Abstract
Domesticated cotton species provide raw material for the majority of the world's textile industry. Two independent domestication events have been identified in allopolyploid cotton, one in Upland cotton (Gossypium hirsutum L.) and the other to Egyptian cotton (Gossypium barbadense L.). However, two diploid cotton species, Gossypium arboreum L. and Gossypium herbaceum L., have been cultivated for several millennia, but their status as independent domesticates has long been in question. Using genome resequencing data, we estimated the global abundance of various repetitive DNAs. We demonstrate that, despite negligible divergence in genome size, the two domesticated diploid cotton species contain different, but compensatory, repeat content and have thus experienced cryptic alterations in repeat abundance despite equivalence in genome size. Evidence of independent origin is bolstered by estimates of divergence times based on molecular evolutionary analysis of f7,000 orthologous genes, for which synonymous substitution rates suggest that G. arboreum and G. herbaceum last shared a common ancestor approximately 0.4–2.5 Ma. These data are incompatible with a shared domestication history during the emergence of agriculture and lead to the conclusion that G. arboreum and G. herbaceum were each domesticated independently.
Keywords: Gossypium, repetitive DNA, molecular evolution, genome size, crop plants
Introduction
Understanding the origins of crop plants and their relationships to wild relatives have long been central concerns of plant biologists. This historical interest, stimulated by the importance of this understanding to crop plant improvement, traces to before the landmark volumes by de Candolle (1883) and Darwin (1868) and remains an area of active research today (Abbo et al. 2012; Hancock 2012; Meyer and Purugganan 2013; Olsen and Wendel 2013). A major challenge in the study of crop plants has been determining the wild ancestors of domesticated species. This difficulty reflects multiple processes, including the often dramatic morphological transformation between progenitor and derivative, possible introgression between crops and wild relatives, cultivation far outside the native range, and rarity or extinction of wild ancestors. Accordingly, the wild ancestors and germplasm relationships of some of our major crop species have remained obscure.
A case in point concerns the two Old World cultivated cotton species, Gossypium arboreum and Gossypium herbaceum (fig. 1). Both species have an ancient history of cultivation, extending back perhaps 5,000 years or more (Chowdhury and Buth 1971). This antiquity of original domestication followed by human-mediated dispersal over vast geographic ranges, extending from Africa through the Levant and Indian subcontinent into the Far East, has generated extensive variability within each species (Hutchinson and Ghose 1937; Silow 1944; Hutchinson et al. 1947; Fryxell 1978). The two species are similar morphologically (Stanton et al. 1994) and with respect to chemical and protein traits (Parks et al. 1975; Wendel et al. 1989). When grown in sympatry, fertile hybrids may arise, although F2 and later generations display “breakdown,” that is, aberrant recombinant phenotypes including some sterility and lethality (Silow 1944; Stephens 1950; Phillips 1961). This indication of genetic differentiation is supported by cytogenetic data, which demonstrate that G. arboreum and G. herbaceum differ by a reciprocal translocation (Gerstel 1953; Brubaker et al. 1999).
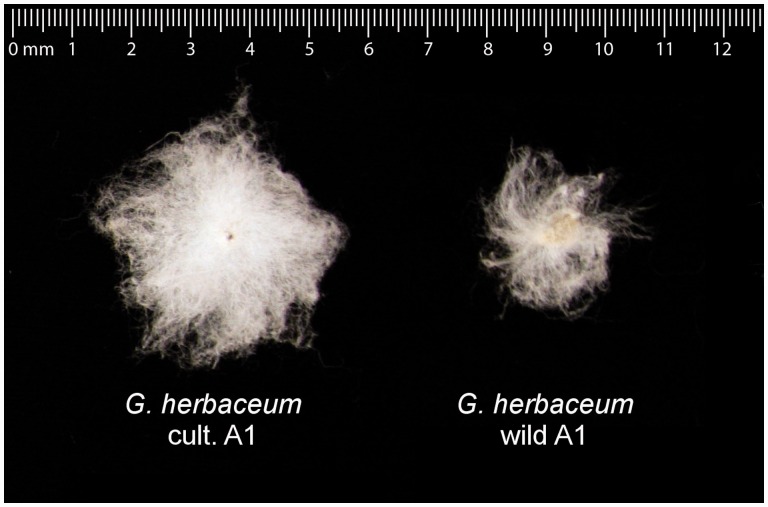
Fig. 1.
Morphological differences between fiber from wild and domesticated A-genome diploid cotton species. Shown are single seeds with single celled trichomes (fiber) from two cotton species, Gossypium herbaceum and Gossypium arboreum. Gossypium arboreum exists only as a cultigen.
Exemplifying the general problem of inference regarding the origins of many crop plants, almost nothing is known about the location and timing of original domestication of either cultivated diploid cotton species. Wild progenitor populations have not been identified with certainty, but a wild and morphologically distinct form of G. herbaceum (G. herbaceum subsp. africanum (Watt) Mauer) occurs in southern Africa (Botswana, Lesotho, and possibly elsewhere) in regions far removed from known historical or present cultivation (Saunders 1961; Fryxell 1978; Vollesen 1987). Its small fruit with seeds bearing sparse, coarse epidermal seed trichomes (“lint” or “cotton”) suggests that G. herbaceum subsp. africanum is a reasonable model of the ancestor of cultivated G. herbaceum (Hutchinson 1954; Fryxell 1978). For G. arboreum, no wild forms have been identified; instead, this species occurs only as a cultigen, with an enormous indigenous range extending from China and Korea westward into northern Africa: its center of diversity lies in India.
