Abstract
Phosphatidylinositol transfer proteins (PITP) are a family of monomeric proteins that bind and transfer phosphatidylinositol and phosphatidylcholine between membrane compartments. They are required for production of inositol and diacylglycerol second messengers, and are found in most metazoan organisms. While PITPs are known to carry out crucial cell-signaling roles in many organisms, the structure, function and evolution of the majority of family members remains unexplored; primarily because the ubiquity and diversity of the family thwarts traditional methods of global alignment. To surmount this obstacle, we instead took a novel approach, using MEME and a parsimony-based analysis to create a cladogram of conserved sequence motifs in 56 PITP family proteins from 26 species. In keeping with previous functional annotations, three clades were supported within our evolutionary analysis; two classes of soluble proteins and a class of membrane-associated proteins. By, focusing on conserved regions, the analysis allowed for in depth queries regarding possible functional roles of PITP proteins in both intra- and extra- cellular signaling.
Keywords: Protein Evolution, Structural Domain, Phylogenetics, Sequence Motif
1. INTRODUCTION
Phosphatidylinositol transfer proteins (PITP) are monomeric, lipid-binding proteins that bind and transfer phosphatidylinositol (PtdIns) and phosphatidylcholine (PtdCho) between membrane compartments (see reviews by: [1,2,3,4,5,6]. Inositol lipids have specialized functions in the regulation of eukaryotic cells, providing a source of second messengers and acting as signaling molecules. Monomeric phospholipids have extremely low solubilities and negligible spontaneous transfer rates between membranes, necessitating protein factors to shield them in the aqueous environment of the cell. Almost all phospholipid exchange activity within the eukaryotic cytosol is accomplished by three groups of proteins: PITP, phosphatidylcholine transfer protein (PCTP), or sterol carrier protein 2. PITP-domain proteins, which are the focus of this study, are found in five classes of proteins: three are soluble and cytosolic, and two are membrane-associated proteins [1,2] (Figure 1).
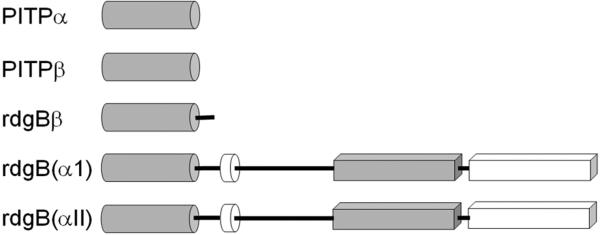
Figure 1.
Gene structure of PITP-domain proteins. Grey cylinder is PITP-like domain, white cylinder is the FFAT region of ER binding, the grey box is the DDHD region, and the white box is the LSN2 region. The the human pitpnm1 protein, the FFAT region is residues 360-365, the DDHD region is 686-879, and the LSN2 domain is residues 1022-1152 [6].
Soluble PITPs are found in virtually all metazoan organisms. These are approximately 77% identical at the amino-acid level. Two isoforms, PITPα and PITP, exist in mammals; they are highly conserved with about 98% sequence identity at the amino acid level. The proteins bind one molecule of PtdIns or PtdCho and are typically about 32 kDa in mass.
PITP-like domains are also detected in the retinal degeneration B (rdgB) class of proteins (see reviews [6,7]). These 160-170 kDa proteins are membrane-associated proteins first identified in Drosophila melanogaster, as mutations in these genes lead to retinal degeneration. The rdgBs contain an N-terminal PITP-like domain (42% sequence identity to PITP isoforms), a FFAT sequence motif [2,8], a DDHD metal binding domain (which has calcium-binding capabilities in the human proteins [9]), and a LNS2 domain. The FFAT motif is a short sequence containing two phenylalanines in an acidic tract that targets the protein to the endoplasmic reticulum [8]. The DDHD domain of 180 residues may be involved in metal binding, but the function of this domain is unknown [6]. The LNS2 domain is believed to be involved in protein: protein interactions, and in the human homologues is a protein tyrosine kinase 2 (PYK2) binding domain [9]. Two homologues of rdgB are found in most mammalian genomes studied to date and are usually called 1) rdgB(I), rdgBαI, or PITPnm1 and 2) rdgB(II), rdgBαII, or PITPnm2. In Drosophila, rdgB is believed to function in the termination of phototransduction and in the establishment and maintenance of rhodopsin levels in photoreceptor cells [10,11]. In humans, PITPnm1 has widespread tissue distribution and can rescue fly rdgB mutant phenotypes [1]; whereas PITPnm2 has a neuronal-specific expression pattern and is unable to functionally rescue fly rdgB mutants [12]. Although Drosophila rdgB possesses the capability to transfer PtdCho and PtdIns in vitro [10,11], the PITP-domains of rdgB and PITPα are not functionally interchangeable [10].
An additional class of PITP-like proteins, rdgBβ, has been identified in mice, humans, and Drosophila. These are 38 kDa, soluble proteins that have sequence similarity most comparable to the N-terminal region of rdgB-class proteins. It shares approximately 40% sequence identity with PITPα/. The purified protein has been shown to possess in vitro PtdIns transfer capabilities [13,14].
Plants and fungi generally do not contain a sequence homologue to PITP, but do possess a functional analog referred to as Sec14p in yeast systems. Sec14p is approximately the same size as PITP, and although there is no detectable amino acid similarity between the two proteins, temperature-sensitive mutants of yeast Sec14p are rescued by rat PITPα and PITP [15,16]. Likewise, Sec14p can successfully substitute for PITP in the PITP-dependent reconstitutions studied to date [17,18,19,20]. Interestingly, the slime mold Dicytostelium discoideum has been shown to contain not only homologues to PITP, called DdPITP1 and DdPITP2, but also a Sec14p relative, called DdSec14 [21].
The experimentally determined three-dimensional structures of rat PITPα-PtdCho [22] and PITP-PtdCho [23], human PITPα-PtdIns [24], and the apo form of mouse PITPα [25] have been reported. The PITP structures share little resemblance to the crystal structure of yeast Sec14p [26]. The PITP structure is composed of three regions (Figure 2(a)): a lipid-binding core, a regulatory loop, and the C-terminal region. The lipid-binding core of PITPα-PtdCho shares a fold with the steroidogenic acute regulatory protein-related transfer (START) domain [22] first observed in human MLN64 [27]. It has been proposed that the START domain may be a common fold adapted for binding lipid molecules. PCTP is a START-domain protein based on structure and sequence identity, whereas PITP is not considered to be a START-domain protein due to a lack of sequence identity.
