Abstract
The bacterial cell surface proteins intimin and invasin are virulence factors that share a common domain structure and bind selectively to host cell receptors in the course of bacterial pathogenesis. The β-barrel domains of intimin and invasin show significant sequence and structural similarities. Conversely, a variety of proteins with sometimes limited sequence similarity have also been annotated as “intimin-like” and “invasin” in genome datasets, while other recent work on apparently unrelated virulence-associated proteins ultimately revealed similarities to intimin and invasin. Here we characterize the sequence and structural relationships across this complex protein family. Surprisingly, intimins and invasins represent a very small minority of the sequence diversity in what has been previously the “intimin/invasin protein family”. Analysis of the assembly pathway for expression of the classic intimin, EaeA, and a characteristic example of the most prevalent members of the group, FdeC, revealed a dependence on the translocation and assembly module as a common feature for both these proteins. While the majority of the sequences in the grouping are most similar to FdeC, a further and widespread group is two-partner secretion systems that use the β-barrel domain as the delivery device for secretion of a variety of virulence factors. This comprehensive analysis supports the adoption of the “inverse autotransporter protein family” as the most accurate nomenclature for the family and, in turn, has important consequences for our overall understanding of the Type V secretion systems of bacterial pathogens.
Keywords: intimin, invasin, beta-barrel, TamA, BamA
INTRODUCTION
The intimin/invasin family of proteins constitutes a set of virulence factors secreted by bacterial pathogens. Both intimin and invasin share a common domain structure and a broad function to promote adherence of the pathogen to host cell surfaces. The founding member of this protein family was identified in enteropathogenic Escherichia coli (EPEC) and initially termed E. coli attaching/effacing protein (EaeA) and is now called the intimin (Jerse et al. 1990). The eaeA gene encoding intimin is found in the locus for enterocyte effacement (LEE), along with the genes encoding numerous virulence factors including the components of a Type III secretion system (McDaniel et al. 1995). Intimin from EPEC was suggested to be an adhesin because EPEC eaeA mutants fail to adhere to epithelial cells. In order to mediate adhesion, intimin docks onto a protein called the Tir (translocated intimin receptor), itself a bacterial protein encoded in the LEE, that is pre-injected into the host cell surface via the type III secretion system (Kenny et al. 1997; Knutton et al. 1998; Batchelor et al. 2000). The intimin–Tir interaction ultimately triggers a re-arrangement of the host cell cytoskeleton into a pedestal structure that enables the intimate association of the pathogen on the host cell surface (Kelly et al. 1999). The structure of the intimin–Tir interaction domain revealed a crucial role for the C-terminal lectin domain of intimin (Kelly et al. 1999; Batchelor et al. 2000; Luo et al. 2000). Intimin has been characterized in other attaching and effacing pathogens such as enterohemorrhagic E. coli (EHEC) and Citrobacter rodentium (Jores et al. 2004; Mundy et al. 2005; Lai et al. 2013; Law et al. 2013).
Following a broadly similar paradigm, species of Yersinia can adhere to and then invade specific host cells via a mechanism that depends on the interaction between extracellular domains in invasin docking to host cell surface integrins (Hamburger et al. 1999). The prototypical invasin is InvA from Yersinia pseudotuberculosis, but a variety of proteins with sometimes limited sequence similarity have also been annotated as “invasins” in genome datasets. The bona fide InvA has a characteristic C-terminal lectin domain that binds to β1-integrins, triggering a rearrangement of the host cell cytoskeleton that ultimately allows entry of the pathogen into the host cell (Isberg and Barnes 2001). The domain organization of invasin is similar to that of intimin, with a highly conserved N-terminal 12-stranded β-barrel domain serving as the most characteristic feature; the β-barrel domains of the intimin from EHEC and the invasin from Y. pseudotuberculosis showed 50% sequence identity (65% sequence similarity) and are structurally superimposable (Isberg et al. 1987; Fairman et al. 2012). The β-barrel domain and the linker region of invasin, and intimin, correspond to a conserved Pfam domain termed the invasin_beta. The passenger domains of intimin and invasin are also structurally related, being composed of a series of three to five bacterial immunoglobulin-like (Big) domains (Bodelón et al. 2013; Leo et al. 2015).
In various lineages of E. coli, intimin has undergone a rapid evolution, forming a large number of variants classified according to their immunological properties as the following: α1, α2, β1, β2 (ξR/β2B), β3, γ1, γ2, δ (δ/β2O), ϵ1, ϵ2 (νR/ϵ2), ϵ3, ϵ4, ϵ5 (ξB), ζ, η1, η2, θ, ι1, ι2 (μR/ι2), κ, λ, μB, νB, o, π, ρ, and σ (Adu-Bobie et al. 1998; Oswald et al. 2000; Tarr and Whittam 2002; Zhang et al. 2002; Blanco et al. 2004; Blanco et al. 2005; Blanco, Blanco, Dahbi, Alonso, et al. 2006; Blanco, Blanco, Dahbi, Mora, et al. 2006; Garrido et al. 2006; Yamamoto et al. 2009). This incredible diversity is functionally relevant, as reflected in the cell types to which each intimin can dock. For example, intimin sub-types γ1/γ2 appear to restrict pathogen binding to the follicle-associated epithelium in Peyer's patches of the ileum (Fitzhenry et al. 2002). The diversity seen in intimin sequences is likely to have been driven by the selective pressure in the host–pathogen relationship, and has supported a notion of broad sequence diversity within the intimin/invasin protein family. In keeping with this notion, recent sequence and structural studies have revealed a wide variety of non-redundant proteins categorized as intimin-related or invasin-related proteins found in diverse species of bacterial pathogens (Tsai et al. 2010; Fairman et al. 2012).
In the assembled intimin and invasin, the C-terminal passenger domain is threaded through the lumen of the N-terminal β-barrel domain (Fairman et al. 2012; Oberhettinger et al. 2015). This topology is reminiscent of that seen in autotransporter proteins and, because of this overall similar architecture, the intimin/invasin family of proteins are also referred to as inverse autotransporters and are categorized as the Type Ve secretion system (Leo et al. 2015). The prefix “inverse” refers to the fact that the autotransporter β-barrel domain is the C-terminal domain, while in intimin and invasin, the β-barrel is an N-terminal domain. The pathway by which autotransporters are assembled depends on two elements of the β-barrel assembly machinery: the BAM complex and the translocation and assembly module (TAM) (Selkrig et al. 2014; van Ulsen et al. 2014; Bernstein 2015). As with the autotransporters, bamA repression studies have revealed that intimin and invasin require the BAM complex for assembly, but whether the TAM plays a synergistic role has not been tested for the inverse autotransporters (Bodelón et al. 2009; Oberhettinger et al. 2012).
Recent work suggests that at least two other virulence-associated bacterial outer membrane proteins share a structural and evolutionary history with intimin and invasin: FdeC protein from extraintestinal pathogenic E. coli (ExPEC) and the Salmonella virulence factor SinH (also known as SivH, Kingsley et al. 2003). The passenger domain of FdeC was reported to have a number of Big domains, similar to those of intimin and invasin (Nesta et al. 2012), while the passenger domain of SinH is more divergent and not annotated at all in Pfam. With the relationships within the intimin/invasin protein family warranting further investigation, we have characterized sequence and structure relationships and show that several other virulence factors also belong to this family. Surprisingly, intimins and invasins represent a very small minority of the sequence diversity in what has been previously the “intimin/invasin protein family”. Analysis of the assembly pathway for the expression of both the classic intimin (EaeA) and what now appears to be the typical representative of the family, FdeC, reveal an involvement of the TAM as a common feature in both cases. FdeC was recently shown to function as an adhesin in ExPEC (Nesta et al. 2012; Easton et al. 2014). While the majority of the sequences in the grouping are most similar to FdeC, a further and widespread group appears instead to be two-partner secretion systems using the inverse autotransporter as the delivery device. This detailed analysis supports the adoption of the “inverse autotransporter protein family” as the most accurate nomenclature for the family, and further supports the characterization of this group of proteins as the Type Ve secretion system (Oberhettinger et al. 2012). This in turn has important consequences for the definition of, and cohesion within, the Type V secretion systems of bacterial pathogens.
