Abstract
Recent studies have revealed that the intestinal microbiota plays an important role in host physiology and pathophysiology in health and disease. One of the major mechanisms by which the gut microbiota influences the host is through its interactions with and effects on the host immune system. In this review, we discuss the reciprocal interactions between the host immune system and the gut microbiota, with a particular focus on individual microbes that impact the host through dramatic and specific interactions with the adaptive immune system. We highlight the idea that the presence or absence of specific immunologically important members of the microbiota can determine disease susceptibility and propose that the identification and characterization of these bacteria in humans will eventually allow us to elucidate the role of microbiota composition in human disease.
Introduction
Humans are constitutively colonized by trillions of bacteria, archaea, fungi and viruses, which are collectively referred to as the ‘microbiota’. These so-called ‘commensal’ microbes inhabit all epithelial surfaces, including the skin, oral cavity, respiratory and gastrointestinal tracts, and the female reproductive tract. The gastrointestinal tract in particular is home to the largest community of bacterial members of the microbiota and is estimated to contain more than 100 trillion bacterial cells. Advances in next-generation sequencing have led to an explosion in our understanding of the diversity and complexity of the microbiota in humans in both health and disease. These studies have revealed that individual humans harbor a unique consortium of bacterial species with recent estimates suggesting that each human gut plays host to approximately 100-150 individual species that can be further divided into approximately 200 strains (1). Notably, it is estimated that the ‘microbiome’, which is the collection of genes encoded by members of the microbiota, contains more than one-hundred times more genes than our own Homo sapiens genome (2). Taken together, these observations have led to the realization that the microbiota can be thought of as a previously-ignored ‘organ’ and that humans should be considered a ‘superorganism’ consisting of a combination of Homo sapiens cells and our microbiota.
Recent studies have revealed that the microbiota plays an important role in host physiology and pathophysiology in health and disease (3-5). The relationship between the host and its microbiota is the result of millions of years of co-evolution and, therefore, is generally mutually beneficial (i.e., symbiotic) (6). However, unhealthy ‘imbalances’ in the microbiota, which are referred to as dysbiosis, have been associated with a multitude of diseases of various etiologies, including inflammatory bowel disease, autoimmunity, obesity, metabolic syndrome, and even neurodevelopmental disorders. One of the major mechanisms by which the microbiota has been shown to influence such diseases is through its chronic interactions with and effects on the host immune system (3).
Impact of the microbiota on the intestinal immune system
The gut microbiota is separated from the inside of the host by a single layer of epithelial cells. This poses a special challenge for the host immune system, which has evolved to recognize microbial non-self as a sign of potential pathogenic infection; therefore, continuous interactions with the microbiota dramatically impact the host intestinal immune system. On the other hand, the immune response to the microbiota also impacts microbial ecology in the intestine and can shape microbiota composition and function. Thus, the host and the microbiota are in constant communication, and reciprocal interactions between these two entities shape both host immunity and microbial ecology. In this review, we will focus on these interactions, with a special focus on antigen-specific responses of the adaptive immune system.
Maturation of the intestinal immune system
The intestinal microbiota plays a crucial role in the development and maturation of the host immune system. This is most clearly demonstrated in germ free mice that have been raised to be completely void of all microbes from birth. These animals exhibit a severely underdeveloped intestinal immune system as compared to conventionally raised mice. For example, germ free mice exhibit dramatically reduced numbers of intra-epithelial lymphocytes, reduced sizes and numbers of Peyer’s patches and cryptopatches, altered crypt structure, and reduced mucus thickness due to a decreased number of goblet cells (7-9). Maturation of the intestinal immune system in germ free mice can be induced through colonization with a variety of different microbes, including a variety of bacterial species as well as ‘commensal’ viruses (8, 10). This maturation is largely dependent on host recognition of the presence of microbial non-self by pattern recognition receptors (PRRs) of the innate immune system (11). These receptors sense microbial colonization through the detection of conserved microbial components termed Pathogen Associated Molecular Patterns (PAMPs), such as bacterial cell wall components (e.g., lipopolysaccharide) and nucleic acids (e.g., viral and bacterial DNA). Since all microbes, including all commensals, contain PAMPs, this type of interaction between the microbiota and the innate immune system can be considered to be relatively generic; that is, any given microbe that can colonize the intestine can trigger such responses and (at least largely) correct the alterations in the intestinal immune system seen in germ free mice.
