Summary
Background
Genetic determinants of stroke, the leading neurological cause of death and disability, are poorly understood and have seldom been explored in the general population. Our aim was to identify additional loci for stroke by doing a meta-analysis of genome-wide association studies.
Methods
For the discovery sample, we did a genome-wide analysis of common genetic variants associated with incident stroke risk in 18 population-based cohorts comprising 84 961 participants, of whom 4348 had stroke. Stroke diagnosis was ascertained and validated by the study investigators. Mean age at stroke ranged from 45·8 years to 76·4 years, and data collection in the studies took place between 1948 and 2013. We did validation analyses for variants yielding a significant association (at p<5 × 10−6) with all-stroke, ischaemic stroke, cardioembolic ischaemic stroke, or non-cardioembolic ischaemic stroke in the largest available cross-sectional studies (70 804 participants, of whom 19 816 had stroke). Summary-level results of discovery and follow-up stages were combined using inverse-variance weighted fixed-effects meta-analysis, and in-silico lookups were done in stroke subtypes. For genome-wide significant findings (at p<5 × 10−8), we explored associations with additional cerebrovascular phenotypes and did functional experiments using conditional (inducible) deletion of the probable causal gene in mice. We also studied the expression of orthologs of this probable causal gene and its effects on cerebral vasculature in zebrafish mutants.
Findings
We replicated seven of eight known loci associated with risk for ischaemic stroke, and identified a novel locus at chromosome 6p25 (rs12204590, near FOXF2) associated with risk of all-stroke (odds ratio [OR] 1·08, 95% CI 1·05–1·12, p=1·48 × 10−8; minor allele frequency 21%). The rs12204590 stroke risk allele was also associated with increased MRI-defined burden of white matter hyperintensity—a marker of cerebral small vessel disease—in stroke-free adults (n=21 079; p=0·0025). Consistently, young patients (aged 2–32 years) with segmental deletions of FOXF2 showed an extensive burden of white matter hyperintensity. Deletion of Foxf2 in adult mice resulted in cerebral infarction, reactive gliosis, and microhaemorrhage. The orthologs of FOXF2 in zebrafish (foxf2b and foxf2a) are expressed in brain pericytes and mutant foxf2b−/− cerebral vessels show decreased smooth muscle cell and pericyte coverage.
Interpretation
We identified common variants near FOXF2 that are associated with increased stroke susceptibility. Epidemiological and experimental data suggest that FOXF2 mediates this association, potentially via differentiation defects of cerebral vascular mural cells. Further expression studies in appropriate human tissues, and further functional experiments with long follow-up periods are needed to fully understand the underlying mechanisms.
Introduction
Stroke is the leading neurological cause of death and disability worldwide.1 A substantial proportion of stroke risk remains unexplained, and contribution of genetic factors is supported by discoveries of common genetic variation associated with stroke risk, identified through large, collaborative, genome-wide association studies (GWAS).2 These studies have estimated the proportion of phenotype variance explained by the genome-wide genotypes to range between 16% and 40% for ischaemic stroke, and between 34% and 73% for intracerebral haemorrhage.3 Most associations so far have been specific to particular ischaemic or haemorrhagic stroke types, although a few risk loci for overall ischaemic stroke have also been reported.2 Overall, the search for stroke loci has been less successful than for other complex phenotypes.4 Potential explanations include heterogeneity of stroke and limited ability to detect genetic variants increasing both stroke risk and severity because of the cross-sectional design of most studies, with hospital-based case ascertainment and non-inclusion of severe strokes with early mortality. Population-based cohort studies, with blood samples drawn at recruitment and prospective incident stroke ascertainment offer the advantage of including participants with severe strokes leading to early death.
We did a genome-wide analysis for common genetic variants associated with an increased risk of incident stroke in population-based cohort studies and validated these results with analysis of association between the identified variants and stroke in the largest available cross-sectional studies. Detailed functional exploration of novel genome-wide significant association was done in zebrafish and mice.
Research in context.
Evidence before this study
We searched PubMed with the search terms “stroke”, “cerebral small vessel disease”, “genetics”, “GWAS”, ”genomics”, and the GWAS catalogue for reports before June, 2015, with no language restrictions; we only included peer-reviewed reports in English. Most of the seven studies found only associations with cardioembolic or large-vessel ischaemic stroke, and no robust genetic association has been reported for other subtypes, especially the very common but poorly understood small-vessel ischaemic stroke. Genetic associations with overall stroke were also scarcely reported, with only a few genetic studies thoroughly investigating incident stroke in a population-based longitudinal setting. Although genome-wide association studies (GWAS) have successfully identified numerous genetic associations with complex diseases, including stroke, biological mechanisms underlying these associations are unknown for most of the variants, precluding clinical applications beyond risk prediction.
Added value of this study
First, discovery analyses were done in 4348 cases and 80 613 controls and findings were validated in an additional 19 816 cases and 50 988 controls. We identified a novel risk locus for stroke that appears to be mediated by small vessel disease. Although small vessel disease is one of the major subtypes of stroke, GWAS have so far not discovered risk loci for small-vessel ischaemic stroke (except for an association with the PRKCH locus identified in a study in Japanese participants, which was not found in European populations). Second, we provide preliminary experimental evidence from zebrafish and mouse models that the recorded statistical association reflects an effect of the nearby transcription factor FOXF2 (a gene predominantly expressed in fetal tissue) on the development of cerebral vasculature. Conditional deletion of Foxf2 in adult mice led to cerebral infarction, reactive gliosis, and microhaemorrhage. In zebrafish, foxf2b−/− mutants showed decreased smooth-muscle cell and pericyte coverage. Third, we show that patients with a rare monogenic ophthalmological condition due to segmental deletions encompassing FOXF2 also exhibit features of cerebral small vessel disease, providing an example of how monogenic conditions can inform the mechanisms of complex diseases.
Implications of all the available evidence
The present findings provide insight into the genetic underpinnings of stroke, especially of the small vessel subtype, with evidence from multiple approaches for a pivotal role of FOXF2, a neural crest expressed transcription factor involved in cerebral vessel development. Cerebral small vessel disease is a major, but poorly understood, cause of stroke in all ethnic groups, and subclinical small vessel disease (which was also associated with the stroke risk variants near FOXF2 in our study) has been associated with progressive functional and cognitive decline, and increased risk of dementia. At present, no mechanism-based treatment is available for small vessel disease, other than management of risk factors. Our findings indicate a possible novel mechanism of stroke and small vessel disease. Further research is warranted to explore whether these findings can be applied to clinical practice.
Methods
Study population for discovery analysis
The GWAS discovery sample comprised 84 961 participants of European origin from 18 community-based prospective cohort studies participating in the Cohorts of Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. All participants did not have stroke at baseline and 4348 developed incident stroke during an average of 10 years (SD 3·6) of follow-up (table 1, appendix pp 8–13). Some, but not all, of the cohorts included in our analysis (representing <1544 incident stroke cases) have been included in published HapMap-based stroke GWAS.5,6
Table 1.
