Abstract
The number of annual cannabinoid users exceeds 100,000,000 globally and an estimated 9 % of these individuals will suffer from dependency. Although exogenous cannabinoids, like those contained in marijuana, are known to exert their effects by disrupting the endocannabinoid system, a dearth of knowledge exists about the potential toxicological consequences on public health. Conversely, the endocannabinoid system represents a promising therapeutic target for a plethora of disorders because it functions to endogenously regulate a vast repertoire of physiological functions. Accordingly, the rapidly expanding field of cannabinoid biology has sought to leverage model organisms in order to provide both toxicological and therapeutic insights about altered endocannabinoid signaling. The primary goal of this manuscript is to review the existing field of cannabinoid research in the genetically tractable zebrafish model—focusing on the cannabinoid receptor genes, cnr1 and cnr2, and the genes that produce enzymes for synthesis and degradation of the cognate ligands anandamide and 2-arachidonylglycerol. Consideration is also given to research that has studied the effects of exposure to exogenous phytocannabinoids and synthetic cannabinoids that are known to interact with cannabinoid receptors. These results are considered in the context of either endocannabinoid gene expression or endocannabinoid gene function, and are integrated with findings from rodent studies. This provides the framework for a discussion of how zebrafish may be leveraged in the future to provide novel toxicological and therapeutic insights in the field of cannabinoid biology, which has become increasingly significant given recent trends in cannabis legislation.
Keywords: Endocannabinoid, Phytocannabinoid, Synthetic Cannabinoid, Cannabis, Marijuana
1. Introduction
Cannabis sativa has been used medicinally and recreationally for thousands of years, and is the most widely abused illicit drug in the world today (Köfalvi, 2008). However, current legislation is trending toward the widespread decriminalization and legalization of this drug. In the United States, 23 states allow legal use of cannabis for medicinal or recreational purposes, and an additional 9 states have legislation pending for the legalization of medical marijuana. These changes in legislation have coincided with increases in the prevalence and frequency of cannabis use. Indeed, recent surveys estimate there are now 17.4 million past-month users in the United States, and reveal that teenagers are more likely to have used cannabis than cigarettes (Substance Abuse and Mental Health Services Administration, 2011, Centers for Disease Control and Prevention, 2012). Similarly, the prevalence of synthetic cannabinoid (sCB) abuse has surged with approximately 11% of high school seniors reporting past-year sCB use in 2012 (Johnston et al., 2013). sCBs are designed to imitate the psychoactive effects of the phytocannabinoids (pCBs) contained in Cannabis sativa, such as the delta-9-tetrahydrocannabinol (Δ9-THC) but typically exhibit a much higher binding affinity for the cognate receptors (Zimmermann et al., 2009). In recent years the Drug Enforcement Agency has begun emergency scheduling sCBs in an attempt to stem their rampant abuse, but a repertoire of novel variants are constantly being developed and marketed to circumvent such legislation (Drug Enforcement Administration, 2014). Accordingly, sCBs have remained one of the most widely abused drugs in the United States. Despite the increasing social acceptance of both pCB and sCB use, there are no existing pharmacological interventions for managing related substance abuse disorders, and little is known about the potential long-term toxicological consequences of cannabinoid (CB) use on public health.
Both pCBs and sCBs exert widespread effects on physiological processes including those involved with Alzheimer’s disease, appetite, anxiety, bone health, cardiovascular function, cancer, epilepsy, gastrointestinal function, immune system function, learning, memory, metabolism, mood, multiple sclerosis, nervous system function, pain perception, and pulmonary function by disrupting the endocannabinoid (eCB) system (Gurney et al., 2014, Hermanson & Marnett, 2011, Idris & Ralston, 2012 Köfalvi, 2008, Scott et al., 2014, Silvestri & Di Marzo, 2013, Witkin et al., 2005). Because these processes are often perturbed in disease states, the therapeutic potential of manipulated eCB signaling rivals the toxicological potential. Therefore, clarifying the role of the eCB system in these functions is critical for both the interpretation of the potential harm resulting from CB use or abuse, and the adaptation of novel therapeutics targeting the eCB system. The eCB system produces lipid-derived eCBs, like anandamide (AEA) and 2-arachidonoylglycerol (2-AG), to endogenously regulate this diverse array of processes (Dipatrizio & Piomelli, 2012). Within the central nervous system, eCBs are typically synthesized locally on-demand in response to stimuli such as neuronal depolarization or elevated intracellular calcium signaling (Di Marzo et al., 1994, Kondo et al., 1998, Stella et al., 1997). These stimuli activate the biosynthetic machinery, such as that contained in the 2-AG signalosome, which are necessary to convert precursor molecules into the respective eCBs (Jung et al., 2007, Jung et al., 2012). Once synthesized the eCBs interact with their cognate CB receptors that are typically presynaptically localized (Ohno-Shosaku et al., 2001). Cannabinoid receptor 1 is the most widely distributed G protein-coupled receptor within the central nervous system, and is predominately coupled to Gi/o- type G proteins (Iliff et al., 2011, Pertwee, 1997) The intracellular signaling cascades initiated by cannabinoid receptor 1 activation typically function to decrease synaptic output by inhibiting voltage-gated calcium channels, by potentiating inward rectifying potassium channels, and possibly by inhibiting mitochondrial activity (Benard et al., 2012, Daniel et al., 2004, Hoffman & Lupica, 2000). Although its distribution is more limited, cannabinoid receptor 2 expression patterns are detected in the central nervous system and its signaling has been shown to regulate appetite and addictive behavior (Onaivi et al., 2008a, Onaivi et al., 2008b). eCB signaling is terminated via the degradation of the short-lived receptor ligands by enzymes like fatty acid amide hydrolase and monoglyceride lipase (Cravatt et al., 1996, Dinh et al., 2002). Although the aforementioned components of eCB signaling are key players in the eCB system, many other putative eCB ligands and eCB receptors have been identified (Abood et al., 2013, Howlett et al., 2002). This includes G protein-coupled receptor 55, which is extensively expressed in the central nervous system and is activated by many of the same eCBs as cannabinoid receptor 1 and cannabinoid receptor 2 (Lauckner et al., 2008). The complexity of how eCB signaling regulates the activity of circuits within the central nervous system is further obscured by the fact that numerous genes have been identified that provide alternate routes of eCB degradation or synthesis, thereby resulting in region specific ligand signaling profiles (Ahn et al., 2008, Di Marzo, 2011). Moreover, the effects of CB receptor signaling depend on the specific cell type involved, which may be associated with opposing outcomes such as the inhibition or excitation of a neural circuit. Additionally, CB signaling has been shown to take place in diverse tissues outside of the central nervous system including the peripheral nervous system, immune system, digestive tract, liver, and muscle (Mouslech & Valla, 2009). The complicated dynamics that define eCB signaling, coupled with the increasing use and development of exogenous CBs that differentially target this system, mandates scientific investigation of the toxicological and therapeutic potential of altered CB signaling. To this end, many groups have sought to use the unique advantages of different model organisms to further explore CB biology.
Although the bulk of CB literature published to date consists of experiments using in vitro or rodent models, zebrafish have recently gained traction as a powerful in vivo model for complementing and expanding upon the findings of these existing studies. Juvenile zebrafish absorb small molecules like pCBs or sCBs across the skin, and feature transparent ex-utero development. These features make the zebrafish well suited for rapidly assessing the effects of exogenous CB exposure. Moreover, zebrafish are highly amenable to genetic manipulation, and are well suited for both reverse and forward genetic screening applications. Therefore zebrafish represent an additional tool for the investigation of individual eCB gene functions, and for identifying novel genetic modifiers of the CB signaling. In addition to the tools available for dissecting the physiology of CB signaling in this model, an increasing repertoire of behavioral assays have become available for investigating the behaviors associated with disrupted CB signaling in both juvenile and adult zebrafish. Herein, we examine how the zebrafish model has been leveraged to elucidate CB biology in the context of both eCB gene expression and eCB gene function.
2. Zebrafish eCB Gene Expression
Phylogenetic analyses have revealed that the eCB system is highly conserved between zebrafish and mammals—a feature not found in common high-throughput invertebrate model organisms such as Saccharomyces cerevisiae, Drosophila melanogaster, or Caenorhabditis elegans (Klee et al., 2012, Mcpartland et al., 2007). Given their highly conserved eCB system and unique suite of features that may be leveraged for research applications, zebrafish represent an effective preclinical model for studying CB signaling in vivo. However, establishing when and where eCB genes are expressed is essential in order to permit later studies of CB signaling at any given point in the zebrafish lifespan. To this end, numerous groups have expanded on the knowledge gained from phylogenetic analyses by providing critical insights regarding the ontogeny of eCB gene expression in this organism. A summary of the known zebrafish eCB gene expression patterns are detailed below, and have been classified with regard to individual eCB genes.
2.1. Cannabinoid Receptor 1 (cnr1)
cnr1 mRNA has been detected in whole-body lysates by quantitative real-time reverse transcriptase polymerase chain reactions (qRT-PCRs) as early as the 3 somite stage, with the transcript exhibiting a progressive increase through the first 15 days postfertilization (dpf) (Migliarini & Carnevali, 2009). One exception to this trend was documented at the 25 somite stage, where a slight decrease in cnr1 transcript occurred. A whole-body western blot analysis using an antibody developed to detect rat CNR1 did not identify any protein product at the 3 somite, or 25 somite stages. A 63 kDa band was, however, detected at hatching time, 7 dpf, and 15 dpf. This band size was consistent with results from experiments that used the antibody to detect CNR1 in rat models. Special consideration has been given to the expression of cnr1 within the central nervous system. In situ hybridization (ISH) has detected cnr1 expression as early as 1 dpf where it is localized to the preoptic area. The expression spreads throughout the telencephalon, hypothalamus, tegmentum, and anterior hindbrain by 2 dpf (Lam et al., 2006, Nishio et al., 2012, Watson et al., 2008). Henceforth, the expression patterns remain similar into adulthood, and share homologies with known mammalian expression patterns (Lam et al., 2006). The 63 kDa band detected by western blots in whole-body lysates has also been detected in adult zebrafish brain homogenates using the same antibody (Migliarini & Carnevali, 2009, Piccinetti et al., 2010). Additionally, this antibody has been used to identify expression patterns within the retina of adult zebrafish via immunocytochemistry (Yazulla & Studholme, 2001). Outside of the central nervous system, cnr1 expression has been examined within the liver and ovary (Migliarini & Carnevali, 2008, Migliarini & Carnevali, 2009). cnr1 transcript expression has been detected in the liver of adult zebrafish (Migliarini & Carnevali, 2008). Fluorescent ISH identified transcript expression throughout the ovary, including within class III and class IV oocytes (Migliarini & Carnevali, 2009). A corresponding western blot analysis detected high levels of the protein in class III oocytes, and low levels in class IV oocytes. No protein product was observed in class V oocytes.
2.2. Cannabinoid Receptor 2 (cnr2)
cnr2 mRNA has been detected by RT-PCR in the adult zebrafish brain, intestine, retina, gills, heart, muscle, pituitary, and spleen (Rodriguez-Martin et al., 2007a). ISH also documented transcript expression in the pituitary gland. However, ISH did not corroborate the expression that had been detected in the brain. The reason for this discrepancy is unknown, and could be due to differences in the sensitivity of the methods employed or due to extremely punctate regional expression patterns.