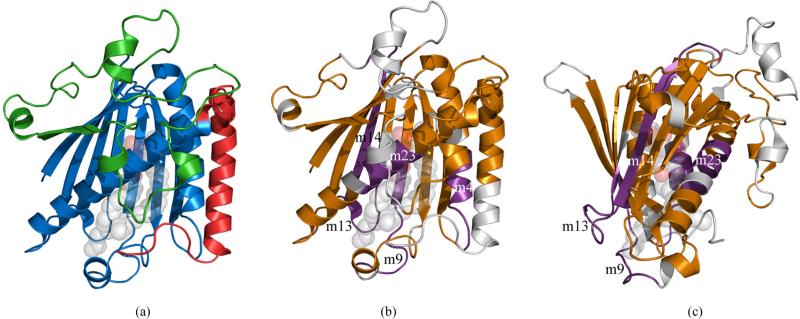
Figure 2.
Cartoon representation of rat PITPα complexed to PtdCho. (a). Structural domains of the class I PITPs displayed on a representative PITP (PDB:1T27). The lipid binding core is shown in blue, the regulatory loop in green, and the C-terminal region in red. The PtdCho molecule is in transparent spheres with standard CPK coloring; (b) Conserved sequence motifs mapped onto the same structure. The four conserved sequence motifs; m4, m9, m13, m14, and m23, representing branch points in the cladogram shown in Figure 3 are in purple and are annotated. The remaining sequence motifs are in gold, regions not present in conserved sequence motifs are colored grey; (c). Image B is rotated 90° counterclockwise around a vertical axis.
Mutations and gene-knockout studies provide insight into the functions of PITP. In mice, a mutation in the PITPα gene causes the vibrator phenotype, which is characterized by a progressive-action tremor, degeneration of brain stem and spinal cord neurons, and juvenile death. PITPβ does not compensate for loss of PITPα [40]. Furthermore, in mice, embryonic stem cells deficient in PITPα or PITPβ reveal differences in physiological function between the two isoforms. PITPβ deficiency leads to catastrophic failure early in embryonic development, and the protein is therefore posited to have an essential housekeeping role in the cell [29]. In contrast, PITPα-deficient embryonic stem cells are not compromised in growth or in bulk phospholipid metabolism; however, PITPα is required for neonatal survival. PITPα deficiency affects regulation of phospholipid transport in the ER, endocrine pancreas function, and glycogen metabolism, due to compromised lipid absorption, homeostatic problems, and severe hypoglycemia [30].
Protein similarity is usually detected by conservation of function as revealed by biochemical analysis, sequence similarity as detected by amino acid pattern rec ognition, or by structural similarity as detected by X-ray crystallography or NMR spectroscopy. The PITP/rdgB proteins possess intriguing patterns of similarity. PITP has functional similarity to Sec14p, but lacks sequence or structure similarity. PITPs have sequence similarity to rdgB and both possess the hallmark ability to transfer phospholipids between the protein and membranes. PITP has been shown to have structural similarity to the START domains, even though it does not share amino acid sequence similarity. Here, we have used a comprehensive evolutionary analysis to synthesize information from sequence analysis and structural comparisons. This approach leads to a cohesive understanding of evolutionary, structural, and functional relationships of the PITP/rdgB protein families, and may suggest that a re-analysis of PITP protein naming conventions is overdue.
2. MATERIALS AND METHODS
2.1. Identification of Sequences
Rat PITPα (gi:8393962) was used as a query sequence in a BLASTp (version 2.2.11, database versions as of May 8, 2005) [31] search. The 134 returned sequences were manually culled to remove duplicated or significantly fragmented sequences, reducing the number to 60 sequences in 26 species (Table 5). The final data set contained both cytosolic PITP and rdgBs of approximately 270-330 residues each, as well as the membrane-associated rdgB/PITPnm proteins, of approximately 1250 residues. Several proteins were determined to be splice variants of existing proteins in the tree; they are annotated in all figures and tables.
Table 5.
Proteins used in phylogenetic analysis.
| gi # | Tracking # | Species | common | Protein designations |
|---|---|---|---|---|
| 55243798 | 19 | Anopheles gambiae | mosquito | |
| 48135931 | 20 | Apis mellifera | honeybee | |
| 39590557 | 16 | Caenorhabditis briggsaae | nematode | |
| 39590636 | 33 | Caenorhabditis briggsaae | nematode | CB09751 b (NCBI-COG: PITP) |
| 17556182 | 32 | Caenorhabditis elegans | ||
| 17554244 | 15 | Caenorhabditis elegans | nematode | PITP DDHD (NCBI-COG: PITP) |
| 48059855 | 58 | Caenorhabditis elegans | nematode | Y71G12B.17 b (NCBI-COG: PITP) |
| 57091327 | 40 | Canus familiaris | dog | similar to PITPα a |
| 41055500 | 49 | Danio rerio | zebrafish | PITP |
| 41055576 | 51 | Danio rerio | zebrafish | similar to PITP |
| 28422482 | 52 | Danio rerio | zebrafish | |
| 8307957 | 56 | Dictyostelium discoideum | slime mold | PITP 2 |
| 8307955 | 57 | Dictyostelium discoideum | slime mold | PITP 1 |
| 24641869 | 17 | Drosophila melanogaster | fruit fly | CG11111-PB, isoform B, rdgB |
| 62484257 | 12 | Drosophila melanogaster | fruit fly | CG17818-PA, rdgBβ |
| 7300495 | 35 | Drosophila melanogaster | fruit fly | CG5269-PA PITP, vib |
| 20151901 | 34 | Drosophila melanogaster | fruit fly | SD01527p, vib |
| 54642914 | 18 | D.pseudoobscura | fly | GA10766-PA, Dpse\GA10766 |
| 19170839 | 60 | Encephalitozoon cuniculi | PITP | |
| 50758480 | 41 | Gallus gallus | chicken | similar to PITPα a |
| 50756343 | 31 | Gallus gallus | chicken | similar to PITP, membrane-associated 2 a |
| 50755397 | 13 | Gallus gallus | chicken | similar to rdgBβ a |
| 53134209 | 47 | Gallus gallus | chicken | |
| 50757849 | 8 | Gallus gallus | chicken | similar splicing variant rdgBβ a |
| 29250063 | 59 | Giardia lamblia | similiar to D. discoideum PITP1 | |
| 18490106 | 21 | Homo sapiens | human | PITPNM1 |
| 24308237 | 27 | Homo sapiens | human | PITPNM2 |
| 5453908 | 38 | Homo sapiens | human | PITPα |
| 6912594 | 43 | Homo sapiens | human | PITPβ |
| 32307140 | 7 | Homo sapiens | human | PITP, rdgBβ 1 |
| 6679337 | 36 | Mus musculus | mouse | PITPα |
| 9790159 | 45 | Mus musculus | mouse | PITPβ |
| 22003862 | 5 | Mus musculus | mouse | rdgB |
| 6679339 | 23 | Mus musculus | mouse | PITPNM1 |
| 47124324 | 25 | Mus musculus | mouse | PITPNM2 |
| 2137007 | 39 | Oryctolagus cuniculus | rabbit | PITPα |
| 55660937 | 42 | Pan troglodytes | chimp | PITP a |
| 55644759 | 54 | Pan troglodytes | chimp | similar to PITPα a |
| 55639157 | 30 | Pan troglodytes | chimp | similar to PITP membrane-associated 2 a |
| 55636463 | 24 | Pan troglodytes | chimp | similar to PITP membrane-associated 1 a |
| 56493706 | 2 | Plasmodium berghei | PITP b | |
| 56509884 | 4 | Plasmodium chabaudi | protein b | |
| 23619433 | 3 | Plasmodium falciparum | PITP b | |
| 23485539 | 1 | Plasmodium yoelii yoelii | PITP 2 | |
| 55731914 | 46 | Pongo pygmaeus | orangutan | protein b |
| 56605814 | 22 | Rattus norvegicus | rat | PITPnm, PI membrane-associated a |
| 62658898 | 26 | Rattus norvegicus | rat | similar KIAA1457, PITPnm2a |
| 8393962 | 37 | Rattus norvegicus | rat | PITP, PITPn |
| 16758568 | 44 | Rattus norvegicus | rat | PITP |
| 62657241 | 6 | Rattus norvegicus | rat | rdgB |
| 56756428 | 55 | Schistosoma japonicum | Unknown, similiar to vib (fly) | |
| 47228496 | 53 | Tetraodon nigroviridis | pufferfish | |
| 47228470 | 28 | Tetraodon nigroviridis | pufferfish | |
| 47223953 | 9 | Tetraodon nigroviridis | pufferfish | |
| 47206861 | 14 | Tetraodon nigroviridis | pufferfish | |
| 47226436 | 29 | Tetraodon nigroviridis | pufferfish | |
| 47937798 | 48 | Xenopus laevis | frog | MGC84500 proteinb |
| 49256286 | 10 | Xenopus laevis | frog | MGC84224 protein |
