Abstract
Background
Mortality for non variceal upper gastrointestinal bleeding (UGIB) is clinically relevant in the first 12–24 hours of the onset of haemorrhage and therefore identification of clinical factors predictive of the risk of death before endoscopic examination may allow for early corrective therapeutic intervention.
Aim
1) Identify simple and early clinical variables predictive of the risk of death in patients with non variceal UGIB; 2) assess previsional gain of a predictive model developed with conventional statistics vs. that developed with artificial neural networks (ANNs).
Methods and results
Analysis was performed on 807 patients with nonvariceal UGIB (527 males, 280 females), as a part of a multicentre Italian study. The mortality was considered “bleeding-related” if occurred within 30 days from the index bleeding episode. A total of 50 independent variables were analysed, 49 of which clinico-anamnestic, all collected prior to endoscopic examination plus the haemoglobin value measured on admission in the emergency department. Death occurred in 42 (5.2%). Conventional statistical techniques (linear discriminant analysis) were compared with ANNs (Twist® system-Semeion) adopting the same result validation protocol with random allocation of the sample in training and testing subsets and subsequent cross-over. ANNs resulted to be significantly more accurate than LDA with an overall accuracy rate near to 90%.
Conclusion
Artificial neural networks technology is highly promising in the development of accurate diagnostic tools designed to recognize patients at high risk of death for UGIB.
Keywords: upper gastrointestinal haemorrhage, non variceal bleeding, 30-day mortality, clinical outcomes, endoscopic therapy, prognostic factors, artificial neural networks
Introduction
Acute upper gastrointestinal haemorrhage (UGIH) remains a common reason for hospital admission, requiring high levels of assistance and health-care expenditures. The incidence of acute UGIH and hospital related 30-day mortality varies widely, partly because of the changing epidemiology in Western countries, in which a number of factors, such as older age, comorbidites, prescriptions for proton pump inhibitors (PPI), low dose aspirin, oral anticoagulants, cyclooxygenase-2 inhibitors, are involved(1–6). Risk assessment is mainly based on endoscopic recognition of high-risk stigmata at ulcer base(7,8) and controversy exists in the literature about the predicting value of clinical, biochemical or therapeutic variables on the risk of death.
Mortality for non variceal UGIB is clinically relevant in the first 12–24 hours of the onset of haemorrhage and therefore identification of clinical factors predictive of the risk of death before endoscopic examination may allow for early corrective therapeutic intervention.
Predictive models, like Blatchford risk score(9) or Rockall risk score(7), have been developed with conventional statistical procedures to quantify mortality risk linked to UGIB.
The application of these methods at individual level has been hampered by the high interdependence and complex interaction among clinical variables involved. Many clinical variables are inter-dependent and may potentially interact with each other with reciprocal enhancement.
Artificial neural networks, at variance with the classical statistical tests, can manage complexity in a brilliant way and have been employed in the prediction of lower GI bleeding with success(10).
The main aim of this prospective multi-centre database study was to describe the ultimate outcome of patients with non variceal UGIH in a contemporary “real-life” setting. Additional analysis assessed the impact of clinical, endoscopic or therapeutic factors on the risk of death in this patient population.
This study has two aims:1) identify simple and early clinical variables predictive of the risk of death in patients with non variceal UGIB; 2) assess predictive gain of a predictive model developed with conventional statistics vs. that developed with artificial neural networks (ANNs).
Methods
The PNED initiative and data collection
A dedicated software including an endoscopic reporting system (Cartella Clinica Endoscopia Digestiva, Bracco, Italy) linked to a project-specific research database was developed. This software was distributed to 23 participating sites across Italy, establishing a network of centers that received emergency admissions from which source data were collected—the Progetto Nazionale Emorragie Digestive (PNED) study group. Fifteen community hospitals and eight tertiary care institutions were included. At each site a “lead” consultant represented the project locally. At each hospital a coordinator identified subjects daily in the accident and emergency department, the wards, the endoscopy unit, the operating theatre, and from blood transfusion records and admission data. Specially trained research assistants collected data directly from the patient’s point of care into case report forms. The coordinator was then responsible for checking and returning a completed form for each patient correctly identified. All data were denominalized and downloaded into a central database on a monthly basis. They were reviewed at a single national location for internal logic of patient flow and biological plausibility. All data queries were resolved within 30 days following original data entry. To help assure the internal validity of the registry, there was an independent data validation of a random subset of all information collected; quality of data were validated on a quarterly basis by randomly comparing 5% of all records to the source data recorded in the hospital charts. Personnel from a clinical research organization trained all research staff at a common start-up meeting and at each initial on-site visit prior to the first patient entry. On that occasion, all endoscopists participating in the registry were invited to review a wide set of video images of different bleeding stigmata in order to estimate inter-rater variability and find as much agreement as possible on the diagnosis of stigmata.
Patient population
Patients were considered for the study if they had clinical evidence of overt UGIH on admission or a history of haematemesis/coffee ground vomiting, melena, hematochezia or a combination of any of the above within 24 hours preceding the admission, or clinical evidence of acute UGIH while hospitalized for any other reason (in-hospital bleeding), independently of their age. UGIH was confirmed only if either haematemesis, melena, or dark, tarry materials on rectal examination was documented and witnessed by nursing or medical staff. Patients were entered in the registry only if an upper GI endoscopy was performed. In case of bleeding from esophago-gastric varices, data were initially computed as patients having an UGIH, but were then excluded from the prospective database.
An audit of all patients presenting over a fixed time period, at each participating institution, was performed to rule out any selection bias in the way in which the study population was enrolled. Patients initially assessed at another hospital for the bleeding episode and subsequently transferred to one of the participating centres were excluded from the analysis.
