Abstract
ASDP is an automated NMR NOE assignment program. It uses a distinct bottom-up topology-constrained network anchoring approach for NOE interpretation, with 2D, 3D and/or 4D NOESY peak lists and resonance assignments as input, and generates unambiguous NOE constraints for iterative structure calculations. ASDP is designed to function interactively with various structure determination programs that use distance restraints to generate molecular models. In the CASD-NMR project, ASDP was tested and further developed using blinded NMR data, including resonance assignments, either raw or manually-curated (refined) NOESY peak list data, and in some cases 15N-1H residual dipolar coupling data. In these blinded tests, in which the reference structure was not available until after structures were generated, the fully-automated ASDP program performed very well on all targets using both the raw and refined NOESY peak list data. Improvements of ASDP relative to its predecessor program for automated NOESY peak assignments, AutoStructure, were driven by challenges provided by these CASD-NMR data. These algorithmic improvements include 1) using a global metric of structural accuracy, the Discriminating Power (DP) score, for guiding model selection during the iterative NOE interpretation process, and 2) identifying incorrect NOESY cross peak assignments caused by errors in the NMR resonance assignment list. These improvements provide a more robust automated NOESY analysis program, ASDP, with the unique capability of being utilized with alternative structure generation and refinement programs including CYANA, CNS, and/or Rosetta.
Keywords: AutoStructure, ASDP, automated structural determination by NMR, CYANA, CNS, Rosetta
Introduction
Automated NOESY peak assignment is a fundamental component of protein NMR structure determination. Several successful programs for interpreting NOESY peaks lists together with resonance assignments are available (Herrmann et al., 2002; Huang et al., 2006; Lee et al., 2011; Nilges, 1995; Nilges et al., 1997). The AutoStructure program (Huang et al., 2006) uses a distinct bottom-up topology-constrained network anchoring approach for NOE interpretation, with 2D, 3D and/or 4D NOESY peak lists and resonance assignments as input. The program generates unambiguous NOE constraints for iterative structure calculations. It is designed to function interactively with various structure determination programs that use distance restraints to generate molecular models, including CYANA (Guntert et al., 1997; Herrmann et al., 2002), CNS (Brunger et al., 1998), and/or distance-restrained Rosetta (Lange et al., 2012; Mao et al., 2014; Raman et al., 2010b; Tejero et al., 2013). AutoStructure has been used for structure determination of more than 370 proteins deposited in the PDB.
In the course of developing AutoStructure, we have also explored a simple metric useful for assessing the accuracy of protein NMR structure models. The RPF-DP score (Huang et al., 2005; Huang et al., 2012) provides a simple approach to compare the short interproton distances in a protein structure model with the network of all potential NOESY cross peak assignments indicated by NOESY peak list and chemical shift resonance assignments. Analysis using NMR -- X-ray pairs, and/or comparisons of decoys generated with various methods with manually-refined NMR structures, demonstrate that RPF-DP scores are highly correlated with structural accuracy (Huang et al., 2012; Mao et al., 2014; Rosato et al., 2012). The DP scores of Rosetta decoys have also been used together with chemical shift data to direct CS-Rosetta calculations, improving the accuracy of models generated from incomplete, sparse NMR data sets (Raman et al., 2010a; Raman et al., 2010b).
CASD-NMR (Critical Assessment of Automated Structure Determination of Proteins from NMR data, www.e-nmr.eu/CASD-NMR), is a community-wide project designed to verify whether unsupervised automated NMR analysis methods can indeed produce structures that closely match those that are refined by manual analysis using the same experimental data (the “reference structures”) (Rosato et al., 2012; Rosato et al., 2009). The concept closely resembles community-wide structure prediction experiments, such as CASP (Moult et al., 1995) and CAPRI (Critical Assessment of Prediction of Interactions) (Janin et al., 2003). However, CASD-NMR utilizes experimental NMR data, presenting special issues in organizing and distributing these data among participants. CASD-NMR is a rolling experiment in which test data sets are released regularly during the course of the experiment. Software developers are invited to test their fully automated protocols on blind data sets and produce structures as if they would directly deposit them into the PDB. The current cycle of CASD-NMR was designed specifically to explore the robustness of automated NOESY resonance assignment programs when provided with “raw”, automatically peak-picked NOESY data, together with largely correct resonance assignment data.
In this paper we describe application of an improved program for automated NOESY peak assignments and restraint generation, ASDP, based on AutoStructure. ASDP was further developed and tested on blinded protein targets of the CASD-NMR experiment. ASDP utilizes the NMR DP score, comparing how well a protein model fits to the experimental NOESY peak list and chemical shift data, to direct the NOESY cross-peak assignment trajectory. Intermediate structures are assessed with the DP metric, and those above a threshold are excluded from use in ruling-in and ruling-out candidate NOESY crosspeak assignments. We have also developed an approach for identifying incorrect NOESY cross peak assignments caused by inaccuracies in the resonance assignment list. These enhancements improve the accuracy of NOESY cross peak assignments and of the resulting final NMR structures.
Materials and Methods
The blinded datasets
ASDP was applied for blind structure determination using 20 different NOESY peak list and chemical shift assignment datasets for 10 protein targets (Table 1). For each dataset, both raw and refined peak lists were provided by the CASD-NMR-2013 organizers. Raw NOESY peak lists were released first. After all structure generation results using these raw NOESY peak lists were submitted to the CASD-NMR site, manually-refined NOESY peak lists were subsequently released as a second test data set. Some of these CASD-NMR targets also had backbone 15N-1H residual dipolar coupling (RDC) data, which were released together with the NOESY peak lists and resonance assignments lists. The last two columns of Table 1 summarize whether 15N-1H RDC data were also used in the ASDP calculation listed in Table 2 along with the raw unrefined peak lists and the manually-refined peak lists.
Table 1.
Statistics on the CASD-NMR benchmark datasets
| Name | PDB | Residues | RMSD ranges | fold | RDCraw | RDCrefine |
|---|---|---|---|---|---|---|
| HR2876B | 2LTM | 107 | 11–107 | Alpha+Beta | Yes | Yes |
| HR2876C | 2M5O | 97 | 16–93 | Alpha+Beta | Yes | Yes |
| HR5460A | 2LAH | 160 | 12–28,32–159 | Alpha | No | Yes |
| HR6430A | 2LA6 | 99 | 12–99 | Alpha+Beta | No | No |
| HR6470A | 2L9R | 59 | 10–59 | Alpha | No | Yes |
| HR8254A | 2M2E | 72 | 553–612 | Alpha | No | No |
| OR135 | 2LN3 | 83 | 3–76 | Alpha+Beta | Yes | Yes |
| OR36 | 2LCI | 134 | 1–48,50–129 | Alpha+Beta | No | Yes |
| StT322 | 2LOJ | 63 | 26–63 | Alpha+Beta | No | No |
| YR313A | 2LTL | 119 | 16–43,45–112,114–116 | Alpha+Beta | Yes | Yes |
Table 2.
Blinded ASDP performance on the 10 CASD benchmark datasets for raw and refined peak lists
| Raw | Refined | |||||||
|---|---|---|---|---|---|---|---|---|
| Name | Energy Refine |
DP | <DP> | RMSD(Å) | Energy Refine |
DP | <DP> | RMSD(Å) |
| HR2876B | Rosetta | 0.798 | 0.787 | 1.32 | Rosetta | 0.923 | 0.903 | 1.23 |
| HR2876C | Rosetta | 0.682 | 0.652 | 1.78 | Rosetta | 0.891 | 0.857 | 0.99 |
| HR5460A | CNS | 0.741 | 0.676 | 1.74 | CNS | 0.853 | 0.808 | 1.70 |
| HR6430A | CNS | 0.853 | 0.820 | 1.38 | CNS | 0.905 | 0.863 | 1.41 |
| HR6470A | CNS | 0.835 | 0.777 | 1.34 | CNS | 0.889 | 0.760 | 1.12 |
| HR8254A | Rosetta | 0.790 | 0.690 | 1.94 | Rosetta | 0.798 | 0.735 | 2.17 |
| OR135 | Rosetta | 0.764 | 0.745 | 1.13 | Rosetta | 0.893 | 0.877 | 0.97 |
| OR36 | CNS | 0.766 | 0.715 | 1.45 | CNS | 0.903 | 0.830 | 1.61 |
| StT322 | Rosetta | 0.635 | 0.490 | 1.46 | Rosetta | 0.771 | 0.656 | 1.45 |
| YR313A | Rosetta | 0.650 | 0.590 | 1.39 | Rosetta | 0.818 | 0.700 | 1.77 |
RPF/DP
RPF/DP is a quality assessment tool for protein NMR structures (Huang et al., 2005; Huang et al., 2012). The algorithms to calculate RPF scores (i.e. Recall, Precision, F-measure) and the DP-score are described elsewhere (Huang et al., 2005). Briefly, Recall measures the percentage of input NOESY peaks that can be explained by the input query structure(s) with a distance cut-off ≤ 5 Å. Precision measures the percentage of 1H–1H distances ≤ 5 Å calculated from the query structure that are observed in the NOESY data. F-measure combines the Recall and Precision scores, and estimates how well the input NMR structure ensemble fits with the input NMR data. DP score is a normalized score of F-measure, which estimates the significance of the F-measure score for the query structure relative to what would be obtained for a random-coil structure fit to the same experimental data. The DP-score is an accuracy predictor of the query structure relative to the NOESY and chemical shift data, ranging from 0 to 1. The RPF/DP also maps the local structure quality measures onto the 3D structure using an online molecular viewer, and onto the NMR spectra, allowing refinement of the structure and/or NOESY peak list data. In summery, the RPF/DP measures the 'goodness-of-fit' of the 3D structure with NMR chemical shift and unassigned NOESY data, and calculates a discrimination power (DP) score, which estimates the differences between the fits of the query structures and random coil structures to these experimental data.
