Abstract
Klebsiella pneumoniae has become one of the most dangerous causative agents of hospital infections due to the acquisition of resistance to carbapenems, one of the last resort families of antibiotics. Resistance is usually mediated by carbapenemases coded for by different classes of genes. A prolonged outbreak of carbapenem-resistant K. pneumoniae infections has been recently described in northeastern Ohio. Most strains isolated from patients during this outbreak belong to MLST sequence type 258 (ST258). To understand more about this outbreak two isolates (strains 140 and 677), one of them responsible for a fatal infection, were selected for genome comparison analyses. Whole genome map and sequence comparisons demonstrated that both strains are highly related showing 99% average nucleotide identity. However, the genomes differ at the so-called high heterogeneity zone (HHZ) and other minor regions. This study identifies the potential value of the HHZ as a potential marker for K. pneumoniae clinical and epidemiological studies.
Keywords: whole genome mapping, optical map, ST-258, carbapenem, KPC
Introduction
Klebsiella pneumoniae is the causative agent of numerous infectious diseases (Woloj, et al. 1986; Daza et al. 2001; Liam, et al. 2001; Huang, et al. 2013; Suzuki et al. 2013; Ramirez et al. 2014a, b; Alcantar-Curiel and Giron 2015; Lin et al. 2015; Qu et al. 2015; Zowawi et al. 2015) and can also act as triggering factor in the initiation and development of ankylosing spondylitis and Crohn’s disease (Ebringer et al. 2007; Rashid and Ebringer 2007; Rashid et al. 2013). K. pneumoniae strains recently acquired genes coding for carbapenemases (Nordmann et al. 2009; Munoz-Price et al. 2013; Xie et al. 2013; Ramirez, et al. 2014b), which increased the severity of the infections by complicating treatments (Hirsch and Tam 2010; van Duin et al. 2014). As a consequence, numerous groups studied aspects of virulence, antibiotic resistance, and epidemiology of carbapenem-resistant K. pneumoniae (CRKp) infections (Lin et al. 2008; Endimiani et al. 2009; Shu, et al. 2009; Hirsch and Tam 2010; Ramirez et al. 2012; Deleo et al. 2014). In one study, a combination of whole genome mapping (WGM) and whole genome sequencing (WGS) led to the identification of a particular chromosome segment, known as high heterogeneity zone (HHZ), later also called region of divergence (rd). The HHZ shows high variability and includes a “hot spot”, also known as attO hot spot, where DNA fragments of a different origin can be inserted, and the capsular polysaccharide biosynthesis gene cluster (CPC) (Lin et al. 2008; Shu et al. 2009; Ramirez et al. 2012; Deleo et al. 2014).
An outbreak of CRKp infection recently occurred in northeastern Ohio with most strains belonging to MLST sequence type 258 (ST258) (van Duin et al. 2014). To characterize KPC-producing strains causing this outbreak we analyzed two isolates that led to different outcomes after treatment. Our results showed high homogeneity at the genomic level, but significant differences occurred at one region within the HHZ.
Results and Discussion
Cases Description
Klebsiella pneumoniae Kb140 is a blood isolate from a 64-year-old man with a history of coronary artery disease, congestive heart failure, peptic ulcer disease, and chronic obstructive pulmonary disease (which was treated with chronic corticosteroids) that was transferred from an outside facility to a referral hospital to be treated for pneumonia (Ramirez et al. 2014b). Despite antibiotic treatment (intravenous tigecycline) and other procedures the patient died on hospital day 8. K. pneumoniae Kb677 was isolated from of urine culture of an 87-year-old woman with a history of localized breast cancer without metastases who was admitted to a referral hospital from the community (Ramirez et al. 2014b). She did not meet CDC/NHSN criteria for urinary tract infection, and was not treated with any antibiotics with in vitro activity against CRKp. She was discharged back to her home after a prolonged hospital stay of 28 days. The antimicrobial susceptibilities of both isolates are summarized in table 1. Significant differences are that Kb140 is susceptible to gentamicin, but not to tobramycin or amikacin; and Kb677 is susceptible to amikacin but not gentamicin or tobramycin.
Table 1.