Materials and Methods
Bioinformatic Analysis
The original dataset for the phylogenetic analysis was retrieved from Fairman et al. (2012), using all sequences with an active UniProt entry (UniProt 2015, http://www.uniprot.org, retrieved 2015 Jun 28). Functionally described sequences (supplementary table S2, Supplementary Material online) were added if they were not already present in the dataset. Alignments were performed using mafft with the -linsi option (Katoh and Standley 2013), the barrel region was extracted according to invasin (P11922; sites 121–501) and, based on this reduced alignment, sites for tree calculation were chosen using the online server TCS (Chang et al. 2015, http://tcoffee.crg.cat/apps/tcoffee/, last accessed August 2, 2015) under the default conditions. The best model was chosen using prottest (Darriba et al. 2011, version 3.1) with the -all-matrices -all-distributions settings resulting in the LG matrix (Le and Gascuel 2008). Trees were calculated using RaXML (Stamatakis 2006, version 7.2.8) with the rapid bootstrap analysis algorithm, the model set as PROTGAMMALGF and 100 bootstrap samples; and MrBayes using the mixed amino acid models calculated for 1 million generations sampling every 500 generations and a burnin of 25% for the calculation of the consensus tree. Mapping and visualization of datasets onto the tree was performed using itol (Letunic and Bork 2011). The alignment and the tree files can be accessed from figshare (https://figshare.com/articles/Conserved_features_in_the_structure_mechanism_and_biogenesis_of_the_inverse_autotransporter_protein_family/3380185).
The domains for displaying the domain architecture were predicted using the Sanger Pfam online server (Finn et al. 2014, http://pfam.xfam.org, last accessed August 12, 2015) with the gathering threshold selected as cutoff. Operon information was retrieved from the ensemble bacteria web resource (Kersey et al. 2012, retrieved 2015 Aug 12) or, if not present there, from GenBank (Benson et al. 2015, http://www.ncbi.nlm.nih.gov/genbank/, retrieved 2015 Aug 12). Pairwise alignment scores were calculated using the needle and water algorithms from the EMBOSS package (Rice et al. 2000, version 6.6.0.0) with the default parameters (gap opening penalty 10.0, gap extension penalty 0.5 for both algorithms); one outlier (D2U3C0) representing a gene duplication was manually reduced to the maximum value of 600. The dataset for the clustering analysis of all invasin_beta domain proteins (fig. 6) was obtained by extracting all accessions assigned as IPR024519 (Pfam domain PF11924 = invasin_beta) from the protein2ipr.dat table provided by InterPro (Mitchell et al. 2015, downloaded 20 May 2015) and retrieving the sequences from UniProt (retrieved on 2015 Jun 27); this sequence selection was reduced by clustering sequences with an id 0.9 cutoff using the uclust software with the usearch -cluster_fast algorithm, sequences were retrieved by using the -centroids option (Edgar 2010), the sequences displayed in figure 3 were reintroduced if they were removed by the clustering and colored according to the groups assigned in figure 3. All clustering analyses were performed using CLANS (Frickey and Lupas 2004) under the default conditions; the network in figure 6 represents a P value cut-off of 1E−10, figure 2 represents a P value cut-off of 1E−35 and figure 9A represents a P value cut-off of 1E−5. To facilitate visualization of sequences in closely related clusters, all clusterings shown are in 2D space.
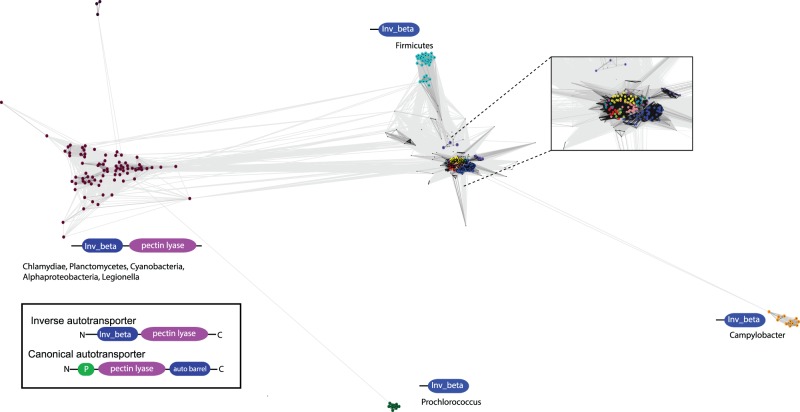
Fig. 6.—
Diversity of invasin_beta β-barrel domain proteins extends beyond the inverse autotransporter family. A protein–protein similarity network of all sequences annotated with an invasin_beta domain (reduced to identity 0.9). The inset frames the majority of sequences which cluster tightly, the colors correspond to the sequence groups as shown in fig. 3. Sequences were observed with both a pectin lyase domain and the invasin_beta translocation device. This is comparable with canonical autotransporters such as pertactin (inset); these Type Va autotransporters often have an additional protease domain indicated by the green icon (“P”).
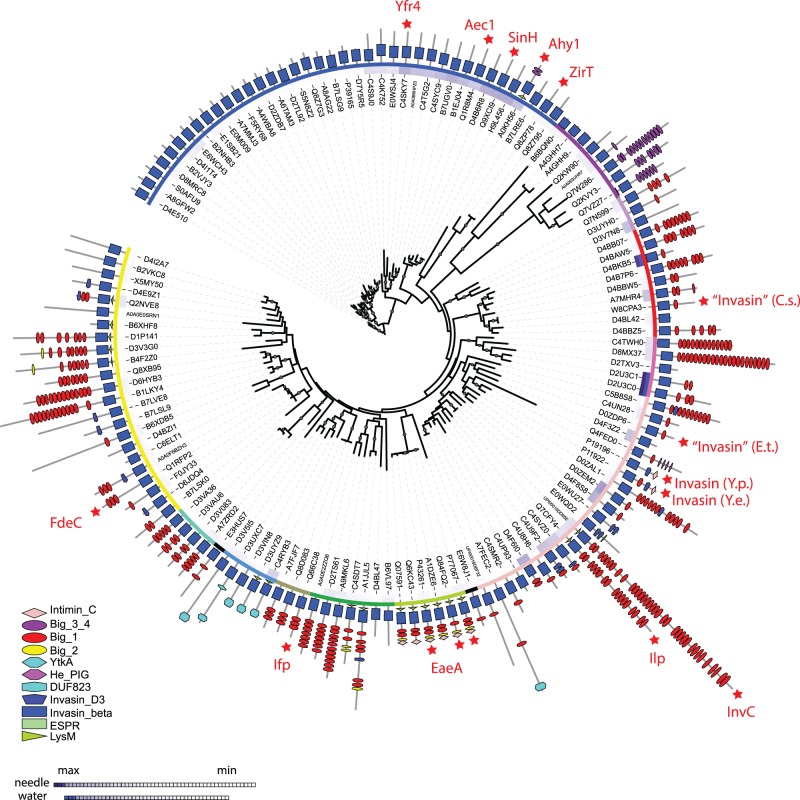
Fig. 3.—
The evolution of the intimin/invasin protein family. The root was set at the proteins without additional adhesion domains for display purposes, assuming a subsequent gain and duplications/new acquisitions of these across the sequence diversity of related proteins; unrooted versions of the tree are shown in fig. 4. The tree was calculated with RaxML using the invasin_beta β-barrel region of the sequences only. Functionally characterized proteins as listed in supplementary table S2 (Supplementary Material online) are indicated with red stars. The domain composition according to Pfam is shown in the outermost circle, with the legend indicating the different respective domain. The two innermost rings indicate similarity values of the proteins sequences with the protein sequences of their respective downstream genes (supplementary table S5, Supplementary Material online) based on pairwise alignments, using either the needle (innermost circle) or the waterman algorithm. The third ring indicates manually defined groups based on their domain composition and branching order, which correspond to the colors used in figs. 4 and 6. The tree was visualized using the itol online server (Letunic and Bork 2011).
