Abstract
Methanogenesis coupled to the Wood–Ljungdahl pathway is one of the most ancient metabolisms for energy generation and carbon fixation in the Archaea. Recent results are sensibly changing our view on the diversity of methane-cycling capabilities in this Domain of Life. The availability of genomic sequences from uncharted branches of the archaeal tree has highlighted the existence of novel methanogenic lineages phylogenetically distant to previously known ones, such as the Methanomassiliicoccales. At the same time, phylogenomic analyses have suggested a methanogenic ancestor for all Archaea, implying multiple independent losses of this metabolism during archaeal diversification. This prediction has been strengthened by the report of genes involved in methane cycling in members of the Bathyarchaeota (a lineage belonging to the TACK clade), representing the first indication of the presence of methanogenesis outside of the Euryarchaeota. In light of these new data, we discuss how the association between methanogenesis and the Wood–Ljungdahl pathway appears to be much more flexible than previously thought, and might provide information on the processes that led to loss of this metabolism in many archaeal lineages. The combination of environmental microbiology, experimental characterization and phylogenomics opens up exciting avenues of research to unravel the diversity and evolutionary history of fundamental metabolic pathways.
Keywords: Archaea, methanogenesis, Wood–Ljungdahl pathway, pathway loss, archaeal ancestor
The Wood–Ljungdahl (WL) pathway is one of the most important metabolisms for energy generation and carbon fixation (Berg 2011). Although its overall scheme is conserved in Archaea and Bacteria, only the carbonyl branch (CBWL) shares homology, whereas the archaeal and bacterial methyl branches (MBWL) involve different C1-carriers, cofactors, electron transporters, and enzymes (Fuchs 2011). Other than carbon fixation, the WL pathway can act in reverse to produce reducing power from the oxidation of organic compounds during organo-heterotrophic growth (Vorholt et al. 1995; Schauder et al. 1988; Hattori et al. 2005). When using the WL pathway for energy generation and carbon fixation, most bacteria produce acetate as an end product (acetogens) whereas most archaea produce methane (CO2-reducing methanogens). The WL pathway has, therefore, been traditionally linked to methanogenesis in the Archaea. Until recently, all known methanogens were known to fall into two classes (Class I and Class II), both belonging to the Euryarchaeota (fig. 1A). Irrespective of the type of methanogenesis performed (CO2-reducing, acetoclasic, and methylotrophic), the representatives of these two classes have been consistently found to share a common set of enzymes for methanogenesis:
The Methyl-Branch of the archaeal type WL pathway (MBWL).
The N5-Methyltetrahydromethanopterin: coenzyme M methyltransferase complex (MTR).
The methyl-coenzyme M reductase complex (MCR).
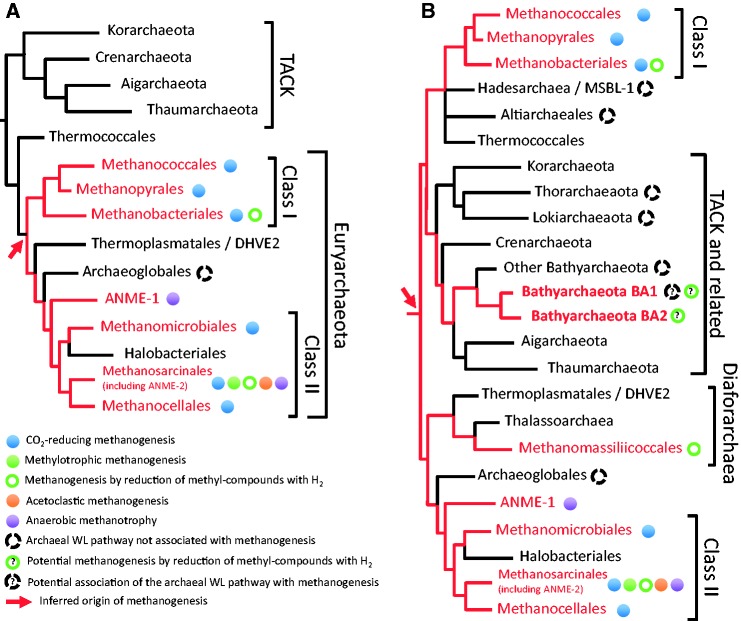
Fig. 1.—
Schematic views of the archaeal phylogeny including complete genomes available before 2012 (A) and currently (B), based on the literature (see text for details). Fast evolving DPANN lineages are not included as their position is unclear. Red arrows indicate the inferred origin of methanogenesis, with further divergence leading to lineages that retained this metabolism based on experimental characterization or the presence of MCR homologues (in red). Colored circles indicate the type of known and predicted pathways in representatives of the lineages according to the descriptive panel.