Antimicrobial Susceptibility of K. pneumoniae Isolates Kb140 and Kb677
| Antibiotic | Kb140 |
Kb677 |
||
|---|---|---|---|---|
| MIC (µg/mL) | Interpretation | MIC (µg/mL) | Interpretation | |
| Amikacin | 32 | I | ≤4 | S |
| Aztreonam | >16 | R | >16 | R |
| Ciprofloxacin | >2 | R | >2 | R |
| Colistin | 0.5 | S | 0.5 | S |
| Doripenema | >2 | R | 2 | I |
| Doxycycline | 16 | R | >16 | R |
| Ertapenem | >4 | R | >4 | R |
| Cefepime | >16 | R | 8 | I |
| Cefotaxime | >32 | R | 32 | R |
| Ceftazidime | >16 | R | >16 | R |
| Gentamicin | ≤1 | S | >8 | R |
| Imipenem | >8 | R | 8 | R |
| Levofloxacin | >8 | R | >8 | R |
| Meropenem | >8 | R | 8 | R |
| Minocycline | 16 | R | >16 | R |
| Piperacillin/ tazobactam | >64/128 | R | >64/128 | R |
| Polymyxin B | 0.5 | S | 0.5 | S |
| Trimethoprim/ sulfamethoxazole | >4/76 | R | >4/76 | R |
| Tigecyclinea | 2 | S | 4 | I |
| Tobramycin | >8 | R | >8 | R |
Values for doripenem and tigecycline are too close (one dilution) to assure that their phenotypes are actually different.
Whole Genome Map Comparison
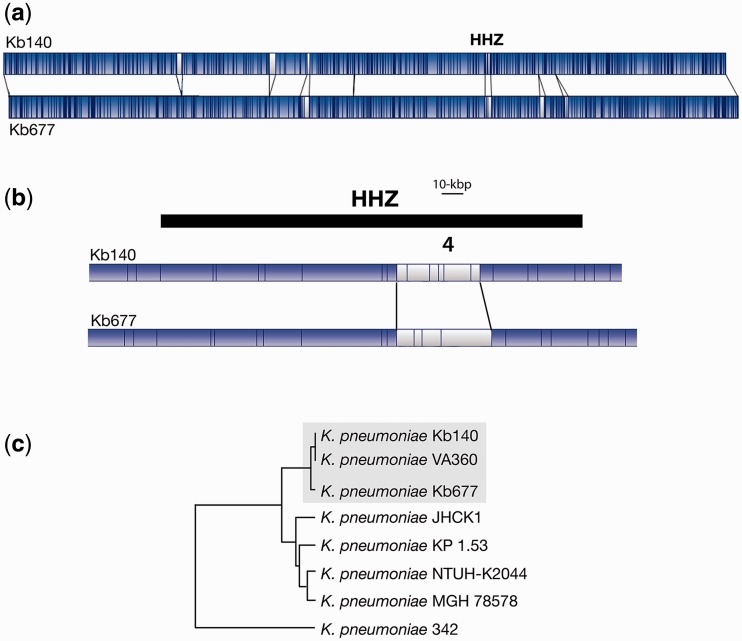
The HHZ can be divided into distinct regions of which 3 and 4 seem to exhibit the highest variability (Ramirez et al. 2012). Region 3 consists of a “hot spot”, also known as attO (Lin et al. 2008), for insertion of DNA fragments. In at least one strain, NTUH-K2044, the insertion in Region 3 is the ICEKp1, a ca. 76-kbp integrative and conjugative element that includes a high-pathogenicity island (Lin et al. 2008). HHZ Region 4 contains the CPC (Ramirez et al. 2012), which shows high variability among strains (Shu et al. 2009). Comparison of WGMs of strains Kb140 and Kb677 revealed that, although not identical, their genomes are very similar (regions in blue, fig. 1a). One of the areas of difference (white in the WGMs) is located within the HHZ (fig. 1a and b), more precisely, at Region 4 (fig. 1b). Further WGM comparisons showed that both strains are closely related to K. pneumoniae VA360, a strain isolated in the same region before the beginning of the surveillance study, strain Kb140 being the most similar (fig. 1c) (Ramirez et al. 2012; Xie et al. 2013).
