Abstract
Plastids and mitochondria have many protein complexes that include subunits encoded by organelle and nuclear genomes. In animal cells, compensatory evolution between mitochondrial and nuclear-encoded subunits was identified and the high mitochondrial mutation rates were hypothesized to drive compensatory evolution in nuclear genomes. In plant cells, compensatory evolution between plastid and nucleus has rarely been investigated in a phylogenetic framework. To investigate plastid–nuclear coevolution, we focused on plastid ribosomal protein genes that are encoded by plastid and nuclear genomes from 27 Geraniales species. Substitution rates were compared for five sets of genes representing plastid- and nuclear-encoded ribosomal subunit proteins targeted to the cytosol or the plastid as well as nonribosomal protein controls. We found that nonsynonymous substitution rates (dN) and the ratios of nonsynonymous to synonymous substitution rates (ω) were accelerated in both plastid- (CpRP) and nuclear-encoded subunits (NuCpRP) of the plastid ribosome relative to control sequences. Our analyses revealed strong signals of cytonuclear coevolution between plastid- and nuclear-encoded subunits, in which nonsynonymous substitutions in CpRP and NuCpRP tend to occur along the same branches in the Geraniaceae phylogeny. This coevolution pattern cannot be explained by physical interaction between amino acid residues. The forces driving accelerated coevolution varied with cellular compartment of the sequence. Increased ω in CpRP was mainly due to intensified positive selection whereas increased ω in NuCpRP was caused by relaxed purifying selection. In addition, the many indels identified in plastid rRNA genes in Geraniaceae may have contributed to changes in plastid subunits.
Keywords: coevolution, plastid, Geraniaceae, ribosome, nonsynonymous substitution
Introduction
Plastids and mitochondria, the cytoplasmic organelles of eukaryotic cells that contain their own genomes, are descendants of free-living prokaryotes (Gray et al. 2001; McFadden 2001). Since the establishment of endosymbiosis, the bulk of plastid genetic material has been transferred to the nuclear genome (Timmis et al. 2004) and nuclear-encoded proteins regulate the expression of genes retained in the organelle genome (Peled-Zehavi and Danon 2007). As a result proper functioning of the plastid requires the import of proteins encoded by nuclear genes and is dependent on coordination between organelle- and nuclear-encoded proteins that assemble plastid-localized multisubunit complexes.
Multisubunit protein complexes comprising proteins encoded by both the organelle and nuclear genomes are critical for cellular function and provide an ideal system to examine intergenomic coevolution (Rand et al. 2004; Sloan 2015; Zhang et al. 2015). In order to maintain proper function, purifying selection is likely the driving force in the evolution of these subunits. However, if mutations arise and become fixed in subunits encoded by one genome, either by drift or positive selection, these alterations may trigger corresponding changes in subunits encoded by the other genome. These coevolutionary changes can be detected by comparisons of the nucleotide substitution rates of subunits encoded in the organelles and nucleus (Lovell and Robertson 2010).
In animal cells, there is evidence of compensatory evolution between mitochondrial and nuclear-encoded genes (Osada and Akashi 2012; Barreto and Burton 2013). In plant cells, studies have found coordinated acceleration in plastid- and nuclear-encoded subunits that are part of the same protein complex (Sloan et al. 2014; Zhang et al. 2015). However, except for the Zhang et al. (2015) study, the evolutionary forces driving this coordinated acceleration in plants have not been investigated within a phylogenetic framework.
The plastid ribosome (70S) is composed of a small subunit (30S) and a large subunit (50S), both of which contain plastid- and nuclear-encoded subunits. In the model plant tobacco (Nicotiana tabacum), the small subunit has 21 proteins that have homologs in Escherichia coli, of which 12 are plastid-encoded and nine are nuclear-encoded (Fleischmann et al. 2011). Similarly, the large subunit has 31 proteins that have homologs in E. coli, of which nine are plastid-encoded and 22 nuclear-encoded (Fleischmann et al. 2011). In addition, there are six nuclear-encoded, plastid-specific ribosomal proteins that have no homology to any bacterial ribosomal protein (Yamaguchi and Subramanian 2000; Yamaguchi et al. 2000).
The pseudogenization or loss of plastid-encoded ribosomal protein genes from plastid genomes has been documented multiple times across angiosperms (Jansen et al. 2007; Ruhlman and Jansen 2014). Endosymbiotic gene transfer is an ongoing evolutionary process (Matsuo et al. 2005; Noutsos et al. 2005; Stegemann and Bock 2006; Kleine et al. 2009) and while rare, the establishment of nuclear expression for a transferred gene could render the plastid-encoded copy redundant. Functional plastid-to-nucleus gene transfers have been associated with pseudogenization and/or loss of plastid-encoded ribosomal protein genes, such as rpl22 in Fagaceae (Jansen et al. 2011) and Fabaceae (Gantt et al. 1991), and rpl32 in Populus (Cusack and Wolfe 2007; Ueda et al. 2007) and Ranunculaceae subfamily Thalictroideae (Park et al. 2015).
In other cases, such as rps16 and rpl23, genes for ribosomal proteins have been lost from the plastid genome with their function substituted by an alternative gene product. The product of plastid-encoded rps16 gene in Medicago and Populus was substituted by a mitochondrial RPS16 that is nuclear-encoded and dual-targeted to both organelles (Ueda et al. 2008). In spinach (Spinacia oleracea), the plastid-encoded rpl23 is a truncated pseudogene, and the eukaryotic cytosolic RPL23 has replaced the product of prokaryotic rpl23 in the plastid ribosome (Bubunenko et al. 1994).
Coordination between the nucleus and organelles is vital for eukaryotic cells, yet molecular evolutionary rates can vary dramatically between the genomes in these compartments (Wolfe et al. 1987; Drouin et al. 2008). The intergenomic dynamics that maintain organismal function despite the contrasting evolutionary trajectories of nuclear and organellar genomes have been previously studied through cytonuclear coadaptation (Budar and Fujii 2012; Greiner and Bock 2013), speciation (Levin 2003), hybrid incompatibility (Jonson 2010), endosymbiotic gene transfer (Brandvain and Wade 2009), and compensatory evolution (Osada and Akashi 2012; Barreto and Burton 2013). Plastid-nuclear compensatory evolution has rarely been investigated due to the low and homogeneous substitution rates in plastid genes. The highly accelerated substitution rates in different lineages and functional groups of genes in Geraniaceae plastid genomes (Guisinger et al. 2008; Weng et al. 2012), however, provide an excellent system for investigating compensatory coevolution in plant cells.
In this study, ribosomal gene sequences were assembled for 25 species of Geraniaceae and seven outgroups from other rosids. We analyzed each species for rates of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions, and dN/dS ratios (ω or omega) for five sets of genes: nuclear-encoded plastid-targeted ribosomal protein genes (NuCpRP), nuclear-encoded cytosol-targeted ribosomal genes (NuCyRP), other nuclear-encoded plastid-targeted genes that are not involved in ribosomes (NuCpOT), plastid-encoded ribosomal protein genes (CpRP) and plastid-encoded photosynthesis genes (CpPS). We found that both NuCpRP and CpRP had significantly higher dN than other nuclear- and plastid-encoded genes, and that both NuCpRP and CpRP had higher ω in Geraniaceae than in the outgroups. Furthermore, our analyses revealed that nonsynonymous substitutions in CpRP and NuCpRP tend to occur along the same branches in the Geraniaceae phylogeny suggesting cytonuclear coevolution between plastid- and nuclear-encoded subunits of the plastid ribosome.
Materials and Methods
Plastid-Encoded Gene Sequences
Seven outgroup and 25 Geraniaceae species were included in the study (fig. 1). Published plastid genome sequences were downloaded from NCBI (supplementary table S1, Supplementary Material online). Sequences for CpRP, CpPS, and ribosomal RNA genes in these species were identified from the available annotations. Previously unpublished Pelargonium and Geranium plastid genomes were de novo assembled following the methods in Weng et al. (2014). Sequences for CpRP and CpPS in assembled contigs were identified using DOGMA (Wyman et al. 2004) with default settings, and submitted to Genbank. The accession numbers for all species in this study are listed in supplementary table S1, Supplementary Material online.