‘Specific’ and ‘intimate’ interactions between the microbiota and the intestinal immune system
In contrast to the generic interactions between the microbiota and the innate immune system described above, select members of the microbiota have been shown to have dramatic and specific effects on the host immune system through their intimate interactions with the host. Such bacteria can be separated into two major categories, depending on their functional effects on the immune system: ‘Inflammatory Commensals’, which primary stimulate inflammatory/effector immune responses, and ‘Immunoregulatory Commensals’, which primarily stimulate immunoregulatory rather than inflammatory responses. It is worth noting that the terminology used to describe these functional subsets of the microbiota is still evolving. Indeed, others have referred to bacteria with largely overlapping features of inflammatory commensals as ‘pathobionts’ to highlight the idea that these bacteria share features with both pathogens and symbionts and can cause pathological outcomes under specific conditions (12). In addition, immunomodulatory bacteria that aren’t overtly associated with disease, or which are thought of as symbiotic rather than parasitic, have been referred to as ‘autobionts’ (13). Regardless of the terms used to describe members of the gut microbiota that impact the immune system, this class of bacteria is defined by the following two critical features that distinguish them from classical pathogens and from other commensals: 1) they are indigenous members of the microbiota; and, 2) they interact with the host in a specific manner that leads to their recognition by the intestinal immune system and stimulation of a specific immune response.
Inflammatory commensals
Since the immune system is critically involved in the development and progression of a variety of diseases, it is perhaps unsurprising that inflammatory commensals also appear to play important roles in the disease development and progression. An important mechanism by which such bacteria appear to impact the health of the host involves the stimulation of chronic Th1 and/or Th17 cell responses. While these responses play a crucial role in host defense against invading bacteria, they can also result in pathological damage to the host when chronically present or improperly controlled. A limited number of bacterial species that drive such T cell responses have been described in detail, with the so-called Segmented Filamentous Bacteria (SFB) and Helicobacter hepaticus being the best characterized members of this group.
SFB colonize the epithelial surface of the small intestine in mice and induce potent Th17 responses (14, 15). These Th17 cells were recently shown to be specific for SFB-derived antigens (16-18). However, in the absence of secondary lymphoid organs, SFB induces non-specific Th17 responses (18). The consequences of these chronic specific and non-specific Th17 responses can be both beneficial and detrimental to the host. For instance, SFB-induced Th17 cells enhance resistance to infection with the enteric murine pathogen Citrobacter rodentium by inducing increased antimicrobial activity in intestinal epithelial cells (15). On the other hand, colonization with SFB results in hypersensitivity to colitis in T-cell dependent models of inflammatory bowel disease and increases the development of Th17-mediated arthritis in susceptible mice (19-21).
Helicobacter hepaticus colonizes the colonic and cecal mucosa of mice and induces both Th1 and Th17 responses. In wild type mice, H. hepaticus does not result in pathological levels of intestinal inflammation. However, H. hepaticus is a potent inducer of colitis in mice lacking the immunoregulatory cytokine IL-10 and in the T cell transfer model of colitis in which CD4+CD45RB high T cells are adoptively transferred into Rag-/- mice (22). A major mechanism by which Helicobacter induces colitis is through induction of the proinflammatory cytokines IL-1β and IL-23 in the colon, which lead to the accumulation of IL-17-secreting innate lymphoid cells (ILCs) and Th17 cells that together mediate the development of colitis (23).