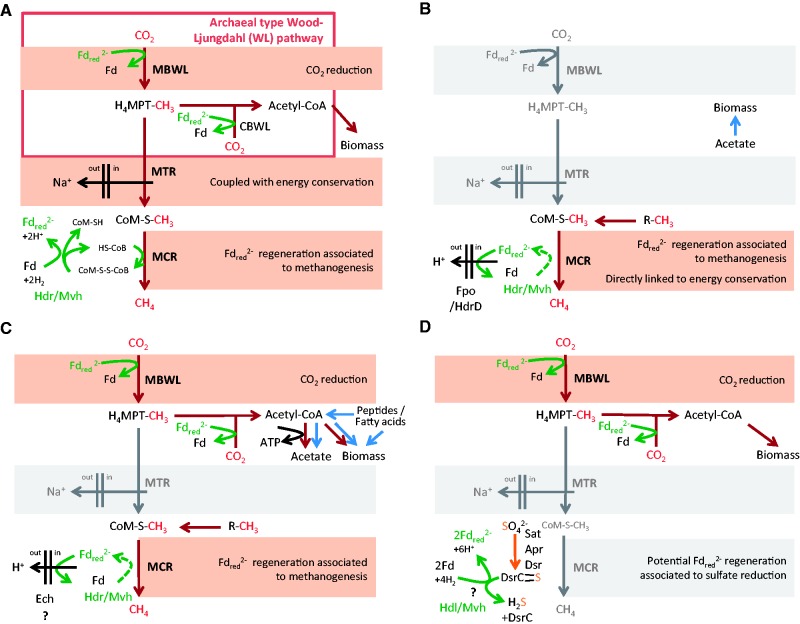
In the case of growth on CO2 and H2, performed by most Class I and II methanogens (CO2-reducing methanogenesis) (fig. 2A), CO2 is sequentially reduced through the MBWL pathway to a methyl-group in a process that requires a reduced low potential ferredoxin (Fdred2−) for CO2 activation. The MTR complex then couples the energetically favorable transfer of a methyl-group from terahydromethanopterin (H4MPT) to coenzyme M (CoM-SH) with the translocation of sodium outside the cytoplasmic membrane, generating an ion motive force exploitable by an ATP synthase for energy conservation. The MCR complex then catalyzes the formation of CH4 and CoM–S–S–CoB (also called heterodisulfide) from CoM–S–CH3 and HS–CoB. In methanogens without cytochromes, a recently described mechanism, called the flavin-based electron bifurcation (Kaster et al. 2011), takes place within the cytoplasmic heterodisulfide reductase (HdrABC)/F420-non reducing hydrogenase (MvhADG) complex (referred to as Hdr/Mvh hereafter) to couple the exergonic reduction of heterodisulfide by H2 to the endergonic reduction of a low potential ferredoxin by H2 (fig. 2A). This Fdred2− is reoxidized at the first step of the MBWL pathway for CO2 reduction. A membrane bound hydrogenase exploits the chemiosmotic gradient generated during methanogensis to produce additional Fdred2− needed for CO2 fixation by the WL pathway (Kaster et al. 2011). It should be noted that a certain variability exists around this scheme. For example, within the MBWL pathway the same step can be carried out by different enzymes (e.g., Mtd/Hmd) (Afting et al. 2000). Moreover, the MBWL pathway and the MTR complex are used in reverse during methylotrophic methanogenesis in Methanosarcinales (Keltjens and Vogels 1993) and are not involved for methanogenesis by reduction of methyl-compounds with H2 (Fricke et al. 2006; Welander and Metcalf 2008).