Fig. 1.—
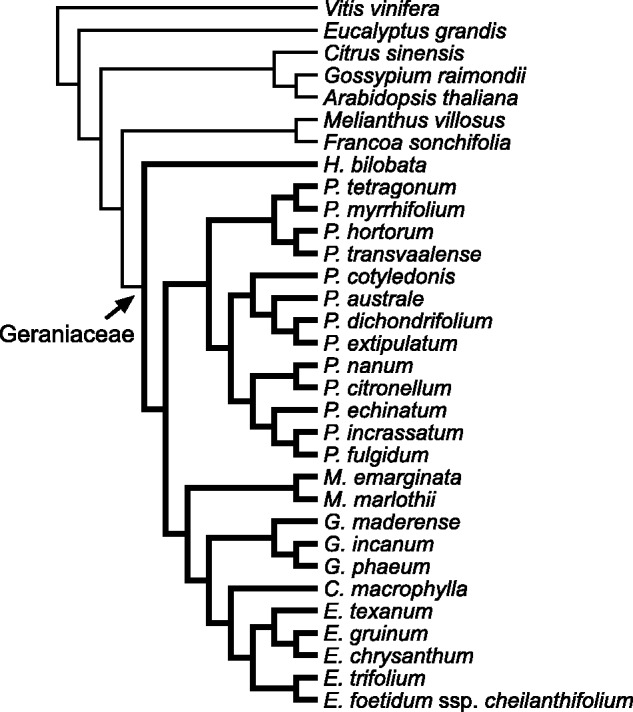
Cladogram of species included in the study. Topology was based on the ML tree generated from concatenated CpRP sequences listed in table 1. Geraniaceae (thick branches) are specified as foreground lineages that can have different ω in branch-site tests implemented in the software of Phylogenetic Analysis by ML, PAML (Yang 2007).
Nuclear-Encoded Gene Sequences
Nuclear genome sequences for Eucalyptus grandis, Vitis vinifera, Gossypium raimondii, Arabidopsis thaliana, and Citrus sinensis were obtained from Phytozome v9.1 (Goodstein et al. 2012). The sequencing and assembly of transcriptomes for Melianthus villosus, Francoa sonchifolia, and 25 Geraniaceae species were described previously (Zhang et al. 2013 and Ruhlman et al. 2015). Nuclear-encoded NuCpRP, NuCpOT, and NuCyRP in A. thaliana were identified from the UniProt database (www.uniprot.org, last accessed on May 18, 2016), Plant Proteomics Data Base (Sun et al. 2009) and the literature (Yamaguchi and Subramanian 2000; Yamaguchi et al. 2000). The corresponding coding sequences for A. thaliana were downloaded from The Arabidopsis Information Resource database (https://www.arabidopsis.org, last accessed on May 18, 2016). Sequences for NuCpRP, NuCpOT, and NuCyRP from 31 species, excluding A. thaliana, were obtained by reciprocal blast searches (tBLASTn and BLASTx default settings in BLAST version 2.2.29+) against downloaded genome or transcriptome assemblies (table 1 and supplementary table S1, Supplementary Material online) using translated A. thaliana genes as queries. Because plant genomes often contain multiple copies of NuCyRP, neighbor-joining trees (Saitou and Nei 1987) were constructed using Geneious Version 6.1.8 (http://www.geneious.com, last accessed on May 18, 2016; Kearse et al. 2012) and the copies congruent with the species tree were selected for rate analyses.
Table 1.
List of Genes included in the Study
| Nuclear-Encoded |
Plastid-Encoded |
||||
|---|---|---|---|---|---|
| NuCyRP (17) | NuCpRP (29) | NuCpOT (15) | CpRP (20) | CpPS (20) | rRNA (4) |
| RPP0C | PSRP-1* | CAO | rpl2 | psaA | rrn23 |
| RPL3A | PSRP-2* | CRTISO | rpl14* | psaB | rrn16 |
| RPL3B | RPL1* | OEP80 | rpl16* | psaC | rrn4.5 |
| RPL4A | RPL3* | PDH-E1-BETA | rpl20 | psaI | rrn5 |
| RPL7A | RPL4* | PSAK* | rpl23 | psaJ | |
| RPL8B | RPL5* | PSAL | rpl32 | psbA | |
| RPL10AA | RPL6* | PSBO2 | rpl33 | psbB | |
| RPL12A | RPL11* | PSBQ-2 | rpl36* | psbC | |
| RPL17A | RPL13* | PSBR | rps2* | psbD | |
| RPL23AA | RPL15 | PSY | rps3* | psbE | |
| RPL26B | RPL17* | SS1 | rps4* | psbF | |
| RPSaA | RPL18-1* | TOC33 | rps7 | psbH | |
| RPS5B | RPL18-2 | TOC64-III | rps8 | psbI | |
| RPS9C* | RPL19* | TSA1 | rps11* | psbJ | |
| RPS13B | RPL21* | TSB1 | rps12 | psbK | |
| RPS15AE | RPL24* | rps14* | psbL | ||
| RPS20C | RPL27* | rps15* | psbM | ||
| RPL28* | rps16 | psbN | |||
| RPL29* | rps18 | psbT | |||
| RPL31 | rps19* | psbZ | |||
| RPL34 | |||||
| RPL35 | |||||
| RPS1* | |||||
| RPS5* | |||||
| RPS6* | |||||
| RPS9* | |||||
| RPS10* | |||||
| RPS13* | |||||
| RPS20* | |||||
NOTE.—Gene names follow the UniProt convention. Numbers in parenthesis are the total number of genes in the gene group. Genes with residues that show positive selection are in bold. Asterisks indicate genes with significantly higher ω in Geraniaceae compared to outgroups.
To identify potential plastid-to-nucleus gene transfer, CpRP sequences from Geraniaceae were translated and reciprocal BLAST searches were performed against transcriptome sequences (tBLASTn and BLASTx default settings in BLAST version 2.2.29+).
The product of the plastid-encoded ribosomal gene, rpl23, has been substituted by a cytosolic homolog of eukaryotic origin in the spinach plastid ribosome (Bubunenko et al. 1994). To examine whether this gene substitution occurred in Geraniaceae, the amino acid sequence of Rpl23 (UniProt id: Q9LWB5) from the spinach plastid ribosome was downloaded from the UniProt database and used to perform BLAST searches against translated transcriptome sequences (tBLASTn default settings in BLAST version 2.2.29+).
Prediction of Transit Peptides
The cleavage site of 5′-transit peptides for nuclear-encoded plastid-targeted genes (NuCpRP and NuCpOT) was predicted in silico using the TargetP web server (http://www.cbs.dtu.dk/services/TargetP/, last accessed on May 18, 2016; Nielsen et al. 1997; Emanuelsson et al. 2000). Transit peptide sequences were excluded from rate and coevolution analyses.
Prediction of Conserved Domains
Conserved domains were predicted by uploading the sequences to the NCBI Conserved Domain Database for comparison (v3.14 with default options and E-value threshold of 0.01; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, last accessed on May 18, 2016; Marchler-Bauer et al. 2015).
Codon Usage Bias Estimation
To estimate the codon usage bias for NuCpRP and NuCyRP, the effective number of codons (Nc) (Wright 1990) was estimated using CodonW (http://codonw.sourceforge.net, last accessed on May 18, 2016; Peden 1999). A t-test was used to assess the difference in Nc between NuCpRP and NuCyRP.