In addition to SFB and Helicobacter, a number of other bacterial species have been shown to exacerbate the development of immunological diseases in various mouse models. However, the specific effects of these bacteria on the immune response (and in particular the T cell response) are less well defined. For example, our group identified Prevotellaceae species that, in the context of intestinal dysbiosis driven by deficiency in the NLRP6 inflammasome, drive chronic intestinal inflammation that is dependent on the chemokine CCL5 and strongly excacerbates DSS-induced colitis (24); mice that are deficient in T-bet and Rag2 spontaneously develop communicable ulcerative colitis that is mediated by Klebsiella pneumoniae and Proteus mirabilis, two members of the Enterobactereceae family (25, 26); high-fat diet caused the outgrowth and induction of colitis by Bilophila wadsworthia in IL-10 deficient mice (27); and finally, Bacteroides fragilis has been shown to induce Th1 and Th17 responses in the gut (28), and can regulate iNKT cell homeostasis (29).
One important question regarding inflammatory commensals is whether these bacteria share any specific features or behaviors that lead them to preferentially trigger chronic innate and adaptive immune responses. While this remains an open question, one intriguing possibility is that these bacteria are uniquely able to invade/colonize normally sterile sites, such as intestinal crypts and the inner mucus layer of the large intestine. Such behavior would naturally be interpreted by the immune system as a signal of potential microbial invasion and would provide a source of antigen for cells of the adaptive immune system. Indeed, many known inflammatory commensals appear to exhibit these types of behaviors: SFB adheres tightly to the epithelium of the terminal ileum (14, 15), Prevotellaceae species inhabit intestinal crypts (24), and B. fragilis colonizes the inner mucus layer in the colon (30).
Immunoregulatory commensals
The co-evolution of the mammalian immune system with its microbiota has led to a relationship that is largely cooperative rather than antagonistic. Therefore it is not surprising that the major effect of the microbiota on the immune system is the induction of immunoregulatory responses, which enforce host-microbiota homeostasis. One of the major immunoregulatory responses to the microbiota involves the induction of intestinal regulatory T cells. For example, colonization of germ free mice with a model microbiota referred to as Altered Schaedler Flora induces the production and recruitment of regulatory T cells in the intestine and these T cells are essential for the maintenance of intestinal homeostasis and resistance to colitis (31). Notably, specific members of the microbiota that are potent inducers of regulatory cells have recently been described. Two of the best-studied Treg-inducing bacteria are members of the genus Clostridium and capsular polysaccharide A (PSA)-producing B. fragilis.
Members of the genus Clostridium are a diverse group of spore-forming bacteria that belong to the phylum Firmicutes. It was recently discovered that a collection of Clostridia species isolated from conventionally reared mice (in particular, Clostridia from clusters IV and XIVa) could potently induce the accumulation of Tregs in the colonic mucosa of mice, and could ameliorate colitis and reduce the induction of Immunoglobulin E (32). Subsequently, a similar approach was taken to identify a group of 17 Clostridium strains from human feces that could mediate Treg accumulation and resistance to colitis when transferred into mice (33). Notably, Clostridia species are major producers of the Short Chain Fatty Acids (SCFA) butyrate, propionate and acetate, which are produced during the processing of dietary fibers. SCFA have recently been shown to mediate immunoregulation through the induction of Tregs, although the mechanism by which they mediate this effect remains controversial (34-36).
In addition to its ability to stimulate Th1 and Th17 responses in the intestine (described above), B. fragilis has also been shown to be a potent inducer of Tregs, which resulted in resistance to colitis in mouse models of disease (37). However, unlike in the case of Clostridia species, which mediate their immunoregulatory functions through the production of microbial metabolites, B. fragilis mediates its immunoregulatory functions due to recognition of its PSA by the innate immune system (37). In particular, it was found that recognition of PSA by TLR2 was critical for the induction of Tregs in the intestine in response to B. fragilis (38).
Regulation of the microbiota by the immune system
It is abundantly clear that the microbiota has a dramatic effect on the immune system; on the other hand, the immune system also plays an important role in controlling and shaping the composition of the microbiota. This immunological-control of the microbiota plays an essential role in maintaining a symbiotic relationship between the host and the microbiota and, therefore, in maintaining intestinal homeostasis and preventing intestinal disease. A variety of innate and adaptive components have been shown to play critical roles in controlling the composition of the intestinal microbiota.
