Abstract
Metaseiulus occidentalis is an eyeless phytoseiid predatory mite employed for the biological control of agricultural pests including spider mites. Despite appearances, these predator and prey mites are separated by some 400 Myr of evolution and radically different lifestyles. We present a 152-Mb draft assembly of the M. occidentalis genome: Larger than that of its favored prey, Tetranychus urticae, but considerably smaller than those of many other chelicerates, enabling an extremely contiguous and complete assembly to be built—the best arachnid to date. Aided by transcriptome data, genome annotation cataloged 18,338 protein-coding genes and identified large numbers of Helitron transposable elements. Comparisons with other arthropods revealed a particularly dynamic and turbulent genomic evolutionary history. Its genes exhibit elevated molecular evolution, with strikingly high numbers of intron gains and losses, in stark contrast to the deer tick Ixodes scapularis. Uniquely among examined arthropods, this predatory mite’s Hox genes are completely atomized, dispersed across the genome, and it encodes five copies of the normally single-copy RNA processing Dicer-2 gene. Examining gene families linked to characteristic biological traits of this tiny predator provides initial insights into processes of sex determination, development, immune defense, and how it detects, disables, and digests its prey. As the first reference genome for the Phytoseiidae, and for any species with the rare sex determination system of parahaploidy, the genome of the western orchard predatory mite improves genomic sampling of chelicerates and provides invaluable new resources for functional genomic analyses of this family of agriculturally important mites.
Keywords: Metaseiulus Typhlodromus Galendromus occidentalis, western orchard predatory mite, genome assembly, Helitron rolling-circle transposons, parahaploid sex determination, Dicer-2 gene duplication
Introduction
The western orchard predatory mite Metaseiulus (= Typhlodromus or Galendromus) occidentalis (Arthropoda: Chelicerata: Arachnida: Acari: Parasitiformes: Phytoseiidae) is an important natural enemy of agricultural pests, and is employed for the biological control of mites that feed on crops including apples, grapes, peaches, almonds, cotton, and strawberries (Hoy 2009). Metaseiulus occidentalis can be laboratory-cultured using the two-spotted spider mite Tetranychus urticae as prey, developing in approximately 1 week from eggs, to larvae with three pairs of legs, and then nymphs, and adults with four pairs. Fully grown females are about 500 microns long and have six holocentric chromosomes, whereas males are about half the size and have only three chromosomes after haploidization in the embryo by heterochromatinization and chromosome elimination, that is, parahaploidy (Nelson-Rees et al. 1980). Eyeless like other phytoseiid mites, its sensory system consists of olfactory sensilla and tactile setae on the tips of the first pair of legs and palps (van Wijk et al. 2006). With no real “head,” these palps, together with the chelicerae (mouthparts) make up the gnathosoma, which in mites and ticks is fused to the rest of the body (the idiosoma) with flexible cuticle. To feed, the chelicerae are inserted into the quarry, delivering paralyzing venoms and salivary digestive enzymes for preoral digestion before ingestion of the liquefied prey (Flechtmann and McMurtry 1992).
Despite being a free-living predator, M. occidentalis belongs to the order Parasitiformes, comprising ticks and both predatory and parasitic mites. With the Acariformes, which includes many plant-parasitic mites, they make up the species-rich Acari that span some 400 Myr with many ancient and rapid radiations making resolution of the phylogeny somewhat contentious (Pepato et al. 2010; Sharma et al. 2014). Genomic sampling of the Parasitiformes lineage remains limited, so far including the draft genome sequence of the Lyme disease-carrying tick Ixodes scapularis (Gulia-Nuss et al. 2015) and low-coverage sequencing of the honeybee parasitic mite Varroa destructor (Cornman et al. 2010). Sequenced Acariformes include the plant-feeding spider mite T. urticae (Grbić et al. 2011), as well as the recently sequenced draft genomes of the American house dust mite Dermatophagoides farinae (Chan et al. 2015) and the scabies-causing itch mite Sarcoptes scabiei (Rider et al. 2015). Additional arachnids with draft genome assemblies include a scorpion (Cao et al. 2013) and two spiders (Sanggaard et al. 2014), and improved species sampling continues with sequencing projects such as those of the i5K initiative (Consortium 2013).
Here we present the principal features from the initial genomic analyses of the 152-Mb draft reference assembly of the M. occidentalis genome, the first for a predatory mite, and for any parahaploid species. Comparisons with sequenced Acari and selected representatives of other major arthropod lineages provide the first glimpses of shared and unique attributes of this genome, the most complete arachnid genome sequenced to date. Genome-wide analyses reveal distinct patterns of transposable element (TE) activity and dynamic intron evolution. Analyses of putative genes involved in chemosensation, phototransduction and circadian rhythms, paralysis and preoral digestion, parahaploidy and epigenetics, development, immunity, and RNA interference (RNAi) focus on key aspects of the unique biology of this agriculturally important predator.
Materials and Methods
Genome Sequencing, Assembly, and Annotation
The carbaryl-organophosphate-sulfur resistant strain of M. occidentalis was subjected to inbreeding for ten generations and the selected approximately 12,000 females were then starved for 4 h prior to DNA extraction. DNA was isolated and 15× coverage was generated using 454 XLR technology, with an additional 30× clone coverage in 3- and 8-kb inserts. The resulting 7 million sequence reads were assembled using the Celera assembler (Myers et al. 2000), omitting small and redundant contigs. Assembly completeness was assessed using both the Core Eukaryotic Genes Mapping Approach, CEGMA (Parra et al. 2007) and Benchmarking Universal Single-Copy Orthologs, BUSCOs (Waterhouse et al. 2013; Simão et al. 2015). Gene annotation was performed using version 4.0 of the National Center for Biotechnology Information (NCBI) Eukaryotic Genome Annotation Pipeline, using a process similar to that described for the honeybee (Elsik et al. 2014), and incorporating multi-life-stage RNA-sequencing (RNA-seq) transcriptome evidence (Hoy et al. 2013). The NCBI pipeline produced two annotation sets: 1) The Gnomon set includes all gene models generated from alignment evidence or a combination of alignments and ab initio, plus additional models derived solely from ab initio prediction; and 2) the RefSeq set includes only those Gnomon gene models based at least in part on alignments or with high-quality matches to a subset of the NCBI nonredundant protein database limited to proteins with conserved domain features or nonhypothetical names. See supplementary material, Supplementary Material online, for additional details on genome sequencing, assembly, and annotation.
Comparative Genomics and Manual Curation
Homology and orthology relations of M. occidentalis genes to those from selected other arthropods and outgroup species were sourced from OrthoDB (Kriventseva et al. 2015), which includes the M. occidentalis Gnomon gene set. In total, 117 single-copy orthologs from M. occidentalis and each of 7 other arthropods, 2 trochozoans, 2 vertebrates, and an outgroup cnidarian were used to build the maximum-likelihood species phylogeny with RAxML (Stamatakis 2014). Analyses of intron evolution employed the Maximum Likelihood Analysis of Intron Evolution suite, MALIN (Csurös 2008), to estimate ancestral intron content and counts of gains and losses along the species phylogeny. The posterior probability estimates were based on the alignments of 1,281 orthologous groups with a total of 12,786 intron sites after quality filtering. Manual curation of selected genes and gene families employed tBLASTn and BLASTp sequence searches, sequence profile searches, conserved domain searches, orthology searches, homology-based sequence clustering, multiple sequence alignment, and phylogenetic tree building approaches. See supplementary material, Supplementary Material online, for additional details on comparative genomics and manual curation of selected genes and gene families.