Structure determination with ASDP
ASDP, the newest version of AutoStructure (Huang et al., 2006), utilizes (i) a topology-based algorithm to build secondary structures, including helices, anti-parallel beta-sheets and parallel beta-sheets, from unassigned NOE data and resonance assignments in the first cycle, and (ii) a bottom-up iterative strategy, beginning from these secondary structure elements, to assign additional NOESY peaks and generate distance restraints. Ambiguous restraint approaches are not used in ASDP analysis. The key feature distinguishing ASDP from its predecessor AutoStructure is the use of the DP score (Huang et al., 2005; Huang et al., 2012) to rank and filter intermediate structures that are used to direct the trajectory of NOESY cross peak assignment process.
Dihedral angle restraints for ASDP are generated from backbone chemical shift data using TALOS+ (Shen et al., 2009). Only the dihedral angles classified as ‘good’ by TALOS+ (reliability score = 10) were used as restraints. The ranges of these dihedral angles were set to their predicted value ± 20 degrees or to twice the standard deviation, whichever was larger. 100 structures were then calculated with the structure generation components of the CYANA program, using distance, dihedral angle, and hydrogen bond restraints provided by the ASDP, together with RDC data when available. Among these 100 structures, the 20 structures with the best combined DP and CYANA target function scores [i.e. (target function/weight) − (DP score), where weight = min(target function of 100 models)*100], were selected and used to rule-in and rule-out potential NOESY cross peaks assignments. This process was carried out for five cycles of NOE analysis. TALOS+ dihedral angle restraints violated in all 20 models were removed, and the ASDP process was repeated. Only one iteration of this overall ASDP protocol is performed to avoid potential over-fitting. All ASDP / CYANA calculations were distributed on 50 cpu processors. Each structure calculation required only minutes to complete. The resulting ensemble of 20 conformers were then energy-refined with the WaterRefCNS protocol (Brunger et al., 1998) with slow cooling steps (tsc) = 0.001 and RDC weight (wrdc1) = 0.2 when applicable (for a detailed protocol see http://www.nmr2.buffalo.edu/nesg.wiki/), or refined with using distance-restrained Rosetta calculations (Mao et al., 2014).
Distance-restrained Rosetta calculations
Restrained Rosetta refinement was done using protocols presented elsewhere (Mao et al., 2014; Tejero et al., 2013). Restraints were converted from CYANA to Rosetta format using the PDBStat program (Tejero et al., 2013). Calculations were done using Rosetta Ver 3.X. Input includes PDB coordinates, chemical shifts, and restraint lists. Fragment libraries for the restrained Rosetta calculations were generated without considerations of chemical shift data.
Automatic residue disorder filter for unrefined raw peak lists
Long segments of residues predicted to be disordered were identified by using a disorder prediction server, DisMeta (Huang et al., 2014), which is based on a consensus analysis of eight disorder prediction and two secondary structure prediction methods. Potential long-range NOE assignments involving disordered residues are excluded from the structure calculations. For StT322, the first 27 residues were predicted to be disordered by DisMeta (Huang et al., 2014).
Automatic noise filter for unrefined raw NOESY peak lists (“ASDP filter”)
The following protocol was applied automatically to filter out noise peaks from the unrefined raw NOESY peak lists:
Step 1. Initial noise peaks were first filtered out from the raw NOESY peak list using preprocessing scripts of ASDP which removed (i) peaks with negative intensities; ii) peaks with no matches against the chemical shift table; iii) possible peaks due to solvent saturation transfer or incomplete solvent suppression in the chemical shift range between 4.4 – 5.2 ppm in the indirect 1H dimension.
Step 2. Run ASDP.
Step 3. Check the DP scores from the ASDP run and identify a “peak intensity cutoff” for further peak filtering. This process was done differently depending on the accuracy of the models as assessed by the DP score.
CASE A: DP score from Step 2 > 0.6
These decoy structures are considered to be reasonably accurate. NOESY peaks were separated into symmetry and non-symmetry classes; symmetry NOESY peaks are pairs of NOESY peaks that are symmetric in the NOESY spectrum, and non-symmetry peaks cannot be confirmed by identification of a symmetric NOESY peak partner. For each of these two groups, the NOESY peaks were sorted based on peak intensities and then equally distributed into 20 equal size bins [i.e. ≥ 0%, ≥ 5%, ≥ 10%, … ≥ 95% of all peaks]. In this notation, the “≥ 20% bin” means removing 20% of peaks with lowest intensities. In each bin, scripts were used to identify false negative (FN) and true positive (TP) peaks, using the RPF recall analysis (Huang et al., 2005); i.e. FN peaks are peaks in the NOESY peak lists which are not satisfied by any structure in the ensemble considering all possible assignments to the peak consistent with the chemical shift assignment list. The FN and TP peaks in each bin were then used to define a “peak intensity cutoff”. The peak intensity cutoff was defined as the highest bin with FN/TP ratio > 1. The optimum peak intensity cutoffs were different for different NOESY peak lists, and also different for the class of peaks that could be validated by identifying a symmetric peak in the NOESY spectrum. The intensity cut offs for non-symmetric peaks typically ranged from ≥ 20% (i.e. the 20% of peaks with lowest intensity were removed) to ≥40% (i.e. the 40% of peaks with lowest intensity were removed). For symmetric peaks in these same NOESY peak lists, cutoff thresholds were typically ≥5% or ≥10% of peaks; for some data sets the algorithm identified an intensity cutoff of ≥0% (i.e. no NOESY peaks were removed). For the raw NOESY data sets of targets HR6430A and HR6470A, none of the raw NOESY peaks were filtered out by this algorithm.
CASE B: DP score from Step 2 < 0.6
In this case the “peak intensity cutoff” was set to ≥10%, ≥15%, ≥20%, …, ≥80% of the total number of peaks (15 bins), sorted again based on NOESY peak intensity. For each bin, we excluded all peaks with intensities below the peak intensity cutoff, ran ASDP, and calculated DP scores. The “peak intensity cutoff” was defined as the bin value generating the highest DP score for the resulting ensemble of NMR models among the 15 bins. Among the 10 CASD-NMR targets, only HR8254A and StT322 NOESY peak lists were filtered using the CASE B protocol. The peak intensity cutoffs were ≥ 70% for HR8254A and ≥ 35% for StT322; i.e. the 70% lowest intensity peaks of HR8254A and the 35% lowest intensity peaks of StT322 were identified as potential noise peaks by this approach.
A DP score < 0.6 indicates that the model does not fit the data, and the resulting structures are not recommended for use in guiding the peak picking process. However, for both targets HR8254A and StT322 that were processed using the peak filtering protocol CASE B, we also tested the CASE A method. The noise/signal (FN/TP) ratios were > 1 for all of the peak intensity cutoff bins. For these NOESY peak lists, no “peak intensity cutoff” can be identified using the CASE A method.
Step 4: All NOESY peaks with intensities below the peak intensity cutoff were removed. The resulting NOESY peak lists were then used for the final ASDP calculations.
RMSD calculations
Backbone (defined as N, Cα, C’, and O atoms) Root Mean Square Deviations (RMSDs) were computed using the fit command as implemented in the PyMol software (Schrodinger; Valafar and Prestegard, 2004). The residue ranges used for RMSD calculations (Table 1) were identified using the FindCore2 algorithm (Snyder et al., 2014; Tejero et al., 2013).