| 62860160 | 11 | Xenopus tropicalis | frog | protein LOC549393 b |
| 38512077 | 50 | Xenopus tropicalis | frog | PITPβ |
indicated as a predicted protein
indicated as a hypothetical or a putative protein
2.2. Phylogenetic Analysis
Unaligned proteins were processed using the MEME program [32] (implemented in the Wisconsin Package, version 10.4) [33], using the “zero or more” setting, which permits anywhere from zero instances of a block to extreme repetition of blocks, thereby permitting the generation of the most robust matrix of sequence motifs possible. The phylogenetic parsimony tree for the sequence motifs was made by PAUP* (version 4.0b) [34]. The consensus tree was imported into MacClade (v 4.08) [35] for further character-state analysis. Character-state changes were traced along tree clades and unambiguous changes were examined. A database of sequence motifs and proteins was created to facilitate this study and the tables from it are available upon request.
In this phylogenetic analysis, the tree root was designated by the branch that incorporated the most sequence motif additions and that led to the most consistent character-state changes along the tree. That is, the basal proteins that rooted the tree had the fewest sequence motifs character-state changes within the tree with this root present. This rooting was borne out by the consistency andwas utilized. This rooting suggested the fewest character state changes and least amount of homoplasy within the tree. The tree root consisted of small PITP proteins from Plasmodium, Giardia, and Encephalitozoon. Sequence motif 14 defined this branch point, proteins from these species lacked the motif, and all other proteins possessed the conserved sequence motif.
3. RESULTS
3.1. Characterization of PITP-Like Proteins
An initial approach to characterizing the PITP family was relatively standard. Rat PITPα was used as the seed for a BLAST search targeting relatively distant protein relatives. BLAST results were imported to Pileup [33] to generate a “first pass” global alignment. Under a variety of different conditions, including weighting end gaps, not weighting end gaps, utilizing low gap opening and extension penalties, high-road and low-road alignment options, alignments were very weak, with notable cases where locally homologous segments identified by BLAST were not aligned in the global matrix. The derived nature of many of the proteins in the PITP family was already apparent; sequence motifs appeared in some classes but not others, rendering standard alignment and analysis methods ineffective. A novel approach was required to identify and analyze the large, divergent PITP family.
3.2. Identification of Conserved Sequence Motifs
When a high-confidence global alignment in Pileup proved impossible, unaligned proteins were processed using MEME [32] which identifies conserved sequence motifs in proteins. The program makes identifications based on prior probabilities of amino acid occurrence. Because it performs ungapped determinations, no preceding global alignment of sequence motifs was necessary, families with highly derived members. Of the 46 conserved sequence motifs detected by MEME, 25 fell within PITP-like domains.
3.3. Phylogenetic Analysis
With sequence motifs identified, two character-state matrices were created. One contained character-state information on the presence or absence of detected motifs, and one contained aligned amino acid data (Tables 1 and 2). These matrices were utilized for the creation of a character state phylogenetic tree via maximum parsimony. The overall shape of the tree was determined by the presence/absence matrix, with the amino acid alignment data utilized to determine internal clade structures. Gaps, i.e., missing sequence motifs were considered a new state, and thus, the difference between having a specific set of amino acids and not having an amino acid could served as an informative character state.
Table 1.
Conserved sequence motif matrix of PITP and rdgB proteins, PITP domain only.
| Sequence motif identifier |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tracking # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 |
| 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 2 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 |
| 3 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| 4 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 59 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 60 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 56 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 15-16 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 17-19, 25, 28-29 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 20 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 21-24 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 |
| 26 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 27 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 30 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| 57 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 5-7 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 8 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 9 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 10 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| 11 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 12 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 58 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 34 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 |
| 53 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 |
| 54 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 |
| 36-39, 42-51 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 40-41, 52 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 55 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 |
| 32 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| 33 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
Table 2.
Conserved sequence motif matrix of rdgB proteins, C-terminal of the PITP-domain.
| Sequence motif identifier |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tracking # | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 |
| 15 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| 17,21-22, 26, 28 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| 18 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 20 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 19 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 24 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 25 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 27 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 29 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| 30 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Twelve most parsimonious trees of differing structures were identified by PAUP, but the strict consensus tree structure suggests the likely evolutionary history of this intriguing family of proteins (Figure 3). The tree revealed that membrane-associated proteins are derivatives of a group of PITP-like genes, and that many genes previously thought to be related to PITPα and PITP via sequence homology fell outside of clades that contain exclusively PITPα or PITP genes.
