Abstract
Neisseria meningitidis is an important cause of meningococcal disease globally. Sequence type (ST)-11 clonal complex (cc11) is a hypervirulent meningococcal lineage historically associated with serogroup C capsule and is believed to have acquired the W capsule through a C to W capsular switching event. We studied the sequence of capsule gene cluster (cps) and adjoining genomic regions of 524 invasive W cc11 strains isolated globally. We identified recombination breakpoints corresponding to two distinct recombination events within W cc11: A 8.4-kb recombinant region likely acquired from W cc22 including the sialic acid/glycosyl-transferase gene, csw resulted in a C→W change in capsular phenotype and a 13.7-kb recombinant segment likely acquired from Y cc23 lineage includes 4.5 kb of cps genes and 8.2 kb downstream of the cps cluster resulting in allelic changes in capsule translocation genes. A vast majority of W cc11 strains (497/524, 94.8%) retain both recombination events as evidenced by sharing identical or very closely related capsular allelic profiles. These data suggest that the W cc11 capsular switch involved two separate recombination events and that current global W cc11 meningococcal disease is caused by strains bearing this mosaic capsular switch.
Keywords: capsule switch, horizontal gene transfer, serogroup W135
Introduction
Polysaccharide capsule is the most important virulence determinant of Neisseria meningitidis, a normal commensal of the human nasopharynx that occasionally causes invasive meningococcal disease (IMD) (Stephens 2009). Even though un-encapsulated meningococci are common in asymptomatic carriage, they are very rarely associated with IMD (Mackinnon et al. 1993). The capsule is the target of most meningococcal vaccines and its biochemical and genetic properties form the basis of classifying meningococci into capsular groups (serogroups). Out of 12 capsular groups, groups A, B, C, W, X and Y cause almost all cases of IMD (Stephens 2009).
Sequence type (ST)-11 clonal complex (cc11), a hypervirulent meningococcal lineage, is a leading cause of IMD across all six continents (Caugant 1998; Harrison et al. 2009). Cc11 is a highly genetically diverse lineage associated with IMD caused by capsular serogroups C, B, W, and less commonly serogroup Y (Caugant 1998; Lucidarme et al. 2015). Serogroup C cc11 strains were associated with endemic IMD cases beginning in the 1960s, caused epidemics in the African “meningitis belt” in the 1970s, and small outbreaks globally (Broome et al. 1983; Caugant 1998; Halperin et al. 2012). Serogroup W cc11 was uncommon among IMD strains in the 1970s–1990s (Mustapha et al. 2016). The first epidemic of W cc11 occurred in Mecca, Saudi Arabia among Hajj pilgrims and their close contacts in 2000 (Taha et al. 2000). Since 2000, W cc11 has emerged as a leading cause of epidemic IMD in the meningitis belt, whereas endemic clusters have emerged in South America, Middle East, Europe, and China in 2000–2015 (Mustapha et al. 2016).
The capsule gene cluster, cps, is a 24-kb genetic island horizontally acquired by N meningitidis that is not present in closely related N. lactamica and N. gonorrhoeae (Dunning Hotopp et al. 2006; Lam et al. 2011). A recent study described cps architecture and gene content (Harrison et al. 2013). All known meningococcal serogroups had cps containing five gene regions involved in capsule synthesis, transport and assembly (cps regions A–E) (Harrison et al. 2013). Serogroups B, C, W and Y have capsules containing sialic acid residues (Swartley et al. 1997; Harrison et al. 2013; Romanow et al. 2014). The transferase gene, located in region A, dictates linkages and thus determines serogroup phenotype (Claus et al. 1997; Harrison et al. 2013). Capsular serogroup diversity among meningococci within the same clonal complex is driven by “capsular switching”—lateral exchange of capsule biosynthetic genes through homologous recombination (Swartley et al. 1997; Kong et al. 2013). For a recombination event to result in change of capsular phenotype, allelic changes must include region A genes, particularly the transferase gene (Swartley et al. 1997; Romanow et al. 2013). Meningococcal capsular switching is relatively common and has been associated with the emergence and persistence of IMD (Swartley et al. 1997; Beddek et al. 2009; Harrison et al. 2010; Castineiras et al. 2012). Capsular switching is also of public health relevance given that majority of meningococcal vaccines target the capsule (Cohn and Harrison 2013).
Recent phylogenomic study of 750 meningococcal cc11 isolates (Lucidarme et al. 2015) revealed a highly complex population structure with extensive genetic diversity among cc11 strains. Serogroup C cc11 strains formed several clusters linked to multiple epidemiological instances of serogroup C disease; B strains were interspersed among serogroup C clusters; whereas serogroup W formed a distinct phylogenetic branch that was not interspersed with either B or C strains. Additionally, W cc11 lineage was genetically heterogeneous, containing diverse phylogenetic clusters associated with the following: (1) the Hajj clone, (2) endemic W cc11 strains unrelated the Hajj clone in Africa, and (3) a distinct South American W cc11 strain in Brazil, Argentina, Chile that has also emerged in United Kingdom (Lucidarme et al. 2015; Mustapha et al. 2015, 2016).
Given the preponderance of serogroup C among cc11 strains, it is regarded by some to be the founder capsule type with serogroups W, B and Y representing capsular switch variants (Kelly and Pollard 2003; Mueller et al. 2006; Mustapha et al. 2016). However, the direction and exact genes involved in capsular switching have not been established. The purpose of this study is to analyze capsular gene sequences from a global collection of strains to characterize the capsular switching events associated with W cc11.