Innate immunity
Toll-like receptors are critical mediators of innate immune recognition of pathogens and commensals, and play a critical role in maintaining intestinal homeostasis (39). TLRs also appear to play a critical role in regulating microbiota composition (40, 41). In particular, multiple studies have demonstrated alterations in the intestinal microbiota specifically in mice lacking TLR5, which senses bacterial flagellin (42). Interestingly, these alterations in the microbiota could confer susceptibility to obesity, metabolic syndrome and colitis; this suggests that TLR5 may be necessary to maintain a ‘healthy’ microbiota composition (42-45). However, some controversy over the role of TLRs in controlling microbiota composition remains since separate studies have demonstrated that alterations in the microbiota in TLR-deficient mice are largely due to ‘microbiota drift’ that occurs when wildtype and knockout breeding colonies are maintained separately over multiple generations (46).
Nod-like receptors (NLRs), which are cytosolic sensors of infection and tissue stress, have also been shown to influence microbiota composition. For example, mice lacking either Nod1 or Nod2 have both been shown to exhibit alterations in their microbiotas (11, 47, 48). Like with TLRs, these studies also remain controversial (49). Finally, our recent studies have demonstrated that NLR family members that mediate activation of the inflammasome exhibit a so-called inflammasome-mediated dysbiosis. Notably, this dysbiosis is transmissible to wildtype mice through co-housing and predisposes mice to the development of obesity, metabolic syndrome, colitis, colorectal cancer, and non-alcoholic fatty liver disease (NAFLD) (24, 50, 51). Interestingly, recent studies have revealed that inflammasome-deficient mice also show defects in mucus production due to goblet cell dysfunction, which may explain their susceptibility to dysbiosis (52).
Taken together, the above-described studies suggest that sensors of the innate immune system play an important role in regulating microbiota composition and maintaining host-microbiota homeostasis in the intestine. In addition, a number of innate effector molecules have also been shown to affect microbiota composition. For example, dysfunctions in paneth cells, which produce large amounts of antimicrobial peptides (AMPs) in small intestinal crypts, or genetically-induced alterations in AMP expression both trigger alterations in the microbiota and dysbiosis (53, 54). Furthermore, defective AMP expression also leads to alterations in the spatial organization of the microbiota; for example, deficiency in the AMP RegIIIgamma leads to bacterial colonization of the inner mucus layer, which is normally sterile (55). Notably, defects in the mucus layer itself also are associated with alterations in the intestinal microbiota (56).
Adaptive immunity
T cells
It has been suggested that the adaptive immune system evolved in part to enable mutualism with a complex microbiota (57). Indeed, mice lacking adaptive immunity exhibit alterations in their microbiotas, which suggests that adaptive immunity plays an important role in regulating microbiota composition (58, 59). T cell deficient mice also exhibit alterations in the microbiota (59, 60). While it is possible that T cells may directly influence microbiota composition (e.g., by inducing AMP expression in paneth cells), evidence for a direct role for T cells is currently lacking. Instead, the main mechanism by which T cells appear to influence microbiota composition is through their role as B cell helpers for the production of secretory IgA (59).
Two types of T helper cells have been reported to be critical for supporting IgA production in the intestine: Tregs and Th17 cells. Multiple studies have shown that Tregs can provide help to B cells in the production of IgA in the intestine. For example, transfer of Tregs into T cell-deficient mice led to the differentiation of T follicular helper cells in the intestine that served as helpers for the production of IgA (61). Furthermore, IgA production in a TCR transgenic mouse with a TCR directed against flagellin was largely dependent on Tregs (62). These data suggest that Tregs are major helpers for T-dependent IgA production against microbiota-derived antigens in the gut. Separate studies showed that Th17 cells are the major helpers for antigen-specific IgA production in response to immunization with the classical IgA-inducing adjuvant cholera toxin (63). This highlights the possibility that, in addition to Tregs, Th17 cells can also provide help for the production of IgA to the microbiota. In this regard, it is notable that SFB, which is a potent inducer of Th17 responses, is also a potent stimulator of IgA (64, 65).