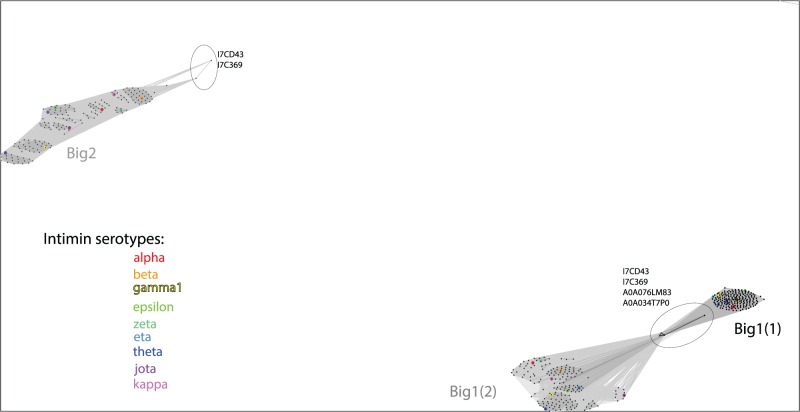
Fig. 2.—
A characterization of the Big domains in intimins by similarity clustering. The most N-terminal Big domain 1 is designated Big1(1) and the subsequent Big domain 1 is Big1(2). These sequences as well as the Big2 domain where present, were extracted from intimin sequences and clustered using the CLANS software. The analysis highlights that little sequence divergence is seen in the domain that would be positioned closest to the outer membrane (Big1(1) domain) and divergence is seen to increase towards the C-terminal lectin domain. Color coding denotes the serotypes, with the CLANS sequence groups corresponding very well with the serotypes. Encircled are the sequences from organisms other than E. coli that sit more diverged from the intimin serotypes of Escherichia species.
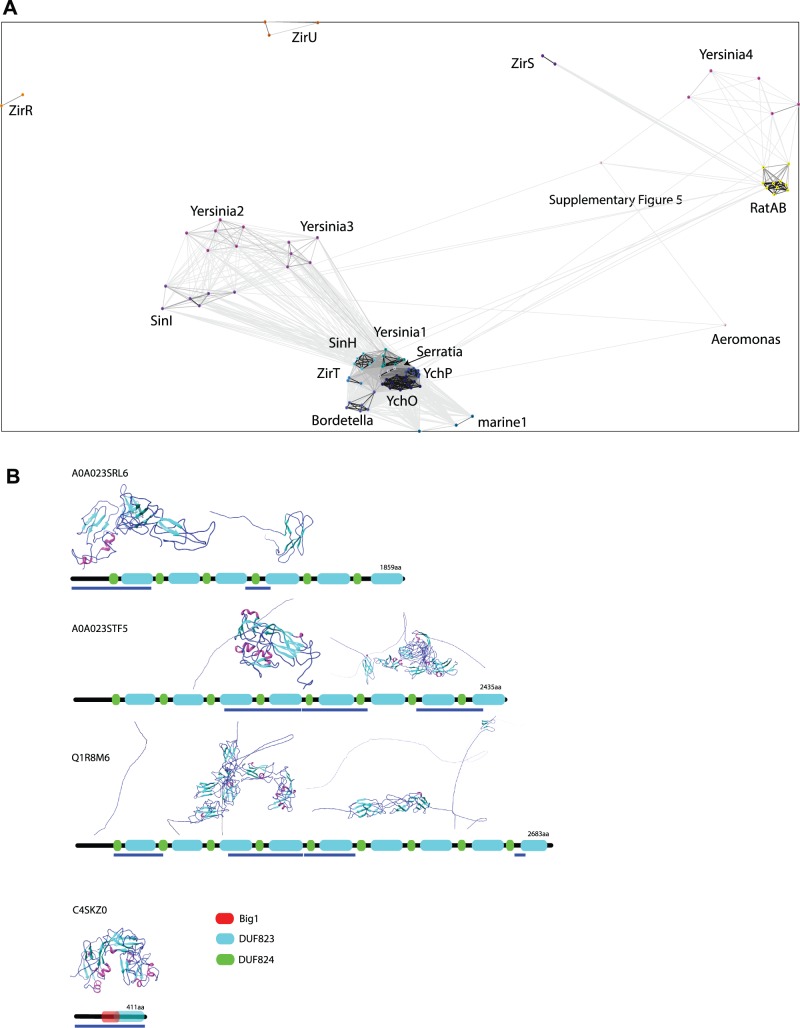
Fig. 9.—
Relationships between Rat-like proteins and Big domain proteins. (A) Protein–protein similarity network of putative two-partner secretion systems with inverse autotransporters and closely related sequences, including the membrane-embedded proteins as well as their putative substrates as shown in fig. 8A, using CLANS. The coloring corresponds to that used in fig. 8A. (B) Domain architecture of selected Rat-like proteins as annotated in the InterPro database (Mitchell et al. 2015, retrieved 2015 Aug 12). All proteins were subjected to Phyre2 (Kelley et al. 2015) in the intensive mode, and the parts of the retrieved structures that were not disordered are shown; the colors show secondary structure elements, and the images were prepared in Chimera (Pettersen et al. 2004). The blue bars below the domain profiles indicate the regions of the protein that were not disordered but are represented by structural elements.
Structure predictions were performed using the online server Phyre2 (Kelley et al. 2015), and structural models were retrieved when using the “intensive” mode for selected sequences in Phyre2, representation of the structures was drawn using Chimera (Pettersen et al. 2004). Secondary structure predictions were performed using Psipred (McGuffin et al. 2000) under the default conditions, using DomPred if the sequences exceeded the length cut-off for PsiPred. Sequences that exceeded the length cut-off for Phyre2 were split according to predicted domain borders as predicted by DomPred. For the clustering analysis of the Big domains, the intimins (supplementary table S1, Supplementary Material online) were reduced to remove identical sequences using uclust as described above, the sequences of the main intimin groups (supplementary table S1, Supplementary Material online) were added subsequently. The alignment was performed using mafft as above, and the Big domains (Big1-1: 560–649, Big1-2: 658–751, and Big2: 752–841) were defined according to Luo et al. (2000) as well as PSI (Haas et al. 2013) Protein Model Portal; http://www.proteinmodelportal.org/query/up/P19809, last accessed August 10, 2015) where no structural information was available, for the sequence P19809 (EPEC intimin). The selected sequence sets were then clustered using CLANS as described above.
Bacterial Strains and Culture Conditions
Supplementary table S4 (Supplementary Material online) summarizes the E. coli strains, plasmids, and primers used in this study. The open-reading frame encoding eaeA from enteropathogenic E. coli (EPEC) E2348/69 and fdeC from uropathogenic E. coli CFT073 were cloned into the plasmid pET-15b (Novagen). The open-reading frame encoding tamA from E. coli K-12 str. MG1655 was cloned into plasmid pACYCDuet-1 (Novagen) for complementation. The region comprising tamA interrupted with a kanamycin resistance casette from BW25113 ΔtamA (Baba et al. 2006) was used to generate an isogenic BL21 Star (DE3) ΔtamA via the linear fragment method (Datsenko and Wanner 2000). Escherichia coli cells were routinely incubated at 37 °C on an orbital platform rotating 200 strokes per minute in LB (Lennox 1955) or M9 Minimal Medium (M9 + S). M9 + S was prepared with 0.2% (w/v) glucose, 1.12 mM thiamine, 1 mM MgSO4, 0.1 mM CaCl2, and M9 Salts (Sambrook and Russell 2001), but an amino acid drop-out mix was used as the nitrogen source (instead of ammonium chloride), where the final concentration of each standard amino acid was 45.4 pg/mL, except leucine, which was 227 pg/mL. A second M9 Minimal Media formulation, but without methionine and cysteine and 1 mM MgCl2 in place of MgSO4, was also used (M9-S). When required, antibiotics were used at the following final concentrations: ampicillin, 100 µg mL−1; kanamycin, 30 µg mL−1; chloramphenicol, 34 µg mL−1.
To measure assembly of either EaeA or FdeC, the method of Stubenrauch et al. (2016) was followed, but optimized with specific details: cells were starved of methionine and cysteine for 30 min in M9-S and then cells were aliquoted into 20% (v/v) glycerol, snap frozen in liquid nitrogen and stored at − 80 °C. Aliquots from the one batch were considered to be technical replicates, whereas aliquots from separate batches were considered to be biological replicates. For the subsequent pulse chase experiment, cells were thawed on ice and resuspended in M9-S (to remove the glycerol). Rifampicin (0.5 mg mL−1) was added to block native RNA polymerase-based transcription. Following a 1-h incubation, T7 RNA polymerase-based transcription was induced with 0.2 mM IPTG for 5 min (at 30 °C). Cells were “pulse” labeled with 22 µCi mL − 1 [35S]-Methionine and [35S]-Cysteine (NEG072, Perkin Elmer) for 40 s and then “chased” (at 30 °C) by resuspension in M9 + S. Where indicated, aliquots were removed and incubated on ice for 10 min with or without 50 μg mL−1 proteinase K (MC500, Promega). Samples were then TCA precipitated, followed by an acetone wash, and were resuspended in SDS-loading buffer. Samples were boiled for 3 min and proteins were separated by SDS-PAGE followed by western transfer to 0.45 µm nitrocellulose or PVDF membrane. Radiation was captured using a storage phosphor screen (GE Health Sciences) and analyzed using Typhoon Trio (320 nm).