Results
Neither Minuscule nor Massive, a Remarkably Complete Mite Genome
Being so tiny, approximately 12,000 females were required to produce sufficient high-quality DNA for construction of a shotgun and two mate-pair sequencing libraries (see Materials and Methods). Maximizing genome homozygosity within this population was therefore critical, and was achieved by subjecting the already inbred carbaryl-organophosphate-sulfur resistant laboratory strain to ten additional generations of sibling matings. These efforts enabled an extremely contiguous assembly of 2,211 scaffolds spanning 152 Mb, with a contig N50 of 200.7 kb and a scaffold N50 of almost 900 kb (table 1). Compared with other sequenced arachnids, this genome is nearly twice as large as the compact T. urticae genome, but much smaller than most others: About half the size of the V. destructor assembly (whose genome is estimated to be even larger at 565 Mb; Cornman et al. 2010) and less than a tenth of the I. scapularis genome (table 1). The dust and scabies mite genomes are estimated to be small like that of T. urticae, but their draft assemblies appear to capture only about half of their total estimated nucleotide content (Chan et al. 2015; Rider et al. 2015). Assessing assembly completeness in terms of expected gene content with 248 Conserved Eukaryotic Genes (CEGs; Parra et al. 2007) and 2,675 arthropod BUSCOs (Waterhouse et al. 2013; Simão et al. 2015) revealed a remarkably complete M. occidentalis genome assembly, the most complete of any arachnid to date (table 1).
Table 1.
Metaseiulus occidentalis Genome Assembly and Gene Set Statistics Compared with Eight Other Arachnids
| Acari: Parasitiformes | Acari: Acariformes | Araneae | Scorpiones | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Metaseiulus occidentalis | Varroa destructor | Ixodes scapularis | Dermatophagoides farinae | Sarcoptes scabiei | Tetranychus urticae | Stegodyphus mimosarum | Acanthoscurria geniculata | Mesobuthus martensii |
| Organism | Western orchard predatory mite | Honey bee parasitic mite | Black-legged deer tick | American house dust mite | Itch mite (scabies) | Two-spotted spider mite | African social velvet spider | Brazilian white-knee tarantula | Chinese scorpion |
| Assembly | GCA_000255335.1 | GCA_000181155.1 | GCA_000208615.1 | GCA_000767015.1 | GCA_000828355.1 | GCA_000239435.1 | GCA_000611955.2 | GCA_000661875.1 | GCA_000484575.1 |
| Assembly method | Celera v.6.1 | Celera v.5.2 | Celera v.4.0 | AllPaths v.39075 | Minia v.1.6088 | Arachne 20071016 | SOAPdenovo v.2 | SOAPdenovo v.2 | Velvet v.1.1.04 |
| Velvet v.1.1.03 | SSPACE | ||||||||
| SOAPdenovo v.1.05 | Standard v.3.0 | ||||||||
| Coverage (×) | 17.7 | 5 | 6 | 436 | 174 | 8a | 70 | 70 | 200 |
| Assembly length (Mb) | 151.7 | 294.1 | 1,765.4 | 53.5 | 56.3 | 90.8 | 2,738.7 | 7,178.4 | 925.5 |
| 1,128.5b | |||||||||
| No. of Contigs | 3,993 | 184,190 | 570,637 | 11,600 | 19,811 | 2,035 | 174,164 | 12,478,692 | 92,408 |
| 18,600c | |||||||||
| Contig N50 (bp) | 200,706 | 2,262 | 2,942 | 8,538 | 11,197 | 212,780 | 40,146 | 541 | 45,228 |
| 11.6 kbc | 17,272d | 277d | 43,135b | ||||||
| No. of scaffolds | 2,211 | na | 369,492 | 515 | na | 640 | 68,653 | na | na |
| Scaffold N50 (bp) | 896,831 | na | 76,228 | 186,342 | na | 2,993,488 | 480,636 | 47,837d | 223,560b |
| No. of protein-coding genes | 18,338e | 11,432g | 20,486h | 16,376i | 10,473 | 18,414a | 27,135 | 73,821k | 32,016b |
| 11,430f | 10,644c | 18,224j | 27,235d | ||||||
| % CEGsl | 98.0/96.8 | 67.7/32.3 | 79.8/41.9 | 97.6/96.4 | 98.0/93.5 | 98.0/95.2 | 61.7/24.2 | 32.7/14.9 | 57.3/24.2 |
| % BUSCOsm(assembly) | 82.3 [6.4], 8.3, 9.9 | 15.2 [0.5], 15.4, 69.5 | 68.9 [2.4], 21.0, 10.1 | 63.8 [3.5], 7.6, 28.6 | 62.0 [3.0], 8.4, 29.6 | 68.8 [5.8], 9.9, 21.3 | 53.7 [4.0], 27.4, 18.9 | 13.7 [1.9], 10.6, 75.7 | 34.4 [4.0], 23.0, 42.7 |
| % BUSCOsm(gene set) | 82.8 [12.0], 10.6, 6.5 | na | 69.4 [6.7], 23.4, 7.2 | na | 54.0 [4.7], 11.4, 34.6 | 69.3 [11.4], 9.6, 21.0 | 64.6 [10.5], 17.3, 18.1 | na | 23.1 [6.7], 16.4, 60.5 |
Note.—Unless otherwise indicated, values were retrieved from the NCBI.
From Grbić et al. (2011).
From Cao et al. (2013).
From Rider et al. (2015).
From Sanggaard et al. (2014).
NCBI Gnomon gene set.
NCBI RefSeq gene set.
Identified open-reading frames with significant sequence similarity to database sequences from Cornman et al. (2010).
IscaW1.4 gene set from VectorBase (www.vectorbase.org).
From Chan et al. (2015).
From EnsemblGenomes (http://metazoa.ensembl.org).
From Sanggaard et al. (2014), transcripts of more than 17 amino acids.
% of 248 CEGMA genes Found/Complete, in the assembly.
% of 2,675 arthropod BUSCOs Complete [Duplicated], Fragmented, Missing, in the assembly or in the gene set.
Protein-coding genes, pseudogenes, and noncoding RNAs were annotated using the NCBI Eukaryotic Genome Annotation Pipeline (see Materials and Methods and supplementary table S1, Supplementary Material online), which identified a total of 18,338 Gnomon protein-coding genes, with a processed subset of 11,430 well-supported RefSeq genes. The gene repertoire is larger than that of many sequenced insects (Waterhouse 2015), but it is similar in size to those of I. scapularis and T. urticae and only about two-thirds those of the spider Stegodyphus mimosarum, and the scorpion Mesobuthus martensii (table 1). Comparative analysis with genes from ten representative arthropod species to estimate numbers of near-universally present orthologs that are potentially missing indicated that the Gnomon gene set is more complete than the gene sets of both I. scapularis and T. urticae (supplementary fig. S1, Supplementary Material online). The predatory mite genome is thus neither as compact as that of the spider mite nor as large as many other arachnids, and represents a remarkably complete assembly and annotation for the study of the currently sparsely sampled Acari.
Paraphyly of the Acari and Orphan Arachnid Genes
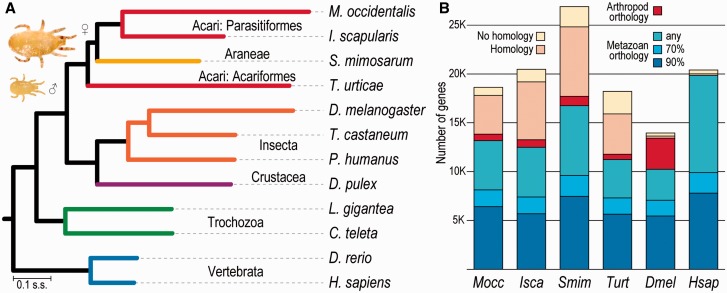
Phylogenomics offers opportunities to address conflicting views of the evolutionary relationships among mites and other arachnids (Pepato et al. 2010; Sharma et al. 2014) using genome-wide sequence data, albeit with a limited sample of fully sequenced genomes with good quality gene annotations. The maximum-likelihood species phylogeny estimated from the concatenated protein sequence alignments of single-copy orthologs (see Materials and Methods) confidently pairs the two Parasitiformes M. occidentalis and I. scapularis (fig. 1A). However, placement of the African social velvet spider Ste. mimosarum as a sister group to these Parasitiformes with the acariform two-spotted spider mite T. urticae as the outgroup results in the paraphyly of the Acari. Complementary phylogenomic analyses with subsets of the species from figure 1A and with orthologs from the less complete gene sets (see table 1) of the M. martensii scorpion and the S. scabiei mite further support these observed relationships (supplementary fig. S2, Supplementary Material online). Depending on the species set and the alignment trimming stringency, these analyses employed some 100 to more than 1,000 single-copy orthologs to build superalignments of between about 55,000 and almost 250,000 amino acids, all resulting in phylogenies with 100% bootstrap support (supplementary material, Supplementary Material online). The relationships are in agreement with recent phylogenomic analyses that also found spiders (Araneae) and ticks to be sister groups (Sanggaard et al. 2014). The molecular phylogeny also highlights the ancient divergence of the two Parasitiformes and their last common ancestor with T. urticae, conservatively estimated to be some 320–360 and 370–420 Ma, respectively (Jeyaprakash and Hoy 2009).