Population characteristics
| Country | Total (n) | All stroke (n) | Ischaemic stroke (n) | Cardioembolic-ischaemic stroke (n) | Non-cardioembolic ischaemic stroke (n)* | Women (%)† | Age at stroke (years) | Age of controls at DNA draw (years) | Follow-up (years) | |
|---|---|---|---|---|---|---|---|---|---|---|
|
Discovery sample (longitudinal population-based cohort studies)
| ||||||||||
| AGES | USA | 2996 | 114 | 99 | NA | NA | 58% | 79·9 (5·5) | 76·4 (5·5) | 3·6 (1·1) |
| ARIC | USA | 8939 | 473 | 416 | 108 | 305 | 53% | 69·3 (7·4) | 54·2 (5·7) | 19·1 (4·6) |
| CHS | USA | 3268 | 563 | 447 | 139 | 308 | 61% | 82·7 (6·2) | 72·3 (5·4) | 13·5 (6·3) |
| FHS | USA | 4369 | 235 | 198 | 57 | 127 | 55% | 73·4 (11·7) | 65·5 (12·7) | 8·0 (3·2) |
| FINRISK CoreExome | Finland | 5202 | 94 | 60 | NA | NA | 55% | 69·9 (9·4) | 45·8 (12·8) | 13·3 (2·7) |
| FINRISK Corogene | Finland | 1887 | 60 | 46 | NA | NA | 49% | 73·7 (9·5) | 55·2 (12·2) | 9·0 (4·0) |
| FINRISK Predict CVD | Finland | 1309 | 352 | 294 | NA. | NA | 53% | 66·5 (9·8) | 46·5 (13·3) | 10·0 (5·2) |
| Health ABC | USA | 1661 | 124 | NA | NA | NA | 47% | 80·1 (4·2) | 73·8 (2·8) | 9·3 (2·9) |
| MESA | USA | 2364 | 49 | 43 | NA | 35 | 52% | 75·1 (8·9) | 62·7 (10·2) | 7·2 (1·4) |
| PROSPER | Netherlands | 4658 | 193 | NA | NA | NA | 53% | 77·8 (3·6) | 75·2 (3·3) | 3·1 (0·7) |
| Rotterdam Study I | Netherlands | 6066 | 821 | 448 | 95 | 353 | 60% | 80·6 (8·1) | 69·2 (9·0) | 13·0 (6·2) |
| Rotterdam Study II | Netherlands | 2080 | 125 | 88 | 17 | 71 | 54% | 75·9 (9·2) | 64·6 (7·9) | 9·7 (2·5) |
| SHIP | Germany | 3112 | 75 | 37 | NA | NA | 52% | 71·8 (10·6) | 48·7 (15·2) | 12·1 (2·5) |
| TWINGENE | Sweden | 6702 | 116 | 95 | NA | NA | 52% | 75·6 (8·8) | 64·9 (8·1) | 3·2 (1·0) |
| ULSAM | Sweden | 1139 | 216 | 171 | 56 | 115 | 0% | 79·9 (4·8) | 71·0 (0·6) | 12·8 (5·2) |
| WGHS | USA | 23 294 | 499 | 402 | 82 | 320 | 100% | 69·6 (9·3) | 54·7 (7·1) | 16·0 (3·2) |
| 3C Study Dijon | France | 3762 | 157 | 125 | 33 | 92 | 62% | 81·5 (6·1) | 72·4 (5·6) | 8·7 (3·1) |
| 3C Study Bordeaux and Montpellier | France | 2153 | 82 | 59 | 15 | 44 | 60% | 81·7 (5·2) | 73·9 (5·1) | 7·8 (2·5) |
| Total | 84 961 | 4348 | 3028 | 602 | 1770 | 67% | 75·8 (8·0) | 63·7 (8·4) | 10·0 (3·6) | |
|
| ||||||||||
|
Validation sample (cross-sectional case-control studies)*
| ||||||||||
| SiGN | USA and Europe | 49 324 | 16 851 | 16 851 | 3427 | 2346/3150‡ | 46% | 66·5 (14·8) | NA‡ | NA |
| METASTROKE | USA and Europe | 9654 | 1729 | 1729 | 276 | 206/159‡ | 36% | 67·0 (10·1) | 60·6 (11·9) | NA |
| HVH1 | USA | 2012 | 681 | 577 | 92 | 62/175‡ | 57% | 68·8 (8·9) | 66·7 (9·1) | NA |
| CADISP | Europe | 9814 | 555 | 555 | 211 | 67/31‡ | 61% | 43·7 (9·9) | NA‡ | NA |
| Total | 70 804 | 19 816 | 19 712 | 4006 | 2681/3515‡ | 47% | NA | NA | NA | |
Data are mean (SD) or n (%). NA=not available. AGES=Age, Gene/Environment Susceptibility-Reykjavik Study. ARIC=Atherosclerosis Risk in Communities Study. CHS=Cardiovascular Health Study. FHS=Framingham Heart Study. ABC=Health, Aging, and Body Composition. MESA=Multi-Ethnic Study of Atherosclerosis. PROSPER=Prospective Study of Pravastatin in the Elderly at Risk. SHIP=Study of Health In Pomerania. ULSAM=Uppsala Longitudinal Study of Adult Men. WGHS=Women’s Genome Health Study. SiGN=Stroke Genetics Network. HVH1= Heart and Vascular Health. CADISP=Cervical Artery Dissections and Ischemic Stroke Patients.
More detailed descriptions of composition of replication studies are shown in the appendix.
Percentage of women calculated from the total (n) column.
Samples are from large-vessel ischaemic stroke or small-vessel ischaemic stroke.
Mean age of controls in SiGN and CADISP study is not available because they were obtained from anonymous genotype databases.
This study was approved by the ethics committees of the participating studies and written informed consent was obtained from all study participants in the original cohort studies that allowed for data use in subsequent studies.
Stroke definition and classification of subtypes
Stroke was defined as a focal neurological deficit of presumed vascular origin with sudden onset and lasting for at least 24 hours, or until death if the participant died less than 24 hours after onset of symptoms. Stroke diagnosis and classification was validated by an expert committee in participating studies (appendix pp 13–19). Strokes were classified as ischaemic stroke (n=3028), intracerebral haemorrhage (n=277), or unknown type based on clinical and imaging criteria; for cohorts in which ischaemic stroke subtypes were available (table 1), ischaemic stroke was subdivided into cardioembolic (n=602) and non-cardioembolic (n=1770) subtypes. Numbers of events were too small to analyse large-vessel ischaemic stroke (n=117) and small-vessel ischaemic stroke (n=87) separately in this discovery dataset. Subarachnoid haemorrhage was not analysed because of its distinct mechanisms and very small number of events. Detailed definitions of stroke types and subtypes are given in the appendix.
Genotyping and imputation
Genotyping platforms and quality control filters are described in the appendix (pp 42–43). All but one study used imputed genotypes based on the 1000GpIv3 “All” reference panel (appendix pp 44–45).
Genome-wide association analyses
Using genome-wide multivariable Cox regression, we tested associations of genetic variants with incident stroke (all stroke, ischaemic stroke, cardioembolic ischaemic stroke, non-cardioembolic ischaemic stroke, and in secondary analyses intracerebral haemorrhage) under an additive genetic model, adjusting for sex and age and, when relevant, for principal components of population stratification, study site, or familial structure (appendix pp 19, 20, 46). Meta-analysis of study-specific association statistics was done by GC and AYC at two sites (University of Bordeaux, Bordeaux, France, and Harvard Medical School, Boston, MA, USA) with inverse-variance weighted meta-analysis with METAL (A software designed to facilitate meta-analysis of large datasets). The quantile–quantile plots and values of the genomic inflation factor λ suggested no systematic inflation of test statistics due to population stratification, cryptic relatedness, or technical arteficts (appendix pp 33, 47). Power of the discovery stage to detect association with various stroke subtypes is presented in the appendix (p 34).
Validation analyses
We selected variants with high imputation accuracy (mean r2>0·80) yielding an association at p<5 × 10−6 significance level with all stroke, ischaemic stroke, cardioembolic ischaemic stroke, or non-cardioembolic ischaemic stroke. 177 variants belonging to 21 loci (linkage disequilibrium r2>0·7 within each locus) were selected. We did in-silico lookups of association results using data from four independent, previously published cross-sectional studies, with mostly hospital-based stroke ascertainment, totalling 19 816 stroke patients (table 1) and 50 988 control participants from the Stroke Genetics Network (SiGN),7,8 METASTROKE,6 Heart and Vascular Health 1 (HVH1),9 and Cervical Artery Dissections and Ischaemic Stroke Patients (CADISP) studies.10 Except for 4963 black and 3371 Hispanic participants (cases and controls) in SiGN, participants in the validation samples were of European ancestry. Validation analyses were done with the same, or most similar, stroke phenotype as in the discovery phase (table 1), with logistic regression under an additive genetic model, followed by inverse-variance weighted meta-analysis of study-specific association statistics (appendix pp 20–25).
After Bonferroni correction for the number of independent loci (r2<0·01 reflecting absence of linkage disequilibrium), p<2·38 × 10−3 was considered significant evidence of replication. We did not correct for the number of stroke phenotypes (all stroke, ischaemic stroke, cardioembolic ischaemic stroke, and non-cardioembolic ischaemic stroke) because they are not independent (cardioembolic and non-cardioembolic ischaemic stroke being subtypes of ischaemic stroke, and ischaemic stroke a subtype of stroke). Only loci reaching genome-wide significance at p<5 × 10−8 in the combined meta-analysis of discovery and validation samples were given further consideration.
Secondary analyses
We examined the association of novel, genome-wide significant (at p<5×10−8), all-stroke risk variants with stroke subtypes in CHARGE and follow-up samples (using TOAST subtyping11 and, in sensitivity analyses, the Causative Classification System [CCS] implemented in SiGN7,8). We examined whether the same variants were associated with burden of white matter hyperintensity, a quantitative MRI-marker of cerebral small vessel disease, in 21 079 participants,12 and with intracerebral haemorrhage in an independent study comprising 1576 patients (682 patients with lobar intracerebral haemorrhage and 894 with deep intracerebral haemorrhage) and 1303 controls of European ancestry.13 We tested whether risk variants with genome-wide or suggestive association for all-stroke or ischaemic stroke in the population-based discovery stage were associated with incident fatal and non-fatal stroke. We also looked for a significant association of these risk variants comparing fatal or non-fatal stroke, which would suggest different genetic influences in the two groups of cases, assuming systematic bias in follow-up does not influence the frequency of the candidate variant in either group.