Because wild forms of G. arboreum are unknown, and because the location of wild G. herbaceum subsp. africanum is geographically disjunct from known historical regions of cultivation of either species, the origin of the two species and their relationships to each other are unclear. Two opposing views have been forwarded, one that stresses the overall similarity of the two species and a second that emphasizes their differences. Hutchinson, a proponent of the first view, proposed G. herbaceum subsp. africanum as a model of the ancestor of both species (Hutchinson and Ghose 1937; Hutchinson 1954; Fryxell 1978). According to Hutchinson’s hypothesis, G. arboreum arose from G. herbaceum early in the history of diploid cotton cultivation, suggesting one cultivated cotton species arose from another. An alternative hypothesis is that G. arboreum and G. herbaceum diverged prior to domestication. Fryxell (1978) and Wendel et al. (1989) among others argue that genetic differences between the two species are too great to have arisen during the relatively brief period in which domesticated cottons have existed. Thus, according to this view, cultivated Old World cottons originated from at least two independent domestication events from two different wild progenitor species.
The purpose of this study was to use genomic data from ongoing resequencing efforts to gain insight into the relative validity of the common progenitor hypothesis versus the different progenitor hypothesis for the two domesticated diploid cotton species. We report results based on whole-genome resequencing (of 13 cotton accessions) and two sources of information derived from these data, that is, types and abundances of repetitive DNA sequences, using a genome skimming approach (Novák et al. 2013) and divergence estimates derived from synonymous substitutions at more than 7,000 confidently aligned orthologous genes between G. arboreum and G. herbaceum. These data collectively provide compelling evidence for independent domestication of the two Old World diploid cotton species, shed new light on processes of genome size evolution, and have relevance to our understanding of the origin of the genomes of the two modern allotetraploid cotton species that presently dominate world cotton commerce (i.e., Gossypium hirsutum and Gossypium barbadense).
Materials and Methods
Plant Samples, DNA Extraction, and Sequencing
We utilized data generated as part of the cotton resequencing project (see supplementary file S1, Supplementary Material online), which includes both wild (A1-73; subsp. africanum) and domesticated accessions of G. herbaceum, as well as multiple accessions of the exclusively domesticated species G. arboreum. Gossypium arboreum and G. herbaceum collectively comprise the A-genome diploid genome clade, the donor of the A genome to allopolyploid (AD-genome, which includes the commercially important G. hirsutum and G. barbadense) cottons at the time of their formation in the mid-Pleistocene (Wendel and Grover 2015). The diploid genomes studied include the D-genome species Gossypium raimondii, which is the best living model of the D-genome ancestor of allopolyploid cotton (Wendel and Grover 2015), and which has a genome size (885 Mb) that is about half as large as that found in both of the A-genome species (G. arboreum and G. herbaceum; 1,700 Mb) studied. In total, 13 accessions of diploid cotton were analyzed (table 1).
Table 1.
Sample and Clustering Details for the 13 Accessions Used in this Analysis
| Species | Reads Per Sample (Number of Samples) | Coverage (%) | Mean Number of Clustered Reads | Genome Sizea (Mb/1C) |
|---|---|---|---|---|
| Gossypium herbaceum (A1) | 175,474 (3) | 1 | 123,766 | 1,667 |
| Gossypium arboreum (A2) | 180,063 (5) | 1 | 132,223 | 1,698 |
| Gossypium raimondii (D5) | 92,632 (5) | 1 | 46,617 | 880 |
Preparation of Sequence Data
Illumina sequencing reads were filtered for quality using default parameters in the program Trimmomatic version 0.33 (Bolger et al. 2014) retaining high-quality reads that were trimmed to 95 bp (from 125 bp) resulting in over 3 million reads per sample. For each sample we took a random 1% genome equivalent (as is typical for analysis with RepeatExplorer), according to genome size estimates at the Kew C-Value Database (Bennett and Leitch 2010; Novák et al. 2013) (accessions used are detailed in table 1 and supplementary file S1, Supplementary Material online). A sample identifier was prefixed to each sequence name, after which we combined genomic samples from all 13 accessions into a single data set for analysis (table 1).
Graph-Based Clustering
The combined sequence reads were analyzed using the RepeatExplorer pipeline (Novak et al. 2010, 2013), which identifies repetitive DNA families in low-pass, next-generation sequence data and has been used successfully in other species (Macas et al. 2007; Swaminathan et al. 2007; Wicker et al. 2009; Hribova et al. 2010; Novak et al. 2010; Renny-Byfield et al. 2011, 2012, 2013; Piednoel et al. 2012; Novák et al. 2013). Briefly, reads are linked based on sequence similarity and a graph-based clustering algorithm groups reads into clusters where reads within a given cluster are more densely connected to each other than they are to other reads in the data set. All resulting clusters were annotated, where possible, using the RepeatMasker (Smit et al. 2010) default library, a custom repeat library consisting of repetitive DNA sequences identified in the recently published G. raimondii genome assembly (Paterson et al. 2012) and previous genomic sequencing data (Grover et al. 2004, 2007, 2008; Hawkins et al. 2006).