Fig. 1.—
Species phylogeny and orthology profiles. (A) The molecular species phylogeny built from aligned protein sequences of single-copy orthologs and rooted with the cnidarian Hydra vulgaris highlights the ancient divergence of the two Parasitiformes, the fast rate of gene-sequence evolution in Metaseiulus occidentalis, and supports the paraphyly of the Acari. All nodes showed 100% bootstrap support. Branch lengths in substitutions per site (s.s.). Inset: Female (♀) M. occidentalis are about 500 microns long, about twice the size of males (♂). (B) Homology assessments across 173 metazoan species from OrthoDB identify ancient orthologs for more than half of M. occidentalis genes (blue fractions: Orthologs in >90% or >70% of metazoans, or in any nonarthropod metazoan) and a small fraction with only arthropod orthologs (red), but about a quarter remain “orphaned” with no clear orthology, reflecting the sparse sampling and ancient divergences within the arachnids. Homology cutoff: e value < 1e-3. Species: Predatory mite Metaseiulus occidentalis Mocc; deer tick Ixodes scapularis Isca; velvet spider Stegodyphus mimosarum Smim; spider mite Tetranychus urticae Turt; fruit fly Drosophila melanogaster Dmel; flour beetle Tribolium castaneum; body louse Pediculus humanus; water flea Daphnia pulex; owl limpet Lottia gigantea; polychaete worm Capitella teleta; zebrafish Danio rerio; human Homo sapiens Hsap.
Rooted with the cnidarian Hydra vulgaris (= H. magnipapillata), the molecular species phylogeny shows the variable rates of gene sequence evolution, particularly among the fast-evolving arthropods, and highlights the fastest rates in the two mites and Drosophila melanogaster (fig. 1A). These conserved orthologs show that amino acid substitutions per site between M. occidentalis and I. scapularis are more than three times more numerous than between Danio rerio and Homo sapiens, underscoring the large evolutionary distance between these two Parasitiformes. Assessing homology and orthology with 172 other metazoan species (see Materials and Methods) identified ancient orthology for approximately 35% of the M. occidentalis Gnomon gene set (fig. 1B). Others are less well maintained across the metazoans (∼36%), or have orthologs only in other arthropods (∼4%). This leaves about a quarter of this mite’s genes with no clear orthology; most of these have homologs in other metazoans, but a small fraction remains with no detectable homology. Other arachnids exhibit similarly high proportions of genes that lack clear orthology (fig. 1B), reflecting the sparse sampling and ancient divergences within this lineage, as previously noted for a tick transcriptome (Gibson et al. 2013), and in contrast to the dipterans and primates, represented by D. melanogaster and H. sapiens, which are well-sampled with many closely related species.
Dynamic Gene Architecture Evolution in a Genome Rich in Helitrons
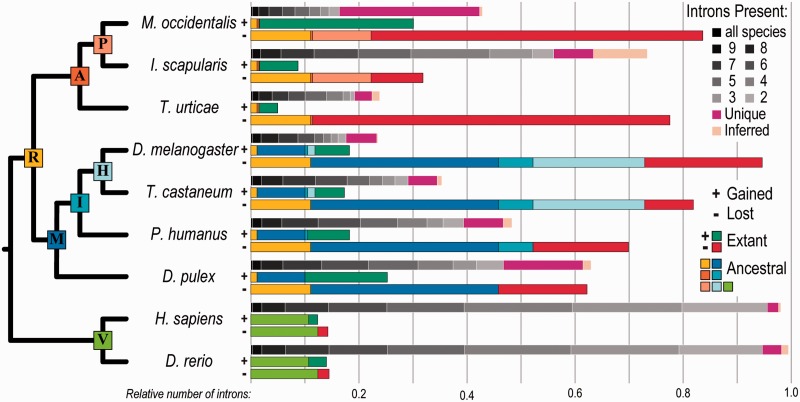
Examining gene architectures of ancient universal orthologs to identify shared and unique intron positions (see Materials and Methods) revealed dramatic intron losses from M. occidentalis genes accompanied by striking numbers of intron gains (fig. 2). The vast majority of these losses and gains occurred since the last common ancestor with ticks, and only D. melanogaster, known for substantial intron losses (Csuros et al. 2011), exhibits more total losses across the phylogeny. At the same time, these mite genes exhibit the most numerous intron gains, exceeding even those of Daphnia pulex, previously noted for extensive intron accumulation (Li et al. 2009). The combination of extensive intron losses with numerous gains leaves this predatory mite with an unusually large proportion of unique, lineage-specific introns (fig. 2). This contrasts the intron-rich gene architectures of the tick, which are more similar to those of a metazoan ancestor than to other arthropods (Gulia-Nuss et al. 2015). The representative vertebrates, fish and human, have the most introns that are shared with any other species, that is, introns that have been stably maintained since the last common metazoan ancestor. Comparing gained versus lost intron sites in M. occidentalis genes revealed an approximately 50:25:25% 0–1–2 phase distribution for both—a ratio observed across many eukaryotes (Csuros et al. 2011) (phase 0: Between codons; phases 1 and 2: After first or second nucleotide in a codon, respectively). However, small but significant biases showed more losses occurred from phase 0 sites (P = 2.0 × 10−07) and more gains occurred at phase 1 sites (P = 1.3 × 10−06), with losses tending to occur more frequently toward the ends of genes (last tenth, P = 1.8 × 10−04; second last tenth, P = 5.1 × 10−05) (supplementary fig. S3, Supplementary Material online). Comparing the lengths of introns unique to M. occidentalis with those that are shared with at least two other species revealed that unique introns are generally shorter (mean 143, 257 bp and median 116, 122 bp; unique, shared, respectively; P = 6.3 × 10−04). Across the examined phylogeny, the relative stability of vertebrate gene architectures contrasts the dynamic changes in intron content throughout the evolution of the arthropods, which appear especially dramatic in M. occidentalis genes that have about half the intron content of I. scapularis and almost double that of T. urticae (fig. 2).
Fig. 2.—
Dynamic Metaseiulus occidentalis intron evolution. Comparing orthologous intron positions in extant species to infer gain and loss events since their last common ancestors reveals dramatic changes in intron content throughout the evolution of arthropods, in contrast to the relative stability of vertebrate gene architectures. Metaseiulus occidentalis has by far the largest proportion of unique introns, driven by the combination of extensive losses and numerous gains. For each species, the three stacked bars show the following: (top bar) shows introns in this species, colored according to the “Introns Present” legend from those found in orthologs from all ten examined species (black) to only two species (gray), or unique (pink) or inferred (almond) to this species; (middle bar, labeled +) shows introns gained in this species color-coded to match the nodes in the adjacent phylogeny where the gains occurred, with gains since the most recent node (extant) shown in bright green; (bottom bar, labeled −) shows lost introns in this species color-coded to match the nodes in the adjacent phylogeny where the losses occurred, with losses since the most recent node (extant) shown in bright red. Introns present in all species are found in the nine organisms labeled on the dendrogram as well as the cnidarian Hydra vulgaris (the outgroup for this analysis). Species: A subset from figure 1. Ancestral nodes on dendrogram: P, Parasitiformes; A, Arachnida; R, Arthropoda; H, Holometabola; I, Insecta; M, Mandibulata; V, Vertebrata. Relative number of introns: Counts normalized by the inferred number of introns in the last common ancestor.