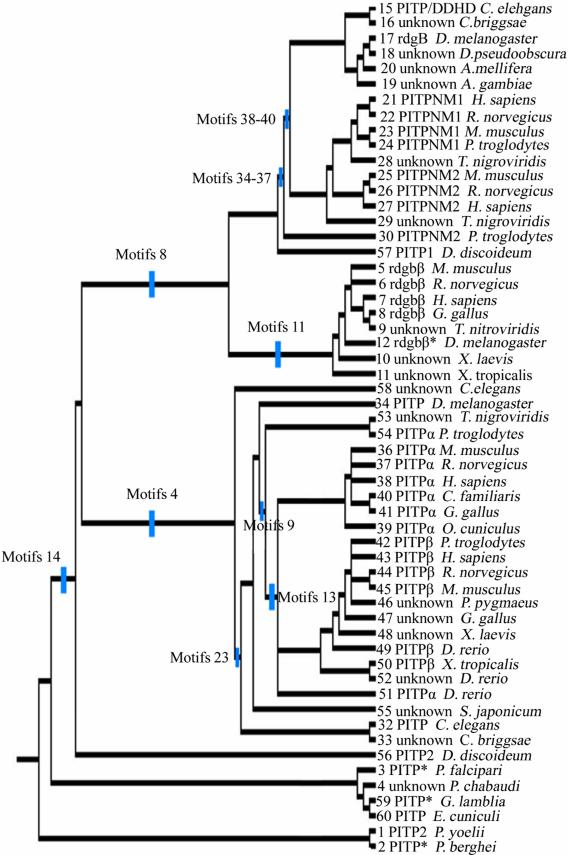
Figure 3.
Phylogenetic tree for 56 PITP family proteins. A strict consensus tree was derived from the presence/absence of sequence motifs as well as the motif sequence, analyzed via parsimony as described in the text. The blue lines show which sequence motifs are uniformly gained within each clade on the tree.
3.4. Identification of Clades Within the Phylogenetic Tree
In order to be cladistically sound, nomenclature must be hierarchical; all proteins in the tree were “PITP-like.” The tree lends itself to consideration of the PITP-like family consisting of three large divisions (the classification nomenclature proposed by Allen-Baume [1] is adapted here): Class I comprised the soluble PITPs, Class IIA were the membrane-associated rdgB proteins (which contain additional domains C-terminal to the PITP-like domain), and Class IIB were the soluble rdgBβ proteins. Further subdivisions along cladistic lines allowed for the naming of the Alpha clade and the Beta clade within Class I, and an ancestral group of PITP-like proteins, predominantly from protista, that occured in several clades rooting the tree. Table 3 lists the nomenclature commonly utilized for the genes and proteins in this study, as well as the HUGO-approved nomenclature.
Table 3.
Gene and protein nomenclature for proteins utilized in this study.
| PITPα | PITP | rdgBβ | rdgB(1) | rdgB(2) | |
|---|---|---|---|---|---|
| Gene name | Pitpna (mouse) Pitpn (rat) Pitpna (human) |
Pitpnb (mouse) Pitpnb (rat) Pitpnb (human) |
pitpnc1 (human) | pitpnm1 (mouse) NIR2 rdgBα1 (fly) DRES9 |
pitpnm2 (mouse) NIR3 rdgBα2 (fly) |
| Gene symbols | Pitpna (mouse) Pitpn (rat) Pitpna (human) |
Pitpnb (mouse) Pitpnb (rat) Pitpnb (human) |
pitpnc1 (human) | pitpnm1 (mouse) | pitpnm2 (mouse) |
| Gene aliases | Pitpn, vib1A | Vib1B | Rd9; RdgB; DRES9; mpt-1; Pitpnm | NIR3; rdgB2; rdgB 2; mKIAA1457 | |
| Protein name | PITPα | PITP | rdgBβ; rdgB 1; PITP, cytoplasmic 1* | PITP membrane-associated 1 | PITP membrane-associated 2 |
| Approved gene symbol | PITPNA | PITPNB | PITPNC1 | PITPNM1 | PITPNM2 |
3.5. Examination of Conserved Sequence Motifs
To understand the functional shifts that have occurred, examination of some of the sequence motifs in each proposed class was necessary. The amino acid sequence of each motif in the human proteins PITPα (Class I), PITPNM1 (Class IIA), and PITPNC1, also called rdgBβ (Class IIB) are given in Table 4.
Table 4.
Conserved sequence motifs in the PITP-domain.
| Motif Identifier | secondary structure element, PITPαa | Sequence rangeb: PITPα PITPnm1 PITPNC1 |
Motif sequenceb: PITPα PITPnm1 PITPNC1 |
|---|---|---|---|
| 1 | sheet 1 (3-11) | 2-9 | 2-VLLKEYRV-9 |
| 1 | 1-8 | 1-MLIKEYHI-8 | |
| 1 | 1-8 | 1-MLLKEYRI-8 | |
| 2 | helix A (14-33) | 12-24 | PVSVDEYQVGQLY |
| 2 | 11-23 | PMSLDEYQVAQLY | |
| 2 | 11-23 | PLTVDEYKIGQLY | |
| 3 | - | -------- | |
| 3 | 24-31 | MIQKKSRE | |
| 3 | 24-31 | MISKHSHE | |
| 4 | helix A (14-33) | 26-36 | VAEASKNETGG |
| 4 | - | ----------- | |
| 4 | - | ----------- | |
| 5 | sheet 2 (39-49) | 37-48 | GEGVEVLVNEPY |
| 5 | 37-48 | GSGVEILANRPY | |
| 5 | 36-47 | GEGVEVVQNEPF | |
| 6 | sheet 3 (55-64) | 55-64 | GQYTHKIYHL |
| 6 | 56-65 | GQYTHKVYHV | |
| 6 | 55-64 | GQFTEKRVYL | |
| 7 | helix B (70-75) | 66-73 | SKVPTFVR |
| 7 | 204-211 | AKIEQFIH | |
| 7 | 198-205 | TRVEQFVH | |
| 8 | - | ----------- | |
| 8 | 69-79 | IPGWFRALLPK | |
| 8 | 68-78 | LPSWARAVVPK | |
| 9 | 76-86 | APEGALNIHEK | |
| 9 | - | ----------- | |
| 9 | - | ----------- | |
| 10 | sheet 4 (84-91) | 87-96 | AWNAYPYCRT |
| 10 | 88-97 | SWNAYPYTRT | |
| 10 | 86-95 | AWNYYPYTIT | |
| 11 | - | -------- | |
| 11 | - | -------- | |
| 11 | 97-104 | YTCSFLPK | |
| 12 | - | -------- | |
| 12 | 98-105 | RYTCPFVE | |
| 12 | - | -------- | |
| 13 | sheet 5 (94-100) | 99-106 | TNEYMKED |
| 13 | - | -------- | |
| 13 | - | -------- | |
| 14 | sheet 6 (108-117) | 107-120 | FLIKIETWHKPDLG |
| 14 | 107-120 | FSIEIETYYLPDGG | |
| 14 | 105-118 | FSIHIETKYEDNKG | |
| 15 | helix C (132-137) | 123-134 | ENVHKLEPEAWK |
| 15 | - | ------------ | |
| 15 | - | ------------ | |
| 16 | - | ----------------------- | |
| 16 | 124-146 | NVFNLSGAERRQRILDTIDIVRD | |
| 16 | - | ----------------------- | |