Mass spectrometry-based identification of SDS-PAGE separated protein fragments was conducted as previously published (Selkrig et al. 2012).
Results
Structure and Assembly of the Intimin Protein EaeA
Fairman et al. (2012) previously investigated a dataset of 146 protein sequences related to prototypical intimin (EaeA) and invasin (InvA) while demonstrating the structural features of the invasin_beta β-barrel domain as a translocation device. In order to determine how prevalent the intimin and invasin proteins are, a large dataset of proteins with an invasin_beta domain (IPR024519; for details on the datasets, see the methods section) was analyzed for the prevalence of classical intimin and invasin sequences. Given the immunological diversity of intimin sequences, we sought to address how prevalent characteristic intimins, and characteristic invasins, are within this set. The domain profile of intimin (fig. 1A) consists of an N-terminal “LysM” (PF01476) domain, the β-barrel domain, and a “passenger domain” formed from four bacterial immunoglobulin-like domains “Big_1” (PF02369) and “Big2” (PF02368), terminating with a lectin domain “Intimin_C”. Even though the domain “Big2” (PF02368) is generally present as a characteristic feature, it is not predicted in some EaeA proteins (e.g., Q6KC43_intimin νB) and was, therefore, not used as a search criterion.
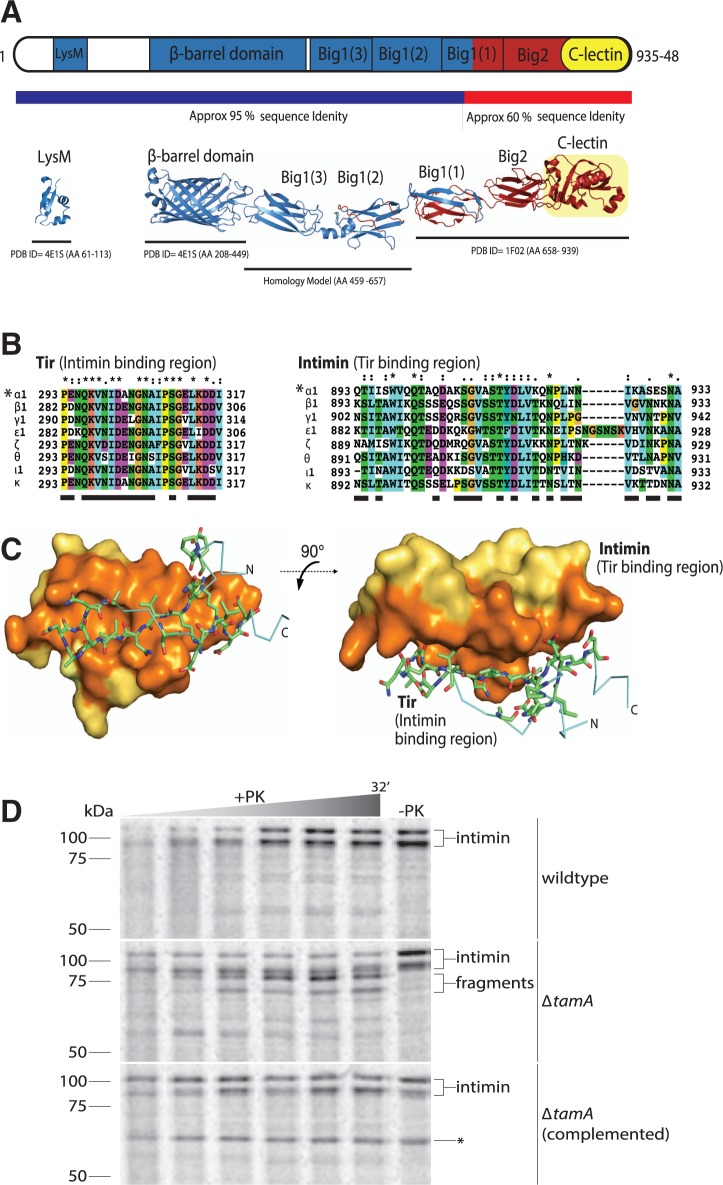
Fig. 1.—
Structure and assembly of the intimin EaeA. (A) The domain architecture of intimin (i.e., EaeA) is represented based on the PDB structures and homology-modeled domains. The predicted overall structure of intact intimin is colored based on sequence conservation between intimin subtypes (blue = 95% sequence identity; red = 60% sequence identity). (B) Multiple sequence alignments of Tir (showing the region that binds intimin) and of the C-lectin domain of intimin α1, β1, γ1, ϵ1, ζ, θ, ι1, and κ (showing the region that binds Tir). Residues are shaded according to the chemical properties, and contact residues identified in the Tir-intimin crystal structure (pdb 1F02) (sequence indicated with *) are underlined (supplementary table S3, Supplementary Material online, has the accession numbers for these intimin and Tir sequences). (C) The Tir:Int complex structure (pdb 1F02) is shown with Tir represented as a stick frame, while the space-filling model of the intimin C-lectin domain is shown in yellow and orange, where orange shading designates residues mediating contact with Tir. (D) Escherichia coli harboring plasmid pCJS30(intimin) were starved of methionine and cysteine, pulse labeled with 35S-Met and -Cys for 40 s, followed by a chase period with excess 32S-Met and-Cys, where aliquots were taken at 10 s, 2, 4, 8, 16, and 32 min. Aliquots were incubated with proteinase K (+PK) or without (−PK) for 10 min on ice, followed by TCA-precipitation, acetone washing, and resuspension in SDS loading dye, before analysis by SDS-PAGE. Middle and upper panels, cells also carry the control plasmid pACYCDuet-1. Lower panel, cells carry the complementing plasmid pCJS69(tamA) (plasmids detailed in supplementary table S4, Supplementary Material online). The labeled protein band corresponding to the ∼60 kDa TamA is indicated as *.
In a search that insisted on the presence of these characteristic domains (and allowed for the presence or absence of any other additional domains), 231 proteins were discovered with this domain profile, and are, therefore, classified here as “intimins”. This set of sequences is equivalent to those detected recently in a similar search (Leo et al. 2015) and encompasses the immunologically diverse intimin variants. The vast majority of these (i.e., 221 sequences) are found in strains of E. coli, in support of the key role of intimins in forming attaching/effacing lesions in E. coli pathovars (Jores et al. 2004; Lai et al. 2013; Law et al. 2013). The remaining 10 protein sequences are found in genera closely related to E. coli: Citrobacter, Edwardsiella, Salmonella, and Shigella (supplementary table S1, Supplementary Material online). Because the β-barrel domains of intimins are structurally conserved (Fairman et al. 2012), a protein-protein similarity network analysis was calculated using the passenger domain elements of the 231 intimin sequences (fig. 2). In these plots, each dot represents a sequence and the distance between each dot is a reflection of their sequence similarity. The relative clustering of the sequences highlights that stronger divergence is seen in the passenger domains, and it increases towards the C-terminus: the Big1(1) domains are very tightly clustered, Big1(2) domains are somewhat less tightly clustered, and the Big2 domains disperse into sub-clusters where each corresponds to one or two serotypes (figs. 1A and 2).
Analysis of the genomes carrying the 231 intimin-encoding sequences revealed a Tir sequence as well in the vast majority of these (supplementary table S1, Supplementary Material online). Intimin binds Tir via its C-lectin domain; a multiple sequence alignment of partner Tir and intimin proteins shows that while the intimin-binding region of Tir proteins is highly conserved, the Tir-binding surface of intimin exhibits considerable sequence variation (fig. 1B). This observation is intriguing and suggests that the broad Tir-interacting surface of intimin (fig. 1C, shaded orange) has diversified to find multiple solutions for binding the narrow, β-hairpin of the Tir-binding surface (fig. 1C). This sequence diversification of intimin in the Tir-binding region, and in its C-terminus more generally, is likely due to pressure from the host immune system.
