Abstract
Many genes in the plastid genomes (plastomes) of plants are organized as gene clusters, in which genes are co-transcribed, resembling bacterial operons. These plastid operons are highly conserved, even among conifers, whose plastomes are highly rearranged relative to other seed plants. We have determined the complete plastome sequence of Sciadopitys verticillata (Japanese umbrella pine), the sole member of Sciadopityaceae. The Sciadopitys plastome is characterized by extensive inversions, pseudogenization of four tRNA genes after tandem duplications, and a unique pair of 370-bp inverted repeats involved in the formation of isomeric plastomes. We showed that plastomic inversions in Sciadopitys have led to shuffling of the remote conserved operons, resulting in the birth of four chimeric gene clusters. Our data also demonstrated that the relocated genes can be co-transcribed in these chimeric gene clusters. The plastome of Sciadopitys advances our current understanding of how the conifer plastomes have evolved toward increased diversity and complexity.
Keywords: plastome, gene cluster, rearrangement, evolution, conifer
Introduction
Due to the loss of many genes in early endosymbiosis, plastomes are reduced compared with their cyanobacterial counterparts (Ku et al. 2015). To date, plastomes have invariably retained a small handful of prokaryotic features, including organization of genes into polycistronic transcription units resembling bacterial operons (Sugiura 1992; Wicke et al. 2011). A hallmark of seed plant plastomes is the presence of two 20- to 30-Kb inverted repeats (IRs) (hereafter referred to as “typical IRs,” including IRA and IRB), which usually contain four ribosomal RNAs. However, a few exceptions have been reported. For example, conifers—the largest gymnosperm group comprising Cupressophyta (cupressophytes) and Pinaceae—have lost a typical IR copy from their plastomes (Raubeson and Jansen 1992). Recent studies have further suggested that cupressophytes and Pinaceae might have lost different IR copies, with the former losing IRA and the latter losing IRB (Wu, Wang, et al. 2011; Wu and Chaw 2014).
Conifer plastomes are also characterized by extensive genomic rearrangements. The plastome of Cryptomeria japonica—the first completed plastome of cupressophytes (Hirao et al. 2008)—experienced at least 12 inversions after its split from the basal gymnosperm clade, cycads, whose plastomes have remained virtually unchanged for 280 million years (Wu and Chaw 2015). The co-existence of four different plastome forms among Pinaceae genera is associated with intra-plastomic recombination mediated by three specific types of short IRs (Wu, Lin, et al. 2011). Furthermore, Cephalotaxus oliveri (Cephalotaxaceae; Yi et al. 2013) and four Juniperus species (Cupressaceae; Guo et al. 2014) harbor isomeric plastomes that deviate from each other by an inversion possibly triggered by a trnQ-containing IR (“trnQ-IR”). Although conifer plastomes are highly rearranged (Wu and Chaw 2014), disruptions in their operons are rare. Until recently, only one case was reported in the plastome of Taxus mairei, in which the S10 operon (trnI-rpoA region) was disrupted into two separate segments by a fragment of approximately 15 Kb (Hsu et al. 2014). However, the impact of such operon disruptions on plastid evolution remains poorly understood.
Cupressophytes comprise five conifer families: Araucariaceae, Cupressaceae, Podocarpaceae, Sciadopityaceae, and Taxaceae. Among them, Sciadopityaceae is the only family with the single species Sciadopitys verticillata (hereafter referred to as Sciadopitys). Sciadopitys is an evergreen tree that can reach 27 m tall. Its spectacular needle-like leaves are arranged in whorls, like an umbrella. Thus, Sciadopitys is commonly called the Japanese umbrella pine. Three genome-based (Chaw et al. 2000) and plastome-based (Rai et al. 2008) phylogenetic studies are congruent in placing Sciadopitys as a sister to Taxaceae and Cupressaceae. Recent molecular dating suggests that Sciadopitys diverged from other cupressophytes more than 200 million years ago (Crisp and Cook 2011). Although Sciadopitys is considered a living fossil endemic to Japan, paleo-biogeographic evidence indicates that its ancestors existed in China during the early and middle Jurassic (Jiang et al. 2012).
The 25 published cupressophyte plastomes available on GenBank (December, 2015) represent four of the five cupressophyte families but no complete plastome is available for Sciadopityaceae. As a part of our continuing efforts to decipher the diversity and evolution of conifer plastomes, we have completed and elucidated the plastome sequence of Sciadopitys. We found that the plastome of Sciadopitys is characterized by several unusual features, including shuffling of the conserved plastid operons and re-organization of plastid genes into new chimeric gene clusters.
