Abstract
We present a novel method to measure the local GC-content bias in genomes and a survey of published fungal species. The method, enacted as “OcculterCut” (https://sourceforge.net/projects/occultercut, last accessed April 30, 2016), identified species containing distinct AT-rich regions. In most fungal taxa, AT-rich regions are a signature of repeat-induced point mutation (RIP), which targets repetitive DNA and decreases GC-content though the conversion of cytosine to thymine bases. RIP has in turn been identified as a driver of fungal genome evolution, as RIP mutations can also occur in single-copy genes neighboring repeat-rich regions. Over time RIP perpetuates “two speeds” of gene evolution in the GC-equilibrated and AT-rich regions of fungal genomes. In this study, genomes showing evidence of this process are found to be common, particularly among the Pezizomycotina. Further analysis highlighted differences in amino acid composition and putative functions of genes from these regions, supporting the hypothesis that these regions play an important role in fungal evolution. OcculterCut can also be used to identify genes undergoing RIP-assisted diversifying selection, such as small, secreted effector proteins that mediate host-microbe disease interactions.
Keywords: fungi, genome evolution, two-speed genome, repeat-induced point mutation, isochore
Introduction
The fungal kingdom contains many highly specialized organisms of interest to the agriculture, food, and medical industries. Specialization and adaptation are keys to the success of many fungi, with accelerated evolution capabilities increasingly recognized as facilitating these processes. For plant pathogens in particular, rapid evolution to overcome chemical control measures and host resistance is essential to their survival. Genome sequences of fungi—small but nonetheless eukaryotic—have revealed many features that challenge the conventional paradigm of genomic stability and contribute to their ability to evolve.
Even before whole genome sequences of fungi were available, genetic variability in the form of dispensable chromosomes (Tzeng et al. 1992) and chromosome length polymorphisms (Zolan 1995) was observed using pulsed-field gel electrophoresis. Saccharomyces cerevisiae was the first fungus to have its genome sequence published (Goffeau et al. 1996). Genomics of the filamentous fungi began some years later with the genome sequences of model organisms Neurospora crassa in 2003 (Galagan et al. 2003) and Aspergillus nidulans in 2005 (Galagan et al. 2005). An avalanche of sequencing projects followed to the point where hundreds of fungal genomes, representing a range of different lifestyles and taxa, are now publically available. These data have enabled further studies of genomic variability and adaptability in fungi relating to multiple mechanisms for sex (Heitman et al. 2013), chromosome length polymorphism (Zolan 1995), horizontal gene transfers (Hane et al. 2011; Gardiner et al. 2012; Sun et al. 2013; Dhillon et al. 2015), repeats and transposable elements (Spanu 2012), conditional dispensability of DNA segments or whole chromosomes (Coleman et al. 2009; Croll and McDonald 2012; Croll et al. 2013), and the conservation and breakdown of synteny and co-linearity (Hane et al. 2011).
The GC-content of genomic DNA has historically been of broad interest in the life sciences. In fungi, broad variation in DNA GC-content (38–63%) was observed from melting-temperature and buoyant-density measurements (Storck 1965) long before whole genome sequence data became available. Genome GC-content is now reported as one of the basic attributes of a genome assembly, confirming wide variation in the GC-content of fungal genomes. For example, Pneumocystis jirovecii—which causes severe lung infections in immunocompromised humans—has a genome GC-content of just 29.5% (Cissé et al. 2013). In contrast, the wood degrading fungus Phanerochaete chrysosporium has a much higher genome GC-content of 57% (Martinez et al. 2004). GC-content variation also exists within genomes, a property that was first observed in warm-blooded vertebrates (Bernardi et al. 1985). The human genome was observed to be composed of a mosaic of long stretches (typically > 300 kb) of DNA homogeneous in base composition (Bernardi 2000). These regions were termed “isochores” and can be grouped into families based on their GC-content. The presence of isochores has been documented in many species, including some fungi (Costantini et al. 2013). However, in fungal genomics the term “isochore” or “AT-isochore” has sometimes been used to refer specifically to sequence regions with markedly depleted GC-content (AT-rich). As this study focuses on these fungal AT-rich regions rather than what is traditionally termed an isochore, we refer to them as “AT-rich regions” from here on. AT-rich regions appear to differ from traditional isochores in their length and suspected origin (Rouxel et al. 2011) and do not necessarily have the homogeneous base composition implicit when using the terminology isochore. Fungal genomes with large proportions of AT-rich regions exhibit a distinctive bimodal pattern of GC-content bias. Observations of higher evolutionary rates in repeat rich genome compartments of filamentous plant pathogens (Raffaele et al. 2010), coupled with evidence of host jumps (Raffaele et al. 2010) and rapid adaptation to crop resistance (Fudal et al. 2009; Van de Wouw et al. 2010), have given rise to the concept of “two-speed” genome evolution. This concept describes a genome in which gene content has been compartmentalized into two types of sequence regions: regions containing the essential or “core” genome and the variable genome, often characterized by a higher density of repetitive elements and in some cases AT-rich sequence.
One mechanism by which AT-rich regions can occur is repeat-induced point mutation (RIP), a process specific to fungi and primarily considered to act as a defense against transposon propagation. RIP was initially observed in the saprobic Ascomycete N. crassa (Selker et al. 1987) and a cytosine methyltransferase homologue (rid) gene was shown to be essential for RIP (Freitag and Williams 2002). RIP has been identified experimentally in Leptosphaeria maculans (Idnurm and Howlett 2003; Fudal et al. 2009; Van de Wouw et al. 2010; Rouxel et al. 2011), Fusarium graminearum (Cuomo et al. 2007), Magnaporthe oryzae (Nakayashiki et al. 1999; Ikeda and Nakayashiki 2002; Dean et al. 2005; Farman 2007), and Podospora anserina (Graïa et al. 2001) with in silico evidence supporting RIP activity in many more species within the subphylum Pezizomycotina (Hane and Oliver 2008, 2010; Clutterbuck 2011; Goodwin et al. 2011) and some species within the Basidiomycota (Horns et al. 2012). RIP occurs during heterokaryon formation prior to meiosis, targeting repetitive DNA above a certain length (Watters et al. 1999) and identity (Cambareri et al. 1991) (400 bp and 80% in N. crassa), with irreversible transitions from cytosine to thymine bases (i.e., C to T). RIP has also been observed to leak beyond repetitive DNA (Irelan et al. 1994) into nearby single copy and often genic regions, in some cases mutating genes with known roles in pathogenicity (Fudal et al. 2009; Van de Wouw et al. 2010). Within genome assemblies, the observable impact of RIP is the depletion of GC-content within, and to a lesser extent nearby, repeats. Over time, the GC-content of RIP-affected sequence becomes distinct from nonRIP affected regions and can be described as “AT-rich”.
Leptosphaeria maculans was the first published fungal genome reported to have a significant proportion of distinctly AT-rich regions, accounting for approximately one-third of the assembly (Rouxel et al. 2011). AT-rich regions within L. maculans were found to have a few genes but many transposons and showed strong evidence of RIP. While it has been suggested that L. maculans is unusual in its high component of AT-rich regions (Raffaele and Kamoun 2012; Lo Presti et al. 2015), several other studies have identified AT-rich regions in fungal genomes including Blastomyces dermatitidis (Clutterbuck 2011) (note: genome unpublished), Passalora fulva (de Wit et al. 2012), multiple Epichloë spp. (Schardl et al. 2013), and Zymoseptoria tritici (Croll et al. 2013; Testa, Oliver, et al. 2015). Furthermore, Clutterbuck (2011) found widespread sequence-based evidence of RIP in the examination of 49 filamentous Ascomycetes (subphylum Pezizomycotina), suggesting that bimodal GC bias and high AT-rich content may be common across this large taxon.
Within the discipline of plant pathology, interest in AT-rich regions has been fuelled by observations of genes encoding avirulence/effector-like proteins within or close to AT-rich regions (e.g., L. maculans genes AvrLm6, AvrLm4-7 and AvrLm1; Gout et al. 2006; Fudal et al. 2009). Effector proteins are typically small, secreted proteins and play an important role in interactions with the host plant. The frequent proximity of effector genes to repetitive regions is well documented (Lo Presti et al. 2015). It has been proposed that pathogenic fungi with effector genes in or near AT-rich RIP hotspots have been selected by evolution as they have an advantageous mechanism by which to rapidly lose or modify these genes, avoid recognition by host defenses and thus repeatedly overcome newly deployed resistance genes (Oliver 2012).
Despite the clear motivations for investigating AT-rich regions, several obstacles limit our understanding of this interesting genome feature. The first is in identifying and defining AT-rich regions. In past studies AT-rich regions within fungi have generally been identified using a “moving window” approach that reports the GC-content within a series of windows over the entire length of the genome. The disadvantage of such an approach is the uncertainty in region boundaries. Another method by which AT-rich regions have been identified is by annotating repeats and recording their GC-content, but this method does not account for repeats that have been degraded (e.g., by RIP) to the point where they are not recognized by repeat-detection software (Hane and Oliver 2010).
Several approaches exist to identify isochore-like regions have been previously described. A highly successful method uses the Jensen–Shannon divergence (Elhaik, Graur, Josic 2010). In isochore studies, genome segments are classified by GC-content into isochore families L1 (< 37% GC), L2 (37–42% GC), H1 (42–47% GC), H2 (47–52% GC) and H3 (> 52% GC) (Oliver et al. 2001; Costantini et al. 2013). AT-rich regions in L. maculans were reported as 34% GC (Rouxel et al. 2011), compared with the 44% GC-content of AT-rich regions within Z. tritici (Testa, Oliver, et al. 2015). This variation in closely related species indicates that broadly applied and arbitrary GC-content categories may not be suitable for investigating AT-rich regions within fungal genomes. This highlights the need for a systematic method to measure GC-content distributions in fungal genomes and define different regions. Furthermore, small-scale studies often differ in their method of identifying and analyzing AT-rich regions, making comparisons difficult. This relates to the second obstacle—that although there have been detailed studies of a few individual fungi and wider surveys of repetitive elements and RIP within fungal genomes, we lack a taxonomically broad survey and understanding of AT-rich regions across the fungi.
In overcoming these obstacles, we present: firstly, a reproducible method and software tool, OcculterCut, which facilitates AT-content analysis in genomes; and secondly, a survey of the AT-content of >500 published fungal genomes. We identify species in which RIP (or other means of mutagenesis with similar effects) has led to significant proportions of AT-rich regions. By extension, this has predicted several species in which RIP-mediated “two-speed” genome evolution is likely to have significantly influenced their roles in plant association. OcculterCut also returns information about the proximity of genes to AT-rich regions, making it a useful tool for identifying genes likely to be under the influence of RIP-mediated “hyper-mutation”, such as candidate avirulence/effector genes or other genes involved in rapid evolution and specialization.
