Abstract
All eukaryotic genomes encode multiple members of the heat shock protein 70 (HSP70) family, which evolved distinctive structural and functional features in response to specific environmental constraints. Phylogenetic analysis of this protein family thus can inform on genetic and molecular mechanisms that drive species-specific environmental adaptation. Here we use the eukaryotic pathogen Leishmania spp. as a model system to investigate the evolution of the HSP70 protein family in an early-branching eukaryote that is prone to gene amplification and adapts to cytotoxic host environments by stress-induced and chaperone-dependent stage differentiation. Combining phylogenetic and comparative analyses of trypanosomatid genomes, draft genome of Paratrypanosoma and recently published genome sequences of 204 L. donovani field isolates, we gained unique insight into the evolutionary dynamics of the Leishmania HSP70 protein family. We provide evidence for (i) significant evolutionary expansion of this protein family in Leishmania through gene amplification and functional specialization of highly conserved canonical HSP70 members, (ii) evolution of trypanosomatid-specific, non-canonical family members that likely gained ATPase-independent functions, and (iii) loss of one atypical HSP70 member in the Trypanosoma genus. Finally, we reveal considerable copy number variation of canonical cytoplasmic HSP70 in highly related L. donovani field isolates, thus identifying this locus as a potential hot spot of environment–genotype interaction. Our data draw a complex picture of the genetic history of HSP70 in trypanosomatids that is driven by the remarkable plasticity of the Leishmania genome to undergo massive intra-chromosomal gene amplification to compensate for the absence of regulated transcriptional control in these parasites.
Keywords: Leishmania, heat shock protein, HSP70, evolution, phylogeny, synteny, copy number variation, gene loss
Introduction
Members of the heat shock protein 70 (HSP70) super family are ubiquitous molecular chaperones that play an essential role in the maintenance of cellular homeostasis in almost all organisms. These proteins are implicated in a variety of essential cellular processes, including protein folding, assembly, or refolding, thereby modulating their activity, interaction, and localization (Boorstein et al. 1994; Mayer and Bukau 2005). Although HSP70 family members have a wide range of diverse functions, their sequence and structure are highly conserved. All HSP70 members show a characteristic N-terminal ATPase domain, and a C-terminal portion containing a conserved substrate-binding domain (Liu and Hendrickson 2007). HSP70 activity and substrate binding is regulated in an adenosine triphosphate (ATP)-dependent fashion, with HSP70-adenosine diphosphate (ADP) showing high substrate affinity that is reduced in its ATP-bound state (Szabo et al. 1994 ; Suh et al. 1999). HSP70 family members that are able to accomplish the full ATP binding and release cycle are defined as “canonical”, whereas atypical HSP70 family members do not follow this functional cycle (Shaner et al. 2006). The latter ones have likely acquired new functions, including nucleotide-exchange activity that synergizes with Hsp40/DnaJ-type co-chaperones to accelerate HSP70 nucleotide cycling (Dragovic et al. 2006; Raviol et al. 2006; Shaner et al. 2006; Steel et al. 2004).
Despite the high degree of inter-species sequence conservation of individual HSP70 family members, the evolution of the HSP70 protein family is very dynamic and often highly adapted to species-specific constraints. This dynamics is documented by substantial variations across eukaryotic organisms in HSP70 gene copy number (Daugaard et al. 2007), the number of phylogenetically distinct sub-families, and the evolution of unique phylogenetic groups or atypical family members (Hughes 1993; Boorstein et al. 1994; Gupta and Singh 1994; Kampinga and Craig 2010; Kominek et al. 2013). Genetic adaptation of the HSP70 protein family is especially well illustrated in unicellular eukaryotes, notably pathogenic protists that often have to adapt to different environments during their infectious cycle. Parasitic protists of the order Trypanosomatida, including Trypanosoma brucei, T. cruzi, and Leishmania spp. (referred to as TriTryp), are particularly interesting organisms to investigate the evolutionary potential of the HSP70 family for several reasons.
First, HSP and chaperone proteins play an essential role in TriTryp adaptation to environmental changes and stress-induced stage differentiation, and thus are important for disease transmission and pathogenesis (Requena et al. 2015). For example Leishmania parasites show a digenetic life cycle, alternating between extracellular promastigotes that develop inside the midgut of phlebotomine sandflies, and intracellular amastigotes that multiply within macrophages of the mammalian host. This stage differentiation is regulated by several environmental factors, notably pH and temperature shifts from pH 7.4/26 °C in the insect vector to pH 5.5/37 °C in the vertebrate host for visceral Leishmania species (Zilberstein and Shapira 1994; Sibley 2011). Using various stress protein inhibitors, including geldanamycin and cyclosporine A, a regulatory role of the Leishmania chaperones HSP90 and cyclophilin 40 in stage development has been uncovered (Wiesgigl and Clos 2001; Yau et al. 2010). Thus stress protein activities are functionally linked to adaptive differentiation, which may have unique consequences for chaperone evolution in these organisms.
Second, trypanosomatids represent a family belonging to the likely early-branching eukaryotic supergroup Excavata (e.g., Hampl et al. 2009; He et al. 2014; Forterre 2015) with unique biological and genetic features that may impact on evolution of the HSP70 family. In particular, regulation of gene expression in these eukaryotes does not follow the paradigm of transcriptional regulation via trans-acting factors that regulate transcript abundance by binding to cis-acting sequence elements. Indeed, TriTryp genomes do not code for regulatory transcription factors and gene expression is largely constitutive (Clayton 2002; Leifso et al. 2007). As a consequence, the need for increased chaperone activity under stress conditions is fulfilled either by increased stability or preferential translation of respective mRNA (Quijada et al. 2000; Zilka et al. 2001; David et al. 2010; Droll et al. 2013), HSP phosphorylation (Hem et al. 2010; Morales et al. 2010), or amplification of HSP genes (Wiesgigl and Clos 2001). Conceivably, the potential regulation of HSP70 protein abundance by gene duplication may have a major impact on the evolution of this family in these protists.
Despite the importance of environmental stress in development and infectivity of pathogenic trypanosomatids, and their unique biology to regulate protein abundance independent of transcriptional regulation, only little is known on how these features shaped the evolution of stress protein families in these early-branching eukaryotes. Here, combining phylogenetic and comparative analyses of the trypanosomatid genomes, a draft genome of early-branching Paratrypanosoma and recently published genome sequences of 204 L. donovani field isolates from the Indian sub-continent (Imamura et al. 2016), we uncover unique, parasite-specific features in both canonical and non-canonical HSP70 family members. We demonstrate genomic expansion of this family in Leishmania, and provide evidence for its adaptation at various taxonomic levels ranging from the trypanosomatid family down to parasite strain. Our data uncover that the trypanosomatid HSP70 protein family is shaped by gene loss and gene birth as a result of the remarkable genome plasticity of these eukaryotic pathogens.
Materials and Methods
Genome Information and Sequencing
A draft genome sequence of Paratrypanosoma confusum (strain CUL13-MS, Flegontov et al. 2013) was produced by sequencing total DNA using Illumina MiSeq system with paired-end (insert size 450 bp) and mate-pair (insert size 2–6 kb) libraries. AUGUSTUS 2.5.5 (Stanke et al. 2006) was used for genome annotation, which was further manually improved. An updated draft genome sequence of Leishmania donovani (BPK282/0cl4, referred to in this manuscript using the internal reference number LdPBQ7IC8 to better distinguish this sequence from the current reference LdBPK282A1) was created using Pacific Biosciences Single Molecule, Real-Time (SMRT) Sequencing (unpublished) with a weighted median length (N50) of 11.7 kb. Final iterative base and indel corrections were performed with BPK282/0cl4 Illumina reads used for the previous assembly (Downing et al. 2011). Sequencing information of 204 field isolates of L. donovani together with corresponding clinical and epidemiological meta-data were published elsewhere (Imamura et al. 2016).
SNPs Analysis
Single-nucleotide polymorphisms (SNPs) were identified using read aligner SMALT v7.4 (sourceforge.net/projects/smalt/files/) and genetic variation identifying tool GenomeAnalysisTK-3.4 (GATK) Unified Genotyper (McKenna et al. 2010). All candidate SNPs were visually checked and false positive SNPs were removed using the Integrative Genomic Viewer (IGV_2_3_47, Thorvaldsdottir et al. 2013). Allele frequency was generated using samtools pilup repetitive regions where reads were mapped identically at multiple locations because GATK did call genetic variants on these regions. Median copy number of the HSP70 genes was calculated based on alignment depth and was calibrated to be one for a single copy gene on a diploid chromosome. When reads were mapped to multiple locations equally, they were randomly assigned to one position.
HSP70 Family Member Search
A total of 16 coding sequences (CDS) were retrieved from the L. major CDS set (as available in GeneDB, Logan-Klumpler et al. 2012) by performing reciprocal BLAST searches (blastp, Altschul et al. 1997) using human HSPA1 as a query. Each of these 16 putative HSP70 members were used as query to perform a PSI-BLAST search (Altschul et al. 1997) against all L. major predicted coding sequence (CDS), but no new homologous sequences were discovered. Finally, all 16 CDS were confirmed to belong to the HSP70 family by comparison with its corresponding Pfam profile (pfam00012; Finn et al. 2015).