Materials and Methods
DNA Extraction
Approximately 2 g of fresh leaves were collected from an individual of Sciadopitys verticillata (voucher chaw 1496) growing in the Floriculture Experiment Center, Taipei, Taiwan. The voucher specimen was deposited in the herbarium of Biodiversity Research Center, Academia Sinica, Taipei (HAST). Total DNA of the leaves was extracted with 2X CTAB buffers (Stewart and Via 1993). The extracted DNA was qualified with a threshold of DNA concentration >300 ng/μl, 260/280 = 1.8–2.0 and 260/230 > 1.7.
Sequencing, Plastome Assembly, and Genome Annotation
Sequencing was conducted on an Illumina MiSeq Sequencing System (Illumina, San Diego, CA) in Yourgene Bioscience (New Taipei City, Taiwan) to yield 300-bp paired-end reads of approximately 4 Gb. De novo assembly of the Sciadopitys plastome was performed using CLC Genomics Workbench 4.9 (CLC Bio, Arhus, Denmark). Plastid genes were predicted using DOGMA (Wyman et al. 2004) and tRNAscan-SE 1.21 (Schattner et al. 2005) with the default option that real tRNA genes should have >= 20 Cove scores. Boundaries of predicted genes were manually adjusted by aligning them with their orthologs of other gymnosperms.
Estimate of Dispersed Repeats and Plastomic Inversions
Repeat sequences were searched by comparing the plastome against itself using NCBI BLASTn with the default settings, followed by manually deleted overlapping or conjoined pairs. To assess the possible scenarios of plastomic inversions in Sciadopitys, the plastome of Cycas taitungensis (NC_009618) with its IRA removed was used for comparison. We identified the syntenic block of genes between Sciadopitys and Cycas using Mauve 2.3.1 (Darling et al. 2004). The resulting matrix of syntenic blocks was utilized to estimate the minimal inversion steps with MGR 2.0.1 (Bourque and Pevzner 2002). The plastome map of Sciadopitys was drawn using Circos 0.67 (Krzywinski et al. 2009).
Detection of Isomeric Plastomes
Primer pairs listed in supplementary table S1 (Supplementary Material online) were used to amplify DNA fragments specific to the two isomeric plastomes in Sciadopitys (i.e., rpl33 + rpoC2 and rpoC1 + rps18 for the presence of A form; rpl33 + rps18 and rpoC1 + rpoC2 for the presence of B form). PCR reactions were conducted with three different numbers of cycles. The conditions were 94 °C for 5 min, followed by 25, 30, or 35 cycles of 94 °C for 20 s, 55 °C for 20 s, and 72 °C for 2 min, and an extension of 72 °C for 10 min.
Detection of RNA Transcripts in Chimeric Gene Clusters
Total RNA was extracted from fresh leaves of Sciadopitys according to a modified RNA isolation protocol (Kolosova et al. 2004). We employed a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham) to synthesize the first-strand cDNA with four specific primers (SpsbN, SatpA, SrpoC2, and Srps18 in supplementary table S1, Supplementary Material online). PCR reactions were conducted with the synthesized cDNA and four pairs of specific primers (atpF-1 + psbT for a 358-bp fragment; psbB-2 + atpA-2 for a 565-bp fragment; rpl33 + rpoC2 for a 687-bp fragment; rpoC1 + rps18 for a 939-bp fragment). The PCR conditions were 94 °C for 5 min, followed by 30 cycles at 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 1 min, and an extension at 72 °C for 10 min.