Fig. 1.—
Genomic comparisons. (a) The K. pneumoniae Kb140 and Kb677 whole genome maps were compared using the MapSolver 3.2.0 software. The blue and white regions represent matching and nonmatching DNA fragments, respectively. The location of the HHZ is indicated. (b) Zoom-in the HHZ region. (c) Map similarity cluster showing K. pneumoniae Kb140 and Kb677 in relation to other completely sequenced K. pneumoniae strains. Dendogram was generated using Unweighted Pair Group Method with Arithmetic Mean. The strains Kb140, Kb670, and the closely related VA360 are boxed.
WGS: Comparison of the HHZs
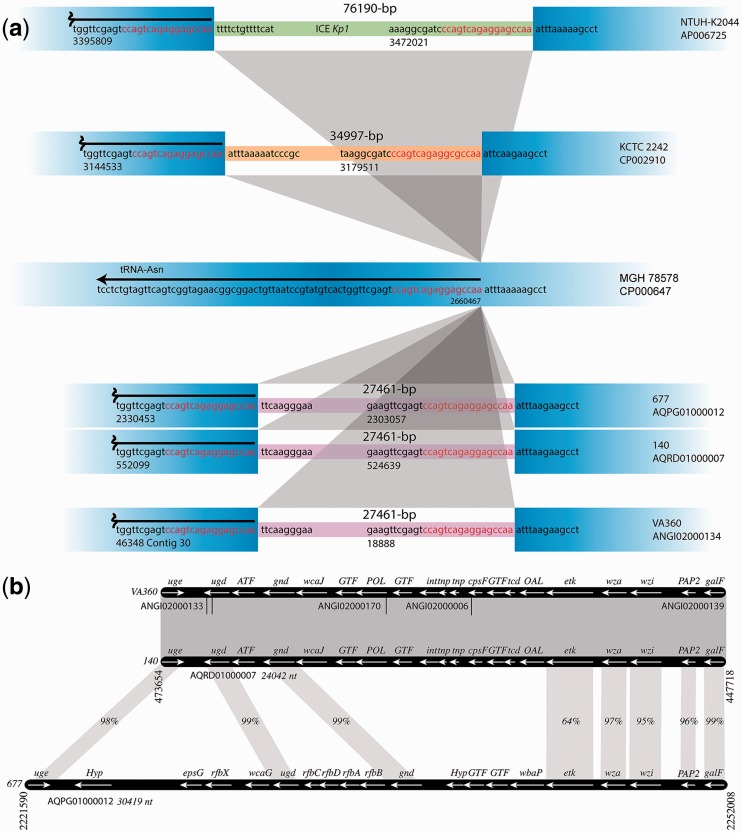
The genomes of strains Kb140 and Kb677 are 5,677,714 and 5,894,762-bp long, respectively. Analysis of the HHZs at the nucleotide level showed that the Region 3 of both strains, as well as that of strain VA360, includes the same DNA fragment, an indication of relatedness. A comparison of the Region 3 of all three strains and those of well-studied strains such as NTUH-K2044, KCTC2242, which include unique DNA fragments, and MGH 78578, which does not include an insert is shown in figure 2a.