Results
W cc11 Reference Strain, M7124 Contains Mosaic Recombinant Sequences
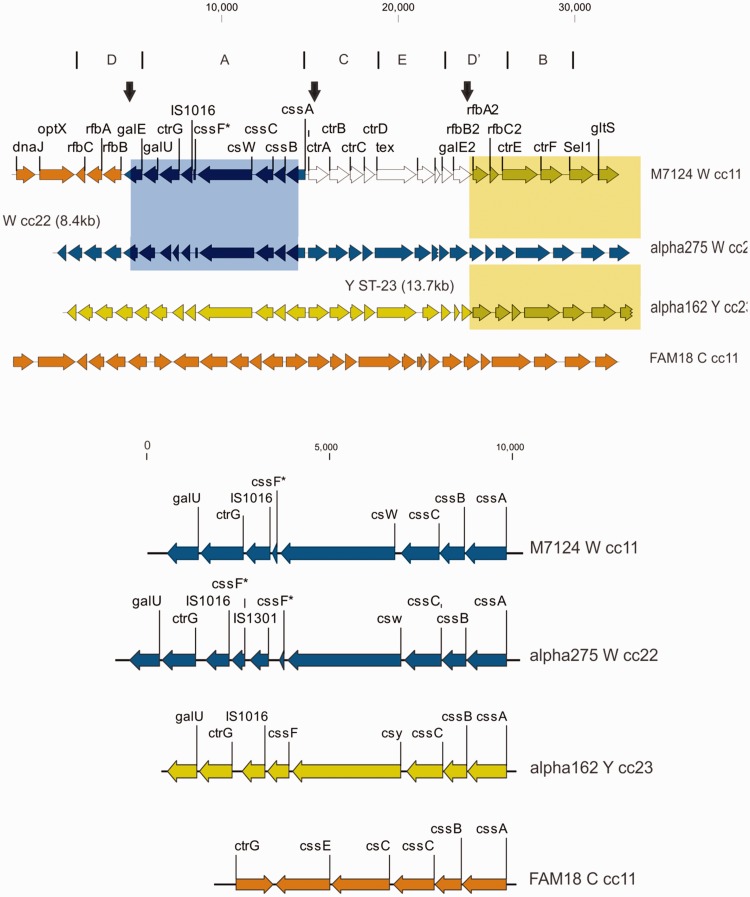
The cps locus of M7124, a previously characterized W ST11 Hajj clone reference strain (Mustapha et al. 2015), is 27 kb in length with genes organized into regions D–A–C–E–D′–B (fig. 1) similar to that described for other meningococcal lineages (Harrison et al. 2013).
Fig. 1.
Top panel: Capsule gene cluster (cps) for W cc11 reference strain M7124 and corresponding genes for reference strains α-275 (W ST-22), α-162 (Y ST-23) and FAM18 (C ST-11). Cps is comprised of five regions A–E; synteny and gene content is divergent within region A (see below) but conserved within regions B–E among listed reference strains. Two recombination events are depicted: (1) an 8.4-kb W cc22 donor fragment (blue rectangle) corresponding to α-275 reference genome. This recombinant segment includes all region A genes; (2) a 13.7-kb Y cc23 donor fragment corresponding to α-162 reference genome (yellow rectangle). This recombinant fragment includes parts of cps region D′, entire region B and adjoining sequences downstream of cps. Downstream end of this recombinant segment, outside of cps, is truncated for clarity; Vertical (black) arrows indicate recombination breakpoints within galE, cssA and rfbA2. Bottom panel: Gene content within cps region A among meningococcal reference strains. cssA-C genes are syntenic in all strains; sialyl transferase gene is divergent with a 1.5-kb csc in serogroup C, whereas the csw and csy genes for serogroups W and Y are both 3.1 kb in size. In addition, W ST-11 further differs from C ST-11 by having truncated O-acetyl transferase, cssF gene (*) fragments, one or more IS elements, reverse orientation of ctrG and presence of the galU gene.
When M7124 cps sequences were compared with other cps reference sequences, a number of abrupt changes in sequence similarity consistent with distinct recombination events were identified. First, M7124 shares very high sequence similarity with the serogroup W cc22 reference strain α-275 over a 8.4-kb segment that includes all cps region A genes (figs. 1 and 2, panel II). Only three nucleotide differences are evident between M7124 and α-275 over the entire 8.4-kb segment (fig. 1). Flanking this segment of high sequence similarity are two areas of sharp divergence in sequence similarity in keeping with recombination breakpoints at nucleotide positions 858 and 804 on galE and cssA genes, respectively (fig. 3A and B). In contrast, there were >1255 and 52 nucleotide differences between M7124 and α-275 within 5-kb upstream and downstream of the respective recombination breakpoints (data not shown). A recombination event with W cc22 as the likely donor strain is supported by alignment of galE and cssA gene sequences (fig. 3A and B). Phylogenetic networks (fig. 2, panel II) of meningococcal reference sequences across cps region A demonstrate that M7124 sequences were phylogenetically indistinguishable from corresponding W cc11 (WUE171) and W cc22 (α-275) strains, providing further evidence of this recombination event. Genes involved in this recombination event (listed from 5′ to 3′) included galE (partial), galU, ctrG, cssF, csw, cscC, cssB and cssA (partial) (fig. 1). The transferase gene, csw acquired within this recombination is responsible for capsular serogroup W phenotype (fig. 1).