Results and Discussion
Loss of IRA from Sciadopitys Plastome
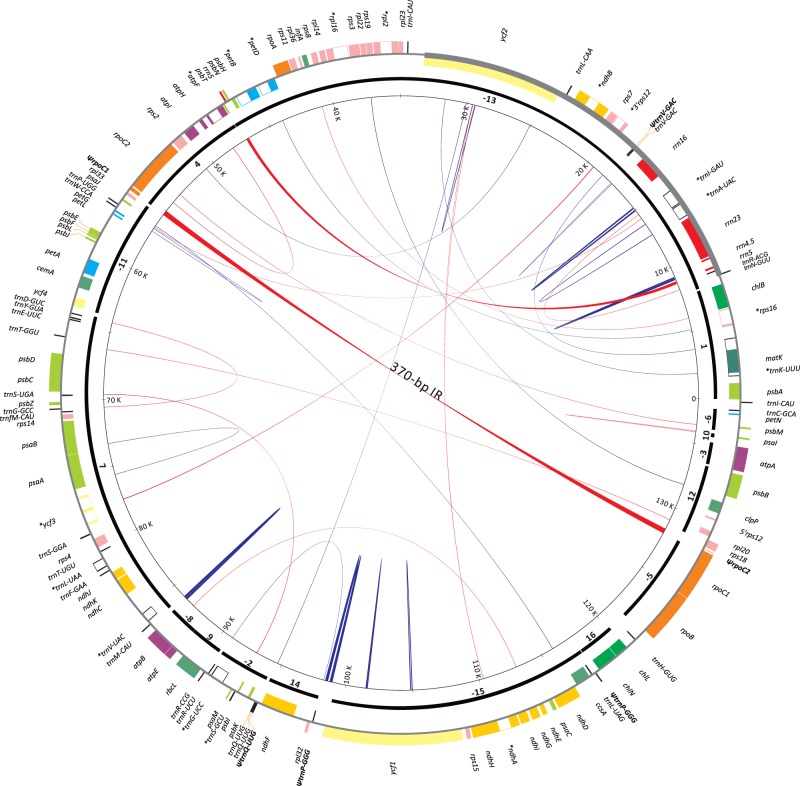
The plastome of Sciadopitys verticillata (AP017299) is illustrated as a circular molecule that consists of 138,309 bp (fig. 1). It is the largest among the known plastomes of Cupressales (including Sciadopityaceae, Taxaceae s. l., and Cupressaceae s. l.). Flanking and adjacent genes of the typical IRs are informative markers for inferring the intact (or retained) IR copy in conifer plastomes. For example, the boundary of IRA or IRB is adjacent to the psbA or S10 operon (i.e., trnI-rpoA region), respectively (Wu, Wang, et al. 2011). In the Sciadopitys plastome, the retained typical IR copy, which encompasses the region from trnN-GUU to ycf2, is adjacent to the S10 operon (fig. 1), indicating that it should be IRB. In other words, the lost IR copy is IRA. This observation reinforces the hypothesis that cupressophytes have lost IRA rather than IRB (Wu, Wang, et al. 2011; Wu and Chaw 2014).
Fig. 1.—
Plastome map of Sciadopitys verticillata. Colored boxes represent genes with counterclockwise (outer boxes) or clockwise (inner boxes) transcriptional directions. Thick and thin grey lines are IRB and single-copy region as compared to those of Cycas, separately. Syntenic blocks of genes between Cycas and Sciadopitys are depicted by thick black bars with Arabic numerals, where pluses or minuses indicate the corresponding syntenic blocks with the same or opposite directions between the two species, respectively. Pairs of dispersed repeats are connected by blue (direct repeats) or red (inverted repeats) lines, with their width proportional to the repeat size. Pseudogenes are bold and marked with a “Ψ.” Intron-containing genes are indicated with an “*”.
The plastome of Sciadopitys contains a total of 121 genes, 83 of which are protein-coding genes and the rest are structural RNA genes (supplementary table S2, Supplementary Material online). Sixteen genes contain introns, but the intron of rpoC1 has been lost. Three genes, rrn5, trnI-CAU, and trnQ-UUG, have two copies. The duplicated rrn5 is located in the region between psbN and psbT (fig. 1). The plastomes of both Agathis dammara and Wollemia nobilis (Araucariaceae) also have a duplicated rrn5, but it is in the region between psbB and clpP (Yap et al. 2015). Among the elucidated cupressophyte plastomes available on GenBank, only these three taxa contain two copies of plastid rrn5. Because Sciadopitys (Sciadopityaceae) and Araucariaceae differ in the locations of their extra rrn5, it is most parsimonious that duplications of rrn5 took place independently in the two families. Moreover, Sciadopitys has lost its plastid accD, which possibly has been transferred to the nucleus (Li et al. 2016).