Functional exploration of novel stroke risk locus
Based on in-silico functional annotation (appendix pp 29–31) and literature review of the novel genome-wide significant stroke risk locus identified by the aforementioned approach, we examined the effect of loss of function of the putative causal gene on brain vasculature and stroke-related phenotypes in humans, mice, and zebrafish (appendix pp 31, 32). Studies into human participants and animal models were approved by the ethics committees of the participating institutions. To complement the genome-wide analysis, we used a rare cohort of patients with Axenfeld-Rieger syndrome with deletions of the novel stroke risk locus to directly determine if loss of this locus resulted in more severe cerebrovascular MRI phenotypes. By extracting data from individual MRI slices, we calculated the volume of white matter hyperintensity in two patients (aged 2 and 32 years) with large segmental deletions encompassing the putative causal gene and two patients (aged 15 and 17 years) with smaller deletions in whom this gene was intact.
The putative causal gene was deleted in adult (12 weeks) conditional (inducible) mouse mutants by Cre-ERT2, an inducible Cre recombinase14 and killed 6 weeks later. Brains of conditional knockout mice and controls were examined by histology (using haematoxylin and eosin or Richardson’s methylene blue–Azure 2 staining) and glial fibrillary acidic protein immuno fluorescence (DAKO Z0334) to search for features of vascular brain injury.
Zebrafish allow live imaging of blood vessel and mural cell interactions to be done with exceptional clarity, using transgenic lines that permit in-vivo visualisation of endothelium and smooth muscle. Expression of the putative causal gene in the brain of zebrafish larvae was assessed by in-situ hybridisation and compared with that of established pericyte markers notch3 and pdgfrβ.15 Function of forkhead transcription factor 2 (Foxf2) was assessed by knockout with transcription activator-like effector nucleases (TALENs) to create targeted nonsense mutations in the DNA-binding domain (appendix p 35). Smooth muscle cell coverage of branches of brain vessels was examined in live transgenic mutant and wild-type zebrafish embryos; these smooth muscle cells in zebrafish were modified to express green fluorescent protein with the α-smooth muscle actin promoter (acta2:GFP) at 4–6 days postfertilisation. Pericyte density was assessed by pdgfrβ in-situ hybridisation.15
Role of the funding source
All funders were involved in the study design but had no role in data collection. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
In the population-based discovery stage (4348 stroke patients vs 80 613 controls), 177 genetic variants in 21 independent loci were associated with incident all-stroke, ischaemic stroke, cardioembolic ischaemic stroke, or non-cardioembolic ischaemic stroke at p<5 × 10−6 significance (appendix pp 48–54). 11 loci showed suggestive association with incident all-stroke or ischaemic stroke at p<5 × 10−6. Ten additional loci were associated with incident cardioembolic ischaemic stroke at p<5 × 10−6 significance, with one locus (the lead single nucleotide polymorphism [SNP] rs72794386 in SLC12A2) showing genome-wide significance (hazard ratio [HR] 1·67, 95% CI 1·39–2·00, p=4·37 × 10−8) and minor allele frequency (MAF) of 10% (table 2, appendix pp 36, 37)
Table 2.
Single nucleotide polymorphism associated with risk of stroke at p<5 × 10−6
| Chromosome: position* |
Function | Gene | Number of variants† |
Minor allele |
Minor allele frequency |
Hazard ratio (95% CI) |
p value‡ | Direction§ | Heterogeneity, I2¶ |
Heterogeneity, p value|| |
Imputation quality** |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
All stroke
| ||||||||||||
| rs6433905 | 2:182138150 | Intergenic | UBE2E3 | 2 | C | 0·08 | 1·21 (1·12–1·31) | 2·54 × 10−6 | +++++++++++++++ | 0 | 0·54 | 0·97 |
| rs12204590 | 6:1337393 | Intergenic | FOXF2 | 6 | A | 0·21 | 1·14 (1·08–1·20) | 2·17 × 10−6 | ++++++++++++−+−++ | 0 | 0·60 | 1·00 |
| rs790919 | 6:154298875 | Intergenic | OPRM1 | 2 | A | 0·44 | 1·12 (1·07–1·17) | 2·44 × 10−6 | +++++++++++++−−++ | 16·2 | 0·26 | 0·96 |
| rs11788316 | 9:13445687 | Intergenic | MPDZ | 4 | T | 0·28 | 1·13 (1·07–1·19) | 2·49 × 10−6 | ++−++++−−++++++−−+ | 0 | 0·84 | 0·96 |
| rs11627959 | 14:35160471 | Intergenic | CFL2 | 4 | A | 0·44 | 0·89 (0·85–0·93) | 2·23 × 10−6 | −−+−−−−−−+−−+−−−− | 0 | 0·87 | 0·93 |
| rs4899120 | 14:64335447 | Intronic | SYNE2 | 1 | T | 0·09 | 1·19 (1·11–1·29) | 4·71 × 10−6 | +++−++++++++−−++ | 24 | 0·18 | 0·98 |
|
| ||||||||||||
|
Ischaemic stroke
| ||||||||||||
| rs62262077 | 3:105014929 | Intergenic | ALCAM | 5 | A | 0·27 | 1·17 (1·10–1·24) | 6·04 × 10−7 | −+++++++++++−+++ | 19·2 | 0·23 | 0·94 |
| rs10037362 | 5:31110857 | Intergenic | CDH6 | 2 | A | 0·07 | 1·27 (1·15–1·40) | 4·41 × 10−6 | −+−++++++++ | 25·7 | 0·20 | 0·97 |
| rs4448595 | 10:21666138 | Intergenic | C10orf114 | 8 | G | 0·16 | 0·83 (0·77–0·90) | 2·50 × 10−6 | −−−−−−−−+−−−−−−− | 0 | 0·80 | 0·98 |
| rs11833579 | 12:775199 | Intergenic | NINJ2 | 2 | A | 0·24 | 1·19 (1·12–1·27) | 5·74 × 10−8 | −+++−++++++−++++ | 18·5 | 0·24 | 0·92 |
| rs77858481 | 13:81142325 | Intergenic | SPRY2 | 1 | G | 0·06 | 1·38 (1·22–1·55) | 2·32 × 10−7 | ++++++++ | 0 | 0·97 | 0·83 |
|
| ||||||||||||
|
Cardioembolic ischaemic stroke
| ||||||||||||
| rs4284256 | 1:157675273 | Intergenic | FCRL3 | 1 | T | 0·18 | 1·41 (1·22–1·64) | 3·13 × 10−6 | −+++++ | 66·5 | 0·01 | 0·96 |
| rs12646447 | 4:111699326 | Intergenic | PITX2 | 102 | C | 0·12 | 1·53 (1·31–1·80) | 1·92 × 10−7 | ++++++ | 0 | 0·44 | 0·99 |
| rs72184 | 5:123754837 | Intergenic | ZNF608 | 1 | G | 0·43 | 1·30 (1·17–1·46) | 2·29 × 10−6 | +++++++++ | 9·1 | 0·6 | 0·90 |
| rs72794386 | 5:127479278 | Intronic | SLC12A2 | 22 | T | 0·10 | 1·67 (1·39–2·00) | 4·37 × 10−8 | +++++ | 0 | 0·87 | 0·97 |
| rs1428155 | 5:151281633 | Intronic | GLRA1 | 2 | C | 0·38 | 1·28 (1·16–1·43) | 3·10 × 10−6 | +++++++++ | 36·2 | 0·13 | 1·00 |
| rs7771564 | 6:22504092 | Intergenic | HDGFL1 | 4 | G | 0·10 | 1·53 (1·28–1·82) | 2·10 × 10−6 | ++++++ | 0 | 0·67 | 0·99 |
| rs1495081 | 8:15314955 | Intergenic | TUSC3 | 1 | C | 0·14 | 1·48 (1·25–1·74) | 3·09 × 10−6 | ++−+++ | 49·9 | 0·08 | 0·88 |
| rs2393938 | 10:44063812 | UTR5 | ZNF239 | 1 | C | 0·12 | 1·45 (1·24–1·70) | 3·47 × 10−6 | ++++++ | 0 | 0·51 | 0·99 |
| rs11021485 | 11:95968208 | Intronic | MAML2 | 1 | A | 0·12 | 1·60 (1·32–1·94) | 1·24 × 10−6 | +++++ | 30·5 | 0·22 | 0·82 |
| rs710009 | 14:59184500 | Intergenic | DACT1 | 4 | G | 0·16 | 1·41 (1·22–1·64) | 3·62 × 10−6 | +++++++ | 0 | 0·84 | 0·98 |
|
| ||||||||||||
|
Non-cardioembolic ischaemic stroke
| ||||||||||||
| rs77744591 | 13:81142325 | Intergenic | SPRY2 | 1 | T | 0·08 | 1·34 (1·18–1·51) | 3·44 × 10−6 | +++++−+ | 0 | 0·97 | 0·93 |
Only associations with the lead single nucleotide polymorphism in each locus are shown, and full set of genetic associations at p<5×10−6 is presented in the appendix (pp 48–54). All results are presented with respect to the minor allele as coded allele. UTR57
Chromosome positions with respect to National Center for Biotechnology Information Build 37 data.
