Abstract
In many plant species, somatic cell differentiation is accompanied by endoreduplication, a process during which cells undergo one or more rounds of DNA replication cycles in the absence of mitosis, resulting in nuclei with multiples of 2C DNA amounts (4C, 8C, 16C, etc.). In some orchids, a disproportionate increase in nuclear DNA contents has been observed, where successive endoreduplication cycles result in DNA amounts 2C + P, 2C + 3P, 2C + 7P, etc., where P is the DNA content of the replicated part of the 2C nuclear genome. This unique phenomenon was termed “progressively partial endoreplication” (PPE). We investigated processes behind the PPE in Ludisia discolor using flow cytometry (FCM) and Illumina sequencing. In particular, we wanted to determine whether chromatin elimination or incomplete genome duplication was involved, and to identify types of DNA sequences that were affected. Cell cycle analysis of root tip cell nuclei pulse-labeled with EdU revealed two cell cycles, one ending above the population of nuclei with 2C + P content, and the other with a typical “horseshoe” pattern of S-phase nuclei ranging from 2C to 4C DNA contents. The process leading to nuclei with 2C + P amounts therefore involves incomplete genome replication. Subsequent Illumina sequencing of flow-sorted 2C and 2C + P nuclei showed that all types of repetitive DNA sequences were affected during PPE; a complete elimination of any specific type of repetitive DNA was not observed. We hypothesize that PPE is part of a highly controlled transition mechanism from proliferation phase to differentiation phase of plant tissue development.
Keywords: cell cycle, DNA replication, EdU, endoreduplication, Ludisia discolor, orchids
Background
Differentiated somatic cells of many plants are often endopolyploid. It has been estimated that this phenomenon affects more than 90% of angiosperm species (Joubes and Chevalier 2000). Endopolyploidy occurs to a different extent in various plant cells and tissues with the exception of embryonic and meristematic cells (Barow and Jovtchev 2007) and is common in economically important tissues and organs including endosperm (Sabelli and Larkins 2009), fruits (Chevalier et al. 2011), cotton fibres (Wilkins et al. 2000), and nitrogen-fixing root nodules (Kondorosi and Kondorosi 2004). The mechanism behind endopolyploidization in plants is endoreduplication, where replication of whole nuclear genome takes place but no mitosis (and cell division) follows (D′Amatto 1989). This leads to proportional (2-fold) increase in nuclear DNA contents from basic 2C content in diploid somatic cell to 4C, 8C, 16C, etc. in successive rounds of endoreduplication. Although biological significance of endoreduplication remains unclear, it has been linked to increased cell size and higher metabolic activity, and seems essential for developmental processes, including cell fate maintenance (De Veylder et al. 2011). Cells with endoreduplicated nuclei do not divide. However, they can be induced to divide by a treatment with growth regulators (D′Amato 1952). In addition to chromosomes with two chromatids observed in dividing 4C cells, cells with chromosomes comprising 4, 8, 16, etc. chromatids lying side-by-side, are observed as a consequence of repeating rounds of DNA replication without chromatid separation (D′Amato 1989).
Unlike regular endoreduplication, where all chromosomes are completely reduplicated and the amount of nuclear DNA is doubled, disproportionate increase of DNA content has been observed in some plant species. Early experiments with orchid protocorms in 1970’s indicated differential amplification of certain genome regions in relation to unequal replication of AT- and GC-rich regions (Nagl 1972, 1977; Schweizer and Nagl 1976; Capesius and Nagl 1978). This phenomenon was called “DNA underreplication,” but since then no detailed research has been conducted, largely due to the lack of suitable experimental methods. The interest was revived after flow cytometry (FCM) was introduced to perform nuclear DNA content estimation in plants. In several orchid species, histograms of DNA content showed unusual profiles that could not be explained by methodological errors (Suda 2004; Suda et al. 2007). Bory et al. (2008) observed a similar phenomenon in Vanilla planifolia and found the replication ratios of consecutive peaks to be 1.43, 1.63, 1.76, and 1.82 instead of two as expected for regular endoreduplication. On the basis of almost perfect linearity between the DNA content and the number of endoreduplication cycles, the authors hypothesized that the same genome part is amplified at each cycle and suggested that each successive peak should be called 2C, 2C + P, 2C + 3P, etc., where P is the DNA content of the replicated part of 2C genome. To describe this phenomenon they coined a new term “progressively partial endoreplication” (PPE). Subsequently, PPE has been revealed in other orchid species (Trávníček et al. 2011, 2015). However, none of the published studies answered the crucial question on the mechanism behind the PPE—whether this unique phenomenon is a result of incomplete genome replication, or excision of DNA following whole-genome replication.
Differences in nuclear DNA content among different cells of one organism were found also in animals. These differences occur predominantly between germline and somatic cells, and are caused by processes called “programmed DNA elimination” (reviewed in Wang and Davis 2014). There have been two ways of programmed DNA elimination described in a wide range of organisms: 1) “chromatin diminution,” where chromosome breaks occur, followed by extrusion of chromatin portions into cytoplasm (Tobler et al. 1992; Smith et al. 2009; Bracht et al. 2013), and 2) “whole chromosome elimination” (Pigozzi and Solari 2005; Del Priore and Pigozzi 2014).
Early studies on chromatin diminution showed that large amounts of repetitive sequences were eliminated (Muller et al. 1982). The advent of massively parallel sequencing methods enabled to study differences in genomic contents between germline and somatic cells in more detail. Wang et al. (2012) compared sequences of somatic and germline cells of the nematode Ascaris suum and found out that not only repetitive sequences, but also single-copy sequences corresponding to protein-coding genes were missing in somatic cells. Similar study in sea lamprey demonstrated that a few thousand genes were absent in somatic cells (Smith et al. 2012). These findings suggest that DNA elimination may serve as “unorthodox” mechanism of ultimate gene regulation (silencing). The programmed DNA elimination may have also been implicated in sex determination and dosage compensation (Pigozzi and Solari 1998; Goday and Esteban 2001; Sanchez 2014).