| 17 | sheet 7 (138-142) | 137-144 | EAVYIDIA |
| 17 | - | -------- | |
| 17 | 134-141 | EVCFIDIA | |
| 18 | 153-160 | DYKAEEDP | |
| 18 | 152-159 | EYKAEEDP | |
| 18 | 149-156 | YYKESEDP | |
| 19 | 163-173 | FKSIKTGRGPL | |
| 19 | 162-172 | YHSVKTGRGPL | |
| 19 | 159-169 | FKSEKTGRGQL | |
| 20 | helix E (177-183) | 175-183 | PNWKQELVN |
| 20 | - | --------- | |
| 20 | - | --------- | |
| 21 | sheet 8 (191-201) | 190-206 | MCAYKLVTVKFKWWGLQ |
| 21 | 187-203 | MCAYKLCKVEFRYWGMQ | |
| 21 | 181-197 | MCSYKLVTVKFEVWGLQ | |
| 22 | helix F (206-232) | 207-214 | NKVENFIH |
| 22 | - | -------- | |
| 22 | - | -------- | |
| 23 | helix F (206-232) | 217-224 | ERRLFTNF |
| 23 | - | -------- | |
| 23 | - | -------- | |
| 24 | helix F (206-232) | 225-244 | HRQLFCWLDKWVDLTMDDIR |
| 24 | 223-242 | HRQAWCWQDEWTELSMADIR | |
| 24 | 216-235 | HRQAFAWVDEWYDMTMDEVR | |
| 25 | helix G (240-261) | 245-253 | RMEEETKRQL |
| 25 | 243-252 | ALEEETARML | |
| 25 | - | ---------- | |
| 26 | - | -------- | |
| 26 | 294-301 | PPGPDASP | |
| 26 | 269-276 | RSAPSSAP |
Secondary structure nomenclature for rat PITP as defined in [22], parenthesis indicate the complete range of the secondary structure element.
Sequences and residue numbering refer to human proteins, PITPα is gi:5453908, tracking #38; PITPnm1 is gi:18490106, tracking #21; PITPNC1 is gi:32307140, tracking #7. PITPNC1 is also called rdgBβ.
Class I: These proteins are cytosolic and contain sequence motifs 1-2, 4-7, 9-10, 13-15, and 17-25 (Figure 4). This class was distinguished from the Class II divisions by sequence motif 8, which was unique to the Class IIA and IIB clades, and sequence motif 4, which was unique to the non-protista Class I PITPs. A striking observation about sequence motif 8 was that it appears to be an amino acid sequence overlap with sequence motif 9, which defines subnodes within Class I. In the structure of rat PITPα, the residues defined by sequence motif 9 incorporate helix B, which is a putative membrane insertion helix [25]. There was no unique conserved sequence motif that distinguishes the functionally divergent alpha and the beta clades.
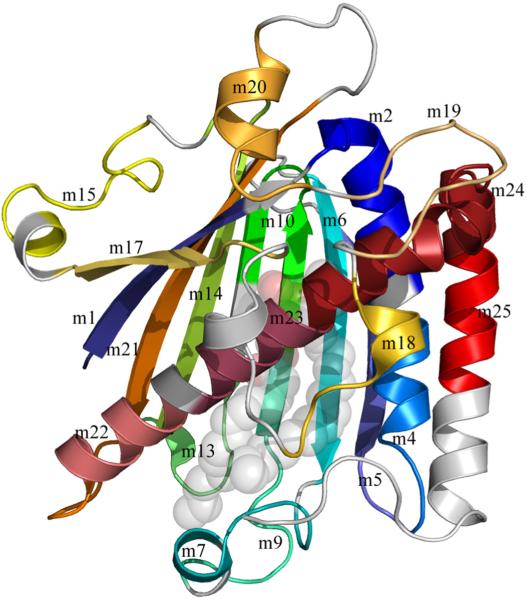
Figure 4.
Sequence motifs mapped to the structure of rat PITPα:PtdCho. The sequence motifs present in class I PITPs are shown in a rainbow spectrum of colors from blue to red and from the N- to the C-terminus. Sequence motifs are annotated ‘m1’ to m25’, the PtdCho molecule is in transparent spheres with standard CPK coloring. The lipid binding core contains sequence motifs 1-2, 4-7, 9-10, 13-15, and 17-25. The regulatory loop contains sequence motifs 15, 17, and 18-20. The C-terminal region contains part of sequence motif 24 and all of motif 25.
Class IIA: These proteins are membrane-associated and typically are 160 kDa. In this analysis they were distinguished by the unique presence of sequence motifs 34-40, which consisted of amino acids residues 831-844, 856-866, 899-920, 937-951, 993-1015, 1022-1038, and 1040-1086 in the human PITPnm1 protein. The division had two major subnodes, one consisting of non-mammalian proteins and containing the canonical rdgB protein from D. melanogaster, and the other subnode containing the mammalian sequences PITPNM1 and PITPNM2. There were 21 conserved sequence motifs outside the PITP domain, with sequence motifs 34-37 observed in all analyzed proteins in this class. In human pitpnm1, the FFAT domain was found at the end of sequence motif 27, with a derived motif sequence of EFFDCLD. The DDHD region contained sequence motifs 30-35, which comprises 35% of the defined DDHD range, and the LSN2 region contained sequence motifs 39 and the N-terminal portion of sequence motif 40, which is 23% of this region. Lev, et al. defined six, short hydrophobic regions based on hydrophathy plots that were originally postulated to be transmembrane domains [9] then later to mediate membrane association [7]. None of the six regions fell in a conserved sequence motif identified in this study On the other hand, sequence motif 26 appeared in multiple repeats throughout the non-PITP domain in sequence motifs 29, 32-33, and 44-45. This is a Ser rich motif of eight amino acids, the significance of this observation is currently unclear.