Pseudogenization of Four tRNA Genes after Tandem Duplications
Four pseudo tRNA genes (ΨtrnV-GAC, ΨtrnQ-UUG, and two copies of ΨtrnP-GGG) were detected in the Sciadopitys plastome (fig. 1). Although the Cove scores for both ΨtrnV-GAC (score = 28.41) and ΨtrnQ-UUG (score = 26.8) were marginal, we predicted them as pseudogenes because their sequences could not form proper cloverleaf structures. Both ΨtrnV-GAC and ΨtrnQ-UUG are near their functional paralogs, implying that pseudogenization of these two genes might have occurred after tandem duplications. In angiosperms, the plastid trnP-GGG likely has been lost for 150 MY (Chaw et al. 2004). In contrast, this tRNA gene is retained and commonly located in the region between trnL and rpl32 in Cycas, Ginkgo, Gnetum, and Pinus (Wu et al. 2007), and other cupressophyte families, such as Araucariaceae (Yap et al. 2015), Podocarpaceae (Vieira Ldo et al. 2014; Wu and Chaw 2014), and Taxaceae s. l. (Yi et al. 2013; Hsu et al. 2014). In the Sciadopitys plastome, two ΨtrnP-GGG copies are separated by a distance of approximately 20-Kb: one is adjacent to trnL-UAG and the other is located near rpl32 (fig. 1). Therefore, the two ΨtrnP-GGG copies might have resulted from tandem duplications, and subsequently, a plastomic inversion of an approximately 20-Kb fragment separated them.
Evolution of the Plastid trnI-CAU in Sciadopitys
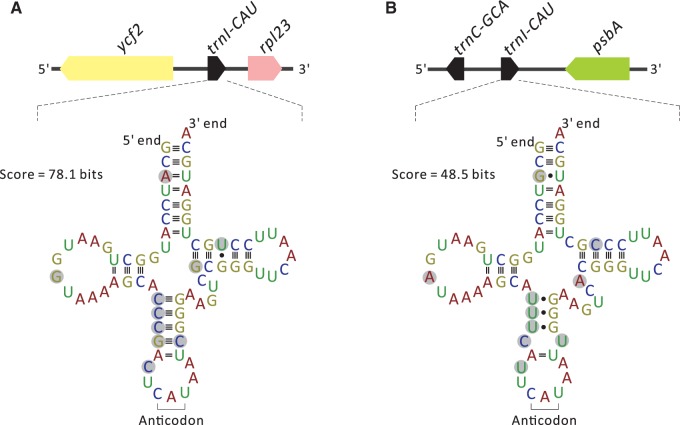
Sciadopitys has two plastid trnI-CAU copies, one between trnC-GCA and psbA, and the other between ycf2 and rpl23 (figs. 1 and 2). In Cryptomeria, one of the two plastid trnI-CAU copies was considered residual from the lost typical IR (Hirao et al. 2008). Indeed, the majority of cupressophyte plastomes have two trnI-CAU copies with the sequence identity higher than 85% (table 1), connoting their homologous origin.
Fig. 2.—
Comparison between the two copies of trnI-CAU in the Sciadopitys plastome. (A) Predicted cloverleaf structure of trnI-CAU located between ycf2 and rpl23 and (B) that of the other located between trnC-GCA and psbA. Pairwise substitutions of nucleotides between the two copies are highlighted in gray.
Table 1.
Presence of trnI-CAU copies in the plastomes of cupressophytes
| Family | Species (GenBank accession) | Copya | Seq. identity (%)b | Score |
|---|---|---|---|---|
| Cupressaceae | Calocedrus formosana (NC_023121) | (+)trnI-CAU | 98.63 | 70.17 |
| (-)trnI-CAU | 75.14 | |||
| Juniperus bermudiana (NC_024021) | (+)trnI-CAU | 98.63 | 70.17 | |
| (-)trnI-CAU | 75.44 | |||
| Cryptomeria japonica (NC_010548) | (+)trnI-CAU | 98.63 | 77.3 | |
| (-)trnI-CAU | 74.56 | |||
| Metasequoia glyptostroboides (NC_027423) | (+)trnI-CAU | 98.63 | 69.43 | |
| (-)trnI-CAU | 74.71 | |||
| Cunninghamia lanceolate (NC_021437) | (+)trnI-CAU | 100 | 75.44 | |
| (-)trnI-CAU | 75.44 | |||
| Taiwania cryptomerioides (NC_016065) | (+)trnI-CAU | 100 | 75.44 | |
| (-)trnI-CAU | 75.44 | |||
| Taxaceae | Taxus mairei (AP014575) | (+)trnI-CAU | 98.63 | 75.44 |
| (-)trnI-CAU | 75.39 | |||
| Amentotaxus formosana (NC_024945) | (+)trnI-CAU | 98.63 | 75.44 | |
| (-)trnI-CAU | 75.67 | |||
| Cephalotaxaceae | Cephalotaxus wilsoniana (NC_016063) | (+)trnI-CAU | — | 77.31 |
| — | ||||
| Sciadopityaceae | Sciadopitys verticillata (AP017299) | (+)trnI-CAU | 86.30 | 48.48 |
| (+)trnI-CAU | 78.15 | |||
| Podocarpaceae | Nageia nagi (NC_023120) | (+)trnI-CAU | — | 75.44 |
| — | ||||
| Podocarpus lambertii (NC_023805) | (+)trnI-CAU | — | 77.3 | |
| — | ||||
| Araucariaceae | Agathis dammara (NC_023119) | (+)trnI-CAU | 97.26 | 75.25 |
| (-)trnI-CAU | 66.46 | |||
| Wollemia nobilis (NC_027235) | (+)trnI-CAU | 94.52 | 75.25 | |
| (-)trnI-CAU | 55.15 |
“+” and “-“ in parentheses denote the transcriptional directions.