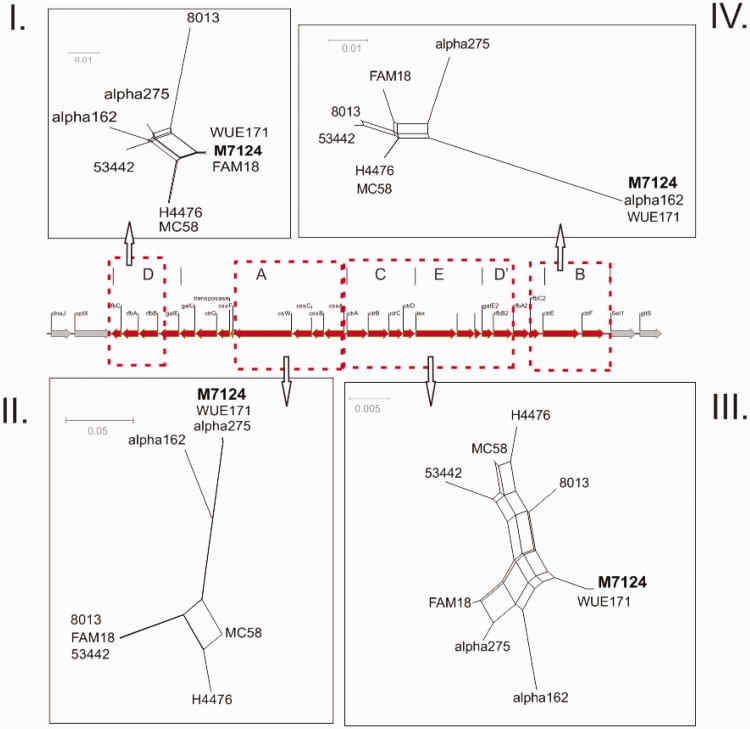
Fig. 2.
Phylogenetic relationships of M7124, W cc11 to cps from meningococcal reference genomes. Aligned rfbC1-rfbB1 genes (region D, panel I) demonstrate very close phylogenetic relationship between W cc11 (M7124 and WUE171) and serogroup C cc11 (FAM18). Phylogeny of aligned region A genes (panel II) shows that W cc11 (M7124 and WUE171) cluster with serogroup W cc22 (α-275). Across regions C, E and upstream parts of region D′ (panel III) W cc11 sequences do not cluster with any of the reference strains. While aligned region B sequences (panel IV) demonstrate clustering between W cc11 and Y cc23 (α-162). WUE171 serogroup W cc11; FAM18, C cc11; MC58, B cc32; H4476, B cc41/44 α-275, W cc22; 8013 C cc18; 53442 C cc4821; and α-162 Y cc23. Dotted red boxes depict margins of sequence alignments used to generate corresponding phylogenetic network.
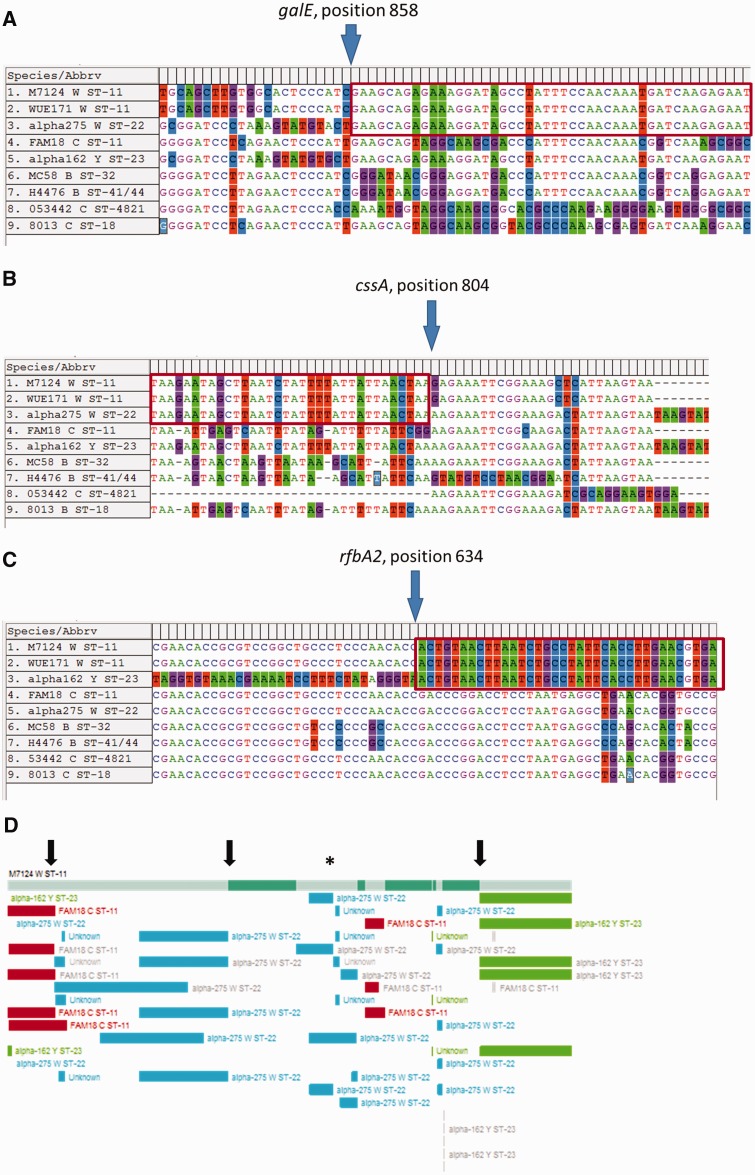
Fig. 3.