Gene Density Analysis
Different sets of CDS were collected from public databanks for six euglenozoans, four apicomplexans, one amoebozoan, five fungi, and two metazoa (see the full taxon list in fig. 3). In order to gather HSP70 family members of each of these selected taxa, the allele sequence of LmjF.28.2770 was used as a query to perform a first BLAST similarity search (blastp) against each CDS sets, and each of the first hits was used as query to perform a PSI-BLAST search. The HSP70 gene density was estimated by the percentage of CDS belonging to the HSP70 family.
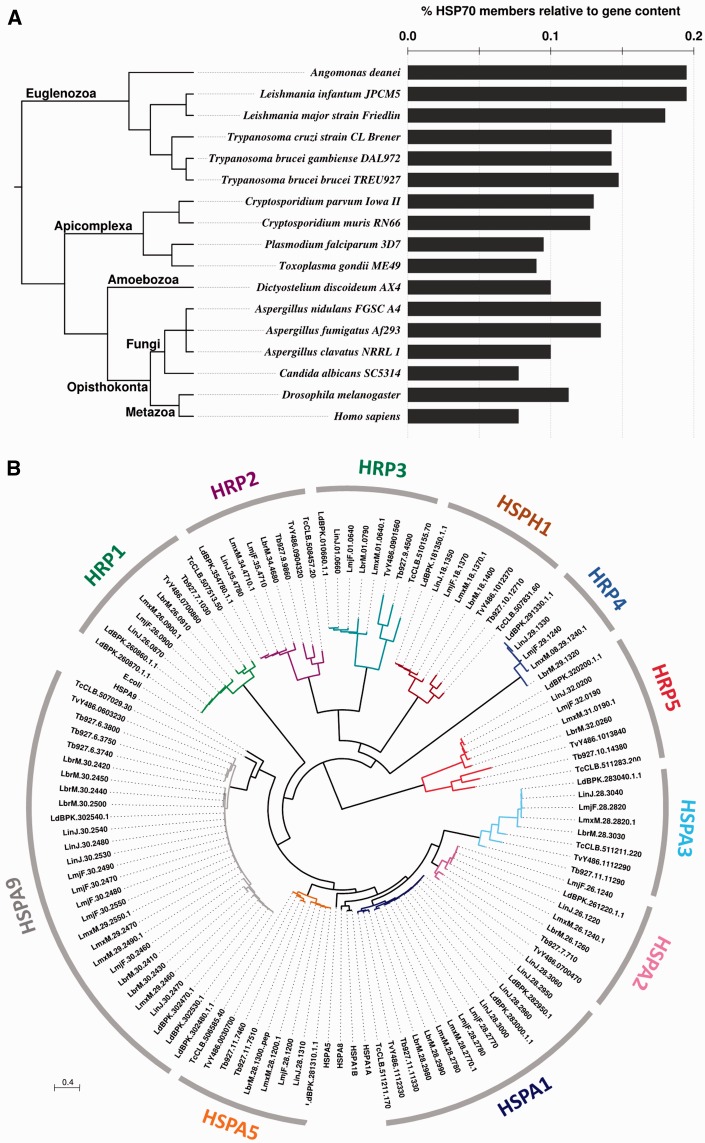
Fig. 3.—
Comparative analyses of the L. major HSP70 family. (A) HSP70 gene coding density. Number of HSP70 members relative to the genome size of selected eukaryotes is shown. The phylogenetic relationship (left) is freely derived from consensus knowledge on eukaryote systematics (Adl et al. 2012; He et al. 2014; Williams 2014). (B) Unrooted ML phylogenetic tree of the TriTryp HSP70 family. E. coli DnaK (acc. no. WP_000516138.1), human HSPA9 (AAH24034.1), HSP1A1A (AAH18740.1), HSPA1B (EAX03529.1), HSPA5 (AAI12964.1), and HSPA8 (AAH07276.2) were included to identify the cluster of potential mitochondrial, cytoplasmic, or ER HSP70 family members. The different colors correspond to different phylogenetic clusters that are labeled according to table 1. Scale bar represents 0.4 amino acid substitutions per site.
Multiple Sequence Alignments and Phylogenetic Analyses
Multiple amino acid sequence alignments were obtained with MAFFT (Katoh and Standley 2013). All phylogenetic analyses were performed on aligned character regions selected with BMGE (Criscuolo and Gribaldo 2010). Tree inferences were performed by PhyML (Guindon and Gascuel 2003) with SPR-based optimal tree searches (Guindon et al. 2010) and amino acid evolutionary model LG + Γ4 + I (Le and Gascuel 2008). In order to minimize the number of irresolutions within phylogenetic trees inferred from very similar amino acid sequences, some specific phylogenetic reconstructions were performed from codon sequence alignments obtained by back-translating multiple amino acid sequence alignments. Aligned character regions were selected by BMGE and phylogenetic trees were inferred by PhyML with evolutionary model GTR + Γ4 + I (Rodríguez et al. 1990). To explore the evolutionary history of HRP4, we have created datasets for phylogenetic analyses from publicly available sequences for HRP4 orthologs and conserved cytosolic Hsp70 (LinJ.28.2960) using blastp at an E-value cut-off of 10−20. All amino acid sequences were validated for the presence of expected domains by Pfam.
Synteny Assessment
Interactive visualization of the syntenic organization of the Leishmania genomes was performed using the SynTView web tool (Lechat et al. 2013). Results are publicly available via a web interface at the following URL http://genopole.pasteur.fr/SynTView/flash/Leishmania/SynWeb.html (last accessed June 18, 2016) for four Leishmania genomes: L. major strain Friedlin (NC_001905, NC_004916, NC_007244-73, NC_007284-87; Ivens et al. 2005; Rogers et al. 2011), L. infantum JPCM5 (NC_009277, NC_009386-420; Peacock et al. 2007; Rogers et al. 2011), L. donovani BPK282A1 (NC_018228-63; Downing et al. 2011), and LdPBQ7IC8.
Results
Characterization of L. major HSP70 Protein Family Members
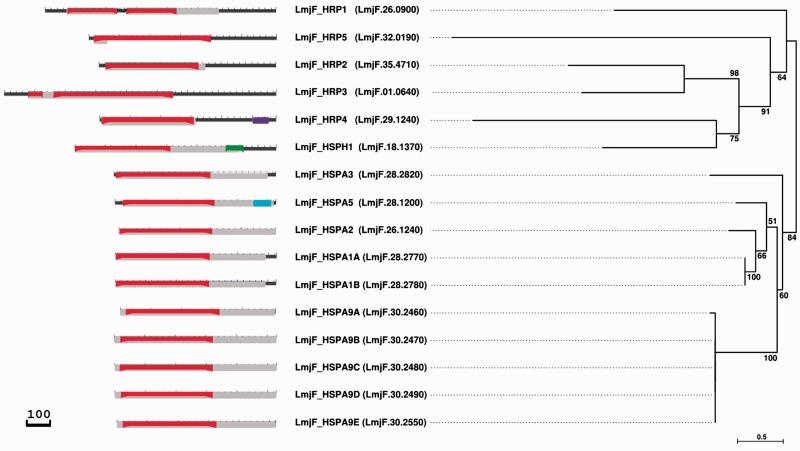
Members of the HSP70 protein family are characterized by conserved signature domains and sequence elements, including an N-terminal nucleotide binding domain of about 45 kDa (NBD) and a C-terminal substrate binding domain of about 25 kDa (SBD), which both are connected through a protease sensitive linker element (Liu and Hendrickson 2007). Using reciprocal BLAST and PSI-BLAST searches, we identified a total of 16 genes within the L. major CDS with homology to either the HSP70 domain (accession numbers pfam00012, PTZ00009, and PTZ00186) or the NBD sub-domain (accession numbers cd10233, cd11733, and cd10170) with E-values of 0–3.6×10−4, and 0–2.6×10−4, respectively (fig. 1, left panel and table 1). Five family members (LmjF.26.0900, LmjF.18.1370, LmjF.35.4710, LmjF.29.1240, and LmjF.32.0190) showed considerable C-terminal sequence extensions that are conserved across Leishmania species (data not shown), likely conferring highly trypanosomatid-specific functions to these proteins. In addition, unlike most eukaryotic HSP70 proteins, three L. major HSP70 members showed additional domains, including a domain of unknown function (DUF3919) characterized by a conserved YLNG motif (LmjF.28.1200), a Hydantoiniase/Oxoprolinase domain (accession number pfam01968) involved in hydrolysis of pyrimidine precursors (Syldatk et al. 1999; LmjF.18.1370), and a tetratricopeptide (TPR) domain (accession number pfam13424) involved in protein–protein interaction (Blatch and Lassle 1999; LmjF.29.1240; see table 1 and fig. 1).
