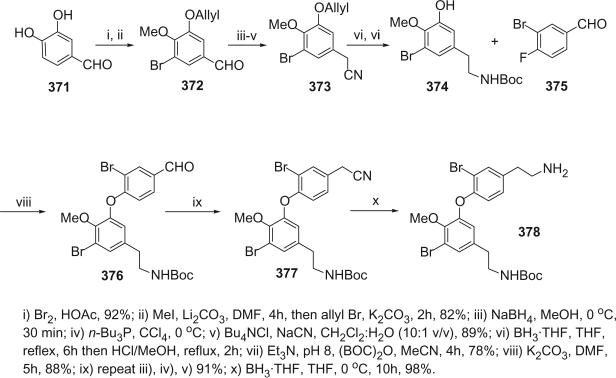
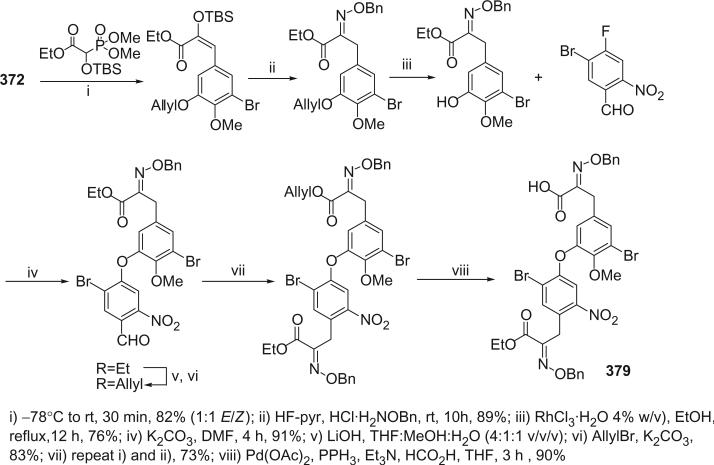
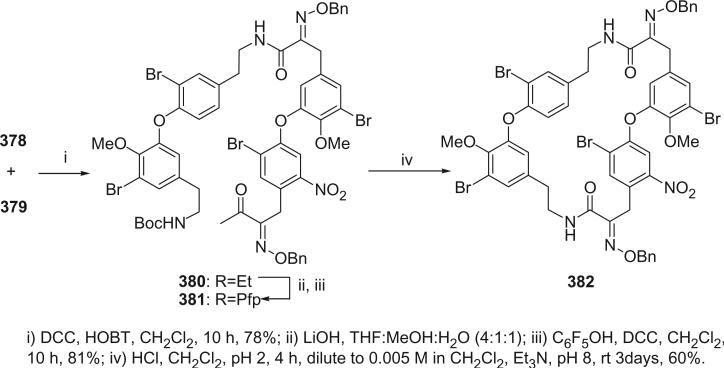
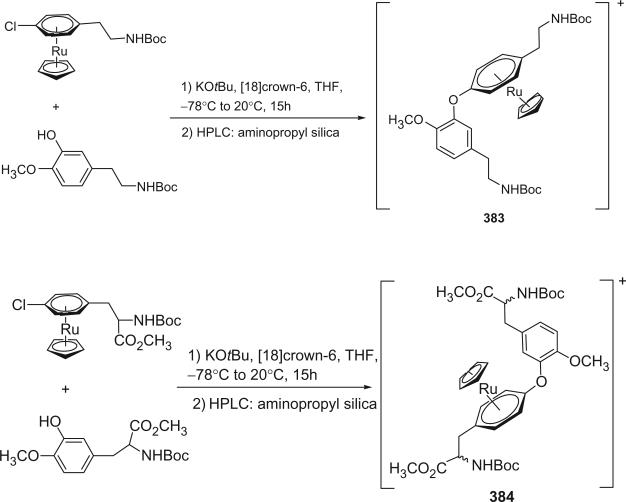
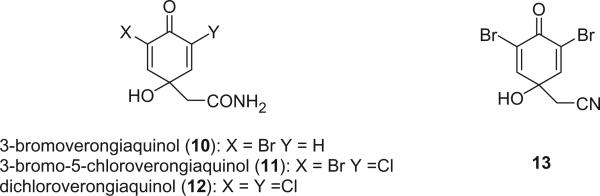I. Introduction
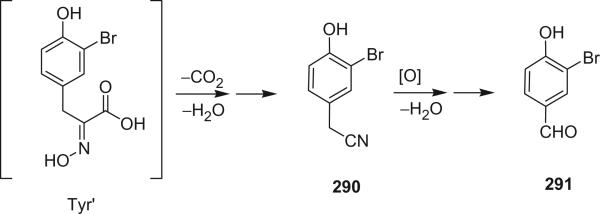
The isolation of bromotyrosine secondary metabolites from marine organisms can be traced back to 1913, when Morner reported the isolation of dibromotyrosine from two coral species (1). There were no reports provided for these secondary metabolites again until 1967, when Sharma and Burkholder isolated 2,6-dibromo-4-acetamide-4-hydroxycyclohexadienone (1) and the dimethoxyketal 2 from two marine sponges Verongia fistularis and V. cauliformis (2–4). Since then, driven by the diverse bioactivities, more and more bromotyrosine-derived marine natural products have been reported. To date, there are over 280 bromotyrosine-derived alkaloids reported from marine invertebrates with a variety of biological activities including: antimicrobial, anticancer, antifouling, antiviral, ATPase regulator, calcium channel modulator, etc.
In this review, we discuss the isolation, structure, physicochemical and spectral data of all bromotyrosine derivatives isolated from marine organisms. The biosynthesis, total synthesis, and bioactivity of the bromotyrosine derivatives are also reviewed. Neither tyrosine derivatives without halogenation, nor indole alkaloids (with or without halogenation), are included in this review. Proteins or peptides containing bromotyrosine units are not included in this review since they are considered as primary metabolites. Cyclopeptides containing halogenated tyrosine units are, however, discussed in this review.
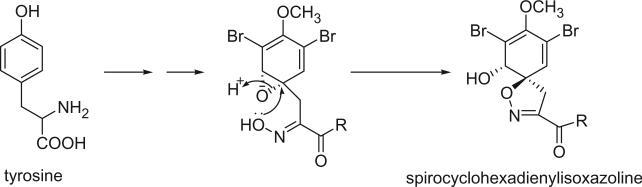
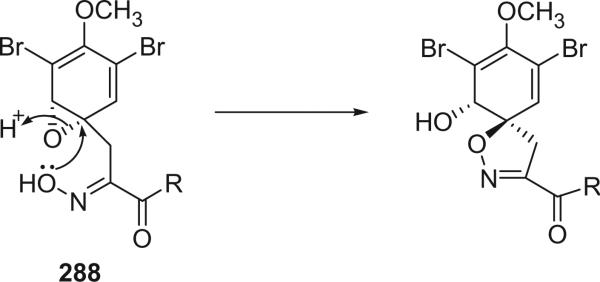
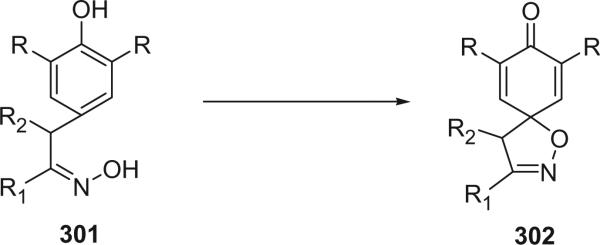
For convenience, the bromotyrosine derivatives are divided into six categories: simple bromotyrosine derivatives, spirocyclohexadienylisoxazolines, spirooxepinisoxazolines, oximes, bastadins, and other structural classes. The simple bromotyrosine derivatives are products of one bromotyrosine undergoing degradation, reduction, hydroxylation, alkylation, or esterification with simple functional groups. In spirocyclohexadienylisoxazoline bromotyrosine derivatives, one or two bromotyrosine units are transformed into a spirocyclohexadienylisoxazoline undergoing an arene oxide biosynthetic pathway. This class of alkaloids generally consists of one to three bromotyrosine-derived units, as well as other functional groups, such as histamine.
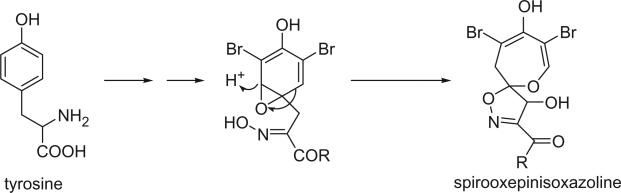
In the spirooxepinisoxazoline bromotyrosine derivatives, one bromotyrosine is transferred into a spirooxepinisoxazoline. There are only eight alkaloids in this class reported to date.
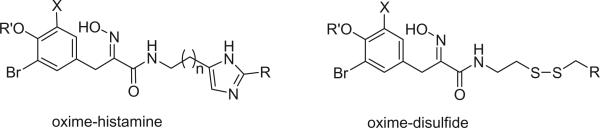
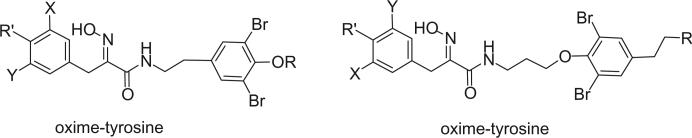
The amine functionality is transferred into an oxime in the oxime class of bromotyrosine derivatives. The geometries of the oxime functionalities were determined to be E in almost every case of this class of compounds. Although the geometries of some alkaloids in this class were not reported, it is easy to assign the E geometries for most of them from the 13C NMR data. There are basically three structural groups in this class of bromotyrosine derivatives. The first group of alkaloids consists of a bromotyrosine oxime and a histamine moiety. The second group of alkaloids has one or two bromotyrosine oximes connected with a disulfide chain, cysteine. The third group of alkaloids has a bromotyrosine oxime connected to a bromotyramine directly or through a three carbon chain.
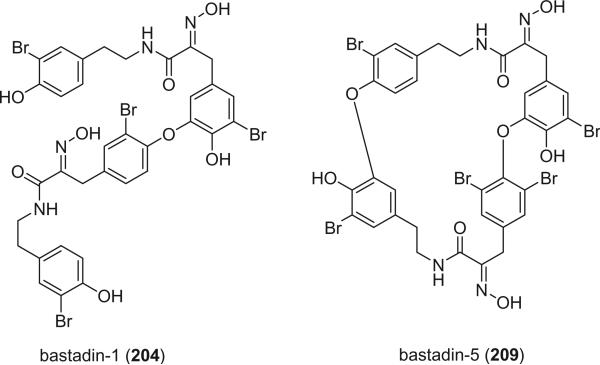
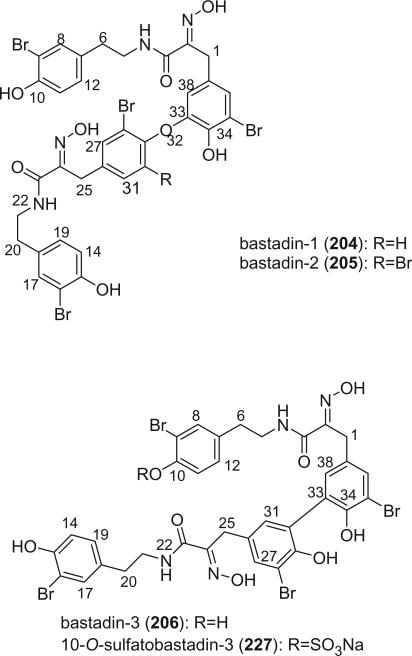
The bastadins are a series of predominantly macrocyclic bromotyrosine derivatives, which are biogenetically derivable from four bromotyrosines by the oxidative phenolic coupling of two tyramine–tyrosine units connected through an amide bond. Until now, there are four acyclic, twenty cyclic bastadins, and sixteen hemibastadins isolated from marine sponges and ascidians. Examples of this class of alkaloids are bastadins-1 (204) and −5 (209).
There are a number bromotyrosine derived compounds not belonging to any of the above structure classes. Geodiamolides, a series of cyclic depsipeptides, are included in this review since they contain halogenated tyrosine. Polycitones and polycitrins are condensation products of substituted bromotyrosine molecules, isolated from ascidians, and are also included in this review. Similar structures including lamellarins are not included due to the absence of halogenation. For the same reason, polyandrocarpamides A-C (253–255), chelonin B (256), and 5-bromochelonin B (257) are included.
II. Isolation and Structure Elucidation
A. SIMPLE BROMOTYROSINE DERIVATIVES
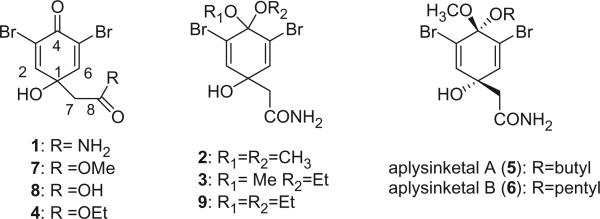
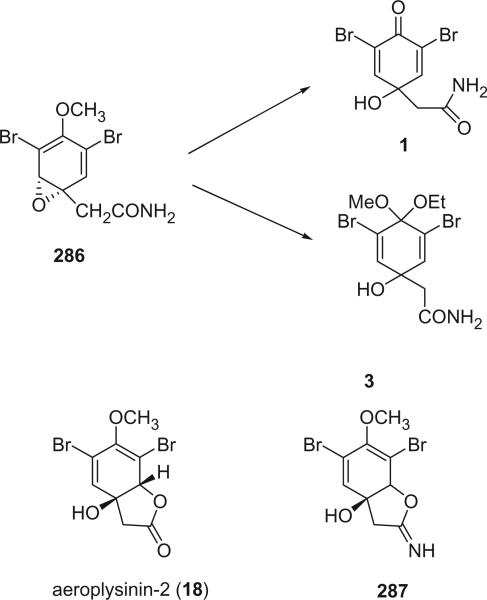
The first two members of this series, 2,6-dibromo-4-acetamide-4-hydroxycyclohexadienone (1) and the dimethoxyketal 2, were isolated from the methanolic extracts of Verongia fistularis and V. cauliformis by Sharma and Burkholder in 1967 (2–4). Anderson and Faulkner isolated a mixed methoxy–ethoxy ketal 3 from Verongia sp. in 1973. Since the 1H NMR spectrum of 3 showed two methoxy signals, indicating that 3 is a mixture of two diastereoisomers, 2 and 3 were considered as artifacts generated during the extraction process (5). Faulkner et al. also reported the dienone 4 from Tylodina fungina (6). Two additional mixed ketals, aplysinketal A (5) and aplysinketal B (6), along with 7 and 8, were obtained from the Mexican sponge Aplysina (Verongia) thiona (7). Unlike 2 and 3, the structure of aplysinketal A (5) was shown to be only one of the two diastereoisomers by X-ray and NMR data. Furthermore, the mixed ketals 5 and 6 would not be expected to be formed without simultaneous formation of the dimethoxy ketal 2, which was never detected. According to these results, aplysinketal A (5) and aplysinketal B (6) are very likely to be natural products. The diethyl ketal 9 was obtained from a Turkish sponge V. aerophoba (8).
Monobromo- (10), bromochloro- (11), and dichloro- (12) dienones were isolated from Aplysina cavernicola (9,10). Both 10 and 11 were isolated as racemic mixtures. The dienone 13 was first obtained as a synthetic product after treatment of aeroplysinin-1 (14) with trifluoroacetic acid (11). Our group isolated 13 from the Jamaican sponge Verongula gigantea as a natural product (12).
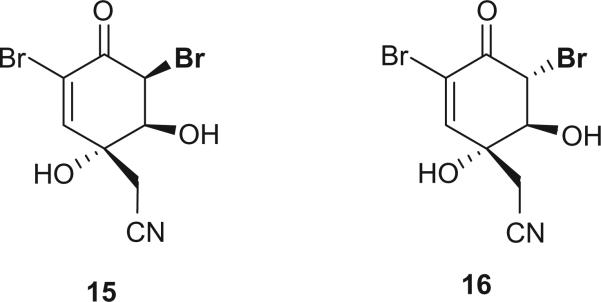
A 3:1 mixture of the two epimeric dibromonitriles, 15 and 16, was isolated from an Australian sponge Aplysina laevis (11). Attempts to resolve the mixture by either normal or reverse-phase HPLC, as well as GC, proved unsuccessful. Treatment of (+)-aeroplysinin-1 (14) with neat trifluoroacetic acid resulted in a good yield of 15 and 16 (3:1 mixture) with a small optical rotation value, which permitted the assignment of the absolute stereochemistries of 15 and 16 as shown. Molecular modeling calculations suggested that the cis isomer 15 was the thermodynamically more stable isomer.
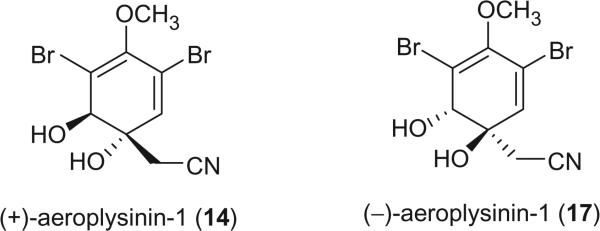
Aeroplysinin-1 (14), the first naturally occurring 1,2-dihydroarene-1,2-diol (13), was initially isolated as a dextrorotatory isomer from V. aerophoba [the species name was later revised to V. cavernicola (72)] (14,15). Fulmor et al. isolated the laevorotatory antipode of aeroplysinin-1 from a closely related sponge Ianthella ardis, for which the absolute configuration was proposed as shown in 17 on the basis of chemical, CD and NMR data (16). The absolute stereochemistries of both antipodes, as shown in 14 and 17, were firmly established by X-ray diffraction analysis (17,18).
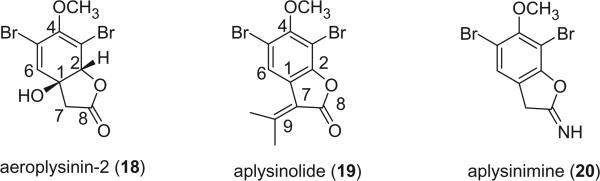
Aeroplysinin-2 (18), the first bromotyrosine derivative with a lactone functionality, was isolated from V. aerophoba in 1972 [the species name was later revised to V. cavernicola (69)] (19), and its structure was established on the basis of proton NMR and chemical methods. The small coupling (0.7 Hz) between the olefinic and methine protons suggested a W relationship for these two protons, indicating a quasi-equatorial orientation for the methine proton and accordingly, a quasi-diaxial orientation for the hydroxyl- and acyloxy-groups. The circular dichroism curve indicated a right-handed helicity for the diene, and therefore confirmed the absolute configuration as depicted in 18. Aplysinolide (19) and aplysinimine (20) were obtained from Aplysina (Verongia) thiona (7).
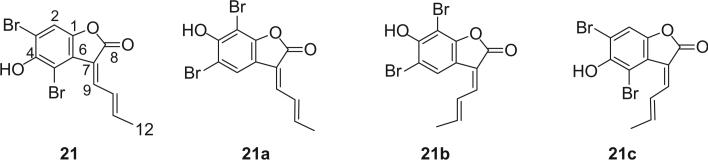
The isolation of aplysinimine (20) is remarkable because this alkaloid could be considered as a possible precursor of other bromo compounds obtained from Aplysina sponges. Aplysinolide (19) has an unusual α,β-unsaturated side chain. Another bromotyrosine derivative containing an α,β-unsaturated side chain is aplysinadiene (21), isolated from Aplysina aerophoba (20). The structure was determined by comparison of the NMR data with the synthesized isomers 21a and 21b. Structure 21c was precluded due to the interaction of the bromine atom with the butenylide side chain (21).
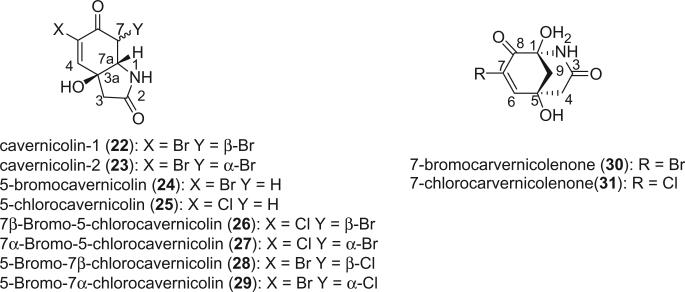
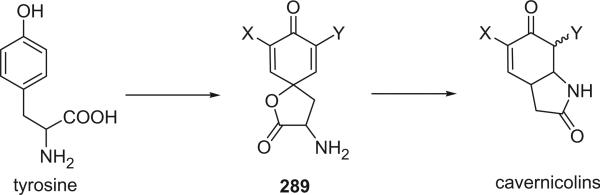
Eight lactams, including cavernicolin-1 (22), cavernicolin-2 (23), 5-bromocavernicolin (24), 5-chlorocavernicolin (25), 7β-bromo-5-chlorocavernicolin (26), 7α-bromo-5-chlorocavernicolin (27), 5-bromo-7β-chlorocavernicolin (28), and 5-bromo-7α-chlorocavernicolin (29), were identified from V. cavernicola (9,22,23). 5-Bromocavernicolin (24) and 5-chlorocavernicolin (25) were the first examples of marine products with low enantiomeric purity. Cavernicolin-1 (22) and cavernicolin-2 (23), 7β-bromo-5-chlorocavernicolin (26) and 7α-bromo-5-chlorocavernicolin (27), 5-bromo-7β-chlorocavernicolin (28) and 5-bromo-7α-chlorocavernicolin (29), could be separated by HPLC, but they quickly equilibrate as a 3:1 mixture (9).
Two δ-lactams, 7-bromocarvernicolenone (30) and 7-chlorocarvernicolenone (31), were also isolated from V. cavernicola (24, 25) and exhibited mild antibacterial activities. The structure and relative stereochemistry of 7-bromocarvernicolenone (30) was confirmed by X-ray diffraction analysis. 7-Bromocarvernicolenone (30) and 7-chlorocarvernicolenone (31) are additional examples of marine natural products having low enantiomeric purity.
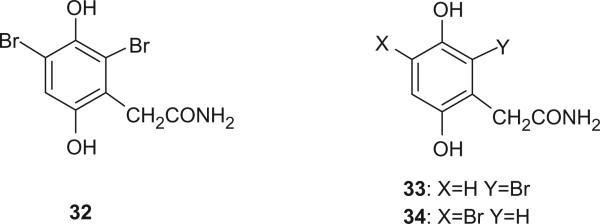
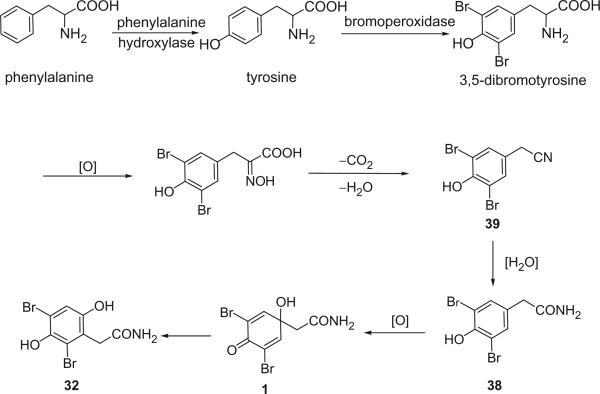
The first, skeletally rearranged dibromotyrosine metabolite, and also the first hydroquinone in this family of marine natural products, 32, whose structure was determined by X-ray crystallography, was isolated from Verongia aurea, along with 33 or 34, as detected by gas chromatography–mass spectrometry of the ether extract (26). It represented a major departure from the normal dibromotyrosine metabolites, in which the aliphatic side chain remained in the para position relative to the hydroxyl group flanked by bromine atoms. An analogy for such a rearrangement of the tyrosine skeleton is available, however, in the conversion of 4-hydroxyphenylpyruvic acid into 2,5-dihydroxyphenylacetic (homogentisic acid), catalyzed by an enzyme classified as a mono-oxygenase (27).
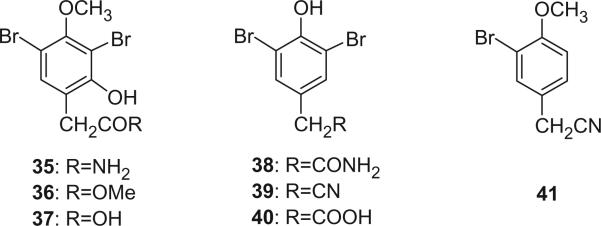
Additional aromatic bromotyrosine derivatives, 35–41, were isolated from Psammaplysilla purpurea, Verongia aerophoba, V. archeri, and Pseudoceratina crassa, respectively (28–33).
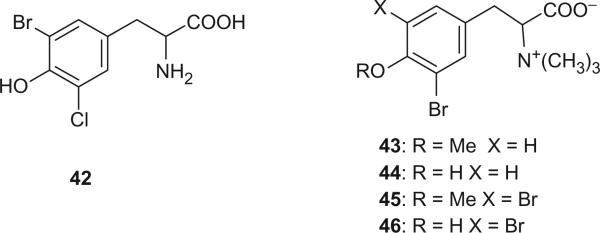
3′-Chloro-5′-bromotyrosine (42) was identified from hydrolysates of a sclero-protein constituting the operculum of the gastropod mollusk Baccinum undatum in 1971 (34). N,N,N-Trimethyl halogenated tyrosines, 43, 44, 45, and 46 were isolated from the Caribbean sponge Pseudoceratina crassa by Fattorusso's group (35). The absolute stereochemistries of 43 and 44 were determined to be L by Gao and Hamann (36).
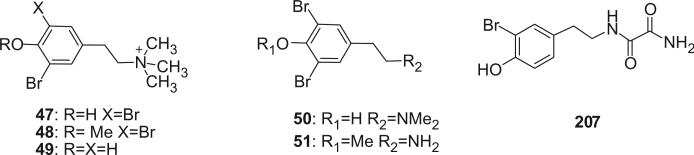
The first tyramine derivative, N,N,N-trimethyl-dibromotyramine (47), was identified from the sponge Verongia fistularis as a dual adrenergic compound in 1978 (37). Compound 48 was obtained as a natural and major bromo compound (1.7% dry weight) from a Caribbean sponge Verongula sp. (38). N,N,N-Trimethyl-3′-bromotyramine (49) and N,N-dimethyl-3′,5′-dibromotyramine (50) were isolated from the marine sponge Verongula gigantea (39). An undescribed ascidian Eudistoma sp. was found to contain 3′,5′-dibromo-4′-methoxyphenethylamine (51), tryptamine and 4-hydroxyphenylacetamide (40). The 3′-bromotyramine amide of oxalic acid amide (207) was obtained from the Papua New Guinea sponge Ianthella basta (41).
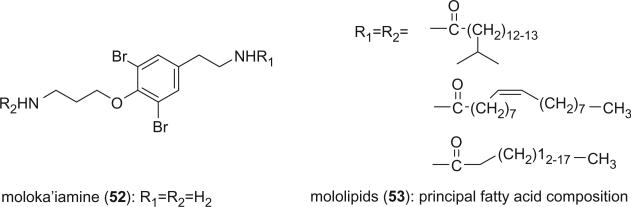
Moloka'iamine (52), which is often represented as a substructure of many bromotyrosine derivatives, was reported as an independent entity from an undescribed Hawaiian Verongid sponge by Hamann and Scheuer (42). Mololipids (53) are a mixture of bisamides of moloka'iamine with long chain fatty acids and were isolated as an anti-HIV agent from this sponge more recently (43). The fatty acids of the mololipid mixture (53) are a homologous series of saturated linear and methyl branched fatty acids ranging from C14 to C20. Included is at least one monounsaturated fatty acid of 18 carbon atoms, with a double bond at C-9. There were no fatty acids with more than one double bond detected. There are also mono-, di-, and trimethyl, internally branched, fatty acids containing 15, 17, and 19 carbons. The positions of the methyl substituents vary in the carbon chain. The 13C NMR data clearly showed that the internal branching of the most abundant fatty acids did not occur at positions α, β, or γ from the carbonyl carbon or the terminal methyl group. There were no data to support or dismiss the possibility that the individual acids may occur randomly on either nitrogen of the core moloka'iamine nucleus.
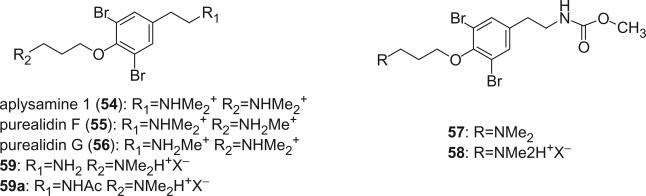
Different N-methylation derivatives of moloka'iamine, aplysamine 1 (54) (44), purealidin F (55), and G (56) (45), were obtained from an Australian sponge Aplysina sp. and the Okinawan sponge Psammaplysilla purea, respectively. 3,5-Dibromo-4-(3-dimethylaminopropoxy)phenethyl carbamic acid methyl ester (57) and its salt 58 were obtained as the first bromotyrosine derivatives containing a carbamate group from an Indian sample of Psammaplysilla purpurea (46), along with 59 (47,48).
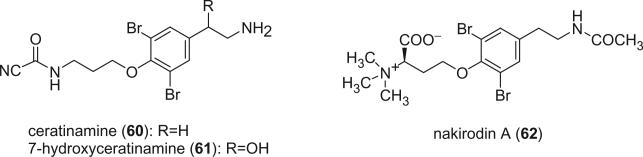
An unprecedented cyanoformyl derivative, ceratinamine (60), was reported from the marine sponge Pseudoceratina purpurea by Fusetani's group in 1996 (49). Ceratinamine (60) was the first report of a cyanoformamide metabolite in natural products. It exhibited antifouling activity against Balanus amphitrite cyprides and cytotoxicity against P388 murine leukemia cells. The second example of a naturally occurring cyanoformamide metabolite, 7-hydroxyceratinamine (61), was isolated from a Micronesian sponge Aplysinella sp. by Fu and Schmitz (50). Nakirodin A (62) was isolated from an Okinawan marine Verongid sponge (51). The absolute configuration was determined to be R by the CD spectrum, of which N,N,N-trimethylhomoserine hydrolyzed from nakirodin A (62) showed a positive Cotton effect at 203.5 nm (Δε +2.0) that was identical to that of the authentic sample of N,N,N-trimethyl-D-homoserine. Although many bromotyrosine alkaloids possess one or more aminopropanol units (52), bromotyrosine alkaloids having an N,N,N-trimethylhomoserine residue, such as nakirodin A (62), are very rare (35,36). The structure of 62 indicated that the aminopropanol units found in many bromotyrosine alkaloids may be biogenetically derived from a homoserine through decarboxylation.
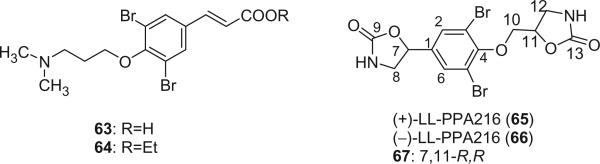
3,5-Dibromo-4-(3′-N,N-dimethylaminopropyloxy)cinnamic acid (63) and its ethyl ester (64) were identified from the Caribbean sponge Pseudoceratina crassa by spectroscopic methods and total synthesis (53). LL-PPA216 (65) was first isolated as dextrorota-tory ([α]D + 8.9°) from the sponge Verongia lacunosa collected off the coast of Puerto Rico by Borders et al. in 1977 (54), and appeared to be the first bromine compound containing 2-oxazolidone rings isolated from a sponge. Makarieva et al. later reported the isolation of the (–)-enantiomer of LL-PPA216 (66, [α]D= −6.5°) from Aplisina sp. collected in Cuba (55). Another isomer 67 was identified from the marine sponge Aplysina aerophoba by Norte et al. in 1988 and its absolute configuration was determined as R, R by X-ray analysis (21). The optical rotation of 67 ([α]D= −33°) is significantly different from that of 65 and 66, indicating that it is a new diastereomer of 65 and 66.
B. SPIROCYCLOHEXADIENYLISOXAZOLINE BROMOTYROSINE DERIVATIVES
1. Bis-Spirocyclohexadienylisoxazolines
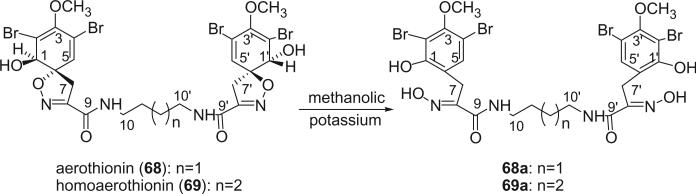
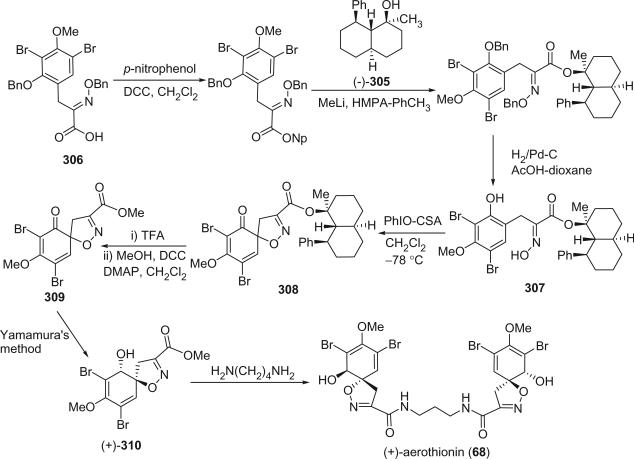
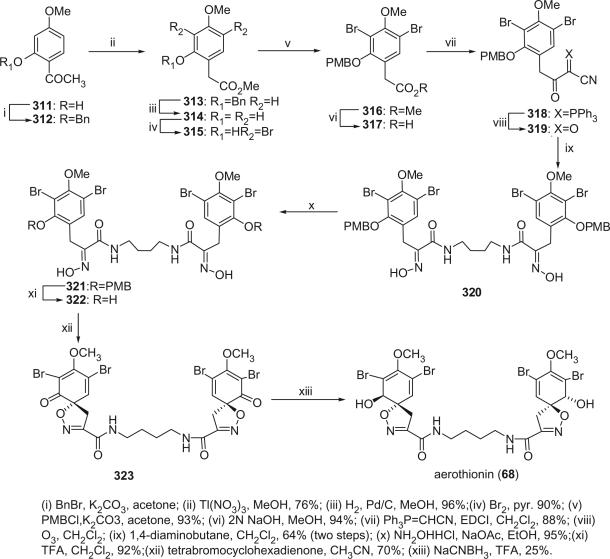
The first spirocyclohexadienylisoxazoline type bromotyrosine derivatives were aerothionin (68) and homoaerothionin (69), which were initially isolated from the marine sponge Verongia aerophoba [the species name was later revised to V. cavernicola (69)] and V. thiona by Fattorusso et al. in 1970 (56–58). The structure of aerothionin, the major component of both sponges (ca. 10% in V. aerophoba), was a result of a collaborative effort between Minale's laboratory and Thomson's laboratory based on proton NMR and chemical methods (59). Treatment of aerothionin with aqueous methanolic potassium converted it quantitatively into the oxime 68a. This reaction was used to identify the spirocyclohexadienylisoxazoline structure. It must be noted that since aerothionin is optically active ([α]D +252°), the asymmetric end units must be identical and not in a mirror-image relationship. The structures of aerothionin and related bisspirocyclohexadienylisoxazoline derivatives were mistakenly drawn as mirror-image relationships in many later publications. 3′,5′-Dibromotyrosine is the probable precursor of the spirocyclohexadienylisoxazoline, and the C4N2 and C5N2 chains are derived from ornithine and lysine, respectively. The absolute stereochemistry was determined by X-ray crystallographic analysis and circular dichroism (60). The 13C NMR chemical shift assignments of C-2 and C-4 were inverted based on the correlation peaks between H-1/H-1′ with C-2/C-2′ and between H-5/H-5′ and C-4/C-4′ in the COLOC NMR experiment (61).
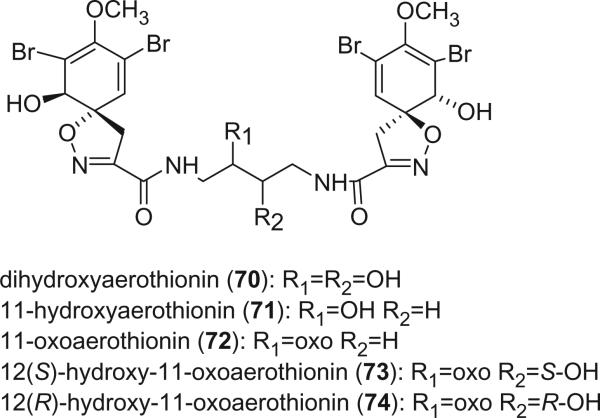
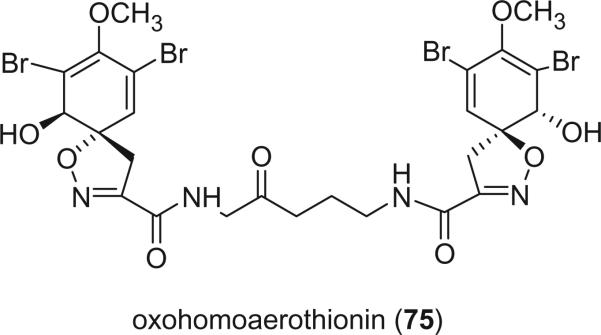
Dihydroxyaerothionin (70) was isolated from the sponge Verongula rigida collected from Sweetings Cay, Bahamas at a depth of 228 feet using an untethered manned submersible. Its structure was determined based on the NMR data and the stereochemistry of the hydroxy group in the central chain was not determined (62). 11-Hydroxyaerothionin (71) was identified from the sponge Pseudoceratina durissima, collected from the Great Barrier Reef, Australia, as an antimicrobial component against Staphylococcus aureus at 100 μg/disk, Bacillus subtilis at 50 μg/disk, and Candida albicans at 50 μg/disk. The relative stereochemistry of the 11-hydroxyl group was not determined (63). 11-Oxoaerothionin (72) was isolated from a Caribbean sponge Aplysina lacunosa (64). It showed pronounced selective cytotoxicity toward the human colon HCT-116 cell line within a limited concentration range (0.01–0.1 μg/mL), in addition to moderate antimicrobial activity against Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. Two epimeric 11-oxo-12-hydroxyaerothionins, 73 and 74, were isolated from the Caribbean sponge Aplysina fistularis forma fulva (Pallas) by Fattorusso's group (61). Their 13C NMR, UV, and IR spectra are identical. However, their 1H NMR spectra have minor differences, suggesting that they differ only in some stereochemical details. Because their CD spectra, which are sensitive to the configuration of the spirocyclohexadiene chromophore, are superimposable, 73 and 74 were concluded to be epimers at C-12. The configurations of 12-hydroxyl groups were determined as 12S for 73 and 12R for 74 using the modified Mosher's method. Oxohomoaerothionin (75) was reported from the sponge Aplysina cavernicola (65).
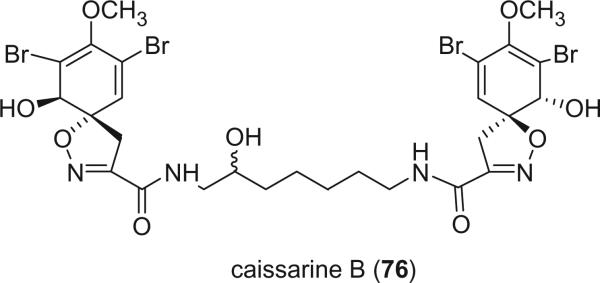
Structurally related to aerothionin and homoaerothionin, caissarine B (76), recently isolated from the Brazilian sponge Aplysina caissara, is the only bromotyrosine-derived alkaloid bearing a 1,7-diamino-3-hydroxyheptane chain, a diamine moiety that has no precedent among natural products (66).
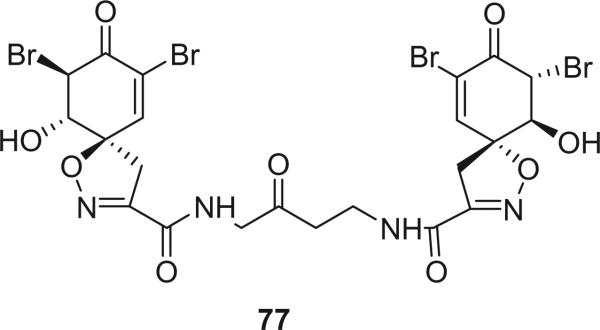
Modification of the spirocyclohexadienylisoxazoline is very rare, which includes compound 77 and calafianin (78). Alkaloid 77, isolated from the sponge Aplysina archeri, was the second example of a bromotyrosine derivative with a spirocyclohexenonylisoxazole ring. Its structure was assigned on the basis of spectroscopic evidence, including 2D-NMR experiments, and the absolute configuration shown in the structure has been suggested using the helicity rule (67).
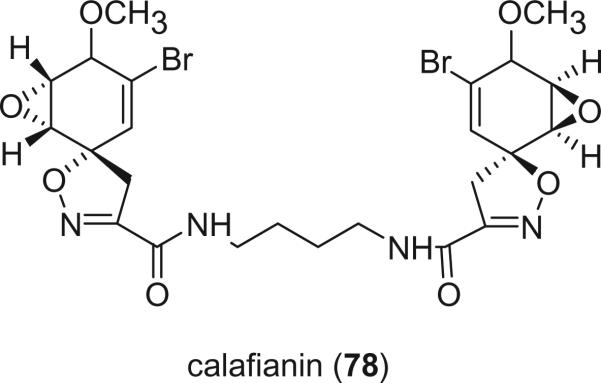
Calafianin (78), isolated from the Mexican sponge Aplysina gerardogreeni, is the only bromotyrosine derivative containing an epoxy group (30).
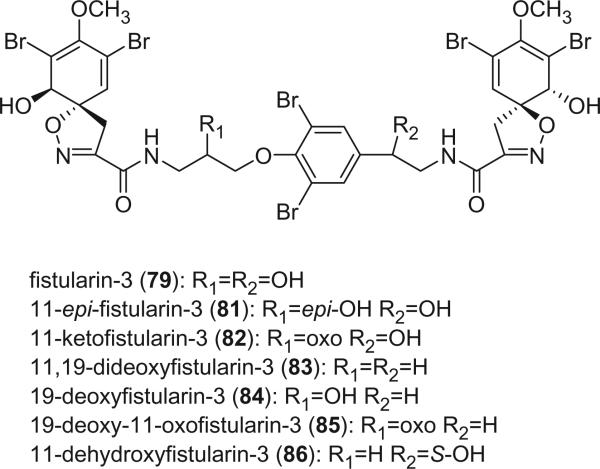
Fistularin-3 (79) was isolated from Aplysina fistularis forma fulva as a cytotoxic component by Gopichand and Schmitz in 1979 (68). Its planar structure was determined as two spirocyclohexadienylisoxazolines connected by a bromotyramine central chain based on proton NMR and alkaline hydrolysis. The relative stereochemistry of the spirocyclohexadienylisoxazoline was the same as aerothionin, based on the proton chemical shifts and coupling constants of H-1/1′ and H-7/7′. The absolute stereochemistry was also determined to be the same as aerothionin based on comparable CD spectrum (36). The configuration of the two chiral centers at C-11 and C-17 remain to be determined. Three stereoisomers of fistularin-3 were reported later. Cimino et al. argued that isofistularin-3 (80), which was isolated from Verongia aerophoba with practically identical UV, IR and optical rotation, was different from fistularin-3 in the stereo-chemistry at one (or more) of the chiral centers based on minor differences when the proton NMR spectra of the acetates of both isofistularin-3 and fistularin-3 were directly compared (69).
König and Wright reported a C-11 stereoisomer of fistularin-3, 11-epi-fistularin-3 (81), from the tropical sponge Agelas oroides (70). Comparison of the 13C and 1H NMR of 81 with those for fistularin-3 revealed small, but significant, differences, notably at C-11 (70.7 ppm in 81 and 69.5 ppm in fistularin-3). All other differences between the two sets of 13C NMR data were in the range of 0.1 to 0.2 ppm. The optical rotation of 81 is +66° compared to +104° (68) and +102° (71) for fistularin-3. The stereochemistry of the spiroisoxazole moiety was deduced as 1R, 1′R, 6S, and 6′S by CD analysis, leaving the configuration of the hydroxyl groups at C-11 and C-17 undetermined. 11-epi-Fistularin-3 was not cytotoxic towards KB-cells (IC50>20 μg/mL) in contrast to fistularin-3 (IC50>4.1 μg/mL) (68) and isofistularin-3 (IC50>4 μg/mL) (69). Aydogmus et al. claimed another stereoisomer of fistularin-3, 1-epi-fistularin-3, based on the minor differences (less than 0.03ppm) of the proton chemical shifts of H-1,1′ and H-7,7′ for 1-epi-fistularin-3 tetraacetate compared to isofistularin-3 tetraacetate (69) and fistularin-3 (68), and an optical rotation of +51.6° (8).
11-Ketofistularin-3 (82) and fistularin-3 were obtained from the sponge Aplysina archeri (71). Both compounds exhibited antiviral activity against feline leukemia virus with ED50 values of 42 μM and 22 μM, respectively. 11,19-Dideoxyfistularin-3 (83) was reported as an antibiotic from the sponge Pseudoceratina durissima, which is more active than aerothionin (68) and homoaerothionin (69) (63). 19-Deoxyfistularin 3 (84) and 19-deoxy-11-oxofistularin 3 (85) were isolated from an undescribed Italian sponge Verongia sp. by Mancini et al. in 1993 (72). 19-Dehydroxyaerothion (86) was isolated from the sponges Aplysina cavernicola (65) and Aplysina fistularis (73). The absolute stereochemistry of the spirocyclohexadienylisoxazole moieties were determined to be the same as aerothionin by the CD spectrum, and the configuration of C-19 was deduced as S by the modified Mosher's method (65).
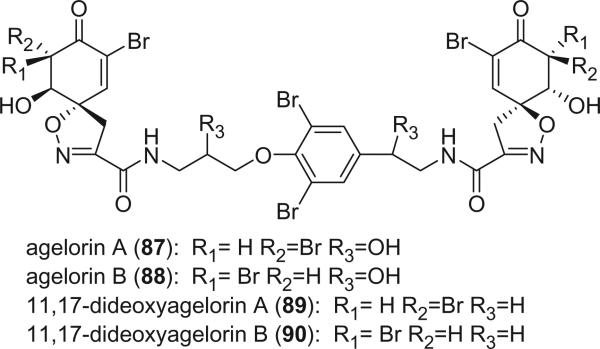
Agelorins A and B (87 and 88) were isolated from the marine sponge Agelas oroides (Agelasidae, Axinellida) (70). The structures contain two units quite similar to the usual spirocyclohexadienylisoxazole fragments differing only by the presence of a cyclohexenone instead of a cyclohexadienyl ring. This structural feature is unique among the bromotyrosine derivatives. The relative stereochemistry of the spirocyclohexenonylisoxazole was determined from the proton NMR and NOESY spectra. A collection of the marine sponge Suberea aff. praetensa from the Gulf of Thailand furnished the bromotyrosine derivatives fistularin-3, agelorin A and B, and the new 11,17-dideoxyagelorin A (89) and B (90) (74).
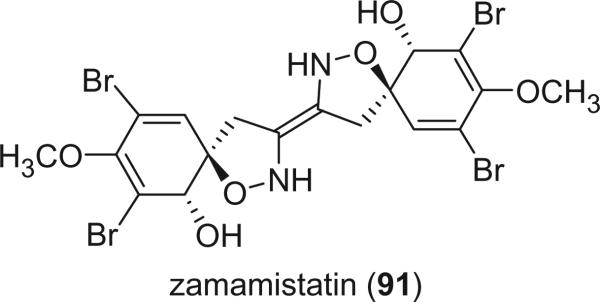
Zamamistatin (91) was isolated from the sponge Pseudoceratina purpurea with significant antibacterial activity against Rhodospirillum salexigens, which has adhering properties, and may be a valuable candidate for novel antifouling agents (75,76). It was determined to be an optically active dimer of spirocyclohexadienylisoxazoline by the careful analysis of the 1D and 2D NMR spectra and its absolute stereochemistry was determined by the modified Mosher's method.
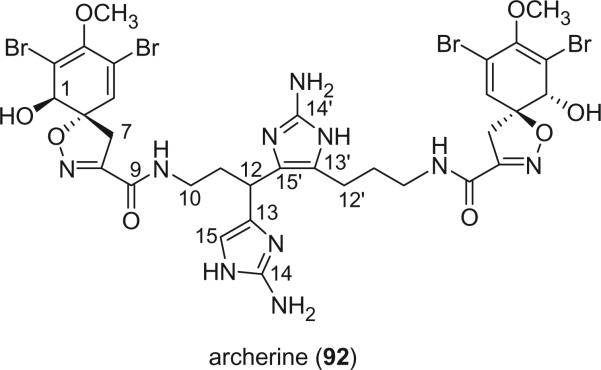
Archerine (92), a novel anti-histaminic bromotyrosine derivative, was identified from the Caribbean marine sponge Aplysina archeri by Fattorusso's group in 2001 (77). Its structure is novel in having the central chain formed by two 2-amino-homohistamine residues through a carbon–carbon bond. The configuration of C-12 remains to be assigned. Histamine or homohistamine are not unusual in bromotyrosine derivatives. However, archerine (92) is the only example having two homohistamine residues. The structure of archerine (92) suggests a biogenetic origin from a [1+1] intermolecular oxidative coupling of two molecules of aerophobin-2 (114), which also exists in this sponge.
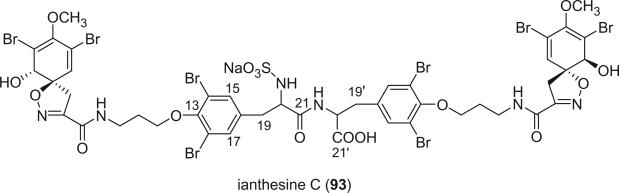
Ianthesine C (93) was isolated as a 3:2 mixture of diastereomers from an Australian marine sponge of the genus Ianthella sp. by Okamoto et al. in 2000 (78). It is a tetrameric bromotyrosine derivative having two spirocyclohexadienylisoxazoline ring systems linked by two bromotyrosines and two C3 units. The configurations of C-20 and C-20′ of these diastereomers are unclear.
2. Mono-spirocyclohexadienylisoxazolines
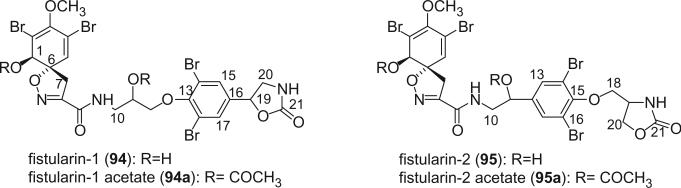
This class of bromotyrosine derivatives contains one spirocyclohexadienylisoxazoline ring system. The first two alkaloids in this class are fistularin-1 (94) and fistularin-2 (95) isolated from the marine sponge Aplysina fistularis forma fulva by Gopichand and Schmitz in 1979 (68). Both compounds contain a spirocyclohexadienylisoxazoline, a bromotyramine, and a C3 unit.
a. Simple Mono-spirocyclohexadienylisoxazolines
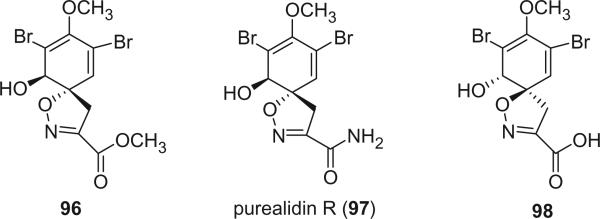
There are only three alkaloids in this group. Alkaloids 96 and 97 were reported by Ciminiello et al. from the Caribbean sponge Verongula sp. (38). Alkaloid 97 was also isolated from the Okinawan sponge Psammaplysilla purea and was named as purealidin R (79). Alkaloid 98 was isolated from a Caribbean sponge Pseudoceratina sp. as a major brominated metabolite (80).
b. Bromotyrosine Mono-spirocyclohexadienylisoxazoline
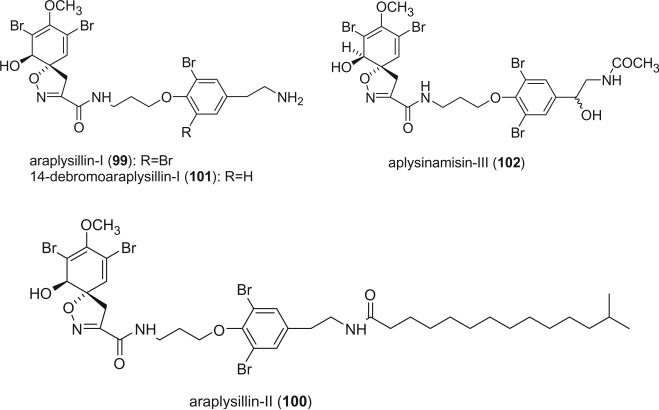
Araplysillins-I (99) and -II (100) were isolated from the sponge Psammaplysilla arabica and were established to be inhibitors of Na,K-ATPase and have antimicrobial activity (81). Two closely related alkaloids, 14-debromoaraplysillin-I (101) and aplysinamisin III (102) were isolated from P. purpurea (82) and Aplysina cauliformis (83), respectively.
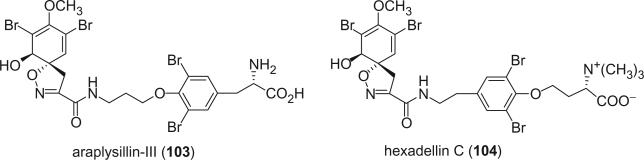
Gao et al. reported araplysillin-III (103) and hexadellin C (104) from the sponge Aiolochroia crassa (36). The absolute configuration of the spiroisoxazoline of both alkaloids was determined by CD spectra. The absolute configuration at C-18 of araplysillin-III (103) was shown to be l by HPLC analysis according to Marfey's procedure. The configuration of the N,N,N-trimethylhomoserine moiety was deduced as l by comparison of the optical rotation with the d- and l- standards.
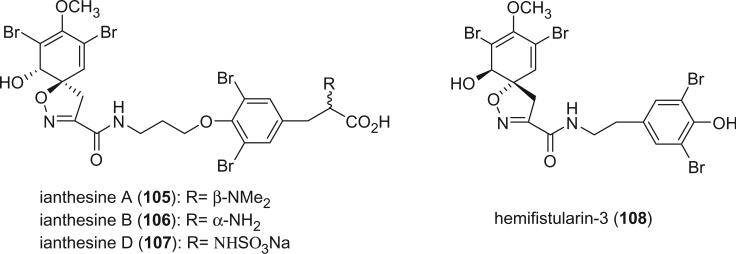
Ianthesines A (105), B (106), and D (107) were isolated from the Australian marine sponge Ianthella sp. as Na,K-ATPase inhibitors with an active range of 50–440 μM (78). The 1S, 6R configuration of the ianthesines was deduced by the negative optical rotation and Cotton effects at 248 and 285 nm. The dibromo-N,N-dimethyltyrosine moiety of ianthesine A (105) was determined as D by chiral HPLC analysis of the hydrobromic acid hydrolysis product of 105. The configuration of the dibromotyrosine moiety of ianthesine B (106) was found to be a mixture of l- and d-forms in the ratio of 7:3, using the same method. Chiral HPLC analysis revealed that ianthesine B (106) is a 7:3 mixture of two diastereomers, 1S, 6R, 20S and 1S, 6R, 20R. The minor diastereomer is an enantiomer of araplysillin-III (103). Hemifistularin-3 (108), which is the right-side portion of 11-oxofistularin-3 (82), was isolated from a new species of sponge in the family Aplysinellidae Bergquist, order Verongida, collected in the Coral Sea (72). Hemifistularin-3 (108) can be obtained by treatment of 11-oxofistularin-3 with methanolic KOH. Since both hemifistularin-3 (108) and 11-oxofistularin-3 (82) were found to exist in the same sponge, it is possible that hemifistularin-3 is a product of the degradation of 11-oxofistularin-3 or an elaborated biogenetic precursor.
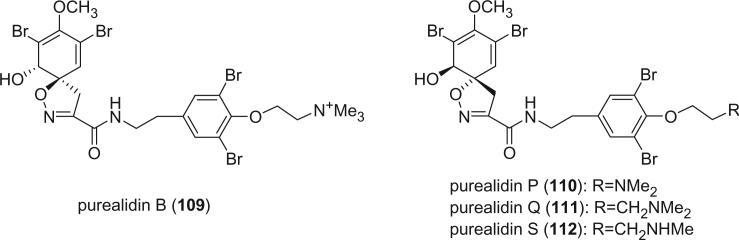
Purealidins B (109), P (110), and Q (111) were isolated from two different collections of an Okinawan sponge Psammaplysilla purea by Kobayashi et al. (79,84,85). The absolute stereochemistry of the spirocyclohexadienylisoxazoline in purealidin B (109) (84), as determined by the CD spectrum, was opposite of that in purealidin P (110) and Q (111) (79). Purealidin S (112) was isolated from the Fijian sponge Druinella sp. (86).
c. Histamine Mono-spirocyclohexadienylisoxazolines
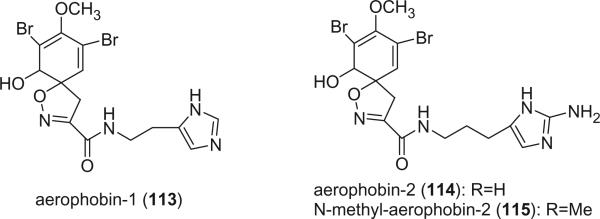
Aerophobin-1 (113) and aerophobin-2 (114) were isolated from the sponge Verongia aerophoba by Cimino et al. in 1983 (69). Their structures consisted of a spirocyclohexadienylisoxazoline and a histamine or 2-amino-homohistamine and were determined by NMR spectroscopy and hydrolysis. A methylation derivative of aerophoin-2, N-methyl-aerophobin-2 (115), was isolated from a Caribbean sponge specimen of Aiolochroia crassa (87).
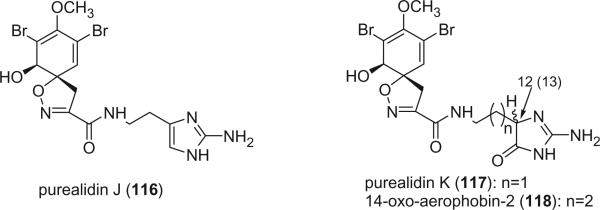
Purealidins J (116) and K (117) were isolated from the Okinawan sponge Psammaplysilla purea by Kobayashi et al. (79). The absolute stereochemistry of the spirocyclohexadienylisoxazoline was determined by the positive Cotton effects at 248 and 184 nm in the CD spectra. Purealidin K was subjected to ozonolysis, followed by oxidation with H2O2, and subsequently acid hydrolysis. Chiral HPLC analysis of the hydrolysate revealed D- and L-2,4-diaminobutyric acids in the ratio of 1:1, suggesting that C-12 of purealidin K is racemic. A similar alkaloid, 14-oxo-aerophobin-2 (118), was identified from the Caribbean sponge Aplysina insularis (88). The stereochemistry of C-13 was not determined.
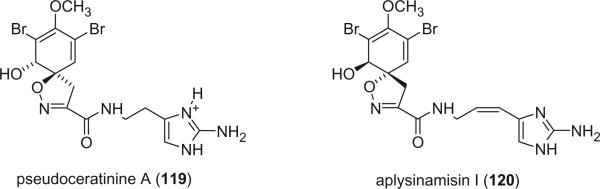
The enantiomer of purealidin J, pseudoceratinine A (119), was reported one year later from Pseudoceratina verrucosa (89). Pseudoceratinine A showed a negative optical rotation and Cotton effects near 250 and 290 nm, which is opposite to that of aerothionin (68), whose absolute stereochemistry was determined by X-ray and CD spectra (60). Aplysinamisine I (120), which can be considered as a 11,12-dehydro product of aerophobin-2 (114), was isolated from the Caribbean sponge Aplysina cauliformis (83).
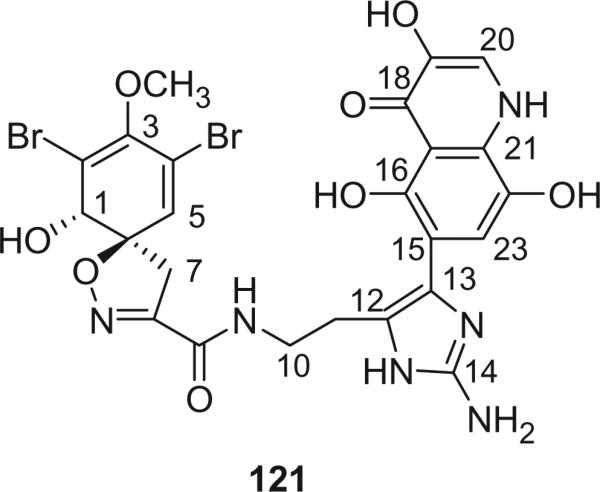
Alkaloid 121 was isolated from an Australian, non-verongid sponge Oceanapia sp. by Bewley's group in 2001 (90). Its structure was elucidated as an unprecedented imidazolyl-quinolinone substructure attached to a spirocyclohexadienylisoxazoline by 1D and 2D NMR, and its absolute configuration was determined to be 1-(S), 6-(R) by comparison of its specific rotation and CD spectrum with those of pseudoceratinine A (119) and purealidin J (116). Bromotyrosine-derived metabolites were initially limited exclusively to sponges of the order Verongida. While a voucher specimen corresponding to the sponge from which 121 was obtained was re-identified as Oceanapia sp., it remains possible that a sample of a verongid sponge was present in the actual collection. Alkaloid 121 was the first example of a natural product that inhibits an enzyme central to a mycothiol-dependent detoxification pathway found in mycobacteria.
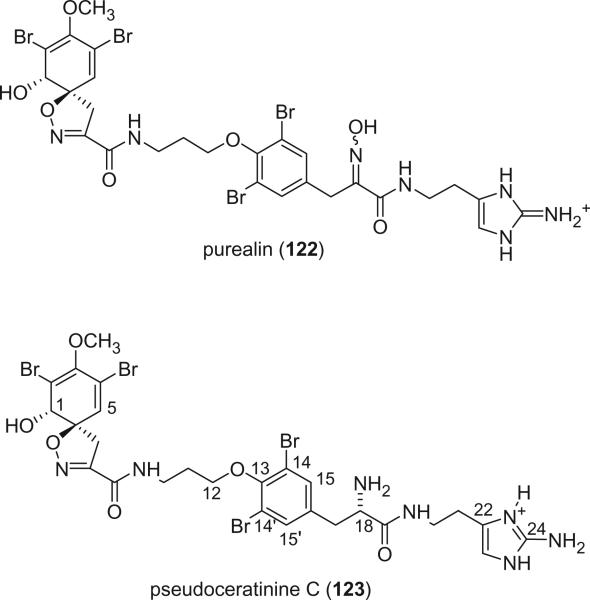
Purealin (122) and pseudoceratinine C (123) are two spirocyclohexadienylisoxazoline derivatives containing both bromotyrosine and 2-amino-histamine in the side chain. Purealin (122) was first isolated from Psammaplysilla purea by Nakamura et al. in 1985 (91). Both alkaloids were reported from Pseudoceratina verrucosa by Benharref and Païs in 1996 (89). The absolute configurations of 1-(S) and 6-(R) of both compounds were deduced by the negative specific rotation and the Cotton effects near 250 and 290 nm. The 18-(S) configuration of pseudoceratinine C (123) was also determined by a positive Cotton effect at 215 nm, corresponding to the S-configuration of the tyrosine residue (89).
d. Linear Side Chain Mono-spirocyclohexadienylisoxazolines
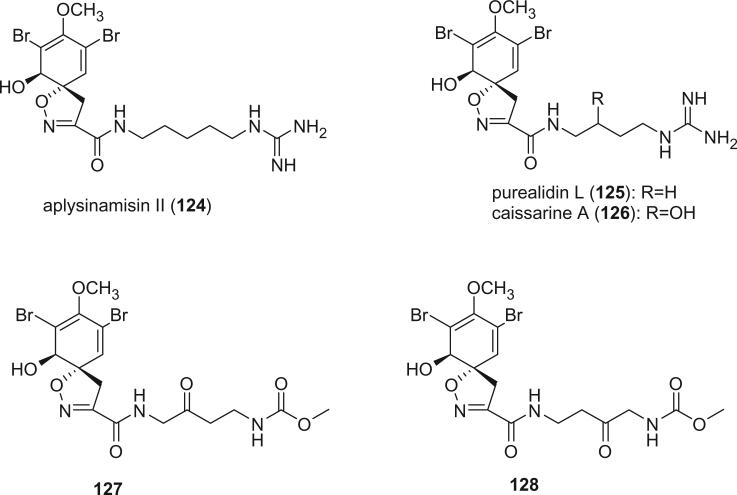
Alkaloids in this class include aplysinamisin II (124) from the Caribbean sponge Aplysina cauliformis (83), purealidin L (125) from Psammaplysilla purea (79), caissarine A (126) from Aplysina caissara (66), and 127 and 128 from Aplysina cauliformis collected in the Bahamas (92).
C. SPIROOXEPINISOXAZOLINE (OXEPIN) BROMOTYROSINE DERIVATIVES
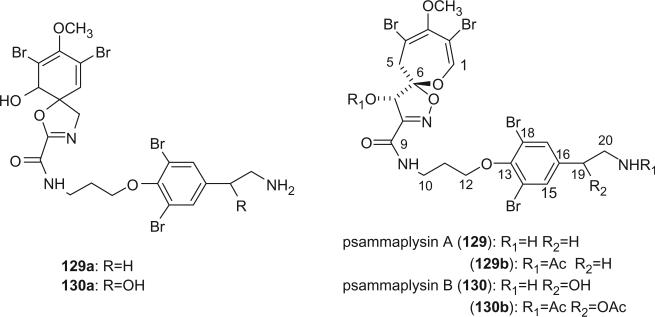
Prompted by the antibiotic activity of the MeOH extract of Psammaplysilla purpurea collected from the southern part of the Gulf of Eilat (off the Red Sea), two spirooxepinisoxazoline type dibromotyrosine derivatives, psammaplysins A and B were first isolated by Kashman's group from P. purpurea in 1982 (93). The freeze-dried sponge was extracted with methanol and the extract was chromatographed on a Sephadex LH-20 column eluted with CHCl3–MeOH 1:1 to yield psammaplysin A and a mixture of psammaplysin A and B. After acetylation, psammaplysins A and B were separated by silica gel column chromatography and eluted with 2% and 6% EtOH in CHCl3. The structures were proposed as having a spiro[4.5]oxazadecane skeleton (129a and 130a) based mainly on the 1H and 13C NMR spectra data and the alkaline degradation of psammaplysin A, which is different from that of aerothionin (58). In 1985, Scheuer's group isolated psammaplysins A and B from the sponge P. purpurea collected in Palau and revised the structures of spiro[4.5]oxazadecane skeleton to a spiro[4.6]dioxazunde-cane (129) on the basis of two-dimensional 13C–13C connectivity and single-crystal X-ray diffraction studies on psammaplysin A acetamide acetate (129b) (94).
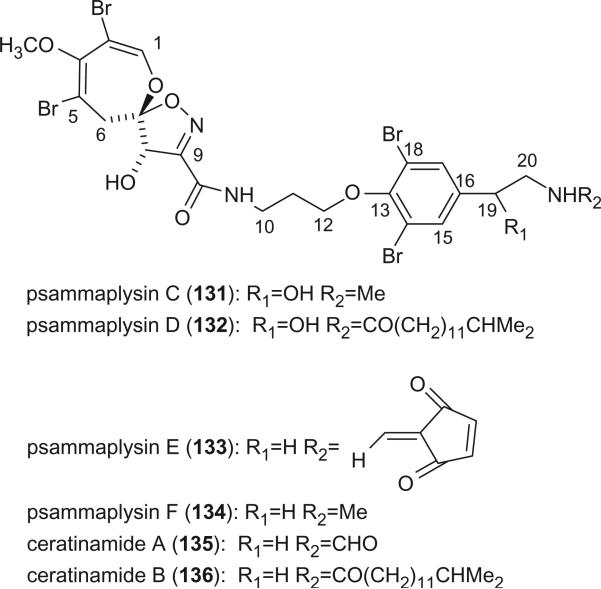
Psammaplysin C (131) was isolated from P. purpurea collected off Makaluva Island in Fiji in 1992, it exhibited in vitro cytotoxicity towards the human colon tumor cell-line HCT 116 with an IC50 of 3 μg/mL (95). Psammaplysin D (132) and E (133) were identified from a new species of Aplysinella (order Verongida) collected from Pingelap Atoll, Micronesia in 1993 (96). Psammaplysin D is the isopentadecanoyl of psammaplysin B, while psammaplysin E has an unprecedented cyclopentenedione, which was not previously encountered in compounds from natural sources. Psammaplysin D displays anti-HIV activity against the Haitian RF strain of HIV-I with a 51% inhibition at 0.1 μg/mL. Psammaplysin E exhibits cytotoxicity against KB and LOVO cells at 5 μg/mL. Psammaplysin F (134) was isolated from an undescribed species of Aplysinella sponge, collected from Chuuk, Micronesia (97). Ceratinamides A (135) and B (136) were isolated from P. purpurea collected from Hachijo-jima in 1996 and were found to exhibit antifouling activity with EC50 values of 0.10 and 2.4 μg/mL (98).
D. OXIMES
1. Oxime-histamines
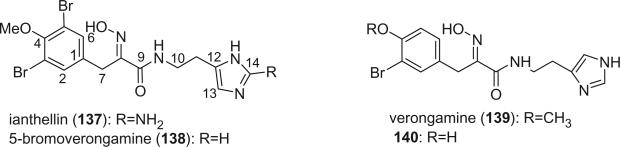
Ianthellin (137) was isolated from the sponge Ianthella ardis, collected in the Bahamas, and was determined to be the major antibiotic and antifungal component (99). 5-Bromoverongamine (138), a novel antifouling tyrosine alkaloid has been isolated from the Caribbean sponge Pseudoceratina sp. (100). Verongamine (139), a specific histamine-H3 antagonist at concentrations as low as 1.0 μg/mL, was isolated from Verongula gigantea (101). Alkaloid 140 was isolated from the Caribbean sponge Pseudoceratina crassa (33).
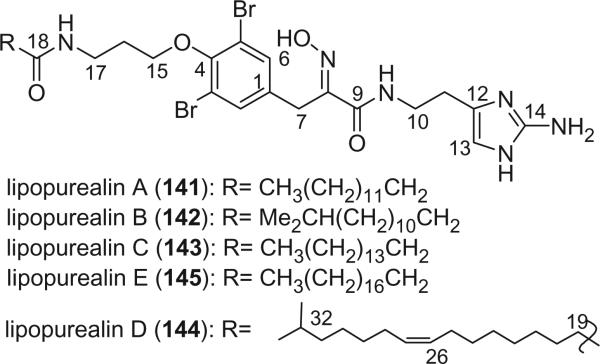
Lipopurealins A (141), B (142), and C (143) are unique metabolites with a long alkyl chain, isolated from the marine sponge Psammaplysilla purea by Kobayashi's group in 1986 (102). Lipopurealins are inhibitors of Na,K-ATPase, and lipopurealin B is most active. Two additional lipopurealins, lipopurealins D (144) and E (145) were isolated from the same sponge in 1995 (103).