Study variables
The following independent variables were included in the electronic form: demographics (age, sex, site and date of endoscopy); historical data (presenting signs or symptoms, any significant comorbidity, the patient’s physical status on presentation using the American Society of Anesthesiologists (ASA) classification(12), relevant past medical history, any concomitant intake of medications in the 7 days preceding the bleeding episode, time elapsed from the onset of bleeding); physical examination findings and laboratory data (haemodynamic data, rectal exam, nasogastric tube use, complete blood count and coagulation parameters).
The outcomes evaluated were the frequency of death, recurrent bleeding and need for surgery. Such outcomes were monitored from the admission to the hospital or the onset of bleeding for in-hospital patients up to 30 days after the endoscopic examination. Both investigators and nurses all worked with the same operational definitions of outcome. A priori definitions for all outcomes were adopted according to established definitions(13). Thyrty-day mortality was the primary investigated outcome; a “bleeding-related” death was defined as any death occurring within 30 days of the index bleeding episode. To ensure the completeness of follow-up information, the study nurses called all patients or their families at 30 days. Furthermore, after PNED had been completed, administrative databases were consulted and all charts of included patients were reviewed for a full 30 days following admission or onset of bleeding while in hospital.
Data analysis
Advanced intelligent systems based on novel coupling of artificial neural networks and evolutionary algorithms have been applied. The results obtained have been compared with those derived from the use of standard neural networks and classical statistical analysis.
In this study we applied supervised ANNs(14), in order to develop a model able to predict with high degree of accuracy the diagnostic class starting from genotype data alone.
Supervised ANNs are networks which learn by examples, calculating an error function during the training phase and adjusting the connection strengths in order to minimize the error function. The learning constraint of the supervised ANNs make their own output coincide with the predefined target. The general form of these ANNs is: y = f(x,w*), where w* constitutes the set of parameters which best approximate the function.
We employed as benchmark linear discriminant analysis (LDA) applied on the same training and testing data sets used for ANNs. For the analysis of LDA, the SAS version 6.04 (SAS Institute, Cary, NC, U.S.A.) using forward stepwise procedure was employed.
Preprocessing methods and experimental protocols
Data preprocessing was performed using two different re-sampling criteria of the global dataset.
Random criterion
We employed the so-called 5 × 2 cross-validation protocol(15). In this procedure the study sample is five-times randomly divided into two sub-samples, always different but containing similar distribution of cases and controls: the training one (containing the dependent variable) and the testing one. During the training phase the ANNs learn a model of data distribution and then, on the basis of such a model, classify subjects in the testing set in a blind way. Training and testing sets are then reversed and consequently 10 analyses for every model employed are conducted.
Optimized criterion: TWIST system
The TWIST system consists in an ensemble of two previously described systems: T&T and IS(16). The T&T system is a robust data resampling technique that is able to arrange the source sample into sub-samples that all possess a similar probability density function. In this way, the data is split into two or more sub-samples in order to train, test and validate the ANN models more effectively. The IS system is an evolutionary wrapper system able to reduce the amount of data while conserving the largest amount of information available in the dataset. The combined action of these two systems allow us to solve two frequent problems in managing Artificial Neural Networks.
Both systems are based on a Genetic Algorithm, the Genetic Doping Algorithm (GenD) developed at Semeion Research Centre(17).
The TWIST system is described in detail in the appendix.
After this processing, the features that were most significant for the classification were selected and at the same time the training set and the testing set were created with a function of probability distribution similar to the one that provided the best results in the classification.
A supervised Multi Layer Perceptron, with four hidden units, was then used for the classification task.
Ethics
The registry was approved by the Institutional Review Board of all participating centers. In addition, all eligible patients were asked to sign a written informed consent.
Results
Study population
A total of 807 cases with complete data set were identified and entered in ANNs analysis. Patient characteristics are outlined in Table 1. A recent history of drugs’ consumption was recorded in a high proprotion of upper GI bleeders, mostly non-steroidal anti-inflammatory drugs (34%) and low dose aspirin (17.5%). One or more co-morbidities were recorded at the time of presentation in 60.6% of cases, with 25% having more than two comorbidities. The median number of comorbid conditions per patient was 1.0 (IQR 1.0–2.0), mainly cardiovascular diseases affecting almost a half of the patients. Mean length of stay was 7.2 ± 6.3 days (median: 4.0, IQR: 2.0–9.0).
Table 1.
Characteristics of the study population.