Fig. 1.—
Identification of L. major HSP70 family members. Domain structure (left) and unrooted ML phylogenetic tree (right) of L. major HSP70 family members (proposed and current gene names are denoted in the middle). Domain structures were built using the NCBI Batch CD-search (Marchler-Bauer et al. 2015) and curated manually respecting the identified domain boundaries denoted in table 1. Black, protein backbone; red, nucleotide binding domain (cd10233, cl17037, cd11733, cd10228, cd10230, and cd10170); gray, HSP70 domain (PTZ00009, pfam00012, PTZ00186, and PTZ00400); purple, TPR domain (pfam00515, cl22897, and pfam13424); cyan, protein domain of unknown function (DUF3919); green, Hydantoinase/oxoprolinase domain (pfam01968). Left and right scale bars represent 100 amino acids and 0.5 amino acid substitutions per site, respectively. Confidence support at the branches of the tree is based on 100 bootstrap replicates.
Table 1.
The L. major HSP70 protein family
| Gene ID | MW | AA | GeneDB annotation | Namea | Domains (E-value) |
|---|---|---|---|---|---|
| LmjF.28.2770 | 71.7 | 658 | HSP70, putative | LmjF_HSPA1A | cd10233, 6-384 (0); cl17037, 6-384 (0); PTZ00009, 1-616 (0); pfam00012, 6-615 (0) |
| LmjF.28.2780 | 71.7 | 658 | HSP70, putative | LmjF_HSPA1B | cd10233, 6-384 (0); cl17037, 6-384 (0); PTZ00009, 1-616 (0); pfam00012, 6-615 (0) |
| LmjF.26.1240 | 70.6 | 641 | HSP70.4 | LmjF_HSPA2 | cd10233, 6-382 (0); cl17037, 6-382 (0); PTZ00009, 1-641 (0); pfam00012, 6-613 (0) |
| LmjF.28.1200 | 71.9 | 658 | BiP | LmjF_HSPA5 | cd10233, 36-408 (0); cl17037, 36-408 (0); PTZ00009, 34-649 (0); pfam00012, 38-642 (0); pfam01968, 208-250 (9e-05); cl00668, 208-250 (9e-05); pfam01968, 208-250 (9.0e-05); cl00668 208-250 (9.0e-05); pfam13057, 568-642 (4.6e-3); cl16063, 568-642 (4.6e-3) |
| LmjF.30.2460 | 68.9 | 635 | HSP70-related protein 1 | LmjF_HSPA9A | cd11733, 26-402 (0); cl17037, 26-402 (0); PTZ00186, 1-633 (0); pfam00012, 29-625 (0) |
| LmjF.30.2470 | 71.9 | 662 | HSP70-related protein 1 | LmjF_HSPA9B | cd11733, 26-402 (0); cl17037, 26-402 (0); PTZ00186, 1-662 (0); pfam00012, 29-625 (0) |
| LmjF.30.2480 | 71.9 | 662 | HSP70-related protein 1 | LmjF_HSPA9C | cd11733, 26-402 (0); cl17037, 26-402 (0); PTZ00186, 1-662 (0); pfam00012, 29-625 (0) |
| LmjF.30.2490 | 71.6 | 660 | HSP70-related protein 1 | LmjF_HSPA9D | cd11733, 26-402 (0); cl17037, 26-402 (0); PTZ00186, 1-660 (0); pfam00012, 29-625 (0) |
| LmjF.30.2550 | 70.6 | 652 | HSP70-related protein 1 | LmjF_HSPA9E | cd11733, 26-402 (0); cl17037, 26-402 (0); PTZ00186, 1-652 (0); pfam00012, 29-625 (0) |
| LmjF.28.2820 | 72.3 | 662 | HSP70, putative | LmjF_HSPA3 | cd10233, 10-395 (1.6e-172); cl17037, 10-395 (1.6e-172); pfam00012, 10-628 (0); PTZ00009, 6-555 (0) |
| LmjF.18.1370 | 91.8 | 823 | HSP110, putative | LmjF_HSPH1 | cd10228, 2-390 (0); cl17037, 2-390 (0); pfam02782, 286-388 (4.4e-06); cl17173, 618-689 (8.5e-3) |
| LmjF.26.0900 | 104.1 | 944 | HSP70-like protein | LmjF_HRP1 | cd10170, 94-538 (3.5e-92); cl17037, 94-538 (3.5e-92); pfam00012, 93-545 (5.7e-69); PTZ00400, 84-547 (1.8e-52) |
| LmjF.35.4710 | 78.8 | 723 | Hypothetical protein | LmjF_HRP2 | cd10230, 28-407 (1.7e-138); cl17037, 28-407 (1.7e-138); pfam00012, 27-436 (3.2e-43); PTZ00186, 5-407 (2.7e-28) |
| LmjF.01.0640 | 117.5 | 1112 | HSP70-like protein | LmjF_HRP3 | cd10230, 99-690 (3.1e-74); cl17037, 99-690 (3.1e-74); pfam00012, 335-689 (1.6e-19); PTZ00186, 399-689 (4.2e-13); |
| LmjF.29.1240 | 78.9 | 722 | Hypothetical protein | LmjF_HRP4 | cd10170, 8-384 (1.2e-29); cl17037, 8-384 (1.2e-29); pfam00012, 8-386 (3.2e-14); PTZ00186, 8-386 (10e-06); pfam00515, 659-692 (2e-4); cl22897, 659-692 (2e-4); pfam13424, 625-691 (3.4e-4) |
| LmjF.32.0190 | 81.5 | 768 | Hypothetical protein | LmjF_HRP5 | cd10170, 23-498 (2.2e-06); cl17037, 23-498 (2.2e-06); pfam00012, 23-73 (2.6e-4); PTZ00186, 20-73 (3.6e-4) |
Proposed name following the nomenclature assigned by the HUGO Gene Nomenclature Committee (http://www.genenames.org/genefamilies/HSP, last accessed June 18, 2016) and used in the NCBI Entrez Gene database for human HSPs. cd10233, Nucleotide-binding domain of HSPA1-A, -B, -L, HSPA-2, -6, -7, -8, and similar proteins (HSPA1-2_6-8-like_NBD); cl17037, nucleotide-binding domain of the sugar kinase/HSP70/actin superfamily (NBD_sugar-kinase_HSP70_actin); PTZ00009, heat shock 70 kDa protein domain; pfam00012, HSP70 domain; pfam01968, Hydantoinase/oxoprolinase domain; cl00668, Hydantoinase_A Superfamily; pfam13057, Protein domain of unknown function (DUF3919); cl16063, protein domain of unknown function (DUF3919); cd11733, nucleotide-binding domain of human HSPA9, Escherichia coli DnaK, and similar proteins (HSPA9-like_NBD); PTZ00186, heat shock 70 kDa precursor protein domain; cd10228, nucleotide-binding domain of 105/110 kDa heat shock proteins including HSPA4 and similar proteins (HSPA4_like_NDB); pfam02782, FGGY family of carbohydrate kinases, C-terminal domain (FGGY_C); cl17173, AdoMet_MTases superfamily; cd10230, nucleotide-binding domain of human HYOU1 and similar proteins (HYOU1-like_NBD); cd10170, nucleotide-binding domain of the HSP70 family (HSP70_NBD); PTZ00400, DnaK-type molecular chaperone; pfam00515, tetratricopeptide repeat domain TPR_1; cl22897, TPR_1 superfamily; pfam13424, tetratricopeptide repeat domain TPR_12. In bold are the domains shown in fig. 1.
We next performed phylogenetic analysis of the L. major HSP70 family members to gain insight into their evolutionary relationship and to propose new allele names in accordance with the recommendations of the HUGO Gene Nomenclature Committee (www.genenames.org/genefamilies/HSP, last accessed June 18, 2016). The tree in fig. 1 displays a strongly supported clade (i.e., 100% bootstrap support) of five highly related genes (see fig. 2) encoded on chromosome 30 (LmjF.30.2460-90 and LmjF.30.2550), which we propose to name LmjF_HSPA9A-E (table 1 and fig. 1) as they cluster together with human mitochondrial HSP70 member HSPA9 with strong bootstrap support (supplementary fig. S1, Supplementary Material online) and are characterized by a C-terminal acidic poly-glutamine stretch diagnostic for mitochondrial HSP70 members (Louw et al. 2010; see fig. 2B). Even though LmjF.30.2460 lacks this signature motif, its phylogenetic relationship to the other members of this cluster defines this gene as an LmjF_HSPA9 homolog.