Despite many studies exploring intron gain/loss mechanisms, they remain poorly understood and, while losses can occur by reverse transcriptase-mediated processes and genomic deletions, gain mechanisms remain elusive but may be facilitated by processes including intron transpositions and transposon insertions (Yenerall et al. 2011; Yenerall and Zhou 2012). Analysis of the TE content of the M. occidentalis genome revealed a comparable total fraction of TEs (6.8%) to that of the D. melanogaster euchromatic genome (6.6%) (supplementary figs. S4 and S5 and table S2, Supplementary Material online). However, although the fruit fly genome is dominated by long terminal repeat (LTR) elements, this genome contains much greater proportions of both cut-and-paste DNA transposons and Helitrons (rolling-circle transposons). In contrast, LTR elements are the most abundant TE in the compact T. urticae genome (Grbić et al. 2011) and Mariner-type DNA transposons are the most common in V. destructor (Cornman et al. 2010). Helitron activity can capture gene fragments and may generate chimeric transcripts or alter gene architectures (Thomas and Pritham 2015), thereby contributing to genomic innovations such as alternative transcripts expressed in maize (Barbaglia et al. 2012) or possible gains or losses of gene features as described in bats (Thomas et al. 2014). Compared with other representative arthropods, this genome harbors the greatest proportion of Helitrons (supplementary fig. S5, Supplementary Material online), suggesting a possible link between elevated rolling-circle transposon activity and dynamic gene architecture evolution in this phytoseiid mite.
Detecting, Disabling, and Digesting Prey
This obligatory predator has no eyes, yet it is able to detect and distinguish between prey species using various sensory receptors to identify silk and chemical cues deposited by spider mites and other prey. The ability to then subdue active prey species and to conduct preoral digestion is critical to this mite’s feeding behavior; investigating genes and gene families likely to be involved in these biological traits therefore offers the best clues to understanding its success.
Chemosensation
Like most arthropods, M. occidentalis obtains considerable information about its environment through chemical cues. Noninsect arthropods employ two major gene families for chemosensation, the gustatory receptors (GRs) (Robertson et al. 2003) and the ionotropic receptors (IRs) (Rytz et al. 2013). The insects, however, possess a third family of chemoreceptors, the odorant receptors (ORs), which is likely to have evolved in ancient insects from the GRs (Missbach et al. 2014). Accordingly, like those of other noninsect arthropods including the crustacean Da. pulex (Peñalva-Arana et al. 2009), the myriapod Strigamia maritima (Chipman et al. 2014), and the spider mite (Grbić et al. 2011), the M. occidentalis genome does not harbor any ORs. Instead, it must rely on the 64 GR (six of which appear to be pseudogenes) and 65 IR genes encoded in its genome (see Materials and Methods and supplementary fig. S6 and table S3, Supplementary Material online) for processing the chemical signals from other individuals, for example, male–female signals, as well as from potential prey. The identified GRs are highly divergent from insect GRs of known function, such as the sugar, fructose, carbon dioxide, and bitter taste receptors; assignment of specific functions therefore requires further experimental evidence beyond the clues from homology. Interestingly, although arthropod GRs generally have about five introns, the majority (50/64) of this mite’s GRs form a large clade of intronless genes, spread across the genome, some as singletons but most in small tandem arrays, suggesting multiple duplications of an intronless ancestral gene. Although functional studies are required to explore their specific biological roles, the IRs form two groups comparable to the “conserved” and “divergent” IRs involved in smell and taste, respectively, in D. melanogaster (Rytz et al. 2013). Like the GRs, the mite genome encodes a group of intronless IRs; however, three IR genes that cluster with this group appear to have independently acquired novel introns. In the second large group most genes have five to eight introns, generally fewer than most insect IRs, but one has only a single intron and another has 14. Thus, gene architecture evolution of these divergent groups of chemoreceptors mirrors the dynamism observed from the analysis of introns from near-universal single-copy orthologs, and although sequence divergence prevents detailed functional inferences, the identified GR and IR gene repertoires suggest a comprehensive chemosensory capacity of this predatory mite.
Light Perception
Although spiders are well-known for their many eyes, this phytoseiid has no apparent eyes or ocelli and thus cannot rely on even weak visual cues for prey detection. Nevertheless, diapause induced by shortening daylengths suggests that this mite is capable of perceiving and responding to light (Hoy 1975). Searches for genes involved in early eye development and retinal differentiation (see Materials and Methods) identified homologs of known regulators (supplementary table S4, Supplementary Material online), including twin of eyeless but not eyeless, the D. melanogaster Pax6-like genes that regulate ocular development (Shaham et al. 2012). Homology searches for canonical genes of the phototransduction cascade (see Materials and Methods) identified all four key components: Photoreceptors, trimeric G-proteins, phospholipases, and transient receptor potential channels (supplementary table S5, Supplementary Material online). However, the two putative photoreceptors both appear to be visual pigment-like peropsins rather than true visual opsins, unlike in spiders that have both opsins and peropsins, which, in the wandering spider Cupiennius salei show different expression patterns in primary and secondary eyes (Eriksson et al. 2013). In addition to mite diapause behaviors, examining oviposition times revealed synchronized periodicity (supplementary fig. S7, Supplementary Material online) that provides further support for the ability to perceive light. This is despite the apparent absence of several key components of the circadian rhythm system—no orthologs of Cycle, Period, or Cryptochrome genes were found (supplementary table S6, Supplementary Material online). In contrast, I. scapularis and T. urticae have orthologs of both Period and Cryptochrome, and although Cycle appears to be missing from the tick, it is present in the spider mite. Thus, despite having no eyes this mite seems to possess key eye developmental genes, as well as all the major molecular components necessary for light perception, and it exhibits behaviors that indicate light-induced responses.
Paralysis and Preoral Digestion
Observations of M. occidentalis feeding confirm the use of preoral digestion common to most predaceous arthropods and reveal symptoms of paralysis in spider mite prey suggesting they immobilize their quarry (Flechtmann and McMurtry 1992). Homology searches with digestive peptidases and some 800 salivary peptides from spiders and other venomous arthropods (see Materials and Methods) identified genes with putative roles in this predator’s feeding (supplementary tables S7 and S8, Supplementary Material online). Two neurotoxin-like genes were identified with matches to agatoxins from funnel-web spiders, which block calcium channels causing paralysis in insects (Wang et al. 2001). A single sphingomyelinase D-like gene matched those from recluse and assassin spiders, which cause lesions in mammals and are potent insecticidal toxins (Zobel-Thropp et al. 2012), compared with at least five in T. urticae, three in Ste. mimosarum and four in I. scapularis. Many toxins are short peptides that appear to have emerged independently in different lineages or diverged rapidly; it is thus not surprising that homology searches revealed few confident matches. In contrast, proteolytic digestive enzymes are more easily recognizable, for example, InterPro protein domain annotations identified 124 M. occidentalis trypsin-like cysteine/serine peptidases; however, this is substantially fewer than in the spider mite and tick genomes (supplementary table S9, Supplementary Material online). Specifically, this predator has only one C13 legumain cysteine peptidase and at least 23 confidently identified C1A cathepsins whereas the spider mite has 19 and 57, respectively (supplementary table S10, Supplementary Material online). Interestingly, all the M. occidentalis C1A peptidases belong to the subfamily of cathepsin L-like proteins, with no cathepsin B-like peptidases, unlike in T. urticae, which has at least 17 (supplementary fig. S8, Supplementary Material online). It is possible that such variations in the repertoires of encoded proteases may reflect the different diets and feeding behaviors of these two distantly related mites.