Number of variants reaching p<5×10−6 in the locus.
p value after genomic control.
Direction refers to direction of effect size with respect to the minor allele in each study contributing to the meta-analysis (in alphabetic order), “+” sign refers to positive values of betas and “−” sign referes to negative values of betas with respect to the minor allele.
Heterogeneity (I2) ranges between 0 and 100, with higher values suggesting more heterogeneity.
p value for heterogeneity was calculated with the Cochran’s Q test.
Mean value of imputation quality across studies.
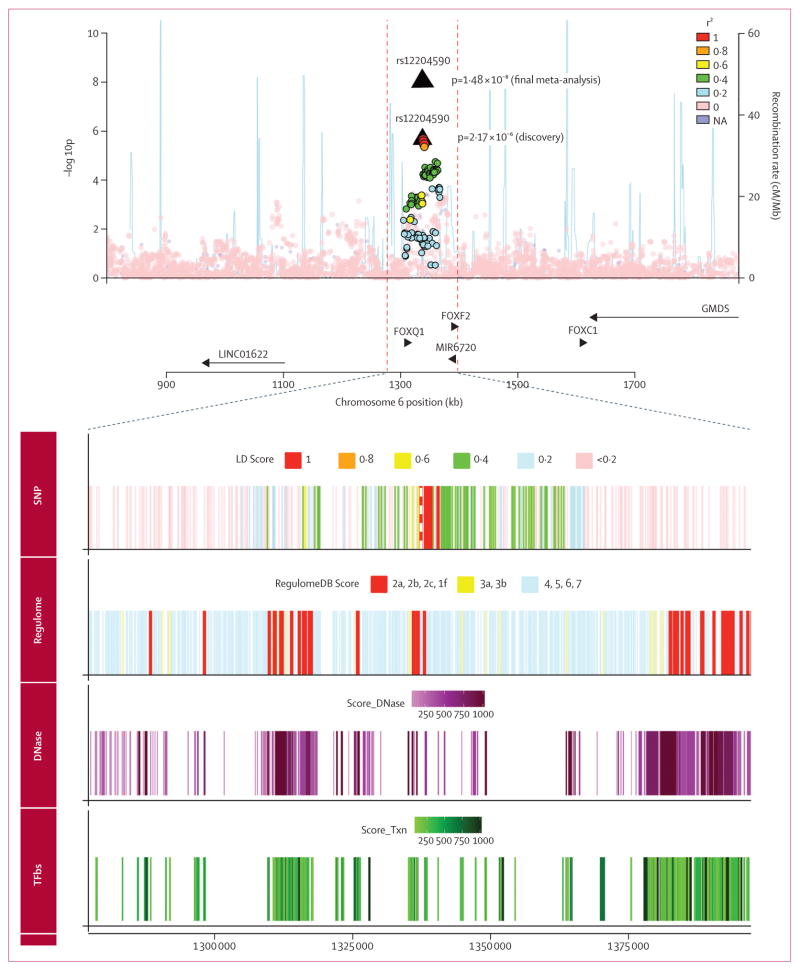
In the cross-sectional validation samples (19 816 stroke patients vs 50 988 control participants), associations for two loci were replicated at p<2·38 × 10−3 significance and reached genome-wide significance (p<5 × 10−8) in the combined analysis (table 3). The first association we found was a previously unreported locus (chr6p25·3, lead SNP rs12204590), located between FOXQ1 and FOXF2, showing association with risk of incident all-stroke (combined odds ratio [OR] 1·08 95% CI 1·05–1·12, p=1·48 × 10−8; MAF 21%; figure 1). Associations in each study are shown in the appendix (p 38). The second locus (chr4q25, near PITX2) is a known risk locus for cardioembolic ischaemic stroke (combined OR 1·37, 1·29–1·46, p=4·72 × 10−23 for incident cardioembolic ischaemic stroke; MAF 12%). The SLC12A2 locus (genome-wide significant in the small cardioembolic ischaemic stroke discovery sample) did not show evidence of replication (p=0·27).
Table 3.
Validation of top loci in independent cross-sectional case-control studies associated with risk of stroke
| Gene | Discovery
|
Validation
|
Combined total
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHARGE
|
SiGN
|
METASTROKE
|
HVH1
|
CADISP
|
Validation meta-analysis |
Meta-analysis
|
|||||||||
| Hazard ratio (95% CI) |
p value | Odds ratio (95% CI) |
p value | Odds ratio (95% CI) |
p value | Odds ratio (95% CI) |
p value | Odds ratio (95% CI) |
p value | Odds ratio (95% CI) |
p value | Odds ratio (95% CI) |
p value | ||
|
All stroke*
| |||||||||||||||
| rs6433905 | UBE2E3 | 1·21 (1·12–1·31) | 2·54 × 10−6 | 1·02 (0·97–1·08) | 0·41 | 1·11 (0·98–1·27) | 0·11 | 1·09 (0·83–1·44) | 0·54 | 0·72 (0·54–0·95) | 0·018 | 1·03 (0·98–1·07) | 0·22 | 1·07 (1·03–1·12) | 8·72 × 10−4 |
| rs12204590† | FOXF2 | 1·14 (1·08–1·20) | 2·17 × 10−6 | 1·07 (1·03–1·11) | 1·02 × 10−3 | 1·07 (0·98–1·16) | 0·13 | 1·03 (0·86–1·24) | 0·73 | 1·08 (0·92–1·26) | 0·36 | 1·06 (1·03–1·09) | 2·15 × 10−4 | 1·08 (1·05–1·12) | 1·48 × 10−8 |
| rs790919 | OPRM1 | 1·12 (1·07–1·17) | 2·44 × 10−6 | 1·00 (0·97–1·03) | 0·88 | 1·01 (0·95–1·09) | 0·70 | 0·88 (0·76–1·02) | 0·10 | 1·06 (0·93–1·21) | 0·37 | 1·00 (0·98–1·03) | 0·80 | 1·03 (1·01–1·05) | 0·013 |
| rs11788316 | FLJ41200 | 1·13 (1·07–1·19) | 2·49 × 10−6 | 0·99 (0·96–1·03) | 0·73 | 1·07 (0·99–1·16) | 0·074 | 1·01 (0·84–1·21) | 0·93 | 0·99 (0·85–1·15) | 0·89 | 1·01 (0·99–1·04) | 0·38 | 1·03 (1·01–1·06) | 7·53 × 10−3 |
| rs11627959 | CFL2 | 0·89 (0·85–0·93) | 2·23 × 10−6 | 0·99 (0·96–1·02) | 0·70 | 1·00 (0·93–1·08) | 0·93 | 1·00 (0·85–1·17) | 0·96 | 1·03 (0·90–1·18) | 0·62 | 1·01 (0·98–1·03) | 0·53 | 0·97 (0·95–0·99) | 0·011 |
| rs4899120 | SYNE2 | 1·19 (1·11–1·29) | 4·71 × 10−6 | 1·02 (0·97–1·07) | 0·50 | 1·00 (0·88–1·14) | 0·98 | 1·05 (0·80–1·37) | 0·73 | 1·16 (0·92–1·46) | 0·21 | 1·02 (0·99–1·07) | 0·23 | 1·06 (1·02–1·10) | 2·00 × 10−3 |
|
| |||||||||||||||
|
Ischaemic stroke‡
| |||||||||||||||
| rs62262077 | ALCAM | 1·17 (1·10–1·24) | 6·04 × 10−7 | 0·99 (0·96–1·02) | 0·52 | 1·03 (0·95–1·12) | 0·46 | 1·03 (0·84–1·26) | 0·78 | 1·02 (0·88–1·18) | 0·82 | 1·01 (0·98–1·03) | 0·63 | 1·03 (1·01–1·06) | 0·015 |
| rs10037362 | CDH6 | 1·27 (1·15–1·40) | 4·41 × 10−6 | 0·98 (0·93–1·03) | 0·40 | 0·99 (0·87–1·12) | 0·84 | 0·93 (0·69–1·26) | 0·65 | 0·86 (0·66–1·13) | 0·27 | 0·97 (0·93–1·02) | 0·22 | 1·01 (0·97–1·05) | 0·55 |
| rs4448595 | NEBL-AS1 | 0·83 (0·77–0·90) | 2·50 × 10−6 | 1·01 (0·97–1·05) | 0·72 | 1·02 (0·93–1·12) | 0·69 | 0·89 (0·72–1·10) | 0·28 | 0·96 (0·81–1·14) | 0·65 | 1·00 (0·97–1·03) | 0·97 | 0·98 (0·95–1·00) | 0·09 |
| rs11833579 | NINJ2 | 1·19 (1·12–1·27) | 5·74 × 10−8 | 0·98 (0·95–1·01) | 0·21 | 0·96 (0·88–1·04) | 0·28 | 1·04 (0·85–1·29) | 0·69 | 0·96 (0·82–1·12) | 0·57 | 0·98 (0·95–1·01) | 0·14 | 1·01 (0·98–1·03) | 0·47 |
| rs77858481 | SPRY2 | 1·38 (1·22–1·55) | 2·32 × 10−7 | 0·99 (0·93–1·06) | 0·75 | 0·90 (0·77–1·05) | 0·17 | 1·02 (0·72–1·44) | 0·93 | 0·91 (0·69–1·20) | 0·48 | 0·98 (0·93–1·03) | 0·50 | 1·03 (0·99–1·08) | 0·17 |
|
| |||||||||||||||
|
Cardioembolic ischaemic stroke§
| |||||||||||||||
| rs4284256 | FCRL3 | 1·41 (1·22–1·64) | 3·13 × 10−6 | 0·96 (0·90–1·04) | 0·31 | 0·93 (0·78–1·12) | 0·45 | 1·03 (0·69–1·55) | 0·88 | 0·98 (0·75–1·28) | 0·90 | 0·96 (0·90–1·01) | 0·13 | 1·02 (0·97–1·09) | 0·41 |
| rs12646447† | PITX2 | 1·53 (1·31–1·80) | 1·92 × 10−7 | 1·39 (1·29–1·50) | 3·15 × 10−18 | 1·17 (0·98–1·41) | 0·083 | 1·62 (1·09–2·42) | 0·018 | 1·04 (0·78–1·37) | 0·80 | 1·36 (1·28–1·44) | 1·89 × 10−23 | 1·37 (1·29–1·46) | 4·72 × 10−24 |