Class IIB: The proteins in this class are historically referred to as rdgBβs and are monomeric, cytosolic proteins of ~330 amino acids. They were distinguished by the unique presence of sequence motif 11, which consists of residues 97-104 in the human PITPNC1 sequence. Sequence motif 11 is interesting; although it was only present in proteins in this division, it overlaped in sequence with motif 12 and motif 13. Sequence motif 12 was only observed in class IIA, whereas sequence motif 13 was unique to Class I, alpha and beta clades. Another notable feature of this division was PITP1 from D. discoideum. It grouped outside the other rdgB. proteins and rooted the class IIA clade.
4. DISCUSSION
4.1. Phylogenetics
Ocaka et al [36] provide an extensive analysis of the chromosomal location of the PITPN genes in humans with a phylogenetic tree rooted with C. elegans PITP as the chosen outgroup. The analysis presented here differs in two respects. First, the original gene-duplication event leading to the evolution of soluble and membrane-associated PITP proteins likely occurred not early in animal evolution, but instead was initiated in some protists. D. discoideum has been shown to have two genes, pitA and pitB, and expression has been demonstrated for both proteins, PITP1 and PITP2 [21]. The phylogeny shown here indicates that PITP1 is a class IIB protein, perhaps representative of an ancestral precursor to class IIA proteins. Second, data presented here indicate that the Class IIB proteins are derived from Class I proteins, and the membrane-associated Class IIA proteins are derived from the class IIB proteins. Previous cladograms and dendrograms have indicated different derivization patterns. Ocaka et al [36] indicate that mammalian PITPNC1 (Class IIB in the present analysis) shares a most recent common ancestor (MRCA) with PITPNA and PITPNB; more so than with any of the PITPNMs. Fullwood et al. present a dendrogram in which the membrane-associated PITPs and the soluble PITPs split from a shared derivation from Class IIB proteins (rdgBβ). An increased sample size, an unbiased tree root, and the use of conserved sequence motifs for global alignment in the present study lead to a different interpretation of the evolution of this protein family.
The phylogenetic tree produced here is indicative of the evolutionary history of these sequence motifs, and not of the proteins as a whole. This is important, because in some proteins, large sequences with no known sequence motif patterns are observed (such as in the Plasmodium yoelli “PITP1” protein) that may be the result of gene conversion, unequal crossing-over, or repeat expansion during the evolution of specific proteins. Such large insertions and deletions in protein sequence are difficult to utilize in any phylogenetic analysis; they are purely derived characters that are phylogenetically un-informative. However, the existence of these derived sequences suggests that either the function or regulation of proteins in this family has changed dramatically, indicative of the type of specific adaptation often seen within protein families.
4.2. Functional Shifts in PITP Proteins
Alignment and subsequent phylogenetic analysis of divergent members of protein families is a problem compounded by shifts in primary, secondary, and tertiary structure that occur during protein evolution and functional diversification. Global alignments of complete protein coding regions often fail when functional units (such as regulatory regions, transmembrane domains, DNA-binding domains, protein-protein interaction domains, etc.) are gained or lost over evolutionary time. This issue complicates the otherwise straightforward phylogenetic analysis of related proteins. Additional complications arise in organismal surveys in which entire genomes are sequenced and annotated by automated algorithms based on homology. BLAST and FASTA searches find protein regions with high local identity, which can suggest functional similarity. However, outside of these identified regions, major changes can occur in protein function that obscure identity and make global protein alignment difficult or impossible. Without corresponding functional data, these genes are often named inconsistently. For example, a gene found in one species with locally high homology to a protein region in another species is often named “Similar to...” or “Species X homologue of Species Y protein”. Because BLAST is a local homology search tool, it is inadequate for identification and annotation of species orthologs. For this reason, many of the genes identified in species genome projects are insufficiently annotated; phylogenetic analysis of all proteins labeled as “Phosphatidylinositol transfer proteins, Beta” for example will yield a spurious alignment and therefore a misleading tree. As demonstrated in this study, analysis of the PITP/rdgB family was complicated by both issues. It should be noted that full-length sequences produce similar phylogenetic trees for alignments of many related sets of proteins (all Class I or Class II proteins, for example).
The resultant phylogenetic tree reveals that the general naming convention for the PITP family needs to be critically examined. Many of the proteins in this family cannot be named exclusively by local sequence identity, as their functions appear to be significantly altered from previously studied proteins. Many annotated genes are named in a way that might not be truly indicative of their function. In other words, sequence homology in local segments has obscured the greater functional diversification within this family. For example, although the membrane-associated proteins (Class IIB) are shown to be derived and monocladistic, the tree root appears to be within the PITP family, suggesting that many other functions are derived characteristics from this single-domain protein. At best, this tree demonstrates the need for better functional and structural classification of PITP-like proteins before final assignment of their names.
An additional feature of the strict consensus tree is the hypothesis it suggests about the evolution of PITP protein family members. First, no PITP proteins are found within prokaryotes or within yeast. Yeast have an analogous enzyme, Sec14p, but it appears to have an independent evolutionary origin. The slime-mold and Plasmodium sequences represent the most primitive species in this PITP family tree. In contrast, the multiplication of forms within mammals is impressive. Humans, mice, and rats have well-characterized genomes and each have a variety of PITP family-member proteins.
4.3. Conserved Sequence Motifs
The use of conserved sequence motifs in a cladistic analysis of the PITP family highlights several regions for consideration. Two regions are of particular interest: one containing sequence motifs 7, 8, and/or 9, and the other containing sequence motifs 11, 12, or 13. These two regions form loops at the surface of the protein near the ends of the acyl chains of the bound phospholipid (Figure 2(b) and 2(c)). Sequence motif 7 is the only motif present that is not found in the same primary sequence space in the three main protein classes. In human PITPα, the amino acid sequence is at residues 66-73, while it occurs at residues 204-211 and 198-205 in human PITPNM1 and PITPNC1, respectively. Sequence motif 8 is not present in class I PITPs, and partially overlaps with sequence motif 9, which is exclusive to the alpha/beta clades of class I. Sequence motif 7 in the class I PITP's partially overlaps with sequence motif 8 of class IIA and IIB. Sequence motif 7 contains the putative helix insertion loop proposed to anchor PITPα to the mem brane during lipid exchange [25]. It is tempting to speculate that this helix has adapted a different conformation or perhaps has migrated across the lipid-binding cavity to near residues 200-208 relative to the PITPα structures.