Fig. 2.—
Different configurations for the associated or independent functioning of the archaeal version of the Wood-Ljungdahl (WL) pathway and methanogenesis. Missing enzymatic complexes and related reactions are shaded in gray. (A) CO2-reducing methanogenesis as present in Class I and Class II methanogens without cytochromes. (B) Methanogenesis by reduction of methyl-compounds using H2 as present in Methanomassiliicoccales. (C) Methanogenesis by reduction of methyl-compounds using H2 as inferred in Bathyarchaeota BA1, and potential link with the WL pathway in absence of MTR. (D) Carbon fixation using the archaeal WL pathway in absence of methanogenesis, and proposal of a mechanism to generate low potential ferredoxin (Fdred2−) during sulphate reduction in the case of Archaeoglobales. Carbon fluxes originating from CO2 or methyl-compounds are shown by red arrows, and carbon fluxes from other sources by blue arrows. Green arrows indicate electron transfers associated with ferredoxins reduction or oxidation. The dotted green arrows in (B) and (C) integrate the electron bifurcation process leading to the generation of an Fdred2− by the Hdr/Mvh complex as described in (A). The reduction of heterodisulfide by Fdred2− in (B) and the reduction of 2H+ by Fdred2− in (C) that are coupled to proton translocation across the membrane are not shown. Abbreviations are as follows: MBWL, methyl-branch of the WL pathway; CBWL, carbonyl-branch of the WL pathway; MTR, N5-methyltetrahydromethanopterin: coenzyme M methyltransferase complex; MCR, methyl-coenzyme M reductase complex; Hdr, cytoplasmic heterodisulfide reductase complex; Mvh, F420-non-reducing hydrogenase complex; Hdl, cytoplasmic heterodisulfide reductase-like complex; Ech, Energy converting hydrogenase complex; Fpo, truncated F420H2 hydrogenase; H4MPT, tetrahydromethanopterin; CoM–S–H, coenzyme M; CoB–S–H, coenzyme B; CoM–S–S–CoB, heterodisulfide; Fd/Fdred2−, oxidized/reduced low potential ferredoxin; R-CH3, methylated compound such as methanol or methylamines; DsrC>S, DsrC trisulfide.
Given the complexity and importance of the methanogenesis pathway, its origin and evolution are key questions. Early phylogenetic analyses showed no evidence for horizontal gene transfer of CO2-reducing methanogenesis among methanogens, suggesting a single origin in Euryarchaeota, after the divergence of Thermococcales (Bapteste et al. 2005) (fig. 1A). A corollary to such unique origin of methanogenesis was that most present-day non-methanogen lineages within the Euryarchaeota would have lost the capacity to gain energy from this metabolism (Brochier et al. 2004). Methanogenic and non-methanogenic archaea form clearly distinct lineages (at least at the level of orders), suggesting that such losses would have been ancient and relatively rare events (fig. 1A). This is notably reflected by the much more important deepness of taxonomic conservation of methanogenesis when compared with other metabolisms (Martiny et al. 2013). In fact, there has been so far no evidence of loss of methanogenesis at small taxonomic scale in Class I and II methanogens. This may be due to the fact that methanogens are not capable to obtain energy from another metabolism, probably a necessary intermediate step toward methanogenesis loss. These aspects distinguish methanogenesis from most other energetic metabolisms that are generally less exclusive within a given clade and more easily transferable. Shifting away from methane-metabolism could take very different directions. For example, Halobacteriales have lost both MTR and MCR complexes as well as the MBWL pathway, and have adapted to radically different conditions (e.g., aerobic environments), helped by an important contribution of lateral gene transfer from Bacteria throughout their evolution (Nelson-Sathi et al. 2012; Becker et al. 2014). In contrast, Archaeoglobales have retained some or all enzymes of the archaeal WL pathway, remnant of their ancestral methane-cycling lifestyle (Vorholt et al. 1995; Bapteste et al. 2005).