| rs72184 | ZNF608 | 1·30 (1·17–1·46) | 2·29 × 10−6 | 1·02 (0·96–1·08) | 0·53 | 0·99 (0·87–1·13) | 0·90 | 0·92 (0·67–1·28) | 0·63 | 1·03 (0·85–1·26) | 0·74 | 1·02 (0·97–1·06) | 0·49 | 1·06 (1·01–1·10) | 0·017 |
| rs72794386 | SLC12A2 | 1·67 (1·39–2·00) | 4·37 × 10−8 | 0·97 (0·89–1·06) | 0·51 | 1·09 (0·87–1·37) | 0·46 | 0·95 (0·56–1·62) | 0·85 | 0·78 (0·54–1·14) | 0·18 | 0·96 (0·90–1·03) | 0·27 | 1·06 (0·99–1·14) | 0·12 |
| rs1428155 | GLRA1 | 1·28 (1·16–1·43) | 3·10 × 10−6 | 0·97 (0·92–1·03) | 0·33 | 0·95 (0·83–1·08) | 0·44 | 0·90 (0·66–1·24) | 0·51 | 0·99 (0·81–1·21) | 0·94 | 0·99 (0·95–1·03) | 0·64 | 1·02 (0·98–1·07) | 0·38 |
| rs7771564 | HDGFL1 | 1·53 (1·28–1·82) | 2·10 × 10−6 | 1·01 (0·92–1·10) | 0·87 | 0·88 (0·70–1·10) | 0·25 | 1·23 (0·77–1·96) | 0·39 | 1·20 (0·89–1·62) | 0·24 | 1·00 (0·93–1·07) | 0·97 | 1·08 (1·01–1·17) | 0·031 |
| rs1495081 | TUSC3 | 1·48 (1·25–1·74) | 3·09 × 10−6 | 1·05 (0·98–1·14) | 0·18 | 0·89 (0·72–1·09) | 0·25 | 1·35 (0·84–2·16) | 0·21 | 1·05 (0·79–1·40) | 0·73 | 1·04 (0·98–1·10) | 0·24 | 1·10 (1·03–1·17) | 5·07 × 10−3 |
| rs2393938 | ZNF239 | 1·45 (1·24–1·70) | 3·47 × 10−6 | 1·02 (0·94–1·10) | 0·68 | 1·01 (0·84–1·22) | 0·88 | 0·95 (0·60–1·52) | 0·84 | 0·96 (0·71–1·29) | 0·76 | 1·00 (0·94–1·07) | 0·95 | 1·07 (1·01–1·15) | 0·029 |
| rs11021485 | MAML2 | 1·60 (1·32–1·94) | 1·24 × 10−6 | 0·94 (0·86–1·03) | 0·17 | 0·77 (0·63–0·95) | 0·015 | 0·51 (0·25–1·04) | 0·063 | 1·06 (0·78–1·43) | 0·73 | 0·92 (0·86–0·99) | 0·017 | 0·99 (0·92–1·07) | 0·82 |
| rs710009 | DACT1 | 1·41 (1·22–1·64) | 3·62 × 10−6 | 1·00 (0·92–1·07) | 0·93 | 0·96 (0·80–1·15) | 0·68 | 1·21 (0·80–1·83) | 0·37 | 1·10 (0·84–1·43) | 0·50 | 1·00 (0·94–1·06) | 0·96 | 1·06 (1·00–1·13) | 0·048 |
|
| |||||||||||||||
|
Non-cardioembolic ischaemic stroke¶
| |||||||||||||||
| rs77744591 | SPRY2 | 1·34 (1·18–1·51) | 3·44 × 10−6 | 1·08 (0·96–1·21) | 0·19 | 1·08 (0·84–1·40) | 0·53 | 1·39 (0·74–2·59) | 0·30 | 0·91 (0·47–1·77) | 0·78 | 1·08 (0·99–1·18) | 0·80 | 1·18 (1·09–1·28) | 3·22 × 10−5 |
| rs77744591 | SPRY2 | 1·34 (1·18–1·51) | 3·44 × 10−6 | 1·12 (1·02–1·24) | 0·023 | 1·06 (0·80–1·40) | 0·68 | 0·83 (0·52–1·31) | 0·42 | 0·73 (0·24–2·22) | 0·56 | 1·11 (1·02–1·20) | 0·16 | 1·18 (1·10–1·27) | 1·08 × 10−5 |
Validationresults from association analyses of ischaemic stroke for SiGN, METASTROKE, and CADISP, and of all stroke for HVH1.
Loci that reach genome-wide significance (p<5 × 10−8) in the combined meta-analysis.
Follow-up results from association analyses of ischaemic stroke for SiGN, METASTROKE, HVH1, and CADISP.
Validation results from association analyses of cardioembolic ischaemic stroke for SiGN, METASTROKE, HVH1, and CADISP (TOAST subtyping).
Validation results from association analyses of large-vessel ischaemic stroke (first line) and small-vessel ischaemic stroke (second line) for SiGN, METASTROKE, HVH1, and CADISP (TOAST subtyping).
Figure 1. Regional association plot of rs12204590 in discovery stage.
Association of rs12204590 and other genotyped or imputed SNPs (circles) in the region with incident all-stroke. Colour variation shows linkage disequilibrium between SNPs (r2) as calculated in 1000 Genomes Project (phase 1, version 3). Blue lines show estimated recombination rates. Coloured tracksseen at bottom of figure were added using the University of California, Santa Cruz genome browser and the RegulomeDB database. SNP track=SNPs encompassing the selected region and red dashed line shows position of top SNP (rs12204590). Regulome track=RegulomeDB scores and variants with lower scores have higher probability of acting as regulatory variants. DNase track=DNase hypersensitive regions assayed in 125 cell types (ENCODE project, Release 3,2014). TFbs track=regions where transcription factors, proteins responsible for modulating gene transcription, bind to DNA as assayed by ChIP-seq assay (ENCODE project, Release 3, 2013).
We also explored association of the chr6p25·3 locus with stroke subtypes. In the discovery sample, lead SNP rs12204590 was associated with incident ischaemic stroke (HR 1·13, 95% CI 1·06–1·20), p=1·64 × 10−4; non-cardioembolic ischaemic stroke: HR 1·12, 1·04–1·22, p=4·35×10−3; and cardioembolic ischaemic stroke: HR 1·10 0·95–1·27, p=0·21). In validation samples, we detected an association with small-vessel ischaemic stroke (OR 1·08, 95% CI 1·02–1·14, p=0·0094) for rs12200309 (in complete linkage disequilibrium with rs12204590), using TOAST subtypes, whereas association with large-vessel and cardioembolic ischaemic stroke was not significant (p>0·35). The association with small-vessel ischaemic stroke was also evident when using CCS-causative subtyping where available (SiGN; OR 1·11, 1·05–1·18, p=0·00029; appendix p 55). chromosome 6p25·3 was also significantly associated with increasing burden of white matter hyperintensity in the general population (most significant p value was p=0·0025, appendix p 56). The HR for association of rs12204590 with incident fatal ischaemic stroke (n=271; HR 1·21, 0·99–1·50, p=0·0684) was higher than with non-fatal ischaemic stroke (n=2300; HR 1·14, 1·06–1·22, p=4·93×10−4), but the difference was not significant (appendix pp 57–59). We did not record any heterogeneity by ethnicity for associations between the chr6p25 locus and stroke risk (appendix p 60).