| Total population (n = 807)
|
||
|---|---|---|
| Mean (median, IQR**) | 95% CI/SD* | |
| Males | 62% | 61.8–67.7 |
| Age (yr) | 68 yrs (71, 53–81) | ± 16 yrs |
| Number of comorbidities | 1.0 (1.0, 0–2) | ± 1.3 |
| ASA score | ||
| 1–2 | 42% | 39.2–44.5 |
| 3 | 45% | 42.5–47.3 |
| 4 | 13% | 11.8–15.7 |
| In-hospital bleeding | 14% | 12–16 |
| Symptoms on presentation | ||
| Melena | 79% | 76.0–81.1 |
| Haematemesis | 28% | 25.0–30.0 |
| Hematochezia | 9% | 7.9–11.5 |
| Timing of endoscopy (hrs) | 8 (10, 2–14) | ± 10 hrs |
| Medications at presentation | ||
| NSAIDs | 34% | 31.1–36.9 |
| Aspirin | 17.5% | 15.3–20.0 |
| Anti-coagulants | 8.2% | 6.7–10.0 |
| Ca++-antagonists | 7.5% | 6.0–9.2 |
| Steroids | 6.7% | 5.5–7.8 |
| Nitrates | 6.3% | 4.9–7.9 |
| Coxib | 3.3% | 2.5–4.0 |
| Initial haemodynamic instability | 27% | 24.7–30.2 |
| Laboratory results | ||
| Initial mean haemoglobin (g/dL) | 9.21 (9.0, 6.9–11.4) | ± 2.8 SD |
| Hematocrit | 31 (30, 24–35) | ± 7 SD |
| INR | 1.4 (1.2, 1.0–1.3) | ± 1.3 SD |
| Source of bleeding at endoscopy | ||
| Duodenal ulcer | 37.8% | 34.9–40.9 |
| Gastric ulcer | 28.3% | 25.6–31.2 |
| Gastroduodenal erosions | 9.1% | 7.4–10.9 |
| Neoplasia | 5.7% | 4.0–6.9 |
| Mallory-Weiss tears | 4.5% | 3.8–5.5 |
| Esophagitis | 3.6% | 3.0–5.0 |
| Dieulafoy’s lesion | 1.4% | 0.9–2.4 |
| Other vascular source | 2.4% | 1.5–3.4 |
| No lesion identified | 7.2% | 5.7–8.9 |
IQR: interquartile range; SD: standard deviation; CI: confidence interval; NSAIDs: non steroidal anti-inflammatory drugs; SRH: stigmata of recent haemorrhage.
Mean daily dose of aspirin 100 ± 25 mg.
ASA score refers to the American Society of Anaesthesiologists classification of a patient’s severity and acuity of disease index.
Outcomes
Death was registered in 42 out of 807 patients (5.2%). The median time to death was 4 days (95% CI 2–6). In the absence of any comorbidity, mortality was 0.7% (1.1% if no severe comorbidity). Mortality rate increased to 8.4% if one severe comorbidity was recorded, and to 23.1% if two or more severe comorbidities were present (p < 0.01). The mean age of the patients who died was 76.6 ± 14.0 yr.
Classification performances with ANNs
Table 2 summarizes the input variables used for modelling. The linear correlation index between the 50 input variables and the target variable ranged between −0.1 to +0.13. Results obtained with LDA werecompared with those obtained with a simple Back Propagation approach with 5 × 2 cross validation protocol (Tables 3 and 4).
Table 2.
input variables.
| male | rofecoxib |
| female | NSAIDs |
| Age | Anticoagulant agents |
| Bleeding in hospital | calcium antagonists |
| previous bleeding | nitroderivates |
| peptic ulcer | heparine |
| gastric surgery | corticosteroids |
| hypertension | antidepressant |
| Obstructive lung disease | syncope |
| kidney failure | hematemesis |
| dialisis | melena |
| ischemic hearth disease | black vomiting |
| cardiac dilatation | red blood vomiting |
| cardiac failure | sistolic blood pressure |
| diabetes | diastolic blood pressure |
| cancer | heart rate |
| cancer site | presence of blood in rectum |
| leukemia | black blood |
| liver cirrosis | red blood vomiting |
| cerebro vascular disease | nasogastric intubation |
| no concomitant diseases | red blood from gastric tube |
| aspirin | black blood from gastric tube |
| ticlopidine | solid material from gastric tube |
| clopidogrel | time from symptoms and hospital |
| celecoxib | hemoglobin value |
Table 3.
Results obtained with Linear Discriminant Analysis (LDA).
| LDA | Sensitivity | Specificity | Overall accuracy |
|---|---|---|---|
| LDA-1AB | 19.1% | 88.8% | 53.9% |
| LDA-1BA | 23.8% | 87.7% | 55.8% |
| LDA-2AB | 52.4% | 83.3% | 67.8% |
| LDA-2BA | 42.9% | 88.0% | 65.4% |
| LDA-3AB | 42.9% | 85.9% | 64.4% |
| LDA-3BA | 61.9% | 77.7% | 69.8% |
| LDA-4AB | 38.1% | 84.9% | 61.5% |
| LDA-4BA | 28.6% | 84.3% | 56.4% |
| LDA-5AB | 42.9% | 84.1% | 63.5% |
| LDA-5BA | 47.6% | 83.3% | 65.4% |
| Average | 40.0% | 84.8% | 62.4% |
Table 4.
Results obtained with Back Propagation artificial neural network.
| ANN | Sensitivity | Specificity | Overall accuracy |
|---|---|---|---|
| FF_Bp(1ab) | 19.1% | 94.5% | 56.8% |
| FF_Bp(1ba) | 23.8% | 96.1% | 59.9% |
| FF_Bp(2ab) | 28.6% | 93.7% | 61.2% |
| FF_Bp(2ba) | 23.8% | 97.4% | 60.6% |
| FF_Bp(3ab) | 33.3% | 96.9% | 65.1% |
| FF_Bp(3ba) | 42.9% | 96.1% | 69.5% |
| FF_Bp(4ab) | 38.1% | 89.6% | 63.8% |
| FF_Bp(4ba) | 23.8% | 97.4% | 60.6% |
| FF_Bp(5ab) | 47.6% | 84.3% | 66.0% |
| FF_Bp(5ba) | 28.6% | 96.3% | 62.5% |
| Average | 31.0% | 94.2% | 62.6% |
The overall predictive accuracy obtained with LDA and standard ANNs ranged from 54% to 70% (average 62.31%) and from 57% to 69.5% (average 62.59%) respectively.