The second region of interest contains sequence motifs 11, 12, or 13. These three motifs are present in approximately the same space in primary sequence, but each is unique to the three family classes. Sequence motif 11 is unique to class IIB, whereas Sequence motif 12 is unique to class IIA, and sequence motif 13 is unique to class I. The role of this loop in the structure or the function of any class of PITP is currently unexplored.
A concern with the novel phylogenetic analysis technique used in this study is the proportion of the sequence data actually used in the evolutionary analysis and whether amino acids that have been structurally and functionally shown to affect phospholipid transfer and signal transduction capabilities are included in the alignment matrices. In human PITPα, 214 of the 270 amino acids (79.3%) are included in the conserved sequence motifs. Most site-directed mutagenic analysis of the role of specific residues in the phospholipid transfer, specificity, and PLC reconstitution capabilities have been done on human or rat PITPα and PITPβ. Residues in rat PITPα that have been studied and shown to play a functional role in PITPs include Thr59, Lys61, Glu86, Asn90, Tyr103, and the double Trp pair at 203-204 [37,38]. These residues are all in conserved sequence motifs. Thr59 and Lys 61 map to sequence motif 6, which is the only sequence motif present in all sequences in this study. Interestingly, sequence motif 9, containing Glu 86, is present only in Class I proteins. Tyr103 has been shown to diminish PLC reconstitution without affecting phospholipid transfer. This residue is a Tyr only in the Class I proteins, and is a conservative substitution to a Phe in the class IIA and class IIB proteins. Examination of these amino acids and sequence motifs from an evolutionary perspective enables a more thorough understanding of their importance in shaping PITP function and structure.
4.4. Functional Specialization across Evolution
Functional shifts within the PITP family are difficult to assess in light of the relative paucity of experimental analysis of different family members. In rat, PITPs are broadly expressed and have been detected in at lease 20 tissues [39]. Unigene expression profiles (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene) for mammalian species indicate a slightly more general and ubiquitous expression for the alpha isoform than the beta isoform. Mouse PITPβ −/− embryonic stem cells are embryonically lethal, indicating essential functions in cell viability [29]. In contrast, mice deficient in PITPα develop normally to term, but fail to thrive neonatally [30]. These observations and the EST expression profiles support the argument that PITP isoforms are not redundant, and have different functions [29]. One implication of the consensus tree is that, given that the ancestral proteins have putative roles in cellular function, e.g., PITP2 in Dictyostelium, they may be more widely expressed than the more derived family members, which appear to have roles only in specific tissues or at specific developmental times. PITPnm1 in mice, for example, is highly expressed primarily in the retina, with weaker expression in the brain and some suggestion (from rat data) of central nervous system expression. Other studies in humans showed PITPnm2 and PITPnm3 expression in brain and retina.
The phylogenetic analysis described here has demonstrated the utility of the use of conserved sequence motifs in detecting evolutionary relationships among proteins in large and/or diverse functions. This method uses short conserved sequence motifs instead of single character amino acids. The presence and absence of motifs becomes an important data point. The use of sequence motifs in phylogenetic analysis as described here should be applicable to the analysis of a wide range of protein families, particularly large, diverse families where re-shuffling of domains occurred over evolutionary time.
ACKNOWLEDGEMENTS
The authors wish to acknowledge Lee Likins, Christine Malcom, Justin Paschall, and Ming Yang for comments and assistance. Funding for this study was provided in part by a University of Missouri Research Board grant and NIH R15HD055668-01A1 (to GJW).
REFERENCES
- 1.Allen-Baume V, Segui B, Cockcroft S. Current thoughts on the phosphatidylinositol transfer protein family. FEBS Letters. 2002;531:74–80. doi: 10.1016/s0014-5793(02)03412-9. [DOI] [PubMed] [Google Scholar]
- 2.Cockcroft S, Carvou N. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim. Biophys. Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Hsuan J, Cockcroft S. The PITP family of phosphatidylinositol transfer proteins. Genome Biol. 2001;2:3011.1–3011.8. doi: 10.1186/gb-2001-2-9-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Routt SM, Bankaitis VA. Biological functions of phosphatidylinositol transfer proteins. Biochem. Cell Biology. 2004;82:254–262. doi: 10.1139/o03-089. [DOI] [PubMed] [Google Scholar]
- 5.Thomas GMH, Pinxteren JA. Phosphatidylinositol transfer proteins: One big happy family or strangers with the same name? Mol. Cell Biol. Res. Comm. 2000;4:1–9. doi: 10.1006/mcbr.2000.0253. [DOI] [PubMed] [Google Scholar]
- 6.Wirtz KWA. Phospholipid transfer proteins in perspective. FEBS Lett. 2006;580:5436–5441. doi: 10.1016/j.febslet.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 7.Trivedi D, Padinjat R. RdgB proteins: functions in lipid homeostasis and signal transduction. Biochim. Biophys. Acta. 2007;1771:692–699. doi: 10.1016/j.bbalip.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lev S, Hernandez J, Martinez R, Chen A, Plowman G, Schlessinger J. Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mole. Cell. Biol. 1999;19:2278–2288. doi: 10.1128/mcb.19.3.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milligan SC, Alb JG, Jr., Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vihtelic TS, Goebl M, Milligan S, O'Tousa JE, Hyde DR. Localization of Drosophila retinal degeneration B, a membrane associated phosphatidylinositol transfer protein. J. Cell Biol. 1993;122:1013–1022. doi: 10.1083/jcb.122.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, Vihtelic TS, Hyde DR, Li T. A neuronal-specific mammalian homolog of the Drosophila retinal degeneration B gene with expression restricted to the retina and dentate gyrus. J. Neurosci. 1999;19:7317–7325. doi: 10.1523/JNEUROSCI.19-17-07317.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fullwood Y, dos Santos M, Hsuan JJ. Cloning and characterization of a novel human phosphatidylinositol transfer protein, rdgBb. J. Biol. Chem. 1999;274:31553–31558. doi: 10.1074/jbc.274.44.31553. [DOI] [PubMed] [Google Scholar]