2.3. Diacylglycerol Lipase, Alpha (dagla)
dagla transcript has been detected in the central nervous system of zebrafish by ISH at 2 dpf. Expression was detected in the telencephalon, hypothalamus, tegmentum, and hindbrain (Watson et al., 2008). This expression pattern was similar to that of cnr1 at this stage, with the exception that dagla transcript was detected throughout both the anterior and posterior hindbrain.
2.4. Fatty Acid Amide Hydrolase (faah)
To date, only one study has examined faah expression in zebrafish. In 2001, Yazulla et al. performed immunocytochemistry with an antibody designed for rat FAAH and detected protein expression in the adult zebrafish retina (Yazulla & Studholme, 2001).
2.5. Abhydrolase Domain Containing 4 (abhd4); Glycerophosphodiester Phosphodiesterase 1 (gde1); Fatty Acid Amide Hydrolase 2a (faah2a); & Monoglyceride Lipase (mgll)
Thisse, B. and Thisse, C. have uploaded data from ISH experiments investigating zebrafish abhd4, gde1, faah2a, and mgll expression patterns to The Zebrafish Model Organism Database available at zfin.org (Thisse et al., 2001). abhd4 expression patterns were assessed in developmental stages ranging from 1-cell to the pec-fin. No spatially restricted expression was observed, however, the authors noted that a low quality probe was used for this experiment. gde1 expression patterns were assessed in developmental stages ranging from 1-cell to the pec-fin, and labeling above the basal expression levels was observed in the yolk syncytial layer. No faah2a expression was observed at stages ranging from 50% epiboly through bud, from 1–4 somite(s) through 10–13 somites, from 14–19 somites, from 20–25 somites through prim-5, from prim-15 through prim-25, or from high-pec through long pec. faah2a expression was, however, observed in the intestinal bulb at 5 dpf. A low–medium quality probe was used for this experiment and it is important to note that the methodologies employed may not detect transcript in the notochord, or throughout the trunk and tail at 5 dpf. The authors evaluated mgll expression using a medium quality probe. mgll expression was observed in the enveloping layer and the periderm at stages ranging from 50% epiboly through bud. mgll expression was observed in the periderm at stages ranging from 1–4 somite(s) through 10–13 somites. mgll expression was observed in the periderm and pronephric duct at stages ranging from 14–19 somites, and from 20–25 somites through prim-5. mgll expression was observed in the diencephalon and pharynx at stages ranging from 20–25 somites through prim-5. mgll expression was observed in the diencephalon, forebrain hindbrain, tegmentum, telencephalon, and ventricular zone at stages ranging from high-pec through long-pec.
2.6. Abhydrolase Domain Containing 6a (abhd6a); Abhydrolase Domain Containing 6b (abhd6b); Abhydrolase Domain Containing 12 (abhd12); Diacylglycerol Lipase, Beta (daglb); & N-acyl Phosphatidylethanolamine Phospholipase D (napepld)
No expression data is currently available for any of these genes that are involved with CB signaling.
3. Zebrafish eCB Gene Function
In addition to the insights from phylogenetic and gene expression analyses, binding assays have confirmed the presence of receptors in the zebrafish brain that may interact with known ligands of mammalian CB receptors. A radiolabeled version of the sCB WIN55212-2 has been shown to bind to targets throughout the hypothalamus, optic tectum, and telencephalon in adult zebrafish brain slices (Connors et al., 2014). Similarly, binding assays have confirmed that the eCB AEA, and the sCBs HU-210, WIN55212-2, and CP55940 interact with receptors in adult brain homogenates (Rodriguez-Martin et al., 2007b). In addition to binding to cognate targets in the zebrafish brain, the CB receptor ligands were shown to activate G protein-coupled receptor signaling via [35S] GTPγS assays. Taken together, these data suggests that the zebrafish eCB system may serve similar functional roles to that of their mammalian counterparts. To this end, numerous experiments have complemented these studies by investigating the functional roles of the zebrafish eCB system (Table 1). Although these biological functions are unquestionably multidisciplinary in nature, for this discussion the corresponding results have been classified as pertaining to addiction, anxiety, development, energy homeostasis and food intake, immune system function, and learning and memory.
Table 1. A Summary of Zebrafish Endocannabinoid Gene Function Studies.
A comprehensive list of existing endocannabinoid gene function studies that have used the zebrafish model. Studies are sorted with regard to the specific gene function investigated, and may be listed more than once. The age of the zebrafish used in each study is provided, along with the pharmacological and genetic approaches that were used to study gene function. A summary of the key experimental endpoints used to assess gene function are included for each study. List of abbreviations: delta-9-tetrahydrocannabinol (Δ9-THC), delta-9-tetrahydrocannabivarin (Δ9-THCV), anandamide (AEA), cannabidiol (CBD), endocannabinoid (eCB), phytocannabinoid (pCB), synthetic cannabinoid (sCB), and not applicable (NA).
| eCB Gene Function Studies |
Zebra fish Ages |
Pharmacolo gical Approaches |
Genetic Approaches |
Key Experimental Endpoints |
|---|---|---|---|---|
|
| ||||
| Addiction | ||||
| Braida et al., 2007 | Adult | SR141716A (sCB) |
NA | Locomotor Activity, & Conditioned Place Preferences |
| Anxiety | ||||
| Barba-Escobedo & Gould, 2012 | Adult | WIN55212-2 (sCB) |
NA | Visual Social Choices |
| Connors et al., 2014 | Adult | WIN55212-2 (sCB) |
NA | Light-Dark Plus Maze Behaviors |
| Ruhl et al., 2014 | Adult | Δ9-THC (pCB) |
NA | Locomotor Activity, & Escape Responses |
| Stewart & Kalueff, 2014 | Adult | Δ9-THC (pCB) |
NA | Assorted Spatiotemporal Behavioral Parameters |
| Development | ||||
| Akhtar et al., 2013 | Juveni le |
Δ9-THC (pCB), AM251 (sCB), CP55940 (sCB), & WIN55212- 2 (sCB) |
NA | Morphology, & Mortality Locomotor Activity, & Visual Motor Responses |
| Migliarini & Carnevali, 2009 | Juveni le |
AM251 (sCB) | NA | Morphology, & Hatching Rates Locomotor Activity |
| Thomas, 1975 | Juveni le |
Δ9-THC (pCB) |
NA | Morphology, & Mortality Spontaneous Tail Muscle Twitches |
| Watson et al., 2008 | Juveni le |
NA |
cnr1 Knockdown (Whole Organism) |
Axonal Outgrowth, & Fasciculation |
|
Energy
Homeostasis & Food intake |
||||
| Migliarini & Carnevali, 2008 | Juveni le, & Adult |
AEA (eCB), & AM251 (sCB) |
NA | CB1, IGF1, IGF2, & SREBP mRNA Expression |
| Nishio et al., 2012 | Juveni le, |
SR141716A (sCB,) & WIN55212- 2 (sCB) |
cnr1 & cnr2 Knockdown (Whole Organism) |
CB1, & CART1–3 mRNA Expression Yolk Sac Size, & Lipid Metabolism |
| Pai et al., 2013 | Juveni le, & Adult |
AEA (eCB), & AM251 (sCB) |
cnr1 Overexpressio n (Hepatic) |
Lipotoxic, & Lipogenic Markers Lipid Metabolism, & Liver Morphology |
| Piccinetti et al., 2010 | Adult | NA | NA | CB1 mRNA, & Protein Expression Food Intake |
| Shimada et al., 2012 | Juveni le |
SR141716A (sCB,) |
cnr1 Knockdown (Whole Organism) |
Locomotor Activity, & Food Intake |
| Silvestri et al., 2015 | Juveni le |
Δ9-THCV (pCB), & CBD (pCB) |
NA | Lipid Metabolism |
|
Immune System
Function |
||||
| Liu et al., 2013 | Juveni le |
JWH-015 (sCB), & WIN55212- 2 (sCB) |
cnr2 Knockout (Whole Organism) |
Alox5, & JNK Signaling Leukocyte Inflammatory Migration |
|
Learning &
Memory |
||||
| Ruhl et al., 2014 | Adult | Δ9-THC (pCB) |
NA | Telencephalic Akt, & Erk Phosphorylation Associative, & Spatial Memory |
3.1. Addiction
Many exogenous CBs are addictive, and the use of these compounds may lead to the subsequent presentation of cannabis use disorder (Galanter et al., 2015). Moreover, numerous pharmacological and genetic studies have demonstrated that CB signaling affects the neurocircuitry that modulates the addictive properties of other drugs of abuse including amphetamine, cocaine, ethanol, heroin, morphine, and nicotine (Maldonado et al., 2006). Accordingly, CB use is not only associated with a risk of cannabis use disorder development but also influences the development of additional substance use disorders. In recent years zebrafish have emerged as a useful tool for modeling aspects of addiction, and for investigating the underlying mechanisms (Stewart et al., 2011). Although zebrafish have not yet been used to annotate the addictive properties of exogenous CBs themselves, a recent study used this organism to identify a potential role for CB signaling in mediating the behavioral effects of salvinorin A—a potent hallucinogen and kappa opioid receptor agonist found in the recreationally used plant Salvia divinorum (Braida et al., 2007). Salvinorin A treatment was shown to have biphasic effects on adult zebrafish swimming behavior, where locomotor activity was increased by 0.1 or 0.2 μg/kg doses and suppressed by 5 or 10 μg/kg doses. A biphasic response was also observed when the rewarding properties of this drug were assessed, where conditioned place preference was induced with 0.2 or 0.5 μg/kg doses and conditioned place aversion was induced with a 80 μg/kg dose. The response to low or high doses of salvinorin A was attenuated when fish were pretreated with a 1 mg/kg dose of the sCB SR141716A. Because SR141716A is a known cannabinoid receptor 1 antagonist, the authors postulated that CB receptor signaling might modulate the observed effects of salvinorin A via cross talk with kappa opioid receptors. The biphasic effects of salvinorin A on locomotor activity and condition place preference/aversion observed in zebrafish are consistent with the results of rodent studies (Braida et al., 2008). Moreover, SR141716A has been shown to similarly block the induction of conditioned place preference following salvinorin A administration in rodent models (Braida et al., 2008). However, it remains to be seen whether such interactions are indeed due to intracellular crosstalk, or some other mechanism. Both cannabinoid receptor 1 and kappa opioid receptor are believed to be linked to common signaling cascades including the protein kinase C signaling pathways, but there is relatively little overlap in the distribution of these receptors in the brain (Braida et al., 2007). Therefore the observed effects could be due to intracellular crosstalk in limited cellular populations with overlapping receptor distributions, or more indirectly through the modification of interconnected circuit activity associated with disparate receptor distributions. Additionally, there remains the possibility that the observed effects could be ascribed to off target drug effects. This is unlikely to take place at cannabinoid receptor 1 because several studies have demonstrated the salvinorin A has little or no affinity at this receptor (Walentiny et al., 2010). However, recent studies suggest that SR141716A may modulate the functioning of numerous opioid receptors including kappa opioid receptor (Seely et al., 2012, Zador et al., 2014, Zador et al., 2015). More studies are needed to clarify the specific mechanisms underlying the observed interactions between drugs that target these neurotransmitter systems.