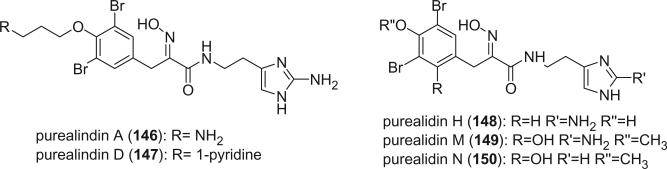
Additional oxime bromotyrosine derivatives containing a histamine group were reported by Kobayashi's group from Psammaplysilla purea, including purealidins A (146), D (147), H (148), M (149), and N (150) (79,103-105). The E-geometries of the oxime in lipopurealins A (141), B (142), C (143), purealidins A (146), and D (147) were assigned based on the chemical shifts of C-7 around δ 28 ppm, which were not assigned in the original literature.
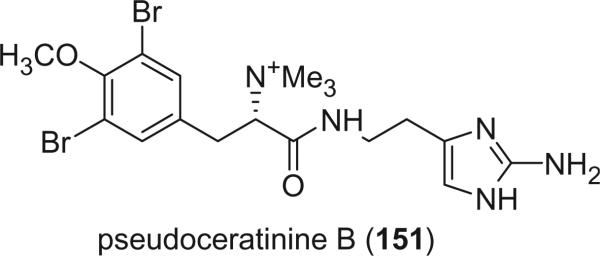
Pseudoceratinine B (151), in which the oxime is reduced and tri-methylated, was isolated from Pseudoceratina verrucosa (89). The absolute configuration of tyrosine was deduced from the positive Cotton effect at 212 nm, corresponding to an S-configuration (106). Pseudoceratinine B (151) is the second bromotyrosine alkaloid containing a non-oxidized amino group.
2. Oxime-Disulfides
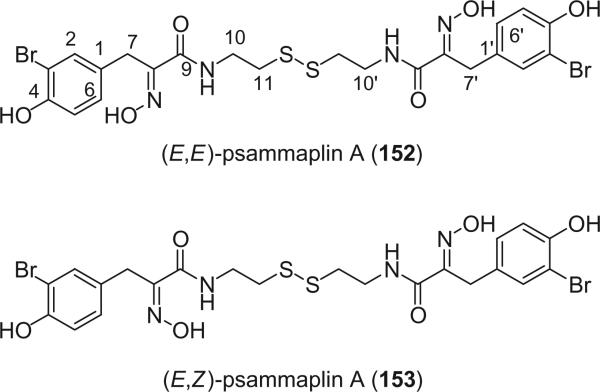
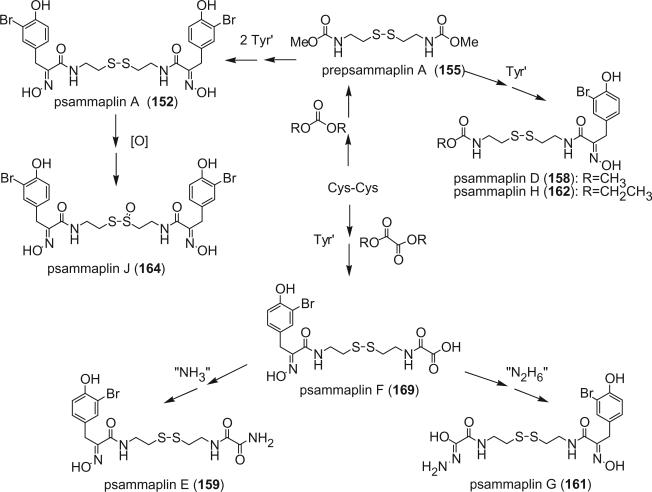
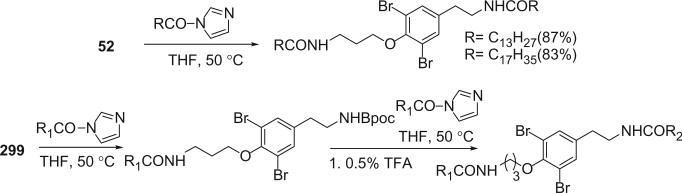
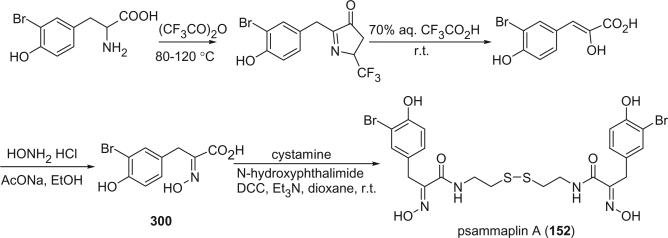
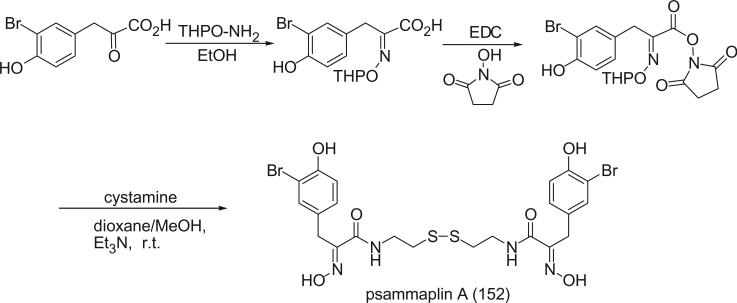
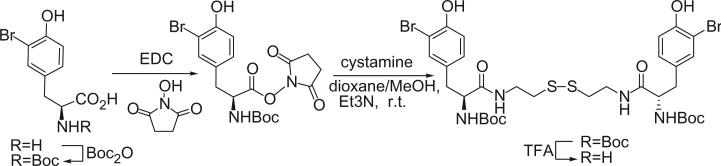
Alkaloids 152 and 153 were the first bromotyrosine derivatives characterized containing a disulfide moiety, and were isolated from an unidentified Verongida sponge by Arabshahi and Schmitz in 1987 (107). Almost at the same time, Quinoa and Crews reported the isolation of 152 from a marine sponge Psammaplysilla sp. collected from Tonga and gave the compound a trivial name of psammaplin A (108). The geometry of the oxime was determined by the carbon chemical shift of the methylene group (C-7) α to the oxime. The chemical shift of C-7 is about 28 ppm for an E-geometry and 35 ppm for a Z-geometry (107).
Scheuer et al. later reported the psammaplin A (152) and the psammaplin A dimer, bisaprasin (154), from the Guam sponge Thorectopsamma xana (109).
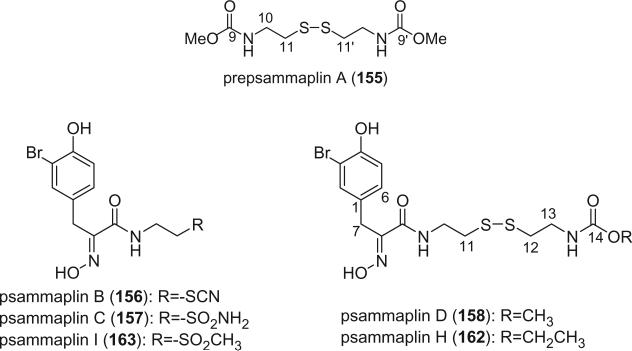
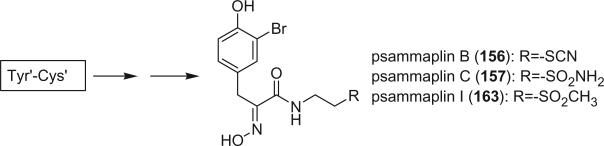
Prepsammaplin A (155) and psammaplins B (156), C (157), and D (158) were isolated from the marine sponge Psammaplysilla purpurea by Jimenez and Crews (110). The isolation of prepsammaplin A (155), a cysteine dimer, indicated that cysteine might be the precursor of the central part of psammaplin A. Interestingly, the R functional groups of psammaplin B (156) and C (157) are unique and do not appear to have counterparts among any known marine sponge amino acid derivatives (110). Additionally, the only precedent of the thiocyanate group, as found in psammaplin B (156), is the sesquiterpene thiocyanate isolated from the sponge Trachyopsis aplysinoides (111).
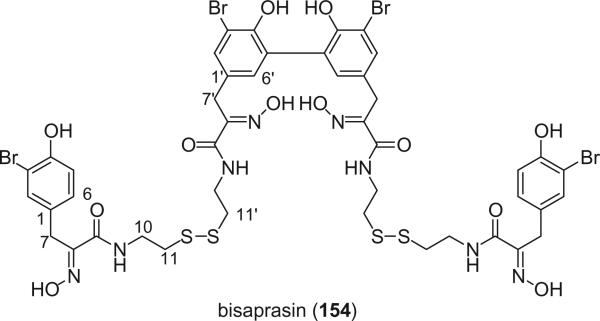
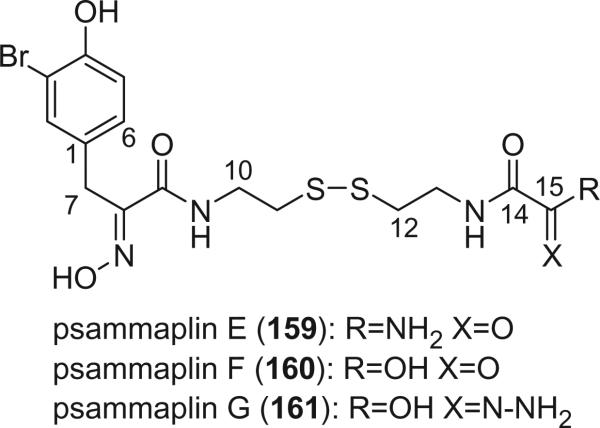
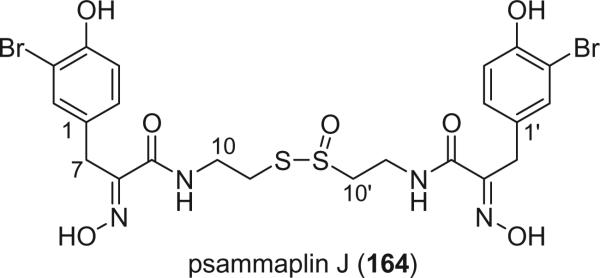
Additional disulfide bromotyrosine derivatives, psammaplin E (159), F (160), G (161), H (162), I (163), and J (164) were isolated from the sponge Psammaplysilla purpurea collected from Papua New Guinea by Crews’ group in 2003 (112). Psammaplin D (158) and H (162) possess a methyl or ethyl carbamate moiety, respectively. The unprecedented functional groups in psammaplin E (159), F (160), G (161), and J (164) were identified by detailed spectroscopic analysis. The N-substituted oxalamide functionality of psammaplin E (159) is rare among marine natural products. The only known marine natural products that possess an oxalamide functional group are igzamide, isolated from the marine sponge Plocamissa igzo (113), and 3-bromotyramine amide, isolated from Ianthella basta (41). Psammaplin F (160) is the first marine natural product containing an oxalamic functionality. The functionality in psammaplin G (161) has no precedent in the natural products literature. Psammaplin A (152), F (160), and bisaprasin (154) are potent histone deacetylase inhibitors with mild cytotoxicity. Psammaplin A (152), G (161), and bisaprasin (154) are potent DNA methyltransferase inhibitors.
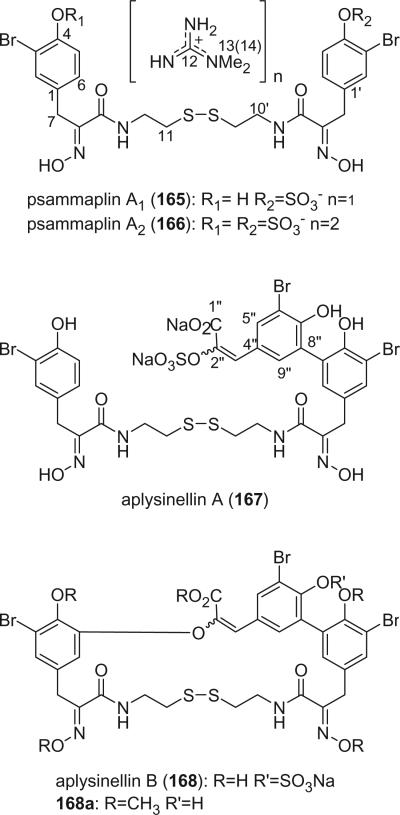
Shin and Paul reported four new psammaplin A analogs, psammaplins A1 (165), A2 (166), aplysinellin A (167), and B (168), from the marine sponge Aplysinella rhax collected from Guam, Palau, and Pohnpei Micronesia (114). Psammaplins A1 (165) and A2 (166) are mono- or bis-N,N-dimethylguanidinium salts of psammaplin A (152) monoor dualsulfate. Organic sulfates with N,N-dimethylguanidinium as counter ions are very rare among sponge metabolites. Suvanine and sulfircin, both sesterterpenoid sulfates from the sponge Ircinia sp., are two examples (115,116). Aplysinellin A (167) possesses a bromotyrosine-derived C9-unit of the cinnamic acid type attached to psammaplin A by a biphenyl linkage, while aplysinellin B (168) is the corresponding cyclic enol ether. Treatment of aplysinellin B (168) afforded compound 168a. Alkaloids 165–168 exhibited inhibitory activity against the growth of the K562 cell-line and against farnesyl protein transferase.
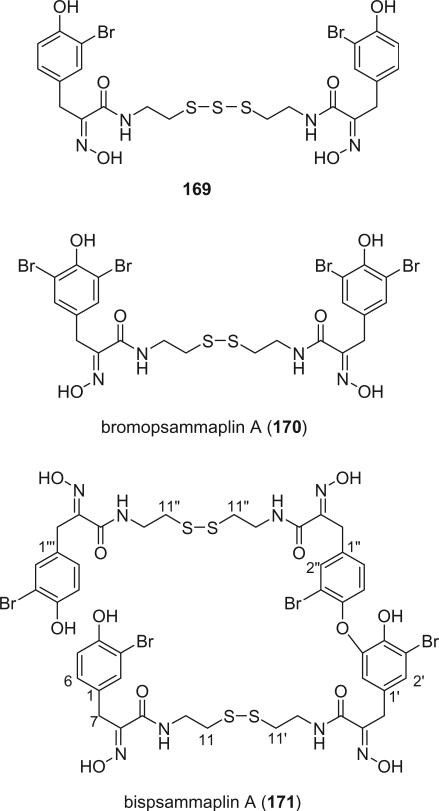
Jung's group reported the isolation of three new psammaplin analogs, 169, (E,E)-bromopsammaplin A (170), and bispsammaplin A (171), along with the known alkaloids (E,E)-psammaplin A (152), (E,Z)-psammaplin A (153), psammaplin D (158), and bisaprasin (154), from an association of two sponges, Jaspis wondoensis and Poecillastra wondoensis collected from Korea (117). Alkaloid 169 is the only bromotyrosine derivative containing a trisulfide moiety. This is the second report of bromotyrosine derivatives isolated from sponges not belonging to the order Verongida. The first example described the isolation of three bromotyrosine derivatives from a non-verongid sponge Oceanapia sp., but it remains possible that a sample of verongid sponge was present in the actual sample analyzed (90).
3. Oxime-Bromotyramines
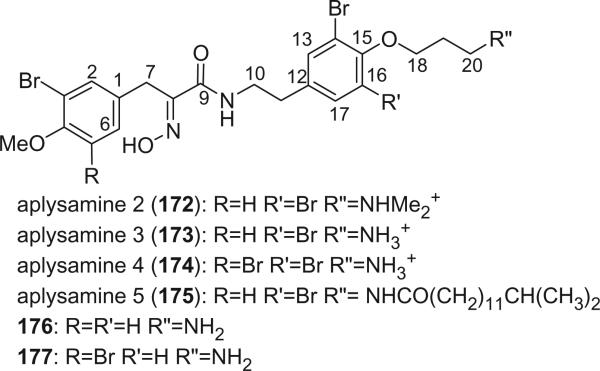
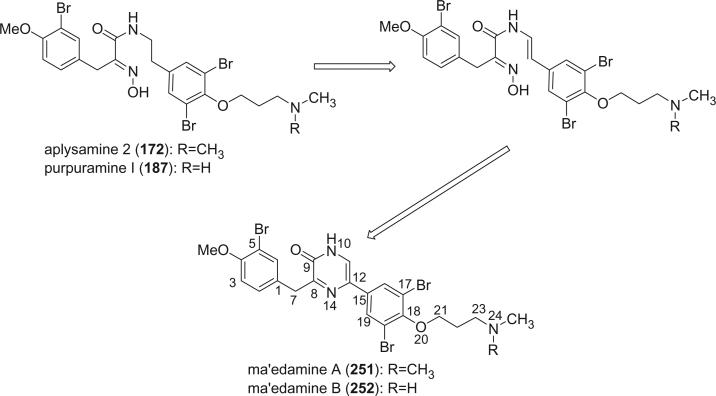
Xynas and Capon reported aplysamine 2 (172) from an Australian marine sponge Aplysina sp. in 1989 (44). Aplysamines 3 (173), 4 (174), and 5 (175) were isolated from the Hawaiian sponge Psammaplysilla purpurea by Scheuer's group (118). All of these alkaloids exhibited cytotoxic activity, while aplysamine 3 and 4 showed mild antibacterial activity against Staphylococcus aureus. Alkaloids 176 and 177 were isolated from the sponge P. purpurea collected in Okinawa by an Indian group (119,120).
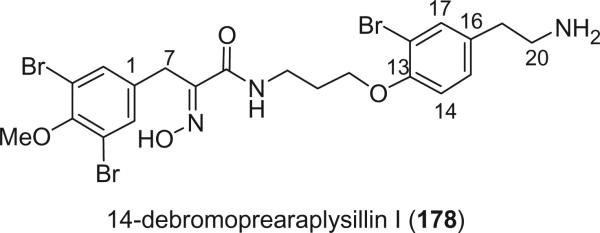
Alkaloid 178 was isolated from P. purpurea collected in the Seychelles by Faulkner's group (82). Since it is the presumed biogenetic precursor of 14-debromoaraplysillin-I (101), the name 14-debromoprearaplysillin I was given. The isolation of spiroisoxazoline 101 and the oxime 178 from the same sponge was the first reported instance in which a spiroisoxazoline has been isolated together with its proposed precursor.
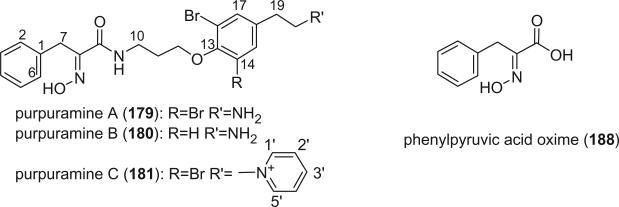
Fusetani et al. reported nine new bromotyrosine-derived metabolites, purpuramines A-I (179–187), from P. purpurea (121). The carboxylic acid oxime units present in the bromotyrosine-derived metabolites were exclusively derived from bromotyrosines, while the oxime function in purpuramines A-I (179–183) is part of a phenylalanine moiety, which is a new variant among verongid metabolites. Phenylpyruvic acid oxime (188), which is a building block for 179–183, was also isolated from the sponge.
Purpuramines A-I (179–187) exhibited antibacterial activity against Staphylococcus aureus, but phenylpyruvic acid oxime (188) was not active. Purpuramine J (189) was isolated from the Fijian sponge Psammaplysilla (Druinella) sp., along with purpuramine I (187), aplysamine 2 (172), and eight other bromotyrosine derivatives (86). Purpuramine J (189) is the first bromotyrosine derivative containing an N-oxide functionality, which is considered rare in marine natural products.
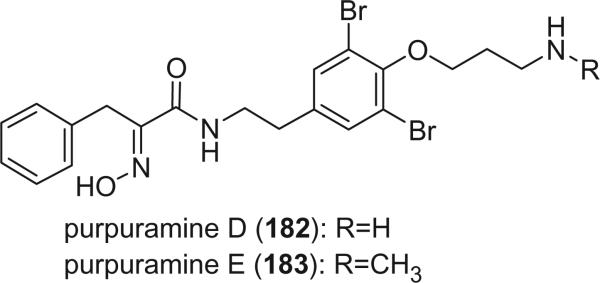
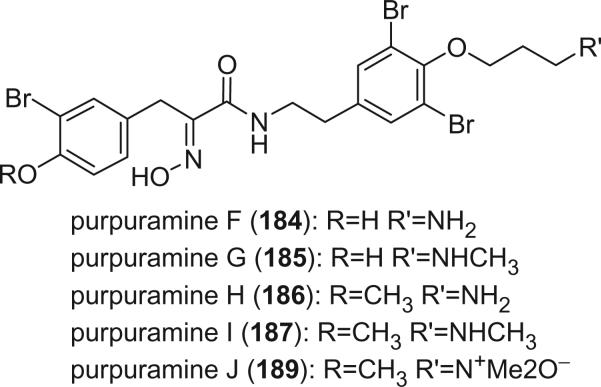
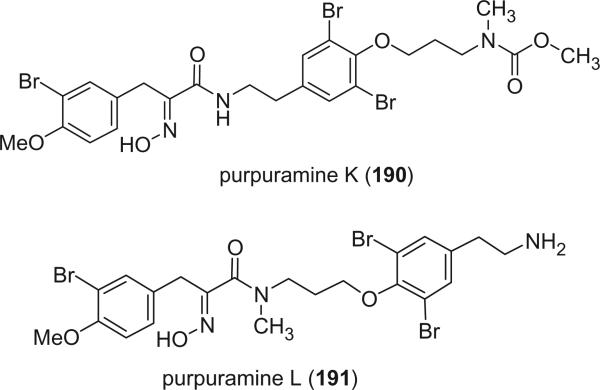
Purpuramines K (190) and L (191), isolated from the Indian sponge Psammaplysilla purpurea, exhibited antimicrobial activity against both Gram positive and Gram negative bacteria (122).
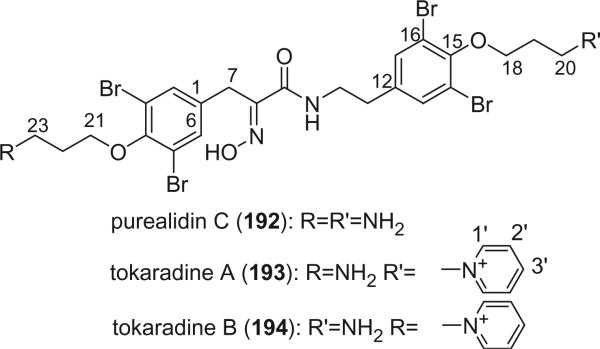
Purealidin C (192) was isolated from the Okinawan sponge P. purea, which exhibited cytotoxic, antifungal, and antimicrobial activities (84). Tokaradines A (193) and B (194), isolated from Pseudoceratina purpurea, are rare examples of marine bromotyrosine-derived metabolites containing an N-substituted pyridinium group. Another example is purpureamine C (181) (123). Tokaradines A (193) and B (194) were seen to be lethal to the crab Hemigrapsus sanguineus (123).
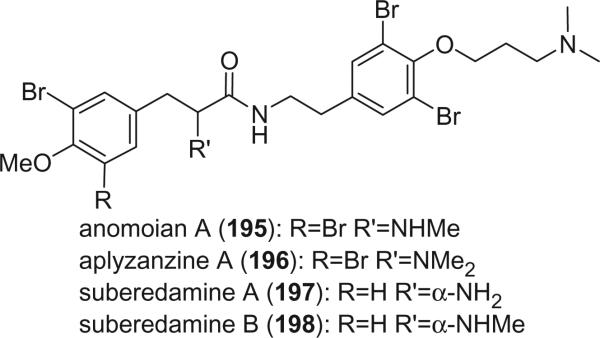
Anomoian A (195) was isolated from an Australian sponge Anomoianthella popeae, belonging to a new genus of the family Ianthellidae (124). Anomoian A is the first bromotyrosine-derived metabolite with a non-oxidized amino group and exhibited strong antimicrobial activity. Aplyzanzine A (196) was isolated from the sponge Aplysina sp. collected from Zanzibar (125). The stereochemistry at C-8 was not determined. Suberedamines A (197) and B (198) were isolated from an undescribed sponge of the genus Suberea (126). The absolute configurations at C-8 of both alkaloids were assigned as S by chiral HPLC analysis of the hydrobromic acid hydrolysate of each compound. Both suberedamines A (197) and B (198) exhibited moderate cytotoxic and antibacterial activity.
4. Other Oxime Structures
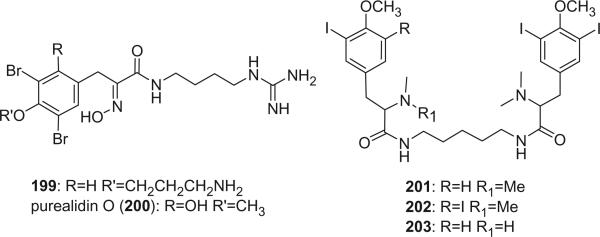
Compound 199 and purealidin O (200) are two oxime-type bromotyrosine-derived metabolites containing an agmatine moiety, isolated from Psammaplysilla purea and Oceanapia sp., respectively (79,90). Alkaloids 201, 202, and 203 were iodinated tyrosine derivatives isolated from the ascidian Aplidium sp. (127).
E. BASTADINS
1. Bastadins
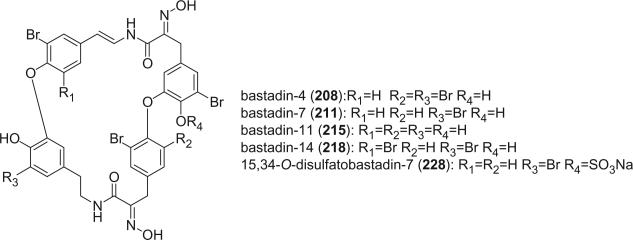
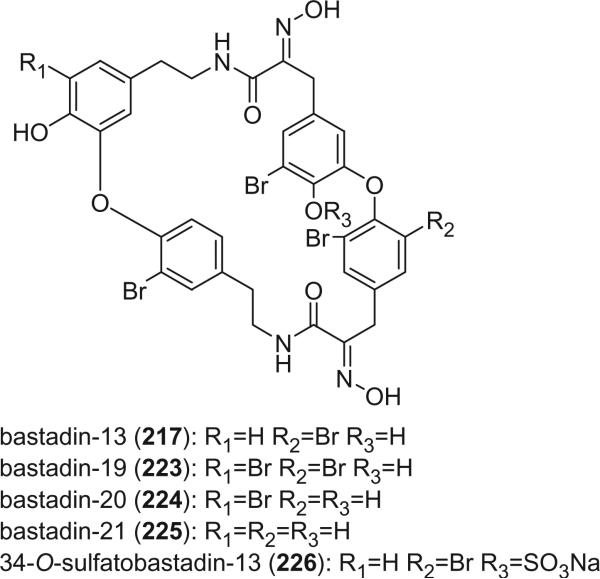
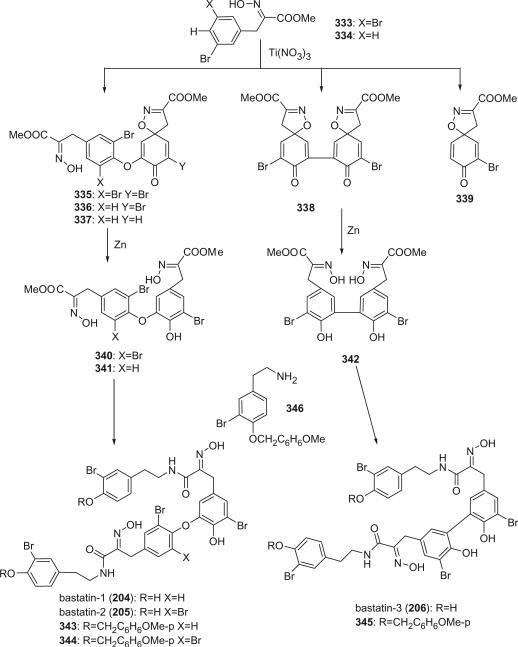
The bastadins are a series of predominantly macrocyclic sponge metabolites, which are biogenetically derivable from four bromotyrosines by oxidative phenolic coupling of two tyramine–tyrosine units connected through an amide bond. To date, there are 24 bastadins isolated from marine sponges. The pioneering studies of Wells and colleagues in the late 1970s led to the isolation and identification of bastadins 1–7 from Ianthella basta collected from Queensland, Australia (128,129). Bastadins-1 (204), 2 (205), and 3 (206), and 10-O-sulfatobastadin-3 (227) (130), are the only known acyclic bastadins. Bastadin-3 (206) and its sulfate (227) (130) both have a biaryl bond connecting two tyramine–tyrosine units, other than the ether bridge present in the remaining bastadins (129).
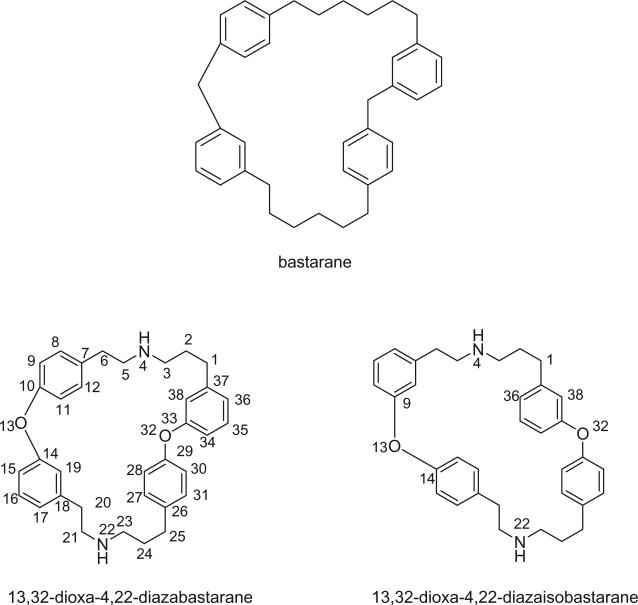
The remaining 20 bastadins possess a macrocyclic system, which was given a name bastarane and numbered as shown (129). According to the oxidative cyclization, there are two structural classes, i.e. 13,23-dioxa-4,22-diazabastarane and 13,32-dioxa-4,22-diazaisobastarane (131).
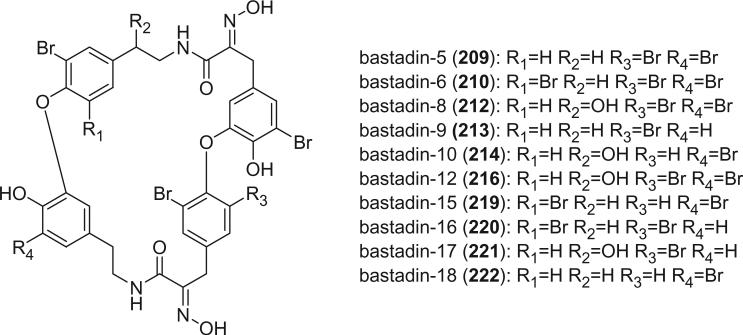
Most of the bastadins are of the 13,23-dioxa-4,22-diazabastarane type, including bastadins-4 (208), −5 (209), −6 (210), −7 (211) (129), −8 (212), −9 (213), −10 (214), −11 (215) (132), −12 (216), −14 (218) (133), −15 (219) (134), −16 (220), −17 (221) (135–137), −18 (222) (138), and 15,34-O-disulfatobstadin-7 (228) (130).
Five bastadins contain the 13,32-dioxa-4,22-diazaisobastarane ring pattern, including bastadins-13 (217) (131), −19 (223) (139), −21 (225) (140), and 34-O-sulfatobastadin-13 (226) (141).
The majority of the bastadins were isolated from the marine sponge Ianthella basta (class Demospongiae, order Verongida, family Inthellidae) with several obtained from I. flabelliformis, I. quadrangulata, Ianthella sp., and Psammaplysilla purpurea. Bastadins 1–7 (204–211) were isolated from the Australian sponge I. basta, as antimicrobial agents against Gram positive organisms in 1980 (128,129). Their structures were determined by spectroscopic methods and alcoholic KOH hydrolysis. X-ray diffraction analyses were performed on a single crystal of bastadin-4 (208) and bastadin-5 permethylate. The X-ray structure showed that the conformations of bastadin-4 and bastadin-5 permethylate are considerably different. The macrocyclic ring of bastadin-5 permethylate appears to be open, whereas the ring of bastadin-4 is more elongated.
The studies of bastadins 8–13 were published simultaneously, unfortunately leading to the assignment of the same number to different bastadins. Scheuer et al. suggested that these alkaloids be renamed in the order of when they were received for publication. Thus, bastadins-8 to −11 of Pordesimo and Schmitz (132) retained their original numbering, bastadin-9 of Miao et al. (142) was renumbered as bastadin-12 (bastadin-8 from Miao et al. coincidentally has the same structure as bastadin-8 of Pordesimo and Schmitz), and bastadin-12 of Butler et al. (131) became bastadin-13. Bastadins-8 to −11 (212–215) were isolated from the sponge I. basta collected in Guam by Pordesimo and Schmitz in 1990 (132). Bastadin-4 (208), −8 (212), and −9 (213) exhibited cytotoxic and anti-inflammatory activities. Bastadins-8 and −12 were isolated from the Papua New Guinean sponge I. basta by Miao et al. (142).
The structure of bastadin-13 (217) was elucidated as a novel alternative oxidative cyclization of bastarane, which was proposed for nomenclature purposes as 13,32-dioxa-4,22-diazaisobastarane, based on detailed spectroscopic analyses and derivatization (131). The co-occurrence of bastadin-13 and bastadin-9 (213) in a single specimen of I. basta collected from Australia introduced another dimension to the structure elucidation of the bastadins, therefore it can no longer be assumed on biosynthetic grounds that all cyclic bastadins possess the same 13,32-dioxa-4,22-diazabastarane ring system (131). Bastadin-14 (218), isolated from Psammaplysilla purpurea collected in Pohnpei, Micronesia, is the only bastadin isolated from a sponge that does not belong to the genus Ianthella. Bastadin-15 was identified from an undescribed marine sponge of the genus Ianthella collected from Australia (134). Bastadin-16 (220) and −17 (221) were isolated from an Indonesian collection of I. basta (135–137). Bastadin-18, along with bastadins-1, −2, −5, −6, −8, and −10, were isolated from the marine sponge I. basta collected in Papua New Guinea (138). In addition, a mixture of bastadins-2 and −5 were also isolated from I. flabelliformis.
Bastadins-8 and −10 were found to be moderate inhibitors of inosine 5′-phosphate dehydrogenase (138). Molinski et al. reported bastadins-19 (223), −20 (224), 15,34-O-disulfatobastadin-7 (228), and 10-O-sulfatobastadin-3 (227) from I. basta, as novel modulators of the skeletal isoform of the ryanodine-sensitive sarcoplasmic reticulum calcium channel by a novel mechanism involving the FKBP12/RyR-1 complex (130,139). Bastadin-21 (225) was isolated from an Australian marine sponge I. quadrangulata (140). Bastadins-6 and −16 were also reported from the Indian sponge Psammaplysilla purpurea, along with other bromotyrosine derivatives (46). The absolute stereochemistry of the C-6 hydroxyl group in bastadins-8 (212), −10 (214), and −12 (216) were determined as S by Pettit et al. using the Mosher–Trost method (143).
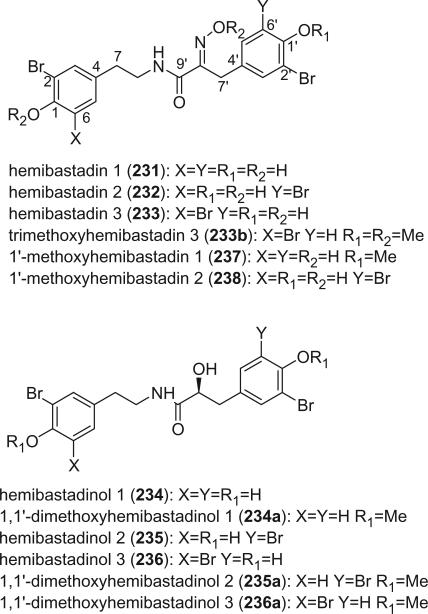
2. Hemibastadins
Two dimers of bromotyrosine, trimethylhemibastadin-1 (229) and trimethylhemibastadin-2 (230), were isolated as their methyl ethers, along with bastadin-9 and bastadin-12 from the sponge I. basta (131). Although trimethylhemibastadin-1 (229) and trimethylhemibastadin-2 (230) were obtained from a methylated fraction, it is not clear whether the natural products themselves were methylated to any extent.
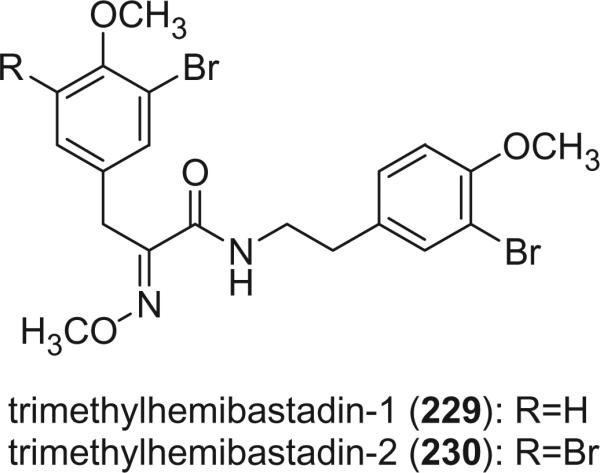
Eight hemibastadins were identified from a scale-up (160 kg wet weight) collection of the sponge I. basta, including hemibastadins 1 (231), 2 (232), and 3 (233), hemibastadinols 1 (234), 2 (235) and 3 (236), and 1′-methoxyhemibastadin 1 (237) and 2 (238) (41). Hemibastadin 2 (232) and 3 (233) were obtained as a mixture (3:1), while hemibastadinol 2 (235) and 3 (236) were an optically active oily mixture (19:1) that effectively resisted separation. The absolute stereochemistry of hemibastadinols 1 (234), 2 (235), and 3 (236) was determined as S using Trost's modification of the Mosher's method (144).
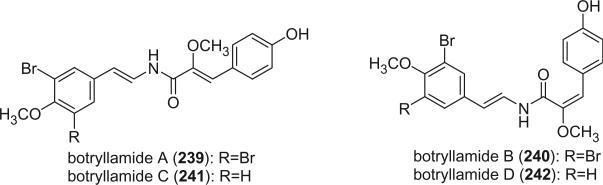
Botryllamides A–D (239–242), which have similar structures as the hemibastadins, but with two additional double bonds, were isolated from the Philippine ascidian Botryllus sp. and the Australian ascidian Botryllus schlosseri (145).
F. OTHER STRUCTURAL CLASSES
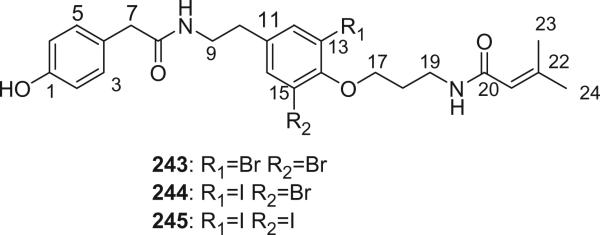
Three novel halogenated metabolites (243–245) were isolated from the Caribbean sponge Iotrochota birotulata, which is taxonomically far removed from the order Verongida (146). Bromotyrosine-derived metabolites have been considered to be characteristic of the sponges in the order of Verongida (147). I. birotulata is the first sponge that elaborates such metabolites which does not belong to the order Verongida. The presence of iodine atoms in alkaloids 244 and 245 is interesting. Iodo-compounds are relatively rare in marine chemistry, and particularly in sponges, even if all known haloperoxidases are effective in oxidizing iodide (148). The biosynthesis of iodinated metabolites seems to be related to the ability of the organism to concentrate iodide from sea water, rather than the presence of a specific peroxidase. Most iodo-metabolites have been isolated from red algae, which are known to contain as high as 0.5% of iodine by wet weight. Significant amounts of iodine (0.12–1.21%), together with comparable quantities of bromine (0.16–2.66%), were reported to exist in the spicule tracts of I. birotulata (149), further supporting the relationship between the presence of iodo-metabolites and high concentrations of iodine in sponge tissues.
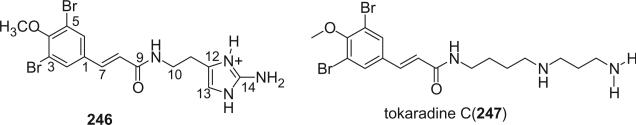
Alkaloid 246 was isolated from the Caribbean sponge Verongula sp. (38). Tokaradine C (247) was isolated from the marine sponge Pseudoceratina purpurea as a toxic constituent against the crab Hemigrapsus sanguineus (123).
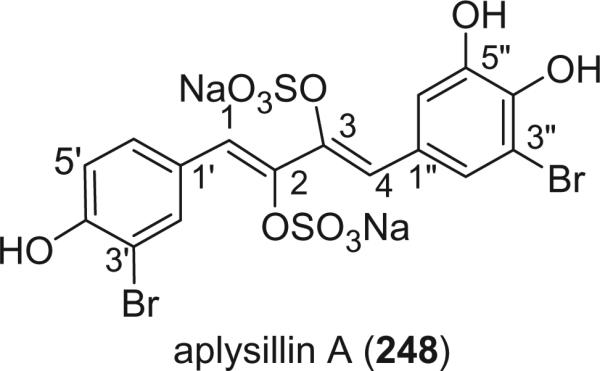
An unusual disulfate ester of a 1,4-diphenyl-1,3-butadiene, aplysillin A (248), was isolated from the sponge Aplysina fistularis fulva, collected at a depth of 369 feet off Sweetings Cay, Grand Bahama Island (150). Aplysillin A weakly inhibited the binding of thrombin to platelet membranes with an IC50 value of 20 μM.
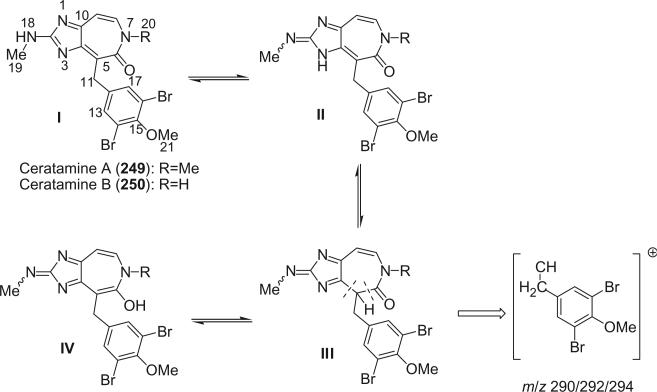
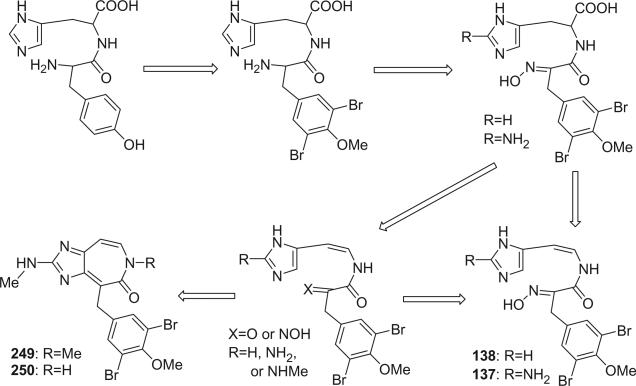
Two novel antimitotic heterocyclic alkaloids, ceratamines A (249) and B (250), were isolated from the marine sponge Pseudoceratina sp. collected in Papua New Guinea (151). Their structures were elucidated by analysis of the spectroscopic data. A number of possible tautomers exist for ceratamine A as shown below. Each of the constitutional isomers II, III, and IV can exist as the E and Z stereoisomers about the C-2/N-18 imine bond. Both 1H and 13C NMR spectra showed evidence for two forms. Scalar coupling observed between the Me-19 and NH-18 resonances provided evidence that the major tautomer observed in the 1H NMR spectrum was I. A significant fragment peak cluster at m/z 290/292/294 (1:2:1) in the EIMS of ceratamine A could formally arise from tautomer(s) III via an α cleavage next to the carbonyl accompanied by cleavage of the bond linking the substituted phenethyl fragment to the imidazole ring carbon as shown (151).
The obvious biogenetic relationship of ceratamines A and B to 5-bromoverogamine (138) (100), also isolated from a Pseudoceratina sp., and ianthelline (137) (99) supports the proposed structures 249 and 250. The putative biogenetic precursors to these alkaloids are histidine and tyrosine as shown.
Two new cytotoxic bromotyrosine alkaloids, ma'edamines A (251) and B (252), with a unique 2(1H)pyrazinone ring were isolated from the Okinawan marine sponge Suberea sp., along with aplysamine 2 (172), purpuramine H (186), and purpuramine I (187) (152). Biogenetically, ma'edamines A (251) and B (252) may be generated from the 11,12-dehydro form of aplysamine-2 (172) and purpuramine I (187) through the formation of a six-membered ring and dehydroxylation.
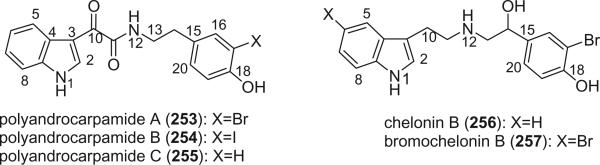
Polyandrocarpamides A–C (253–255), which are derived from tryptophan and tyrosine subunits, were isolated from the marine ascidian Polyandrocarpa sp. (153). Structurally related alkaloids, the chelonins, were reported from the marine sponge Chelonaplysilla sp., of which chelonin B (256) and 5-bromochelonin B (257) contain a bromotyrosine (154).
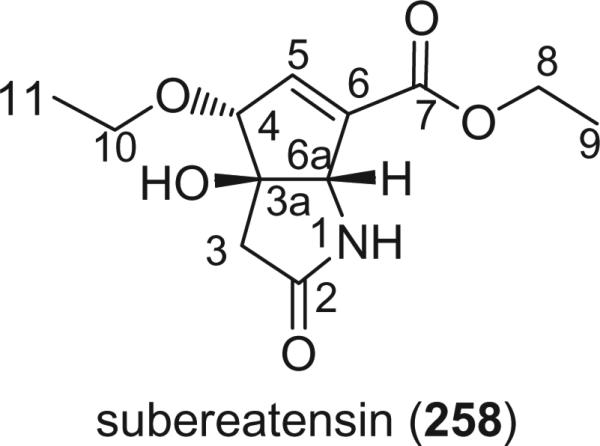
Examination of the marine sponge Suberea aff. praetensa (Row) from the Gulf of Thailand furnished, in addition to the known cavernicolins, an unusual rearranged tyrosine metabolite subereatensin (258) (155).
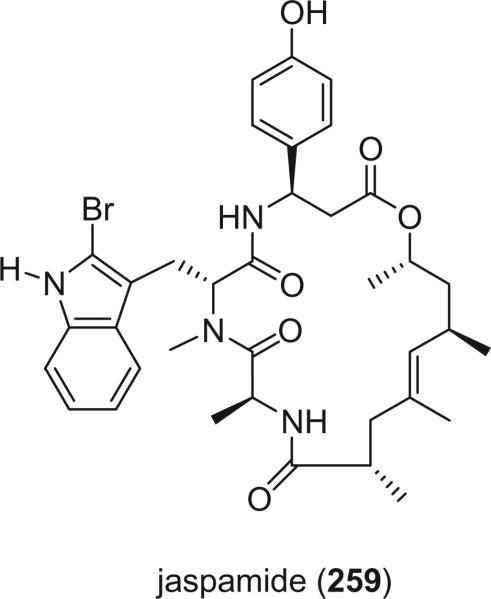
Geodiamolides and jaspamides are closely related cyclic depsipeptides isolated from a variety of tropical marine sponges. Jaspamide (259), also named jasplakinolide, obtained independently from Jaspis sp. collected in Palau (156) and Fiji (157), was the first member of this group of depsipeptides to be reported.
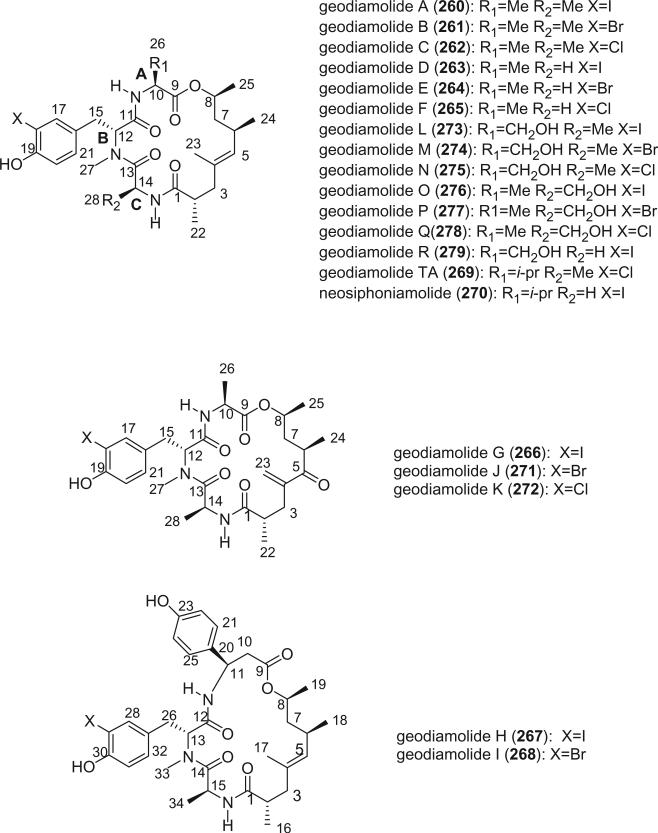
Shortly thereafter, geodiamolides A (260) and B (261) were isolated from the Caribbean sponge Geodia sp. (158). Subsequently, geodiamolides C–G (262–265, 266) were reported from the sponge Cymbastela (Pseudaxinyssa) sp. collected in Papua New Guinea (159,160); geodiamolides H (267) and I (268) were reported from a Geodia sp. (161); geodiamolide TA (269) was reported from the South African sponge Hemiastrella minor (162); neosiphoniamolide A (270) was reported from the New Caledonian sponge Neosiphonia superstes (163); and geodiamolides J–P (271–277) and R (279) were isolated from a recollection of the sponge Cymbastela sp. from Papua New Guinea (164). Although no chloro analogue of geodiamolide O (276) was isolated, it is likely to exist as a natural product, and therefore Andersen et al. reserved the name geodiamolide Q (278) for the hypothetical structure in anticipation of its future discovery (164).
The structures of the geodiamolides were determined by 1D and 2D NMR spectroscopy, and the absolute stereochemistries of geodiamolides A (260) and H (267) were determined by X-ray crystallography (158,161). There are now 19 known members of the geodiamolide family of cyclic depsipeptides. Variations have been observed in all three amino acid positions, as well as the polyketide portion of the molecule. Amino acid A variants include alanine (260, 261, 262–265, 276, 277), serine (273–275, 267), β-tyrosine (267, 268), and valine (269, 270); amino acid B variants include only the C-18 halogen atom, with iodine (260, 263, 266, 267, 273, 276, 278, 270), bromine (261, 264, 268, 271, 274, 277), and chlorine (262, 264, 272, 275, 269) all being observed; amino acid C variants include alanine (260, 261, 262, 267, 268, 273–275, 269), glycine (263–265), and serine (276, 277). The jaspamide/geodiamolide family of metabolites occurs across a taxonomically distant group of sponge species (156,158–160,162,163). To account for this observation, it has been suggested that microorganisms associated with the sponges produce these metabolites (162). The chondramides, which are jaspamide analogues from cultures of various strains of Chondromyces myxobacteria (165,166), strongly support the hypothesis of a microbial origin for the jaspamide/geodiamolides.
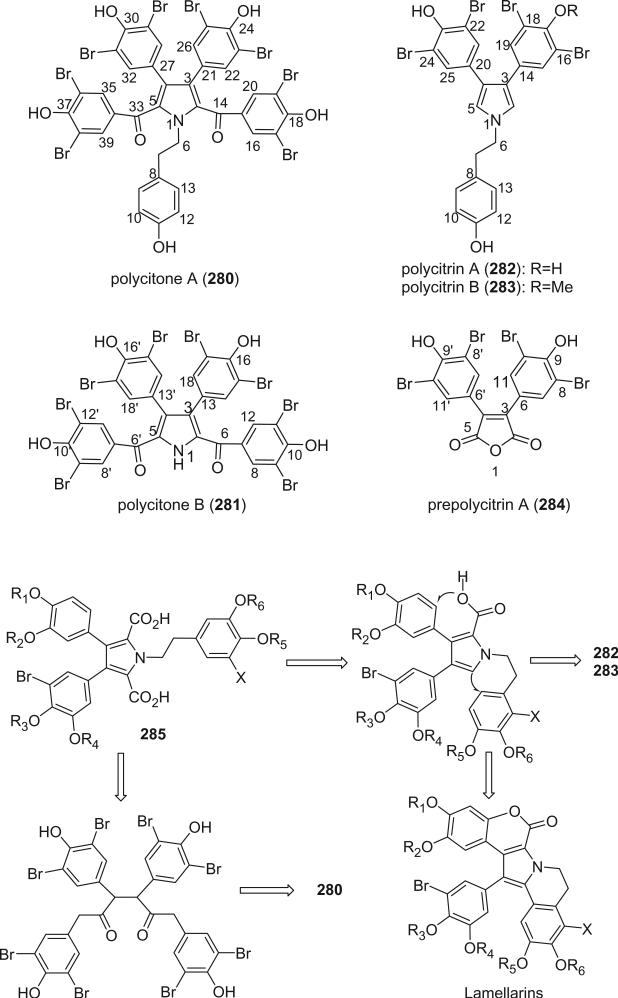
Polycitone A (280) and B (281), polycitrin A (282) and B (283), and prepolycitrin A (284) were reported from the Indo-Pacific ascidians Polycitor sp. and P. africanus by Kashman's group (167,168). Their structures were established by spectroscopic and chemical methods. The structure of polycitone A (280) was confirmed by X-ray crystallography (167).
The structures of polycitone A (280) and B (281) and polycitrin A (282) and B (283) were close to the lamellarins isolated from the mollusk Lamellaria sp. (169) and an ascidian of the genus Didemnum (170,171). Lammellarins are not included in this review because of the absence of a halogen in their structures. A possible biogenetic relationship between the lamellarins, polycitone A (280), and polycitrin A (282) and B (283) is shown below. The lamellarins may be derived from a precursor 285, which is a condensation product of three, suitably substituted tyrosine molecules.
III. Spectroscopic Data
As seen from the previous section, bromotyrosine derivatives exhibit a variety of chemical structures. The structure elucidation of bromotyrosine derivatives was based on chemical methods and the contemporary spectroscopic methods, which include 1D proton and carbon NMR, 2D homo- and heteronuclear correlations, as well as different mass spectroscopic methods. Some of the structures were verified by X-ray diffraction analysis. The physicochemical properties are listed in Table I, including appearance, molecular formula, MS, MP, [α]D, UV, IR, and CD. The proton and carbon NMR data are collected in Table II. All of the 13C-NMR resonance assignments for C-2 and C-4 of the spirocyclohexadienylisoxazoline type of compounds were revised based on Fattorusso's (61) COLOC NMR experiment, and our HMQC and HMBC experiment on a number of mono- and bis-spirocyclohexadienylisoxazoline bromotyrosine derivatives (12). The chemical shift of C-2 is at around 114 ppm and that of C-4 is around 122 ppm. The relative stereochemistry of the hydroxyl group at C-1 and the oxygen atom in the spirocyclohexadienylisoxazoline ring can be determined by comparison of the chemical shifts of H-1 and H-7 with synthesized cis and trans isomers (217). For the trans configuration, H-1 resonances at 4.2 ppm, H-7 resonances at 3.1 (d, J = 18 Hz) and 3.8 (d, J = 18 Hz) while in cis configuration δ 4.5 (H-1) and 3.42 (2H, H-7) were observed. The absolute configuration of aerothionin (68) was determined by X-ray crystallographic analysis and the CD spectrum (60). The stereochemistry of the spirocyclohexadienylisoxazoline ring in other alkaloids can be conventionally deduced by comparison of the CD spectrum with that of aerothionin.
TABLE I.
Physical-Chemical Properties of Bromotyrosine Derivatives.
