Abstract
The heme uptake pathway (hmu) of Corynebacterium diphtheriae utilizes multiple proteins to bind and transport heme into the cell. One of these proteins, HmuT, delivers heme to the ABC transporter HmuUV. In this study, the axial ligation of the heme in ferric HmuT is probed by examination of wild-type HmuT and a series of conserved heme pocket residue mutants, H136A, Y235A, and M292A. Characterization by UV-visible, resonance Raman, and magnetic circular dichroism spectroscopies indicate that H136 and Y235 are the axial ligands in ferric HmuT. Consistent with this assignment of axial ligands, ferric WT and H136A HmuT are difficult to reduce while Y235A reduces readily in the presence of dithionite. Raman frequencies of the FeCO distortions in WT, H136A, and Y235A HmuT–CO complexes provide further evidence for the axial ligand assignments. Additionally, the se frequencies provide insight into the nonbonding environment of the heme pocket. Ferrous Y235A and the Y235A–CO complex reveal that the imidazole of H136 exists in two forms, one neutral and one with imidazolate character, consistent with a hydrogen-bond acceptor on the H136 side of the heme. The ferric fluoride complex of Y235A reveals the presence of at least one hydrogen-bond donor on the Y235 side of the heme. Hemoglobin utilization assays showed that the axial Y235 ligand is required for heme uptake in HmuT.
Graphical Abstract
Iron is required for infection in essentially all bacterial pathogens (1). In vertebrate infections, the most abundant source of iron is heme (iron protoporphyrin IX), which comes primarily from hemoglobin. Bacteria have developed sophisticated approaches to transport heme into the cytoplasm. These pathways have been the focus of many recent reviews (1–11) with particular emphasis on the structures of heme transport proteins (12–14), Gram-positive bacteria (15), including Bacillus anthracis (16) and Staphylococcus aureus (17–20), and Gram-negative bacteria (21).
To date, pathogenic bacterial heme uptake pathways that have been characterized in detail have involved ATP-binding cassette (ABC) transporters. These transmembrane systems utilize the energy yield from ATP hydrolysis to pump various compounds across cellular membranes (22–24). An ABC transporter comprises two transmembrane modules and two ATPase subunits. Import ABC transporters are commonly found in prokaryotic systems and have an associated substrate binding protein [in this instance a heme binding protein (HBP)] that brings the substrate to the ABC transporter (14).
A number of HBP’s have been structurally characterized. Pseudomonas aeruginosa PhuT (25) and Shigella dysenteriae ShuT (25;26) both contain a conserved tyrosine that binds the heme in a pentacoordinate fashion. Yersinia pestis HmuT (YpHmuT) utilizes a His/Tyr ligation and binds heme both as a monomer and as a π-stacked dimer (27;28). S. aureus IsdE (29) and Streptococcus pyogenes SiaA/HtsA (30;31) both form hexacoordinate heme complexes having His/Met axial ligation.
A number of other membrane-bound proteins involved in the capture of host heme are known to have tyrosine axial ligands. The S. aureus Isd (Iron-regulated surface determinant) pathway employs a series of proteins for heme uptake and transfer including IsdA (29), IsdB (32), IsdC (33), and IsdH (34). Each of these proteins has one to three NEAT (near-iron transport) domains (17;35). The NEAT domains that bind heme [IsdA-N1 (29;36;37), IsdB-N2 (38), IsdC-N2 (33;37), and IsdH-N3 (34;38)] do so using tyrosine as an axial ligand (17;39;40). The x-ray structure of IsdB-N2 indicates that this protein can bind heme with an axial methionine in addition to the conserved tyrosine (32). X-ray structures of other NEAT proteins, such as IsdX1 and IsdX2-N5 from B. anthracis, also show the heme to be bound by an axial tyrosine (41–43). All of these Isd heme-binding NEAT domains have a second Tyr four residues downstream of the ligand on the same β-strand. The side chain of the second Tyr serves as a hydrogen-bond (H-bond) donor to the phenolate oxygen atom of the axial Tyr ligand, whose bent coordination geometry enforces a π-stacking interaction with the porphyrin ring. Recently, a NEAT domain, Hbp2-N2, has been described in which a tyrosine from the β-7 strand is the axial ligand, rather than one in the β-8 YXXXY motif (44).
Bis-histidine motifs are found in HmuY from Porphyromonas gingivalis (45) as well as in membrane-spanning heme receptors (7). Other motifs in the heme receptor class are also known; HmuR from Neisseria meningitidis has a five-coordinate tyrosine ligation (46) while PhuR from P. aeruginosa has been shown to have His/Tyr ligation (47). In still another variant on heme binding motifs in heme uptake pathways, Shp from S. pyogenes has bis-Met axial ligation (48).
The variety of heme binding motifs in heme uptake pathways leads to an interest in further characterization of other proteins associated with heme transfer. One such pathway is found in the pathogen Corynebacterium diphtheriae, a Gram-positive bacterium which is the causative agent of diphtheria (49–53), a well-known upper respiratory tract disease that carries a high mortality rate in humans (54). Diphtheria is still common in developing countries due to low vaccination rates (55;56). C. diphtheriae requires iron for survival and for virulence (49;57–59).
C. diphtheriae acquires heme via an ABC-type heme binding protein transporter system (57). A variety of heme sources can be used including hemoglobin (Hb), hemoglobin/haptoglobin, and myoglobin (Mb) (60). The heme utilization (hmu) operon includes hmuT (the HBP /substrate binding protein), hmuU (the permease) and hmuV (the ATPase), which form an ABC transport system (61;62). The htaA gene is located immediately upstream of the hmuTUV locus, and it is followed by a promoter region and the htaC gene. Downstream of the hmuV gene is a promoter region and the htaB gene. The hmuTUV and htaA genes form a single operon, while htaB and htaC are transcribed independently (63). HtaA and HtaB are proposed to be anchored to the cytoplasmic membrane through a C-terminal hydrophobic region. Both proteins are exposed to the bacterial surface, suggesting that these heme binding proteins may function as heme receptors (62). It has been shown that HtaA passes heme to HtaB (64). The next protein in the pathway is the HBP HmuT, which donates heme to the HmuUV transporter. Once the heme is brought into the cell, the heme oxygenase HmuO catalyzes the O2–dependent degradation of the heme, which releases its iron for further use in cellular functions (65;66).
Sequence alignment studies of CdHmuT with other Corynebacterium species reveal two conserved tyrosines: Tyr235 and Tyr349 as well as a conserved histidine (His136) and methionine (Met292). Homology modeling shows that Tyr235 is probably an axial ligand and that either His136 or Met292 could also bind as a sixth ligand. CdHmuT is unique among its homologs in that the proposed axial His and Tyr ligands are reversed with respect to their positions in the structurally characterized proteins ShuT, PhuT and YpHmuT. That is, the tyrosines in all proteins in heme uptake pathways known to date come from the N-terminal part of the sequence, but CdHmuT is predicted to have the tyrosine from the C-terminal part of the sequence. As this prediction indicates a key role for tyrosine in heme transfer, it was important to establish the axial ligands in CdHmuT. Herein we report the biophysical characterization of CdHmuT and a number of its mutants. UV-visible, resonance Raman (rR), and magnetic circular dichroism (MCD) studies combine to give a picture of this novel heme uptake protein.
Materials and Methods
Bacterial strains and media
E. coli strains DH5α and TOP10 (Invitrogen) were used for routine cloning and plasmid maintenance, while XL-1 Gold (Stratagene) was used in the mutagenesis experiments. E. coli BL21(DE3) (Novagen) was used for protein expression. Corynebacterium ulcerans strain CU77 was previously described (Schmitt & Drazek, 2001) and carries a point mutation that results in premature termination of the hmuT gene (Schmitt, unpublished observation). Chromosomal DNA from C. diphtheriae strain 1737 (Popovic et al., 1996) was used as the source DNA for PCR. Luria-Bertani (LB) medium was used for culturing of E. coli, and Heart Infusion Broth containing 0.2% Tween 80 (HIBTW) was used for growth of C. ulcerans strains. Bacterial stocks were maintained in 20% glycerol at −80°C. Antibiotics were added to LB medium at 50 μg/ml for kanamycin and to HIBTW for C. ulcerans cultures at 2 μg/ml for chloramphenicol. HIBTW was made low iron by the addition of ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) at 12 μg/ml. Modified PGT (mPGT) is a semi-defined low iron media that has been previously described (Tai et al., 1990). Antibiotics, EDDA, Tween 80, were obtained from Sigma Chemical Co. and hemoglobin (human) was purchased from MP BioMedical.
Plasmid construction
The HmuT expression construct was developed using the pET28a expression vector (Novagen). A PCR-derived DNA fragment containing the C. diphtheriae hmuT coding region was initially cloned into the pCR-Blunt II-TOPO vector (Invitrogen). The DNA fragment harboring the hmuT gene was subsequently ligated into the NcoI-EcoRI sites in pET28a and the expression plasmid was then transformed into BL21(DE3). The cloned hmuT gene in the pET28a vector lacked the 20 - amino acid N-terminal secretion signal and contained an N-terminal Strep-tag, which was used for protein purification. The following primers were used in the PCR: hmuTF; 5′-CC ATGGCA AGC TGG AGC CAC CCG CAG TTC GAA AAG GGT GTC CAG GGC ACA TAT-3′; hmuTR; 5′-GAATTC CTA TAC CTG TGG GTC ATAC-3′: underlined sequences indicate restriction sites and the sequence in italics encodes the 8-amino acid Strep-tag.
Site-directed mutagenesis and hemoglobin-iron utilization assays
Site directed mutants were made using the QuikChange Lightning kit (Stratagene) according to the manufacturer’s instructions. Briefly, 125 ng of each primer containing the targeted base change and 50 ng of plasmid template were used in the QuikChange reaction. Methylated template DNA was removed from the reaction by digestion with DpnI restriction endonuclease, and mutagenized DNA was recovered by transformation into XL1-Gold competent cells. The presence of the base changes was confirmed by sequence analysis. Plasmids used for site-directed mutagenesis were pET28a containing the cloned Strep-tag-hmuT gene, and plasmid pCD842, which harbors the hmuT gene on the E. coli-Corynebacterium shuttle vector pCM2.6 (67). The hemoglobin utilization assay has been described previously (66).
Expression and purification of CdHmuT
HmuT was expressed and purified from BL21DE3 (pEThmuT) cells. The N-terminal leader sequence was deleted and replaced with a Strep-tag. The native construct started at residue 21 (Gly) and extended through the native stop codon. The culture was prepared in a Luria-Bertini (LB) medium containing 50 μg/mL kanamycin. Inoculation was done with an overnight pre-culture and cells were grown at 37 °C with shaking at 220 rpm. When the OD600 of the culture reached 0.5 – 0.6, protein expression was induced by adding isopropyl β-D-1- thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM. The culture was incubated for 3 h at 27 °C. Cells were harvested by centrifugation at 8000 x g. The cell pellet was resuspended in lysis solution (100 mM Tris-Cl, 150 mM NaCl, pH 8.0) containing a protease inhibitor cocktail (Roche Complete Mini, EDTA-free, following the manufacturer protocol). The cells were broken using a cell disrupter or sonication. The lysate was then centrifuged at 8000 x g, and the supernatant was syringe-filtered with a 0.45 μm filter.
All of the following purification steps were conducted at 4 °C using fast protein liquid chromatography and all buffer solutions were pH 8.0 unless specified otherwise. The protein sample was loaded onto a Strep-Tactin Superflow column (5 mL, IBA BioTAGnology) equilibrated with buffer A (100 mM Tris-Cl, 150 mM NaCl, pH 8.0). Unbound material was washed out with 5 column volumes (CV) of buffer A. HmuT was eluted with 10 CV of buffer B containing 100 mM Tris-Cl, 150 mM NaCl, 2.5 mM desthiobiotin, pH 8.0 applied via a linear gradient. The purities of the fractions were evaluated using sodium dodecyl sulfate polyacrylamide gel electrophoresis. Native PAGE did not show dimers, indicating that the protein is monomeric in solution. Minor differences in the optical spectra were observed as a function of the buffer type.