Fig. 2.—
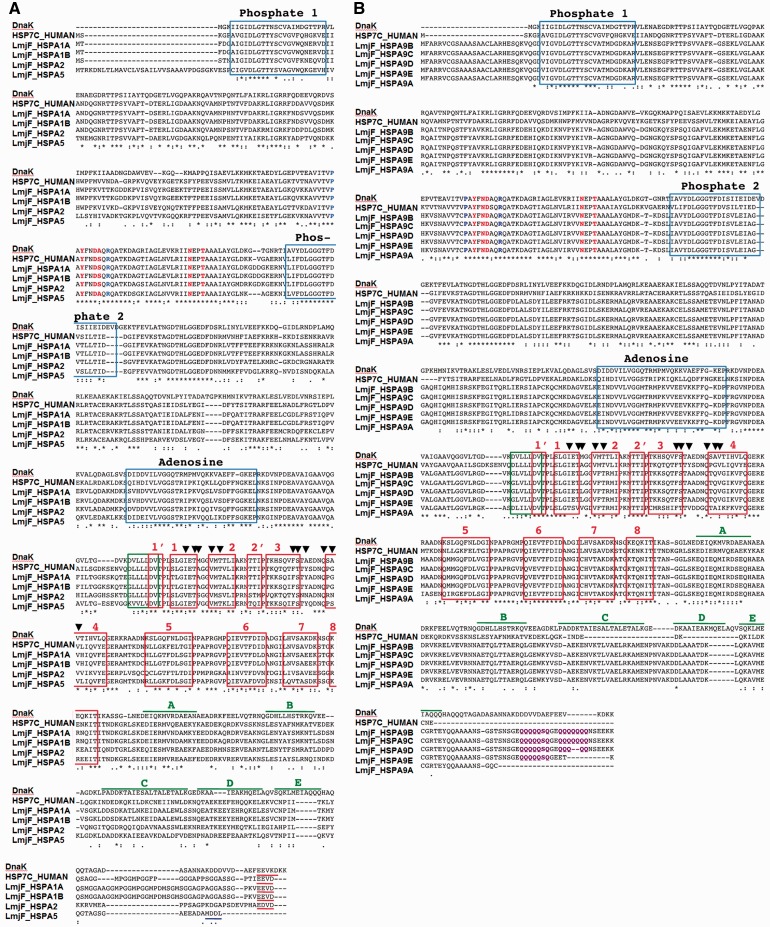
Multiple sequence alignment of cytoplasmic (A) and mitochondrial (B) canonical L. major HSP70 members. Sequences of the L. major HSP70 family members were aligned with E. coli DnaK (acc. no. EDX37800) and human HSC70 (acc. no. P11142). Blue boxes, sequence elements implicated in nucleotide binding (phosphate-1, phosphate-2, and adensoine); green box, linker; red boxes, sub-domains of the beta-sheet; blue, residues involved in allosteric switching and inter-domain function (P143 and R151 in E. coli DnaK; Vogel, et al. 2006); red, residues interacting with the DnaJ domain of HSP40 (Y145, N147, D148, N170, and T173 in E. coli DnaK; Gassler et al. 1998; Suh et al. 1998); purple, acidic Q stretch characteristic of mitochondrial HSP70; black arrow heads, residues in contact with the substrate (Mayer et al. 2000); red underlined, terminal EEV motif; blue underlined, terminal ER retention signal (Pelham 1989); green lines, lid sub-domains of DnaK. Adapted from Shonhai, et al. 2007.
The tree in fig. 1 also reveals four HSP70 family members (LmjF.28.2770, LmjF.28.2780, LmjF.28.1200, and LmjF.26.1240) that are grouped together. Despite the corresponding clade being only moderately supported (i.e., 51% bootstrap support caused by a phylogenetic signal that is likely too weak to lead to a significant bootstrap support for this deep part of the tree; see also supplementary fig. S1, Supplementary Material online), they show high conservation of NBD, linker, SBD, signature domains involved in phosphate and adenosine binding, and residues that interact with HSP40 or are implicated in substrate binding or allosteric regulation of inter-domain function (fig. 2A). Based on the presence of a conserved C-terminal EEVD motif and clustering with the canonical human cytosolic HSP70 members HSPA1A (supplementary fig. S1, Supplementary Material online), we propose to name LmjF.28.2770, LmjF.28.2780, and LmjF.26.1240, respectively, LmjF_HSPA1A, LmjF_HSPA1B, and LmjF_HSPA2 (table 1 and fig. 1). Clustering with human HSP5A (supplementary fig. S1, Supplementary Material online) and the presence of a conserved MDDL ER-retention motif (fig. 2A) reveals LmjF.28.1200 as the Leishmania ortholog of BiP (Folgueira and Requena 2007), which is consequently named LmjF_HSPA5 (table 1 and fig. 1). Another conserved L. major HSP70 family member includes LmjF.28.2820 (51% identity to the LmjF_HSPA1 HSP70 domain; see supplementary table S1, Supplementary Material online) that we propose to name LmjF_HSPA3 (table 1, and fig. 1 and supplementary fig. S2, Supplementary Material online).
Finally, five L. major HSP70 family members show substantial divergence to the canonical HSP70 domain (upper clade in fig. 1 and supplementary fig. S1, Supplementary Material online), including LmjF.26.0900, LmjF.35.4710, LmjF.01.0640, LmjF.29.1240, and LmjF.32.0190, which we propose to name, respectively, Leishmania HSP70-related protein (HRP) 1, 2, 3, 4, and 5 (supplementary figs. S4–S8, Supplementary Material online, table 1 and supplementary table S1, Supplementary Material online). These five sequences made up a strongly supported clade (i.e., 84 bootstrap support), with one additional member, LmjF.18.1370 (table 1 and fig. 1 and supplementary fig. S3, Supplementary Material online), that is currently annotated as putative HSP110 and is proposed to be named LmjF_HSPH1 (table 1 and fig. 1) as it groups in proximity with members of the human HSP70-related sub-family HSPH (supplementary fig. S1, Supplementary Material online).
The Leishmania HSP70 Family Shows Evolutionary Expansion Compared to Other Eukaryotes and is Largely Conserved across Trypanosomatids
Given the small size of the L. major reference genome of 32.8 Mb, the high number of HSP70 family members may indicate evolutionary expansion, likely as an adaptation to the various forms of stress encountered during the parasitic life cycle. Comparison of the HSP70 gene density (number of HSP70 genes normalized to total gene number, see supplementary table S2, Supplementary Material online) supports this hypothesis, as judged by a higher density in trypanosomatids compared with most other eukaryotes, including apicomplexan parasites or pathogenic fungi (fig. 3A). This expansion is reflected for example by the amplification and functional specialization of the mitochondrial HSP70 members as detailed below (see fig. 5C). Surprisingly, despite exposure to similar environmental challenges during infection, the number of HSP70 orthologs between trypanosomatids is significantly different, suggesting specific evolution of the HSP70 family down to the species level. We further investigated species-specific HSP70 evolution across the order Trypanosomatida by phylogenetic analysis.
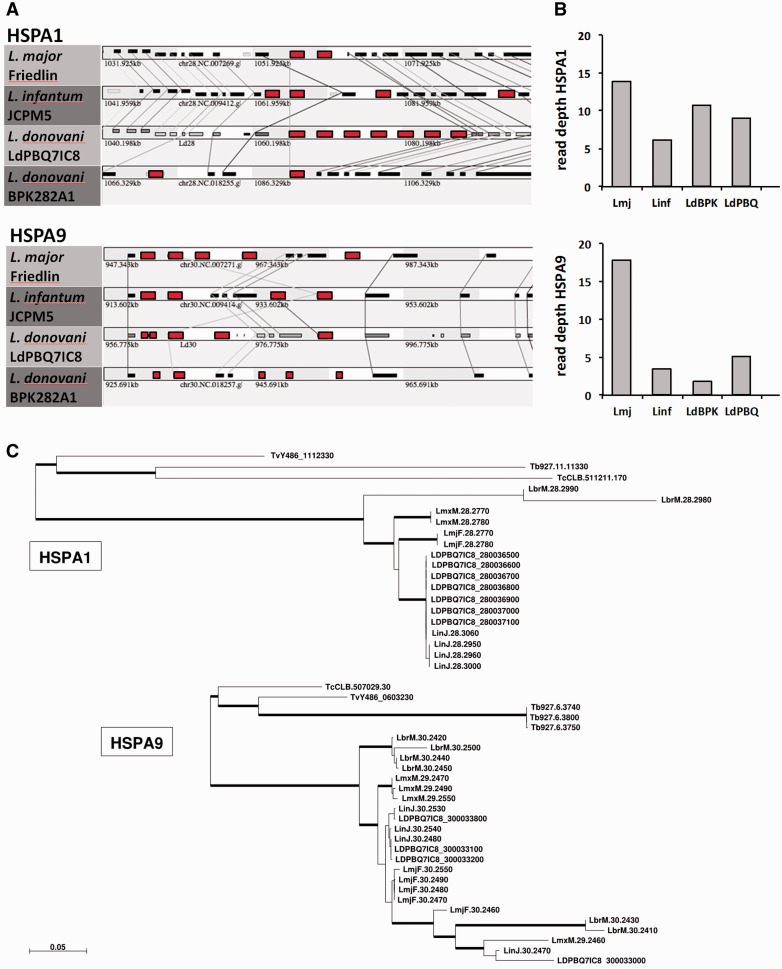
Fig. 5.—
Species-specific copy number variation of the Leishmania HSPA1 and HSPA9 sub-families. (A) Synteny analysis. Synteny view of the HSPA1 (upper panel, colored in red) and HSPA9 loci (lower panel, colored in red) across the different Leishmania species indicated. Homologous genes are connected by lines. (B) Read depth analysis. The cumulative number of read counts that map to the annotated HSPA1 and HSPA9 genes in the various reference strains is plotted. Lmj, L. major Friedlin; Linf, L. infantum JCPM5; LdBPK, L. donovani LdBPK282A1; LdBPQ, L. donovani LdPBQ7IC8. (C) ML phylogenetic trees of HSPA1 and HSPA9 sequences. Thick branches correspond to bootstrap-based confidence support > 70%. Scale bar represents 0.05 nucleotide substitutions per site. Of note, same ML trees were inferred when using codon evolutionary models (Gil et al. 2013).