Materials and Methods
Collecting Genomes for Survey
Published fungal genome sequences were obtained from multiple sources, a full list of which is provided in the Supplementary Material online (supplementary table S1, Supplementary Material online). As assembled genome sequence(s) could potentially be contaminated with AT-rich mitochondrial sequences, a filtering process was carried out on all genomes surveyed. Firstly, a database of fungal mitochondrial sequences (mtDNAs) was downloaded from NCBI’s Organelle Genome resource (NCBI Resource Coordinators 2014) (supplementary table S6, Supplementary Material online—accessions of sequences used in mtDNA screen set). BLASTn (Altschul et al. 1990) was used to search for alignments between scaffolds in the surveyed genomes and the mtDNA database (e ≤ 1e-10 and ≥ 75% identity). Scaffold coverage of mtDNA matches was calculated via BEDtools (version 2.17) (Quinlan and Hall 2010) coverageBed and scaffolds were removed if 50% or more of the scaffold length was covered by alignments (see supplementary table S7, Supplementary Material online, matches to mtDNA screen). A bash script for mtDNA filtering has been included in the OcculterCut release files (available from https://sourceforge.net/projects/occultercut, last accessed April 30, 2016).
The lifestyle of the surveyed species was documented in order to reveal possible links between AT-rich regions and fungal lifestyle. The broad lifestyle categories used were saprobe, pathogen, and symbiont. Symbionts were separated into plant symbionts, which account for the majority of symbionts surveyed, and other symbionts. Pathogens were separated into plant pathogens and animal pathogens, and plant pathogens were further separated into obligate biotrophs, nonobligate biotrophs, hemibiotrophs, and necrotrophs as in (Spanu 2012). Sources of information about fungal lifestyle are cited in supplementary table S1, Supplementary Material online. We note that in plant pathology the reliability of such lifestyle categories has become a contentious issue for many species, and in such cases, we aimed to identify the recent consensus in the literature.
Identifying AT-Rich Regions with OcculterCut
The GC-contents of genome assemblies included in this survey were evaluated using a procedure that we have presented as a software tool, OcculterCut. The steps employed by OcculterCut in segmenting genome sequence and identifying AT-rich and GC-equilibrated regions are described below.
Genome Segmentation
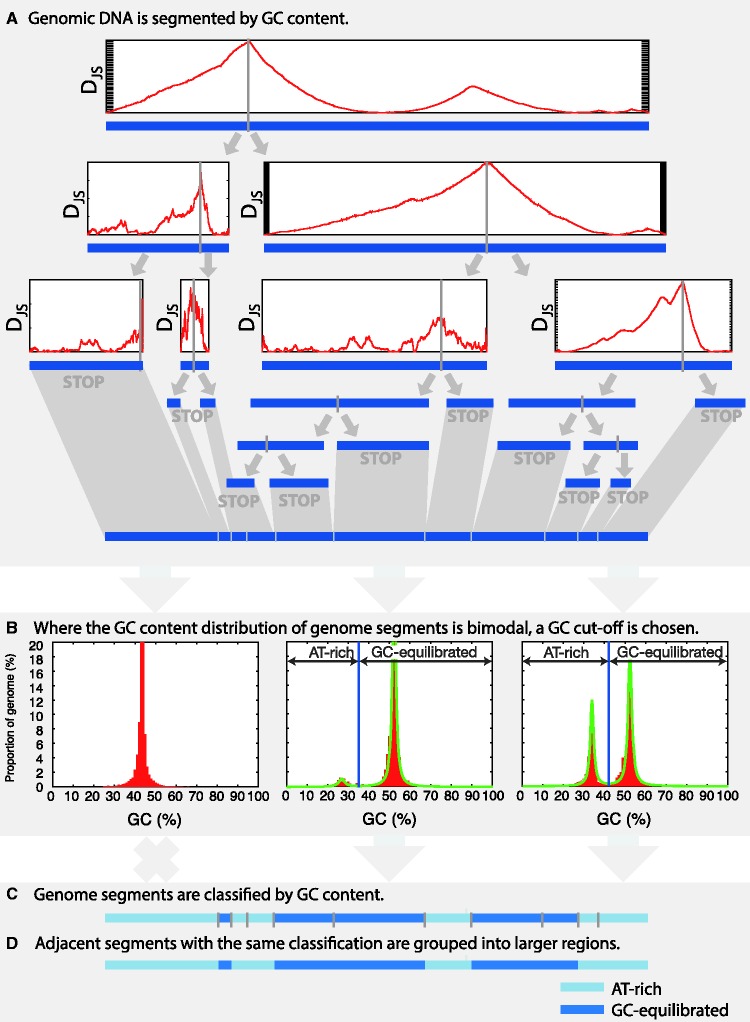
Assembled genome sequence is segmented into regions of differing GC-content using the Jensen–Shannon divergence (DJS), based on the methods described in past isochore studies (Bernaola-Galván et al. 1996; Elhaik, Graur, Josic 2010; Elhaik, Graur, et al. 2010). A border is moved along the query sequence and at each position the Jensen–Shannon divergence is calculated. At the position where the Jensen–Shannon divergence maximized, the sequence is split into two proposed subsequences, and the split is retained providing certain conditions are met (fig. 1A). This process continues recursively until a proposed split is rejected. The conditions that decide whether a split is rejected are based on the size of the segments to the left and right of a potential split and whether the potential split would result in two adjacent segments with a statistically significant difference in GC-contents. In this study, the minimum segment length of 1 kb was set. To assess the statistical significance of the difference between GC-contents either side of the split, the left-hand and right-hand segments are divided into nonoverlapping 300 bp sections and the GC-content of each section is recorded. Similar to Oliver et al. (2004), a t-test is then used to decide whether the GC-contents of the left-hand sections differ significantly from the GC-contents of the right-hand sections.
Fig. 1.
The basic steps employed by OcculterCut in annotating AT-rich and GC-equilibrated genome regions. (A) DNA sequence is recursively split into segments of differing GC-content. The point at which a sequence is split into two segments is chosen to be where the Jensen–Shannon divergence statistic (DJS on the y-axis of cartoon plots in (A) is maximized. (B) AT-rich and GC-equilibrated genome segments are identified, and a GC-content cut-off is selected. (C) Genome segments are categorized as either AT-rich or GC-equilibrated, and the genome segments are grouped into broader regions.
Segment Classification
The genome segmentation process results in genome sequence designated into segments of differing GC-content. In assessing the AT-rich region content of a genome the next step is to determine whether AT-rich genome segments are present and, if present, to categorize segments and as either AT-rich or GC-equilibrated. This requires an assessment of the GC-content distribution of the genome segments. The GC-content of each genome segment, including un-segmented contigs <1 kb, and the proportion of the genome taken up by that particular segment (segment length/genome assembly size) is recorded. These data can be visualized as a plot of the GC-content of the genome segments against the proportion of the genome (see examples in figs. 1B and 2). A mixture of two Cauchy distributions is fit to the data. That is, the data are assumed to be of the form:
where 0 ≤ ω ≤ 1 and describes the weight of each peak, x01and x02describe the x-axis position (GC-content) of each peak and γ1 and γ2 relate to the peak widths. Examples of Cauchy distributions fit to genome GC-content distributions are shown in figure 1B. The Cauchy distribution mixture model of the GC-content distribution (as described above) is fit to the genome segment GC-content data by determining the values ω, x01, γ1, x02, and γ2 using expectation maximization. Whether genome segments can be grouped into AT-rich and GC-equilibrated regions and the GC-content cut-off used to define each region type are based on this Cauchy distribution mixture model.
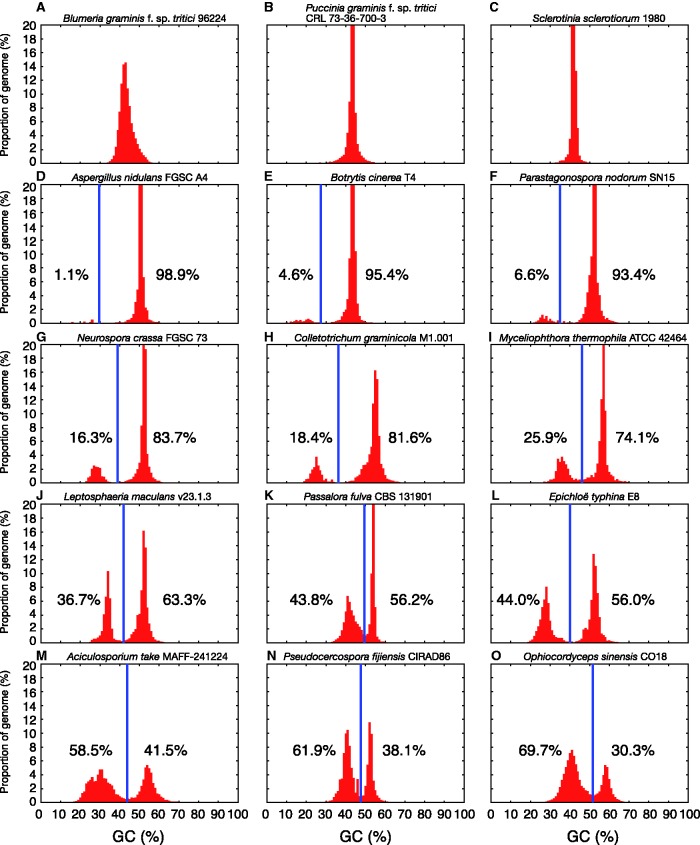
Fig. 2.
Example GC-content plots of 15 surveyed fungal genomes are displayed, arranged from (A) to (O) ordered in increasing AT-rich region composition. Diversity in the GC-content of peaks, their shape, spacing, and height can be observed. Vertical blue lines show the GC cut-off chosen by OcculterCut and used to classify genome segments into distinct AT-rich and GC-equilibrated region types. The percentage values shown either side of the vertical blue lines indicate the percentage of the genome classified as AT-rich (left) and GC-equilibrated (right).