In order to be accessible to Tir, the entire passenger domain of intimin is secreted as a part of the assembly of intimin into the outer membrane. The Omp85 protein BamA is responsible for the assembly of β-barrel proteins into bacterial outer membranes, and repression of bamA expression inhibits intimin assembly (Bodelón et al. 2009; Oberhettinger et al. 2012). Given the recent finding that repressing bamA expression also depletes the level of TamA (Dunstan et al. 2015), we sought to use ΔtamA mutants of E. coli to determine whether the TAM plays a synergistic role with the BAM complex in the assembly of intimin. To monitor assembly, a rifampicin-blocking technique Stubenrauch et al. (2016) was employed to selectively label intimin with 35S-labeled amino acids in vivo. A 32-min time course of intimin expression in wild-type E. coli showed the expression of the expected 98 kDa protein as well as a form migrating at ∼115 kDa (fig. 1D). This unusual migration pattern is due to the tight-folding of surface-exposed intimin, which is only partially unfolded under conditions of SDS-PAGE (Bodelón et al. 2009). The protein is unaffected by the addition of proteinase K (PK), given that the profile is equivalent in the “−PK” or “+PK” samples. Strikingly, in the ΔtamA mutant strain, the majority of intimin exist in a form that is susceptible to cleavage by proteinase K, with the “+PK” treatment generating a ladder of fragments of intimin from the surface of the cells. That this cleavage is the result of defective assembly of intimin due to the absence of TamA was confirmed by complementation of the ΔtamA mutant strain with a plasmid containing tamA, which rescued this phenotype (fig. 1D). This finding suggests that the TAM assists in the assembly of members of two distinct, yet topologically equivalent protein families: autotransporters (Selkrig et al. 2012, 2015) and inverse autotransporters (fig. 1D).
The Intimin/Invasin Protein Family
For phylogenetic analyses, the original dataset from Fairman et al. was retrieved, using all sequences with an active UniProt entry (see the Methods section). Functionally described sequences (supplementary table S2, Supplementary Material online) were added if they were not already present in the dataset. Phylogenetic analysis of the inverse autotransporter family suggests a monophyletic group of sequences where the invasin_beta β-barrel domain is appended to a very short C-terminal extension (figs. 3 and 4), which could represent the origin of the superfamily where other domains where subsequently gained. Given (i) the small proportion of sequences corresponding to bona fide intimins and invasins, (ii) that the invasin_beta domain appears to occur independently of any particular passenger domain, and (iii) to move away from any misnomer in the literature, we hereafter refer to the overall set of proteins as the inverse autotransporter protein family (Leo et al. 2015).
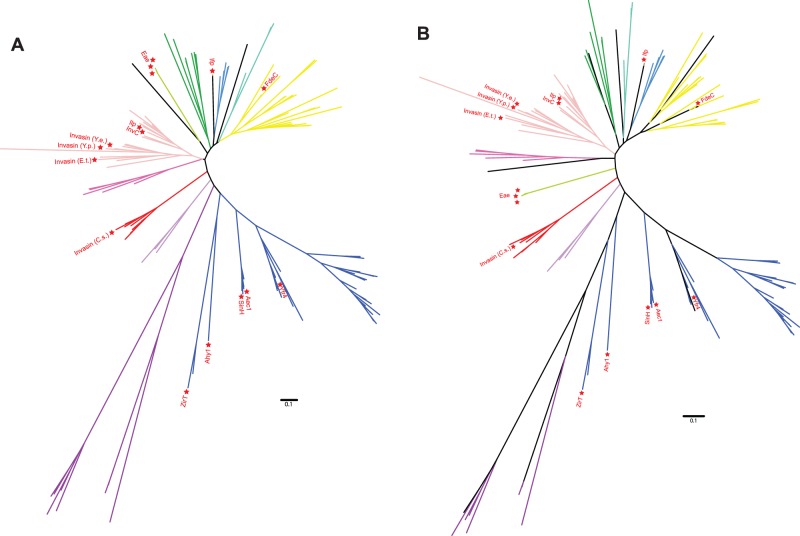
Fig. 4.—
Phylogenetic relationships of the major groups in the intimin/invasin superfamily as discussed in this study. (A) The unrooted representation of the tree in fig. 3 is shown, and the branches are colored according to the groups as assigned and shown as ring 3 in fig. 3. Functionally characterized proteins are indicated for additional orientation. This figure highlights the monophyly of some, but not all, groups as discussed in this study. (B) The same alignment was analyzed under a Bayesian algorithm (MrBayes, for details see the Methods section), not only highlighting the monophyly of the discussed groups but also showing the low confidence of the ordering of the deep branches.
Phylogenetic analysis combined with the domain profiles of the sequences demonstrates that intimins and invasins form distinct, monophyletic branches with only few members (figs. 3 and 4). Analysis of the domain profiles for all these proteins underscores that intimins and invasins are an exception with respect to their domain architecture too. Despite the immunologically relevant sequence diversity mapped in the Big domains of the intimin serotypes (fig. 2), all the intimin sequences form a tight, distinct group divorced from other sequences (fig. 4). The canonical invasin, InvA, is also part of a conserved monophyletic group of sequences that takes up relatively little of the sequence space of the inverse autotransporter family (figs. 3 and 4). In addition to the characteristic β-barrel domain, invasin has domains recognized by Pfam as “Intimin_C” (PF07979) and “Invasin_D3” (PF09134). Of all sequences encoding inverse autotransporter β-barrel domain, only 22 out of 2,468 proteins are annotated with this domain profile even when allowing for other, additional domains to be present; all these sequences belong to the genus Yersinia (supplementary table S1, Supplementary Material online).
Figure 3 also shows that the majority of the inverse autotransporter sequences have domain architectures distinct from intimin and invasin, but that these are all instead similar to that of the functionally characterized adhesin, FdeC. FdeC is a protein of 150 kDa, with an extensive series of Big domains extending from the bacterial surface (Nesta et al. 2012; fig. 5A). Using selective 35S-labeling of FdeC in E. coli to monitor the assembly of FdeC in vivo revealed that it is assembled in a time-dependent manner, in the “−PK” control, the size of FdeC is consistent with the predicted size of 150 kDa (fig. 5B). Treatment of bacteria expressing FdeC with proteinase K generated 35S-labeled fragments of ∼100 kDa and ∼50 kDa. In gel, tryptic digestion and mass spectrometry revealed that the ∼50 kDa fragment includes the N-terminal β-barrel domain, and the ∼100 kDa fragment comprises the rest of the C-terminal passenger domain (fig. 5A). As judged by comparison with the intensity of the “−PK” control lane, the majority of the FdeC expressed in this system is cleaved into these two fragments of the protein (fig. 5B).
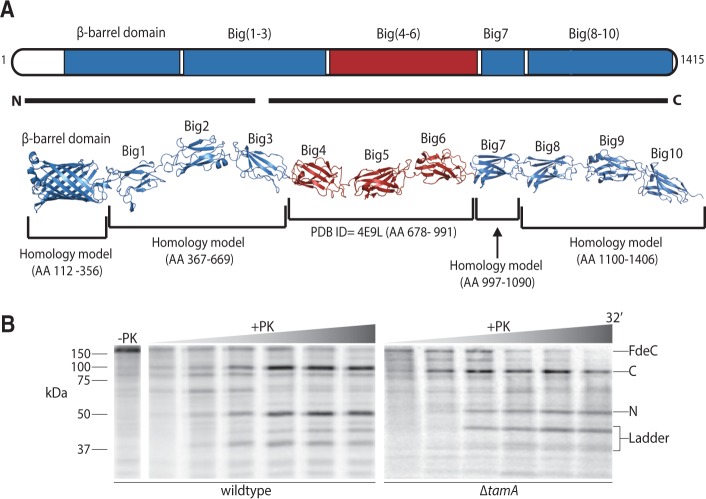
Fig. 5.—
The assembly of FdeC is enhanced by the TAM. (A) The domain architecture of FdeC is represented based on PDB structures and homology-modeled domains. The predicted overall structure of intact FdeC is colored based on the sequence conservation between FdeC subtypes (blue = 95% sequence identity; red = 60% sequence identity). The black lines “N” and “C” encompass the peptides recovered from the 50 kDa fragment and the 100 kDa fragment derived by proteinase K as described in the main text. (B) Escherichia coli harboring plasmid pCJS50(fdeC) were pulse labeled as in D. Mass spectrometry defined the identity of the 50 kDa N-terminal fragment of FdeC (N), and the 100 kDa C-terminal fragment of FdeC (C).
Two observations were made when following the assembly reaction for FdeC in ΔtamA mutants, lacking the TamA subunit of the TAM. First, the amount of FdeC assembled per unit time is less in the mutants, as judged by the relative intensity protein fragments in the wild-type and ΔtamA extracts (fig. 5B). Second, very little of the ∼50 kDa β-barrel domain fragment is observed, with a relative increase in the amount of other, smaller fragments (fig. 5B, “ladder”) seen in the assays. This is consistent with increased protease sensitivity in the surface-exposed loops of the β-barrel domain in the mutants, enabling different-sized fragments of FdeC to be generated in the mutants. Thus, in terms of biogenesis of members of the protein family, the TAM stimulates the expression of a specialist isoform (intimin) and the prototypical form (FdeC) of inverse autotransporters.