Magnetic circular dichroism spectroscopy
Magnetic circular dichroism (MCD) spectra were measured with a magnetic field strength of 1.41 T by using a JASCO J815 spectrophotometer. This instrument was equipped with a JASCO MCD-1B electromagnet and interfaced with a Silicon Solutions PC through a JASCO IF-815-2 interface unit. Data acquisition and manipulation using Cary or Jasco software has been previously described (68). To ensure homogeneity of ferric oxidation for the various mutants, ferricyanide was used to fully oxidize the heme center, followed by desalting chromatography. The resulting spectra were compared to data from other heme-containing proteins with known binding site structures and optical spectra. All spectral measurements for all proteins were carried out with a 0.2 cm quartz cuvette at 4 °C in 50 mM phosphate buffer (either pH 6.5 or 10).
Resonance Raman spectroscopy
Resonance Raman (rR) spectra were collected using the 441.6-nm emission line from a HeCd laser or either 406.7 nm or 413.1 nm emission from a Kr+ laser. Spectra were recorded at ambient temperature using the 135° backscattering geometry with the laser be am focused to a line on a spinning 5 mm NMR tube. Toluene, DMSO, and CH2Br2 were used as external standards for spectral calibration. UV-visible spectra were recorded before and after rR experiments to verify that the samples were not altered by exposure to the laser beam. The final concentrations of all Fe(III) samples were between 25 and 80 μM protein in 100 mM buffer solution. The buffers used were CHES, pH 10.0, Tris-Cl, pH 8.8 or 8.0, sodium phosphate buffer, pH 7.0 or 5.8, MES, pH 5.1, and sodium acetate buffer, pH 5.0. The D2Oand H218O samples of the Y235A mutant were made by diluting a concentrated sample of the protein into CHES buffer made with either D2O or H218O at pD 10 or pH 10, respectively. Resonance Raman spectra were collected with laser powers between 9 and 12 mW at the sample.
The ferric fluoride adduct of Y235A HmuT was prepared by titration with 0.8 M NaF solution in 0.1 M sodium phosphate buffer at pH 5.8. The final protein and NaF concentrations were 80 μM and 330 mM, respectively. The laser power was 9.7 mW for the 406.7 nm Kr+ excitation and 4.6 mW for the 441.6 nm HeCd excitation.
Ferrous HmuT(Y235A) samples (36 μM) were prepared under anaerobic conditions in 0.1 M Tris-Cl pH 8.2 by adding an 86-fold excess of sodium dithionite in buffered solution. Laser power for the ferrous samples was 4 to 8 mW. Ferrous carbonyl adducts (36 to 75 μM) were made by reducing the proteins with a 70-to 180-fold excess of buffered sodium dithionite under an atmosphere of natural abundance CO or 13CO (99 atom % 13C) in 0.1 M Tris-Cl pH 8.8. Laser power for the heme carbonyl samples was held between 2 and 4.5 mW to minimize photolysis.
Results
Heme ligation in ferric HmuT
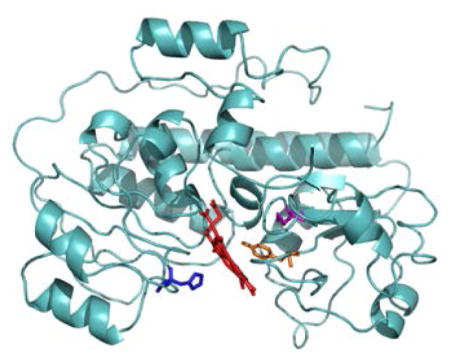
An alignment of the HmuT amino acid sequences of various Corynebacterium species showed that two tyrosine residues (Y235 and Y349) as well as H136 and M292 were highly conserved in this group of bacteria (Figure S1). A homology model of HmuT was created by I-TASSER(69) with YpHmuT as one of the templates (Figure 1). The two structures were similar overall with a root mean square difference (RMSD) between the model and YpHmuT of 0.673 Å. Alignment of the CdHmuT sequence with four HBP’s for which crystal structures have been solved: PhuT from P. aeruginosa (25), ShuT from S. dysenteriae (25), IsdE from S. aureus (70) and YpHmuT(27) indicated that it was probable that CdHmuT would have a Tyr as one axial ligand, and a Met or His as the second (Figure S2). However, CdHmuT was predicted to be unique in that the axial ligands would be reversed (N-terminal His and C-terminal Met or Tyr) with respect to structurally characterized homologous hexacoordinate heme proteins (N-terminal Tyr and C-terminal His for YpHmuT and N-terminal Met and C-terminal His for IsdE). Based on this homology model and sequence alignment, the H136A, Y235A, and M292A mutants were created to probe the heme axial ligation.
Figure 1.
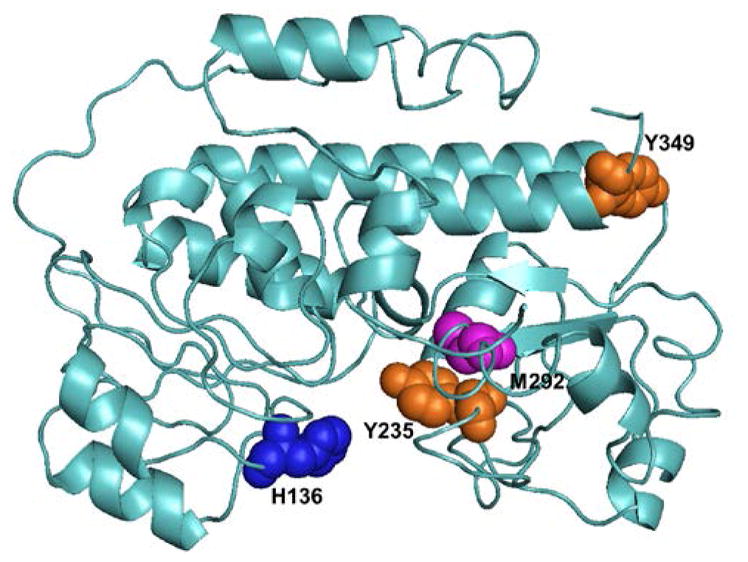
I-TASSER homology modeling of CdHmuT created using PyMOL (119). Shown are the locations of H136, Y235, M292, and Y349.
Conserved residues and the biological function of HmuT
To determine if H136A, Y235A, and M292A are important for the biological function of HmuT, the ability of the cloned hmuT genes to complement a Hb-iron utilization defect was assessed. It was previously shown that the CU77 mutant strain of C. ulcerans HmuT (CuHmuT) was defective for HmuT activity (57). The cloned hmuT gene from either Corynebacterium species could fully restore the wild-type phenotype to this strain. Complementation studies with each of the H136A, Y235A, and M292A CdHmuT mutants revealed that only Y235A was unable to restore growth fully (Figure S3). Thus, Y235 is essential for the heme-iron utilization function of HmuT.
Spectroscopy of wild-type CdHmuT
The UV-visible spectrum of ferric wild-type (WT) CdHmuT showed a Soret peak at 407 nm and four peaks in the α,β region at 492, 546, 569, and 616 nm (Figure 2). The charge transfer band at 616 nm is characteristic of a high-spin species, and the two α,β bands at 569 and 546 nm are consistent with the presence of a low-spin species. Hence, these data suggest an equilibrium mixture of high-spin (HS) and low-spin (LS) WT CdHmuT. This UV-visible spectrum is strikingly similar to that observed for S. marcescens HasA (SmHasA) (406, 494, 537, 568 and 618 nm) which is known to have His/Tyr axial ligation and exists in a thermal spin equilibrium (71).
Figure 2.
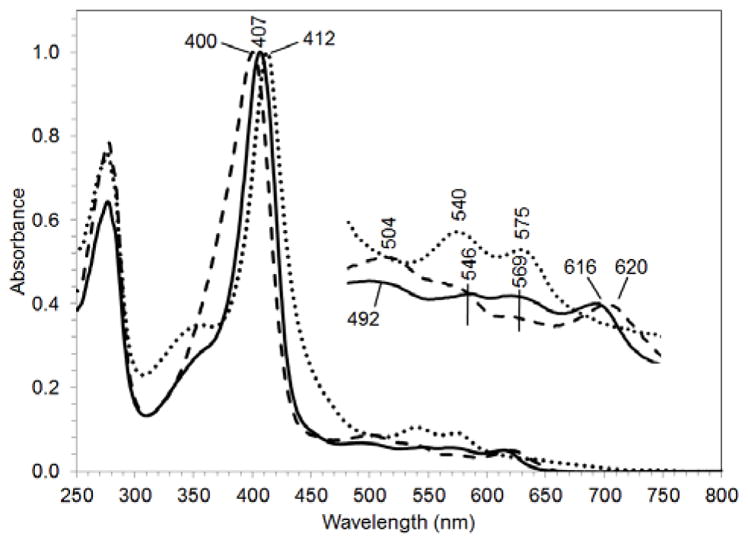
UV-visible spectra of the Fe(III) forms of WT CdHmuT (solid line), H136A (dashed line), and Y235A (dotted line) normalized at the Soret. The samples were taken in 50 mM Tris-Cl at pH7.0.
Ligation of the heme was further probed by the use of MCD. Initial comparisons of the spectra of WT CdHmuT with models having His-only or His/Met axial ligation did not give entirely similar spectral profiles (data not shown). The UV-visible and MCD spectra were then compared with two five-coordinate Tyr-ligated proteins, bovine liver catalase (BLC) (72–74) and the H93Y mutant of Mb (75). Figure S4 shows agreement in the Soret region and moderate agreement in the α,β region of the UV-visible spectrum. However, the three species are different in the MCD spectrum. Towards the blue, Mb-H93Y and HmuT are similar, but BLC is quite different. Towards the red, the peaks and troughs observed for HmuT are similar to, but shifted from, Tyr-ligated BLC, and there is no agreement with Mb-H93Y. These results lead to the conclusion that WT CdHmuT does not adopt a five-coordinate, tyrosine-ligated geometry.
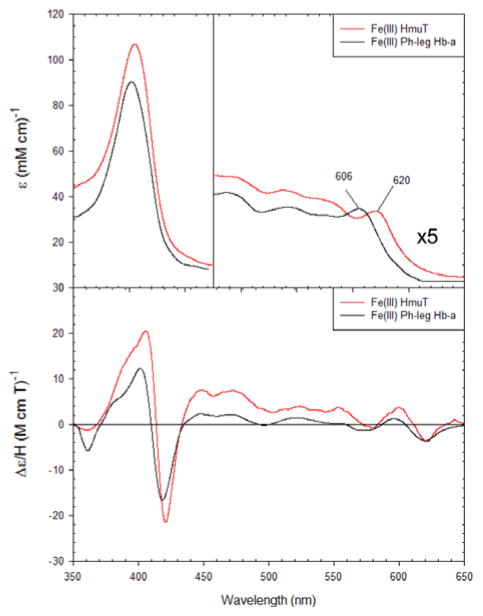
WT CdHmuT was then compared to a model for the His/Tyr ligand set: leghemoglobin a with exogenous phenol (76). This complex adopts a six-coordinate heme binding structure with a histidine axial ligand and a phenol in the transposition. Figure 3 shows that both the UV-visible and MCD spectra were in agreement with those of the WT CdHmuT supporting the conclusion that CdHmuT is a His/Tyr protein.
Figure 3.