3.2. Anxiety
Besides substance use disorders, disrupted CB signaling has been associated with features of other complex neuropsychiatric disorders such as anxiety (Kedzior & Laeber, 2014). Typically exposure to a low dose of a CB receptor agonist induces anxiolytic behavioral responses, while exposure to a high dose triggers anxiogenic behavioral responses (Viveros et al., 2005). The effects of CB signaling on anxiety-associated behavior do depend on the specific CB tested, however, and a myriad of additional factors such as differences in drug administration routes, environments, or basal anxiety levels (Patel & Hillard, 2006, Viveros et al., 2005). Recently, several groups have adapted behavioral paradigms using adult zebrafish to compliment and advance the knowledge gained from existing rodent studies regarding the effects of CB signaling in modulating anxiety-associated behaviors. In 2014, Stewart and Kalueff established a novel tank test and tracked several spatiotemporal behavioral parameters of zebrafish locomotion following a 20 min treatment with either a 30 mg/L or 50 mg/L dose of the pCB Δ9-THC (Stewart & Kalueff, 2014). The 30 mg/L dose reduced the total distance traveled and the velocity of the fish, and increased the duration of slow movement relative to controls. Both concentrations decreased the transitions to and time spent in the upper half of the tank, and increased the latency to enter the upper half of the tank. The attenuation of swimming in the upper half of the tank suggests that the tested doses of Δ9-THC may exert anxiogenic behavioral effects in zebrafish, in addition to the observed reduction of locomotor activity. In contrast, Ruhl et al. did not observe any anxiogenic responses from zebrafish treated with 100 nM Δ9-THC in an escape response chamber test (Ruhl et al., 2014). Although this discrepancy could be due to the different behavioral paradigms used in these two experiments, it more is likely due to differences in the selected doses that are approximately an order of magnitude apart. This notion is in agreement with several mammalian studies where low doses of Δ9-THC have been shown to elicit either no behavioral effects or an anxiolytic response, while high doses of Δ9-THC have been shown to trigger anxiogenic responses and reductions in locomotor activity (Viveros et al., 2005).
The effects of the sCB WIN55212-2 on anxiety-associated behavior have also received consideration using the zebrafish model (Barba-Escobedo & Gould, 2012). Visual choice tests similar to rodent three-chamber sociability paradigms were used to investigate the potential anxiolytic effects of cannabinoid signaling in relation to shoaling behavior. Exposure to 1 mg/L WIN55212-2 was shown to increase shoaling behavior, thereby demonstrating anxiolytic effects in this visual-social interaction test. Subsequently, Connors et al. investigated the potential anxiolytic effects of WIN55212-2 on zebrafish behavior using an aquatic light-dark plus maze test (Connors et al., 2014). In this paradigm, dietary exposure to 1 μg/day WIN55212-2 significantly decreased the latency to enter the light arms and increased the time spent dwelling in them. These results are consistent with previous findings that demonstrated anxiolytic effects of WIN55212-2 administration in the rodent elevated-plus maze paradigm (Naderi et al., 2008, Patel & Hillard, 2006). However, in contrast to the findings from the zebrafish visual-choice test, a 0.1 mg/kg dose of WIN55212-2 did not increase sociability in rodents (Gould et al., 2012). The cause of this incongruity remains to be seen and could be due to a host of the previously mentioned factors, or organismal differences. Nonetheless it is worth noting that discrepancies are commonly observed in this area of research, and that these studies collectively indicate that CB signaling in zebrafish may similarly function to modulate anxiety-associated behavior.
3.3. Development
A functional eCB system is detected in the early stages of embryonic neural development, and has been shown to mediate components of it including axon guidance and synaptogenesis (Gomez et al., 2008). Disruptions of the molecular constituents in the eCB system during key developmental time points, such as those due to exogenous CB exposure, have been implicated in a number of adverse health consequences. Altered CB signaling during pregnancy is correlated with impaired embryonic implantations, premature births, and low birth weights (Park et al., 2004). Moreover, exposure to exogenous CBs during adolescent development has been associated with lasting cognitive impairments and an increased risk for developing psychoses (Malone et al., 2010, Meier et al., 2012). Although these studies have provided valuable insights to the role of the eCB system during development and risks of CB exposure at this time, there remains a dearth of evidence establishing causative links between developmental CB exposure and long-term toxicological effects. Given that CB signaling has acute effects on pulmonary, cardiovascular, gastrointestinal, and nervous system functioning, it is imperative to further discern the roles of CB signaling in the context of development (Gurney et al., 2014). The role of CB signaling in zebrafish development was first documented in a report published in 1975 that investigated the effects of 1 ppm – 10 ppm Δ9-THC exposure during embryogenesis, beginning at the late high blastula stage (Thomas, 1975). The administration of this pCB at a dose of 1 ppm did not result in any toxicological or teratological effects at any stage throughout 9 days post-hatching. Conversely, at a dose of 2 ppm, morphological defects were observed in the distal trunk. Exposure to 5 ppm or 10 ppm Δ9-THC led to a reduction of spontaneous tail twitches and death after 24 h posttreatment. Following this study, another group documented the developmental defects resulting from exposure to the sCB AM251 during embryogenesis (Migliarini & Carnevali, 2009). In this experiment, no morphological defects were observed following up to 96 h of chronic exposure to either 10 nM or 20 nM AM251. Both doses, however, significantly reduced hatching rates at 72 h posttreatment and swimming rates at 96 h posttreatment. More recently, Akhtar et al. annotated the developmental effects of exposure to the pCB Δ9-THC, and the sCBs CP55940 and WIN55212-2 (Akhtar et al., 2013). In this study, the authors first characterized the LC50 and morphological defects associated with several CBs at 5 dpf, following 4 dpf of drug exposure. The LC50 identified for each drug tested was: 0.01 mM (Δ9-THC), 0.049 mM (CP55940), and 0.003 mM (WIN55212-2). Although no morphological defects were found following exposure to any of the tested concentrations of WIN55212-2 or CP55940, Δ9-THC treatment caused pericardial edema, yolk sac edema, and a curvature of the rostro-caudal axis at doses of 0.3 mg/L or higher.
Subsequently, the effects of acute and chronic CB exposure on the visual motor response to a dark challenge were examined in 5 dpf zebrafish (Akhtar et al., 2013). A biphasic response was observed where low doses of CBs potentiated the distance moved in response to the dark challenge and high doses attenuated it following 1 h, 4 h, or 12 h of treatment. In contrast to the acute effects of CB exposure, a chronic 96 h treatment had little effect on the response to the dark challenge at 5 dpf. The observed acute effects were rescued with the co-administration of AM251, which could attenuate the locomotor response to the dark challenge when administered by alone either acutely or chronically. The suppressive effect of acute AM251 treatment on locomotor activity of developing zebrafish is consistent with the results of rodent experiments (Sink et al., 2010). Similarly, the biphasic effects of acute Δ9-THC, WIN55212-2, or CP55940 treatment on the locomotor activity are analogous to the effects that have been observed following their administration in adult zebrafish and rodent models. However, while chronic treatments with Δ9-THC, WIN55212-2, or CP55940 were shown to cause no effect on the locomotor response of 5 dpf zebrafish, the results of rodent studies suggest that the effects of chronic CB treatment during development are complicated to interpret. Factors such as the specific CB used, the CB doses tested, the development time point when CBs were administered, the developmental time point when behavioral tests were performed, the specific behavioral test used, and a multitude of additional influences may alter the specific outcome (Schneider, 2009). Therefore, although these compounds exerted no effect in the experiment by Akhtar et al., there is reason to believe that additional experimentation could reveal significant effects if changes in the experimental design were implemented. It is also important to note that the teratogenic effects of CB exposure observed in the zebrafish studies, including alterations in gross morphology or mortality, are inconsistent with the data from epidemiologic studies. Although teratogenic effects have been observed in studies with other model organisms, these effects are typically achieved using doses of CBs that are significantly higher than the doses used by humans for recreational purposes (Park et al., 2004). Nonetheless, more epidemiological studies may be warranted given the increasing prevalence of cannabis containing elevated concentrations of CBs, the rising popularity of alternate CB administration routes such as methods that utilize extracts, and the growing use of potent sCBs.
In addition to studies that used pharmacological approaches, one group used morpholino-mediated gene knockdown to investigate the role of CB signaling during zebrafish development (Watson et al., 2008). Injections of antisense morpholinos directed against two sequences in cnr1 were shown to cause abnormal axonal outgrowth and fasciculation. Neurofilament staining of fish at the prim-12 stage revealed that an injection with a morpholino directed against a cnr1 sequence beginning at the start codon disrupted axonal track formation in the anterior commissure, while an injection with a morpholino directed against a downstream cnr1 sequence disrupted fasciculation in both the anterior and posterior commissure. At 72 h, fish injected with either morpholino demonstrated similar defects in neural populations including the Mauthner neurons, the reticulospinal neurons, and the tracks of neurons comprising the medial longitudinal fasciculi. The observed results are consistent with rodent studies where Cnr1 mutants have been shown to exhibit disrupted fasciculation in a number of tracts in the developing brain (Mulder et al., 2008, Wu et al., 2010). It is possible that such disruptions could underlie the persistent behavioral deficiencies that are associated with prenatal CB exposure.
3.4. Energy Homeostasis & Food Intake
CB signaling is known to play a crucial role in regulating energy homeostasis and food intake centrally via the nervous system, as well as through peripheral tissues (Silvestri & Di Marzo, 2013). Zebrafish have been used to provide novel insights regarding the regulatory role of CB signaling at both of these levels. In 2008, Migliarini et al. showed that zebrafish treated for 2 h with a bath application of the eCB 10 nM AEA had increased whole-body cnr1 transcript levels at hatching time, as well as at 7 dpf and 15 dpf (Migliarini & Carnevali, 2008). These responses were attenuated using a 2 h pretreatment with a 10nM dose of the sCB AM251. The increase in whole-body cnr1 transcript levels following 2 h 10 nM AEA treatment was accompanied by increased expression of several genes implicated in growth and lipid metabolism including sterol regulatory element binding protein, insulin-like growth factor 1, and insulin-like growth factor 2. Similarly, a dose dependent increase in expression of these genes was observed in the liver of adult zebrafish following a 2 h treatment with either 10 nM or 100 nM AEA. This suggests that CB signaling in the peripheral tissues of zebrafish has a role in metabolic-regulation that is conserved with rodents (Osei-Hyiaman et al., 2005b). To further examine this potential role, a transgenic line was recently developed to study the effects of hepatic cnr1 overexpression (Pai et al., 2013). Overexpression of cnr1 in the liver resulted in elevated hepatic expression of SREBP-1c, a transcription factor involved in lipogenesis, as well as a host of genes involved with the synthesis, transport, and storage of fatty acids. Oil Red O staining revealed that this overexpression leads to the accumulation of lipids in the liver, and hepatic steatosis in 6 dpf zebrafish. A 72 h systemic administration of 10 nM AEA resulted in lipid accumulation and hepatic steatosis in control groups that did not overexpress cnr1 by 6 dpf, while 25 nM AM251 treatment rescued these phenotypes in the cnr1 overexpressing fish. The pathological phenotypes resulting from hepatic cnr1 overexpression in juveniles were also present in adult zebrafish, and eventually lead to hepatic hypoplasia and lipotoxicity. These findings are in agreement with experiments using rodent models, which implicated cannabinoid receptor 1 in the modulation of obesity-associated liver pathology (Gary-Bobo et al., 2007). Moreover, Cnr1 knockout mice are protected from the dyslipidemia and steatosis induced by high-fat diets (Osei-Hyiaman et al., 2005b).