The Inverse Autotransporter Superfamily of Proteins
We sought to determine whether the β-barrel domain characteristic of inverse autotransporters has been acquired in other bacterial lineages. Using the invasin_beta domain as a diagnostic criterion, a dataset of proteins was retrieved from UniProt (for details on the datasets, see the Methods section) and was analyzed using a protein–protein similarity network (fig. 4). Groups of more distantly related sequences were highlighted by the clustering analysis. These include proteins from distinct Phyla such as the Chlamydiae and Cyanobacteria, but only distinct taxa within these Phyla; a comparative set of diversity was also detected in a study focused on the distribution of the LysM domain in the inverse autotransporter superfamily (Leo et al. 2015). Such a distribution is suggestive of horizontal gene transfer (HGT) events. Some of these sequence groups mirror the “invasin_beta-only” domain architecture also found in the conserved part of the inverse autotransporter protein family (fig. 3), but further support for HGT-based acquisition comes from the observation that a defined group of the more diverse sequences have a C-terminal domain defined as a “pectin lyase” (IPR011050), a domain common to virulence factors like pertactin, delivered via a classic autotransporter mechanism (fig. 6, inset). This observation adds strength to the hypothesis that the inverse autotransporter β-barrel domain is functionally equivalent to the classic autotransporter domain (Oberhettinger et al. 2015), with it being a device that can deliver equivalent virulence factors to the bacterial cell surface.
ZirT and Other Inverse Autotransporters with Co-Encoded Substrates
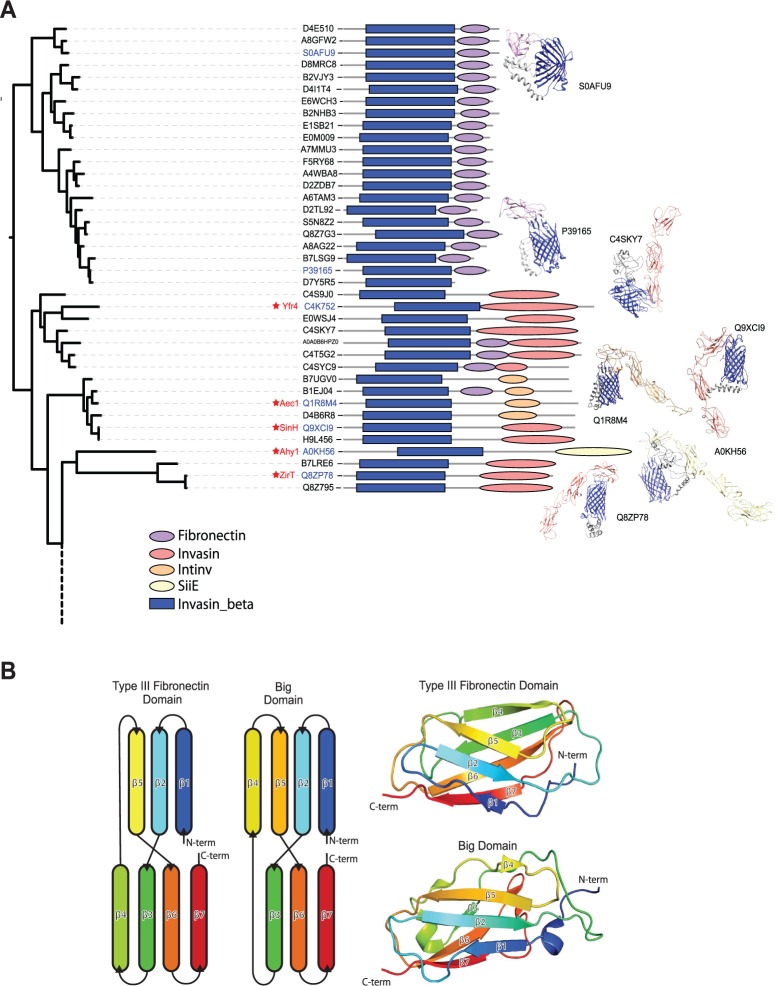
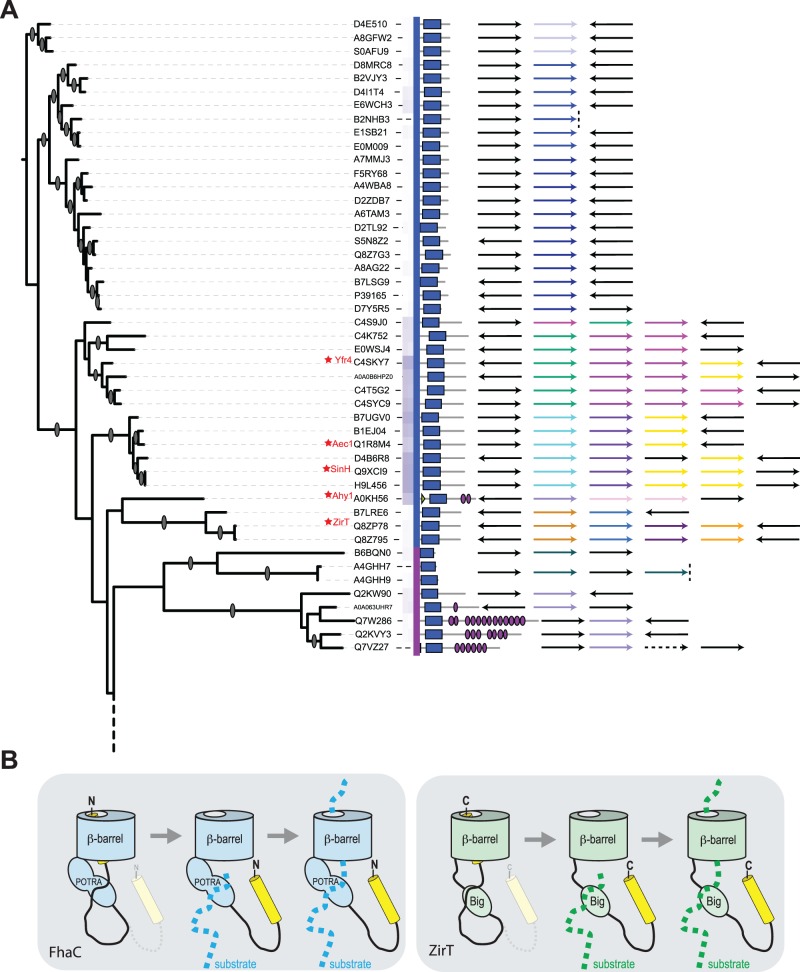
According to Pfam predictions, a substantial set of proteins shown in figure 3 do not have Big domains. Many of these have only a short C-terminal extension, in some cases, with structural characteristics of other bacterial immunoglobulin super-family (IgSF) domains (Bodelón et al. 2013) such as the fibronectin III domain (fig. 7A). The secondary structure elements in a fibronectin domain comprise a β-sandwich domain wherein three anti-parallel β-strands on one side are intimately packed against four anti-parallel β-strands on the opposite side of the sandwich. This overall β-sandwich structure is the same as that found in Big domains (fig. 7B) and all the other IgSF domains found in bacteria (Bodelón et al. 2013). This highly adaptable fold is generally used for protein–protein interactions, particularly in the periplasm where a number of chaperones and disulphide exchange folding factors have an IgSF structure that serves in chaperoning protein substrates that will either reside in the periplasm or be translocated across the outer membrane (Bodelón et al. 2013). Pfam aims to identify homologous domains based on sequence signatures, and while their ancestry might distinguish these domains as either “Big” or “fibronectin”, these are both equivalent in three-dimensional structure (Bodelón et al. 2013). This further commonality binds together the inverse autotransporter protein family. Analysis of the operons (fig. 8A) and the proteins they encode (fig. 9A) shows an organization reminiscent of the well-characterized ZirTS secretion system (Gal-Mor et al. 2008; Prehna et al. 2012). Notably, SinH from Salmonella and Yfr4 from Yersinia (fig. 8A) are inverse autotransporter proteins which share sequence similarity with the protein encoded downstream; this observation was previously documented for SinH, where the downstream protein SinI lacks a β-barrel domain, but has substantial sequence similar to the IgSF domains of SinH (Kingsley et al. 2003). In the case of ZirT, the downstream proteins ZirS and ZirU lack a β-barrel domain, but have sequence similar to the IgSF domains of ZirT (Prehna et al. 2012). It has been experimentally demonstrated that the products of the downstream genes (i.e., ZirS and ZirU) are substrates that are secreted via the ZirT β-barrel domain (Gal-Mor et al. 2008; Prehna et al. 2012).