Results
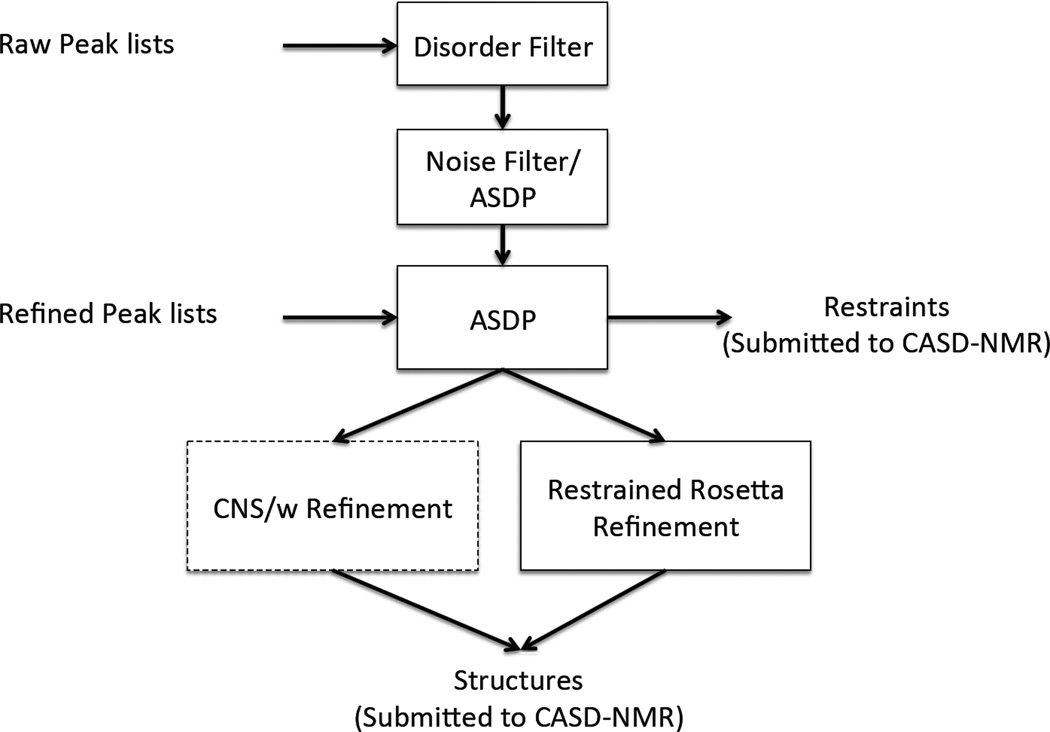
ASDP results for 10 blinded CASD-NMR datasets, each with raw and refined NOESY peak lists
The ASDP protocols used for the CASD-NMR-2013 experiments are summarized in Figure 1. The performance of ASDP with 10 blind datasets, using both raw and refined NOESY peaks lists, are summarized in Table 2 and Figure 2. NOESY peak lists were assigned using ASDP, and the structures were generated from these restraints using CYANA. The results structures were then refined with CNS in explicit water solvent (CNSw) (Brunger et al., 1998) or with restrained Rosetta (Mao et al., 2014). The DP score was calculated two ways. The value reported as the “DP value” in Table 2 is based on interproton distances averaged across the ensemble of 20 conformers, which is the conventional method of computing the DP score (Huang et al., 2005; Huang et al., 2012). In addition, a <DP> score was computed by determining the DP score for each of the 20 models in the ensemble, and then averaging these values. These are equivalent metrics, though the <DP> score is generally smaller than the DP score based on the average distance. RMSD, DP, and <DP> scores for structures generated from the raw unrefined peak list (after noise filtering) were computed by comparing models against these same unrefined peak lists; the scores for structures generated using manually-refined NOESY peak lists were computed by comparing models against the corresponding refined NOESY peak lists. The differences in RMSDs and DP scores for raw (after noise filtering) and refined peak lists are small, demonstrating that the peak editing methods used in ASDP are robust in removing random noise from these peak lists.
Fig. 1.
The ASDP protocols used for the CASD-NMR experiments. For unrefined raw peak list, the peak lists were filtered by both a Disorder Filter and a Noise Filter, followed by ASDP run together with dihedral angle constraints from Talos+ (Shen et al., 2009). For refined peak lists, no filters were applied to the peak lists. All structures were energy refined before submitted to the CASD using either CNSw (Brunger et al., 1998) or restrained Rosetta refinement (Mao et al., 2014). These resulting restraints and structures were submitted to the CASD-NMR server.
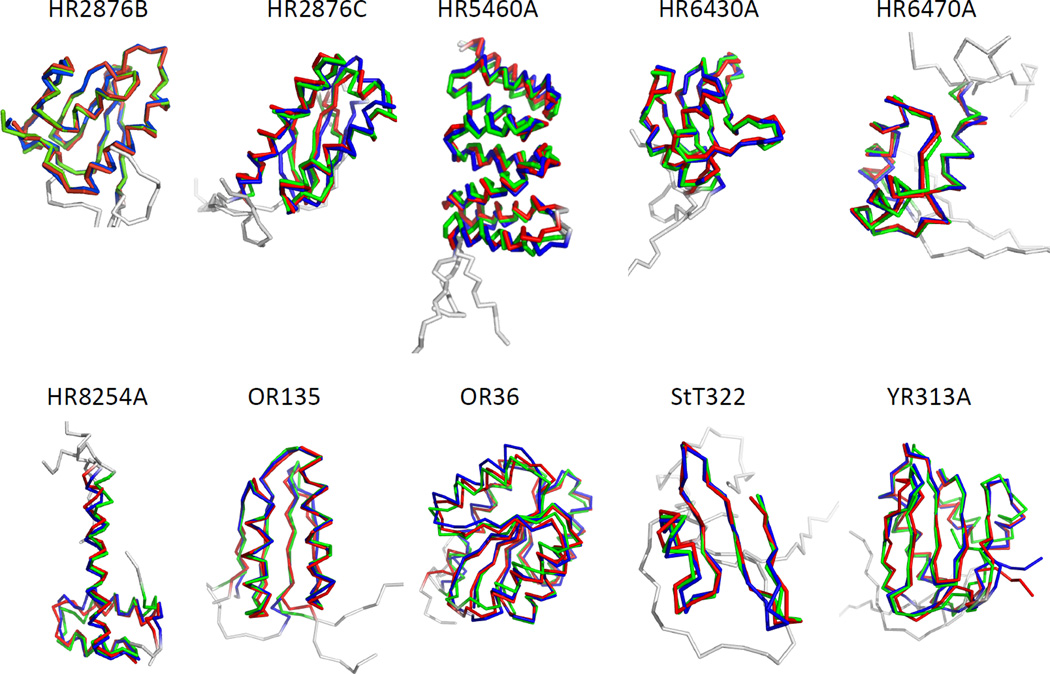
Fig. 2.
Superimposed diagrams for manually-refined models deposited in PDB (red), generated by ASDP using raw peak lists (blue), and generated by ASDP using refined peak lists (green) for all ten CASD-NMR targets. Flexible or disordered regions excluded from RMSD calculations are shown in gray.
Impact of using DP scores for guiding model selection when using unrefined NOESY peak lists
The DP score is a global measure of how well the model of the structure fits to the NOESY peak and chemical shift NMR data. Using the DP score for model selection can guide the ASDP program to generate globally-optimized models, and to utilize these models in its algorithms to rule-in and rule-out additional NOESY cross peak assignments. In particular, using the global DP score to distinguish more accurate from less accurate intermediate structures will reduce the use of inaccurate intermediate structures for assigning NOESY cross peaks. In order to assess this assumption, we also tested the performance of ASDP without using DP scores for intermediate model selection. The final DP, <DP>, and RMSD scores reported in Table 3 were calculated using the unrefined (raw) NOESY peak lists after automatic noise filtering using the preprocessing scripts of ASDP. Models were generated with or without DP filtering of intermediate structures, using ASDP for NOESY cross peak assignment and restraint generation, and Cyana for structure generation with the resulting restraints, with no further energy refinement. In eight of ten cases, trajectories using the DP score for model selection significantly improved the accuracy of the resulting structures (Table 3). These results demonstrate that the ASDP algorithm incorporating the DP filtering of intermediate structures is more robust to the noise peaks present in the NOESY peak lists than the corresponding algorithm that does not use the DP score for filtering out inaccurate decoy structures.
Table 3.