Another type of somatic DNA content alterations found in animals is called “DNA underreplication” and is commonly associated with polytene chromosomes of Diptera (Rudkin 1969). Johnston et al. (2013) used FCM to study Drosophila melanogaster thorax cells and observed the ratio of 4C/2C amounts not two but lower (1.75 and 1.83 for wild type and mutant strains, respectively), and also found age-dependency of this effect. Yarosh and Spradling (2014) used massively parallel sequencing on polytene larval salivary gland cells and identified many underreplicated chromosome regions. Similarly, Nordman et al. (2011) using array-based comparative genomic hybridization found sites of underreplication in euchromatic, nonrepetitive regions of multiple polytene tissues of D. melanogaster.
While DNA elimination and DNA underreplication have been observed in many groups of eukaryotes, ranging from protozoa to vertebrates, PPE in plants seems to be restricted to the Orchidaceae family. This conclusion is supported by the vast number of FCM studies over the past few decades which failed to reveal PPE in any other plant family, incl. closely related Boryaceae, Hypoxidaceae, and Lanariaceae (our personal observations). Trávníček et al. (2015) found that both PPE and regular endoreduplication occur in orchids, but always only one type in a particular species. The authors suggested that PPE must be developmentally regulated as its extent was species-specific. The observed differences in the extent of PPE, based on the replicated fraction (P) of 2C genome, ranged from 21.1% in Cynorkis guttata to 81.1% in Paphiopedilum callosum. Akin to regular endoreduplication, PPE was found in a majority of differentiated cells, but the proportion of different nuclei classes varied among tissues and organs.
In this work, we aimed to unravel the mechanism of PPE. In particular, we wanted to determine whether chromatin elimination or incomplete genome duplication was involved, and to identify types of DNA sequences that were affected. As a model for our study, we chose Ludisia discolor (Ker Gawl.) A. Rich. belonging to Orchidaceae subfamily Orchidoideae, tribe Cranichidae, subtribe Goodyerinae. The species possesses relatively small genome (2C = 2.20 pg = 2.15 Gb) (Trávníček et al. 2015) allowing to keep the sequencing costs reasonably low, and has suitable difference in DNA content between 2C and 2C + P nuclei (P = 1.31 pg). We have employed a multidisciplinary approach combining molecular cytogenetics, next-generation sequencing (NGS) and FCM. FCM analysis of nuclear DNA content and EdU (5-ethynyl-2′-deoxyuridine) incorporation into newly synthesized DNA is a powerful approach to study proliferation patterns on cellular, chromosome, and chromatin levels, changes in DNA replication upon different stresses or estimation of average length of the S-phase (reviewed in Bass et al. 2014). Combination of FCM sorting and NGS of individual chromosomes has proven to be instrumental in unlocking complex genomes of important plant species (reviewed in Doležel et al. 2012, 2014), and we have used the same approach to purify defined subpopulations of cell nuclei and sequence their DNA. Here, we report that the nature of PPE in L. discolor is incomplete genome replication.
Materials and Methods
Plant Material
To ensure genetic purity of the material, we used a large clump of L. discolor of clonal origin obtained from a private collection. This clone is usually named L. discolor var. alba hort. (similar to L. discolor var. dawsoniana nom. nud.—syn. Haemaria discolor var. dawsoniana (H.Low ex Rchb.f.) B.S.Williams but lacking red pigment in leaves). The clone is maintained in Prague Botanical Garden living collection (accession number 2015.07344). For sequencing, we used young plantlets obtained from protocorms (specific formation found in orchids, originating from germinating embryo which is nourished heterotrophicaly using mycorrhiza and which later creates foundations of future plant organs) originating from self-pollination of the same mother plant. For FCM analysis of DNA content of L. discolor we used ovaries of nonpollinated flowers and young leaves of Pisum sativum cv. Ctirad as an internal reference standard. Somatic tissue of young plantlets and ovaries of nonpollinated flowers yielded sufficient numbers of both 2C and 2C + P nuclei, in contrast to fully developed leaves of mature plants where 2C nuclei may be virtually absent (Trávníček et al. 2015). Seeds of Hordeum vulgare cv. Morex were germinated and used for cell cycle analysis. Leaves of Arabidopsis thaliana were used for comparison of endoreduplication profiles.
DNA Content Analysis
DNA content analysis was performed according to Doležel et al. (2007). Briefly, 50 mg of young leaves tissues of L. discolor and P. sativum were chopped simultaneously using a razor blade in 0.5 ml of Otto I solution (1M citric acid, 0.5% (v/v) Tween 20). The crude homogenate was filtered through 50 µm pore size nylon mesh and the nuclei were stained by adding 1 ml of Otto II solution (0.4M Na2HPO4.12H2O) containing propidium iodide and RNase A (both at 50 µg/ml final concentration). FCM analysis was performed using CyFlow Space flow cytometer (Sysmex Partec, Goerlitz, Germany) equipped with green laser (532 nm, 100 mW) and 2C nuclear DNA content of L. discolor was determined by comparing its 2C peak position on histograms of PI fluorescence intensity with the position of 2C peak of P. sativum (2C = 9.09 pg DNA; Doležel et al. 1998), which served as an internal reference standard. Three plants were measured and the measurements were repeated on three different days and mean 2C amount was calculated. Genome size of L. discolor was calculated using formula: 1 pg DNA = 0.978 × 109 bp (Doležel al. 2003). To show an example of a regular endoreduplication, we performed FCM analysis of nuclei of A. thaliana leaves in the same way as in case of L. discolor. We adjusted instrument settings so that the peak corresponding to 2C nuclei of A. thaliana was positioned on the same channel (100) as 2C nuclei of L. discolor.