The UV-visible and MCD comparison spectra for Fe(III) WT CdHmuT at pH 6.5 with Fe(III) phenol-bound leghemoglobin a. The samples were taken in 50 mM phosphate buffer. Spectra were slightly dependent on buffer conditions. The spectrum of phenol-bound leghemoglobin a was replotted from (76).
Soret-excited resonance Raman (rR) spectra of WT CdHmuT were recorded over the pH range of 5.0 to 10.0 (Figure S5). Two bands are observed in the 1470 – 1510 cm−1 region of the rR spectrum. The ν3 core size marker band frequencies observed in this range are indicative of heme spin state and coordination number. The pair of ν3 bands observed at 1475 and 1504 cm−1 indicates that the protein contains a mixture of six-coordinate high-spin (6cHS) and six-coordinate low-spin (6cLS) hemes. This is consistent with their UV-visible spectra shown in Figure 2. The rR spectra were independent of pH, i.e., the identities of the axial ligands and populations of the 6cHS and 6cLS hemes were pH independent. Thus, the HS/LS equilibrium is not governed by any acid-base speciation of the heme or the protein over this pH range.
Spectroscopy of M292A CdHmuT
The UV-visible (Figure S6) and rR (Figure 4) spectra for the WT and M292A proteins, as well as the dependence of the rR spectrum on pH (Figure S7), were almost identical. In the MCD, as well, the M292A spectra were remarkably similar to the WT protein, with only slight differences in absorbance maxima (Figure S6). The overall similarity between the spectral signatures of the WT and M292A proteins suggest that Met is not an axial ligand.
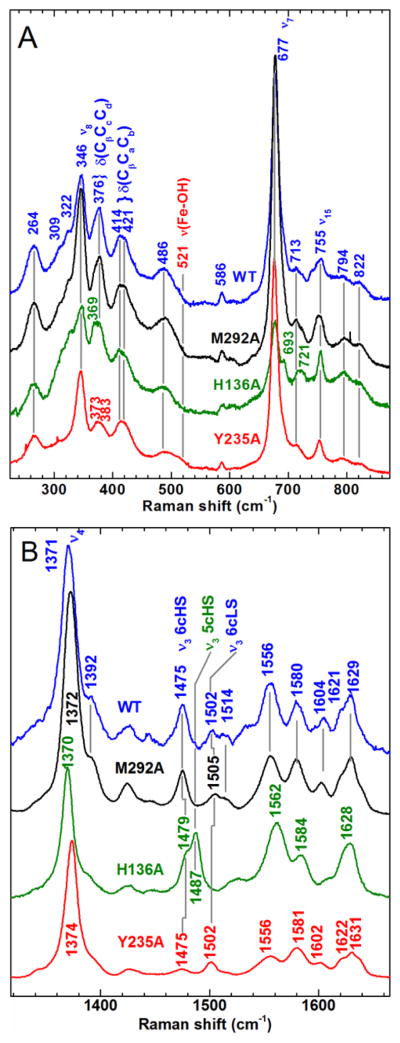
Figure 4.
Comparison of the Soret-excited rR spectra of WT CdHmuT, M292A, H136A, and Y235A. Protein concentrations were 80, 70, 25 and 36 μM, respectively. All samples were prepared in 50 mM Tris-Cl at pH 7.0. The spectra were recorded with 406.7-nm excitation. A) Low frequency and B) high frequency spectra of WT CdHmuT (blue), M292A (black), H136A (green), and Y235A (red).
Spectroscopy of H136A CdHmuT
H136A CdHmuT gave a broad Soret band that was blue shifted to 400 nm and three defined bands in the α,β region, including one at 620 nm (Figure 2). The H136A mutant was compared with three tyrosine models: ShuT (26), BLC (72–74), and Mb-H93Y (75) (Figure S8). In the UV-visible spectra, the Soret peaks for all four proteins were similar, with maxima located from 400 to 407 nm. In the visible region, the characteristic high-spin peak located past 600 nm was evident for H136A HmuT, BLC, and ShuT and blue-shifted for H93Y Mb. The peaks and troughs in the visible region of the MCD spectrum are evidence for tyrosinate ligation. Together, the data indicate a ferric five-coordinate tyrosinate-bound heme.
The Soret-excited rR spectrum of H136A HmuT is significantly different from that of WT; it comprises ν3 bands at 1479 and 1487 cm−1, indicating the presence of 6cHS and five-coordinate high-spin (5cHS) hemes, respectively (Figure 4). The presence of only HS species reflects the weakened axial ligand field in the absence of His136 (77;78). The bands at 721 and 693 cm−1 are assigned to γ5 (symmetric pyrrole ring fold, A2u in D4h) and γ15 (symmetric pyrrole ring fold, B2u in D4h), respectively, by analogy to Mb (79–81). Intensification of the bands arising from these modes, which are normally Raman forbidden, reveals lowering of the porphyrin symmetry. Pentacoordination of the heme iron center, as anticipated for H136A mutation and as indicated by the core-size marker band frequencies in Figure 4B, is expected to drive the heme iron out of the mean porphyrin plane toward the Tyr235 side chain. This type of coordination typically increases the extent of porphyrin doming. The A2u and B2u symmetries of γ5 and γ15 correlate with A1 and B2 in the approximate C4v symmetry of a domed conformation. Since A1 and B2 modes of a C4v metalloporphyrin are Raman allowed, the intensification of scattering by both γ5 and γ15 is consistent with the heme in H136A being more domed than the WT protein.
The presence of 5cHS heme in H136A, which is absent in the WT protein, strongly suggests that His136 is a heme ligand in the WT protein. The rR spectrum of H136A does not exhibit a pH dependence over the range of 5.0 to 10.0, as seen in Figure S9, suggesting that neither the 5cHS or the 6cHS H136A species binds hydroxide under alkaline conditions. The inability to form a hydroxide complex has also been reported for HasA(H32A), wherein tyrosinate is the lone amino acid ligand to the heme (82). In fact, there are very few examples of Fe(III) porphyrinates having two anionic axial RO− ligands, including OH−, because large overall positive charges are necessary to stabilize such axial ligand sets (83;84). Thus, the rR spectra are consistent with an equilibrium mixture of 5cHS Fe–Tyr− and 6cHS H2O–Fe–Tyr− axial ligation.
Spectroscopy of Y235A CdHmuT
Y235A CdHmuT had a Soret band that was red-shifted by 7 nm compared to the native protein and a significant shoulder near 350 nm, which was absent from the native and other mutants (Figure 2). The α,β region had bands at 540 and 575 nm and no charge transfer band, consistent with a predominantly LS heme.
Figure 5 compares the UV-visible and MCD spectra of this mutant in the ferric state at pH 10 with two His/OH− complexes, alkaline Hb at pH 10 (85) and horseradish peroxidase (HRP) at pH 12.5 (86). The peaks and troughs in both the Soret and visible regions are located at comparable wavelengths and exhibit similar relative intensities. These similarities indicate that Y235A adopts a His/OH− ligation set at high pH.
Figure 5.
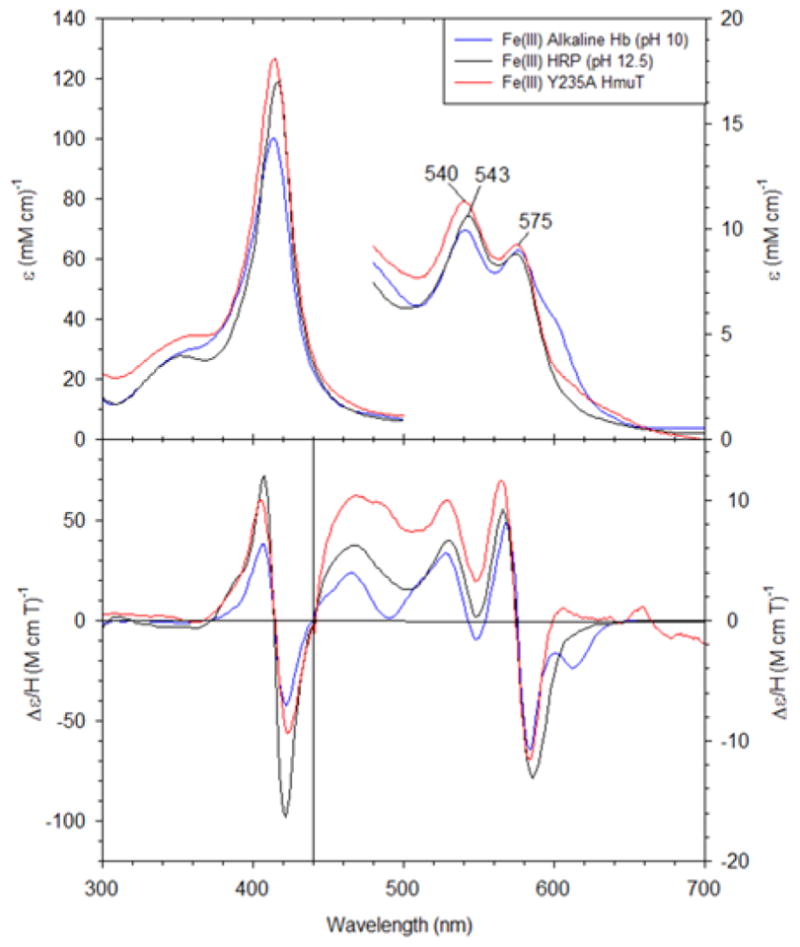
The UV-visible and MCD spectra for Fe(III) Y235A CdHmuT at pH 10 with Fe(III) alkaline Hb (pH 10) and Fe(III) HRP (pH 12.5). The samples were prepared in 50 mM phosphate buffer. The spectra of alkaline Hb and HRP were replotted from (85) and (86), respectively.
The heme speciation of Y235A was found to be sensitive to pH in the Raman spectra. Y235A was shown to shift from a mixture of 6cHS (1473 cm−1) and 6cLS (1501 cm−1) at pH 10.0 to all 6cHS (1477 cm−1) at pH 5.0 (Figure 6). Based on changes in the relative rR intensities as a function of pH, the pKa of the acidic form is near 6.
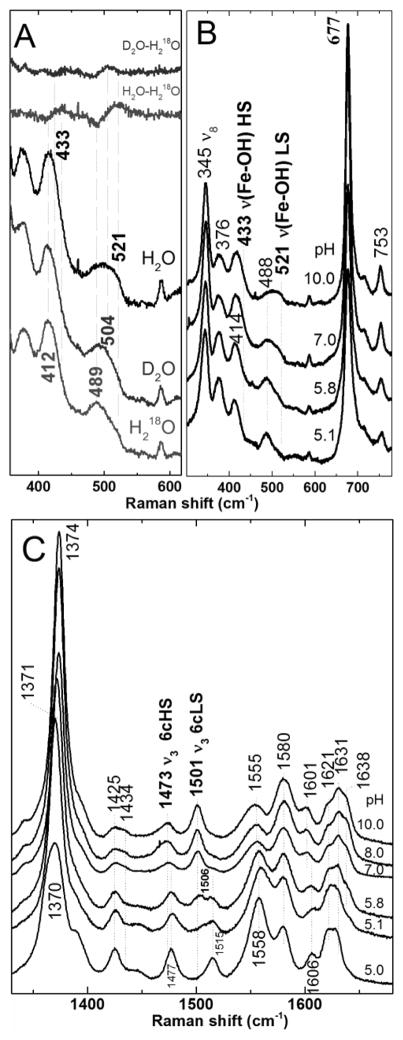
Figure 6.
The pH dependence of ferric Y235A monitored by 406.7 nm-excited rR spectra (11 mW power at sample). A) Isotopologs of Y235A at pH 10 prepared in H2O, D2O, and H218O. Difference spectra of D2O-H218O and H2O-H218O shown at the top of the figure were generated by subtraction of the respective parent spectra at the bottom of the figure. B) Low frequency and C) high frequency spectra of ferric Y235A as a function of pH. Samples were between 25 and 60 μM.