Homologous sequences of the L. major HSP70 members were retrieved for various Leishmania and Trypanosoma species, and phylogenetic analysis was performed including canonical human and bacterial HSP70 sequences, leading to overall 11 distinct groups (i.e., HSPA1, HSPA2, HSPA3, HSPA5, HSPA9, HRP1, HRP2, HRP3, HRP4, HRP5, and HSPH1; see fig. 3B). Most HSP70 members in these groups show a closer relationship across the various Leishmania and Trypanosoma species than to other HSP70 family members inside the same species, suggesting that duplication events at the origin of these genes occurred in a common trypanosomatid ancestor. In contrast, the L. major orthologs for human HSPA1A/B and HSPA9, which include two and five largely identical genes, respectively, cluster out in a species-specific manner (see also fig. 5C and supplementary fig. S1, Supplementary Material online).
Despite the high conservation of the various HSP70 family members across the trypanosomatids, there are several interesting differences that inform on the dynamic evolution of this protein family in these closely related protists. First, the genomes of Trypanosoma spp. do not code for an HRP4 ortholog, which was either acquired in Leishmania spp. after the evolutionary separation of both trypanosomatid genera, or selectively lost in the Trypanosoma lineage. Second, the number of canonical cytoplasmic and mitochondrial HSP70 members varies considerably among trypanosomatids, suggesting species-specific evolution that may be driven by different environmental constraints. Below, we investigate in more detail these two important evolutionary aspects.
Leishmania HRP4 is a Co-chaperone with Unusual Domain Structure Lost from the Trypanosoma Genome
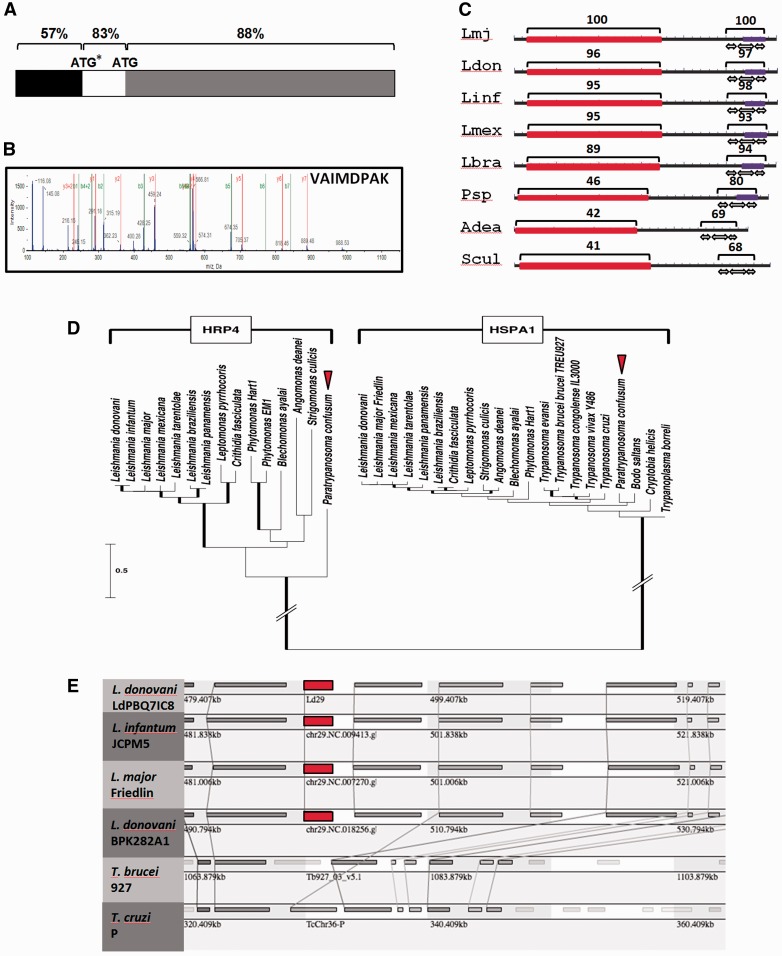
The Leishmania HSP70 family member HRP4 attracted our attention for further bioinformatics assessment due to its unusual domain structure and its absence in the related genus Trypanosoma. An initial analysis by multiple sequence alignment revealed an N-terminal sequence extension of 153 nucleotides unique to the otherwise highly conserved L. tarentolae HRP4 homolog (supplementary fig. S10, Supplementary Material online). We predicted a similar extension of 171 nucleotides to be also part of the L. donovani HRP4 coding sequence based on our own manual ORF analysis performed with the genome sequence surrounding this locus (data not shown), and the high sequence conservation of 83% in this region compared to 57% of the putative 5′ UTR (fig. 4A and supplementary fig. S11, Supplementary Material online). Re-interrogation of our previous proteomics analysis (Hem et al. 2010) indeed identified a diagnostic peptide for this longer L. donovani HRP4 CDS (fig. 4B), thus confirming its expression and revealing mis-annotation of HRP4 (with the exception of L. tarentolae) in all current Leishmania public genome releases that lack this N-terminal part of the protein.
Fig. 4.—
Absence of HRP4 in the Trypanosoma spp. genome is due to chromosomal deletion. (A) Re-annotation of the L. donovani HRP4 ORF. Schematic representation of the L. donovani HRP4 CDS region. The percent nucleotide identity is indicated for 5′ UTR (black), the putative additional coding sequence of 171 nucleotides of L. donovani HRP4 (white), and the currently annotated ORF (gray). The numbers indicate the percent of nucleotide identity to the L. tarentolae HRP4 CDS region (see supplementary fig. S11, Supplementary Material online). ATG, currently mis-annotated start codon; ATP*, correct start codon identified in this study. (B) Validation of the HRP4 ORF by proteomics analysis. The spectrum of the peptide VAIMDPAK that is diagnostic for the additional 57 amino acids of the longer ORF is shown as identified by LC-MS-MS analysis of L. donovani LdBob (Hem et al. 2010). (C) HRP4 domain structure. The domain structure for HRP4 was built using the NCBI Batch CD-search (Marchler-Bauer et al. 2015) and curated manually. The number indicates % identity to the L. major HRP4 protein across the domains indicated. Black, protein backbone; red, nucleotide binding domain (cd10170); purple, TPR domain (pfam13424); white arrows, TPR repeats. (D) ML phylogenetic tree of HRP4 and HSPA1 sequences. Thick branches correspond to bootstrap-based confidence support > 70%. Scale bar represents 0.5 amino acid substitutions per site. Of note, the long-branch joining the two clades was shortened for better reading. (E) Synteny analysis of HRP4. Synteny view of the HRP4 locus (red) and surrounding genes across the indicated Leishmania and Trypanosoma species. Homologous genes are connected by lines. The HRP4 gene is indicated in red.
We next investigated the unusual domain structure of HRP4, which is composed of a divergent, N-terminal, HSP70-related NBD, and a C-terminal TPR domain (fig. 1 and supplementary S7, Supplementary Material online and table 1) that can confer interaction with HSP70 and HSP90 (Blatch and Lassle 1999). Both domains are equally well conserved across different Leishmania species (fig. 4C and supplementary fig. S12, Supplementary Material online), but show a distinct evolutionary pace when compared to the HRP4 orthologs in the related parasitic plant trypanosomatid Phytomonas spp., and parasitic insect trypanosomatids Angomonas deanei and Strigomonas culicis. The respective TPR domains of these organisms show between 68 and 80% sequence identity to L. major HRP4, whereas the sequence identity of the NBD ranges only from 41 to 46% (fig. 4C), suggesting a stronger selection pressure on the maintenance of protein-protein interaction rather than conservation of the NBD.