Categorization of genome segments as AT-rich or GC-equilibrated is not carried out in all cases; many genomes do not have AT-rich regions and the plot of the GC-content is unimodal (figs. 1B and 2A–C). In some cases, the plot of the GC-content may suggest multiple peaks, but these overlap too much to allow the segments to be classified reliably as from one or the other. The decision to categorize the segments is therefore based on the %GC separation between the two peaks (the difference between x01 and x02), the existence of a local minimum in the Cauchy distribution mixture model between the two peaks, and the confidence with which segments can be classified. The minimum GC-content peak separation is set at a default 5%. This filters out cases where the GC-content distribution is unimodal or the peak separation of is too small to be of interest. Segment classification is always carried out where peak separation is ≥10% GC and a minimum in the Cauchy distribution mixture model can be identified between the two peaks. In cases where the peak separation is between 5% and 10%, region grouping is only carried out where a minimum can be identified between the peaks and 75% of the segments in each group can be classified with 75% or better confidence.
Where classification of genome segments is carried out, segments are classified as AT-rich or GC-equilibrated according to whether they have a GC-content above or below a cut-off GC value set as the local minimum between the two peaks in the Cauchy distribution mixture model (fig. 1B). Adjacent genome segments with the same classification are then merged into single, larger regions (fig. 1C and D). To allow the user to explore different grouping of genome segments, the Cauchy fit-to-data can be disabled in favor of the user specifying two or more GC-content intervals on which to group the genome segments.
Outputs
The presented software, OcculterCut, automates the described genome segmentation and segment classification steps and outputs a number of files containing the results of these steps. A brief description of the content of the OcculterCut outputs is given here and detailed description of output file names and content is given in the instruction manual that accompanies OcculterCut.
A General Feature Format (GFF) (Wellcome Trust Sanger Institute 2015) containing the genomic coordinates of genome segments resulting from the GC-content segmentation is output. This is accompanied by a text file containing a list of GC-content intervals (from 0% to 100% GC in 1% intervals) and the proportion of the genome covered by genome segments with a GC-content within each interval. These data can be plotted with GC-content on the x-axis and the proportion of the genome on the y-axis to produce a plot of the GC-content distribution (see examples in fig. 2). These data are returned for all genomes, regardless of whether the genome is bimodal or not.
In cases where the genome is found to be bimodal, the parameters of the Cauchy mixture fit-to-data (see the segment classification section) are returned in a text file along with the GC-content cut-off used to define AT-rich and GC-equilibrated regions. A GFF containing the genomic coordinates of AT-rich and GC-equilibrated regions is also included (fig. 1C). The user can choose to supply a GFF of gene locations when running OcculterCut, in which case a summary of the distance from each gene to the closest AT-rich region or scaffold end is returned. This feature may be particularly useful to phytopathogen researchers interested in identifying effector gene candidates. Single, di- and tri-nucleotide frequencies from AT-rich and GC-equilibrated regions are also returned as text files, allowing the user to assess sequence biases.
Analyzing AT-Rich Regions and Their Gene Content
In assemblies found to have an AT-rich region component ≥5%, AT-rich and GC-equilibrated regions were compared. This included looking at the length of each of the region types, di-nucleotide frequencies within each region type, and gene content. Additional analysis of the difference between coding sequences from genes within AT-rich and GC-equilibrated regions was conducted on a subset of species. For this purpose, incomplete coding sequences (lacking a methionine at the start), coding sequences containing an in-frame stop codon, and coding sequences <90 nucleotides (corresponding to a protein length <30 amino acids) were excluded from the analysis. This was done in an attempt to remove false positive or incorrect coding sequences from the analysis that may bias results. An additional filtering step was carried out to ensure the coding sequences being analyzed were not false positive predictions made on transposons sequences. TBLASTn (Camacho et al. 2009) was used to search for alignments between the Repbase (Jurka et al. 2005) fungal database and translated protein sequences from each genome. Where one or more of the alignments had e-value(s) <10 −5 and covered 50% or more of the query protein sequence, the protein sequence and corresponding nucleotide sequence were removed from the sets used for analysis. Finally, analysis was carried out only where 50 or more genes with coding sequences meeting the above criteria could be identified in the AT-rich regions of a particular genome assembly. For calculations of gene density the full set of annotations was included regardless of the above criteria.
When comparing the amino acid composition of proteins from genes within and outside AT-rich regions, the frequency of each amino acid was calculated for each gene from the different sets. A Mann–Whitney U test (P= 0.05) was used to compare the distributions of frequency values between the different sets. Codon frequencies for each amino acid were calculated within and outside AT-rich regions on the full set of coding sequence rather than a per gene basis.
Hmmscan (Eddy 2011) was used to identify Pfam domains in proteins within and outside AT-rich regions (automatic cut-off determined using the parameter—cut-ga) using the Pfam library version 28.0 (Finn et al. 2014). A Fisher’s exact test (two sided, implemented in R) was then used to compare the number proteins found to contain a particular Pfam domain in relation to the number of proteins found to contain a different Pfam domain in protein sets within and outside AT-rich regions. An additional comparison was made between proteins with a Pfam domain hit and with no Pfam domain hit. A P value of 0.05 was used, with a Bonferroni correction for the total number of tests for significance carried out for each species.
The number of secreted genes, as predicted by SignalP v. 4.1 (Petersen et al. 2011), was recorded for the sets of genes within and outside AT-rich regions for each species. A Fisher’s exact test (two sided, P value 0.05, implemented in R) was used to compare the number of proteins that were predicted to be secreted in relation to the number of proteins that were not predicted to be secreted in protein sets within and outside AT-rich regions.
Results
OcculterCut—A Tool for AT-Rich Region Analysis
OcculterCut was used to apply GC-content genome segmentation (see fig. 1 and Materials and Methods) to over 500 published fungal genomes (see supplementary table S1, Supplementary Material online, for list of genomes). Plotting the proportion a given genome accounted for by genome segments with a GC-content within 1% intervals (from 0% to 100%) offered a way of visualizing the GC-content distribution of each genome (see examples in fig. 2). Such plots of the surveyed genomes revealed diversity in peak GC-content(s) and distribution shapes and spreads (fig. 2). Heat map summary plots of the GC-content of segmented genomes of the species surveyed are displayed next to dendograms generated according to taxonomic classifications in figures 3 and 4. In most cases, GC-content distributions could be classified as unimodal (having a single peak, fig. 2A–C) or bimodal (two peaks, see fig. 2D–O). OcculterCut carried out this classification automatically (see Materials and Methods and fig. 1B and C, examples shown in fig. 2), and in genomes where distinctly AT-rich segments were present, the genome segments were grouped into broader AT-rich and GC-equilibrated region types.
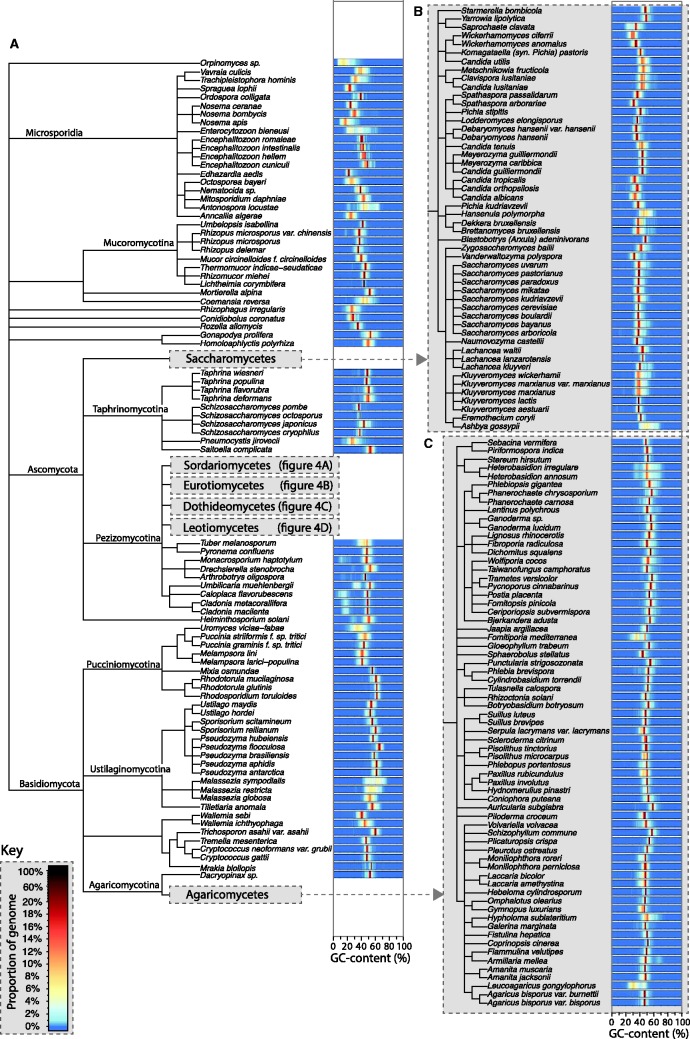
Fig. 3.
(A) Clade trees showing the taxonomic relationship between the fungal species included in this study. To the right of each species’ name, there is a bar displaying a heat map plot of the GC-content distribution of segments of each genome, where different colors represent varying genome proportions (see key). Classes containing high numbers of sequenced genomes have been displayed separately in panels (B) (Saccharomycetes) and (C) (Agaricomycetes), and in figure 4 (Sordariomycetes, Eurotiomycetes, Dothideomycetes and Leotiomycetes).
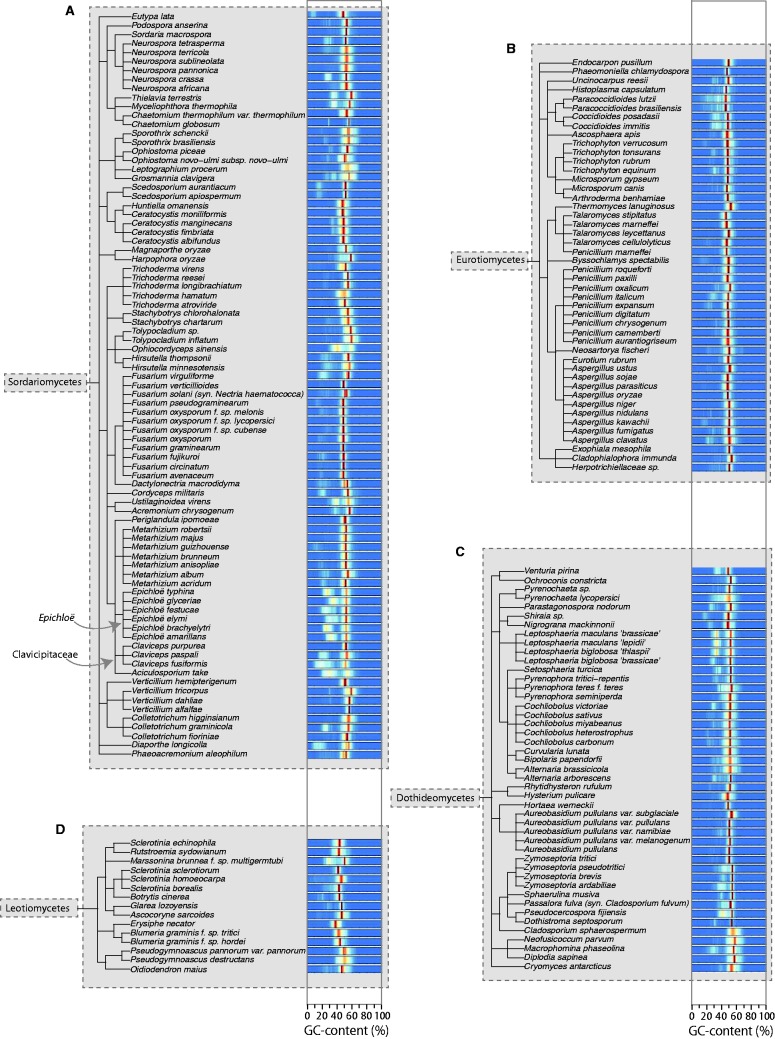
Fig. 4.