To determine whether the alkaline species comprising the HS/LS equilibrium are hydroxide complexes, the 2H and 18O isotopologs were generated in D2O and H218O, and their Soret-excited rR spectra recorded (Figure 6A). Two isotope-sensitive bands were observed. The band at 521 cm−1 in H2O shifted to 504 and 489 cm−1 in D2O and H218O, respectively, and is assigned to the νFe─OH mode of LS Y235A–OH. The second isotope-sensitive band appeared at 433 cm−1 and shifted to 412 cm −1 in H218O and is tentatively assigned to the νFe─OH mode of HS hemin–OH component of the mixture. The difference feature for the HS νFe─OH observed in the D2O – H218O difference spectrum has considerably less amplitude than that of the LS νFe─OH making the shift due to deuterium in the HS complex difficult to measure. Y235A is similar to Mb, Hb, and various other heme protein hydroxides (87–91) that exist as HS/LS mixtures. Since the alkaline form of Y235A HmuT is a hydroxide complex, it is likely that the 6cHS species that dominates below pH 6 is an aqua complex.
Heme environment in CdHmuT
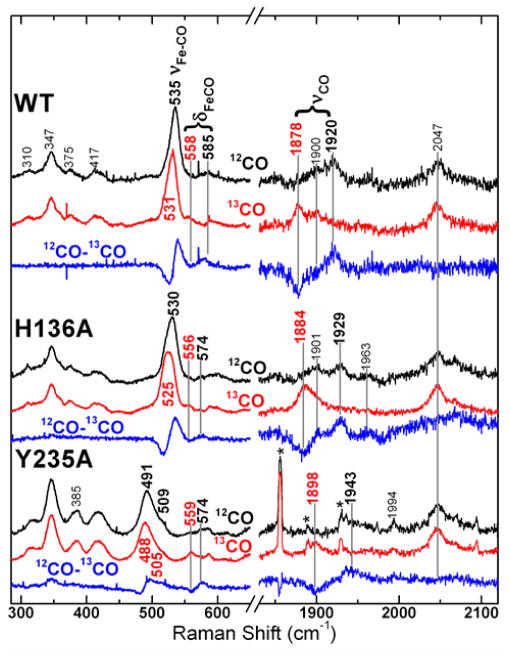
The carbon monoxide (CO) complex of ferrous heme is a particularly sensitive probe of the coordination and electrostatic character of its protein environment. The C–O and Fe–C stretching frequencies reflect the extent of FeCO π-backbonding, which is sensitive to FeCO geometry, the trans ligand, and nonbonded interactions between the CO and the distal heme pocket. Useful insight into the distal electrostatic landscape and the σ-donor strength of the trans ligand are derived from the position of a heme carbonyl complex on the νFe–CO/νC–O correlation plot (92–94). The Fe–13CO isotopologs of WT CdHmuT and heme pocket mutants were used to identify rR bands corresponding to the Fe–CO stretching, Fe–CO bending and C–O stretching modes. The Soret-excited rR spectra are shown in Figure 7. Three 13C-sensitive bands were observed for the WT and H136A CdHmuT proteins while four were observed for the Y235A mutant. The spectrum of WT reveals isotope-sensitive bands at 535, 585, and 1920 cm−1 which shift to 531, 558, and 1878 cm−1, respectively, in the HmuT–13CO spectrum. They are assigned to the νFe–C, δFeCO, and νC–O modes, respectively. For the H136A mutant, the isotope–sensitive bands shifted from 530, 574, and 1929 cm−1 to 525, 556, and 1884 cm−1, respectively. Thus, its νFe-C frequency was 5 cm−1 lower than those of WT, and its νC–O frequency was 9 cm −1 greater, consistent with a weakened π-backbonding characteristic of the trans-Tyr ligand in this mutant. The largest differences were observed for the Y235A mutant with two νFe–C bands occurring at 491 and 509 cm −1, a δFeCO frequency of 574 cm−1, and νC-O band at 1943 cm−1 which shift to 488, 505, 559, and 1898 cm−1, respectively, with 13C. The two νFe-C frequencies were assigned by peak fitting the iron-carbon stretching region of the 12CO and 13CO spectra and generating a simulated difference spectrum; both are shown in Figure S10.
Figure 7.
Resonance Raman spectra of the ferrous carbonyls of WT CdHmuT, H136A, and Y235A recorded using 413.1-nm excitation. Natural abundance HmuT–CO (black), HmuT–13CO (red) and difference (blue) spectra are shown for each protein. Spectra of WT and H136A were recorded at pH 8.8 and that of Y235A at pH 8.2. The asterisks in the carbonyl stretching region of the Y235A spectrum mark plasma emission lines from the Kr+ laser.
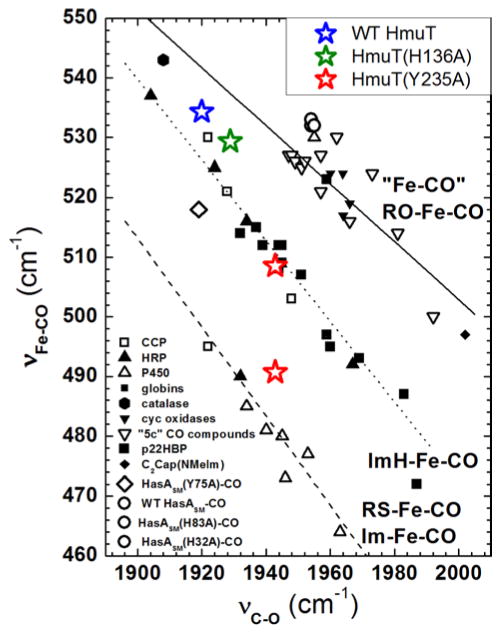
The inverse correlation between νFe–C and νC–O frequencies is plotted for CdHmuT and its heme pocket mutants along with a number of heme–CO proteins and model complexes for comparison in Figure 8. The WT HmuT–CO point falls on the νFe–C/νC–O correlation plot in a position consistent with its proximal ligand being a weaker donor than His, suggesting that WT HmuT–CO contains a weak proximal Fe(II)–O bond, such as Fe(II)–Tyr or Fe(II)–OH2 (95).
Figure 8.
Backbonding correlation plot of νFe-CO versus νC-O for ferrous carbonyls of heme proteins showing the dependences of their positions on axial ligation and distal pocket properties. WT (blue), H136A (green), Y235A (red) are shown as stars on the plot. Catalase, hexagon; HasA(Y75A), ◇; HasA(WT), ○; HasA(H83A), ○; HasA(H32A), ○ (105;120). The dashed line is the least squares line for six-coordinate Fe–CO adducts in which the proximal ligand is thiolate or imidazolate; the dotted line is the least squares line for Fe–CO adducts with proximal histidine (neutral imidazole) (91;96;120) (and references therein); and the solid line represents a compilation of “five-coordinate” model complexes (93) (and references therein) and heme proteins which the ligand trans to CO is coordinated through an oxygen atom (98).
Distal H-bond donors to the bound CO ligand and positive charge enhance π-backbonding. Both of these interactions weaken the C–O bond while strengthening the Fe–C bond, placing points high and to the left on the imidazole correlation line (e.g., νFe–C 520 cm−1 and νC–O 1935 cm−1) (96). Off-axis Fe–C–O distortion and negative charge weaken backbonding which, absent other factors such as hydrogen bonding, is characterized by positions to the low and to the right on the correlation line. Thus, the positions of WT HmuT–CO and H136A–CO would be interpreted as due to a positive charge near and/or H-bond donation to the bound CO.
The bottom set of spectra in Figure 7 reveal that Y235A HmuT–CO has two conformers whose positions on the νFe–C/νC–O correlation plot are distinct from each other and from WT and H136A HmuT–CO. One conformer is located on the imidazole line, indicating neutral histidine axial ligation; its position relative to other proteins on the imidazole correlation line is consistent with a modest distal H-bonding interaction. In contrast, the second conformer is slightly above the imidazolate line suggesting that the proximal histidine (His136) has some imidazolate character, perhaps due to its interaction with an H-bond acceptor in the proximal pocket.
Ferric Y235A CdHmuT–fluoride
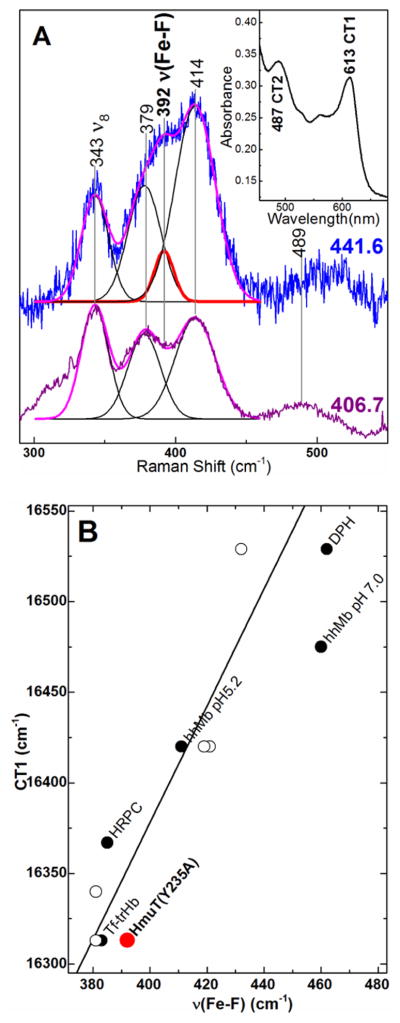
Fluoride complexes of heme proteins are sensitive probes of the distal H-bonding environment (97;98). The energy of the charge transfer band at 600 – 620 nm (CT1) together with the Fe–F stretching frequency constitutes a sensitive probe of H-bond strength between a distal H-bond donor and the bound F− ligand. Low νFe─F frequencies correlate with red-shifted CT1 bands in complexes having strong hydrogen bonds. The νFe─F mode of Y235A HmuT–F was identified by exciting into its CT2 band (450 – 460 nm) with 441.6-nm light. Relative enhancement of scattering by the νFe─F mode with 441.6-nm excitation is considerably greater than with Soret excitation; peak fitting of the 441.6-nm excited rR spectrum shown in Figure 9 revealed that the νFe─F band occurs at 392 cm−1. The Y235A HmuT–F CT1 band was observed at 613 nm (16,313 cm−1) (Figure 9A inset). Correlation of its νFe─F and the CT1 energy places Y235A CdHmuT low on the correlation plot in Figure 9B which is consistent with strong hydrogen bonding between the bound F− ligand and the distal pocket.
Figure 9.
Characterization of Y235A–F by correlation of the FeIII–F stretching frequency and CT1 energy. A) Low frequency window of the rR spectra of Y235A–F using Raman excitation into the CT2 (441.6 nm) and Soret (406.7 nm) bands. Protein was 80 μM in 100 mM sodium phosphate buffer in 330 mM sodium fluoride, pH 5.8. Laser power at the sample was 4.6 mW with 441.6-nm excitation and 9.7 mW with 406.7-nm excitation. Peak fitting analyses of both spectra are overlaid on the original spectra with the calculated FeIII–F stretching band shown in red; calculated ν8 and propionate and vinyl bending bands are shown in black; the overall fit is shown in magenta. Inset: Visible spectrum of Y235A–F rR sample. B) Correlation plot of νFe-F frequency and the CT1 energy. Y235A is shown in red. Other points are from Nicoletti and coworkers (98). Open circles are for mutants of truncated Hb from Thermobifida fusca (Tf-trHb) with varying number of hydrogen bonds between the distal pocket and the fluoride (97;98).