The absence of an HRP4 ortholog in Trypanosoma spp. raises interesting questions on the evolutionary history of this protein, i.e., whether it has been specifically acquired by a common ancestor of Leishmania, Phytomonas, Leptomonas, Crithidia, Angomonas, and Strigomonas, or whether it has been lost in Trypanosoma spp. We used the draft genome of the early branching trypanosomatid Paratrypanosoma confusum and phylogenetic analysis to distinguish between these possibilities. Despite the large number of clades that are weakly supported by the bootstrap analysis, the phylogenetic subtree of the slowly evolving and, therefore, highly conserved trypanosomatid HSPA1 orthologs recapitulated the recently published evolutionary relationship between these organisms (Flegontov et al. 2013) clustering Leishmania, Angomonas and Strigomonas together, whereas Paratrypanosoma creates its own well-separated basal clade (fig. 4D). Phylogenetic analysis of HRP4 across trypanosomatids confirmed the absence of this protein in Trypanosoma spp., but identifies an early emerging HRP4 ortholog in Paratrypanosoma. This finding suggests that HRP4 was likely present in the common ancestor of all trypanosomatids and thus has been lost by members of the genus Trypanosoma for unknown reasons. Synteny analysis further confirmed the loss of HRP4 and an adjacent gene (LmjF.29.1250, coding for a hypothetical protein) in the otherwise syntenic chromosomal region of T. brucei and T. cruzi (fig. 4E, data accessible via URL http://genopole.pasteur.fr/SynTView/flash/Leishmania/SynWebTryp.html, last accessed June 18, 2016).
Species-specific Copy Number Variation of the Leishmania HSPA1 and HSPA9 sub-families
Our phylogenetic analysis of the trypanosomatid HSP70 family revealed a significant difference in the gene number between Leishmania species, notably for the cytoplasmic and mitochondrial HSP70 gene arrays HSPA1 on chromosome 28 and HSPA9 on chromosome 30, respectively, suggesting that both loci may be under species-specific selection. We more closely investigated the evolution of these genes in their respective genomic contexts analyzing the synteny between all HSP70 loci in L. major, L. infantum, and the L. donovani reference genome LdBPK282A1 using the SynTView web tool (Lechat et al. 2013). The repetitive regions of the first published draft genome of L. donovani (LdBPK282A1; Downing, et al. 2011) were not fully assembled because the length of 454 and Illumina reads was too short for reconstruction of repetitive regions (Downing et al. 2011). Reads from BPK282/0cl4 (referred here to as LdPBQ7IC8) generated by Pacific Biosciences SMRT sequencing (unpublished) were able to reduce over 2,500 existing gaps to 20 and achieved better annotations for previously mis-assembled regions. Our analysis documents synteny across all species and all HSP70 family members (data not shown but accessible via URL http://genopole.pasteur.fr/SynTView/flash/Leishmania/SynWeb.html, last accessed June 18, 2016), but revealed substantial gene copy number variations for the HSPA1 and HSPA9 gene arrays across the different Leishmania species (fig. 5A and supplementary table S3, Supplementary Material online) and confirms important differences in copy number of these genes to the L. major genome database (Folgueira et al. 2007). To more accurately estimate the number of gene copies, we exploited recently published HTseq data (Rogers et al. 2011). Read depth analysis uncovered even more dramatic changes in copy number across various Leishmania spp., with L. major showing the highest rate of gene amplification with estimated 14 and 17 copies for HSPA1 and HSPA9, respectively (fig. 5B). No CNV was observed for all other HSP70 members, which are all encoded by single copy genes.
The availability of a high-quality L. donovani genome sequence allowed us to investigate in more detail the evolutionary dynamics of these gene arrays between related species using phylogenetic analysis. Re-drawing the phylogenetic tree shown in fig. 3B using this new sequence information did not change the overall clustering of the HSP70 family members (supplementary fig. S9, Supplementary Material online). However, an increased resolution of the HSPA1 and HSPA9 clades was obtained. All HSPA1 genes cluster in an intra-species-specific manner (i.e., they form species-specific clusters), suggesting the occurrence of gene duplication after Leishmania speciation (fig. 5C). In contrast, the HSPA9 genes of the different species cluster in an interspersed way, drawing a more complex picture of gene duplication events that occurred before Leishmania speciation (e.g., LmjF30.2460 and its orthologs), and thereafter (all other genes).
The L. donovani HSPA1 Array may be a Hot Spot of Environment-genotype Interactions
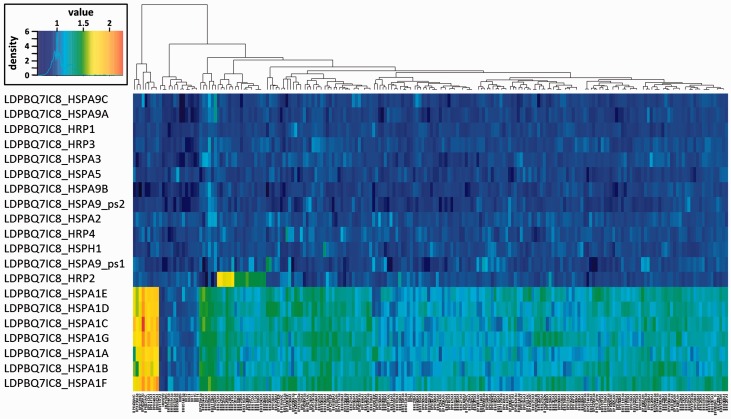
The highly dynamic structure of the Leishmania HSPA1 and HSPA9 gene loci across Leishmania species primed us to investigate if these gene arrays may even have evolved in a strain-specific manner. We investigated this possibility drawing from recently published HTseq data of 204 L. donovani field isolates from the Indian sub-continent (Imamura et al. 2016) and using the PacBio-sequenced isolate LDPBQ7IC8 as a reference. As indicated by the heat map shown in fig. 6, most single-copy HSP70 family members show little or no variation in gene copy number. One exception is represented by the HRP2 gene locus that shows duplication in one branch of the Indian strains as part of a large sub-telomeric amplification in chromosome 35 reported elsewhere (Downing et al. 2011). A highly dynamic evolutionary pattern was revealed for the HSPA1 gene array on chromosome 28, which characterizes three clusters of field isolates: (i) strains from Bangladesh with a locus of about seven gene copies similar to the LDPBQ7IC8 reference, (ii) strains from highlands in Nepal that show a strong expansion of this array with up to 14 genes, and (iii) the majority of the strains originating from the lowlands in India and Nepal (further called core population) and showing moderate expansion with likely 8–10 genes. In contrast to the HSPA1 locus, the HSPA9 array is stable across the entire sample set, demonstrating that only cytoplasmic but not mitochondrial HSP70 genes may show dynamic, strain-specific evolution.
Fig. 6.—
Heat map of gene copy number variation of HSP70 family members in LdBPK field isolates. The normalized read depth for the genes coding for the indicated HSP70 members is plotted against 204 L. donovani field isolates (Imamura et al. 2016), Leishmania infantum JPCM5 and L. donovani LV9. The color scale at the upper right shows normalized depth calibrated to correspond to one gene copy for a single copy gene on a diploid chromosome. The dendrogram shown at the top of the heat map was established based on euclidean distance of the depth of HSP70 genes.
We determined single-nucleotide polymorphisms (SNPs) to investigate if individual HSPA1 and HSPA9 gene copies show non-synonymous nucleotide substitutions as a sign of divergent evolution. In the core population, the genes for all HSP70 family members were well conserved and only one non-synonymous (nsyn) SNP site shared by three strains was found in LdPBQ_HSPA5 at position 452,103, and one synonymous (syn) SNP site was identified in the pseudogene LdPBQ_HSPA9_ps1 at position 962,090 (supplementary table S4, Supplementary Material online). In contrast, the 12 strains originating from the highlands contained 44 SNP sites, including 13 nsyn and 31 syn sites. None of LdPBQ_HSPA1 and LdPBQ_HSPA9 genes had nsyn SNPs thus ruling out onset of divergent evolution in these highly related field isolates. LdPBQ_HRP3 in these 12 strains had complex genetic variants, including a cluster of six SNPs and a nine bp deletion (supplementary table S4, Supplementary Material online). Other SNPs in this group of 12 strains are described in supplementary table S4, Supplementary Material online.
Discussion
The HSP70 superfamily contains some of the most conserved proteins in eukaryotes. Canonical HSP70 members share sequence elements and functional residues essential for nucleotide binding and interaction with peptide substrates, nucleotide exchange factors, and co-chaperones. At the same time, all eukaryotes have evolved non-canonical HSP70 members through gene duplication events and divergent evolution that resulted in unique, non-conserved sequence elements and highly species-specific interactions and functions. Combining phylogenetic and comparative analyses of the TriTryp genomes, L. donovani field isolates, and early-branching Paratrypanosoma, we provide novel insight into the evolutionary dynamics of the Leishmania HSP70 protein family, and uncover unique, parasite-specific features in both its canonical and non-canonical members that are discussed in detail below.