Substitution Rate Estimation
The CODEML program implemented in PAML v4.7 (Yang 2007) was used to estimate dN and dS for each gene. Nucleotide sequences were aligned by Multiple Alignment using Fast Fourier Transform (MAFFT) (Katoh et al. 2002) and gaps were excluded for rate estimations (cleandata = 1 in CODEML control file). The codon frequencies were determined by the F3 x 4 model. Transition/transversion ratios and ω were estimated with initial values of 2 and 0.4, respectively. The constraint species tree was constructed by maximum likelihood (ML) on the RAxML web server (Stamatakis et al. 2008) using a data set of concatenated plastid ribosomal protein genes (table 1). The free-ratio model (m1) allowing branch-specific ω was specified to estimate dN and dS for each branch. The Kruskal–Wallis rank sum tests were used to test whether branch-specific ω values were significantly different from each other. Likelihood ratio tests (LRTs) between two branch-models were conducted in PAML v4.7 (Yang 2007) and tested whether genes in Geraniaceae have elevated ω. The null model (m0) specified one ω shared across the entire tree, whereas the alternative model (m2) allowed the Geraniaceae lineages to have different values of ω. The same LRT scheme for detecting significant difference of dN and dS between Geraniaceae and outgroups was conducted using a custom batch script with MG94xHKY85 codon model in HyPhy (Kosakovsky Pond and Muse 2005), in which the null model specified a shared dN or dS across the tree whereas the alternative model allowed the Geraniaceae lineages to have different dN or dS. The dN and dS values for the Geraniaceae and outgroup branches were summed and binned by gene type (NuCpRP, NuCpOT, NuCyRP, CpRP, and CpPS). Wilcoxon rank sum tests were used to test rate differences between gene types and P values were adjusted by Bonferroni correction.
Detection of Positive Selection
LRTs were used to evaluate whether sites were under positive selection in Geraniaceae lineages. Two branch-site models were specified in the CODEML program in PAML (Yang 2007). The alternative model specified four different types of codon sites: sites under purifying selection across the phylogeny (ω < 1), neutral sites across the phylogeny (ω = 1), sites under purifying selection in outgroups but with positive selection (ω > 1) in Geraniaceae, and sites that are neutral in outgroups but with positive selection in Geraniaceae. The settings for the alternative model in CODEML control file were model = 2, NSsites = 2, fix_omega =0. The null model fixed the ω equal to one in Geraniaceae lineages. The settings for the null model in CODEML control file were model =2, NSsites =2, fix_omega =1, omega =1. Posterior probabilities for sites under positive selection were computed by Bayes Empirical analysis (Yang et al. 2005) included in the CODEML analysis.
Detection of Relaxed Selection
The RELAX program (Wertheim et al. 2015) implemented in HyPhy (Kosakovsky Pond and Muse 2005) was used to test whether the increased ω in Geraniaceae was due to relaxed purifying selection. Outgroup and Geraniaceae lineages were used as the reference and test branches, respectively. The selection intensity parameter (k) was introduced in the RELAX program to describe the relationship of ω in the test and the reference branches. The null model constrained k equal to one, whereas the alternative model set k as a free parameter. A LRT was used to test whether k significantly deviated from one. Rejection of the null model and both ω and k smaller than one indicated that purifying selection was relaxed in Geraniaceae relative to outgroups.
Identification of Contact Residues in the Plastid Ribosome
Protein structures for small and large subunits (PDB ids: 3BBN and 3BBO, Sharma et al. 2007) of the spinach plastid ribosome were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB, http://www.rcsb.org/, last accessed on May 18, 2016). Amino acid residues between two proteins that are within 10Å were defined as contact residues. Bhattacherjee et al. (2015) showed that slightly deleterious mutations that destabilize structure within a protein and compensatory mutations that re-establish stable protein folding tend to co-occur within 10Å radius in 3D space. Nucleotide sequences of NuCpRP and CpRP from Geraniaceae and outgroups were translated and aligned with spinach ribosomal proteins using MAFFT to identify contact residues across the alignment.
Coevolution Analysis
For the analysis of coevolution, we tested 1) whether the nonsynonymous substitutions in CpRP and NuCpRP tend to occur along the same branches and 2) whether the nonsynonymous substitutions in CpRP and NuCpRP tend to occur sequentially on the tree more often than the null expectation. The posterior probability of ancestral codons for each node along the phylogenetic tree was estimated by the CODEML program in PAML (Yang 2007) using the F3 x 4 codon model. The most probable ancestral codons were recorded and branch-specific nonsynonymous substitutions for each codon were determined.
We analyzed every combination of two proteins from CpRP and NuCpRP. Each protein pair comprised a leading and a trailing protein. The terms “leading” and “trailing” protein did not imply physical interaction, but instead, simply describe the first and second protein in any given protein pair comparison. Every CpRP and NuCpPR can be either a leading or a trailing protein. For example, in Rpl2-Rpl14 comparison, Rpl2 is leading protein and Rpl14 is trailing protein, whereas in Rpl14-Rpl2 comparison, Rpl14 is leading and Rpl2 is trailing protein. To evaluate co-occurring substitutions, we systematically compared each pair of variable codons from a given protein pair. The number of codons that shared nonsynonymous substitutions at one, two, and tree branches were counted. This value was defined as the type I coevolution statistic. To evaluate sequential substitution, we tested whether the nonsynonymous substitutions in the leading protein on the phylogenetic tree were followed by nonsynonymous substitutions in the trailing protein more quickly than the null expectation (Kryazhimskiy et al. 2011). First, the number of consecutive nonsynonymous substitution pairs between the leading and the trailing proteins was calculated. Consecutive nonsynonymous substitutions were defined as a pair of nonsynonymous substitutions that occur consecutively in the same lineage (supplementary fig. S1, Supplementary Material online). Second, the number of branches between each consecutive nonsynonymous substitution pair was determined. Third, the number of branches between all consecutive nonsynonymous substitutions was summed and divided by the total number of consecutive nonsynonymous substitution pairs. This average was defined as the type II coevolution statistic between two proteins.
A null expectation of the coevolution statistics between two proteins was generated by simulation (Shapiro et al. 2006). One thousand codon sequences were simulated by the EVOLVER program in PAML (Yang 2007) using the estimated codon frequency, transition/transversion ratio, and branch specific ω from the original codon sequence of CpRP and NuCpRP. The null distribution of the coevolution statistics between a pair of proteins was the combination of the statistics between leading protein and simulated trailing protein and between the simulated leading protein and trailing protein. A one-tail significance of observed coevolution statistics was computed against the generated null normal distribution and P values were adjusted by false discovery rate using R (R Core Team 2015).
To address the effects of physical interaction on ribosomal protein coevolution, the positions from each sequence alignment that were identified as contact residues were extracted. The coevolution analyses described above were repeated on the alignments that included contact residues only.
Our method differs from the Osada and Akashi (2012) approach in two ways. First, we reported the most probable ancestral codon per node instead of every possible reconstructed codon. Due to the combinatorial explosion problem, including multiple reconstructed codons per node is not suitable for computing the coevolution statistics in a phylogeny of more than a dozen species. Second, in our analysis statistical significance was tested by sequence simulation rather than randomization, the former was able to explore the potential sequence space that a given gene might have evolved into.
Insertion and Deletion (Indel) Analysis for Plastid Ribosomal RNA Genes
Four plastid ribosomal RNA genes (rrn23, rrn16, rrn5, and rrn4.5) were extracted from all 32 species (supplementary table S1, Supplementary Material online) and aligned using MAFFT (Katoh et al. 2002). Indels were analyzed using the simple indel coding algorithm (Simmons and Ochoterena 2000) implemented in SeqState (Müller 2005).
Results
For all 32 taxa included in the study, 40 plastid-encoded genes (20 CpRP and 20 CpPS) were obtained from annotations of published plastid genomes or identified in DOGMA for de novo assembled plastomes (table 1 and supplementary table S1, Supplementary Material online). Using reciprocal blast searches between translated gene sequences from A. thaliana and published nuclear genomes or de novo assembled transcriptomes, 61 nuclear-encoded (29 NuCpRP, 17 NuCyRP, and 15 NuCpOT) genes were obtained for all other species (table 1 and supplementary tables S1 and S2, Supplementary Material online). The alignments for all individual genes are available in supplementary data file S1, Supplementary Material online.
Identification of rpl23 Gene Transfer or Substitution Events
The plastid-encoded rpl23 in Geranium maderense is truncated leaving only 99 bp at the 5′-end (compared with 324 bp in A. thaliana). The absence of the 3′-sequence encoding the functional domain of Rpl23 (Marchler-Bauer et al. 2015) suggests that this may be a pseudogene. To investigate a potential plastid-to-nucleus functional transfer, we performed a reciprocal blast search between the translated plastid-encoded Rpl23 and the translated nuclear transcriptome of G. maderense (Zhang et al. 2013) and evaluated the top hits for the presence of a plastid transit peptide using TargetP (Emanuelsson et al. 2000). None of the top hits from reciprocal BLAST searches were predicted to contain a plastid transit peptide.