B cells: Immunoglobulin A
IgA is unique among antibody isotypes in that it is primarily secreted into the intestinal lumen where it can bind and coat members of the intestinal microbiota. This positions IgA as the major antigen-specific mechanism by which the immune system can directly interact with and influence the luminal microbiota. There are two non-mutually exclusive mechanisms by which IgA has been proposed to affect the composition of the microbiota and support host-microbiota homeostasis: 1) by restricting the growth or inflammatory effects of commensals; and 2) by enforcing maintenance of a diverse and ‘healthy’ microbiota composition.
The potential role of IgA in shaping the microbiota was first noted in mice lacking Activation Induced Cytidine Deaminase (AID), which are unable to undergo class switch recombination (CSR) or somatic hypermutation (SHM) (66). In particular, it was found that these mice displayed an outgrowth of SFB, which could be corrected via reconstitution of intestinal IgA production (67). Thus, it appears that IgA responses to SFB restrict the growth of this species. Notably, the ‘quality’ of the IgA response to the microbiota also appears to be important for IgA-mediated regulation of the microbiota since mice displaying defects in T cell help for IgA production in the gut also display altered IgA coating of the intestinal microbiota and dysbiosis (68, 69). In addition, mice with defective T-dependent IgA responses due to T cell-intrinsic deletion of MyD88 also showed outgrowth of specific gut microbes and hypersensitivity to colitis (70). Finally, IgA is also important for shaping the composition of the microbiota during development as the absence of an IgA response leads to a failure to supress Proteobacteria during microbiota maturation (71).
Microbiota-specific IgA can also promote host microbiota mutualism by reducing the inflammatory response to the microbiota (72). This reduced inflammatory response is proposed to be mediated largely ‘immune exclusion’, which involves the exclusion of microorganisms from directly contacting the mucosal epithelium (73). An additional mechanism by which IgA reduces inflammatory responses to the microbiota is through direct effects of IgA coating on bacterial gene expression; for example, the IgA response to bacterial flagellin can mediate downregulation of flagellin gene expression, which naturally reduces inflammatory responses to these bacteria (74). Finally, in addition to restricting the growth or inflammatory potential of specific indigenous microbes, it has been hypothesized that high affinity, T cell-dependent IgA is also important for enforcing maintenance of a diverse and ‘healthy’ microbiota composition. This hypothesis is based on the observation that T cell-deficient mice display a dysbiosis that is characterized by reduced complexity (60). However, the precise mechanisms by which IgA mediates this effect remain to be determined.
Exploiting IgA to identify inflammatory and immunomodulatory commensals: IgA-SEQ
We recently hypothesized that bacterial IgA-coating could be used to identify specific members of the microbiota that selectively interact with and impact the intestinal immune response and disease susceptibility (75). To test this hypothesis, we developed a technique to quantify taxa-specific levels of IgA coating in fecal samples (IgA-SEQ). In support of this approach, we found that known inflammatory commensals, including Helicobacter sp., Prevotellaceae and SFB, were uniquely highly coated with IgA in dysbiotic mice. Furthermore, we were able to use this technique to identify specific members of the human gut microbiota in IBD patients that conferred susceptibility to colitis onto germ-free mice. These data suggest that functional categorization of the microbiota based on IgA-coating will be useful to identify novel immunologically important members of the microbiota in both mice and humans.
Future Perspective
The intestinal microbiota has a dramatic impact on intestinal and extra-intestinal immunity in both health and disease. Notably, recent studies have demonstrated that not all commensals are equal in terms of their impact on the host immune system; indeed, specific commensals can exert dramatic effects on host immunity and disease susceptibility. We propose that these immunologically important commensals can be split into two broad categories based on their overall effects on the immune system: inflammatory commensals and immunoregulatory commensals. The identification and characterization of such immunologically important commensals in humans will eventually allow us to elucidate the role of microbiota composition in determining disease susceptibility and progression.