| 1 (3) | 5 (7) | 6 (7) | 14 (15) | 17 (16) | |
|---|---|---|---|---|---|
| Appearance | Crystal | Colorless crystals | Colorless crystals | Crystal | Crystal |
| Molecular formula | C8H7Br2NO3 | C9H9Br2NO3 | C9H9Br2NO3 | ||
| MS m/z | 380, 382, 384 [M–MeO]+ (8, 14, 8), 338, 340, 342, [M–BuO]+ (40, 78, 39), 306, 308, 310 [M–MeOH–BuO]+ (14, 27, 18) | 394, 396, 398 [M–MeO]+ (1, 1.5, 1), 338, 340, 342 [M–PenO]+ (21, 40, 20), 306, 308, 310[M–MeOH–PenO]+ (12.5, 25, 13) | 341, 339, 337 [M]+, 323, 321, 319 [M–H2O]+, 240, 242 [M–H2O–Br]+ | ||
| MP (°C) | 193–195 | 194–195 | 166–168 | 120–121 | 112–116 |
| [α]D | +186° (MeOH) | −189° (c 0.5, acetone) | |||
| UV λmax (MeOH) nm | 257 (8000) | 204 (9912) | 203 (12134) | 231 (3220), 284 (4915) | |
| IR νmax (KBr) cm−1 | 3445, 3420, 3125, 1700, 1675, 1660, 1650 | 3410, 3200, 1660 | 3390, 3185, 1668 | 3380, 2265, 1635, 1585 |
| 15:16 3:1(11) | 18 (19) | 19 (7) | 20 (7) | 21 (20) | |
|---|---|---|---|---|---|
| Appearance | Oil | Yellow crystals | |||
| Molecular formula | C8H7Br2NO3 | C9H8Br2O4 | C12H10Br2O3 | C24H24Br4N4O10 | C24H24Br4N4O10 |
| MS m/z | 323, 325, 327 [M]+, 306, 308, 310 (2), 278, 280, 282 (3), 201, 203, 205 (100) | 360, 362, 364 (49, 100, 48), 345, 347, 349 (M–Me, 15, 25, 14), 331, 333, 335 (M-29, 5, 9, 4), 317, 319, 321 (M-43, 36, 70, 35). | |||
| MP (°C) | 106–108 | 142–144 | |||
| [α]D | +30° (c 5.7, MeOH) | +22° (c 3, MeOH) | +152.5° (c 0.92, MeOH) | +160.7° (c 0.92, MeOH) | |
| UV λmax (MeOH) nm | 254 (4100), 249 (4000) | 281 (4900) | 207 (6717), 327 (3732) | 280 (10600), 231 (17400) | 281 (10500), 230 (17500) |
| IR νmax (KBr) cm−1 | 3443, 2263, 1710, 1611 | 3400, 1785 | CHCl3 1780 | 3402, 1728, 1665 | 3350, 1732, 1662 |
| 22:23 (3:1 mixture) (22) | 24 (23) | 25 (9) | 26, 27, 28, 29 mixture (9) | 31 (25) | 30 (24) | |
|---|---|---|---|---|---|---|
| Appearance | Oily mixture | Sticky semi-solid | Sticky semi-solid | Colorless needles | ||
| Molecular formula | C8H7Br2NO3 | C8H8BrNO3 | C8H8ClNO3 | C8H7BrClNO3 | ||
| MS m/z | VG-ZAB, EI 328 (2), 326 (4), 324 (2) (M+1), 327 (6), 325 (12), 323 (6) (M) [calcd for C8H779Br2NO3 322.8790; found 322.8766 ± 0.005], 246 (52) (M-Br), 229 (5), 227 (6) (244-OH), 218 (7), 216 (7) (244-CO), 204 (40), 202 (42) (244-ketone), 176 (24), 174 (24) (202-CO), 95 (17) (174-Br), 43 (100) (O=C=NH) | EIMS: 245 (2), 175 (5), 174 (6) | EIMS 203 (3), 201 (9, M+), 186 (1), 184 (3, M+–17), 161 (15), 159 (46, M+–CH2CO), 134 (5), 132 (15), 133 (5), 131 (15, M+–C2H2O2), 96 (21), 70 (95), 43 (100, OCNH+) | EIMS 283 (4), 281 (14), 279 (11) (four M+) | EIMS 219 (1), 217 (3, M+), 203 (12), 201 (35), 191 (2), 189 (7), 174 (4), 172 (11), 159 (33), 157 (100), 182 (13), 136 (15), 123 (19), 59 (52), 44 (46), 43 (35), 39 (40) | MS 264, 262 (2.7 each, (M+1)+), 263, 261 (1.4 each, M+), 247, 245 (7 each, ((M+1)–17)+), 246, 244 (1.5 each, M+–17), 235, 223 (23 each, M+–28), 218, 216 (19 each HR: 215.9613 ±0.01 C7H7BrNO2), 204, 205 (5 each, 244–42), 203, 201 (16 each, 244–243) 182 (70 HR: 182.0480 ±0.01 C7H6NO2), 59 (100), 53 (48), 44 (61), 43 (59) |
| MP (°C) | 183–185 | 165–170 | ||||
| [α]D | +0.036° (c 0.084, MeOH) | |||||
| UV λmax (MeOH) nm | 255 (4000), 315 sh (100) | 250 (6000), 315 (35) | 238 (4100) | 250 (2500), 312 (60) | 242 (2200) | 254 (2600) |
| IR νmax (KBr) cm−1 | (Nujol) 3400–3200, 1700, 1620 | (Nujol) 3300, 1680, 1605 |
| 43 (35) | 44 (35) | 45 (35) | 46 (35) | 67 (21) | 51 (40) | |
|---|---|---|---|---|---|---|
| Appearance | Amorphous white solid | Amorphous solid | Amorphous solid | Amorphous solid | Solid | Colorless solid |
| Molecular formula | C12H15Br2O3N | C13H17Br2O3N | C13H18BrO3N | C12H16BrO3N | C13H12Br2N2O5 | C9H11Br2NO |
| MS m/z | HRMS 378.9406 | HRMS 333.8839 [M+–NMe3] | HRMS 255.9720 [M+–NMe3] | HRMS 301.0323 | HRMS 433.9094 MS 434, 436, 438 (M+) 348, 350, 352 (M+–C3H4O3N), 318, 320, 322 (M+–C4H6O3N) | HRFAB: 307.9279 |
| MP (°C) | 220–222 | |||||
| [α]D | −1.35° | −8.33° | −9.00° | −15.00° | −33° (c 1.1, MeOH) | |
| UV λmax (MeOH) nm | (H2O) 287 (1260), 307 (1995), 281 (1250) | (H2O) 277 (1263) | (H2O) 277 (1260) | (H2O) 287 (1257), 307 (1994), 281 (1254) | (EtOH) 274 (524), 282 (491) | 224 (3.91), 284 (3.08) |
| IR νmax (KBr) cm−1 | (KBr matrix) 3446, 1576 | (KBr matrix) 3446, 1576 | (KBr matrix) 3446, 1576 | (KBr matrix) 3446, 1576 | (CHCl)3 3670, 3000, 1760, 1595 |
| 48 (38) | 207 (41) | 52 (42) | 54 (44) | 55 (45) | |
|---|---|---|---|---|---|
| Appearance | Amorphous solid | Colorless amorphous solid | Off-white powder | Pale yellow semicrystalline solid | Colorless oil |
| Molecular formula | C12H18NBr2O | C10H11BrN2O3 | C11H17Br2N2O | C15H24Br2N2O | C14H22N2OBr2 |
| MS m/z | FABMS 350, 352, 354 | EI 288 (6), 286 (6), 200 (99), 199 (13), 198 (100), 187 (32), 185 (32), 120 (20), 101 (10), 77 (11) HREIMS 287.9935 | EIMS 354.9 (M++H+4,1), 352.9 (M++H+2,3), 350.9752 (M++H, 2), 324.9 (57), 322.9 (100), 320.9 (61), 294.9 (18), 292.9 (16), 273.0 (88), 271.0 (91), 264.8 (30), 262.8 (16), 81.9 (66), 80.9 (27), 79.9 (68), 78.9 (28) | EI 406, 408, 410 (M+–2HCl, 3%; found: 406.025; C15H2479Br2N2O requires 406.0255), 165 (5), 121 (54; found: 121.065; C8H9O requires 121.0653), 85 (64), 58 (100) FAB 407, 409, 411 ([M+–2HCl]+1) | EIMS 396, 394, 392 (1:2:1), 352, 349, 347 (1:2:1), 324, 322, 320 (1:2:1), 95, 89, 69, 53, 51, 44 HREIMS 392.0081 |
| MP (°C) | 122–123 | ||||
| [α]D | |||||
| UV λmax (MeOH) nm | (EtOH) 277 (1260) | (CH3OH) 212 (7762), 281 (1380), 205 (18900) | 206 (28000), 284 (600) | 293 (400), 448 (440) | (MeOH, HCl salt) 218 (12000), 274 (800), 285 (1000) |
| IR νmax (KBr) cm−1 | (Dry film) 2578, 1635 | (NaCl film) 3387, 3310, 1651 | (NaCl) 3000, 1592, 1473, 1459, 1385, 1261, 865, 739 | 3000, 2940, 1632, 1454, 1256, 1043 | (KBr, HCl salt) 3420, 2950, 1635, 1455, 1260, 1035 |
| 56 (45) | 57 (46) | 58 (46) | 60 (49) | 59a (47) | |
|---|---|---|---|---|---|
| Appearance | Colorless oil | Brown solid | White solid | Colorless solid | Colorless crystalline compound |
| Molecular formula | C14H22N2OBr2 | C15H23Br2N2O3 | C15H23Br2N2O3 | C13H15Br2N3O2 | C15H22N2O2Br2 |
| MS m/z (int) | EIMS 396, 394, 392 (1:2:1), 353, 351, 349 (1:2:1), 315, 313 (1:1), 87, 69, 58, 44 HREIMS 394.0053 | FABMS 437, 439, 441 (1:2:1) HRFABMS 437.0083 | FABMS 437, 439, 441 (1:2:1) HRFABMS 437.0090 | FAB 404, 406, 408 (1:2:1) (Δ −1.6 mmu) | FABMS 421, 423, 425 (1:2:1) HRFABMS 421.010534 |
| MP (°C) | 160–162 | 170–173 | 148 | ||
| [α]D | |||||
| UV λmax (MeOH) nm | (MeOH, HCl salt) 214 (14000), 276 (8000), 288 (1100) | 207 (37800), 220 (12600), 276 (850 | (MeOH, HCl salt) 280 (944), 275 (950), 223 (5769) | ||
| IR νmax (KBr) cm−1 | (KBr, HCl salt) 3420, 2950, 1635, 1455, 1260, 1035 | 3300, 2720, 1720, 1620, 1480 | 3350, 2700, 1725, 1610 | (Film) 3200, 3040, 2950, 1680, 1450, 1200, 1130 | 3328, 2700, 1680, 1580, 1480, |
| 61 (50) | 62 (51) | 63 (53) | 64 (53) | 65 (54) | 67 (21) | |
|---|---|---|---|---|---|---|
| Appearance | Amorphous solid | Colorless oil | Colorless flakes | White crystals | Colorless crystals | Solid |
| Molecular formula | C13H16Br2N3O3 | C7H25N2O4Br2 | C16H21Br2NO3 | C14H18Br2NO3 | C13H12N2O5Br2 | C13H12Br2N2O5 |
| MS m/z | FABMS 420, 422, 424 (1:2:1) HRFABS 419.9568 | ESIMS 479, 481, 483 (1:2:1, [M+H]+), 501, 503, 505 (1:2:1, [M+Na]+) HRFABMS 479.0176 | EIMS 437 (0.7), 435 (1.4), 356 (1), 354 (1), 167 (1), 149 (1), 86 (8), 84 (2), 73 (3), 71 (3), 58 (100) HREIMS 432.9860 | FABMS 410 (46), 408 (94), 406 (61), 325 (11), 323 (24), 321 (14), 307 (8), 305 (19), 303 (10), 244 (12), 58 (100) HRFABMS 407.9641 | 433.9109 | HRMS 433.9094 MS 434, 436, 438 (M+) 348, 350, 352 (M+–C3H4O3N), 318, 320, 322 (M+–C4H6O3N) |
| MP (°C) | 67 | 193–194 | 222–225 | 220–222 | ||
| [α]D | +3.0° (c 0.8, MeOH) | +35° (c 0.1, MeOH) | +8.9° (c 0.87, MeOH) | −33° (c 1.1, MeOH) | ||
| UV λmax (MeOH) nm | 283 (880) | 230 (11200), 282 (9500) | 223 (13700), 267 (11000) | 275 (480) | (EtOH) 274 (524), 282 (491) | |
| IR νmax (KBr) cm−1 | (neat) 3365, 3280, 1675, 1453, 1206, 1138 | 3411, 1630, 1057 | (Neat, NaCl) 2920, 2840, 2805, 2758, 1712, 1635, 1195, 950 | (Neat, NaCl) 3700–2900, 2940, 2870, 1635, 1565, 930 | 1748 (sh. 1722, 1703) | (CHCl3) 3670, 3000, 1760, 1595 |
| 68 (58) | 70 (62) | 71 (63) | 72 (64) | 73 (61) | 74 (61) | 75 (65) | |
|---|---|---|---|---|---|---|---|
| Appearance | Plates | White powder | Colorless glass | White powder | |||
| Molecular formula | C24H26Br4N4O8 | C24H27Br4N4O10 | C24H26Br4N4O9 | C24H24Br4N4O9 | C24H24Br4N4O10 | C24H24Br4N4O10 | C25H26Br4N4O9 |
| MS m/z | HRFABMS 850.84 13 (Δ 1.9 mmu) | HRFABMS 831.8235 (Δ 0.1 mmu) | FABMS 841, 843, 845, 847, 849 | ||||
| MP (°C) | 134–137 (dec.) | 162–164 | 174.6–176.6 (dec) | ||||
| [α]D | +252° (acetone) | −64.2° (c 0.1, MeOH) | +189° (c 0.15) | +181.15° (c 2.17, DMSO) | +152.5° | +160.7° | |
| UV λmax (MeOH) nm | EtOH) 234 (4.16), 284 (4.13) | 282 (9400), 225 (19000), 202 (34000) | 284 (11100), 233 (19750), 205 (18900) | (DMSO) 284 (11500), 262 (11600) | 280.5 (10600), 231 (17400) | 281 (10500), 230.5 (17500) | 284 (10500), 232 (19000) |
| IR νmax (KBr) cm−1 | 3335, 3160, 1675, 1660, 1580, 1550 | 3600–3000, 1660, 1600, 1530 | 3400, 3350 (OH, NH), 1650 (CONH) | 3600–3000, 1713, 1665, 1558, 1552, 1435, 1293, 1271, 1131, 1100, 1025 | 3402, 1728, 1665 | 3350, 1732, 1662 | (KBr matrix) 3450, 1715, 1665 |
| CD | N/A | N/A | N/A | N/A | N/A | N/A | 285, +52000 250, +60,200 |
| 76 (66) | 77 (67) | 78 (30) | 79 (25) | 81 (70) | |
|---|---|---|---|---|---|
| Appearance | Colorless glassy solid | NA | Colorless solid | Amorphous white solid | Amorphous white solid |
| Molecular formula | C27H32Br4N4O9 | C22H20Br4N4O9 | C22H20Br2N4O8 | C31H30Br6N4O11 | C31H30Br6N4O11 |
| MS m/z | FABMS (thioglycerol+MeOH) 875 (1), 873 (2), 871 (4) [M]+, 869 (2), 867 (1), 861 (1.5), 859 (3.5), 857 (5), 855 (3.5), 853 (1.5), 322 (1.5), 320 (2.5), 318 (1.5), 297 (2.5), 295 (4.5). 293 (2.5), 281 (18), 279 (37), 277 (18), 70 (100) HRFABMS 871.97028, found 871.97930 [M]+ | FABMS 801, 803, 805, 807, 809 | FABMS 629 | NA | FABMS 1141, 1139, 1137, 1133 ([M+Na]+, 39, 64, 100, 80, 48) |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | +104° (c 1.67, MeOH) | +65.2° (c, 1.04, acetone) |
| UV λmax (MeOH) nm | 234 (9000), 283 (4300) | 250 (7850) | 260 (0.63), 230 (0.78), 210 (0.90) | 283 (10387), 223 (26545) | (EtOH) 233 (13500), 283 (2650) |
| IR νmax (KBr) cm−1 | 3382, 2357, 1665, 1549, 1106, 603 | (KBr matrix) 3360, 1713, 1705, 1660, 1600 | NA | 3395, 1655, 1602, 1550 | 3350, 2920, 1655, 1545 |
| CD | NA | 252, +1.6 | 344.1 (2.5), 270.3 (13.8), 233.6 (24.1), 199.0 (−25.0) | NA | NA |
| 82 (71) | 83 (63) | 84 (72) | 85 (72) | 86 (65) | |
|---|---|---|---|---|---|
| Appearance | Pale yellow gummy solid | Unstable yellow power | Powder | NA | |
| Molecular formula | C31H29Br6N4O11 | C31H30Br6N4O9 | C31H28Br6N4O10 | C31H30Br6N4O10 | |
| MS m/z | HRFABMS 1112.6897 (Δ 0.6 mmu) | EIMS 351 (2), 349 (4), 347 (2), 336 (2), 334 (4), 332 (2), 323 (4), 321 (9), 319, (5), 308 (6), 306 (12), 304 (6), 267 (6), 265 (12), 263 (6) | FABMS 1091, 1093, 1095, 1097, 1099, 1101, 1103 | ||
| MP (°C) | NA | NA | NA | NA | |
| [α]D | +130° (c 0.1, MeOH) | +98.5° (c 0.10, MeOH) | +136° (c 0.2, acetone) | NA | |
| UV λmax (MeOH) nm | 283 (9900), 242(14000), 225 (27000) | 284(10400), 257(16000), 224(26000) | 280 (3400), 205 (18500) | 284 (11000), 232 (19000) | |
| IR νmax (KBr) cm−1 | 3600–3200, 1720, 1660, 1590, 1525, | 3450, 3350 (OH, NH), 1645 (CONH) | (Nujol) 3400 br, 1725, 1650, 1590, 1530 | (KBr matrix) 3450, 1715, 1665 | |
| CD | NA | NA | NA | 285, +56500 250, +65200 |
| 87 (70) | 88 (70) | 89:90 (74) | 91(75) | |
|---|---|---|---|---|
| Appearance | Amorphous off-white powder | Amorphous white powder | Noncrystalline mixture | |
| Molecular formula | C29H26N4O11Br6 | C29H26N4O11Br6 | C29H26N4O9Br6 | C18H18Br4N2O6 |
| MS m/z | FABMS 1092, 1090, 1088, 1086, 1084, 1082, (1, 1.8, 1.8, 2, 1.8, 1.8, 1) [M+], 707 (0.8), 705 (1), 703 (0.8), 427 (10), 425 (18), 423 (10) | FABMS 1092, 1090, 1088, 1086, 1084, 1082 (<1) [M+], 707 (1), 705 (1), 703 (1), 427 (6), 425 (10), 423 (6) | FABMS 1054.68209 [M+H]+ | ESIMS 696.7766 [M+Na]+ |
| MP (°C) | N/A | N/A | N/A | |
| [α]D | −17.1° (c 1.26, acetone) | +50.0° (c 0.27, acetone) | +248° (c 0.012, CHCl3) | |
| UV λmax (MeOH) nm | (EtOH) 220 (12600), 250 (7740) | (EtOH) 215 (12570), 250 (7940) | ||
| IR νmax (KBr) cm−1 | 3360, 2930, 1750, 1660, 1600, 1540, 1260, 1095 | 3350, 2920, 1700, 1665, 1605, 1540, 1255, 1095, 910 |
| 92 (77) | 93 (78) | 94 (68) | 95 (68) | |
|---|---|---|---|---|
| Appearance | Brown amorphous solid | Yellow powder | Amorphous white solid | Crystalline |
| Molecular formula | C32H36Br4N10O8 | C44H43Br8N6NaO16S | C22H21Br4N3O8 | C26H25Br4N3O10 |
| MS m/z | FABMS 1005, 1007, 1009, 1011, 1013 HRFABMS 1008.9488 | FABMS 1583 [M–Na]+, 1503 [M–SO3Na] | NA | NA |
| MP (°C) | NA | 200 (dec.) | NA | 168–171 |
| [α]D | +111.4° (c 0.07, MeOH) | −96° (c 0.86, DMSO) | +93.5° (c 1.2, MeOH) | +122.7° (c 0.44, CHCl3) |
| UV λmax (MeOH) nm | 221 (19600, 279 (9500) | 231 (2690), 283 (11900) | 284 (5681), 230 (15217) | NA |
| IR νmax (KBr) cm−1 | (KBr matrix) 3450, 1665 | 3700–2500, 1660, 1584, 1541, 1257, 1218, 1183, 1047, 990 | 3360, 1750, 1660, 1600, 1545 | 3360, 1750, 1740, 1660, 1600, 1545 |
| CD | 245, +60000 285, +50000 |
252 (−16.0), 289 (−16.9) |
| 97 (79) | 98 (80) | 99 (81) | 101 (82) | |
|---|---|---|---|---|
| Appearance | Colorless oil | Amorphous white solid | White solid | Glass |
| Molecular formula | C10H10Br2N2O4 | C10H9Br2NO5 | C21H23Br4N3O5 | C21H24Br3N3O5 |
| MS m/z | EI: 380, 382, 384 (1:2:1) | FABMS 938, 940, 942, 944, 946 (1:4:6:4:1) [M+H]+ | FABMS 714, 716, 718, 720, 722 (1:4:6:4:1) [M+H]+ | HRFABMS 635.9360 |
| MP (°C) | 40–42 | 140–142 | NA | |
| [α]D | +86° | −38° (c 0.73, CHCl3) | −70° (c 0.7, MeOH) | +21° (c 0.47, CHCl3–MeOH 1:1) |
| UV λmax (MeOH) nm | 228 (8000), 290 (2000) | 270 (11650) | 283 (9392) | (CHCl3) 240 (23000), 287 (16700) |
| IR νmax (KBr) cm−1 | 3400, 2940, 1675, 1135, 1120 | NA | NA | (CHCl3–MeOH 1:1) 3690, 3630, 3420, 3020, 2945, 1600, 1570, 1535, 1500, 1465, 1255, 1015 |
| CD | 248 (+4.0), 284 (+4.0) |
| 102 (83) | 100 (81) | 102 (36) | 104(36) | 105 (78) | |
|---|---|---|---|---|---|
| Appearance | Colorless semisolid | Amorphous white solid | Colorless solid | Colorless solid | Colorless fine crystals |
| Molecular formula | C23H25Br4N3O7 | C36H51Br4N3O6 | C22H24Br4N3O7 | C25H29Br4N3O7 | C24H27Br4N3O7 |
| MS m/z | HRFABMS 797.82940 FABMS 802 (19), 800 (67), 798 (95), 796 (67), 794 (21), 776 (28), 700 (9), 681 (14), 661 (4), 612 (33), 532 (38), 510 (100), 482 (85), 464 (42), 437 (65), 413 (48) | FABMS 938, 940, 942, 944, 946 (1:4:6:4:1) [M+H]+ | ESI-FTMS 761.8307 (Δ +10.3 mmu) | ESI-FTMS 803.8777 (Δ −27.3 mmu) [M+H]+ | FABMS (Thiogylcerol matrix) 786, 788, 790, 792, 794 (1:4:6:4:1) [M+H] HRFABMS 785.8661, found 785.8649 [M+H] |
| MP (°C) | NA | 40–42 | NA | NA | 154–156 |
| [α]D | +69.0° (c 6.4, MeOH) | −38° (c 0.73, CHCl3) | +96.3° (c 0.19, MeOH) | +102° (c 0.067, MeOH) | −118° (c 1.02, MeOH) |
| UV λmax (MeOH) nm | 282 (5200), 208 (145000) | 270 (11650) | (EtOH) 208 (8100), 284 (1200) | (EtOH) 207 (8300), 284 (1200) | 210 (29500), 283 (7390) |
| IR νmax (KBr) cm−1 | 3692–3026, 2933, 2853, 1649, 1643, 1545, 1400, 1384, 1281, 1095, 1044, 988, 918, 868, 765, 739, 704 | NA | (NaCl) 3251 br, 2935, 1658, 1543, 1456, 1257, 989 | (NaCl) 3255 br, 3058, 2933, 1771, 1631, 1542, 1456, 1257, 1024, 987 | 3700–2300, 1660, 1630, 1540, 1260, 1045 |
| 106 (78) | 107 (78) | 108 (72) | 109 (85) | 110 (79) | |
|---|---|---|---|---|---|
| Appearance | Colorless fine crystals | Colorless powder | Powder | Colorless amorphous solid | Colorless oil |
| Molecular formula | C22H23Br4N3O7 | C22H22Br4N3O7S | C18H16Br4N2O6 | C24H30Br4N3O5 | C23H27Br4N3O5 |
| MS m/z | FABMS (Glycerol matrix) 758, 760, 762, 764, 766 (1:4:6:4:1) [M+H] HRFABMS 757.8348, found 757.8359 [M+H] | FABMS (Glycerol matrix) 836, 838, 840, 842, 844 (1:4:6:4:1) [M–Na]− | FABMS (Glycerol–MeOH–H+) 676.8 (0.2%, as the centre of a cluster of ions) [MH+] FABMS (3-nitrobenzyl alcohol) 698.6 (1.0% as the centre of a cluster of ions) [MNa+], 658.6 (1.1% as a quintet) [MH+–H2O] | FABMS 764, 762, 760, 758, 756 [M+], 748, 746, 742, 740, 684, 682, 680, 678, 602, 600, 658 HRFABMS 759.8892 | FABMS 742, 744, 746, 748, 750 (1:4:6:4:1) [M++H] HRFABMS 745.8721 [M+4+H]+, found 745.8776 |
| MP (°C) | 154–157 | 190 | 73–75 | NA | NA |
| −97° (c 0.58, MeOH) | −69° (c 0.19, MeOH) | +110° (c 0.2, acetone) | −4.5° (c 1.3, MeOH) | +6.6° (c 0.75, MeOH) | |
| UV λmax (MeOH) nm | 207 (45400), 283 (7300) | 206 (53200), 220 (25000 sh), 282 (6760) | 281 (6900), 207 (39200) | 220 (10000), 284 (1000) | 277 (1700), 284 (1400) |
| IR νmax (KBr) cm−1 | 3700–2300, 1660, 1635, 1540, 1260, 1045 | 3700–2500, 1662, 1596, 1541, 1258, 1217, 1046 | (Nujol) 3350 br, 1660, 1590, 1540, 1270, 1220, 1095 | 3450, 2980, 2880, 1690, 1470, 1400, 1220, 1150 | 3400, 2940, 2845, 1670, 1520, 1470, 1135, 1120 |
| 111 (79) | 112 (86) | 113 (69) | 114 (69) | 116 (79) | 117(79) | |
|---|---|---|---|---|---|---|
| Appearance | Colorless oil | Colorless oil | NA | NA | Colorless oil | Colorless oil |
| Molecular formula | C23H27Br4N3O5 | C22H25Br4N3O5 | C15H16Br2N4O4 | C16H19Br2N5O4 | C15H17Br2N5O4 | C15H17Br2N5O5 |
| MS m/z | FABMS 742, 744, 746, 748, 750 (1:4:6:4:1) [M++H] HRFABMS 745.8721 [M+4+H]+, found 745.8729 | ESIMS 726.85, 728.85,730.85, 732.86,734.85 [M+H]+ HRESIMS 728.8676 (Δ 0.8 mmu) [M+H]+ | FABMS 475, 477, 479 [MH+] | FABMS 504, 506, 508 [MH+] | FABMS 490, 492, 494 (1:2:1) [M++H] HRFABMS 491.9705 [M+2+H]+, found 491.9682 | FABMS 506, 508, 510 (1:2:1) [M++H] HRFABMS 507.9654 [M+2+H]+, found 507.9672 |
| MP (°C) | NA | NA | NA | NA | NA | NA |
| [α]D | +9.1° (c 0.39, MeOH) | NA | +187° (c 2.0, MeOH) | +139° (c 1.9, MeOH) | +24° (c 0.98, MeOH) | +26° (c 0.38, MeOH) |
| UV λmax (MeOH) nm | 277 (1700), 284 (1400) | 280 (3.28) | NA | NA | 277 (1700), 284 (1400) | 231 (6300), 284 (2400) |
| IR νmax (KBr) cm−1 | 3400, 2940, 2850, 1675, 1470, 1200, 1135, 1120 | 1736, 1655, 1591, 1542, 1458, 1390, 1257, 1044, 737 | NA | NA | 3400, 2930, 2845, 1680, 1540, 1430, 1200, 1135 | 3400, 2920, 2850, 1670, 1540, 1430, 1200, 1135, 1125 |
| 119 (89) | 120 (83) | 121 (90) | 122 (91) | 123 (89) | |
|---|---|---|---|---|---|
| Appearance | NA | Colorless oil | NA | Colorless amorphous solid | NA |
| Molecular formula | C15H18Br2N5O4 | C16H18Br2N5O4 | C24H22O8N6Br2 | C27H29Br4N7O7 · HCl | C27H32Br4N7O6 |
| MS m/z | FABMS 490, 492 494 [MH+] | HRFABMS 503.97073 FABMS 506 (53), 504 (100), 502 (55), 307 (5), 154 (50) | FABMS 681, 683, 685 (1:2:1) HRFABMS 680.9940 | FDMS 880, 882, 884, 886, 888 (1:4:6:4:1) | FABMS 866, 868, 870, 872, 874 [MH+] |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]d | −158° (c 1.0, MeOH) | +121.9° (c 5.7, MeOH) | NA | −85° (c 2.10, MeOH) | −10° (c 1.0, MeOH) |
| UV λmax (MeOH) nm | 220 (4.47), 284 (4.08) | 266 (13100), 226 (13700) | NA | NA | 220 (4.54), 228 (4.11) |
| IR νmax (KBr) cm−1 | 3400, 1680, 1543, 1437, 1381, 1250 | 3654–3000, 2937, 1660, 1595, 1543, 1435, 1273, 1219, 1047, 1025, 997, 824, 765 | NA | NA | 3375, 1680, 1543, 1456, 1387, 1250 |
| 124 (83) | 125 (79) | 126 (66) | 127 (92) | 128(92) | |
|---|---|---|---|---|---|
| Appearance | Colorless semisolid | Colorless oil | Colorless glassy solid | NA | NA |
| Molecular formula | C16H24Br2N5O4 | C15H21Br2N5O4 | C15H7D7 Br2N5O5 | C16H19Br2N3O7 | C16H19Br2N3O7 |
| MS m/z | HRFABMS 510.01640 FABMS 512 (25), 510 (50), 508 (25), 451 (7), 273 (22), 219 (12), 154 (100) | FABMS 494, 496, 498 (1:2:1) [M++H] HRFABMS 496.0018 [M+4+H]+, found 496.0035 | FABMS (Thioglycerol) 518 [M+7D–7H]+ (13), 513 [M+2D–2H]+ (4), 496 (8), 478 (6), 295 (23), 279 (22), 157 (60), 71 (100) HRFABMS 518.20911 [M+7D–7H]+ | ESIMS 524, 526, 528 (1:2:1) [M+H]+ | ESIMS 524, 526, 528 [M+H]+ |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | +47.0° (c 7.9, MeOH) | +27° (c 0.18, MeOH) | NA | NA | NA |
| UV λmax (MeOH) nm | 284 (2500), 218 (4600) | 228 (8600), 290 (3400) | 232 (9100), 283 (4250) | 229 (18500), 280 (10600) | 227 (19000), 281(10500) |
| IR νmax (KBr) cm−1 | 3588–3050, 3024, 2782, 1658, 1402, 1024, 992 | 3400, 2920, 1660, 1520, 1470, 1210 | 3500–3000, 2928, 2860, 1690–1630, 1405, 1100, 605 | 3450, 1730, 1715, 1665 | 3450, 1730, 1715, 1665 |
| 129 (94) | 130 (94) | 131 (95) | 132 (96) | |
|---|---|---|---|---|
| Appearance | Colorless glass | Colorless glass | Glass | Colorless oil |
| Molecular formula | C21H23Br4N3O6 | C21H23Br4N3O7 | C22H25Br4N3O7 | C36H51Br4N3O8 |
| MS m/z | EIMS 406 (0.4), 404 (0.4), 402 (0.4), 3.62 (1), 360 (2), 358 (2), 356 (1), 323 (1), 321 (2), 319 (2), 281 (2), 280 (7), 279 (6), 278 (17), 277 (4), 276 (4), 268 (1), 267 (4), 266 (2), 265 (6), 264 (1), 263 (7), 248 (5), 246 (3), 200 (6), 198 (4), 58 (100). CIMS 407, 405, 403, 361, 359, 281, 279, 277, 275, 249, 247, 233, 231, 221, 219, 193, 191, 166, 164, 125, 123, 121 | EIMS 523 (9), 521 (27), 519 (26), 517 (8) overlapping 2 Br clusters, 477 (3), 475 (4), 473 (2) 2 Br cluster, 441 (6), 439 (13), 437 (6) 2 Br cluster, 423 (5), 421 (11), 419 (4) 2 Br cluster, 407 (3), 405 (4), 403 (2) 2 Br cluster, 393 (11), 391 (39), 358 (2), 356 (6), 354 (2), 283 (8), 281 (16), 279 (12), 278 (9), 276 (20), 274 (18), 272 (8) 2 Br cluster, 249 (94), 247 (97) 1 Br cluster, 235 (16), 233 (14) 1 Br cluster, 221 (24), 219 (27) 1 Br cluster, 207 (11), 205 (24), 203 (16) 2 Br cluster, 196 (9), 194 (100), 192 (17), 179 (9), 177 (18), 175 (8), 169 (4), 167 (19), 155 (45), 127 (38), 125 (43), 113 (9), 111 (12). CIMS 441, 439, 437 (2 Br), 407, 405, 403 (2 Br), 340, 338, 336 (2 Br), 249, 247 (1 Br), 221, 219 (1 Br). HRFDMS 749.8308 (M+H, C21H2479Br281Br2N3O7, calcd 749.83068) | HRFABMS 763.8499 | HRFABMS 974.0468 [M+H] |
| [α]D | −65.2° (c 0.52, MeOH) | −60.2° (c 0.63, MeOH) | −57.1° (c 0.014, MeOH) | −71.4° (c 2.8, acetone) |
| UV λmax (MeOH) nm | 229 (12700), 262 (7100 sh), 276 (4650 sh), 283 (3200 sh) | 218 (28600), 256 (7000 sh), 262 (6400 sh), 277 (4000 sh), 282 (3000) | 218 (17600), 255 (6700 sh), 279 (3000 sh) | 210 (50900), 224 (27600 sh), 258 (10400 sh) |
| IR νmax (KBr) cm−1 | (CHCl3) 3420, 2950, 1675 s, 1660, 1540–1530, 1460, 1260, 1150, 1120, 910 | (CHCl3) 3420, 2940, 1670 s, 1620, 1590, 1530 s, 1450, 1250, 1140, 1110, 900 | (KBr smear) 3362, 2925, 2851, 1668, 1652, 1558, 1540, 1456, 1258, 1197, 1146, 1118, 1044, 1024 | (CHCl3) 3430, 3390, 2910, 2830, 1660, 1570, 1445, 1135, 1105, 1030, 980, 950, 890 |
| 133 (96) | 134 (97) | 135 (98) | 136 (98) | |
|---|---|---|---|---|
| Appearance | Bright yellow oil | NA | Colorless solid | Colorless solid |
| Molecular formula | C27H25Br4O8 | C22H25O6Br4N3 | C22H23Br4N3O7 | C35H51Br4N3O7 |
| MS m/z | FABMS 994.4 [M+H+C4H10O2S2], 840.0 [M+H]. HRFABMS 993.8563 [M+H+C4H10O2S2] | FABMS [M+H]+ 752 (22), 750 (57), 748 (100), 746 (71), 744 (23), 393 (26) | FABMS (positive, glycerol matrix) 758, 760, 762, 764, 766 [M+H]+, 780, 782, 784, 786, 788 [M+Na]+ HRFABMS 783.8152 (calcd for C22H2379Br281Br2N3O7Na, Δ +2.6 mmu) | FABMS (Positive, NBA matrix) 954, 956, 958, 960, 962 [M+H]+, 976, 977, 980, 982, 984 [M+Na]+ HRFABMS 980.0320 (calcd for C36H5179Br281Br2N3O7Na, Δ −0.4 mmu) |
| MP (°C) | NA | NA | NA | |
| [α]D | −80.3° (c 0.3, acetone) | −62.3° (c 1.2, MeOH-CH2Cl2) | −89.7° (c 0.146, MeOH) | −53.5° (c 0.263, acetone) |
| UV λmax (MeOH) nm | 208 (53400), 224 (33300 sh), 298 (25000) | NA | (EtOH) 207 (64200), 218 (28900 sh), 255 (11000 sh) | (EtOH) 207 (51100), 219 (22700 sh), 255 (9000 sh) |
| IR νmax (KBr) cm−1 | (CHCl3) 3380, 2910, 1700, 1640, 1610, 1445, 1145, 1105, 1035, 985, 950, 880 | (Neat) 3450, 3300, 1660 s, 1620, 1590, 1260 | (Film) 3300, 2930, 1660, 1530, 1450, 1380, 1260, 1110, 760 | (Film) 3320, 2920, 2850, 1650, 1540, 1450, 1250, 1120 |
| 137 (99) | 138 (100) | 139 (101) | 140 (33) | 141 (102) | |
|---|---|---|---|---|---|
| Appearance | NA | Oil | Yellow semisolid, oil | NA | Colorless amorphous solid |
| Molecular formula | C15H17N5O3Br2 | C15H16N4O3Br2 | C15H18N4O3Br | C14H15BrN4O3 | C31H48N6O4Br2 |
| MS m/z | EIMS 477, 475, 473 | EIMS 351, 149, 247 (2.5, 5, 2.5), 335, 333, 331 (2.5, 5, 2.5), 307, 305, 303 (50, 100, 50), 292, 290, 288 (20, 40, 20), 195 (22), 181 (31), 137 (30), 81 (80) | FABMS 381 (92), 383 (100) [M+H]+, 403 (36), 405 (38) [M+Na]+ HRFABMS 381.0573 | FAMS 367, 369 | SIMS 727, 729, 731 (1:2:1) |
| MP (°C) | 113–115 | NA | NA | NA | 94–95 |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | NA | 207.5 (44140) | 388 (630), 289 (sh 2660), 280 (3040), 206 (33600) | 387 (620), 289 (2600), 278 (3020), 207 (31000) | 284 (970) |
| IR νmax (KBr) cm−1 | 1538, 1372, 1107 | 3271, 1652, 1471, 1260 | 3390 br, 3231 br, 2936, 2837, 1655, 1495, 1254, 1054 | 3151, 1675 | 2930, 2850, 1675, 1520, 1450, 1250 |
| 142 (102) | 143 (102) | 144 (103) | 145 (103) | |
|---|---|---|---|---|
| Appearance | Colorless amorphous solid | Colorless amorphous solid | Colorless amorphous solid | Colorless amorphous solid |
| Molecular formula | C32H50N6O4Br2 | C33H52N6O4Br2 | C34H53N6O4Br2 | C36H59N6O4Br2 |
| MS m/z | SIMS 741, 743, 745 (1:2:1) | SIMS 755, 757, 759 (1:2:1) | FABMS 767, 769, 771 (1:2:1) [M+H]+ HRFABMS 767.2540 | FABMS 797, 799, 801 (1:2:1) [M+H]+ HRFABMS 767.2956 |
| MP (°C) | 93–95 | 108–110 | NA | NA |
| [α]D | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 284 (930) | 284 (910) | 284 (1100), 274 (1400) | 284 (710), 274 (1000) |
| IR νmax (KBr) cm−1 | 2930, 2850, 1675, 1540, 1455, 1260 | 2930, 2860, 1675, 1520, 1450 | 3400, 2910, 1675, 1625, 1535, 1450, 1250, 1120 | 3400, 2840, 1675, 1620, 1535, 1450, 1200, 1130 |
| 146 (104) | 147 (105) | 148 (103) | 149 (79) | 150 (79) | |
|---|---|---|---|---|---|
| Appearance | Colorless amorphous solid | NA | Colorless oil | Colorless oil | Colorless oil |
| Molecular formula | C17H23O3N6Br2 | C22H25N6O3Br2 | C14H15Br2N6O3 | C15H18Br2N5O4 | C15H17Br2N4O4 |
| MS m/z | FABMS (Positive glycerol matrix) 559, 557, 555 (1:2:1) [M+K]+, 521, 519, 517 (1:2:1) [M+H]+, 505, 503, 501 (1:2:1) [M+H–NH2]+, 441, 439 (1:2:1) [M–Br]+ HRFABMS 517.0241 | FABMS 579, 581, 583 (1:2:1) HRFABMS 581.0366 (Δ 3.1 mmu) | 459.9631 | FABMS 490, 492, 494 (M++H, 1:2:1) HRFABMS 491.9705 | FABMS 475, 477, 479 (M++H, 1:2:1) HRFABMS 476.9596 |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 277 (520) | 286 (800), 269 (shoulder), 260 (2900), 217 (15300) | 277 (1700), 284 (1400) | 277 (1700), 284 (1400) | 235 (2600), 290 (2800) |
| IR νmax (KBr) cm−1 | (Film) 3400, 1680, 1540, 1435, 1200, 1140 | 3400, 1680, 1200, 1140 | 3400, 2920, 2845, 1680, 1520, 1470, 1200, 1135, 1120 | 3400, 2920, 1680, 1520, 1470, 1200, 1135, 1120 | 3400, 2920, 2845, 1680, 1520, 1135, 1120 |
| 151 (89) | 152 (107) | 153 (107) | 154 (109) | 155 (110) | |
|---|---|---|---|---|---|
| Appearance | NA | White powder | White powder | Colorless foam | Oil |
| Molecular formula | C18H26Br2N5O2 | C22H24Br2N4O6S2 | C22H24Br2N4O6S2 | C44H46Br4N8O12S4 | C8H17N2O4S2 |
| MS m/z | 502.0432 | FABMS 689 (5), 687 (15), 685 (7) [M++Na], 667 (62), 665 (100), 663 (50) [M++H] HRFABMS 664.9560 | NA | NA | EIMS 268 (18) [M+], 193 (60), 134 (60) [M+–SCH2CH2NHCOOMe], 102 (100) [M+–SSCH2CH2NHCOOMe] CIMS (isobutane) 269 (100) [M++H], 237 (18), 197 (5), 134 (80), 102 (8) HRFABMS 269.0626 (Δ 0.4 mmu) |
| MP (°C) | NA | 172–174 | NA | NA | NA |
| [α]D | +17° (c 1.0, MeOH) | NA | NA | 0° (c 1.0, MeOH) | NA |
| UV λmax (MeOH) nm | 285 (3.17), 330 (2.69) | (EtOH) 277 (4875) | NA | 212 (110,000), 290 (14,100) | NA |
| IR νmax (KBr) cm−1 | 3406, 1680, 1556, 1475, 1262 | 3348, 1657, 1637, 1536, 1025, 679 | 3300 br, 1670, 1540 | 3400–3100 br, 2900, 1670, 1570, 1450, 1200, 1025 | 3630, 3547, 2960, 1724, 1630, 1525, 1255, 1052 |
| 156 (110) | 157 (110) | 158 (110) | 159 (112) | 160 (112) | |
|---|---|---|---|---|---|
| Appearance | Oil | Oil | Oil | NA | NA |
| Molecular formula | C12H13N3O3BrS | C11H15N3O5BrS | C15H20N3O5BrS2 | C15H19N4O5S2Br | C15H18N3O6S2Br |
| MS m/z | EIMS 357/359 (3) [M+], 328/330 (5) [M+–HCN–2H], 298/300 (5) [M+–HSCN], 281/283 (7), 211/213 (100) [C8H6NOBr], 59 (30) [HSCN]. CIMS (isobutane) 358/360 (21) [M++H], 329/331 (29) [M+–HCN–H], 299/301 (35) [M+–HSCN+H], 283/285 (25). HRFABMS 357.9856 (Δ 0.5 mmu) | EIMS 379/381 (4) [M+], 363/365 (2) [M+–NH2], 211/213 (100) [C8H6NOBr]. CIMS (isobutane) 380/382 (32) [M++H], 364/366 (20) [M+–NH2+H], 301/303 (15) [M+–SO2NH2+2H]. HRFABMS 379.9913 (Δ 0.3 mmu) | EIMS 465/467 (5) [M+], 390/392 (1) [M+–NHCOOMe–H], 331/333 (22) [M+–SCH2CH2NHCOOMe], 299/301 (37) [M+–SSCH2CH2NHCOOMe], 226/230 (40), 211/213 (31) [C8H6NOBr]. CIMS (isobutane) 466/468 (10) [M++H], 375/377 (3), 333/335 (8), 299/301 (35). HREIMS 465.0030 (Δ 0.2 mmu) | HRFABMS 479.0094 [M+H]+ (Δ −3.5 mmu) | HRFABMS 501.9724 [M+H]+ (Δ 0.6 mmu) |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 280 (5450) | NA | NA | 206 (18070), 284 (1858) | 206 (25935), 280 (2057) |
| IR νmax (KBr) cm−1 | 3627, 3541, 2257, 2157, 1678, 1634, 1442, 1378, 1040 | 3300 br, 1670, 1540 | 3630, 3547, 2960, 1724, 1630, 1525, 1255, 1052 | (Film) 3500–3100 br, 3327, 1668, 1539, 1420, 1358, 1287, 1221, 1010, 985 | (Film) 3600–2600 br, 1647, 1527, 1424, 1390, 1287, 1219, 1047, 1019, 985 |
| 161 (112) | 162 (112) | 163 (112) | 164 (112) | 165 (114) | |
|---|---|---|---|---|---|
| Appearance | NA | NA | NA | NA | Pale yellow amorphous solid |
| Molecular formula | C15H20N5O5S2Br | C16H22N3O5S2BrNa | C12H15N2O5S2Br | C22H24N4O7S2Br2 | C25H34Br2N7O6S2 |
| MS m/z | HRFABMS 494.0055 [M+H]+ (Δ 3.4 mmu) | HRFABMS 502.0102 [M+Na]+ (Δ 2.0 mmu) | HRFABMS 400.9795 [M+Na]+ (Δ 1.2 mmu) | HRFABMS 700.9396 [M+Na]+ (Δ 4.5 mmu) | (+)-HRFABMS 752.0381 for C25H34Br2N7O6S2 (Δ −2.3 mmu) (−)-HRFABMS 742.8976 for C22H23Br2N4O9S2 (Δ −0.3 mmu) ESIMS 743 |
| MP (°C) | NA | NA | NA | NA | 76–80 |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | NA | NA | NA | NA | 204 (4.73), 281 (3.88) |
| IR νmax (KBr) cm−1 | (Film) 3600–2600 br, 1666, 1631, 1596, 1531, 1437, 1408, 1354, 1290, 1220, 1145, 1043. 1025, 996 | (Film) 3500–3000 br, 1660, 1531, 1419, 1384, 1360, 1260, 1231, 1037, 985 | (Film) 3600–3000 br, 1654, 1531, 1501, 1420, 1361, 1284, 1043, 973 | 3600–2900 br, 1660, 1625, 1531, 1502, 1449, 1425, 1361, 1287, 1284, 1213, 1149, 1044 | 3400 br, 2925, 1655, 1530, 1485, 1250, 1230, 1040 |
| 166 (114) | 167 (114) | 168 (114) | 168a (114) | 169 (117) | |
|---|---|---|---|---|---|
| Appearance | Pale yellow amorphous solid | Yellow amorphous solid | Yellow amorphous solid | White solid | White amorphous solid |
| Molecular formula | C25H32Br2N7O12S4 | C31H27Br3N4O13S3 | C31H26Br3N4O13S3 | C36H37Br3N4O10S2 | C22H24N4O6S3Br2 |
| MS m/z | HRFABMS 909.9285 | (+)–FABMS 1068.8/1066.8, 1046.8/1044.8, 944.8/942.8 [M-SO3+Na+2H]+ (−)-FABMS 1044.9/1042.9 [M+2Na–H]−, 1022.9/1020.9 [M+Na]− (+)–HRFABMS 1068.7794 [M+3Na]+, 1046.8165 [M+2Na+H]+ | (+)–FABMS 1044.7/1042.7, 1022.6/1020.6 [M+Na+H]+, 942.8/940.8 [M–SO3+Na+H]+ (−)-FABMS 998.7/996.8 ESIMS 998.7/996.8 (+)–HRFABMS 1044.8008 [M+2Na]+ | HRFABMS 990.9545 [M+H]+ | ESIMS 695 (46), 697 (100), 699 (60) [M+H]+, 717 (35), 719 (74), 721 (45) [M+Na]+, 466 (19), 468 (20), 363 (19), 365 (22) MALDI-TOFMS 718.9037 [M+Na]+ |
| MP (°C) | 155–160 | 191–194 | 236–239 | 136–138 | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 204 (4.60), 281 (3.98) | 204 (4.83), 291 (4.11) | 205 (4.88), 280 (3.93) | NA | NA |
| IR νmax (KBr) cm−1 | 3400 br, 2930, 1660, 1530, 1485, 1230, 1040 | 3400 br, 2925, 1660, 1565, 1270, 1230, 1040 | 3400 br, 2925, 1655, 1540, 1385, 1270, 1235, 1045 | NA | NA |
| 170 (117) | 171 (117) | 172 (44) | 173 (118) | 174 (118) | |
|---|---|---|---|---|---|
| Appearance | White amorphous solid | Yellow oil | Pale tan semicrystalline solid | White semicrystalline solid | White semicrystalline solid |
| Molecular formula | C22H23N4O6S2Br3 | C44H46N8O12S4Br4 | C23H28Br3N3O4 | C21H24Br3N3O4 | C21H23Br4N3O4 |
| MS m/z | ESIMS 741 (25), 743 (81), 745 (100), 747 (35) [M+H]+ MALDI-TOFMS 766.8495 [M+Na]+ | ESIMS 1323 (8), 1325 (38), 1327 (52), 1329 (43), 1331 (15) [M+H]+, 1345 (13), 1347 (54), 1349 (100), 1351 (77), 1353 (25) [M+Na]+ MALDI-TOFMS 1348.9407 [M+Na]+ | EIMS 647, 649, 651, 653, (M+–HCl. 5%), 631, 633, 635, 637 (4), 562, 564, 566, 568 (9), 484, 486, 488 (5), 404, 406 (2), 225, 227 (35), 58 (100) FABMS 648, 650, 652, 654 ([M+–HCl]+1) | FABMS 619.9380 [M+H]+ | FABMS 699.8480 [M+H]+ |
| MP (°C) | NA | NA | 87–88.5 | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | NA | NA | 294 (3286), 266 (3850), 260 (4000), 250 (8300), 215 (26100) | 222 (16700), 280 (3100) | 222 (17400), 274 (2100) |
| IR νmax (KBr) cm−1 | NA | 3369, 1655, 1534, 1490, 1426, 1228, 1016, 669 | 3427, 2922, 1632, 1454, 1256, 1053 | 3350, 2940, 1655, 1620, 1530, 1490, 1455 | 3400, 2990, 1670, 1630, 1530, 1460 |
| 175 (118) | 176 Acetate (119) | 177 Acetate (119) | 178 (82) | 179 (121) | |
|---|---|---|---|---|---|
| Appearance | Colorless gum | NA | NA | Glass | NA |
| Molecular formula | C36H52Br3N3O5 | C21H25Br2N3O4 | C21H24Br3N3O4 | C21H24Br3N3O4 | C20H23Br2N3O3 |
| MS m/z | HRFABMS 844.1459 [M+H]+ | FABMS 588 (4), 586 (7), 584 (4) [M+H]+, 572 (5), 570 (8), 568 (5), 492 (4), 490 (4), 343 (11), 341 (10), 301 (9), 299 (9), 228 (19), 226 (21), 201 (47), 199 (57) | FABMS 668 (2), 666 (4), 664 (4), 662 (2) [M+H]+, 652 (2), 650 (4), 648 (4), 646 (2), 586 (2), 584 (1), 572 (3), 570 (4), 568 (3), 492 (1), 490 (1), 343 (8), 341 (7), 308 (2), 306 (2), 304 (3), 281 (8), 279 (13), 277 (7) | EIMS 409 (13), 407 (33), 405 (13), 307 (20), 305 (45), 303 (23), 292 (18), 290 (29), 288 (16), 187 (40), 185 (54), 183 (38), 181 (32) HRFABMS 619.9377 | HRFABMS 514.0178 [M+H]+ |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 214 (18500), 222 (17200), 282 (2700) | NA | NA | (CHCl3) 240 (6750), 280 (3500) | 280 (834) |
| IR νmax (KBr) cm−1 | 3400, 2910, 1660, 1645, 1490, 1445 | NA | NA | [CHCl3–MeOH (1:1)] 3410, 3015, 2930, 1670, 1605, 1530, 1495, 1470, 1420 | (Film) 1658, 1540, 1458, 1420, 1258, 1200, 1010 |
| 180 (121) | 181 (121) | 182 (121) | 182 (121) | 183 (121) | |
|---|---|---|---|---|---|
| Appearance | NA | NA | NA | NA | NA |
| Molecular formula | C20H24BrN3O3 | C25H26Br2N3O3 | C20H23Br2N3O3 | C21H25Br2N3O3 | C20H22Br3N3O4 |
| MS m/z | HRFABMS 434.1086 [M+H]+ | HRFABMS 578.0306 [M]+ | HRFABMS 512.0141 [M+H]+ | HRFABMS 526.0292 [M+H]+ | HRFABMS 609.9254 [M+H]+ |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 280 (641) | 250 (5600) | 282 (512) | 282 (600) | 282 (2600) |
| IR νmax (KBr) cm−1 | (Film) 1680, 1540, 1498, 1458, 1200, 1138, 840, 800 | (Film) 1670, 1200, 1130, 800 | (Film) 1678, 1200, 1138, 840, 800 | 1678, 1204, 1140, 840, 800 | 1678, 1204, 1140, 800 |
| 185 (121) | 186 (121) | 187 (121) | 188 (121) | 189 (86) | 190 (122) | |
|---|---|---|---|---|---|---|
| Appearance | NA | NA | NA | White powder | Colorless oil | White solid |
| Molecular formula | C21H24Br3N3O4 | C21H24Br3N3O4 | C22H26Br3N3O4 | C9H9NO3 | C23H28Br3N3O5 | C24H28N3O6Br3 |
| MS m/z | HRFABMS 619.9391 [M+H]+ | HRFABMS 619.9349 [M+H]+ | HRFABMS 639.9457 [M+H]+ | NA | ESIMS 663.97, 665.96, 667.98, 669.96 [M+H]+ HRESIMS 663.9763 [M+H]+ (Δ 2.3 mmu) | FABMS 692, 694, 696, 698, (1:2:2:1) [M+H]+ HRFABMS 694.2150 [M+H]+ (Δ −2.2 mmu) |
| MP (°C) | NA | NA | NA | NA | 190–195 | |
| [α]D | NA | NA | NA | NA | NA | |
| UV λmax (MeOH) nm | 282 (2800) | 280 (2000) | 280 (1800) | 280 (3.26) | 217 (3.65), 280 (4.78) | |
| IR νmax (KBr) cm−1 | 1680, 1400, 1200, 800 | 1678, 1202, 1138, 842, 800, 722 | 1670, 1200, 1135, 838, 798, 721 | 1694, 1650, 1600, 1480, 1452, 1430, 1282, 1200, 1132, 1120, 1072, 1020, 920, 890, 740, 688 | 2937, 2852, 1743, 1656, 1533, 1493, 1452, 1253, 1047, 739 | 3404, 2926, 1675, 1494 |
| 191 (122) | 192 (84) | 192 (123) | 194 (123) | 195 (124) | |
|---|---|---|---|---|---|
| Appearance | White solid | Colorless amorphous solid | Yellow amorphous solid | Yellow amorphous solid | White powder |
| Molecular formula | C22H26N3O4Br3 | C23H29O4N4Br4 | C28H31Br4N4O4 | C28H31Br4N4O4 | C24H31N3Br4O3 · HCl |
| MS m/z | FABMS 633, 635, 637, 639 (1:2:2:1) [M+H]+. HRFABMS 635.7964 (Δ −2.8 mmu) | FABMS 771, 769, 767, 765, 763 [M+Na]+, 749, 747, 745, 743, 741 [M+H]+, 669, 667, 665, 663 HRFABMS 744.8868 | FABMS (glycerol matrix) 803, 805, 807, 809, 811 [M]+; 645, 647, 649; 466, 468, 470; 411, 413, 415; 347, 349, 351 HRFABMS 806.9037 [M]+ (Δ 0.0 mmu) | FABMS 803, 805, 807, 809, 811 [M]+; 645, 647, 649; 466, 468, 470; 426. 428. 430; 409, 411, 413 HRFABMS 806.9007 [M]+ (Δ −3.1 mmu) | DEIMS 726, 728, 730, 732, 734 (1.7); 448, 450, 452 (25:50:25); 403, 405, 407 (10:15:10); 320, 322, 324 (50:100:50) |
| MP (°C) | 175–178 | NA | NA | NA | 200 |
| [α]D | NA | NA | NA | NA | +5.1° (c 0.013, MeOH) |
| UV λmax (MeOH) nm | 214.5 (5.12), 280.0 (4.02) | 210 (23000), 285 (1600) | 216 (21800), 258 (4000 sh) | 214 (20600), 258 (3800 sh) | 283 (7800), 276 (9400), 245 (12300) |
| IR νmax (KBr) cm−1 | 3350, 1670, 1200, 1137, 720 | 3400, 3100, 1680, 1400, 1200, 1130 | NA | NA | (CHCl3) 3350, 1645, 1535, 1515, 1470, 1450, 1240, 980 |
| 196 (125) | 197 (125) | 198 (125) | 199 (90) | |
|---|---|---|---|---|
| Appearance | Pale orange oil | Colorless amorphous solid | Colorless, amorphous solid | NA |
| Molecular formula | C25H33Br4N3O3 | C23H31N3O3Br3 | C24H33N3O3Br3 | C17H26N6O3Br2 |
| MS m/z | CIMS 740 (22), 742 (70), 744 (100), 746 (64), 748 (20) [MH+]; 696 (1), 698 (5), 700 (7), 702 (4), 704 (1) [MH+–NMe2]; 662 (4), 664 (10), 666 (8), 668 (2) [MH+–Br]; 582 (3), 584 (6), 586 (3) [MH+–Br2]; 462 (3), 464 (12), 466 (12), 468 (3), 334 (15), 336 (26), 338 (15) [C11H14Br2NO+]; 309 (12), 311 (21), 313 (8), 118 (22). EIMS 696 (1), 698 (3), 700 (7), 702 (3), 704 (1) [MH+–NMe2]; 462 (4), 464 (9), 466 (7), [C17H26Br2N3O2+]; 377 (5), 379 (6), 381 (3) [C12H15Br2N2O2+]; 334 (52), 336 (99), 338 (50) [C11H14Br2NO+]; 256 (20), 258 (15) [C11H14BrNO+]; 84 (17), 58 (100) [CH2NMe2+] | FABMS 634, 636, 638, 640 (1:3:3:1) [M+H]+ HRFABMS 633.9944 | FABMS 648, 650, 652, 654 (1:3:3:1) [M+H]+ HRFABMS 648.0092 | FABMS 521, 523, 525 (1:2:1) HRFABMS [MH+] 521.0501 |
| MP (°C) | NA | 64–67 | 79–81 | NA |
| [α]D | 0° (c 5.0, CHCl3) | +21° (c 1.0, MeOH) | +16° (c 1.0 MeOH) | NA |
| UV λmax (MeOH) nm | NA | 281 (2400) | 281 (2900) | NA |
| IR νmax (KBr) cm−1 | 1036, 1211, 1221, 1259, 1473, 1545, 1678, 2450, 2969, 3023, 3222 | 3425, 1655 | 3425, 1655 | NA |
| 200 (79) | 201 (127) | 202 (127) | 203 (127) | |
|---|---|---|---|---|
| Appearance | Colorless oil | White needles | White needles | Colorless gum |
| Molecular formula | C5H22Br2N5O4 | C29H41I3N4O4 | C29H40I4N4O4 | C28H39I3N4O4 |
| MS m/z | FABMS 494, 496, 498 (1:2:1) [M++H] HRFABMS 496.0018 | FABMS 891 (100) [MH+], 643 (18), 517 (10), 430 (93), 391 (12), 304 (93). EIMS 890 (Not observed) [M], 643 (10), 517 (10), 430 (25), 304 (35), 254 (56), 142 (100), 127 (92) | FABMS 1017 (87) [MH +], 643 (18), 430 (100), 391 (12), 307 (45). EIMS 1016 (Not observed) [M], 643 (30), 430 (50), 304 (100), 254 (10) | FABMS 877 (100) [MH +], 629 (13), 503 (17), 430 (100), 329 (10), 290 (37). EIMS 876 (Not observed) [M], 629 (45), 503 (15), 430 (100), 304 (12), 290 (11), 142 (40), 127 (15) |
| MP (°C) | NA | 135–137 | 114–116 | NA |
| [α]D | NA | −0.23° (c 0.51) | −0.20° (c 0.35) | −0.74° (c 0.24) |
| UV λmax (MeOH) nm | 235 (2600), 287 (700) | 211 (25000), 223 (24000), 273 (5400) | 207 (32000), 221 (35000), 275 (8400) | 210 (40000), 222 (39000), 278 (9200) |
| IR νmax (KBr) cm−1 | 3400, 2920, 2850, 1680, 1635, 1135, | (CHCl3) 3368, 2935, 2883, 2855, 1672, 1600, 1518, 1500, 1491, 1479, 1463, 1447, 1441, 1416, 1278, 1272, 1254, 1050, 1020, 996 | (CHCl3) 3368, 2938, 2882, 2855, 1674, 1614, 1602, 1522, 1464, 1416, 1264, 996 | (CHCl3) 3367, 2940, 2861, 1671, 1522, 1492, 1463, 1264, 1260, 1255, 1050, 996, 925 |
| 204 (129) | 205 (129) | 206 (129) | 227 (129) | 208 (129) | |
|---|---|---|---|---|---|
| Appearance | Colorless foam | Colorless foam | Pale yellow foam | White powder | Yellow needles |
| Molecular formula | C34H30Br4N4O8 | C34H29Br5N4O8 | C34H30Br4N4O8 | C34H29Br4N4O11Sna | C34H25Br5N4O8 |
| MS m/z | 424 (23), 423 (11), 422 (48), 421 (6), 420 (24), 398 (24), 397 (10), 396 (13), 3959 (11), 394 (8), 316 (26), 314 (25), 303 (27), 301 (25), 262 (16), 243 (9), 241 (9), 201 (17), 200 (68), 199 (19), 198 (70), 188 (26), 187 (100), 186 (31), 185 (95), 120 (25), 119 (10), 107 (19), 105 (21), 103 (15), 102 (12), 91 (18), 89 (22), 88 (10), 82 (49), 81 (17), 80 (43), 79 (16), 78 (28), 77 (76), 76 (27), 75 (25), 74 (10), 65 (15), 64 (17), 63 (43), 62 (20), 58 (11), 53 (29), 52 (21), 51 (82), 50 (33) | No molecular ion; 504 (3), 502 (10), 500 (10), 498 (3), 200 (100), 198 (100), 187 (87), 185 (87) | 426 (2), 425 (6), 424 (11), 423 (10), 422 (18), 421 (4), 420 (8), 399 (6), 398 (8), 397 13), 396 (8), 395 (9), 243 (10), 201 (24), 200 (84), 199 (26), 198 (80), 189 (12), 188 (54), 187 (100), 186 (54), 185 (100), 120 (20), 108 (12), 107 (30), 105 (14), 91 (12), 89 (10), 82 (56), 81 (27), 80 (62), 79 (22), 78 (28), 77 (64), 51 (60) | MALDI FTMS [M+Na2+] 1062.8130 | 504 (4), 502 (11), 500 (11), 498 (4), 424 (2), 422 (6), 420 (7), 418 (2), 396 (6), 394 (7), 392 (3), 342 (12), 340 (12), 82 (100), 81 (27), 80 (100), 79 (27) Found: 497.8219, C16H979Br3N2O2 requires 497.8214 |
| MP (°C) | NA | NA | NA | NA | 250 (dec) |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 220 (4.80) 282 (4.30), 288 sh (3.93) | 220 sh (4.70), 280 (3.93), 193 sh (3.87) | 220 (4.77), 285 (3.94) | 209 (84400), 279 (5550) | 210 (4.84), 288 (4.00), 315 (4.00) |
| IR νmax (KBr) cm−1 | NA | NA | NA | (ZnSe film) 3400–3000, 2921, 2852, 1660, 1531, 1486, 1424, 1262, 1234, 1180, 989 | (KBr disc) 3600, 2800, 1657, 1633, 1490, 1250 |
| 209 Tetramethyl ether (129) | 210 (129) | 211 (129) | 228 (130) | 212 (132) | |
|---|---|---|---|---|---|
| Appearance | Colorless prisms | White powder | Off-white foam | Yellow solid | White film |
| Molecular formula | C38H35Br5N4O8 | C34H26Br6N4O8 | C34H26Br4N4O8 | C34H25Br4N4O14S2Na2 | C34H27Br5N4O9 |
| HR-MS m/z | 1073.8310 | NA | NA | MALDI FTMS [M+K3+] 1208.6481 | EIMS 504 (7.0), 502 (18.7), 500 (18.7), 498 (5.8), 342 (15.8), 340 (21.1), 199 (7.6), 199 (7.0) |
| MP (°C) | 262–264 | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 220 sh, 275 | 220 sh (4.69), 281 (3.47) | 290 (4.18), 315 (4.19) | 207 (82300), 321 (12400) | 208 (5.1), 280 (3.9) |
| IR νmax (KBr) cm−1 | 1670, 1565, 1520, 1495, 1455, 1043 | 3500–2800, 1665, 1658, 1630, 1500, 1450, 1420, 1280, 1245, 1020, 980 | 3600–2900, 1663, 1635, 1490, 1455, 1430, 1250, 1015, 1000, 990 | (ZnSe film) 3300–2860, 1670, 1653, 1571, 1523, 1482, 1418, 1277, 1240, 1051, 1026, 1005 | (Film on NaCl plate) 3340, 1700, 1660, 1590, 1530, 1490, 1450, 1420, 1360, 1280, 1240, 990 |
| 212 Tetramethyl ether (132) | 213 (132) | 213 Tetramethyl ether (132) | 214 (132) | 215 (132) | |
|---|---|---|---|---|---|
| Appearance | White powder | White powder | NA | Colorless oil | Whitish film |
| Molecular formula | C37H33Br5N4O9 | C34H28O8N4Br4 | NA | C34H28O9N4Br4 | C34H26O8N4Br4 |
| MS m/z | FABMS [p-nitrobenzyl alcohol/magic bullet (dithiothreitol/dithioerythritol) matrix] 1076.9 (33.2), 1074.9 (52.5), 1072.8 (24.7), 1070.8 (19.9). HRFABMS 1110.8194 (C38H35N4O979Br481BrNa requires 1110.8198), 1112.8194 (C38H35N4O979Br381Br2Na requires 1112.8178), 1114.8035 (C38H35N4O979Br281Br3Na requires 1114.8157), 1116.7491 (C38H35N4O979Br81Br4Na requires 1116.8137) | FABMS (Magic bullet matrix) 942 (1), 940 (2), 938 (2), 936 (1) [M+] | FABMS 1014 (7.1) [M+Na]+, 1016 (21.1), 1018 (28.3), 1020 (26.3), 1022 (1.2). HREIMS 511.8354 (34.4), 513.8460 (95.8), 515.88416 (100), 517.8290 (37.5) | NA | NA |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | NA | 208 (5.1), 280 (4.0) | NA | 210 (4.7), 277 (3.7) | 208 (4.3), 285 (3.4), 331 (3.5) |
| IR νmax (KBr) cm−1 | NA | (Film) 3420–3200, 1710, 1660, 1620, 1585, 1450, 1420, 1360, 1230 | NA | (Film) 3340 br, 1698, 1662, 1590, 1545, 1483, 1421, 1290, 1240 | (Film) 3300 br, 1718, 1662, 1646, 1544, 1495, 1479, 1447, 1418, 1284, 1243 |
| 216 Methyl ether (142) | 217 (131) | 217 Methyl ether (131) | 226 (141) | |
|---|---|---|---|---|
| Appearance | White amorphous solid | White powder | White powder | NA |
| Molecular formula | C38H35N4O9Br5 | C34H28Br4N4O8 | C38H36Br4N4O8 | C34H27Br4N4O11S |
| MS m/z | 1096 (0.5), 1094 (2.0), 1092 (4.1), 1090 (4.2), 1088 (2.4), 1086 (0.7), 1078 (1.0), 1076 (3.7), 1074 (7.2), 1072 (6.5), 1070 (3.4), 1068 (0.8), 1065 (1.1), 1063 (2.3), 1061 (5.6), 1059 (5.3), 1057 (3.0), 1055 (0.8), 1047 (1.1), 1045 (3.2), 1043 (5.6), 1041 (5.0), 1039 (2.9), 1037 (0.7), 1015 (1.3), 1013 (2.2), 1011 (2.7), 1009 (2.1), 1007 (0.9), 648 (0.4 ), 632 (0.8), 630 (0.6), 602 (3.1), 600 (5.7), 598 (2.8), 592 (1.2), 590 (1.6), 571 (8.0), 569 (13.9), 567 (7.6), 551 (2.0), 549 (5.1), 547 (4.9), 545 (2.3), 536 (2.5), 534 (3.6), 532 (4.6), 530 (3.3), 438 (21.1), 436 (40.4), 434 (20.8), 412 (12.9), 411 (14.1), 410 (19.6), 409 (14.0), 408 (10.0), 342 (20.5), 340 (18.3), 318 (5.6), 316 (13.8), 314 (10.4), 276 (22.5), 261 (8.8), 169 (9.2), 115 (9.4), 82 (41.5), 80 (42.1), 31 (73.6), 29 (100.0). HREIMS 1089.8312. CIMS (Showed one extra Br) [M+Br]+ 1178 (122.7), 1176 (37.3), 1174 (72.2), 1172 (92.9), 1170 (56.4), 1168 (21.8), 1166 (1.2), 1096 (21.0), 1094 (52.0), 1092 (74.2), 1090 (54.0), 1088 (22.6), 1086 (2.4) | EI: 520 (4), 518 (4), 516 (3), 396 (9), 394 (16), 392 (8), 342 (23), 340 (24), 317 (8), 316 (7), 315 (10), 57 (100) | EI: 1000 (4), 998 (14), 996 (20), 994 (13), 992 (4), 969 (4), 967 (10), 965 (14), 963 (9), 518 (20), 516 (530, 514 (54), 512 (18420 (16), 419 (16), 417 (21), 376 (33), 375 (53), 73 (52), 333 (31), 332 (97), 331 (31), 330 (100) | HRFABMS: 1018.8064 [M–Na]− |
| MP (°C) | NA | 177–179 | 107–109 | NA |
| [α]D | +2.7° (c 0.77, CH2Cl2) | 0.0° (c 1.52, CH2Cl2) | NA | |
| UV λmax (MeOH) nm | NA | 209 (81100), 283 (6200) | 209 (76400), 276 (7450) | 280 (4800), 204 (74000) |
| IR νmax (KBr) cm−1 | (Film) 3327, 2932, 2857, 1669, 1487 | 3413, 1666 | NA |
| 218 (133) | 219 (134) | 220 (135) | 221 (135) | |
|---|---|---|---|---|
| Appearance | NA | White amorphous solid | NA | NA |
| Molecular formula | C38H25Br5N4O8 | C34H27N4O8Br5 | C34H27Br5N4O8 | C34H28Br4N4O9 |
| MS m/z | EIMS 424 (2.4), 422 (4.5), 420 (2.5), 316 (3.4), 314 (2.6), 303 (2.7), 301 (3.1) HREIMS 421.9089 | HRFABMS 1016.7785 [M+H]+ | HRFABMS 1018.7792 [M+H]+ | HRFABMS 956.8606 [M+H]+ |
| MP (°C) | NA | NA | NA | NA |
| [α]D | NA | Negative, too small to be measured | NA | NA |
| UV λmax (MeOH) nm | 284 (4.3), 292 (4.3), 314 (4.4) | NA | (Nujol) 280 (3.6) | (Nujol) 278 (4.1) |
| IR νmax (KBr) cm−1 | (Nujol) 3500–3300, 1655, 1580, 1530, 1500, 1455, 1425, 1280, 1245, 1230, 1180 | NA | 3500–3300, 1650, 1630, 1480, 1240, 1225 | 3600–3100, 1660, 1640, 1490, 1470, 1285, 1220 |
| 222 (138) | 223 (139) | 224 (130) | 225 (140) | 225 Tetramethyl ether (140) | |
|---|---|---|---|---|---|
| Appearance | Colorless amorphous solid | Colorless amorphous powder | Colorless solid | Amorphous white solid | Pale yellow gum |
| Molecular formula | C34H28Br4N4O8 | C34H27Br5N4O8 | C34H28Br4N4O8 | C34H29N4O8Br3 | C38H37N4O8Br3 |
| MS m/z | FABMS [M++1] 613 (4.9), 581 (1.8), 461 (5.3), 427 (1.5) HRFABMS 940.8705 | FABMS 1018.8 | MALDI FTMS [M+Na+] 958.8588 | ESIMS [M+Na]+ 887 (40), 885 (100), 883 (96), 881 (32) HRESIMS 886.9394, 884.9406, 882.9412, 880.9431 | HRESI: 920 (24), |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | NA | NA | 209 (84400), 279 (5550) | 279 (3.4), 3.83 (2.99) | 205 (4.71) |
| IR νmax (KBr) cm−1 | NA | NA | (ZnSe film) 3400–3000, 2921, 2852, 1660, 1531, 1486, 1424, 1262, 1234, 1180, 989 | 3422, 2925, 1743, 1654, 1697, 1490, 1380, 1235, 1044 | 3054, 2970, 2954, 2854, 1735, 1676, 1376, |
| 231 (41) | 237 (41) | 232:233 3:1 (41) | 238 (41) | |
|---|---|---|---|---|
| Appearance | Colorless oil | Colorless oil | Colorless solid | |
| Molecular formula | C17H16Br2N2O4 | C18H18Br2N2O4 | C17H15Br3N2O4 | C18H18Br3N2O4 |
| MS m/z | EIMS 474 (10), 472 (19), 470 (10), 458 (7), 456 (16), 454 (7), 256 (9), 214 (23), 213 (91), 212 (33), 211 (84), 201 (26), 200 (97), 199 (27), 198 (98), 188 (13), 187 (95), 186 (15), 185 (100), 132 (43), 120 (31), 77 (42). HREIMS 469.9437 (C17H1681Br2N2O4 calcd 473.9430), 471.9445 (C17H1679Br81BrN2O4 calcd 471.9457), 469.9467 (C17H1679Br2N2O4 calcd 473.9477) | EIMS 488 (6), 486 (13), 484 (6), 472 (6), 470 (11), 468 (6), 288 (12), 286 (12), 228 (21), 227 (97), 226 (20), 225 (100), 201 (38), 200 (30), 199 (38), 198 (27), 187 (50), 185 (53), 146 (52), 120 (18), 103 (33), 77 (38). HREIMS 487.9615 (C18H1881Br2N2O4 calcd 487.9593), 485.9609 (C18H1879Br81BrN2O4 calcd 485.9614), 483.9633 (C18H1879Br2N2O4 calcd 485.9634) | EIMS 554 (4), 552 (14), 550 (14), 548 (5), 293 (33), 291 (67), 289 (34), 267 (24), 265 (50), 263 (24), 213 (41), 212 (51), 211 (24), 198 (76), 187 (91), 185 (100), 132 (28), 120 (25), 77 (49). HREIMS 553.8549 (C17H1581Br3N2O4 calcd 553.8522), 551.8536 (C17H1579Br81Br2N2O4 calcd 551.8542), 549.8562 (C17H1579Br281BrN2O4 calcd 549.8562), 547.8570 (C17H1579Br3N2O4, calcd 547.8582) | EIMS 568 (2), 566 (6), 564 (7), 562 (2), 552 (2), 550 (7), 548 (8), 546 (2), 307 (30), 305 (61), 303 (30), 292 (14), 290 (25), 288 (13), 281 (10), 279 (17), 277 (9), 271 (7), 269 (23), 267 (10), 201 (15), 200 (96), 199 (18), 198 (100), 187 (44), 185 (48), 183 (17), 181 (15), 143 (18), 120 (18), 77 (17). HREIMS 567.8672 (C18H1881Br3N2O4 calcd 567.8679), 565.8701 (C18H1879Br81Br2N2O4 calcd 565.8699), 563.8715 (C18H1879Br281BrN2O4 calcd 563.8719), 561.8716 (C18H1879Br3N2O4 calcd 561.8739) |
| UV λmax (MeOH) nm | 210 (4.53), 281 (3.88) | 213 (4.53), 280 (3.87) | NA | 213 (4.50), 281 (3.87) |
| IR νmax (KBr) cm−1 | (NaCl film) 3383, 3000, 1659, 1537, 1495, 1422, 1283, 1256 | (NaCl film) 3385, 3283, 1657, 1537, 1497, 1283, 1256 | (NaCl film) 3393, 1657, 1476 | (NaCl film) 3300, 1661, 1537, 1497, 1472, 1422, 1260, 993 |
| 234 (41) | 234a (41) | 235 (41) | 235a:236a (41) | |
|---|---|---|---|---|
| Appearance | Colorless solid | Colorless solid | Mixture with hemibastadinol-3 | Colorless solid |
| Molecular formula | C17H17Br2NO4 | C19H21Br2NO4 | C17H16Br3NO4 | C19H20Br3NO4 |
| MS m/z | EIMS 461 (>1), 459 (0.3), 457 (>1), 443 (6), 441 (13), 439 (6), 244 (30), 242 (35), 241 (18), 227 (14), 225 (15), 201 (19), 200 (98), 199 (21), 198 (100), 187 (28), 185 (28), 120 (20), 107 (23), 77 (18) HREIMS 460.9477 (C17H1781Br2NO4 calcd 460.9494), 458.9483 (C17H1779Br81BrNO4 calcd 458.9514), 456.9507 (C17H1779Br2NO4 calcd 456.9534) | HRFABMS [M+H] 485.9897 (C19H2279Br2NO4, calcd 485.9915) | EIMS 541 (<1), 539 (< 1), 537 (<1), 535 (<1), [M+–H2O] 523 (2), 521 (7), 519 (6), 517 (2), 244 (9), 242 (10), 201 (15), 200 (100), 199 (16), 198 (100), 187 (16), 185 (17), 120 (20), 77 (13). HREIMS 540.8558 (C17H1681Br3NO4 calcd 540.8572), 538.8586 (C17H1679Br81Br2NO4 calcd 538.8593), 536.8631 (C17H1679Br281BrNO4 calcd 536.8612), 534.8605 (C17H1679Br3NO4 calcd 534.8633), [M+–H2O] 522.8446 (C17H1481Br3NO3 calcd 522.8464), 518.8497 (C17H1479Br281BrNO3 calcd 518.8504), 516.8509 (C17H1479Br3NO3 calcd 516.8524) | HRFABMS [M+H], 563.9019 (C19H2079Br3NO4, calcd 563.9020) |
| MP (°C) | NA | NA | NA | NA |
| [α]D | −31° (c 1.83, MeOH) | −23° (c 0.66, MeOH) | −24° (c 0.10, MeOH) | NA |
| UV λmax (MeOH) nm | 208 (4.43), 281 (3.69) | 208 (4.45), 281 (3.60) | NA | NA |
| IR νmax (KBr) cm−1 | (Film) 3381, 1643, 1541, 1495, 1420, 1289 | (Film) 3387, 1651, 1497, 1279, 1256, 1057 | (Film) 3381, 1643, 1539, 1478, 1279 | 3393, 2930, 1651, 1537, 1497, 1472, 1258 |
| 236 (41) | 239 (145) | 240 (145) | 241 (145) | |
|---|---|---|---|---|
| Appearance | Mixture with hemibastadinol-2 | White needles | Colorless gum | White needles |
| Molecular formula | C17H16Br3NO4 | C19H17Br2NO4 | C19H17Br2NO4 | C19H18BrNO4 |
| MS m/z | EIMS 541 (<1), 539 (<1), 537 (<1), 535 (<1), [M+–H2O] 523 (2), 521 (7), 519 (6), 517 (2), 244 (9), 242 (10), 201 (15), 200 (100), 199 (16), 198 (100), 187 (16), 185 (17), 120 (20), 77 (13) HREIMS 540.8558 (C17H1681Br3NO4 calcd 540.8572), 538.8586 (C17H1679Br81Br2NO4 calcd 538.8593), 536.8631 (C17H1679Br281BrNO4 calcd 536.8612), 534.8605 (C17H1679Br3NO4 calcd 534.8633), [M+–H2O] 522.8446 (C17H1481Br3NO3 calcd 522.8464), 518.8497 (C17H1479Br281BrNO3 calcd 518.8504), 516.8509 (C17H1479Br3NO3 calcd 516.8524) | EIMS 484.9 (3), 482.9 (6), 480.9 (3), 177 (17), 149 (20), 134 (100), 106 (25), 78 (15), 77 (13) HREIMS 480.9526 | EIMS 484.9 (4), 482.9 (8), 480.9 (4), 177 (21), 149 (21), 134 (100), 106 (19), 78 (10), 77 (9) HREIMS 480.9526 | EIMS 405 (7), 403 (6), 229 (7), 227 (7), 177 (11), 149 (16), 78 (13), 77 (15) HREIMS 403.418 |
| MP (°C) | NA | 169–171 | NA | 173–175 |
| [α]D | −24° (c 0.10, MeOH) | NA | NA | NA |
| UV λmax (MeOH) nm | NA | (EtOH) 205 (10900), 220 (10300), 225 sh (9900), 325 (15900) | (EtOH) 206 (10800), 214 (9600), 230 sh (9400), 328 (14100) | (EtOH) 204 (13200), 227 sh (10600), 329 (14100) |
| IR νmax (KBr) cm−1 | (NaCl film) 3381, 1643, 1539, 1478, 1279 | (CHCl3) 3410, 3018, 2927, 2855, 1735, 1650, 1607, 1513, 1496, 1468, 1423, 1265, 1171, 1087, 909 | (CHCl3) 3409, 3018, 1700, 1650, 1607, 1513, 1468, 1423, 1250, 1171, 1087, 1001, 949 | (CHCl3) 3409, 3018, 2927, 2855, 1730, 1693, 1653, 1607, 1513, 1486, 1463, 1441, 1259, 1171, 1089, 1054, 1019 |
| 242 (145) | 243 (146) | 244 (146) | 245 (146) | 247 (123) | |
|---|---|---|---|---|---|
| Appearance | Colorless gum | NA | NA | NA | Yellow amorphous solid |
| Molecular formula | C19H18BrNO4 | C24H28Br2N2O4 | C24H28BrIN2O4 | C24H28I2N2O4 | C17H25Br2N3O2 |
| MS m/z | EIMS 405 (8), 403 (7), 229 (8), 227 (7), 177 (12), 149 (18), 135 (9), 134 (100), 106 (21), 78 (13), 77 (16) HREIMS 403.0418 | FABMS 571, 569, 567 [M+H]+ | FABMS 617, 615 [M+H]+ | FABMS 663 [M+H]+ | FABMS 803, 805, 807, 809, 811 [M]+, 466, 468, 470, 426, 428, 430, 409, 411, 413 HRFABMS 806.9007 [M]+ |
| MP (°C) | NA | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | (EtOH) 203 (11100), 224 sh (9100), 326 (11700) | NA | NA | NA | 214 (20600), 258 sh (3800) |
| IR νmax (KBr) cm−1 | (CHCl3) 3413, 3018, 2935, 1710, 1650, 1607, 1513, 1466, 1425, 1248, 1168, 1087, 1001, 949 | (Neat) 3286, 1635, 1542, 1515, 1455, 1256 | (Neat) 3283, 1628, 1537, 1514, 1443, 1260 | (Neat) 3276, 1629, 1538, 1514, 1435, 1256 | NA |
| 246 (38) | 248 (150) | 249 (151) | 250 (151) | 251 (152) | 252 (152) | |
|---|---|---|---|---|---|---|
| Appearance | Amorphous solid | NA | Small yellow crystals | Small yellow crystals | Yellow amorphous solid | Yellow amorphous solid |
| Molecular formula | C15H17N4O2Br2 | C14H10O11S2Br2 | C17H16Br2N4O2 | C16H14Br2N4O2 | C23H24N3O3Br3 | C22H22N3O3Br3 |
| MS m/z | FABMS 443, 445, 447 | NA | HREIMS 467.9624 [M+] | HREIMS 453.9460 [M+] | FABMS 628, 630, 632, 634 (1:3:3:1) [M+H]+ HRFABMS 631.9403 [M+H]+ (Δ −0.2 mmu) | FABMS 614, 616, 618, 620 (1:3:3:1) [M+H]+ HRFABMS 615.9280 (M+H)+ (Δ +1.8 mmu) |
| MP (°C) | NA | NA | 236 | 242 | NA | NA |
| [α]D | NA | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | (EtOH) 226 (12200), 298 (32000) | NA | NA | NA | 285 (4000), 350 (1000) | 285 (5000), 350 (1200) |
| IR νmax (KBr) cm−1 | (Dry film) 1690 | (Film) 3454 (br d), 1631, 1257 (br d), 1059, 1018 | NA | NA | 3422, 1680 | 3422, 1680 |
| 253 (153) | 254 (153) | 255 (153) | 256 (154) | 257 (154) | |
|---|---|---|---|---|---|
| Appearance | Needles | White amorphous solid | White amorphous solid | White solid | White gum |
| Molecular formula | C18H15N2O3Br | C18H15N2O3I | C18H16N2O3 | C19H21N2O2Br | C19H20N2O2Br2 |
| MS m/z | HRFABMS [M++H] 387.0313 | HREIMS 434.0146 | HREIMS 308.1159 | HREIMS 370.0681 [M–H2O]+ | HREIMS 465.9891 [M+] |
| MP (°C) | 178–179 | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA | NA |
| UV λmax (MeOH) nm | (MeOH) 324 (8400), 290 (5000), 274 (9600), 267 (9900), 255 (10300), 205 (41400) (MeOH+NaOH), 309 (9800), 272 (8600), 266 (9900), 247 (1600), 207 (58500) | (MeOH) 325 (6600), 274 (8200), 267 (sh), 254 (9100), 230 (sh), 205 (50000) (MeOH+NaOH), 311 (8200), 274 (7600), 267 (sh), 247 (13200), 203 (79100) | (MeOH) 324 (4600), 285 (sh), 272 (5700), 266 (6100), 254 (6400), 205 (19200) (MeOH+NaOH), 218 (4600), 272 (5300), 266 (6100), 246 (8300), 205 (21400) | 288 (sh), 280 (6400), 221 (30600), 208 (29000) | (DMSO) 299 (sh), 287 (2500), 279 (sh), 221 (30600), 208 (29000) |
| IR νmax (KBr) cm−1 | (CHCl3) 3010, 1680, 1635, 1500, 1420, 1215, 1120 | (neat NaCl) 2930, 1680, 1640, 1505, 1420 | NA | (CHCl3) 3470, 3330 br, 1605, 1500, 1460, 1445, 1420, 1285, 1260, 1055 | (CHCl3) 3470, 3330 br, 1605, 1500, 1460, 1445, 1420, 1285, 1260, 1055 |
| 258 (155) | 259 (164) | 260 (158) | 261 (158) | 262 (159) | |
|---|---|---|---|---|---|
| Appearance | Gum | NA | Colorless prisms | Colorless crystals | Colorless glass |
| Molecular formula | C12H17NO5 | C36H45IN4O6Br | C28H40IN3O6 | C28H40BrN3O6 | C28H40N3O6Cl |
| MS m/z | FABMS 256 (M+H+, 100) HRFABMS 256.11858, 256.11850 | NA | FABMS 642 (100) [MH+], 516 (50), 393 (15), 276 (30), 217 (50), 150 (50) | FABMS 594 (95) [MH+], 516 (60), 345 (30), 267 (30), 228 (80), 150 (100) | HREIMS 549.2603, 551.2562 (Δ −0.2, −1.4 mmu) |
| MP (°C) | NA | NA | 217–218 | 203–204 | NA |
| [α]D | +25.51° (c 0.002, MeOH) | +35° (c 3.62, CHCl3) | +53° (c 0.04, CHCl3) | +101° (c 0.04, CHCl3) | NA |
| UV λmax (MeOH) nm | NA | 281 (5400), 290 (4100) | 219 (13800), 284 (3200), 292 (3000) | 214 (15400), 281 (2700), 290 (2500) | NA |
| IR νmax (KBr) cm−1 | NA | (CDCl3) 3400–3100, 1710, 1660, 1630 | (CHCl3) 1725, 1670, 1655, 1630 | (CHCl3) 3510, 3410, 1725, 1670, 1655 (sh), 1630 | NA |
| 263 (159) | 264 (159) | 265 (159) | 266 (164) | 267 (161) | |
|---|---|---|---|---|---|
| Appearance | Colorless glass | NA | NA | Colorless glass | NA |
| Molecular formula | C27H38N3O6I | C27H38N3O6Br | C27H38N3O6Cl | C28H38N3O7I | C34H44O7N3I |
| MS m/z | HREIMS 627.1798 (Δ −0.9 mmu) | HREIMS 579.1955, 581.1909 (Δ +1.1, −1.6 mmu) | HREIMS 535.2449, 537.2442 (Δ 0.0, +2.2 mmu) | HREIMS 655.1760 (ΔM, 0.6 mmu) | EIMS 773 (48) [M]+, 706 (10), 608 (15), 552 (5), 460 (54), 413 (17), 321 (24), 276 (100), 250 (19), 162 (73), 109 (46), HREIMS 733.2199 |
| MP (°C) | NA | NA | NA | NA | 186–189 |
| [α]D | NA | NA | NA | NA | +19.1° (c 0.17, CHCl3) |
| UV λmax (MeOH) nm | NA | NA | NA | NA | 215 (12500), 280 (2800) |
| IR νmax (KBr) cm−1 | NA | NA | NA | 3313, 1732, 1675, 1635 | 3495, 1724, 1670 |
| 268 (161) | 271 (164) | 272 (164) | 273 (164) | 274 (164) | |
|---|---|---|---|---|---|
| Appearance | NA | Colorless glass | Colorless glass | Colorless glass | Colorless glass |
| Molecular formula | C34H44O7N3Br | C28H39N3O7Br | C28H39N3O7Cl | C28H40N3O7I | C28H39N3O7Br |
| MS m/z | EIMS 685 (38) [M]+, 657 (10), 460 (52), 413 (24), 273 (26), 228 (100), 162 (87), 109 (59) HREIMS 685.2374 | HRDCIMS 610.19509, 608.19713 (C28H39N3O781Br, C28H39N3O779Br); found 610.19431, 608.19508 (Δ −1.49, −3.38 ppm) | HRDCIMS 565.23688, 563.23981 (C28H39N3O737Cl, C28H39N3O735Cl); found 565.23956, 563.24011 (Δ −4.7, −0.5 ppm) | HREIMS 658.19892, found 658.19767 (Δ −1.91 ppm) | HREIMS 611.20294, 609.20496 (C28H40N3O781Br, C28H40N3O779Br); found 611.20371, 609.20519 (Δ −1.3, −0.4 ppm) |
| MP (°C) | 168–170 | NA | NA | NA | NA |
| [α]D | +39.3° (c 0.17, CHCl3) | NA | NA | NA | NA |
| UV λmax (MeOH) nm | 214 (14000), 280 (3000) | NA | NA | NA | NA |
| IR νmax (KBr) cm−1 | (CHCl3) 3500, 1725, 1670 | NA | NA | NA | NA |
| 275 (164) | 276 (164) | 277 (164) | 278 (164) | |
|---|---|---|---|---|
| Appearance | Colorless glass | Colorless glass | Colorless glass | Colorless glass |
| Molecular formula | C28H39N3O7Cl | C28H40O7N3I | C28H39N3O7Br | C27H38O7N3I |
| MS m/z | HREIMS 567.25250, 265.25549 (C28H40N3O737Cl, C28H40N3O7735Cl); found 567.25184, 565.25403 (Δ 1.2, 2.6 ppm) | HREIMS 658.19892, found 658.19880 (Δ −0.19 ppm) | HREIMS 611.20294, 609.20496 (C28H40N3O781Br, C28H40N3O7779Br); found 611.20374, 609.20515 (Δ −1.3, −0.3 ppm) | HREIMS 643.17548, found 643.17539 (Δ 0.1 ppm) |
| MP (°C) | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA |
| UV λmax (MeOH) nm | NA | NA | NA | NA |
| IR νmax (KBr) cm−1 | NA | NA | NA | NA |
| 269(164) | 270 (164) | 280 (167) | 281 (168) | 282 (167) | |
|---|---|---|---|---|---|
| Appearance | Colorless glass | Colorless amorphous solid | Yellowish needles | Yellow oil | Yellowish, fluorescent oil |
| Molecular formula | C30H44N3O6I | C38H21Br8NO7 · (CH3)2CO | C30H13Br8NO6 | C24H15Br4NO5 | |
| MS m/z | HRMS 670.2335 [MH+] | FABMS 656 [M+H]+ | NA | EIMS 1123 (20) [M+], 596 (55), 526 (55), 448 (15), 337 (20), 279 (55) | HRMS 719.0348 |
| MP (°C) | NA | NA | 285 | NA | NA |
| [α]D | +30° (c 0.027, CHCl3) | +5.2° | NA | NA | NA |
| UV λmax (MeOH) nm | NA | 219 (13270), 284 (4000), 292 (3000) | 36 (17000), 300s (11700) | NA | 400 (2000), 280 (5250) (MeOH/KOH) 515 (2800), 400 (2800), 300 (5000) |
| IR νmax (KBr) cm−1 | (Neat) 3420, 1720, 1670, 1655, 1630 | NA | (CHCl3) 3500, 2900, 1645, 1580, 1514, 1300 | (Neat) 3500, 1645, 1582, 1515 | 3500, 2919, 1699, 1400 |
| 283 (167) | 284 (168) | 229 (41) | 230 (41) | |
|---|---|---|---|---|
| Appearance | Yellowish fluorescent oil | Yellow oil | White solid | White solid |
| Molecular formula | C25H17Br4NO5 | C16H6Br4O5 | C20H22Br2N2O4 | C20H21Br3N2O4 |
| MS m/z | HRMS 732.0619 [M+H]+ | HREIMS 597.6915 | EIMS 516 (1), 514 (2), 512 (1), 258 (2), 257 (2), 256 (3), 255 (9), 228 (10), 227 (64), 226 (14), 225 (65), 215 (10), 214 (38), 213 (11), 212 (46), 201 (61), 199 (65), 149 (48), 146 (40), 57 (100) HREIMS 511.9948 | EIMS 596 (0.4), 594 (2), 592 (2), 590 (0.4), 565 (<<1) 563 (1), 561 (1), 559 (<<1) 366 (0.3), 364 (1), 362 (0.4), 338 (0.6), 336 (2), 334 (0.7), 308 (1), 307 (10), 306 (4), 305 (20), 304 (4), 303 (10), 302 (1), 292 (5), 290 (10), 288 (5), 281 (6), 279 (14), 277 (7), 257 (2), 255 (2), 224 (2), 223 (3), 222 (3), 221 (2), 215 (15), 214 (97), 213 (16), 212 (100), 201 (30), 199 (34), 185 (4), 183 (11), 181 (8), 134 (16), 105 (17), 77 (39) HREIMS 589.9049 |
| MP (°C) | NA | NA | NA | NA |
| [α]D | NA | NA | NA | NA |
| (MeOH) nm | NA | NA | ||
| (KBr) cm"1 | 3500, 1702, 1698, 1400 | (Neat) 3460, 1761, 1721, 1475 | NA | NA |
TABLE II.