Fig. 7.—
Structural models of inverse autotransporter passenger domains. (A) The protein sequences without predicted Big domains (fig. 3) were subjected to structure prediction with Phyre2 (Kelley et al. 2015). Results (see supplementary table S7, Supplementary Material online) are illustrated as icons. Most of the sequences appear to conform to fibronectin-like domains (purple), others predict to fold into Big domain folds (orange/red). Structural models built with Phyre2 were edited in Chimera (Pettersen et al. 2004). (B) Structural comparison of the human type III fibronectin domain and a Big domain from FdeC shows that both domains possess an equivalent β-sandwich fold. A schematic representation of the β-sheets from each domain (left) illustrates the common β-sheet topology, while a cartoon representation (right) of Type III Fibronectin repeat 9 (pdb: 1FNF, residues 1327–1417) and FdeC region B Big1 domain (pdb: 4E9L, residues 678–793) shows the β-sandwich fold.
Fig. 8.—
Inverse autotransporters and co-expressed substrate proteins. (A) The operon structures of a subset of inverse autotransporter candidates. The blue and green arrows (first column in the legend) show the inverse autotransporter proteins as given in the tree, whereas the other arrows illustrate the genetic environment (see supplementary table S6, Supplementary Material online). Black arrows indicate genes assessed as unlikely to be transcriptionally linked, dashed lines represent contig ends, and dashed arrows represent pseudogenes. (B) A model for two-partner secretion employing the inverse autotransporter barrel. FhaC is an archetypal two-partner secretion system (Jacob-Dubuisson et al. 2013). It employs an Omp85-type β-barrel domain, fused to a pair of substrate-binding POTRA domains (Clantin et al. 2007) and an N-terminal helix which sits in the barrel lumen, but which is displaced by the presence of the substrate protein FHA (Maier et al. 2015). By analogy, secretion of the substrates ZirS and ZirU by the inverse autotransporter ZirT might rely on reversible binding of a C-terminal helix in the lumen of the β-barrel domain. In its resting state, the C-terminus of ZirT would be oriented towards the cell surface, and a C-terminal epitope would be accessible to exogenously added protease, as observed by Gal-Mor et al. (2008).
Some of the operons shown in figure 8A also include proteins similar to the Rat-like proteins (RatA and RatB) from Salmonella enterica serovar Typhimurium (Kingsley et al. 2003). Further analysis of the Rat-like protein sequences showed that while the N-terminal region is highly divergent, the C-terminus shows sequence and predicted structure similar to the Big domains of intimin and invasin (fig. 9B). Based on the ZirT:ZirS translocator:substrate relationship (Gal-Mor et al. 2008; Prehna et al. 2012), and given their IgSF domain structures, we hypothesize that the Rat-like proteins encoded in these operons would also be substrates for the inverse autotransporter. In this regard, we note that the Rat-like protein from Yersinia was previously suggested to be a substrate for the protein translocase Yfr4 (Tsai et al. 2010). Our sequence analysis now makes clear that Yfr4 is a member of the inverse autotransporter family (fig. 8A).
Discussion
Oberhettinger et al. (2012) suggested the intimin/invasin family should be classified as the Type Ve secretion system, and coined the term inverse autotransporter. While this terminology has not yet been picked up widely in the literature, it is a highly appropriate classification and we suggest it be expanded to include four sub-families: intimin, invasin, FdeC-type adhesins, and two-partner inverse-autotransporters. The diagnostic features of the family are an N-terminal β-barrel domain that functions as the delivery device, and C-terminal domains that are predicted to have an IgSF fold, many of which are recognized by sequence profiling as either “Big” or “fibronectin” domains.
Analyses of the sequence and putative structures of the diverse members of the inverse autotransporter family gave insight into its evolutionary history. Almost exclusively restricted to the Gammaproteobacteria, these proteins show a highly disperse taxonomic distribution; in accordance with the sort of pattern now being recognized for classical virulence factors. The best studied members of the inverse autotransporter family, intimin and invasin, are two highly specialized forms almost exclusively restricted to Escherichia spp. and Yersinia spp., respectively. In the case of the intimin, it seems highly likely that this key component of the attaching/effacing lesion has rapidly evolved under pressure from the host immune system to generate serotypes with important functional differences within a relatively narrow range of overall sequence diversity. Despite being able to modulate diverse tissue tropism and evade immune surveillance to some extent, within the context of the overall inverse autotransporter protein family, the serotypes of intimin are effectively a single protein isoform and are rightly considered EaeA homologs.
Here we discovered that the majority of proteins in the inverse autotransporter family display a domain architecture-like FdeC, suggesting that they will function as adhesins. Expression of the adhesin FdeC on the bacterial cell surface is triggered by the interaction of E. coli with human cells (Nesta et al. 2012). From analysis by protein crystallography and structural modeling, FdeC carries 10 Big domains, and is predicted to extend substantially from the outer surface of the outer membrane (Nesta et al. 2012). FdeC provides adhesive properties important in two modes of attachment for E. coli: bacteria displaying FdeC on their surface are better able to interact with each other to form biofilms (Easton et al. 2014), and FdeC on ExPEC was shown to bind specifically to collagen and to promote colonization of kidney tissue surfaces (Easton et al. 2014).
The range in passenger domain size seen in inverse autotransporters, based on variation in the number of Big domains, is remarkable. It is yet another feature reminiscent of the autotransporters, where passenger domains can differ dramatically in length depending on the number of β-elements present in the predominantly β-helical structure of the passenger domain (Celik et al. 2012). Conceptually, this again ties the inverse autotransporter proteins of the Type Ve protein secretion system together with the autotransporters of the Type Va protein secretion system.
We suggest a further group of inverse autotransporters function as protein translocation devices for various protein substrates. A biochemical assessment of ZirT demonstrated that an epitope-tag on the C-terminus of the protein can be accessed by exogenously added protease, and it was, therefore, suggested that the entire C-terminal domain, composed of several Big domains, was extracellular (Gal-Mor et al. 2008; Prehna et al. 2012). In contradistinction to this finding, two proteins, ZirS and ZirU, have been shown to depend on ZirT as a protein translocase, delivering them across the bacterial outer membrane (Gal-Mor et al. 2008; Prehna et al. 2012). This conundrum has remained a Gordian knot, since structural studies show clearly that the channel in the β-barrel domain is blocked once it is occupied by even one α-helix of the C-terminal domain (Fairman et al. 2012).
Given how other devices within the Type V protein secretion system function, we suggest an alternative topology for proteins such as ZirT and SinH (fig. 8B). In our model, ZirT and the related proteins Aec1, SinH, Yfr4, etc. function dynamically and wholly equivalent to classic, FhaC-type two-partner secretion systems. The β-barrel domain of ZirT and other inverse autotransporter translocases are assembled into the outer membrane by the β-barrel assembly machinery. The C-terminal passenger domain would fold in the periplasm, with the final segment of this domain folding back into the channel of the β-barrel domain. This is equivalent to the structurally defined topology of the two-partner secretion system FhaC (Clantin et al. 2007; Jacob-Dubuisson et al. 2013; Maier et al. 2015) and would represent the resting state for the translocation device. This proposition is consistent with observations that diverse proteins with IgSF folds are found in the periplasm, and especially given that several of these function to bind and chaperone other IgSF-domain proteins to assist their biogenesis: either to complete their folding in the periplasm or to promote their translocation across the outer membrane (Bodelón et al. 2013).
According to this model, the Big domains of ZirT would serve as substrate-binding domains equivalent to the POTRA domains of FhaC. Big domains are known to function in protein-protein interactions, many of these being in the periplasm (Bodelón et al. 2013). Binding of the substrates ZirS and ZirU to the Big domains of ZirT would initiate a withdrawal of the C-terminal domain from the ZirT channel, and enable the translocation of the substrates through the outer membrane. By this mechanism, the secretion of multiple substrates such as ZirS and ZirU (Gal-Mor et al. 2008; Prehna et al. 2012) and other proteins contained within a secretion system operon (fig. 8A) can be accommodated. A repetitive mechanism such as this is the simplest explanation for how some of the inverse autotransporter operons shown in figure 8A might secrete as many as four related substrate proteins. A further attraction of this model is that it brings together the various arms of the Type V secretion system based on their mechanism: autotransporters (Type Va), inverse-autotransporters (Type Ve), and two-partner secretion systems (Type Vb), with two-partner inverse-autotransporters.