In addition to the studies that focused on the peripheral metabolic roles of CB signaling, considerable attention has been focused on the putative central roles as well. The systemic administration of the sCB SR141716A caused an increase in yolk sac size at several stages of development, suggesting that CB signaling modulates metabolism because the yolk sac is the principle source of energy for embryonic and larval zebrafish (Jones et al., 2008, Nishio et al., 2012). This effect could be rescued with the injection of a morpholino directed against cnr1, but not with an injection of a morpholino directed against cnr2 (Nishio et al., 2012). Oil Red O staining indicated that the increase yolk sac size was accompanied by decreased lipid accumulation in the telencephalon and around the eye of fish treated with SR141716A. In contrast, this fat accumulation was increased in fish treated with a 2 μM dose of the sCB WIN55212-2. To gain insights regarding the potential mechanisms of these effects the authors investigated the expression of cocaine- and amphetamine- related transcript (cart) genes, which are potential modifiers of CB signaling, in the brain of 2 dpf zebrafish. cnr1 and cart3 were shown to be coexpressed in both the hypothalamus and the anterior hindbrain—two brain structures implicated in appetite regulation. The expression of cart genes were shown to be dependent on CB signaling because the expression of these genes were disrupted following injection of a morpholino directed against cnr1, with the cart3 transcript exhibiting a significant down regulation in 2 dpf fish. Subsequently, the authors investigated the effects of fasting on the expression of cart3 and observed a down regulation of its expression in the brains of both juvenile, and adult zebrafish. Interestingly, this down regulation was only observed in the brain regions where cart3 was coexpressed with cnr1, and could be reversed with the administration of SR141716A. Collectively, these data suggests that zebrafish cnr1 has a central role in relaying information concerning food intake, and does so in a cart3 dependent manner. It is important to note that the names for zebrafish cart genes have changed since the publication of these findings. That aside, these findings serve to advance the hypothesis that cocaine- and amphetamine- associated transcript genes may be downstream targets of CB signaling that mediate the regulation of food intake (Cota et al., 2003, Osei-Hyiaman et al., 2005a).
In addition to directly studying potential relationships of central Cnr1 signaling and food intake, the role of hormones involved in the maintenance of anorexic and orexic signaling has also been researched in this context (Piccinetti et al., 2010). It was demonstrated that adult zebrafish treated with a 100 nM or 1 μM dose of melatonin had decreased food intake relative to controls. The decreased food intake correlated with altered transcript levels in the brain of genes involved with anorexigenic and orexigenic signaling, including cnr1. Western blotting revealed that the decreased cnr1 transcript coincided with decreased levels of protein in the brain, providing further evidence that CB signaling plays a role in metabolic regulation at the central level. The role of CB signaling in regulating larval zebrafish food intake has been further tested using a novel feeding assay with fluorescently-labeled paramecium (Shimada et al., 2012). An injection with a morpholino directed against cnr1 was shown to decrease both food intake, and locomotor activity in this behavioral assay. These effects were also observed in a dose dependent manner following the systemic administration of 0.1 μM 0.3 μM or 1 μM SR141716A, providing further evidence that Cnr1 signaling regulates appetite. This notion is consistent with previous findings where the mutation of Cnr1 or the administration of SR141716A was shown to reduce food intake mice (Wiley et al., 2005). More recently, Silvestri et al. examined the effects of systemic pCB administration on yolk sac metabolism in 3 dpf zebrafish (Silvestri et al., 2015). AdipoRed staining revealed that a 5 μM dose of cannabidiol (CBD) increased lipolysis at 24 h, 48 h, and 72 h posttreatment. Likewise, a 5 μM dose of delta-9-tetrahydrocannabivarin (Δ9-THCV) increased yolk sac lipid mobilization at 48 h, and 72 h posttreatment. Although it remains to be seen whether these effects are due to peripheral or central actions, they are also consistent with the results of rodent studies where these compounds have been shown to decrease hepatic triglyceride levels (Silvestri et al., 2015, Wargent et al., 2013).
3.5. Immune System Function
While cannabinoid receptor 1 is the most highly expressed CB receptor in the central nervous system, cannabinoid receptor 2 has been shown to have 10 – 100 fold higher expression levels in hematopoietic stem cell-derived constituents of the immune system (Galiegue et al., 1995). Accordingly, there is significant interest in understanding how cannabinoid receptor 2 regulates immune system functioning during disease-relevant processes such as inflammatory migration. In 2013 Liu et al. performed a chemical screen with juvenile transgenic zebrafish and observed that the administration of WIN55212-2, a cannabinoid receptor 1 and cannabinoid receptor 2 agonist, inhibited the migration of fluorescently labeled leukocytes to the site of a tail wound in a time-dependent manner (Liu et al., 2013). Similarly to other model systems the cnr2 transcript levels were shown to be over 10-fold higher than cnr1 transcript levels in the leukocytes, and the administration of the cannabinoid receptor 2 agonist JWH-015 inhibited the leukocyte migration in a dose- and time-dependent manner. These effects of JWH-015 were not observed in cnr2 −/− fish, and untreated mutants exhibited an increase in leukocyte migration to the post-injury tail wound site relative to the wild-type controls—an effect that was rescued with the injection of a morpholino directed against alox5. Subsequently it was demonstrated that tail injury resulted in increased phosphorylation of JNK and elevated whole-body transcript levels of alox5. These responses were also attenuated with the administration of JWH-015. When a transgenic line with fluorescently labeled leukocytes was crossed to a line expressing constitutively active JNK in leukocytes, the decreased leukocyte migration to the tail wound following JWH-015 treatment was rescued. Following these observations, the authors investigated the potential mechanisms that linked cnr2 activity to alox5 transcription in the injury model. After identifying three AP-1 sites upstream of the alox5 transcriptional start site, an embryo-chromatin immunoprecipitation assay was used to reveal that phosphorylated c-Jun bound to two of the AP-1 sites in response to tail-injury. This binding was blocked with the administration of either JWH-015, or the JNK inhibitor SP600125. Furthermore, it was demonstrated that constitutive JNK upregulated alox5 expression in leukocytes, and could partially rescue the JWH-015 induced reduction in alox5 transcript in leukocytes following tail injury. Taken together, these data suggests that cannabinoid receptor 2 activation inhibits leukocyte migration to a wound site by suppressing the JNK-mediated activation of c-Jun, thereby decreasing alox5 expression. This model is consistent with the results of previous studies that have demonstrated a role for both cannabinoid receptor 2 and arachidonate 5-lipoxygenase in modulating leukocyte migration (Csoka et al., 2009, Newton et al., 1994, Tschop et al., 2009). Moreover, the results of several independent studies have suggested that cannabinoid receptor 2 activation may inhibit the phosphorylation of JNK, and that interactions may occur between cannabinoid receptor 2 and arachidonate 5-lipoxygenase (Klegeris et al., 2003, Massi et al., 2008, Olea-Herrero et al., 2009). Using several genetically modified zebrafish models Liu et al. have expanded upon these studies by establishing several direct links in this putative intracellular signaling cascade, thereby unveiling a novel mechanism for CB signaling in regulating the inflammatory response to injury.
3.6. Learning & Memory
Several lines of evidence have indicated that the lateral and medial regions of the zebrafish dorsal pallium are functionally homologous to the mammalian hippocampus and amygdala respectively, and similarly express CB receptors (Lam et al., 2006, Mueller et al., 2011). The mammalian hippocampus and teleostian lateral dorsal pallium serve a critical role in spatial information memory processing, while the mammalian amygdala and teleostian medial dorsal pallium are more heavily involved with emotional associative memory (Broglio et al., 2005). In 2014 Ruhl et al. annotated the effects of Δ9-THC on intracellular signaling in the pallium of adult zebrafish, and the associated effects on memory function (Ruhl et al., 2014). A 1 h bath application of 100 nM Δ9-THC increased the amount of phosphorylated Erk in the telencephalon, but did not have a significant effect on the levels of phosphorylated Akt. The observed effect on phosphorylated Erk was shown to be specific to the lateral pallium rather than the medial pallium, suggesting that the treatment may result in corresponding disruptions in spatial memory. To test this the authors used a behavioral paradigm where fish learned to navigate an aquatic arena using an ego-allocentric strategy in order to locate a hidden food reward. The administration of 100nM Δ9-THC on day 19 of the training protocol significantly increased the distance traveled and the latency to find the hidden food, relative to several days in the protocol before and after day 19 when no drug was administered. The increased distance traveled and latency was similar to, and not significantly different from, earlier time points in the protocol that may have involved memory acquisition and consolidation. In contrast, no effects were observed on associative memory following the administration of 100nM Δ9-THC in a color discrimination behavioral paradigm. Higher doses of Δ9-THC, including 300 nM and 3 μM were also tested but resulted in impaired locomotor activity. These results are similar to rodent studies where no impairments were observed in analogous tests of associate memory following the acute administration of Δ9-THC at a dose that did not impair locomotor activity (Jentsch et al., 1997, Mishima et al., 2001). Discrepancies have, however, been observed in rodent tests of spatial memory. Acute doses of Δ9-THC were shown to impair performance a radial arm maze, but not in the water maze (Mishima et al., 2001). Ruhl et al. speculate that these differences could be a result of the motivation behind the behaviors in each paradigm, which are a positive reward in the radial arm maze and a negative reward in the water maze (Ruhl et al., 2014). The positive, food reward used in the radial arm maze is similar to the food reward used in the aquatic arena, which suggests that the eCB system may have similar functions in the learning and memory processes in teleostian brain structures.