The genomic region where the variant associated with increased stroke risk and adjacent linked variants (linkage disequilibrium r2>0·50) are located seems to include enhancers (regions of DNA that activate transcription of nearby genes). This genomic region also includes DNase I hypersensitive regions, a marker of open chromatin associated with active cis-regulatory elements important for transcription of nearby genes (figure 1, appendix pp 61, 62). Two SNPs (rs7750826 and rs2006798, r2>0·75 with rs12204590) had RegulomeDB scores of 2b, suggesting a probable role in regulating gene expression (combination of transcription factor binding site and DNase peak and footprint; figure 1, appendix pp 63, 64). The genomic region that includes the lead variant and variants in linkage disequilibrium also includes two protein-coding genes, FOXQ1 and FOXF2. The same region also includes a microRNA (MIR6720). However, if we expand the regional plot to a 1 Mb region around the lead variant, the region also includes two other protein coding genes (FOXC1 and GMDS) and a long non-coding RNA (LINC01622; figure 1). We did an extensive search of publicaly available expression quantitative trait loci (eQTL)16 and miRNA databases,17 examined mRNA expression of FOXF2 and adjacent genes in the dorsolateral prefrontal cortex of 508 participants enrolled in the Religious Orders Study and the Rush Memory and Aging Project,18 and mined large sets of epigenomic data from the International Human Epigenome Consortium (appendix pp 65–67). eQTL or methylation quantitative trait loci in this region were absent except for a long non-coding RNA (LOC285768) in the human brain (p=5·25×10−7 for rs7746700, average in ten brain regions in the BRAINEAC database). However, the different distribution of histone modifications associated with active genes in cells expressing FOXF2 or FOXQ1 suggested that these variants are likely to lie within the regulatory region of FOXF2 (appendix pp 39, 40). Moreover, we noted the lowest CpG methylation levels (indicating highest activity) at this locus in the fetal brain compared with any other tissue.19
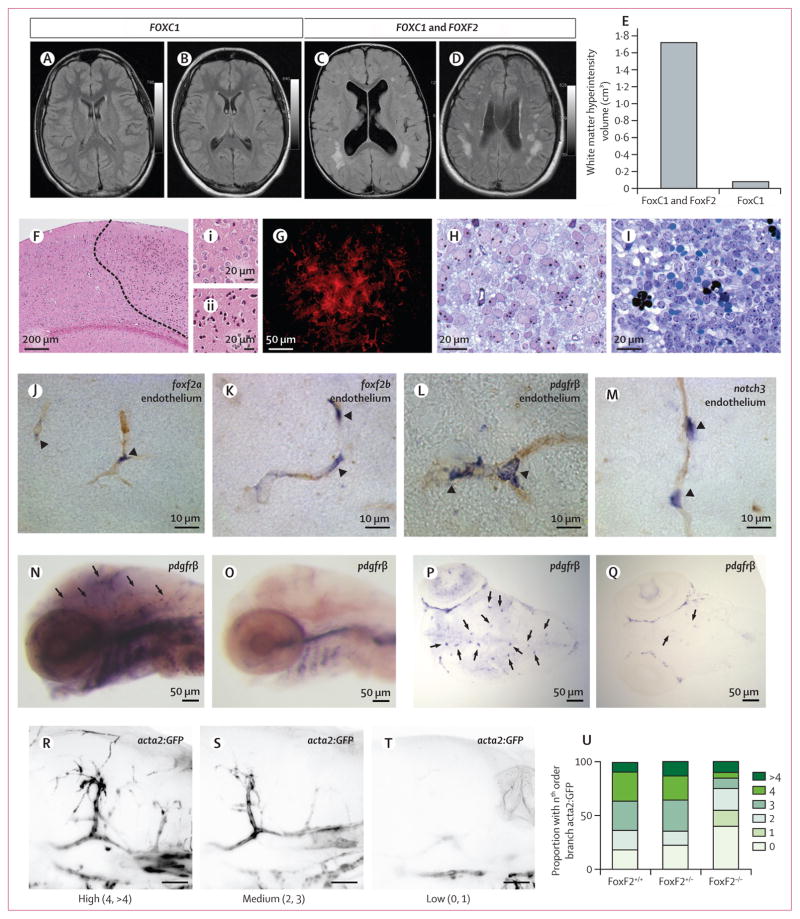
We have previously described that patients with Axenfeld-Rieger syndrome, a rare heterogeneous condition with maldevelopment of the ocular anterior segment attributable to mutation or copy number variation of FOXC1 (adjacent to FOXF2 on chr6p25), have increased burden of MRI markers of cerebral small vessel disease.20 Within this cohort of people with FOXC1-attributable Axenfeld-Rieger syndrome, we identified two patients with 300 kb segmental deletions encompassing both FOXC1 and FOXF2 and found that they had extensive, confluent white matter hyperintensity, with more than ten-times larger volume of white matter hyperintensity than two patients with segmental deletions of FOXC1 only (30 kb), although the small number of patients does not allow a formal statistical comparison to be made. All patients were younger than 35 years (range 2–32 years) and lacked vascular risk factors (figure 2A–E, appendix p 68). White matter hyperintensities are normally absent or negligible in this age range in the general population.21
Figure 2. Cerebrovascular phenotype of FOXF2 and FOXC1 deletions in humans and FoxF2 knockout mice, and expression of foxf2a and foxf2b in zebrafish.
(A–D) In two patients with segmental deletion encompassing FOXC1, white matter hyperintensities are noted in periventricular region (A,B) and subcortical regions (B). In two patients with segmental deletion of both FOXC1 and FOXF2 (C,D), mean white matter hyperintensities volume is increased by more than ten times (E), in subcortical and periventricular regions (see appendix p 68 for white matter hyperintensities volumes in each of the four patients). (F–I) Cerebral cortex of conditional Foxf2 knockout mouse showing ischaemic infarction and haemorrhagic tissue. (F) Area with condensed eosinophilic cytoplasm and pyknotic nuclei (to the right dashed line) that indicates recent ischaemic infarction. (Fi) Normal tissue and (Fii) tissue with ischaemic infarction at higher magnification. (G) Glial fibrillary acidic protein immunofluorescence of area with reactive astrogliosis in cerebral cortex of Foxf2 conditional knockout mouse. (H) Cerebral cortex from control mouse showing normal neuronal tissue and intact capillaries. (I) Haemorrhagic area of cerebral cortex from Foxf2 conditional knockout mouse. Extravascular erythrocytes seen both as intact cells (homogeneous greenish blue) and lysed cells (black). (J–M) RNA in situ hybridisation (purple) of larval zebrafish brains shows expression of foxf2a (J) and foxf2b (K) in presumptive pericytes compared with known pericyte markers pdgfrβ (L) and notch3 (M; purple) around capillaries in 1-month old larval zebrafish (brown). (N-Q) foxf2−/− mutants have reduced expression of pericyte marker pdgfrβ in 4-day postfertilisation embryonic cerebral vasculature. (R–U) Loss of foxf2b results in reduction of the smooth-muscle marker acta2:GFP coverage of blood vessels in wild type (n=11), foxf2+/− (n=31), and Foxf2−/− (n=20) embryonic cerebral vasculature. (R–T) Examples of high, medium, and low branch order coverage scored from 0 to the 4th order branch in vessel coverage presented as percentages of total embryo counts (U). Arrow heads in images J–Q refer to positions showing pericytes.
To understand the mechanism at the tissue level we used histology of the brains from six mice 6 weeks after Foxf2 inactivation and found areas of neurons with pyknotic nuclei and eosinophilic cytoplasm (figure 2F–Fii), suggestive of ischaemic infarction, in five of the brains. Patches with elevated levels of glial fibrillary acidic protein in astrocytes (figure 2G) indicated reactive gliosis. Increased magnification revealed a few instances of microhaemorrhage with extravascular erythrocytes (figure 2I). By contrast, brains from control mice contained only occasional and scattered neurons that showed signs of apoptosis or isolated astrocytes with increased glial fibrillary acidic protein immuonoreactivity and no visible haemorrhagic lesions (figure 2H). Mice in which Foxf2 had been deleted had significantly higher mortality than control mice. Usually the animals were found dead, but some had to be euthanised after showing behaviour indicative of brain damage, such as circling or lopsided gait.