Fig. 2.—
HHZ Regions 3 and 4. (a) Schematic diagram of the Region 3 within the HHZs of several K. pneumoniae strains. The DNA sequences surrounding the hot spot and inserted fragments are represented in blue. The inserted regions are shown in thinner lines with different colors to indicate that the sequences are different. The sequences inserted in strains Kb140, Kb677, and VA360 are identical. The coordinates are those found in GenBank. The hot spot sequences, also known as attO (Lin et al. 2008), are red and the sequences immediately adjacent to the hot spots are black. In the case of strain NTUH-K2044 the inserted sequenced is the ICEKp1 pathogenicity island. (b) Schematic comparison of the Region 4, which consists of the CPC, within the HHZs of K. pneumoniae strains Kb140, Kb677, and VA360. Grayed regions represent high homology. In those cases where there is no 100% identity, the identity percentage is shown. In the case of strain VA360, the Region 4 was put together using different contigs, which are identified by their accession numbers. Coordinate numbers are those in GenBank. Potential function of genes (Kelly and Whitfield 1996; Whitfield 2006; Shu et al. 2009): uge, UDP-galacturonate 4-epimerase; ugd, UDP-glucose dehydrogenase; ATF, acyl transferase; gnd, gluconate-6-phosphate dehydrogenase; wcaJ, undecaprenyl-phosphate glycosyltransferase; GTF, glycosyl transferase; POL, polysaccharide biosynthesis; int, integrase-like; tnp, transposase-like; cpsF, CMP-N-acetylneuraminic acid synthetase; tcd, glycosyltransferase; OAL, O-antigen ligase; etk, tyrosine-protein kinase; wza, polysaccharide export lipoprotein; wzi, integral outer membrane protein; PAP2, PAP2 superfamily; galF, modulator of GalU to elevate the cellular concentration of UDP-glucose; Hyp, hypothetical; epsG, putative capsular polysaccharide biosynthesis protein; rfbX (wzx), flippase; wcaG, GDP-4-keto-6-deoxy-D-mannose 3,5-epimerase/reductase; rfbAB, ATP-binding cassette transporter; rfbC, involved in polysaccharide synthesis (epimerase), rfbD, involved in polysaccharide transport; wbaP, membrane protein that helps transfer a galactose residue from UDP-Gal to undecaprenol diphosphate to form Gal-p-UndP.
Region 3 of strains Kb140, Kb677, and VA360 includes elements coding for functions involved in conjugation, a type III restriction endonuclease and the cognate methylase as well as a P4-like integrase and other hypothetical genes with unknown function (table 2). The integrase is found in numerous genomes of Gram-negatives (supplementary fig. 1S, Supplementary Material online) and its amino acid sequence shares 50% identity and 68% similarity with the ICEKp1 integrase (Lin et al. 2008) (supplementary fig. S2, Supplementary Material online). As it is the case in ICEKp1, the integrase could mediate insertion and excision of this DNA fragment. The Region 3 of strains Kb140, Kb677, and VA360 was also found in sixteen CRKp strains (supplementary fig. S3a, Supplementary Material online) (Snitkin et al. 2012).
Table 2.
Summary of main ORFs found in Region 3
| Putative function |
Coordinates |
||
|---|---|---|---|
| Kb140 | Kb677 | VA360 | |
| AQRD01000007 | AQPG01000012 | ANGI02000134 | |
| Hypothetical protein—DNA-binding family protein | c (528973.529680) | c (2307327.2308034) | c (23222.23929) |
| tRNA_anti-like family protein | 531400.531966 | 2309754.2310320 | 25649.26215 |
| Putative DNA-directed RNA polymerase | 531981.532532 | 2310335.2310886 | 26230.26781 |
| TIR domain protein | c (532862.533281) | c (2311216.2311635) | c (27111.27530) |