4. Toxicological & Therapeutic Considerations
Phylogenetic analyses of the zebrafish eCB system have demonstrated that it is highly conserved with the mammalian counterpart, and ontogenetic analyses have revealed that eCB gene expression begins early in zebrafish development. In addition to the conservation of eCB genes themselves, many of the zebrafish eCB gene expression patterns also appear to be homologous. For instance, cnr1 expression in the zebrafish brain exhibits homologies with rodent distribution patterns as early as 2 dpf, which are maintained into adulthood (Lam et al., 2006). The expression of cnr2 in peripheral tissues of zebrafish is expected, and the discrepancies regarding expression in the brain have similarly occurred in rodent studies (Onaivi et al., 2008b, Rodriguez-Martin et al., 2007a). The expression of dagla, a key gene involved with the biosynthesis of the eCB 2-AG, is detected in the 2 dpf zebrafish brain and has a significant overlap with cnr1 expression (Watson et al., 2008). Similarly, in rodent models Dagla and Cnr1 are expressed in the brain during development and have overlapping expression patterns, which are also present in adulthood (Ahn et al., 2008). In contrast, Daglb also has high levels of neural expression early in development but its expression in the brain decreases in later stages, suggesting that a development switch occurs between these two diacylglycerols involved with 2-AG biosynthesis (Ahn et al., 2008). Whether or not similar transitions in expression exist during the ontogeny of the zebrafish eCB system remains to be seen. It is important to note that while some genes are conserved and do seem to display homologous expression patterns, some expression patterns may deviate due to differences that exist between organisms. For instance, rodent models have a gene encoding N-acylethanolamine acid amidase but do not have a gene encoding fatty acid amide hydrolase 2 (Tai et al., 2012, Wei et al., 2006). Zebrafish do not have a gene encoding N-acylethanolamine acid amidase but do feature a gene encoding fatty acid amide hydrolase 2, although the gene was affected by a duplication event resulting in genes designated as faah2a and faah2b (Mcpartland et al., 2007). In contrast, humans have genes encoding both fatty acid amide hydrolase 2 and N-acylethanolamine acid amidase (Klee et al., 2012). In addition to the inconsistencies that exist with regard to gene conservation, the fundamental physiological differences between fish, rodents and humans are also likely to account for incongruities in eCB gene expression and function between these organisms. Thus it is critical that investigations of eCB gene function use an array of preclinical models to ensure that a holistic understanding of the eCB system emerges. As previously discussed, zebrafish have already been used to study CB biology in diverse contexts including addiction, anxiety, development, energy homeostasis and food intake, immune system function, and learning and memory. Herein we contemplate how future studies may continue to capitalize on the unique advantages of the zebrafish model to further advance our understanding of the toxicological and therapeutic potential of the eCB system.
One useful application of zebrafish is pharmacological screening in a whole vertebrate system, which is ideal for studying more complex pharmacological dynamics not assessed in in vitro models such as the effects of drug bioavailability and metabolites in a multiorgan system. Zebrafish are able to absorb small molecules from the surrounding water at all stages of development and the fecundity of adult mating pairs, which can produce clutches of several hundred offspring per week, readily enables high-throughput screening in juvenile fish. Adult zebrafish themselves also provide cost effective model for molecular screening due to their size and the three dimensional nature of their housing, which allows 10 or more adult fish to survive in the same amount space required for one mouse. These features are particularly relevant to CB biology because the development, and identification of pharmacological modifiers of the eCB system has outpaced our ability to characterize the physiological and behavioral effects of these compounds in any meaningful way. To date, dozens of pCBs have been identified in Cannabis sativa and hundreds of novel sCBs have been developed. An expanding list of putative eCBs has also emerged following the initial discoveries of AEA and 2-AG. The understanding of how these numerous compounds affect an organism has been obscured by the fact that exposure often involves various combinations and concentrations of CBs, differences in frequencies and durations of CB use, and administrations of CBs under different physiological conditions. The advantages of the zebrafish model could be readily employed in order to provide insights about the effects resulting from the countless permutations of exogenous CB treatment conditions.
With regard to the toxicological effects of exogenous CBs, zebrafish could be used to annotate the potential deleterious effects of the pCBs contained in cannabis, or the plethora of recreationally used sCBs. Juvenile zebrafish could be used to rapidly screen for drastic effects such as alterations in gross morphology or mortality, or to investigate subtle tissue-specific effects such as alterations in cellular populations or functions. Juvenile zebrafish are also frequently used to study behavioral endpoints associated with altered physiology, providing another powerful tool for assessing the potentially harmful effects of CB exposure. Such paradigms could be adapted to model the effects CB exposure on simple behavioral states such as basal locomotor activity, or on more complex behavioral states including responses associated with anxiety or depression. In addition to testing the effects of individual CBs or combinations of CBs, the high-throughput zebrafish model will allow for an investigation of how these compounds interact with other pharmacological modifiers often administered in combination with CBs, including drugs of abuse such as ethanol or nicotine. Any observed alterations in physiology or behavior could then be linked to specific molecular signaling cascades, ideally with the use of comprehensive analyses such as RNA sequencing. Taken together, this approach provides critical information about existing CBs, and would provide a powerful tool for quickly predicting the potential threat of newly identified or developed CBs based on the unique molecular, physiological, and behavioral signatures.
Conversely, zebrafish could be also used to identify novel therapeutic strategies involving CB signaling. Due to the diverse scope of physiological processes that involve eCB signaling, the potential therapeutic applications of modified eCB signaling are just as diverse as the potential toxicological effects. Exogenous CBs could be screened using disease models to identify compounds that corrected the pathological phenotypes of interest. Similarly to toxicological applications, juvenile zebrafish would enable a rapid assessment of the potential therapeutic effects in both physiological and behavioral paradigms. Such compounds would likely include CBs that preferentially bind to receptors other than cannabinoid receptor 1 or that target enzymes involved with eCB metabolism, and that do not have the addictive properties or psychoactive effects characteristic of recreationally used CBs. Moreover, this model organism would permit for high-throughput screening of compounds that could attenuate or prevent any deleterious effects of CB use including behaviors associated with cannabis use disorder, which currently has no approved pharmacological treatment options. Although juvenile zebrafish are excellent models for high-throughput pharmacological analyses, the fact that development is still occurring may causes discrepancies from experiments performed in their adult counterparts. Therefore, the results of any studies performed early in the zebrafish lifecycle that do not pertain to development itself should be corroborated in adult models. Additionally, features unique to the adult zebrafish model, including the more sophisticated repertoire of behavioral assays, could be leveraged to provide further insights about pharmacological manipulations of the eCB system.
Although it is undoubtedly critical to study the effects of exogenous CB exposure, there is still a need to advance the understanding of the endogenous system that is targeted by these compounds. Elucidating the contributions of individual eCB genes to physiological and behavioral functions will help enable a mechanistic understanding of disease states, and a more targeted approached to the development of corresponding therapeutics. However, one problem that has hampered our understanding of the eCB gene function is the promiscuous signaling dynamics that are characteristic of this system. In addition to cannabinoid receptor 1 and cannabinoid receptor 2, eCBs bind to many non-CB receptors such as transient receptor potential cation channels, peroxisome proliferator-activated receptors, and numerous orphan G protein-coupled receptors. Together, these receptors comprise a complex multipartite signaling system where a host of ligands bind to multiple receptors with differential affinities (Alexander & Kendall, 2007). Furthermore, there are multiple enzymatic pathways that are responsible for the anabolism and catabolism of individual eCBs. Accordingly, eCB signaling in a given region of interest is not only contingent on the array of cognate receptors, but also the available ligand pool that results from the specific profile of enzymes involved with eCB metabolism in that area (Ahn et al., 2008, Di Marzo, 2011). These promiscuous signaling dynamics are also conserved in the exogenous pCBs and sCBs that target this system, and therefore limit the insights about eCB gene function that can be gained from pharmacological manipulations of this system. Consequently, a targeted manipulation of the genome itself is required in order to provide definitive insights about individual eCB gene functions.
Zebrafish are ideal for such quantitative trait analyses, in part, because they are both high-throughput and genetically amenable. In recent years, developments in genome editing techniques have enabled precise, and highly efficient modification of the zebrafish genome (Blackburn et al., 2013). Custom nuclease technology such as transcription activator-like effector nucleases (TALENs) and clustered regularly interspersed short palindromic repeat/Cas9 (CRISPR/Cas9) may be used for targeted mutagenesis of loci via non-homologous end joining, or the incorporation of new sequences using small single stranded oligonucleotide donors for homology directed repair (Blackburn et al., 2013, Campbell et al., 2013). This technology has provided an accessible methodology for studying the roles of individual genes, or variations in genes, in the zebrafish model. With regard eCB biology, establishing zebrafish lines with mutations in individual genes will be indispensable for studying their function in this organism. These lines would allow researchers to definitively establish functional roles for a given gene, which may have been previously obscured by the potential off-target effects associated with pharmacological investigations. Moreover, the high-throughput nature of zebrafish allows for a whole panel of mutant lines to be rapidly tested in parallel, providing a consistent and comprehensive assessment of gene functions in any given context of interest. Additionally, genome engineering technology will enable conditional and tissue-specific investigations of eCB gene function because the ability to integrate small single stranded oligonucleotides through homology directed repair has provided a means for the site-specific introduction of sequences used in recombinase technology, such as loxP sites (Bedell et al., 2012). In addition to studies related to recombinase technology, specific sequence modifications could be used to investigate a host of other aspects of CB biology including the development of single nucleotide polymorphism models and lines featuring epitope tagged genes products.
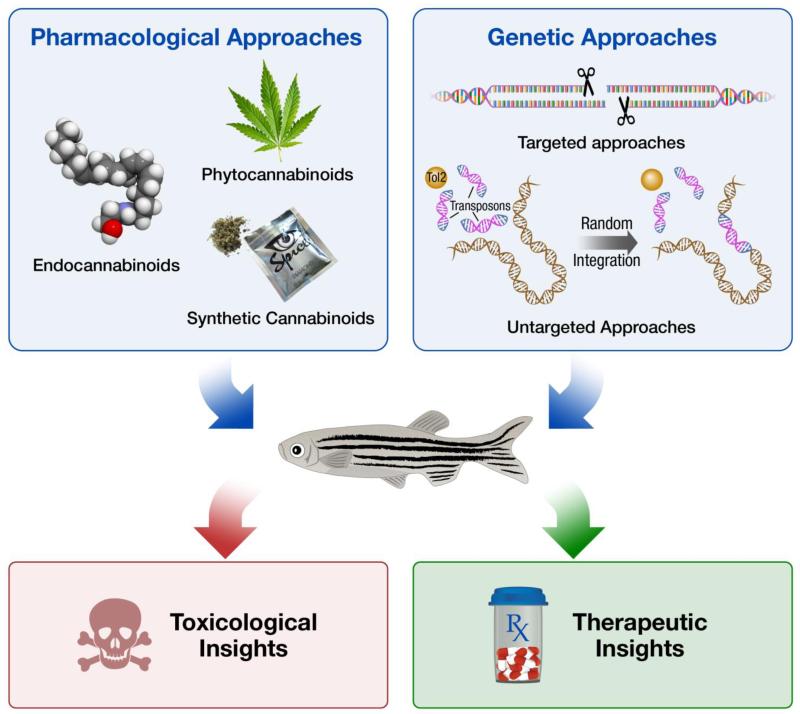
Although zebrafish can significantly advance the understanding of CB biology with the use of pharmacological or genome editing approaches, their greatest contributions to the field will likely result from studies enabled through combined approaches (Figure 1). The genetic amenability of zebrafish have allowed for the development of numerous transgenic lines that enable the visualization of everything from axon patterning to stress response dynamics (Higashijima et al., 2000, Krug et al., 2014). The transparent embryos and larvae obtained from such lines could provide an invaluable tool for screening the effects of CB exposure in a living organism, which may be either toxicological or therapeutic depending on the specific paradigm employed. Similar screens could be performed in models created with custom nuclease technology, including loss of function mutants or single nucleotide polymorphism lines. The development of assays with such preclinical models could be used to advance individualized medicine by enabling the identification of novel pharmacological interventions that are optimized for a given genotype. In addition to reverse genetic approaches, paradigms could be developed around the response to specific CBs and used for forward genetic applications. Extensive libraries of mutants, such as gene-break transposon lines could be rapidly screened in these paradigms to identify novel genetic modifiers of CB signaling (Clark et al., 2011). Additionally, lines with fluorescently tagged gene products could be used for studies of gene expression dynamics in a living organism. Ultimately, it is the combination of pharmacological and genetic approaches that is necessary to advance the understanding of potential toxicological effects of disrupted eCB signaling, and to develop therapeutic strategies that involve this system. Accordingly, it is quite apparent that the zebrafish model is poised to make a splash in the burgeoning field of CB research.