Clade trees showing the taxonomic relationship between fungal species belonging the classes Sordariomycetes (A), Eurotiomycetes (B), Dothideomycetes (C) and Leotiomycetes (D) within the subphylum Pezizomycotina. These fit into the broader taxonomic tree displayed in figure 3A. To the right of each species’ name, there is a bar displaying a heat map plot of the GC-content of segments of each genome, where different colors represent varying proportions of the genome with each percentage GC (see fig. 3 key).
The majority (∼63%) of surveyed genomes had unimodal GC-content distributions, with variation in the mode, peak width and shape observed between species (fig. 2A–C). GC-content plots of genomes known from previous studies to have distinct AT-content patterns showed two clearly separate peaks (fig. 2, L. maculans [J], P. fulva [K]). Additional genomes with bimodal GC-content distributions (see fig. 2D–O) confirm the efficacy of the GC-content genome segmentation method in distinguishing these regions. Where the GC-content of a segmented genome was bimodal (fig. 2D–O), OcculterCut selected a local minima between the peaks to divide the distribution and group genome segments into broader categories: AT-rich and GC-equilibrated. The selected GC-content cut-offs, shown as vertical blue lines in figure 2D–O and listed in supplementary table S2, Supplementary Material online, varied from species to species. Harpophora oryzae (R5-6-1) (Xu et al. 2014) had the highest GC-content AT-rich region component at 48% and Monacrosporium haptotylum (CBS 200.50) (Meerupati et al. 2013) the lowest at 12.3% (fig. 4). This wide range of 35.7% strongly supports the need to define AT-rich and GC-equilibrated regions case-by-case, based on comparisons within each genome, rather than by broadly applying pre-defined GC-content cut-offs.
There was a broad range in the proportion of each genome containing AT-rich regions—from ∼1% to AT-rich region components over 50%. AT-rich regions are not present in all genomes with a high repeat content, with known examples of repeat rich genomes showing clearly unimodal GC-content distributions (e.g., Blumeria spp. and Puccinia spp. genomes fig. 2A and B and supplementary table S1, Supplementary Material online). A total of 79 genome assemblies from 61 distinct species were identified as being composed of ≥ 5% AT-rich regions (supplementary table S1, Supplementary Material online). Although L. maculans is perhaps the best known case of a fungal genome interspersed with AT-rich regions (Gout et al. 2006; Fudal et al. 2009; Van de Wouw et al. 2010; Rouxel et al. 2011; Ohm et al. 2012), we note 14 genomes with higher AT-rich region contents than L. maculans and many more with comparable AT-rich region content (supplementary table S2, Supplementary Material online). In four species, AT-rich regions were in higher proportions than GC-equilibrated regions: Leucoagaricus gongylophorus (Ac12) (Aylward et al. 2013) with 76.6%, Ophiocordyceps sinensis (CO18) (Hu et al. 2013) with 69.7%, Aciculosporium take (MAFF-241224) (Schardl et al. 2013) with 58.5%, and Fomitiporia mediterranea (LYAD-421 SS1) (Floudas et al. 2012) with 57.8%. Despite the dominant component of each of these genomes being AT-rich, where gene annotations were available for these species (L. gongylophorus, A. take, and F. mediterranea) the majority of the gene content was still attributed to GC-equilibrated genome regions.
In six of the surveyed 500+ genomes, manual inspection of the GC-content plots of genomes motivated their removal from the set of AT-rich region genomes. The GC-content plots of Armillaria mellea (DSM 3731) (Collins et al. 2013), Nosema bombycis (CQ1) (Pan et al. 2013), and Lachancea kluyveri (NRRL Y-12651) (Cliften et al. 2003) showed evidence of segments of DNA, amounting to small components of the overall genome, with a higher GC-content than the rest of the genome. Armillaria mellea and Candida orthopsilosis (MCO456) (Pryszcz et al. 2014) contained small percentages of 0–1% GC-content segments that may be an artefact of assembly. Malassezia sympodialis (ATCC 42132) (Gioti et al. 2013) (fig. 3A, see Ustilaginomycotina for M. sympodialis GC-content heat map) appeared to have an unusually broad, albeit unimodal, GC-content distribution. In response to these examples, we have included a feature to allow the manual input of one or more GC-content boundaries for the categorization of genome segments into genome regions. This feature allows unusual and exceptional cases of genome GC-content distribution to be studied.
Taxonomic Distribution of Genomes with AT-Rich Regions
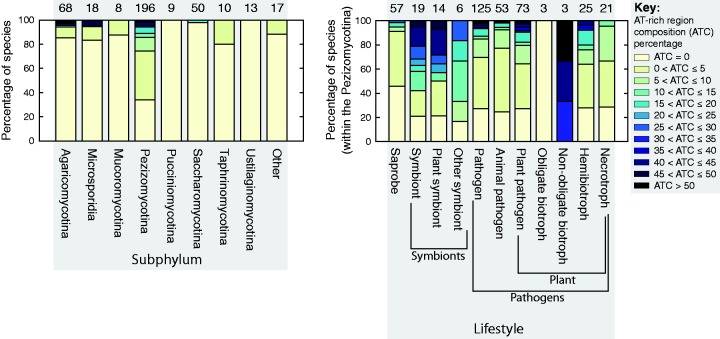
Figure 5A shows the percentage of genomes with various AT-rich genome contents for each subphylum surveyed, showing that this genome type is most common within the Pezizomycotina. The majority of genomes with ≥ 5% AT-rich region content are from the subphylum Pezizomycotina with only six exceptions: four Agaricomycetes (L. gongylophorus, F. mediterranea, Paxillus rubicundulus, Moniliophthora roreri), one Microsporidia (Enterocytozoon bieneusi), and one Saccharomycotina (Saprochaete clavata). Heat-map plots of GC-content distributions arranged alongside dendograms generated according to taxonomic classifications (figs. 3 and 4) also show this trend. Additionally, clusters of species with AT-rich regions are clearly visible in figures 3 and 4 (e.g., the family Clavicipitaceae within the class Sordariomycetes).
Fig. 5.
A summary of the number of species surveyed for each subphylum is shown (left-hand plot), where within each bar the colours describe the proportion of surveyed genomes within each subphyla with an AT-rich content in a particular range (see key far right). Subphyla with less than five surveyed species have been grouped into the entry “Other”. A similar plot (right-hand plot) is shown where species (from within the Pezizomycotina) have been classified according to their lifestyle.
AT-Rich Regions and Fungal Lifestyle
Classifications of fungal species into broad lifestyle categories (saprobe, pathogen, and symbiont) are displayed next to each surveyed species in supplementary table S1, Supplementary Material online. Pathogens and symbionts are further classified based on whether they have a plant host, and plant pathogens are additionally classified into obligate biotrophs, nonobligate biotrophs, hemibiotrophs, and necrotrophs. The distribution of fungal lifestyles in the different subphylum varied. The percentage of AT-rich genomes with various AT-rich genome contents are displayed according to their lifestyle categories in figure 5B. Within 196 surveyed Pezizomycotina species, 125 (∼64%) were pathogens, compared with 9 out of 68 (∼13%) of the Agaricomycotina species and 10 out of 50 (20%) of the Saccharomycotina species. As discussed in the previous section, AT-rich bimodal genomes were found more commonly in the Pezizomycotina subphylum. To prevent these taxonomic biases from influencing our assessment of AT-rich regions in relation to fungal lifestyle, the results shown in figure 5B are restricted to Pezizomycotina species.
Attributes of AT-Rich Regions Compared with GC-Equilibrated Regions
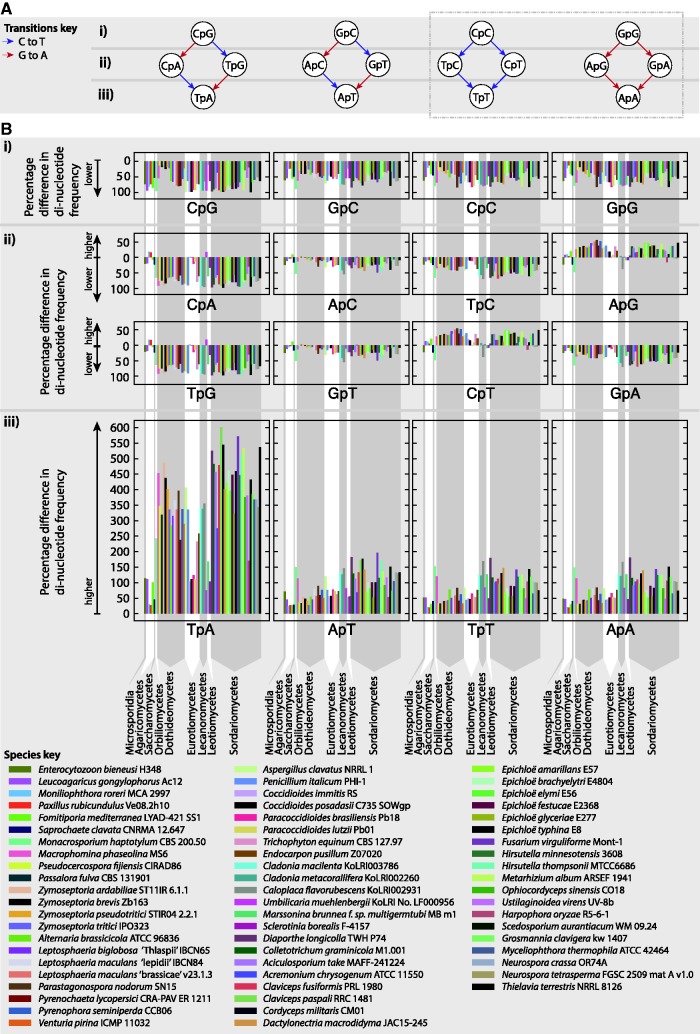
The subset of genomes with a ≥ 5% proportion of AT-rich regions was further analyzed (see Materials and Methods) to assess the role of RIP in AT-rich region formation and compare AT-rich regions between species. For each of these species, the frequency of each of the 16 possible dinucleotides was calculated from AT-rich region genome sequences and GC-equilibrated genome sequences (supplementary table S3, Supplementary Material online). Dinucleotide frequencies have commonly been employed in previous studies to ascertain the genomic impact of RIP (Hane and Oliver 2008; Clutterbuck 2011). The percentage differences between dinucleotide frequencies within AT-rich and GC-equilibrated regions are shown in a series of plots (fig. 6), with each different colored bar representing a different species. In all cases, the dinucleotide pairs TpA, ApT, TpT and ApA were higher in AT-rich regions, as expected and reflecting their lower GC-content. There was however a much larger difference in the frequency of the TpA dinucleotide, the primary product of RIP in the Pezizomycotina (Clutterbuck 2011) (fig. 6A), when compared with the other low GC dinucleotides ApT, TpT and ApA (fig. 6Biii). This is a strong indicator of RIP activity in these species.