Estimates based on comparison of the two trnI-CAU copies within each species.
In Sciadopitys plastome, both copies of trnI-CAU are able to fold into cloverleaf structures, but they differ in prediction scores. The gene located between ycf2 and rpl23 has a score of 78.1 bits (fig. 2A), much higher than the score of the other gene (score = 48.5 bits; fig. 2B). Ten nucleotide substitutions (highlighted in gray in fig. 2) were detected between the two trnI-CAU copies, including three mismatches and four U•G abnormal pairings in the stems of the low-scoring trnI-CAU (fig. 2B). Although trnI-CAU is essential for plastid biology (Alkatib et al. 2012), why two copies of trnI-CAU are required remains to be investigated. The elucidated cupressophyte plastomes, such as Cephalotaxus, Nageia, and Podocarpus, contain only one copy of trnI-CAU (table 1). Therefore, whether the low-scoring trnI-CAU of Sciadopitys is functionally redundant and subjected to relaxed structural constraint is worthy of further investigation.
Presence of Two Isomeric Plastomes in Sciadopitys
Recent studies of conifer plastomes revealed that dispersed short IRs can trigger rearrangements to generate isomeric forms. In Pinaceae, a shift between different plastomic forms is often associated with homologous recombination (HR) mediated by the short IRs of approximately 949 bp (Tsumura et al. 2000; Wu, Lin, et al. 2011). The short IRs that contain trnQ-UUG (trnQ-IR) can also promote the formation of isomeric plastomes in Cephalotaxus (Yi et al. 2013) and Juniperus (Guo et al. 2014).
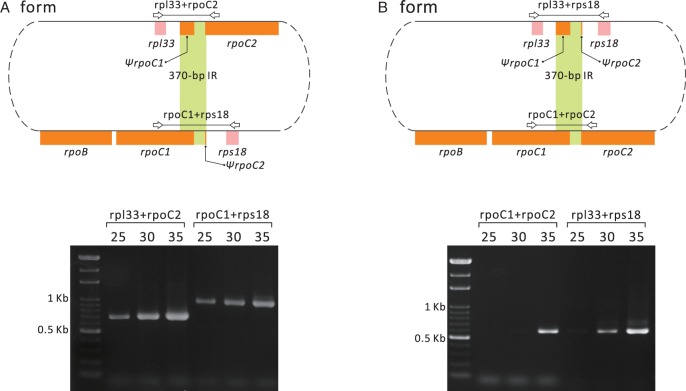
Thirty-seven pairs of dispersed repeats were detected in the Sciadopitys plastome. Among them, the longest IR pair is 370 bp and contains the sequences of 3′rpoC1 and 5′rpoC2 (fig. 1). In the Sciadopitys plastome, only this 370-bp IR pair is longer than the 250-bp “trnQ-IR” of Juniperus (Guo et al. 2014). Hence, if the 370-bp IR is able to mediate HR in Sciadopitys, we would expect the presence of two plastomic forms, as depicted in figure 3. The plastome form illustrated in figure 1 is designated as the A form, and the other is the B form. We have verified the presence of both the A and B forms by amplicons of four specific DNA fragments across the 370-bp IR in the PCR with 35 cycles (fig. 3). However, the amount of the PCR amplicons differs between the two forms. With 25 PCR cycles, the two specific amplicons of the A form are evident, but those of the B form are undetectable (fig. 3). These results suggest that A form is predominant in Sciadopitys plastome populations, in agreement with our assembly results.
Fig. 3.—
Co-existence of two isomeric plastomes in Sciadopitys. The A form is the plastome map obtained from our genome assembly and is shown in fig. 1. The B form differs from the A form by an inversion of the rpoC2–rps18 (or rpl33–rpoC1) fragment. Light green areas are the 370-bp IRs involved in homologous recombination that allows for conversion between the two forms. Paired open arrows are primers specific for the PCR amplification of each form. The corresponding PCR amplicons are shown, and the numbers above each lane of gel photos denote the PCR cycles conducted.