Cell Cycle Analysis
Cell cycle analyses were performed on roots obtained from fresh stem cuttings of L. discolor which were immersed in tap water for 3–4 weeks until the roots achieved 1–2 cm in length. For comparison, we conducted cell cycle analysis on roots of young seedlings of H. vulgare cv. Morex as a representative of a species with regular mitotic cycle. Actively growing roots were incubated in tap water containing 10 µM EdU (Click-iT EdU Alexa Fluor 488 kit; Life Technologies, Eugene) for 3 h at room temperature. Suspensions of intact nuclei were prepared according to Doležel et al. (1992), with modifications. Briefly, roots were cut approximately 0.5 cm from apex and incubated in 2% (v/v) formaldehyde solution for 30 min at 5 °C. The roots were then washed three times in Tris buffer for 5 min at 5 °C. Root apices were excised and chopped with a razor blade in 1 ml LB01 buffer on a Petri dish. The crude homogenate was filtered through a 50 µm pore size mesh. The suspension of nuclei was then centrifuged at 400 × g for 10 min at 4 °C. The supernatant was discarded and the pellet resuspended in 500 µl Click-iT reaction cocktail (prepared according manufacturer’s instructions) and incubated for 30 min in the dark at 25 °C. Next, the nuclei were pelleted again and the pellet was resuspended in 500 µl LB01 buffer containing DAPI (0.2 µg/ml final concentration). FCM experiments were carried out on FACSAria II SORP flow cytometer and sorter (BD Biosciences, Santa Clara) equipped with two lasers (488 nm and 355 nm) and optical detectors with appropriate optical filters for simultaneous detection of DNA content (DAPI fluorescence) and nascent DNA containing incorporated EdU (Alexa Fluor 488 fluorescence).
Nuclei Sorting and Preparation of DNA for Sequencing
Suspensions of intact nuclei were prepared from leaves and stems of young plantlets in the same way as described above, except that all EdU labeling steps were omitted. For Illumina sequencing, batches of 200,000 nuclei representing 2C and 2C + P populations were flow sorted using into 1.5 ml PCR tubes containing 100 µl ET buffer (1 mM Tris, 75 mM EDTA, pH 8.0). At least ten batches for each fraction were sorted in each of the two sequencing experiments to obtain enough DNA. Preparation of nuclear DNA was done according to Šimková et al. (2008), with modifications. Briefly, aliquots of 200,000 intact nuclei were incubated with 0.35% SDS (Sodium dodecyl sulfate) and 0.35 µg/µl proteinase K for 20 h at 50 °C, and DNA was purified using Microcon YM100 column (Millipore, Bedford).
Illumina Sequencing
DNA libraries for sequencing were prepared separately from 2C and 2C + P DNA samples with TruSeq DNA PCR-Free Library Preparation Kits (Illumina, San Diego). Two micrograms of DNA dissolved in 100 μl deionized water were fragmented with Bioruptor Plus (Diagenode, Denville) with the following settings: 5 cycles, 30/90 s (On/Off). DNA was sheared to 500–700 bp long fragments, purified, end-repaired, adenylated, size selected to 500–600 bp, and ligated with adapters according to manufacturer’s protocol (Illumina). Libraries were diluted 1:10,000 and their concentration was estimated by real-time PCR using KAPA Library Quantification Kit (Bio-Rad, Hercules), pooled and diluted to final concentration of 19 pM. Samples were paired-end sequenced on an Illumina MiSeq instrument for 301 cycles using the MiSeq Reagent Kit v3 (Illumina). Two separate Illumina libraries were created for each nuclei fraction and sequenced independently with the aim to increase the information value of the sequence data analysis.
Sequence Data Analysis
Illumina reads were trimmed for adapters, length of 200 bp and for quality using FASTX-toolkit [-q 20 -p 90] (http://hannonlab.cshl.edu/fastx_toolkit/index.html (last accessed June 14, 2016). Characterization of repeat families was performed using the Repeat Explorer pipeline (Novák et al. 2013). Random data sets representing 0.5× coverage of the L. discolor nuclear genome were retrieved from the Illumina data obtained from 2C and 2C + P nuclei, respectively, and used for reconstruction of repetitive elements using the graph-based method according to Novák et al. (2010). This pipeline uses computational processes which are demanding and therefore it is not possible to use large amounts of data. The effectiveness as well as correctness of this computing process was shown in a number of studies (Novák et al. 2010; Piednoël et al. 2012; Kelly et al. 2015). The assembled sequences within each individual cluster were characterized based on the homology searches and other tools useful for repeat characterization. The assembled sequences within individual clusters were annotated by different sources including similarity searches using RepeatMasker, BLASTX and BLASTN programs against public databases as well as database of domains derived from plant mobile elements (http://repeatexplorer. umbr.cas.cz, last accessed June 14, 2016). Tandem organized repeats were identified using Dotter (Sonnhammer and Durbin 1995).
Databases of Illumina reads as well as assembled contigs specific to different types of repetitive DNA elements identified de novo in the sequencing data were established and are publicly available (http://olomouc.ueb.cas.cz/projects/Ludisia_discolor, last accessed June 14, 2016) and raw data were submitted to Sequence Read Archive (accession code: SRP074524).