Fig. 6.
Possible changes to dinucleotide pairs are described in a series of graphs (A). Dinucleotides are represented by nodes, and arrows represent possible changes resulting from C to T (blue) and G to A (red) transitions. Dinucleotides are organized into bands marked (i), (ii), and, (iii) based on the number of mutable sites they have (2, 1 and 0, respectively). Below each graph, corresponding plots (B) of percentage differences between dinucleotide frequencies within and outside AT-rich regions are shown for species with 5% or greater AT-rich region content. Percentage differences above y = 0 correspond to higher values within AT-rich regions and below y = 0 correspond to lower values within AT-rich regions (see axis labels). Each species is represented by a different color as shown in the species key and arranged according to taxonomy. Vertical grey bands group species by class, with class names marked at the bottom of (B).
While analysis and comparison of the lengths of AT-rich regions can be impeded by relative differences in assembly quality, we were able to observe variation in the size of AT-rich regions accurately in higher quality genomes. Coccidioides immitis (RS) (Sharpton and Stajich 2009), Thielavia terrestris (NRRL 8126) (Berka et al. 2011), Myceliophthora thermophile (ATCC 42464) (Berka et al. 2011), N. crassa (OR74A) (Galagan et al. 2003), Z. tritici (IPO323) (Goodwin et al. 2011), Cordyceps militaris (CM01) (Zheng et al. 2011), and L. maculans “brassicae” (v23.1.3) (Rouxel et al. 2011) all have assemblies with <50 scaffolds and we consider these to be of “good quality”. Among these, L. maculans had the longest average AT-rich region length at 31.7 kb and Z. tritici the shortest at 10.7 kb. GC-equilibrated regions were also longer within L. maculans than within Z. tritici, indicating that the Z. tritici genome is interrupted with AT-rich regions more frequently than L. maculans, despite having a smaller overall AT-rich genome component. Similarly N. crassa, M. thermophila and C. immitis have comparable average AT-rich region lengths, with differing overall AT-rich region composition and correspondingly different higher GC-region average lengths.
Gene Content
Mutations accumulated during AT-rich region formation could influence the properties of coding sequences in those regions. All observed AT-rich regions are gene-sparse when compared with GC-equilibrated regions (table 1). Indeed, many of the surveyed genomes contained only a very small number of genes within AT-rich regions, making meaningful comparisons between sets of genes within and outside AT-rich regions difficult. After quality filtering the sets of genes (see Materials and Methods), 19 species were identified with >50 genes within AT-rich regions and thus suitable for further analysis (supplementary table S4, Supplementary Material online). Comparisons between coding sequences within AT-rich and GC-equilibrated regions confirmed differences in amino acid usage, codon usage, and putative function between these sets as well as identifying common trends across the selected species.
Table 1.
Attributes of Coding Sequences from Genome Regions within AT-Rich and GC-Equilibrated Regions
| Species | AT-Rich Region Gene Content |
GC-Equilibrated Gene Content |
Genome Source | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Density (Genes Per Mb) | No. Genes (Total) | No. Genes (Filtered)a | Average Length (Amino Acids) | No. Secreted | Per cent Secreted | Gene Density (Genes Per Mb) | No. Genes (Total) | No. Genes (Filtered)a | Average Length (Amino Acids) | No. Secreted | Per cent Secreted | ||
| Enterocytozoon bieneusi (H348) | 865 | 1,616 | 1,549 | 313.2 | 40 | 2.58 | 1,010 | 2,016 | 1,341 | 184.3 | 36 | 2.68 | JGI (Akiyoshi et al. 2009) |
| Leucoagaricus gongylophorus (Ac12) | 1.9 | 148 | 66 | 186.8 | 3 | 4.54 | 222 | 5,272 | 3,610 | 421.6 | 194 | 5.37 | JGI (Aylward et al. 2013) |
| Moniliophthora roreri (MCA2997) | 188 | 1,473 | 1,050 | 248.9 | 33 | 3.14 | 353 | 15,677 | 15,128 | 433.0 | 1,660 | 10.97 | NCBI (Meinhardt et al. 2014) |
| Fomitiporia mediterranea (LYAD-421 SS1) | 53.2 | 1,947 | 1,614 | 332.1 | 112 | 6.93 | 351 | 9,386 | 8,836 | 464.9 | 681 | 7.70 | JGI (Floudas et al. 2012) |
| Pseudocercospora fijensis (CIRAD86) | 25.7 | 1,179 | 1,039 | 392.1 | 70 | 6.73 | 421 | 11,928 | 10,514 | 439.2 | 752 | 7.15 | JGI |
| Passalora fulva (CBS 131901) | 19.8 | 528 | 93 | 152.4 | 11 | 11.70 | 395 | 13,589 | 13,380 | 449.7 | 1,134 | 8.47 | JGI (de Wit et al. 2012) |
| Leptosphaeria maculans (v23.1.2) | 11.3 | 186 | 181 | 141.1 | 28 | 15.46 | 432 | 12,283 | 12,272 | 422.8 | 1,042 | 8.49 | JGI (Rouxel et al. 2011) |
| Coccidioides immitis (RS) | 65.2 | 319 | 249 | 140.8 | 3 | 1.20 | 399 | 9,591 | 9,537 | 445.4 | 537 | 5.63 | JGI (Sharpton and Stajich 2009) |
| Paracoccidioides lutzii (Pb01) | 34.4 | 202 | 193 | 195.5 | 8 | 4.14 | 330 | 8,934 | 8,841 | 448.7 | 386 | 4.36 | Broad (Desjardins et al. 2011) |
| Marssonina brunnea f. sp. Multigermtubi (Mb m1) | 15.4 | 303 | 251 | 220.2 | 49 | 19.52 | 302 | 9,724 | 9,634 | 505.1 | 922 | 9.57 | JGI (Zhu et al. 2012) |
| Aciculosporium take (MAFF-241224) | 17.9 | 616 | 223 | 89.2 | 7 | 3.05 | 364 | 8,899 | 8,213 | 443.6 | 532 | 6.47 | NCBI (Schardl et al. 2013) |
| Claviceps fusiformis (PRL 1980) | 93.9 | 2,338 | 272 | 85.6 | 9 | 3.20 | 324 | 8,955 | 8,479 | 450.6 | 671 | 7.91 | NCBI (Schardl et al. 2013) |
| Epichloë festucae (E2368) | 37.8 | 400 | 130 | 76.3 | 3 | 2.23 | 355 | 8,573 | 8,032 | 477.0 | 613 | 7.63 | NCBI (Schardl et al. 2013) |
| Epichloë glyceriae (E277) | 37.8 | 765 | 388 | 162.3 | 15 | 3.81 | 408 | 12,755 | 10,329 | 456.0 | 754 | 7.29 | NCBI (Schardl et al. 2013) |
| Ustilaginoidea virens (UV-8b) | 10.6 | 144 | 135 | 270.9 | 5 | 3.70 | 322 | 8,282 | 8,271 | 467.8 | 607 | 7.33 | NCBI (Berka et al. 2011) |
| Myceliophthora thermophila (ATCC42464) | 15.9 | 160 | 64 | 169.9 | 2 | 3.12 | 312 | 8,950 | 8,613 | 487.8 | 722 | 8.38 | JGI (Zhang et al. 2014) |
| Neurospora crassa (FGSC 73) | 41.1 | 249 | 56 | 112.4 | 3 | 5.35 | 341 | 11,723 | 11,190 | 447.6 | 910 | 8.13 | JGI (Baker et al. 2015) |
| Neurospora tetrasperma (FGSC 2509) | 71.3 | 254 | 62 | 225.3 | 6 | 9.67 | 308 | 10,938 | 10,287 | 466.6 | 805 | 7.82 | JGI (Ellison et al. 2011) |
| Neurospora tetrasperma (FGSC 2508) | 38.5 | 160 | 64 | 229.0 | 4 | 6.25 | 292 | 10,220 | 9,741 | 484.8 | 797 | 8.18 | JGI (Ellison et al. 2011) |
Note.—The number of genes, average gene length, and number of genes predicted to be secreted is shown for each species. Species listed are those with 50 or more protein sequences from each region type (AT-rich and GC-equilibrated) after quality filters were applied to remove incomplete genes and transposons (see Materials and Methods and supplementary table S4, Supplementary Material online).
a>Number of genes (filtered) and subsequent values relate post-quality filtering gene sets (see Materials and Methods).