With the exception of Archaeoglobales, the archaeal WL pathway has long been thought to be associated to methanogenesis only, and thus specifically linked to the MTR and MCR complexes. This view has been widely altered over the last 3 years, as genomic data from previously poorly characterized archaeal lineages have become available (fig. 1B). A first unconventional case showed up with the discovery of a seventh order of methanogens, the Methanomassiliicoccales, phylogenetically unrelated to Class I or Class II methanogens, but rather belonging to the recently proposed superclass Diaforarchaea (Borrel et al. 2013; Petitjean et al. 2015) (fig. 1B). Methanomassiliicoccales are distinguished from all previously known methanogens by the complete lack of the archaeal MBWL pathway and the MTR complex (fig. 2B) (Borrel et al. 2013, 2014; Lang et al. 2015; Söllinger et al. 2016). For methanogenesis, they use methyltransferases and corrinoid proteins allowing the transfer of methyl-groups from methanol, methylated-amines, and dimethyl sulfide to HS-CoM (fig. 2B). As the archaeal MBWL pathway and MTR complex are missing, methanogenesis in Methanomassiliicoccales is restricted to the reduction of methyl-compounds with H2, validated by physiological characterization (Dridi et al. 2012; Brugère et al. 2014; Lang et al. 2015). As in Class I and II methanogens without cytochromes, here Fdred2− are also likely generated by the Hdr/Mvh complex through flavin-based electron bifurcation (Borrel et al. 2014). Energy conservation potentially involves an additional coupling between ferredoxin and heterodisulfide, where the Fdred2− generated by Hdr/Mvh reduces a second heterodisulfide (Borrel et al. 2014; Lang et al. 2015; Kröninger et al. 2016). This reaction is likely operated by the association of a truncated F420H2 hydrogenase (Fpo) and a second heterodisulfide reductase (HdrD), and is coupled to the generation of a chemiosmotic gradient (fig. 2B) exploitable by an ATP synthase (Lang et al. 2015; Kröninger et al. 2016). This process represents, a novel way for coupling methanogenesis to energy conservation.
In parallel with the growing availability of genomic data from an ever-wider spectrum of archaeal diversity, large-scale phylogenomic analyses are sensibly changing our view on the early evolution of the third Domain of Life. For example, a recent analysis has proposed a novel root for the archaeal tree lying within the Euryarchaeota (Raymann et al. 2015; fig. 1B). According to this novel topology, the first divergence in archaeal diversification would have separated two clusters, one containing Methanococcales, Thermococcales, and the TACK clade, and the other containing all other Euryarchaeota (fig. 2B). Because both clusters contain methanogenic lineages, this result suggests that the last common ancestor of Archaea was a methanogen, pushing further back in time the origin of this important metabolism, in agreement with its antiquity (Liu et al. 2012). This also implies even more losses of methanogenesis than currently assumed. Moreover, the hypothesis of a methanogenic ancestor for a cluster that also includes the TACK opened up the possibility for the existence of additional lineages capable of methane metabolism in this clade and related lineages.
This prediction has been met by the recent report of the presence of methane metabolism in the Bathyarchaeota (formerly known as Miscellaneous Crenarchaeota Group) (Evans et al. 2015; fig. 1B). This diversified phylum is associated with the TACK and is composed of versatile archaea that can gain energy at least from fermentation, as inferred by the analysis of the first available genomes from uncultured members (Lloyd et al. 2013; Evans et al. 2015; Sara Lazar et al. 2015; He et al. 2016). Stunningly, among those genomes, two (BA1 and BA2) contain genes encoding the MCR complex, and, therefore, likely represent the first methane-cycling archaea not affiliated with Euryarchaeota (Evans et al. 2015). Moreover, in unrooted phylogenies of MCR subunits, Bathyarchaeota sequences are very divergent with respect to Euryarchaeota sequences, suggesting vertical inheritance of these genes in BA1 and BA2, strengthening an ancient origin of methanogenesis (Evans et al. 2015). This suggests even more independent losses of methanogenesis during archaeal evolution (fig. 1B).