Recombination breakpoints within M7124 capsule. (A) Sequence alignment of galE genes showing recombination breakpoint at position 858 relative to W cc22 strain, α-275. (B) Sequence alignment of cssA genes demonstrating breakpoint at position 804 relative to α-275. (C) Sequence alignment of rfbA2 and rfbC2 showing recombination breakpoint within rfbA2 position 634. (D) Recombination breakpoints detected by at least one of seven methods included in RDP v4.56. The topmost horizontal bar represents M7124 cps sequence; three black arrows correspond to locations of breakpoints identified in (A–C) above; asterisk marks location of multiple small (0.1–1.5 kb) putative mosaic recombinant segments that were not supported by visual examination of aligned sequences (see main text). Topmost horizontal bar represents W cc11; colored boxes represent donor sequences from Y cc23 (green), W cc22 (blue) and C cc22 (red).
To further investigate whether W cc22 was the potential donor lineage, we assessed the prevalence of the recombinant sequence among 55 W ST-22 genome sequences in the PubMLST database (http://pubmlst.org/neisseria/). Eighty percent (44/55) of W ST-22 strains shared a common cssB-C, csw and ctrG allelic profile differing from M7124 by only two nucleotide substitutions within csw. These data demonstrate that this upstream recombination is predominant among W cc22 strains.
Second, M7124 shares a 13.7-kb region of very high sequence similarity with the serogroup Y cc23 strain, including 4.5 kb of cps and 9.2 kb downstream of the cps cluster (fig. 1). There was 100% nucleotide sequence identity between M7124 and Y cc23 reference strain α-162 over this region. This region of 100% sequence identity contrasts with the significant sequence divergence observed within the sequences flanking the 13.7-kb region. The upstream breakpoint corresponds to position 634 of rfbA2 gene (figs. 1, 2, and 3C), whereas a downstream breakpoint is located outside the cps cluster at the 5′ end of pykA gene (data not shown). The entire region B (ctrE and ctrF) and parts of region D′ (rfbA2 and rfbC2) were part of this recombinant segment (figs. 1 and 2) which also extends beyond cps and includes the genes encoding a sodium/glucose transport protein (gltS), a pyruvate kinase (pykA), tetratricopeptide repeat (sel1), and several hypothetical proteins. Phylogenetic network analysis of aligned region B among reference cps sequences demonstrates that M7124, WUE171 (W cc11) and α-162 (Y cc23) formed a phylogenetic cluster that was distinct from other meningococcal reference strains (fig. 2, panel IV).
We also compared M7124 ctrE and ctrF genes to 201 Y cc23 genome sequences from the PubMLST database. Over 88% (177/201) of all Y cc23 isolates shared identical ctrE and ctrF gene sequences with M7124 demonstrating that this allelic combination is predominant among Y cc23 strains. Taken together these data support the presence of a 13.7-kb recombinant segment that extends downstream of cps with Y cc23 as the most likely donor lineage.
All three recombination break points within cps were confirmed by automated detection of recombination using RDP v4.56 (fig. 3D) (Martin et al. 2010). In addition, one-third 3.0-kb recombinant fragment within cps region C involving ctrB–D genes was suggested by RDP analysis. This putative recombinant fragment was ruled out following visual inspection of aligned sequences that found 39 nucleotide differences between M7124 and the putative donor, α-275 (W ST-22).
M7124 Recombination Pattern Is Highly Conserved among Global W cc11 Strains Isolated from 1970 to 2014
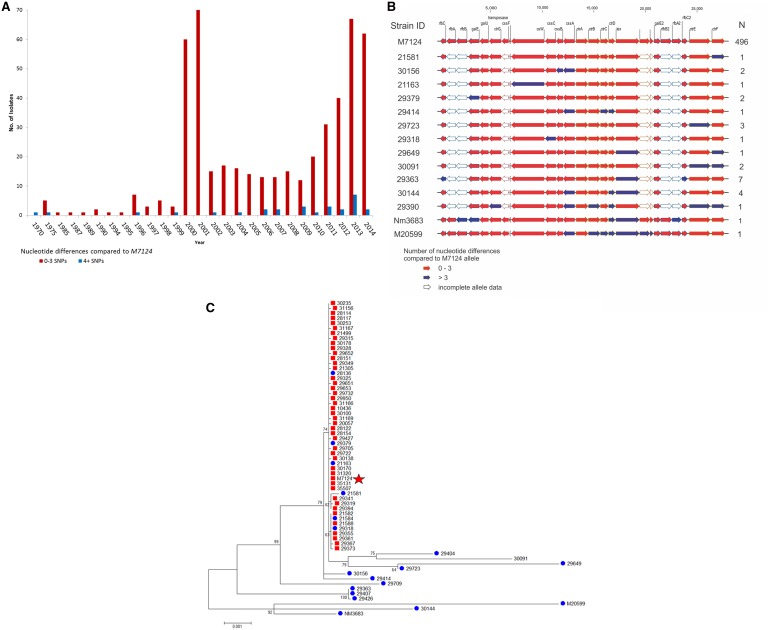
To assess what proportion of W cc11 strains contained this M7124 mosaic recombination, sequence comparison of 16 cps alleles (ctrA–G, cssA–C, csw, tex, galE, galE2, rfbC and rfbC2) from 524 W cc11 strains isolated from 21 different countries was performed (supplementary table, Supplementary Material online). The majority of W cc11 strains (63.9%; 334/523) shared identical cps allelic profiles with M7124, whereas 31.0% (162/523) differed by ≤3 point mutations across all 16 cps genes. Thus, the majority (95%, 496/523) of sequenced W cc11 strains representing a global collection isolated from 1970 to 2014 shared the same cps gene structure as M7124 (fig. 4A and B). A phylogeny of contiguous cps gene sequences shows a single cluster highly related to M7124 (fig. 4C, red boxes). While some strains with >3 SNPs relative to M7124 belonged to the same cluster, the majority were distinct from M7124 (fig. 4C, blue dots).