In zebrafish, foxf2a and foxf2b (FOXF2 orthologs) are expressed in the cerebral endothelium in pericytes, similar to expression in pericytes in mice (figure 2J,K). Expression occurrs in a pattern similar to those of established pericyte markers pdgfrβ and notch3 (figure 2L,M). We made two zebrafish knockout lines, foxf2bca22 and foxf2bca23, with nonsense mutations in the first exon of foxf2b that would result in a translation block before the essential DNA-binding domain (appendix p 35). Both alleles had identical phenotypes. foxf2b mutants had decreased expression of the brain pericyte marker pdgfrβ (figure 2N–Q), which is indicative of pericyte maturation defects and decreased acta2-positive smooth muscle cell coverage on large cerebral vessels, which in turn is suggestive of smooth-muscle defects (figure 2R–U, appendix p 41). In homozygous mutants, acta2 was visible up to second order of cerebral vessel branching or less versus fourth or higher order branch in wild-type or heterozygous embryos.
When considering risk loci for ischaemic stroke reported by previous cross-sectional studies,2 we found that seven of the eight published loci were associated with incident stroke in the discovery stage of the present population-based GWAS, predominantly in the same subtype as the original study (p value range=0·047–7·82 × 10−5, appendix p 69). One known risk locus for ischaemic stroke (PITX2) also showed association with incident intracerebral haemorrhage (p=0·0031), and one known risk locus for intracerebral haemorrhage (PMF1-BGLAP), also a known risk locus for increasing white matter hyperintensity burden,12 was associated with incident ischaemic stroke (p=0·00064), both in the same direction (appendix p 31).
Discussion
In a large population-based GWAS meta-analysis of incident stroke, with validation in the largest available cross-sectional stroke GWAS, we identified a novel genome-wide significant association of common variants in the chr6p25·3 region, near FOXF2, with risk of stroke. Associations predominated with small-vessel ischaemic stroke, and significant association was noted with burden of white matter hyperintensity; in addition, patients with rare segmental deletions of FOXF2 also showed extensive burden of white matter hyperintensity. These findings suggest an effect of this locus on cerebral small vessel disease; however, the mechanism by which this transcription factor results in cerebral small vessel disease and stroke is unclear. To investigate the possibility of a role of FOXF2 in stroke, we did experimental studies in two animal species to examine its role in cerebral vessel development and stability. We showed areas of infarction and microhaemorrhages in brains of conditional Foxf2 mutant mice. We showed in zebrafish that foxf2b was expressed in brain pericytes (as in mice14) and that reduction in foxf2b function led to differentiation defects of both pericytes and smooth muscle cells in the developing cerebral vasculature.
Converging evidence from the present study and previous publications suggests an important role of FOXF2 in cerebrovascular disease. In mice, we previously showed that Foxf2 is required for brain pericyte differentiation and blood–brain barrier development, with Foxf2−/− embryos showing thickened endothelium, perivascular oedema, thinning of the vascular basal lamina, and a leaky blood–brain barrier.14 We had also described that Foxf2 inactivation in adult mice results in endothelial thickening and blood–brain barrier breakdown,14 an important mediator of cerebral small vessel disease, and increased mortality.22 In the conditional Foxf2 mutant, we analysed mice brains 6 weeks after Foxf2 inactivation and we found signs of brain infarction, with reactive astrogliosis and microhaemorrhages. Of note in previous experiments, Foxf2−/− mouse embryos developed intracranial haemorrhage,14 whereas areas of microhaemorrhage were scarce in conditional Foxf2 mutant brains. We were also unable to show an association of the chr6p25.3 locus with intracerebral haemorrhage in the largest available collaborative genetic association study, although power might have been limited by the size of the study (appendix p 70). In zebrafish, foxf2b knockout led to disruption of cerebral vasculature with decreased pericyte density and smooth muscle cell coverage (figure 2). Foxf2 is first expressed in the neural crest and in mice regulates pathways involved in mural cell (pericyte and vascular smooth muscle cell) differentiation, including the pdgfβ and serum response factor pathways.14,23 We did not see haemorrhage in the zebrafish foxf2 mutant model during embryonic stages. We note that we knocked out only one of two foxf2 genes in zebrafish, and even though mural cell markers have changed expression in foxf2b mutants, there might be genetic compensation from the foxf2a gene that could make the phenotype less severe. However, we have not found expression changes of foxf2a in zebrafish having mutant foxf2b. We cannot exclude haemorrhage occurring at juvenile or adult stages that we have not been able to examine. In summary, our data suggest that association of FOXF2 and stroke might arise from differentiation defects of cerebral vascular mural cells. To demonstrate the cellular requirement for Foxf2 by expression under a vascular mural cell promoter for prevention of stroke and mural cell phenotypes in mutants is an important future experiment, but is beyond the scope of this study.
Forkhead transcription factors are involved in various developmental and biological processes, and tend to be distributed in clusters on the genome.24,25 The evolutionarily conserved chr6p25 cluster comprises FOXQ1, FOXF2, and FOXC1. Our lead stroke risk variants lie between FOXQ1 (22·4 kb) and FOXF2 (52·7 kb). By contrast with the compelling experimental evidence for a central role of FOXF2 in cerebrovascular disease, FOXQ1 has not been implicated in cerebrovascular phenotypes; mutant mice for this gene show mainly altered hair differentiation and gastric mucin secretion.26,27 The third gene in the cluster, FOXC1 (225 kb downstream of FOXF2 and 273·3 kb downstream of stroke risk variants), is also expressed in the brain vasculature and influences vessel morphogenesis28 and arteriovenous specification.29 MRI analysis of patients with FOXC1-attributable Axenfeld-Rieger syndrome revealed MRI features of cerebral small vessel disease (including increased burden of white matter hyper intensity), and genetic variants downstream of FOXC1 were associated with burden of white matter hyper intensity in the general population.20 These previously described variants are independent (r2<0·017) from the stroke risk variants near FOXF2 described here.20,30 These findings, together with the ten-times increased burden of white matter hyperintensity in patients with segmental deletion of both FOXC1 and FOXF2 versus FOXC1 alone, suggest an independent role of FOXF2 in cerebral small vessel disease. Differences in the roles of foxf2 and foxc1a and foxc1b are also seen in the zebrafish model.31 Knockdown of zebrafish foxc1a/b leads to embryonic cerebral hemorrhage in embryos,31 whereas knockout of foxf2b at the same developmental stage does not. Thus, although the two genes are closely related, there are indications that their roles in vascular mural cells might be distinct.
Intriguingly, PITX2 (chr4q25), a known risk locus for cardioembolic ischaemic stroke and atrial fibrillation32,33 as well as being genome-wide significant for cardioembolic ischaemic stroke in the present sample, encodes a neural crest–expressed transcription factor that physically interacts with FOXC1 and harbours causal mutations for Axenfeld-Rieger syndrome.20 Variants near PITX2 were associated with burden of white matter hyperintensity.20,30 In this study, common variants near PITX2 were also associated with risk of intracerebral haemorrhage, of which the main mechanism in the general population is small vessel disease.34 These findings suggest that FOXF2, FOXC1, and PITX2 could perhaps contribute to cerebrovascular disease via partly shared pathways, of relevance in cerebral small arteries.
The small number of incident strokes (n=4300), particularly stroke subtypes (n=602 for cardioembolic ischaemic stroke, whereas small-vessel occlusion, large-vessel ischaemic stroke, and other stroke subtypes had to be merged into a single category) might have hampered our ability to detect additional associations. The study was also underpowered (<80% power) to detect associations with effect sizes smaller than OR 1·10 and allele frequency lower than 5% (appendix p 34). Although the results of the described animal studies suggest that FOXF2 is the causal gene underlying the observed genetic association with stroke and small vessel disease at chr6p25, functional annotation of the identified risk variants is limited, with an absence of eQTL despite an extensive search of publicly available and other databases, possibly reflecting tissue specificity or primarily developmental effects. These effects are supported by a higher expression of FOXF2 in fetal brain than adult brain, and lower methylation levels of the stroke risk locus in fetal tissues than adult tissues. Another limitation is that we only explored common variants and not rare variants in this region. In addition, we have used an additive genetic model only, which is the most powerful approach when the underlying genetic model is unknown, but we cannot exclude that associations with genetic risk loci following a recessive or dominant model might have been missed. Nevertheless, we confirmed and extended the range of associations for previously discovered stroke risk loci in a population-based sample. For the first time, to our knowledge, we describe shared genetic variation underlying both ischaemic stroke (chr4q25) and intracerebral haemorrhage (chr1q22), in agreement with some monogenic strokes having both ischaemic and haemorrhagic phenotypes, mostly with underlying small vessel disease.35–37 The association we previously described between chr12p13 and incident stroke in a smaller, overlapping sample was also the most significant association with ischaemic stroke in the present population-based GWAS,5 showing a stronger association with incident fatal stroke versus non-fatal stroke, which suggests an effect on stroke survival (appendix pp 58, 59).