Figure 1. Strategies for Providing Toxicological, & Therapeutic Insights About Cannabinoid Biology Using Zebrafish.
Cannabinoid biology may be elucidated in the zebrafish using a tandem of pharmacological and genetic approaches. Pharmacological approaches should include investigations of endocannabinoids, phytocannabinoids, and synthetic cannabinoids. Genetic approaches should include the use of targeted genome editing approaches such as a custom nuclease technology, as well as untargeted genome editing approaches such as transposon systems. The use of pharmacological and genetic approaches with the zebrafish model will reveal novel toxicological and therapeutic insights about cannabinoid biology.
Highlights.
An overview of fundamental cannabinoid biology is provided.
Endocannabinoid gene expression and function studies using zebrafish are reviewed.
The results of zebrafish and rodent cannabinoid studies are integrated.
The use of zebrafish to advance the field of cannabinoid biology is discussed.
The toxicological and therapeutic potentials of cannabinoid biology are examined.
Acknowledgments
The National Institutes of Health (Grant: DA032194), Mayo Graduate School, and Mayo Foundation for Medical Education and Research provided funding for our work. The Mayo Clinic Division of Media Support Services assisted with the development of Figure 1. The National Institutes of Health (Grant: RR15402-01) provided funding for the work referenced in this manuscript that has been published on The Zebrafish Model Organism Database available at zfin.org (Thisse et al., 2001).
Abbreviations
- Δ9-THC
delta-9-tetrahydrocannabinol
- Δ9-THCV
delta-9-tetrahydrocannabivarin
- μ
micro
- 2-AG
2-arachidonoylglycerol
- abhd4
abhydrolase domain containing 4 (zebrafish gene)
- abhd6a
abhydrolase domain containing 6a (zebrafish gene)
- abhd6b
abhydrolase domain containing 6b (zebrafish gene)
- abhd12
abhydrolase domain containing 12 (zebrafish gene)
- AEA
anandamide
- alox5
arachidonate 5-lipoxygenase (zebrafish gene)
- cart
cocaine- and amphetamine- related transcript (zebrafish genes)
- CBD
cannabidiol
- CB
cannabinoid
- Cnr1
cannabinoid receptor 1 (rodent gene)
- CNR1
cannabinoid receptor 1 (rodent protein)
- cnr1
cannabinoid receptor 1 (zebrafish gene)
- Cnr1
cannabinoid receptor 1 (zebrafish protein)
- cnr2
cannabinoid receptor 2 (zebrafish gene)
- CRISPR/Cas9
clustered regularly interspersed short palindromic repeat/Cas9
- Dagla
diacylglycerol lipase, alpha (rodent gene)
- dagla
diacylglycerol lipase, alpha (zebrafish gene)
- Daglb
diacylglycerol lipase, beta (rodent gene)
- daglb
diacylglycerol lipase, beta (zebrafish gene)
- dpf
days postfertilization
- eCB
endocannabinoid
- FAAH
fatty acid amide hydrolase (rodent protein)
- faah
fatty acid amide hydrolase (zebrafish gene)
- faah2a
fatty acid amide hydrolase 2a (zebrafish gene)
- faah2b
fatty acid amide hydrolase 2b (zebrafish gene)
- g
grams
- gde1
glycerophosphodiester phosphodiesterase 1 (zebrafish gene)
- h
hours
- ISH
in situ hybridization
- k
kilo
- M
molar
- L
liters
- LC50
lethal concentration 50
- m
milli
- min
minutes
- mgll
monoglyceride lipase (zebrafish gene)
- n
nano
- napepld
n-acyl phosphatidylethanolamine phospholipase d (zebrafish gene)
- pCB
phytocannabinoid
- ppm
parts per million
- qRT-PCR
quantitative real-time reverse transcriptase polymerase chain reaction
- RNA
ribonucleic acid
- sCB
synthetic cannabinoid
- TALEN
transcription activator-like effector nuclease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- Abood ME, Sorensen RG, Stella N. Endocannabinoids : actions at non-CB1/CB2 cannabinoid receptors. Springer; New York: 2013. [Google Scholar]
- Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chemical reviews. 2008;108:1687–1707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MT, Ali S, Rashidi H, van der Kooy F, Verpoorte R, Richardson MK. Developmental effects of cannabinoids on zebrafish larvae. Zebrafish. 2013;10:283–293. doi: 10.1089/zeb.2012.0785. [DOI] [PubMed] [Google Scholar]
- Alexander SP, Kendall DA. The complications of promiscuity: endocannabinoid action and metabolism. British journal of pharmacology. 2007;152:602–623. doi: 10.1038/sj.bjp.0707456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Escobedo PA, Gould GG. Visual social preferences of lone zebrafish in a novel environment: strain and anxiolytic effects. Genes, brain, and behavior. 2012;11:366–373. doi: 10.1111/j.1601-183X.2012.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard G, Massa F, Puente N, Lourenco J, Bellocchio L, Soria-Gomez E, Matias I, Delamarre A, Metna-Laurent M, Cannich A, Hebert-Chatelain E, Mulle C, Ortega-Gutierrez S, Martin-Fontecha M, Klugmann M, Guggenhuber S, Lutz B, Gertsch J, Chaouloff F, Lopez-Rodriguez ML, Grandes P, Rossignol R, Marsicano G. Mitochondrial CB(1) receptors regulate neuronal energy metabolism. Nature neuroscience. 2012;15:558–564. doi: 10.1038/nn.3053. [DOI] [PubMed] [Google Scholar]
- Blackburn PR, Campbell JM, Clark KJ, Ekker SC. The CRISPR system--keeping zebrafish gene targeting fresh. Zebrafish. 2013;10:116–118. doi: 10.1089/zeb.2013.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braida D, Limonta V, Capurro V, Fadda P, Rubino T, Mascia P, Zani A, Gori E, Fratta W, Parolaro D, Sala M. Involvement of kappa-opioid and endocannabinoid system on Salvinorin A-induced reward. Biological psychiatry. 2008;63:286–292. doi: 10.1016/j.biopsych.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Braida D, Limonta V, Pegorini S, Zani A, Guerini-Rocco C, Gori E, Sala M. Hallucinatory and rewarding effect of salvinorin A in zebrafish: kappa-opioid and CB1-cannabinoid receptor involvement. Psychopharmacology. 2007;190:441–448. doi: 10.1007/s00213-006-0639-1. [DOI] [PubMed] [Google Scholar]
- Broglio C, Gomez A, Duran E, Ocana FM, Jimenez-Moya F, Rodriguez F, Salas C. Hallmarks of a common forebrain vertebrate plan: specialized pallial areas for spatial, temporal and emotional memory in actinopterygian fish. Brain research bulletin. 2005;66:277–281. doi: 10.1016/j.brainresbull.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Campbell JM, Hartjes KA, Nelson TJ, Xu X, Ekker SC. New and TALENted genome engineering toolbox. Circulation research. 2013;113:571–587. doi: 10.1161/CIRCRESAHA.113.301765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Youth Risk Behavior Surveillance - United States, 2011. Morbidity and Mortality Weekly Report. 2012;61 [PubMed] [Google Scholar]
- Clark KJ, Balciunas D, Pogoda HM, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, Ni J, Nielsen AL, Patowary A, Scaria V, Sivasubbu S, Xu X, Hammerschmidt M, Ekker SC. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nature methods. 2011;8:506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors KA, Valenti TW, Lawless K, Sackerman J, Onaivi ES, Brooks BW, Gould GG. Similar anxiolytic effects of agonists targeting serotonin 5-HT1A or cannabinoid CB receptors on zebrafish behavior in novel environments. Aquatic toxicology. 2014;151:105–113. doi: 10.1016/j.aquatox.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D, Marsicano G, Tschop M, Grubler Y, Flachskamm C, Schubert M, Auer D, Yassouridis A, Thone-Reineke C, Ortmann S, Tomassoni F, Cervino C, Nisoli E, Linthorst AC, Pasquali R, Lutz B, Stalla GK, Pagotto U. The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. The Journal of clinical investigation. 2003;112:423–431. doi: 10.1172/JCI17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- Csoka B, Nemeth ZH, Mukhopadhyay P, Spolarics Z, Rajesh M, Federici S, Deitch EA, Batkai S, Pacher P, Hasko G. CB2 cannabinoid receptors contribute to bacterial invasion and mortality in polymicrobial sepsis. PloS one. 2009;4:e6409. doi: 10.1371/journal.pone.0006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel H, Rancillac A, Crepel F. Mechanisms underlying cannabinoid inhibition of presynaptic Ca2+ influx at parallel fibre synapses of the rat cerebellum. The Journal of physiology. 2004;557:159–174. doi: 10.1113/jphysiol.2004.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzo V. Endocannabinoid signaling in the brain: biosynthetic mechanisms in the limelight. Nature neuroscience. 2011;14:9–15. doi: 10.1038/nn.2720. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz JC, Piomelli D. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPatrizio NV, Piomelli D. The thrifty lipids: endocannabinoids and the neural control of energy conservation. Trends in neurosciences. 2012;35:403–411. doi: 10.1016/j.tins.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drug Enforcement Administration . Schedules of Controlled Substances: Temporary Placement of Four Synthetic Cannabinoids Into Schedule 1. no. 9. Vol. 79. Department of Justice, Federal Register; 2014. pp. 1776–1780. [PubMed] [Google Scholar]
- Galanter M, Kleber HD, Brady K. The American Psychiatric Publishing textbook of substance abuse treatment. American Psychiatric Publishing; Washington, DC: 2015. [Google Scholar]
- Galiegue S, Mary S, Marchand J, Dussossoy D, Carriere D, Carayon P, Bouaboula M, Shire D, Le Fur G, Casellas P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. European journal of biochemistry / FEBS. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- Gary-Bobo M, Elachouri G, Gallas JF, Janiak P, Marini P, Ravinet-Trillou C, Chabbert M, Cruccioli N, Pfersdorff C, Roque C, Arnone M, Croci T, Soubrie P, Oury-Donat F, Maffrand JP, Scatton B, Lacheretz F, Le Fur G, Herbert JM, Bensaid M. Rimonabant reduces obesity-associated hepatic steatosis and features of metabolic syndrome in obese Zucker fa/fa rats. Hepatology. 2007;46:122–129. doi: 10.1002/hep.21641. [DOI] [PubMed] [Google Scholar]
- Gomez M, Hernandez M, Fernandez-Ruiz J. Cannabinoid signaling system: does it play a function in cell proliferation and migration, neuritic elongation and guidance and synaptogenesis during brain ontogenesis? Cell adhesion & migration. 2008;2:246–248. doi: 10.4161/cam.2.4.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould GG, Seillier A, Weiss G, Giuffrida A, Burke TF, Hensler JG, Rock C, Tristan A, McMahon LR, Salazar A, O’Connor JC, Satsangi N, Satsangi RK, Gu TT, Treat K, Smolik C, Schultz ST. Acetaminophen differentially enhances social behavior and cortical cannabinoid levels in inbred mice. Progress in neuro-psychopharmacology & biological psychiatry. 2012;38:260–269. doi: 10.1016/j.pnpbp.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney SMR, Scott KS, Kacinko SL, Presley BC, Logan BK. Pharmacology, Toxicology, and Adverse Effects of Synthetic Cannabinoid Drugs. 2014;26 [PubMed] [Google Scholar]