Improvement provided by combining target function and DP scores for model selection with filtered raw peak lists1
| Target Function only | Target Function + DP | |||||
|---|---|---|---|---|---|---|
| Name | DP | <DP> | RMSD(Å) | DP | <DP> | RMSD(Å) |
| HR2876Braw | 0.799 | 0.775 | 1.26 | 0.802 | 0.780 | 1.44 |
| HR2876Craw | 0.698 | 0.669 | 1.74 | 0.680 | 0.653 | 2.09 |
| HR5460Araw | 0.702 | 0.640 | 3.88 | 0.741 | 0.684 | 2.05 |
| HR6430Araw | 0.843 | 0.810 | 1.78 | 0.860 | 0.831 | 1.46 |
| HR6470Araw | 0.872 | 0.779 | 1.88 | 0.882 | 0.811 | 1.34 |
| HR8254Araw | 0.774 | 0.672 | 2.90 | 0.802 | 0.721 | 2.25 |
| OR135raw | 0.732 | 0.675 | 2.07 | 0.757 | 0.739 | 1.21 |
| OR36raw | 0.749 | 0.708 | 2.60 | 0.761 | 0.729 | 1.91 |
| StT322raw | 0.624 | 0.518 | 2.23 | 0.629 | 0.544 | 1.75 |
| YR313Araw | 0.643 | 0.596 | 1.37 | 0.655 | 0.598 | 1.34 |
Structures were assessed prior to energy refinement.
Impact of restrained CNS or Rosetta energy refinement
In this work, ASDP used Cyana structure generation methods for rapid calculations of models from restraints generated by ASDP, followed by restrained energy optimization using CNSw in explicit solvent or Rosetta. ASDP used CNSw for restrained energy refinement on 8 NMR data sets for 4 targets released in this cycle of CASD-NMR. During the CASD-NMR experiments, Mao et al developed a new restrained Rosetta refinement tool (Mao et al., 2014). Therefore, we replaced CNSw refinement with Rosetta refinement for the remaining 12 data sets provided for 6 targets (Table 2). Restrained CNSw or Rosetta energy refinement generally improved the RMSD and DP scores. This is illustrated by comparing the DP, <DP> and RMSD scores for the energy-refined structures generated using raw NOESY peak lists (Table 2) with the corresponding scores in Table 3 for structures generated using the same NOESY peak list data and protocols excluding energy refinement (i.e. “Target Function+DP” protocol of Table 3). For 9 of the 20 peak lists, we carried out both CNSw and restrained Rosetta energy refinement. Except for the refined peak list of HR8254A (discussed below), restrained Rosetta refinement generated models with either similar or smaller RMSD and higher DP scores than CNSw refinement, relative to the manually-refined reference structure (data not shown).
Restrained Rosetta refinement provides better sampling of the conformational space consistent with NMR data
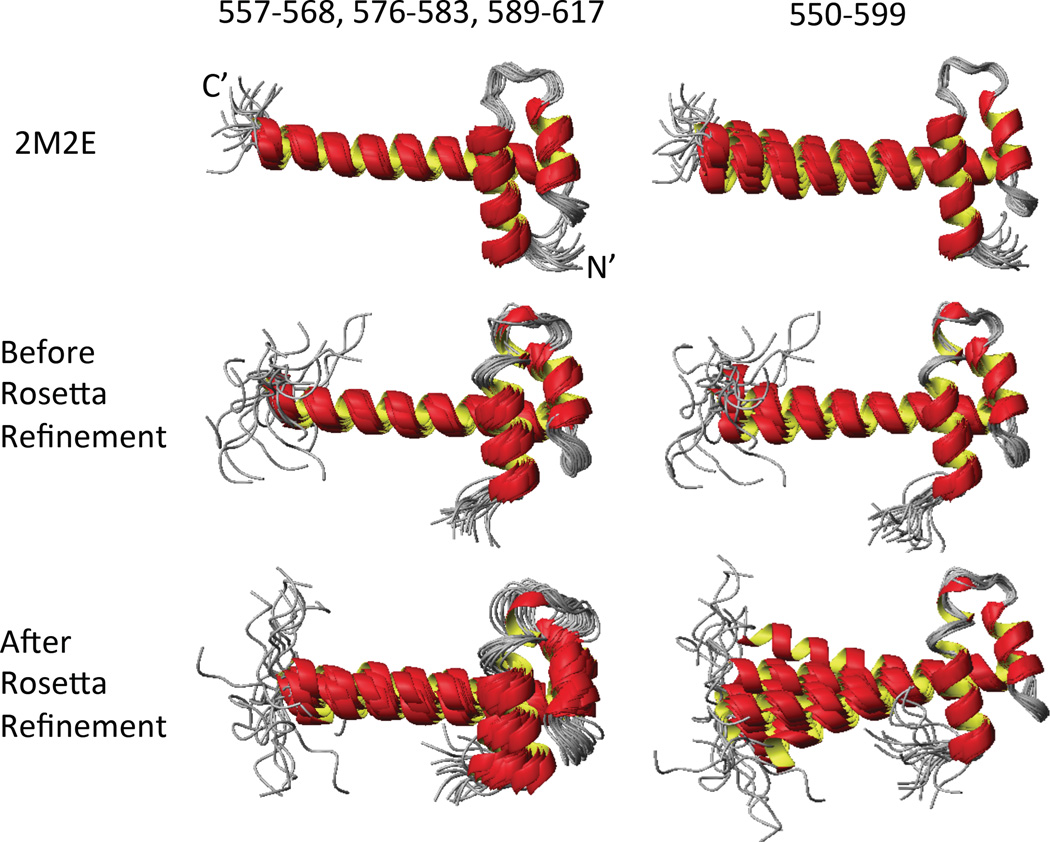
We also compared the backbone RMSD within the ensemble for targets before and after energy refinement. Figure 3 shows superimposed ribbon diagrams using different regions for three ensembles of target HR8254A: PDB ID 2M2E: DP=0.827, <DP>=0.752; ASDP structures before Rosetta refinement: DP = 0.802, <DP>=0.721; and ASDP structures after Rosetta refinement : DP=0.798, <DP>=0.735. The refined peak lists were used for these ASDP structure calculations. These ensembles have similar DP scores before and after Rosetta refinement; i.e. they are equally good fits to the NMR data. However, superimpositions (with the first 50 residues) show that helix-3 of target HR8254A has a wider range of conformations relative to the first two helices in the restrained-Rosetta refined structures. The similar DP scores between these two ASDP ensembles demonstrate that for this particular target the Rosetta refinement provides a broader sampling of the distribution of conformations that are equally consistent with the NOESY peak lists. RDC data was not available for target HR8254A, but would be helpful to resolve the uncertainty of the helix-3 tilt angle. We also carried out CNSw refinement for the models generated with the manually-refined NOESY peak list. The RMSD for the CNSw-refined structure was 1.48 Å, DP = 0.778, <DP> = 0.719. The DP scores of CNSw refined structures are essentially the same (slightly lower) than the DP scores of the Rosetta refined structures, while the RMSD of the CNSw-refined structures is much lower than the RMSD of the Rosetta refined structures. For this target, restrained Rosetta refinement provides broader sampling of the conformational space consistent with the NMR data, resulting in larger RMSDs of the Rosetta models relative to the manually-refined structure of HR8254A.
Fig. 3.
Superimposed ribbon diagrams using different regions for three HR8254A ensembles: 2M2E, ASDP structures before Rosetta refinement, and ASDP structure after Rosetta refinement. The ASDP structures were calculated using refined peaks list. Left – Residues 557–568,576–583 and 589–617 were used for superimposition. Right – Residues 550–599 were used for superimposition. The N- and C-termini of the ensemble in the upper left are labeled.
Assessment of NOESY peak filtering protocols
In this work, two protocols were used for automatic NOESY peak list filtering, referred to as CASE A and CASE B, as outlined in the Methods Section. Two targets (HR8254A and StT322) could not be addressed with CASE A peak filtering, and required the CASE B NOESY peak filtering. In the overall evaluation of CASD-NMR 2013 (Rosato et al, accompanying paper in this issue of JBNMR), these two targets were identified as particularly challenging; only a few of the participants in CASD-NMR 2013 provided any result at all using the raw NOESY peak lists for HR8254A and StT322. For these two targets, ASDP was successful in obtaining reasonably accurate structures using the CASE B filtering method. However, this rough filtering method appears to remove weak real peaks, which if preserved could improve the accuracy of ASDP structures.