The plastomes of Cupressaceae and Taxaceae possess a trnQ-IR (Guo et al. 2014). Nonetheless, such a trnQ-IR is absent from the plastome of Sciadopitys. The plastomes of Araucariaceae (Yap et al. 2015) have an IR pair that is approximately 600-bp long and contains the gene rrn5. Such the length of IRs could potentially trigger HR; however, the presence of associated isomeric plastomes has not been experimentally demonstrated in Araucariaceae. Including the unique 370-bp IR of Sciadopitys, it is apparent that in cupressophytes, the presence of isomeric plastomes is overwhelming and associated with diverse short IRs.
Birth of Four Chimeric Gene Clusters
We identified a total of 16 syntenic blocks between Cycas and Sciadopitys plastomes (fig. 1 and supplementary fig. S1, Supplementary Material online). In addition to the loss of IRA from the Sciadopitys plastome mentioned above, eight plastomic inversions were detected to distinguish Sciadopitys from Cycas (supplementary fig. S1, Supplementary Material online). Since Cycas was proposed to retain the ancestral gene order of seed plant plastomes (Jansen and Ruhlman 2012), these eight inversions should have occurred after cupressophytes split from cycads.
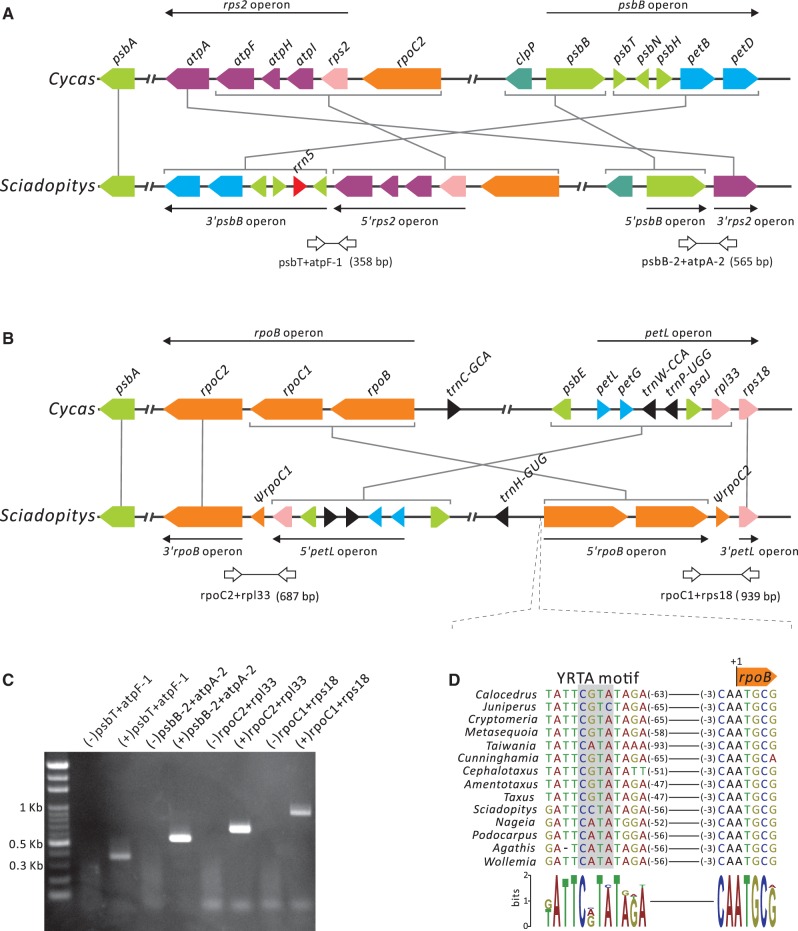
In Sciadopitys, plastomic inversions have also disrupted four operons that are generally conserved among seed plants. These disrupted operons are rps2–atpI–atpH–atpF–atpA (hereafter, rps2 operon), psbB–psbT–psbH–petB–petD (psbB operon), rpoB–proC1–rpoC2 (rpoB operon), and petL–petG–psaJ–rpl33–rps18 (petL operon) (fig. 4A and B). On one hand, recombination between rps2 and psbB operons is associated with inversion 8 (supplementary fig. S1, Supplementary Material online), creating the rps2–petD and psbB–atpA gene clusters (fig. 4A). On the other hand, inversion 4 recombined the rpoB and petL operons and then generated the petL–rpoC2 and rpoB–rps18 gene clusters (fig. 4B). Most genes in each of the four chemic gene clusters have the same transcriptional direction (fig. 4A and B). Therefore, we postulated that genes in these chemic gene clusters might be co-transcribed. To verify this, we performed RT-PCR assays with specific primers designed from genes near the junction between different operon-derived segments.