By which mechanisms methanogenesis could be lost is unknown, but it should involve a transitory state where energy is also gained by an alternative energetic metabolism. In this regard, BA1 and BA2 are exceptionally valuable as they represent the only archaea that likely handle both methane metabolism and an alternative energetic metabolism (i.e., fermentation). If fermentation processes are fully independent from methanogenic ones, BA1 and BA2 could be much closer to lose methanogenesis than any other known methanogen. Interestingly, recently sequenced SG8-32-3 and AD8-1 genomes share 87–89% sequence identity with BA1 and BA2 based on 16S rRNA, but lack MCR and MTR gene homologues, implying that they are not capable of obtaining energy through methanogenesis (Sara Lazar et al. 2015). A variable presence of methane-metabolism at such shallow phylogenetic distance has never been observed previously.
Analysis of the BA1 and BA2 draft genomes (91.6% and 93.8% completeness, respectively) allowed to infer their potential methane-related metabolic capabilities (Evans et al. 2015). Because of the lack of the MTR complex in both genomes, along with the presence of sequences with homology to corrinoid proteins, MtaA/MtbA methyltransferases and novel methyltransferases, it was proposed that methanogenesis in these Bathyarchaeota could occur via reduction of methyl compounds by H2, similar to Methanomassiliicoccales (Evans et al. 2015). Interestingly, the analysis of BA1 revealed yet another variation on the theme of methanogenesis and the archaeal WL pathway. In fact, BA1 harbors both the WL pathway and the MCR complex, without the MTR complex that links the two in Class I and Class II methanogens (fig. 2C; Evans et al. 2015). For energy conservation, an Hdr/Mvh complex could produce Fdred2− through flavin-based electron bifurcation, as described in other methanogens, and an energy-converting hydrogenase (Ech) could reduce protons with those Fdred2− to generate a chemiosmotic gradient. However, no evidence for the presence of an ATP synthase that could exploit this gradient was found in BA1 and BA2 genomes or in their source metagenomes (Evans et al. 2015). Therefore, an alternative hypothesis may be proposed, where the Fdred2− generated by methanogenesis from H2-dependent reduction of methyl compounds might be used for CO2 reduction in the archaeal WL pathway (fig. 2C). The produced acetyl-CoA could be both integrated into biomass and converted into acetate for ATP generation (fig. 2C). In other words, the reducing power produced by methanogenesis from H2-dependent reduction of methyl compounds would be used in reductive acetogenesis for carbon fixation and energy conservation.
Might this hypothetical alternative coupling of methanogenesis and the archaeal WL pathway be more fragile than the one that has a connection through the MTR complex? And would this make it easier to replace methanogenesis by another source of reducing power for the archaeal WL pathway? Alternative energetic coupling for carbon fixation with the archaeal WL pathway has long been restricted to Archaeoglobales (fig. 2D). This case might be no longer unique as the presence of the archaeal WL pathway in the absence of MCR and MTR complexes has now been reported from an increasing number of archaeal lineages (fig. 1B), within the Bathyarchaeota (Sara Lazar et al. 2015; He et al. 2016), the Altiarchaeales (Probst et al. 2014), the Hadesarchaea/MSBL-1 (Baker et al. 2016; Mwirichia et al. 2016), the Lokiarchaeota (Sousa et al. 2016), and the Thorarchaeota (Seitz et al. 2016). As suggested for Archaeoglobales, the presence of the archaeal WL pathway in these lineages might be the remnant of a previous association with methanogenesis. The studies describing these novel archaea have discussed the direction of the WL pathway (CO2-reduction to acetyl-CoA or oxidation of acetyl-CoA to CO2). However, mechanisms to generate low potential reduced ferredoxin for CO2-reduction by the archaeal WL pathway in the absence of methanogenesis remain to be elucidated. It was shown that during sulfate reduction, Archaeoglobus fulgidus generates two disulfide bonds on the DsrC protein (forming a DsrC trisulfide) that are reduced by a membrane bound heterodisulfide reductase-like enzyme for energy conservation (Santos et al. 2015). Interestingly, Archaeoglobales representatives also possess a cytoplasmic complex very similar to the Hdr/Mvh complex present in methanogens (Mander et al. 2004). The hypothesis could thus be made that the required for carbon fixation might be generated through an electron bifurcation mechanism akin to the process taking place in methanogens, with DsrC trisulfide replacing CoM–S–S–CoB (fig. 2D). In the other lineages of non-methanogens bearing the archaeal WL pathway (fig. 1B), homologues of HdrABC and sometimes MvhADG/FrhAB/Hyd hydrogenases have been identified and might be involved in the processes of generation, with potentially new types of energetic coupling that remain to be fully explored. Alternatively, the archaeal WL pathway might also be used in reverse in these novel archaeal lineages to produce reducing power from the oxidation of organic compounds, as observed in Archaeoglobales growing organo-heterotrophically (Klenk et al. 1997).