Results and Discussion
Cell Cycle Analysis
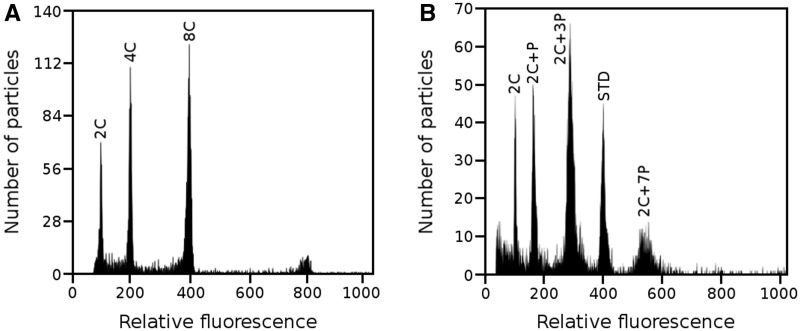
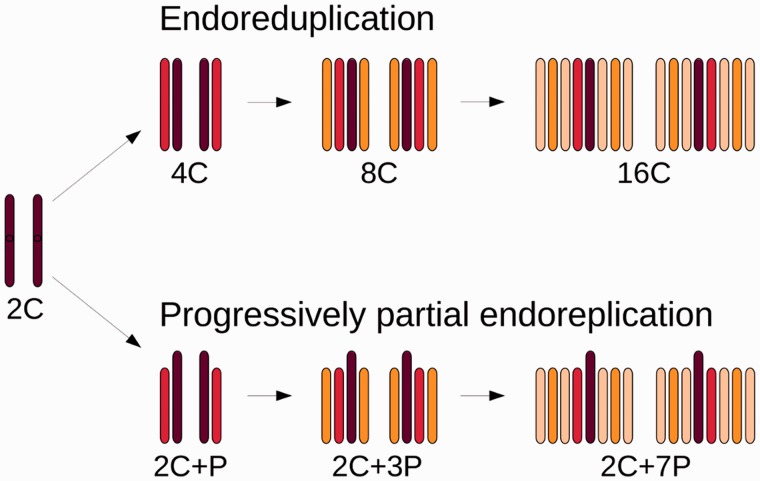
To confirm the occurrence of PPE in L. discolor, we performed DNA content analysis of propidium iodide-stained nuclei using FCM (fig. 1). Panel A shows example of regular endoreduplication found in leaves of A. thaliana. Note that the replication ratio between consecutive peaks is two, thus indicating that the whole DNA complement was replicated in each cycle. On the other hand, the histograms of L. discolor nuclei isolated from ovaries of nonpollinated flowers (panel B) show replication ratios between consecutive peaks 2C : 2C + P:2C + 3P:2C + 7P to be 1.00:1.59:1.76:1.89. The DNA content of individual peaks therefore perfectly corresponds to the number of endoreduplication cycles, which is in agreement with the results of Bory et al. (2008) obtained in V. planifolia. A simplified model visualizing the anticipated process of PPE is shown in figure 2. The model implies that in each consecutive round of endoreduplication, only the same defined portion of the original (complete) 2C genome is replicated.
Fig. 1.
Histogram of relative fluorescence intensity obtained after the analysis of propidium iodide-stained nuclei. (A) Nuclei of A. thaliana leaves showing regular endoreduplication. (B) Example of PPE observed in nuclei isolated from an ovary of nonpollinated flower of L. discolor. 2C peak of the internal reference standard (P. sativum cv. Ctirad) is marked as STD. Note that the histograms in panels A and B were obtained using different instrument settings and hence the positions of peaks on x axes do not reflect genome sizes.
Fig. 2.
Scheme of endoreduplication and PPE. Newly replicated chromatids are drawn in red in the first round of replicating cycle (4C or 2C + P), in orange in the second round of replicating cycle (8C or 2C + 3P), and light orange represents replicated chromatids in the third round of replicating cycle (16C or 2C + 7P).
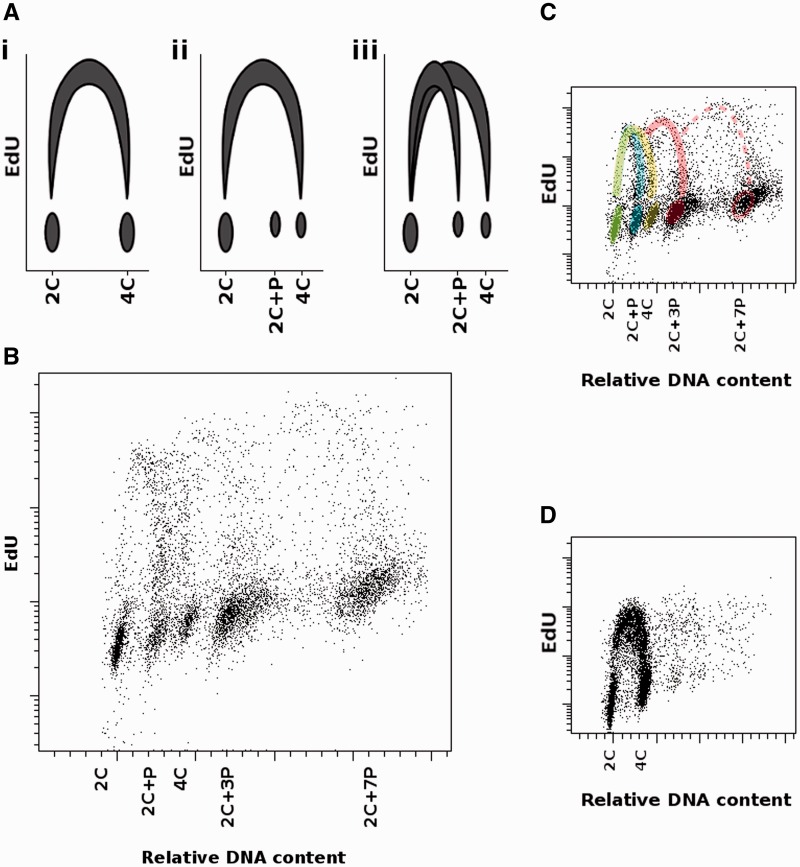
Panel A of the figure 3 shows possible theoretical scenarios of cell cycle progression as observed by FCM analysis of nuclei stained for DNA content and the amount of newly replicated DNA. The first graph (i) depicts regular mitotic cycle where DNA content is duplicated from 2C to 4C. Two other graphs illustrate two different ways which could lead to PPE: complete nuclear DNA replication followed by removal of some DNA sequences (graph ii), and incomplete nuclear replication which results in two horseshoe-like distributions, one spanning 2C and 2C + P nuclei and the second ranging from 2C to 4C nuclei, which represents complete DNA replication in meristematic cells (graph iii). The results obtained after biparametric FCM analysis of root cell nuclei labeled with EdU are shown in panels B–D of the figure 3. Despite the low number of dividing cells in roots of L. discolor, patterns of several cell cycles are visible (panels B and C). The first cycle corresponds to PPE (blue), while the second cycle ending above the population of 4C nuclei belongs to meristematic cells (yellow). Successive endoreplication cycles (red) having origin in 2C + P and 2C + 3P populations, respectively, are also visible. For comparison, cell cycle of barley meristematic cells with a regular pattern is also shown (panel D). These results suggest that partial DNA replication is the most probable mechanism behind PPE. Moreover, as tissue differentiation in plants is often accompanied by endoreduplication cycles, we predict that PPE might be a hallmark of transition of cells from proliferation to differentiation phase of plant development. Similar exit from the mitotic cycle into