1H and 13C NMR Data of Bromotyrosine Derivatives.
| 1 (176) |
5 (7) |
6 (7) |
14 (14) |
15 (11) |
||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 1H | 1H | 13C (12) | 1H | 13C | |
| acetone-d6 | acetone-d6 | acetone-d6 | acetone-d6 | CD3CN | acetone-d6 | acetone-d6 | acetone-d6 | |
| 1 | 72.8 | 75.8 | 74.2 | |||||
| 2 | 7.59 s | 121.5 | 7.20 s | 7.14 s | 4.10 br | 87.8 | 7.30 d, 2.2 | 146.6 |
| 3 | 153.2 | 106.4 | 123.7 | |||||
| 4 | 174.5 | 150.1 | 183.0 | |||||
| 5 | 153.2 | 118.1 | 5.72 d, 2.2 | 57.1 | ||||
| 6 | 7.59 | 121.5 | 7.20 s | 7.14 s | 6.34 | 134.9 | 4.44 ddd, 2.2, 2.2, 5.6 | 78.9 |
| 7 | 2.75 | 46.0 | 2.58 s | 2.57 s | 2.74 s | 41.7 | 3.11 s | 28.4 |
| 8 | 173.0 | 171.9 | 116.9 | |||||
| NH | 2.97 | 6.68, 6.77 s | 6.65, 6.76 s | |||||
| OMe | 3.07 s | 3.10 s | 3.70 s | 60.0 | ||||
| OH | 5.88 s | 5.85 s | 2.28 | 5.96 d, 5.6, 5.93 s | ||||
| 1′ | 3.25 t | 3.22 t | ||||||
| 2′ | 1.55 m | 1.50 m | ||||||
| 3′ | 1.41 m | 1.50 m | ||||||
| 4′ | 0.98 t | 1.50 m | ||||||
| 5′ | 0.98 t | |||||||
| 16 (11) |
18 (19) |
19 (7) |
20 (7) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C (12) | 1H | 13C | 1H | |
| acetone-d6 | acetone-d6 | CDCl3 | acetone-d6 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 75.5 | 78.2 | 122.4 | ||||
| 2 | 7.52 s | 151.7 | 5.20 d, 0.7 | 113.4 | 162.0 | ||
| 3 | 122.7 | 147.9 | 111.8 | ||||
| 4 | 183.0 | 120.4 | 150.8 | ||||
| 5 | 5.04 d, 11.2 | 56.1 | 133.2 | 117.5 | |||
| 6 | 4.28 dd, 11.2, 5.0 | 78.4 | 6.36 d, 0.7 | 73.9 | 7.65 s | 125.8 | 7.48 s |
| 7 | 3.15 dq, 14.0 | 28.4 | 2.88 d, 18 | 26.3 | 100.7 | 3.67 s | |
| 8 | 116.9 | 117.5 | 165.2 | ||||
| 9 | 154.3 | ||||||
| NH | 5.82 s | ||||||
| 10 | 2.39 s | 23.7 | |||||
| 11 | 2.59 s | 25.2 | |||||
| OMe | 3.79 s | 59.6 | 3.92 s | 3.85 s | |||
| OH | 5.98 d, 5.0, 5.70 s | 3.38 | |||||
| 21 (20) |
22 (22) |
23 (22) |
24 (23) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CDCl3 | CDCl3 | acetone-d6 | DMSO-d6 | acetone-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | |
| 1 | 147.2 | 7.6 br | 7.6 br | 7.1 br | ||||
| 2 | 7.30 s | 109.1 | 173.9 | 173.5 | 172.4 | |||
| 3 | 113.7 | 2.92 d, 16.8 2.48 d, 16.8 |
42.3 | 2.75 d, 16.8 2.58 d, 16.8 |
44.1 | 2.60 d, 16.8 2.69 d, 16.8 |
43.4 | |
| 3a | 75.5 | 74.2 | 73.9 | |||||
| 4 | 146.4 | 7.44 s | 150.4 | 7.33 d, 0.6 | 149.2 | 7.28 br | 149.7 | |
| 5 | NA | 119.0 | 119.2 | 122.2 | ||||
| 6 | NA | 183.7 | 183.9 | 188.6 | ||||
| 7 | 103.8 | 5.16 d, 9.8 | 57.4 | 5.28 d, 4.2 | 52.7 | 2.82 dd, 16.4, 4.9 3.06 dd, 16.4, 4.9 |
39.5 | |
| 7a | 4.22 dd, 9.8, 1.6 | 68.3 | 4.46 brd, 4.2 | 63.4 | 4.14 brt | 58.4 | ||
| 8 | 165.9 | |||||||
| 9 | 8.21 d, 11.4 | 148.9 | ||||||
| 10 | 7.74 ddq, 14.3, 11.4, 1.6 | 144.7 | ||||||
| 11 | 6.59 dq, 14.3, 7.4 | 128.1 | ||||||
| 12 | 2.07 dd, 7.4, 1.6 | 19.8 | ||||||
| OH | 5.5 br | 5.6 br | 5.4 br | |||||
| 25 (9) |
26 (9) |
27 (9) |
28 (9) |
29 (9) |
|||
|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 1H | 1H | |
| acetone-d6 | acetone-d6 | acetone-d6 | DMSO-d6 | acetone-d6 | acetone-d6 | acetone-d6 | |
| 1 | 5.5 br | 5.5 br | 5.6 br | 5.6 br | |||
| 2 | 176.3 | 173.6 | |||||
| 3 | 2.70 s | 45.0 | 2.90 d, 16.9 2.50 d, 16.9 |
42.4 | 2.74 d, 16.9 2.59 d, 16.9 |
2.90 d, 16.9 2.48 d, 16.9 |
2.74 d, 16.9 2.56 d, 16.9 |
| 3a | 74.8 | 74.4 | |||||
| 4 | 7.03 d, 0.9 | 145.4 | 7.21 s | 146.3 | 7.07 br | 7.43 s | 7.32 br |
| 5 | 133.0 | 127.4 | |||||
| 6 | 183.1 | 183.4 | |||||
| 7 | 3.04, 2.80 d, 16.4 | 41.0 | 5.15 d, 9.8 | 58.0 | 5.24 d, 4.1 | 5.06 d, 10.4 | 5.26 d, 3.8 |
| 7a | 4.14 ddd, 6.0, 5.1, 0.9 | 60.8 | 4.19 brd, 9.8 | 67.9 | 4.8 brd, 4.1 | 4.03 brd, 10.4 | 4.48 brd, 3.8 |
| 30 (24) |
31 (25) |
43 (35) |
44 (35) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | DMSO-d6 | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 80.9 | 82.7 | 128.8 | 136.4 | ||||
| 2 | 7.46 s | 134.2 | 7.30 s | 135.1 | ||||
| 3 | 168.3 | 171.4 | 113.0 | 119.0 | ||||
| 4 | 2.64 br | 43.1 | 2.66 dd, 18.0, 1.7 2.55 dd, 18.0, 1.7 |
44.0 | 153.4 | 154.8 | ||
| 5 | 69.5 | 69.5 | 113.0 | 119.0 | ||||
| 6 | 7.43 d, 2.2 | 156.9 | 7.16 d, 2.2 | 153.2 | 7.46 s, 3.12 t, 12.0 | 134.2 | 7.0 s, 3.12 t, 12.0 | 135.1 |
| 7 | 118.8 | 129.5 | 3.29 | 32.8 | 3.29 | 32.9 | ||
| 8 | 187.8 | 188.9 | 8.73 dd, 12.0, 3.5 | 81.3 | 3.74 dd, 12.0, 3.5 | 81.0 | ||
| 9 | 2.43 d, 12.5 2.31 dd, 12.5, 2.2 |
46.6 | 2.44 dd, 12.4, 1.7 2.32 dd, 12.4, 2.2 |
47.6 | 170.6 | 170.6 | ||
| OMe | 3.81 s | 61.0 | ||||||
| N(Me)3 | 3.28 | 52.7 | 3.30 s | 52.7 | ||||
| 45 (35) |
46 (35) |
51 (40) |
||||
|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | |
| D2O | CD3OD | D2O | CD3OD | DMSO-d6 | DMSO-d6 | |
| 1 | 130.5 | 128.0 | 136.7 | |||
| 2 | 7.22 d, 2.0 | 135.3 | 7.20 | 134.9 | 7.58 s | 133.1 |
| 3 | 112.8 | 111.4 | 117.6 | |||
| 4 | 156.8 | 155.8 | 152.2 | |||
| 5 | 6.66 d, 8.5 | 113.9 | 6.65 d, 8.5 | 117.8 | 117.6 | |
| 6 | 6.95 dd, 8.5, 2.0; 2.80 t, 12.0 | 131.0 | 6.89 dd, 8.5, 2.0; 2.80 t, 12.0 | 130.7 | 7.58 s | 133.1 |
| 7 | 2.95a | 33.7 | 3.01a | 33.2 | 2.82 t, 7.4 | 31.4 |
| 8 | 3.42 dd, 12.0, 3.7 | 81.9 | 3.52 dd, 12.0, 3.7 | 81.6 | 3.06 t, 7.4 | 39.4 |
| 9 | 170.5 | 170.9 | ||||
| OMe | 3.54 s | 57.0 | 3.76 | 60.3 | ||
| N(Me)3 | 2.96 s | 53.2 | 3.02 s | 52.7 | ||
| 48 (38) |
207 (41) |
52 (42) |
54 (44) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | CD3OD | CD3OD | |
| 1 | 136.4 | 132.8 | 137.1 | |||||
| 2 | 7.69 s | 134.84 | 7.01 dd, 2.0, 8.2 | 130.0 | 7.30 s | 133.1 | 7.62 s | 134.4 |
| 3 | 119.21 | 6.81 d, 8.2 | 117.3 | 117.5 | ||||
| 4 | 154.46 | 154.0 | 150.8 | |||||
| 5 | 119.21 | 110.8 | 117.5 | |||||
| 6 | 7.69 s | 134.84 | 7.33 d, 2.0 | 134.3 | 7.30 s | 133.1 | 7.62 s | |
| 7 | 3.17 ddd, 17.5, 12.5, 5.5 | 29.04 | 2.72 t, 7.4 | 35.0 | 2.70 t | 31.3 | 3.02 t, 8.0 | 35.2 |
| 8 | 3.67 ddd, 17.5, 12.5, 5.5 | 67.54 | 3.41 t, 7.4 | 42.2 | 2.92 t | 39.6 | 3.50 t, 8.0 | 41.3 |
| 9 | 161.8 | 3.84 t | 70.4 | 4.12 t, 5.5 | 71.7 | |||
| 10 | 164.0 | 1.97 m | 27.6 | 2.30 tt, 5.5, 5.5 | 26.4 | |||
| 11 | 3.07 t | 36.4 | 3.22 t, 5.5 | 56.9 | ||||
| OMe | 3.85 s | 61.42 | ||||||
| N(Me)3 | 3.32 s | 54.11 | ||||||
| NH(Me)2 | 2.91, 2.96 s | 43.7, 43.6 | ||||||
| 55 HCl salt (45) |
56 HCl salt (45) |
59a (47) |
57 (46) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | NA | CD3OD | NA | CD3OD | CD3OD | CDCl3 | CDCl3 | |
| 1 | 140.4 | 137.6 | ||||||
| 2 | 7.62 s | 7.59 s | 7.40 s | 134.3 | 7.35 s | 132.9 | ||
| 3 | 118.7 | 118.2 | ||||||
| 4 | 152.2 | 151.6 | ||||||
| 5 | 118.7 | 118.2 | ||||||
| 6 | 7.62 s | 7.59 s | 7.40 s | 134.3 | 7.35 s | 132.9 | ||
| 7 | 3.01 t, 8.1 | 3.23 t, 8.1 | 2.72 t, 7.5 | 35.1 | 2.65 t, 7.0 | 35.0 | ||
| 8 | 3.25 t, 8.1 | 2.94 t, 8.1 | 3.35 t, 7.5 | 41.4 | 3.40 q, 6.8 | 41.9 | ||
| 9 | 4.13 t, 5.5 | 4.12 t, 5.5 | 4.12 t, 7.5 | 71.1 | 4.05 t, 6.5 | 71.1 | ||
| NH | 4.70 brt | |||||||
| 10 | 2.24 tt, 5.5, 8.4 | 2.30 tt, 5.5, 8.4 | 2.38 m | 26.4 | 2.05 m | 27.1 | ||
| 11 | 3.34 t, 8.4 | 3.52 t, 8.4 | 3.51 t, 7.5 | 57.0 | 2.75 t, 7.0 | 56.0 | ||
| OMe | 3.70 s | 52.1 | ||||||
| N(Me)2 | 2.93 s | 2.96 s | 3.0 s | 43.7 2C | 2.32 s | 44.4 | ||
| NH2(Me) | 2.77 s | 2.72 s | ||||||
| NHAc | 2.01 s | 22.6 | ||||||
| CO | 173.3 | 156.8 | ||||||
| 58 (46) |
60 (49) |
61 (50) |
62 (51) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | DMSO-d6-TFA | CD3OD | |
| 1 | 140.4 | 136.8 | 141.8 | 141.2 | ||||
| 2 | 7.38 s | 134.3 | 7.59 s | 133.2 | 7.66 s | 131.6 | 7.50 s | 135.2a |
| 3 | 118.7 | 117.5 | 119.4 | 119.6 | ||||
| 4 | 152.1 | 151.1 | 154.0 | 153.3 | ||||
| 5 | 118.7 | 117.5 | 119.4 | 119.6 | ||||
| 6 | 7.38 s | 134.3 | 7.59 s | 133.2 | 7.66 s | 131.6 | 7.50 s | 135.3a |
| 7 | 2.70 t, 7.0 | 34.9 | 7.82 t, 7.2 | 31.4 | 4.86 1H, m | 69.2 | 2.65 t, 7.0 | 31.6 |
| 8 | 3.20 q, 6.8 | 42.8 | 3.07 t, 7.2 | 39.3 | 3.16 dd, 3.5, 13 | 46.9 | 3.32 m | 42.8 |
| 9 | 4.05 t, 6.5 | 71.1 | 3.95 t, 5.7 | 70.6 | 4.04 t, 6 | 71.6 | 3.98, 4.07, m | 71.1 |
| NH | 9.92 br | |||||||
| 10 | 2.30 m | 26.4 | 1.99 tt, 6.1, 5.7 | 28.6 | 2.09 m | 30.1 | 2.35, 2.70, m | 30.3 |
| 11 | 3.50 t, 7.0 | 57.1 | 3.43 t, 6.1 | 36.8 | 3.56 t, 7 | 38.4 | 4.35 dd, 2.1, 2.7 | 77 |
| 12 | 143.0 | 145.2 | 165b | |||||
| 13 | 112.4 | 113.1 | 174.1 | |||||
| 14 | 1.79 s | 23.8 | ||||||
| OMe | 3.65 s | 52.5 | ||||||
| N(Me)3 | 3.25 s | 53.6 | ||||||
| NH(Me)2 | 3.00 s | 44.0 | ||||||
| CO | 159.5 | |||||||
| NH3 | 7.79 br | |||||||
| 63 (53) |
64 (53) |
65 (54) |
67 (21) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CDCl3 | CDCl3 | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | CDCl3 | CDCl3 | |
| 1 | 133.0 | 136.6 | 140.6 | 140.0 | ||||
| 2 | 7.63 s | 132.0 | 7.70 s | 132.7 | 7.75 s | 131.9 | 7.75 s | 130.8 |
| 3 | 118.9 | 119.4 | 119.3 | 118.2 | ||||
| 4 | 154.0 | 154.0 | 153.5 | 142.3 | ||||
| 5 | 118.9 | 119.4 | 119.3 | 118.2 | ||||
| 6 | 7.63 s | 132.0 | 7.70 s | 132.7 | 7.75 s | 131.9 | 7.75 s | 130.8 |
| 7 | 7.46 d, 16.0 | 141.0 | 7.21 d, 15.9 | 128.1 | 5.60 dd, 8.7, 4.5 | 5.65 dd, 8.7, 9.2 | 53.6a | |
| 8 | 6.04 d, 16.0 | 120.1 | 6.44 d, 15.9 | 137.8 | 3.38 d, 9.3 3.89 d, 9.0 |
3.71 dd, 9.3, 11.5 4.30 dd, 7.1, 8.5 |
41.5 | |
| 9 | 166.3 | 174.0 | 160.2c | 158.4b | ||||
| 10 | 4.06 t, 6.2 | 72.2 | 4.10 t, 5.6 | 71.4 | 4.16 d, 7.1 | 4.15 dd, 4.5, 4.2 | 53.0 | |
| 11 | 2.12 tt, 7.1, 6.2 | 28.2 | 2.25 tt, 7.8, 6.5 | 26.7 | 4.96 d, 8.7 | 4.96 dd, 7.1, 7.1 | 54.1a | |
| 12 | 2.72 t, 7.1 | 56.1 | 3.40 t, 7.8 | 56.8 | 3.61 d, 7.1 | 3.52 dd, 9.0, 8.5 3.65 dd, 8.7, 8.5 |
47.0 | |
| 13 | 43.8 | 159.8c | 157.8b | |||||
| 14 | 43.8 | |||||||
| NMe2 | 2.38 s | 45.5 | 2.83 s | |||||
| OEt | 4.23 d, 7.1 | 60.7 | ||||||
| OEt | 1.30 t, 7.1 | 14.3 | ||||||
| 68 (58) |
69 (58) |
70 (62) |
71 (63) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H (820) | 13C (6485) | 1H | 1H | 13C | 1H | 1H | 13C | 13C | |
| acetone-d6 | CD3OD | CDCl3 + DMSO-d6 | 1%TFA-D in DMSO-d6 | 1%TFA in DMSO-d6 | CDCl3 + CD3OD 4:1 | acetone-d6 | CDCl3 + CD3OD 4:1 | CDCl3 | |
| 1,1′ | 4.18 d, 8 | 75.5 | 4.16 s | 3.92 s | 73.6 | 4.10 s 4.11 s | 4.16 d, 7 | 73.9 | 73.8 |
| 2,2′ | 114.2 | 113.1 | 113.5 | 113.1 | |||||
| 3,3′ | 149.3 | 147.2 | 147.9 | 147.7 | |||||
| 4,4′ | 122.7 | 120.8 | 121.8, 121.7 | 121.4, 121.3 | |||||
| 5,5′ | 6.50 s | 133.2 | 6.28 s | 6.56 s | 131.3 | 6.17 s | 6.51 s | 130.8 | 130.6 |
| 6,6′ | 92.6 | 90.3 | 91.9, 91.8 | 91.9, 91.8 | |||||
| 7,7′ | 3.84 d, 18 3.14 d, 18 |
40.1 | 3.87 d, 18.5 3.02 d, 18.5 |
3.61 d, 17.8 3.19 d, 17.8 |
42.5 | 3.70, 3.71 d, 18 2.85 d, 18 |
3.82, 3.83, d, 18, 3.12 d, 18 | 39.7 | 38.7 |
| 8,8′ | 155.5 | 154.5 | 154.1 | 153.9 | |||||
| 9,9′ | 161.6 | 159.0 | 160.3 | 160.0 | |||||
| NH | 7.58 bt | 7.60 b | 8.10, 2H t, 6.9 | 7.56 brt, 6 7.81 brt, 6 |
|||||
| 10 | 3.34 m | 38.4 | 3.35 m | 3.48 m 3.14 m |
39.0 | 3.46 dd,14, 3 3.32 dd,14, 7 |
3.29 m, 3.52 m | 36.4 | 36.2 |
| 11 | 1.60 m | 26.1 | 1.69 m | 3.43 m | 71.0 | 3.60 m | 3.79 m | 68.1 | 68.0 |
| 12 | 26.1 | 1.73, 1.61 m | 1.97, 1.60 m | 45.3 | 45.0 | ||||
| 13 | 38.4 | 3.55, 3.37 m | 3.43 2H m | 33.8 | 33.6 | ||||
| OMe | 3.72 s | 60.4 | 3.73 s | 3.63 s | 59.6 | 3.59 s | 3.71 s | 60.3 | 60.0 |
| OH | 5.37 d, 8 | 5.46, 5.47 d, 7 | |||||||
| 72 (64) |
74 (61) |
73 (61) |
75 (65) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1,1′ | 3.94 d, 8.5 3.88 d, 8.5 |
73.6 | 4.15 d, 0.9 4.13 d, 0.9 |
75.5 | 4.14 d, 0.9 4.13 d, 0.9 |
75.5 | 4.16 s 4.14s |
75.5 |
| 2,2′ | 113.1 | 114.1 | 114.1 | 114.2 | ||||
| 3,3′ | 147.1 | 149.3 | 149.3 | 149.3 | ||||
| 4,4′ | 120.9 120.8 | 122.8 | 122.8 | 122.8 | ||||
| 5,5′ | 6.60 s 6.57 s |
131.2 | 6.46d, 0.9 6.47d, 0.9 |
132.2 133.2 |
6.47 d, 0.9 6.45 d, 0.9 |
132.2 133.2 |
6.46s 6.47s |
132.3 |
| 6,6′ | 90.5, 90.2 | 92.6, 92.5 | 92.6, 92.5 | 92.6, 92.4 | ||||
| 7,7′ | 3.60 d,18.2 3.19 d,18.2 |
39.7 39.5 |
3.14, 3.13 d, 18.3 3.82, 3.81 d, 18.3 |
40.1 40.0 |
3.15, 3.12 d, 18.3 3.82, 3.81 d, 18.3 |
40.1 40.0 |
3.15, 3.13 d, 18 3.82, 3.80 d, 18 |
40.2 40.0 |
| 8,8′ | 154.4, 154.1 | 155.1, 154.9 | 155.1 154.9 | 155.3, 154.9 | ||||
| 9,9′ | 159.1, 158.9 | 161.9, 161.8 | 161.9 161.8 | 161.8 | ||||
| NH | 8.66, 8.48 t, 5.5 | |||||||
| 10 | 4.02 d, 5.7 | 48.5 | 4.43, 4.42 d, 18.8 | 49.9 | 4.43, 4.43 d, 18.8 | 49.9 | 4.12 s | 49.2 |
| 11 | 204.4 | 207.8 | 207.8 | 206.2 | ||||
| 12 | 2.70 2H t, 6.8 | 38.6 | 4.41 dd, 6.3, 4.9 | 75.7 | 4.40 dd, 6.3, 4.9 | 75.7 | 2.60 t, 7 | 37.57 |
| 13 | 3.34 2H m | 33.8 | 3.66 dd, 13.9, 4.9 3.63 dd, 13.9, 6.3 |
43.4 | 3.66 dd, 13.9, 4.9 3.63 dd, 13.9, 6.3 |
43.4 | 1.88 dt, 7, 7 | 24.1 |
| 14 | 3.32 t, 7 | 39.6 | ||||||
| OMe | 3.63, 3.60 3H s | 59.6 | 3.76 3H s | 60.4 | 3.76 3H s | 60.4 | 3.75 3H s | 60.4 |
| OH | 6.41, 6.35 d, 8.5 | |||||||
| 76 (66) |
77 (67) |
78 (30) |
||||
|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | acetone-d6 | acetone-d6 | DMSO-d6 | DMSO-d6 | |
| 1,1′ | 3.93 s | 74.1 | 4.38 dd, 5, 12 4.40 dd, 5, 12 |
74.7 74.8 | 4.13 dd, 2.6b, 3.7 | 56.9 |
| 2,2′ | 113.5 | 5.08 d, 12; 5.06 d, 12 | 57.1 | 3.93 d, 3.5 | 53.0 | |
| 3,3′ | 147.6 | 183.5 | 186.0 | |||
| 4,4′ | 121.2 | 122.5, 122.4 | 122.7 | |||
| 5,5′ | 6.57 s | 131.7 | 7.63 s; 7.67 s | 149.1, 149.2 | 7.49 d, 2.6 | 143.7 |
| 6,6′ | 7.47 dd, 8.6, 1.5 | 90.7 | 91.7, 91.4 | 84.0 | ||
| 7,7′ | 3.12 d, 18 3.62 d, 18 |
40.0 | 3.31, 3.29 d, 18 3.85, 3.86 d, 18 |
38.2 38.1 |
3.68 d, 17.8 3.61 d, 17.9 |
43.6 |
| 8,8′ | 155.0 | 154.5, 154.3 | 154.9 | |||
| 9,9′ | 159.3 | 159.8, 159.6 | 158.3 | |||
| 10 | 3.08 m, 3.15 m | 45.7 | 4.22 d, 6 | 49.2 | 3.19 br | 38.6 |
| 11 | 3.55 m, 3.61 m | 67.4, 68.9 | 204.7 | 1.50 br | 26.4 | |
| 12 | 1.28 m, 1.40 m | 32.3 | 2.87 t, 6 | 39.6 | ||
| 13 | 1.50 m, 1.60 m | 25.5 | 3.58 dt, 6, 6 | 34.8 | ||
| 14 | 3.28 m, 3.22 m | 36.6 | ||||
| 15 | 1.45 m, 1.65 m | 34.4 | ||||
| 16 | 3.15 m | 39.4 | ||||
| OMe | 3.65 m | 60.1 | ||||
| NH | 8.41 t, 6, 8.45 t, 5 | 7.90 t, 6 | 8.63 t, 5.7 | |||
| NH | 8.27 t, 6; 8.21 t, 5 | 7.66 t, 6 | ||||
| OH | 6.36 s | 6.03 d, 5; 6.08 d, 5 | ||||
| 79 (71) |
81 (70) |
82 (71) |
||||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| pyridine-d5 | DMSO-d6 (36) | acetone-d6 | pyridine-d5 | |||||
| 1,1′ | 4.61, 4.58 d, 7.9 | 74.7 | 3.93, 3.91 d, 7.9 | 73.6 | 4.19 m | 75.0 | 4.65, 4.64 s | 74.7 |
| 2,2′ | 115.2 | 113.2 | 113.7 | 115.2 | ||||
| 3,3′ | 148.1 | 147.2 | 148.6 | 148.1 | ||||
| 4,4′ | 121.8 | 121.1 | 122.0 | 121.9 | ||||
| 5,5′ | 6.62, 6.63 s | 132.4 | 6.58, 6.56 s | 131.3 131.2 |
6.49, 6.51 d, 1.0 | 132.0 132.1 |
6.62, 6.58 s | 132.3 |
| 6,6′ | 91.9 | 90.4 | 91.8 | 91.9 | ||||
| 7,7′ | 3.47, 3.44 d, 18.2 4.40, 4.43 d, 18.2 |
40.3 | 3.61, 3.59d, 18.8 3.24, 3.19 d, 18.8 |
39.4 | 3.16, 3.19 d, 18.5 3.85, 3.83 d, 18.5 |
39.839.9 | 3.40, 3.47 d, 18.2 4.44, 4.45 d, 18.2 |
40.3 40.2 |
| 8,8′ | 155.2, 155.3 | 154.5 | 154.9, 155.0 | 155.2, 155.8 | ||||
| 9,9′ | 160.5 | 159.2, 159.1 | 160.4, 160.5 | 160.5, 160.6 | ||||
| NH | 9.34, 9.71 t,5.8 | 8.43, 8.39, d 5.8 | 7.66, 7.71 t, 5.9 | 9.80, 9.71 t, 5.8 | ||||
| 10 | 4.25, 3.98 m | 44.0 | 3.47, 3.29 m | 42.6 | 3.55, 3.76 m | 43.4 | 4.97 dd, 18.0, 5.8 | 47.49 |
| 11 | 4.76 m | 69.5 | 4.06 m | 68.1 | 4.25 m | 69.7 | 201.3 | |
| 12 | 4.34 dd, 8.8, 5.5; 4.26 m | 76.1 | 3.89, 3.82 m | 75.4 | 4.05 m | 75.7 | 4.82s | 76.1 |
| 13 | 152.3 | 151.3 | 152.5 | 152.2 | ||||
| 14 | 118.4 | 117.3 | 118.3 | 118.1 | ||||
| 15 | 7.92s | 131.1 | 7.57 s | 130.5 | 7.65 s | 131.3 | 7.91s | 131.1 |
| 16 | 143.5 | 142.7 | 142.9 | 144.2 | ||||
| 17 | 7.92s | 131.1 | 7.57 s | 130.5 | 7.65 s | 131.3 | 7.91s | 131.1 |
| 18 | 118.4 | 117.3 | 118.3 | 118.1 | ||||
| 19 | 5.29 dd, 4.3, 5.8 | 69.5 | 4.68 m, 5.2 | 69.4 | 4.90 dd, 4.3, 7.5 | 71.3 | 5.30 dd, 7.5, 4.6 | 70.7 |
| 20 | 3.97 m 3.81 ddd, 15.3, 5.8, 4.3 |
48.2 | 3.33 d, 5.3 | 46.4 | 3.48 m 3.61 m | 47.5 | 3.97 ddd, 5.8, 13.3, 4.6 3.81 ddd, 5.8, 13.3, 7.5 |
48.0 |
| OMe | 3.63 s | 59.9 | 3.64 s | 59.7 | 3.71, 3.71 s | 60.2 | 3.63 s | |
| 11-OH | 7.23 d, 5.2 | 5.29 d, 5.4 | 5.41 br s | |||||
| 1,1′-OH | 8.62, 8.61 d, 7.9 | 6.39, 6.37 d, 7.9 | 8.60 br s | |||||
| 17-OH | 7.77 d, 4.3 | 5.75 d, 4.6 | 5.43 br s | |||||
| 83 (63) |
84 (72) |
85 (72) |
86 (65) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| (CD3)2CO | CDCl3 | (CD3)2CO | (CD3)2CO | (CD3)2CO | (CD3)2CO | CD3OD | CD3OD | |
| 1,1′ | 4.15d, 6 4.16d, 6 |
73.9 73.8 | 4.23 dd, 8.1, 0.9 4.18 dd, 8.1, 0.9 |
75.1, 75.2 | 4.23, 4.18 dd, 8.1, 0.9 | 75.1, 75.2 | 4.12 s 4.11 s |
75.4 |
| 2,2′ | 112.7 | 113.8 | 113.9 | 114.2 | ||||
| 3,3′ | 147.9 | 148.8 | 148.7 | 149.3 | ||||
| 4,4′ | 121.4 | 122.1 | 122.1 | 122.7 | ||||
| 5,5′ | 6.50, 6.51 d, 1 | 130.9 | 6.56, 6.52 d, 0.9 | 132.3 | 6.57, 6.52 d, 0.9 | 132.3 | 6.46, 6.44 s | 132.2 |
| 6,6′ | 91.8, 91.6 | 91.6, 91.8 | 92.0, 91.6 | 92.5 | ||||
| 7,7′ | 3.83, 3.18 d, 18 3.81, 3.15 d, 18 |
38.9 38.8 |
3.86, 3.22 d, 18.1 3.84, 3.18 d, 18.1 |
40.0 40.1 |
3.86, 3.22 d, 18.1 3.84, 3.16 d, 18.1 |
40.0 40.1 | 3.81, 3.14 d, 18 3.79, 3.09 d, 18 |
40.1, 40.0 |
| 8,8′ | 154.1, 154.0 | 155.1, 155.2 | 154.8, 155.2 | 155.3, 155.1 | ||||
| 9,9′ | 159.3 | 160.0, 160.4 | 160.2, 160.0 | 161.7, 161.6 | ||||
| NH | 7.78, 7.73 t, 6 | |||||||
| 10 | 3.53 q, 6 | 37.2 | 3.50, 3.70 m | 43.5 | 4.60 d, 6.0 | 47.5 | 3.62 t, 6.5 | 37.9 |
| 11 | 2.11 m | 29.2 | 4.25 m | 69.7 | 200.9 | 2.15 t, 6.5 | 30.6 | |
| 12 | 4.07 t, 6 | 71.7 | 4.03 m | 75.8 | 4.75 s | 76.5 | 4.10 t, 6.5 | 71.6 |
| 13 | 151.3 | 152.0 | 151.2 | 153.6 | ||||
| 14 | 118.1 | 118.3 | 118.1 | 119.0 | ||||
| 15 | 7.50 s | 132.9 | 7.53 s | 134.1 | 7.57 s | 134.2 | 7.64 s | 131.7 |
| 16 | 137.4 | 139.9 | 140.5 | 143.1 | ||||
| 17 | 7.50 s | 132.9 | 7.53 s | 134.1 | 134.2 | 7.64 s | 131.7 | |
| 18 | 118.1 | 118.3 | 118.1 | 119.0 | ||||
| 19 | 2.86 t, 7 | 34.2 | 2.88 t, 7.0 | 34.7 | 2.91 t, 7.0 | 34.7 | 4.79 dd, 4.5, 7.5 | 72.2 |
| 20 | 3.60 td, 7, 6 | 40.4 | 3.58 td, 7.0, 6.0 | 40.9 | 3.58 q, 6.9 | 40.9 | 3.51, 3.46 | 47.6 |
| OMe | 3.71 s, 6H | 60.1 | 3.78, 3.73 s | 60.2 | 3.74, 3.72 s | 60.2 | 3.76 s | 60.4 |
| OH | 5.46, 5.45 d, 6 | |||||||
| 87 (70) |
88 (70) |
89 (74) |
90 (74) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| acetone-d6 | DMSO-d6 | acetone-d6 | acetone-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | |
| 1,1′ | 4.39, 4.40 d, 11.3 | 74.6 | 4.56 2H m | 72.3 | 4.18 d, 11.4 | 73.3 | 4.29 br | 72.01 |
| 2,2′ | 5.06, 5.07 d, 11.3 | 57.0, 57.1 | 5.27 2H m | 54.8 | 5.19 d, 11.3 | 57.8 | 5.35 | 55.84 |
| 3,3′ | 183.7 | 183.6 | 183.8 | 183.39 | ||||
| 4,4′ | 122.6 | 124.9 | 121.3 | 122.75 | ||||
| 5,5′ | 7.61 s 7.64 s |
149.2 149.3 | 7.46 d, 1.0 7.49 d, 0.9 |
146.2 146.4 |
7.68 s 7.65 s |
149.1 149.0 |
7.54 s | 145.35 |
| 6,6′ | 91.7 | 90.8 | 90.5 | 89.38 | ||||
| 7,7′ | 3.26, 3.30 d, 18.2 3.88, 3.86 d, 18.2 |
38.4 | 3.38, 3.35 d, 18.2 3.87, 3.90 d, 18.2 |
41.4 | 3,65, 3.63 d, 18 3.13, 3.10 d, 18 |
38.0 | 3.65, 3.63 d, 18 3.33, 3.30 d, 18 |
38.03 39.79 |
| 8,8′ | 154.6, 154.7 | 155.4, 155.5 | 153.9, 153.9 | 154.57, 153.92 | ||||
| 9,9′ | 160.2 | 160.2 | 158.9 | 158.78 | ||||
| NH | 7.65 br 7.67 br |
7.66 br 7.68 br |
8.67 dd, 11.8, 5.7 8.64 dd, 11.8, 5.7 |
8.67 8.64 |
||||
| 10 | 3.53, 3.81 m | 43.4 | 3.53, 3.80 m | 43.6 | 3.42 m | 36.3 | 3.42 m | 36.31 |
| 11 | 4.26 m | 69.7 | 4.25 m | 69.9 | 2.00 q, 6 | 29.6 | 2.00 q, 6 | 29.62 |
| 12 | 4.04, 4.08 m | 75.8 | 4.04, 4.08 m | 76.0 | 3.95 dd, 6, 4 | 71.2 | 3.95 dd, 6, 4 | 71.24 |
| 13 | 152.6 | 152.7 | 150.8 | 150.75 | ||||
| 14,18 | 118.4 | 118.4 | 117.4 | 117.38 | ||||
| 15,17 | 7.67 s | 131.4 | 7.67 s | 131.5 | 7.53 s | 133.0 | 7.53 s | 133.04 |
| 16 | 143.1 | 143.3 | 133.9 | 133.88 | ||||
| 19 | 4.91 dd, 4.4, 7.5 | 71.0 | 4.91 ddd, 4.2, 4.2, 7.5 | 71.4 | 2.76 t, 7 | 33.2 | 2.76 t, 7 | 33.20 |
| 20 | 3.48, 3.65 m | 47.4 | 3.51, 3.62 m | 47.4 | 3.42 m | 34.0 | 3.42 m | 39.79 |
| OH | 5.97 d, 5.6 5.99 d, 5.6 |
4.45 d, 5.2; 5.00 d, 4.2 5.92 d, 5.6; 5.92 d, 5.6 |
6.77 br | 6.77 br | ||||
| 91 (75) |
92 (77) |
93 (3:2 diastereomeric mixture) (78) |
|||||
|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | ||
| CDCl3 | CDCl3 | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | ||
| 1,1′ | 4.44 d, 5.1 | 77.4 | 4.13, 4.14 s | 75.4, 75.5 | 1,1′ | 3.96 br | 73.7 |
| 2,2′ | 112.3 | 114.2 | 2,2′ | 113.0 | |||
| 3,3′ | 148.1 | 149.3 | 3,3′ | 147.1 | |||
| 4,4′ | 121.3 | 122.8 | 4,4′ | 120.6 | |||
| 5,5′ | 6.42 | 131.6 | 6.46 s | 132.3 | 5,5′ | 6.54 s | 131.3 |
| 6,6′ | 74.3 | 92.4, 92.5 | 6,6′ | 90.3 | |||
| 7,7′ | 2.81 d, 16 2.85 d, 16 |
25.5 | 3.11, 3.14 d, 18.4 3.79, 3.83 d, 18.4 |
40.3 | 7,7′ | 3.64 d, 17.5 3.17 d, 17.5 |
39.4 |
| 8,8′ | 116.6 | 155.3, 155.4 | 8,8′ | 154.3 | |||
| 9,9′ | 161.4, 161.6 | 9,9′ | 158.9 | ||||
| NH | 2.52 br | NH | 8.35 m, 7.90 brd, 6.4; 8.06 br | ||||
| 10,10′ | 3.28, 3.29 m | 39.1, 40.0 | 10,10′ | 3.40 q, 6.6 | 36.2 | ||
| 11,11′ | 2.07, 2.23 m; 1.80 q, 6.8 | 33.9, 30.1 | 11,11′ | 2.00 q, 6.6 | 29.4 | ||
| 12,12′ | 3.96 t, 7.4; 2.48 t, 6.8 | 33.5, 23.1 | 12,12′ | 3.96 m | 71.2 | ||
| 13,13′ | 135.9, 126.1 | 13,13′ | 150.6, 150.7, 150.8 | ||||
| 14,14′ | 150.7, 149.5 | 14,14′, 18,18′ | 116.7, 116.8, 116.9 | ||||
| 15,15′ | 6.36 s | 111.3, 126.7 | 15,17 | 7.52 s, 7.44 s | 133.5 | ||
| OH | 2.45 d, 5.1 | 16 | 138.0, 138.2 | ||||
| OMe | 3.77 s, 3H | 60.3 | 3.75 s | 60.4 | 19a | 2.94 m, 2.79 m | 36.7, 37.0 |
| 19b | 2.79 m, 3.01 dd, 4.7, 13.6 | ||||||
| 20 | 3.87 m, 3.89 m | 57.9, 57.3 | |||||
| 21 | 171.8, 172.5 | ||||||
| OH | 6.24 br | ||||||
| 20-NH | 4.91 br, 4.52 br | ||||||
| OMe | 3.66 s | 59.5 | |||||
| 15′,17′ | 7.38 s, 7.47 s | 133.5 | |||||
| 19′a | 2.95 m, 2.64 dd, 7.5, 14 | 36.7, 36.2 | |||||
| 19′b | 2.86 dd, 6.2, 13.4; 2.81 m | ||||||
| 20′ | 4.26 m, 4.24 m | 54.5, 54.5 | |||||
| 21′ | 172.0, 172.3 | ||||||
| 94 (68) |
94a (68) |
95a (68) |
97 (79) |
99 (81) |
101 (82) |
||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 1H | 1H | 13C | 1H | 13C | 1H | 13C | |
| CDCl3 | CDCl3 | CDCl3 | DMSO-d6 | CD3OD | CD3OD | CD3OD | CDCl3 | CDCl3 | |
| 1 | 4.21 s | 5.87 s | 5.86 s | 3.92 d, 6.2 | 75.5 | 4.12s | 74.1 | 4.32 s | 60.1 |
| 2 | 114.2 | 113.5 | 113.4 | ||||||
| 3 | 149.3 | 148.1 | 147.7 | ||||||
| 4 | 122.7 | 121.9 | 121.1 | ||||||
| 5 | 6.57 s | 6.34 s | 6.32 s | 6.58 s | 132.3 | 6.34 s | 130.9 | 6.30 s | 131.1 |
| 6 | 92.6 | 91.9 | 91.7 | ||||||
| 7 | 3.23 d, 18.2 3.87 d, 18 |
3.08 d, 18 3.47 d, 18 |
3.06 d, 18 3.45 d, 18 |
3.18 d, 18.2 3.60 d, 18.2 |
40.0 | 3.03 d, 18.3 3.80 d, 18.3 |
39.3 | 3.86 d, 18.5 2.94 d, 18.5 |
38.7 |
| 8 | 155.2 | 154.2 | 154.0 | ||||||
| 9 | 163.6 | 159.9 | 159.4 | ||||||
| 10 | 3.40–3.60 m | 3.80 m 3.96 m |
3.57 m 3.70–3.95 m |
3.59 t | 37.7 | 3.55 m | 38.0 | ||
| 11 | 4.27 m | 5.28 q, 4 | 5.76 dd, 7, 4 | 2.10 tt | 29.5 | 2.10 m | 28.9 | ||
| 12 | 4.11 m | 4.19 dd, 10, 4 4.24 dd, 10, 4 |
4. 06 t | 71.3 | 4.12 t, 5.7 | 73.4 | |||
| 13 | 7.50 s | 151.6 | 153.4 | ||||||
| 14 | 118.5 | 134.4 | |||||||
| 15 | 7.72 m | 7.54 s | 7.43 s | 133.3 | 7.39 d, 2 | 133.4 | |||
| 16 | 4.22 d, 4.5 | 137.0 | 112.2 | ||||||
| 17 | 5.68 t, 8 | 5.54 t, 8 | 5.03 m | 7.43 s | 133.3 | 7.09 dd, 8.4, 2 | 128.8 | ||
| 18 | 3.56, 4.08 t, 8 | 3.50, 4.00 t, 8 | 3.70–3.95 m | 118.5 | 6.83 d, 8.4 | 113.3 | |||
| 19 | 2.74 t | 35.9 t | 2.67 t, 6.8 | 37.3 | |||||
| 20 | 2.95 bt | 42.2 t | 2.94 t, 7.2 | 42.9 | |||||
| OMe | 3.74 s, 3H | 3.74 s, 3H | 3.77 s, 3H | 3.65 s | 59.6 | 3.71 s | 60.2 | 3.74 s | |
| 1-OH | 6.35 d, 6.2 | ||||||||
| NH | 7.10 t, 6.5 5.80 br |
6.84 brt 5.33 br |
7.58 br 7.82 br |
7.33 t | 7.00 brt | ||||
| 102 (83) |
100 (81) |
103 (36) |
104 (36) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | |
| 1 | 4.08 s | 75.4 | 4.08 s | 3.95 s | 73.7 | 3.93 d, 8.3 | 73.7 |
| 2 | 114.1 | 113.4 | 113.3 | ||||
| 3 | 149.2 | 147.2 | 147.3 | ||||
| 4 | 122.7 | 121.0 | 121.1 | ||||
| 5 | 6.37 s | 132.1 | 6.41 s | 6.55 s | 131.4 | 6.56 s | 131.3 |
| 6 | 92.6 | 90.4 | 90.4 | ||||
| 7 | 3.74, 3.03 d, 18.0 | 40.1 | 3.78, 3.08 d, 18.3 | 3.21, 3.67 d, 18.2 | 39.5 | 3.17, 3.60 d, 18.1 | 39.5 |
| 8 | 155.1 | 154.6 | 154.5 | ||||
| 9 | 161.6 | 159.2 | 159.1 | ||||
| 10 | 3.48 m | 37.9 | 3.58 t | 3.41 bdd | 36.4 | 3.36 m | 39.9 |
| 11 | 2.02 tt, 6.9, 13.8 | 30.8 | 2.12 tt | 1.99 q, 6.6 | 29.5 | 2.74 t, 6.9 | 33.3 |
| 12 | 4.01 t, 6 | 72.3 | 4.04 t | 3.93 m | 71.3 | 139.0 | |
| 13 | 153.6 | 151.2 | 7.49 s | 133.2 | |||
| 14 | 118.9 | 117.5 | 117.3 | ||||
| 15 | 7.57 s | 131.7 | 7.44 s | 7.55 s | 133.9 | 151.1 | |
| 16 | 142.8 | 136.8 | 117.3 | ||||
| 17 | 7.57 s | 131.7 | 7.44 s | 7.55 s | 133.9 | 7.49 s | 133.2 |
| 18 | 118.9 | 117.5 | 3.94, 4.08 m | 71.3 | |||
| 19 | 4.75 t | 71.64 | 2.73 t | 2.90 m; 3.08 bd | 35.3 | 2.23, 2.38 m | 27.8 |
| 20 | 3.42 brt, 7.8 | 47.62 | 3.37 dt | 3.56 m | 55.0 | 3.56 m | 74.8 |
| 21 | 173.3 | 2.12m | 169.9 | 167.1 | |||
| 22 | 1.93 s | 22.7 | 1.25 s | ||||
| 23-32 | 1.25 s | ||||||
| 33 | 1.54m | ||||||
| OMe | 3.69 s | 60.4 | 3.72 s | 3.63 s | 59.8 | 3.63 s | 59.8 |
| Me2 | 0.87 d | ||||||
| NH | 7.31 t, 5.67 t | 8.57 t, 5.6 | 8.57 t, 5.8 | ||||
| +NMe3 | 3.15 s | 51.0 | |||||
| 105 (78) |
106 (78) |
107 (78) |
108 (72) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | acetone-d6 | ||
| 1 | 4.10 s | 75.4 | 4.07 s | 75.5 | 3.94 br | 73.7 | 4.18 dd, 7.8, 0.9 | 75.2 |
| 2 | 114.2 | 114.2 | 113.1 | 113.9 | ||||
| 3 | 149.2 | 149.3 | 147.2 | 148.8 | ||||
| 4 | 122.7 | 122.8 | 120.7 | 122.2 | ||||
| 5 | 6.40 s | 132.3 | 6.41 s | 132.3 | 6.56 s | 131.4 | 6.52 d, 0.9 | 132.3 |
| 6 | 92.4 | 92.4 | 90.3 | 91.8 | ||||
| 7 | 3.09, 3.79 d, 18.2 | 40.2 | 3.09, 3.77 d, 18.2 | 40.2 | 3.63, 3.20 d, 18.2 | 39.4 | 3.83, 3.17 d, 18.2 | 40.1 |
| 8 | 155.3 | 155.3 | 154.5 | 155.1 | ||||
| 9 | 161.4 | 161.6 | 159.0 | 160.4 | ||||
| 10 | 3.57 t, 7.0 | 38.0 | 3.58 t, 7.0 | 38.0 | 3.40 dt, 5.8, 6.5 | 36.3 | 3.58 ddd, 13.5, 6.4, 5.8 3.45 ddd, 13.5, 7.3, 5.8 |
47.7 |
| 11 | 2.09 m | 30.6 | 2.11 m | 30.6 | 1.99 quint, 6.5 | 29.5 | 4.84 ddd, 7.3, 6.4, 3.8 | 71.4 |
| 12 | 4.02 t, 6.0 | 72.2 | 4.07 t, 6.0 | 72.2 | 3.95 m | 71.2 | 138.5 | |
| 13 | 153.3 | 153.7 | 150.4 | 7.57 s | 130.9 | |||
| 14 | 119.1 | 119.4 | 116.5 | 111.4 | ||||
| 15 | 7.57 s | 134.9 | 7.55 s | 135.0 | 7.53 s | 134.0 | 150.7 | |
| 16 | 136.9 | 136.6 | 138.8 | 111.4 | ||||
| 17 | 7.57 s | 134.9 | 7.55 s | 135.0 | 7.53 s | 134.0 | 7.57 s | 130.9 |
| 18 | 119.1 | 119.4 | 116.5 | |||||
| 19 | 3.09 dd, 8.8, 13.9 3.17 dd, 5.2, 13.9 |
33.7 | 2.98 dd, 8.1, 14.7 3.20 dd, 4.7, 14.7 |
36.8 | 2.98 dd, 4.0, 13.0 2.86 dd, 4.0, 13.0 |
36.3 | ||
| 20 | 3.74 dd, 5.2, 8.8 | 72.6 | 3.75 dd, 4.7, 8.1 | 57.1 | 3.82 br | 57.2 | ||
| 21 | 171.4 | 173.0 | 174.3 | |||||
| OMe | 3.72 s | 60.4 | 3.65 s | 59.6 | 3.72 s | 60.2 | ||
| NH-20 | Not detected | 7.69 brt, 5.8 | ||||||
| OH-1 | 6.32 br | 5.48 d, 7.8 | ||||||
| OH-11 | 5.02 d, 3.8 | |||||||
| OH-15 | 3.79 s | |||||||
| NH-9 | 8.46 t, 5.8 | |||||||
| 109 (84) |
110 (79) |
111 (79) |
112 (86) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | DMSO-d6 | CD3OD | DMSO-d6 | CD3OD | CD3OD | CD3OD | |
| 1 | 3.91 s | 73.5 | 3.92 d, 8.1 | 75.5 | 3.92 d, 8.2 | 75.5 | 4.04 s | 75.2 |
| 2 | 113.0 | 114.2 | 114.2 | 113.8 | ||||
| 3 | 147.1 | 149.4 | 149.3 | 148.9 | ||||
| 4 | 120.8 | 122.8 | 122.8 | 122.5 | ||||
| 5 | 6.58 s | 131.2 | 6.59 s | 132.3 | 6.57 s | 132.3 | 6.36 s | 132.0 |
| 6 | 90.2 | 92.5 | 92.4 | 92.0 | ||||
| 7 | 3.20 d, 18.1 3.62 d, 18.1 |
39.1 | 3.23 d, 18.0 3.62 d, 18.0 |
40.1 | 3.18 d, 18.2 3.60 d, 18.2 |
41.4 | 3.73 d, 18.2 3.01 d, 18.2 |
39.0 |
| 8 | 154.4 | 155.3 | 155.2 | 154.8 | ||||
| 9 | 158.9 | 161.6 | 161.6 | 161.2 | ||||
| 10 | 3.38 m | 36.1 | 3.41 m | 37.9 | 3.36 m | 40.1 | 3.42 t, 7.6 | 40.1 |
| 11 | 1.99 m | 29.3 | 2.01 m | 30.6 | 2.76 t, 6.9 | 35.1 | 2.76 t, 7.6 | 34.7 |
| 12 | 3.95 t, 6.3 | 71.3 | 3.97 t, 6.3 | 72.3 | 140.3 | 139.8 | ||
| 13 | 151.3 | 153.7 | 7.54 s | 134.5 | 7.45 s | 134.1 | ||
| 14 | 117.5 | 119.5 | 118.8 | 118.4 | ||||
| 15 | 7.68 s | 132.9 | 7.64 s | 134.4 | 152.5 | 151.9 | ||
| 16 | 135.8 | 136.5 | 118.8 | 118.4 | ||||
| 17 | 7.68 s | 132.9 | 7.64 s | 134.5 | 7.54 s | 134.6 | 7.45 s | 134.1 |
| 18 | 117.5 | 153.7 | 71.2 | 4.08 t, 5.6 | 71.4 | |||
| 19 | 3.05 m | 26.9 | 2.93 t, 8.1 | 30.3 | 2.15 m | 26.4 | 2.14 t, 6.0 | 28.0 |
| 20 | 3.50 m | 64.2 | 3.35 m | 59.2 | 3.40 m | 57.2 | 3.30 t, 6.6 | 48.8 |
| OMe | 3.64 s | 59.5 | 3.65 s | 60.4 | 3.64 s | 60.4 | 3.69 s | 60.1 |
| OH-1 | 6.36 m | 6.36 d, 8.1 | 6.34 d, 8.2 | |||||
| NMe | 2.69 s | 32.0 | ||||||
| NH-9 | 8.57 t, 5.8 | 8.57 t, 5.8 | 8.58 t, 5.7 | |||||
| NMe3-18 | 3.09 s | 52.3 | 2.78 s | 43.6 | 2.83 s | 43.7 | ||
| 113 (69) |
113 acetate (69) |
114 (69) |
114 acetate (69) |
115 (87) |
116 (79) |
|||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD/CDCl3 | CD3OD | CD3OD | CD3OD/CDCl3 | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | |
| 1 | 4.26 s | 74.7 | 4.14s | 73.4 | 4.13 d, 0.8 | 75.5 | 3.91 d, 6.4 | 73.6 |
| 2 | 108.5 | 107.5 | 114.1 | 113.0 | ||||
| 3 | 151.5 | 148.9 | 149.3 | 147.1 | ||||
| 4 | 122.4 | 121.8 | 122.8 | 120.8 | ||||
| 5 | 6.35 s | 132.3 | 6.35 s | 130.5 | 6.45 d, 0.9 | 132.2 | 6.58 s | 131.2 |
| 6 | 90.6 | 89.7 | 92.4 | 90.2 | ||||
| 7 | 3.77 d, 17.5 3.93 d, 17.5 |
40.7 | 3.20 d, 17.5 3.85 d, 17.5 |
40.0 | 3.12 d, 18.3 3.82 d, 18.3 |
40.1 | 3.19 d, 18.6 3.61 d, 18.6 |
39.2 |
| 8 | 155.5 | 153.9 | 155.2 | 154.3 | ||||
| 9 | 160.9 | 159.4 | 161.7 | 158.9 | ||||
| 10 | 3.67 t, 7 | 40.3 | 3.43 t, 7 | 38.6 | 3.38 t, 6.8 | 39.5 | 3.37 brt, 6.4 | 37.3 |
| 11 | 2.99 t, 7 | 27.4 | 2.01 t, 7 | 21.9 | 1.90 q, 7.1 | 22.9 | 2.61 t, 6.4 | 24.2 |
| 12 | 134.6 | 2.67 t, 7 | 27.8 | 2.60 t, 7.1 | 28.9 | 124.1 | ||
| 13 | 6.85 s | 117.7 | 130.8 | 128.5 | 6.62 br | 109.4 | ||
| 14 | 7.59 s | 135.9 | 6.47 s | 108.4 | 6.62 s | 110.3 | 146.9 | |
| 15 | 146.9 | 149.5 | ||||||
| OMe | 3.86 s | 60.6 | 3.79 s | 60.3 | 3.77 s | 60.4 | 3.64 s | 59.6 |
| OH-1 | 6.37 d, 6.4 | |||||||
| NMe | 2.98 s | 29.5 | ||||||
| OCOCH3 | NA | 171.1, 20.5 | NA | 170.0, 20.7 | ||||
| NH-9 | 8.63 t, 5.6 | |||||||
| NH-12 | 11.71 br | |||||||
| NH-13 | 12.13 br | |||||||
| NH-14 | 7.40 s | |||||||
| 117 (79) |
118 (88) |
119 (89) |
120 (83) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | 13C | |
| DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | CD3OD | |
| 1 | 3.92 d, 6.3 | 73.6 | 4.13 s | 75.5 | 4.08 s | 75.6 | 3.92 br | 73.5 | 75.5 |
| 2 | 113.1 | 114.1 | 114.5 | 113.0 | 114.1 | ||||
| 3 | 147.2 | 149.3 | 149.0 | 147.0 | 149.2 | ||||
| 4 | 120.9 | 122.8 | 123.1 | 120.6 | 122.7 | ||||
| 5 | 6.58 s | 131.3 | 6.46 s | 132.3 | 6.40 s | 132.6 | 6.56 s | 131.2 | 132.2 |
| 6 | 90.4 | 92.3 | 92.9 | 90.1 | 92.4 | ||||
| 7 | 3.19 d, 18.0 3.62 d, 18.0 |
39.5 | 3.13 d, 18.3 3.81 d, 18.3 |
40.2 | 3.09 d, 18.0 3.75 d, 18.0 |
39.5 | 3.62 Abq, 18.0 3.20 Abq, 18.0 |
39.3 | 40.2 |
| 8 | 154.3 | 155.3 | 155.6 | 154.5 | 155.3 | ||||
| 9 | 157.9 | 161.6 | 162.1 | 158.4 | 161.3 | ||||
| 10 | 3.35 m | 35.0 | 3.34-3.37 m | 39.9 | 3.51 t, 7.0 | 40.5 | 4.16 brt, 5.1 | 37.9 | 39.5 |
| 11 | 1.86, 1.96 m | 30.2 | 1.65 m | 29.8 | 2.76 t, 7.0 | 26.1 | 5.24 dtb | 121.0 | 123.5 |
| 12 | 4.30 dd, 4.9, 7.8 | 56.9 | 1.87, 1.65 m | 25.6 | 126.3 | 6.06 d, 11.4 | 121.8 | 122.3 | |
| 13 | 174.8 | 4.09 dd, 5.2, 5.2 | 61.9 | 6.55 s | 111.3 | 115.1 | 130.8 | ||
| 14 | 159.2 | 190.6 | 149.7 | 150.0 | 151.5 | ||||
| 15 | 171.4 | 6.52 s | 131.2 | 118.1 | |||||
| OMe | 3.65 s | 59.7 | 3.77 s | 60.4 | 3.72 s | 59.6 | 3.63 s | 29.5 | 60.4 |
| OH-1 | 6.36 d, 6.3 | 6.55 br | 6.56 | ||||||
| NH-15 | 5.28 br, 5.28 br, 9.01 t, 5.4 | ||||||||
| NH2 | |||||||||
| NH-9 | 8.63 t, 5.1 | 8.75 br | |||||||
| NH-12 | 12.5 br | 11.70 s | |||||||
| NH-13 | 9.64 br | 12.30 s | |||||||
| NH-14 | 8.96 s | 7.45 s | |||||||
| 121 (90) |
122 (91) |
123 (89) |
124 (83) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1Ha | 13C | 1H | 13C | |
| CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | |
| 1 | 3.89 d, 0.6 | 73.5 | 3.98 br | 73.5 | 4.10 s | 75.4 | 3.88 br | 73.4 |
| 2 | 113.1 | 113.0 | 114.1 | 113.0 | ||||
| 3 | 147.1 | 147.0 | 149.3 | 147.1 | ||||
| 4 | 120.8 | 120.7 | 122.7 | 120.6 | ||||
| 5 | 6.56 d, 0.6 | 131.2 | 6.58 br | 131.2 | 6.40 s | 132.3 | 6.53 s | 131.2 |
| 6 | 90.1 | 90.2 | 92.5 | 90.1 | ||||
| 7 | 3.12 d, 18.0 3.60 overlapped |
39.4 | 3.24 d, 18 3.71 d, 18 |
39.4 | 3.10 d, 18.0 3.78 d, 18.0 |
40.2 | 3.62 d, 18.0 3.20 d, 18.0 |
39.5 |
| 8 | 154.4 | 154.4 | 155.3 | 154.4 | ||||
| 9 | 158.9 | 158.9 | 161.1 | 158.8 | ||||
| 10 | 3.40 overlapped | 38.1 | 3.43 dt, 6, 7 | 36.1 | 3.60 t, 7.0 | 38.3 | 3.10 m | 38.5 |
| 11 | 2.75 m | 24.6 | 2.02 tt, 7 | 29.3 | 2.12m | 30.6 | 1.42 m | 28.2c |
| 12 | 120.4 | 3.97 t, 7 | 71.2 | 4.02 t, 7.0 | 72.3 | 1.24 m | 23.3 | |
| 13 | 118.7 | 150.7 | 154.1 | 1.42 m | 28.0c | |||
| 14,14′ | 146.2 | 117.1 | 119.2 | 3.10 m | 40.7 | |||
| 15,15′ | 103.1 | 7.46 s | 132.8 | 7.52 s | 135.0 | 157.2 | ||
| 16 | 149.32 | 136.1 | 136.0 | |||||
| 17 | 112.2 | 3.78 s | 27.8 | 3.07, m | 38.0 | |||
| 18 | 173.2 | 150.7 | 4.07 m | 56.0 | ||||
| 19 | 137.7 | 163.1 | 170.1 | |||||
| 20 | 7.70 s | 124.2 | 3.42 dt, 6, 7 | 37.3 | 3.45 m | 39.3 | ||
| 21 | 128.9 | 2.66 t, 7 | 24.3 | 2.65 m | 25.7 | |||
| 22 | 137.0 | 124.1 | 125.9 | |||||
| 23 | 6.96 | 113.8 | 6.59 s | 109.0 | 6.55 s | 110.6 | ||
| 24 | 146.7 | 149.9 | ||||||
| OMe | 3.69 | 59.6 | 3.67 s | 59.5 | 3.72 s | 60.4 | 3.58 s | 59.6 |
| OH-1 | 6.42 br | 6.48 br | 6.50 br | 6.56 br | ||||
| OH | 10.4, 11.8, 14.4 br | |||||||
| NH2-18 | 4.18 br | |||||||
| NH | 11.95, 9.0 br | 7.85 brt (NH-14) | ||||||
| NH | 7.35 br (NH-15) | |||||||
| NH2 | 7.27 br | 7.38 br | 7.35 br | 7.35 br | ||||
| NH-9 | 8.62 t, 5.7 | 8.57 t, 6 | 8.60 t, 6.0 | 8.46 brt, 5.4 | ||||
| NH-19 | 8.15 t, 6 | 8.70 t, 6.0 | ||||||
| NH-22 | 11.74 br | 11.80 s | ||||||
| NH-23 | 12.16 br | 12.35 s | ||||||
| 125 (79) |
126 (66) |
127 (92) |
128 (92) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | CD3OD | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | |
| 1 | 3.91 d, 7.9 | 76.3 | 4.10 s | 76.4 | 3.93 d, 8.6 | 73.6 | 3.91 d, 8.2 | 73.6 |
| 2 | 114.9 | 115.1 | 113.1 | 113.0 | ||||
| 3 | 150.1 | 150.2 | 147.2 | 147.1 | ||||
| 4 | 123.5 | 123.7 | 120.9 | 120.8 | ||||
| 5 | 6.57 s | 133.0 | 6.40 s | 133.2 | 6.60 s | 131.2 | 6.57 s | 131.2 |
| 6 | 93.2 | 93.4 | 90.5 | 90.2 | ||||
| 7 | 3.19 d, 18.2 3.62 d, 18.2 |
40.1 | 3.11 d, 18 3.78 d, 18 |
41.0 | 3.21 d, 18.4 3.62 d, 18.4 |
39.3 | 3.18 d, 18.3 3.61 d, 18.3 |
39.5 |
| 8 | 159.5 | 156.0 | 154.1 | 154.3 | ||||
| 9 | 162.5 | 162.8 | 159.1 | 158.9 | ||||
| 10 | 3.16 m | 40.5 | 3.54 d, 4 | 44.7 | 3.99 d, 5.5 | 48.6 | 3.34 dt, 6.5, 5.9 | 33.8 |
| 11 | 1.47 m | 28.3 | 4.45 m | 78.4 | 205.8 | 2.67 t, 6.5 | 38.3 | |
| 12 | 1.45 m | 28.0 | 1.78, 2.05 m | 25.7 | 2.61 t, 6.8 | 39.8 | 205.7 | |
| 13 | 3.09 m | 42.9 | 3.35 m | 40.2 | 3.16 dt, 6.8, 4.8 | 35.2 | 3.83 d, 6.2 | 49.9 |
| 14 | 156.1 | 157.5 | 157.0 | 157.0 | ||||
| OMe | 3.64 s | 61.2 | 3.72 s | 61.3 | 3.64 s | 59.8 | 3.65 s | 59.6 |
| OH-1 | 6.34 d, 7.9 | 6.42 d, 8.6 | 6.33 d, 8.2 | |||||
| OMe-14 | 3.50 s | 51.3 | 3.55 s | 51.5 | ||||
| NH-9 | 8.54 t, 5.8 | 8.64 t, 5.5 | 8.43 t, 5.9 | |||||
| NH-13 | 7.44 br | 7.09 t, 4.8 | 7.34 t, 6.2 | |||||
| NH-14 | 7.1–7.3 4H, br | |||||||
| 131 (95) |
132 (96) |
133 (96) |
129 (94) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 1H | 13C | |
| CD3OD | CD3OD | acetone-d6 | acetone-d6 | acetone-d6 | acetone-d6 | CD3OD | CD2Cl2 | DMSO-d6 | |
| 1 | 7.13 s | 146.7 | 7.17 s | 146.4 | 7.17 s | 147.3 | 7.13 s | 7.01 s | 146.3 |
| 2 | 104.5 | 103.6 | 103.5 | 104.3 | |||||
| 3 | 149.9 | 149.3 | 149.3 | 149.4 | |||||
| 4 | 104.4 | 104.1 | 104.1 | 103.9 | |||||
| 5 | 3.38 d, 16.0 3.06 d, 16.0 |
38.3 | 3.45 d, 16.0 3.15 d, 16.5 |
37.5 | 3.44 d, 16.2 3.13 d, 16.2 |
37.4 | 3.38 d, 16 3.05 d, 16 |
3.37 d, 16 3.08 d, 16 |
38.2 |
| 6 | 120.9 | 119.9 | 119.9 | 120.4 | |||||
| 7 | 4.98 s | 80.4 | 5.07 d,7.0 | 80.2 | 5.07 d, 7.2 | 80.1 | 4.97 s | 5.05 s | 80.2 |
| 8 | 158.7 | 158.5 | 158.4 | 158.2 | |||||
| 9 | 160.7 | 159.1 | 159.1 | 160.2 | |||||
| CONH | 7.84 t, 5.8 | 8.64 br | 5.48 s | 7.15 brt, 6 | |||||
| NH | 7.26 brt, 5.3 | 6.02 d,7.2 | 5.40 br | ||||||
| 10 | 3.61 t, 6.0 | 38.0 | 3.64 q, 6.5 | 37.5 | 3.63 q, 6.6 | 37.9 | 3.61 t, 7.0 | 3.68 t, 6.0 | 39.7 |
| 11 | 2.14 dd, 6.0, 6.5 | 30.5 | 2.15 q, 6.0 | 30.4 | 2.14 q, 6.6 | 30.4 | 2.13 dd, 6.0, 7.0 | 2.10 2H | 29.9 |
| 12 | 4. 08 t, 6.0 | 72.2 | 4.10 t, 6.3 | 72.0 | 4.08 t, 6.2 | 72.0 | 4. 06 t, 6.0 | 4.08 t, 6.0 | 71.9 |
| 13 | 154.2 | 152.6 | 152.5 | 153.0 | |||||
| 14 | 119.4 | 118.4 | 118.6 | 119.1 | |||||
| 15 | 7.67 s | 1316 | 7.60 d,0.5 | 131.4 | 7.55 s | 134.3 | 7.48 s | 7.38 s | 134.1 |
| 16 | 141.5 | 143.6 | 138.6 | 136.9 | |||||
| 17 | 7.67 s | 131.6 | 7.60 d,0.5 | 131.4 | 7.55 s | 134.3 | 7.48 s | 7.38 s | 134.1 |
| 18 | 119.4 | 118.4 | 118.6 | 119.1 | |||||
| 19 | 4.92 dd, 3.0, 10 | 68.6 | 4.78 q, 5.3 | 72.3 | 3.01 t, 7.2 | 32.4 | 2.76 dd, 7.5, 7.0 | 2.66 t, 7.0 | 32.9 |
| 20 | 3.21 dd, 3.0, 13 3.09 dd, 10, 13 |
56.4 | 3.49 dt, 13.8, 5.1 3.37 dt, 14.0, 6.0 |
47.9 | 3.75 q, 7.0 | 51.2 | 2.96 dd, 7.5, 7.0 | 2.90 t, 7.0 | 41.5 |
| 21 | 174.8 | 7.34 d, 14.4 | 149.8 | ||||||
| 22 | 2.14 t, 7.0 | 39.7 | 99.4 | ||||||
| 23 | 26.6 | 197.4 | |||||||
| 24 | 6.64 d, 6.3 | 142.6 | |||||||
| 25 | 1.28 br(H23-H32) | 27.8 (C24-C31) | 6.73 d, 6.3 | 142.4 | |||||
| 26 | 194.1 | ||||||||
| 32 | 28.6 | ||||||||
| 33 | 1.55 m | 28.1 | |||||||
| 34,35 | 0.85 d, 6.5 | 22.9 | |||||||
| OMe | 3.64 s | 59.3 | 3.64 s | 59.0 | 3.65 s | 60.0 | 3.64 s | 3.66 s | 59.3 |
| NMe | 2.73 s | 33.9 | |||||||
| 7-OH | 5.99 d, 7.0 | 7.86 brt, 5.4 | |||||||
| 19-OH | 5.19 dd ,4.5 | ||||||||
| 130 (94) |
135 (98) |
136 (98) |
134 (97) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1Ha | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | acetone-d6 | acetone-d6 | CDCl3 | CDCl3 | |
| 1 | 7.13 s | 146.7 | 7.14s | 146.8 | 7.17 s | 147.3 | 7.00 s | 145.4 |
| 2 | 104.3 | 104.4 | 103.5 | 103.5 | ||||
| 3 | 149.8 | 149.9 | 149.3 | 148.5 | ||||
| 4 | 104.5 | 104.6 | 104.1 | 104.6 | ||||
| 5 | 3.38, 3.07 d | 38.2 | 3.39, 3.07 d, 16.1 | 38.3 | 3.44, 3.13 d, 16.2 | 37.4 | 3.38, 3.09 d, 16 | 37.1 |
| 6 | 120.8 | 120.9 | 119.9 | 121.3 | ||||
| 7 | 4.97 s | 80.4 | 4.99 s | 80.4 | 5.07 d,7.2 | 80.1 | 5.03 s | 78.8 |
| 8 | 158.7 | 158.8 | 158.4 | 156.7 | ||||
| 9 | 160.6 | 160.7 | 159.1 | 158.8 | ||||
| CONH | 8.64 br | 7.26 s | ||||||
| NH | 6.02 d, 7.2 | 3.68 | ||||||