As judged by clustering with well-characterized bacterial and human HSP70 members, three highly conserved monophyletic groups were identified in Leishmania spp. that correspond to cytoplasmic (HSPA1 and HSPA2), ER (HSPA5), and mitochondrial HSP70 (HSPA9; see fig. 3B and supplementary fig. S1, Supplementary Material online), some of which have been confirmed previously for their predicted sub-cellular localization by immuno-fluorescence and immuno-EM analyses (Searle and Smith 1993; Searle et al. 1993; Jensen et al. 2001; Campos et al. 2008; Týč et al. 2015). However, despite their conservation (e.g., over 70% of sequence identity between the Leishmania and human HSPA1 HSP70 domain; see supplementary table S1, Supplementary Material online), there are several important features that distinguish the parasite canonical HSP70 proteins from their orthologs in other eukaryotes. First, all canonical members are characterized by non-conserved, N- and/or C-terminal sequence extensions that likely confer trypanosomatid-specific interactions or localization. This is further supported by the presence of divergent, C-terminal consensus sequence motifs in some of these proteins, including the motif EDVD in Leishmania HSPA2 that does not correspond to the canonical EEVD motif known to interact with co-chaperones via their TPR domain (Li et al. 2009). The same applies to the MDDL motif in Leishmania HSPA5 that differs from the canonical KDDL motif conferring ER localization (Pidoux and Armstrong 1992; Bangs et al. 1996), or extensive poly-glutamine stretches in Leishmania HSPA9A and B absent in bacterial and human orthologs.
Second, both canonical HSPA1 and HSPA9 show a significant increase in gene copy number in Leishmania spp. compared with other eukaryotes. Even though the quantification of exact copy number variation of duplicated regions remains difficult for current sequencing techniques, our data indicate that the copy number of these genes is significantly underestimated in current public databases, which, for example, may attain up to 14 and 17 gene copies for L. major HSPA1 and HSPA9, respectively. The presence of abundant gene arrays is a main characteristic of the Leishmania genome structure, believed to increase gene expression in the absence of transcriptional control (Ivens et al. 2005; Peacock et al. 2007; Rogers et al. 2011). Often gene amplification is selected for in response to environmental stress. For example, drug resistance in Leishmania has been correlated to gene amplification (Downing et al. 2011; Leprohon et al. 2015), and Leishmania antimony tolerance has been linked to increased HSP70 abundance (Brochu et al. 2004). Analyzing copy number variation (CNV) across L. donovani HSP70 members in 204 field isolates from the Indian sub-continent (Imamura et al. 2016) uncovered a surprising diversity in the gene copy number of HSPA1 ranging from seven to at least 14 copies (per haploid genome). Interestingly, there was a geographical/environmental structure in those results, with seven copies in isolates from Bangladesh, 8–10 in lowland Indo-Nepalese isolates and 14 in isolates from Nepalese highlands (belonging to the ISC1 group described by Imamura et al. 2016). Furthermore, part of this expansion is rather recent, as lowland Indo-Nepalese isolates and those from Bangladesh were estimated to have diverged around the year 1850 (Imamura et al. 2016). Given the importance of cytoplasmic HSP70 to confer resistance to oxidative stress in Leishmania (Miller et al. 2000), its reported link to treatment failure (Torres et al. 2013) and antimony resistance (Brochu et al. 2004), it is interesting to speculate that this rapid intra-chromosomal amplification may reflect a strain-specific adaptation to unknown environmental insults. The absence of non-synonymous SNP in the HSPA1 gene copies suggests that this change was driven by gene dosage adaptation. Although increased HSPA1 copy number does not correlate with antimony-resistance (data not shown), our observation nevertheless suggests that this locus may be a target of environment-genotype interaction with potential epidemiological relevance, and suggests CNV has an important signal for biomarker discovery in Leishmania. However, we cannot rule out that the dynamics of this locus represents random drift. In contrast, the HSPA9 gene array—whose products are indispensable for the maintenance and replication of kinetoplast DNA (Týč et al. 2015)—shows stable copy number across the 204 field isolates, thus serving as an internal control and demonstrating that this locus is likely not target of environment-genotype interactions.
Finally, despite their high-sequence conservation and hence likely recent amplification, various members of the HSPA1 and HSPA9 gene arrays show regulatory or structural differences that point towards a recent onset of divergent evolution and the birth of two novel HSP70 sub-families in Leishmania spp. For example, one gene of the HSPA1 array in L. major and L. amazonensis shows temperature-induced expression due to the presence of a unique 3′ UTR that is not shared by the other, constitutively expressed copies (Folgueira et al. 2005). Likewise, individual members of the HSPA9 array show significant sequence divergence in their C-terminal domain, and are differentially regulated, thus likely carrying out distinct mitochondrial functions (Campos et al. 2008; Týč et al. 2015). Our phylogenetic analyses allow insight into the evolutionary history of these arrays. Members of the HSPA1 and (with some exceptions) HSPA9 sub-families largely cluster in a species-specific manner with good bootstrap support, pointing towards an evolutionary scenario of convergent amplification, where a single gene in the common ancestor was duplicated independently in various Leishmania species. However, the very small inter-species sequence divergence, and the conservation of regulatory and structural features across orthologs seems more compatible with a scenario of gene amplification occurring in the common ancestor of all Leishmania spp., likely triggered by the wide array of functions fulfilled by the HSP70 proteins that may then have further evolved in a species-specific fashion by divergent evolution.
In contrast to the HSPA1 and HSPA9 gene arrays, all other Leishmania HSP70 members are encoded by single-copy genes that are more closely related across Leishmania species than to paralogs inside the same species. This inter-species phylogenetic clustering reveals evolution of these HSP70 members through an ancient gene duplication and divergent evolution in an ancestral trypanosomatid that predates Leishmania speciation. As judged by the conservation of functional domains, aside HSPA1 and HSPA9 two additional canonical family members (i.e., HSPA2 and HSPA3) likely retain ATP-dependent chaperone function. However, these functions seem adapted to the parasite-specific biology given the divergence of functional residues involved in substrate and NEF binding. In contrast, the substantial degeneration of the HSP70 domain of the non-canonical family members HSPH1 and HRP1-5 indicates that these proteins are probably not directly involved in protein folding but have evolved novel functions that may include NEF or co-chaperone activities as suggested for non-canonical HSP70 members in other eukaryotes (Shaner et al. 2006). Although all these atypical HSP70 proteins find a highly conserved ortholog in the genus Trypanosoma, highlighting their ancestral origin, no HRP4 ortholog is present in the T. brucei or T. cruzi genomes. The presence of HRP4 orthologs in all Leishmania species and the early-branching Paratrypanosoma reveals the secondary loss of this gene from the Trypanosoma genome by chromosomal deletion.
In conclusion, we provide here a unique insight into the evolutionary dynamics of the Leishmania HSP70 protein family and reveal features of this superfamily that are specific at various taxonomic levels, including the trypanosomatid family, the Leishmania genus, the Leishmania species, and even the Leishmania strain. Intra-chromosomal amplification and subsequent divergent evolution as documented here for the Leishmania HSPA9 gene locus may represent an important source for genetic diversity in the absence of frequent genetic recombination in this largely asexual organism. The re-annotation of this family, the proposition of a new nomenclature following official recommendations, and the discovery of CNV as an evolutionary mechanism that drives Leishmania species- and even strain-specific adaptation of the this family sheds important new light on the evolutionary potential of eukaryotic HSP70 proteins and sets the stage for future functional analyses elucidating the biological and epidemiological significance of these essential chaperones.
Supplementary Material
Supplementary tables S1–S4 and figures S1–S12 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Sophie Veillault for editorial help, Simonetta Gribaldo for discussion, and Mariette Matondo from the Institut Pasteur Proteomics platform for proteomics data analysis. This work was supported by the French Government’s Investissements d’Avenir programme: Laboratoire d’Excellence ‘Integrative Biology of Emerging Infectious Diseases’ (Grant no. ANR-10-LABX-62-IBEID to G.F.S.); KALADRUG-R (EU/FP7-222895 to J.C.D.), the Belgian Development Cooperation (FA3 II VL control and FA3 project 95502 to J.C.D.), the Belgian Science Policy Office (TRIT, P7/41 to J.C.D.), the Flemish Fund for Scientific Research (G.0.B81.12 to J.C.D.), the INBEV-Baillet Latour foundation, and EWI (GEMINI and SINGLE grants to ITM SOFI-B to J.C.D.); the Czech Grant Agency (Grant 14-23986S to J.L.); a fellowship from the Pasteur–Paris University (PPU) International PhD program and the Institut Carnot Pasteur Maladies Infectieuses (to S.D.), and a Bourse Fin de thèse scientifique from the Fondation de Recherche Médicale (contract FDT20150532765 to S.D.).