the endocycle during the S-phase occurs in D. melanogaster salivary gland cells (Nordman et al. 2011) where underreplication of specific chromosomal regions has been observed, especially in centromeric, telomeric, and intercalary heterochromatin. Our results might also indicate that the differentiation/endoreplication program in orchid species with PPE does not start from G2-phase of cell cycle (4C nuclei), but already from G1-phase (2C), when the destiny of a cell is determined and the cell enters a modified S-phase during which some parts of the genome are not replicated.
Fig. 3.
Cell cycle analysis using FCM. (A) Simplified models of different possible cell cycle progressions. (i) Regular cell cycle, where cells double their nuclear DNA content during the transition from G1-phase through S-phase to G2-phase. ii) Cells replicate their whole DNA content but afterwards some DNA sequences are eliminated. Population with 4C DNA content represents meristematic cells. (iii) Two types of cell cycle occur, one representing PPE cells (2C + P), the other meristematic cells (4C). x axis: nuclear DNA content; y axis: the extent of EdU incorporation into newly synthetized DNA.(B–D) Cell cycle analysis in root cells of orchid L. discolor (B, C) and barley Hordeum vulgare (D) using FCM. The roots were pulse-labeled with EdU. x axis represents relative DNA content measured as intensity of DAPI fluorescence (linear scale). y axis shows the extent of EdU incorporation as measured by Alexa Fluor 488 fluorescence intensity (log scale). Different populations represent 2C nuclei, 2C+P nuclei, 4C nuclei, and endopolyploid nuclei (2C+3P, 2C+7P). (B) Overall view showing different classes of nuclei. (C) The same view with different cell cycles highlighted—PPE (blue), mitotic (yellow), successive endoreplication cycles (red). The green color is caused by the overlap of initial phases of PPE and mitotic cycle. (D) Example of a typical “horseshoe” pattern found in barley as a representative of species with regular cell cycle and complete DNA replication.
DNA Sequence Composition of 2C and 2C + P Nuclei
To find out whether PPE in L. discolor is characterized by excluding specific types of DNA sequences from replication, 2C and 2C + P nuclei were purified by flow sorting and their DNA was sequenced by Illumina technology. The two sequencing reactions produced more than 30 and 28 million of paired-end reads from 2C and 2C + P nuclei, respectively. After trimming to quality and length, 31,221,377 and 29,322,104 reads were obtained for 2C and 2C + P nuclei, respectively, corresponding to approximately 16× coverage of the L. discolor nuclear genome (1C = 1075.8 Mb; Trávníček et al. 2015). In order to avoid a random bias, two sequencing libraries were created from each nuclei fraction and sequenced in two independent sequencing runs.
Illumina sequence reads of both sequencing reactions were trimmed to quality and length and used to characterize the most abundant repetitive DNA elements and create database of repetitive DNA sequences specific to L. discolor. RepeatExplorer pipeline (Novák et al. 2014) was used to identify and classify orthologous repeat families. The pipeline enables to analyze intra- and interspecific similarities as well as to reconstruct and quantify major repeat families not only within one species but also between several species; hereafter called comparative analysis (Novák et al. 2014). In our study, the comparative analysis was used to evaluate overall differences in DNA sequence composition and abundance between 2C and 2C + P nuclei fractions and, at the same time, to analyze intra- and interspecific similarities between the two individual sequencing reactions.