- Hermanson DJ, Marnett LJ. Cannabinoids, endocannabinoids, and cancer. Cancer metastasis reviews. 2011;30:599–612. doi: 10.1007/s10555-011-9318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AF, Lupica CR. Mechanisms of cannabinoid inhibition of GABA(A) synaptic transmission in the hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:2470–2479. doi: 10.1523/JNEUROSCI.20-07-02470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, Mechoulam R, Pertwee RG. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacological reviews. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- Idris AI, Ralston SH. Role of cannabinoids in the regulation of bone remodeling. Frontiers in endocrinology. 2012;3:136. doi: 10.3389/fendo.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff HA, Lynch DL, Kotsikorou E, Reggio PH. Parameterization of Org27569: an allosteric modulator of the cannabinoid CB1 G protein-coupled receptor. Journal of computational chemistry. 2011;32:2119–2126. doi: 10.1002/jcc.21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch JD, Andrusiak E, Tran A, Bowers MB, Jr., Roth RH. Delta 9-tetrahydrocannabinol increases prefrontal cortical catecholaminergic utilization and impairs spatial working memory in the rat: blockade of dopaminergic effects with HA966. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 1997;16:426–432. doi: 10.1016/S0893-133X(97)00018-3. [DOI] [PubMed] [Google Scholar]
- Johnston LD, O’Malley PM, Bachman JG, Schulenberg JE. In: Monitoring the Future national survey results on adolescent drug use: Overview of key findings, 2012. T.U.o.M., editor. Institute for Social Research; Ann Arbor: 2013. [Google Scholar]
- Jones KS, Alimov AP, Rilo HL, Jandacek RJ, Woollett LA, Penberthy WT. A high throughput live transparent animal bioassay to identify non-toxic small molecules or genes that regulate vertebrate fat metabolism for obesity drug development. Nutrition & metabolism. 2008;5:23. doi: 10.1186/1743-7075-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KM, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D. A key role for diacylglycerol lipase-alpha in metabotropic glutamate receptor-dependent endocannabinoid mobilization. Molecular pharmacology. 2007;72:612–621. doi: 10.1124/mol.107.037796. [DOI] [PubMed] [Google Scholar]
- Jung KM, Sepers M, Henstridge CM, Lassalle O, Neuhofer D, Martin H, Ginger M, Frick A, DiPatrizio NV, Mackie K, Katona I, Piomelli D, Manzoni OJ. Uncoupling of the endocannabinoid signalling complex in a mouse model of fragile X syndrome. Nature communications. 2012;3:1080. doi: 10.1038/ncomms2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzior KK, Laeber LT. A positive association between anxiety disorders and cannabis use or cannabis use disorders in the general population--a meta-analysis of 31 studies. BMC psychiatry. 2014;14:136. doi: 10.1186/1471-244X-14-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee EW, Schneider H, Clark KJ, Cousin MA, Ebbert JO, Hooten WM, Karpyak VM, Warner DO, Ekker SC. Zebrafish: a model for the study of addiction genetics. Human genetics. 2012;131:977–1008. doi: 10.1007/s00439-011-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klegeris A, Bissonnette CJ, McGeer PL. Reduction of human monocytic cell neurotoxicity and cytokine secretion by ligands of the cannabinoid-type CB2 receptor. British journal of pharmacology. 2003;139:775–786. doi: 10.1038/sj.bjp.0705304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köfalvi A. Cannabinoids and the brain. Springer; New York: 2008. [Google Scholar]
- Kondo S, Kondo H, Nakane S, Kodaka T, Tokumura A, Waku K, Sugiura T. 2-Arachidonoylglycerol, an endogenous cannabinoid receptor agonist: identification as one of the major species of monoacylglycerols in various rat tissues, and evidence for its generation through CA2+-dependent and -independent mechanisms. FEBS letters. 1998;429:152–156. doi: 10.1016/s0014-5793(98)00581-x. [DOI] [PubMed] [Google Scholar]
- Krug RG, 2nd, Poshusta TL, Skuster KJ, Berg MR, Gardner SL, Clark KJ. A transgenic zebrafish model for monitoring glucocorticoid receptor activity. Genes, brain, and behavior. 2014;13:478–487. doi: 10.1111/gbb.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, Rastegar S, Strahle U. Distribution of cannabinoid receptor 1 in the CNS of zebrafish. Neuroscience. 2006;138:83–95. doi: 10.1016/j.neuroscience.2005.10.069. [DOI] [PubMed] [Google Scholar]
- Lauckner JE, Jensen JB, Chen HY, Lu HC, Hille B, Mackie K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2699–2704. doi: 10.1073/pnas.0711278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Fan HB, Jin Y, Ren CG, Jia XE, Wang L, Chen Y, Dong M, Zhu KY, Dong ZW, Ye BX, Zhong Z, Deng M, Liu TX, Ren R. Cannabinoid receptor 2 suppresses leukocyte inflammatory migration by modulating the JNK/c-Jun/Alox5 pathway. The Journal of biological chemistry. 2013;288:13551–13562. doi: 10.1074/jbc.M113.453811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado R, Valverde O, Berrendero F. Involvement of the endocannabinoid system in drug addiction. Trends in neurosciences. 2006;29:225–232. doi: 10.1016/j.tins.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Malone DT, Hill MN, Rubino T. Adolescent cannabis use and psychosis: epidemiology and neurodevelopmental models. British journal of pharmacology. 2010;160:511–522. doi: 10.1111/j.1476-5381.2010.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi P, Valenti M, Vaccani A, Gasperi V, Perletti G, Marras E, Fezza F, Maccarrone M, Parolaro D. 5-Lipoxygenase and anandamide hydrolase (FAAH) mediate the antitumor activity of cannabidiol, a non-psychoactive cannabinoid. Journal of neurochemistry. 2008;104:1091–1100. doi: 10.1111/j.1471-4159.2007.05073.x. [DOI] [PubMed] [Google Scholar]
- McPartland JM, Glass M, Matias I, Norris RW, Kilpatrick CW. A shifted repertoire of endocannabinoid genes in the zebrafish (Danio rerio) Molecular genetics and genomics : MGG. 2007;277:555–570. doi: 10.1007/s00438-007-0207-3. [DOI] [PubMed] [Google Scholar]
- Meier MH, Caspi A, Ambler A, Harrington H, Houts R, Keefe RS, McDonald K, Ward A, Poulton R, Moffitt TE. Persistent cannabis users show neuropsychological decline from childhood to midlife. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2657–2664. doi: 10.1073/pnas.1206820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliarini B, Carnevali O. Anandamide modulates growth and lipid metabolism in the zebrafish Danio rerio. Molecular and cellular endocrinology. 2008;286:S12–16. doi: 10.1016/j.mce.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Migliarini B, Carnevali O. A novel role for the endocannabinoid system during zebrafish development. Molecular and cellular endocrinology. 2009;299:172–177. doi: 10.1016/j.mce.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Mishima K, Egashira N, Hirosawa N, Fujii M, Matsumoto Y, Iwasaki K, Fujiwara M. Characteristics of learning and memory impairment induced by delta9-tetrahydrocannabinol in rats. Japanese journal of pharmacology. 2001;87:297–308. doi: 10.1254/jjp.87.297. [DOI] [PubMed] [Google Scholar]
- Mouslech Z, Valla V. Endocannabinoid system: An overview of its potential in current medical practice. Neuro endocrinology letters. 2009;30:153–179. [PubMed] [Google Scholar]
- Mueller T, Dong Z, Berberoglu MA, Guo S. The dorsal pallium in zebrafish, Danio rerio (Cyprinidae, Teleostei) Brain research. 2011;1381:95–105. doi: 10.1016/j.brainres.2010.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Aguado T, Keimpema E, Barabas K, Ballester Rosado CJ, Nguyen L, Monory K, Marsicano G, Di Marzo V, Hurd YL, Guillemot F, Mackie K, Lutz B, Guzman M, Lu HC, Galve-Roperh I, Harkany T. Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8760–8765. doi: 10.1073/pnas.0803545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderi N, Haghparast A, Saber-Tehrani A, Rezaii N, Alizadeh AM, Khani A, Motamedi F. Interaction between cannabinoid compounds and diazepam on anxiety-like behaviour of mice. Pharmacology, biochemistry, and behavior. 2008;89:64–75. doi: 10.1016/j.pbb.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Newton CA, Klein TW, Friedman H. Secondary immunity to Legionella pneumophila and Th1 activity are suppressed by delta-9-tetrahydrocannabinol injection. Infection and immunity. 1994;62:4015–4020. doi: 10.1128/iai.62.9.4015-4020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio S, Gibert Y, Berekelya L, Bernard L, Brunet F, Guillot E, Le Bail JC, Sanchez JA, Galzin AM, Triqueneaux G, Laudet V. Fasting induces CART down-regulation in the zebrafish nervous system in a cannabinoid receptor 1-dependent manner. Molecular endocrinology. 2012;26:1316–1326. doi: 10.1210/me.2011-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno-Shosaku T, Maejima T, Kano M. Endogenous cannabinoids mediate retrograde signals from depolarized postsynaptic neurons to presynaptic terminals. Neuron. 2001;29:729–738. doi: 10.1016/s0896-6273(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Olea-Herrero N, Vara D, Malagarie-Cazenave S, Diaz-Laviada I. Inhibition of human tumour prostate PC-3 cell growth by cannabinoids R(+)-Methanandamide and JWH-015: involvement of CB2. British journal of cancer. 2009;101:940–950. doi: 10.1038/sj.bjc.6605248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Carpio O, Ishiguro H, Schanz N, Uhl GR, Benno R. Behavioral effects of CB2 cannabinoid receptor activation and its influence on food and alcohol consumption. Annals of the New York Academy of Sciences. 2008a;1139:426–433. doi: 10.1196/annals.1432.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gong JP, Patel S, Meozzi PA, Myers L, Perchuk A, Mora Z, Tagliaferro PA, Gardner E, Brusco A, Akinshola BE, Hope B, Lujilde J, Inada T, Iwasaki S, Macharia D, Teasenfitz L, Arinami T, Uhl GR. Brain neuronal CB2 cannabinoid receptors in drug abuse and depression: from mice to human subjects. PloS one. 2008b;3:e1640. doi: 10.1371/journal.pone.0001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Hyiaman D, Depetrillo M, Harvey-White J, Bannon AW, Cravatt BF, Kuhar MJ, Mackie K, Palkovits M, Kunos G. Cocaine- and amphetamine-related transcript is involved in the orexigenic effect of endogenous anandamide. Neuroendocrinology. 2005a;81:273–282. doi: 10.1159/000087925. [DOI] [PubMed] [Google Scholar]