| 10 | 3.62 t | 38.0 | 3.62 t, 6.8 | 38.0 | 3.63 q, 6.6 | 37.9 | 3.68 m | 37.0 |
| 11 | 2.10 pentet | 30.5 | 2.13 tt, 6.0, 6.8 | 30.6 | 2.14 q, 6.6 | 30.4 | 2.08 m | 29.3 |
| 12 | 4.07 t | 72.1 | 4.07 t, 6.0 | 72.1 | 4.08 t, 6.2 | 72.0 | 4.05 t, 5.5 | 70.8 |
| 13 | 153.6 | 152.9 | 152.5 | 151.0 | ||||
| 14 | 119.2 | 119.0 | 118.6 | 118.0 | ||||
| 15 | 7.62s | 134.3 | 7.48s | 134.3 | 7.55 s | 134.3 | 7.34 s | 132.8 |
| 16 | 143.2 | 139.7 | 138.6 | 138.6 | ||||
| 17 | 7.62 s | 131.5 | 7.48 s | 134.3 | 7.55 s | 134.3 | 7.34 s | 132.8 |
| 18 | 119.4 | 119.0 | 118.6 | 118.0 | ||||
| 19 | 2.83 dd | 72.7 | 2.77 t, 7.1 | 35.0 | 3.01 t, 7.2 | 32.4 | 2.73 t, 7.0 | 34.3 |
| 20 | 2.97 dd | 18.4 | 3.44 t, 7.1 | 40.0 | 3.75 q, 7.0 | 51.2 | 2.82 t, 7.0 | 52.1 |
| 21 | 7.80 s | 163.8 | 7.34 d, 14.4 | 149.8 | ||||
| 22 | 99.4 | |||||||
| 23 | 197.4 | |||||||
| 24 | 6.64 d, 6.3 | 142.6 | ||||||
| 25 | 6.73 d, 6.3 | 142.4 | ||||||
| 26 | 194.1 | |||||||
| NCH3 | 2.44 s | 35.8 | ||||||
| OMe | 3.65 s | 59.3 | 3.65 s | 59.3 | 3.65 s | 60.0 | 3.67 s | 59.0 |
| 7-OH | 7.86 brt, 5.4 | 4.1 s | ||||||
| 137 (99) |
138 (100) |
139 (101) |
140 (33) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 13C | 1H | 13C | 13C | 1H | 13C | |
| CD3OD | DMSO-d6 | CD3OD | 0.2% TFA-H in CD3OD | CD3OD | CD3OD | CD3OD + 5% CD3CO2D | 0.2% TFA-H in CD3OD | |||
| 1 | 126.0 | 137.7 | 112.1 | 112.1 | 130.6 | |||||
| 2 | 7.46 s | 7.44 s | 134.4 | 7.46 s | 134.8 | 7.42 d, 2.1 | 134.8 | 134.7 | 7.45 d, 2.2 | 134.3 |
| 3 | 118.5 | 118.8 | 131.8 | 131.7 | 110.4 | |||||
| 4 | 148.6 | 154.2 | 155.9 | 155.9 | 153.6 | |||||
| 5 | 118.5 | 118.8 | 6.89 d, 8.4 | 113.1 | 113.1 | 6.81 d, 8.5 | 117.1 | |||
| 6 | 7.46 s | 7.44 s | 134.4 | 7.46 s | 134.8 | 7.17 dd, 8.4, 2.1 | 130.5 | 130.4 | 7.06 dd, 8.5, 2.2 | 130.4 |
| 7 | 3.81 s | 3.75 s | 28.8 | 3.82 | 29.1 | 3.79 2YH, s | 28.7 | 28.7 | 3.78 s | 28.7 |
| 8 | 152.1 | 152.4 | 153.0 | 152.9 | 153.1 | |||||
| 9 | 165.6 | 165.6 | 165.7 | 165.9 | 166.1 | |||||
| 10 | 38.9 | 3.49 t, 7 | 40.6 | 3.47 t, 7.1 | 40.3 | 39.2 | 3.57 t, 7.0 | 39.0 | ||
| 11 | 25.7 | 2.79 t, 7 | 27.9 | 2.77 t, 7.1 | 27.8 | 26.1 | 2.94 t, 7.0 | 25.7 | ||
| 12 | 3.47 t | 3.36 m | 137.2 | 134.0 | NA | 133.4 | 132.8 | |||
| 13 | 2.69 t | 2.58 t | 110.7 | 6.84 br | 118.0 | 6.80 br | NA | 117.6 | 7.25 br | 117.5 |
| 14 | 153.8 | 7.62 br | 136.2 | 7.55 br | 136.1 | 135.0 | 8.70 br | 134.6 | ||
| OMe | 3.81 s | 3.75 s | 61.0 | 3.80 s | 61.3 | 3.80 s | 56.7 | 56.7 | ||
| NH | 6.50 s | 6.50 s | ||||||||
| 141 (102) |
142 (102) |
143 (102) |
144 (103) |
145 (103) |
||||
|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 137.2 | 138.2 | 138.1 | |||||
| 2 | 7.48 s | 7.48 s | 134.4 | 7.48 s | 7.46 s | 135.3 | 7.46 s | 135.3 |
| 3 | 118.7 | 119.6 | 119.6 | |||||
| 4 | 152.8 | 153.7 | 153.6 | |||||
| 5 | 118.7 | 119.6 | 119.6 | |||||
| 6 | 7.48 s | 7.48 s | 134.4 | 7.48 s | 7.46 s | 135.3 | 7.46 s | 135.3 |
| 7 | 3.83 br | 3.82 br | 28.8 | 3.83 br | 3.82 s | 28.9 | 3.82 s | 28.9 |
| 8 | 151.9 | 152.9 | 152.9 | |||||
| 9 | 165.4 | 166.3 | 166.4 | |||||
| 10 | 3.47 t, 7 | 3.48 t, 7 | 38.9 | 3.47 t, 7 | 3.45 t, 6.8 | 40.2 | 3.45 t, 6.8 | 39.9 |
| 11 | 2.70 t, 7 | 2.71 t, 7 | 25.8 | 2.71 t, 7 | 2.66 t, 6.8 | 24.5 | 2.67 t, 6.8 | 24.5 |
| 12 | 126.0 | 126.2 | 126.4 | |||||
| 13 | 6.51 s | 6.51 s | 110.7 | 6.51 s | 6.41 s | 112.0 | 6.46 s | 111.7 |
| 14 | 148.5 | 148.3 | 148.5 | |||||
| 15 | 4.01 t, 7 | 4.01 t, 7 | 72.3 | 4.02 t, 7 | 4.00 t, 6.1 | 73.1 | 4.00 t, 6.1 | 73.1 |
| 16 | 2.04 quin, 7 | 2.04 quin, 7 | 30.9 | 2.04 quin, 7 | 2.03 m | 31.1 | 2.03 tt, 6.1, 6.8 | 31.1 |
| 17 | 3.44 t, 7 | 3.44 t, 7 | 37.7 | 3.44 t, 7 | 3.43 t, 7.3 | 38.5 | 3.43 t, 66.8 | 39.0 |
| 18 | 176.3 | 177.2 | 177.2 | |||||
| 19 | 2.19 t, 7 | 2.20 t, 7 | 37.2 | 2.19 t, 7 | 2.18 t, 7.3 | 38.0 | 2.18 t, 7.6 | 38.0 |
| 20 | 1.61 quin, 7 | 1.61 quin, 7 | 27.1 | 1.61 quin, 7 | 1.60 m | 27.4 | 1.60m | 34.7–31.2 |
| 21–24 | 1.28 m | 1.28 m | 30.2 | 1.28 m | 1.35–1.27 m | 31.9–31.1 | 1.33–1.26 m | 34.7–31.2 |
| 25 | 1.28 m | 1.28 m | 30.7 | 1.28 m | 2.03 m | 27.9 | 1.33–1.26 m | 34.7–31.2 |
| 26 | 1.28 m | 1.28 m | 30.7 | 1.28 m | 5.33 t, 5.4 | 131.7 | 1.33–1.26 m | 34.7–31.2 |
| 27 | 1.28 m | 1.28 m | 31.0 | 1.28 m | 5.33 t, 5.4 | 131.6 | 1.33–1.26 m | 34.7–31.2 |
| 28 | 1.28 m | 1.28 m | 28.5 | 1.28 m | 2.03 m | 27.9 | 1.33–1.26 m | 34.7–31.2 |
| 29 | 1.28 m | 1.28 m | 40.2 | 1.28 m | 1.35–1.27 m | 31.9–31.1 | 1.33–1.26 m | 34.7–31.2 |
| 30 | 1.28 m | 1.51 m | 29.0 | 1.28 m | 1.35–1.27 m | 31.9–31.1 | 1.33–1.26 m | 34.7–31.2 |
| 31 | 0.90 t, 7 | 0.87 d, 7 | 23.1 | 1.28 m | 1.35–1.27 m | 31.9–31.1 | 1.33–1.26 m | 34.7–31.2 |
| 32 | 0.87 d, 7 | 23.1 | 1.28 m | 1.51 m, 6.8 | 30.0 | 1.33–1.26 m | 34.7–31.2 | |
| 33 | 0.88 t, 7 | 0.86 d, 6.8 | 23.8 | 1.33–1.26 m | 34.7–31.2 | |||
| 34 | 0.86 d, 6.8 | 23.8 | 1.33–1.26 m | 29.6 | ||||
| 35 | 1.33–1.26 m | 27.9 | ||||||
| 36 | 0.89 t, 6.8 | 15.2 | ||||||
| 146 (104) |
147 (105) |
148 (103) |
149 (79) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 + 1 d 1N HCl | |
| 1 | 136.4 | 136.5 | 131.4 | 122.0 | ||||
| 2 | 7.45 s | 132.9 | 7.45 s | 132.8 | 7.32 s | 132.5 | 152.8 | |
| 3 | 117.4 | 117.0 | 111.9 | 107.6 | ||||
| 4 | 150.5 | 150.5 | 149.2 | 152.7 | ||||
| 5 | 117.4 | 117.0 | 111.9 | 105.5 | ||||
| 6 | 7.45 s | 132.9 | 7.45 s | 132.8 | 7.32 s | 132.5 | 7.26 s | 132.2 |
| 7 | 3.75 s | 27.7 | 3.75 s | 27.9 | 3.69 s | 27.7 | 3.70 s | 27.0 |
| 8 | 150.9 | 150.8 | 151.5 | 150.3 | ||||
| 9 | 163.1 | 163.1 | 163.4 | 164.7 | ||||
| CONH | 8.13 t, 5.9 | 8.55 t, 5.9 | ||||||
| NOH | 12.03 s | 12.09 s | 11.93 s | 12.19 s | ||||
| NH-9 | 8.15 t, 6 | 8.17 t, 5.4 | ||||||
| NH-12 | 11.56 br | 11.55 br | ||||||
| NH-13 | 11.77 br | 11.96 br | 11.98 br | 11.19 br | ||||
| NH-14 | 12.16 br | 12.33 br | 7.33s | 7.33 s | ||||
| NH2 | 7.83 br | 7.49 s | ||||||
| 10 | 3.36 dt, 6 | 37.3 | 3.36 dt, 5.4, 6.4 | 37.3 | 3.36 dt, 5.9, 6.8 | 37.5 | 3.40 dt, 5.9, 6.9 | 37.6 |
| 11 | 2.59 t, 6 | 24.4 | 2.61 t, 6.4 | 24.4 | 2.59 t, 6.8 | 24.6 | 2.63 t, 6.9 | 24.7 |
| 12 | 124.3 | 124.2 | 124.5 | 124.2 | ||||
| 13 | 6.57 s | 109.2 | 6.58 br | 109.2 | 6.57 s | 109.4 | 6.60 s | 109.3 |
| 14 | 146.9 | 147.1 | 147.0 | 146.7 | ||||
| 15 | 3.98 t, 6 | 70.4 | 4.01 5.9 | 70.1 | ||||
| 16 | 2.02 m | 27.9 | 2.50 2H overlapped | 31.0 | ||||
| 17 | 3.06 m | 36.5 | 4.89 t, 7.3 | 58.5 | ||||
| 1′,5′ | 9.20 d, 5.9 | 145.1 | ||||||
| 2′,4′ | 8.18 dt, 5.9, 7.8 | 128.0 | ||||||
| 3′ | 8.61 t, 7.8 | 145.6 | ||||||
| OMe | 3.74 s | 60.0 | ||||||
| OH | 9.77 br | 10.48 br | ||||||
| 150 (79) |
151 (89) |
|||
|---|---|---|---|---|
| 1H | 13C | 1H | 13C | |
| DMSO-d6 | CD3OD | DMSO-d6 | DMSO-d6 + 1 d 1N HCl | |
| 1 | 122.7 | 134.0 | ||
| 2 | 155.0 | 7.55 s | 135.2 | |
| 3 | 108.8 | 118.9 | ||
| 4 | 154.3 | 154.8 | ||
| 5 | 107.4 | 118.9 | ||
| 6 | 7.26 s | 134.6 | 7.55 s | 132.5 |
| 7 | 3.70 s | 25.5 | 3.30 m | 32.0 |
| 8 | 151.5 | 4.25 dd, 12.0, 4.0 | 75.8 | |
| 9 | 167.3 | 166.2 | ||
| CONH | 8.59 t, 5.9 | 9.20 t, 6.0 | ||
| NOH | 12.20 s | |||
| NH-12 | 11.70 s | |||
| NH-13 | 14.1 br | 12.25 s | ||
| NH-14 | 7.25 br | |||
| 10 | 3.48 dt, 5.9, 6.7 | 39.2 | 3.30 m | 38.7 |
| 11 | 2.85 t, 6.7 | 25.8 | 2.45 m | 25.2 |
| 12 | 132.2 | 125.3 | ||
| 13 | 7.40 s | 117.7 | 6.42 s | 110.5 |
| 14 | 134.9 | 148.5 | ||
| OMe | 3.74 s | 60.8 | 3.82 s | 61.0 |
| NMe | 3.32 s | 53.2 | ||
| OH | 10.45 br | |||
| 152 (107) |
153 (107) |
154 (109) |
||||||
|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 13C | 1H | 13C | 1H | 13C | |
| CDCl3/CD3OD | pyridine-d5 | CD3OD | DMSO-d6 | DMSO-d6 | CDCl3/CD3OD | DMSO-d6 | CD3OD | |
| 1,1′ | 132.1 | 128.8 | 128.2 | Signals were not assigned | ||||
| 2,2′ | 7.62 d, 2 | 7.76 d, 2 | 136.0 | 132.9 | 7.40 d, 2 7.48 d, 2 |
132.5, 139.7 | 8.08 t, 6; 805, t, 6; 7.29, d, 2; 7.00, dd, 8.3, 2; 6.83, d, 8.3; 6.73, d, 1.9; 6.65, d, 1.9; 3.67, s; 3.61, s; 3.44 dt, 7, 6; 3.41 dt, 7, 6; 2.81 t, 7; 2.79 t, 7 | 166.1, 165.8, 154.7, 153.6, 153.5, 153.0, 134.4, 133.1, 132.1, 130.6, |
| 3,3′ | 112.1 | 108.8 | 108.8, 108.9 | |||||
| 4,4′ | 155.4 | 152.3 | 152.6, 152.9 | |||||
| 5,5′ | 6.85 d, 8 | 6.84 d, 8 | 118.7 | 116.2 | 6.83 d, 8 6.87 d, 8 | 115.9 | ||
| 6,6′ | 7.15 dd, 8, 2 | 7.26 dd, 8, 2 | 131.9 | 129.1 | 7.10 dd, 8, 2 7.17 dd, 8, 2 |
128.9, 129.1 | 117.0, 114.4, 110.5, 39.7 | |
| 7,7′ | 3.87 s | 4.01 s | 30.3 | 26.7 | 3.68, 3.87 s | 27.5, 35.7 | 39.6, 38.5, 28.9, 28.7 | |
| 8,8′ | 154.7 | 151.8 | 151.1, 151.6 | |||||
| 9,9′ | 167.5 | 161.9, 163.1 | ||||||
| 10,10′ | 3.63 t, 7 | 3.48 q, 7 | 41.2 | 38.1 | 3.50, 3.65 m | 37.6, 38.0 | ||
| 11,11′ | 2.87 t, 7 | 2.65 t, 7 | 40.1 | 37.0 | 2.95 m | 36.6, 36.7 | ||
| OH | 12, 14 br | |||||||
| NH | 8.56 t, 7 | 8.08, 8.05 t, 6 | ||||||
| 155 (110) |
156 (110) |
157 (110) |
158 (110) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3CN | CD3CN | CD3CN | CD3CN | CD3CN | CD3CN | CD3CN | CD3CN | |
| 1 | 130.9 | 130.8 | 130.9 | |||||
| 2 | 7.08 dd, 2.1, 8.4 | 130.4 | 7.07 dd, 1.8, 8.4 | 130.4 | 7.07 dd, 1.8, 8.4 | 130.4 | ||
| 3 | 6.83 d, 8.4 | 117.2 | 6.82 d, 8, 4 | 117.1 | 6.82 d, 8.4 | 117.1 | ||
| 4 | 153.0 | 152.8 | 152.8 | |||||
| 5 | 109.1 | 109.9 | 109.9 | |||||
| 6 | 7.36 d, 2.1 | 134.1 | 7.35 d, 1.8 | 134.0 | 7.35 d, 2.1 | 134.1 | ||
| 7 | 3.76 s | 28.5 | 3.75 s | 28.5 | 3.76 d, 2.1 | 28.5 | ||
| 8 | 153.4 | 153.2 | 153.4 | |||||
| 9 | 154.2 | 164.5 | 164.2 | 164.1 | ||||
| 10 | 3.35 q, 6.3 | 40.7 | 3.57 q, 6.3 | 39.7 | 3.65 q, 6.3 | 35.1 | 3.50 q, 6.6 | 39.1 |
| 11 | 2.77 t, 6.6 | 38.9 | 3.10 t, 6.6 | 34.1 | 3.21 t, 6.3 | 54.5 | 2.79 t, 6.6 | 39.0 |
| 12 | 2.76 t, 6.3 | 38.2 | ||||||
| 13 | 3.33 q, 6.3 | 40.6 | ||||||
| 14 | 158.2 | |||||||
| OMe | 3.57 s | 52.4 | 3.56 s | 52.4 | ||||
| NH-9 | 5.77 br | 7.37 br | 7.47 t, 5.4 | 7.29 t, 5.4 | ||||
| NH-13 | 5.77 br | 5.82 br | ||||||
| CN | 111.9 | |||||||
| NH2 | 5.49 br | |||||||
| 159 (112) |
160 (112) |
161 (112) |
162 (112) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | CD3OD | CD3OD | CD3OD | |
| 1 | 130.2 | 130.7 | 130.7 | 130.6 | ||||
| 2 | 7.06 dd, 8.0, 2.0 | 130.5 | 7.05 dd, 8.0, 2.0 | 130.5 | 7.01 dd, 8.0, 2.0 | 130.5 | 7.06 dd, 8.0, 2.5 | 130.5 |
| 3 | 6.75 d, 8.0 | 117.4 | 6.75 d, 8.0 | 117.2 | 6.83 d, 8.0 | 117.2 | 6.75 d, 8.5 | 117.2 |
| 4 | 154.5 | 153.9 | 153.8 | 153.9 | ||||
| 5 | 111.0 | 110.6 | 110.6 | 110.6 | ||||
| 6 | 7.35 d, 2.0 | 134.6 | 7.35 d, 2.0 | 134.6 | 7.28 d, 2.5 | 134.6 | 7.36 d, 2.5 | 134.6 |
| 7 | 3.78 s | 28.8 | 3.78 s | 28.8 | 3.69 s | 28.2 | 3.78 s | 28.8 |
| 8 | 153.4 | 153.3 | 153.2 | 153.2 | ||||
| 9 | 166.0 | 166.0 | 166.0 | 166.0 | ||||
| 10 | 3.54 t, 6.0 | 39.9 | 3.54 t, 6.0 | 39.8 | 3.42 t, 6.5 | 39.7 | 3.54 t, 7.0 | 39.8 |
| 11 | 2.84 t, 6.0 | 38.8 | 2.84 t, 6.0 | 38.6 | 2.83 t, 6.5 | 38.6 | 2..82 t, 7.0 | 39.4 |
| 12 | 2.84 t, 6.0 | 38.2 | 2.84 t, 6.0 | 37.9 | 2.83 t, 6.5 | 37.9 | 2.78 t, 7.0 | 38.5 |
| 13 | 3.53 t, 6.0 | 39.8 | 3.54 t, 6.0 | 40.1 | 3.42 t, 6.5 | 40.1 | 3.36 t, 7.0 | 41.0 |
| 14 | 162.1 | 161.9 | 161.9 | 159.2 | ||||
| 15 | 164.0 | 159.2 | 159.2 | |||||
| OCH2CH3 | 4.05 q, 7.0, 1.21 t, 7.0, | 61.9, 15.1 | ||||||
| 164 (112) |
165 (114) |
166 (114) |
167 (114) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 129.2 | 130.6 | 136.1 | 130.0 | ||||
| 2 | 7.35 d, 2.0 | 133.0 | 7.36 d, 2.0 | 134.4 | 7.49 d, 2.0 | 134.6 | 7.34 d, 2.0 | 134.4 |
| 3 | 109.0 | 110.4 | 116.5 | 110.8 | ||||
| 4 | 152.3 | 153.7 | 149.5 | 154.3 | ||||
| 5 | 6.75 d, 8.5 | 115.6 | 6.75 d, 8.3 | 117.0 | 7.47 d, 8.3 | 123.3 | 6.73 d, 8.3 | 117.3 |
| 6 | 7.05 dd, 8.5, 2.0 | 129.0 | 7.06 dd, 8.3, 2.0 | 130.3 | 7.21 dd, 8.3, 2.0 | 130.0 | 7.03 dd, 8.3, 2.0 | 130.3 |
| 7 | 3.78 s | 27.2 | 3.78 s | 28.7 | 3.86 s | 29.0 | 3.77 s | 28.7 |
| 8 | 151.6 | 153.0 | 152.5 | 153.0 | ||||
| 9 | 164.6 | 165.8 | 165.6 | 165.9 | ||||
| 10 | 3.62 m | 33.5 | 3.51 t, 6.8 | 39.7b | 3.52 t, 6.8 | 39.6 | 3.49 t, 6.8 | 39.7c |
| 11 | 3.51 m | 39.3 | 2.80 t, 6.8 | 38.6a | 2.80 t, 6.8 | 38.5 | 2.78 t, 6.8a | 38.6b |
| 12 | 158.6 | 158.6 | ||||||
| 13 | 3.02 s | 38.3 | 3.01 s | 38.3 | ||||
| 14 | 3.02 s | 38.3 | 3.01 s | 38.3 | ||||
| 1′ | 129.2 | 130.6 | 136.1 | 127.9 | ||||
| 2′ | 7.35 d, 2.0 | 133.0 | 7.49 d, 2.0 | 134.6 | 7.49 d, 2.0 | 134.6 | 7.31 d, 2.0 | 132.5 |
| 3′ | 109.0 | 116.5 | 116.5 | 114.6 | ||||
| 4′ | 152.3 | 149.6 | 149.5 | 155.0 | ||||
| 5′ | 6.75 d, 8.5 | 115.6 | 7.47 d, 8.8 | 123.3 | 7.47 d, 8.3 | 123.3 | 131.7 | |
| 6′ | 7.05 dd, 8.5, 2.0 | 129.0 | 7.21 dd, 8.3, 2.0 | 130.0 | 7.21 dd, 8.3, 2.0 | 130.0 | 7.35 d, 2.0 | 132.2 |
| 7′ | 3.78 s | 27.2 | 3.86 s | 29.0 | 3.86 s | 29.0 | 3.86 s | 29.0 |
| 8′ | 151.6 | 152.5 | 152.5 | 153.8 | ||||
| 9′ | 164.6 | 165.6 | 165.6 | 166.1 | ||||
| 10′ | 3.22, 3.32 m | 55.0 | 3.51 t, 6.8 | 39.6b | 3.52 t, 6.8 | 39.6 | 3.51 t, 6.8 | 39.6c |
| 11′ | 3.29 m | 32.2 | 2.80 t, 6.8 | 38.5a | 2.80 t, 6.8 | 38.5 | 2.77 t, 6.8a | 38.5b |
| 1″ | 172.7 | |||||||
| 2″ | 142.5 | |||||||
| 3″ | 6.93 s | 123.8 | ||||||
| 4″ | 124.1 | |||||||
| 5″ | 8.22 d, 2.2 | 134.3 | ||||||
| 6″ | 116.4 | |||||||
| 7″ | 160.2 | |||||||
| 8″ | 130.8 | |||||||
| 9″ | 7.65 d, 2.2 | 134.2 | ||||||
| 168 (114) |
168a (114) |
169 (117) |
170 (117) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CDCl3 | CDCl3 | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 129.6 | 133.1 | 130.3 | 130.8 | ||||
| 2 | 6.81 d, 2.0 | 126.3 | 6.96 d, 1.4 | 126.0 | 7.35 d, 2.0 | 134.6 | 7.36 d, 2.0 | 134.5 |
| 3 | 110.5 | 117.6 | 110.0 | 110.2 | ||||
| 4 | 143.7 | 144.3 | 153.2 | 153.7 | ||||
| 5 | 147.5 | 151.2 | 6.74 d, 8.5 | 116.8 | 6.74 d, 8.5 | 117.0 | ||
| 6 | 6.18 d, 2.0 | 115.0 | 6.17 d, 1.4 | 114.3 | 7.05 dd, 8.5, 2.0 | 130.0 | 7.06 dd, 8.0, 2.0 | 130.6 |
| 7 | 3.72 s | 28.3 | 3.71 s | 25.6 | 3.79 s | 28.0 | 3.76 s | 28.7 |
| 8 | 153.1 | 151.7 | 152.8 | 152.8 | ||||
| 9 | 166.2 | 163.1 | 165.8 | 166.0 | ||||
| 10 | 3.51 md | 40.0 | 3.48 br | 38.7 | 3.60 t, 7.0 | 39.0 | 3.51 t, 7.0b | 39.6 |
| 11 | 2.87 md | 41.1 | 2.48 br | 38.8 | 2.99 7.0 | 37.8 | 2.81 7.0c | 38.5 |
| 1′ | 130.3 | 132.9 | 130.3 | 128.3 | ||||
| 2′ | 7.32 d, 2.0 | 134.6 | 7.43 d, 1.8 | 134.7 | 7.35 d, 2.0 | 134.6 | 7.35 s | 133.0 |
| 3′ | 113.1 | 117.1 | 110.0 | 112.0 | ||||
| 4′ | 151.2 | 153.1 | 153.2 | 151.0 | ||||
| 5′ | 128.4 | 132.0 | 6.74 d, 8.5 | 116.8 | 112.0 | |||
| 6′ | 6.78, d, 2.0 | 131.3 | 6.65 d, 1.8 | 129.6 | 7.05 dd, 8.5, 2.0 | 130.0 | 7.35 s | 133.0 |
| 7′ | 3.68 br sd | 28.7 | 3.63 s | 28.6 | 3.79 s | 28.0 | 3.78 s | 28.7 |
| 8′ | 152.9 | 151.1 | 152.8 | 153.3 | ||||
| 9′ | 165.4 | 162.7 | 165.8 | 166.0 | ||||
| 10′ | 3.43 t, 6.8 | 39.6 | 3.54 t, 6.1 | 39.0 | 3.60 t, 7.0 | 39.0 | 3.50 t, 7.0b | 39.6 |
| 11′ | 2.87 t, 6.8 | 38.4 | 3.48 br | 38.8 | 2.99 7.0 | 37.8 | 2.80 7.0c | 38.5 |
| 1″ | 171.4 | 166.2 | ||||||
| 2″ | 146.8 | 140.0 | ||||||
| 3″ | 6.96 s | 120.9 | 6.50 s | 108.5 | ||||
| 4″ | 134.5 | 132.6 | ||||||
| 5″ | 8.42 d, 2.0 | 135.7 | 8.12 d, 1.8 | 134.6 | ||||
| 6″ | 118.2 | 117.9 | ||||||
| 7″ | 150.4 | 148.1 | ||||||
| 8″ | 135.5 | 133.8 | ||||||
| 9″ | 7.59 d, 2.0 | 134.1 | 7.67 d, 1.9 | 132.1 | ||||
| 1″-Me | 3.92 s | 53.5 | ||||||
| 8,8′-Me | 3.91 s, 3.85 s | 63.0, 62.9 | ||||||
| 4,4′-Me | 3.61 s, 3.58 s | 61.1, 60.5 | ||||||
| NH | 7.71 br; 7.23 t, 6.3 | |||||||
| 171 (117) |
163 (112) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | ||||
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |||||||||
| 1 | 130.8 | 1′ | 128.7 | 1″ | 129.3 | 1‴ | 130.8 | 129.1 | |||||
| 2 | 7.35 d, 2.0 | 133.3 | 2′ | 7.14 d, 2.0 | 128.2 | 2″ | 7.54 d, 2.0 | 134.0 | 2‴ | 7.35 d, 2.0 | 133.3 | 7.05 dd, 8.5, 2.0 | 129.0 |
| 3 | 109.5 | 3′ | 110.9 | 3″ | 113.6 | 3‴ | 109.5 | 6.75 d, 8.5 | 115.6 | ||||
| 4 | 152.8 | 4′ | 145.3 | 4″ | 152.3 | 4‴ | 152.8 | 152.4 | |||||
| 5 | 6.74 d, 8.5 | 116.0 | 5′ | 145.3 | 5″ | 6.75 d, 8.5 | 119.6 | 5‴ | 6.74 d, 8.5 | 116.0 | 109.0 | ||
| 6 | 7.05 dd, 8.5, 2.0 | 129.2 | 6′ | 6.75 d, 2.0 | 118.3 | 6″ | 7.20 dd, 8.5, 2.0 | 129.6 | 6‴ | 7.05 dd, 8.5, 2.0 | 129.2 | 7.35 d, 2.0 | 133.0 |
| 7 | 3.78 s | 27.5 | 7′ | 3.70 s | 27.6 | 7″ | 3.88 s | 27.9 | 7‴ | 3.78 s | 27.5 | 3.78 s | 27.2 |
| 8 | 152.0 | 8′ | 151.7 | 8″ | 151.5 | 8‴ | 152.0 | 151.6 | |||||
| 9 | 164.7 | 9′ | 164.6 | 9″ | 164.6 | 9‴ | 164.7 | 164.5 | |||||
| 10 | 3.51 t, 7.0 | 38.5 | 10′ | 3.54 t, 7.0a | 38.5 | 10″ | 3.47 t, 7.0a | 38.5 | 10‴ | 3.51 t, 7.0 | 38.5 | 3.62 dt, 6.5, 2.0 | 31.9 |
| 11 | 2.80 t, 7.0 | 37.4 | 11′ | 2.81 t, 7.0b | 37.4 | 11″ | 2.76 t, 7.0a | 37.4 | 11‴ | 2.80 t, 7.0 | 37.4 | 2.96 m | 55.8 |
| 12 | 3.75 s | 54.1 | |||||||||||
| 172 (44) |
173 (118) |
174 (118) |
175 (118) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 113.1 | 112.2 | 137.4 | ||||
| 2 | 7.43 d, 2.2 | 134.7 | 7.17 dd, 2.2, 8.5 | 130.4 | 7.47 s | 134.5 | 7.14 dd, 2.0, 8.5 |
| 3 | 130.3 | 6.88 d, 8.5 | 113.2 | 113.2 | 6.88 d, 8.5 | ||
| 4 | 155.8 | 155.9 | 152.1 | ||||
| 5 | 6.88 d, 8.5 | 112.1 | 131.8 | 118.6 | |||
| 6 | 7.17 dd, 2.2, 8.5 | 131.1 | 7.42 d, 2.0 | 134.7 | 7.47 s | 134.5 | 7.47 d, 2.0 |
| 7 | 3.79 s | 28.7 | 3.79 s | 28.7 | 3.81 s | 28.8 | 3.79 s |
| 8 | 152.9 | 153.0 | 152.2 | ||||
| 9 | 165.8 | 165.8 | 165.5 | ||||
| 10 | 3.41 t, 7.0 | 41.3 | 3.42 t, 7.3 | 41.3 | 3.43 t, 7.2 | 41.3 | 3.42 t, 7.2 |
| 11 | 2.73 t, 7.0 | 35.2 | 2.75 t, 7.3 | 35.2 | 2.75 t, 7.2 | 35.2 | 2.73 t, 7.2 |
| 12 | 140.3 | 140.3 | 140.3 | ||||
| 13,17 | 7.42 s | 134.4 | 7.43 s | 134.4 | 7.44 s | 134.4 | 7.41 s |
| 14,16 | 118.7 | 118.7 | 118.7 | ||||
| 15 | 152.1 | 152.2 | 152.1 | ||||
| 18 | 4.05 t, 5.5 | 71.7 | 4.08 t, 5.7 | 71.6 | 4.06 t, 5.8 | 71.6 | 3.99 t, 6.2 |
| 19 | 2.25 tt, 5.5, 7.6 | 26.4 | 2.19 tt, 5.5, 7.5 | 29.0 | 2.18 tt, 5.8, 7.8 | 29.0 | 2.03 tt, 6.2, 6.5 |
| 20 | 3.46 t, 7.6 | 56.9 | 3.29 t, 7.5 | 39.0 | 3.29 t, 7.8 | 39.0 | 3.41 t, 6.5 |
| 22 | 2.17 t, 7.3 | ||||||
| 23 | 1.60 m | ||||||
| 24–32 | 1.28 16H, m 1.16 m | ||||||
| 33 | 1.51 m | ||||||
| 34,35 | 0.86 d, 6.5 | ||||||
| Ome | 3.81 s | 56.7 | 3.82 s | 56.7 | 3.81 s | 61.0 | 3.82 s |
| NH(CH3)2 | 2.93 s | 43.7 | |||||
| 176 acetate (119) |
177 acetate (119) |
178 (82) |
179 (121) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 13C | 1H | 13H | |
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | CD3OD | ||
| 1 | 132.6 | 135.6 | 138.2 | ||||
| 2 | 7.56 d, 2 | 7.50 s | 133.5 | 7.48 s | 133.4 | 7.25 d, 7.5 | 130.1 |
| 3 | 117.8 | 117.7 | 7.20 dd, 7.3, 7.5 | 129.3 | |||
| 4 | 153.1 | 153.8 | 7.12 t, 7.3 | 127.2 | |||
| 5 | 6.80 d, 8 | 117.8 | 117.7 | 7.20 dd, 7.3, 7.5 | 129.3 | ||
| 6 | 7.2–7.5 m | 7.50 s | 133.5 | 7.48 s | 133.4 | 7.25 d, 7.5 | 130.1 |
| 7 | 3.85 s | 3.84 s | 27.9 | 3.84 s | 28.0 | 3.91 s | 29.9 |
| 8 | 152.5 | 150.9 | 153.2 | ||||
| 9 | 162.8 | 163.4 | 166.0 | ||||
| 10 | 3.4–3.7 m | 3.4–3.6 m | 39.9 | 3.60 dt, 6.1, 5.8 | 37.3 | 2.60 t, 6.7 | 37.7 |
| 11 | 2.76 t, 7 | 2.74 t, 6 | 34.0 | 2.06 m | 28.4 | 2.05 tt, 6.2, 6.7 | 30.7 |
| 12 | 135.3 | 4.06 t, 5.8 | 67.9 | 4.10 t, 6.2 | 72.2 | ||
| 13 | 7.2–7.5 m | 7.38 d, 2 | 152.4 | 153.6 | |||
| 14 | 111.9 | 114.0 | 119.5 | ||||
| 15 | 151.2 | 7.32 d, 1.8 | 133.3 | 7.51 s | 134.4 | ||
| 16 | 6.80 d, 8 | 6.76 d, 8 | 113.0 | 111.5 | 137.3 | ||
| 17 | 7.02 dd, 8, 2 | 7.02 dd, 8, 2 | 129.2 | 7.02 dd, 8.3, 1.8 | 128.8 | 7.51 s | 134.4 |
| 18 | 4.20 t, 6 | 4.14 t, 6 | 68.4 | 6.78 d, 8.6 | 133.2 | 119.5 | |
| 19 | 1.9–2.2 m | 1.9–2.2 m | 28.1 | 2.68 t, 6.5 | 37.0 | 2.88 t, 7.5 | 33.3 |
| 20 | 3.4–3.7 m | 3.4–3.6 m | 38.6 | 2.96 t, 6.5 | 42.5 | 3.13 t, 7.5 | 41.5 |
| Ome | 3.86 s | 3.86 s | 60.6 | 3.82 s | 60.5 | ||
| NH(CH3)2 | |||||||
| NH | 6.60 brt, 6 | 6.64 t, 6 | 7.10 brt, 6.1 | ||||
| NH | 6.32 m | 6.44 m | |||||
| NOH | 9.80 br | 10.64 br | |||||
| OCOCH3 | 1.94 s | 1.94 s | 23.1 | ||||
| NHCOCH3 | 171.3 | ||||||
| 180 (121) |
181 (121) |
182 (121) |
183 (121) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | CD3OD | CD3OD | CD3OD | |
| 1 | 138.1 | 138.1 | 138.1 | 137.1 | |||||
| 2 | 7.25 d, 7.5 | 130.0 | 7.25 d, 7.5 | 130.0 | 7.21 m | 7.23 m | 130.0 | 7.21 m | 130.0 |
| 3 | 7.20 dd, 7.3, 7.5 | 129.3 | 7.20 dd, 7.3, 7.5 | 129.3 | 7.21 m | 7.14 m | 129.3 | 7.21 m | 129.3 |
| 4 | 7.15 t, 7.3 | 127.2 | 7.12 t, 7.3 | 127.2 | 7.13 m | 7.14 m | 127.2 | 7.14 m | 127.2 |
| 5 | 7.20 dd, 7.3, 7.5 | 129.3 | 7.20 dd, 7.3, 7.5 | 129.3 | 7.21 m | 7.14 m | 129.3 | 7.21 m | 129.3 |
| 6 | 7.25 d, 7.5 | 130.0 | 7.25 d, 7.5 | 130.0 | 7.21 m | 7.23 m | 130.0 | 7.21 m | 130.0 |
| 7 | 3.90 s | 30.2 | 3.91 s | 29.9 | 3.88 s | 3.78 s | 29.9 | 3.88 s | 29.9 |
| 8 | 153.4 | 153.3 | 153.2 | 153.2 | |||||
| 9 | 166.1 | 166.0 | 166.0 | 166.0 | |||||
| 10 | 3.43 t, 6.7 | 37.7 | 3.51, t, 6.8 | 37.7 | 3.42 t, 7.2 | 3.32 m | 41.3 | 3.42 t, 7.1 | 41.4 |
| 11 | 2.00 tt, 6.1, 6.7 | 30.0 | 2.04 tt, 6.1, 6.8 | 30.7 | 2.73 t, 7.2 | 2.73 t, 6.8 | 35.2 | 2.74 t, 7.1 | 35.2 |
| 12 | 4.02 t, 6.1 | 68.3 | 3.99 t, 6.1 | 72.3 | 140.3 | 139.1 | |||
| 13 | 155.9 | 153.9 | 7.45 s | 7.48 s | 134.4 | 7.45 s | 134.4 | ||
| 14 | 113.4 | 119.6 | 118.7 | 118.7 | |||||
| 15 | 7.47 d, 1.8 | 134.4 | 7.44 s | 134.4 | 152.2 | 152.2 | |||
| 16 | 131.6 | 136.1 | 118.7 | 118.7 | |||||
| 17 | 7.18 d, 1.8, 8.4 | 130.0 | 7.44 s | 134.4 | 7.45 s | 7.48 s | 134.4 | 7.45 s | 134.4 |
| 18 | 6.95 d, 8.4 | 115.0 | 119.6 | 4.09 t, 5.7 | 3.98 t, 5.9 | 71.6 | 4.10 t, 5.7 | 71.4 | |
| 19 | 2.88 t, 7.6 | 33.3 | 3.27 t, 7.3 | 36.6 | 2.19 tt, 5.7, 7.3 | 2.06 m | 29.0 | 2.22 tt, 5.7, 7.2 | 33.8 |
| 20 | 3.13 t, 7.6 | 41.9 | 4.83 t, 7.3 | 63.3 | 3.30 t, 7.3 | 3. 60 m | 38.9 | 3.32 t, 7.2 | 48.2 |
| 21,25 | 8.88 t, 5.6 | 146.2 | |||||||
| 22,24 | 8.08 t, 5.6, 7.1 | 129.5 | |||||||
| 23 | 8.59 t, 7.1 | 147.2 | |||||||
| NH-9 | 8.00 t, 5.8 | ||||||||
| NOH-8 | 11.77 s | ||||||||
| NMe-20 | 2.76 s | 27.1 | |||||||
| 184 (121) |
185 (121) |
186 (121) |
187 (121) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 130.3 | 130.6 | 131.8 | 131.8 | ||||
| 2 | 7.04 dd, 2.0, 8.3 | 130.6 | 7.03 dd, 1.7, 8.3 | 130.3 | 7.18 dd, 1.8, 8.5 | 130.4 | 7.17 dd, 2.0, 8.4 | 130.4 |
| 3 | 6.75 d, 8.3 | 117.1 | 6.75 d, 8.3 | 117.1 | 6.90 d, 8.5 | 113.2 | 6.89 d, 8.4 | 113.2 |
| 4 | 153.7 | 153.7 | 155.9 | 155.9 | ||||
| 5 | 110.5 | 110.5 | 112.2 | 112.2 | ||||
| 6 | 7.43 d, 2.0 | 134.4 | 7.34 d, 1.7 | 134.4 | 7.42 d, 1.8 | 134.4 | 7.42 d, 2.0 | 134.7 |
| 7 | 3.76 s | 28.7 | 3.75 s | 28.7 | 3.79 s | 28.7 | 3.79 s | 28.7 |
| 8 | 153.2 | 153.1 | 153.0 | 153.0 | ||||
| 9 | 166.2 | 165.8 | 165.8 | 165.8 | ||||
| NMe-20 | 2.76 s | 27.7 | 2.76 s | 27.7 | ||||
| 10 | 3.41 t, 7.1 | 41.4 | 3.42 t, 7.0 | 41.3 | 3.43 t, 7.2 | 41.3 | 3.42 t, 7.1 | 41.3 |
| 11 | 2.74 t, 7.1 | 35.2 | 2.73 t, 7.0 | 35.2 | 2.75 t, 7.2 | 35.2 | 2.74 t, 7.1 | 35.2 |
| 12 | 140.3 | 140.3 | 140.3 | 140.3 | ||||
| 13 | 7.43 s | 134.4 | 7.43 s | 134.4 | 7.43 s | 134.4 | 7.43 s | 134.4 |
| 14 | 118.7 | 118.7 | 118.7 | 118.7 | ||||
| 15 | 152.2 | 152.1 | 152.2 | 152.2 | ||||
| 16 | 118.7 | 118.7 | 118.7 | 118.7 | ||||
| 17 | 7.43 s | 134.4 | 7.43 s | 134.4 | 7.44 s | 134.4 | 7.43 s | 134.4 |
| 18 | 4.07 t, 5.7 | 71.6 | 4.06 t, 5.7 | 71.5 | 4.08 t, 5.7 | 71.6 | 4.05 t, 5.6 | 71.6 |
| 19 | 2.18 tt, 5.7, 7.2 | 29.0 | 2.21 tt, 5.7, 7.2 | 33.8 | 2.18 tt, 5.7, 7.6 | 29.0 | 2.21 tt, 5.6, 7.6 | 33.8 |
| 20 | 3.30 t, 7.2 | 39.0 | 3.34 t, 7.2 | 48.2 | 3.30 t, 7.6 | 38.9 | 3.30 t, 7.6 | 38.2 |
| OMe | 3.82 s | 56.7 | 3.82 s | 56.7 | ||||
| 188 (121) |
189 (86) |
190 (122) |
191 (122) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | |
| 1 | 138.0 | 131.4 | 130.4 | 131.7 | ||||
| 2 | 7.23 m | 130.0 | 7.40 d, 2.0 | 134.4 | 7.38 d, 2.0 | 133.0 | 7.44 d. 2.0 | |
| 3 | 7.23 m | 129.3 | 111.8 | 110.2 | 112.5 | |||
| 4 | 7.16 m | 127.3 | 155.4 | 153.8 | 155.7 | |||
| 5 | 7.23 m | 129.3 | 6.85 d, 8.0 | 112.8 | 6.97 d, 8.0 | 112.6 | 6.78 d, 8.0 | 113.2 |
| 6 | 7.23 m | 130.0 | 7.13 dd, 2.4, 8.5 | 130.0 | 7.12 dd, 2.0, 8.0 | 129.1 | 7.02 dd, 2.0, 8.0 | 130.4 |
| 7 | 3.91 s | 31.0 | 3.76 s | 28.2 | 3.72 s | 27.8 | 3.76 s | 28.1 |
| 8 | 152.2 | 152.6 | 151.8 | 152.7 | ||||
| 9 | 166.9 | 165.4 | 163.2 | 165.5 | ||||
| 10 | 3.39 t, 7.2 | 41.0 | 3.35 t, 7.0 | 41.1 | 3.15 t, 7.2 | 48.7 | ||
| 11 | 2.70 t, 6.9 | 34.8 | 2.73 t, 7.0 | 34.0 | 2.15 t, 7.2 | 29.0 | ||
| 12 | 139.8 | 139.0 | 4.08 t, 6.0 | 71.5 | ||||
| 13 | 7.39 s | 134.0 | 7.46 s | 132.9 | 152.2 | |||
| 14 | 118.4 | 117.2 | 118.9 | |||||
| 15 | 152.0 | 150.7 | 7.36 s | 134.3 | ||||
| 16 | 118.0 | 117.2 | 139.9 | |||||
| 17 | 7.39 s | 134.0 | 7.46 | 132.9 | 7.36 s | 134.3 | ||
| 18 | 4.03 t, 5.7 | 70.9 | 3.91 t, 6.0 | 71.0 | 118.9 | |||
| 19 | 2.37 q, 6.4 | 25.1 | 2.03 t, m | 28.0 | 2.72 t, 7.8 | 35.5 | ||
| 20 | 3.61 dt, 8.4, 7.6 | 69.0 | 3.38 t, 6.8 | 40.6 | 3.45 t, 7.8 | 41.5 | ||
| 21 | 156.5 | |||||||
| 22 | 3.57 s | 52.3 | ||||||
| 23 | ||||||||
| OMe | 3.79 br | 56.0 | 3.79 s | 56.2 | 3.79 s | 57.0 | ||
| NOMe2 | 3.24 br | 58.0 | ||||||
| NH | 7.99 t, 6.0 | |||||||
| NMe | 2.83 s | 41.4 | 2.60 s | 34.4 | ||||
| 192 (84) |
193 (123) |
194 (123) |
195 (124) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | CD3OD + TFA | CDCl3 | CDCl3 | ||
| 1 | 136.4 | 137.9 | 138.0 | 135.7 | ||||
| 2 | 7.48 s | 132.9 | 7.49 s | 134.4 | 7.47 s | 134.6 | 7.41 br | 134.2 |
| 3 | 117.2 | 117.8 | 118.5 | 118.6 | ||||
| 4 | 150.5 | 152.2 | 152.3 | 152.6 | ||||
| 5 | 117.2 | 117.8 | 118.5 | 118.6 | ||||
| 6 | 7.48 s | 132.9 | 7.49 s | 134.4 | 7.47 s | 134.6 | 7.41 br | 134.2 |
| 7 | 3.74 s | 27.7 | 3.82 s | 30.7 | 3.81 s | 30.7 | 3.07 dd, 13, 6, 2.95 dd, 13, 9 | 37.2 |
| 8 | 150.8 | 151.9 | 151.9 | 3.70 dd, 9, 6 | 63.9 | |||
| 9 | 163.0 | 165.5 | 165.4 | 169.4 | ||||
| 10 | 3.34 m | 49.7 | 3.41 t, 7.1 | 41.5 | 3.42 t, 7.1 | 41.5 | 3.53 ddd, 14, 8, 6, 3.40 dt, 14, 7 | 36.7 |
| 11 | 2.27 t | 33.3 | 2.74 t, 7.1 | 35.2 | 2.74 t, 7.1 | 35.2 | 1.90 m | 30.0 |
| 12 | 139.1 | 140.3 | 140.2 | 3.91, 3.89 m | 71.1 | |||
| 13 | 7.45 s | 132.9 | 7.43 s | 134.6 | 7.43 s | 134.4 | 153.9 | |
| 14 | 117.1 | 118.7 | 118.7 | 118.9 | ||||
| 15 | 150.4 | 152.4 | 152.3 | 7.46 br | 133.6 | |||
| 16 | 117.1 | 118.7 | 118.7 | 134.7 | ||||
| 17 | 7.45 s | 132.9 | 7.43 s | 134.6 | 7.43 s | 134.4 | 7.46 br | 133.6 |
| 18 | 3.97 t | 70.3 | 4.10 t, 5.4 | 70.8 | 4.08 t, 5.8 | 71.6 | 118.9 | |
| 19 | 2.07 m | 27.7 | 2.60 tt, 5.4, 6.9 | 32.5 | 2.20 tt, 5.8, 7.7 | 29.0 | 3.01 m | 30.2 |
| 20 | 3.04 m | 36.5 | 4.99 t, 6.9 | 60.9 | 3.29 t, 7.7 | 38.9 | 3.20 m | 58.9 |
| 21 | 3.97 t | 70.4 | 4.80 t, 5.7 | 71.6 | 4.10 t, 5.6 | 70.9 | ||
| 22 | 2.07 m | 27.7 | 2.19 tt, 5.7, 7.7 | 29.0 | 2.60 tt, 5.6, 7.1 | 32.5 | ||
| 23 | 3.04 m | 36.5 | 3.28 t, 7.7 | 38.9 | 4.98 t, 7.1 | 60.8 | ||
| NOH | 12.05 s | |||||||
| 1′,5′ | 9.13 d, 5.8 | 146.4 | 9.13 d, 5.8 | 146.4 | ||||
| 2′,4′ | 8.14 dd, 5.8, 7.7 | 129.5 | 8.13 dd, 5.8, 7.7 | 129.5 | ||||
| 3′ | 8.60 t, 7.7 | 147.1 | 8.59 t, 7.7 | 147.1 | ||||
| NH | 8.12 t | |||||||
| NH2 | 7.86 br | |||||||
| NMe | 2.82 s | 43.4 | ||||||
| NMe2 | 2.52 s | 33.1 | ||||||
| OMe | 3.77 s | 60.9 | ||||||
| 196 (125) |
197 (126) |
198 (126) |
199 (90) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD + CDCl3 | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | ||
| 1 | 137.7 | 129.8 | 129.4 | 136.5 | ||||
| 2 | 7.31 s | 133.2 | 7.47 d, 1.9 | 136.1 | 7.44 d, 1.1 | 136.1 | 7.49 s | 132.9 |
| 3 | 117.6 | 113.6 | 113.6 | 117.2 | ||||
| 4 | 152.3 | 157.9 | 157.9 | 150.5 | ||||
| 5 | 117.6 | 7.04 d, 8.5 | 114.5 | 7.04 d, 8.4 | 114.4 | 117.2 | ||
| 6 | 7.31 s | 133.2 | 7.20 dd, 1.9, 8.5 | 131.7 | 7.18 dd, 1.1, 8.4 | 131.8 | 7.49 s | 132.9 |
| 7 | 2.71 dd, 4.5, 13.8 2.94 dd, 8.8, 13.5 |
31.6 | 3.08 dd, 7.8, 14.0 2.98 dd, 7.3, 14.0 |
38.3 | 3.12 dd, 5.7, 13.6 3.05 dd, 8.6, 13.6 |
37.4 | 3.82 s | 28.1 |
| 8 | 3.14 dd, 4.5, 8.8 | 69.8 | 4.00 t, 7.3 | 56.5 | 3.90 m | 64.9 | 151.0 | |
| 9 | 170.8 | 170.3 | 168.9 | 165.7 | ||||
| 10 | 3.29 dt, 2.8, 7.0 | 39.8 | 3.56 m 3.32 dd, 6.6, 13.2 |
42.3 | 3.47 m 3.38 m |
42.1 | 3.27 m | 38.9 |
| 11 | 2.57, 2.54 m | 34.2 | 2.74 t, 7.2 | 35.8 | 2.73, 2.63 m | 35.8 | 1.56 m | 27.1 |
| 12 | 137.9 | 140.9 | 140.8 | 1.56 m | 27.2 | |||
| 13 | 7.23 s | 132.8 | 7.51 s | 135.1 | 7.47 s | 135.1 | 3.17 m | 41.8 |
| 14 | 117.7 | 119.7 | 119.7 | 156.8 | ||||
| 15 | 150.9 | 153.2 | 153.2 | 4.09 t, 5.5 | 70.7 | |||
| 16 | 117.7 | 119.7 | 119.7 | 1.26 m | 33.6 | |||
| 17 | 7.23 s | 132.8 | 7.51 s | 135.1 | 7.47 s | 135.1 | 3.25 m | 39.5 |
| 18 | 3.96 t, 5.5 | 69.7 | 4.13 t, 5.6 | 71.9 | 4.12 t, 5.5 | 71.9 | ||
| 19 | 2.18 m | 25.4 | 2.32 tt, 5.6, 7.9 | 27.2 | 2.32 tt, 5.5, 7.5 | 27.2 | ||
| 20 | 3.16 m | 55.4 | 3.54 t, 7.9 | 57.8 | 3.54 t, 7.4 | 57.6 | ||
| 21 | 3.74 s | 60.4 | ||||||
| 22,23 | 2.26 s | 41.5 | ||||||
| 24,25 | 2.67 s | 42.9 | ||||||
| NH | 8.67 brt | |||||||
| OMe | 3.91 s | 57.6 | 3.90 s | 57.6 | ||||
| NMe2 | 3.00 s | 44.5 | 3.00 s | 44.4 | ||||
| NMe | 2.62 s | 33.3 | ||||||
| 200 (79) |
201 (127) |
202 (127) |
203 (127) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 123.7 | 133.7 | 139.1 | 130.9 | ||||
| 2 | 155.2 | 7.63 d, 2.1 | 139.8 | 7.65 s | 140.5 | 7.61 d, 2.1 | 140.0 | |
| 3 | 108.2 | 85.7 | 90.3 | 86.1 | ||||
| 4 | 153.6 | 156.4 | 157.2 | 157.2 | ||||
| 5 | 108.2 | 6.72 d, 8.3 | 110.7 | 90.3 | 6.76 d, 8.3 | 111.0 | ||
| 6 | 7.31 s | 135.4 | 7.22 dd, 2.1, 8.3 | 130.4 | 7.65 s | 140.5 | 7.14 dd, 2.1, 8.3 | 130.3 |
| 7 | 3.70 | 26.7 | 2.78 dd, 4.9, 13.5 3.05 dd, 7.5, 13.5 |
31.5 | 2.73 dd, 4.6, 13.8 3.02 dd, 7.9, 13.8 |
31.0 | 2.63 dd, 8.7, 13.5 3.05 dd, 4.3, 13.5 |
37.1 |
| 8 | 152.6 | 3.13 dd, 4.9, 7.5 | 70.9 | 3.14 dd, 4.6, 7.9 | 69.8 | 3.13, dd, 8.7, 4.3 | 65.1 | |
| 9 | 167.8 | 171.5 | 170.3 | 171.6 | ||||
| 10 | 3.18 m | 40.7 | 3.18 q, 6.6 | 38.8 | 3.20 q, 6.6 | 39.0 | 3.22 quin, 7.2 | 38.7 |
| 11 | 1.44 m | 29.0 | 1.44 m | 29.2 | 1.45 m | 29.0 | 1.46 m | 28.9 |
| 12 | 1.42 m | 28.4 | 1.22 m | 24.1 | 1.22 quin, 7.2 | 24.1 | 1.24 m | 24.0 |
| 13 | 3.09 m | 42.9 | 1.44 m | 29.2 | 1.45 m | 29.0 | 1.46 m | 28.9 |
| 14 | 156.4 | 3.18 q, 6.6 | 38.9 | 3.20 q, 6.6 | 39.0 | 3.22 quin, 7.2 | 38.9 | |
| 15 | 171.2 | 170.3 | 170.2 | |||||
| 16 | 3.14 dd, 4.6, 7.8 | 70.4 | 3.14 dd, 4.6, 7.9 | 69.8 | 3.14 dd, 4.4, 7.9 | 69.6 | ||
| 17 | 2.72 dd, 4.6, 13.7 3.01 dd, 7.8, 13.7 |
30.5 | 2.73 dd, 4.6, 13.8 3.02 dd, 7.9, 13.8 |
31.0 | 2.73 dd, 4.4, 13.8 3.02 dd, 7.9, 13.8 |
31.0 | ||
| 18 | 139.9 | 139.1 | 139.0 | |||||
| 19,23 | 7.65 s | 140.4 | 7.65 s | 140.5 | 7.65 s | 140.5 | ||
| 20,22 | 90.1 | 90.3 | 90.2 | |||||
| 21 | 157.0 | 157.2 | 157.3 | |||||
| 4-OMe | 3.74 | 61.7 | 3.81 s | 56.3 | 3.82 s | 60.7 | 3.81 s | 56.4 |
| 21-OMe | 3.84 s | 60.6 | 3.82 s | 60.7 | 3.86 s | 60.7 | ||
| 8-NMe | 2.33 s | 42.1 | 2.30 s | 41.9 | 2.28 s | 34.6 | ||
| 18-NMe | 2.30 s | 42.2 | 2.30 s | 41.9 | 2.30 s | 41.8 | ||
| NH2 | 7.1–7.3 br | |||||||
| 9-NH 13-NH | 7.56 t, 5.9, 7.43 br | |||||||
| OH | 10.68, 10.12 br | |||||||
| 204 (129) |
205 (129) |
206 (129) |
|
|---|---|---|---|
| 1H | 1H | 1H | |
| acetone-d6 | acetone-d6 | acetone-d6 | |
| 1 | 3.65 sH | 3.54 sG | 3.72 s |
| 4 | 7.96 tF | 7.89 t, 6E | 8.00 t, 7 |
| 5 | 3.32 qE | 3.25 dt, 6, 7D | 3.29 q, 7 |
| 6 | 2.62 tD | 2.67 t, 7C | 2.62 t, 7 |
| 8 | 7.29 d, 1.9A | 7.28 d, 1.9A | 7.32 d, 1.9A |
| 11 | 6.86 d, 8.2B | 6.85 d, 8 | 6.85 d, 8.2 |
| 12 | 6.96 dd, 1.9, 8.2C | 6.96 dd, 1.9, 8B | 6.98 dd, 1.9, 8.2 |
| 16 | 6.87 d, 8.2B | 6.85 d, 8 | 6.85 d, 8.2 |
| 17 | 6.99 dd, 1.9, 8.2C | 6.99 dd, 1.9, 8B | 6.98 dd, 1.9, 8.2 |
| 19 | 7.31 d, 1.9A | 7.30 d, 1.9A | 7.32 d, 1.9A |
| 20 | 2.65 tD | 2.60 t, 7C | 2.62 t, 7 |
| 21 | 3.27 qE | 3.33 dt, 6, 7D | 3.29 q, 7 |
| 22 | 8.01 tF | 8.11 t, 6E | 8.00 t, 7 |
| 25 | 3.76 sH | 3.83 sG | 3.72 s |
| 27 | 7.51 d, 1.7 | 7.57 s | 7.29 d, 1.9A |
| 30 | 6.75 d, 8.5 | ||
| 31 | 7.15 dd, 1.7, 8.5 | 7.57 s | 6.93 d, 1.9 |
| 36 | 7.13 d, 1.7 | 6.97 d, 1.9 | 7.29 d, 1.9A |
| 38 | 6.64 d, 1.7 | 6.21 d, 1.9 | 6.93 d, 1.9 |
| OH-15 | 10.02 s | 10.02s | 10.02 s |
| OH-10 | 10.02 s | 10.02s | 10.02 s |
| OH-29 | 8.94 s | ||
| OH-34 | 9.85 s | 10.02s | 8.94 s |
| NOH-24 | 11.94 sG | 11.75 sF | 11.84s |
| NOH-2 | 11.83 sG | 12.08 sF | 11.84s |
| 208 (129) |
208 Tetramethyl ether (129) |
210 (129) |
211 (129) |
|
|---|---|---|---|---|
| 1H | 1H | 1H | 1H | |
| acetone-d6 | acetone-d6 | acetone-d6 | acetone-d6 | |
| 1 | 3.52 sC | signals were not assigned | 3.57 sE | 3.50 sD |
| 4 | 10.24 d, 10.2 | 8.00 t, 6C | 10.29 d, 10 | |
| 5 | 7.31 dd, 10.2, 14.5 | 7.50 2H s | 3.30 m | 7.34 dd, 10, 15 |
| 6 | 6.44 d, 14.5 | 7.45 d, 2 | 2.72 m | 6.42 d, 15 |
| 8 | 7.63 d, 1.9 | 7.16 d, 2 | 7.65 sF | 7.67 d, 1.9 |
| 11 | 6.92 d, 8.5 | 7.10 d, 2 | 6.99 d, 8.8 | |
| 12 | 7.48 dd, 1.9, 8.5 | 6.80 d, 2 | 7.65 sF | 7.45 dd, 1.9, 8.8 |
| 171 | 6.55 d, 1.9 | 6.24 d, 2 | 6.23 d, 1.7BA | 6.43 d, 2.0AB |
| 191 | 7.13 d, 1.9A | Other aromatic signals are overlapped. | 7.08 d, 1.7AB | 7.20 d, 2.0AB |
| 20 | 2.73 m | 2.72 m | 2.64 t, 6 | |
| 21 | 3.35 m | 3.30 m | 3.24 q, 6 | |
| 22 | 7.85 t, 6 | 4.02, 3.98, 3.94, 3.52, 3H each, s, OCH3 | 8.13 t, 6C | 7.85, 6 |
| 25 | 3.64 sC | 3.66 sE | 3.71 sD | |
| 27 | 7.51 s | 7.64 sF | 7.44 d, 1.9 | |
| 30 | 6.66 d, 8.3 | |||
| 31 | 7.51 s | 7.64 sF | 7.10 dd, 1.9, 8.3 | |
| 36 | 7.08 d, 1.9A | 7.04 d, 1.9AB | 7.09 d, 1.5BA | |
| 38 | 6.09 d, 1.9 | 6.16 d, 1.9BA | 6.40 d, 1.5BA | |
| OH | 9.85 sB | 10.00 s | 9.89 s | |
| OH-34 | 10.10 sB | 10.00 s | 9.89 s | |
| NOH | 12.00 s | 11.92 sD | 12.09 sC | |
| NOH-2 | 12.00 s | 11.71 sD | 11.99 sC |
| 228 (130) |
212 (132) |
212 Tetramethyl ether (132) |
213 (132) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | CDCl3 | NA | DMSO-d6 | DMSO-d6 | |
| 1 | 3.74 s | 27.8 | 3.57 br | 27.3 | 3.79 br | 3.54 br | 27.6 | |
| 2 | 151.2e | 150.9 | 151.4 | |||||
| 3 | 161.2 | 162.8 | 162.9 | |||||
| 4 | 10.26 d, 10 | |||||||
| 5 | 7.32 dd, 10, 14 | 124.1 | 3.40, 3.00 m | 47.3 | 3.72, 3.38 m | 3.18 q | 40.9 | |
| 6 | 6.40 d, 14 | 111.4 | 4.63 m | 70.4 | 4.88 m | 2.62 t | 34.0 | |
| 7 | 134.6 | 140.6 | 135.4 | |||||
| 8 | 7.62 d, 2 | 129.6 | 7.69 d, 2.0 | 130.2 | 7.69 d, 2.0 | 7.49 d, 2.1 | 132.8 | |
| 9 | 114.9 | 112.0 | 111.8 | |||||
| 10 | 151.3e | 151.8 | 152.4 | |||||
| 11 | 7.01 d, 8.5 | 122.7 | 6.94 d, 8.4 | 119.5 | 6.97 d, 8.0 | 6.75 d, 8.4 | 120.3 | |
| 12 | 7.42 dd, 8.5, 2 | 125.6 | 7.27 dd, 8.4, 2.0 | 126.5 | 7.28 dd, 8.0, 2.0 | 7.10 dd, 8.4, 2.1 | 125.7 | |
| 14 | 151.4 | 144.7 | 148.0 | |||||
| 15 | 139.6 | 143.8 | 143.0 | |||||
| 16 | 118.8 | 110.6 | 6.85 d, 8.1 | 116.5 | ||||
| 17 | 7.12 d, 2 | 127.2 | 7.13 d, 2.0 | 128.1 | 7.14 d, 2.0 | 6.81 dd, 8.1, 1.8 | 128.9 | |
| 18 | 137.1 | 131.4 | 130.6 | |||||
| 19 | 6.39 d, 2 | 116.6 | 6.60 d, 2.0 | 118.0 | 6.65 d, 2.0 | 6.73 d, 1.8 | 118.0 | |
| 20 | 2.71 t, 6 | 33.0 | 2.68 t, 6.0 | 33.4 | 2.75 m | 2.67 t, 5.7 | 34.0 | |
| 21 | 3.27 m | 38.7 | 3.40 q | 39.2 | 3.46, 3.55 | 3.41 q | 39.4 | |
| 22 | 7.85 t, 6.0 | 6.76 t, 6.0 | ||||||
| 23 | 163.2 | 163.2 | 163.0 | |||||
| 24 | 151.0 | 150.4 | 150.4 | |||||
| 25 | 3.43 s | 29.0 | 3.62 br | 28.7 | 3.78 br | 3.52 br | 28.8 | |
| 26 | 133.9 | 137.6 | 137.7 | |||||
| 27 | 7.38 d, 2 | 132.6 | 7.56 s | 133.5 | 7.52 s | 7.59 s | 133.1 | |
| 28 | 112.8 | 117.1 | 117.2 | |||||
| 29 | 151.5 | 145.9 | 145.9 | |||||
| 30 | 6.70 d, 8.5 | 120.2 | 117.1 | 117.2 | ||||
| 31 | 7.02 dd, 2, 8.5 | 129.8 | 7.56 s | 133.5 | 7.52 s | 7.59 s | 133.1 | |
| 33 | 151.4 | 144.7 | 144.8 | |||||
| 34 | 140.5 | 141.9 | 141.9 | |||||
| 35 | 119.0 | 109.7 | 109.8 | |||||
| 36 | 7.23 d, 2 | 128.1 | 7.07 d, 2.0 | 126.8 | 7.16 d, 2.0 | 7.05 d, 1.8 | 126.8 | |
| 37 | 133.6 | 127.9 | 128.1 | |||||
| 38 | 6.40 d, 2 | 117.9 | 6.24 d, 2.0 | 112.9 | 6.28 d, 2.0 | 6.22 d, 1.8 | 112.8 | |
| OMe | 4.02 s, 4.01 s, 3.97 s, 3.70 s | |||||||
| OH | 5.62 d, 4.2 | 6.87 t, 6.0 | ||||||
| NH-4 | 7.77 t, 6.0 | 7.97 t, 6.0 | ||||||
| NH | 8.00 t, 6.0 | 8.03 t, 5.7 | ||||||
| NOH | 11.94, 12.13 s | 11.83 br, 12.0 br | 11.73, 11.96 br | |||||
| ArOH | 9.8 br | 10.02 br | ||||||
| 213 Tetramethyl ether(132) |
214 (132) |
215 (132) |
215 Tetramethyl ether(132) |
|||
|---|---|---|---|---|---|---|
| 1H | 1H (132) | 13C (143) | 1H | 13C | 1H | |
| CDCl3 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | CDCl3 | |
| 1 | 3.67 br | 3.61 br | 27.6 | 3.64 br | 27.3 | |
| 2 | 151.2 | 151.3 | ||||
| 3 | 162.9 | 161.4 | ||||
| 4 | ||||||
| 5 | 3.30 q | 47.0 | 7.25 dd, 14.6, 10.3 | 123.6 | 7.34 dda | |
| 6 | 4.62 m | 70.2 | 6.39 d, 14.6 | 110.9 | 6.17 d, 14.7 | |
| 7 | 141.0 | 133.3 | ||||
| 8 | 7.44 d, 2.0 | 7.64 d, 1.7 | 130.6 | 7.52 d, sm | 130.5 | 7.54 d, 2.7 |
| 9 | 113.3 | 112.5 | ||||
| 10 | 151.7 | 152.3 | ||||
| 11 | 6.85 d, 8.0 | 6.91 d, 8.5 | 120.0 | 6.73 d, 8.7 | 119.9 | 6.90 d, 8.4 |
| 12 | 7.03 dd, 8.3, 2.0 | 7.21 dd, 8.0, 2.0 | 126.8 | 7.43 dd, 8.7, 2.1 | 124.1 | 7.38 dda |
| 14 | 145.0 | 146.7 | ||||
| 15 | 143.4 | 143.0 | ||||
| 16 | 6.97 d, 8.3 | 110.7 | 6.87 d, 8.0 | 116.7 | 6.96 d, 8.4 | |
| 17 | 6.91 dd, 8.3, 2.0 | 7.12 d, 2.1 | 127.7 | 6.82 dd, 8.0, sm | 125.8 | 6.90 dda |
| 18 | 131.7 | 130.6 | ||||
| 19 | 6.66 d, 2.0 | 6.45 d, 1.8 | 117.4 | 6.72 d, sm | 119.1 | 6.71 d, 2.1 |
| 20 | 2.61 t, 6.0 | 33.5 | 2.70 bt | 33.5 | ||
| 21 | 3.38 q | 40.6 | 3.43 q | 38.5 | ||
| 23 | 163.1 | 162.7 | ||||
| 24 | 150.8 | 150.1 | ||||
| 25 | 3.86 br | 3.61 br | 28.5 | 3.53 br | 28.9 | |
| 26 | 134.5 | 137.6 | ||||
| 27 | 7.53 s | 7.49 d, 2.0 | 133.5 | 7.51 s | 133.5 | 7.46 s |
| 28 | 113.3 | 116.9 | ||||
| 29 | 151.2 | 145.9 | ||||
| 30 | 6.78 d, 8.5 | 119.9 | 116.9 | |||
| 31 | 7.53 s | 7.07 dd, 8.5, sm | 129.6 | 7.51 s | 133.5 | 7.46 s |
| 33 | 145.0 | 144.8 | ||||
| 34 | 143.4 | 141.9 | ||||
| 35 | 110.5 | 110.1 | ||||
| 36 | 7.19 d, 2.0 | 7.15 d, 1.9 | 126.8 | 7.07 d, 1.9 | 126.8 | 7.12 d, 2.0 |
| 37 | 127.6 | |||||
| 38 | 6.27 d, 2.0 | 6.47 d, 1.9 | 117.4 | 6.14 d, 1.9 | 112.4 | 6.25 d, 2.0 |
| OMe | 3.90, 3.49, 4.05, 4.00, Each s | 4.01 s; 3.93 s; 3.92 s | ||||
| OH | 5.64 d, 4.4 | |||||
| NH-4 | 6.79 t, 6.0 | 7.78 t, 6.0 | 10.2 d, 10.3 | 8.30 d, 11.4 | ||
| NH-22 | 6.61 t, 6.0 | 7.94 t, 5.8 | 7.78 t, 6.0 | 6.58 t, 5.4 | ||
| NOH | 11.87 br, 11.86 br | 11.98 br, 11.97 br | ||||
| ArOH | 9.78 br | 10.04 br, 9.38 br | ||||
| CH2 | 3.53 q, 6.3; 3.46 q; 2.75 t; 2.73 t | 3.73 br; 3.65 br; 3.58 q, 5.4; 2.82 t, 5.4 | ||||
| 216 (142) |
216 Tetramethyl ether (142) |
227 (130) |
217 (131) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 1H | 1H | 1H | 13C | 1H | 13C | |
| CDCl3 | CDCl3 | CDCl3/C6D6 1:2 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | |
| 1 | 3.72 d, 12.9 3.78 d, 12.9 |
3.80 s | 3.73 s | 3.69 s | 27.8 | 3.40 s | 27.8 |
| 2 | 152.0 | 151.2 | |||||
| 3 | 163.1 | 162.8 | |||||
| 4 | 7.08 t, 6.1b | 6.73 t, 6.1b | 6.26 t, 6.1b | 7.89 t, 5.5 | |||
| 5 | 3.41 m 3.63 m | 3.38 m | 2.94 m 3.32 m | 3.30 m | 39.9 | 3.20 dt, 6.6, 6.6 | 40.5 |
| 6 | 4.80 t, 6.1 | 4.86 dd, 3.2, 7.5 | 4.37 brd, 7.3 | 2.68 t, 7 | 33.7 | 2.54 t, 6.6 | 34.0 |
| 7 | 135.4 | 130.8 | |||||
| 8 | 7.58 br | 7.65 br | 7.30–7.60 br | 7.36 d, 2 | 132.3 | 6.53 d, 1.8 | 120.4 |
| 9 | 113.8 | 143.0 | |||||
| 10 | 148.9 | 146.9 | |||||
| 11 | 7.47 d, 8.5 | 121.2 | 6.86 d, 8.1 | 117.2 | |||
| 12 | 7.58 br | 7.65 br | 7.30–7.60 br | 7.09 dd, 2, 8.5 | 128.6 | 6.81 dd, 1.8, 8.1 | 125.2 |
| 14 | 109.0 | 152.4 | |||||
| 15 | 152.1 | 111.9 | |||||
| 16 | 6.76 d, 8 | 116.2 | 7.48 d, 1.8 | 133.3 | |||
| 17 | 6.13 d, 1.8c | 7.08 d, 1.9c | 6.86 d, 1.9c | 6.88 dd, 2, 8 | 128.7 | 135.4 | |
| 18 | 131.4 | 7.02 dd, 1.8, 8.4 | 129.4 | ||||
| 19 | 7.03 d, 1.8c | 6.15 d, 1.9c | 6.30 d, 1.9c | 7.25 d, 2 | 132.5 | 6.58 d, 8.4 | 117.7 |
| 20 | 2.66 m | 2.69 m | 2.19 m | 2.62 t, 7 | 33.5 | 2.68 t, 6.6 | 33.9 |
| 21 | 3.36 m | 3.38 m | 3.10, 2.94 m | 3.30 m | 39.8 | 3.37 obscured | 39.9 |
| 22 | 7.14 t, 6.1b | 7.07 t, 6.1b | 6.70 t, 6.0b | 7.87 t, 5.5 | |||
| 23 | 163.0 | 163.3 | |||||
| 24 | 152.0 | 151.3 | |||||
| 25 | 3.89 d, 13.0 3.81 d, 13.0 |
3.84 d, 13.0 3.78 d, 13.0 |
3.69 s | 3.69 s | 27.8 | 3.70 s | 28.5 |
| 26 | 131.4 | 138.0 | |||||
| 27 | 7.52 d, 1.8 | 7.52 d, 2.0 | 7.60 d, 2.0 | 7.22 br | 132.3 | 7.46 s | 133.2 |
| 28 | 112.4 | 117.3 | |||||
| 29 | 152.2 | 146.2 | |||||
| 30 | 6.78 d, 8.4 | 6.65 d, 8.4 | 6.64 d, 8.4 | 128.2 | 117.3 | ||
| 31 | 7.14 dd, 2.0, 8.4 | 7.14 dd, 2.0, 8.4 | 7.13 dd, 2.0, 8.4 | 6.96 d, 2 | 130.7 | 7.46 s | 133.2 |
| 33 | 128.2 | 144.6 | |||||
| 34 | 152.2 | 141.9 | |||||
| 35 | 112.3 | 110.1 | |||||
| 36 | 7.26 d, 1.8a | 7.29 d, 2.0a | 7.28 d, 2.0a | 7.22 br | 132.3 | 6.93 d, 2.0 | 126.2 |
| 37 | 131.4 | 128.4 | |||||
| 38 | 6.64 d, 1.8a | 6.70 d, 2.0a | 6.78 d, 2.0a | 6.97 d, 2 | 130.7 | 6.02 d, 2.0 | 113.2 |
| OMe-10 | 4.05 s 4.01 s 3.89 s 3.87 s |
4.00 s 3.80 s 3.65 s 3.58 s |
|||||
| OH | 3.21 br | 2.70 br | 10.00 s | ||||
| OH-10 | 9.39 s | ||||||
| OH-24 | 8.90 br | ||||||
| OH-34 | 8.90 br | 9.97 s | |||||
| NH-4 | 7.82 brt, 6.6 | ||||||
| NH-22 | 8.16 brt, 6.6 | ||||||
| NOH-24 | 11.75 s | 11.93 s | |||||
| NOH-2 | 11.69s | 11.66 s | |||||
| ArOH | 10.73 br | ||||||
| OMe-34 | 11.27 br | ||||||
| 217 Methyl ether(131) |
226 (141) |
218 (133) |
218a (133) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CDCl3 | DMSO-d6 | DMSO-d6 | CDCl3 | |||||
| 1 | 3.48 s | 29.0 | 3.41 | 27.9 | 3.70 s | 27.5 | 3.85 br | 28.3 |
| 2 | 150.7 | 150.7 | 151.5 | 150.9 | ||||
| 3 | 161.9 | 162.6 | 161.4 | 159.6 | ||||
| 4 | 6.58 brt, 6.1 | 7.72 t, 5.8 | 10.31 d, 10.2 | 8.44 d, 8.4 | ||||
| 5 | 3.47 obscured | 40.8 | 3.22 dt, 5.8, 7.0 | 40.3 | 7.45 dd, 14.3, 10.2 | 126.0 | 7.46 dd, 14.6, 8.4 | 124.8 |
| 6 | 2.71 brt, 6.0 | 34.5 | 2.54 t, 7.0 | 33.8 | 6.41 d, 14.3 | 110.0 | 6.11 d, 14.6 | 110.0 |
| 7 | 132.1 | 130.7 | 137.6 | 136.9 | ||||
| 8 | 6.68 d, 1.5 | 121.2 | 6.50 d, 2. | 120.1 | 7.77 s | 129.5 | 7.57 s | 129.8 |
| 9 | 144.3 | 143.0 | 118.1 | 118.5 | ||||
| 10 | 149.5 | 146.7 | 145.6 | 146.4 | ||||
| 11 | 6.96 m | 113.6 | 6.85 d, 8.5 | 117.0 | 118.1 | 118.5 | ||
| 12 | 6.96 m | 125.2 | 6.81 dd, 2.0, 8.5 | 125.0 | 7.77 s | 129.5 | 7.57 s | 129.8 |
| 14 | 153.1 | 152.3 | 145.0 | 150.4 | ||||
| 15 | 112.9 | 111.9 | 141.6 | 144.3 | ||||
| 16 | 7.45 d, 2.2 | 134.4 | 7.47 d, 2.0 | 133.1 | 110.1 | 118.1 | ||
| 17 | 133.7 | 135.4 | 7.00 d, 1.5 | 126.4 | 7.03 d, 2.0 | 126.7 | ||
| 18 | 6.94 dd, 2.2, 8.4 | 128.6 | 7.00 dd, 2.0, 8.3 | 129.3 | 130.8 | 135.5 | ||
| 19 | 6.64 d, 8.4 | 117.8 | 6.57 d, 8.3 | 117.9 | 6.18 d, 1.5 | 111.7 | 6.15 d, 2.0 | 112.5 |
| 20 | 2.78 brt, 6.0 | 34.5 | 2.66 t, 6.5 | 33.9 | 2.67 t, 6.5 | 32.8 | 2.72 t, 6.5 | 34.5 |
| 21 | 3.55 dt, 5.8, 6.0 | 40.5 | 3.37 dt, 6.5, 6.3 | 39.8 | 3.19 q, 6.3 | 38.3 | 3.36 bq, 6.5 | 39.4 |
| 22 | 6.76 brt, 5.8 | 8.10 t, 6.3 | 7.80 t, 5.7 | 6.71 t, 6.5 | ||||
| 23 | 162.0 | 163.1 | 162.9 | 162.3 | ||||
| 24 | 150.8 | 151.3 | 150.9 | 150.4 | ||||
| 25 | 3.89 s | 28.7 | 3.69 s | 28.4 | 3.59 s | 28.4 | 3.75 br | 29.2 |
| 26 | 137.4 | 134.4 | 132.5 | |||||
| 27 | 7.51 s | 136.6 | 7.42 s | 133.1 | 7.46 br | 133.7 | 7.47 d, 2.2 | 134.1 |
| 28 | 118.1 | 117.2 | 112.8 | 112.3 | ||||
| 29 | 146.8 | 146.6 | 150.9 | 152.1 | ||||
| 30 | 118.1 | 117.2 | 6.64 d, 8.3 | 119.1 | 6.47 d, 8.4 | 117.1 | ||
| 31 | 7.51 s | 136.6 | 7.42 | 113.1 | 7.14 dd, 8.3, 1.9 | 130.3 | 7.12 dd, 8.4, 2.2 | 129.9 |
| 33 | 149.6 | 150.3 | 144.8 | 148.9 | ||||
| 34 | 144.2 | 138.6 | 143.8 | 147.5 | ||||
| 35 | 117.9 | 119.1 | 110.9 | 117.9 | ||||
| 36 | 7.16 d, 2.0 | 127.3 | 6.93 s | 125.9 | 7.20 d, 1.9 | 126.0 | 7.32 d, 2.2 | 129.9 |
| 37 | 133.0 | 133.6 | 128.3 | 133.0 | ||||
| 38 | 6.09 d, 2.0 | 114.7 | 5.98 s | 113.5 | 6.40 br | 117.3 | 6.86 d, 2.0 | 121.3 |
| OMe-10 | 3.81 s | 56.3 | ||||||
| C=NOH | 3.48 s | 62.4 | 11.68, 11.88 | |||||
| OMe-34 | 4.01 s | 63.3 | 3.83 s | 61.1 | ||||
| NOMe-2 | 4.04 s | 61.0 | 4.05 s | 61.0 | ||||
| NOMe-24 | 4.10 s | 63.7 | ||||||
| OMe-15 | 4.03 s | 63.1 | ||||||
| 10-OH | 9.35 | |||||||
| 219 (134) |
220 (135) |
221 (135) |
222 (138) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 13C | 1H | 13C | |
| CDCl3 + 1 drop DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | CD3OD | CD3OD | |
| 1 | 3.71 s | 3.55 s | 27.4 | 3.52 s | 27.4 | 3.67 s | 28.7 |
| 2 | 11.65 s | 151.4 | 11.77 s | 151.0 | 153.3 | ||
| 3 | 163.0 | 162.9 | 166.0 | ||||
| 4 | 6.76 t, 6.2 | 8.06 t, 6.0 | 7.77 t, 6.3 | ||||
| 5 | 3.51 m | 3.28 m | 40.4 | 2.92, 3.38 m | 47.7 | 3.30 m | 41.5 |
| 6 | 2.82 m | 2.71 t, 6.6 | 33.9 | 4.58 m, 5.53 d, 4.4 | 70.5 | 2.55 t, 7 | 35.4 |
| 7 | 139.8 | 139.3 | 138.6 | ||||
| 8 | 7.38 s | 7.59 s | 133.2 | 7.62 d, 1.9 | 129.8 | 7.39 d, 2.0 | 135.1 |
| 9 | 117.7 | 111.4 | 115.6 | ||||
| 10 | 146.6 | 153.1 | 152.4 | ||||
| 11 | 117.7 | 6.73 d, 8.5 | 117.4 | 6.82 d, 8.3 | 122.2 | ||
| 12 | 7.38 s | 7.59 s | 133.2 | 7.13 dd, 8.5, 1.9 | 126.2 | 7.06 dd, 8.3, 2.1 | 130.9 |
| 14 | 144.1 | 142.5 | 145.3 | ||||
| 15 | 144.3 | 146.9 | 146.6 | ||||
| 16 | 6.81 d, 8.2 | 116.6 | 6.83 d, 8.2 | 116.5 | 111.6 | ||
| 17 | 7.04 d, 1.9 | 6.67 dd, 8.2. 1.8 | 122.9 | 6.81 dd, 8.2, 1.9 | 126.1 | 7.04 d, 2.0 | 128.5 |
| 18 | 129.5 | 130.6 | 132.7 | ||||
| 19 | 6.14 d, 1.9 | 6.17 d, 1.8 | 112.5 | 6.76 d, 1.9 | 112.5 | 6.35 d, 2.0 | 118.0 |
| 20 | 2.67 m | 2.67, 6.6 | 33.3 | 2.65 t, 6.3 | 34.1 | 2.66 t, 6.5 | 35.5 |
| 21 | 3.34 m | 3.29 m | 38.8 | 3.40 m | 39.2 | 3.34 m, 7 | 41.7 |
| 22 | 7.16 t, 6.4 | 7.94 t, 6.0 | 7.95 t, 6.3 | ||||
| 23 | 163.3 | 163.0 | 166.6 | ||||
| 24 | 11.86 s | 150.7 | 11.91 s | 150.4 | 152.6 | ||
| 25 | 3.88 s | 3.68 s | 28.7 | 3.51 d, 12.9 3.56 d, 12.9 |
28.7 | 3.72 s | 29.3 |
| 26 | 137.8 | 137.6 | 135.8 | ||||
| 27 | 7.49 d, 2.0 | 7.63 s | 133.7 | 7.58 s | 133.6 | 7.42 d, 2.0 | 135.1 |
| 28 | 117.2 | 117.2 | 115.2 | ||||
| 29 | 146.1 | 146.0 | 153.0 | ||||
| 30 | 6.87 d, 8.3 | 117.2 | 117.2 | 6.67 d, 8.4 | 121.2 | ||
| 31 | 7.14 dd, 2.0, 8.3 | 7.63 s | 133.7 | 7.58 s | 133.6 | 6.94 dd, 8.5, 2.1 | 130.3 |
| 33 | 144.8 | 144.6 | 145.1 | ||||
| 34 | 142.0 | 141.8 | 147.0 | ||||
| 35 | 109.8 | 109.7 | 111.7 | ||||
| 36 | 7.22 d, 2.0 | 7.07 d, 1.6 | 127.0 | 7.03 d, 1.9 | 126.8 | 7.13 d, 1.9 | 129.5 |
| 37 | 128.2 | 128.0 | 129.7 | ||||
| 38 | 6.52 d, 2.0 | 6.15 d, 1.6 | 112.8 | 6.24 d, 1.9 | 113.1 | 6.46 d, 1.9 | 118.6 |
| OH-15 | 9.25 s | 9.36 s | |||||
| OH-34 | 9.99 s | 9.98 s | |||||
| NOHe | 11.48 br | ||||||
| Ar-OH | 10.17 br | ||||||
| 223a (139) |
224 (130) |
225 (140) |
225 Tetramethyl ether(140) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | CDCl3 | CDCl3 | |
| 1 | 3.72 s | 28.3 | 3.63 s | 28.1 | 3.58 br | 27.8 | 3.62 br | 29.1 |
| 2 | 150.9 | 151.3 | 151.4 | 151.0 | ||||
| 3 | 163.0 | 162.9 | 162.8 | 162.0 | ||||
| 4 | 7.78 t, 6.8 | 6.75 t, 6 | 7.92 brt | 6.62 brt | ||||
| 5 | 3.36 m | 39.6 | 3.39 m | 40.4 | 3.31 m | 39.7 | 3.47 q, 6 | 40.9 |
| 6 | 2.68 t, 6.8 | 34.0 | 2.62 t, 6.5 | 34.4 | 2.61 m | 33.9 | 2.72 6 | 34.6 |
| 7 | 136.8 | 131.6 | 130.6 | 132.0 | ||||
| 8 | 6.76 d, 1.9 | 133.3 | 7.06 d, 2 | 127.5 | 6.62 d, 2 | 120.0 | 6.69 d, 2 | 121.1 |
| 9 | 113.2 | 110.3 | 142.9 | 145.0 | ||||
| 10 | 151.2 | 144.7 | 146.7 | 149.5 | ||||
| 11 | 119.9 | 143.2 | 6.96 d, 8 | 116.9 | 6.93 d, 8 | 125.0 | ||
| 12 | 7.09 dd, 8.3, 1.9 | 129.5 | 6.44 d, 2 | 117.2 | 6.89 dd, 8, 2 | 124.9 | 6.93 dd, 2, 8 | 113.5 |
| 14 | 145.1 | 151.4 | 152.2 | 153.2 | ||||
| 15 | 143.4 | 114.1 | 111.9 | 113.0 | ||||
| 16 | 7.41 d, 2 | 133.7 | 7.58 d, 2 | 133.1 | 7.43 d, 2 | 133.6 | ||
| 17 | 7.10 d, 1.9 | 127.4 | 136.8 | 135.5 | 135.0 | |||
| 18 | 131.8 | 7.06 dd, 2, 8 | 129.3 | 7.10 dd, 8, 2 | 129.2 | 6.94 dd, 2, 8 | 128.8 | |
| 19 | 7.50 d, 1.9 | 117.4 | 6.84 d, 8 | 120.4 | 6.69 d, 8 | 117.8 | 6.66 d, 8 | 118.0 |
| 20 | 2.49 t, 6.8 | 33.2 | 2.77 t, 6.5 | 34.6 | 2.79 m | 33.6 | 2.78 t, 6 | 34.5 |
| 21 | 3.16 m | 40.0 | 3.54 m | 40.2 | 3.50 m | 39.5 | 3.58 q, 6 | 40.5 |
| 22 | 8.19 t, 6.8 | 6.97 t, 6 | 8.09 brt | 6.67 brt | ||||
| 23 | 162.7 | 163.4 | 163.3 | 162.5 | ||||
| 24 | 151.1 | 151.9 | 151.7 | 151.5 | ||||
| 25 | 3.36 s | 27.6 | 3.88 s | 28.3 | 3.79 br | 28.4 | 3.87 br | 28.7 |
| 26 | 128.3 | 134.5 | 134.5 | 135.0 | ||||
| 27 | 6.89 d, 1.9 | 125.8 | 7.54 d, 2 | 134.2 | 7.17 dd, 8, 2 | 129.5 | 7.11 dd, 2, 8 | 129.4 |
| 28 | 110.0 | 114.1 | 6.92 d, 8 | 120.2 | 6.77 8 | 120.9 | ||
| 29 | 141.7 | 151.1 | 150.9 | 151.2 | ||||
| 30 | 144.5 | 6.83 d, 8.5 | 120.2 | 113.3 | 114.2 | |||
| 31 | 5.94 d, 1.9 | 112.9 | 7.13 dd, 2, 8.5 | 129.3c | 7.53 d, 2 | 133.4 | 7.54 d, 2 | 134.5 |
| 32 | ||||||||
| 33 | 117.0 | 142.8 | 145.1 | 153.5 | ||||
| 34 | 146.1 | 144.6 | 143.4 | 146.1 | ||||
| 35 | 117.0 | 109.8 | 110.6 | 114.5 | ||||
| 36 | 7.46 s | 133.0 | 7.20 d, 2 | 128.1 | 7.14 d, 2 | 127.2 | 7.21 d, 2 | 128.3 |
| 37 | 137.7 | 129.3 | 128.9 | 133.2 | ||||
| 38 | 7.46 s | 133.0 | 6.53 d, 2 | 117.8 | 6.44 2 | 117.2 | 6.48 d, 2 | 119.0 |
| OMe-10 | 3.83 s | 56.3 | ||||||
| OH-10 | 11.91, 11.68 s | 6.70 br | 9.41 s | |||||
| OH-24 | 11.87 s | |||||||
| OH-34 | 7.65 br | 9.89 s | ||||||
| NOH-24 | 11.24 sd | |||||||
| NOH | 9.8 br | 10.89 sd | 11.77 s | |||||
| OMe-34 | 3.92 s | 61.0 | ||||||
| NOMe-2 | 3.65 s | 62.8 | ||||||
| NOMe-24 | 3.99 s | 63.0 | ||||||
| 231 (41) |
237 (41) |
232 (41) |
||||||
|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | pyridine-d5 | CD3OD | pyridine-d5 | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 153.9 | 154.1 | 153.8 | 154.1 | ||||
| 2 | 110.7 | 111.0 | 110.7 | 110.9 | ||||
| 3 | 7.29 d, 2.0 | 7.53 s | 134.3 | 133.8 | 7.29 d, 2.0 | 134.2 | 7.30 d, 2.0 | 134.5 |
| 4 | 133.1 | 132.2 | 133.0 | 133.3 | ||||
| 5 | 6.95 dd, 2.0, 8.3 | 7.05 br | 130.1 | 129.6 | 6.95 dd, 2.0, 8.3 | 130.0 | 6.96 dd, 2.0, 8.3 | 130.2 |
| 6 | 6.77 d, 8.3 | 7.05 br | 117.3 | 117.2 | 6.77 d, 8.3 | 117.2 | 6.78 d, 8.3 | 117.5 |
| 7 | 2.67 t | 2.87 t, 6.5 | 35.3 | 35.0 | 2.67 t, 7.2 | 35.3 | 2.68 t, 7.3 | 35.1 |
| 8 | 3.38 t, 7.3 | 3.67 dt, 6.5, 6.5 | 42.0 | 41.5 | 3.40 t, 7.2 | 41.9 | 3.39 t, 7.3 | 42.0 |
| 1′ | 153.8 | 154.2 | 155.9 | 151.0 | ||||
| 2′ | 110.5 | 111.1 | 112.2 | 112.2 | ||||
| 3′ | 7.35 d, 2.0 | 8.06 d, 2.1 | 134.5 | 134.5 | 7.43 d, 2.3 | 134.8 | 7.38 s | 134.2 |
| 4′ | 130.7 | 130.4 | 131.6 | 132.6 | ||||
| 5′ | 7.03 dd, 2.0, 8.3 | 7.55 dd, 2.1, 8.3 | 130.4 | 130.3 | 7.14 dd, 2.3, 8.4 | 130.4 | 7.38 s | 134.2 |
| 6′ | 6.76 d, 8.3 | 7.12 d, 8.3 | 117.1 | 117.2 | 6.89 d, 8.4 | 113.2 | 112.2 | |
| 7′ | 3.77 s | 4.32 s | 28.7 | 28.9 | 3.80 s | 28.7 | 3.77 s | 28.3 |
| 8′ | 153.3 | 153.1 | 153.1 | 152.9 | ||||
| 9′ | 165.8 | 164.6 | 165.7 | 165.9 | ||||
| NH | 8.60 t, 6.5 | |||||||
| OMe-1′ | 3.82 s | 56.7 | ||||||
| 238 (41) |
233 (41) |
233b (41) |
234 (41) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 1H | 1H | 13C | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CDCl3 | CD3OD | pyridine-d5 | CD3OD | CD3OD | |
| 1 | 153.9 | 154.0 | 153.9 | 154.1 | |||||
| 2 | 110.7 | 112.3 | 110.7 | 111.0 | |||||
| 3 | 7.30 d, 2.0 | 134.2 | 7.31 s | 133.9 | 7.42 s | 7.28 d, 1.9 | 7.56 obscured | 134.2 | 133.8 |
| 4 | 133.0 | 135.0 | 133.0 | 132.2 | |||||
| 5 | 6.97 dd, 2.0, 8.3 | 130.0 | 7.31 s | 133.9 | 7.42 s | 6.92 dd, 1.9, 8.3 | 7.08 br | 130.0 | 129.4 |
| 6 | 6.79 d, 8.3 | 117.2 | 112.3 | 6.79 d, 8.3 | 7.08 br | 117.3 | 117.2 | ||
| 7 | 2.68 t, 7.3 | 35.2 | 2.68 t, 7.3 | 34.8 | 2.76 t, 7.0 | 2.57 m | 2.82 m | 35.4 | 35.2 |
| 8 | 3.40 t, 7.3 | 42.0 | 3.39 t, 7.3 | 41.7 | 3.51 dt, 7.0, 7.0 | 3.26 m, 3.38 m | 3.69 m | 41.5 | 40.9 |
| 1′ | 153.8 | 151.0 | 154.0 | 154.1 | |||||
| 2′ | 118.6 | 110.7 | 110.4 | 110.7 | |||||
| 3′ | 7.47 s | 134.5 | 7.36 d, 2.0 | 134.8 | 7.38 d, 2.2 | 7.34 d, 1.9 | 7.82 d, 1.9 | 135.2 | 134.8 |
| 4′ | 137.4 | 130.9 | 131.6 | 131.7 | |||||
| 5′ | 7.47 s | 134.5 | 7.04 dd, 2.0, 8.3 | 130.5 | 7.08 dd, 2.2, 8.4 | 7.02 dd, 1.9, 8.3 | 7.32 dd, 1.9, 8.3 | 131.0 | 130.6 |
| 6′ | 118.6 | 6.76 d, 8.3 | 117.3 | 6.83 d, 8.4 | 6.79 d, 8.3 | 7.12 d, 8.3 | 116.9 | 116.8 | |
| 7′ | 3.82 s | 28.8 | 3.77 s | 28.6 | 3.81 s | 2.72 dd, 7.2, 14.0 2.90 dd, 4.0, 14.0 |
3.18 dd, 7.8, 13.5 3.47 dd, 3.5, 13.5 |
40.6 | 40.6 |
| 8′ | 152.1 | 153.5 | 4.13 dd, 4.0, 7.2 | 4.73 dd, 3.5, 7.8 | 73.7 | 73.4 | |||
| 9′ | 165.3 | 166.2 | 176.2 | 174.3 | |||||
| NH | 6.76 brt, 7.0 | 8.39 t, 5.6 | |||||||
| NOMe | 3.99 s | ||||||||
| OMe-1 | 3.84 s | ||||||||
| OMe-1′ | 3.80 s | 61.0 | 3.87 s | ||||||
| 234a (41) |
235 (41) |
235a (41) |
236 (41) |
|||
|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 1H | 13C | |
| CDCl3 | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 153.8 | 150.8 | ||||
| 2 | 110.7 | 112.2 | ||||
| 3 | 7.35 d, 2.1 | 7.28 d, 2.0 | 134.2 | 7.36 d, 2.0 | 7.31 s | 133.6 |
| 4 | 133.1 | 131.9 | ||||
| 5 | 7.04 dd, 2.1, 8.4 | 6.92 dd, 2.0, 8.0 | 129.9 | 7.05 dd, 2.0, 8.3 | 7.31 s | 133.6 |
| 6 | 6.92 d, 8.4 | 6.80 d, 8.0 | 117.3 | 112.2 | ||
| 7 | 2.60 m | 2.53 ddd, 7.0, 8.0, 14.0 2.60 ddd, 6.5, 8.5, 14.0 |
35.5 | 2.59 m | 35.1 | |
| 8 | 3.25 ddd, 7.4, 7.4, 15.0 3.41 ddd, 6.5, 7.9, 13.3 |
3.23 ddd, 6.5, 8.0, 15.0 3.40 ddd, 7.0, 8.5, 15.0 |
41.5 | 3.24 ddd, 6.9, 8.0, 14.6 3.42 ddd, 6.3, 8.3, 14.6 |
41.2 | |
| 1′ | 150.9 | 153.9 | ||||
| 2′ | 111.8 | 110.4 | ||||
| 3′ | 7.42 d, 2.1 | 7.34 s | 134.6 | 7.45 s | 7.34 d, 2.0 | 135.2 |
| 4′ | 132.9 | 131.6 | ||||
| 5′ | 7.15 dd, 2.1, 8.4 | 7.34 s | 134.6 | 7.45 s | 7.02 dd, 2.0, 8.0 | 130.9 |
| 6′ | 6.92 d, 8.4 | 111.8 | 6.92 d, 8.3 | 6.80 d, 8.0 | 116.9 | |
| 7′ | 2.77 dd, 6.9, 14.0 2.93 dd, 4.1, 14.0 |
2.75 dd, 4.0, 14.0 2.88 dd, 7.0, 14.0 |
41.5 | 2.80 dd, 6.8, 14.0 2.93 dd, 4.1, 14.0 |
40.6 | |
| 8′ | 4.15 dd, 4.1, 6.9 | 4.14 dd, 4.0, 7.0 | 73.3 | 4.17 dd, 4.1, 6.8 | 73.7 | |
| 9′ | 175.8 | 176.2 | ||||
| OMe-1 | 3.82 s | 3.80 s | ||||
| OMe-1′ | 3.83 s | 3.83 s | ||||
| 236a (41) |
239 (145) |
240 (145) |
241 (145) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | NA | CDCl3 | CDCl3 | acetone-d6 | acetone-d6 | CDCl3 | CDCl3 | |
| 1 | 152.5 | 152.8 | 155.0 | |||||
| 2 | 118.3 | 118.8 | 112.3 | |||||
| 3 | 7.40 s | 7.48 s | 129.4 | 7.42 s | 130.1 | 7.55 d, 2.0 | 130.7 | |
| 4 | 135.0 | 137.3 | 132.5 | |||||
| 5 | 7.40 s | 7.48 s | 129.4 | 7.42 s | 130.1 | 7.27 dd, 8.4, 2.0 | 126.4 | |
| 6 | 118.3 | 118.8 | 6.85 d, 8.4 | 112.3 | ||||
| 7 | 2.59 m | 6.08 d, 14.6 | 110.7 | 5.95 d, 14.6 | 110.5 | 6.14 d, 14.6 | 113.5 | |
| 8 | 3.24 ddd, 6.9, 8.0, 14.6 3.42 ddd, 6.3, 8.3, 14.6 |
7.53 dd, 11.1, 14.6 | 124.0 | 7.46 dd, 11.6, 14.6 | 125.9 | 7.49 dd, 11.4, 14.6 | 123.8 | |
| 1′ | 156.9 | 157.2 | 158.9 | |||||
| 2′ | 6.88 d, 8.4 | 115.8 | 6.78 d, 8.7 | 115.4 | 6.86 d, 8.6 | 116.4 | ||
| 3′ | 7.42 d, 2.0 | 7.59 d, 8.4 | 132.0 | 7.29 d, 8.7 | 131.3 | 7.60 d, 8.6 | 132.5 | |
| 4′ | 125.2 | 126.4 | 125.8 | |||||
| 5′ | 7.16 dd, 2.0, 8.3 | 7.59 d, 8.4 | 132.0 | 7.29 d, 8.7 | 131.3 | 7.60 d, 8.6 | 132.5 | |
| 6′ | 6.92 d, 8.3 | 6.88 d, 8.4 | 115.8 | 6.78 d, 8.7 | 115.4 | 6.86 d, 8.6 | 116.4 | |
| 7′ | 2.80 dd, 6.8, 14.0 2.93 dd, 4.1, 14.0 |
7.13 s | 122.3 | 6.14 s | 109.9 | 7.12 s | 121.3 | |
| 8′ | 4.14 dd, 4.1, 6.8 | 145.3 | 148.2 | 147.8 | ||||
| 9′ | 162.0 | 162.0 | 162.4 | |||||
| NH | 8.45 d, 11.1 | 8.23 d, 11.6 | 8.37 d, 11.4 | |||||
| OMe-1 | 3.83 s | 3.87 s | 60.7 | 3.85 s | 60.9 | 3.89 s | 56.6 | |
| OMe-1′ | 3.82 s | |||||||
| OMe-8′ | 3.68 s | 59.5 | 3.78 s | 56.1 | 3.68 s | 59.7 | ||
| OH-1′ | 5.85 br | 5.75 br | 5.95 br | |||||
| 242 (145) |
243 (146) |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 1H | 13C | 13C | ||
| DMSO-d6 | acetone-d6 | DMSO-d6 | CDCl3 | acetone-d6 | CDCl3 | acetone-d6 | ||
| 1 | 153.9 | 1 | 156.4 | 157.1 | ||||
| 2 | 111.0 | 2 | 6.77 d, 8.1 | 6.75 d, 8.5 | 115.9 | 116.1 | ||
| 3 | 7.55 d, 2.0 | 7.49 d, 2.0 | 129.5 | 3 | 6.92 d, 8.1 | 7.04 d, 8.5 | 130.7 | 130.9 |
| 4 | 130.6 | 4 | 124.6 | 127.5 | ||||
| 5 | 7.34 m | 7.23 dd, 8.6, 2.1 | 125.7 | 5 | 6.92 d, 8.1 | 7.04 d, 8.5 | 130.7 | 130.9 |
| 6 | 7.01 d, 8.7 | 6.81 d, 8.6 | 112.9 | 6 | 6.77 d, 8.1 | 6.75 d, 8.5 | 115.9 | 116.1 |
| 7 | 6.21 d, 14.8 | 6.04 d, 14.6 | 111.8 | 7 | 3.47 s | 3.33 s | 42.9 | 43.1 |
| 8 | 7.34 m | 7.41 dd, 14.6, 11.7 | 122.5 | 8 | 171.7 | 171.5 | ||
| 1′ | 156.0 | NH-9 | 5.01 br | 6.94 br | ||||
| 2′ | 6.64 d, 8.5 | 6.74 d, 8.6 | 114.9 | 10 | 3.39 q, 6.2 | 3.42 t, 6.5 | 40.0 | 40.8 |
| 3′ | 7.11 d, 8.5 | 7.26 d, 8.6 | 129.4 | 11 | 2.68 t, 6.2 | 2.74 t, 6.5 | 33.6 | 34.8 |
| 4′ | 124.9 | 12 | 137.5 | 139.9 | ||||
| 5′ | 7.11 d, 8.5 | 7.26 d, 8.6 | 129.4 | 13 | 7.18 s | 7.42 s | 133.0 | 134.1 |
| 6′ | 6.64 d, 8.5 | 6.74 d, 8.6 | 114.9 | 14 | 124.8 | 118.4 | ||
| 7′ | 5.93 s | 6.13 s | 105.9 | 15 | 151.8 | 152.3 | ||
| 8′ | 148.4 | 16 | 124.8 | 118.4 | ||||
| 9′ | 161.7 | 17 | 7.18 s | 7.42 s | 133.0 | 134.1 | ||
| NH | 10.40 d, 10.0 | 8.22 d, 11.7 | 18 | 4.04 t, 7.5 | 4.04 t, 6.4 | 71.9 | 72.3 | |
| OMe-1 | 3.82 s | 3.87 s | 56.2 | 19 | 2.13 m | 2.06 submerged | 31.9 | 32.2 |
| OMe-8′ | 3.66 s | 3.78 s | 55.6 | 20 | 3.54 q, 6.2 | 3.45 t, 6.5 | 36.1 | 36.6 |
| OH-1′ | 9.38 br | 5.75 br | 21 | 5.80 br | 7.10 br | |||
| 22 | 167.9 | 167.5 | ||||||
| 23 | 5.62 br | 5.70 br | 124.8 | 119.9 | ||||
| 24 | 151.8 | 149.8 | ||||||
| 25 | 1.85 br | 1.96 br | 27.2 | 26.5 | ||||
| 26 | 2.13 br | 2.13 br | 20.1 | 19.6 | ||||
| OH-1 | 8.37 s | |||||||
| 244 (146) |
245 (146) |
247 (123) |
246 (38) |
|||
|---|---|---|---|---|---|---|
| 1H | 1H | 1H | 13C | 1H | 13C | |
| CDCl3 | CDCl3 | CD3OD + TFA | CD3OD + TFA | CD3OD | CD3OD | |
| 1 | 135.3 | 136.4 | ||||
| 2 | 6.77 d, 8.1 | 6.77 d, 8.1 | 7.78 s | 132.9 | 7.82 s | 133.0 |
| 3 | 6.92 d, 8.1 | 6.92 d, 8.1 | 119.5 | 119.5 | ||
| 4 | 156.3 | 154.5 | ||||
| 5 | 6.92 d, 8.1 | 6.92 d, 8.1 | 119.5 | 119.5 | ||
| 6 | 6.77 d, 8.1 | 6.77 d, 8.1 | 7.78 s | 132.9 | 7.82 s | 133.0 |
| 7 | 3.47 s | 3.47 s | 7.38 d, 15.8 | 138.2 | 7.42 d, 15.5 | 138.4 |
| 8 | 6.56 d, 15.8 | 123.8 | 6.57 d, 15.5 | 124.0 | ||
| 9 | 3.39 q, 6.2 | 3.39 q, 6.2 | 168.0 | 167.5 | ||
| 10 | 2.67 t, 6.2 | 2.65 t, 6.2 | 3.34 t, 7.7 | 39.5 | 3.56 t, 6.6 | 39.3 |
| 11 | 1.65 tt, 6.9, 7.7 | 27.5 | 2.77 t, 6.6 | 25.9 | ||
| 12 | 7.42 d, 1.8 | 7.45 s | 1.75 tt, 6.9, 8.1 | 24.4 | 125.9 | |
| 13 | 3.06 t, 8.1 | 48.5 | 6.59 s | 110.9 | ||
| 14 | 3.12 t, 7.7 | 45.7 | 146.4 | |||
| 15 | 2.07 tt, 7.7, 8.1 | 25.3 | ||||
| 16 | 7.21 d, 1.8 | 7.45 s | 3.05 t, 8.1 | 37.8 | ||
| 17 | 4.02 t, 7.5 | 4.00 t, 7.5 | ||||
| 18 | 2.15 m | 2.16 m | ||||
| 19 | 3.55 q, 6.2 | 3.56 2H | ||||
| 21 | 5.63 br | 5.64 br | ||||
| 23 | 1.85 s | 1.85 s | ||||
| 24 | 2.13 s | 2.13 s | ||||
| OMe | 3.87 s | 61.2 | 3.91 s | 61.3 | ||
| NH-8 | 5.01 br | 5.01 br | ||||
| NH-19 | 5.81 br | 5.82 br | ||||
| 248 (150) |
249 (151) |
250 (151) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 13C | 1H | 13C | 1H | 13C | ||
| CD3OD | DMSO-d6 | CD3OD | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | DMSO-d6 | ||
| 1 | 6.71 s | 6.66 s | 118.3 | 115.5 | 1 | ||||
| 2 | 144.3 | 144.7 | 2 | 175.6 | 175.3 | ||||
| 3 | 144.3 | 144.5 | 3 | ||||||
| 4 | 6.65 s | 6.60 s | 118.6 | 116.1 | 4 | 160.5 | 161.2 | ||
| 1′ | 129.7 | 128.3 | 5 | 121.3 | 121.3 | ||||
| 2′ | 7.94 d, 2.1 | 7.90 d, 1.9 | 135.7 | 133.6 | 6 | 164.1 | 164.7 | ||
| 3′ | 110.4 | 108.8 | 7 | 10.55 br | |||||
| 4′ | 154.6 | 152.4 | 8 | 7.73 d, 10.0 | 142.9 | 7.14 t, 9.5 | 137.9 | ||
| 5′ | 6.83 d, 8.4 | 6.83 d, 8.7 | 116.7 | 115.5 | 9 | 6.42 d, 10.0 | 100.4 | 6.42 d, 9.5 | 101.2 |
| 6′ | 7.72 dd, 8.4, 2.1 | 7.55 dd, 8.7, 1.9 | 131.5 | 130.0 | 10 | 169.7 | 170.8 | ||
| 1″ | 129.1 | 128.9 | 11 | 4.23 s | 35.0 | 4.17 s | 33.9 | ||
| 2″ | 7.32 d, 1.7 | 7.55 d, 1.7 | 127.0 | 124.3 | 12 | 140.2 | 140.1 | ||
| 3″ | 110.0 | 109.4 | 13 | 7.67 s | 133.2 | 7.66 s | 133.1 | ||
| 4″ | 143.9 | 141.9 | 14 | 116.7 | 116.7 | ||||
| 5″ | 146.5 | 145.1 | 15 | 151.4 | 151.4 | ||||
| 6″ | 7.48 d, 1.7 | 7.04 d, 1.7 | 116.5 | 116.4 | 16 | 116.7 | 116.7 | ||
| OH-4′ | 10.12 br | 17 | 7.67 s | 133.2 | 7.66 s | 133.1 | |||
| OH-4″ | 9.00 br | 18 | 8.69 | 8.69 | |||||
| OH-5″ | 9.52 br | 19 | 3.07 d, 3.7 | 29.2 | 3.07 d, 3.7 | 28.9 | |||
| 20 | 3.56 s | 43.7 | |||||||
| 21 | 3.71 s | 60.3 | 3.72 s | 60.3 | |||||
| 251 (152) |
252 (152) |
253 (153) |
||||
|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | |
| 1 | 133.0 | 133.0 | ||||
| 2 | 7.60 d, 1.7 | 136.0 | 7.61 d, 1.7 | 136.1 | 8.67 s | 139.5 |
| 3 | 113.6 | 114.0 | 114.0 | |||
| 4 | 157.0 | 157.1 | 127.9 | |||
| 5 | 7.00 d, 7.0 | 114.0 | 7.00 d, 7.0 | 114.1 | 8.29 m | 122.9 |
| 6 | 7.33 dd, 1.7, 7.0 | 131.5 | 7.34 dd, 1.7, 7.0 | 131.6 | 7.25 m | 123.8 |
| 7 | 4.10 s | 39.8 | 4.20 s | 39.9 | 7.25 m | 124.8 |
| 8 | 160.8 | 160.8 | 7.47 m | 113.1 | ||
| 9 | 157.7 | 157.8 | 138.0 | |||
| 10 | 183.3 | |||||
| 11 | 7.83 s | 124.4 | 7.86 s | 124.5 | 165.8 | |
| 12 | 131.5 | 131.6 | ||||
| 13 | 3.51 t, 7.1 | 41.8 | ||||
| 14 | 137.5 | 137.6 | 2.79 t, 7.1 | 35.2 | ||
| 15 | 8.07 s | 130.9 | 8.12 s | 131.7 | 132.9 | |
| 16 | 120.2 | 120.3 | 7.38 d, 2.0 | 134.3 | ||
| 17 | 153.7 | 153.7 | 110.8 | |||
| 18 | 120.2 | 120.3 | 154.0 | |||
| 19 | 8.07 s | 131.0 | 8.12 s | 131.7 | 6.82 dd, 8.2 | 117.3 |
| 20 | 7.06 dd, 8.2, 2.0 | 130.1 | ||||
| 21 | 4.19 s, 5.6 | 72.1 | 4.21 s, 5.6 | 72.5 | ||
| 22 | 2.35 tt, 5.6, 7.9 | 27.2 | 2.30 tt, 5.6, 7.4 | 34.7 | ||
| 23 | 3.57 t, 7.9 | 57.9 | 3.43 t, 7.4 | 49.4 | ||
| 38 | 3.88 s | 57.5 | 3.88 s | 57.6 | ||
| OMe | ||||||
| NMe2 | 3.02 s | 44.5 | ||||
| NMe | 2.83 s | 28.6 | ||||
| 254 (153) |
255 (153) |
256 (154) |
257 (154) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | CD3OD | DMSO-d6 | DMSO-d6 | |
| 1 | Signals were not assigned | Signals were not assigned | ||||||
| 2 | 8.65 s | 139.6 | 8.67 s | 139.5 | 2.75 d, 5.7; 2.76 d, 7.3; 2.96 4H, br; 3.82 s; 4.66 dd, 7.3, 5.7; 6.86 d, 8.5; 8.5; 6.98 ddd, 7.3, 7.1, 1.0; 7.04 ddd, 7.7, 7.3, 1.1; 7.14 dd, 8.5, 2.0; 7.32 dd 7.7, 1.1; 7.48 d, 2.0; 7.52 dd, 7.1, 1.1. | 26.0 50.5 | 2.63 br d, 5.9; 2.79 4H, br; 3.81 s; 5.25 br; 6.98 d, 8.4; 7.13 dd, 8.5, 1.6; 7.14 br; 7.23 dd, 8.4, 1.7; 7.18 d, 8.5; 7.48 d, 1.7; 7.66 d, 1.6; 11.04 br. | 24.7 |
| 3 | 114.0 | No | 50.5 | 49.4 | ||||
| 4 | 127.9 | 127.8 | 56.7 | 56.1 | ||||
| 5 | 8.28 m | 122.9 | 8.29 m | 123.0 | 57.5 | 56.7 | ||
| 6 | 7.25 m | 123.8 | 7.25 s | 123.8 | 72.0 | 69.9 | ||
| 7 | 7.25 m | 124.8 | 7.25 m | 124.8 | 112.3 | 110.2 | ||
| 8 | 7.45 m | 113.1 | 7.46 m | 113.1 | 112.4 | 110.9 | ||
| 9 | 138.0 | 138.0 | 113.0 | 112.0 | ||||
| 10 | 183.3 | 182.9 | 113.0 | 112.1 | ||||
| 11 | 166.0 | 165.7 | 119.2 | 113.3 | ||||
| 13 | 3.49 t, 7.2 | 41.8 | 3.51 t, 7.7 | 42.1 | 119.6 | 120.6 | ||
| 14 | 2.76 t, 7.2 | 35.2 | 2.80 t, 7.7 | 35.6 | 122.4 | 123.2 | ||
| 15 | 131.0 | 131.0 | 123.6 | 124.4 | ||||
| 16 | 7.59 d, 2.0 | 140.5 | 7.08 d, 2.0 | 130.8 | 127.4 | 126.4 | ||
| 17 | 84.9 | 6.72 d, 8.5 | 116.3 | 128.6 | 129.1 | |||
| 18 | 157.1 | 157.0 | 131.8 | 130.3 | ||||
| 19 | 7.07 dd, 8.1 | 115.7 | 6.72 dd, 8.5 | 116.3 | 138.9 | 134.9 | ||
| 20 | 7.07 dd, 8.1, 2.0 | 133.3 | 7.08 dd, 8.5 | 130.8 | 138.2 156.7 |
137.9 154.2 |
||
| 258 (155) |
259 (164) |
260 (158) |
261 (158) |
|||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| DMSO-d6 | DMSO-d6 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 175.1a | 170.9 | 170.7 | |||||
| 2 | 174.2 | 2.50 m | 40.1 | 2.32 m | 42.4 | 2.32 m | 42.4 | |
| 3 | 2.37 d, 17.4 2.19 d, 17.4 |
43.4 | 2.38 dd, 15.7, 10.8 1.89 d, 15.7 | 40.7 | 2.04 dd, 15, 4 2.16 d, 15 |
43.3 | 2.03 dd, 14, 4 2.16 d, 14 | 43.4 |
| 3a | 81.6 | |||||||
| 4 | 4.1–4.2 m | 83.2 | 131.1 | 129.6 | 129.9 | |||
| 5 | 6.75 d, 2.5 | 142.0 | 4.75 d, 7.1 | 127.8 | 4.93 d, 8 | 131.6 | 4.94 d, 8 | 131.7 |
| 6 | 137.8 | 2.23 m | 29.2 | 2.16 m | 29.0 | 2.16 m | 29.0 | |
| 6a | 67.4 | |||||||
| 7 | 163.4 | 1.32 m | 43.3 | 1.34 m, 1.59 m | 43.7 | 1.35 m, 1.60 m | 43.8 | |
| 8 | 4.1–4.2 m | 60.6 | 4.62 m | 70.8 | 4.91 m | 71.0 | 4.88 m | 71.0 |
| 9 | 1.24 t, 7 | 14.0 | 174.4a | 168.8 | 168.8 | |||
| 10 | 3.69 dq, 9.2, 7 3.55 dq, 9.2, 7 |
65.9 | 2.65 dd, 4.7, 15.0 | 39.7 | 4.75 dq, 8, 6.5 | 49.0 | 4.73 dq, 8, 6.5 | 49.0 |
| 11 | 1.13 t, 7 | 15.4 | 5.26 dd, 4.7, 8.4 | 49.0 | 175.1 | 175.1 | ||
| 12 | 170.5a | 5.21 dd, 9, 8 | 56.7 | 5.20 dd, 9, 8 | 56.7 | |||
| 13 | 5.85 dd, 6.4, 10.2 | 55.5 | 174.4 | 174.4 | ||||
| 14 | 168.9 | 4.46 dq, 8, 7 | 45.9 | 4.47 dq, 8, 7 | 45.9 | |||
| 15 | 4.75 m | 45.8 | 2.95 dd, 15, 9 3.15 dd, 15, 8 |
32.7 | 2.95 dd, 15, 93.18 dd, 15, 8 | 32.5 | ||
| 16 | 1.12 d, 6.8 | 20.3 | 133.0 | 133.1 | ||||
| 17 | 1.56 s | 18.5 | 7.29 d, 2 | 132.2 | 7.30 d, 2.0 | 132.1 | ||
| 18 | 0.81 d, 6.5 | 21.9 | 85.1 | 110.0 | ||||
| 19 | 1.05 d, 6.3 | 19.0 | 154.5 | 151.4 | ||||
| 20 | 133.6 | 6.87 d, 9 | 116.1 | 6.95 d, 9 | 116.0 | |||
| 21 | 6.94 d, 8.3 | 127.1 | 7.05 dd, 9, 2 | 129.4 | 7.07 dd, 9, 2 | 129.5 | ||
| 22 | 6.66 d, 8.3 | 115.6 | 1.14 d, 6.5 | 20.4 | 1.13 d, 6.5 | 20.4 | ||
| 23 | 155.7 | 1.49 s | 17.7 | 1.49 s | 17.7 | |||
| 24 | 6.66 d, 8.3 | 115.6 | 0.86 d, 6.5 | 18.2 | 0.84 d, 6.5 | 18.2 | ||
| 25 | 6.94 d, 8.3 | 127.1 | 1.24 d, 6.5 | 20.6 | 1.22 d, 6 | 20.5 | ||
| 26 | 3.38 dd, 6.3, 15.2 3.24 dd, 10.5, 15.2 |
23.2 | 1.02 d, 6.5 | 18.7 | 1.02 d, 6.5 | 18.8 | ||
| 27 | 109.0 | 2.97 s | 30.7 | 2.96 s | 30.6 | |||
| 28 | 111.1 | 1.34 d, 7 | 18.8 | 1.33 d, 7 | 18.8 | |||
| 29 | 131.3 | |||||||
| 30 | 7.24 d, 7.3 | 118.1 | ||||||
| 31 | 7.13 dd, 7.3, 7.7 | 122.3 | ||||||
| 32 | 7.10 dd, 7.3, 7.7 | 120.9 | ||||||
| 33 | 7.56 brd, 7.3 | 110.6 | ||||||
| 34 | 136.1 | |||||||
| 35 | 2.98 s | 30.8 | ||||||
| 36 | 0.70 d, 6.9 | 17.8 | ||||||
| OH | 5.36 br | 6.27 s | 6.29 s | |||||
| NH | 8.14 br | 6.63, 8.70 br | 6.59 d, 8 | 6.59 d, 8 | ||||
| NH | 7.65 d, 8.4 | 6.52 d, 8 | 6.50 d, 8 | |||||
| 262 (159) |
263 (159) |
264 (159) |
265 (159) |
||||
|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | |
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 170.8 | 170.5 | 170.5 | ||||
| 2 | 2.32 m | 42.3 | 2.42 m | 42.3 | 2.42 m | 42.3 | 2.4 m |
| 3 | 2.03 dd, 13.8, 3.6, 2.16 | 43.2 | 2.1 m | 43.2 | 2.1 m | 43.2 | 2.1 m |
| 4 | 129.0 | 130.6 | noa | ||||
| 5 | 4.93 d, 8.8 | 129.5 | 4.99 d, 8.3 | 131.5 | 4.99 d, 8.7 | 131.4 | 4.99 d, 8.7 |
| 6 | 2.16 m | 28.9 | 2.21 m | 29.1 | 2.21 m | 29.1 | 2.21 m |
| 7 | 1.36 m, 1.60 m | 43.6 | 1.40 m, 1.69 m | 43.5 | 1.40 m, 1.69 m | 43.5 | 1.40 m, 1.69 m |
| 8 | 4.88 m | 70.9 | 4.86 m | 71.4 | 4.86 m | 71.4 | 4.86 m |
| 9 | 168.7 | 168.2 | |||||
| 10 | 4.48 dq, 7.7, 7.1 | 49.0 | 4.51 dq, 8.0, 7.1 | 48.9 | 4.52 dq, 8.0, 7.1 | 48.9 | 4.52 dq, 8.0, 7.1 |
| 11 | 175.1 | 175.8 | noa | ||||
| 12 | 5.21 dd, 9.0, 7.3 | 56.6 | 5.08 dd, 9.0, 6.8 | 57.6 | 5.09 dd, 9.0, 6.8 | 57.5 | 5.11 dd, 9.0, 6.8 |
| 13 | 174.5 | 169.9 | 169.9 | ||||
| 14 | 4.75 dq, 6.3, 6.3 | 45.8 | 3.77 dd, 17.6, 3.8 4.16 dq, 17.6, 1 | 42.2 | 3.77 dd, 17.9, 3.2 4.16 dq, 17.9, 4.2 |
42.0 | 3.77 dd, 18, 3.24.16 dq, 18, 4.2 |
| 15 | 2.92 dd, 14.7, 9.0 3.17 dd, 14.7, 7.3 |
32.7 | 2.81 dd, 14, 6.8 3.25 dd, 14, 9 |
32.2 | 2.83 dd, 14, 6.8 3.26 dd, 14, 9 |
32.4 | 2.84 dd, 14, 6.7 3.26 dd, 14, 9 |
| 16 | 133.0 | 133.4 | NA | ||||
| 17 | 7.15 d, 1.7 | 131.5 | 7.15 d, 1.7 | 138.4 | 7.32 d, 2.0 | 132.2 | 7.18 d, 2.0 |
| 18 | 119.8 | noa | noa | ||||
| 19 | 150.2 | noa | noa | ||||
| 20 | 6.93 d, 8.3 | 116.3 | 6.90 d, 8.3 | 115.1 | 6.94 d, 8.3 | 116.2 | 6.93 d, 8.3 |
| 21 | 7.01 dd, 8.3, 1.7 | 128.8 | 7.08 dd, 8.3, 1.7 | 130.7 | 7.06 dd, 8.3, 2 | 129.7 | 7.01 dd, 8.3, 2 |
| 22 | 1.15 d, 6.7 | 18.7 | 1.16 d, 6.7 | 18.5 | 1.16 d, 6.7 | 18.5 | 1.16 d, 6.7 |
| 23 | 1.51 3H | 17.6 | 1.53 d, 1 | 18.2 | 1.54 d, 1.2 | 18.2 | 1.5 d, 1.0 |
| 24 | 0.88 d, 6.6 | 20.4 | 0.90 d, 6.6 | 20.4 | 0.90 d, 6.7 | 20.4 | 0.91 d, 6.7 |
| 25 | 1.24 d, 6.3 | 20.5 | 1.25 d, 6.2 | 20.8 | 1.25 d, 6.2 | 20.7 | 1.25 d, 6.2 |
| 26 | 1.35 d, 7.1 | 18.2 | 1.30 d, 7.1 | 18.2 | 1.30 d, 7.1 | 18.2 | 1.30 d, 7.1 |
| 27 | 2.97 s | 30.5 | 2.94 s | 29.8 | 2.97 s | 29.7 | 2.93 s |
| 28 | 1.09 d, 6.7 | 18.7 | |||||
| OH | 5.5 s | 5.3 s | 5.46 s | 5.46 s | |||
| NH | 6.56 d, 7.7 | 6.60 d, 8 | 6.60 d, 8 | 6.60 d, 8 | |||
| NH | 6.46 d, 6.3 | 6.45 dd, 3.8, 1 | 6.44 dd, 3.8, 1 | 6.44 dd, 3.8, 1 | |||
| 266 (164) |
267 (161) |
268 (161) |
271 (164) |
|||
|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 13C | 1H | |
| CDCl3 | CDCl3 + 5% DMSO-d6 | CDCl3 + 5% DMSO-d6 | CDCl3 + 5% DMSO-d6 | CDCl3 + 5% DMSO-d6 | CDCl3 | |
| 1 | 175.0 | 174.9 | ||||
| 2 | 2.46 m | 2.55 m | 40.1 | 2.56 m | 40.1 | 2.45 m |
| 3 | 2.55 dd, 4, 12 2.14 t, 12 |
2.36, 15.2, 11.4 1.95, 15.2, 2.0 |
40.9 | 2.35, 15.2, 11.4 1.94, 15.2, 2.0 |
40.9 | 2.54 dd, 4, 13 2.14, 12 |
| 4 | 133.6 | 133.6 | ||||
| 5 | 4.82, 9.6 | 128.4 | 4.82 9.6 | 128.3 | ||
| 6 | 2.95 | 2.28 m | 29.2 | 2.28 m | 29.2 | nsa |
| 7 | 1.82 ddd, 3, 9, 15 1.61 ddd, 3, 11, 15 |
1.39, 13.6, 11.1, 4.7 1.18, 13.5, 9.4, 4.5 |
43.4 | 1.38, 13.6, 11.1, 4.7 1.19, 13.5, 9.4, 4.5 |
43.4 | 1.82 m, 1.60 m |
| 8 | 5.11 m | 4.64 m | 70.4 | 4.64 m | 70.4 | 5.11 m |
| 9 | 170.5 | 170.5 | ||||
| 10 | 4.51 dq, 7, 7 | 2.68, 15.5, 4.5 2.59, 15.5, 6.6 |
40.2 | 2.69, 15.5, 4.5 2.60, 15.5, 6.6 |
40.0 | 4.50 dq, 7, 7 |
| 11 | 5.25 m | 48.8 | 5.24 m | 48.8 | ||
| 12 | 5.06 dd, 8, 9 | 168.4 | 168.4 | 5.06 dd, 8, 9 | ||
| 13 | 5.38, 9.8, 6.7 | 56.8 | 5.40, 9.8, 6.7 | 56.7 | ||
| 14 | 4.72 dq, 7 | 174.1 | 174.1 | 4.71 dq, 7, 8 | ||
| 15 | 3.12 dd, 8, 15 2.90 dd, 9, 15 | 4.75 m | 46.0 | 4.24 m | 45.0 | 3.13 dd, 8, 15 2.90 dd, 9, 15 |
| 16 | 1.16, 7.0 | 20.2 | 1.16, 7.0 | 20.2 | ||
| 17 | 7.45 d, 1 s | 1.60, 1.0 | 18.5 | 1.60, 1.0 | 18.5 | 7.25 d, 2 |
| 18 | 0.86, 7.0 | 21.9 | 0.86, 7.0 | 21.9 | ||
| 19 | 1.09, 7.0 | 19.1 | 1.08, 7.0 | 19.1 | ||
| 20 | 6.88 d, 8 | 130.8 | 130.7 | 6.91 d, 8 | ||
| 21 | 7.04 dd, 1, 8 | 7.00, 8.5 | 127.1 | 7.01, 8.5 | 127.1 | 7.02 dd, 2, 8 |
| 22 | 1.15 d, 6 | 6.77, 8.5 | 115.6 | 6.87, 8.5 | 115.6 | 1.14 d, 7 |
| 23 | 5.90 s 5.78 s |
156.6 | 156.6 | 5.90 s 5.78 s |
||
| 24 | 1.09 d, 7 | 6.77, 8.5 | 115.6 | 6.87, 8.5 | 115.6 | 1.09 d, 7 |
| 25 | 1.28 d, 6 | 7.00, 8.5 | 127.1 | 7.01, 8.5 | 127.1 | 1.28 d, 6 |
| 26 | 1.32 d, 7 | 3.18, 14.5, 5.7 2.86, 14.5, 9.9 |
31.7 | 3.19, 14.5, 5.7 2.88, 14.5, 9.9 |
31.9 | 1.31 d, 7 |
| 27 | 2.97 s | 129.6 | 129.2 | 2.96 s | ||
| 28 | 1.04 d, 7 | 7.48, 2.3 | 138.7 | 7.26, 2.3 | 132.7 | 1.03 d, 7 |
| 29 | 84.2 | 109.7 | ||||
| 30 | 155.2 | 152.6 | ||||
| 31 | 6.79, 8.5 | 115.1 | 6.85, 8.5 | 116.4 | ||
| 32 | 6.96, 8.5, 2.3 | 129.8 | 6.93, 8.5, 2.3 | 128.8 | ||
| 33 | 2.94 s | 30.3 | 2.92 s | 30.3 | ||
| 34 | 1.08, 7.0 | 18.2 | 1.06, 7.0 | 18.1 | ||
| OH | 9.12 s | 8.85 s | ||||
| OH | 8.68 s | 8.69 s | ||||
| NH-10 | 6.35 d, 7 | 7.46, 8.3 | 7.47, 8.3 | 6.21 d, 7 | ||
| NH-14 | 6.19 d, 7 | 6.78, 8.5 | 6.77, 8.5 | 6.36 d, 8 | ||
| 272 (164) |
273 (164) |
274 (164) |
275 (164) |
276 (164) |
|||
|---|---|---|---|---|---|---|---|
| 1H | 1H | 13C | 1H | 1H | 1H | 13C | |
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | |
| 1 | 175.5 | 177.6 | |||||
| 2 | 2.46 m | 2.26 m | 42.4 | 2.28 m | 2.27 m | 2.40 m | 42.2 |
| 3 | 2.54 d, 12 NA |
2.17 m 1.98 dd, 3, 14 |
42.3 | 2.17 m 1.98 dd, 3, 14 |
2.16 m 1.98 dd, 2, 12 |
2.14 m 2.04 m |
43.2 |
| 4 | 132.9 | 147 | |||||
| 5 | 4.87 d, 9 | 131.3 | 4.87 d, 9 | 4.87 d, 9 | 4.93 d, 9 | 131.2 | |
| 6 | NA | 2.18 m | 29.3 | 2.18 m | 2.16 m | 2.16 m | 28.7 |
| 7 | NA NA |
1.63 ddd, 6, 8, 14 1.39 ddd, 4, 8, 14 |
43.7 | 1.64 m 1.37 m |
nsb 1.37 m |
1.56 m 1.37 m |
43.9 |
| 8 | 5.11 m | 4.94 m | 71.9 | 4.94 m | 4.93 m | 4.87 m | 70.6 |
| 9 | 169.8 | 170.4 | |||||
| 10 | 4.51 m | 4.43 dq, 7, 7 | 53.3 | 4.43 m | 4.43 m | 4.5 dq, 7, 8 | 48.8 |
| 11 | 168.9 | 168.4 | |||||
| 12 | 5.06 m | 5.26 dd, 7, 9 | 56.9 | 5.27 dd, 7, 9 | 5.28 dd, 7, 9 | 5.16 dd, 8, 9 | 57.1 |
| 13 | 174.6 | 171.3 | |||||
| 14 | 4.71 m | 4.71 dq, 4, 9 | 45.8 | 4.71 quint, 7 | 4.72 quint, 6 | 4.71 m | 52.8 |
| 15 | 3.14m 2.90 m |
3.19 dd, 7, 15 2.89 dd, 9, 15 |
32.5 | 3.22 dd, 7, 15 2.91 dd, 9, 15 |
3.22 dd, 7, 15 2.93 dd, 9, 15 |
3.17 dd, 8, 15 2.88 dd, 9, 15 |
32.2 |
| 16 | 130.3 | 130.1 | |||||
| 17 | 7.12s | 7.46 d, 2 | 138.2 | 7.27 d, 2 | 7.13 d, 2 | 7.46 d, 2 | 138 |
| 18 | 85 | 85.3 | |||||
| 19 | 154.0 | 154.1 | |||||
| 20 | 6.91 d, 8 | 6.86 d, 8 | 115.1 | 6.91 d, 8 | 6.91 d, 8 | 6.88 d, 8 | 114.9 |
| 21 | 6.98 d, 8 | 7.03 dd, 2, 8 | 130.4 | 7.03 dd, 2, 8 | 7.03 dd, 2, 8 | 7.05 dd, 2, 8 | 130.1 |
| 22 | 1.14 d, 7 | 1.12 d, 7 | 19 | 1.12 d, 7 | 1.13 d, 7 | 1.16 d, 7 | 18.8 |
| 23 | 5.90 s 5.79 s | 1.45 s | 17.8 | 1.45 s | 1.45 s | 1.49 s | 17.7 |
| 24 | 1.09 d, 7 | 0.86 d, 7 | 20.5 | 0.86 d, 7 | 0.87 d, 7 | 0.88 d, 7 | 20.4 |
| 25 | 1.28 d, 6 | 1.24 d, 6 | 20.8 | 1.23 d, 6 | 1.23 d, 6 | 1.23 d, 6 | 20.5 |
| 26 | 1.31 d, 7 | 3.97 dd, 4, 11 3.81 dd, 4, 11 |
63.1 | 3.97 dd, 4, 11 3.82 dm, 4, 11 |
3.97 dd, 4, 11 3.82 dm, 4, 11 |
1.32 d, 7 | 18.1 |
| 27 | 2.96 s | 2.97 | 30.7 | 2.97 s | 2.97 s | 3.03 s | 30.9 |
| 28 | 1.03 d, 7 | 1.08 d, 7 | 18.5 | 1.09 d, 7 | 1.09 d, 7 | 3.54 br | 65.2 |
| NH-10 | 4.51 m | 7.00 d, 8 | 6.99 d, 8 | 6.98 d, 7 | 6.45 d, 7 | ||
| NH-14 | 6.37 d, 8 | 6.44 d, 6 | 6.43 d, 6 | 6.44 d, 6 | 6.67 br | ||
| 277 (164) |
278 (164) |
269 (162) |
270 (163) |
280 (167) |
||||
|---|---|---|---|---|---|---|---|---|
| 1H | 1H | 1H | 13C | 1H | 13C | 1H | 13C | |
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | CDCl3 | DMSO-d6 | ||
| 1 | 175.2 | 169.7 | ||||||
| 2 | 2.41 m | 2.39 m | 2.23 m | 42.1 | 2.47 m | 42.0 | 132.0 | |
| 3 | 2.10 m, 2.04 m | 2.10 m, 2.05 m | 1.98 m, 2.06 m | 42.5 | 2.10 m | 43.3 | 136.3 | |
| 4 | 133.5 | 130.5 | 136.3 | |||||
| 5 | 4.93 d, 9 | 4.97 d, 9 | 4.86 d, 8.5 | 132.0 | 4.99 d, 8.5 | 131.6 | 132.0 | |
| 6 | 2.15 m | 2.19 m | 2.06 m | 29.0 | 2.21 m | 29.1 | 4.57 t, 6.0 | 47.2 |
| 7 | 1.56 m, 1.37 m | 1.70 m, 1.41 m | 1.30 m, 1.60 m | 43.6 | 1.78 m, 1.40 m | 43.3 | 2.97 t, 6.0 | 37.1 |
| 8 | 4.87 m | 4.88 m | 4.78 sextet, 7 | 71.7 | 4.84 sextet, 6.4 | 71.5 | 132.8 | |
| 9 | 170.0 | 168.5 | 6.89 d, 8.0 | 134.7 | ||||
| 10 | 4.45 m | 4.53 dq, 7, 8 | 4.25 dd, 6, 9 | 58.6 | 4.43 dd, 8.7, 7.0 | 58.4 | 6.58 d, 8.0 | 120.4 |
| 11 | 169.5 | 175.9 | 161.2 | |||||
| 12 | 5.17 dd, 7, 10 | 5.11 dd, 7, 9 | 5.18 t, 7 | 57.2 | 5.13 dd, 9.8, 6.4 | 57.8 | 6.58 d, 8.0 | 120.4 |
| 13 | 174.7 | 169.5 | 6.89 d, 8.0 | 134.7 | ||||
| 14 | 4.79 m | 4.15 dd, 4, 18 | 4.67 quin, 6.5 | 46.8 | 4.15 dd, 18.3, 4.1 3.19 dd, 18.3, 3.4 |
41.9 | 189.7 | |
| 15 | 3.18 dd, 7, 15 2.91 dd, 10, 15 |
3.24 dd, 9, 14 2.81 dd, 7, 14 |
2.82 dd, 14.5, 8 3.08 dd, 14.5, 8 |
32.2 | 3.25 dd, 13.2, 9.0 2.80 dd, 13.2, 6.1 |
32.7 | 132.8 | |
| 16 | 130.5 | 133.2 | 7.62 4H, s | 138.9 | ||||
| 17 | 7.27 d, 2 | 7.50 d, 2 | 7.42 d, 2 | 138.8 | 7.53 d, 2.0 | 138.6 | 116.1 | |
| 18 | 86.2 | 85.5 | 159.9 | |||||
| 19 | 154.5 | 154.0 | 116.1 | |||||
| 20 | 6.92 d, 8 | 6.89 d, 8 | 6.82 d, 8 | 115.1 | 6.90 d, 8.5 | 115.2 | 7.62 4H, s | 138.9 |
| 21 | 7.03 dd, 2, 8 | 7.07 dd, 2, 8 | 7.01 d, 8 | 131.0 | 7.10 dd, 8.5, 2.0 | 130.9 | 136.0 | |
| 22 | 1.16 d, 7 | 1.14 d, 7 | 1.09 d, 6.5 | 18.5 | 1.17 d, 6.8 | 18.8 | 7.00 4H, s | 139.3 |
| 23 | 1.49 s | 1.49 s | 1.46 s | 17.7 | 1.56 s | 17.8 | 116.2 | |
| 24 | 0.86 d, 6 | 0.89 d, 7 | 0.79 d, 6.5 | 20.5 | 0.90 d, 6.8 | 20.5 | 154.6 | |
| 25 | 1.23 d, 6 | 1.26 d, 6 | 1.16 d, 6.5 | 20.6 | 1.25 d, 6.4 | 20.8 | 116.2 | |
| 26 | 1.32 d, 7 | 3.91 dm, 8, 3.75 m | 1.98 m | 32.0 | 1.99 m | 31.5 | 7.00 4H, s | 139.3 |
| 27 | 3.03 s | 2.93 s | 2.97 s | 30.5 | 2.96 s | 29.7 | 136.0 | |
| 28 | 3.54 br | 1.04 d, 6.5 | 18.8 | 0.79 d, 6.5 | 18.1 | 7.00 4H, s | 139.3 | |
| 29 | 0.75 d, 6.5 | 17.8 | 0.75 d, 6.5 | 18.1 | 116.2 | |||
| 30 | 0.77 d, 6.5 | 154.6 | ||||||
| 31 | 116.2 | |||||||
| 32 | 7.00 4H, s | 139.3 | ||||||
| 33 | 189.7 | |||||||
| 34 | 132.8 | |||||||
| 35,39 | 7.62 4H, s | 138.9 | ||||||
| 36,38 | 116.1 | |||||||
| 37 | 159.9 | |||||||
| NH-10 | 6.46 d, 8 | 6.6 d, 8 | 6.42 d, 8.8 | |||||
| NH-14 | 6.68 br | 6.45 dd, 4, 1 | 6.64 t, 3.4 | |||||
| 282 (167) |
283 (167) |
281 (168) |
284 (168) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | ||
| CDCl3 | CDCl3 | CDCl3 | CDCl3 | CD3OD | CDCl3 | CDCl3 + CD3OD | |||
| 2 | 170.0 | 170.0 | 2,5 | 131.2 | 166.6 | ||||
| 3 | 122.6 | 122.6 | 3,4 | 128.0 | 135.2 | ||||
| 4 | 122.6 | 122.4 | 6,6′ | 186.0 | 124.9 | ||||
| 5 | 170.0 | 170.0 | 7,7′ | 129.0 | 7.75 | 136.2 | |||
| 6 | 3.58 t, 6 | 40.0 | 3.85 t, 6 | 40.0 | 8,8′ | 8.00 s | 136.1 | 114.2 | |
| 7 | 2.90 t, 6 | 34.0 | 2.90 t, 6 | 34.0 | 9,9′ | 114.0 | 153.7 | ||
| 8 | 129.5 | 129.5 | 10,10′ | 162.6 | 114.2 | ||||
| 9,13 | 7.10 d, 8 | 130.0 | 7.10 d, 8 | 128.9 | 11,11′ | 114.0 | 7.75 | 136.2 | |
| 10,12 | 6.85 d, 8 | 115.5 | 6.85 d, 8 | 115.0 | 12,12′ | 8.00 s | 136.1 | ||
| 11 | 155.0 | 153.1 | 13,13′ | 131.2 | |||||
| 14 | 132.9 | 132.5 | 14,14′ | 7.40 | 135.9 | ||||
| 15,19 | 7.60 s | 133.3 | 7.63 | 133.0 | 15,15′ | 112.2 | |||
| 16,18 | 110.2 | 109.0 | 16,16′ | 153.5 | |||||
| 17 | 152.8 | 154.0 | 17,17′ | 112.2 | |||||
| 20 | 132.9 | 132.5 | 18,18′ | 7.40 | 135.9 | ||||
| 21,25 | 7.60 s | 133.3 | 7.62 | 132.8 | NH | 8.31 s | |||
| 22,24 | 110.2 | 118.2 | |||||||
| 23 | 152.8 | 156.2 | |||||||
| 26 | 3.94 s | 61.0 | |||||||
NA: not reported
Interchangeable
Broad signal
Exchangeable.