To explore the possible substitution of Rpl23 by a nuclear homolog, we queried the translated G. maderense transcriptome assembly with the amino acid sequence of the spinach Rpl23, which has a eukaryotic-cytosolic origin (Bubunenko et al. 1994). Of the two transcripts identified, one was predicted to contain a plastid transit peptide (0.771) (fig. 2). The conserved 50S and 60S ribosomal protein L23 domains were identified in the C-terminus in both translated transcripts (fig. 2). The two transcripts shared 81% and 71% amino acid sequence identity in the 50S and 60S ribosomal protein L23 domains, respectively.
Fig. 2.—
Amino acid sequence alignment of two putative nuclear-encoded rpl23 transcripts in Geranium maderense. The 5′-plastid transit peptide and the cleavage site indicated by the black bar were predicted by TargetP (Nielsen et al. 1997; Emanuelsson et al. 2000). The conserved domains of the 50S and 60S ribosomal proteins L23 indicated by gray bars were predicted by NCBI Conserved Domain Database (Marchler-Bauer et al. 2015). The BLOSUM62 protein substitution matrix with a threshold of one was used to assess similarity between residues. The identical and 60–99% similarity residues were highlighted in black and gray, respectively.
Both Plastid-Encoded and Plastid-Targeted Ribosomal Protein Genes in Geraniaceae Show Elevated dN and ω
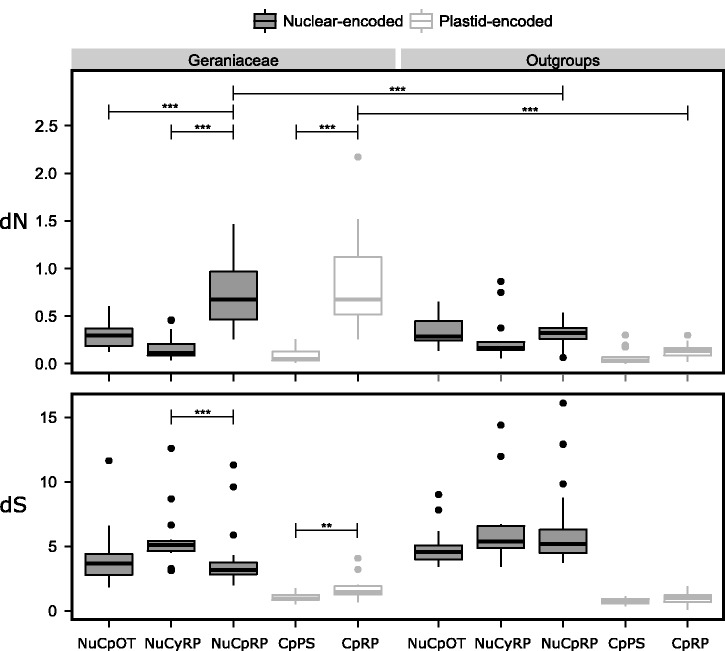
The value of dN and dS was estimated for each gene by the free-ratio model (m1). The sum of branch lengths of dN and dS trees for Geraniaceae and outgroups were binned by gene and are shown in boxplots (fig. 3). In Geraniaceae, dN for NuCpRP was significantly higher than all other nuclear-encoded genes (P < 0.001). Similarly among plastid-encoded genes, CpRP had significantly higher dN than CpPS (P < 0.001). In the outgroups, there were no significant differences in dN either between nuclear-encoded genes or between plastid-encoded genes. Comparisons between Geraniaceae and outgroups using Wilcoxon rank sum tests and LRTs both showed that Geraniaceae NuCpRP and CpRP had significantly higher dN (P < 0.001) (fig. 3 and supplementary table S3, Supplementary Material online).
Fig. 3.—
Comparison of dN and dS values across different gene groups in Geraniaceae and outgroups. Asterisks indicate P < 0.01 (**) and P < 0.001 (***) after Bonferroni correction. NuCpRP: nuclear-encoded plastid targeted ribosomal genes, NuCyRP: nuclear-encoded cytosol-targeted ribosomal genes, NuCpOT: nuclear-encoded plastid-targeted nonribosomal genes, CpRP: plastid-encoded ribosomal genes, CpPS: plastid-encoded photosynthetic genes.
In Geraniaceae comparison of dS among nuclear-encoded genes showed that NuCpRP had significantly lower dS than NuCyRP (P < 0.001) but there was no significant difference between NuCpRP and NuCpOT. Among plastid-encoded genes, CpRP showed significantly higher dS than CpPS (P < 0.01) (fig. 3). There was no significant difference in dS between nuclear-encoded genes and between plastid-encoded genes in outgroups.
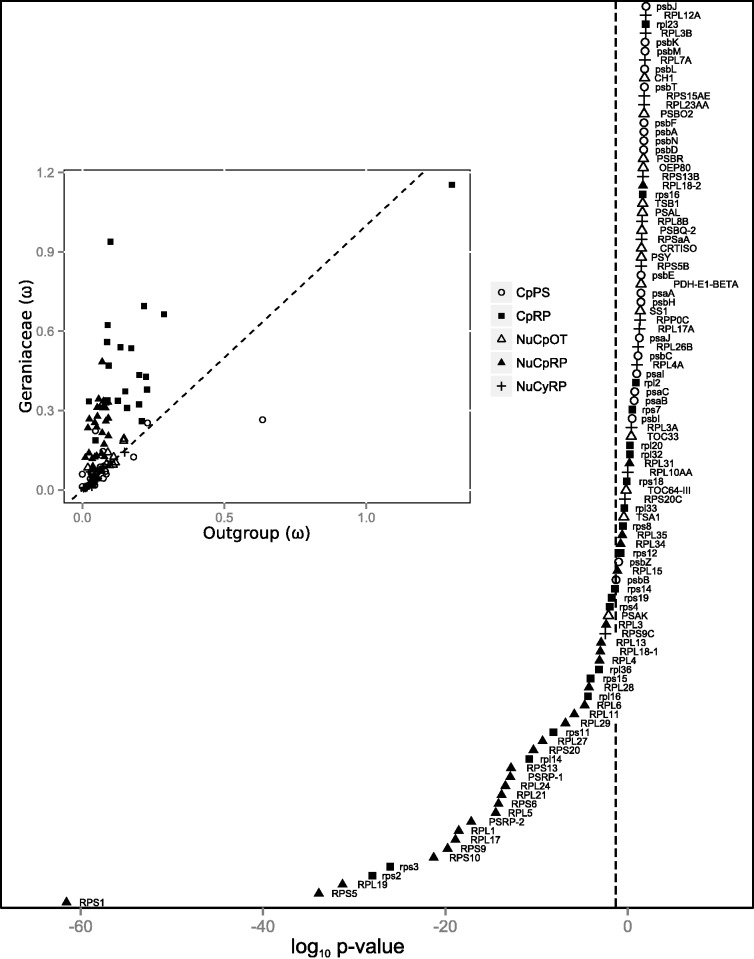
The estimate of ω under the m2 model, which allows Geraniaceae and outgroups to have different ω values, was plotted for each gene (fig. 4 inset). Most points on the graph for NuCpRP and CpRP were above the diagonal dashed line showing that ω values were higher in Geraniaceae than outgroups. To test whether the deviation was significant, LRTs were conducted. The m2 model was tested against the null model (m0) where a single ω was shared across the entire phylogeny. The logarithmic P values of the LRTs were ranked, Bonferroni corrected and plotted (fig. 4). At a cutoff of 5%, 36 genes had significantly higher ω in Geraniaceae than in outgroups (genes appearing to the left of the vertical dashed line in fig. 4), including 24 NuCpRP, 10 CpRP, one NuCyRP, and one NuCpOT (table 1). The estimate of ω for CpRP and NuCpRP under the free-ratio model (m1), which allows branch-specific ω, was plotted on the phylogeny (supplementary fig. S2, Supplementary Material online). The branch-specific ω values for CpRP were significantly different from each other (P < 0.001). The branch-specific ω values for NuCpRP were also significantly different from each other (P < 0.001).