Ferrous Y235A CdHmuT
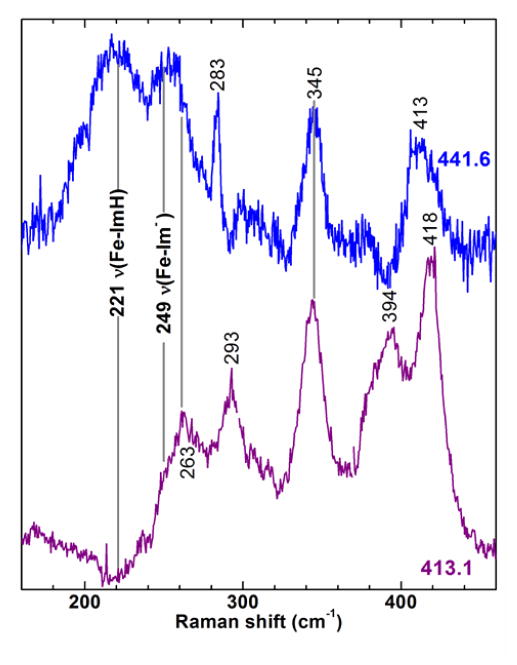
Unlike WT and H136A CdHmuT, which are slow to reduce with aqueous buffered S2O42−, Y235A CdHmuT was readily converted to a 5cHS ferrous heme (ν4, 1354 cm−1; ν3, 1467 cm−1) upon reaction with S2O42− (spectra not shown). The νFe─His frequency for 5cHS ferrous hemes has been shown to be significantly enhanced with 441.6 nm laser excitation (99) and two νFe─His modes for ferrous HmuT-Y235A are tentatively assigned to bands at 221 and 249 cm−1 based on comparison of their relative enhancements in the 441.6 and 413.1-nm excited spectra in Figure 10. Observation of two νFe─His modes is consistent with two proximal pocket conformers in Y235A CdHmuT, which are distinguished by the extent of imidazolate character of the proximal His ligand. These are likely the same conformers responsible for the two νFe–C frequencies (491 and 509 cm −1) in the corresponding carbonyl spectrum shown in Figure 7 and Figure S10.
Figure 10.
Comparison of the low frequency RR window of ferrous Y235A spectra obtained with 413.1-nm (violet) and 441.6-nm (blue) excitation. Laser powers at the sample were 4.0 mW and 4.6 mW, respectively. The solutions were 38 μM in protein and 100 mM in Tris-Cl, pH 8.8.
Discussion
His/Tyr ligand set identified for HmuT:heme complex
Ferric species
Sequence alignment, homology modeling, and UV-visible, MCD, and Raman spectroscopies all lead to the conclusion that WT CdHmuT contains a six-coordinate active site with a tyrosine bound to one axial position of the heme and a histidine bound to the other. Mutation studies validated these findings, with M292A showing spectra very similar to the wild-type protein, indicating that this methionine is not an axial ligand.
H136A HmuT exhibited both UV-visible and MCD spectral features of other five-coordinate tyrosine-ligated proteins such as BLC(74), Mycobacterium avium ssp. paratuberculosis (MAP) (74), Plexaura homomalla coral allene oxide synthase (cAOS) (73), S. dysenteriae ShuT (26), and the S. aureus Isd system: IsdA-N1 (29;36;37), IsdB-N2 (38), IsdC-N2 (33;37), and IsdH-N3 (34;38). The ~620 nm charge transfer band appears to be characteristic of a tyrosine bound to the heme.
The Y235A mutant showed loss of the charge transfer band in the UV-visible spectrum, consistent with removal of the axial tyrosine. Both the UV-visible and MCD spectra of Y235A were not comparable with other known 5c His-bound heme proteins, but rather with 6c His/OH− species, such as HRP(86) or alkaline hemoglobin (85). It was concluded that the ferric iron was still ligated to H136, but with the axial position vacated by the mutated Y235 occupied by a hydroxide ligand. This His/OH− motif was seen not only at pH 10, but also at pH 6.5. pKa values of water trans to histidine in ferric heme proteins vary from > 10 down to at least 6.8 (see examples in Supplementary Table 1). The homology model indicates that the Arg237 side chain is found in the heme pocket on the side opposite the axial histidine. Thus, the low pKa for Y235A may be attributable to the interaction of water with this cationic side chain, which could serve as the H-bond donor to the axial Tyr235 ligand. Consistent with this proposal, in almost all tyrosine-heme proteins studied to date, the axial tyrosine is hydrogen-bonded to a second residue, common examples include Tyr, Arg and His (see examples in Supplementary Table 2). For heme uptake proteins, the number of residues between the tyrosine and its hydrogen bonding partner (located on the C-terminal side in known examples) can be as few as one or as many as seven.
For heme proteins having abound water ligand, a decrease in the Fe–OH stretching frequency relative to Mb and Hb has been attributed to strong hydrogen bond donation to the hydroxide. For example, in hemoglobin from M. tuberculosis (HbN) a 35 cm −1 decrease in HS Fe–OH stretching frequency has been attributed to a strong interaction between the bound hydroxide and a distal tyrosine side chain (90). Very strong hydrogen bonding in alkaline HRP similarly gives rise to the LS νFe─OH frequency of 503 cm−1, 47 cm−1 lower than the LS νFe─OH Mb frequency of 550 cm−1(87). For Y235A CdHmuT, the LS and HS Fe–OH stretching frequencies are 29 and 58 cm−1 lower, respectively, than those reported for Mb. This strongly suggests Tyr235 has a hydrogen bonding partner that upon removal of Tyr235 its normal H-bond partner serves as a H-bond donor to the hydroxide ligand. The presence of a strong hydrogen bonding partner on the Tyr235 side of the heme is further supported by the position of the Y235A HmuT–F on the νFe─F frequency/CT1 energy empirical correlation plot (Figure 9B). On the correlation plot, it lies very close to the Thermobifida fusca (Tf-trHb) fluoride adduct which has two strong hydrogen bonds between the fluoride and the distal heme pocket (98).
Ferrous Species
The ferrous spectra of the wild-type and the H136A mutant could not be reduced under standard reductive conditions, e.g., sodium dithionite, unless in the presence of CO. This contrasts with reduction of Y235A, which is readily effected. These observations are consistent with His/Tyr heme ligation. Related observations have been made for HasA(100), which has a very low reduction potential (−550 mV versus SHE) (101). Upon mutation of its axial tyrosine, it is also readily reduced by dithionite (100).
Proteins and engineered mutants that can bind CO with either a trans His or Tyr have a histidine in some instances and an oxygen ligand (tyrosine or water) in others. For example, hemoglobin Saskatoon, with histidine and tyrosine as axial ligands, forms the His–Fe–CO species (102). Sperm whale HisE7Tyr (with a Tyr–Fe–His motif in the ferric form) also forms a His–Fe–CO complex (103). Other hemoglobins with axial tyrosines (e.g., Boston, Hyde Park and Saskatoon) form His–Fe–CO species, presumably utilizing the distal histidine (102). In contrast, HasA, with a His/Tyr ligand set, gives an O–Fe–CO species upon reduction in the presence of CO(100). The CO complex of human heme oxygenase H25Y, with a tyrosine axial ligand in the ferric form, gives an H2O–Fe–CO species (104). Catalase, with an axial tyrosine, is thought to form the Tyr–Fe–CO species (105).
The νFe–CO band in the Soret-excited rR spectrum of catalase–CO has been reported to be more intense than the totally symmetric ν7 band (105). This signature was correlated with the anionic character of the proximal Tyr whose coordination to heme iron is stabilized by H-bonding to a His residue. Although the νFe–CO bands for WT and H136A HmuT CO complexes are not more intense than their ν7 bands, they are quite intense relative to the ν Fe–CO band of Y235A HmuT–CO. This is taken as further evidence for a charge neutral Tyr–Fe(II)–CO species in H136A HmuT and WT HmuT.
Unlike the wild-type and H136A, Y235A was reduced by aqueous S2O42− in the absence of CO. Based on the two Fe–His stretching frequencies of 221 and 249 cm−1, ferrous Y235A HmuT has one conformer with Fe–His proximal bond of strength comparable to that of Mb and Hb (218 – 224 cm−1) (106;107) and a second conformer with significant imidazolate character in the proximal histidine similar to that observed for peroxidases (i.e., νFe–Im− occurs at 244 cm−1 for HRP)(108).
Why tyrosine?
Although not common in heme proteins (109), the His/Tyr ligand set has also been observed in S. marcescens HasA(110), P. aeruginosa HasA(111;112) and PhuR (47), E. coli CcmE (113;114), Paracoccus denitrificans MauG (115), and Y. pestis HmuT (27). Of these, HasA, PhuR, CcmE and HmuT are all involved in heme transfer. MauG, in contrast, appears to use the tyrosinate axial ligand to stabilize a high oxidation center in the mechanistic pathway of this protein (116). The His/Tyr ligand set is also known in hemoglobin variants such as Hb M Saskatoon (102) and has been created by site-directed mutagenesis of sperm whale Mb (HisE7Tyr) (103). S. aureus IsdA binds heme through His83 and hemin through Tyr166 (36;37;70).
Tyrosine alone is also an axial ligand in a number of proteins in heme uptake pathways characterized to date, including S. dysenteriae ShuT (25), P. aeruginosa PhuT (25;26), Y. pestis HasA (117), N. meningitidis HmbR (46) and the heme uptake proteins from the S. aureus Isd system [IsdA (29), IsdB (32), IsdC (33), and IsdH (34)] as well as IsdX1 and IsdX2-N5 of B. anthracis (41;42). Sequence alignment of the Isd proteins revealed these tyrosine residues are conserved among species, and their role in heme binding is significant.
It is possible that modulation of H-bond donation to the axial tyrosine plays a role in triggering heme release and transfer. In some instances, the hydrogen bonding residue participates in an extended H-bonding network that also involves the heme propionates (17). Disruption of the hydrogen bonding network in these cases could have cooperative effects on the structure of the protein, its affinity for hemin, and potentially on the kinetics and mechanism of hemin transfer (71;82;100).
Tyrosine may also be employed as an axial ligand to ensure the hemin remains in its ferric form (100). Tyrosinate-bound heme proteins are characterized by low reduction potentials, consistent with stabilization of the Fe(III) center by the negative charge of the tyrosinate. For example, the midpoint reduction potential of SmHasA was reported to be −550 mV (101), nearly 0.5 V more negative than the potential of −60 mV for the hemin|heme couple (118). The redox potential indicates that tyrosine binds more strongly with the Fe(III) of hemin than with the Fe(II) of heme. Assuming that the His/Tyr axial ligation in HmuT imposes a similarly negative potential as in HasA, estimation of this difference in binding free energy is facilitated by the thermodynamic cycle shown in Scheme 1. Based on this cycle, (ΔGIII–ΔGII) = nF(Eb°–Ef°) = −4.7×104 J·mol−1 where ΔGIII and ΔGII are the free energies of apoHmuT complexation with hemin (FeIIIPPIX) to give HmuTIII and heme (FeIIPPIX) to yield HmuTII, respectively. Eb° and Ef° are the reduction potentials of bound (HmuTIII) and free hemin, respectively. Thus, formation of HmuTIII is favored by an estimated 47 kJ·mol−1 over HmuT II. This would strongly favor binding of hemin over heme, thereby favoring uptake of Fe(III) by any bacterium having a Tyr−-based HBP that delivers hemin to the ABC permease. This discrimination could help guard against the damaging effects of Fenton-type chemistry from buildup of free heme in the reducing environment of the cell.
Scheme 1.