Exchangeable.
Exchangeable.
Coupling constants were not reported in original paper.
Interchangeable signals.
Interchangeable signals.
Interchangeable signals.
a Interchangeable signals.
Interchangeable signals.
Interchangeable signals.
Shape of signal was changed in temperature-variable experiment in DMSO-d6.
May be interchanged
May be interchanged
c May be interchanged
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
May be interchanged.
Coupling constants were not reported.
may be interchanged.
may be interchanged.
may be interchanged.
not observed due to limited sample size
The 1H and 13C NMR spectra of the spirooxepinisoxazoline type of alkaloids are significantly different from those of the spirocyclohexadieneisoxazoline type of alkaloids. The chemical shift of H-1 of the spirooxepinisoxazoline type at 7.3 ppm is 1.0 ppm lower than that of the corresponding proton (H-5) in the spirocyclohexadieneisoxazolines. The carbon-13 chemical shifts of C-1 and C-6 (the spiro-carbon) for the spirooxepinisoxazo-line type are at 145 and 102 ppm, which are 15 and 12 ppm lower than those of the corresponding carbons (C-5, C-6) in the spirocyclohexadieneisoxazoline type of bromotyrosine derivatives.
The geometry of the oxime can be determined by the chemical shift of C-7, where 25.3–27.0 ppm indicated an E-geometry, while 35 ppm indicated a Z-geometry (107).
Bastadins are often spectroscopically similar to one another. 1H and 13C NMR alone are insufficient to discriminate bastarane and isobastarane isomers (130). For example, the structure of bastadin 5 (209) is almost impossible to distinguish from bastadin 19 (223) by 1D NMR, without recourse to 2D heteronuclear experiments. Molinski et al. reported that using “fingerprinting” of the MeO 1H NMR signals of the permethyl derivatives of bastadins provided a partial solution to the problem (130). The chemical shift pattern of the MeO signals measured in CDCl3 can be matched reliably against those of previously reported bastadins tetra-O-methyl ethers (see Table III). For example, the 1H NMR of the three isomers, bastadin 5, bastadin 15, and bastadin 19, are very similar in CD3OD or DMSO-d6. However, the 1H NMR signals of the MeO of the corresponding tetramethyl ethers are readily distinguishable in CDCl3.
TABLE III.
Selected 1H-NMR Data of Bastadin Tetra-O-methyl Ethers (130).
| Parent bastadins | −4 (208) | −5 (209) | −6 (210) | “Isobastadin 6” | −8 (212) | −9 (213) | −11 (215) | −12 (216) | −13 (217) | −14 (218) | 15 (219) | 19 (223) | 20 (224) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δ of MeO groups (CDCl3) | 4.01 | 4.02 | 4.06 | 4.08 | 4.02 | 4.05 | 4.01 | 4.05 | 4.04 | 4.10 | 4.03 | 4.04 | 4.00 |
| 4.01 | 3.98 | 4.03 | 4.04 | 4.01 | 4.00 | 4.01 | 4.01 | 4.01 | 4.05 | 4.01 | 4.01 | 3.92 | |
| 4.01 | 3.94 | 4.02 | 4.02 | 3.97 | 3.90 | 3.93 | 3.89 | 3.81 | 4.03 | 3.89 | 3.91 | 3.91 | |
| 4.01 | 3.52 | 3.61 | 3.92 | 3.70 | 3.49 | 3.92 | 3.87 | 3.48 | 3.83 | 3.73 | 3.61 | 3.72 |
IV. Biosynthesis of Bromotyrosine Derivatives
When the first bromotyrosine derivative, the dienone 1, was isolated (3), the biogenetic precursor of this antibiotic was easily envisioned from tyrosine, which, after bromination, may be transformed into 3,5-dibromo-4-hydroxy-phenylacetaldehyde through a pathway already suggested in the literature (172). From the inspection of the structures, Minale et al. (19) and Moddy et al. (58) suggested that the Verongia bromo-metabolites are most likely biogenetically derived from 3,5-dibromo-tyrosine and, presumably, the central C4N2 and C5N2 chains of aerothionin (68) and homoaerothionin (69) are derived from ornithine and lysine, respectively.
A. BIOGENESIS OF ISOSPIROXAZOLINE
Based on the acid-catalyzed addition of methanol to 1,4-dimethylbenzene oxide which gave 4-methoxy-1,4-dimethyl-2,5-cyclohexadienol (173), Anderson and Faulkner (5) proposed that an analogous 1,4-addition of solvent to an arene oxide 286 could result in the formation of dienone 1 or the corresponding ketal 3 (5). Although arene oxides have been proposed as intermediates in the biosynthetic oxidation of aromatic compounds, there is no evidence of their existence as natural products. The coexistence of the lactone aeroplysinin-2 (18) with dienone 1 and the ketal 3 suggested that the imino-ether 287 should also be considered as a possible precursor of these alkaloids.
The spiro systems could arise in various ways, including nucleophilic attack by an oxime function on an arene oxide as shown in 288 (59). Following suggestions that nitriles may be derived in vivo from α-amino acids (174), the oxime 288 could be speculated as a precursor of the nitrile (±)-aeroplysinin-1 (14 and 17).
Quite surprisingly, the first attempt to demonstrate the conversion of tyrosine into bromotyrosine derivatives was unsuccessful (175). In that experiment, Verongia aerophoba failed to incorporate radioactivity from {U–14C}-l-tyrosine into aerothionin (68), aeroplysinin-1 (14) and the dienone 1. Inactive aerothionin (68) was also isolated when the sponge was fed with {U–14C}-l-ornithine and {CH3–14C}methionine. However, the sponge utilized these amino acids for the synthesis of fatty acids. A very slow rate of biosynthesis might account for these results.
By using liposome-enclosed precursors and modified culture conditions, which allowed the sponge to survive in the laboratory for as long as 2 weeks, Rinehart's group demonstrated the conversion of phenylalanine and tyrosine to the dienone 1, as well as the rearranged product dibromohomogetisamide (32), by the sponge Aplysina fistularis in 1981 (176). In addition to implying that the sponge can convert phenylalanine to tyrosine (177), the comparable radioactivity found in 1 and 32 supports the hypothesis (26) that 32 is formed from 1 via a skeletal rearrangement analogous to the mammalian catabolism of tyrosine to homogentisic acid. Although the enzymatic mechanism of the side chain migration is still unclear (178), the conversion of 4-hydroxy-2,5-cyclohexadienone-4-acetic acid to homogentisic acid in aqueous alkali has been demonstrated (179). Double labeling studies also revealed that the sponge can convert the side chain in phenylalanine to the acetamide side chain in 32 without deamination. In addition to corroborating the known occurrence of bromophenol nitriles and oximes in Verongid sponges, this work supported the following biosynthetic pathway:
Moreover, it has been demonstrated that α-oximino acids undergo facile dehydration/decarboxylation to give nitriles in vitro (180) and in vivo (181), that 1 was converted to 32 (182), and that (4-hydroxyphenyl)pyruvic acid oxime was isolated from Hymeniacidon sanguinea (183), further supporting the proposed mechanism.
Carney and Rinehart reported the results of additional feeding experiments in the marine sponge Aplysina fistularis in 1995 (184). {U–14C}-l-tyrosine, {U–14C}-l-3-bromotyrosine, and {U–14C}-l-3,5-dibromotyrosine were incorporated into both the dienone 1 and aeroplysinin-1 (14), and {methyl-14C}-methionine was specifically incorporated into the O-methyl group of aeroplysinin-1. In contrast to expectations, tyrosine was incorporated more efficiently than 3-bromotyrosine, which is in turn incorporated more efficiently than 3,5-dibromotyrosine. This may result from the bulky bromines interfering with the permeability of the precursors across cell membranes. It was surprising that {methyl-14C}-l-methyltyrosine, {methyl-14C}-l-3,5-dibromo-O-methyltyrosine, 3-bromo-4-hydroxybenzyl cyanide, and 3,5-dibromo-4-hydroxybenzyl cyanide were not incorporated into the dienone 1 and aeroplysinin-1 (14). Both 3-bromo-4-hydroxybenzyl cyanide and 3,5-dibromo-4-hydroxybenzyl cyanide have been identified as metabolites of A. fistularis (185), and their involvement as a potential precursor to the dienone 1 seems reasonable. The possible reasons for the lack of incorporation into 1 and 14, aside from the possibility of their not being part at the biosynthetic path, are poor transport of the halogenated nitriles across cell membranes, or poor solubility of the nitriles in sea water (184).
Although, from the above feeding experiments, it is clear that the Verongida bromotyrosine derivatives are biogenetically derived from tyrosine, it is still a matter of debate whether these alkaloids are produced via an arene oxide, or through phenol oxidative coupling (186). Based on the isolation of the racemic verongiaquinols, (±)-3-bromoverongiaquinol (10) and (±)-3-bromo-5-chloroverongiaquinol (11), and the low enantiomeric pure cavernicolins, 5-bromocavernicolin (24) (23) and 5-chlorocavernicolin (25), D'Ambrosio et al. (9) proposed a phenol oxidative (187) route based upon tyrosine precursors for their formation, since the proposal of an arene epoxide as a biogenetic precursor demands enantiomerically pure 11 and 10. In fact, natural products derived from phenol oxidations occur in nature in both optically active forms or as racemates, or even in nearly racemic forms (188). One way to rationalize the formation of the cavernicolins in full respect of the classical ortho–para orientation rules for the oxidative coupling of phenolic compounds (188) is to postulate a racemic or nearly racemic spirolactone 289 as the intermediate derived from the halotyrosine precursor (9). There is, in fact, ample precedent for cyclohexadienone spirolactone in oxidative couplings of phenols (188). Hydrolysis of the lactone, followed by conjugate attack of the amino acid N-atom and decarboxylative oxidation may then be seen to lead to the cavernicolins (9).
7-Bromocavernicolenone (30) and 7-chlorocavernicolenone (31) may be biogenetically derived from 5-bromodopa undergoing a similar pathway (24,25).
Anodic oxidation of 3′,5′-dibromo-4′-methoxyphenylalanine methyl ester led to the cavernicolin model compounds as four possible stereoisomers (189,190). This may constitute a new model for the biogenesis of the carvernicolins as an alternative to a spirolactone route from oxidation of amino-protected tyrosines
B. BIOGENESIS OF THE PSAMMAPLYSINS
Biogenesis of the psammaplysins may proceed through an oximino epoxide as shown in the scheme below (94). The epoxide ring opening leads to ring enlargement and results in the spiro[4.6]dioxazundecane. The benzene oxide-oxepin pathway, first adumbrated on theoretical grounds by Vogel and Günther, (191) was experimentally demonstrated through the biosynthesis of aranotin by Neuss et al. (192,193).
C. BIOSYNTHESIS OF PSAMMAPLIN TYPE OF COMPOUNDS
Crews et al. proposed a biogenetic pathway, based on the isolation of prep-sammaplin A (155), to rationalize the formation of psammaplin A (152) and other bromotyrosine derivatives isolated from Pseudoceratina purea by his group (110, 112). A straightforward dimerization of a rearranged cysteine (Cys′=HS–CH2–CH2–NHCOOMe) could generate prepsammaplin A (155). Condensation of the dimer 155 with either one or two functionalized tyrosine groups (Tyr′=the bromo oxime derivative) could rationalize the formation of psammaplins A (152), D (158), and H (162), respectively. Likewise, cysteine can lead to psammaplin F (160), which is envisioned to be the precursor of psammaplin E (159) and psammaplin G (161). Oxidation of psammaplin A (152) (possibly by a cytochrome P450 reaction) provides a connection to psammaplin J (164). The two esterified psammaplins D (158) and H (162) may be artifacts of the isolation derived from the acid formed via condensation of cysteine and tyrosine.
Similarly, condensation of the functional tyrosine (Tyr′) and the rearranged cysteine (Cys′) could generate psammaplins B (156), C (157), and I (163).
Modified tyrosine (Tyr′) is also the logical precursor of alkaloids 290 and 291.
V. Synthesis
The diverse and unique chemical structures, and the variety of biological activities of bromotyrosine derivatives, make them ideal synthetic targets for synthetic and natural products chemists. Studies on the synthesis of bromotyrosine can be traced back to 1912 (194). In former studies, degradation, chemical transformation to known compounds, or even total synthesis were conducted for the purpose of verifying the structures. Later studies were focused on the total synthesis of more complex structures, such as the spirocyclohexadienylisoxazoline and bastadin ring systems.
A. SYNTHESIS OF SMALL BROMOTYROSINE DERIVATIVES
Synthesis of functionalized cyclohexadienones, dienols, and dienediols was studied, which may be useful for the synthesis of some bromotyrosine derived dienones or ketals (195). Andersen and Faulkner reported the synthesis of aeroplysinin-1 (14). 3,5-Dibromo-2-hydroxy-4-methoxyphenylacetonitrile (292) was oxidized with excess lead tetraacetate in acetic acid at 25°C for 18 h to obtain the dienone 293 in 35% yield. Reduction of 293 with sodium borohydride in absolute ethanol at 0°C gave three products: isoaeroplysinin-1 (294) (40%), the corresponding monoacetate 295 (18%), and 2-deoxyaeroplysinin-1 (296) (22%). The failure to obtain a trans-diol was attributed to the influence of the acetoxy function. The dienone 293 was therefore transformed to the corresponding keto alcohol 297 by transesterification in methanol containing p-toluenesulfonic acid. Reduction of the keto alcohol 297 with sodium borohydride in absolute ethanol at 0°C for 10 min gave aeroplysinin-1 in 60% yield (14). This synthesis of aeroplysinin-1 (14) and isoaeroplysinin-1 (294) constitutes a novel approach to the synthesis of arene glycols and has the added advantage in that both cis- and trans- glycols can be prepared stereospecifically (196).
Ganem et al. synthesized ceratinamine (60), moloka'iamine (52), and mololipids (53), together with several analogues, for antifouling, anti-HIV and cytotoxicity studies (197–199).
Mololipids can be produced smoothly by the direct dualacylation of moloka'iamine (52) with myristoyl, stearoyl, and oleoyl imidazoles or the step-wise acylation of a protected precursor with different fatty acyl groups (198).
Hoshino et al. reported a convenient method to synthesize psammaplin A (152) (200). As for a synthetic strategy, direct coupling of the phenolic oxime-acid 300 with cystamine was considered, because 300 could be readily prepared from 3′-bromotyrosine. After several attempts to couple 300 directly with cystamine, a mixture of 300 (1.0 eq.) and free cystamine (0.5 eq.) in dioxane containing one equivalent of Et3N, DCC and N-hydroxyphthalimide was stirred at room temperature for 12 h to afford, after purification using column chromatography, psammaplin A (152) in 67% yield.
Inspired by the structure of psammaplin A (152), a combinatorial scrambling strategy for the construction of heterodimeric disulfide analogues was developed and applied to the construction of a 3828-membered library starting from 88 homodimeric disulfides (201). The disulfide motif was utilized as a readily exchangeable linkage (202), which would allow rapid construction of a heterodimeric analogue library suitable for defining structure–activity relationships (203). It is well-precedented that disulfide bonds will readily undergo facile exchange reactions with other disulfides in high yield under mild conditions (204). For example, if two homodimeric disulfides, A-SS-A and B-SS-B, are mixed under basic conditions in the presence of a suitable catalyst they will undergo rapid exchange reactions to afford a statistical mixture of three disulfides: A-SS-A, A-SSB, and B-SS-B in a ratio of 1:2:1, respectively. Hoshino's synthesis of psammaplin A (200) was modified as shown in the following scheme to synthesize 44 homodimeric psammaplin A analogues.
Amino acid homodimeric disulfide analogues were prepared using the following scheme.
Additional sulfonamide analogues and amide analogues were synthesized by standard techniques. Finally, 43 homodimeric disulfide building blocks were obtained from commercial sources in order to enhance the library's structural diversity. The disulfide exchange reaction in DMSO/H2O (3:1) by adding catalytic amounts of dithiothreitol was reproducible and led to a statistical mixture. Solution phase combinatorial synthesis of the psammaplin A (152) analogue library was conducted in 88 96-well plates. The disulfide products could be screened directly from the reaction mixture since the catalyst, dithiothreitol, and the byproduct trans-1,2-dithiane-4,5-diol did not show any detectable antibacterial activity at a concentration which was 50-fold greater than the concentration at which either of these compounds would be in the screen mixture. After screening, six structurally distinct psammaplin analogues from this library demonstrated higher antibacterial activity than psammaplin A (201).
B. SYNTHESIS OF SPIROCYCLOHEXADIENYLISOXAZOLINE
The unique spirocyclohexadienylisoxazoline ring system of the bromotyrosine derivatives, such as aerothionin (68), aerophobin-1 (69), and fistularin 3 (79), and their wide range of bioactivities make them attractive targets for synthesis. A number of studies on the formation of the spiroisoxazoline through intramolecular oxidative cyclization of a phenolic oxime have been reported (205). Forrester et al. examined the reactions of oxime 301 with different oxidizing reagents including lead tetra-acetate, potassium ferricyanide, silver oxide, sodium periodate, Fremy's salt, and manganese tris(acetylacetonate) (206, 207). Of these, the manganese reagent was found to be the most effective oxidant, and gave the spiroisoxazoline 302 in fair yields of 19–62%. Oxidation with lead tetra-acetate did give the corresponding spiroisoxazolines, but in a much lower yield of 10%, while potassium ferricyanide, silver oxide, sodium periodate, and Fremy's salt failed to effect cyclization of the phenolic oximes. In a subsequent study, the same researchers showed that oxidation of 301 (R=H, R1=CO2Me) with bromine water resulted in both electrophilic bromination and spirocyclization to give 302 (R=Br, R1=CO2Me) in 65% yield, while use of NBS with 301 (R = t-Bu, R1 = Me) gave 302 (R = t-Bu, R1 = Me) in 72% yield (208). Similarly, Boehlow et al. reported that addition of one equivalent of NBA to the oxime 301 (R = H, R1 = CO2Et) in THF at −60°C gave the monobromide 301 (R = 3-mono-Br, R1 = CO2Et, 60%). Addition of two equivalents of NBA to the oxime 301 (R = H, R1 = CO2Et) in THF or DMF at 0°C gave the dibromide 301 (R = 3,5-di-Br, R1 = CO2Et, 69%), and three equivalents at 0°C cleanly produced the spiroisoxazoline 302 (R = 3,5-di-Br, R1 = CO2Et, 74%) (209). Treatment of the oxime 301 (R = H, R1 = CO2Et) with Furia's bromoperoxidase enzyme mimic (210) of NH4VO3, H2O2, and KBr in water/chloroform resulted in the formation of both the monobromide 301 (R = 3-mono-Br, R1 = CO2Et) and dibromide 301 (R = 3,5-di-Br, R1 = CO2Et) without forming the spiroisoxazoline (209). Yamamura reported that oxidation of 301 (R = Br, R1 = CO2Me) with thallium (III) nitrate in methanol gave a mixture of products, which included 7–11% of 302 (R = Br, R1 = CO2Me) (211). Intriguingly, the same transformation could be carried out in quantitative yield by anodic oxidation (211). Kacan et al. reported phenyliodine (III) bis(trifluoroacetate) as an efficient reagent for the intramolecular oxidative cyclization. A series of oximes 301 (R = H, Br, R1 = Me, Et, CMe3, C6H5, CO2Et, 4-BrC6H4) reacted with phenyliodine (III) bis(trifluoroacetate) in acetonitrile at 0 °C smoothly and rapidly to give the corresponding isoxazolines 302 in good to excellent yield (212). Application of this procedure to an ortho-phenolic oxime resulted in the formation of the [4 + 2] dimer of the initially formed spiroisoxazoline.
Cyclization of the ortho-phenolic oxime has also been studied. Forrester et al. found that of all the oxidants they examined, only tetrabromocyclohexa-2,5-dienone would convert 303 (R = 2,4-di-tBu, R1 = Me) into 304 (R = 2,4-di-tBu, R1 = Me) in 20% yield (208). An equally low yield of 27% was obtained by Yamamura, on oxidation of 303 (R = 2,4-di-Br, 3-OMe, R1 = COOMe) to 304 (R = 2,4-di-Br, 3-OMe, R1 = COOMe) with thallium trifluoroacetate in trifluoroacetic acid (213). Application of the above phenyliodine (III) bis(trifluoroacetate) to the ortho-phenolic oxime resulted in the direct formation of the [4 + 2] dimer of the initially formed spiroisoxazoline (212). Independently, Murakata et al. found that the hypervalent iodine compound phenyliodium diacetate (PIDA) was an efficient cyclization reagent of o-phenolic oxime-esters and oxime-amides (214). The cyclization of various oximes 303 (R = mono-, di-bromo, H, OMe, R1 = OMe, OtBu, NH(CH2)3OMe) in acetonitrile at 0°C proceeded smoothly to afford the spiroisoxazoline in good yield.
Asymmetric oxidative cyclization of o-phenolic oxime-esters was also studied by Murakata et al. (215). A novel, optically active tertiary alcohol (–)-305 was synthesized as a chiral auxiliary. The ortho-phenolic oxime 306 was first transformed to the oxime ester 307 in three steps (see the following scheme). The reaction of 307 with PhIO in the presence of camphor sulfonic acid (CSA) in CH2Cl2 at −78°C proceeded smoothly to afford the spiroisoxazoline 308 in 83% yield with 70–80% estimated diastereomeric excess by the 1H NMR spectrum. The chiral auxiliary in 308 was removed by treatment with TFA at room temperature. Methylation using DCC and MeOH gave methyl ester (–)-309 ([α]D −56.4°) in 74% ee with the S-configuration. Optically active spiroisoxazoline 309 was reduced with Zn(BH4)2 to give cyclohexadienylisoxazoline (+)-310, amidation of which with butanediamine afforded (+)-aerothionin ([α]D + 166°) with the S-configuration.
With the key spiroisoxazoline synthon in hand, total synthesis of some spirocyclohexadienylisoxazoline bromotyrosine derivatives was achieved. Forrester et al. reported that reducing tetradehydroaerothionin using NaBH4 did not yield the natural trans, trans-aerothionin, but cis,cis-aerothionin (216). Yamamura et al. reduced the spiroisoxazoline 309 using excess Zn(BH4)2 in CH2Cl2–Et2O (3:2) to give the corresponding cis- and trans-isomers 310 in 29% and 40% yields, which were easily separated on preparative TLC (217, 213). Mixing trans- 310 with 1,4-butanediamine, 1,5-pentanediamine, or histamine, at room temperature overnight afforded aerothionin (68), homoaerothionin (69), and aerophobin-1 (113) in 18, 4.4, and 82% yields, respectively.
Wasserman et al. developed an efficient methodology for the formation of key α-keto amido residues common to many members of this family of natural products (218–221). Using this method, the α,β-diketo nitrile 319 was synthesized from the oxidation of the ylide 318 (222). The generation of 318 from 2-hydroxy-4-methoxyacetophenone (311) was accomplished according to the sequence outlined in the following scheme by protecting the phenolic hydroxyl as the benzyl ether 312, rearrangement with Ti(NO3)3 to 313, deprotection to 314, and bromination to form dibromide 315. Following reprotection to 316 and hydrolysis of the ester to the acid 317, coupling with Ph3P=CHCN in the presence of EDCI yielded 318. After the ylide 318 was oxidized by O3 to the labile diketonitrile 319, generation of the bis-α-ketoamide 320 took place on treatment of 319 with 1,4-diaminobutane in CH2Cl2. Conversion of 320 to the oxime 321 and deprotection provided the substrate 322, which underwent the oxidative cyclization with tetrabromocyclohexadienone to afford 323 in 71% yield. Reduction of 323 with NaCNBH3 in TFA gave the desired trans,trans-aerothionin (68) (222).
Murakata et al. synthesized the cyclohexadienonespiroisoxazoline amides 324, which could be useful as intermediates for the synthesis of araplysillin I (99) and II (100) (223). The synthesis of these metabolites was efficiently achieved using two routes. The first route is direct cyclization of o-phenolic oxime-amide 325 with phenyliodonium diacetate (PIDA) to produce 324. The second is the amidation of the cyclohexadienonespiroisoxazoline-acid 326 to afford 324.
Based on the enantioselective cycloaddition of the optically pure cyclopentadiene (4) with spirolactone (224,225), Winterfeldt et al. synthesized enantiomerically pure spiroxazoline 332, which is the crucial intermediate en route to the agelorins (87, 88) and their analogues (226). Face selectivity was successfully achieved in the cycloaddition of cyclopentadiene 327 and spiroxazoline 328 to form the enantiopure 329. This may be due to the lower spatial demand of an oxygen atom than a methylene group. Flash-hydroxylation of 329 with ruthenium tetraoxide afforded cis-diol 329a, which underwent pyrolysis after it was transformed into the acetal 330 to generate the spirocyclohexenone 331. Subsequently, hydrolysis afforded the enantiopure intermediate 332, representing the agelorins (87, 88) chromophore (226).
In a continuing report, several enantiopure spiroisoxazoline amides were prepared using the above method and tested on an isoxazoline-splitting enzyme, which is involved in an injury-induced defense reaction of the sponge Aplysina cauliformis (227). The results indicated that the bromoatoms in the cyclohexenone moiety are important for enzyme binding, while the presence of the N–H bond of a monoalkylamide turned out to be mandatory for ring fission.
Boehlow et al. summarized the approach to the synthesis of some spirocyclohexadienylisoxazoline and oxime bromotyrosine derivatives and synthesized two oxime bromotyrosine derivatives verongamine (139) and purealidin N (150) (205).
C. SYNTHESIS OF BASTADINS
Yamamura's group reported the total synthesis of bastadins 1, 2, 3, and 6 (228,229,230). Biomimetic oxidation of methyl 3,5-dibromo-4-hydroxyphenylpyruvate oxime (333) was carried out using Ti(NO3)3 to afford 7–11% of spiroisoxazole 339 and 37–44% of the dimeric spiroisoxazole 335. Reduction of 335 gave the biphenyl ether 340, as a key intermediate for bastadins-2 synthesis, in almost quantitative yield (228). Similarly, on oxidation with thallium trifluoroacetate in trifluoroacetic acid containing a small amount of CH2Cl2, methyl 3-bromo-4-hydroxyphenylpyruvate oxime (334) was converted into three spiroisoxazoles [339 (7%), 336 (6%), and 337 (5%)] and a plausible compound 338, which was directly reduced with Zn powder to the biphenyl compound 342 in 8% overall yield. Zn reduction of 336 afforded the biphenyl ether 341 in 48% yield. The biphenyl ether (341, 340, and 342) was reacted with excess amounts of 3-bromotyramine p-methoxybenzyl ether 346 (60°, 3–4 days) to give the desirable diamide (343, 344, and, 345), which gave the corresponding bastadins-1, −2, and −3 after deprotection (230).
Bastadin-6 can be produced by phenolic oxidation of the corresponding acyclic precursor, bastadin-2 (229). Bis-p-methoxybenzyl bastadin-2 (344) was converted to tribenzyl bastadin-2 (347), which was further treated with Br2 to afford dibromobastadin-2 tribenzyl ether (348). Under the same procedure, 348 was oxidized with Ti(NO3)3 to afford two macrocyclic dienones (349 and 350). Compound 349 was reduced with Zn to give the tribenzyl ether 351. Finally, 351 was subjected to hydrogenlysis, using Pd black to afford bastadin-6 (229). Bastadin-6 trimethyl ether was produced in a similar procedure (231).
Guo et al. developed a chemoenzymatic strategy for the synthesis of bastadins-2, −3, and −6 (232). The requisite dimeric dityrosine and isodityrosine were successfully prepared by C–C and C–O oxidative phenolic coupling of mono- and dihalogenated derivatives of tyrosine and tyramine using horseradish (233) and soybean peroxidases. By carefully controlling the experimental conditions, the required synthons were prepared in synthetically useful yields without the exhaustive protection and deprotection of the sensitive functional groups. This mode of oxidative coupling may represent the biogenetic synthetic route for bastadins, which is, the isodityrosine and isodityromine are formed via the coupling of dihalogenated tyrosine and tyramine derivatives.
Couladouros and Moutsos reported a general synthetic route for the synthesis of the eastern and western parts of bastadins, which can be used for construction of bastadins 4–16 (234–236). The brominated biaryl ethers are synthesized using the iodonium salt method. The eastern segment 352 was synthesized within 18 steps in 15.5% overall yield. The western segment 353 and 354, which can be used to construct bastadins-8 (212), −10 (214), −12 (216), and −17 (221), was synthesized in 13 steps (see the following scheme).
In order to utilize the hydroxyl group as a general precursor for all functionalities present at C-5 and C-6 of the bastadins, 355 was transformed after desilylation either to alkene 356 or alkane 357 using DBU or NaBH4, respectively, providing the necessary intermediates for the construction of bastadins-4 to −7 (208–211), −9 (213), −11 (215), and −14 to −16 (218–220).
Having the western and eastern parts of the molecule, the synthesis of the cyclic skeleton of bastadins was attempted. In order to study the efficiency of macrolactamization, four protected amino acids (358, 359, 360, and 361) were used as advanced intermediates. Several experiments revealed that these amino acids could be synthesized in good yields through the coupling of imidazolide 364 with primary amines 353 or 354. Subsequently, these open bastadin precursors could be selectively brominated at Y2 position with TBS (237). The terminal amino and carboxylic acid were unmasked followed by in situ cyclization using EDC and N-hydroxybenzotriazole to afford a mixture of bastadin precursor 362 and its desilylated analog 363. The mixture was treated with TBAF providing compound 393 in 67% yield from the precursor 361. Compound 363 is protected bastadin-12, which possesses the unsymmetrical bromination pattern and asymmetric hydroxyl group.
Bailey and Molinski presented a convergent strategy that allows the rapid assembly of isodityrosine-isodityrosine cyclic analogues of bastadin-5 in four steps from a common precursor, dialdehyde 365, by exploiting a tandem Erlenmeyer condensation – macrolactamization (238). Dialdehyde 365, prepared by Ullmann coupling of commercially available 3-hydroxy-4-methoxybenzaldehyde with 4-bromobenzaldehyde, was treated with N-benzoylglycine to obtain 366. Diamine 367 was readily prepared from 365 by double Henry condensation-elimination followed by reduction of the resultant bis-β-nitrostyrene. Heating 366 and 367 in pyridine (50°C) resulted in the regioisomeric 28-membered macrocycles bastarane 368 and isobastarane 369 as a 1:1 mixture with a 30% combined yield. Hydrogenation of the mixture of 368 and 369 provides the isodityrosine cyclic peptide 370 as a mixture of isomers in 20–26% overall yield.
In order to gain control of the regiochemistry of the ring closure, Bailey and Molinski proposed an intermolecular SNAr strategy to synthesize the unsymmetrical bastadin-5 analog 382 (239). Bromination of catechol 371 followed by two sequential directed alkylations gave the differentially protected benzaldehyde 372, which was converted into phenylacetonitrile 373 in three steps. Reduction of 373 and simultaneous removal of the O-allyl protecting group was achieved in one step to give the expected phenethylamine, which was immediately protected as the N-Boc compound 374. Intermolecular SNAr substitution of 374 with 3-bromo-4-fluorobenzaldehyde (375) afforded aldehyde 376 in high yield (88%). Repetition of the homologation sequence on 376 (reduction-halide displacement-cyanide displacement) afforded 377 followed by nitrile reduction to give the monoprotected western diamine 378 (55% from 375).
The eastern hemisphere intermediate 379 was also prepared from 372, using a Horner–Emmons strategy for step-wise extension of each carboxyaldehyde group to the corresponding α-ketoxime (see the following scheme). The western diamine 378 and the eastern acid 379 were coupled to give the protected teracycle 380 in 78% yield. Compound 380 was transformed to the activated pentafluorophenol (Pfp) ester 381, which underwent the macrolactamization upon removal of the N-Boc group to give the product 382. The overall yield of 382 from 371 was 16% (16 steps).
Molinski et al. also reported the reduction of bastadin-4 (208) to bastadin-5 (209) using cationic hydrogenation (Et3SiH, TFA, 60%) (240). Specific deuteration at H-5 of bastadin-5 was conducted following this method.
Leone-Stumpf and Lindel synthesized the ruthenium labeled western (383) and eastern (384) diaryl ether and tripeptide (385) of bastadin-5, and for the first time established an HPLC method using aminopropyl-functionalized silica to purify the [CpRu]+ complexes (241,242).
The free amino group of 4-O-methyldopamine and 4-O-methyltyramine react as nucleophiles in a chemoselective manner with [RuCp]+-complexed p-substituted chloroarenes. As a consequence, it is not necessary to use protecting groups for the synthesis of peptides with alternating amide and diaryl ether bonds.
The total synthesis of polycitone A (280) and B (281) (243), and the cyclodepsipeptides, jaspamide (259) (244), geodiamolide A (260) (245), and geodiamolide B (261) (246) were also reported. Since these are not typical bromotyrosine derivatives, the details of the syntheses are not included here.
VI. Bioactivity
Bromotyrosine alkaloids provide a unique diversity in chemical structure and in bioactivity. Since the first bromotyrosine derivative, the dienone 1, was isolated as an antibiotic, a large number of bromotyrosine derivatives have been found to have diverse activities, which include antibacterial, antifungal, anticancer, antiviral, antifouling, Na,KATPase inhibitor, etc. The majority of the dibromotyrosine-derived natural products from Verongida have been reported to possess significant antimicrobial and cytotoxic activity (247). The presence of antibacterial compounds in marine sponges has been reported as a general phenomenon (248), and has been suggested to reflect a defensive strategy of these sedentary, filter-feeding animals (249).
A. ANTIMICROBIAL ACTIVITY
Bromotyrosine derivatives were first isolated in the process of searching for antibiotics from marine sources. The dienone 1, aeroplysinin-1 (14), and aerothionin (68) are the earliest antimicrobial bromotyrosine derivatives (2,14,58). The antibacterial and antifungal activities of the bromotyrosine alkaloids are summarized in Table IV and Table V. Bromotyrosine derivatives exhibited a broad antimicrobial spectrum, which include Gram-positive bacteria such as Streptococcus faecalis, Streptococcus pyogenes, Staphylococcus aureus, Bacillus subtilis, Micrococcus luteus, Sarcina lutea, Gram-negative bacteria such as Escherichia coli, Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Alcaligena faecalis, and fungi such as Candida albicans.
TABLE IV.
Antibacterial Activity of Bromotyrosine Derivatives.
| Alkaloids | Antimicrobial activitya | Ref. |
|---|---|---|
| 3-Bromoverongiaquinol (10) | Active against S. f., B. s.; not active against A. f., P. v. | 9 |
| 3-Bromo-5-chloroverongiaquinol (11) | Active against S. l., A. f., P. v. | 9 |
| (–)-Aeroplysinin-1 (17) | MIC 20–100 μg/mL against S. a., B. s., M. l., E. c., P. a. | 16 |
| (+)-Aeroplysinin-1 (14) | MIC 20–100 μg/mL against S. a., B. s., M. l., E. c., P. a. | 16 |
| 31 | ||
| 7-Bromocavernicolenone (30) | Active against P. c.; not active against B. s. | 24 |
| 38 | 3000 μg/disk shows inhibition zone against E. c. 18–22 mm | 31 |
| Aeroplysinin-2 (18) | 3000 μg/disk shows inhibition zone against E. c. <15 mm | 31 |
| Moloka'iamine (52) | Inhibit the growth of B. m., P. n., A. m., V. a., F. m. at 10 μg /disk | 49 |
| Aplysamine-1 (54) | Not active against S. a., B. s., S. l., E. c., S. sp. | 44 |
| Aplysamine 2 (172) | Not active against S. a., B. s., S. l., E. c., S. sp. | 44 |
| Ceratinamine (60) | Inhibit the growth of B. m., P. n., A. m., V. a., F. m. at 10 μg/disk | 49 |
| Aerothionin (68) | Inhibit the growth of S. a. at 100 μg/disk, B. s. at 50 μg/disk | 63 |
| Homoaerothionin (69) | Inhibit the growth of S. a. at 100 μg/disk, B. s. at 50 μg/disk | 63 |
| 11,19-Dideoxy-fistularin 3 (83) | Inhibit the growth of S. a. at 25 μg/disk, B. s. at 10 μg/disk | 63 |
| 11-Hydroxyaerothionin (71) | Inhibit the growth of S. a. at 100 μg/disk, B. s. at 50 μg/disk | 63 |
| 11-Oxoaerothionin (72) | MIC 30 μg/mL against S. a., P. a.; 10 μg/mL against E. c. | 64 |
| 11-Epi-fistularin-3 (81) | Active against B. s., M. l.; not active against E. c. | 70 |
| Agelorin A (87) | Active against B. s., M. l.; not active against E. c. | 70 |
| Agelorin B (88) | Active against B. s., M. l.; not active against E. c. | 70 |
| Zamamistatin (91) | Active against R. s. | 75 |
| Aplysinamisine I (120) | Inhibit the growth of S. a., E. c., P. a. at 50–100 μg/disk | 83 |
| Aplysinamisine II (124) | Inhibit the growth of S. a., E. c., P. a. at 50–100 μg/disk | 83 |
| Aplysinamisine III (102) | Inhibit the growth of S. a., E. c., P. a. at 50–100 μg/disk | 83 |
| Purealidin B (109) | MIC 62.5 μg/mL against S. a., 15.6 μg/mL against B. s., 3.9 μg/mL against S. l. | 84 |
| Purealidin C (192) | MIC 62.5 μg/mL against S. a., 15.6 μg/mL against S. l. | 84 |
| Araplysillin-I (99) | 250 μg/disk shows inhibition zone against S. a. 12 mm | 81 |
| Araplysillin-II (100) | 250 μg/disk shows inhibition zone against S. a. 7 mm | 81 |
| Anomoian A (195) | Inhibit the growth of S. a. at 10 μg/disk, B. s. at 5 μg/disk | 124 |
| Bromochelonin B (257) | Inhibit the growth of B. s. at 100 μg/disk | 154 |
| Purpuramine K (190) | 25 μg/disk shows inhibition zone against S. a. 10, P. a. 10; B. s. 8; C. v. 7; K. a. 10 mm | 122 |
| Purpuramine L (191) | 25 μg/disk shows inhibition zone against S. a., B. s. 14; P. a. 16; B. sp 12 mm | 122 |
| Psammaplin A (152) | Active against S. a., B. s. | 109 |
| Bisaprasin (154) | Active against S. a., B. s. | 109 |
| Psammaplin D (158) | Inhibit the growth of S. a., T. m. at 100 μg/disk. | 110 |
| Bastadin-1 (204) | MIC: S. a. 6.25–12.5, E. c. 100, N. g. 50–100, E. f. 12.5–25 μg/disk | 41 |
| Bastadin-2 (205) | MIC: S. a. 6.25–12.5, N. g. 50–100, E. f. 50–100 μg/disk | 41 |
| Bastadin-3 (206) | MIC: S. a. 1.56–3.12, N. g. 0.78–1.56, E. f. 3.12–6.25 μg/disk | 41 |
| Bastadin-4 (208) | MIC: S. a. 12.5–25, N. g. 12.5–25, E. f. 50–100 μg/disk | 41 |
| Bastadin-5 (209) | MIC: S. a. 12.5–25, N. g. 50–100, E. f. 50–100 μg/disk | 41 |
| Bastadin-6 (210) | MIC: S. a. 6.25–12.5, E. f. 12.5–25 μg/disk | 41 |
| 207 | MIC: N. g. 50–100 μg/disk | 41 |
| Hemibastadin 1 (231) | MIC: S. a. 1.56–3.12, N. g. 12.5–25, E. f. 50–100 μg/disk | 41 |
| 1′-(Methyloxy) hemibastadins 1 (237) | MIC: S. a. 6.25–12.5, N. g. 6.25–12.5 μg/disk | 41 |
| Hemibastadins 2 and 3 (232, 233) | MIC: S. a. 6.25–12.5, N. g. 6.25–12.5, E. f. 50–100 μg/disk | 41 |
| Hemibastadinol 1 (234) | MIC: N. g. 50–100 μg/disk | 41 |
| Hemibastadinols 2 and 3 (235, 236) | MIC: N. g. 50–100 μg/disk | 41 |
| Bastadin-13 (217) | MIC 6 μg/mL against B. s. | 41 |
Streptococcus faecalis (S. f.); Staphylococcus aureus (S. a.); Bacillus subtilis (B. s.); Micrococcus luteus (M. l.); Sarcina lutea (S. l.); Bacillus marinus (B. m.); Bacillus sphaericus (B. sp.). Escherichia coli (E. c.); Pseudomonas aeruginosa (P. a.); Neisseria gonorrhoeae (N. g.); Alcaligena faecalis (A. f.); Chromobacterium violaceum (C. v.); Klebsiella aerogenes (K. a.); Proteus vulgaris (P. v.); Vibrio alginolyticus (V. a.); Pseudomonas cichorii (P. c.); Pseudomonas nautical (P. n.); Alteromonas macleodii (A. m.); Flavobacterium marinotypicum (F. m.); Rhodospirillum salexigens (R. s.); Serratia sp. (S. sp.); Escherichia faecalis (E. f.).
TABLE V.
Antifungal activity of Bromotyrosine Derivatives.
| Alkaloids | Antifungal activity | Ref. |
|---|---|---|
| Aplysamine-1 (54) | No activity | 44 |
| Aplysamine-2 (172) | ||
| Ceratinamine (60) | No activity against Candida albicans, Penicillium chrysogenum, Mortierelia ramanniana at 10 μg/ disk | 49 |
| Aerothionin (68) | Active against C. albicans at 50 μg/ disk | 63 |
| Homoaerothionin (69) | Active against C. albicans at 50 μg/ disk | 63 |
| 11-Hydroxyaerothionin (71) | Active against C. albicans at 50 μg/ disk | 63 |
| 11,19-Dideoxyfistularin-3 (83) | Active against C. albicans at 25 μg/ disk | 63 |
| 77 | MIC 64 μg/ml on Criptococcus neoformans ATCC90113 | 67 |
| 11-Epi-fistularin-3 (81) | Not active toward Penicillium oxalicum | 70 |
| Agelorin A (87) | Not active toward Penicillium oxalicum | 70 |
| Agelorin B (88) | Not active toward Penicillium oxalicum | 70 |
| Purealidin C (192) | Modest activity against C. albicans, Crypotococcus neoformans, and Paecilomyces variotii | 84 |
| Anomoian A (195) | Inhibit the growth of C. albicans at 25 μg/disk | 124 |
| Geodiamolide A (260) | Active against C. albicans (MIC: 31.3 μg/ml) | 158 |
| Geodiamolide B (261) | Active against C. albicans (MIC: 31.3 μg/ml) | 158 |
| Psammaplin D (158) | Inhibit the growth of Trichopyhton mentagrophytes at 100 μg/disk. | 110 |
The active compounds distribute in all of the structural subclasses. The simple bromotyrosine derivatives that were reported to possess antibacterial activity include the dienone 1, aeroplysinin-1 (14), aeroplysinin-2 (18), 3-bromoverongiaquinol (10), 3-bromo-5-chloroverongiaquinol (11), aplysamine 1 (54), moloka'iamine (52), ceratina-mine (60), 7-bromocavernicolenone (30), 38, and 51. The first bromotyrosine derivative, the dienone 1, was reported as a broad-spectrum antibiotic (2). Both dextrorotatory and levorotary aeroplysinin-1 are active against Gram-positive and Gram-negative bacteria (14,16). The MICs (MBCs) of the dienone 1 and aeroplysinin-1 (14) ranged from 12.5–25 μg/mL against Bacillus subtilis, Staphylococcus aureus, and Escherichia coli (250). (+)-Aeroplysinin-1 showed greater than 65% inhibition of the growth of phytopathogenic fungus Phytophthora infestans (potatoes late blight) (251).
Although the bis-spirocyclohexadienylisoxazoline bromotyrosine derivatives, aerothionin (68), homoaerothionin (69), 11-hydroxyaerothionin (71), and 11,19-dideoxyfistularin 3 (83), all had in vitro antimicrobial activity, 11,19-dideoxyfistularin 3 is the most active. Aerothionin (68), homoaerothionin (69), and 11-hydroxyaerothionin (71) all inhibited the growth of Staphylococcus aureus at 100 μg/disk, Bacillus subtilis at 50 μg/disk, and Candida albicans at 50 μg/disk, whereas 11,19-dideoxyfistularin 3 (83) inhibited the growth of S. aureus at 25 μg/disk, B. subtilis at 10 μg/disk, and C. albicans at 25 μg/disk (63). Aerothionin (68) was reported to be active against a group of monoresistant variants of Mycobacterium tuberculosis H37Rv at 12.5 μg/mL (252). It was also active against a group of multidrug resistant TB clinical isolates with MIC values between 6.5 and 25 μg/mL and active against three of nine nontuberculosis mycobacteria (252).
Both 11-oxo-12-hydroxyaerothionin (73) and 11-hydroxyaerothionin (71) induced 60 and 70% inhibition, respectively, of Mycobacterium tuberculosis growth at 12.5 μg/mL, while 11-oxoaerothionin (72) induced no inhibition at all (253). Single spirocyclohexadienylisoxazoline bromotyrosine derivatives, aplysinamisine I (120), aplysinamisine Π (124), aplysinamisine III (102), purealidin B (109), araplysillin-I (99), and araplysillin-II (100), showed moderate antimicrobial activity (81,83,84). Some of the oxime type bromotyrosine derivatives showed potent antimicrobial activity. Anomoian A (195), a reduced oxime, exhibited strong antimicrobial activity against Staphylococcus aureus at 10 μg/disk, Bacillus subtilis at 5 μg/disk, and Candida albicans at 25 μg/disk.
Purpuramine L (191) is highly active against Staphylococcus aureus, Bacillus subtilis, and Chromobacterium violaceum, moderately against Bacillus sphaericus, K. aerogenes, and P. aeruginosa (122). Purpuramine K (190) is moderately active against all the organisms. Interestingly, Gram-positive bacteria are highly susceptible to purpuramine L when compared to the positive control kanamycin (122). Psammaplin A (152) was found to possess antimicrobial activity (109,254,255). It also selectively inhibited the growth of methicillin-resistant Staphylococcus aureus (MRSA) (256). The minimal inhibitory concentration of psammaplin A against twenty-one MRSAs ranged from 0.78 to 6.25 μg/mL while that of ciprofloxacin was 0.39–3.13 μg/mL. Psammaplin A (152) could not bind to the penicillin binding protein, but inhibited the DNA synthesis and the DNA gyrase activity with the respective 50% (DNA synthesis) and 100% (DNA gyrase) inhibitory concentrations of 2.83 and 100 μg/mL. These results indicated that psammaplin A inhibited the growth of bacteria probably by inhibiting DNA gyrase (256). Since the significant antimicrobial activity of psammaplin A (152) was disclosed, the antibacterial activities against 40 drug-resistant Staphylococcus aureus strains were determined for psammaplin D (158), 169, bromopsammaplin A (170), bispsammaplin A (171), and bisaprasin (154) (117). Alkaloid 169 showed the highest potency, which is higher than that of meropenem against several strains (see Table VI.) (117). Bromopsammaplin A (170) and bisaprasin (154) exhibited strong to moderate activity against all these strains, while psammaplin D (158) and bispsammaplin A (171) did not show significant activity against all strains.
TABLE VI.
Antibacterial Activity against Different Staphylococcus Strainsa (MIC, μg/mL).
| Strains | 158 | 169 | 170 | 171 | 154 | Meropenem |
|---|---|---|---|---|---|---|
| S. aureus KIST 1b | > 25.0 | 3.1 | 12.5 | > 50.0 | 12.5 | 25.0 |
| S. aureus KIST 2b | > 25.0 | 3.1 | 12.5 | > 50.0 | 6.3 | 3.1 |
| S. aureus KIST 3b | > 25.0 | 6.3 | 12.5 | > 50.0 | 6.3 | 50.0 |
| S. aureus KIST 4b | > 25.0 | 3.1 | 12.5 | > 50.0 | 6.3 | 50.0 |
| S. aureus 003c | > 25.0 | 3.1 | 12.5 | > 50.0 | 12.5 | 3.1 |
| S. aureus 004c | > 25.0 | 3.1 | 12.5 | > 50.0 | 6.3 | 3.1 |
| S. aureus Y-80-12-1109d | > 25.0 | 6.3 | 12.5 | > 50.0 | 50.0 | 50.0 |
| S. aureus Y-80-12-1999d | > 25.0 | 6.3 | 12.5 | > 50.0 | 50.0 | 50.0 |
| S. aureus Y-80-12-844d | > 25.0 | 25.0 | 25.0 | > 50.0 | 25.0 | 50.0 |
| S. epidermidis 178e | > 25.0 | 0.8 | 3.2 | > 50.0 | 6.3 | 0.8 |
| S. epidermidis 291e | > 25.0 | 0.8 | 3.2 | > 50.0 | 12.5 | 1.6 |
A total of 40 strains were employed for testing, and only the strains to which 169 exhibited equipotency to or higher potency than meropenem were registered.
Strains were obtained from KIST (Korea Institute of Science and Technology).
Strains were obtained from LG Chemical, Korea.
Strains were obtained from Yon-Sei Medical Center, Korea.
Ofloxacin-resistant strains.
Bastadins-1 to- 7 (204–211) were isolated because of the potent activity against Gram-positive and Gram-negative bacteria, but data were not provided (129). Bastadin-13 showed a minimum inhibitory concentration of 6 μg/mL against Bacillus subtilis, while 34-sulfatobastadin-13 showed no activity at 50 μg/mL, suggesting the free phenolic group at C-34 is required for the antimicrobial activity (141). Pettit et al. evaluated the ability of bastadins-1 to 6 (204–210), hemibastadins (231–233, 237), hemibastadinols (234, 235, 236), and amide 51 to inhibit growth of Gram-positive bacteria, Gram-negative bacteria, and two fungi (Table IV) (41). Except for bastadin-6, all of these alkaloids inhibited the growth of the Gram-negative pathogen Neisseria gonorrhoeae. Most of the compounds also inhibited the growth of the Gram-positive opportunists Enterococcus faecalis and Staphylococcus aureus. At up to 100 μg/disk, these alkaloids exhibited no antimicrobial activity against Esherichia coli or the fungi Candida albicans and Cryptococcus neoformans (41).
Antibiotic activity of sponge metabolites has frequently been demonstrated in the past (257–259). However, most of these studies were conducted with terrestrial rather than marine bacteria. For example, antibiotic activity of aeroplysinin-1 (14) and dienone 1 was proven using terrestrial Gram-positive and Gram-negative bacteria (250). Proksch et al. confirmed this activity using several marine bacteria such as Alteromonas, Moraxella, or Vibrio sp. (see Table VII) (260). The EC50 values of aeroplysinin-1 (14) and the dienone 1 towards Photobacterium phophoreum, which is a well-established model for the analysis of water-borne toxins (261,262), are comparable to those of pentachlorophenol (EC50: 1.3 μM), lead (EC50: 1.9 mM) or cadmium ions (EC50: 71 μM) which are known for their pronounced toxicity. At the same time, aeroplysinin-1 and the dienone 1 are also active against microalgae (260).
TABLE VII.
Antibiotic Activity against Several Marine Bacteria.
| Diameter of inhibitory zone (mm) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Compounds | Isofistularin-3 (80) | Aerophobin-2 (114) | Aeroplysinin-1 (14) | Dienone (1) | ||||
| Dose (μg/disk) | 100 | 100 | 100 | 50 | 5 | 100 | 50 | 5 |
| Alteromonas sp.* NCIMB 224 | – | – | 11 | 7 | – | 10 | 9 | – |
| Cytophage/Flexibacter sp.* NCIMB 251 | – | – | 30 | n.d. | n.d. | 28 | n.d. | n.d. |
| Moraxella sp.* NCIMB 308 | – | – | 22 | 20 | – | 19 | 16 | 7 |
| Pseudomonas fluorescens* NCIMB 129 | – | – | 8 | – | – | 9 | 7 | 7 |
| Serratia plymuthica * | – | – | 11 | 8 | – | 14 | 11 | 7 |
| Vibrio sp.* | – | – | 28 | 24 | 12 | 24 | 21 | 8 |
| Vibrioanguillarum* NCIMB 407 | – | – | 27 | 24 | 11 | 34 | 21 | 7 |
| Plancoccus citraus+ NCIMB 1495 | – | – | 14 | n.d. | n.d. | 14 | n.d. | n.d. |
n.d.: not determined
Gram positive bacteria
Gram negative bacteria.
B. ANTICANCER ACTIVITY
Anticancer active bromotyrosine derivatives are summarized in Table VIII. Many of the bromotyrosine derivatives exhibited cytotoxicity, emphasizing the ecological relevance of this type of compounds in the sponge's chemical defense. The small molecules, aeroplysinin-1 (14) and the dienone 1, which are from the enzymatic degradation of larger bromotyrosine derivatives such as isofistularin-3 (80), aerophobin-2 (114), fistularin-1 (94), and other alkaloids, are more toxic than the parent compounds, suggesting a wound defense reaction (250,263,264).
TABLE VIII.
Cytotoxicity of Bromotyrosine Derivatives
| Alkaloids | Cytotoxicity | Ref. |
|---|---|---|
| Psammaplin A (152) | IC50: K562 0.4 mM. | 114 |
| IC50: P388 0.3 μg/mL; | 108 | |
| ED50: A549 0.57; SK-OV-3 0.14; SK-MEL-2 0.13; XF498 0.57; HCT15 0.68 μg/mL | 117 | |
| Psammaplin D (158) | ED50: A549 0.80; SK-OV-3 0.17; SK-MEL-2 0.20; XF498 0.60; HCT15 1.23 μg/mL | 117 |
| Bromopsammaplin A (170) | ED50: A549 1.34; SK-OV-3 1.38; SK-MEL-2 0.90; XF498 0.92; HCT15 3.31 μg/mL | 117 |
| Bispsammaplin A (171) | ED50: A549 1.53; SK-OV-3 1.52; SK-MEL-2 1.02; XF498 1.10; HCT15 3.35 μg/mL | 117 |
| Bisaprasin (154) | ED50: A549 3.40; SK-OV-3 2.78; SK-MEL-2 2.94; XF498 2.44; HCT15 6.0 μg/mL | 117 |
| Psammaplin A1 (165) | K562: LC50 1.9 mM | 114 |
| Psammaplin A2 (166) | K562: LC50 4.2 mM | 114 |
| Aplysinellin A (167) | K562: LC50 10.7 mM | 114 |
| Aplysinellin B (168) | K562: LC50 7.1 mM | 114 |
| Moloka'iamine (52) | MIC: KB 50 μg/mL; LOVO 10 μg/mL; | 42 |
| IC50: P-388 5 μg/mL; A-549 10 μg/mL; HT-29 5 μg/mL; CV-1 10 μg/mL | 49 | |
| Mololipids (53) | > 100 μM (–) | 43 |
| Ceratinamine (60) | P388 IC50 3.4 μg/mL | 49,98 |
| N,N,N-Trimethyl-dibromotyramine (47) | P388 ED50 20 μg/mL | 37 |
| 11-Oxoaerothionin (72) | HCT 116 (0.01-0.1 μg/mL) | 64 |
| 11-Epi-fistularin-3 (81) | IC50: KB > 20; BCI 5.9; ZR-75-1 4.5 μg/mL | 70 |
| 11-Oxofistularin-3 (82) | 100% inhibition of KB in culture at 7 μg/mL | 72 |
| Isofistularin-3 (80) | KB: 4 μg/mL active | 69 |
| Fistularin-3 (79) | ED50: KB 4.1; PS 4.3; LE 1.3 μg/mL | 68 |
| 11-Deoxyfistularin-3 (86) | LD50: CF-7 17 μg/mL; X-17, Hela, Hep-2, RD, Lovo > 50 μg/mL | 73 |
| Aplysinamisine I (120) | IC50: MCF-7 > 50; CCRF-CEM > 50 μg/mL; HCT116 (-). | 83 |
| Aplysinamisine II (124) | IC50: MCF-7 > 50; CCRF-CEM > 50; HCT116 10 μg/mL. | 83 |
| Aplysinamisine III (102) | IC50: MCF-7 30; CCRF-CEM 6; HCT116 10 μg/mL | 83 |
| Purealidin N (150) | IC50: K1210 0.07; KB cells 0.074 μg/mL | 79 |
| Purealidin P 110 | IC50: K1210 2.8; KB: 7.6 μg/mL | 79 |
| Purealidin Q (111) | IC50: K1210 0.95; KB 1.2 μg/mL | 79 |
| Purealidin J (116) | > 10 μg/mL. | 79 |
| Purealidin K (117) | > 10 μg/mL | 79 |
| Purealidin L (125) | > 10 μg/mL | 79 |
| Purealidin M (149) | > 10 μg/mL | 79 |
| Purealidin O (200) | > 10 μg/mL | 79 |
| Purealidin R (97) | > 10 μg/mL | 79 |
| Aerophobin-1 (113) | > 10 μg/mL | 79 |
| Purealidin C (192) | IC50: KB 3.2; K1210 2.4 μg/mL | 84 |
| Purealidin B (109) | – | 84 |
| Purealidin A (146) | IC50: L1210 1.1 μg/mL | 104 |
| Fistularin-1 (94) | ED50: 21-35 μg/ml against KB, PS, LE | 68 |
| 128 | IC50: HeLa 50 μg/mL | 92 |
| Jaspamide (259) | ED50: larynx epithelial carcinoma 0.32 μg/mL, human embryonic lung 0.01 μg/mL | 157 |
| Geodiamolide A (260) | L1210: 0.0032 (ED50). ED50: U373 0.016, HEY 0.043 μg/mL | 158,159,169 |
| Geodiamolide B (261) | L1210: 0.0026 (ED50). | 158,159 |
| Geodiamolide C (262) | L1210: 0.0025 (ED50). | 159 |
| Geodiamolide D (263) | L1210: 0.0039 (ED50). | 159 |
| Geodiamolide E (264) | L1210: 0.014 (ED50). | 159 |
| Geodiamolide F (265) | L1210: 0.006 (ED50). | 159 |
| Geodiamolide G (266) | ED50: U373 7.7, HEY 8.6 μg/mL | 160 |
| Geodiamolide H (267) | TGI: HOP92 0.118, SF-268 0.153, OV Car-4 0.0186, A498 0.0948, UO-31 0.185, MDA-MB-23/ATCC 0.433 μM | 161 |
| Geodiamolide I (268) | not active | 161 |
| Bastadin-4 (208) | P-388 ED50: 2.0 μg/mL | 132 |
| Bastadin-8 (212) | P-388 ED50: 3.6 μg/mL, L1210 ED50: 5 μg/mL | 132, 142 |
| Bastadin-9 (213) | P-388 ED50: 2.7 μg/mL | 132 |
| Bastadin-12 (216) | L1210 ED50: 5 μg/mL | 142 |
| Bastadin-14 (218) | IC50 2 μg/mL against A-549, HT-29, P-388 | 133 |
| Botryllamide D (242) | HCT116: 17 μg/mL | 145 |
| Ma'edamine A (251) | IC50: L1210 4.3 μg/mL; KB 5.2 μg/mL | 152 |
| Ma'edamine A (252) | IC50: L1210 3.9 μg/mL; KB 4.5 μg/mL | 152 |
| Psammaplysin A (129) | HCT116 IC50 6 μg/mL | 95 |
| Psammaplysin B (130) | HCT116 IC50 6 μg/mL | 95 |
| Psammaplysin C (131) | HCT116 IC50 3 μg/Ml | 95 |
| Psammaplysin E (133) | active at 5 μg/mL against KB and LoVo cells | 96 |
The anticancer activity of aeroplysinin-1 (14) was the most studied among all bromotyrosine derivatives. Kreuter et al. evaluated the in vitro and in vivo anticancer activity of (+)-aeroplysinin-1 (14) (265,266). Aeroplysinin-1 displayed pronounced cytostatic activity against L5178y mouse lymphoma cells (ED50: 0.5 μM), Friend erythroleukemia cells (ED50: 0.7 μM), human breast carcinoma cells (ED50: 0.3 mM), and human colon carcinoma cells (ED50: 5.0 mM), but not against the related normal cells in vitro. The ED50 values of aeroplysinin-1 for these cell lines are significantly lower than those obtained in cultures with non-transformed lymphocytes. The inhibitory activity of aeroplysinin-1 in the L5178y cell system is higher than other cytostatic agents including 9-β-d-arabinofuranosyladenine, ED50 29 μM (267), 1-β-d-arabinofuranosylthymine, ED50 9.8 μM (268), bleomycin, ED50 0.9 μM (269), and distamycin, ED50 13.1 μM (270). Aerothionin caused a preferential inhibition of [3H]thymidine incorporation rates in L5178y mouse lymphoma cells when compared with murine spleen lymphocytes in vitro. These results showed that a higher concentration of the alkaloid is required to influence the incorporation rates in lymphocytes than in L5178y cells; e.g., at a concentration of 2.9 mM the incorporation rates in lymphocytes are reduced only by 40–50%, while those in L5178y cells are diminished by almost 100%. The same differential effect in vitro was found with the following epithelial cells: 14.7 μM of the compound was required to inhibit normal human fibroblasts to 50%, but only 2.9 μM in the assays with human malignant keratinocytes or malignant melanoma cells was needed to observe the same inhibitory effect.