We subsequently calculated the number of reads from each sample contributing to each cluster by counting the number of reads with each sample identifier. This allowed us to assess the number of reads, nucleotides (as each read is 95 bp long), and fraction of the genome for all clusters in each sample. Because many of the clusters have annotations, we summed the number of reads, nucleotides, and fraction of the genome associated with each annotation for each sample. Following this, we pooled samples by species and calculated the mean and standard error for each unique annotation.
Statistical Analysis of Cluster Abundance
Analyzing a standard 1% of the genome per sample allows us to estimate the absolute abundance of each cluster in all samples such that the number of reads and/or bp are directly comparable among species. We used a Generalized Linear Model (GLM) to examine differential abundance of the largest 1,000 clusters in the three species in a manner similar to how RNA-seq count data are used to assess differential gene expression between samples.
Using the GLM, we performed contrasts, with the R (version 3.1.2) package contrast (version 0.19), to assess whether mean abundance of each cluster is statistically different among species. All P values were subsequently corrected using the method of Benjamini and Hochberg (1995).
Hierarchical Clustering of Samples Based on Repetitive DNA Content
Using the largest 1,000 clusters, we assessed the similarity of repeat content on a per sample basis using hierarchical clustering, based on Euclidean distance. We used multidimensional scaling to place each sample in two-dimensional space, using the cmdscale function implemented in R (R Development Core Team 2010), and highlighted each cluster using the ordihull and ordispider functions of the R package vegan (version 2.2-1).
Estimation of Synonymous Substitution Rate
We generated gene sequences of two accessions each of the two A-genome diploid species (A1-155 and A1-97 for G. herbaceum; A2-1011 and A2-34 for G. arboreum). These pseudomolecules were produced using a single-nucleotide polymorphism data set generated using extensive EST and genomic resequencing data (Page et al. 2013). Short read alignments to the G. raimondii reference genome, generated in Page et al. (2013), allowed us to identify single nucleotide polymorphism between species. Since the consensus sequences were generated from the same annotation coordinates, the consensus sequences were not aligned in the traditional sense. Consensus sequences were formatted for input into BioPerl using ClustalW (Larkin et al. 2007) and nonsynonymous and synonymous substitution rates were estimated with a Jukes–Cantor substitution model. Two estimates of divergence (using upper and lower bounds of rate estimates) time between G. herbaceum and G. arboreum were obtained from the substitution rate at neutral loci using the formula T = K/2r, where K equals divergence amount and r corresponds to the rate of divergence for a small sampling of nuclear genes from woody plants (1.5 × 10 − 8 and 2.6 × 10 − 9 substitutions/site/year), as discussed in Senchina et al. (2003).
Results
Clustering of Next-Generation Sequences from Three Species of Gossypium
To characterize and quantify the repetitive content of diploid Gossypium genomes, we sampled multiple 1% genome equivalents from each species (table 1). The complete data set was subjected to clustering using the RepeatExplorer pipeline (Novak et al. 2010, 2013), producing approximately 60,000 clusters ranging from a minimum of only two reads to over 27,000 (a summary of the RepeatExplorer run is provided in supplementary file S2, Supplementary Material online). Sequence similarity searches to a custom repeat library resulted in 65% of clusters being annotated. Not surprisingly, however, the distribution of annotations was heavily skewed in favor of the larger clusters (e.g., 92% of the 1,000 largest, and 100% of the 100 largest clusters were annotated). We grouped clusters based on shared annotation and calculated the total number of Mb/1C that could be attributed to each annotation type (fig. 2; supplementary file S3, Supplementary Material online). As the majority of the data relevant to genome size evolution resides in the largest clusters, our bioinformatic and statistical approaches used the portion of the data most pertinent to question of genome size and independent origins of G. herbaceum and G. arboreum.
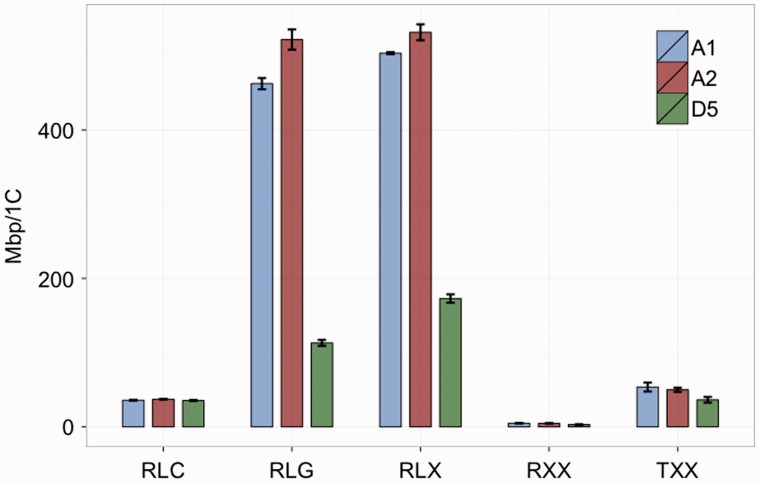
Fig. 2.