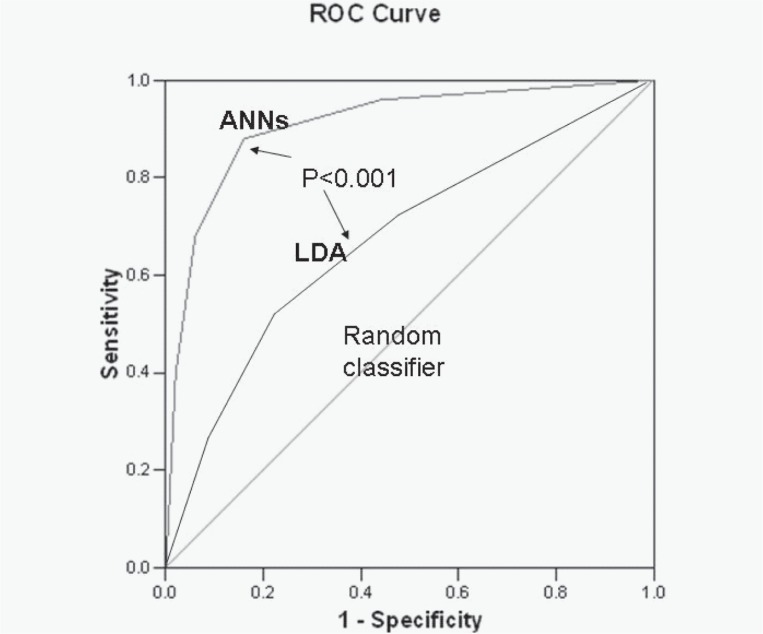
With the TWIST approach, every experiment was conducted in a blind and independent manner in two directions: training with sub-sample A and blind testing with sub-sample B vs training with sub-sample B and blind testing with sub-sample A. The results in variables selection from the best 11 applications of TWIST procedures are reported in Table 5. This advanced intelligent system, through the final selection of a subgroup of 17 variables which resulted most often selected along eleven independent applications (at least 10 times), provided the highest predictive performance with a sensitivity ranging from 81.48% to 93.33% (average 89.18%), and a specificity ranging from 80.85% to 88.24% (average 82.98%) and with an overall accuracy ranging from 81.17 to 89.03% (average 86.04%) (Table 6). The resulting ROC curve is reported in Figure 1, with a comparison with ROC curve obtained with LDA.
Table 5.
Variables selection in TWIST system application. In yellow variables selected in each round of TWIST applications; in purple variables selected in at least 10 TWIST applications; in green variables selected by LDA.
| Num | INPUT VARIABLES | LDA | Tw1 | Tw2 | Tw3 | Tw4 | Tw5 | Tw6 | Tw7 | Tw8 | Tw9 | Tw10 | Tw11 | SUM | Euristic |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | male | 3 | |||||||||||||
| 2 | female | 11 | |||||||||||||
| 3 | Age | 10 | |||||||||||||
| 4 | Bleeding in hospital | 11 | |||||||||||||
| 5 | previous bleeding | 1 | |||||||||||||
| 6 | peptic ulcer | 1 | |||||||||||||
| 7 | gastric surgery | 1 | |||||||||||||
| 8 | hypertension | 1 | |||||||||||||
| 9 | Obstructive lung disease | 11 | |||||||||||||
| 10 | kidney failure | 0 | |||||||||||||
| 11 | dialisis | 8 | |||||||||||||
| 12 | ischemic hearth disease | 1 | |||||||||||||
| 13 | cardiac dilatation | 8 | |||||||||||||
| 14 | cardiac failure | 7 | |||||||||||||
| 15 | diabetes | 0 | |||||||||||||
| 16 | cancer | 11 | |||||||||||||
| 17 | cancer site | 10 | |||||||||||||
| 18 | leukemia | 9 | |||||||||||||
| 19 | liver cirrosis | 11 | |||||||||||||
| 20 | cerebro vascular disease | 2 | |||||||||||||
| 21 | no concomitant diseases | 1 | |||||||||||||
| 22 | aspirin | 5 | |||||||||||||
| 23 | ticlopidine | 11 | |||||||||||||
| 24 | clopidogrel | 10 | |||||||||||||
| 25 | celecoxib | 9 | |||||||||||||
| 26 | rofecoxib | 10 | |||||||||||||
| 27 | NSAIDs | 11 | |||||||||||||
| 28 | Anticoagulant agents | 1 | |||||||||||||
| 29 | calcium antagonists | 1 | |||||||||||||
| 30 | nitroderivates | 10 | |||||||||||||
| 31 | heparine | 6 | |||||||||||||
| 32 | corticosteroids | 3 | |||||||||||||
| 33 | antidepressant | 4 | |||||||||||||
| 34 | syncope | 4 | |||||||||||||
| 35 | hematemesis | 1 | |||||||||||||
| 36 | melena | 1 | |||||||||||||
| 37 | black vomiting | 11 | |||||||||||||
| 38 | red blood vomiting | 11 | |||||||||||||
| 39 | sistolic blood pressure | 11 | |||||||||||||
| 40 | diastolic blood pressure | 5 | |||||||||||||
| 41 | heart rate | 2 | |||||||||||||
| 42 | presence of blood in rectum | 11 | |||||||||||||
| 43 | black blood | 11 | |||||||||||||
| 44 | red blood vomiting | 2 | |||||||||||||
| 45 | nasogastric intubation | 0 | |||||||||||||
| 46 | red blood from gastric tube | 7 | |||||||||||||
| 47 | black blood from gastric tube | 9 | |||||||||||||
| 48 | solid material from gastric tube | 3 | |||||||||||||
| 49 | time from symptoms and hospital | 0 | |||||||||||||
| 50 | hemoglobin value | 9 | |||||||||||||
| Total variables selected | 10 | 26 | 25 | 25 | 24 | 24 | 28 | 30 | 34 | 28 | 19 | 27 | 17 |
Table 6.