Fig. 4.
Summary of W cc11 cps allelic profiles. (A) Graphic depiction of 524 cps allelic profiles relative to M7124. Red arrows: genes with ≤3 nucleotide differences; blue arrows: genes with >3 nucleotide differences; white boxes: genes with incomplete sequence data. Strain ID: representative strain within each cps allelic profile. N: number of isolates. (B) Distribution of W cc11 cps allelic profile relative to M7124 over time; Red bars: strains with ≤3 nucleotide differences; blue bars: strains with >3 nucleotide differences. (C) Maximum likelihood tree depicting phylogenetic relationship between selected W cc11 cps sequences. Red squares: strains with ≤3 nucleotide differences relative to M7124 (red star); blue dots: strains with ≥4 nucleotide differences.
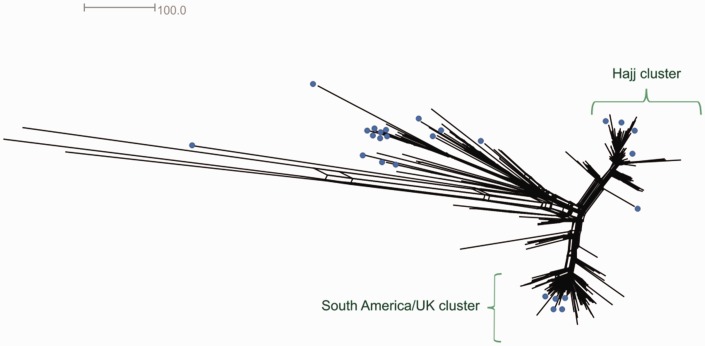
Twenty-eight isolates (28/523, 5.4%) with nonM7124 cps gene structure were further examined. All 28 isolates had one or more cps genes that exhibited cps allelic shift, defined as >3 nucleotide differences within a single gene (fig. 4a, blue boxes). Thirteen isolates (46.4%, 13/28) had allelic shift in a single cps gene, whereas the remaining 16 isolates had two to seven genes with allelic shift (fig. 4a,blue rectangles). Also, seven isolates collected from South Africa in 2006–2013 each had three genes with allelic shift and shared a common cps allelic combination suggesting geographically limited clonal spread (fig. 4A and supplementary table, Supplementary Material online). Twenty-two remaining strains with one or more allelic shifts were geo-temporally and phylogenetically diverse without any predominant allelic profile (fig. 4B and C and supplementary table S1, Supplementary Material online). Interestingly, all 28 isolates with cps allelic shift contained at least one of the two recombinant sequences identified in M7124. Core genome phylogenetic network of 524 W cc11 isolates demonstrated that strains with cps allelic shift were interspersed among strains with M7124 cps type (fig. 5, blue dots) in keeping with sporadic rather than clonal pattern of allelic shift.
Fig. 5.
Core genome phylogenetic network of 524 W cc11 strains reconstructed using neighbor-net algorithm on SplitsTree v4. Twenty-eight strains with cps allelic shift relative to M7124 are represented by blue dots.
There was no statistically significant difference in proportion of M7124 allelic type among W cc11 strains isolated in 1970–1999 compared with 2000–2014 (87.9% vs. 95.1%, P = 0.09). Likewise, excluding United Kingdom and South Africa strains does not change the proportion of isolates with M7124 cps allelic type (96.2% vs. 94.7% P = 0.6). These data suggest that observed dominance of M7124 cps allelic type among W cc11 isolates is unlikely to be a result of clonal expansion of the Hajj clone in 2000 or an artifact caused by the large proportion of isolates from United Kingdom and South Africa.
Discussion
In this study, we present detailed characterization of the capsule gene locus, the primary meningococcal virulence determinant within the context of globally emergent hypervirulent serogroup W cc11 lineage. Our results suggest that W cc11 acquired the capsule polymerase (csw) gene along with other region A genes from a W cc22 strain and capsule translocation genes from a strain belonging to the Y cc23 lineage. Also, an overwhelming majority of W cc11 strains had cps allelic profiles very closely related to M7124 suggesting a shared recombinant capsule structure from a common ancestor. Strains with one or more divergent alleles were a very small minority but were observed sporadically from 1970 to 2014. These data are consistent with recent genome sequencing studies showing that all W cc11 strains belonged to a single lineage that is phylogenetically distinct from serogroup C and B cc11 lineages (Lucidarme et al. 2015). Also, these data raise the possibility of a multi-step C to W switch involving a serogroup Y strain. Detailed characterization of global C, B and Y capsular genes is needed to address these questions.
We propose that the Hajj clone and a vast majority of W cc11 strains acquired an 8.4-kb recombinant fragment from serogroup W cc22 lineage. This recombination event includes transferase gene (csw) that mediates formation of (α2→6) sialic acid and glucose heteropolymers characteristic of serogroup W capsule (Harrison et al. 2013). The observed 8.4 kb recombinant fragment in this study is consistent with the finding of a 9-kb allelic exchange of cps regions resulting in a serogroup B to C ST-32 capsular switch among isolates from a serogroup B epidemic in Oregon (Swartley et al. 1997); and a 12-kb recombination involving the entire cps region A and flanking genes within region D and C leading to a switch from serogroup A ST-7 to C in China (Wang et al. 2010). These studies highlight that even though allelic exchange within a single gene (sialyl-transferase) may be sufficient to cause capsular switching, large recombination events affecting several genes are common.