Bar plot showing the abundance of the most common repeat types in the genomes of three Gossypium species. Species are color coded and indicated using genome designations (A1, Gossypium herbaceum; A2, Gossypium arboreum; and D5, Gossypium raimondii). Standard error bars are shown. Annotation abbreviations are as follows: RLG, Ty3/Gypsy retroelements; RLC, Ty1/Copia retroelements; RLX, unknown LTR retroelements; RXX, retroelement unknown superfamily; TXX, unknown DNA transposon; AT, AT-rich simple repeat.
In some cases, our pipeline was unable to distinguish Ty3/Gypsy- and Ty1/Copia-derived clusters, and there is reasonable fraction of long terminal repeat (LTR) retroelements of unknown type (RLX; fig. 2; supplementary file S3, Supplementary Material online). These are derived from degraded copies of the Ty3/Gypsy and Ty1/Copia and are likely in roughly the same proportions as those LTR retroelements we could distinguish. Other than the non-specific LTR retroelement annotation, Ty3/Gypsy retroelements (RLG) are the next largest category, comprising between 133 and 522 Mb/1C of the genome, depending on the species. Not surprisingly, G. raimondii (D5), with the smallest genome (880 Mb/1C), had the fewest Ty3/Gypsy retroelements (fig. 2; supplementary file S3, Supplementary Material online), accounting for approximately 13% of the genome. The two Old World diploid cottons, G. herbaceum (A1) and G. arboreum (A2), each had a larger complement of Ty3/Gypsy retroelements, in congruence with their larger genome sizes relative to G. raimondii (fig. 2; supplementary file S3, Supplementary Material online). As expected, Ty1/Copia retroelements are significantly less abundant when compared to Ty3/Gyspy, comprising only approximately 36 Mb of the genomes in all three species (fig. 2; supplementary file S3, Supplementary Material online).
In interspecific comparisons of the 1,000 largest clusters, we found a variable number with significant deviation in abundance between the three species analyzed (tables 2 and 3). Between 149 and 342 of the 1,000 largest clusters exhibited evidence of divergence in abundance, depending on the species and clusters being compared.
Table 2.
Two-Way Analysis of Variance of Cluster Abundance among the Largest 1,000 Clusters in Four Gossypium Species
| df | Sum of Squares | Mean Sq | F Value | P | |
|---|---|---|---|---|---|
| Cluster (C) | 999 | 6.18 × 108 | 618,949 | 279.56 | <0.00005 |
| Species (S) | 2 | 2.03 × 107 | 10,193,026 | 4603.81 | <0.00005 |
| Cluster:species (CxS) | 1998 | 2.69 × 108 | 134,759 | 60.87 | <0.00005 |
| Residuals | 10,000 | 2.21 × 107 | 2,214 |
Table 3.
Statistically Significant Differences in Cluster (Using a GLM, See Materials and Methods) Abundance between Species of Gossypium
| Comparison | Clusters with Differential Abundancea |
|---|---|
| A1: A2 | 149 |
| A1: D5 | 297 |
| A2: D5 | 342 |
aOf the largest 1,000 clusters.
Variation in Repeat Content among A-Genome Diploids
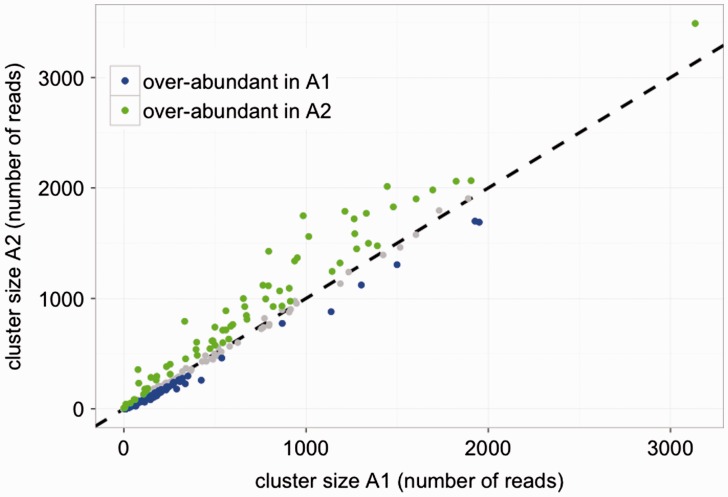
We used a GLM to estimate the effect of species on cluster abundance. Subsequently, we used contrasts to individually compare the abundance of each cluster between species. This revealed 149 of the top 1,000 clusters had statistically different abundance in the two sister species G. herbaceum and G. arboreum (A1 and A2, respectively; table 3). Examining the clusters with significant difference revealed that the overall variation is attributable to some clusters being highly represented in G. arboreum and others being over-represented in G. herbaceum (fig. 3). Importantly, the largest clusters are typically more highly abundant in G. arboreum, whereas the smaller set of clusters are generally more highly in G. herbaceum.