Results obtained with different kind of supervised neural networks applied to the 17 variables selected with TWIST system.
| ANN | train - test order | Sensitivity | Specificity | Overall accuracy |
|---|---|---|---|---|
| SelfSABp 3 | ab | 93.3% | 84.7% | 89.0% |
| TasmSABp 4 | ab | 93.3% | 83.2% | 88.3% |
| SelfDABp 1 | ab | 93.3% | 81.9% | 87.6% |
| SelfSABp 4 | ab | 86.7% | 88.2% | 87.5% |
| FF_Bp 5 | ab | 93.3% | 81.5% | 87.4% |
| SelfSABp 2 | ba | 93.3% | 81.2% | 87.3% |
| TasmDABp 1 | ba | 88.9% | 83.2% | 86.0% |
| SelfSABp 1 | ba | 86.7% | 83.2% | 84.9% |
| FF_Bp 3 | ba | 81.5% | 80.9% | 81.2% |
| SelfSABp 1 | ba | 81.5% | 80.9% | 81.2% |
| Average | 89.2% | 82.9% | 86.0% |
Notes: In the column under ANN are listed different kind of Artificial Neural Networks developed by Semeion Research Centre:
SelfSABp 3: self-recurrent-static-adaptive No.3.
TasmSABp 4: Temporal Associative Subjective Memory back propagation No.4.
SelfDABp 1: self-recurrent dynamic-adaptive No.1.
SelfSABp 3: self-recurrent-static-adaptive No.4.
FF_Bp 5: Feed forward Back propagation No.5.
SelfSABp 2: self-recurrent-static-adaptive No.2.
TasmSABp 4: Temporal Associative Subjective Memory dynamic adaptive No.1.
SelfDABp 1: self-recurrent dynamic-adaptive No.1.
SelfSABp 3: self-recurrent-static-adaptive No.4.
FF_Bp 5: Feed forward Back propagation No.5.
In column train-test order ab = train on subset a and test on subset b; ba = train on subset b and test on subset a.
Figure 1.
ROC curve obtained with TWIST system (ANNs) and with Linear discriminant analysis(LDA). The respective AUC are: 0.87 and 0.65 (P < 0.005).
The following variables resulted to be selected both by LDA and TWIST system: age, cancer, cancer site, nitroderivates while other 4 variables were never selected by all models: gastric surgery hypertension, diabetes, time from symptoms to hospital.
Discussion
Acute UGIH remains a common medical problem that has significant associated morbidity, 30-day mortality, and health care resource use. The PNED data indicate that ulcers are by far the most common cause of nonvariceal UGIH, accounting for 66% of all diagnoses. More than a half of the patients in the present study were taking at least one ulcerogenic drug, mainly NSAID’s, whereas the use of low dose aspirin was much lower than reported in North America (19–20)and this should be taken into account in terms of generalizability of results. An important feature of the PNED study is that it represents a consortium of practice sites that use a structured endoscopy reporting system to collect information in a centralised endoscopic database. Such data are useful because they reflect “real world” endoscopic practice from a wide-range of practice settings and minimize patient selection or potential referral bias. Of the 23 participating centers, in fact, only eight were tertiary institutions, unlike the Canadian RUGBE study in which tertiary institutions accounted for two-thirds of the participating centers and 62% of patients were enrolled at six centers(21).
This results obtained with artificial neural networks analysis allow the following considerations. First of all, the comparison of results obtained with three different analytical approaches (classical statistics, standard neural networks and advanced artificial neural networks), points out the need to employ systems that are really able to handle the disease complexity instead of treating the data with reductionist approaches that are unable to detect multiple variables interaction effects in predisposing to the adverse outcome. The possibility to derive high diagnostic accuracy from limited and selected information using these new analytical tools, open the possibility to develop application software to be used in hospital ward for individual risk stratification.
Because patients in trials, as in clinical practice, have many attributes that can affect the likelihood of treatment being beneficial or harmful, exploring each of these attributes “one variable at a time” (e.g. male vs female, old vs young) risks spurious false-positive subgroup results from chance fluctuations. Furthermore, although patients have simultaneous multiple characteristics that can affect the likelihood of the outcome and the effect of therapy, one-variable-at-a-time comparisons are fundamentally limited because they compare groups that vary only on a single factor, usually resulting in the subgroups being more similar than different (21).
Second point, artificial neural networks, at variance with the classical statistical tests, can manage complexity even with relatively small samples and to the subsequent unbalanced ratio between variables and records. In this connection, it is important to note that adaptive learning algorithms of inference, based on the principle of a functional estimation like artificial neural networks, overcome the problem of dimensionality. An important obstacle in approaching in conventional manner the biological basis of a rare events like death for UGIB, is related to the difficulty to find an homogeneous sample population large enough to be analysed for a wide number of clinical variants. Our study identified a number of prognostic clinical factors independently associated with the risk of 30-day mortality after an acute UGI bleed. The relevant predictors were female gender, age, bleeding in hospital, obstructive lung disease, cancer, cancer site, liver cirrhosis, ticlopidine, clopidogrel, rofecoxib, NSAIDs, nitroderivates, black vomiting, red blood vomiting, systolic blood pressure, presence of blood in rectum, black blood.