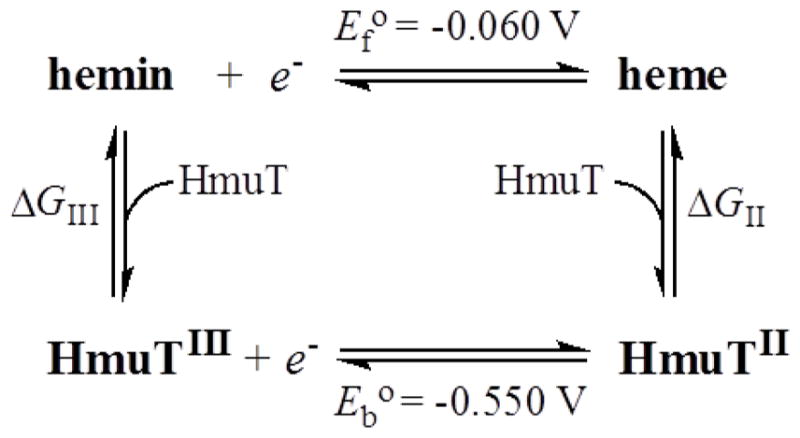
HmuT joins a growing number of extracellular and cell-surface HBPs that use the H-bond assisted axial Tyr ligand motif to bind and stabilize hemin. These proteins share high affinities for hemin and cognate hemin-accepting proteins that, ostensibly, use the free energy of protein-protein complexation to destabilize their hemin-bound states, thereby facilitating hemin transfer to the acceptor protein (10).
Conclusions
Multiple lines of spectroscopic evidence have revealed the hemin axial ligand set in HmuT from C. diphtheriae to be His136/Tyr235 from the N - and C-terminal domains of the protein, respectively. The same axial ligand set is found in YpHmuT, but with the His and Tyr ligands arising from the C-and N-terminal domains of the proteins, respectively. This highlights the variety of binding motifs used by heme binding proteins in bacteria. Solution speciation of the ferric form of CdHmuT is dominated by the 1:1 complex, which, like HasA, exists as a thermal spin state equilibrium between 6cHS and 6cLS complexes. Ferric CdHmuT is slow to reduce with S2O42− in the presence of CO with His136 being replaced by CO upon reduction. The position of the trans-Tyr carbonyl complex on the neutral O- bound ligand line of the π-backbonding correlation plot suggests that, as in HasA–CO, the Tyr-based phenol ligand is a charge neutral heme carbonyl. Thus, HmuT reinforces the emerging theme of extracellular and cell surface heme-binding proteins that use H-bond assisted axial Tyr ligands to stabilize hemin-bound states that require a heme-accepting partner for release and transfer of the hemin substrate. Moreover, the axial Tyr ligand (Tyr235 in CdHmuT) is required for full hemin uptake function in C. diphtheriae.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by National Institutes of Health Grants AI072719 (to K.R.R.), GM26730 (to J.H.D.) and GM094039 (to G.L.R.) and the Research Corporation (to D.W.D.).
We thank Jonathan M. Burgos for technical assistance and Emily Johnson and Dr. Masanori Sono for helpful discussions. We also like to thank the Georgia State University Molecular Basis of Disease Fellowship program for financially supporting E. Bennett.
ABBREVIATIONS
- ABC transporter
ATP-binding cassette transporter
- BLC
bovine liver catalase
- CdHmuT
Corynebacterium diphtheriae HmuT
- HBP
heme binding protein
- Mb
myoglobin
- Hb
hemoglobin
- HRP
horseradish peroxidase
- MCD
magnetic circular dichroism
- rR
resonance Raman spectroscopy
- LS
low spin
- HS
high spin
- WT
wild-type
Footnotes
Supporting Information. Additional MCD and resonance Raman studies of HmuT and its mutants, sequence alignments, and tables of pKa values and axial ligand hydrogen bonding partners in other heme proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15:328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Anzaldi LL, Skaar EP. Overcoming the heme paradox: Heme toxicity and tolerance in bacterial pathogens. Infect Immun. 2010;78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelis P, Andrews SC. Iron Uptake and Homeostasis in Microorganisms. Caister Academic Press; Norfolk, UK: 2010. [Google Scholar]
- 4.Mayfield JA, Dehner CA, Dubois JL. Recent advances in bacterial heme protein biochemistry. Curr Opin Chem Biol. 2011;15:260–266. doi: 10.1016/j.cbpa.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilks A, Barker KD. Mechanisms of heme uptake and utilization in bacterial pathogens. In: Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science with Applications to Chemistry, Physics, Materials Science, Engineering, Biology and Medicine, Vol 15: Biochemistry of Tetrapyrroles. World Scientific; Hacksensack, NJ: 2011. pp. 357–398. [Google Scholar]
- 6.Gruss A, Borezée-Durant E, Lechardeur D. Environmental heme utilization by heme-auxotrophic bacteria. In: Poole RK, editor. Advances in Bacterial Respiratory Physiology. Academic Press; London, England: 2012. pp. 69–124. [DOI] [PubMed] [Google Scholar]
- 7.Smith AD, Wilks A. Extracellular heme uptake and the challenges of bacterial cell membranes. Curr Top Membr. 2012;69:359–392. doi: 10.1016/B978-0-12-394390-3.00013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DR, Rivera M. Heme uptake and metabolism in bacteria. Met Ions Life Sci. 2013;12:279–332. doi: 10.1007/978-94-007-5561-1_9. [DOI] [PubMed] [Google Scholar]
- 9.Farrand AJ, Skaar EP. Heme and infectious diseases. In: Ferreira GC, Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science with Applications to Chemistry, Physics, Materials Science, Engineering, Biology and Medicine, Vol 26: Heme Biochemistry. World Scientific; Hackensack, NJ: 2014. pp. 317–377. [Google Scholar]
- 10.Rodgers KR, Lukat-Rodgers GS. Biophysical perspectives on the aquisition, transport, and trafficking of heme in bacteria. In: Ferreira GC, Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science with Applications to Chemistry, Physics, Materials Science, Engineering, Biology and Medicine. World Scientific; Hackensack, N.J: 2014. pp. 251–309. [Google Scholar]
- 11.Wilks A, O’Neill MJ. Extracellular heme uptake and metabolism in bacterial pathogenesis. In: Ferreira GC, Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science with Applications to Chemistry, Physics, Materials Science, Engineering, Biology and Medicine, Vol 26: Heme Biochemistry. World Scientific; Hackensack, NJ: 2014. pp. 267–315. [Google Scholar]
- 12.Krewulak KD, Vogel HJ. Structural biology of bacterial iron uptake. Biochim Biophys Acta. 2008;1778:1781–1804. doi: 10.1016/j.bbamem.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 13.Tong Y, Guo M. Bacterial heme-transport proteins and their heme-coordination modes. Arch Biochem Biophys. 2009;481:1–15. doi: 10.1016/j.abb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu BC, Vogel HJ. A structural and functional analysis of type III periplasmic and substrate binding proteins: Their role in bacterial siderophore and heme transport. Biol Chem. 2011;392:39–52. doi: 10.1515/BC.2011.012. [DOI] [PubMed] [Google Scholar]
- 15.Nobles CL, Maresso AW. The theft of host heme by Gram-positive pathogenic bacteria. Metallomics. 2011;3:788–796. doi: 10.1039/c1mt00047k. [DOI] [PubMed] [Google Scholar]
- 16.Honsa ES, Maresso AW. Mechanisms of iron import in anthrax. Biometals. 2011;24:533–545. doi: 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 17.Grigg JC, Ukpabi G, Gaudin CFM, Murphy MEP. Structural biology of heme binding in the Staphylococcus aureus Isd system. J Inorg Biochem. 2010;104:341–348. doi: 10.1016/j.jinorgbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haley KP, Skaar EP. A battle for iron: Host sequestration and Staphylococcus aureus acquisition. Microbes Infect. 2012;14:217–227. doi: 10.1016/j.micinf.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiedemann MT, Heinrichs DE, Stillman MJ. Multiprotein heme shuttle pathway in Staphylococcus aureus: Iron-regulated surface determinant cog-wheel kinetics. J Am Chem Soc. 2012;134:16578–16585. doi: 10.1021/ja305115y. [DOI] [PubMed] [Google Scholar]
- 21.Runyen-Janecky LJ. Role and regulation of heme on acquisition in gram-negative pathogens. Front Cell Infect Microbio. 2013;3:55. doi: 10.3389/fcimb.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.George AM, Jones PM. Perspectives on the structure-function of ABC transporters: The switch and constant contact models. Prog Biophys Mol Biol. 2012;109:95–107. doi: 10.1016/j.pbiomolbio.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Rice AJ, Park A, Pinkett HW. Diversity in ABC transporters: Type I, II and III importers. Crit Rev Biochem Mol Biol. 2014;49:426–437. doi: 10.3109/10409238.2014.953626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Beek J, Guskov A, Slotboom DJ. Structural diversity of ABC transporters. J Gen Physiol. 2014;143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho WW, Li HY, Eakanunkul S, Tong Y, Wilks A, Guo ML, Poulos TL. Holo-and apo-bound structures of bacterial periplasmic heme-binding proteins. J Biol Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 26.Eakanunkul S, Lukat-Rodgers GS, Sumithran S, Ghosh A, Rodgers KR, Dawson JH, Wilks A. Characterization of the periplasmic heme-binding protein ShuT from the heme uptake system of Shigella dysenteriae. Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 27.Mattle D, Zeltina A, Woo JS, Goetz BA, Locher KP. Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J Mol Biol. 2010;404:220–231. doi: 10.1016/j.jmb.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Woo JS, Zeltina A, Goetz BA, Locher KP. X-ray structure of the Yersinia pestis heme transporter HmuUV. Nature Struc Mol Biol. 2012;19:1310–1315. doi: 10.1038/nsmb.2417. [DOI] [PubMed] [Google Scholar]
- 29.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy MEP. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 30.Sook BR, Block DR, Sumithran S, Montañez GE, Rodgers KR, Dawson JH, Eichenbaum Z, Dixon DW. Characterization of SiaA, a streptococcal heme-binding protein associated with a heme ABC transport system. Biochemistry. 2008;47:2678–2688. doi: 10.1021/bi701604y. [DOI] [PubMed] [Google Scholar]
- 31.Ran Y, Liu M, Zhu H, Nygaard TK, Brown DE, Fabian M, Dooley DM, Lei B. Spectroscopic identification of heme axial ligands in HtsA that are involved in heme acquisition by Streptococcus pyogenes. Biochemistry. 2010;49:2834–2842. doi: 10.1021/bi901987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaudin CF, Grigg JC, Arrieta AL, Murphy ME. Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry. 2011;50:5443–5452. doi: 10.1021/bi200369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J Biol Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe M, Tanaka Y, Suenaga A, Kuroda M, Yao M, Watanabe N, Arisaka F, Ohta T, Tanaka I, Tsumoto K. Structural basis for multimeric heme complexation through a specific protein-heme interaction - The case of the third NEAT domain of IsdH from Staphylococcus aureus. J Biol Chem. 2008;283:28649–28659. doi: 10.1074/jbc.M803383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade MA, Ciccarelli FD, Perez-Iratxeta C, Bork P. NEAT: A domain duplicated in genes near the components of a putative Fe(3+) siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 2002;3:RESEARCH0047. doi: 10.1186/gb-2002-3-9-research0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeiren CL, Pluym M, Mack J, Heinrichs DE, Stillman MJ. Characterization of the heme binding properties ofStaphylococcus aureus IsdA. Biochemistry. 2006;45:12867–12875. doi: 10.1021/bi0607711. [DOI] [PubMed] [Google Scholar]