Clustering of individual sequences by graph-based method running under RepeatExplorer (Novák et al. 2010) was done under high stringency to avoid reconstruction of hybrid clusters (mixed repeat families). Only those sequences which shared low variation (90% similarity over at least 55% of their length) were included in individual clusters representing different families of repetitive DNA sequences. Reconstruction of repetitive DNA sequences and evaluation of differences in sequence composition and abundance between 2C and 2C + P nuclei was done separately on reads from the two Illumina runs and corresponding to 0.5× coverage of L. discolor nuclear genome fractions. Graph-based clustering resulted in creation of clusters (groups) of mutually connected sequences representing orthologous repeat families. These clusters contained approximately 84% of sequenced reads and 307 largest clusters corresponding to at least 0.01% of the Illumina data set were used for further characterization.
Classification of the largest clusters obtained after comparative analysis showed that Ty3/gypsy and Ty1/copia long terminal repeat-retrotransposons were the most abundant repeats in the genome. Ty3/gypsy elements prevailed and were mainly represented by the Chromoviridae lineage, whereas Ty1/copia family was represented mainly by the Maximus-SIRE lineage (table 1). Tandem organized repeats identified using Dotter (Sonnhammer and Durbin 1995) represented approximately 3.5% of the genome, and sequences similar to DNA transposons and LINE elements were rare, accounting only for approximately 1.0% of the sequence data. Classification of the largest clusters resulted in identification of sequences homologous to Caulimoviridae, DNA pararetroviruses. Apart from sequences showing homology to known repeats, approximately 40% of the analyzed reads (table 1) were not homologous to previously characterized DNA sequences and may represent unknown DNA repeats, or noncoding parts of repetitive DNA elements. The newly identified and characterized DNA repeats will be useful for repeat masking of L. discolor and closely related species. Large database of repetitive DNA elements can also serve as a source of molecular markers. Finally, some repetitive DNA sequences can be used as cytogenetic probes to study molecular organization of nuclei or chromosomes in L. discolor and closely related orchid species. While nuclear genomes of some orchid species were characterized also at chromosome level using FISH with probes for rDNA and repetitive DNA sequences (D'emerico et al. 2001; Begum et al. 2009; Lan and Albert 2011; Lee et al. 2011), this information is missing in L. discolor and closely related species.
Table 1.
Proportion of Repetitive DNA Sequences Identified In Illumina Data of Ludisia Discolor
| Repeat | Lineage/class | Alternative Names | Proportion in 2C Nuclei [%] | Proportion in 2C+P Nuclei [%] | |
|---|---|---|---|---|---|
| LTR retroelements | Ty1/copia | Maximus-SIRE | 8.86 | 7.55 | |
| Angela | 2.58 | 2.35 | |||
| TAR | Tont | 0.32 | 0.32 | ||
| Tork | Tnt | 0.37 | 0.37 | ||
| Ale | Hopscotch | 0.15 | 0.14 | ||
| Ivana-Oryoco | 0.13 | 0.13 | |||
| Total Ty1/copia | 12.41 | 10.86 | |||
| Ty3/gypsy | Chromoviridae | 15.67 | 14.61 | ||
| Ogre-Tat | 1.7 | 1.62 | |||
| Athila | 0.21 | 0.20 | |||
| Total Ty3/gypsy | 17.58 | 16.43 | |||
| Other | |||||
| Caulimoviridea | 0.69 | 0.62 | |||
| LINE | 1.10 | 1.01 | |||
| DNA transposons | 0.93 | 0.90 | |||
| rDNA sequences | 0.10 | 0.09 | |||
| Tandem repeats | 3.58 | 3.60 | |||
| Unclassified repeats* | 7.83 | 6.61 | |||
| Nonannotated sequences | 39.46 | 42.36 | |||
| Chloroplast DNA | 1.38 | 1.17 |
Note.—LTR, long terminal repeat. Unclassified repeats contain sequences of unclassified LTR retrotransposons and unknown repeats.
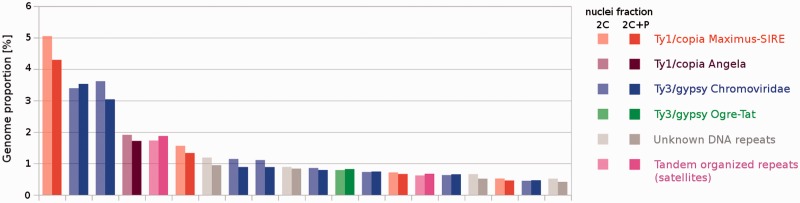
We hypothesized that a particular group of DNA sequences were either partially or completely excluded during PPE and that these sequenced could be revealed by comparing copy numbers in 2C and 2C + P sequence data sets. However, comparative analysis and characterization of sequences within the largest clusters (accounting for at least 0.01% of sequence data) did not show substantial reduction in copy number for any specific repetitive element. Thus, our results indicate that incomplete replication in L. discolor does not concern a specific group of DNA repeats. Twenty largest clusters containing highly similar repetitive DNA sequences specific for 2C and 2C + P nuclei, respectively, are shown in figure 4.
Fig. 4.
Genome proportion of the most abundant DNA sequence types from 20 largest clusters obtained after graph-based analysis of 2C and 2C + P nuclei. The height of columns represents per cent of reads in the genome of L. discolor. Classification of the clusters is marked with colors.