The fact that most deaths occur in elderly patients with severe comorbidities should prompt us to switch our focus to improve management of this selected high-risk subgroup, to be included into specifically designed trials. Furthermore, major preventive steps should be taken in order to reduce the risk of NSAIDs’ related haemorrhage, particularly in the highest risk population.
Risk stratification in patients with UGIH is essential for optimal management of bleeders, both for triage of those at high risk to inpatient care and for identification of patients at low risk of adverse outcome who can be safely managed as outpatients(23,24,25).
Predictive models, like Blatchford risk score or Rockall risk score, have been developed with conventional statistical procedures to quantify mortality risk linked to UGIB.
The application of these methods at individual level has been hampered by the high interdependence and complex interaction among clinical variables involved.
However, as pointed out in a recent review on the topic by Das et al. (25), at present there is not an ideal risk score. For acute bleeders, urgent EGD should not be an essential part of the initial risk score; however, EGD should be an important component of the risk score at a subsequent point in patient care. Hence, the need to categorise the patient’s risk profile with the aid of accurate and user-friendly clinical predictors that typically are available during initial patient triage.
There are several limitations to the present registry. The adopted study design is not an experimental one, and therefore not as rigorous as that of a randomized controlled trial. The demonstrated associations should be seen as suggestive, but require prospective independent validation, which is actually under way. Observational databases can be useful adjuncts to randomised controlled trials to determine whether efficacy under controlled condition in referral specialists units can be translated into effective treatment in routine clinical practice. Methodological limitations can nonetheless threaten the internal validity of a registry: completeness of follow-up, ascertainment of outcomes, possible patient selection bias and inadequate adjustment for confounders when attempting at identifying predictors of outcome. Like others(21), we attempted to address these possible shortcomings by establishing conservative and a priori definitions for all study variables including outcomes, by training all research staff in a standardized fashion, by enforcing strict data verification and validation protocols and by ensuring complete 30-day follow-up.
In conclusion, death after an acute non variceal UGIH occurs mostly among elderly patients with severe comorbidity or those with failure of endoscopic intention to treatment. These factors should be taken into account in a struggle toward a further reduction of overall 30-day mortality. Future studies should reconsider early resort to surgery, especially for young and surgically fit patients, to reduce the risk of death during the first 24 hours of the bleeding episode. Also, the use of preventive strategies to reduce the bleeding risk especially in patients with advanced age, neoplasia, renal failure and liver cirrhosis deserves appropriate investigation. Improving the ultimate outcome of patients with non-variceal UGIH will take an integrated approach by a team focused on treating the patient and not just the source of bleeding.
The results of this study illustrate that ANNs can be added to the list of computational methods that may provide answers to some questions about complex biological process.
Acknowledgments
The PNED initiative was a collaborative effort supported by the Italian Society for Digestive Endoscopy (SIED) and the Italian Association of Hospital Gastroenterologists (AIGO). We acknowledge the great deal of work performed by medical and nursing staff in each of the participating units.
Appendix
The PNED investigators’ group includes Riccardo Marmo, Ospedale Curto, Polla; Livio Cipolletta, Gianluca Rotondano, and Maria A. Bianco, Ospedale Maresca, Torre del Greco; Lucio Capurso, Maurizio Koch and Angelo Dezi, ACO San Filippo Neri, Rome; Angelo Pera and Rodolfo Rocca, Ospedale Mauriziano Umberto I, Torino; Fausto Barberani and Sandro Boschetto, Ospedale San Camillo De Lellis, Rieti; Alfredo Pastorelli and Elena Sanz Torre, Ospedale Bel Colle, Viterbo; Sergio Brunati and Renato Fasoli, Ospedale Cantù, Abbiategrasso; Ivano Lorenzini and Ugo Germani, AO Umberto I, Ancona; Giorgio Minoli and Giorgio Imperiali, Ospedale Valduce, Como; Giovanni Gatto and Mariano Amuso, AO Villa Sofia, Palermo; Massimo Proietti and Anna Tanzilli, Ospedale Del Prete, Pontecorvo; Walter Piubello, Maria Tebaldi and Fabrizio Bonfante, Ospedale di Desenzano, Desenzano del Garda; Renzo Cestari and Domenico Della Casa, AO Ospedali Civili, University of Brescia, Brescia; Paolo Michetti and Paola Romagnoli, Ospedali Galliera, Genova; Omero Triossi and Andrea Buzzi, Ospedale Santa Maria delle Croci, Ravenna; Alessandro Casadei and Claudio Cortini, AO Morgagni, Forlì; Giorgio Chiozzini and Lisa Girardi, Ospedale Umberto I, Mestre; Luciano Allegretta and Salvatore Tronci, Ospedale Santa Caterina Novella, Galatina; Giovanni Aragona and Francesco Giangregorio, Ospedale Civile, Piacenza; Sergio Segato and Giuseppe Chianese, AO Ospedale Circolo e Fondazione Macchi, Varese; Andrea Nucci and Francesca Rogai, AO Ospedale Careggi, Firenze; Giampiero Bagnalasta and Claudio Leoci, Ospedale Civile, Manerbio; Giovanni Di Matteo and Paolo Giorgio, IRCCS Ospedale De Bellis, Castellana Grotte; Marco Martorano, Ospedale dell’Immacolata, Sapri; Mario Salvagnini, Ospedale San Bortolo, Vicenza.
Appendix: TWIST system
TWIST system is an ensemble of two algorithms: T&T and I.S.