In order to assess the robustness of the two peak filtering methods protocols (i.e., CASE A and CASE B), we calculated RPF scores for all 10 CASD-NMR reference structures (down loaded from the PDB) against (i) unfiltered raw NOESY peak lists, (ii) raw NOESY peak lists processed using the ASDP filtering protocols, and (iii) the manually-refined NOESY peak lists (Table 4). This study demonstrates that, as expected, the DP scores for the reference structures calculated with the manually-refined NOESY peak lists are > 0.73; the reference structures fit well to these refined NOESY peak lists. Only 2 of the raw NOESY peak list data sets (HR6430A, DP = 0.857 and HR6470A, DP = 0.849) have DP scores > 0.73. Indeed, based on the protocol CASE A, no filtering was required for the NOESY peak lists of these two targets. For the NOESY peak lists of 6 targets filtered using the protocol CASE A, 5 out of 6 have DP scores against the reference structures > 0.73. This analysis, demonstrates that NOESY peak filtering protocol CASE A can generally generate good quality NOESY peak lists. However, for targets HR8254A and StT322, which could not be filtered using protocol CASE A, the DP scores of the reference structure against NOESY peak lists filtered with protocol CASE B are 0.736 and 0.553, respectively, indicating that these NOESY peak lists filtered with protocol CASE B could potentially be improved. Indeed, while both automated NOESY peak list filtering protocols CASE A and CASE B provide good quality NMR structures (Table 1), comparisons of DP scores for the reference structures against (i) automatically filtered NOESY peak lists and (ii) manually-refined NOESY peak lists (Table 4) suggests that these data sets will be useful for developing improved NOESY peak list filtering algorithms.
Table 4.
RPF results for reference PDB structures with raw, filtered and refined NOESY peak lists
| Raw | Raw with noise filter | Manual | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Recall | Precision | DP | Recall | Precision | DP | Recall | Precision | DP |
| HR2876Ba | 0.645 | 0.940 | 0.590 | 0.862 | 0.875 | 0.787 | 0.989 | 0.963 | 0.919 |
| HR2876Ca | 0.737 | 0.951 | 0.645 | 0.802 | 0.904 | 0.707 | 0.959 | 0.976 | 0.891 |
| HR5460Aa | 0.680 | 0.884 | 0.592 | 0.859 | 0.840 | 0.736 | 0.965 | 0.956 | 0.860 |
| HR6430Ab | 0.958 | 0.955 | 0.857 | 0.958 | 0.955 | 0.857 | 0.986 | 0.973 | 0.932 |
| HR6470Ab | 0.975 | 0.934 | 0.849 | 0.975 | 0.934 | 0.849 | 0.993 | 0.960 | 0.898 |
| HR8254Ac | 0.434 | 0.972 | 0.394 | 0.901 | 0.849 | 0.736 | 0.965 | 0.936 | 0.827 |
| OR135a | 0.848 | 0.931 | 0.721 | 0.926 | 0.871 | 0.743 | 0.971 | 0.972 | 0.889 |
| OR36a | 0.706 | 0.911 | 0.579 | 0.898 | 0.853 | 0.733 | 0.990 | 0.966 | 0.904 |
| StT322c | 0.454 | 0.894 | 0.325 | 0.858 | 0.754 | 0.553 | 0.963 | 0.840 | 0.748 |
| YR313Aa | 0.627 | 0.913 | 0.456 | 0.832 | 0.857 | 0.642 | 0.988 | 0.947 | 0.848 |
The NOESY peak intensity filtering used protocol CASE A.
No NOESY peak intensity filtering was required.
The NOESY peak intensity filtering could not use protocol CASE A and instead used protocol CASE B.
Effects of simulated errors in resonance assignments
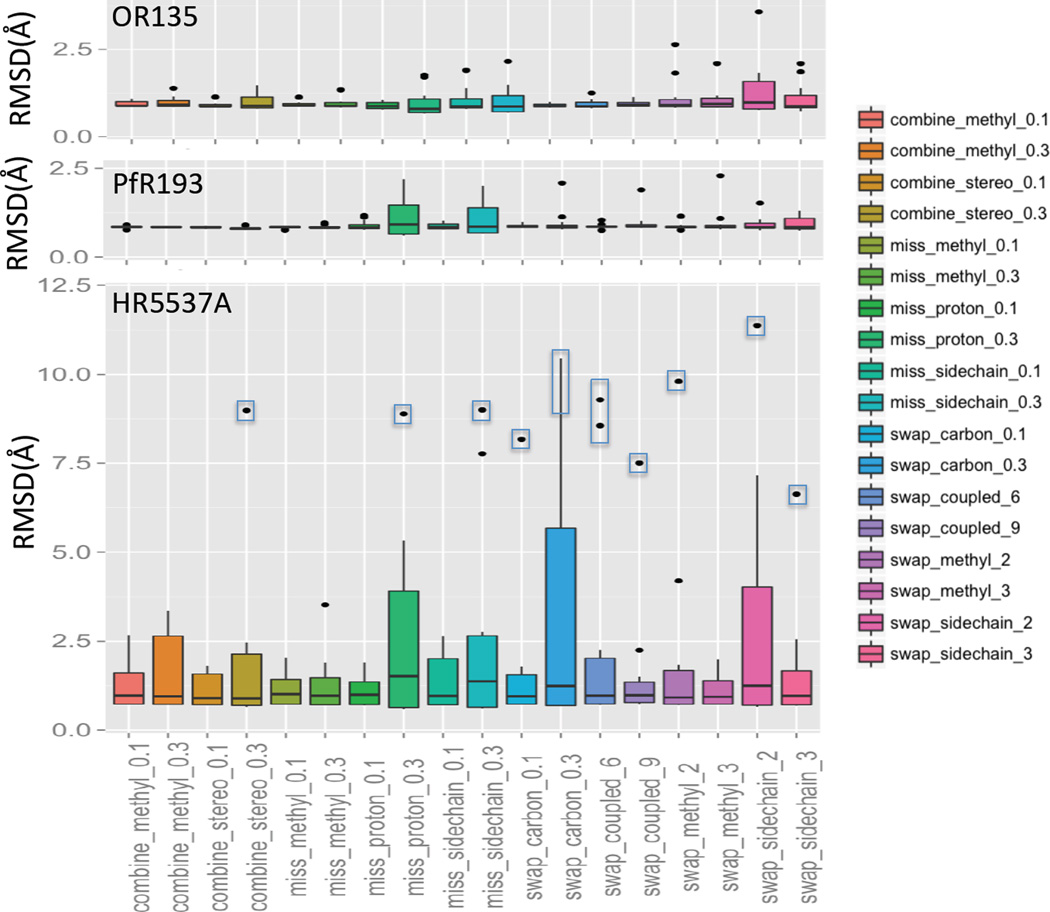
The resonance assignment table may contain some degree of incomplete and/or incorrect chemical shift assignments. Though rare, swapped or combined chemical shift assignments also can happen. Zhang et al. (Zhang, in preparation) have systematically simulated various input errors in the resonance assignment tables and tested the robustness of AutoNOE (Zhang et al., 2014), Cyana (Herrmann et al., 2002) and ASDP (Huang et al., 2005; Huang et al., 2006) to such errors. In these sensitivity tests, various random errors in the input resonance assignment table were simulated (Zhang et al, in preparation), including cases where 1) chemical shift assignments were combined, 2) chemical shift assignments were incomplete, and 3) chemical shift assignments were swapped (see Figure 4 legend for details).
Fig. 4.
The RMSD statistics of structures generated with ASDP for three data sets with various types of simulated chemical shift errors. The class of simulation is described along the X-axis. Six random errors were simulated for each class. The boxed dots were outliers used to test the two-run ASDP method. The reference structures for RMSD calculations are 2LN3 for OR135 (83 residues, alpha-beta fold), 2KL6 for PfR193A (114 residues, beta fold), and 2KK1 for HR5537A (135 residues, alpha fold). Secondary structure regions annotated in the X-ray PDB files were used to compute superimpositions for RMSD calculations. Random chemical shift assignment combination includes combining the methyl carbon and proton resonances of Leu, Ile and Val by 10% and 30% (combine_methyl_0.1, combine_methyl_0.3) and combining diastereo specifically assigned protons by 10% and 30% (combine_stereo_0.1, combine_stereo_0.3). Random resonance assignments incompleteness includes methyl groups were removed by 10% and 30% (miss_methyl_0.1, miss_methyl_0.3), protons were removed by 10% and 30% (miss_proton_0.1, miss_proton_0.3), and the whole side chain atoms were removed by 10% and 30% (miss_sidechain_0.1, miss_sidechain_0.3). Random chemical shift assignment swapping includes swapping between similar carbon assignments (i.e. with the same atom names) by 10% and 30% (swap_carbon_0.1 and swap_carbon_0.3), swapping between carbon-proton coupled assignments by 6 and 9 pairs (swap_coupled_6 and swap_coupled_9), swapping between methyl group assignments by 2 and 3 pairs (swap_methyl_2, swap_methyl_3), and swapping between the whole sidechain atom assignments for the same residue type by 2 and 3 pairs (swap_sidechain_2, swap_sidechain_3). These simulations of incomplete and/or incorrect NMR assignment tables were generated by Z. Zhang and O. Lange (Zhang et al, manuscript in preparation).