We also identified an additional recombination event involving genes associated with capsule translocation (ctrE, ctrF) from Y cc23 donor lineage. This recombination event did not involve capsule synthesis genes in region A and therefore had no corresponding change in capsular serogroup. However, even without an obvious change in capsular phenotype, such allelic exchanges outside region A could affect meningococcal virulence by altering capsule transport and modification (Tzeng et al. 2005). For example, ctrE is up-regulated during meningococcal invasion of human cells (Spinosa et al. 2007) so recombination within this gene has the potential to enhance virulence through enhanced intracellular survival.
The very high degree of sequence conservation among W cc11 capsule genes markedly contrasts with the substantial global and temporal variability in W cc11 disease incidence patterns (Mustapha et al. 2016), a lineage associated with both sporadic disease (Molling et al. 2001; Zhou et al. 2013) and epidemics (Taha et al. 2000; MacNeil et al. 2014). Meningococci are associated with extensive genetic diversity through homologous recombination, phase variation and point mutations (Holmes et al. 1999; Hao et al. 2011; Kong et al. 2013). In fact, our previous study demonstrated that the epidemic W cc11 strain, “Hajj clone,” and its descendants acquired virulence gene alleles outside of the cps cluster, through a set of unique recombination events affecting factor H binding protein (fHbp), nitric oxide reductase (nor) and nitrite reductase (aniA) genes (Mustapha et al. 2015). Taken together, these studies suggest that although a unique, mosaic, recombinant capsule locus is a common feature across most hypervirulent W cc11 strains, recombination across other genomic loci mediate changes in strain virulence over time.
To our knowledge, these data represent the first description of allelic coupling across a large number of cps genes. Such extensive allelic coupling was unexpected given that the horizontal transfer of a single cps region A gene allele (csw) from W cc22 was sufficient to cause phenotypic change from an ancestral C to W serogroup (Claus et al. 1997; Harrison et al. 2013; Swartley et al. 1997). Significance of the second recombination from Y cc23 could be to offset any fitness cost associated with the first (W cc22) recombination event. This is supported by the observation that W cc11 represents a rare example where a capsular switch strain has persisted for decades and spread to several continents, as compared with a majority of capsular switch strains that do not persist in the long term (Harrison et al. 2010; Barroso et al. 2013; He et al. 2014; Zhu et al. 2015). Dominance of single cps allelic type is consistent with the “genocloud” concept in which a set of mutually co-adapted sets of genes persist despite presence of a few, mostly transient, escape variants (Gupta et al. 1996; Zhu et al. 2001). In general, DNA uptake sequences (Duffin and Seifert 2010) and restriction modification systems (Budroni et al. 2011; Tibayrenc and Ayala 2015) are known to affect the rate, extent and donor specificity of recombination in Neisseria. However, the precise role of these factors in capsular switching is yet to be established.
A limitation of our study is that we were unable to identify intermediate N. meningitidis isolates that had only one of the two recombinant sequences. These intermediate strain(s) may have lacked the biologic fitness to survive for several decades (Zhu et al. 2001). Also, our method of assigning donor lineages is, by design, conservative in that it only assigns events when there is a match that spans at least 2 kb and if the identified recombinant allele is present in a majority of putative donor lineage isolates in the Neisseria genome database. This approach could miss smaller recombination fragments that may have given rise to novel, mosaic, genes matching neither the donor nor the ancestral allele. In addition, a vast majority of Neisseria genome sequences, including most of the 524 W cc11 strains in this study, were isolated from invasive disease cases. Therefore, some of the genetic diversity among carriage strains could possibly have been missed.
In summary, we have demonstrated that the W cc11 lineage likely arose through recombination from ancestral serogroup W cc22 and serogroup Y cc23 donor strains. Remarkably, strains with this mosaic recombination have persisted as the dominant global W cc11 strain from 1970 to 2014 despite emergence of a phylogenetically distinct epidemic Hajj clone (Mustapha et al. 2016) and genetically diverse endemic case clusters globally.
Material and Methods
Study Isolates
A total of 528 serogroup W cc11 genome sequences with corresponding capsule gene allele designations were identified from PubMLST (www.pubmlst.org/neisseria, last accessed September, 2015), a database that captures Neisseria genetic diversity by assigning allele numbers for all Neisseria genes. Four isolates (19369, 19377, 19379, and 19381) were excluded because they had missing data for a majority of cps alleles, whereas cps allele designations for 524 isolates were included in the study (supplementary table, Supplementary Material online). All but 1 of the 524 W ST-11 strains were isolated from patients with IMD. M7124, identified in Saudi Arabia during the Hajj 2000 epidemic, is a well characterized reference strain for the W cc11 lineage (GenBank accession number, CP009419) sequenced using single molecule, real-time technology (SMRT, www.pacb.com) into a single contiguous sequence (contig) and is used in this study as W cc11 cps reference genome (Mustapha et al. 2015).
Identification of Recombination Donors and Breakpoints for M7124
To determine the organization of cps genes cps sequences were extracted for the M7124 and compared with eight previously described cps sequences for reference meningococcal strains (Harrison et al. 2013) using SplitsTree v4 (Huson and Bryant) phylogenetic networks and visual inspection of sequence alignments. These strains were selected because they represent the best characterized cps gene sequences with no assembly gaps or missing data. Also, potential recombination donor lineages were assessed by querying Neisseria genome sequences in PubMLST and GenBank databases to determine closest matches outside of the W cc11 lineage.