Fig. 4.—
Plot of log P value for each gene from LRTs. Two models were compared to determine P value. The null model constrained a single value for ω across the entire phylogeny, and the alternative model relaxed the constraint by allowing Geraniaceae and outgroups to have different ω. The vertical dashed line shows the Bonferroni corrected 5% significance cutoff. Genes appearing to the left of the vertical dashed line have significantly higher ω in Geraniaceae compared to outgroups. The inset scatter plot shows the ω values in Geraniaceae versus outgroups. The dashed diagonal line is the 1:1 line showing ω is equal in Geraniaceae and outgroups. Gene group acronyms are the same as in figure 3.
Plastid-Encoded Ribosomal Protein Genes Show Positive Selection
The branch-site test, which allows detection of codon-specific positive selection (ω > 1) in prespecified lineages (Zhang et al. 2005), was applied to all genes included in the study using PAML (Yang 2007). The Geraniaceae and outgroup lineages were specified as “foreground” and “background,” respectively (fig. 1). Positive selection was not detected among nuclear-encoded genes (NuCyRP, NuCpRP, and NuCpOT). For plastid-encoded genes (CpPS and CpRP), the branch-site test did not detect positive selection in CpPS, but five out of 20 CpRP showed evidence of sites under positive selection in Geraniaceae (table 2). Among the five genes, rps2 had the largest number of sites showing positive selection with posterior probability greater than 0.9 (34 sites). Among all sites that showed positive selection, six (one, two, and three in rps4, rps2, and rps7, respectively) involved residues within 10Å of a residue in another ribosomal complex protein (asterisk labeled sites in table 2).
Table 2.
Candidate Sites under Positive Selection with ω > 1
| Gene | Encoded genome | -lnL | -lnL | ω | Selected Sitesa | |
|---|---|---|---|---|---|---|
| Model 2a | Model 2b | |||||
| rpl32 | Plastid | 1520.600 | 1532.556 | 3.272 | 18I (0.931) | 47V (1.00) |
| 25W (0.919) | 49K (1.00) | |||||
| 46F (0.997) | ||||||
| rps2 | Plastid | 4319.073 | 4339.983 | 2.193 | 5Y (0.973) | 80A (0.974) |
| 9D (0.946 | 112E (0.967) | |||||
| 13M (0.959) | 114R (0.951) | |||||
| 14 M (0.995) | 116H (0.983) | |||||
| 17G (0.974) | 117K (0.924) | |||||
| 20F (0.998) | 118F (0.965) | |||||
| 23G (1.00) | 123T (0.986) | |||||
| 37A (0.979) | 124E (1.00) | |||||
| 39G (1.00) | 127G (0.983) | |||||
| 41G (0.944) | 141S (0.99) | |||||
| 42I (1.00) | 148G (0.972) | |||||
| 45I (0.908) | 154T (0.985) | |||||
| 52R (0.986) | 166Q (0.951) | |||||
| 53F (0.993)* | 167E (1.00) | |||||
| 67R (0.919) | 168E (0.996) | |||||
| 70Q (1.00) | 169Y (0.994)* | |||||
| 79K (0.996) | 222E (0.978) | |||||
| rps4 | Plastid | 3789.815 | 3814.208 | 4.749 | 13R (1.00) | 30S (0.999) |
| 15R (1.00) | 149E (0.94) | |||||
| 28S (0.988) | 172C (0.951) | |||||
| 29R (0.999) * | ||||||
| rps7 | Plastid | 2368.431 | 2397.530 | 5.945 | 14S (1.00)* | 112P (0.995) |
| 68G (0.962) | 125V (0.999) | |||||
| 81G (1.00)* | 128A (0.999) | |||||
| 85H (0.914)* | 133D (0.902) | |||||
| 93S (0.955) | ||||||
| rps18 | Plastid | 3025.523 | 3048.144 | 4.296 | 5L (1.00) | 14P (0.992) |
| 6T (1.00) | 74Q (0.962) | |||||
| 8S (0.972) | 82S (0.949) | |||||
Posterior probabilities of ω > 1 are shown in the parentheses. Sequence positions and amino acid residues are based on the spinach plastid ribosome structure (PDB ids: 3BBN and 3BBO). Residues lying within 10Å of a residue in another ribosomal protein are indicated with an asterisk.
Nuclear-Encoded Ribosomal Protein Genes Show Relaxed Purifying Selection
The selection intensity parameter (k) estimated by RELAX for NuCpRP ranged from 0.39 to 17.94 with mean of 4.47, whereas for CpRP it ranges from 0.22 to 13.99 with mean of 1.52 (supplementary table S4, Supplementary Material online). Twenty-one NuCpRP (72%) had k significantly lower than one, whereas ten CpRP (50%) had k significantly higher than one (supplementary table S4, Supplementary Material online). The selection intensity parameter (k) was significantly lower in NuCpRP than CpRP (P < 0.01, supplementary fig. S3, Supplementary Material online).
Cytosol-Targeted Ribosomal Protein Genes Have Stronger Codon Usage Bias than Plastid-Targeted Genes
Overall the effective number of codons (Nc) was significantly higher in NuCpRP than NuCyRP indicating greater codon usage bias in NuCyRP. In Geraniaceae the mean Nc was 51.38 for NuCpRP and 46.98 for NuCyRP (P < 0.05). In outgroups, the mean Nc was 53.34 for NuCpRP and 48.97 for NuCyRP (P < 0.05).
Contact Residues in Plastid Ribosomal Proteins Did Not Show Higher Conservation
On average, 24.3% of the residues of CpRP and NuCpRP were within 10Å of a residue in another ribosomal complex protein, whereas 50.3% were within 10Å of a nucleotide in rRNAs (supplementary table S5, Supplementary Material online). Pair-wise amino acid sequence identities were computed for CpRP and NuCpRP (supplementary table S6, Supplementary Material online). The pair-wise identity for CpRP ranged from 68.6% to 86.1% when the entire sequence was considered, from 53.8% to 92.0% for protein contact residues only and from 72.8% to 87.7% for protein and rRNA contact residues. The pair-wise identity for NuCpRP ranged from 72.3% to 92.7% for entire protein sequences, from 61.3% to 100% for protein contact residues, and from 73.4% to 93.5% for protein and rRNA contact residues. The protein contact residues did not show higher sequence identity than the entire sequence (P = 0.38). Also, the protein and rRNA contact residues combined did not show higher sequence identity than the entire sequence (P = 0.06).
Plastid-Encoded and Plastid-Targeted Ribosomal Protein Genes Are Coevolving
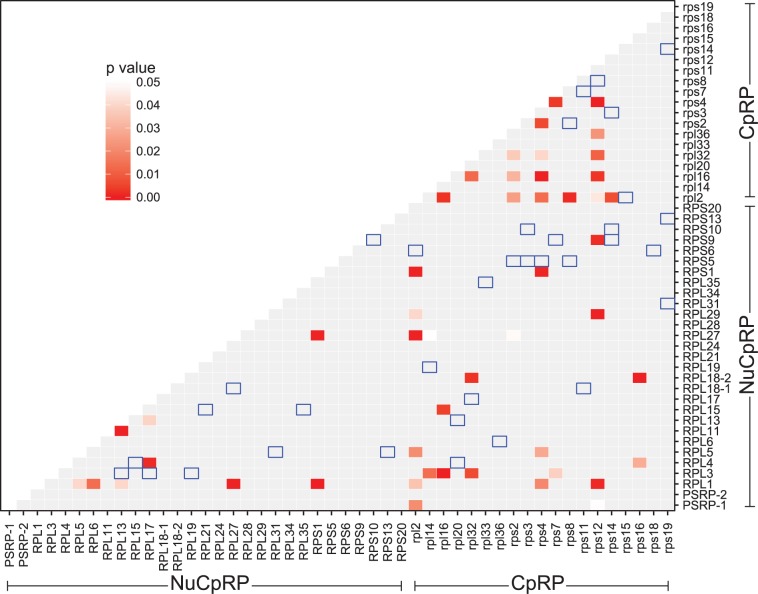
The type I coevolution statistic, defined as the number of codon pairs that share nonsynonymous substitutions along the same branch, was computed. To test whether these statistics were significantly larger than expected, the one-tail significance was computed against a null normal distribution generated through sequence simulation (see Materials and Methods). When analyzing the number of codon pairs that had nonsynonymous substitutions along one or two branches, none of the protein pairs showed significant results. When the threshold was increased to three branches, 17 CpRP-CpRP pairs, 9 NuCpRP-NuCpRP pairs, and 23 CpRP-NuCpRP pairs had significantly more nonsynonymous substitutions occurring along the same branches on the Geraniaceae lineages (fig. 5, table 3, and supplementary data file S2, Supplementary Material online). Among the protein pairs that had significantly more nonsynonymous substitutions along three branches, the largest number (12) were within Rpl2; five other CpRP (Rpl16, Rpl32, Rps2, Rps4, Rps12) and two NuCpRP (RPL1 and RPL27) involved more than five protein pairs. The protein pairs that tend to have nonsynonymous substitutions at the same branches did not have residues within 10Å distance (fig. 5, none of the blue open squares coincide with red squares).