| Tetratricopeptide repeat family protein | c (533283.534548) | c (2311637.2312902) | c (27532.28797) |
| MobA/MobL family protein | c (535635.537155) | c (2313989.2315509) | c (29884.31404) |
| Hypothetical protein | 538002.538484 | 2316356.2316838 | 32251.32733 |
| Conserved hypothetical protein | 539844.540359 | 2318198.2318713 | 34093.34608 |
| TraD | 540459.540782 | 2318813.2319136 | 34708.35031 |
| Type IV pilin | 541350.541919 | 2319704.2320273 | 35599.36168 |
| PilV—tail fiber protein | 542123.543682 | 2320477.2322036 | 36372.37931 |
| Type III restriction endonuclease subunit R | c (544145.547105) | c (2322499.2325459) | c (38394.41354) |
| DNA methyltransferase | c (547115.548965) | c (2325469.2327319) | c (41364.43214) |
| SpnT | 549159.550547 | 2327513.2328901 | 43408.44796 |
| Phage integrase | c (550619.551893) | c (2328973.2330247) | c (44868.45566)* |
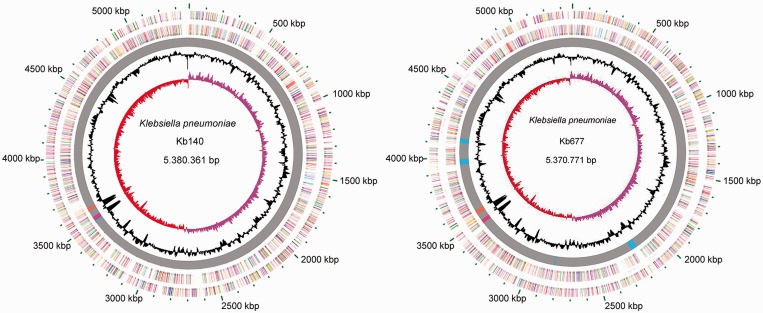
A comparison of the HHZ Region 4 among strains Kb140, Kb677, and VA360 showed that while the CPCs were identical in strains Kb140 and VA360, they were significantly different from that of strain Kb677 (fig. 2b). BLAST analyses of both versions of Region 4 indicated that only ten completed genomes include a CPC identical to that of strain Kb140 (supplementary fig. S3b, Supplementary Material online). Conversely, the CPC from strain Kb677 was found in 11 strains, 3 of them the KPNIH10, 30660/NJST258_1, and 30684/NJST258_2 that also have identical versions of HHZ Region 3 (supplementary fig. S3c, Supplementary Material online). There is a conspicuous GC% difference in value at the HHZ with respect to the rest of the genome at the locations corresponding to the Regions 3 and 4 (see fig. 3).
Fig. 3.—
Circular genome representation of the Kb140 and Kb677 genomes. Ring 1: ORF distribution, plus strand. Ring 2: ORF distribution, negative strand. ORFs are color coded based on COG classifications. Ring 3: Common regions and unique regions are represented in gray or colors, respectively. Pink and orange correspond to HHZ Regions 3 and 4. Ring 4: GC content. Ring 5: GC skew, calculated in Artemis.
WGS: Multiple Genome Alignment and Genomic Features
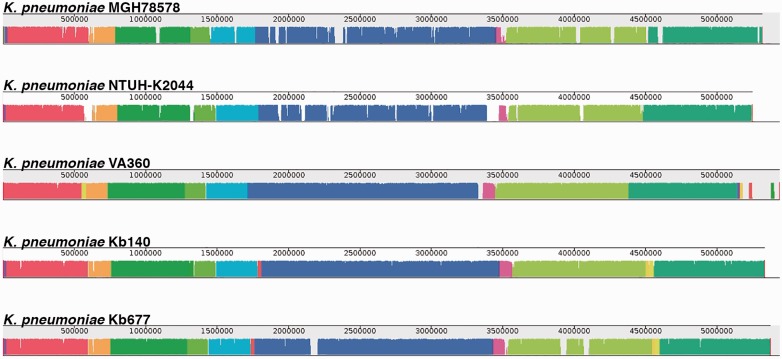
The nucleotide sequences of both strains showed 99% average identity. MAUVE alignment including the K. pneumoniae VA360, NTUH-K2044, and MGH 78578 genomes identified seven locally collinear blocks (LCBs) and no inversions (fig. 4). A 5,757-nucleotides deletion was observed in the Kb677 genome. The deleted fragment includes genes coding for homologs to a two-component regulatory system, a heat shock protein, a putative LuxR regulator, the competence damage-inducible protein A, and a hypothetical protein. A 2,096-nucleotide deletion coding for the 23S ribosomal RNA was identified in the Kb140 genome. Furthermore, 5 and 14 unique regions were found in the Kb140 and Kb677 genomes, respectively. Three of them are prophage-like sequences and considerably larger than the rest (table 3 and fig. 3).
Fig. 4.—
Alignment of K. pneumoniae strains genomes using MAUVE. Each genome’s panel contains a scale showing the sequence coordinates, colored blocks that represent regions of the genome sequence that aligned to part of the other genomes, and a single black horizontal center line where the blocks that lie above are in the forward orientation (in this case there are no blocks in reverse orientation, then the lines look to be at the bottom across the genome).
Table 3.