ASDP results for the OR135 data set from CASD-NMR-2013 using these simulated inaccurate chemical shift data are summarized in Figure 4 (top panel). ASDP was also tested using two additional datasets (HR5537A and PfR193) (Figure 4, middle and lower panels). In these tests, RMSD results are relative to corresponding X-ray crystal reference structures, including 2LN3 for OR135 (83 a.a, alpha-beta fold), 2KL6 for PfR193A (114 a.a, beta fold), and 2KK1 for HR5537A (135 a.a., alpha fold).
ASDP utilizes a topology-based algorithm to build secondary structures including anti-parallel and parallel beta-sheets from the unassigned back-bone NOEs, and Cα and Cβ resonance assignments in the first cycle before building any 3D structural model. This approach makes it less sensitive against errors in side-chain resonance assignments for beta only (PfR193) and beta-alpha (OR135) proteins, than for all-helical proteins (HR5537A) (Figure 4); all-helical proteins are more likely to be influenced by missing or scrambled assignments of sidechains than beta and beta-alpha proteins. In contrast, for beta-sheet containing proteins, tertiary structure is determined to a large extent by NOEs involving correct backbone proton assignments. Detailed comparison of the performance of ASDP with other NOESY assignments programs will be presented elsewhere (Zhang et al, in preparation).
Impact of resonance assignment errors on DP scores
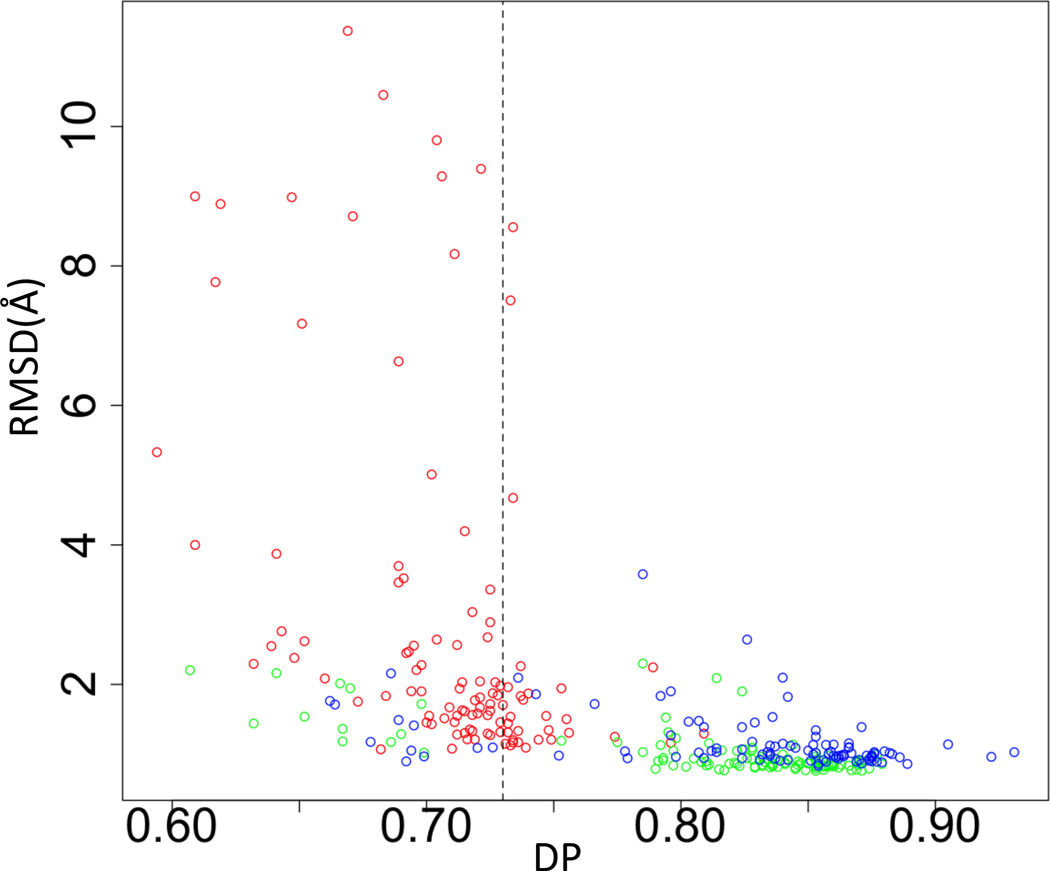
DP scores (Huang et al., 2005; Huang et al., 2012) were calculated for all the models generated with ASDP for data sets with simulated random resonance assignment errors (Figure 5). As in previous work (Huang et al., 2005; Huang et al., 2012), a strong correlation is observed between DP scores and structural accuracy measured by RMSD to reference structures (Figure 5). We have previously demonstrated that a DP cutoff of ~ 0.73 generally separates good from poor structures (Huang et al., 2005; Huang et al., 2012). The data in Figure. 5 demonstrate that even with incorrect chemical shift assignments, the DP cut off of > ~ 0.73 corresponds to a structural accuracy RMSD < ~ 2 Å. Accordingly, even when provided with inaccurate and/or incomplete resonance assignment data, highly inaccurate structures can be identified by the DP score, providing critical feed back that can be used to improve the quality of the input data.
Fig. 5.
The correlation between DP and RMSD for datasets with various simulated chemical shift errors. The value DP=0.73 is shown as vertical dashed line. Blue – OR135 results. Green – PfR193 results. Red – HR5537A results.
Effect of automated editing the input peak lists
In the first cycle of NOESY peak assignments, ASDP will uniquely assign only a very small fraction of long-range NOEs. Chemical shift assignment errors, like the simulated resonance assignment errors used in this study, can result in a small number of incorrect long-range NOE assignments, and very inaccurate structure can sometimes be generated. Indeed, for several of the simulated incorrect resonance assignment data sets, particularly for the all-helical HR5537A target, ASDP generated some highly inaccurate structures, with backbone RMSDs relative to the corresponding X-ray crystal structure > 5 Å (Figure 4. bottom panel). Fortunately, these erroneous restraints, which are mostly generated in the initial stages of ASDP analysis, can be detected and removed from iterative NOE analysis, because they tend to be strongly violated (>10 Å) in the final structures.
In our experience, the most severe incorrect NOESY cross peak assignments often result in interproton distance restraint violations greater than > 10 Å in the final structure. The corresponding NOESY cross peaks can thus be identified and removed for a second independent ASDP run. Using the resulting structures to clean up the initial set of distance restraints provides more accurate structures for subsequent restrained-energy optimization.
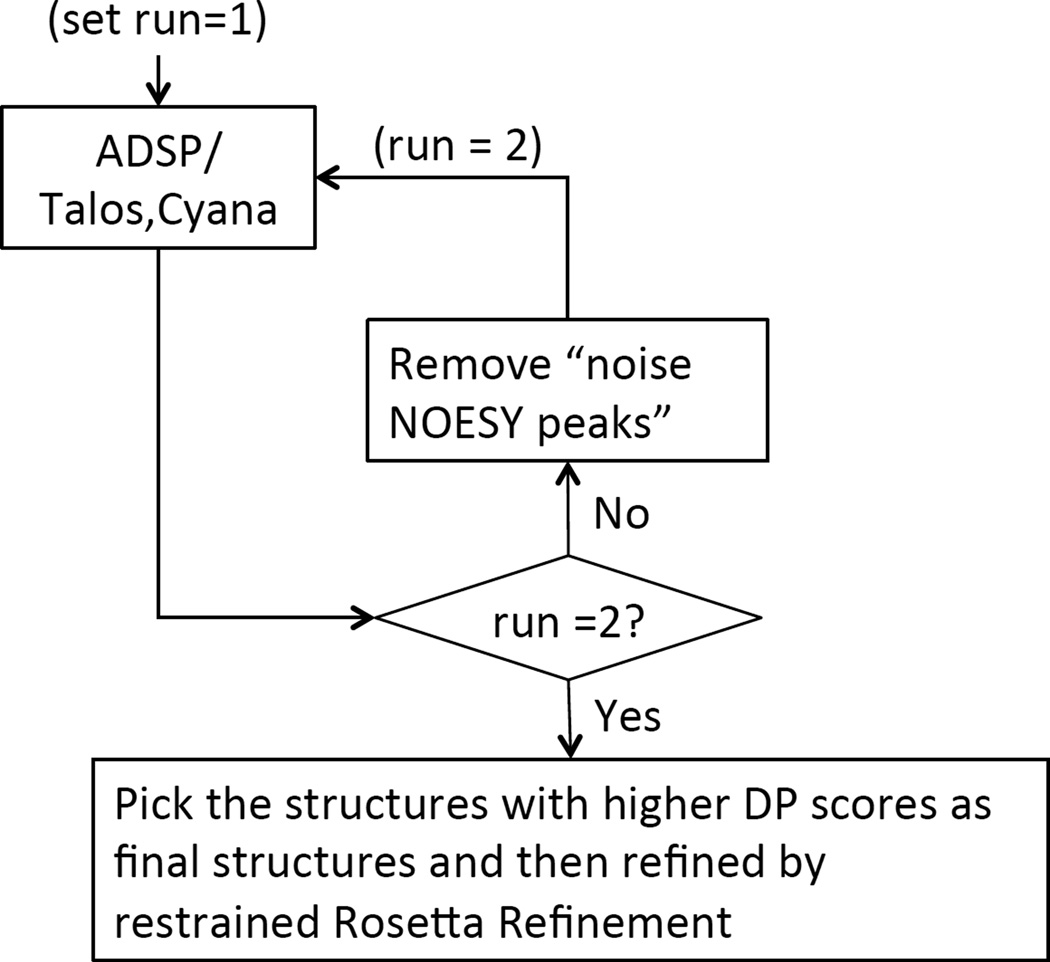
Driven by these data and the challenge of the CASD-NMR project, a new module was implemented in ASDP to automatically detect erroneous restraints resulting from inaccurate resonance assignments. This “two-run ASDP protocol” is illustrated in Figure 6. “Noise NOESY cross peaks” are defined as all NOESY cross peaks for which the corresponding restraint is violated by > 10 Å in all 20 conformers from the final cycle of first run. These “noise” NOESY cross peaks are removed for a second run of ASDP. The DP scores of the two individual runs are compared and the structures from the run with higher DP score are selected as the final ASDP results for further restrained Rosetta refinement.