The classical association of methanogenesis with the archaeal WL pathway appears to be less and less the rule (e.g., MCR without WL/MTR and WL without MTR/MCR), and potentially more flexible than previously thought. First, the larger phylogenetic distribution of methanogens without WL/MTR underlines the increasing importance of methanogenesis based on the reduction of methyl compounds by H2, whose environmental relevance might have been underestimated, as well as its potential antiquity. This is further strengthened by the recent report of this metabolic conformation in a proposed novel class of methanogens named “Candidatus Methanofastidiosa” (formerly known as WSA2) (Nobu et al. 2016). Second, among the growing number of archaeal lineages that harbor the WL pathway in the absence of MTR and MCR complexes, it might be wondered whether − generation processes alternative to the ones driven by methanogenesis represent recent adaptations, or if some of them might have been already present early in evolution.
Future genomic and metabolic exploration of still uncharacterized archaeal lineages, notably those with potential for methane metabolism (Lever & Teske 2015; Lloyd 2015; Lever 2016), promises exciting information on the diversity and evolution of methanogenesis, its connection with the WL pathway, and its loss through different mechanisms and transitory states linked to alternative ways for energy conservation.
Acknowledgments
G.B. is a recipient of a Roux-Cantarini fellowship from the Institut Pasteur. P.S.A. is supported by a PhD fellowship from Paris Diderot University and funds by the PhD Program “Frontières du Vivant (FdV) – Programme Bettencourt.
Literature Cited
- Afting C, Kremmer E, Brucker C, Hochheimer A, Thauer RK. 2000. Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch Microbiol. 174:225–232. doi: 10.1007/s002030000197. [DOI] [PubMed] [Google Scholar]
- Bapteste E, Brochier C, Boucher Y. 2005. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea 1:353–363. doi: 10.1155/2005/859728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EA, et al. 2014. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet. 10:e1004784. doi:10.1371/journal.pgen.1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA. 2011. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, et al. 2013. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol. 5:1769–1780. doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, et al. 2014. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Forterre P, Gribaldo S. 2004. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 5:R17. doi: 10.1186/gb-2004-5-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugère JF, et al. 2014. Archaebiotics: proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes 5:5–10. doi: 10.4161/gmic.26749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. 2012. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol. 62:1902–1907. doi: 10.1099/ijs.0.033712-0. [DOI] [PubMed] [Google Scholar]
- Evans PN, et al. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- Fricke WF, et al. 2006. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G. 2011. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol. 65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- Hattori S, Galushko AS, Kamagata Y, Schink B. 2005. Operation of the CO dehydrogenase/acetyl coenzyme A pathway in both acetate oxidation and acetate formation by the syntrophically acetate-oxidizing bacterium Thermacetogenium phaeum. J Bacteriol. 187:3471–3476. doi: 10.1128/JB.187.10.3471-3476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, et al. 2016. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol. doi: 10.1038/nmicrobiol.2016.35. [DOI] [PubMed] [Google Scholar]
- Kaster A-K, Moll J, Parey K, Thauer RK. 2011. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc Natl Acad Sci U S A. 108:2981–2986. doi: 10.1073/pnas.1016761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keltjens JT, Vogels GD. 1993. Conversion of methanol and methylamines to methane and carbon dioxide In: Ferry JG, editor. Methanogenesis: ecology, physiology, biochemistry & genetics. New York: Springer Science & Business Media; p. 253–303. [Google Scholar]