Fig. 3.
Scatter plot of cluster abundance in Gossypium herbaceum (A1) and Gossypium arboreum (A2). Clusters that exhibit statistically significant difference in abundance between the two species are color coded as indicated. Clusters that do not exhibit and statistical difference are indicated in gray.
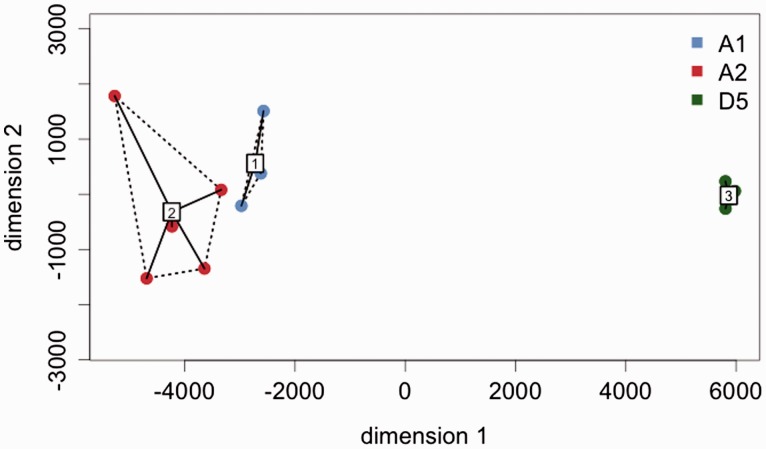
Using the largest 1,000 clusters, we assessed the similarity of repeat content on a per sample basis using hierarchical clustering (fig. 4). This analysis reveals, as expected, that samples cluster by species, with G. herbaceum and G. arboreum being more closely related to each other than either is to G. raimondii. Of particular relevance is that G. herbaceum and G. arboreum are distinct in the first clustering dimension (fig. 4).
Fig. 4.
Cotton samples grouped by repeat content. Using the largest 1,000 clusters we assessed the similarity of repeat content on a per sample basis using hierarchical clustering, based on Euclidean distance. We identified natural groups (gated and numbered) using the ordihull and ordispider functions of the R package vegan. Importantly, Gossypium herbaceum (A1) and Gossypium arboreum (A2) are distinct in the first dimension.
Estimation of Divergence Time Based on Synonymous Substitution Rates for >7,000 Genes
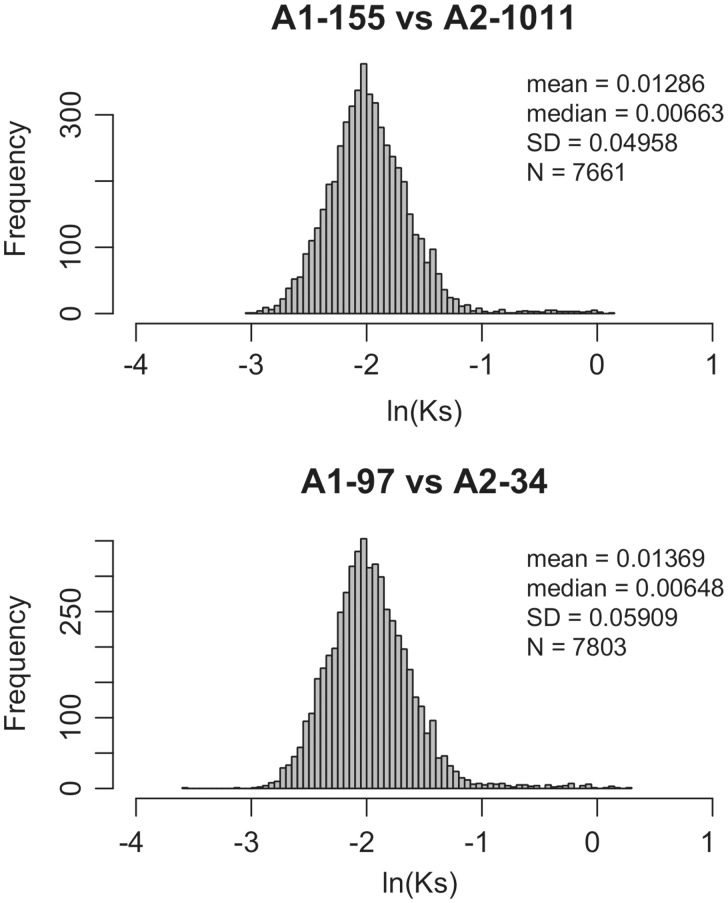
We estimated genome-wide synonymous substitution (Ks) rates for approximately >7,000 orthologous genes of G. herbaceum and G. arboreum for which alignments and inferences of orthology were deemed unambiguous (fig. 5). Depending on the accessions compared, the mean Ks varied from 0.0127 to 0.0137, (average = 0.0132). Assuming a range of reasonable mutation rates, between 1.5 × 10 − 8 and 2.6 × 10 − 9 substitutions per site per year (see Senchina et al. 2003), estimates of divergence time for G. herbaceum and G. arboreum ranged from 400,000 to 2.5 Myr.