Fig. 6.
The two-run ASDP flow chart. The NOE peaks that are inconsistent with the protein structure models from ASDP are identified after run1. These “noise” data are then removed from the input data, and ASDP calculations are repeated. The DP scores of structures from run 1 and run 2 are compared and the ones with higher DP scores are picked as the final structures for further restrained Rosetta refinement.
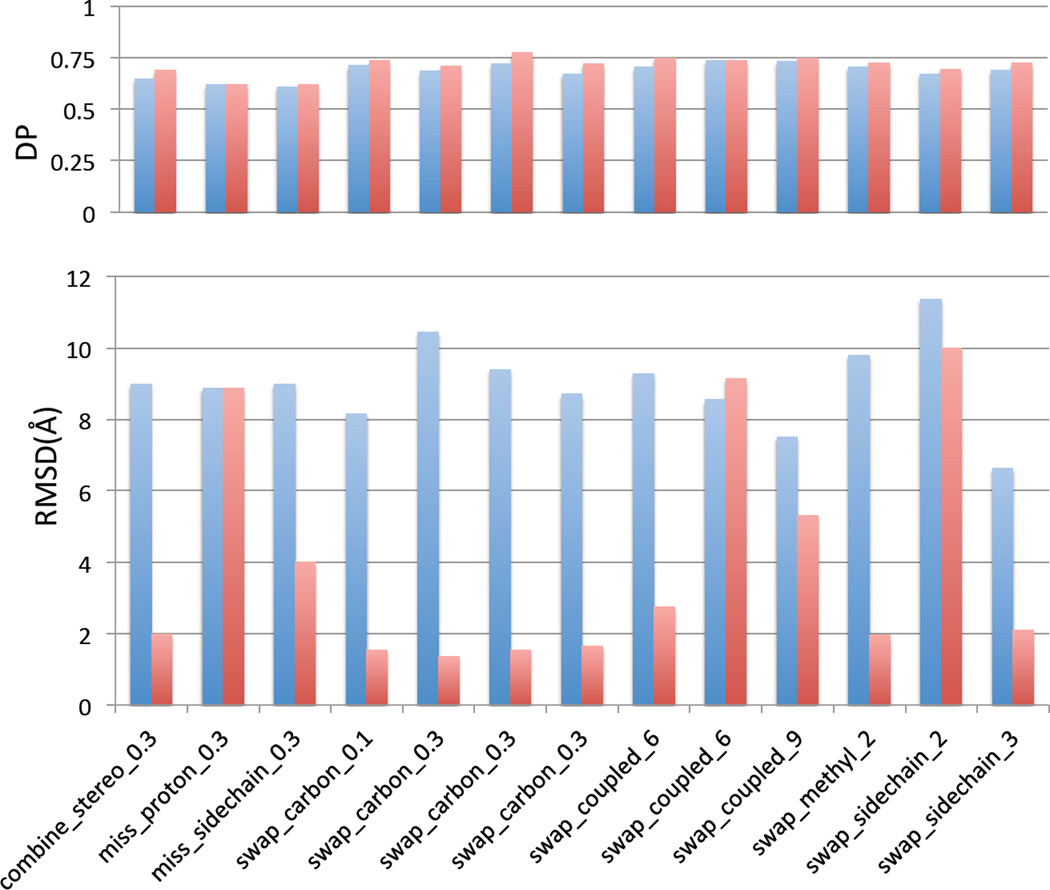
To test this “two run protocol”, we selected 13 outlier structures generated using the simulated inaccurate chemical shift list (marked as boxed dots in Figure 4 HR5537A panel). As demonstrated in the results presented in Figure 7, the two-run ASDP protocol resulted in more accurate structures, with smaller RMSDs compared to X-ray crystal reference structures, for 10 of the 13 outliers. The protocol allowed for identification and exclusion of NOESY cross peaks involving misassigned resonances, providing information which could be potentially used to correct these resonance assignments. For the remaining 3 outliers tested, the assignment inaccuracy was not sufficient to generate consistent violations of > 10 Å in the final structure, and other methods to identify these kinds of errors in resonance assignments still need to be developed. In any case, it is clearly important to identify potential errors in NOESY peak lists and resonance assignments, and to prepare high quality input data in order to generate high-quality structures..
Fig. 7.
Comparison of one-run and two-run ASDPs for outlier structures produced with simulated chemical shift errors. Blue – the DP and RMSD for one-run ASDP. Red – the DP and RMSD for two-run ASDP.
Discussion
We have improved the robustness and accuracy of automated NMR structure determination with AutoStructure by 1) using a global optimization DP score to guide the iterative NOESY cross peak assignment process, 2) identifying input noise and automatically filtering input NOESY peak lists, and 3) identifying incorrect NOESY cross peak assignments caused by errors in resonance assignments and excluding these from the structure determination process. This improved version of the program has been renamed ASDP.
A DP-score-based noise filter was implemented to identify weak noise peaks in the raw NOESY peak lists. We have tested this ASDP “peak intensity filter” with CASD-NMR blinded NOESY data sets, using all 10 raw peak lists. The small differences in RMSDs and DP scores of structures generated from “raw” NOESY peak lists processed with this noise filter compared with structures generated using manually-refined NOESY peak lists demonstrates that the peak editing methods used by ASDP are robust in eliminating noise in these peak lists, thus providing accurate structures in a fully automated analysis.
We have previously reported a high correlation between accuracy (i.e. RMSD to the manually-refined structure) and DP scores in comparing various kinds of decoy structures with manually-refined NOESY peak lists (Huang et al., 2005; Huang et al., 2012). In this work, we also observe good correlation between structural accuracy and DP scores (or <DP> scores) when using raw peak lists processed automatically with the preprocessing scripts of ASDP. These results demonstrate the robustness of the DP score for structure validation, even with different qualities of NOESY peak list data. The results further suggest that the DP score measurement can be potentially applied directly to NOESY spectra (FIDs), followed by automated peak picking and noise filtering, as described in this study.
Using the data in Table 4, we also assessed the degree to which our NOESY peak filtering protocols, CASE A and CASE B, may remove real peaks from the NOESY peak list. The Recall score of the RPF_DP metric is a measure of the percentage of peaks in the NOESY peak list that are consistent with the protein structure model. Noise peaks are generally not consistent with the reference structure, and their presence in the peak list will reduce the Recall score measured against the reference structure. The Precision score is a measure of the percentage of short distances in the protein structure model which are not represented in the NOESY peak list. If real peaks are eliminated from the NOESY peak list by a filtering process, this will reduce the precision score, measured against the reference structure. For the 6 data sets processed with CASE A, the peak intensity filtering significantly increased the Recall, demonstrating removal of noise peaks. The corresponding Precision scores decreased slightly (or not at all), demonstrating good preservation of real peaks. However, both the Recall and Precision scores are generally higher for the manually-refined NOESY peak lists than the peak lists filtered using the CASE A method. For the 2 data sets which could not be processed with CASE A, and were processed with CASE B (HR8254A and StT322), the CASE B filtering significantly increased the Recall scores, from 0.43–0.45 to 0.86–0.90 (Table 4), demonstrating significant reduction in noise peaks. However, the corresponding Precision scores also drop significantly (from 0.89 – 0.97 to 0.75 – 0.85). The manually-refined NOESY peak lists have significantly higher Recall and Precision scores for these two reference structures. The CASE B filtering successfully removes noise peaks, but also reduces the number of real peaks in the NOESY peak list. These CASD-2013 data sets provide good test cases for the development of more robust NOESY peak filtering tools.