Our method of assessing recombination and assigning donor lineage is, by design, conservative based on the following criteria. A strain was considered a potential donor for M7124 capsular genes if: (1) there is a match that spans at least 2 kb, (2) less than one SNP difference per 1000 base pairs between donor and recombinant gene sequences, and (3) the identified recombinant gene allele is present in a majority of putative donor lineage isolates on the Neisseria genome databases. Recombination breakpoints were identified from sequence alignments as points of abrupt change in sequence similarity between M7124 and a potential donor strain (Swartley et al. 1997). Recombination break points identified by visual inspection of cps sequence alignments were re-assessed using Recombination Detection Program (RDP) v4.56 (Martin et al. 2010). Putative recombination breakpoints identified by ≥1 of 7 algorithms (RDP, Chimaera, Bootscan, 3Seq, GENECONV, MaxChi and SiScan) within cps sequence alignment of four meningococcal strains, specifically M7124, C ST-11 (FAM18), Y ST-23 (α-162), and W ST-22 (α-275), were compared with visually detected break points.
Comparison of cps Alleles within W cc11 Lineage
M7124 cps gene sequences were compared with 524 W cc11 strains with genome sequence data isolated from 21 different countries. Majority (93.5%, 489/523) were isolated in 2000–2014 with the remaining 34 isolated in 1970–1999. Most isolates were from United Kingdom (330/523, 63.1%), South Africa (116/523, 22.2%) or the African “meningitis belt” (34/523, 6.5%; supplementary table, Supplementary Material online) (Lapeyssonnie 1968). Allelic variants for 16 cps genes (ctrA-G, cssA-C, csw, tex, galE, galE2, rfbC and rfbC2) that had complete sequences for ≥80% of the study isolates were downloaded. Allele comparisons of rfbA and rfbB genes was not performed due to missing sequence data among 91% (479/524) and 79% (416/524) of total isolates, respectively. Also, gene elements not curated by PubMLST including insertion sequences, putative fragments in close proximity to insertion sequences (specifically, cssF gene fragments) and two pseudo-genes (putative methyl transferases) within region E were excluded from further analyses. Over 70% of isolates (368/524) had complete sequence data for all 16 cps genes, 22.3% (117/524) had missing data for one or two genes, whereas 39 (7.3%) had missing data for three to five genes. galE2 (99/524, 19.9%), and galE (94/524, 17.9%) genes had the highest frequency of missing data (supplementary table, Supplementary Material online).
Isolates that had either an identical allelic profile to M7124 or had one to three nucleotide differences across all nonmissing cps genes were considered the same cps gene structure as M7124. Isolates with >3 nucleotide differences across all cps genes were further examined to identify whether nucleotide differences clustered into one or more genes. Any gene that differed from M7124 by >3 nucleotide was defined as an “allelic shift” suggestive of a unique recombination in that particular gene in a given isolate compared with M7124. Presence and total number of allelic shifts were assessed for each isolate.
Phylogenetic tree of all 524 W cc11 isolates was obtained using the core genome MLST (cgMLST) protocol from the Neisseria genome PubMLST database (http://pubmlst.org/neisseria/) and visualized on SplitsTree v4 (fig. 5). This method generates a neighbornet phylogenetic network based on allelic profiles across 1605 universally present meningococcal genes (Huson and Bryant 2006; Bratcher et al. 2014). Cps sequences were then concatenated and aligned for a subset of 65 isolates selected to capture capsular allelic diversity within W cc11 (fig. 3). All 29 isolates with ≥4 nucleotide differences across cps gene alleles and 36 isolates, representative of the geo-temporal diversity of isolates within zero to three nucleotide differences from M7124, were selected for this phylogenetic analysis. At least one isolate was selected from every cps allelic combination and strains that were isolated >5 years apart or from different countries were included even if they shared allelic profile with another included strain. MEGA v5.2 (Tamura et al. 2011) and CLC genomics workbench v8.0 (www.clcbio.com) were used to generate sequence alignments and maximum likelihood phylogenetic trees under the under the Hasegawa, Kishino and Yano model of evolution, Γ-distribution of substitution rates and invariant sites (HKY + Γ+I) with 500 bootstrap replicates.