In summary, we identified common variants near FOXF2 associated with increased stroke susceptibility (especially of the small vessel subtype) and extensive subclinical small vessel disease. This association is particularly interesting because GWAS have not yet discovered risk loci for small-vessel ischaemic stroke (except for an association with the PRKCH locus identified in a study of Japanese participants, which was not found in European populations).6,38 Brain imaging data from patients with rare segmental deletions encompassing FOXF2, and functional experiments across evolutionarily separated species, suggest an important role of FOXF2 in cerebrovascular disease—especially cerebral small vessel disease—possibly by affecting differentiation of cerebral vascular mural cells. Cerebral small vessel disease is a major, but poorly understood, cause of stroke in all ethnic groups, and subclinical small vessel disease has been associated with progressive functional and cognitive decline and increased risk of dementia. At present, no mechanism-based treatment for small vessel disease is available, other than management of vascular risk factors. Our findings provide promising grounds for follow-up, pointing to a possible mechanism of stroke and small vessel disease. Further research is warranted to explore whether these findings can have implications for clinical practice.
Supplementary Material
Acknowledgments
Funding NIH, NINDS, NHMRC, CIHR, European national research institutions, Fondation Leducq.
For information about the funding for each section, please see the appendix (pp 63–73).
Footnotes
Contributors
SD, SS, WTL, SJC, DIC, PC, MAI, and LJL jointly supervised research. GC, CRA, AYC, MF, AR, JCB, and ASH contributed equally. SD, SS, WTL, SJC, PC, JR, and BBW conceived and designed the experiments. GC, AYC, MF, JCB, ASH, MS, AVS, HHHA, SHC, SLP, ST, MEGa, AM, AT, SG, TMB, CeB, XJ, FX, YL, MdH, TP, CCW, SB, RM, QW, MT, and YK. did statistical analysis and other data analysis. MAI, LJL, KMR, CT, OJL, CMvD, EI, THM, VS, JR, BBW, COS, ALD, SSR, TBH, BMP, TK, VG, ML, TP, CCW, PLDJ, DAB, MD, CRF, EV, HSM, AB, BGW, DSK, MEGr, RFG, MAN, JIR, OLL, YK, JDa, RC, GBB, US, MRPM, HJG, ADJ, MMS, NBA, DJS, JWJ, IF, AJMdC, AGU, MLP, PJK, AHo, DW, AS, FR, JMo, JJ-C, APM, CML, LLi, LLa, NLP, MT, PKM, AHa, J-FD, ClB, PMRi, JEB, AP, DL, MK, STE, HJA, QW, J-ML, SW-S, ORB, JAJ, CJ, DKA, PMRo, TR, RLS, RPG, OM, AL, VT, JMe, RS, JWC, AMN, CLMS, PS, CL, NLS, KR, SRH, RM, JCH, SB, MdH, JDe, YL, SR, FX, CLS, KLW, OK, XJ, JSV, CeB, TMB, SG, AT, AM, MEGa, ST, SLP, SHC, HHHA, AVS, MS, ASH, JCB, AR, MF, AYC, and CRA revised the draft critically for important intellectual content. SJC, OJL, AMN, AR, and CRA. did the experiments. SD, SS, WTL, SJC, DIC, PC, and GC wrote the Article.
Declaration of interests
We declare no competing interests.
References
- 1.Johnston SC, Mendis S, Mathers CD. Global variation in stroke burden and mortality: estimates from monitoring, surveillance, and modelling. Lancet Neurol. 2009;8:345–54. doi: 10.1016/S1474-4422(09)70023-7. [DOI] [PubMed] [Google Scholar]
- 2.Falcone GJ, Malik R, Dichgans M, Rosand J. Current concepts and clinical applications of stroke genetics. Lancet Neurol. 2014;13:405–18. doi: 10.1016/S1474-4422(14)70029-8. [DOI] [PubMed] [Google Scholar]
- 3.Bevan S, Traylor M, Adib-Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–67. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 4.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 5.Ikram MA, Seshadri S, Bis JC, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–28. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–62. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meschia JF, Arnett DK, Ay H, et al. Stroke Genetics Network (SiGN) study: design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke. 2013;44:2694–702. doi: 10.1161/STROKEAHA.113.001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pulit SL, McArdle PF, Wong Q, et al. Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 2015;15:174–84. doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klungel OH, Heckbert SR, Longstreth WT, Junior, et al. Antihypertensive drug therapies and the risk of ischemic stroke. Arch Intern Med. 2001;161:37–43. doi: 10.1001/archinte.161.1.37. [DOI] [PubMed] [Google Scholar]
- 10.Debette S, Kamatani Y, Metso TM, et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015;47:78–83. doi: 10.1038/ng.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams HP, Junior, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 12.Verhaaren BF, Debette S, Bis JC, et al. Multi-ethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ Cardiovasc Genet. 2015;8:398–409. doi: 10.1161/CIRCGENETICS.114.000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo D, Falcone GJ, Devan WJ, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–21. doi: 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyahi A, Nik AM, Ghiami M, et al. Foxf2 is required for brain pericyte differentiation and development and maintenance of the blood-brain barrier. Dev Cell. 2015;34:19–32. doi: 10.1016/j.devcel.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Arnold CR, Lamont RE, Walker JT, et al. Comparative analysis of genes regulated by Dzip1/iguana and hedgehog in zebrafish. Dev Dyn. 2015;244:211–23. doi: 10.1002/dvdy.24237. [DOI] [PubMed] [Google Scholar]
- 16.Ramasamy A, Trabzuni D, Guelfi S, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L, Chibnik LB, Srivastava GP, et al. Association of brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72:15–24. doi: 10.1001/jamaneurol.2014.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziller MJ, Gu H, Muller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French CR, Seshadri S, Destefano AL, et al. Mutation of FOXC1 and PITX2 induces cerebral small-vessel disease. J Clin Invest. 2014;124:4877–81. doi: 10.1172/JCI75109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim BS, Illes J, Kaplan RT, et al. Incidental findings on pediatric MR images of the brain. Am J Neuroradiol. 2002;23:1674–77. [PMC free article] [PubMed] [Google Scholar]
- 22.Wardlaw JM, Smith C, Dichgans M. Mechanisms of sporadic cerebral small vessel disease: insights from neuroimaging. Lancet Neurol. 2013;12:483–97. doi: 10.1016/S1474-4422(13)70060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolte C, Ren X, Tomley T, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem. 2015;290:7563–75. doi: 10.1074/jbc.M114.609487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development and disease. Trends Genet. 2003;19:339–44. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 25.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 26.Hong HK, Noveroske JK, Headon DJ, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29:163–71. doi: 10.1002/gene.1020. [DOI] [PubMed] [Google Scholar]
- 27.Verzi MP, Khan AH, Ito S, Shivdasani RA. Transcription factor foxq1 controls mucin gene expression and granule content in mouse stomach surface mucous cells. Gastroenterology. 2008;135:591–600. doi: 10.1053/j.gastro.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegenthaler JA, Choe Y, Patterson KP, et al. Foxc1 is required by pericytes during fetal brain angiogenesis. Biol Open. 2013;2:647–59. doi: 10.1242/bio.20135009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fish JE, Wythe JD. The molecular regulation of arteriovenous specification and maintenance. Dev Dyn. 2015;244:391–409. doi: 10.1002/dvdy.24252. [DOI] [PubMed] [Google Scholar]
- 30.Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann Neurol. 2011;69:928–39. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.French CR, Seshadri S, Destefano AL, et al. Mutation of FOXC1 and PITX2 induces cerebral small-vessel disease. J Clin Invest. 2014;124:4877–81. doi: 10.1172/JCI75109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellenguez C, Bevan S, Gschwendtner A, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–33. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–57. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 34.Greenberg SM. Small vessels, big problems. N Engl J Med. 2006;354:1451–53. doi: 10.1056/NEJMp068043. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Q, Yang D, Ombrello AK, et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. N Engl J Med. 2014;370:911–20. doi: 10.1056/NEJMoa1307361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sibon I, Coupry I, Menegon P, et al. COL4A1 mutation in Axenfeld-Rieger anomaly with leukoencephalopathy and stroke. Ann Neurol. 2007;62:177–84. doi: 10.1002/ana.21191. [DOI] [PubMed] [Google Scholar]
- 37.Lanfranconi S, Markus HS. COL4A1 mutations as a monogenic cause of cerebral small vessel disease: a systematic review. Stroke. 2010;41:e513–18. doi: 10.1161/STROKEAHA.110.581918. [DOI] [PubMed] [Google Scholar]
- 38.Kubo M, Hata J, Ninomiya T, et al. A nonsynonymous SNP in PRKCH (protein kinase C eta) increases the risk of cerebral infarction. Nat Genet. 2007;39:212–17. doi: 10.1038/ng1945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.