Protein NMR structure determination is essentially restraint-based modeling, an area of active development in the protein structure prediction community (Monastyrskyy et al., 2014; Moult et al., 2014). Modeling methods are broadly categorized in two broad classes: “knowledge-based” methods which rely on sampling from protein conformations observed in experimental protein structures available in the Protein Data Bank, and “physics-based methods”, which utilize empirical (or even ab initio force) fields, in molecular mechanics calculations. These distinctions are not mutually-exclusive, as most knowledge-based modeling methods also utilize physics-based force fields, and physics-based methods also utilize information from small molecule, or even protein structure data bases. Comparisons of this spectrum of methods for protein NMR structure refinement is an active area of research (Mao et al., 2014; Tejero et al., 2013).
In this work, we compared a largely knowledge-based method, restrained-refinement of CASD-NMR structures generated with Rosetta utilizing polypeptide fragments generated from protein structures available in the PDB, with a physics-based method, restrained refinement with CNSw, using simulated annealing of short molecular dynamics trajectories in explicit solvent. Restrained CNSw or Rosetta energy refinement generally improves the RMSD (accuracy to the manually-refined structure) and DP (fit of the model to the NOESY and chemical shift assignment data) scores. Restrained Rosetta energy refinement generally generated models with smaller RMSD relative to the manually-refined reference structure, and higher DP scores relative to NOESY peak lists, compared with CNSw. Similar improved performance of restrained Rosetta compared with CNSw energy refinement has been reported in our previous papers using NMR – X-ray pairs available for the same protein targets (Mao et al., 2014; Tejero et al., 2013). However, this does not demonstrate that knowledge-based methods are generally superior to physics-based methods for protein NMR structure refinement, as this field is still evolving, and improvements in protocols for both physics-based and knowledge-based protein structure modeling with NMR data can be anticipated in the coming years.
We also tested the robustness of ASDP to simulated errors in resonance assignments. Considering the underlying algorithms of ASDP, all-helical proteins are more likely to be influenced by missing or scrambled assignments of side chains than beta and alpha-beta proteins. However, the new “two-run ASDP protocol” can identify structural inconsistencies caused by some inaccuracies in the NMR resonance assignments. While further development of methods to detect incorrect resonance assignments are needed, these results demonstrate that such two-run protocols can identify some of the kinds of errors that result from misassignment of NMR resonances.
ASDP exhibited reliable performance on all of the 10 CASD-NMR targets released, using both raw and manually-refined peak lists. Other members of the Montelione laboratory contributed to the CASD-NMR project by generously providing the NOESY data sets and manually-refined reference structures for eight of these CASD-NMR targets; i.e. all protein targets except HR8254A and StT322. In order to ensure blind tests of the ASDP protocol, a special data handling process was set up in the Montelione laboratory, so that information about NOESY peak list data and the manually-refined reference structures were not shared by the subgroup doing the blinded, fully-automated ASDP calculations, until the reference structures were released by the CASD-NMR-2013 organizers. As these ASDP calculations were carried out in an automated fashion, they can be repeated using the CASD-NMR-2013 data and the released ASDP software.
It is clearly important to identify potential errors in NOESY peak lists and resonance assignment table and to prepare high quality input data in order to generate high quality structures. Meanwhile, it is also useful to develop automatic tools, which help users to identify potential noise peaks in NOESY spectra and/or incorrect resonance assignments. For example, a unique feature of DP score is that it can detect such errors in the input data by studying the false positive and false negative error reports, which direct the user to the specific NOESY peaks which are inconsistent with protein structure models. These features of the DP analysis provide feedback to the user that is useful to improve the quality of the input data (Huang et al., 2005; Huang et al., 2012). Additional user-friendly tools are under development to further utilize the false positive and false negative error reports generated by NMR DP analysis.
Acknowledgments
We thank all of the members of the Northeast Structural Genomics Consortium who generated and archived NMR data used in the CASD-NMR project, particularly scientists in the laboratories of C. Arrowsmith, M. Kennedy, G.T. Montelione, T. Szyperski, and J. Prestegard. We also thank Z. Zhang and O. Lange for providing the simulated incorrect resonance assignment tables used for testing. This work was supported by a grant from the National Institutes of Health Protein Structure Initiative grant U54-GM094597 to GTM, and by the Jerome and Lorraine Aresty Charitable Foundation.
Footnotes
Software Availability
ASDP v1.0 and associated scripts are available at http://www-nmr.cabm.rutgers.edu/NMRsoftware/asdp/Home.html. The C++/perl source codes are also available at the same site.
References
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallographica Section D, Biological crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. Journal of molecular biology. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. Journal of molecular biology. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Acton TB, Montelione GT. DisMeta: a meta server for construct design and optimization. Methods in molecular biology. 2014;1091:3–16. doi: 10.1007/978-1-62703-691-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Rosato A, Singh G, Montelione GT. RPF: a quality assessment tool for protein NMR structures. Nucleic acids research. 2012;40:W542–W546. doi: 10.1093/nar/gks373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, Vakser I, Wodak SJ Critical Assessment of, P.I. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- Lange OF, Rossi P, Sgourakis NG, Song Y, Lee HW, Aramini JM, Ertekin A, Xiao R, Acton TB, Montelione GT, et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci U S A. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Kim JH, Westler WM, Markley JL. PONDEROSA, an automated 3D-NOESY peak picking program, enables automated protein structure determination. Bioinformatics. 2011;27:1727–1728. doi: 10.1093/bioinformatics/btr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Tejero R, Baker D, Montelione GT. Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures. J Am Chem Soc. 2014;136:1893–1906. doi: 10.1021/ja409845w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrskyy B, D'Andrea D, Fidelis K, Tramontano A, Kryshtafovych A. Evaluation of residue-residue contact prediction in CASP10. Proteins. 2014;82(Suppl 2):138–153. doi: 10.1002/prot.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)--round x. Proteins. 2014;82(Suppl 2):1–6. doi: 10.1002/prot.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995;23:ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- Nilges M. Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. Journal of molecular biology. 1995;245:645–660. doi: 10.1006/jmbi.1994.0053. [DOI] [PubMed] [Google Scholar]
- Nilges M, Macias MJ, O'Donoghue SI, Oschkinat H. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin. Journal of molecular biology. 1997;269:408–422. doi: 10.1006/jmbi.1997.1044. [DOI] [PubMed] [Google Scholar]
- Raman S, Huang YJ, Mao B, Rossi P, Aramini JM, Liu G, Montelione GT, Baker D. Accurate automated protein NMR structure determination using unassigned NOESY data. J Am Chem Soc. 2010a;132:202–207. doi: 10.1021/ja905934c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, et al. NMR structure determination for larger proteins using backbone-only data. Science. 2010b;327:1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato A, Aramini JM, Arrowsmith C, Bagaria A, Baker D, Cavalli A, Doreleijers JF, Eletsky A, Giachetti A, Guerry P, et al. Blind testing of routine, fully automated determination of protein structures from NMR data. Structure. 2012;20:227–236. doi: 10.1016/j.str.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Guntert P, Herrmann T, et al. CASD-NMR: critical assessment of automated structure determination by NMR. Nature methods. 2009;6:625–626. doi: 10.1038/nmeth0909-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. The PyMOL Molecular Graphics System [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DA, Grullon J, Huang YJ, Tejero R, Montelione GT. The expanded FindCore method for identification of a core atom set for assessment of protein structure prediction. Proteins. 2014;82(Suppl 2):219–230. doi: 10.1002/prot.24490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejero R, Snyder D, Mao B, Aramini JM, Montelione GT. PDBStat: a universal restraint converter and restraint analysis software package for protein NMR. J Biomol NMR. 2013;56:337–351. doi: 10.1007/s10858-013-9753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Porter J, Tripsianes K, Lange OF. Robust and highly accurate automatic NOESY assignment and structure determination with Rosetta. J Biomol NMR. 2014;59:135–145. doi: 10.1007/s10858-014-9832-4. [DOI] [PubMed] [Google Scholar]
- Zhang Z, X F, Huang YJ, Tripsiances K, Montelione G, Lange OF. Effect of incorrect chemical shift assignments on automated NOE assignments and NMR structure calculation. (in preparation) [Google Scholar]