- 37.Pluym M, Muryoi N, Heinrichs DE, Stillman MJ. Heme binding in the NEAT domains of IsdA and IsdC of Staphylococcus aureus. J Inorg Biochem. 2008;102:480–488. doi: 10.1016/j.jinorgbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Tiedemann MT, Muryoi N, Heinrichs DE, Stillman MJ. Characterization of IsdH (NEAT domain 3) and IsdB (NEAT domain 2) in Staphylococcus aureus by magnetic circular dichroism spectroscopy and electrospray ionization mass spectrometry. J Porph Phthalo. 2009;13:1006–1016. [Google Scholar]
- 39.Tiedemann MT, Muryoi N, Heinrichs DE, Stillman MJ. Iron acquisition by the haem-binding Isd proteins in Staphylococcus aureus: Studies of the mechanism using magnetic circular dichroism. Biochem Soc Trans. 2008;36:1138–1143. doi: 10.1042/BST0361138. [DOI] [PubMed] [Google Scholar]
- 40.Grigg JC, Mao CX, Murphy ME. Iron-coordinating tyrosine is a key determinant of NEAT domain heme transfer. J Mol Biol. 2011;413:684–698. doi: 10.1016/j.jmb.2011.08.047. [DOI] [PubMed] [Google Scholar]
- 41.Ekworomadu MT, Poor CB, Owens CP, Balderas MA, Fabian M, Olson JS, Murphy F, Balkabasi E, Honsa ES, He C, Goulding CW, Maresso AW. Differential function of Lip residues in the mechanism and biology of an anthrax hemophore. PLoS Path. 2012;8 doi: 10.1371/journal.ppat.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honsa ES, Owens CP, Goulding CW, Maresso AW. The near-iron transporter (NEAT) domains of the anthrax hemophore IsdX2 require a critical glutamine to extract heme from methemoglobin. J Biol Chem. 2013;288:8479–8490. doi: 10.1074/jbc.M112.430009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honsa ES, Maresso AW, Highlander SK. Molecular and evolutionary analysis of NEAr-iron Transporter (NEAT) domains. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malmirchegini GR, Sjodt M, Shnitkind S, Sawaya MR, Rosinski J, Newton SM, Klebba PE, Clubb RT. Novel mechanism of hemin capture by Hbp2, the hemoglobin-binding hemophore from Listeria monocytogenes. J Biol Chem. 2014;289:34886–34899. doi: 10.1074/jbc.M114.583013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojtowicz H, Wojaczynski J, Olczak M, Kroliczewski J, Latos-Grazynski L, Olczak T. Heme environment in HmuY, the heme-binding protein of Porphyromonas gingivalis. Biochem Biophys Res Commun. 2009;383:178–182. doi: 10.1016/j.bbrc.2009.03.148. [DOI] [PubMed] [Google Scholar]
- 46.Mokry DZ, Nadia-Albete A, Johnson MK, Lukat-Rodgers GS, Rodgers KR, Lanzilotta WN. Spectroscopic evidence for a 5-coordinate oxygenic ligated high spin ferric heme moiety in the Neisseria meningitidis hemoglobin binding receptor. Biochim Biophys Acta. 2014;1840:3058–3066. doi: 10.1016/j.bbagen.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith AD, Modi AR, Sun SF, Dawson JH, Wilks A. Spectroscopic determination of distinct heme ligands in outer-membrane receptors PhuR and HasR of Pseudomonas aeruginosa. Biochemistry. 2015;54:2601–2612. doi: 10.1021/acs.biochem.5b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aranda R, Worley CE, Liu M, Bitto E, Cates MS, Olson JS, Lei BF, Phillips GN. Bis-methionyl coordination in the crystal structure of the heme-binding domain of the streptococcal cell surface protein Shp. J Mol Biol. 2007;374:374–383. doi: 10.1016/j.jmb.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt MP. Utilization of host iron sources by Corynebacterium diphtheriae: Identification of a gene whose product is homologous for eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. The pathology of diphtheria. J Infect Dis. 2000;181:S116–S120. doi: 10.1086/315551. [DOI] [PubMed] [Google Scholar]
- 51.Holmes RK. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis. 2000;181(Suppl 1):S156–S167. doi: 10.1086/315554. [DOI] [PubMed] [Google Scholar]
- 52.Burgos JM, Schmitt MP. The ChrA response regulator in Corynebacterium diphtheriae controls hemin-regulated gene expression through binding to the hmuO and hrtAB promoter regions. J Bacteriol. 2012;194:1717–1729. doi: 10.1128/JB.06801-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trost E, Blom J, Soares SD, Huang IH, Al-Dilaimi A, Schroder J, Jaenicke S, Dorella FA, Rocha FS, Miyoshi A, Azevedo V, Schneider MP, Silva A, Camello TC, Sabbadini PS, Santos CS, Santos LS, Hirata R, Mattos-Guaraldi AL, Efstratiou A, Schmitt MP, Hung TT, Tauch A. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J Bacteriol. 2012;194:3199–3215. doi: 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Besa NC, Coldiron ME, Bakri A, Raji A, Nsuami MJ, Rousseau C, Hurtado N, Porten K. Diphtheria outbreak with high mortality in northeastern Nigeria. Epidemiol Infect. 2014;142:797–802. doi: 10.1017/S0950268813001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mattos-Guaraldi A, Moreira L, Damasco P, Hirata R., Junior Diphtheria remains a threat to health in the developing world: An overview. Mem Inst Oswaldo Cruz. 2003;98:987–993. doi: 10.1590/s0074-02762003000800001. [DOI] [PubMed] [Google Scholar]
- 56.Wagner KS, White JM, Lucenko I, Mercer D, Crowcroft NS, Neal S, Efstratiou A. Diphtheria in the postepidemic period, Europe, 2000–2009. Emerg Infect Dis. 2012;18:217–225. doi: 10.3201/eid1802.110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drazek ES, Hammack CA, Schmitt MP. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol Microbiol. 2000;36:68–84. doi: 10.1046/j.1365-2958.2000.01818.x. [DOI] [PubMed] [Google Scholar]
- 58.Schmitt MP, Drazek ES. Construction and consequences of directed mutations affecting the hemin receptor in pathogenic Corynebacterium species. J Bacteriol. 2001;183:1476–1481. doi: 10.1128/JB.183.4.1476-1481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitt MP. Iron acquisition and iron-dependent gene expression in Corynebacterium diphtheriae. In: Burkovski A, editor. Corynebacterium diphtheriae and Related Toxigenic Species: Genomics, Pathogenicity and Applications. 2014. pp. 95–121. [Google Scholar]
- 60.Allen CE, Schmitt MP. Utilization of host iron sources by Corynebacterium diphtheriae: Multiple hemoglobin-binding proteins are essential for the use of iron from the hemoglobin/haptoglobin complex. J Bacteriol. 2014;195:2413–2414. doi: 10.1128/JB.02413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerdeno-Tarraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, Besra GS, Churcher C, James KD, De Zoysa A, Chillingworth T, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, Jagels K, Moule S, Quail MA, Rabbinowitsch E, Rutherford KM, Thomson NR, Unwin L, Whitehead S, Barrell BG, Parkhill J. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 2003;31:6516–6523. doi: 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allen CE, Schmitt MP. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J Bacteriol. 2009;191:2638–2648. doi: 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kunkle CA, Schmitt MP. Analysis of the Corynebacterium diphtheriae DtxR regulon: Identification of a putative siderophore synthesis and transport system that is similar to the Yersinia high-pathogenicity island-encoded yersiniabactin synthesis and uptake system. J Bacteriol. 2003;185:6826–6840. doi: 10.1128/JB.185.23.6826-6840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allen C, Schmitt MP. Novel hemin binding domains in the Corynebacterium diphtheriae HtaA protein interact with hemoglobin and are critical for heme iron utilization by HtaA. J Bacteriol. 2011;193:5374–5385. doi: 10.1128/JB.05508-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bibb LA, Kunkle CA, Schmitt MP. The ChrA-ChrS and HrrA-HrrS signal transduction systems are required for activation of the hmuO promoter and repression of the hemA promoter in Corynebacterium diphtheriae. Infect Immun. 2007;75:2421–2431. doi: 10.1128/IAI.01821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunkle CA, Schmitt MP. Comparative analysis of hmuO function and expression in Corynebacterium species. J Bacteriol. 2007;189:3650–3654. doi: 10.1128/JB.00056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitt MP, Holmes RK. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene Dtxr in C. diphtheriae C7 strains. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pond AE, Roach MP, Thomas MR, Boxer SG, Dawson JH. The H93G myoglobin cavity mutant as a versatile template for modeling heme proteins: Ferrous, ferric, and ferryl mixed-ligand complexes with imidazole in the cavity. Inorg Chem. 2000;39:6061–6066. doi: 10.1021/ic0007198. [DOI] [PubMed] [Google Scholar]
- 69.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Heme coordination by Staphylococcus aureus IsdE. J Biol Chem. 2007;282:28815–28822. doi: 10.1074/jbc.M704602200. [DOI] [PubMed] [Google Scholar]
- 71.Caillet-Saguy C, Turano P, Piccioli M, Lukat-Rodgers GS, Czjzek M, Guigliarelli B, Izadi-Pruneyre N, Rodgers KR, Delepierre M, Lecroisey A. Deciphering the structural role of histidine 83 for heme binding in hemophore HasA. J Biol Chem. 2008;283:5960–5970. doi: 10.1074/jbc.M703795200. [DOI] [PubMed] [Google Scholar]
- 72.Browett WR, Stillman MJ. Magnetic circular dichroism studies of bovine liver catalase. Biochim Biophys Acta. 1979;577:291–306. doi: 10.1016/0005-2795(79)90033-3. [DOI] [PubMed] [Google Scholar]
- 73.Abraham BD, Sono M, Boutaud O, Shriner A, Dawson JH, Brash AR, Gaffney BJ. Characterization of the coral allene oxide synthase active site with UV-visible absorption, magnetic circular dichroism, and electron paramagnetic resonance spectroscopy: Evidence for tyrosinate ligation to the ferric enzyme heme iron. Biochemistry. 2001;40:2251–2259. doi: 10.1021/bi002121h. [DOI] [PubMed] [Google Scholar]
- 74.Bandara DMI, Sono M, Bruce GS, Brash AR, Dawson JH. Coordination modes of tyrosinate-ligated catalase-type heme enzymes: Magnetic circular dichroism studies of Plexaura homomalla allene oxide synthase, Mycobacterium avium ssp. paratuberculosis protein-2744c, and bovine liver catalase in their ferric and ferrous states. J Inorg Biochem. 2011;105:1786–1794. doi: 10.1016/j.jinorgbio.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pond AE, Roach MP, Sono M, Rux AH, Franzen S, Hu R, Thomas MR, Wilks A, Dou Y, Ikeda-Saito M, Ortiz de Montellano PR, Woodruff WH, Boxer SG, Dawson JH. Assignment of the heme axial ligand(s) for the ferric myoglobin (H93G) and heme oxygenase (H25A) cavity mutants as oxygen donors using magnetic circular dichroism. Biochemistry. 1999;38:7601–7608. doi: 10.1021/bi9825448. [DOI] [PubMed] [Google Scholar]
- 76.Sievers G, Gadsby PM, Peterson J, Thomson AJ. Magnetic circular dichroism spectra of soybean leghaemoglobin a at room temperature and 4.2 K. Biochim Biophys Acta. 1983;742:637–647. doi: 10.1016/0167-4838(83)90284-4. [DOI] [PubMed] [Google Scholar]
- 77.Spiro TG, Strekas TC. Resonance Raman spectra of heme proteins-Effects of oxidation and spin state. J Am Chem Soc. 1974;96:338–345. doi: 10.1021/ja00809a004. [DOI] [PubMed] [Google Scholar]