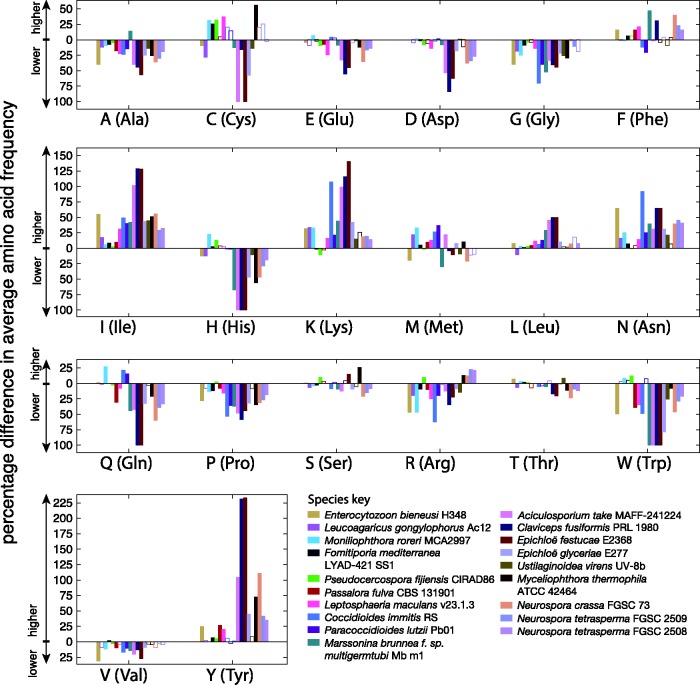
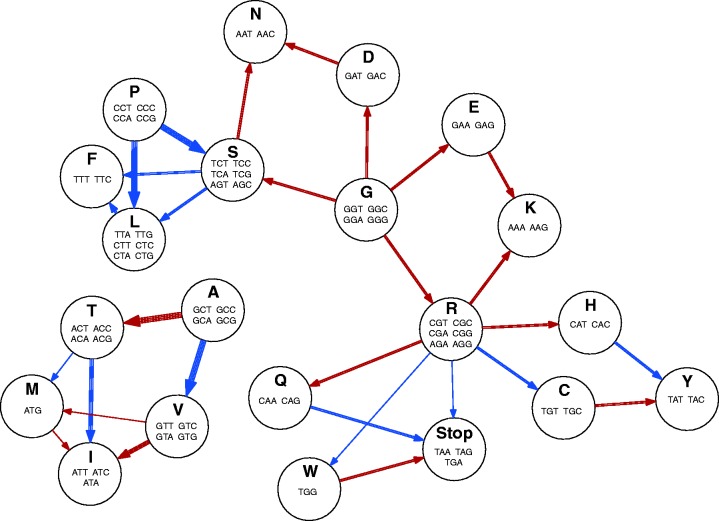
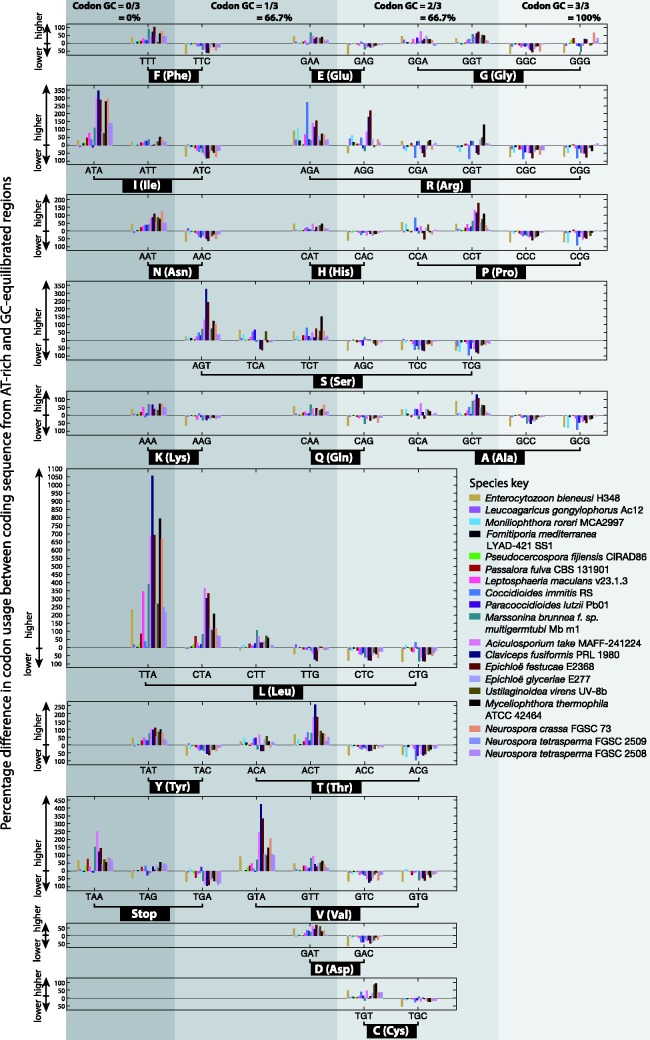
Percentage differences in the average amino acid content of coding sequences from genes within AT-rich regions compared with those in GC-equilibrated regions are shown per species in figure 8. Statistically significant differences (Mann–Whitney U tests, P value ≤ 0.05) in amino acid content were found in all species surveyed, with common trends across different species observable (fig. 8). If these differences occurred due to coding sequences from GC-equilibrated regions being subjected to nonsynonymous RIP-like C to T and G to A transitions, we can form expectations about how the resulting coding sequences in AT-rich regions would differ in amino acid composition. As an aid to understanding this, possible nonsynonymous changes (dN) are summarized in figure 7. In addition to illustrating how C to T and G to A transitions can alter one amino acid to another, this network highlights nodes which are absolute start and end points. Glycine, alanine and proline (G, A, and P) may undergo C to T and G to A changes to other amino acids, but can never be created by these mutations. As such, coding sequences subjected to C to T and G to A transitions are expected to have lower levels of these amino acids. Conversely, phenylalanine, tyrosine, lysine, isoleucine and asparagine (F, Y, K, I, and N) could be expected to increase in number. This appears as expected in the plots shown in figure 8. Both nonsynonymous and synonymous mutations within coding sequences can disrupt codon usage. The proportion of each codon encoding each of the amino acids was compared within and outside AT-rich regions within figure 9. In most cases codon usage in coding sequences within AT-rich regions favored codon choices higher in A and T than coding sequences in GC-equilibrated regions. The particularly strong percentage increases in codon usage were seen in codons containing TpA dinucleotides, which have been observed to be a typical product of RIP in noncoding regions.
Fig. 8.
Percentage differences between average amino acid content of genes within and outside AT-rich regions are shown. Different colors correspond to different fungal species as per the key (bottom right). Bars that have a solid fill color show where the difference in amino acid frequency is statistically significant (Mann–Whitney U test, P= 0.05). Percentage differences above y = 0 correspond to higher values within AT-rich regions and below y = 0 correspond to lower values within AT-rich regions (see axis labels).
Fig. 7.
Possible amino acid changes that can results from nonsynonymous C to T (blue) or G to A (red) transitions. The circular graph nodes represent the amino acids, and the edges represent a mutation resulting in a nonsynonymous change. An edge comprised of multiple lines indicates where multiple different codons of an amino acid can undergo the same amino acid change.
Fig. 9.
Percentage differences between codon usage frequencies in coding sequences within and outside AT-rich regions are shown. Percentage differences above y = 0 correspond to higher values within AT-rich regions and below y = 0 correspond to lower values within AT-rich regions (see axis labels). Each species is represented by different color as per the species key. Codons are grouped according to the amino acid they encode (see brackets and labels on the x-axis). The horizontal position of each plot is based on the GC-content of the codons, with low GC-content codons positioned to the left and higher GC-content codons to the right. Vertical grey bands and labels at the top of the figure show the codon GC-content.
In all but one case (E. bieneusi), genes of the analyzed species within AT-rich regions were of a shorter average length than those outside (table 1). This may be due to mutations causing nonsynonymous changes to certain codons encoding arginine, tryptophan, and glutamine to stop codons (fig. 7), thus shortening the open reading frame (Hane et al. 2015). This possibility is supported by trends of lower levels of these amino acids in proteins within AT-rich regions (fig. 8). Higher levels of secreted proteins in AT-rich regions were previously noted in studies of L. maculans (Rouxel et al. 2011), however, we did not find this was a common theme among the species listed in table 1. Only L. maculans and M. brunnea f. sp. multigermtubi were found to have a significantly enriched (by Fisher’s exact test, P value ≤ 0.05) complement of secreted proteins within AT-rich regions. Indeed, we note several species were significantly depleted in secreted proteins within their AT-rich regions (C. fusiformis, E. festucae, E. glyceriae, C. immitis, and M. roreri). A summary of enriched and depleted Pfam domains in genes/proteins within AT-rich regions compared with those outside (supplementary table S5, Supplementary Material online) did not highlight any notable enriched Pfam domains for most species. However, most species were enriched in proteins lacking conserved Pfam domains (by Fisher’s exact test, P ≤ 0.05)
Discussion
Links between RIP, bimodal genomes, transposon activity, and the evolution of fungal genomes have been made previously. Presented here is a facile method for determining the presence of AT-rich genome regions and a systematic survey of published fungal genomes. We show that far from unusual or unique, bimodal genomes are common in the fungal kingdom. Furthermore, for the first time, we have been able to compare how this genome type manifests in a diverse set of species and compare the gene content of AT-rich regions between species.
AT-Rich Region Formation
Our results are consistent with the hypothesis that AT-rich regions are generally formed by transposon invasion followed by mutation by RIP. The higher frequency of species with AT-rich genome regions within the Pezizomycontina (fig. 5) conforms to the taxonomic distribution of RIP, as previously reported by Clutterbuck (2011) and supports the role of RIP in the formation of these regions. The observed TpA dinucleotide bias in most species with AT-rich regions surveyed provides further evidence of this. Clusters of some closely related species harboring AT-rich regions may indicate that AT-rich regions either developed prior to speciation or that closely related species share a common predisposition for the development of AT-rich regions.
Several species known to be RIP competent were not found to have bimodal genomes. For example, in silico evidence supports RIP activity in Penicillium roqueforti (Ropars et al. 2012), and whereas small amounts of AT-rich genome segments are visible in the GC-content distribution, there is not a distinct AT-rich peak in the GC-content distribution. Previously reported in silico evidence also supports RIP-like activity in obligate biotrophs Melampsora laricis-populina and Puccinia graminia (Horns et al. 2012), from the subphylum Pucciniomycotina, albeit with different dinucleotide biases to those typically seen within the Pezizomycotina. Both these species show unimodal distributions. Therefore, while RIP appears to be necessary for the formation of AT-rich regions, evidence of RIP activity within a genome does not necessarily mean the genome is bimodal. The evidence of RIP can also be due to historical RIP and RIP may no longer be an active process. The presence of repeats is also required for AT-rich region formation though RIP activity, however without subsequent RIP these sequences are not necessarily distinguishable from the rest of the genome by their GC-content.