Sex Determination and Development of a Parahaploid Mite with Completely Atomized Hox Genes
Also known as pseudoarrhenotoky or paternal genome elimination, parahaploidy is a rare genetic system found in some mites, beetles, mealybugs, and scale insects (Sánchez 2014). An improved understanding of this embryonic process could help control programs by biasing predatory mite production toward females, which consume more prey than males. The adults exhibit the reduced body-plan segmentation characteristic of mites and ticks, which in acariform mites has been linked to changes in Hox gene expression (Barnett and Thomas 2013) or Hox gene loss (Grbić et al. 2011). As a distantly related mite with similarly reduced segmentation, characterizing M. occidentalis Hox genes offers clues to understanding their roles in body-plan development.
Parahaploidy and Epigenetics
Paternally derived chromosomes are heterochromatinized and eliminated in M. occidentalis embryos that develop into haploid adult males (Nelson-Rees et al. 1980) through a process that allows the mother to control the sex ratios of her offspring (Nagelkerke and Sabelis 1998). As little is known about the molecular players in this unusual genetic system, the identification of candidate genes involved in sex determination focused on searches both for canonical components of signaling cascades as well as on genes involved in epigenetic DNA modifications that precede chromosome elimination in similar systems (see Materials and Methods). Studies in insects have revealed substantial variations in the components of sex-determination systems and their regulatory circuits (Gempe and Beye 2011), so it is not surprising that several elements of the D. melanogaster system appear to be missing from M. occidentalis (supplementary tables S11–S14, Supplementary Material online). Nevertheless, orthologs of the key upstream splicing factor Sex lethal and at least one of its targets, transformer-2, are present, as well as the downstream target transcription factor doublesex (Pomerantz et al. 2014). Although the transformer-2 ortholog did not exhibit any sex-biased gene expression in M. occidentalis, two doublesex-like genes displayed male-biased expression, but without any evidence of alternative splicing (Pomerantz and Hoy 2015a). Although such signals may be involved in controlling sex determination, the process of heterochromatinization itself is likely governed by sex-specific epigenetic marks, as revealed by studies of the parahaploid mealybug Planococcus citri (Bongiorni et al. 2009). DNA methylation is associated with processes such as genomic imprinting and X chromosome inactivation, but searches for DNA methytransferases (Dnmts) in the M. occidentalis genome revealed only the widely conserved Dnmt2 gene and no Dnmt1 or Dnmt3 homologs (supplementary fig. S9, Supplementary Material online). Dnmt2 in fact functions primarily as a tRNA methytransferase, and species with only Dnmt2 are therefore unlikely to be able to rely on DNA methylation for epigenetic regulation (Raddatz et al. 2013). In contrast, the repertoires of histone deacetylases and their cofactors are similar to those of other arthropods (supplementary table S15, Supplementary Material online), suggesting that this epigenetic regulatory system is fully functional. These comparisons point to candidate genes that can now be targeted for detailed characterizations; however, it seems that DNA methylation may not hold the key to unlocking the secrets underlying the unusual system of sex determination in this parahaploid mite.
Completely Atomized Hox Genes
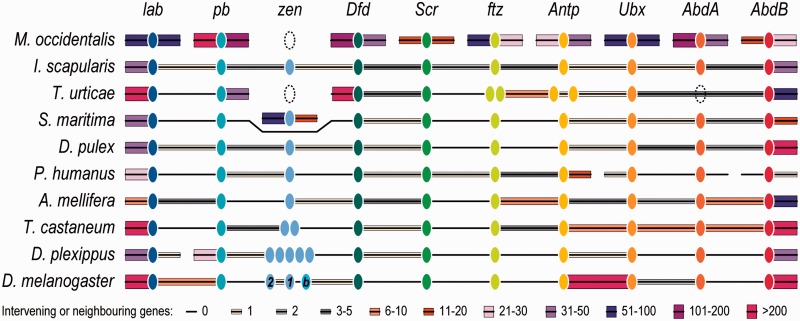
Regardless of their sex, embryonic development leads to the adult form with only two body segments, the gnathosoma fused with flexible cuticle to the idiosoma. This reduced segmentation of mites and ticks is dramatically different from the body plans of insects, crustaceans, and myriapods, and is presumably achieved through segmental fusions during early development. Segment identities and development along the anteroposterior axis are determined by the expression patterns of Hox genes, a widely conserved subset of homeobox-containing transcription factors typically found physically clustered within genomes (Duboule 2007). Of the ten Hox genes present in the arthropod ancestor, searching the M. occidentalis genome (see Materials and Methods) identified orthologs for all except Hox3/zerknüllt/zen and no gene duplications (fig. 3), unlike in the scorpion M. martensii where the complete Hox gene cluster appears to have been duplicated (Di et al. 2015). The zen gene is also missing from the spider mite, but it is present in I. scapularis and has been identified in an oribatid mite (Telford and Thomas 1998) and other chelicerates. Most strikingly, M. occidentalis Hox genes are completely atomized with each gene on a different scaffold, unlike in other examined arthropods where they are generally found to be collinear with no or few intervening genes (fig. 3). They are located on scaffolds ranging in size from 220 kb with 43 genes to 3,231 kb with 413 genes (supplementary fig. S10 and table S16, Supplementary Material online), supporting the conclusion that their atomization is not an artefact of genome assembly fragmentation. Hox genes generally form canonical “organized” clusters but some may be described as “split,” “disorganized,” or “atomized” (Duboule 2007); however, complete Hox gene atomization such as in the tunicate sea squirt Oikopleura dioica (Seo et al. 2004) or the cephalopod Octopus bimaculoides (Albertin et al. 2015), and now for the first time in any arthropod, appears to be a rare occurrence indeed.
Fig. 3.—
Complete atomization of Metaseiulus occidentalis Hox genes. In stark contrast to M. occidentalis, the genomic organization of ten Hox genes from two other arachnids and representative species from seven arthropod lineages shows their generally well-maintained collinearity with no or few intervening genes. This complete atomization of Hox genes is the first reported for any arthropod and highlights the turbulent and dynamic evolutionary history of this predatory mite’s genome. Hox gene orthologs in each species are depicted as color-matched ellipses with dotted ellipses indicating putatively missing genes. Connecting black lines show their scaffold or chromosome collinearity with the number of intervening or neighboring genes indicated by boxes of different heights and colors: Thin gray boxes for 1, 2, or 3–5 intervening or neighboring genes; thicker brown boxes for 6–10 or 11–20 genes; thicker purple boxes for 21–30, 31–50, 51–100, or 101–200 genes; and the thickest bright pink boxes for more than 200 genes. Hox genes: lab, labial; pb, proboscipedia; zen, zerknüllt; Dfd, Deformed; Scr, Sex combs reduced; ftz, fushi tarazu; Antp, Antennapedia; Ubx, Ultrabithorax; AdbA, abdominal A; AbdB, Abdominal B; and labeled in D. melanogaster, 1, zerknüllt; 2, zerknüllt-related; and b, bicoid. Species: A subset from figure 1, plus the centipede Strigamia maritima; the honeybee Apis mellifera; and the monarch butterfly Danaus plexippus.
Immunity without Imd, and RNAi with Duplicated Dicers
In the wild, M. occidentalis is found in low-density populations (less than 0.1/leaf) where diseases appear uncommon. In contrast, infections with microsporidia and other pathogens occur in dense colonies and can seriously impede rearing for mass production in biological control programs (Bjørnson 2008). Understanding this mite’s immune-defense capacities, including the RNAi system, is necessary to improve productivity at mass-rearing facilities.