Fig. 4.—
Birth of chimeric gene clusters in the Sciadopitys plastome. (A) Shuffling between rps2 and psbB operons and (B) between rpoB and petL operons. Syntenic genes in the corresponding operons of Cycas are used as references. Operons and their transcriptional directions are indicated by solid arrows. Syntenic blocks of genes are connected with gray lines. Paired open arrows are primers for amplifying cDNA fragments across junctions between two recombined operons. The expected sizes of amplicons are shown in parentheses. (C) RT-PCR analysis for detecting the transcripts comprising the genes originated from different operons. Primer pairs used for RT-PCR assays are shown above the gel panel, with minus and plus signs (in parentheses) denoting the use of RNA (negative control) and cDNA (experimental set) as templates, respectively. (D) YRTA motif of the NEP promoter upstream of rpoB.
As shown in figure 4C, our RT-PCR results indicate that (1) there was no DNA contamination in the assayed RNA because all the negative controls failed to yield any product; and (2) the expected size of amplicons was clearly detected in all experimental sets. These data suggest that shuffling between different operons could lead to the birth of new co-transcription units in plastids.
Disruptions of conserved plastid operons have been only reported in a few taxa, such as Vigna (Perry et al. 2002), Trifolium (Cai et al. 2008), Trachelium (Haberle et al. 2008), some genera of Geraniaceae (Guisinger et al. 2011), Taxus (Hsu et al. 2014), and Sciadopitys (this study). These taxa also have highly rearranged plastomes. Except for Vigna and Sciadopitys, none of the above taxa has experienced recombination between operons. In the Vigna plastome, recombination between two homologous operons, S10A and S10B, has led to the re-organization of genes in the operons (Perry et al. 2002). Nonetheless, novel chimeric gene clusters created by shuffling between heterologous operons (fig. 4) are documented for the first time in the present study.
Evolutionary Impacts of Novel Chimeric Gene Clusters
The chimeric gene clusters of Sciadopitys provide two novel insights into the evolution of plastomes. First, other than the gene cluster rpoB–rpoC1–rps18, the remaining three chimeric gene clusters do not alter their upstream regions, as the neighboring genes of their 5′ regions are the same as those of Cycas (fig. 4A and B). This finding suggests that the promoter sequences of these gene clusters have not been altered after the associated inversions taken place. Figure 4D shows that the upstream sequence of rpoB harbors an YRTA motif of the nuclear-encoded RNA polymerase (NEP) promoter (Shiina et al. 2005). Furthermore, genes of different origins are able to be co-transcribed in the chimeric gene cluster (fig. 4C). Therefore, we cannot rule out the possibility that the pre-existing promoters are adopted for transcription of the genes in these chimeric gene clusters.
Second, shuffling between rpoB and petL operons (fig. 4B) has relocated rpoC2 to join the segment of the 5′petL operon whose transcription is associated with the plastid RNA polymerase (PEP) promoter (Finster et al. 2013). RpoC2 codes for one of the core units of PEP (Hu and Bogorad 1990). If the chimeric gene cluster petL–petG–psaJ–rpl33–rpoC2 is exclusively transcribed by PEP, we would not expect any transcript of this gene cluster in Sciadopitys. Nonetheless, its associated transcript was observed in figure 4C. Two possibilities might account for the presence of this transcript: (1) the isomeric plastome of the B form (fig. 3) that contains an intact rpoB operon provides RPOC2 proteins; (2) an alternative promoter has evolved to perform transcription because many plastid genes are transcribed by both the NEP and PEP promoters (Börner et al. 2015).
Conclusion
The plastome of Sciadopitys is characterized by several unusual features, such as the loss of the typical IRA copy, the duplication and pseudogenization of four tRNAs, extensive genomic inversions, the presence of isomeric plastomes, and chimeric gene clusters derived from shuffling of remote operons. All these characteristics highlight the fact that the evolution of plastomes may be more complicated than previously thought. The highly rearranged plastome of Scidaopitys advances our understanding of the dynamics, complexity, and evolution of plastomes in conifers.