A number of additional factors that could affect AT-rich genome formation are not investigated in this survey. RIP occurs around the time of meiosis and as such the sexual cycle of the fungi is relevant to the presence of RIP affected sequences. We note that species thought to be predominantly asexual—Verticillium dahliae, Magnaporthe oryzae, Fusarium oxysporum, and most Metarhizium spp. (Taylor et al. 1999; Hu et al. 2014)—contained relatively low proportions of AT-rich regions (supplementary table S1, Supplementary Material online). Time between sexual cycles (and RIP cycles) could relate to how much repeats propagate throughout the genome prior to RIP deactivation. Reproductive frequency (both sexual and asexual) could also influence selection, as the burden of replicating a large genome increases with more frequent reproductive cycles. It is beyond the scope of the current study to gather detailed data on each species, not least because many sequenced species have not yet been studied in detail. Other factors affecting AT-rich region formation that are not investigated in this survey could relate to the efficiency of RIP and the level of activity of other transposon defence mechanisms such as DNA methylation (Miura et al. 2001) and RNA interference (Buchon and Vaury 2006; Cerutti and Casas-mollano 2006; Chung et al. 2008). The genomes we see are those that have survived and as such selection plays a part in the amount of AT-rich sequence we see.
Links to Evolution
Transposon activity contributes to genetic variability and diversification. RIP is primarily considered to be a fungal defence mechanism against the proliferation of repeats. In this survey, we saw a range of AT-rich region genome contents—i.e., the overall proportion of the genome comprised of AT-rich regions—from genomes comprised of ∼1% AT-rich regions, to extreme cases where AT-rich regions dominated the genome. Most examples of genomes identified with AT-rich regions were at the lower end of the spectrum where RIP has prevented the spread of repeats and could be thought of as evidence of selection favoring genome stability. At the higher end of the spectrum, we see some genomes have been extensively invaded by repeats prior to their deactivation by RIP, resulting bloated genomes with large components of AT-rich regions. Transposon proliferation and subsequent RIP has the potential to give a burst of diversification, provided both by the transposons and the RIP mutations, followed by a return to relative stability. Within the surveyed Pezizomycotina, bimodal genomes were similarly common within the genomes of saprobes, pathogens, and symbionts. However, in the genomes of symbionts and pathogens there was a shift toward higher components of AT-rich regions (fig. 5B).
An important consideration in our interpretation of these data is the bias in the subset of fungi that have been sequenced. Fungal species are not selected for study at random, but rather because of their industrial relevance or experimental tractability. For example, model organisms such as saprobic Pezizomycotina species N. crassa and A. nidulans were selected for stability. Many plant pathogens are sequenced because of their economic relevance and as such this group is biased toward pathogens that are particularly successful and thus more frequently employing effective evolutionary strategies. Further biases exist where sequencing has been motivated by the desire to conduct comparative genomics research, manifesting in clusters of genome sequences of closely related organisms.
It is not yet possible to know if a truly random sampling of the fungal kingdom would show the same trends seen here, however there are some reasons why high components of AT-rich regions might be more common in symbionts and pathogens. In both the symbionts and the pathogens, it appears to be species with a plant host that are largely responsible for this trend (fig. 5B). Both plant symbionts and pathogens need to evade host defences and are therefore under different selection pressures to saprobes. For example, plant pathogens frequently have an evolutionary history of host jumps and overcoming host resistance or control measures. Many of these fungi are cosmopolitan—they travel the world and are exposed to a variety of host resistance genes and fungicides—and only the fit survive. Genome dynamism in these species is key to their survival. This could explain why bursts of diversification that could come about with AT-rich region formation would be a successful evolutionary strategy in plant pathogens and thus a common theme to their genomes.
The GC-content distributions of the eight surveyed obligate biotroph plant pathogen species (Blumeria graminis f. sp. hordei, Blumeria graminis f. sp. tritici, Erysiphe necator, M. larici-populina, Melampsora lini, P. graminis f. sp. tritici, Puccinia striiformis f. sp. tritici and Uromyces viciae-fabae) were clearly unimodal. Considering the taxonomic distribution of RIP and that just three of the nine are Pezizomycotina species (B. graminis f. sp. hordei, B. graminis f. sp. tritici and E. necator), we cannot say whether unimodal genomes are common theme in obligate biotrophs. Only three nonobligate biotroph species from the Pezizomycotina were surveyed (A. take, P. fulva, and U. virens with 58.5%, 43.8%, and 34.6% AT-rich region composition, respectively) and as such we cannot reliably compare nonobligate biotrophs with the other plant pathogen groupings. Obligate biotrophs are known to have large, repeat-bloated genomes (Spanu 2012) with the set of assemblies included in this survey ranging in size from a large 82 Mb (B. graminis f. sp. tritici 96224; Wicker et al. 2013) to an enormous 210 Mb (U. viciae-fabae I2; Link et al. 2014). In fact, the genome of B. graminis f. sp. tritici was estimated to be 120 Mb (Wicker et al. 2013) and U. viciae-fabae between 330 and 379 Mb (Link et al. 2014), however due to difficulties assembling repetitive DNA the assemblies are significantly smaller. It has previously been suggested that in the evolution of obligate biotrophs a relaxation of constraints on transposon activity was advantageous as it allowed genetic variability. Notably, the genes necessary for RIP are absent from Blumeria spp., despite being common among other Pezizomycotina. Future sequencing projects may shed light on whether this is a common trend in obligate biotrophs and clarify any differing trends in the nonobligate biotrophs.
Although we have larger sets of hemibiotrophic and necrotrophic species within the Pezizomycotina that are more suitable for comparison (25 and 21, respectively), the distinction between these groups is contentious. Many of these pathogens have at one time or another been referred to in the published literature as both necrotrophs and hemibiotrophs. Hemibiotrophs have a longer latent phase than necrotrophs, but the exact length of latent phase that distinguishes these two states is not clearly defined. Within the surveyed Pezizomycotina, higher AT-rich region components were more common among the hemibiotrophic plant pathogens than the necrotrophs (fig. 5B, see supplementary table S1, Supplementary Material online, for individual species). Three species out of the 25 surveyed hemibiotrophic Pezizomycotina (L. maculans “brassicae”, Marssonina brunnea f. sp. multigermtubi, and Pseudocercospora fijiensis) had bimodal genomes with >30% AT-rich region content, whereas the highest AT-rich region content seen among the 21 necrotrophic Pezizomycotina was 11.8% (Macrophomina phaseolina MS6; Islam et al. 2012). These differences could relate to the length the latent phase of infection, although it is also possible that additional or unknown characteristics not documented in this survey may explain these differences.
As discussed in our introduction, coding sequences can be affected by RIP mutations either by “leakage” of RIP into neighboring nonrepeat regions or by being themselves duplicated and thus directly targeted by RIP. Our results support this, showing evidence that proteins within AT-rich regions have an amino acid composition consistent with them evolving under these conditions. The differences in amino acid frequency in coding sequences within AT-rich regions could support the idea that AT-rich regions contribute to the evolution of proteins with novel sequence and function. On the other hand, some nonsynonymous changes may be tolerated if they occur without affecting protein function. This may occur where amino acids are altered to those with similar properties.
Bioinformatic Challenges and Consequences
We note that several of the genomes with the highest AT-rich component also have high numbers of scaffolds (supplementary table S2, Supplementary Material online). This is likely due to the difficulty in assembling repetitive sequences, possibly compounded by amplification biases toward GC-equilibrated regions when using some sequencing platforms (Elhaik, Graur, Josic 2010; Ross et al. 2013). We placed no restrictions on genome assembly quality when selecting genomes for this survey, and it is likely that some low coverage short read assemblies have failed to capture AT-rich regions. We refer to the reader to the recent paper by Thomma et al. (2015) which highlights the benefits of generating more complete genomes and closing gaps in assemblies. As genomic resources are improved it will be interesting to see how our understanding of repetitive genome regions and AT-rich regions improves.
As demonstrated, AT-rich regions can have a distinctly lower GC-content than other genome regions and have different dinucleotide compositional biases. Results showing different amino acid usage and codon biases in AT-rich regions demonstrate that coding sequences in these regions also differ from the rest of the genome. These factors all affect gene prediction, which in many cases relies on patterns in short (typically 5 or 6 nucleotide) sequences of DNA. Ab initio gene prediction training is typically carried out on a training set of genes, which may result in parameters that are a poor match to sequences in AT-rich regions. Existing fungal specific approaches to gene prediction (Reid et al. 2014; Testa, Hane, Ellwood, et al. 2015) may offer a platform for addressing these issues.
Leveraging AT-Rich Region Annotation to Advance Fungal Bioinformatics and Crop Protection
This study has revealed the true extent of AT-rich regions across the fungal kingdom, with special emphasis on the Pezizomycotina. Theoretical analysis and analysis of the gene content of AT-rich regions has highlighted a tell-tale amino acid bias in RIP affected proteins. This phenylalanine, tyrosine, lysine, isoleucine and asparagine enriched and glycine, alanine and proline depleted (FYKIN-enriched, GAP-depleted) signature could be used to screen for effectors. Searching for FYKIN-enriched GAP-depleted proteins has the potential to locate candidate effectors that have evolved under RIP conditions but no longer have an AT-rich genomic context.
Knowledge of the locations of AT-rich regions within fungal genomes also allows additional metadata to be associated with gene loci, describing their genomic context and potential to accumulate RIP mutations through leakage. There are only a handful established examples of pathogenicity-related avirulence genes in fungi that conform to this pattern, therefore this method may yet highlight many new candidate pathogenicity genes. To this end, we present the software OcculterCut (available from https://sourceforge.net/projects/occultercut, last accessed April 30, 2016) that is capable of replicating the analyses presented herein as well as reporting gene annotations within and near AT-rich regions. Finally, current gene prediction methods typically rely on training based on the overall gene set or existing homologs, both of which are ill suited to accurate prediction of highly unique gene sets residing in AT-rich regions and/or with specialized roles in plant pathogen interactions. We anticipate that this new knowledge will open up avenues for the prediction of nonstandard genes, particularly fungal effector-like genes, which will be the subject of our continued investigations.
Supplementary Material
Supplementary tables S1–S7 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
An Australian Postgraduate Award (APA), funded by the Australian Federal Government, and a Grains Research Scholarship, funded by the Grains Research & Development Corporation (GRS10564), supported the studentship of A.C.T. Many thanks to Megan Meates for giving the article a final read through and polish. This work was funded by the Grains Research & Development Corporation.