The Immune Repertoire
Classical arthropod immune responses include phagocytosis, melanization, and the production of antimicrobial peptides in response to signals induced by pathogen recognition, and have been characterized principally with studies in insects (Buchon et al. 2014; Barribeau et al. 2015). Surveying the M. occidentalis genome (see Materials and Methods) identified homologs of key immune-related genes including recognition proteins and signaling pathway members (supplementary table S17, Supplementary Material online). Only a single gene encoding a peptidoglycan recognition protein (PGRP) was identified; T. urticae also has only one PGRP, whereas I. scapularis has at least four, and none of these Acari appears to have any Gram-negative bacteria-binding proteins (GNBPs). Other recognition-related genes include those encoding a fibrinogen-related protein, at least two thioester-containing proteins and four phagocytosis-related Dscam-like proteins; counts of which are generally lower than in other chelicerates (Palmer and Jiggins 2015). The Toll pathway appears largely intact: A single toll-activating spaetzle gene and six toll-like receptor genes; the signal transducers Myd88 and Pelle (but no Tube-like gene, which is also absent from I. scapularis and T. urticae [Palmer and Jiggins 2015]); the nuclear factor-κB (NF-κB) transcription factor dorsal and its inhibitor cactus, are all present. In contrast, neither Imd nor the NF-κB transcription factor of the Imd pathway, relish, could be identified, and the absence of additional pathway members further suggests that canonical Imd signaling does not occur. Although homologs of the JAK/STAT pathway receptor domeless and transcription factor Stat92E were identified, no clear homolog of the janus kinase hopscotch was found. Finally, like I. scapularis (Smith and Pal 2014), this predatory mite lacks genes related to the melanization (phenoloxidase) pathway.
RNAi Pathways
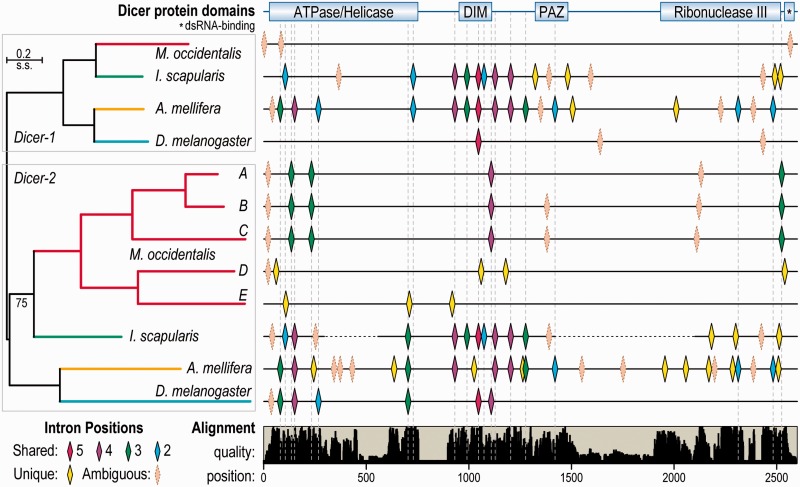
Orally delivered double-stranded RNAs (dsRNAs) induce robust, prolonged, and systemic gene knockdowns in M. occidentalis (Wu and Hoy 2014a, 2014b; Pomerantz and Hoy 2015b; Wu and Hoy 2015). Candidate genes likely involved in facilitating these responses were identified (see Materials and Methods) with searches for homologs of genes of the RNAi or microRNA (miRNA) pathways as well as dsRNA uptake and transport processes (supplementary tables S18 and S19, Supplementary Material online). Most genes implicated in dsRNA uptake and transport were identified, including clathrin heavy chain which, when knocked down, severely impaired RNAi responses (Wu and Hoy 2014a), suggesting this mite uses a receptor-mediated endocytosis pathway for dsRNA uptake. The identification of at least three RNA-dependent RNA polymerase (RDRP) genes, which are also found in other arachnids and Str. maritima but not in other arthropods, suggests RDRP-based dsRNA amplification may occur. The presence of both Dicer-1 and Dicer-2 homologs suggests that, like Drosophila, distinct dicer proteins are used for processing miRNAs and small interfering RNAs (siRNAs). However, although homologs of the miRNA precursor processors Drosha and Pasha, as well as Dicer-1’s partner for miRNA processing, Loquacious, were all identified, Dicer-2’s partner, R2D2, appears to be absent. Most strikingly, the M. occidentalis genome encodes at least five distinct full-length copies of Dicer-2, located on four different scaffolds, with three to seven introns each (fig. 4). These divergent gene copies vary in sequence identity from 66% to 31% (amino acids) and 66–43% (nucleotides). Four of these Dicer-2 genes matched transcripts from a multi-life-stage RNA-seq transcriptome (Hoy et al. 2013) (supplementary table S20, Supplementary Material online), providing support for their functional relevance. Such duplications of Dicer-2 genes appear to be rare events: A single duplication was found in Da. pulex (Mukherjee et al. 2013; Palmer and Jiggins 2015), and surveying more than 80 arthropod species identified no other confidently distinct and full-length gene duplications. Comparing their gene architectures reveals the paucity of introns in M. occidentalis Dicers, especially when compared with the intron-rich genes of I. scapularis and Apis mellifera, and their uniqueness—with only a single confidently aligned intron position that is shared with D. melanogaster Dicer-2 (fig. 4). Such multiple duplications of a normally single-copy gene were also noted for the gene encoding the muscle myosin heavy chain protein, which also showed very different intron–exon architectures, as well as mutually exclusive spliced exons (Kollmar and Hatje 2014). The multiple duplications of Dicer-2 genes highlight how the dynamic evolutionary history of this genome has led to the creation and maintenance of several gene copies, which exhibit dramatic changes in their intron content.
Fig. 4.—
The expanded set of Metaseiulus occidentalis Dicer-2 genes. The phylogeny of Dicer-1 and Dicer-2 genes from four distantly related arthropods (left) with five distinct copies of Dicer-2 from M. occidentalis showing the locations of shared and unique intron positions along the length of the multiple protein sequence alignment (right). Dashed vertical lines mark the positions of unambiguous shared intron positions along the length of the alignment, which are colored according to the number of genes in which they are found (5 pink, 4 purple, 3 green, 2 blue). Confidently identified unique intron positions are shown in yellow, and ambiguous (poorly aligned regions) intron positions are shown in almond. Dramatic gains and losses of introns in M. occidentalis genes have left only a single intron position in three Dicer-2 genes that is shared with any other Dicers—Dicer-2 from Drosophila melanogaster. Examining these gene structures highlights the paucity of introns in M. occidentalis genes, especially compared with the intron-rich orthologs from Ixodes scapularis and Apis mellifera. Dicer-1 and Dicer-2 genes in the phylogeny are color-matched by species, and the only node with less than 100% bootstrap support is labeled “75.” The positions of Dicer protein domains are indicated (top) as well as the relative quality of the alignment (bottom). Dashed lines for I. scapularis Dicer-2 denote missing sequence information due to gaps in the genome assembly. DIM, dicer dimerization domain; PAZ, domain named after Piwi, Argonaut and Zwille proteins.
Discussion
The relatively compact M. occidentalis genome assembly of 152 Mb represents the first sequenced phytoseiid, a family of predatory mites used to control pests such as the two-spotted spider mite. The small genomes of these two mites are in stark contrast to many other chelicerates (table 1), prompting speculation that their rapid life cycles, both averaging about 1 week from egg to adult, compared with their longer-lived relatives may have influenced the evolution of selfish genetic element activity and favored smaller genomes. Phylogenomic analyses highlighted rapid rates of gene-sequence evolution and the ancient divergence of M. occidentalis from other sequenced arachnids, as well as disputing the monophyly of the Acari (fig. 1A). These findings with new genomic data support the previously recognized possibility of two distinct ancestral arachnid lineages having converged on very similar mite-like morphologies (Krantz 2009). Comparative analyses of annotated genes with those of other animals identified a conserved core of orthologs, but the limited sampling and ancient divergences within the arachnids mean that there are many currently “orphaned” genes with no clear orthology (fig. 1B). Some of these taxonomically restricted genes (TRGs) are likely to be involved in lineage-specific adaptations, as proposed for thousands of TRGs identified in ants (Simola et al. 2013). Examining intron–exon structures of genes from the conserved core of orthologs revealed dramatic intron losses accompanied by many intron gains that have led to the majority of intron positions being unique to M. occidentalis (fig. 2). Although no direct evidence exists, analysis of repetitive DNA content revealed an abundance of Helitron rolling circle transposons, making it tempting to speculate that the elevated Helitron activity may be linked to the dynamic history of gene architecture evolution in this mite. Future genomic sampling of the arachnids and other chelicerates will help to explore the roles of TEs in the evolution of gene architectures and genome sizes, to trace the evolutionary histories of the “orphaned” arachnid genes, as well as to bring a more detailed resolution to the apparent paraphyly of the Acari.