Supplementary Material
Supplementary table S1-S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge the Monash Bioinformatics Platform for computing resources, and thank Alex Bateman for critical comments on the manuscript. This work was supported through an National Health & Medical Research Council (NHMRC) Program Grant (APP1092262, to T.L., R.A.S., and G.D.). T.L. is an Australian Research Council (ARC) Australian Laureate Fellow, E.H. is an ARC Laureate Postdoctoral Fellow, R.G. is a Sir Henry Wellcome Postdoctoral Fellow (Grant no. 106077/Z/14/Z) and A.W.P. is a NHMRC Senior Research Fellow. C.J.S. is supported by a Monash Research Scholarship. Author contributions: E.H., C.J.S., R.G., and T.L. designed research; E.H., C.J.S., N.P.C. and R.G., performed research; E.H., C.J.S., R.G., N.P.C., A.W.P., R.A.S., G.D. and T.L. analyzed data; and E.H., R.A.S., G.D. and T.L. wrote the paper.
Literature Cited
- Adu-Bobie J, et al. 1998. Detection of intimins a, b, g, and d, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 36:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2: 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor M, et al. 2000. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. Embo J. 19:2452–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, et al. 2015. GenBank. Nucleic Acids Res. 43:D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein HD. 2015. Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol Microbiol. 97:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M, et al. 2004. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-x). J Clin Microbiol. 42:645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M, et al. 2005. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-h2). BMC Microbiol. 5:23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco M, Blanco JE, Dahbi G, Alonso MP, et al. 2006. Identification of two new intimin types in atypical enteropathogenic Escherichia coli. Int Microbiol. 9:103–110. [PubMed] [Google Scholar]
- Blanco M, Blanco JE, Dahbi G, Mora A,et al. 2006. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (mB and xR/b2B). J Med Microbiol. 55:1165–1174. [DOI] [PubMed] [Google Scholar]
- Bodelón G, Marin E, Fernández LA. 2009. Role of periplasmic chaperones and BamA (YaeT/Omp85) in folding and secretion of intimin from enteropathogenic Escherichia coli strains. J Bacteriol. 191:5169–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelón G, Palomino C, Fernández LA. 2013. Immunoglobulin domains in Escherichia coli and other enterobacteria: from pathogenesis to applications in antibody technologies. FEMS Microbiol Rev. 37:204–250. [DOI] [PubMed] [Google Scholar]
- Celik N, et al. 2012. A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS One 7:e43245.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JM, Di Tommaso P, Lefort V, Gascuel O, Notredame C. 2015. TCS: a web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res. 43:W3–W6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, et al. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317:957–961. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan RA, et al. 2015. Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol Microbiol. 97:616–629. [DOI] [PubMed] [Google Scholar]
- Easton DM, et al. 2014. The intimin-like protein FdeC is regulated by H-NS and temperature in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 80:7337–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Fairman JW, et al. 2012. Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 20:1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzhenry RJ, et al. 2002. Intimin type influences the site of human intestinal mucosal colonisation by enterohaemorrhagic Escherichia coli O157:H7. Gut 50:180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas A. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Gibson DL, Baluta D, Vallance BA, Finlay BB. 2008. A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog. 4:e1000036.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido P, et al. 2006. STEC-EPEC oligonucleotide microarray: a new tool for typing genetic variants of the LEE pathogenicity island of human and animal Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) strains. Clin Chem. 52:192–201. [DOI] [PubMed] [Google Scholar]
- Haas J, et al. 2013. The Protein Model Portal – a comprehensive resource for protein structure and model information. Database (Oxford) 2013: bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger ZA, Brown MS, Isberg RR, Bjorkman PJ. 1999. Crystal structure of invasin: a bacterial integrin-binding protein. Science 286:291–295. [DOI] [PubMed] [Google Scholar]
- Isberg RR, Barnes P. 2001. Subversion of integrins by enteropathogenic Yersinia. J Cell Sci. 114:21–28. [DOI] [PubMed] [Google Scholar]
- Isberg RR, Voorhis DL, Falkow S. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769–778. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Guerin J, Baelen S, Clantin B. 2013. Two-partner secretion: as simple as it sounds? Res Microbiol. 164:583–595. [DOI] [PubMed] [Google Scholar]
- Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A. 87:7839–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores J, Rumer L, Wieler LH. 2004. Impact of the locus of enterocyte effacement pathogenicity island on the evolution of pathogenic Escherichia coli. Int J Med Microbiol. 294:103–113. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly G, et al. 1999. Structure of the cell-adhesion fragment of intimin from enteropathogenic Escherichia coli. Nat Struct Biol. 6:313–318. [DOI] [PubMed] [Google Scholar]
- Kenny B, et al. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, et al. 2012. Ensembl genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 40:D91–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, et al. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun. 71:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, et al. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. Embo J. 17:2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Rosenshine I, Leong JM, Frankel G. 2013. Intimate host attachment: enteropathogenic and enterohaemorrhagic Escherichia coli. Cell Microbiol. 15:1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RJ, Gur-Arie L, Rosenshine I, Finlay BB. 2013. In vitro and in vivo model systems for studying enteropathogenic Escherichia coli infections. Cold Spring Harb Perspect Med. 3:a009977.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Lennox ES. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190–206. [DOI] [PubMed] [Google Scholar]
- Leo JC, Oberhettinger P, Schutz M, Linke D. 2015. The inverse autotransporter family: intimin, invasin and related proteins. Int J Med Microbiol. 305:276–282. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, et al. 2000. Crystal structure of enteropathogenic Escherichia coli intimin–receptor complex. Nature 405:1073–1077. [DOI] [PubMed] [Google Scholar]
- Maier T, et al. 2015. Conserved Omp85 lid-lock structure and substrate recognition in FhaC. Nat Commun. 6:7452.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 92:1664–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. [DOI] [PubMed] [Google Scholar]
- Mitchell A, et al. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43:D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. 2005. Citrobacter rodentium of mice and man. Cell Microbiol. 7:1697–1706. [DOI] [PubMed] [Google Scholar]
- Nesta B, et al. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. MBio 3: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhettinger P, Leo JC, Linke D, Autenrieth IB, Schutz MS. 2015. The inverse autotransporter intimin exports its passenger domain via a hairpin intermediate. J Biol Chem. 290:1837–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhettinger P, et al. 2012. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS One 7:e47069.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald E, et al. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun. 68:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, et al. 2004. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Prehna G, et al. 2012. The zinc regulated antivirulence pathway of Salmonella is a multiprotein immunoglobulin adhesion system. J Biol Chem. 287:32324–32337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. 2001. In: Molecular cloning: a laboratory manual. New York: Cold Spring Habor Laboratory Press. p. A2.2. [Google Scholar]
- Selkrig J, Leyton DL, Webb CT, Lithgow T. 2014. Assembly of beta-barrel proteins into bacterial outer membranes. Biochim Biophys Acta. 1843:1542–1550. [DOI] [PubMed] [Google Scholar]
- Selkrig J, et al. 2012. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol. 19:506–510. S501. [DOI] [PubMed] [Google Scholar]
- Selkrig J, et al. 2015. Conserved features in TamA enable interaction with TamB to drive the activity of the translocation and assembly module. Sci Rep. 5:12905.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Stubenrauch C, et al. 2016. Effective assembly of fimbriae in Escherichia coli depends on the TAM nanomachine. Nature Microbiol. DOI: 10.1038/NMICROBIOL.2016.64. [DOI] [PubMed] [Google Scholar]
- Tarr CL, Whittam TS. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J Bacteriol. 184:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JC, Jr., et al. 2010. The bacterial intimins and invasins: a large and novel family of secreted proteins. PLoS One 5:e14403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. 2015. UniProt: a hub for protein information. Nucleic Acids Res; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ulsen P, Rahman S, Jong WS, Daleke-Schermerhorn MH, Luirink J. 2014. Type V secretion: from biogenesis to biotechnology. Biochim Biophys Acta. 1843:1592–1611. [DOI] [PubMed] [Google Scholar]
- Yamamoto D, et al. 2009. Invasiveness as a putative additional virulence mechanism of some atypical Enteropathogenic Escherichia coli strains with different uncommon intimin types. BMC Microbiol. 9:146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WL, et al. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol. 40:4486–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.