Literature Cited
- Akiyoshi DE, et al. 2009. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 5:e1000261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Lipmann DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Aylward FO, et al. 2013. Leucoagaricus gongylophorus produces diverse enzymes for the degradation of recalcitrant plant polymers in leaf-cutter ant fungus gardens. Appl Environ Microbiol. 79:3770–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SE, et al. 2015. Draft genome sequence of Neurospora crassa strain FGSC’73. Genome Announcements 3:2012–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka RM, et al. 2011. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 29:922–927. [DOI] [PubMed] [Google Scholar]
- Bernaola-Galván P, Román-Roldán R, Oliver J. 1996. Compositional segmentation and long-range fractal correlations in DNA sequences. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Top. 53:5181–5189. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 2000. Isochores and the evolutionary genomics of vertebrates. Gene 241:3–17. [DOI] [PubMed] [Google Scholar]
- Bernardi G, et al. 1985. The mosaic genome of warm-blooded vertebrates. Science 228:953–958. [DOI] [PubMed] [Google Scholar]
- Buchon N, Vaury C. 2006. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96:195–202. [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinform. 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri EB, Singer MJ, Selker EU. 1991. Recurrence of repeat-induced point mutation. Genetics April(127):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Casas-mollano JA. 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 50:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-J, Okamura K, Martin R, Lai EC. 2008. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 18:795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cissé OH, Pagni M, Hauser PM. 2013. De novo assembly of the pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P, et al. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71–77. [DOI] [PubMed] [Google Scholar]
- Clutterbuck AJ. 2011. Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet Biol. 48:306–326. [DOI] [PubMed] [Google Scholar]
- Coleman JJ, et al. 2009. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5:e1000618.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C, et al. 2013. Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: toward a new infection model system. J. Proteome Res. 12:2552–2570. [DOI] [PMC free article] [PubMed]
- Costantini M, Alvarez-Valin F, Costantini S, Cammarano R, Bernardi G. 2013. Compositional patterns in the genomes of unicellular eukaryotes. BMC Genomics 14:755.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, McDonald BA. 2012. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 8:e1002608.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Zala M, McDonald BA. 2013. Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet. 9:e1003567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, et al. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317:1400–1403. [DOI] [PubMed] [Google Scholar]
- de Wit PJGM, et al. 2012. The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8:e1003088.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ. 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986. [DOI] [PubMed] [Google Scholar]
- Desjardins CA, et al. 2011. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 7:e1002345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon B, et al. 2015. Horizontal gene transfer and gene dosage drives adaptation to wood colonization in a tree pathogen. Proc Natl Acad Sci U S A. 112:3451–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhaik E, Graur D, Josic K. 2010. Comparative testing of DNA segmentation algorithms using benchmark simulations. Mol Biol Evol. 27:1015–1024. [DOI] [PubMed] [Google Scholar]
- Elhaik E, Graur D, Josić K, Landan G. 2010. Identifying compositionally homogeneous and non-homogeneous domains within the human genome using a novel segmentation algorithm. Nucleic Acids Research 38:e158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, et al. 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189:55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman ML. 2007. Telomeres in the rice BLAST fungus Magnaporthe oryzae: the world of the end as we know it. FEMS Microbiol Lett. 273:125–132. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floudas D, et al. 2012. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. [DOI] [PubMed] [Google Scholar]
- Freitag M, Williams RL, Kothe GO, Selker EU. 2002. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. PNAS 99:8802–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudal I, et al. 2009. Repeat-induced point mutation (RIP) as an alternative mechanism of evolution toward virulence in Leptosphaeria maculans. Mol Plant Pathol. 22:932–941. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. [DOI] [PubMed] [Google Scholar]
- Galagan JE, et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, et al. 2012. Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8:e1002952.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioti A, et al. 2013. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio. 4:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, et al. 1996. Life with 6000 genes. Science 274:546–567. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, et al. 2011. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7:e1002070.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout L, et al. 2006. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 60:67–80. [DOI] [PubMed] [Google Scholar]
- Graïa F, et al. 2001. Genome quality control: RIP (repeat-induced point mutation) comes to Podospora. Mol Microbiol. 40:586–595. [DOI] [PubMed] [Google Scholar]
- Hane JK, Oliver RP. 2008. RIPCAL: a tool for alignment-based analysis of repeat-induced point mutations in fungal genomic sequences. BMC Bioinform. 9:478.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane JK, Oliver RP. 2010. In silico reversal of repeat-induced point mutation (RIP) identifies the origins of repeat families and uncovers obscured duplicated genes. BMC Genomics 11:655.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane JK, et al. 2011. A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 12:R45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane JK, Williams AH, Taranto AP, Solomon PS, Oliver RP. 2015. Genetic transformation systems in fungi. Fungal Biol. 2:55–68. [Google Scholar]
- Heitman J, Sun S, James TY. 2013. Evolution of fungal sexual reproduction. Mycologia 105:1–27. [DOI] [PubMed] [Google Scholar]
- Horns F, Petit E, Yockteng R, Hood ME. 2012. Patterns of repeat-induced point mutation in transposable elements of basidiomycete fungi. Genome Biol Evol. 4:240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, et al. 2013. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin Sci Bull. 58:2846–2854. [Google Scholar]
- Hu X, et al. 2014. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc Natl Acad Sci U S A. 111:16796–16801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Howlett BJ. 2003. Analysis of loss of pathogenicity mutants reveals that repeat-induced point mutations can occur in the Dothideomycete Leptosphaeria maculans. Fungal Genet Biol. 39:31–37. [DOI] [PubMed] [Google Scholar]
- Ikeda K-i, et al. 2002. Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Mol Microbiol. 45:1355–1364. [DOI] [PubMed] [Google Scholar]
- Irelan JT, Hagemann AT, Selker EU. 1994. High frequency repeat-induced point mutation (RIP) is not associated with efficient recombination. Genetics 138:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, et al. 2012. Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13:493.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110:462–467. [DOI] [PubMed] [Google Scholar]
- Link T, Seibel C, Voegele RT. 2014. Early insights into the genome sequence of Uromyces fabae. Front Plant Sci. 5:587.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, et al. 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol. 66:513–545. [DOI] [PubMed] [Google Scholar]
- Martinez D, et al. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 22(6):695–700. [DOI] [PubMed] [Google Scholar]
- Meerupati T, et al. 2013. Genomic mechanisms accounting for the adaptation to parasitism in nematode-trapping fungi. PLoS Genet. 9:e1003909.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt LW, et al. 2014. Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases. BMC Genomics 15:164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A, et al. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212–214. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Nishimoto N, Ikeda K., Tosa Y, Mayama S. 1999. Degenerate MAGGY elements in a subgroup of Pyricularia grisea: a possible example of successful capture of a genetic invader by a fungal genome. Mol Gen Genet. 261:958–966. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators 2014. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 43:6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm RA, et al. 2012. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8:e1003037.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JL, Bernaola-Galván P, Carpena P, Román-Roldán R. 2001. Isochore chromosome maps of eukaryotic genomes. Gene 276:47–56. [DOI] [PubMed] [Google Scholar]
- Oliver JL, Carpena P, Hackenberg M, Bernaola-Galván P. 2004. IsoFinder: computational prediction of isochores in genome sequences. Nucleic Acids Res. 32:W287–W292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver R. 2012. Genomic tillage and the harvest of fungal phytopathogens. New Phytol. 196:1015–1023. [DOI] [PubMed] [Google Scholar]
- Pan G, et al. 2013. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics 14:186.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8(10):785–786. [DOI] [PubMed] [Google Scholar]
- Pryszcz LP, Németh T, Gácser A, Gabaldén T. 2014. Genome comparison of Candida orthopsilosis clinical strains reveals the existence of hybrids between two distinct subspecies. Genome Biol Evol. 6:1069–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Kamoun S. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol. 10:417–430. [DOI] [PubMed] [Google Scholar]
- Raffaele S, et al. 2010. Genome evolution following host jumps in the irish potato famine pathogen lineage. Science 330:1540–1544. [DOI] [PubMed] [Google Scholar]
- Reid I, et al. 2014. SnowyOwl: accurate prediction of fungal genes by using RNA-Seq and homology information to select among ab initio models. BMC Bioinform. 15:229.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J, et al. 2012. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. PloS One 7:e49665.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MG, et al. 2013. Characterizing and measuring bias in sequence data. Genome Biol. 14:R51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel T, et al. 2011. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat Commun. 2:202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, et al. 2013. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet. 9:e1003323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Cambareri EB, Jensen BC, Haack KR. 1987. Rearrangement of duplicated DNA in specialized cells of neurospora. Cell 51:741–752. [DOI] [PubMed] [Google Scholar]
- Sharpton TJ, et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19:1722–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD. 2012. The genomics of obligate (and nonobligate) biotrophs. Annu Rev Phytopathol. 50:91–109. [DOI] [PubMed] [Google Scholar]
- Storck R. 1965. Nucleotide composition of nucleic acids of fungi. J Bacteriol. 91(1):227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B-F, et al. 2013. Multiple interkingdom horizontal gene transfers in pyrenophora and closely related species and their contributions to phytopathogenic lifestyles. PloS One 8:e60029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Fisher MC. 1999. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol. 37:197–246. [DOI] [PubMed] [Google Scholar]
- Testa A, Oliver R, Hane J. 2015. Overview of genomic and bioinformatic resources for Zymoseptoria tritici. Fungal Genet Biol. 79:13–16. [DOI] [PubMed] [Google Scholar]
- Testa AC, Hane JK, Ellwood SR, Oliver , Richard P. 2015. CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics 16:170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, et al. 2015. Mind the gap; seven reasons to close fragmented genome assemblies. Fungal Genet Biol. 90:24–30. [DOI] [PubMed] [Google Scholar]
- Tzeng TH, Lyngholm LK, Ford CF, Bronson CR. 1992. A restriction fragment length polymorphism map and electrophoretic karyotype of the fungal maize pathogen Cochliobolus heterostrophus. Genetics. 130(1):81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wouw AP, et al. 2010. Evolution of linked avirulence effectors in Leptosphaeria maculans is affected by genomic environment and exposure to resistance genes in host plants. PLoS Pathog. 6:e1001180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters MK, Randall , Thomas A, Margolin BS, Selker EU, Stadier DR. 1999. Action of repeat-induced point mutation on both strands of a duplex and on tandem duplications of various sizes in neurospora. Genetics 153(2):705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Sanger Institute. 2015. GFF (General Feature Format) specifications document. [cited 2016 Apr 30]. Available from: http://www.sanger.ac.uk/resources/software/gff/.
- Wicker T, et al. 2013. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat Genet. 45:1092–1096. [DOI] [PubMed] [Google Scholar]
- Xu X-H, et al. 2014. The rice endophyte Harpophora oryzae genome reveals evolution from a pathogen to a mutualistic endophyte. Sci Rep. 4:5783.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. 2014. Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat Commun. 5:3849.. [DOI] [PubMed] [Google Scholar]
- Zheng P, et al. 2011. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 12:R116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, et al. 2012. Sequencing the genome of Marssonina brunnea reveals fungus-poplar co-evolution. BMC Genomics 13:382.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan ME. 1995. Chromosome-length polymorphism in fungi. Microbiol Rev. 59:686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.