Despite the large evolutionary distances with other sequenced arthropods, orthology delineation and homology searches successfully identified many genes implicated in processes of particular interest to phytoseiid biology. In this blind mite, chemosensation presumably plays an especially important role in prey detection, facilitated by the GR and IR families of chemoreceptors, which are similar in size to those of other arthropods with approximately 64 genes each, and exhibit dynamic patterns of intron evolution. Once located, prey may be immobilized by agatoxin-like and sphingomyelinase D-like proteins, and other venoms that are yet to be discovered. Although this mite has fewer cysteine/serine peptidases than the spider mite or the tick, many of these proteolytic enzymes are likely important components of the saliva that must be injected to perform preoral digestion. Future sialome transcriptomic studies will help identify components active in preoral digestion, as well as offering the opportunity to characterize putative secreted venoms of this predator. Despite lacking eyes, it appears to have a complete phototransduction pathway, albeit with peropsins rather than true visual opsins, and light–dark oviposition periodicity and photoperiodic induction of diapause support the ability to perceive light. Key components of the circadian-rhythm system appear to be absent, suggesting concomitant losses of vision and biological clock genes similar to that observed in centipedes (Chipman et al. 2014). In contrast, most eye-development-related transcription factors have been maintained, perhaps suggesting a rewiring of the regulatory network to drive the expression of molecules required for light detection in other cells.
As the first sequenced species with the rare and poorly understood system of parahaploidy, M. occidentalis provides a unique opportunity to explore this process. The presence of key components of arthropod sex-determination pathways suggests that parts of the signaling cascades are likely maintained, but they may also involve cross-talk with heterochromatinization signals. Although such interactions remain to be determined, it appears possible that cascades regulated by sex-biased gene expression may form part of the signaling processes during parahaploid sex determination. These signals may be in the form of sex-specific epigenetic marks, but interestingly, of the three subfamilies of DNA methyltransferases this mite only has Dnmt2, making methylation an unlikely driver of heterochromatinization of paternally derived chromosomes in male embryos. Development leads to a body plan with reduced segmentation common to mites and ticks, and in these phytoseiid mites it is intriguing to contemplate the possibility that relaxation of the normally tightly coordinated expression of Hox genes in such a dynamically evolving genome has allowed them to become completely dispersed across the genome (fig. 3). It will be interesting for future studies to examine how this dispersal may have affected nearby regulatory sequences that control Hox gene expression. This rare example of the complete “atomization” of the Hox genes is the first reported for any arthropod, and further highlights the turbulent and dynamic evolutionary history of this genome.
Keeping dense colonies of these mass-reared predators free from disease is critical for maximizing their production for control programs. This mite appears to have the capacity for Toll and JAK/STAT immune signaling, albeit with some likely deviations from the canonical models. However, no GNBPs were identified (like in other arachnids [Bechsgaard et al. 2015]), and the absence of Imd pathway and melanization genes suggests immune-related melanization as well as Imd-mediated responses to Gram-positive bacteria may be absent. In the pea aphid, the absence of functional Imd responses has been linked to adaptations for obligate symbioses with bacteria (Douglas et al. 2011), but although some M. occidentalis populations carry a Cardinium endosymbiont associated with nonreciprocal reproductive incompatibility (Wu and Hoy 2012), these mites have no known obligate endosymbionts. In contrast, the RNAi system seems particularly robust, and gene duplications have created five distinct full-length copies of Dicer-2 (fig. 4). Multiple duplications of Dicer-2 appear unique to M. occidentalis, and may exemplify a rare case of a gene having escaped the constraints of evolution under “single-copy control” (Waterhouse et al. 2011). This, and the presence of RDRP genes, adds an interesting layer of complexity to the challenge of characterizing siRNA pathway functions in this predator.
The annotated M. occidentalis genome contributes valuable new genomic resources to investigate the biology and evolution of the currently sparsely sampled Acari. Comparative analyses revealed a remarkably dynamic and turbulent evolutionary history of the genes and genome of this tiny predator, in stark contrast to the Ixodes deer tick. This is exemplified by its completely atomized Hox genes—unique among arthropods to date, its superdynamic evolution of gene intron–exon architectures, and five copies of the normally single-copy RNA processing Dicer-2 gene. These genomic resources facilitate detailed examination of genes putatively involved in detecting, disabling, and digesting prey, parahaploid sex determination and development, as well as in immune and RNAi responses, and provide new opportunities for future functional genomics and taxonomic analyses of the Phytoseiidae. Importantly, investigating the large fraction of genes for which homology offers few confident hypotheses on gene function will be prioritized to comprehensively explore the genetic basis and molecular mechanisms underlying the biology of this agriculturally important predator.
Supplementary Material
Supplementary figures S1–S10 and tables S1–S20 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank the anonymous reviewers for their constructive comments. This work was supported by the Davies, Fischer and Eckes endowment in biological control to M.A.H.; Marie Curie International Outgoing Fellowship (PIOF-GA-2011-303312 to R.M.W.); the Swiss National Science Foundation (31003A-125350, 31003A-143936 to E.M.Z.); and the National Human Genome Research Institute (U54 HG003272 to R.A.G.).