- Osei-Hyiaman D, DePetrillo M, Pacher P, Liu J, Radaeva S, Batkai S, Harvey-White J, Mackie K, Offertaler L, Wang L, Kunos G. Endocannabinoid activation at hepatic CB1 receptors stimulates fatty acid synthesis and contributes to diet-induced obesity. The Journal of clinical investigation. 2005b;115:1298–1305. doi: 10.1172/JCI23057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai WY, Hsu CC, Lai CY, Chang TZ, Tsai YL, Her GM. Cannabinoid receptor 1 promotes hepatic lipid accumulation and lipotoxicity through the induction of SREBP-1c expression in zebrafish. Transgenic research. 2013;22:823–838. doi: 10.1007/s11248-012-9685-0. [DOI] [PubMed] [Google Scholar]
- Park B, McPartland JM, Glass M. Cannabis, cannabinoids and reproduction. Prostaglandins, leukotrienes, and essential fatty acids. 2004;70:189–197. doi: 10.1016/j.plefa.2003.04.007. [DOI] [PubMed] [Google Scholar]
- Patel S, Hillard CJ. Pharmacological evaluation of cannabinoid receptor ligands in a mouse model of anxiety: further evidence for an anxiolytic role for endogenous cannabinoid signaling. The Journal of pharmacology and experimental therapeutics. 2006;318:304–311. doi: 10.1124/jpet.106.101287. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacology & therapeutics. 1997;74:129–180. doi: 10.1016/s0163-7258(97)82001-3. [DOI] [PubMed] [Google Scholar]
- Piccinetti CC, Migliarini B, Olivotto I, Coletti G, Amici A, Carnevali O. Appetite regulation: the central role of melatonin in Danio rerio. Hormones and behavior. 2010;58:780–785. doi: 10.1016/j.yhbeh.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin I, Herrero-Turrion MJ, Marron Fdez de Velasco E, Gonzalez-Sarmiento R, Rodriguez RE. Characterization of two duplicate zebrafish Cb2-like cannabinoid receptors. Gene. 2007a;389:36–44. doi: 10.1016/j.gene.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martin I, Marron Fernandez de Velasco E, Rodriguez RE. Characterization of cannabinoid-binding sites in zebrafish brain. Neuroscience letters. 2007b;413:249–254. doi: 10.1016/j.neulet.2006.11.057. [DOI] [PubMed] [Google Scholar]
- Ruhl T, Prinz N, Oellers N, Seidel NI, Jonas A, Albayram O, Bilkei-Gorzo A, von der Emde G. Acute administration of THC impairs spatial but not associative memory function in zebrafish. Psychopharmacology. 2014;231:3829–3842. doi: 10.1007/s00213-014-3522-5. [DOI] [PubMed] [Google Scholar]
- Schneider M. Cannabis use in pregnancy and early life and its consequences: animal models. European archives of psychiatry and clinical neuroscience. 2009;259:383–393. doi: 10.1007/s00406-009-0026-0. [DOI] [PubMed] [Google Scholar]
- Scott KA, Dalgleish AG, Liu WM. The combination of cannabidiol and Delta9-tetrahydrocannabinol enhances the anticancer effects of radiation in an orthotopic murine glioma model. Molecular cancer therapeutics. 2014;13:2955–2967. doi: 10.1158/1535-7163.MCT-14-0402. [DOI] [PubMed] [Google Scholar]
- Seely KA, Brents LK, Franks LN, Rajasekaran M, Zimmerman SM, Fantegrossi WE, Prather PL. AM-251 and rimonabant act as direct antagonists at mu-opioid receptors: implications for opioid/cannabinoid interaction studies. Neuropharmacology. 2012;63:905–915. doi: 10.1016/j.neuropharm.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Hirano M, Nishimura Y, Tanaka T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PloS one. 2012;7:e52549. doi: 10.1371/journal.pone.0052549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri C, Di Marzo V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell metabolism. 2013;17:475–490. doi: 10.1016/j.cmet.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Silvestri C, Paris D, Martella A, Melck D, Guadagnino I, Cawthorne M, Motta A, Di Marzo V. Two non-psychoactive cannabinoids reduce intracellular lipid levels and inhibit hepatosteatosis. Journal of hepatology. 2015;62:1382–1390. doi: 10.1016/j.jhep.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Sink KS, Segovia KN, Sink J, Randall PA, Collins LE, Correa M, Markus EJ, Vemuri VK, Makriyannis A, Salamone JD. Potential anxiogenic effects of cannabinoid CB1 receptor antagonists/inverse agonists in rats: comparisons between AM4113, AM251, and the benzodiazepine inverse agonist FG-7142. European neuropsychopharmacology : the journal of the European College of Neuropsychopharmacology. 2010;20:112–122. doi: 10.1016/j.euroneuro.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella N, Schweitzer P, Piomelli D. A second endogenous cannabinoid that modulates long-term potentiation. Nature. 1997;388:773–778. doi: 10.1038/42015. [DOI] [PubMed] [Google Scholar]
- Stewart A, Wong K, Cachat J, Gaikwad S, Kyzar E, Wu N, Hart P, Piet V, Utterback E, Elegante M, Tien D, Kalueff AV. Zebrafish models to study drug abuse-related phenotypes. Reviews in the neurosciences. 2011;22:95–105. doi: 10.1515/RNS.2011.011. [DOI] [PubMed] [Google Scholar]
- Stewart AM, Kalueff AV. The behavioral effects of acute Delta(9)-tetrahydrocannabinol and heroin (diacetylmorphine) exposure in adult zebrafish. Brain research. 2014;1543:109–119. doi: 10.1016/j.brainres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration . Results from the 2010 National Survey on Drug Use and Health: Summary of National Findings. Rockville, MD: 2011. NSDUH Series H-41, HHS Publication No. (SMA) 11-4658. [Google Scholar]
- Tai T, Tsuboi K, Uyama T, Masuda K, Cravatt BF, Houchi H, Ueda N. Endogenous molecules stimulating N-acylethanolamine-hydrolyzing acid amidase (NAAA) ACS chemical neuroscience. 2012;3:379–385. doi: 10.1021/cn300007s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pfumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. ZFIN online publication. 2001 [Google Scholar]
- Thomas RJ. The toxicologic and teratologic effects of delta-9-tetrahydrocannabinol in the zebrafish embryo. Toxicology and applied pharmacology. 1975;32:184–190. doi: 10.1016/0041-008x(75)90209-4. [DOI] [PubMed] [Google Scholar]
- Tschop J, Kasten KR, Nogueiras R, Goetzman HS, Cave CM, England LG, Dattilo J, Lentsch AB, Tschop MH, Caldwell CC. The cannabinoid receptor 2 is critical for the host response to sepsis. Journal of immunology. 2009;183:499–505. doi: 10.4049/jimmunol.0900203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveros MP, Marco EM, File SE. Endocannabinoid system and stress and anxiety responses. Pharmacology, biochemistry, and behavior. 2005;81:331–342. doi: 10.1016/j.pbb.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Walentiny DM, Vann RE, Warner JA, King LS, Seltzman HH, Navarro HA, Twine CE, Jr., Thomas BF, Gilliam AF, Gilmour BP, Carroll FI, Wiley JL. Kappa opioid mediation of cannabinoid effects of the potent hallucinogen, salvinorin A, in rodents. Psychopharmacology. 2010;210:275–284. doi: 10.1007/s00213-010-1827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargent ET, Zaibi MS, Silvestri C, Hislop DC, Stocker CJ, Stott CG, Guy GW, Duncan M, Di Marzo V, Cawthorne MA. The cannabinoid Delta(9)-tetrahydrocannabivarin (THCV) ameliorates insulin sensitivity in two mouse models of obesity. Nutrition & diabetes. 2013;3:e68. doi: 10.1038/nutd.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S, Chambers D, Hobbs C, Doherty P, Graham A. The endocannabinoid receptor, CB1, is required for normal axonal growth and fasciculation. Molecular and cellular neurosciences. 2008;38:89–97. doi: 10.1016/j.mcn.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. A second fatty acid amide hydrolase with variable distribution among placental mammals. The Journal of biological chemistry. 2006;281:36569–36578. doi: 10.1074/jbc.M606646200. [DOI] [PubMed] [Google Scholar]
- Wiley JL, Burston JJ, Leggett DC, Alekseeva OO, Razdan RK, Mahadevan A, Martin BR. CB1 cannabinoid receptor-mediated modulation of food intake in mice. British journal of pharmacology. 2005;145:293–300. doi: 10.1038/sj.bjp.0706157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin JM, Tzavara ET, Nomikos GG. A role for cannabinoid CB1 receptors in mood and anxiety disorders. Behavioural pharmacology. 2005;16:315–331. doi: 10.1097/00008877-200509000-00005. [DOI] [PubMed] [Google Scholar]
- Wu CS, Zhu J, Wager-Miller J, Wang S, O’Leary D, Monory K, Lutz B, Mackie K, Lu HC. Requirement of cannabinoid CB(1) receptors in cortical pyramidal neurons for appropriate development of corticothalamic and thalamocortical projections. The European journal of neuroscience. 2010;32:693–706. doi: 10.1111/j.1460-9568.2010.07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazulla S, Studholme KM. Neurochemical anatomy of the zebrafish retina as determined by immunocytochemistry. Journal of neurocytology. 2001;30:551–592. doi: 10.1023/a:1016512617484. [DOI] [PubMed] [Google Scholar]
- Zador F, Kocsis D, Borsodi A, Benyhe S. Micromolar concentrations of rimonabant directly inhibits delta opioid receptor specific ligand binding and agonist-induced G-protein activity. Neurochemistry international. 2014;67:14–22. doi: 10.1016/j.neuint.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Zador F, Lenart N, Csibrany B, Santha M, Molnar M, Tuka B, Samavati R, Klivenyi P, Vecsei L, Marton A, Vizler C, Nagy GM, Borsodi A, Benyhe S, Paldy E. Low dosage of rimonabant leads to anxiolytic-like behavior via inhibiting expression levels and G-protein activity of kappa opioid receptors in a cannabinoid receptor independent manner. Neuropharmacology. 2015;89:298–307. doi: 10.1016/j.neuropharm.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Zimmermann US, Winkelmann PR, Pilhatsch M, Nees JA, Spanagel R, Schulz K. Withdrawal phenomena and dependence syndrome after the consumption of “spice gold”. Deutsches Arzteblatt international. 2009;106:464–467. doi: 10.3238/arztebl.2009.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]