- Klenk H-P, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370. http://dx.doi.org/10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Kröninger L, Berger S, Welte C, Deppenmeier U. 2016. Evidence for the involvement of two different heterodisulfide reductases in the energy conserving system of Methanomassiliicoccus luminyensis. Febs J. 283:472–483. doi: 10.1111/febs.13594. [DOI] [PubMed] [Google Scholar]
- Lang K, et al. 2015. New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of ‘Candidatus Methanoplasma termitum’. Appl Environ Microbiol. 81:1338–1352. doi: 10.1128/AEM.03389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever MA. 2016. A new era of methanogenesis. Trends Microbiol. 24:84–86. doi: 10.1016/j.tim.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Lever MA, Teske AP. 2015. Diversity of methane-cycling archaea in hydrothermal sediment investigated by general and group-specific PCR primers. Appl Environ Microbiol. 81:1426–1441. doi: 10.1128/AEM.03588-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beer LL, Whitman WB. 2012. Methanogens: a window into ancient sulfur metabolism. Trends Microbiol. 20:251–258. doi: 10.1016/j.tim.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Lloyd K. 2015. Beyond known methanogens. Science 350:384. doi: 10.1126/science.aad4066. [DOI] [PubMed] [Google Scholar]
- Lloyd KG, et al. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. doi: 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- Mander GJ, Pierik AJ, Huber H, Hedderich R. 2004. Two distinct heterodisulfide reductase-like enzymes in the sulfate-reducing archaeon Archaeoglobus profundus. Eur J Biochem. 271:1106–1116. doi: 10.1111/j.1432-1033.2004.04013.x. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Treseder K, Pusch G. 2013. Phylogenetic conservatism of functional traits in microorganisms. Isme J. 7:830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, et al. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 109:6–11. doi: 10.1073/pnas.1209119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobu MK, Narihiro T, Kuroda K, Mei R, Liu W-T. 2016. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. Isme J. doi: 10.1038/ismej.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, López-García P, Moreira D, Brochier-Armanet C. 2015. Extending the conserved phylogenetic core of archaea disentangles the evolution of the third domain of life. Mol Biol Evol. 32:1242–1254. doi: 10.1093/molbev/msv015. [DOI] [PubMed] [Google Scholar]
- Probst AJ, et al. 2014. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat Commun. 5:5497. doi: 10.1038/ncomms6497. [DOI] [PubMed] [Google Scholar]
- Raymann K, Brochier-Armanet C, Gribaldo S. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci. 112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AA, et al. 2015. A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350:1541–1545. doi: 10.1126/science.aad3558. [DOI] [PubMed] [Google Scholar]
- Sara Lazar C, et al. 2015. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol. 18:1200–1211. doi: 10.1111/1462-2920.13142. [DOI] [PubMed] [Google Scholar]
- Schauder R, Preuß A, Jetten M, Fuchs G. 1988. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum – 2. Demonstration of the enzymes of the pathway and comparison of CO dehydrogenase. Arch Microbiol. 151:84–89. doi: 10.1007/BF00444674. [Google Scholar]
- Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. 2016. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. Isme J. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllinger A, et al. 2016. Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat. FEMS Microbiol Ecol. 92:fiv149. doi: 10.1093/femsec/fiv149. [DOI] [PubMed] [Google Scholar]
- Sousa FL, et al. 2016. Lokiarchaeon is hydrogen dependent. Nat. Microbiol. 14–16. doi: 10.1038/nmicrobiol.2016.34. [DOI] [PubMed] [Google Scholar]
- Vorholt J, Kunow J, Stetter KO, Thauer RK. 1995. Enzymes and coenzymes of the carbon monoxide dehydrogenase pathway for autotrophic CO2 fixation in Archaeoglobus lithotrophicus and the lack of carbon monoxide dehydrogenase in the heterotrophic A. profundus. Arch Micro biol. 163:112–118. doi: 10.1007/BF00381784. [Google Scholar]
- Welander PV, Metcalf WW. 2008. Mutagenesis of the C1 oxidation pathway in Methanosarcina barkeri: new insights into the Mtr/Mer bypass pathway. J Bacteriol. 190:1928–1936. doi: 10.1128/JB.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]