Unique Region of K. pneumoniae Kb140 and Kb677
| Strain | Unique region | Length size (bp) | Description |
|---|---|---|---|
| Kb140 | 1 | 866 | Second copy of DNA polymerase |
| 2 | 5245 | Genes related to nucleotide metabolism | |
| 3 | 1699 | Putative regulator genes and efflux pump protein | |
| 4 | 1255 | Efflux pump | |
| 5 | 1186 | Transposase | |
| Kb677 | 1 | 1687 | Transposase and hypothetical proteins |
| 2 | 1199 | Efflux pump protein | |
| 3 | 800 | Partial transposase sequences gene | |
| 4 | 46988 | Prophage insertion into the outer membrane protein W | |
| 5 | 6571 | Genes related to carbohydrate metabolism | |
| 6 | 1366 | Putative transposase | |
| 7 | 41536 | Phage-like proteins | |
| 8 | 1961 | Transposase, integrase and hypothetical protein | |
| 9 | 36458 | Phage-like proteins | |
| 10 | 1667 | Genes related to cell wall biogenesis/degradation and DNA repair | |
| 11 | 1326 | Putative transposase |
In both genomes 73% of the genes were assigned to a functional category including carbohydrate and amino acid metabolism, transcription and energy production and conversion (table 4). As expected, there was no significant difference in functional categories between the five strains (table 4).
Table 4.
COGs Between K. pneumoniae Strains
| COG function | MGH 78578 | NTUH-K2044 | VA360 | Kb140 | Kb677 |
|---|---|---|---|---|---|
| Information storage and processing | |||||
| [J] Translation, ribosomal structure and biogenesis | 210 | 203 | 227 | 224 | 222 |
| [A] RNA processing and modification | 1 | 1 | 1 | 1 | 1 |
| [K] Transcription | 489 | 481 | 504 | 493 | 496 |
| [L] Replication, recombination and repair | 187 | 196 | 233 | 238 | 242 |
| [B] Chromatin structure and dynamics | 1 | 1 | 1 | 1 | 1 |
| Cellular processes and signaling | |||||
| [D] Cell cycle control, cell division, chromosome partitioning | 42 | 41 | 42 | 40 | 40 |
| [V] Defense mechanisms | 64 | 57 | 64 | 67 | 65 |
| [T] Signal transduction mechanisms | 192 | 190 | 174 | 171 | 169 |
| [M] Cell wall/membrane/envelope biogenesis | 251 | 251 | 293 | 289 | 294 |
| [N] Cell motility | 69 | 59 | 53 | 53 | 51 |
| [U] Intracellular trafficking and secretion | 113 | 110 | 113 | 113 | 112 |
| [O] Post-translational modification | 151 | 156 | 192 | 183 | 183 |
| Metabolism | |||||
| [C] Energy production and conversion | 308 | 304 | 370 | 365 | 363 |
| [G] Carbohydrate transport and metabolism | 586 | 593 | 653 | 644 | 643 |
| [E] Amino acid transport and metabolism | 613 | 606 | 594 | 584 | 600 |
| [F] Nucleotide transport and metabolism | 97 | 100 | 113 | 112 | 112 |
| [H] Coenzyme transport and metabolism | 190 | 191 | 205 | 203 | 203 |
| [I] Lipid transport and metabolism | 135 | 132 | 159 | 157 | 159 |
| [P] Inorganic ion transport and metabolism | 418 | 418 | 395 | 423 | 393 |
| [Q] Secondary metabolites biosynthesis | 122 | 127 | 78 | 77 | 77 |
| Poorly characterized | |||||
| [R] General function prediction only | 680 | 667 | 674 | 665 | 667 |
| [S] Function unknown | 382 | 370 | 370 | 369 | 373 |
Conclusions
The importance of K. pneumoniae as a pathogen has significantly grown in recent years (Nordmann et al. 2009). This is, at least in part, due the rise of strains that are becoming resistant to most antibiotics used in standard treatments. In particular, the relatively recent acquisition of carbapenemase genes led to dissemination of CRKp strains that are the hardest to treat (Nordmann, et al. 2009; Munoz-Price et al. 2013; Xie et al. 2013; Ramirez et al. 2014b). These facts led us and other research groups to carry out comparative genomics studies that may help understanding the epidemiology and dissemination of CRKp strains.