Fig. 5.
Distribution of synonymous substitutions (Ks) between orthologs from Gossypium arboreum and Gossypium herbaceum. Alignment of over >7,000 genes in each comparison allowed the mean and median substitution rate between species to be estimated.
Discussion
The Repetitive Landscape of the Cotton Genome
Here we used low-coverage next-generation sequencing to analyze the global repeat composition within and among three cotton (Gossypium) species, and subsequently applied the annotated repetitive profiles as evidence, in conjunction with estimates of divergence time, to assess the likelihood that the two Old World cultivated cottons, G. arboreum and G. herbaceum, had independent origins from different wild progenitors.
In total, we annotated between 348 and 1,146 Mb/1C, depending on the species (fig. 2; supplementary file S3, Supplementary Material online), with the three cotton genomes being between 40% and 68% repetitive, in line with estimates from other plants (SanMiguel et al. 1998; Kumar and Bennetzen 1999; Wicker et al. 2001). As expected from previous genomic analyses in cotton and other plant species (Hawkins et al. 2006; Hribova et al. 2010; Renny-Byfield et al. 2011, 2012, 2013; Paterson et al. 2012), Ty3/Gypsy LTR-retroelements (RLG) account for the majority of cotton genomes. Our range in RLG estimates is, however, notably higher than estimates employing methodology that is similar to that which we used here (Macas et al. 2007; Hribova et al. 2010; Novak et al. 2010, 2013; Renny-Byfield et al. 2011, 2013); this may be due, in part, to the inclusion of a cotton-specific repeat database in our analysis. Not surprisingly, G. raimondii (880 Mb/1C; the smallest genome analyzed) had the smallest absolute number of repeats while G. arboreum had the greatest absolute number of repeats.
The results reported here are broadly consistent with earlier work but in detail contrast with the initial estimates reported by Hawkins et al. (2006). Hawkins et al. reported that Ty1/Copia-like sequences were more abundant than Ty3/Gypsy elements in G raimondii. Their analysis was based on cloned, whole-genome shotgun sequences, which then were matched to the NCBI database, which at that time was relatively poor in terms of repeat content, as the authors noted. The use of a custom database of cotton repeats and RepeatMasker libraries, as in this study, allows for more accurate annotation. In this respect we note the proportions of each TE superfamily follow the same pattern as reported for the G. raimondii reference genome (Paterson et al. 2012), with the values reported here being consistently lower. For example, the reference annotation identifies 53.2% of the genome to be retroelement derived, whereas our estimate is 36.91%. Similarly, Ty3/Gypsy and Ty1/Copia retroelements account for 18.8% and 5.9% of the genome according to the reference annotation, whereas we report 12% and 4%, respectively. For DNA transposons, the reference annotation reports 1.5% of the genome, whereas our analysis suggests 0.9%.
Additionally, sequencing of the G. arboreum (A2) genome provides estimates of repeat abundance similar to those reported here. For example, the G. arboreum genome assembly consists of 5.5% Ty1/Copia elements, whereas we report 2.2%. Similarly, our analysis and that reported by Li et al. (2014) indicate Ty3/Gypsy retroelements are far more common than Ty1/Copia, although perhaps not surprisingly whole-genome assembly identifies a larger proportion of Ty3/Gypsy when compared with the clustering detailed here (∼56% vs. ∼30%). All things considered, RepeatExplorer performed well and, at least in terms of the cotton genome, seems to produce repetitive DNA content estimations, whose proportions are in broad agreement with high quality, fully assembled genomes.
Interestingly, Hawkins et al. (2006) reported that in G. herbaceum (A1) Ty3/Gypsy-like retroelements predominated, in contrast to observations in G. arboreum, where Ty1/Copia were reported as more abundant. The results presented in Hawkins et al. (2006) are therefore in agreement with data presented here for G. herbaceum. The recent publication of the G. arboreum genome sequence revealed a notable proliferation of Gorge-Ty3/Gypsy. Furthermore, Ty3-Gyspy-like sequences were more common in the genome of G. arboreum when compared to their Ty1/Copia counterparts (Li et al. 2014), in line with our analysis. Annotation of the G. arboreum genome sequence, however, indicated that 68% of the genome is composed of repetitive DNA, a value very close to our estimate (∼67.5%; fig. 2; supplementary file S3, Supplementary Material online).
A sizeable fraction of each genome was attributable to LTR retroelements of unknown origin. One likely explanation of these is mutational degeneration, rendering difficult particular assignments to source retroelement families (fig. 2; supplementary file S3, Supplementary Material online). We also identified a number of other repeat classes in our data set but almost all of these were in low abundance in all three species (fig. 2; supplementary file S3, Supplementary Material online). A significant proportion of the data for each of the four genomes is composed of relatively low-copy repeat families, with relatively few clusters containing more than 5,000 reads (data not shown).