T&T
The “Training and Testing” algorithm (T&T) is based on a population of n ANNs managed by an evolutionary system. In its simplest form, this algorithm reproduces several distribution models of the complete dataset DΓ (one for every ANN of the population) in two subsets ( the Training Set and , the Testing Set). During the learning process each ANN, according to its own data distribution model, is trained on the subsample and blind-validated on the subsample .
The performance score reached by each ANN in the testing phase represents its “fitness” value (i.e. the individual probability of evolution). The genome of each “network-individual” thus codifies a data distribution model with an associated validation strategy. The n data distribution models are combined according to their fitness criteria using an evolutionary algorithm. The selection of “network-individuals” based on fitness determines the evolution of the population; that is, the progressive improvement of performance of each network until the optimal performance is reached, which is equivalent to the better division of the global dataset into subsets. The evolutionary algorithm mastering this process, named “Genetic Doping Algorithm” (GenD for short), was created at Semeion Research Centre has similar characteristics to a genetic algorithm [6–10] but it’s able to maintain an inner instability during the evolution, carrying out a natural increase of biodiversity and a continuous “evolution of the evolution” in the population. The elaboration of T&T is articulated in two phases:
-
Preliminary phase: in this phase an evaluation of the parameters of the fitness function that will be used on the global dataset is performed. During this phase an inductor is configured, which consists of an Artificial Neural Network with an algorithm (A) Back Propagation standard. For this inductor the optimal configuration to reach the convergence is stabilized at the end of different training trials on the global dataset DΓ; in this way the configuration that most “suits” the available dataset is determined: the number of layers and hidden units and some possible generalizations of the standard learning law. The parameters thus determined define the configuration and the initialization of all the individual-networks of the population and will then stay fixed in the following computational phase. Basically, during this preliminary phase there is a fine-tuning of the inductor that defines the fitness values of the population’s individuals during evolution.
The accuracy of the ANN performance with the testing set will be the fitness of that individual (that is, of that hypothesis of distribution into two halves of the whole dataset).
Computational phase: the system extracts from the global dataset the best training and testing sets. During this phase the individual-network of the population is running, according to the established configuration and the initialization parameters. From the evolution of the population, managed by the GenD algorithm, the best distribution of the global dataset D Γ into two subsets is generated, starting from the initial population of possible solutions . Preliminary experimental sessions are performed using several different initialization and configuration of the network in order to achieve the best partition of the global dataset.
I.S
Parallel to T&T run I.S. (Input Selection), an adaptive system, which is also based on the evolutionary algorithm GenD, and which is able to evaluate the relevance of the different variables of the data-set in an intelligent way. Therefore it can be considered on the same level as a feature selection technique.
From a formal point of view, I.S. is an artificial organism based on the GenD algorithm and consists of a population of ANN, in which each one carries out a selection of the independent variables on the available database. The elaboration of I.S., as for T&T, is developed in two phases:
Preliminary phase: during this phase an inductor is configured to evaluate the parameters of the fitness function. This inductor is a standard Back-Propagation ANN. The parameters configuration and the initialization of the ANNs are carried out with particular care to avoid possible over-fitting problems that can be present when the database is characterized by a high number of variables that describe a low quantity of data. The number of epochs E0 necessary to train the inductor is determined through preliminary experimental tests.
Computational phase: the inductor is active, according to the stabilized configuration and the fixed initialization parameters, to extract the most relevant variables of the training and testing subsets. Each individual-network of the population is trained on the training set and tested on the testing set .
The evolution of the individual-network of the population is based on the algorithm GenD. In the I.S. approach the GenD genome is built by n binary values, where n is the cardinality of the original input space. Every gene indicates if an input variable is to be used or not during the evaluation of the population fitness. Through the evolutionary algorithm, the different “hypotheses” of variable selection, generated by each ANNs of the population, change over time, at each generation: this leads to the selection of the best combination of input variables. As in the T&T systems the genetic operators crossover and mutation are applied on the ANNs population; the rates of occurrence for both operators are self-determinated by the system in adaptive way at each generation.
When the evolutionary algorithm no longer improves its performance, the process stops, and the best selection of the input variables is employed on the testing subset.
In order to improve the speed and the quality of the solutions that have to be optimized, the GenD algorithm makes the evolutionary process of the artificial populations more natural and less centered on the individual liberalism culture.