Fig. 5.—
Matrix of P values for the type I coevolution statistics. The type I coevolution statistic for a protein pair is defined as the number of codons that share nonsynonymous substitutions on three branches. False discovery rate-adjusted P values smaller than 5% are highlighted in red gradient. The nonsignificant P values are shown as gray squares. The blue open squares indicate the protein pairs that have residues within 10Å distance.
Table 3.
Summary of Coevolution Analysis
| Gene group | Gene | Intergenomic Protein Paira | Intragenomic Protein Pairb | Total |
|---|---|---|---|---|
| CpRP | rpl2* | 6 | 6 | 12 |
| rps4* | 3 | 6 | 9 | |
| rps12* | 4 | 5 | 9 | |
| rpl16 | 2 | 5 | 7 | |
| rpl32 | 2 | 4 | 6 | |
| rps2* | 1 | 4 | 5 | |
| rpl14* | 2 | 0 | 2 | |
| rps7* | 1 | 1 | 2 | |
| rps16 | 2 | 0 | 2 | |
| rpl36 | 0 | 1 | 1 | |
| rps8* | 0 | 1 | 1 | |
| rps14 | 0 | 1 | 1 | |
| NuCpRP | RPL1 | 3 | 5 | 8 |
| RPL27 | 3 | 2 | 5 | |
| RPL3 | 4 | 0 | 4 | |
| RPS1 | 2 | 2 | 4 | |
| RPL5 | 2 | 1 | 3 | |
| RPL13 | 0 | 3 | 3 | |
| PSRP-1 | 2 | 0 | 2 | |
| RPL4 | 1 | 1 | 2 | |
| RPL17 | 0 | 2 | 2 | |
| RPL18-2 | 2 | 0 | 2 | |
| RPL29 | 2 | 0 | 2 | |
| RPL6 | 0 | 1 | 1 | |
| RPL11 | 0 | 1 | 1 | |
| RPL15 | 1 | 0 | 1 | |
| RPS9 | 1 | 0 | 1 | |
| Total protein pair | 23 (CpRP-NuCpRP) | 17 (CpRP-CpRP) | 49 | |
| 9 (NuCpRP-NuCpRP) | ||||
Note.—The number of proteins that share significant type I coevolution statistics are shown.
An intergenomic protein pair comprises one CpRP and one NuCpRP.
An intragenomic protein pair comprises two proteins from the same genome. Asterisks indicate core plastid ribosomal protein genes identified in Maier et al. (2013).
The estimated type I coevolution statistic based on alignments that only include contact residues are shown in supplementary figure S4, Supplementary Material online. Seven protein pairs had significantly more nonsynonymous substitutions occurring along the same branches when both protein and rRNA contact residues were included in the analysis (upper diagonal in supplementary fig. S4, Supplementary Material online). Six protein pairs had significantly more nonsynonymous substitutions at protein contact residues occurring along the same branches (lower diagonal in supplementary fig. S4, Supplementary Material online), and one of them (Rpl14-RPL29) has residues within 10Å distance.
The type II coevolution statistic, defined as the average number of branches separating consecutive nonsynonymous substitutions between two proteins in the same lineage, was computed (supplementary fig. S1, Supplementary Material online). To test whether the calculated coevolution statistics were significantly smaller than expectation, one-tail significance was computed against a null normal distribution generated through sequence simulation (see Materials and Methods). None of the protein pairs showed significant type II coevolution statistics (supplementary data file S2, Supplementary Material online). The analysis based on alignments that only include contact residues yielded no significant results (data not shown).
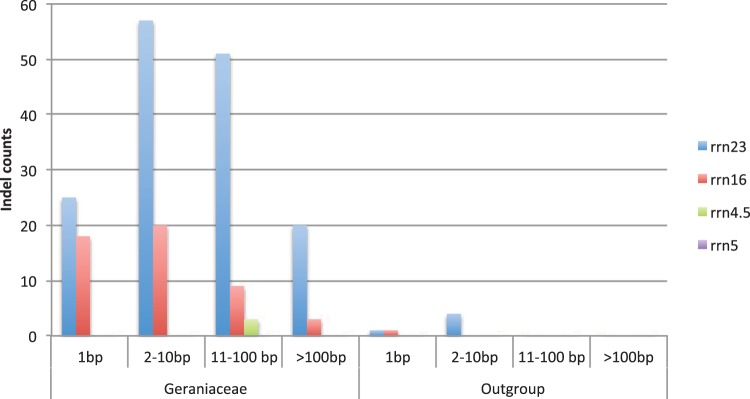
Indels Are Abundant in Geraniaceae Ribosomal RNA Genes
Indels in rRNA genes in Geraniaceae and outgroups were binned by length (fig. 6). Indels longer than 11 bp were absent in the outgroups but abundant in Geraniaceae. There were a total of 206 indels in Geraniaceae compared to only six in the outgroups. Geraniaceae had many more indels in 23S and 16S rRNA genes than in outgroups. Indels in 4.5S rRNA were only present in Geraniaceae but absent from the outgroups. No indels were present in 5S rRNA gene in either group.
Fig. 6.—
Indel distribution in plastid ribosomal RNA genes. Indels in rRNA genes were enumerated and plotted. Indels were identified using the simple indel coding algorithm (Simmons and Ochoterena 2000) and are binned by length.
Discussion
A Nuclear-Encoded Cytosolic Copy Substitutes for the Plastid-Encoded Rpl23 in G. maderense
A pseudogene in the plastid could suggest a recent plastid-to-nucleus gene transfer (Gantt et al. 1991; Cusack and Wolfe 2007; Ueda et al. 2007; Jansen et al. 2011) or gene substitution event (Bubunenko et al. 1994). To investigate whether gene transfer or substitution was associated with the truncated plastid-encoded rpl23 gene in G. maderense, we performed two BLAST searches against the translated G. maderense transcriptome. First, using the native truncated sequence as a query we did not find evidence for a plastid-to-nucleus gene transfer. Second, using the spinach Rpl23 amino acid sequence as the query we identified two transcripts. One of them contained a predicted plastid transit peptide suggesting the nuclear-encoded 60S Rpl23 has substituted for the plastid-encoded copy (fig. 2). In the spinach plastid ribosome, the product of the eukaryotic cytosolic rpl23 gene has also replaced the product of prokaryotic rpl23 (Bubunenko et al. 1994). The protein substitution reported here is the second independent case of Rpl23 substitution. The substitution event provides a plausible explanation why the plastid-encoded ribosomal protein genes have been pseudogenized in G. maderense. Another published plastid genome from Geranium, G. palmatum, also has a truncated rpl23 gene (Guisinger et al. 2011). The Rpl23 substitution event might be shared by G. maderense and G. palmatum given their close phylogenetic relationship (Park S, Park S, Choi KS, Aedo C, Jansen RK, unpublished data). Transcriptome or nuclear genomic data for G. palmatum is needed for further investigation.