Variation in Repetitive DNA Content among Cotton Species
We demonstrate here the first statistical assessment of genome-wide differences in repeat content between closely related Gossypium species. Using a GLM we investigated variation in cluster abundance among the three cotton species analyzed, with a two-way analysis of variance revealing that all factors and interactions are significant (table 2). Additionally, our results provide data on repeat abundance using statistical methods, a practice that is relatively uncommon (Macas and Neumann 2007; Renny-Byfield et al. 2011, 2012, 2013; Piednoel et al. 2012).
For most species comparisons there were a relatively large number of clusters exhibiting evidence of differential abundance (table 3). There were approximately equal number of clusters with statistically significant differences in comparisons between G. herbaceum (A1) and G. raimondii (D5), and G. arboreum (A2) and G. raimondii (D5), an expected result given that G. raimondii is equally divergent from both G. herbaceum and G. arboreum.
Repeat Content, Genic Divergence, and the Question of Parallel Domestication of Two Different A-Genome Cottons
This high level of divergence between the two closely related species G. herbaceum (A1) and G. arboreum (A2) was somewhat unexpected, given their overall similarity in genome size and in other traits (Wendel et al. 1989). Our GLM analysis indicates that 149 of the top 1,000 clusters showed differential abundance following contrast analysis (table 3 and fig. 3), despite the minimal difference in genome size between the species (∼10 to 80 Mb). These observations serve to highlight the ever-changing repeat landscapes of plant genomes; stasis in genome size need not reflect genomic quiescence, even between two closely related genomes. The example presented here clearly demonstrates this point; that is, despite their overall and remarkable similarity, at the repeat content level, the genomes of G. herbaceum and G. arboreum are easily distinguished. Such divergence would be extraordinary, perhaps implausibly so, if these two species had an ancestor-descendant relationship following a single domestication event some 5,000 years ago. Moreover, if G. arboreum had been derived from domesticated G. herbaceum, as suggested in some of the older literature, then one might expect the former to be nested within the latter in a hierarchical clustering analysis; instead, however, there is a separation of the two species into distinct groups in the first dimension after multidimensional scaling (fig. 4), as is the case with allozymes (Wendel et al. 1989).
A key conclusion reached here is that, despite negligible divergence in genome size, the two A-genome cotton species contain variable proportions of repeat families. This observation suggests that they are distinct species with separate evolutionary histories, as opposed to conjoined domesticates, one derived from the other. These data are congruent with a molecular divergence data set derived from >7,000 orthologous genes from G. arboreum and G. herbaceum, which indicate that these two species last shared a common ancestor approximately 400,000 to 2.5 Ma, prior to the origin of agriculture and possibly the origin of modern humans. Collectively, we view these various sources of genomic data as providing compelling support for the hypothesis that the two species were independently domesticated from different wild progenitors, rather than having been derived, one from the other (G. arboreum from G. herbaceum), following a single domestication event (see Introduction). We note that this evidence in support of parallel domestication is consistent with the observation of F2 breakdown following interspecific hybridization (Stephens 1950), the chromosomal translocation that distinguishes the two species (Gerstel 1953; Brubaker et al. 1999), and allozyme data (Wendel et al. 1989) and microsatellite markers (Hinze et al. 2015), which, remarkably, were used a quarter of a century ago to derive a divergence time estimate of 1,400,000 ± 450,000 years. A corollary implication is that the differences that distinguish G. arboreum from G. herbaceum did not arise during agricultural times, but instead were present in their respective ancestors.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation Plant Genome Program and Cotton Inc. (to J.F.W.) and the U.S. Department of Agriculture (ARS 6402-21310-003-24S and 6402-21310-003-18S to D.G.P.), and Cotton Incorporated (awards to J.F.W. and D.G.P.).
Supplementary Material
Supplementary files S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Literature Cited
- Abbo S, Lev-Yadun S, Gopher A. 2012. Plant domestication and crop evolution in the Near East: on events and processes. Crit Rev Plant Sci. 31:241–257. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate–a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 57:289–300. [Google Scholar]
- Bennett MD, Leitch IJ. 2010. Angiosperm DNA C-values database. Available from: http://data.kew.org/cvalues/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker C, Paterson A, Wendel J. 1999. Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome 42:184–203. [Google Scholar]
- Chowdhury K, Buth G. 1971. Cotton seeds from the Neolithic in Egyptian Nubia and the origin of Old World cotton. Biol J Linnean Soc. 3:303–312. [Google Scholar]
- Darwin C. 1868. The variation of animals and plants under domestication. London: John Murray.
- de Candolle A. 1883. Origine des plantes cultivées. Paris: Germer-Baillière.