- 78.Strekas TC, Spiro TG. Resonance Raman evidence for anomalous heme structures in cytochrome c′ from Rhodopseudomonas palustris. Biochim Biophys Acta. 1974;351:237–245. doi: 10.1016/0005-2795(74)90186-x. [DOI] [PubMed] [Google Scholar]
- 79.Hu SZ, Smith KM, Spiro TG. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J Am Chem Soc. 1996;118:12638–12646. [Google Scholar]
- 80.Smulevich G, Hu SZ, Rodgers KR, Goodin DB, Smith KM, Spiro TG. Heme-protein interactions in cytochrome c peroxidase revealed by site-directed mutagenesis and resonance Raman spectra of isotopically labeled hemes. Biospectroscopy. 1996;2:365–376. [Google Scholar]
- 81.Heering HA, Jansen MAK, Thorneley RNF, Smulevich G. Cationic ascorbate peroxidase isoenzyme II from tea: Structural insights into the heme pocket of a unique hybrid peroxidase. Biochemistry. 2001;40:10360–10370. doi: 10.1021/bi0106033. [DOI] [PubMed] [Google Scholar]
- 82.Caillet-Saguy C, Piccioli M, Turano P, Lukat-Rodgers G, Wolff N, Rodgers KR, Izadi-Pruneyre N, Delepierre M, Lecroisey A. Role of the iron axial ligands of heme carrier HasA in heme uptake and release. J Biol Chem. 2012;287:26932–26943. doi: 10.1074/jbc.M112.366385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reed RA, Rodgers KR, Kushmeider K, Spiro TG, Su YO. Iron-hydroxide stretching resonance Raman bands of a water-soluble sterically hindered porphyrin. Inorg Chem. 1990;29:2881–2883. [Google Scholar]
- 84.Rodgers KR, Reed RA, Spiro TG, Su YO. Fe-OH bond strength in high- and low-spin bis-hydroxides of a model heme from resonance Raman spectroscopy - Implications for heme proteins. New J Chem. 1992;16:533–535. [Google Scholar]
- 85.Sono M, Dawson JH. Extensive studies of the heme coordination structure of indoleamine 2,3-dioxygenase and of tryptophan binding with magnetic and natural circular-dichroism and electron paramagnetic resonance spectroscopy. Biochim Biophys Acta. 1984;789:170–187. doi: 10.1016/0167-4838(84)90202-4. [DOI] [PubMed] [Google Scholar]
- 86.Pond AE, Sono M, Elenkova EA, McRee DE, Goodin DB, English AM, Dawson JH. Magnetic circular dichroism studies of the active site heme coordination sphere of exogenous ligand-free ferric cytochrome c peroxidase from yeast: Effects of sample history and pH. J Inorg Biochem. 1999;76:165–174. doi: 10.1016/s0162-0134(99)00112-9. [DOI] [PubMed] [Google Scholar]
- 87.Asher SA, Schuster TM. Resonance Raman examination of axial ligand bonding and spin-state equilibria in metmyoglobin hydroxide and other heme derivatives. Biochemistry. 1979;18:5377–5387. doi: 10.1021/bi00591a019. [DOI] [PubMed] [Google Scholar]
- 88.Feis A, Marzocchi MP, Paoli M, Smulevich G. Spin state and axial ligand bonding in the hydroxide complexes of metmyoglobin, methemoglobin, and horseradish peroxidase at room and low temperatures. Biochemistry. 1994;33:4577–4583. doi: 10.1021/bi00181a019. [DOI] [PubMed] [Google Scholar]
- 89.Lukat-Rodgers GS, Rexine JL, Rodgers KR. Heme speciation in alkaline ferric FixL and possible tyrosine involvement in the signal transduction pathway for regulation of nitrogen fixation. Biochemistry. 1998;37:13543–13552. doi: 10.1021/bi981439v. [DOI] [PubMed] [Google Scholar]
- 90.Yeh SR, Couture M, Ouellet Y, Guertin M, Rousseau DL. A cooperative oxygen finding hemoglobin from Mycobacterium tuberculosis- Stabilization of heme ligands by a distal tyrosine residue. J Biol Chem. 2000;275:1679–1684. doi: 10.1074/jbc.275.3.1679. [DOI] [PubMed] [Google Scholar]
- 91.Streit BR, Blanc B, Lukat-Rodgers GS, Rodgers KR, DuBois JL. How active-site protonation state influences the reactivity and ligation of the heme in chlorite dismutase. J Am Chem Soc. 2010;132:5711–5724. doi: 10.1021/ja9082182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Role of the axial ligand in heme-CO backbonding; DFT analysis of vibrational data. Inorg Chim Acta. 2000;297:11–17. [Google Scholar]
- 93.Spiro TG, Wasbotten IH. CO as a vibrational probe of heme protein active sites. J Inorg Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 94.Spiro TG, Soldatova AV, Balakrishnan G. CO, NO and O2 as vibrational probes of heme protein interactions. Coord Chem Rev. 2013;257:511–527. doi: 10.1016/j.ccr.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Linder DP, Silvernail NJ, Barabanschikov A, Zhao JY, Alp EE, Sturhahn W, Sage JT, Scheidt WR, Rodgers KR. The diagnostic vibrational signature of pentacoordination in heme carbonyls. J Am Chem Soc. 2014;136:9818–9821. doi: 10.1021/ja503191z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smulevich G, Mauro JM, Fishel LA, English AM, Kraut J, Spiro TG. Cytochrome c peroxidase mutant active site structures probed by resonance Raman and infrared signatures of the CO adducts. Biochemistry. 1988;27:5486–5492. doi: 10.1021/bi00415a015. [DOI] [PubMed] [Google Scholar]
- 97.Droghetti E, Nicoletti FP, Bonamore A, Sciamanna N, Boffi A, Feis A, Smulevich G. The optical spectra of fluoride complexes can effectively probe H-bonding interactions in the distal cavity of heme proteins. J Inorg Biochem. 2011;105:1338–1343. doi: 10.1016/j.jinorgbio.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 98.Nicoletti FP, Droghetti E, Boechi L, Bonamore A, Sciamanna N, Estrin DA, Feis A, Boffi A, Smulevich G. Fluoride as a probe for H-bonding interactions in the active site of heme proteins: The case of Thermobifida fusca hemoglobin. J Am Chem Soc. 2011;133:20970–20980. doi: 10.1021/ja209312k. [DOI] [PubMed] [Google Scholar]
- 99.Bangcharoenpaurpong O, Schomacker KT, Champion PM. A Resonance Raman investigation of myoglobin and hemoglobin. J Am Chem Soc. 1984;106:5688–5698. [Google Scholar]
- 100.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel heme ligand displacement by CO in the soluble hemophore HasA and its proximal ligand mutants: Implications for heme uptake and release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 101.Izadi N, Henry Y, Haladjian J, Goldberg ME, Wandersman C, Delepierre M, Lecroisey A. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry. 1997;36:7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 102.Nagai M, Yoneyama Y, Kitagawa T. Unusual carbon monoxide bonding geometry in abnormal subunits of hemoglobin M Boston and hemoglobin M Saskatoon. Biochemistry. 1991;30:6495–6503. doi: 10.1021/bi00240a021. [DOI] [PubMed] [Google Scholar]
- 103.Egeberg KD, Springer BA, Martinis SA, Sligar SG, Morikis D, Champion PM. Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry. 1990;29:9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- 104.Liu Y, Moenne-Loccoz P, Hildebrand DP, Wilks A, Loehr TM, Mauk AG, Ortiz de Montellano PR. Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry. 1999;38:3733–3743. doi: 10.1021/bi982707s. [DOI] [PubMed] [Google Scholar]
- 105.Hu SH, Kincaid JR. Resonance Raman studies of the carbonmonoxy form of catalase-Evidence for and effects of phenolate ligation. FEBS Lett. 1992;314:293–296. doi: 10.1016/0014-5793(92)81492-5. [DOI] [PubMed] [Google Scholar]
- 106.Kitagawa T, Nagai K, Tsubaki M. Assignment of the Fe-nitrogen (His F8) stretching band in the resonance Raman spectra of deoxymyoglobin. FEBS Lett. 1979;104:376–378. doi: 10.1016/0014-5793(79)80856-x. [DOI] [PubMed] [Google Scholar]
- 107.Nagai K, Kitagawa T. Differences in Fe(II)-Nε (His-F8) stretching frequencies between deoxyhemoglobins in the two alternative quaternary structures. Proc Natl Acad Sci U S A. 1980;77:2033–2037. doi: 10.1073/pnas.77.4.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Teraoka J, Kitagawa T. Structural implication of the heme-linked ionization of horseradish peroxidase probed by the Fe-histidine stretching Raman line. J Biol Chem. 1981;256:3969–3977. [PubMed] [Google Scholar]
- 109.Reedy CJ, Elvekrog MM, Gibney BR. Development of a heme protein structure-electrochemical function database. Nucleic Acids Res. 2008;36:D307–D313. doi: 10.1093/nar/gkm814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjzek M. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nature Struct Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 111.Alontaga AY, Rodriguez JC, Schonbrunn E, Becker A, Funke T, Yukl ET, Hayashi T, Stobaugh J, Monne-Loccoz P, Rivera M. Structural characterization of the hemophore HasAp from Pseudomonas aeruginosa: NMR spectroscopy reveals protein-protein interactions between holo-HasAp and hemoglobin. Biochemistry. 2009;48:96–109. doi: 10.1021/bi801860g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kumar R, Matsumura H, Lovell S, Yao HL, Rodriguez JC, Battaile KP, Moenne-Loccoz P, Rivera M. Replacing the axial ligand tyrosine 75 or its hydrogen bond partner histidine 83 minimally affects hemin acquisition by the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry. 2014;53:2112–2125. doi: 10.1021/bi500030p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Couture M, Das TK, Lee HC, Peisach J, Rousseau DL, Wittenberg BA, Wittenberg JB, Guertin M. Chlamydomonas chloroplast ferrous hemoglobin-Heme pocket structure and reactions with ligands. J Biol Chem. 1999;274:6898–6910. doi: 10.1074/jbc.274.11.6898. [DOI] [PubMed] [Google Scholar]
- 114.Das TK, Couture M, Lee HC, Peisach J, Rousseau DL, Wittenberg BA, Wittenberg JB, Guertin M. Identification of the ligands to the ferric heme of Chlamydomonas chloroplast hemoglobin: Evidence for ligation of tyrosine-63 (B10) to the heme. Biochemistry. 1999;38:15360–15368. doi: 10.1021/bi991237e. [DOI] [PubMed] [Google Scholar]
- 115.Jensen LM, Sanishvili R, Davidson VL, Wilmot CM. In crystallo post-translational modification within a MauG/pre-methylamine dehydrogenase complex. Science. 2010;327:1392–1394. doi: 10.1126/science.1182492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abu Tarboush N, Shin S, Geng JF, Liu AM, Davidson VL. Effects of the loss of the axial tyrosine ligand of the low-spin heme of MauG on its physical properties and reactivity. FEBS Lett. 2012;586:4339–4343. doi: 10.1016/j.febslet.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kumar R, Lovell S, Matsumura H, Battaile KP, Moenne-Loccoz P, Rivera M. The hemophore HasA from Yersinia pestis (HasAyp) coordinates hemin with a single residue, Tyr75, and with minimal conformational change. Biochemistry. 2013;52:2705–2707. doi: 10.1021/bi400280z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reedy CJ, Kennedy ML, Gibney BR. Thermodynamic characterization of ferric and ferrous haem binding to a designed four-α-helix protein. Chem Commun. 2003:570–571. doi: 10.1039/b211488g. [DOI] [PubMed] [Google Scholar]
- 119.DeLano WL. The PyMOL Molecular Graphics System, Version 1.7.4. Schrödinger, LLC; 2015. http://www.pymol.org. [Google Scholar]
- 120.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel heme ligand displacement by CO in the soluble hemophore HasA and its proximal ligand mutants: Implications for heme uptake and release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.