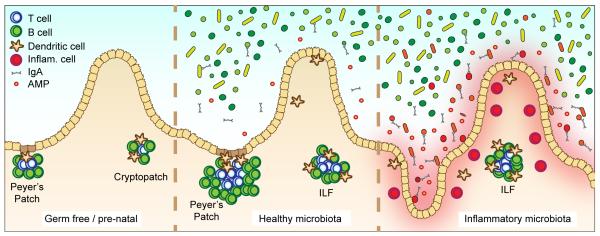
Figure 1. Interactions between the microbiota and the intestinal immune system.
Germ free and prenatal mice (left) contain underdeveloped and reduced numbers of Peyer’s patches and isolated lymphoid follicles (ILF). Colonization of the intestinal tract with a ‘healthy’ microbiota (middle) stimulates the induction and expansion of lymphoid tissues, and the secretion of antimicrobial peptides (AMP) and IgA. Colonization with inflammatory commensals (right), which are often located within normally sterile sites (such as the inner mucus layer) and induce strong IgA responses, induces chronic inflammatory responses that are characterized by an influx of inflammatory cells such as Th17 and Th1 T cells, inflammatory monocytes and neutrophils.
References
- 1.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell host & microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Science translational medicine. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 7.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. The American journal of clinical nutrition. 2001;73:1131s–1141s. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 8.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in immunology. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Macpherson AJ, McCoy KD. Standardised animal models of host microbial mutualism. Mucosal immunology. 2014 doi: 10.1038/mi.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 12.Chow J, Tang HQ, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov II, Honda K. Intestinal Commensal Microbes as Immune Modulators. Cell host & microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto Y, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecuyer E, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Qiu J, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepankova R, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflammatory bowel diseases. 2007;13:1202–1211. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 21.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song-Zhao GX, Maloy KJ. Experimental mouse models of T cell-dependent inflammatory bowel disease. Methods in molecular biology (Clifton, N.J.) 2014;1193:199–211. doi: 10.1007/978-1-4939-1212-4_18. [DOI] [PubMed] [Google Scholar]
- 23.Coccia M, et al. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. The Journal of experimental medicine. 2012;209:1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 29.An D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang JY, Lee SM, Mazmanian SK. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe. 2011;17:137–141. doi: 10.1016/j.anaerobe.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 34.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 36.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 38.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Larsson E, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell host & microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069–1080. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chassaing B, Ley RE, Gewirtz AT. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology. 2014;147:1363–1377. e1317. doi: 10.1053/j.gastro.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. The Journal of experimental medicine. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. The Journal of clinical investigation. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petnicki-Ocwieja T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson SJ, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut microbes. 2013;4:222–231. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu B, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature immunology. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salzman NH, Bevins CL. Dysbiosis--a consequence of Paneth cell dysfunction. Seminars in immunology. 2013;25:334–341. doi: 10.1016/j.smim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sommer F, et al. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PloS one. 2014;9:e85254. doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. Host adaptive immunity alters gut microbiota. The ISME journal. 2015;9:770–781. doi: 10.1038/ismej.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kato LM, Kawamoto S, Maruya M, Fagarasan S. The role of the adaptive immune system in regulation of gut microbiota. Immunological reviews. 2014;260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 60.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 61.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 62.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirota K, et al. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nature immunology. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klaasen HL, et al. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infection and immunity. 1993;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infection and immunity. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fagarasan S, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maruya M, Kawamoto S, Kato LM, Fagarasan S. Impaired selection of IgA and intestinal dysbiosis associated with PD-1-deficiency. Gut microbes. 2013;4:165–171. doi: 10.4161/gmic.23595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawamoto S, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 70.Kubinak JL, et al. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell host & microbe. 2015;17:153–163. doi: 10.1016/j.chom.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mirpuri J, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 73.Corthesy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Frontiers in immunology. 2013;4:185. doi: 10.3389/fimmu.2013.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cullender TC, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell host & microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]