- Fryxell PA. 1978. The natural history of the cotton tribe (Malvaceae, tribe Gossypieae). College Station: Texas A & M University Press. [Google Scholar]
- Gerstel D. 1953. Chromosomal translocations in interspecific hybrids of the genus Gossypium. Evolution 234–244. [Google Scholar]
- Grover CE, Hawkins JS, Wendel JF. 2008. Phylogenetic insights into the pace and pattern of plant genome size evolution. In: Volff, editor. Genome dynamics. Basel (Switzerland): Karger Publishers; p. 57–68. [DOI] [PubMed] [Google Scholar]
- Grover CE, Kim H, Wing RA, Paterson AH, Wendel JF. 2004. Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14:1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover CE, Kim H, Wing RA, Paterson AH, Wendel JF. 2007. Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium). Plant J. 50:995–1006. [DOI] [PubMed] [Google Scholar]
- Hancock JF. 2012. Plant evolution and the origin of crop species. Cambridge (MA): CABI. [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. 2006. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 16:1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix B, Stewart JM. 2015. Estimation of the nuclear DNA content of Gossypium species. Annals of Botany 7:233–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinze LL, et al. 2015. Molecular characterization of the Gossypium Diversity Reference Set of the US National Cotton Germplasm Collection. Theor Appl Genet. 128:313–327. [DOI] [PubMed] [Google Scholar]
- Hribova E, et al. 2010. Repetitive part of the banana (Musa acuminata) genome investigated by low-depth 454 sequencing. BMC Plant Biol. 10:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. 1954. New evidence on the origin of the Old World cottons. Heredity 8:225–241. [Google Scholar]
- Hutchinson JB, Ghose R. 1937. The classification of the cottons of Asia and Africa. Indian J Agric Sci. 7: 233–257.
- Hutchinson JB, Silow R, Stephens S. 1947. The evolution of Gossypium and the differentiation of the cultivated cottons. London: Oxford University Press. New York.
- Kumar A, Bennetzen JL. 1999. Plant retrotransposons. Ann Rev Genet. 33:479–532. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Li F, et al. 2014. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 46:567–572. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P. 2007. Ogre elements—a distinct group of plant Ty3/gypsy-like retrotransposons. Gene 390:108–116. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navratilova A. 2007. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD. 2013. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 14:840–852. [DOI] [PubMed] [Google Scholar]
- Novak P, Neumann P, Macas J. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. 2013. RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next generation sequence reads. Bioinformatics 29(6): 792–793. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Wendel JF. 2013. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu Rev Plant Biol. 64:47–70. [DOI] [PubMed] [Google Scholar]
- Page JT, et al. 2013. Insights into the evolution of cotton diploids and polyploids from whole-genome re-sequencing. G3-Genes Genomes Genet. 3:1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page JT, Liechty ZS, Huynh MD, Udall JA. 2014. BamBam: genome sequence analysis tools for biologists. BMC Res Notes. 7:829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks C, Williams D, Dreyer D. 1975. Symposium on Biological Systematics, genetics and origin of cultivated plants. 7. Application of flavonoid distrubution to taxanomic problems in genus Gossypium. Bull Torrey Bot Club. 102:350–361. [Google Scholar]
- Paterson AH, et al. 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427. [DOI] [PubMed] [Google Scholar]
- Phillips L. 1961. The cytogenetics of speciation in Asiatic cotton. Genetics 46:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piednoel M, et al. 2012. Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol Biol Evol. 29:3601–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2010. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Renny-Byfield S, et al. 2011. Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol Biol Evol. 28:2843–2854. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, et al. 2012. Independent, rapid and targeted loss of a highly repetitive DNA sequence derived from the paternal genome donor in natural and synthetic Nicotiana tabacum. PLoS One 7:e36963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renny-Byfield S, et al. 2013. Diploidization and genome size change in allopolyploids is associated with differential dynamics of low- and high-copy sequences. Plant J. 74:829–839. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. 1998. The paleontology of intergene retrotransposons of maize. Nat Genet. 20:43–45. [DOI] [PubMed] [Google Scholar]
- Saunders JH. 1961. The wild species of Gossypium and their evolutionary history. Empire Cotton Growing Corporation. London: Oxford University Press. [Google Scholar]
- Senchina DS, et al. 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol Biol Evol. 20:633–643. [DOI] [PubMed] [Google Scholar]
- Silow R. 1944. The genetics of species development in the Old World cottons. J Genet. 46:62–77. [Google Scholar]
- Smit A R., Hubley P, Green 2010. RepeatMasker Open-3.0.
- Stanton MA, Stewart JM, Pervical AE, Wendel JF. 1994. Morphological diversity and relationships in the A-genome cottons, Gossypium arboreum and G. herbaceum. Crop Sci. 34:519–527. [Google Scholar]
- Stephens S. 1950. The internal mechanism of speciation in Gossypium. Bot Rev. 16:115–149. [Google Scholar]
- Swaminathan K, Varala K, Hudson ME. 2007. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 8:132–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollesen K. 1987. The native species of Gossypium (Malvaceae) in Africa, Arabia and Pakistan. Kew Bull. 337–349. [Google Scholar]
- Wendel JF, Grover CE. 2015. Taxonomy and evolution of the cotton genus. In: Fang D, Percy R, editors. Cotton, Agronomy Monograph 24. Madison (WI): ASA-CSSA-SSSA; p. 25–44. [Google Scholar]
- Wendel JF, Olson PD, Stewart JM. 1989. Genetic diversity, introgression, and independent domestication of old world cultivated cottons. Am J Bot. 76:1795–1806. [Google Scholar]
- Wicker T, et al. 2001. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26:307–316. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. 2009. A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 59:712–722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.