Supplementary Material
Supplementary figure S1 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Shu-Mei Liu for filling sequence gaps. This work was supported by research grants from the Ministry of Science and Technology, Taiwan (MOST 103-2621-B-001-007-MY3), and from the Investigator’s Award of Academia Sinica (2011–2015) to S.-M.C. Special thanks are due to one anonymous reviewer for critical reading and helpful comments. The authors also like to thank Dr. Robert Jansen for valuable suggestions that improved the manuscript.
Literature Cited
- Alkatib S, Fleischmann TT, Scharff LB, Bock R. 2012. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res. 40:6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA. 2002. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 12:26–36. [PMC free article] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV. 2015. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta. 1847:761–769. [DOI] [PubMed] [Google Scholar]
- Cai Z, et al. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67:696–704. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH. 2004. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 58:18–11. [DOI] [PubMed] [Google Scholar]
- Chaw SM, Parkinson CL, Cheng Y, Vincent TM, Palmer JD. 2000. Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc Natl Acad Sci U S A. 97:4086–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp MD, Cook LG. 2011. Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytol. 192:997–1009. [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finster S, Eggert E, Zoschke R, Weihe A, Schmitz-Linneweber C. 2013. Light-dependent, plastome-wide association of the plastid-encoded RNA polymerase with chloroplast DNA. Plant J. 76:849–860. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28:583–600. [DOI] [PubMed] [Google Scholar]
- Guo W, et al. 2014. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol Evol. 6:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 66:350–361. [DOI] [PubMed] [Google Scholar]
- Hirao T, Watanabe A, Kurita M, Kondo T, Takata K. 2008. Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol. 8:70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CY, Wu CS, Chaw SM. 2014. Ancient nuclear plastid DNA in the yew family (taxaceae). Genome Biol Evol. 6:2111–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Bogorad L. 1990. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc Natl Acad Sci U S A. 87:1531–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA. 2012. Plastid genomes of seed plants In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Netherlands: Springer; p. 103–126. [Google Scholar]
- Jiang ZK, Wang YD, Zheng SL, Zhang W, Tian N. 2012. Occurrence of Sciadopitys-like fossil wood (Conifer) in the Jurassic of western Liaoning and its evolutionary implications. Chin Sci Bull. 57:569–572. [Google Scholar]
- Kolosova N, et al. 2004. Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechniques 36:821–824. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C, et al. 2015. Endosymbiotic origin and differential loss of eukaryotic genes. Nature 524:427–432. [DOI] [PubMed] [Google Scholar]
- Li J, et al. 2016. Evolution of short inverted repeat in cupressophytes, transfer of accD to nucleus in Sciadopitys verticillata and phylogenetic position of Sciadopityaceae. Sci Rep. 6:20934.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AS, Brennan S, Murphy DJ, Kavanagh TA, Wolfe KH. 2002. Evolutionary re-organisation of a large operon in adzuki bean chloroplast DNA caused by inverted repeat movement. DNA Res. 9:157–162. [DOI] [PubMed] [Google Scholar]
- Rai HS, Reeves PA, Peakall R, Olmstead RG, Graham SW. 2008. Inference of higher-order conifer relationships from a multi-locus plastid data set. Botany 86:658–669. [Google Scholar]
- Raubeson LA, Jansen RK. 1992. A rare chloroplast DNA structure mutation is shared by all conifers. Biochem Syst Ecol. 20:17–24. [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. 2005. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol. 244:1–68. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Jr, Via LE. 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–750. [PubMed] [Google Scholar]
- Sugiura M. 1992. The chloroplast genome. Plant Mol Biol. 19:149–168. [DOI] [PubMed] [Google Scholar]
- Tsumura Y, Suyama Y, Yoshimura K. 2000. Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga. Mol Biol Evol. 17:1302–1312. [DOI] [PubMed] [Google Scholar]
- Vieira Ldo N, et al. 2014. The complete chloroplast genome sequence of Podocarpus lambertii: genome structure, evolutionary aspects, gene content and SSR detection. PLoS One 9:e90618.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Chaw SM. 2014. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol J. 12:344–353. [DOI] [PubMed] [Google Scholar]
- Wu CS, Chaw SM. 2015. Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biol Evol. 7:2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 3:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Liu SM, Chaw SM. 2007. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol. 24:1366–1379. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Yi X, Gao L, Wang B, Su YJ, Wang T. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol. 5:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap JY, et al. 2015. Complete chloroplast genome of the wollemi pine (Wollemia nobilis): structure and evolution. PLoS One 10:e0128126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.