As mentioned above, two sequencing libraries were created from 2C and 2C + P nuclei fractions each and sequenced independently. The reads were trimmed to 100 bp and comparative analysis was done on data representing 0.32× genome coverage. The results of comparative analysis showed consistency between the replicates and only small differences (up to 2%; supplementary table S1, Supplementary Material online) in copy number of repetitive DNA elements in both fractions of analyzed nuclei (2C and 2C + P) were observed (supplementary table S1, Supplementary Material online). On the other hand, higher variation (1.4- to 5.9-fold difference between 2C nuclei replicates and 1.5- to 6.2-fold difference in 2C + P nuclei replicates; supplementary table S1, Supplementary Material online) was observed in the abundance of tandem organized repeats. These observations are similar to those of Macas et al. (2015) who found the highest variability between the replicate data sets for clusters containing satellite DNA. The variation could be an artifact caused by mechanical fragmentation of genomic DNA followed by size-fractionation, and also due to variation between sequencing runs. On the other hand, previous studies demonstrated that relative abundance of different repeat types estimated using experimental methods and by in silico analysis of NGS data provided comparable results (Macas et al. 2007; Klemme et al. 2013; Novák et al. 2014).
Our findings indicate that PPE in L. discolor is not due to the removal of a significant fraction of particular classes of DNA repeats, as observed in protozoa and some animals (Smith et al. 2012; Wang et al. 2012). Presence of two types of cell cycle, one representing PPE cells (2C + P), the other meristematic cells (4C), together with no obvious decrease in relative abundance of individual DNA repeat types in 2C + P nuclei as compared with 2C nuclei, indicate that PPE process in L. discolor may be similar to DNA underreplication in polytene chromosomes of D. melanogaster salivary gland cells (Nordman et al. 2011). In that case, most of euchromatin regions are fully replicated except for pericentromeric regions containing satellite DNA sequences. Recent studies based on NGS showed that underreplicated zones in polytene chromosomes of D. melanogaster closely correspond to regions of repressive chromatin, sparse replication origins, and silent genes (Yarosh and Spradling 2014). Thus, it is feasible that sparse replication origins in L. discolor genome may play an important role in PPE.
The lack of any genomic data for L. discolor and the reference genome sequence in particular, precluded unambiguous identification of underreplicated regions in L. discolor genome in this work. The lack of reference genome sequence also hampered the analysis of single and low copy sequence regions and genes, and thus we were unable to verify if also genic regions were involved. Nevertheless, even with a reference genome sequence available, the analysis will be complicated by a fact that the underreplicated nuclei contain two copies of completely replicated genome (corresponding to 2C DNA amount). A solution could be creation of reference genome sequence specific for the 2C nuclei and then sequencing at high coverage (∼60×) the nuclei with higher levels of endoreduplication (2C + 3P, 3C + 7P) and hence higher representation of incompletely replicated parts of nuclear genome.
Conclusion
This study provides the first insights into the phenomenon called PPE, which seems to occur only in some orchid species. Based on the results obtained, that is, the presence of two cell cycles and the lack of apparent sequence-specific elimination of repetitive DNA elements, we suggest that the mechanism behind PPE is the incomplete replication of nuclear DNA. Together with the precise control of the extent of DNA underreplication, our results indicate that PPE is a highly controlled process accompanying cell and tissue differentiation. However, little is known about the evolutionary and/or ecological role of PPE and further work is needed to fully understand the causes and consequences of PPE occurring in the largest plant family (Orchidaceae).
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Mr. B. Vondruš (Explantex, České Budějovice, Czech Republic) for providing the Ludisia discolor plant. The authors also thank Dr Jana Čížková for help with FCM analyses and Dr Z. Milec for drawing model in figure 3. They also thank Ms. Z. Dubská, Ms. R. Šperková, and Ms. H. Tvardíková for excellent technical assistance. The computing was supported by the National Grid Infrastructure MetaCentrum (grant No. LM2010005 under the program Projects of Large Infrastructure for Research, Development, and Innovations). This work was funded by the Czech Science Foundation (grant award P506/12/1320) and the Ministry of Education, Youth and Sports of the Czech Republic (grant award LO1204 from the National Program of Sustainability I).
Literature Cited
- Barow M, Jovtchev G. 2007. Endopolyploidy in plants and its analysis by flow cytometry In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. Weinheim (Germany: ): Wiley-VCH; p. 349–372. [Google Scholar]
- Bass HW, et al. 2014. A maize root tip system to study DNA replication programmes in somatic and endocycling nuclei during plant development. J Exp Bot. 65:2747–2756. [DOI] [PubMed] [Google Scholar]
- Begum R, Alam SS, Menzel G, Schmidt T. 2009. Comparative molecular cytogenetics of major repetitive sequence families of three Dendrobium species (Orchidaceae) from Bangladesh. Ann Bot. 104:863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bory S, et al. 2008. Natural polyploidy in Vanilla planifolia (Orchidaceae). Genome 51:816–826. [DOI] [PubMed] [Google Scholar]
- Bracht JR, et al. 2013. Genomes on the edge: programmed genome instability in ciliates. Cell 152:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capesius I, Nagl W. 1978. Molecular and cytological characteristics of nuclear DNA and chromatin for Angiosperm systematics: DNA diversification in the evolution of four orchids. Plant Syst Evol. 129:143–166. [Google Scholar]
- Chevalier C, et al. 2011. Elucidating the functional role of endoreduplication in tomato fruit development. Ann Bot. 107:1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato F. 1952. New evidence on endopolyploidy in differentiated plant tissues. Caryologia 4:121–144. [Google Scholar]
- D’Amato F. 1989. Polyploidy in cell differentiation. Caryologia 42:183–211. [Google Scholar]
- D'emerico S, Galasso I, Pignone D, Scrugli A. 2001. Localization of rDNA loci by fluorescent in situ hybridization in some wild orchids from Italy (Orchidaceae). Caryologia 51:31–36. [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. 2011. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16:624–634. [DOI] [PubMed] [Google Scholar]