Literature Cited
- Albertin CB, et al. 2015. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524:220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaglia AM, et al. 2012. Gene capture by Helitron transposons reshuffles the transcriptome of maize. Genetics 190:965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett AA, Thomas RH. 2013. Posterior Hox gene reduction in an arthropod: ultrabithorax and Abdominal-B are expressed in a single segment in the mite Archegozetes longisetosus. EvoDevo. 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barribeau SM, et al. 2015. A depauperate immune repertoire precedes evolution of sociality in bees. Genome Biol. 16:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard J, et al. 2015. Comparative genomic study of arachnid immune systems indicates loss of β-1,3-glucanase-related proteins and the immune deficiency pathway. J Evol Biol. 29(2):277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørnson S. 2008. Natural enemies of mass-reared predatory mites (family Phytoseiidae) used for biological pest control. Exp Appl Acarol. 46:299–306. [DOI] [PubMed] [Google Scholar]
- Bongiorni S, et al. 2009. Epigenetic marks for chromosome imprinting during spermatogenesis in coccids. Chromosoma 118:501–512. [DOI] [PubMed] [Google Scholar]
- Buchon N, Silverman N, Cherry S. 2014. Immunity in Drosophila melanogaster–from microbial recognition to whole-organism physiology. Nat Rev Immunol. 14:796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, et al. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun. 4:2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, et al. 2015. The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J Allergy Clin Immunol. 135:539–548. [DOI] [PubMed] [Google Scholar]
- Chipman AD, et al. 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 12:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium i5K 2013. The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 104:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman SR, et al. 2010. Genomic survey of the ectoparasitic mite Varroa destructor, a major pest of the honey bee Apis mellifera. BMC Genomics 11:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csurös M. 2008. Malin: maximum likelihood analysis of intron evolution in eukaryotes. Bioinformatics 24:1538–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csuros M, Rogozin IB, Koonin EV. 2011. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol. 7:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Z, et al. 2015. Genome-wide analysis of homeobox genes from Mesobuthus martensii reveals Hox gene duplication in scorpions. Insect Biochem Mol Biol. 61:25–33. [DOI] [PubMed] [Google Scholar]
- Douglas AE, Bouvaine S, Russell RR. 2011. How the insect immune system interacts with an obligate symbiotic bacterium. Proc Biol Sci. 278:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. 2007. The rise and fall of Hox gene clusters. Development 134:2549–2560. [DOI] [PubMed] [Google Scholar]
- Elsik CG, et al. 2014. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson BJ, Fredman D, Steiner G, Schmid A. 2013. Characterisation and localisation of the opsin protein repertoire in the brain and retinas of a spider and an onychophoran. BMC Evol Biol. 13:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechtmann CHW, McMurtry JA. 1992. Studies on how phytoseiid mites feed on spider mites and pollen. Int J Acarol. 18:157–162. [Google Scholar]
- Gempe T, Beye M. 2011. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 33:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AK, Smith Z, Fuqua C, Clay K, Colbourne JK. 2013. Why so many unknown genes? Partitioning orphans from a representative transcriptome of the lone star tick Amblyomma americanum. BMC Genomics 14:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulia-Nuss M, et al. 2015. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy MA. 1975. Effect of temperature and photoperiod on the induction of diapause in the mite Metaseiulus occidentalis. J Insect Physiol. 21:605–611. [DOI] [PubMed] [Google Scholar]
- Hoy MA. 2009. The predatory mite Metaseiulus occidentalis: mitey small and mitey large genomes. Bioessays 31:581–590. [DOI] [PubMed] [Google Scholar]
- Hoy MA, et al. 2013. Transcriptome sequencing and annotation of the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae): a cautionary tale about possible contamination by prey sequences. Exp Appl Acarol. 59:283–296. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. 2009. First divergence time estimate of spiders, scorpions, mites and ticks (subphylum: Chelicerata) inferred from mitochondrial phylogeny. Exp Appl Acarol. 47:1–18. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Hatje K. 2014. Shared gene structures and clusters of mutually exclusive spliced exons within the metazoan muscle myosin heavy chain genes. PLoS One 9:e88111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz GW. 2009. Introduction In: Krantz GW, Walter DE, editors. A manual of acarology. 3rd ed Lubbock (TX: ): Texas Tech Univ. Press. p. ; 3–4. [Google Scholar]
- Kriventseva EV, et al. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 43:D250–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tucker AE, Sung W, Thomas WK, Lynch M. 2009. Extensive, recent intron gains in Daphnia populations. Science 326:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C, et al. 2014. Evolution of insect olfactory receptors. Elife 3:e02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K, Campos H, Kolaczkowski B. 2013. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol. 30:627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, et al. 2000. A whole-genome assembly of Drosophila. Science 287:2196–2204. [DOI] [PubMed] [Google Scholar]
- Nagelkerke CJ, Sabelis MW. 1998. Precise control of sex allocation in pseudo-arrhenotokous phytoseiid mites. J Evol Biol. 11:649–684. [Google Scholar]
- Nelson-Rees WA, Hoy MA, Roush RT. 1980. Heterochromatinization, chromatin elimination and haploidization in the parahaploid mite Metaseiulus occidentalis (Nesbitt) (Acarina: Phytoseiidae). Chromosoma 77:263–276. [DOI] [PubMed] [Google Scholar]
- Palmer WJ, Jiggins FM. 2015. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol. 32(8):2111–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Pepato AR, da Rocha CE, Dunlop JA. 2010. Phylogenetic position of the acariform mites: sensitivity to homology assessment under total evidence. BMC Evol Biol. 10:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva-Arana D, Lynch M, Robertson H. 2009. The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol. 9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz AF, Hoy MA. 2015a. Expression analysis of Drosophila doublesex, transformer-2, intersex, fruitless-like, and vitellogenin homologs in the parahaploid predator Metaseiulus occidentalis (Chelicerata: Acari: Phytoseiidae). Exp Appl Acarol. 65:1–16. [DOI] [PubMed] [Google Scholar]
- Pomerantz AF, Hoy MA. 2015b. RNAi-mediated knockdown of transformer-2 in the predatory mite Metaseiulus occidentalis via oral delivery of double-stranded RNA. Exp Appl Acarol. 65:17–27. [DOI] [PubMed] [Google Scholar]
- Pomerantz AF, Hoy MA, Kawahara AY. 2014. Molecular characterization and evolutionary insights into potential sex-determination genes in the western orchard predatory mite Metaseiulus occidentalis (Chelicerata: Arachnida: Acari: Phytoseiidae). J Biomol Struct Dyn. 33(6):1239–1253. [DOI] [PubMed] [Google Scholar]
- Raddatz G, et al. 2013. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci U S A. 110:8627–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD, Morgan MS, Arlian LG. 2015. Draft genome of the scabies mite. Parasit Vectors. 8:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100 (2 Suppl):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz R, Croset V, Benton R. 2013. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol. 43:888–897. [DOI] [PubMed] [Google Scholar]
- Sanggaard KW, et al. 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 5:3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, et al. 2004. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nature 431:67–71. [DOI] [PubMed] [Google Scholar]
- Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. 2012. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 31:351–376. [DOI] [PubMed] [Google Scholar]
- Sharma PP, et al. 2014. Phylogenomic interrogation of arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol. 31:2963–2984. [DOI] [PubMed] [Google Scholar]
- Simola DF, et al. 2013. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 23:1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith AA, Pal U. 2014. Immunity-related genes in Ixodes scapularis–perspectives from genome information. Front Cell Infect Microbiol. 4:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L. 2014. Sex-determining mechanisms in insects based on imprinting and elimination of chromosomes. Sex Dev. 8:83–103. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Thomas RH. 1998. Of mites and zen: expression studies in a chelicerate arthropod confirm zen is a divergent Hox gene. Dev Genes Evol. 208:591–594. [DOI] [PubMed] [Google Scholar]
- Thomas J, Phillips CD, Baker RJ, Pritham EJ. 2014. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol. 6:2595–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Pritham EJ. 2015. Chapter 40: Helitrons, the eukaryotic rolling-circle transposable elements In: Craig NL, et al. , editor. Mobile DNA III. 3rd ed Washington DC: American Society of Microbiology Press. p. ; 893–926. [DOI] [PubMed] [Google Scholar]
- van Wijk M, Wadman WJ, Sabelis MW. 2006. Morphology of the olfactory system in the predatory mite Phytoseiulus persimilis. Exp Appl Acarol. 40:217–229. [DOI] [PubMed] [Google Scholar]
- Wang XH, et al. 2001. Discovery and structure of a potent and highly specific blocker of insect calcium channels. J Biol Chem. 276:40306–40312. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM. 2015. A maturing understanding of the composition of the insect gene repertoire. Curr Opin Insect Sci. 7:15–23. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. 2013. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41:D358–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Zdobnov EM, Kriventseva EV. 2011. Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi. Genome Biol Evol. 3:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Hoy MA. 2012. Cardinium is associated with reproductive incompatibility in the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae). J Invertebr Pathol. 110:359–365. [DOI] [PubMed] [Google Scholar]
- Wu K, Hoy MA. 2014a. Clathrin heavy chain is important for viability, oviposition, embryogenesis and, possibly, systemic RNAi response in the predatory mite Metaseiulus occidentalis. PLoS One 9:e110874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Hoy MA. 2014b. Oral delivery of double-stranded RNA induces prolonged and systemic gene knockdown in Metaseiulus occidentalis only after feeding on Tetranychus urticae. Exp Appl Acarol. 63:171–187. [DOI] [PubMed] [Google Scholar]
- Wu K, Hoy MA. 2015. Cloning and functional characterization of two BTB genes in the predatory mite Metaseiulus occidentalis. PLoS One 10:e0144291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenerall P, Krupa B, Zhou L. 2011. Mechanisms of intron gain and loss in Drosophila. BMC Evol Biol. 11:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenerall P, Zhou L. 2012. Identifying the mechanisms of intron gain: progress and trends. Biol Direct. 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel-Thropp PA, Kerins AE, Binford GJ. 2012. Sphingomyelinase D in sicariid spider venom is a potent insecticidal toxin. Toxicon 60:265–271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.