- 14.Takano N, Owada Y, Suzuki R, Sakagami H, Shimosegawa T, Kondo H. Cloning and characterization of a novel variant (mM-rdgB b1) of mouse M-rdgBs, mammalian homologs of Drosophila retinal degeneration B gene proteins, and its mRNA localization in mouse brain in comparison with other M-rdgBs. J Neurochem. 2003;84:829–839. doi: 10.1046/j.1471-4159.2003.01591.x. [DOI] [PubMed] [Google Scholar]
- 15.Skinner HB, Alb JG, Jr., Whitters EA, Helmkamp GM, Jr., Bankaiitis VA. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO. 1993;12:4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka S, Hosaka K. Cloning of a cDNA encoding a second phosphatidylinositol transfer protein of rat brain by complementation of the yeast sec14 mutation. J. Biochem (Tokyo) 1994;115:981–984. doi: 10.1093/oxfordjournals.jbchem.a124448. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham E, Tan SK, Swigart P, Hsuan J, Bankaitis V, Cockcroft S. The yeast and mammalian isoforms of phosphatidylinositol transfer protein can all restore phospholipase C-mediated inositol lipid signaling in cytosol-depleted RBL-2H3 and HL-60 cells. Proc. Natl. Acad. Sci. USA. 1996;93:6589–6593. doi: 10.1073/pnas.93.13.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay JC, Martin TFJ. Phosphatidylinositol transfer protein is required for ATP-dependent priming of Ca2+-activated secretion. Nature. 1993;366:572–575. doi: 10.1038/366572a0. [DOI] [PubMed] [Google Scholar]
- 19.Jones SM, Alb JGJ, Phillips SE, Bankaitis VA, Howell KE. A phosphatidylinositol 3-kinase and phosphatidylinositol transfer protein act synergistically in formation of constitutive transport vesicles from the trans-Golgi network. J. Biol. Chem. 1998;273:10349–10354. doi: 10.1074/jbc.273.17.10349. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi M, de Vries KJ, Frank R, Snoek G, Bankaitis V, Wirtz K, Huttner WB. A role for phosphatidylinositol transfer protein in secretory vesicle formation. Nature. 1995;377:544–547. doi: 10.1038/377544a0. [DOI] [PubMed] [Google Scholar]
- 21.Swigart P, Insall R, Wilkins A, Cockcroft S. Purification and cloning of phosphatidylinositol transfer proteins from Dictyostelium discoideum: homo-logues of both mammalian PITPs and Saccharomyces cerevisiae Sec14p are found in the same cell. Biochem. J. 2000;347:837–843. [PMC free article] [PubMed] [Google Scholar]
- 22.Yoder MD, Thomas LM, Tremblay JM, Oliver RL, Yarbrough LR, Helmkamp GM., Jr. Structure of a multifunctional protein: mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J. Biol. Chem. 2001;276:9246–9252. doi: 10.1074/jbc.M010131200. [DOI] [PubMed] [Google Scholar]
- 23.Vordtriede PB, Doan CN, Tremblay JM, Helmkamp GMJ, Yoder MD. Structure of PITPb in complex with phosphatidylcholine: Comparison of structure and lipid transfer to other PITP isoforms. Biochemistry. 2005;44:14760–14771. doi: 10.1021/bi051191r. [DOI] [PubMed] [Google Scholar]
- 24.van Tiel CM, Schouten A, Snoek GT, Gros P, Wirtz KWA. The structure of phosphatidylinositol transfer protein a reveals sites for phospholipid binding and membrane association with major implications for its function. FEBS. 2002;531:69–73. doi: 10.1016/s0014-5793(02)03403-8. [DOI] [PubMed] [Google Scholar]
- 25.Schouten A, Agianian B, Westerman J, Kroon J, Wirtz KWA, Gros P. Structure of apophosphatidylinositol transfer protein a provides insight into membrane association. EMBO J. 2002;21:2117–2121. doi: 10.1093/emboj/21.9.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 27.Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc. Natl. Acad. Sci. USA. 2002;99:6949–6954. doi: 10.1073/pnas.052140699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton BA, Smith DJ, Mueller KL, Kerrebrock AW, Bronson RT, van Berkel V, Daly MJ, Kruglyak L, Reeve MP, Nemhauser JL, Hawkins TL, Rubin EM, Lander ES. The vibrator mutation causes neurodegeneration via reduced expression of PITPa: positional complementation cloning and extragenic suppression. Neuron. 1997;18:711–722. doi: 10.1016/s0896-6273(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 29.Alb JG, Jr., Phillips SE, Rostand K, Cui X, Pinxteren J, Cotlin L, Manning T, Guo S, York JD, Sontheimer H, Collawn JF, Bankaitis VA. Genetic ablation of phosphatidylinositol transfer protein function in murine embryonic stem cells. Mol. Biol. Cell. 2002;13:739–754. doi: 10.1091/mbc.01-09-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alb JGJ, Cortese JD, Phillips SE, Albin RL, Nagy TR, Hamilton BA, Bankaitis VA. Mice lacking phosphatidylinositol transfer protein-a exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 2003;278:33501–33518. doi: 10.1074/jbc.M303591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Bailey TL, Elkan CP. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press; Menlo Park, CA: 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. pp. 28–36. [PubMed] [Google Scholar]
- 33.Accelrys I. Wisconsin Package v.10.3. Accelrys, Inc; San Diego, CA.: 2005. [Google Scholar]
- 34.Swofford DL. PAUP*. Phylogenetic analysis using parsimony and other methods. Version 4. Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- 35.Maddison DR, Maddison WP. MacClade v.4.08. Sinauer Associates; Sunderland, Massachusetts: 2005. [Google Scholar]
- 36.Ocaka L, Spalluto C, Wilson DI, Hunt DM, Halford S. Chromosomal localization, genomic organization and evolution of the genes encoding human phosphatidylinositol transfer protein membrane- associated (PITPNM) 1, 2 and 3. Cytogenet Genome Res. 2005;108:293–202. doi: 10.1159/000081519. [DOI] [PubMed] [Google Scholar]
- 37.Alb JG, Jr., Gedvilaite A, Cartee RT, Skinner HB, Bankaitis VA. Mutant rat phosphatidylinositol/phosphatidylcholine transfer proteins specifically defective in phosphatidylinositol transfer: Implications for the regulation of phospholipid transfer activity. Proc. Natl. Acad. Sci. USA. 1995;92:8826–8830. doi: 10.1073/pnas.92.19.8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tilley SJ, Skippen A, Murray-Rust J, Swigart PM, Stewart A, Morgan CP, Cockcroft S, McDonald NQ. Structure-function analysis of phosphatidylinositol transfer protein alpha bound to human phosphatidylinositol. Structure. 2004;12:317–326. doi: 10.1016/j.str.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Venuti SE, Helmkamp GMJ. Tissue distribution, purification and characterization of rat phosphatidylinositol transfer protein. Biochim Biophys Acta. 1988;946:119–128. doi: 10.1016/0005-2736(88)90464-6. [DOI] [PubMed] [Google Scholar]
- 40.Lev S. The role of the Nir/rdgB protein family in membrane trafficking and cytoskeleton remodeling. Exp. Cell Res. 2004;297:1–10. doi: 10.1016/j.yexcr.2004.02.033. [DOI] [PubMed] [Google Scholar]