The Plastid Ribosome Evolves Faster in Geraniaceae
Our analyses showed that in Geraniaceae both CpRP and NuCpRP have accelerated dN, relative to other plastid-encoded and nuclear-encoded genes, respectively (fig. 3). The acceleration of dN but not dS in CpRP and NuCpRP indicates that the increased substitution rates are independent of background mutation rates in these genes. However, the observation that dS was consistently higher in nuclear-encoded genes than plastid-encoded genes is congruent with the higher mutation rates in the nuclear genomes of land plants (Wolfe et al. 1987; Drouin et al. 2008; Zhang et al. 2016). Other phenomena, such as positive selection or relaxation of functional constraints, could be responsible for the observed acceleration in dN.
Nonsynonymous substitutions could accumulate through strong positive selection or relaxed purifying selection. To detect positive selection, we used branch-site models that allowed ω to vary across sites in a protein and across branches in the phylogeny (Yang et al. 2005; Zhang et al. 2005). The ω is an estimate of the strength of selection; a value greater than one suggests positive selection and less than 1 indicates purifying selection (Nielsen 2005). The branch-site tests indicated that none of the NuCpRP genes were under positive selection, whereas six CpRP genes showed evidence of positive selection in some amino acid residues (tables 1 and 2). Further, ω values estimated by branch-models were significantly higher for CpRP and NuCpRP in Geraniaceae than in outgroups (fig. 4 inset) suggesting that the strength of purifying selection is relaxed for these genes in Geraniaceae. Accelerated dN has also been observed in plastid-encoded RNA polymerase genes in Geraniaceae (Guisinger et al. 2008; Weng et al. 2012; Blazier et al. 2016) and it was suggested that the acceleration was related to the relaxation of functional constraints.
Codon usage bias can affect ω estimates. Because it inversely correlates with synonymous substitution rates (Sharp and Li 1987; Zhang et al. 2002), strong codon usage bias would inflate the value of ω. However, this is opposite to the pattern observed in Geraniaceae, where NuCyRP genes have stronger codon usage bias and significantly lower ω than NuCpRP. The difference in ω between NuCyRP and NuCpRP would be larger if codon usage bias were taken into account.
Cytonuclear Coevolution in Plastid Ribosomes
Amino acid substitutions in proteins within a multisubunit complex might drive changes in other proteins that assemble in the same complex. Cytonuclear coevolution has been observed in multisubunit complexes that include proteins encoded by both nuclear and mitochondrial genomes (Osada and Akashi 2012; Barreto and Burton 2013; Zhang and Broughton 2013) and by both nuclear and plastid genomes (Sloan et al. 2014; Zhang et al. 2015). In animal cells, Osada and Akashi (2012) provided evidence for compensatory evolution between mitochondrial- and nuclear-encoded subunits in the cytochrome c oxidase (COX) complex in a phylogenetic context. They showed that the fixation of slightly deleterious mutations in mitochondrial-encoded subunits in the COX complex has driven compensatory changes in nuclear-encoded subunits. However, the driving forces for cytonuclear coevolution have not been well characterized in plants.
To investigate cytonuclear coevolution between plastid ribosomal subunits that are encoded in the nucleus and plastids, we used an ML method to reconstruct the ancestral sequences and tested whether the nonsynonymous substitutions of two interacting proteins occurred concurrently or sequentially in the phylogeny, as would be expected under a model of coevolution. Our analyses revealed that 49 protein pairs between plastid ribosome subunits tend to have nonsynonymous substitutions along the same branches in the Geraniaceae phylogeny (fig. 5), a signal suggesting coevolution between these proteins. The three plastid encoded proteins (Rps4, Rps12 and Rpl2) that coevolved with the largest number of nuclear encoded proteins (fig. 5 and table 3) are included in the core set of plastid ribosomal protein genes identified in Maier et al. (2013). This core set of ribosomal protein genes has been retained in plastids and mitochondria across multiple independent eukaryotic lineages. The constraint imposed by ribosome assembly was suggested as the selection pressure driving this conservation (Maier et al. 2013). The essential roles played by the core ribosomal proteins in ribosome assembly (Kaczanowska and Ryden-Aulin 2007) might explain the strong coevolution signals revealed by our analyses.
Although physical interaction between residues in a multisubunit protein structure has been predicted as a driver for coevolutionary changes (Schmidt et al. 2001; Osada and Akashi 2012), our analyses did not find evidence favoring this hypothesis. The coevolving plastid ribosomal protein pairs identified based on complete gene sequences did not share residues within 10Å distance (fig. 5 and supplementary fig. S4, Supplementary Material online). If the physical interaction between ribosomal proteins is driving coevolution, the number of protein pairs with significantly more nonsynonymous substitutions at contact residues occurring along the same branches should have increased. Instead, we found the number of significant protein pairs decreased when analyzing alignments that only include contact residues (supplementary fig. S4, Supplementary Material online). In addition, the sequence identity between complete gene sequences and contact residues were not significantly different (supplementary table S6, Supplementary Material online) suggesting the absence of biased selective constraints on residues at contact sites in plastid ribosomes in Geraniaceae. Compensatory coevolution may be occurring between residues that are not physically proximate, as has been demonstrated for noncontact residues in ribosomal proteins in bacteria (Maisnier-Patin et al. 2007). Given the complexity of the translation machinery in plastids (Marín-Navarro et al. 2007; Peled-Zehavi and Danon 2007), additional nuclear-encoded regulatory factors may coevolve with ribosomal proteins. Further studies that include nuclear genes encoding regulatory factors are needed.
Unlike the observations of nuclear and mitochondrial genomes in animals, where it is argued that the fixation of deleterious mutations in the mitochondrion causes selective pressures that drive compensatory changes in nuclear genomes (Gabriel et al. 1993; Lynch 1996; Neiman and Taylor 2009), our analyses did not find evidence that nonsynonymous substitutions in CpRP tend to occur on branches subsequent to nonsynonymous substitutions in NuCpRp, or vice versa, in Geraniaceae (supplementary data file S2, Supplementary Material online). This result suggests that the driving forces for the increased dN in plastid ribosomal protein genes might not come from plastid ribosomal subunits themselves. Ribosomal RNAs, which provide the foundation for ribosome assembly and structure, could drive changes in ribosome subunits. Geraniaceae ribosomal RNAs indeed had a large number of indels of different lengths (fig. 6), which could cause substantial structural changes in the ribosome and consequently amino acid substitutions in CpRP and NuCpRP.
The cytonuclear coevolution in Geraniaceae plastid ribosomes could also be due to relaxation of purifying selection on the ribosome resulting in accelerated substitutions in ribosomal protein genes, and subsequent selective pressure for compensatory changes. Purifying selection in the majority of NuCpRP in Geraniaceae was relaxed (supplementary fig. S3 and table S4, Supplementary Material online). Relaxation of selection on nuclear genes often occurs after genome or gene duplication (Ohno 1970; Zhang 2003). Whole-genome duplication, either through auto- or allopolyploidy, has occurred numerous times across flowering plants (Jiao et al. 2011). Substantial variation in genome size, chromosome number, and size and ploidy level in Geraniaceae (Bakker et al. 2004; Weng et al. 2012) suggests that genome duplication has occurred in the family. After genome duplication, plastid-targeted genes have strong tendency to be retained as single copy genes suggesting gene losses among duplicated plastid-targeted genes (De Smet et al. 2013). Gene loss after genome duplication could lead to the retention of single copy genes that are divergent among species (reciprocal gene loss; Scannell et al. 2006), leading to the detection of highly accelerated substitution rates in plastid-targeted genes, and potentially to plastome-genome incompatibilities as proposed by Zhang et al. (2015).
Studying the sequences of plastid ribosomal proteins in Geraniaceae showed strong coevolution signals between plastid- and nuclear-encoded subunits that have accelerated nonsynonymous substitution rates. Increased positive selection, relaxed purifying selection, and sequence variation in rRNA genes all contributed to the accelerated nonsynonymous rates in ribosome subunits.