Here, we compared the genomes of two strains isolated during an outbreak of CRKp infection in northeastern Ohio. Although strains Kb677 and Kb140 differ in the carbapenemase gene they carry, only minor differences were detected between their genomes, in general due to insertions and deletions. This was not surprising because as others and we have previously observed comparing other K. pneumoniae strains (Deleo et al. 2014; Ramirez et al. 2014b; Sabirova et al. 2016), the genomes of this bacterium seem to be quite homogeneous. However, in spite of the similarities between both genomes, strains Kb677 and Kb140 show significant differences at the HHZ. This region, also known as “rd”, was shown to be one of the most variable in K. pneumoniae isolates (Deleo et al. 2014). These results underscore the potential importance of the HHZ. This study shows that two strains isolated during the same outbreak could be quickly differentiated by their HHZs. We propose that this zone of the K. pneumoniae genome can be used as a signature for epidemiology analysis.
Materials and Methods
Bacterial Strains
Klebsiella pneumoniae Kb140 and Kb677 belong to ST-258 possess a different rep-PCR type (type-A for Kb140 and type-B for Kb677). Strain Kb140 harbors blaKPC-2 while Kb677 harbors blaKPC-3. Their antimicrobial susceptibilities were determined by broth microdilution according to CLSI methods and interpretations (Clinical-Laboratory-Standard-Institute 2012).
Genomic Analyses
The available scaffolds for the strains Kb140 and Kb677 (accession number AQRD01000000 and AQPG01000000, respectively) were ordered and oriented with the MAUVE Contig Mover (Darling et al. 2010), using the K. pneumoniae NTUH-K2044 genome as reference (accession number AP006725). Genomes were aligned with the open-source MAUVE aligner version 2.3.1 using the progressive algorithm. The alignments were generated using the default settings (http://darlinglab.org/mauve/mauve.html, last accessed June 8, 2016). Conserved segments that appear to be internally free from genome rearrangements, known as LCBs, were identified using MAUVE. Functional categories groups for genes have been predicted by comparison with the Clusters of Orthologous Groups (COGs) (Tatusov et al 1997). The prediction of coding sequences (CDS) was made by Rast version 4.0 software and confirmed by BLAST (Aziz et al. 2008). tRNA genes were identified using tRNAscan-SE (Lowe and Eddy 1997). Circos software was used to represent both genomes showing Open reading frame (ORF) sorted by COGs, Unique region, Region III and IV of HHZ and GC content and skew. WGMs generated at OpGen Technologies, Inc. (Gaithersburg, MD) were also used to perform the first comparison and analyzed comparing the AflII restriction maps using the MapSolver software (version 3.2.0).
Supplementary Material
Supplementary figures S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Funding
This study was supported by Public Health Service grant 2R15AI047115-04 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to M.E.T.), R01AI063517 and R01AI10056 (to R.A.B.), the U.S. Department of Energy Joint Genome Institute through the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231, and Wellcome Trust Program Grant WT083469 (to D.J.S.). The Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program award number 1I01BX001974, and the Geriatric Research Education and Clinical Center VISN 10 supported R.A.B. A.R. was supported by the LA Basin Minority Health and Health Disparities International Research Training Program (MHIRT) 5T37MD001368.
Literature Cited
- Alcantar-Curiel MD, Giron JA. 2015. Klebsiella pneumoniae and the pyogenic liver abscess: implications and association of the presence of rpmA genes and expression of hypermucoviscosity. Virulence 6:407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical-Laboratory-Standard-Institute. 2012. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved standard - Ninth edition. M07–A09.