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley and sited at the University of Oxford (Jolley and Maiden 2010). The development of this site has been funded by the Wellcome Trust and European Union. This publication also made use of the Meningitis Research Foundation Meningococcus Genome Library (www.meningitis.org/research/genome) developed by the Health Protection Agency, the Wellcome Trust Sanger Institute and the University of Oxford as a collaboration. A.P.S.d.L. reports travel grants and personal fees from Novartis, personal fees from Sanofi Pasteur, travel grants from GSK, outside the submitted work. Other co-authors have no interests to declare. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Literature Cited
- Barroso DE, et al. 2013. Neisseria meningitidis ST-11 clonal complex bearing capsule serogroups B, C, or W in Brazil. J Infect. 66:547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddek AJ, Li MS, Kroll JS, Jordan TW, Martin DR. 2009. Evidence for capsule switching between carried and disease-causing Neisseria meningitidis strains. Infect Immun. 77:2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. 2014. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics 15:1138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome CV, et al. 1983. Epidemic group C meningococcal meningitis in Upper Volta, 1979. Bull World Health Organ. 61:325–330. [PMC free article] [PubMed] [Google Scholar]
- Budroni S, et al. 2011. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A. 108:4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castineiras TM, et al. 2012. Capsular switching in invasive Neisseria meningitidis, Brazil. Emerg Infect Dis. 18:1336–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant DA. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. Apmis 106:505–525. [PubMed] [Google Scholar]
- Claus H, Vogel U, Muhlenhoff M, Gerardy-Schahn R, Frosch M. 1997. Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol Gen Genet. 257:28–34. [DOI] [PubMed] [Google Scholar]
- Cohn AC, Harrison LH. 2013. Meningococcal vaccines: current issues and future strategies. Drugs 73:1147–1155. [DOI] [PubMed] [Google Scholar]
- Duffin PM, Seifert HS. 2010. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J Bacteriol. 192:4436–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC, et al. 2006. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology 152:3733–3749. [DOI] [PubMed] [Google Scholar]
- Gupta S, et al. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat Med. 2:437–442. [DOI] [PubMed] [Google Scholar]
- Halperin SA, et al. 2012. The changing and dynamic epidemiology of meningococcal disease. Vaccine 30(Suppl 2):B26–B36. [DOI] [PubMed] [Google Scholar]
- Hao W, et al. 2011. Extensive genomic variation within clonal complexes of Neisseria meningitidis. Genome Biol Evol. 3:1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison LH, et al. 2010. Population structure and capsular switching of invasive Neisseria meningitidis isolates in the pre-meningococcal conjugate vaccine era—United States, 2000-2005. J Infect Dis. 201:1208–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison LH, Trotter CL, Ramsay ME. 2009. Global epidemiology of meningococcal disease. Vaccine 27(Suppl 2):B51–B63. [DOI] [PubMed] [Google Scholar]
- Harrison OB, et al. 2013. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis. 19:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, et al. 2014. CC4821 serogroup W meningococcal disease in China. Int J Infect Dis. 29:113–114. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Urwin R, Maiden MC. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol Biol Evol. 16:741–749. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254–267. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, Pollard AJ. 2003. W135 in Africa: origins, problems and perspectives. Travel Med Infect Dis. 1:19–28. [DOI] [PubMed] [Google Scholar]
- Kong Y, et al. 2013. Homologous recombination drives both sequence diversity and gene content variation in Neisseria meningitidis. Genome Biol Evol. 5:1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT, Claus H, Frosch M, Vogel U. 2011. Sequence analysis of serotype-specific synthesis regions II of Haemophilus influenzae serotypes c and d: evidence for common ancestry of capsule synthesis in Pasteurellaceae and Neisseria meningitidis. Res Microbiol. 162:483–487. [DOI] [PubMed] [Google Scholar]
- Lapeyssonnie L. 1968. [Comparative epidemiologic study of meningococcic cerebrospinal meningitis in temperate regions and in the meningitis belt in Africa. Attempt at synthesis]. Med Trop (Mars). 28:709–720. [PubMed] [Google Scholar]
- Lucidarme J, et al. 2015. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect. 71(5):554–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon FG, et al. 1993. Demonstration of lipooligosaccharide immunotype and capsule as virulence factors for Neisseria meningitidis using an infant mouse intranasal infection model. Microb Pathog. 15:359–366. [DOI] [PubMed] [Google Scholar]
- MacNeil JR, et al. 2014. Neisseria meningitidis serogroup W, Burkina Faso, 2012. Emerg Infect Dis. 20:394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, et al. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molling P, Backman A, Olcen P, Fredlund H. 2001. Comparison of serogroup W-135 meningococci isolated in Sweden during a 23-year period and those associated with a recent Hajj pilgrimage. J Clin Microbiol. 39:2695–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JE, Borrow R, Gessner BD. 2006. Meningococcal serogroup W135 in the African meningitis belt: epidemiology, immunity and vaccines. Expert Rev Vaccines. 5:319–336. [DOI] [PubMed] [Google Scholar]
- Mustapha MM, et al. 2015. Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine 2:1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha MM, Marsh JW, Harrison LH. 2016. Global epidemiology of capsular group W meningococcal disease (1970-2015): multifocal emergence and persistence of hypervirulent sequence type (ST)-11 clonal complex. Vaccine 34:1515–1523. [DOI] [PubMed] [Google Scholar]
- Romanow A, et al. 2013. Biochemical and biophysical characterization of the sialyl-/hexosyltransferase synthesizing the meningococcal serogroup W135 heteropolysaccharide capsule. J Biol Chem. 288:11718–11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanow A, et al. 2014. Dissection of hexosyl- and sialyltransferase domains in the bifunctional capsule polymerases from Neisseria meningitidis W and Y defines a new sialyltransferase family. J Biol Chem. 289:33945–33957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinosa MR, et al. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect Immun. 75:3594–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS. 2009. Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 27(Suppl 2):B71–B77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartley JS, et al. 1997. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci U S A. 94:271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha MK, et al. 2000. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356:2159.. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M, Ayala FJ. 2015. How clonal are Neisseria species? The epidemic clonality model revisited. Proc Natl Acad Sci U S A. 112:8909–8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, et al. 2005. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect Immun. 73:1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, et al. 2010. Genetic study of capsular switching between Neisseria meningitidis sequence type 7 serogroup A and C strains. Infect Immun. 78:3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, et al. 2013. Spread of Neisseria meningitidis serogroup W clone, China. Emerg Infect Dis. 19:1496–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, et al. 2015. Sequence type 4821 clonal complex serogroup B Neisseria meningitidis in China, 1978-2013. Emerg Infect Dis. 21:925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, et al. 2001. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc Natl Acad Sci U S A. 98:5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.