Supplementary Material
Supplementary figures S1–S4, tables S1–S6, and data files S1–S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Support was provided by the National Science Foundation (IOS-1027259 to R.J.K. and T.A.R.). The authors thank Jin Zhang for helpful discussions, the Texas Advanced Computing Center (TACC) at the University of Texas for access to supercomputers and TEX-LL for serving as a repository for voucher specimens.
Literature Cited
- Bakker FT, Culham A, Hettiarachi P, Touloumenidou T, Gibby M. 2004. Phylogeny of Pelargonium (Geraniaceae) based on DNA sequences from three genomes. Taxon 53:17–28. [Google Scholar]
- Barreto FS, Burton RS. 2013. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 30:310–314. [DOI] [PubMed] [Google Scholar]
- Bhattacherjee A, Mallik S, Kundu S. 2015. Compensatory mutations occur within the electrostatic interaction range of deleterious mutations in protein structure. J Mol Evol. 80:10–12. [DOI] [PubMed] [Google Scholar]
- Blazier JC, et al. 2016. Divergence of RNA polymeraseα subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci Rep. 6(24595):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Wade MJ. 2009. The functional transfer of genes from the mitochondria to the nucleus: the effects of selection, mutation, population size and rate of self-fertilization. Genetics 182(4):129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubunenko MG, Schmidt J, Subramanian AR. 1994. Protein substitution in chloroplast ribosome evolution: a eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J Mol Biol. 240:28–41. [DOI] [PubMed] [Google Scholar]
- Budar F, Fujii S. 2012. Cytonuclear adaptation in plants. Adv Bot Res. 63:99–126. [Google Scholar]
- Cusack BP, Wolfe KH. 2007. When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet. 23:270–272. [DOI] [PubMed] [Google Scholar]
- De Smet R, et al. 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci U S A. 110:2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49:827–831. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, et al. 2011. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23:3137–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W, Lynch M, Burger R. 1993. Muller’s ratchet and mutational meltdowns. Evolution 47:1744–1757. [DOI] [PubMed] [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD. 1991. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. Embo J. 10:3073.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(D1):D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. 2001. The origin and early evolution of mitochondria. Genome Biol. 2:1018–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Bock R. 2013. Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 35:354–365. [DOI] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2008. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci U S A. 105:18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28:583–600. [DOI] [PubMed] [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104:19369.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Saski C, Lee S-B, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Jonson NA. 2010. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 26(7):317–325. [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Ryden-Aulin MR. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 71:477–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. 2009. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 60:115–138. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies In: Nielsen R, editor. Statistical methods in molecular evolution. (Statistics for biology and health). New York: Springer; p. 125–181. [Google Scholar]
- Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB. 2011. Prevalence of epistasis in the evolution of influenza a surface proteins. PLoS Genet. 7:e1001301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. 2003. The cytoplasmic factor in plant speciation. Syst Bot. 28:5–11. [Google Scholar]
- Lovell SC, Robertson DL. 2010. An integrated view of molecular coevolution in protein-protein interactions. Mol Biol Evol. 27:2567–2575. [DOI] [PubMed] [Google Scholar]
- Lynch M. 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol Biol Evol. 13:209–220. [DOI] [PubMed] [Google Scholar]
- Maier U-G, et al. 2013. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol. 5:2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S, Paulander W, Pennhag A, Andersson DI. 2007. Compensatory evolution reveals functional interactions between ribosomal proteins S12, L14 and L19. J Mol Biol. 366:207–215. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Navarro J, Manuell AL, Wu J, Mayfield SP. 2007. Chloroplast translation regulation. Photosynth Res. 94:359–374. [DOI] [PubMed] [Google Scholar]
- Matsuo M, Ito Y, Yamauchi R, Obokata J. 2005. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast-nuclear DNA flux. Plant Cell 17:665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. 2001. Primary and secondary endosymbiosis and the origin of plastids. J Phycol. 37:951–959. [Google Scholar]
- Müller K. 2005. SeqState–primer design and sequence statistics for phylogenetic DNA data sets. Appl Bioinformatics. 4:65–69. [DOI] [PubMed] [Google Scholar]
- Neiman M, Taylor DR. 2009. The causes of mutation accumulation in mitochondrial genomes. Proc Biol Sci. 276:1201–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6. [DOI] [PubMed] [Google Scholar]
- Nielsen R. 2005. Molecular signatures of natural selection. Annu Rev Genet. 36:197–218. [DOI] [PubMed] [Google Scholar]
- Noutsos C, Richly E, Leister D. 2005. Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 15:616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer; p. 160. [Google Scholar]
- Osada N, Akashi H. 2012. Mitochondrial–nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome c oxidase complex. Mol Biol Evol. 29:337–346. [DOI] [PubMed] [Google Scholar]
- Park S, Jansen RK, Park S. 2015. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden JF. 1999. Analysis of codon usage. [PhD thesis]. University of Nottingham. [cited 2016 May 18]. Available from: http://codonw.sourceforge.net.
- Peled-Zehavi H, Danon A. 2007. Translation and translational regulation in chloroplasts In: Bock R, editor. Cell and molecular biology of plastids. Vol. 19. Topics in current genetics. Berlin/Heidelberg: Springer; p. 249–281. [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [cited 2016 May 18]. Available from: https://www.R-project.org.
- Rand DM, Haney RA, Fry AJ. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 19:645–653. [DOI] [PubMed] [Google Scholar]
- Ruhlman TA, et al. 2015. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15:100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman TA, Jansen RK. 2014. The plastid genomes of flowering plants In: Maliga P, editor. Chloroplast biotechnology. Vol. 1132 Totowa, NJ: Humana Press; p. 3–38. [cited 2016 May 18]. Available from: http://link.springer.com/10.1007/978-1-62703-995-6_1. [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440:341–345. [DOI] [PubMed] [Google Scholar]
- Schmidt TR, Wu W, Goodman M, Grossman LI. 2001. Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol Biol Evol. 18:563–569. [DOI] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Pybus OG, Holmes EC. 2006. A phylogenetic method for detecting positive epistasis in gene sequences and its application to RNA virus evolution. Mol Biol Evol. 23:1724–1730. [DOI] [PubMed] [Google Scholar]
- Sharma MR, et al. 2007. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci U S A. 104:19315–19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 4:222–230. [DOI] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analysis. Syst Biol. 49:369–381. [PubMed] [Google Scholar]
- Sloan DB. 2015. Using plants to elucidate the mechanisms of cytonuclear co-evolution. New Phytol. 205:1040–1046. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Triant DA, Wu M, Taylor DR. 2014. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol Biol Evol. 31:673–682. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 75:758–771. [DOI] [PubMed] [Google Scholar]
- Stegemann S, Bock R. 2006. Experimental reconstruction of functional gene transfer from the tobacco plastid genome to the nucleus. Plant Cell 18:2869–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, et al. 2009. PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 37(Suppl 1):D969–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5(2):123–135. [DOI] [PubMed] [Google Scholar]
- Ueda M, et al. 2007. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 402:51–56. [DOI] [PubMed] [Google Scholar]
- Ueda M, et al. 2008. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol Biol Evol. 25:1566–1575. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31:645–659. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Ruhlman TA, Gibby M, Jansen RK. 2012. Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae). Mol Phylogenet Evol. 64:654–670. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32:820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 84:9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. 1990. The effective number of codons used in a gene. Gene 87:23–29. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3235–3255. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR. 2000. The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J Biol Chem. 275:28466–28482. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR. 2000. The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J Biol Chem. 275:28455–28465. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22:1107–1118. [DOI] [PubMed] [Google Scholar]
- Zhang F, Broughton RE. 2013. Mitochondrial–nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol Evol. 5:1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18:292–298. [Google Scholar]
- Zhang J, et al. 2016. Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biol Evol. 8:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Mower JP, Jansen RK. 2013. Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biol. 13:228.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Sabir J, Blazier JC, Jansen RK. 2015. Coordinated rates of evolution between interacting plastid and nuclear genes in Geraniaceae. Plant Cell 27:563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vision TJ, Gaut BS. 2002. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol Biol Evol. 19:1464–1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.