- Del Priore L, Pigozzi MI. 2014. Histone modifications related to chromosome silencing and elimination during male meiosis in Bengalese finch. Chromosoma 123:293–302. [DOI] [PubMed] [Google Scholar]
- Doležel J, Bartoš J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry 51:127–128. [DOI] [PubMed] [Google Scholar]
- Doležel J, et al. 1998. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot. 82:17–26. [Google Scholar]
- Doležel J., et al. 2012. Chromosomes in the flow to simplify genome analysis. Funct Integr Genomics. 12:397–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J., et al. 2014. Advances in plant chromosome genomics. Biotechnol Adv. 32:122–136. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2:2233–2244. [DOI] [PubMed] [Google Scholar]
- Doležel J, Sgorbati S, Lucretti S. 1992. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol Plantarum. 85:625–631. [Google Scholar]
- Goday C, Esteban MR. 2001. Chromosome elimination in sciarid flies. BioEssays 23:242–250. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Schoener M, McMahon DP. 2013. DNA underreplication in the majority of nuclei in the Drosophila melanogaster thorax: evidence from Suur and flow cytometry. J Mol Biol Res. 3:47–54. [Google Scholar]
- Joubes J, Chevalier C. 2000. Endoreduplication in higher plants. Plant Mol Biol. 43:735–745. [DOI] [PubMed] [Google Scholar]
- Kelly LJ, et al. 2015. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 208:596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemme S, et al. 2013. High-copy sequences reveal distinct evolution of the rye B chromosome. New Phytol. 199:550–558. [DOI] [PubMed] [Google Scholar]
- Kondorosi E, Kondorosi A. 2004. Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 567:152–157. [DOI] [PubMed] [Google Scholar]
- Lan T, Albert VA. 2011. Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum, a lady's slipper orchid. BMC Plant Biol. 11:126.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YI, Chang FC, Chung MC. 2011. Chromosome affinities in interspecific hybrids reflect phylogenetic distances among lady's slipper orchids (Paphiopedilum). Ann Bot. 108:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J., et al. 2015. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS One 10:e0143424.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navrátilová A. 2007. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8:427.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F., et al. 1982. Nucleotide sequence of satellite DNA contained in the eliminated genome of Ascaris lumbricoides. Nucleic Acids Res. 10:7493–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl W. 1972. Evidence of DNA amplification in orchid Cymbidium in vitro. Cytobios 5:154 145. [Google Scholar]
- Nagl W. 1977. Nuclear structures during the cell cycle In: Rost TI, Gifford EM, editors. Mechanisms and control of cell division. Stroudsburg (US: ): Dowden Hutchinson and Ross, Inc; p. 147–193. [Google Scholar]
- Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL. 2011. Developmental control of the DNA replication and transcription programs. Genome Res. 21:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, et al. 2014. Genome-wide analysis of repeat diversity across the family Musaceae. PLoS One 9:e98918.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Macas J. 2010. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29:792–793. [DOI] [PubMed] [Google Scholar]
- Piednoël M, et al. 2012. Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol Biol Evol. 29:3601–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigozzi MI, Solari AJ. 1998. Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata. Chromosome Res. 6:105–113. [DOI] [PubMed] [Google Scholar]
- Pigozzi MI, Solari AJ. 2005. The germ-line-restricted chromosome in the zebra finch: recombination in females and elimination in males. Chromosoma 114:403–409. [DOI] [PubMed] [Google Scholar]
- Rudkin GT. 1969. Non-replicating DNA in Drosophila. Genetics 61:227–238. [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. 2009. The contribution of cell cycle regulation to endosperm development. Sex Plant Reprod. 22:207–219. [DOI] [PubMed] [Google Scholar]
- Sanchez L. 2014. Sex-determining mechanisms in insects based on imprinting and elimination of chromosomes. Sex Dev. 8:83–103. [DOI] [PubMed] [Google Scholar]
- Schweizer D, Nagl W. 1976. Heterochromatin diversity in Cymbidium and its relationship to differential DNA replication. Exp Cell Res. 98:411–423. [DOI] [PubMed] [Google Scholar]
- Šimková H, et al. 2008. Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genomics 9:294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. 2009. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci U S A. 106:11212–11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Baker C, Eichler EE, Amemiya CT. 2012. Genetic consequences of programmed genome rearrangement. Curr Biol. 22:1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–G10. [DOI] [PubMed] [Google Scholar]
- Suda J. 2004. An employment of flow cytometry into plant biosystematics. PhD thesis, MS., Depon. Prague: Dept. of Botany, Faculty of Science Charles University.
- Suda J, Kron P, Husband BC, Trávníček P. 2007. Flow cytometry and ploidy: Applications in plant systematics, ecology and evolutionary biology In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. Weinheim (Germany: ): Wiley-VCH; p. 103–130. [Google Scholar]
- Tobler H, Etter A, Muller F. 1992. Chromatin diminution in nematode development. Trends Genet. 8:427–432. [DOI] [PubMed] [Google Scholar]
- Trávníček P, et al. 2011. Remarkable coexistence of multiple cytotypes of the fragrant orchid (Gymnadenia conopsea agg.): evidence from flow cytometry. Ann Bot. 107:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trávníček P, et al. 2015. Challenges of flow-cytometric estimation of nuclear genome size in orchids, a plant group with both whole-genome and progressively partial endoreplication. Cytometry A. 87A:958–966. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. 2012. Silencing of germline-expressed genes by DNA elimination in somatic cells. Dev Cell. 23:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Davis RE. 2014. Programmed DNA elimination in multicellular organisms. Curr Opin Genet Dev. 27:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins TA, Rajasekaran K, Anderson DM. 2000. Cotton biotechnology. Crit Rev Plant Sci. 19:511–550. [Google Scholar]
- Yarosh W, Spradling AC. 2014. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 28:1840–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.