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleo FR, et al. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 111:4988–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza R, Gutierrez J, Piedrola G. 2001. Antibiotic susceptibility of bacterial strains isolated from patients with community-acquired urinary tract infections. Int J Antimicrob Agents 18:211–215. [DOI] [PubMed] [Google Scholar]
- Ebringer A, Rashid T, Tiwana H, Wilson C. 2007. A possible link between Crohn's disease and ankylosing spondylitis via Klebsiella infections. Clin Rheumatol. 26:289–297. [DOI] [PubMed] [Google Scholar]
- Endimiani A, et al. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother. 63:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch EB, Tam VH. 2010. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 65:1119–1125. [DOI] [PubMed] [Google Scholar]
- Huang HY, Wu YH, Kuo CF. 2013. Klebsiella pneumoniae sepsis with unusual cutaneous presentation of generalized pustulosis. Clin Exp Dermatol. 38:626–629. [DOI] [PubMed] [Google Scholar]
- Kelly RF, Whitfield C. 1996. Clonally diverse rfb gene clusters are involved in expression of a family of related D-galactan O antigens in Klebsiella species. J Bacteriol. 178:5205–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liam CK, Lim KH, Wong CM. 2001. Community-acquired pneumonia in patients requiring hospitalization. Respirology 6:259–264. [DOI] [PubMed] [Google Scholar]
- Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. 2008. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol. 190:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Pan YJ, Lin TL, Fung CP, Wang JT. 2015. Transfer of CMY-2 ephalosporinase from Escherichia coli to virulent Klebsiella pneumoniae causing a recurrent liver abscess. Antimicrob Agents Chemother. 59:5000–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM Eddy SR.. 1997. Nucleic Acids Res. 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price LS, et al. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 13:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 9:228–236. [DOI] [PubMed] [Google Scholar]
- Qu TT, et al. 2015. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis. 15:161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MS, Traglia GM, Lin DL, Tran T, Tolmasky ME. 2014a. Plasmid-mediated antibiotic resistance and virulence in gram-negatives: the Klebsiella pneumoniae paradigm. Microbiol Spectr. 2: PLAS-0016-2013. [DOI] [PubMed] [Google Scholar]
- Ramirez MS, et al. 2014b. Genome sequences of two carbapenemase-resistant Klebsiella pneumoniae ST258 isolates. Genome Announc. 2:e00558–e00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MS, et al. 2012. Multidrug-resistant (MDR) Klebsiella pneumoniae clinical isolates: a zone of high heterogeneity (HHZ) as a tool for epidemiological studies. Clin Microbiol Infect. 18:E254–E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid T, Ebringer A. 2007. Ankylosing spondylitis is linked to Klebsiella—the evidence. Clin Rheumatol. 26:858–864. [DOI] [PubMed] [Google Scholar]
- Rashid T, Wilson C, Ebringer A. 2013. The link between ankylosing spondylitis, Crohn's disease, Klebsiella, and starch consumption. Clin Dev Immunol. 2013:872632.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirova JS, Xavier BB, Coppens J, Zarkotou O, Lammens C, Janssens L, Burggrave R, Wagner T, Goossens H, Malhotra-Kumar S. 2016. Whole-genome typing and characterization of blaVIM19-harbouring ST383 Klebsiella pneumoniae by PFGE, whole-genome mapping and WGS. J Antimicrob Chemother. 71:1501–1509. [DOI] [PubMed] [Google Scholar]
- Shu HY, et al. 2009. Genetic diversity of capsular polysaccharide biosynthesis in Klebsiella pneumoniae clinical isolates. Microbiology 155:4170–4183. [DOI] [PubMed] [Google Scholar]
- Snitkin ES, et al. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 4:148ra116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, et al. 2013. Septic arthritis subsequent to urosepsis caused by hypermucoviscous Klebsiella pneumoniae. Intern Med. 52:1641–1645. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. 1997. Science. 278:631–637. [DOI] [PubMed] [Google Scholar]
- van Duin D, et al. 2014. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother. 58:4035–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 75:39–68. [DOI] [PubMed] [Google Scholar]
- Woloj M, Tolmasky ME, Roberts MC, Crosa JH. 1986. Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob Agents Chemother. 29:315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, et al. 2013. Genome sequences of two Klebsiella pneumoniae isolates from different geographical regions, Argentina (Strain JHCK1) and the United States (Strain VA360). Genome Announc. 1:e00168–e00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zowawi HM, et al. 2015. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat Rev Urol. 12:570–584. [DOI] [PubMed] [Google Scholar]