Literature Cited
- Adl SM, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol. 59:429–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs JD, Brouch EM, Ransom DM, Roggy JL. 1996. A soluble secretory reporter system in Trypanosoma brucei—studies on endoplasmic reticulum targeting. J Biol Chem. 271:18387–18393. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. 1994. Molecular evolution of the HSP70 multigene family. J Mol Evol. 38:1–17. [DOI] [PubMed] [Google Scholar]
- Brochu C, Haimeur A, Ouellette M. 2004. The heat shock protein HSP70 and heat shock cognate protein HSC70 contribute to antimony tolerance in the protozoan parasite Leishmania. Cell Stress Chaperones 9:294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RM, et al. 2008. Distinct mitochondrial HSP70 homologues conserved in various Leishmania species suggest novel biological functions. Mol Biochem Parasitol. 160:157–162. [DOI] [PubMed] [Google Scholar]
- Clayton CE. 2002. Life without transcriptional control? From fly to man and back again (vol 21, pg 1881, 2002). EMBO J. 21:3917–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Rohde M, Jaattela M. 2007. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581:3702–3710. [DOI] [PubMed] [Google Scholar]
- David M, et al. 2010. Preferential translation of Hsp83 in Leishmania requires a thermosensitive polypyrimidine-rich element in the 3' UTR and involves scanning of the 5' UTR. RNA 16:364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T, et al. 2011. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 21:2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. 2006. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25:2519–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droll D, et al. 2013. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog 9:e1003286.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2015. HMMER web server: 2015 update. Nucleic Acids Res. 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegontov P, et al. 2013. Paratrypanosoma is a novel early-branching trypanosomatid. Curr Biol. 23:1787–1793. [DOI] [PubMed] [Google Scholar]
- Folgueira C, Canavate C, Chicharro C, Requena JM. 2007. Genomic organization and expression of the HSP70 locus in New and Old World Leishmania species. Parasitology 134:369–377. [DOI] [PubMed] [Google Scholar]
- Folgueira C, et al. 2005. The translational efficiencies of the two Leishmania infantum HSP70 mRNAs, differing in their 3'-untranslated regions, are affected by shifts in the temperature of growth through different mechanisms. J Biol Chem. 280:35172–35183. [DOI] [PubMed] [Google Scholar]
- Folgueira C, Requena JM. 2007. A postgenomic view of the heat shock proteins in kinetoplastids. FEMS Microbiol Rev 31:359–377. [DOI] [PubMed] [Google Scholar]
- Forterre P. 2015. The universal tree of life: an update. Front Microbiol. 6:717.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassler CS, et al. 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 95:15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil M, Zanetti MS, Zoller S, Anisimova M. 2013. CodonPhyML: fast maximum likelihood phylogeny estimation under codon substitution models. Mol Biol Evol. 30:1270–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Singh B. 1994. Phylogenetic analysis of 70-Kd heat-shock protein sequences suggests a chimeric origin for the eukaryotic cell-nucleus. Curr Biol. 4:1104–1114. [DOI] [PubMed] [Google Scholar]
- Hampl V, et al. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 106:3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, et al. 2014. An alternative root for the eukaryote tree of life. Curr Biol. 24:465–470. [DOI] [PubMed] [Google Scholar]
- Hem S, et al. 2010. Identification of Leishmania-specific protein phosphorylation sites by LC-ESI-MS/MS and comparative genomics analyses. Proteomics 10:3868–3883. [DOI] [PubMed] [Google Scholar]
- Hughes AL. 1993. Nonlinear relationships among evolutionary rates identify regions of functional divergence in Heat-Shock Protein-70 genes. Mol Biol Evol. 10:243–255. [DOI] [PubMed] [Google Scholar]
- Imamura H, et al. 2016. Evolutionary genomics of epidemic visceral Leishmaniasis in the Indian subcontinent. eLife e12613.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, et al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ATR, Curtis J, Montgomery J, Handman E, Theander TG. 2001. Molecular and immunological characterisation of the glucose regulated protein 78 of Leishmania donovani. BBActa-Protein Struct M 1549:73–87. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 11:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominek J, Marszalek J, Neuveglise C, Craig EA, Williams BL. 2013. The complex evolutionary dynamics of Hsp70s: a genomic and functional perspective. Genome Biol Evol. 5:2460–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Lechat P, Souche E, Moszer I. 2013. SynTView—an interactive multi-view genome browser for next-generation comparative microorganism genomics. BMC Bioinformatics 14:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. 2007. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasit 152:35–46. [DOI] [PubMed] [Google Scholar]
- Leprohon P, Fernandez-Prada C, Gazanion E, Monte-Neto R, Ouellette M. 2015. Drug resistance analysis by next generation sequencing in Leishmania. Int J Parasitol Drugs Drug Resist 5:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Qian XG, Sha BD. 2009. Heat shock protein 40: structural studies and their functional implications. Protein: Peptide Lett. 16:606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QL, Hendrickson WA. 2007. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan-Klumpler FJ, et al. 2012. GeneDB—an annotation database for pathogens. Nucleic Acids Res. 40:D98–D108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw CA, Ludewig MH, Mayer J, Blatch GL. 2010. The Hsp70 chaperones of the Tritryps are characterized by unusual features and novel members. Parasitol Int 59:497–505. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 62:670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, et al. 2000. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 7:586–593. [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, et al. 2000. Inducible resistance to oxidant stress in the protozoan Leishmania chagasi. J Biol Chem. 275:33883–33889. [DOI] [PubMed] [Google Scholar]
- Morales MA, et al. 2010. Phosphoproteome dynamics reveal heat-shock protein complexes specific to the Leishmania donovani infectious stage. Proc Natl Acad Sci USA. 107:8381–8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock CS, et al. 2007. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 39:839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. 1989. Heat-shock and the sorting of Luminal Er proteins. EMBO J. 8:3171–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Armstrong J. 1992. Analysis of the Bip gene and identification of an Er retention signal in Schizosaccharomycespombe. EMBO J. 11:1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada L, Soto M, Alonso C, Requena JM. 2000. Identification of a putative regulatory element in the 3'-untranslated region that controls expression of HSP70 in Leishmania infantum. Mol Biochem Parasitol. 110:79–91. [DOI] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. 2006. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25:2510–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena JM, Montalvo AM, Fraga J. 2015. Molecular chaperones of Leishmania: central players in many stress-related and -unrelated physiological processes. Biomed Res Int 3013:26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez F, Oliver JL, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitution. J Theor Biol. 142:485–501. [DOI] [PubMed] [Google Scholar]
- Rogers MB, et al. 2011. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 21:2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle S, Mccrossan MV, Smith DF. 1993. Expression of a mitochondrial stress protein in the protozoan parasite Leishmania major. J Cell Sci. 104:1091–1100. [DOI] [PubMed] [Google Scholar]
- Searle S, Smith DF. 1993. Leishmania major—characterization and expression of a cytoplasmic stress-related protein. Exp Parasitol. 77:43–52. [DOI] [PubMed] [Google Scholar]
- Shaner L, Sousa R, Morano KA. 2006. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry 45:15075–15084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonhai A, Maier AG, Przyborski JM, Blatch GL. 2007. Intracellular protozoan parasites of humans: the role of molecular chaperones in development and pathogenesis Protein Pept Lett. 18(2):143–57. [DOI] [PubMed] [Google Scholar]
- Sibley LD. 2011. Invasion and intracellular survival by protozoan parasites. Immunol Rev 240:72–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. 2004. Coordinated activation of Hsp70 chaperones. Science 303:98–101. [DOI] [PubMed] [Google Scholar]
- Suh WC, Lu CZ, Gross CA. 1999. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J Biol Chem. 274:30534–30539. [DOI] [PubMed] [Google Scholar]
- Suh WC, et al. 1998. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 95:15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syldatk C, May O, Altenbuchner J, Mattes R, Siemann M. 1999. Microbial hydantoinases—industrial enzymes from the origin of life? Appl Microbiol Biot 51:293–309. [DOI] [PubMed] [Google Scholar]
- Szabo A, et al. 1994. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 91:10345–10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres DC, Ribeiro-Alves M, Romero GAS, Davila AMR, Cupolillo E. 2013. Assessment of drug resistance related genes as candidate markers for treatment outcome prediction of cutaneous leishmaniasis in Brazil. Acta Trop 126:132–141. [DOI] [PubMed] [Google Scholar]
- Týč J, Klingbeil MM, Lukes J. 2015. Mitochondrial heat shock protein machinery hsp70/hsp40 is indispensable for proper mitochondrial DNA maintenance and replication. mBio 6: e02425–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M, Mayer MP, Bukau B. 2006. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 281:38705–38711. [DOI] [PubMed] [Google Scholar]
- Wiesgigl M, Clos J. 2001. Heat shock protein 90 homeostasis controls stage differentiation in Leishmania donovani. Mol Biol Cell 12:3307–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA. 2014. Evolution: rooting the eukaryotic tree of life. Curr Biol. 24:R151–R152. [DOI] [PubMed] [Google Scholar]
- Yau WL, et al. 2010. Cyclosporin a treatment of Leishmania donovani reveals stage-specific functions of cyclophilins in parasite proliferation and viability. PloS Neglect Trop D 4: e729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberstein D, Shapira M. 1994. The role of pH and temperature in the development of Leishmania parasites. Annu Rev Microbiol. 48:449–470. [DOI] [PubMed] [Google Scholar]
- Zilka A, Garlapati S, Dahan E, Yaolsky V, Shapira M. 2001. Developmental regulation of heat shock protein 83 in Leishmania. J Biol Chem. 276:47922–47929. [DOI] [PubMed] [Google Scholar]