References
- 1.Longstreth GF. Epidemiology of hospitalization for acute upper gastrointestinal hemorrhage: a population-based study. Am J Gastroenterol. 1995;90:206–10. [PubMed] [Google Scholar]
- 2.Yavorski RT, Wong RK, Maydonovitch C, et al. Analysis of 3,294 cases of upper gastrointestinal bleeding in military medical facilities. Am J Gastroenterol. 1995;90:568–73. [PubMed] [Google Scholar]
- 3.Rockall TA, Logan RF, Devlin HB, et al. Incidence of and 30-day mortality from acute upper gastrointestinal haemorrhage in the United Kingdom. BMJ. 1995;311:222–6. doi: 10.1136/bmj.311.6999.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higham J, Kang JY, Majeed A. Recent trends in admissions and 30-day mortality due to peptic ulcer in England: increasing frequency of haemorrhage among older subjects. Gut. 2002;50:460–4. doi: 10.1136/gut.50.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vreeburg EM, Snel P, de Bruijne JW, et al. Acute upper gastrointestinal bleeding in the Amsterdam area: incidence, diagnosis, and clinical outcome. Am J Gastroenterol. 1997;92:236–43. [PubMed] [Google Scholar]
- 6.Esrailian E, Gralnek IM. Nonvariceal upper gastrointestinal bleeding: epidemiology and diagnosis. Gastroenterol Clin North Am. 2005;34:589–605. doi: 10.1016/j.gtc.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Rockall TA, Logan RF, Devlin HB, et al. Risk assessment after acute upper gastrointestinal haemorrhage. Gut. 1996;38:316–21. doi: 10.1136/gut.38.3.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laine L, Peterson WL. Bleeding Peptic Ulcer. N Engl J Med. 1994;331:717–27. doi: 10.1056/NEJM199409153311107. [DOI] [PubMed] [Google Scholar]
- 9.Blatchford O, Murray WR, Blatchford M. A risk score to predict need for treatment for upper-gastrointestinal haemorrhage. Lancet. 2000;356(9238):1318–21. doi: 10.1016/S0140-6736(00)02816-6. [DOI] [PubMed] [Google Scholar]
- 10.Das A, Ben-Menachem T, Cooper GS, Chak A, Sivak MV, Jr, Gonet JA, Wong RC. Prediction of outcome in acute lower gastrointestinal haemorrhage based on an artificial neural network: internal and external validation of a predictive model. Lancet. 2003;362:1261–6. doi: 10.1016/S0140-6736(03)14568-0. [DOI] [PubMed] [Google Scholar]
- 11.Owens W, Felts J, Spitznagel E. ASA physical status classifications: a study of consistency of ratings. Anesthesiology. 1978;49:239–43. doi: 10.1097/00000542-197810000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Cooper GS, Chak A, Lloyd LE, et al. The accuracy of diagnosis and procedural codes for patients with upper GI haemorrhage. Gastrointest Endosc. 2000;51:423–6. doi: 10.1016/s0016-5107(00)70442-1. [DOI] [PubMed] [Google Scholar]
- 13.Rumelhart DE, Mc Clelland JL. Parallel Distributed Processing: Explorations in the Microstructure of Cognition Volume 1: Foundations. MIT Press; Cambridge, MA: 1986. [Google Scholar]
- 14.Dietterich TG. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Computation. 1998;10:1885–924. doi: 10.1162/089976698300017197. [DOI] [PubMed] [Google Scholar]
- 15.Buscema M, Grossi E, Intraligi M, Garbagna N, Andriulli A, Breda M. An Optimized Experimental Protocol Based on Neuro-Evolutionary Algorithms. Application to the Classification of Dyspeptic Patients and to the Prediction of the Effectiveness of Their Treatment. Artificial Intelligence in Medicine. 2005;34:279–305. doi: 10.1016/j.artmed.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Buscema M. Genetic Doping Algorihm (GenD): theory and applications. Expert Systems. 2004;21:63–79. [Google Scholar]
- 17.Buscema Massimo, Capriotti Massimiliano, Bergami Francesca, Babiloni Claudio, Rossini Paolo, Grossi Enzo. The Implicit Function as Squashing TimeModel: A Novel Parallel Nonlinear EEG Analysis Technique Distinguishing Mild Cognitive Impairment and Alzheimer’s Disease Subjects with High Degree of Accuracy. Computational Intelligence and Neuroscience. 2007:1–15. doi: 10.1155/2007/35021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis JD, Bilker WB, Brensinger C, Farrar JT, Strom BL. Hospitalization and 30-day mortality rates from peptic ulcer disease and GI bleeding in the 1990s: relationship to sales of nonsteroidal anti-inflammatory drugs and acid suppression medications. Am J Gastroenterol. 2002;97:2540–9. doi: 10.1111/j.1572-0241.2002.06037.x. [DOI] [PubMed] [Google Scholar]
- 19.Peura DA, Lanza FL, Gostout CJ, Foutch PG. The American College of Gastroenterology Bleeding Registry: preliminary findings. Am J Gastroenterol. 1997;92:924–8. [PubMed] [Google Scholar]
- 20.Barkun A, Sabbah S, Enns R, et al. Endoscopic hemostasis and proton pump inhibition are associated with improved outcomes in a real-life setting. Am J Gastroenterol. 2004;99:1238–46. doi: 10.1111/j.1572-0241.2004.30272.x. [DOI] [PubMed] [Google Scholar]
- 21.David M, Kent MD, Rodney MS, A Hayward. Limitations of Applying Summary Results of Clinical Trials to Individual Patients. The Need for Risk Stratification JAMA. 2007;298:1209–12. doi: 10.1001/jama.298.10.1209. [DOI] [PubMed] [Google Scholar]
- 22.Barkun A, Bardou M, Marshall JK. Nonvariceal Upper G.I. 2003. Bleeding Consensus Conference Group. Consensus recommendations for managing patients with nonvariceal upper gastrointestinal bleeding. Ann Intern Med. 139:843–57. doi: 10.7326/0003-4819-139-10-200311180-00012. [DOI] [PubMed] [Google Scholar]
- 23.Cebollero-Santamaria F, Smith J, Gioe S, et al. Selective outpatient management of upper gastrointestinal bleeding in the elderly. Am J Gastroenterol. 1999;94:1242–7. doi: 10.1111/j.1572-0241.1999.01073.x. [DOI] [PubMed] [Google Scholar]
- 24.Cipolletta L, Bianco MA, Rotondano G, et al. Outpatient management for low-risk nonvariceal upper gastrointestinal bleeding: a randomized controlled trial. Gastrointest Endosc. 2002;55:1–5. doi: 10.1067/mge.2002.119219. [DOI] [PubMed] [Google Scholar]
- 25.Das A, Wong RC. Prediction of outcome of acute GI hemorrhage: a review of risk scores and predictive models. Gastrointest Endosc. 2004;60:85–93. doi: 10.1016/s0016-5107(04)01291-x. [DOI] [PubMed] [Google Scholar]