Aeroplysinin-1 (14) also displayed antileukemic activity in vivo using the L5178y cell/NMRI mouse system. Administration of a dose of 50 mg/kg for five consecutive days, the life span T/C (%) value was determined to be 338. Preliminary toxicology studies revealed an acute LD50 of 202 mg/kg, a subacute LD50 of 150 mg/kg, and a subacute LD10 133.3 mg/kg. A daily dose of 30 mg/kg for 30 consecutive days neither caused any toxicity nor a change of body weight compared to the controls. The animals were sacrificed for autopsy 40 days after the last injection. In no case were any morphological abnormalities detected in the peritoneal cavity. Using the umu-test system, which has been demonstrated to detect many types of DNA-damaging agents, such as base change mutagens, frameshift mutagens or oxidative mutagens (271), aeroplysinin-1 turned out to be neither a direct nor an indirect mutagen. The excellent T/C value of aeroplysinin-1 is the combined result of the antitumor effect and low toxicity. Following the arbitrary activity rating proposed (272), aeroplysin-1 was classified to the markedly active (+ + → + + +) to highly active anticancer agents (+ + +).
In the continuing paper, Kreuter et al. reported the inhibition of intrinsic protein tyrosine kinase activity of the EGF-receptor kinase complex from human breast cancer cells by (+)-aeroplysinin-1 (14) (273). Aeroplysinin-1, which possesses a close structure-relationship to tyrosine, blocks the epidermal growth factor (EGF) dependent proliferation of both MCF-7 and ZR-75-1 human breast cancer cells, and inhibited the ligand-induced endocytosis of the EGF receptor in vitro. Aeroplysinin-1 was found to inhibit the tyrosine-specific phosphorylation of lipocortin-like proteins, which have been established as major substrates of the EGF receptor-associated protein–tyrosine kinase (274), by a highly purified preparation of the EGF receptor protein–tyrosine kinase complex, isolated from MCF-7 cells. Treatment of aeroplysinin-1 in the concentration range of 0.25–0.5 μM resulted in MCF-7 cells losing their ability for EGF-mediated cell response and cell death occurred within 36–72 h. At a 10-fold higher concentration, aeroplysinin-1 did not reveal cytostatic activity in normal human fibroblasts. From these data, aeroplysinin-1 was concluded to be a promising compound in a rational chemotherapy (273). Based on the potent anticancer activity, production of aeroplysinin-1 by the in vitro culturing of the sponge Verongia aerophoba was also studied (275).
In other studies, aeroplysinin-1 (14) and the related dienone 1 exhibited cytotoxicity against Ehrlich ascites tumor (EAT) cells and HeLa tumor cells in the microculture tetrazolium (MTT) and clonogenic assays (276). Both alkaloids were able to cause growth inhibition, as well as cell death, in these cell lines. When the cells were depleted of glutathione by pretreatment with buthionine sulfoximine, they were significantly more sensitive toward aeroplysinin-1 (14) and the dienone 1 in the MTT assay. A dose-enhancement factor as high as 11.8 was found in EAT cells after a 2-h incubation with the dienone 1. These results suggested that in both tumor cell lines, glutathione may play an important role in the defense against the cytotoxic action of aeroplysinin-1 (14) and the dienone 1, and a free-radical mechanism might be involved in the cytotoxicity of both compounds (277,278). Using electron paramagnetic resonance, the formation of free radicals from aeroplysinin-1 (14) and the dienone 1 were measured in a culture medium with tumor cells. When dissolved in pure H2O, 1 yielded an EPR spectrum, but 14 failed to do so. Thus, metabolic activation by living cells may be required to obtain free radicals from aeroplysinin-1 (14). Structures 416 and 417 are possibilities for the semiquinone radicals that originate from 14 and 1. Semiquinone radicals are known to be very toxic (279).
From the above results it may be concluded that free radicals are, at least in part, responsible for the cytotoxic effects of aeroplysinin-1 (14) and the dienone 1 in vitro. Although aeroplysinin-1 (14) and the dienone 1 were less effective than the clinically used anticancer drug cisplatin, their chemical structures may be of interest as leads for future drug development. The different reactivity of aeroplysinin-1 (14) and the dienone 1 fits very well in the defensive role of these alkaloids for Aplysina aerophoba, when the following hypothesis is used. As soon as a predator attacks this sponge, aeroplysinin-1 (14) is formed due to enzymatic degradation of the larger precursors. Aeroplysinin-1 will generate free radicals after metabolic activation by the predator's cell, which results in an antifeedant effect. Simultaneously, the dienone 1 is released, which immediately yields free radicals in the water surrounding the sponge, thereby shielding the sponge from further predation or attack (276).
Recent studies revealed that (+)-aeroplysinin-1 (14) has a strong inhibitory antiangiogenic activity (280,281). Since the earliest hypothesis that tumor growth was dependent on angiogenesis (282), it has become clear that interfering with and/or preventing angiogenesis is an attractive therapeutic approach to the treatment of angiogenesis-dependent diseases (283,284). In a variety of experimental systems, representing the sequential events of the angiogenic process, aeroplysinin-1 treatment of endothelial cells resulted in strong inhibitory effects. Aeroplysinin-1 inhibited the growth of endothelial cells in culture with an IC50 of 2.1 μM and induced endothelial cell apoptosis. Capillary tube formation on Matrigel was completely abrogated by addition of aeroplysinin-1 at the low micromolar range. Aeroplysinin-1 also exhibited a clear inhibitory effect on the migration capabilities of endothelial cells. Zymographic assays showed that aeroplysinin-1 treatment produced a decrease in the concentration of matrix metalloproteinase-2 and urokinase in conditioned medium from endothelial cells. Finally, aeroplysinin-1 exhibited a dose-dependent inhibitory effect on the in vivo chorioallantoic membrane assay, showing potent apoptosis-inducing activity in the developing endothelium. The in vivo inhibition of angiogenesis by aeroplysinin-1 was confirmed by the Matrigel plug assay. Together, all of these data indicate that aeroplysinin-1 is a compound that interferes with key events in angiogenesis, making it a promising drug for further evaluation in the treatment of angiogenesis-related pathologies (280).
Most of the bis-spirocyclohexadienylisoxazoline type of bromotyrosine derivatives tested exhibited moderate cytotoxicity against different cancer cell lines. These include fistularin-3 (79) (68), 11-oxoaerothionin (72) (64), 11-epi-fistularin-3 (81) (70), 11-oxofistularin (82) (72), isofistularin-3 (80) (72), and 11-deoxyfistularin-3 (86) (73). 11-Oxoaerothionin (72) is the most potent agent and showed pronounced selective cytotoxic activity toward the human colon HCT cell line at a limited concentration range of 0.01–0.1 μg/mL (64).
Purealidins, which are single spirocyclohexadienylisoxazoline or oxime type of bromotyrosine derivatives, exhibited moderate to strong cytotoxicity, among which purealidin N (150) is very potent against K1210 and KB cells with IC50 values of 0.07 and 0.074 μg/mL, respectively (79,84,104).
The oxime disulfides, psammaplin A (152), psammaplin D (158), bromopsammaplin A (170), bispsammaplin A (171), and bisaprasin (154), were reported to have significant cytotoxicity against human lung A549, ovarian SK-OV-3, skin SK-MEL-2, CNS XF498, and colon HCT15 solid tumor cell lines (Table VIII). Psammaplin A (152) exhibited the highest potency among the five compounds (117). Inspired by its HDAC and DNMT enzyme inhibitory actions and unique structural features, the antiproliferative properties of psammaplin A (152) were thoroughly explored (112). Encouraging in vitro results were found as 152 inhibited cells grown in monolayer and in soft agar at the following levels (IC50 values in mM): A549 lung tumor at 1.35 and 2.5, and MDA-MB-435 breast tumor at 1.15 and 2.0, respectively. These positive results were also observed in vivo, as psammaplin A (152) inhibited tumor growth in the A549 lung xenograph mouse model, while also maintaining low toxicity (285). Very recently, psammaplin A (152) was found to inhibit mammalian aminopeptidase N (APN) that plays a key role in tumor cell invasion and angiogenesis, with an IC50 value of 18 μM in a noncompetitive manner (286). Interestingly, psammaplin A potently inhibited the proliferation of several cancer and endothelial cells and this effect was dependent on the cellular amount of APN expression. Finally, psammaplin A suppressed the invasion and tube formation of endothelial cells stimulated by basic fibroblast growth factor. These data demonstrated that psammaplin A is a new inhibitor of APN and might be developed as a novel anti-angiogenic agent (286).
Geodiamolides are a group of depsipeptides with very strong cytotoxicity. Geodiamolides A (260), B (261), C (262), D (263), E (264), and F (265) exhibited very potent cytotoxicity against L1210 cell line with an ED50 range of 0.0025–0.039 μg/mL (159). Comparison of the cytotoxicities of jaspamide (259) (157) and geodiamolide A (260) to F (265) (158,159) shows that significant variation in the three amino acid residues causes only minor changes in the levels of cytotoxicity exhibited by the peptides. By contrast, geodiamolide G (266), with a modified polyketide fragment, is significantly less cytotoxic than the analogue geodiamolide A (260) (160). Surprisingly, geodiamolide I (268) was completely devoid of activity (161).
Oxepin-type bromotyrosine derivatives, including psammaplysin A (129), B (130), and C (131) and bastadins-4 (208), −8 (212), −9 (213), −12 (216), and −14 (218) showed moderate cytotoxicities (95,132,133,142).
C. ANTIFOULING
Biofouling causes serious problems in the shipping business, in aquaculture and in the cooling systems of power stations. The biofouling of ship hulls increases drag, and consequently increases fuel costs. The biofouling of fishing nets in marine aquaculture prevents a smooth flow of sea water, followed by a serious decline in the number of fish due to an insufficient oxygen supply. Metallic compounds, such as copper (I) oxide and bis(tributyltin)oxide (TBTO), have previously been used as antifouling agents. Today, however, the use of TBTO in antifouling paints is restricted in order to prevent environmental pollution. Therefore, the development of environmentally benign anti-fouling agents is essential for resolving this global problem (287). Soft-bodied benthic invertebrates are believed to have chemical defenses against predators and the overgrowth of other benthic organisms (288). Therefore, metabolites of these invertebrates are potential “environmentally-friendly” antifouling agents (289).
The Verongida sponges provide excellent examples for the use of metabolites in chemical defense (260,290). The marine sponge Verongia aerophoba represents an example for a wound-induced chemical defense that relies on an enzymatic conversion of inactive storage compounds into an active defense metabolite (260). Following disrupture of the compartmentation (e.g. by wounding) both major components, isofistularin-3 (80) and aerophobin-2 (114), are enzymatically converted into aeroplysin-1 (14), which in turn gives rise to the dienone 1 (291). Aeroplysin-1 (14) and dienone 1 were shown to have pronounced inhibitory activity against a number of marine organisms (bacteria, microalgae, and molluscs), which may be associated with biofouling (260). This may account for the remarkably clean surfaces of the Verongida sponges.
A bioassay system suitable for screening crude extracts against biofilm formation was developed by Yamada et al. (292). Interestingly, most sponges examined using this method contained anti-biofilm compounds, among which aeroplysin-1 (14) exhibited activity with an IC50 value of 0.66 μg/cm2.
Ceratinamide A (135), ceratinamide B (136), psammaplysin A (129), psammaplysin E (133), ceratinamine (60), and moloka'iamine (52) isolated from the marine sponge Pseudoceratina purpurea, were found to have antifouling activity (settlement and metamorphosis inhibitory activity) against cyprid larvae of the barnacle Balanus amphitrite with ED50 values of 0.10, 2.4, 0.27, 4.8, 5.0, and 4.3 μg/mL, respectively (49,98). Ceratinamide A (135) and psammaplysin A (129) were particularly potent in the assay. Ceratinamide B (136), psammaplysin A (129), and ceratinamine (60) were lethal to the larvae at a concentration of 30 μg/mL, while other metabolites were not toxic at this concentration. Thus, ceratinamide A (135) is a promising antifouling agent. Interestingly, psammaplysin A (129) induced larval metamorphosis of the ascidian Halocynthia roretzi (98). Ceratinamine (60) and moloka'iamine (52), and 3,5-dibromo-4-methoxy-β-phenethylamine (51), together with several analogues, have been synthesized (197,199). 3,5-Dibromo-4-methoxy-β-phenethylamine (51) and its two analogues, octanoic amide (418) and octylamine (419) strongly inhibited the settlement of barnacle cyprids, with IC50 values of 0.07, 0.2, and 0.008 μg/mL, respectively.
Rhodospirillum salexigens is a marine bacterium, which has adhering properties to form bacteria biofilm. Zamamistatin (91) exhibited significant antibacterial activity against R. salexigens with an inhibition zone of 21 mm at 1.6 μg/disk, and may be a valuable candidate for novel antifouling agents (75,76). 5-Bromoverongamine (138) was also reported to exhibit antifouling activity (100).
Due to the noteworthy absence of fouling observed on sponges of the order Verongida, a number of natural and synthetic dibromotyramine derivatives, including moloka'iamine, were selected as potential zebra mussel (Dreissena polymorpha) anti-foulants in our laboratory (293). The zebra mussel (ZM) re-attachment assays yielded an antifouling activity with an EC50 value of 10.4 μM for moloka'iamine (52). Hemifistularin-3 (108) had significant activity in a preliminary study against ZM attachment at a single high point concentration of 30 μM, while fistularin-3 (79) and aerophobin-2 (114) showed no antifouling activity against ZMs.
D. ENZYME ACTIVITY
Purealin (122) inhibited the activity of myosin Ca-ATPase and Na,K-ATPase (see Table IX) (91). However, the activity of myosin K,EDTA-ATPase was enhanced by purealin. Purealin is the first natural product which activates myosin K,EDTA-ATPase (91). A number of papers have been published about the activity of purealin (122) on ATPase in different modes (294–299). Purealin (122) modulates the ATPase activities of dephosphorylated gizzard myosin by enhancing the stability of myosin filaments against the disassembling action of ATP (294). Purealin blocks the sliding movement of sea urchin flagellar axonemes by selective inhibition of half the ATPase activity of axonemal dyneins. Purealin-sensitive APTase activity of the dynein arms plays an essential role in generating the sliding movement of flagellar axonemes (295). Purealin activates skeletal muscle actomyosin ATPase and myosin EDTA-ATPase that enhanced the superprecipitation of actomyosin (296). Purealin binds to the myosin portion involved in actin–myosin interaction and increases the actin-activated ATPase activity of myosin (298). Purealin acts as a calmodulin antagonist in reconstituted actomyosin from chicken gizzard, resulting in the inhibition of light chain phosphorylation and the actin-activated ATPase activity of myosin (299).
TABLE IX.
Enzyme Activity of Some Bromotyrosine Derivatives.
| Alkaloids | Activity (IC50) |
Ref. | ||||
|---|---|---|---|---|---|---|
| Tyrosine kinase (mM) | Na,K-ATPase | Myosin K,EDTA-ATPase | Farnesyl protein transferase | AP-N (mM) | ||
| Psammaplin D (158) | 2.8 | 110 | ||||
| Purealidin A (146) | 22% Inhibition at 10−4M | – | 104 | |||
| Purealin (122) | Activate | 91 | ||||
| Lipopurealin-A (141) | 30 M (brain) 20 M (kidney) 100 M (heart) |
102 | ||||
| Lipopurealin-B (142) | 6 M (brain) 10 M (kidney) > 100 M (heart) |
Inhibit | 102 | |||
| Lipopurealin-C (143) | 60 M (brain) 20 M (kidney) > 100 M (heart) |
102 | ||||
| Araplysillin-I (99) | 0.5 mM (brain) | 81 | ||||
| Araplysillin-II (100) | 1 mM (brain) | 81 | ||||
| Psammaplin A (152) | 7.0 mM | 70.9 | 114 | |||
| Bisaprasin (154) | 4.2 mM | 30.2 | 114 | |||
| Psammaplin A1 (165) | 3.0 mM | 114 | ||||
| Psammaplin A2 (166) | 4.4 mM | 114 | ||||
| Aplysinellin A (167) | 85.2 mM | 2.4 | 114 | |||
| Aplysinellin B (168) | 25.1 mM | 114 | ||||
Lipopurealin-A (141), lipopurealin-B (142), and lipopurealin-C (143) exhibited inhibitory activities on Na,K-ATPase from porcine brain and dog kidney, lipopurealin-B being the most potent inhibitor. In cardiac Na,K-ATPase all three alkaloids showed only weak activity. In addition, myosin K,EDTA-ATPase was markedly activated by purealin, whereas the enzyme was inhibited by lipopurealin-B (102). Purealidin A (146) showed weak inhibitory activity (22% inhibition at 10−4 M) on Na,K-ATPase, and had no effect on myosin K,EDTA-ATPase. These results suggest that the acryl part of purealin (122), lacking in purealidin A (146), is important for the activity of these ATPase inhibitors (104). Ianthesines B (106), C (93), and D (107) exhibited inhibitory activity against dog kidney Na,K-ATPase with IC50 values of 440, 50, and 280 μM, respectively, while ianthesine A (105) was inactive (>2.5 mM) (78). The amino group of the terminal α-amino acid (in 105, 106, and 93) or the dipeptide moiety (in 93) seems to be important for the activity, because the activity decreased in the order, (93), (107), NH2 (105), and NMe2 (105). Araplysillins-I (99) and -II (100) inhibited the purified porcine brain Na,K-ATPase with IC50 values of 0.5 mM and 1 mM, respectively, which is more significant than the parent alkaloids purealin (122) and lipopurealins (142) (81).
Psammaplin A (152), bisaprasin (154), psammaplin A1 (165), psammaplin A2 (166), aplysinellin A (167), and aplysinellin B (168) exhibited inhibitory activity against farnesyl protein transferase with IC50 values of 7.0, 4.2, 3.0, 4.4, 85.2, and 25.1 mM, respectively. In addition, alkaloids 152, 154, and 167 exhibited inhibitory activity against leucine aminopeptidase (AP-N) with IC50 values of 70.9, 30.2, and 2.4 mM, respectively, while other derivatives tested were inactive (IC50>100 mM).
Recently, psammaplins were reported as potent histone deacetylase (HDAC) and DNA methyltransferase (DNMT) inhibitors (see Table X) (112). Eleven psammaplins (see Table X), isolated from the sponge Pseudoceratina purpurea, were tested in the histone deacetylase (HDAC) enzyme assay at concentrations ranging from 16 nM to 10 μM, and the data obtained were compared to that of two standards, trichostatin A (300) and trapoxin A (301). There were three psammaplin derivatives whose IC50 values were in a range of very potent activity (≤10 nM) and these included the following: psammaplin A (152) IC50 = 4.2 nM, bisaprasin (154) IC50 = 10.7 nM, and psammaplin F (160) IC50 = 8.6 nM. In contrast, psammaplins B (156), C (157), D (158), G (161), H (162), and J (164) were only considered moderately active (IC50 values >40 nM), while the other two psammaplins E (159) and I (163) were weakly active (IC50 values >100 nM). These trends are mirrored in the activation of p21 promoter regions in cell-based assays (see Table X). The factors responsible for the structure–activity relationship (SAR) variations based on potency are clearly complex. It would appear that the structural features required for the best HDAC inhibition activity include the disulfide spacer linked to a hydroxyimino amide capped on each end by modified tyrosine residues. Deletion of one of the tyrosine end groups was not tolerated, but in one case it could be replaced by an oxalamic acid group (i.e. 160).
TABLE X.
Enzyme-Based Histone Deacetylase (HDAC) and DNA Methyltransferase (DNMT) Inhibition Data, and Cell-Based p21 Promoter Activity Data, for the Psammaplins (112).
| Alkaloids | HDAC enzyme inhibition IC50 (nM)a | cell-based fold induction activity AC50 (μM)b | max fold induction (22 MFI is produced at 0.5 μM) | DNMT enzyme inhibition IC50 (nM) |
|---|---|---|---|---|
| Psammaplin A (152) | 4.2 ± 2.4 | 7.5 | 15 | 18.6 |
| Psammaplin B (156) | 48 ± 12 | 15.0 | 7 | nt |
| Psammaplin C (157) | nt | nt | nt | nt |
| Psammaplin D (158) | 44 ± 10 | > 15.0 | 5 | > 30.0 |
| Psammaplin E (159) | 327 ± 39 | > 15.0 | 2 | > 30.0 |
| Psammaplin F (160) | 2.1 ± .4 | 0.7 | 18 | > 30.0 |
| Psammaplin G (161) | 18 ± 8 | 7.5 | 7 | 12.8 |
| Psammaplin H (162) | 79 ± 22 | 3.8 | 7 | > 30.0 |
| Psammaplin I (163) | 299 ± 70 | 2.6 | 9 | > 30.0 |
| Psammaplin J (164) | 20 ± 17 | 3.6 | 8 | nt |
| Bisaprasin (154) | 9 ± 5 | 0.7 | 9 | 3.4 |
| Trichostatin A | 14 ± 1 | 0.2 | 7.2 | nt |
| Trapoxin A | 9 ± 3 | 0.005 | 28.5 | nt |
Compounds were titrated in a 1:5 dilution series (10 μM, 2 μM, 400 nM, 80 nM, and 16 nM); averages from duplicate trials are shown, except for known HDAC inhibitors, trichostatin A and trapoxin A.
Concentration of compound required to produce 50% p21 promoter activity by psammaplin A (152).
Eight psammaplin analogues were then assessed in the DNA methyltransferase assay and the results are presented in Table X. Unfortunately, scarcity of material prevented the testing of three of the analogues, psammaplins B (156), C (157), and J (164). Only one alkaloid, bisaprasin (154), was observed to be very potent (≤10 nM) and two other alkaloids, psammaplin A (152) and G (161), were also potent. Interestingly, two of these, psammaplin A (152) and bisaprasin (154), were also potent HDAC inhibitors. Although the data set is limited, it would appear that the unifying feature for dual activity against both HDAC and DNMT is the presence of the disulfide spacer linked to a hydroxyimino amide capped on each end by aromatic moieties.
Recent findings implicate an interesting relationship between DNMT and HDAC as epigenetic modifiers in the silencing of tumor supressor genes, but to date no single compound has been shown to exhibit dual activity for both of these distinct targets. Psammaplin A (152) and bisaprasin (154) are the first single compounds to inhibit both of these enzymes. Furthermore, while psammaplin F (160) is selective for HDAC, psammaplin G (161) is selective for DNMT. The unique structural differences between these various psammaplins make them relevant for further SAR studies. In summary, psammaplin A (152) and bisaprasin (154), are micromolar inhibitors of both HDAC and DNMT and may be useful as biomolecular probes of epigenetic gene regulation pathways. Finally, the logical next step has been taken by the Novartis group to design a synthetic compound, NVP-LAQ824 (302), based in part on the trichostatin A, tripoxin B, and psammaplin A (152) pharmacophores, and this HDAC inhibitor has been advanced into a Phase I anticancer clinical trial (303).
E. ANTIVIRAL ACTIVITY
Several bromotyrosine alkaloids have been reported to possess antiviral activities (see Table XI). These include moloka'iamine (52), mololipids (53), fistularin-3 (79) 11-ketofistularin-3 (82), and psammaplysin D (132).
TABLE XI.
Antiviral Activity of Bromotyrosine Derivatives
| Alkaloids | Antiviral activity | Ref. |
|---|---|---|
| Moloka'iamine (52) | Mv 1 Lu/HSV II: 10 μg/mL 92% reduced; CV-1/HSV-1 and BHK/VSV not active | 42 |
| Mololipids (53) | HIV-1 ED50 52.2 μM | 43 |
| Fistularin-3 (79) | Feline leukemia [ED50:22 μM (4.8 μg/200 μl)]. ED50: AZT 0.10 μM, ED50: ddCyd 15 μM. Not active against FeLV at 100 μg/200 μl | 71 |
| 11-Ketofistularin-3 (82) | Feline leukemia [ED50:42 μM (9.3 μg/200 μl)]. ED50: AZT 0.10 μM, ED50: ddCyd 15 μM. Not active against FeLV at 100 μg/200 μl | 71 |
| Psammaplysin D (132) | 51% inhibition at 0.1 μg/mL against Haitian RF stran of HIV-1 | 96 |
Moloka'iamine (52) and the naturally occurring mixture mololipids (53) showed moderate anti-HIV activity. Moloka'iamine (52) showed an inhibition of 90% at 10 μg/mL against HSV-II (42). Mololipids (53) exhibited selective activity against HIV-1 with an ED50 of 52.2 μM without cytotoxicity against human peripheral blood mononuclear cells (IC50>100 μg/mL), suggesting that this series of anti-HIV activity lipids has potential for future studies (43). A series of pure synthetic mololipids was screened for gp120 binding by analyzing the interaction of mololipid monolayers with recombinant gp120 at an air– water interface (198). None of these interacted significantly with gp120, suggesting that the action of mololipids seems unlikely by impairing HIV–glycolipid interactions on the plasma cell membrane.
Fistularin-3 (79) and 11-ketofistularin (82) showed activity against feline leukemia virus with an ED50 value of 22 μM (4.8 μg/200 μl) and 42 μM (9.3 μg/200 μl), respectively (71). The highest concentrations tested for cytotoxicity against FeLV were 100 μg/200 μl, and neither compound was toxic at this dosage. The alkaloids were less active than 3′-azido-3′-deoxythymidine (AZT, ED50 0.10 μM), but were comparable to 2′,3′-dideoxycytidine (ddCyd ,ED50 15 μM) in these assays. Fistularin-3 (79) also exhibited anti-HIV-1 activity with an EC50 value of 6.9 μM (311).
Reverse transcriptase (RT) plays a critical role in the early steps of the life of human immunodeficiency virus (HIV) (304), and for over a decade has been one of the major targets of AIDS therapy. Polycitone A (280) was found to be a potent general inhibitor of retroviral reverse transcriptases and cellular DNA polymerases (305). Polycitone A exhibited potent inhibitory capacity of both RNA- and DNA-directed DNA polymerases. It inhibits retroviral reverse transcriptases (RTs) of human immunodeficiency virus type 1 (HIV), murine leukemia virus (MLV) and mouse mammary tumor virus (MMTV)] as efficiently as cellular DNA polymerases of both DNA polymerases α and β and the prokaryotic Klenow fragment of Escherichia coli DNA polymerase I. The mode and mechanism of inhibition of the DNA-polymerase activity associated with HIV-1 RT by polycitone A (280) have been studied. The results suggest that the inhibitory capacity of the DNA polymerase activity is independent of the template-primer used. The RNase H function is hardly affected by this inhibitor. Polycitone A has been shown to interfere with DNA primer extension, as well as with the formation of the RT-DNA complex. Steady-state kinetic studies demonstrate that this inhibitor can be considered as an allosteric inhibitor of HIV-1 RT. The target site on the enzyme may be also spatially related to the substrate binding site, since this inhibitor behaves competitively with respect to dTTP with poly(rA) oligo(dT) as a template primer. Chemical transformation of the five phenol groups of polycitone A by methoxy groups decreased the ability to inhibit the DNA polymerase function by 40-fold. Furthermore, this analog lacks the ability to inhibit DNA primer extension as well as the formation of the RT-DNA complex (305).
F. CALCIUM CHANNEL REGULATOR
Bastadins were found to be modulators of skeletal muscle FKBP12/calcium channel complex, which selectively modulated the skeletal isoform of the ryanodine-sensitive sarcoplasmic reticulum (SR) calcium channel by a novel mechanism involving the FK506 binding protein (FKBP12)/ryanodine receptor skeletal isoform (RyR-1) complex (139). Bastadins-5 (209), −7 (211), and −19 (223) showed marked differences in potency and efficacy toward activation of the binding of [3H]ryanodine. In physiological salt, bastadin-5 (5 μM) increases the [3H]ryanodine binding capacity of SR membranes 5-fold, by stabilizing the high affinity conformation of RyR-1 for ryanodine without shifting the affinity of the activator site for Ca2+ or altering the response to caffeine or adenine nucleotides. Bastadin-5 decreases the inhibitory potency of Mg2+ 7-fold and high concentrations of (>100 μM) Ca2+ nearly 5-fold. Bastadin-5 inhibits Ca2+ uptake into SR vesicles and enhances Ca2+-induced Ca2+ release 8-fold. Bastadin-5 increases single-channel open dwell time, τ1 and τ2, 65- and 92-fold, respectively, without changing unitary conductance for Cs+ (450 picosiemens) or open probability. The most significant finding is that the unique actions of bastadin-5 on [3H]ryanodine binding and Ca2+ transport are antagonized by the immunosuppressant FK506. FK506 alone weakly enhances the binding of [3H]ryanodine, compared to bastadin-5. However, FK506 diminishes bastadin 5-induced changes in [3H]ryanodine binding and Ca2+ transport without altering the efficacy of adenine nucleotides. Unlike FK506, bastadin-5 does not directly promote the dissociation of FKBP12 from the RyR-1 membrane complex. However, it markedly enhances the release of FKBP12 induced by FK506. These results suggest that the bastadin-5 effector site is a novel modulatory domain on FKBP12. Bastadins represent a new class of compounds that will allow insight into the functional interactions between FKBP12 and RyR-1 (139).
Utilizing single channel analysis and measurements of Ca2+ flux across the sarcoplasmic reticulum, bastadin-10 (214) was identified as the structural congener responsible for dramatically stabilizing the open conformation of the RyR channel, possibly by reducing the free energy associated with closed to open channel transitions (ΔG*c → o) (306). The stability of the channel open state induced by bastadin-10 sensitized the channel to activation by Ca2+ to such an extent that it essentially obviated regulation by physiological concentrations of Ca2+ and relieved inhibition by physiological Mg2+. These actions of bastadin-10 were produced only on the cytoplasmic face of the channel, were selectively eliminated by pretreatment of channels with FK506 or rapamycin, and were reconstituted by human recombinant FKBP12. The actions of bastadin-10 were found to be reversible. A structure–activity model is proposed by which substitutions on the Eastern and Western hemispheres of the bastarane macrocycle may confer specificity toward the RyR1–FKBP12 complex to stabilize either the closed or open channel conformation. These results indicate that RyR1–FKBP12 complexes possess a novel binding domain for phenoxycatechols and raise the possibility of molecular recognition of an endogenous ligand (306). Bastadins are now used as tools to study the calcium channel (307,308).
G. OTHER ACTIVITIES
Searching for a selective histamine-H3 antagonist led to the isolation of verongamine (139) (101). Verongamine binds with an IC50 of 0.5 μM to the H3-receptor isolated from guinea pig brain membranes (309). Verongamine also demonstrated H3-antagonist activity in the electrical field stimulated (EFS) contracted guinea pig ileum. (310). In this assay, EFS-induced contractions are inhibited by the H-receptor agonist (R)-α-methyl histamine (RAM), and the effect of RAM can be blocked by H3-antagonists. Verongamine at 1 μg/mL blocked the effect of RAM. For comparison, aerophobin-1 (113) gave an H3-receptor binding IC50 of 9.0 μM, and showed no activity in the guinea pig ileum assay. Additionally, verongamine at 1 μg/mL has no effect against the inhibition of EFS-induced ileum contractions by the α2-adrenoceptor agonist clonidine. These results indicate verongamine (139) is a specific H3-receptor antagonist (101). Although bromotyrosine derivatives have been reported to possess a variety of bioactivities, the detection of histamine-H3 activity is unprecedented among sponge metabolites and marine natural products in general.
Archerine (92) was assessed for antihistaminic activity on histamine-induced contractions of guinea pig isolated ileum. The histamine agonist (10−8–10−4 M) caused a concentration-dependent contraction of the isolated organ. Archerine (92) at the concentration of 1.2 × 10−4 M completely abolished the 1 μM response of histamine, while a milder effect was observed at the concentrations 1.2 × 10−5 M and 1.2 × 10−6 M. This inhibition was removed by washing the tissue with fresh medium, indicating that the antagonism produced by archerine (92) is reversible. This result is particularly interesting if we consider that archerine (92) appears not to possess all the structural features which are currently used as guidelines for the synthesis of antihistamine drugs (77).
Intravenous injection of 100 mcg/kg of N,N,N-trimethyl-dibromotyramine (47) in an anesthetized dog caused a small, transient hypertensive response (20 mm, 25 s), followed by a similar short-lived hypotensive reaction (20mm, 30 s). Large doses of 47 (1 mg/kg) caused a marked and relatively sustained rise in blood pressure (120 mm, 4.8 min), typical of that induced by 1 μg/kg of epinephrine. This pressor effect was blocked by an alpha adrenergic antagonist, tolfazoline (12 mg/kg), thus characterizing 47 as an alpha adrenergic agent (37). When the pressor effect of 47 was blocked by tolazoline, a fall in blood pressure was observed, indicating a beta-adrenoreceptor mediated response. This depressive effect was blocked by propanolol (2 mg/kg), a beta-adrenergic blocker, thereby identifying a dual adrenergic effect of 47 on blood pressure. Synthetic and natural 47 caused identical responses when administered consecutively in identical doses to the same dog. Thus the possibility that the dual adrenergic activity might be due to a trace impurity seems to be ruled out (37).
N,N,N-Trimethyl-dibromotyramine (47) had stimulant properties on the central nervous system. Spontaneous motor activity in mice was increased 41% at a 1 mg/kg i.p. dose. A preliminary study on the smooth muscle preparations indicated that 47 had a non-specific spasmogenic response at 10 μg/mL bath concentrations. Both atropine and antihistamine could partially antagonize this response. Although the mechanism of this action is difficult to explain at present, the quaternary structural features of 47 may possibly contribute to its spasmogenic activity in smooth muscles (37).
Ceratamines A (249) and B (250) both had IC50 values of 10 μg/mL in the cell-based antimitotic assay. They are the first two representatives of a novel family of antimitotic heterocyclic alkaloids. The imidazo[4,5,d]azepine core heterocycle in the ceratamines appears to have no precedent at any oxidation level among known natural products or synthetic compounds (151).
Aplysillin A (248) weakly inhibited the binding of [125I]-thrombin to platelet membranes with an IC50 value of 20 μM (150).
VII. Conclusions
The abundance of metabolites involving the halogenation of tyrosine is an occurrence which is highly unique to the marine environment. Both the broad range of structural classes, as well as the diversity of bioactivity associated with this structural group, suggests that this group of molecules will continue to find their way into clinical applications. From the point of view of the development of novel drug leads, these molecules have a unique potential for the introduction of new drug leads and scaffolds for the generation of new drugs. This is clearly due, in part, to the good drug-like properties for this group of secondary metabolites, which, combined with the unique and abundant structural variations, suggests that the opportunites for applying these molecules to issues associated with human health has just begun.
References
- 1.Morner CT. Z. Physiol. Chem. 1913;88:138. [Google Scholar]
- 2.Sharma GM, Burkholder PR. J. Antibiotics, Ser. A. 1967;20:200. [PubMed] [Google Scholar]
- 3.Sharma GM, Burkholder PR. Tetrahedron Lett. 4147. 1967 doi: 10.1016/s0040-4039(01)89710-0. [DOI] [PubMed] [Google Scholar]
- 4.Sharma GM, Vigand B, Burkholder PR. J. Org. Chem. 1970;35:2823. doi: 10.1021/jo00833a084. [DOI] [PubMed] [Google Scholar]
- 5.Anderson RJ, Faulkner DJ. Tetrahedron Lett. 1973:1175. [Google Scholar]
- 6.Faulkner DJ, Ireland CM. NATO Conference Series IV: Marine Sciences 1 (Mar. Nat. Prod. Chem.) CAN. 1977;2388:19210. [Google Scholar]
- 7.Kernan MR, Cambie RC, Bergquist PR. J. Nat. Prod. 1990;53:720. [Google Scholar]
- 8.Aydogmus Z, Ersoy N, Imre S. Turk. J. Chem. 1999;23:339. [Google Scholar]
- 9.D'Ambrosio M, Guerriero A, Pietra F. Helv. Chim. Acta. 1984;67:1484. [Google Scholar]
- 10.D'Ambrosio M, Guerriero A, De Clauser R, De Stanchina G, Pietra F. Experientia. 1983;39:1091. [Google Scholar]
- 11.Capon RJ, MacLeod JK. J. Chem. 1987;40:341. [Google Scholar]
- 12.Peng J, Hamann MT. Unpublished material [Google Scholar]
- 13.Daly JW, Jerina DM, Witkop D. Experientia. 1972;28:1129. doi: 10.1007/BF01946135. [DOI] [PubMed] [Google Scholar]
- 14.Fattorusso E, Minale L, Sodano G. J. Chem. Soc., Chem. Commun. 1970;751 doi: 10.1039/p19720000016. [DOI] [PubMed] [Google Scholar]
- 15.Fattorusso E, Minale L, Sodano G. J. Chem. Soc., Perkin Trans. 1972;11:16. doi: 10.1039/p19720000016. [DOI] [PubMed] [Google Scholar]
- 16.Fulmor W, Van Lear GE, Morton GO, Mills RO. Tetrahedron Lett. 1970;4551 doi: 10.1016/s0040-4039(00)89414-9. [DOI] [PubMed] [Google Scholar]
- 17.Mazzarella L, Puliti R. Gazz. Chim. Ital. 1972;102:39. [Google Scholar]
- 18.Cosulich DB, Lovell FM. J. Chem. Soc., Chem. Commun. 1971;397 [Google Scholar]
- 19.Minale L, Sodano G, Chan WR, Chen AM. J. Chem. Soc., Chem. Comm. 1972;674 [Google Scholar]
- 20.Norte M, Fernandez JJ. Tetrahedron Lett. 1987;28:1693. [Google Scholar]
- 21.Norte M, Rodriguez ML, Fernandez JJ, Eguren L, Estrada DM. Tetrahedron. 1988;44:4973. [Google Scholar]
- 22.D'Ambrosio M, Guerriero A, Traldi P, Pietra F. Tetrahedron Lett. 1982;23:4403. [Google Scholar]
- 23.Guerriero A, D'Ambrosio M, Traldi P, Pietra F. Naturwissenschaften. 1984;71:425. [Google Scholar]
- 24.D'Ambrosio M, Mealli C, Guerriero A, Pietra F. Helv. Chim. Acta. 1985;68:1453. [Google Scholar]
- 25.D'Ambrosio M, Geurreiro A, Pietra F. Comp. Biochem. Physiol. 1986;83:309. [Google Scholar]
- 26.Krejcarek GE, White RH, Hager LP, McClure WO, Johnson RD, Rinehart KL, McMillan JA, Paul IC, Shaw PD, Brusca RC. Tetrahedron Lett. 1975;507 [Google Scholar]
- 27.Yasunohu K, Tanaka T, Knox WE, Mason HS. Fed. Proc. Am. Soc. Exp. Biol. 1985;17:340. [Google Scholar]
- 28.Chang CWJ, Weinheimer AJ. Tetrahedron Lett. 4005. 1977 [Google Scholar]
- 29.Kelecom A, Kannengiesser GJ. Anais da Academia Brasileira de Ciencias. 1979;51:639. [Google Scholar]
- 30.Encarnacion RD, Sandoval E, Malmstrom J, Christophersen C. J. Nat. Prod. 2000;63:874. doi: 10.1021/np990489d. [DOI] [PubMed] [Google Scholar]
- 31.Chib JS, Stempien MF, Mierzwa RA, Ruggieri GD, Nigrelli RF. J. Pharm. Sci. 1978;67:264. doi: 10.1002/jps.2600670238. [DOI] [PubMed] [Google Scholar]
- 32.Venkateswarlu Y, Chavakula R. J. Nat. Prod. 1995;58:1087. [Google Scholar]
- 33.Ciminiello P, Fattorusso E, Magno S, Pansini M. J. Nat. Prod. 1995;58:689. doi: 10.1021/np990343e. [DOI] [PubMed] [Google Scholar]
- 34.Hunt S, Breuer SW. Biochim. Biophys. Acta. 1971;252:401. doi: 10.1016/0304-4165(71)90021-3. [DOI] [PubMed] [Google Scholar]
- 35.Albrizio S, Ciminiello P, Fattorusso E, Magno S, Pansini M. Tetrahedron. 1994;50:783. [Google Scholar]
- 36.Gao H-F, Kelly M, Hamann MT. Tetrahedron. 1999;55:9717. [Google Scholar]
- 37.Hollenbeak KH, Schmitz FJ, Kaul PN, Kulkarni SK. Drugs Food Sea: Myth or Reality 81. 1978 [Google Scholar]
- 38.Ciminiello P, Fattorusso E, Magno S, Pansini M. J. Nat. Prod. 1994;57:1564. doi: 10.1021/np990343e. [DOI] [PubMed] [Google Scholar]
- 39.Ciminiello P, Dell'Aversano S, Fattorusso E, Magno S, Pansini M. J. Nat. Prod. 2000;63:263. doi: 10.1021/np990343e. [DOI] [PubMed] [Google Scholar]
- 40.Wagoner RMV, Jompa J, Tahir A, Ireland CM. J. Nat. Prod. 1999;62:794. doi: 10.1021/np9805589. [DOI] [PubMed] [Google Scholar]
- 41.Pettit GR, Butler MS, Williams MD, Filiatrault MJ, Pettit RK. J. Nat. Prod. 1996;59:927. doi: 10.1021/np960249n. [DOI] [PubMed] [Google Scholar]
- 42.Hamann MT, Scheuer PJ, Kelly-Borges M. J. Org. Chem. 1993;58:6565. [Google Scholar]
- 43.Ross SA, Weete JD, Schinazi RF, Wirtz SS, Tharnish P, Scheuer PJ, Hamann MT. J. Nat. Prod. 2000;63:501. doi: 10.1021/np980414u. [DOI] [PubMed] [Google Scholar]
- 44.Xynas R, Capon RJ. Aust. J. Chem. 1989;42:1427. [Google Scholar]
- 45.Tsuda M, Shigemori H, Ishibashi M, Kobayashi J. J. Nat. Prod. 1992;55:1325. [Google Scholar]
- 46.Venkateswarlu Y, Venkatesham U, Rao MR. J. Nat. Prod. 1999;62:893. doi: 10.1021/np980419r. [DOI] [PubMed] [Google Scholar]
- 47.Venkateswarlu Y, Rao MR, Venkatesham U. J Nat. Prod. 1998;61:1388. doi: 10.1021/np980032o. [DOI] [PubMed] [Google Scholar]
- 48.Rao MR, Venkatesham U, Venkateswarlu Y. Ind. J. Chem. 1999;38B:1301. [Google Scholar]
- 49.Tsukamoto S, Kato H, Hirota H, Fusetani N. J. Org. Chem. 1996;61:2936. doi: 10.1021/jo9602884. [DOI] [PubMed] [Google Scholar]
- 50.Fu X, Schmitz FJ. J. Nat. Prod. 1999;62(7):1072. doi: 10.1021/np9900425. [DOI] [PubMed] [Google Scholar]
- 51.Tsuda M, Endo T, Watanabe K, Fromont J, Kobayashi J. J. Nat. Prod. 2002;65:1670. doi: 10.1021/np020210k. [DOI] [PubMed] [Google Scholar]
- 52.Faulkner DJ. Nat. Prod. Rep. 2001;18:1. doi: 10.1039/b006897g. and references therein. [DOI] [PubMed] [Google Scholar]
- 53.Kassuklke KE, Faulkner DJ. Tetrahedron. 1991;47:1809. [Google Scholar]
- 54.Borders B, Morton GO, Wetzel ER. Tetrahedron Lett. 1974;2709 [Google Scholar]
- 55.Makarieva TN, Stonik VA, Alcolado P, Elyakov YB. Comp. Biochem. Physiol. 1981;68B:481. [Google Scholar]
- 56.Fattorusso E, Minale L, Sodano G, Moody K, Thomson RH. J. Chem. Soc., Chem. Commun. 1970;752 [Google Scholar]
- 57.Fattorusso E, Minale L, Moody K, Sodano G, Thomson RH. Gazz. Chim. Ital. 1971;101:61. [Google Scholar]
- 58.Moody KRH, Thomson E, Fattorusso L, Minale GS. J. Chem. Soc., Perkin Trans. 1. 1972;10:18. doi: 10.1039/p19720000016. [DOI] [PubMed] [Google Scholar]
- 59.Minale L. Pure Appl. Chem. 1976;48:7. [Google Scholar]
- 60.A McMillan J, Paul IC, Goo YM, Rinehart KL, Krueger WC, Pschigoda LM. Tetrahedron Lett. 1981;22:39. [Google Scholar]
- 61.Ciminiello P, Costantino V, Fattorusso E, Magno S, Mangoni A, Pansini M. J. Nat. Prod. 1994;57:705. [Google Scholar]
- 62.Gunasekera M, Gunasekera SP. J. Nat. Prod. 1989;52:753. [Google Scholar]
- 63.Kernan MR, Cambie RC, Bergquis PR. J. Nat. Prod. 1990;53:615. [Google Scholar]
- 64.Acosta AL, Rodriguez AD. J. Nat. Prod. 1992;55:1007. doi: 10.1021/np50085a031. [DOI] [PubMed] [Google Scholar]
- 65.Ciminiello P, Fattorusso E, Forino M, Magno S, Pansini M. Tetrahedron. 1997;53:6565. [Google Scholar]
- 66.Saeki BM, Granato AC, Berlinck RGS, Magalhaes A, Schefer AB, Ferreira AG, Pinheiro US, Hajdu E. J. Nat. Prod. 2002;65:796. doi: 10.1021/np0105735. [DOI] [PubMed] [Google Scholar]
- 67.Ciminiello P, Dell'Aversano C, Fattorusso E, Magno S, Carrano L, Pansini M. Tetrahedron. 1996;52:9863. [Google Scholar]
- 68.Gopichand Y, Schmitz FJ. Tetrahedron Lett. 3921. 1979 [Google Scholar]
- 69.Cimino G, De S, De Rosa S, De Stefano S, Self R, Sodano G. Tetrahedron Lett. 1983;24:3029. [Google Scholar]
- 70.König GM, Wright AD. Heterocycles. 1993;36:1351. [Google Scholar]
- 71.Gunasekera SP, Cross SS. J. Nat. Prod. 1992;55:509. doi: 10.1021/np50082a020. [DOI] [PubMed] [Google Scholar]
- 72.Mancini I, Guella G, Laboute P, Debitus C, Pietra F. J. Chem. Soc., Perkin Trans. 1993;1:3121. [Google Scholar]
- 73.Compagnone RS, Avila R, Suarez AI, Abrams OV, Rangel HR, Arvelo F, Pina IC, Merentes E. J. Nat. Prod. 1999;62:1443. doi: 10.1021/np9901938. [DOI] [PubMed] [Google Scholar]
- 74.Kijjoa A, Watanadilok R, Sonchaeng P, Silva AMS, Eaton G. Z. Naturforsch. 2001;56c:1116. doi: 10.1515/znc-2001-11-1232. [DOI] [PubMed] [Google Scholar]
- 75.Takada N, Watanabe R, Suenaga K, Yamada K, Ueda K, Kita M, Uemura D. Tetrahedron Lett. 2001;42:5265. [Google Scholar]
- 76.Uemura D, Takada A, Suenaga K, Yamada K, Watanabe R. Jpn. Kokai Tokkyo Koho. 2002;8 CODEN: JKXXAF JP 2002338554 A2 20021127. Application: JP 2001-144150 20010515. [Google Scholar]
- 77.Ciminiello P, Dell'Aversano C, Fattorusso E, Magno S. Eur. J. Org. Chem. 55. 2001 [Google Scholar]
- 78.Okamoto Y, Ojika M, Kato S, Sakagami Y. Tetrahedron. 2000;56:5813. [Google Scholar]
- 79.Kobayashi J, Honma K, Sasaki T, Tsuda M. Chem. Pharm. Bull. 1995;43:403. [Google Scholar]
- 80.Aiello A, Fattorusso E, Menna M, Pansini M. Biochem. Syst. & Ecol. 1995;23:377. [Google Scholar]
- 81.Longeon A, Guyot M, Vacelet J. Experientia. 1990;46:548. [Google Scholar]
- 82.James DM, Kunze HB, Faulkner DJ. J. Nat. Prod. 1991;54:1137. doi: 10.1021/np50076a040. [DOI] [PubMed] [Google Scholar]
- 83.Rodríguez AD, Pina IC. J. Nat. Prod. 1993;56:907. doi: 10.1021/np50096a014. [DOI] [PubMed] [Google Scholar]
- 84.Kobayashi J, Tsuda M, Agemi K, Shigemori H, Ishibashi M, Sasaki T, Mikami Y. Tetrahedron. 1991;47:6617. [Google Scholar]
- 85.Kobayashi J, Tsuda M, Agemi K, Honma K, Shigemori H, Ishibashi M, Sasaki T. Peptide Chem. 1996;1995:197. [Google Scholar]
- 86.Tabudravu JN, Jaspars M. J. Nat. Prod. 2002;65:1798. doi: 10.1021/np020275n. [DOI] [PubMed] [Google Scholar]
- 87.Assmann M, Wray V, Van Soest RWM, Proksch P. Z. Naturforsch. 1998;53:398. [Google Scholar]
- 88.Fendert T, Wray V, Van Soest RWM, Proksch P. Z. Naturforsch. 1999;54c:246. [Google Scholar]
- 89.Benharref A, Nat MJ. Prod. 1996;59:177. [Google Scholar]
- 90.Nicholas GM, Newton GL, Fahey RC, Bewley CA. Organic Lett. 2001;3:1543. doi: 10.1021/ol015845+. [DOI] [PubMed] [Google Scholar]
- 91.Nakamura H, Wu H, Kobayashi J, Nakamura Y, Ohizumi Y, Hirata Y. Tetrahedron Lett. 1985;26:4517. [Google Scholar]
- 92.Ciminiello P, Dell'Aversano C, Fattorusso E, Magno S, Pansini M. J. Nat. Prod. 1999;62:590. doi: 10.1021/np9805138. [DOI] [PubMed] [Google Scholar]
- 93.Kashman Y, Groweiss A, Carmely S, Kinamoni Z, Czarkie D, Rotem M. Pure Appl. Chem. 1982;54:1995. [Google Scholar]
- 94.Roll DM, Chang CWJ, Scheuer PJ, Gray GA, Shoolery JN, Matsumoto GK, Van Duyne GD, Clardy J. J. Am. Chem. Soc. 1985;107:2916. [Google Scholar]
- 95.Copp BR, Ireland CM, Barrows LR. J. Nat. Prod. 1992;55:822. doi: 10.1021/np50084a021. [DOI] [PubMed] [Google Scholar]
- 96.Ichiba T, Scheuer PJ, Kelly-Borges M. J. Org. Chem. 1993;58:4149. [Google Scholar]
- 97.Liu S, Fu X, Schmitz FJ, Kelly-Borges M. J. Nat. Prod. 1997;60:614. doi: 10.1021/np970070s. [DOI] [PubMed] [Google Scholar]
- 98.Tsukamoto S, Kato H, Hirota H, Fusetani N. Tetrahedron. 1996;52:8181. [Google Scholar]
- 99.Litaudon M, Guyot M. Tetrahedron Lett. 1986;27:4455. [Google Scholar]
- 100.Thirionet I, Daloze D, Braekman JC. Nat. Prod. Lett. 1998;12:209. [Google Scholar]
- 101.Mierzwa R, King A, Conover MA, Tozzi S, Puar MS, Patel M, Coval SJ. J. Nat. Prod. 1994;57:175. doi: 10.1021/np50103a029. [DOI] [PubMed] [Google Scholar]
- 102.Wu H, Nakamura H, Kobayashi J, Ohizumi Y, Hirata Y. Experientia. 1986;42:855. doi: 10.1007/BF01941300. [DOI] [PubMed] [Google Scholar]
- 103.Kobayashi J, Honma K, Tsuda M, Kosaka T. J. Nat. Prod. 1995;58:467. [Google Scholar]
- 104.Ishibashi M, Tsuda M, Ohizumi Y, Sasaki T, Kobayashi J. Experientia. 1991;47:299. doi: 10.1007/BF01958166. [DOI] [PubMed] [Google Scholar]
- 105.Tsuda M, Shigemori H, Ishibashi M, Kobayashi J. Tetrahedron Lett. 1992;33:2597. [Google Scholar]
- 106.Sakota N, Okita K, Matsui Y. Bull. Soc. Jap. 1970;43:1138. [Google Scholar]
- 107.Arabshahi L, Schmitz FJ. J. Org. Chem. 1987;52:3584. [Google Scholar]
- 108.Quinoa E, Crews P. Tetrahedron Lett. 1987;28:3229. [Google Scholar]
- 109.Rodriguez AD, Akee RK, Scheuer PJ. Tetrahedron Lett. 1987;28:4989. [Google Scholar]
- 110.Jimenez C, Crews P. Tetrahedron. 1991;47:2097. [Google Scholar]
- 111.He H, Faulkner JD, Shumsky JS, Hong K, Clardy J. J. Org. Chem. 1989;54:2511. [Google Scholar]
- 112.Pina IC, Gautschi JT, Wang G, Sanders ML, Schmitz FJ, France D, Cornell-Kennon S, Sambucetti LC, Remiszewski SW, Perez LB, Bair KW, Crews P. J. Org. Chem. 2003;68:3866. doi: 10.1021/jo034248t. [DOI] [PubMed] [Google Scholar]
- 113.Dumdei E, Andersen RJ. J. Nat. Prod. 1993;56:792. [Google Scholar]
- 114.Shin J, Lee H-S, Seo Y, Rho J-R, Cho KW, Paul VJ. Tetrahedron. 2000;56:9071. [Google Scholar]
- 115.Manes LV, Crews P, Kernan MP, Faulkner DJ, Fronczek FR, Gandour RD. J. Org. Chem. 1988;53:570. [Google Scholar]
- 116.Wright AE, McCarthy PJ, Schulte GK. J. Org. Chem. 1989;54:3472. [Google Scholar]
- 117.Park Y, Liu Y, Hong J, Lee C, Cho H, Kim D-K, Im KS, Jung JH. J. Nat. Prod. 2003;66:1495. doi: 10.1021/np030162j. [DOI] [PubMed] [Google Scholar]
- 118.Jurek J, Yoshida WY, Scheuer PJ, Kelly-Borges M. J. Nat. Prod. 1993;56:1609. [Google Scholar]
- 119.Pakrashi SC, Achari B, Dutta PK, Chakrabarti AK, Giri C, Saha S, Basa S. Tetrahedron. 1994;50:12009. [Google Scholar]
- 120.Pakrashi SC, Achari B, Dutta PK, Chakrabarti AK, Giri C, Saha S, Basa S. Tetrahedron. 1994;50:12783. [Google Scholar]
- 121.Yagi H, Matsunaga S, Fusetani N. Tetrahedron. 1993;49:3749. [Google Scholar]
- 122.Goud TV, Srinivasulu M, Reddy VLN, Reddy AV, Rao TP, Kumar DS, Murty US, Venkateswarlu Y. Chem. & Pharm. Bull. 2003;51:990. doi: 10.1248/cpb.51.990. [DOI] [PubMed] [Google Scholar]
- 123.Fusetani N, Masuda Y, Nakao Y, Matsunaga S, Van Soest RWM. Tetrahedron. 2001;57:7507. [Google Scholar]
- 124.Kernan MR, Cambie RC, Bergquist PR. J. Nat. Prod. 1990;53:720. [Google Scholar]
- 125.Evan T, Rudi A, Ilan M, Kashman Y. J. Nat. Prod. 2001;64:226. doi: 10.1021/np000383e. [DOI] [PubMed] [Google Scholar]
- 126.Tsuda M, Sakuma Y, Kobayashi J. J. Nat. Prod. 2001;64:980. doi: 10.1021/np010077g. [DOI] [PubMed] [Google Scholar]
- 127.Carroll AR, Bowden BF, Coll JC. Aust. J. Chem. 1993;46:825. [Google Scholar]
- 128.Kazlauskas R, Lidgard RO, Murphy PT, Wells RJ. Tetrahedron Lett. 1980;21:2277. [Google Scholar]
- 129.Kazlauskas R, Lidgard RO, Murphy PT, Wells RJ, Blount JF. Aust. J. Chem. 1981;34:765. [Google Scholar]
- 130.Franklin MA, Penn SG, Lebrilla CB, Lam TH, Pessah IN, Molinski TF. J. Nat. Prod. 1996;59:1121. doi: 10.1021/np960507g. [DOI] [PubMed] [Google Scholar]
- 131.Butler MS, Lim TK, Capon RJ, Hammond LS. Aust. J. Chem. 1991;44:287. [Google Scholar]
- 132.Pordesimo EO, Schmitz FJ. J. Org. Chem. 1990;55:4704. [Google Scholar]
- 133.Carney JR, Scheuer PJ, Kelly-Borges M. J. Nat. Prod. 1993;56:153. doi: 10.1021/np50091a025. [DOI] [PubMed] [Google Scholar]
- 134.Dexter AF, Garson MJ, Hemling ME. J. Nat. Prod. 1993;56:56. [Google Scholar]
- 135.Park SK, Jurek J, Carney JR, Scheuer PJ. J. Nat. Prod. 1994;57:407. [Google Scholar]
- 136.Park SK, Park H, Scheuer PJ. Bull. Kor. Chem. Soc. 1994;5:534. [Google Scholar]
- 137.Park SK, Ryu JK, Scheuer PJ. Bull. Kor. Chem. Soc. 1995;16:677. [Google Scholar]
- 138.Jaspars M, Rali T, Laney M, Schatzman RC, Diaz MC, Schmitz FJ, Pordesimo EO, Crews P. Tetrahedron. 1994;50:7367. [Google Scholar]
- 139.Mack MM, Molinski TF, Buck ED, Pessah IN. J. Biol. Chem. 1994;269:23236. [PubMed] [Google Scholar]
- 140.Coll JC, Kearns PS, Rideout JA, Sankar V. J. Nat. Prod. 2002;65:753. doi: 10.1021/np010520n. [DOI] [PubMed] [Google Scholar]
- 141.Gulavita NK, Wright AE, McCarthy PJ, Pomponi SA, Kelly-Borges M, Chin M, Sills MA. J. Nat. Prod. 1993;56:1613. doi: 10.1021/np50099a026. [DOI] [PubMed] [Google Scholar]
- 142.Miao S, Andersen RJ, Allen TM. J. Nat. Prod. 1990;53:1441. doi: 10.1021/np50072a007. [DOI] [PubMed] [Google Scholar]
- 143.Pettit GR, Butler MS, Bass CG, Doubek DL, Williams MD, Schmidt JM, Pettit RK, Hooper JNA, Tackett LP, Filliatrault MJ. J. Nat. Prod. 1995;58:680. doi: 10.1021/np50119a005. [DOI] [PubMed] [Google Scholar]
- 144.Trost BM, Belletire JL, Godleski S, McDougal PG, Balkovec JM, Baldwin JJ, Christy ME, Ponticello GS, Varga SL, Springer JP. J. Org. Chem. 1986;51:2370. [Google Scholar]
- 145.Mcdonald LA, Swersey JC, Ireland CM, Carroll AR, Coll JC, Bowden BF, Fairchild CR, Cornell L. Tetrahedron. 1995;51:5237. [Google Scholar]
- 146.Costantino V, Fattorusso E, Mangoni A, Pansini M. J. Nat. Prod. 1994;57:1552. [Google Scholar]
- 147.Bergwuist RP, Wells RJ. In: Marine Natural Products: Chemical and Biological Perspectives. Scheuer PJ, editor. V. Academic Press; New York: 1983. pp. 12–22. [Google Scholar]
- 148.Neidleman SL, Geigeert J. Biohalogenation: Principles, Basic Roles, and Applications. Ellis Horwood Ltd.; Chichester, UK: 1986. pp. 46–47. [Google Scholar]
- 149.Kaestner A. Invertebrate Zoology. I. Wiley Interscience; New York: 1967. p. 24. [Google Scholar]
- 150.Gulavita NK, Pomponi SA, Wright AE. J. Nat. Prod. 1995;58:954. doi: 10.1021/np50120a023. [DOI] [PubMed] [Google Scholar]
- 151.Manzo E, Van Soest R, Matainaho L, Roberge M, Andersen RJ. Org. Lett. 2003;5:4591. doi: 10.1021/ol035721s. [DOI] [PubMed] [Google Scholar]
- 152.Hirano K, Kubota T, Tsuda M, Watanabe K, Fromont J, Kobayashi J. Tetrahedron. 2000;56:8107. [Google Scholar]
- 153.Lindquist N, Fenical W. Tetrahedron Lett. 1990;31:2521. [Google Scholar]
- 154.Bobzin SC, Faulkner DJ. J. Org. Chem. 1991;56:4403. [Google Scholar]
- 155.Kijjoa A, Watanadilok R, Sonchaeng P, Sawangwong P, Pedro M, Nascimento MSJ, Silva AMS, Eaton G, Herz W. Z. Naturforsch. 2002;57c:732. doi: 10.1515/znc-2002-7-831. [DOI] [PubMed] [Google Scholar]
- 156.Zabriskie TM, Klocke JA, Ireland CM, Marcus AH, Molinski TF, Faulkner DJ, Xu C, Clardy JC. J. Am. Chem. Soc. 1986;108:3123. [Google Scholar]
- 157.Crews P, Manes LV, Boehler M. Tetrahedron Lett. 1986;27:2797. [Google Scholar]
- 158.Chan WR, Tinto WF, Manchand PS, Todaro LJ. J. Org. Chem. 1987;52:3091. [Google Scholar]
- 159.de Silva ED, Andersen RJ, Allen TM. Tetrahedron Lett. 1990;31:489. [Google Scholar]
- 160.Coleman JE, de Silva ED, Kong F, Andersen RJ, Allen TM. Tetrahedron. 1995;51:10653. [Google Scholar]
- 161.Tinto WF, Lough AJ, McLean S, Reynolds WF, Yu M, Chan WR. Tetrahedron. 1998;54:4451. [Google Scholar]
- 162.Talpir R, Benayahu Y, Kashman Y, Pannell L, Schleyer M. Tetrahedron Lett. 1994;35:4453. [Google Scholar]
- 163.D'Auria MV, Paloma LG, Minale L, Zampella A, Debitus C, Perez J. J. Nat. Prod. 1995;58:121. doi: 10.1021/np50115a017. [DOI] [PubMed] [Google Scholar]
- 164.Coleman JE, Soest RV, Andersen RJ. J. Nat. Prod. 1999;62:1137. doi: 10.1021/np990155o. [DOI] [PubMed] [Google Scholar]
- 165.Jansen R, Kunze B, Reichenbach H, Hoefle G. Liebigs Ann. 1996:285. [Google Scholar]
- 166.Kunze B, Jansen R, Sasse F, Hoefle G, Reichenbach H. J. Antibiot. 1995;48:1262. doi: 10.7164/antibiotics.48.1262. [DOI] [PubMed] [Google Scholar]
- 167.Rudi A, Goldberg I, Stein Z, Frolow F, Benayahu Y, Schleyer M, Kashman Y. J. Org. Chem. 1994;59:999. [Google Scholar]
- 168.Rudi A, Evan T, Aknin M, Kashman Y. J. Nat. Prod. 2000;63:832. doi: 10.1021/np9905158. [DOI] [PubMed] [Google Scholar]
- 169.Andersen RJ, Faulkner DJ, He CH, Van Duyne GD, Clardy J. J. Am. Chem. Soc. 1985;107:5492. [Google Scholar]
- 170.Lindquist N, Fenical W, Van Duyne GD, Clardy J. J. Org. Chem. 1988;53:4570. [Google Scholar]
- 171.Carroll AR, Bowden BF, Coll JC. Aust. J. Chem. 1993;46:489. [Google Scholar]
- 172.Leete E. Techniques of Organic Chemistry. Part I. XI. Wiley Interscience Publishers; New York: 1963. p. 437. [Google Scholar]
- 173.Kasperek GJ, Bruice TC, Yagi H, Kaubisch N, Jerina DM. J. Am. Chem. Soc. 1972;94:7876. [Google Scholar]
- 174.Stowe BB. Progress in the Chemistry of Organic Natural Products. Vol. 17. Springer; Vienna: 1959. p. 248. [Google Scholar]
- 175.De Rosa M, Minale L, Sodano G. Comp. Biochem. Physiol. 1973;45:883. doi: 10.1016/0305-0491(73)90149-1. [DOI] [PubMed] [Google Scholar]
- 176.Tymiak AA, Rinehart KL. J. Am. Chem. Soc. 1981;103:6763. [Google Scholar]
- 177.Weiss UU, Edward JM. The Biosynthesis of Aromatic Compounds. Wiley; New York: 1980. p. 144. [Google Scholar]
- 178.Nozaki M. Top. Curr. Chem. 1975;78:145. doi: 10.1007/BFb0048193. [DOI] [PubMed] [Google Scholar]
- 179.Saito I, Chujo Y, Shimazu H, Yamane M, Matsuura T, Cahnmann HJ. J. Am. Chem. Soc. 1975;97:5272. doi: 10.1021/ja00851a042. [DOI] [PubMed] [Google Scholar]
- 180.Ahmad A, Spenser ID. Can. J. Chem. 1961;39:1340. [Google Scholar]
- 181.Tapper BA, Conn EE, Butler GW. Arch. Biochem. Biophys. 1967;119:593. doi: 10.1016/0003-9861(67)90500-0. [DOI] [PubMed] [Google Scholar]
- 182.Bicchierini N, Cavazza M, Nucci L, Pergola F, Pietra F. Tetrahedron Lett. 1991;32:4039. [Google Scholar]
- 183.Cimino G, DeStefano S, Minale L. Experientia. 1975;31:756. [Google Scholar]
- 184.Carney JR, Rinehart KL. J. Nat. Prod. 1995;58:971. doi: 10.1021/np50121a001. [DOI] [PubMed] [Google Scholar]
- 185.Goo YM. Diss. Abstr. Int. B. 1980;41:569. [Google Scholar]
- 186.Cavazza M, Guella G, Nucci L, Pergola F, Bicchierini N, Pietra F. J. Chem., Soc. Perkin Trans. 1993;1:3117. [Google Scholar]
- 187.a Rieker A. In: “Methoden der Organischen Chimie (Houben-Weyl)” Band VII-3b. Thieme Verlag G, editor. Stuttgart; 1979. p. 524. [Google Scholar]; b Coffy S, editor. “Rodd's Chemistry of Carbon Compounds”, Part III A Supplement. Elsevier; msterdam: 1983. p. 174. [Google Scholar]
- 188.a Barton DHR. Proc. Chem. Soc. 1963:293. [Google Scholar]; b Scott AI. Quart. Rev. 1965;19:1. [Google Scholar]; c Natori S. In: Natural Products Chemistry. Nakanishi K, Goto T, Ito S, Natori S, Nozoe S, editors. Vol. 2. Kodanska Ltd., Academic Press; New York: 1975. p. 182. [Google Scholar]; d Geissman TA, Crout G. Organic Chemistry of Secondary Plant Metabolism. Freeman, Cooper & Company; San Francisco: 1969. p. 383. [Google Scholar]
- 189.Cavazza M, Guella G, Nucci L, Pergola F, Bicchierini N, Pietra F. J. Chem. Soc., Perkin Trans. 1993;1:3117. [Google Scholar]
- 190.Cavazza M, Guella G, Nucci L, Pergola F, Bicchierini N, Pietra F. J. Chem. Soc., Perkin Trans. 1995;1:1199. [Google Scholar]
- 191.Vogel E, Günther H. Angew. Chem., Int. Ed. Engl. 1967;6:385. [Google Scholar]
- 192.Neuss N, Nagarajan R, Molly BB, Huckstep LL. Tetrahedron Lett. 1968:4467. [Google Scholar]
- 193.Brannon DRMJA, Molloy BB, Day WA. Biochem. Biophys. Res. Commun. 1971;43:588. doi: 10.1016/0006-291x(71)90654-1. [DOI] [PubMed] [Google Scholar]
- 194.Johnson TB, Bengis R. J. Am. Chem. Soc. 1912;34:1061. [Google Scholar]
- 195.Fisher A, Henderson GN. Tetrahedron Lett. 1983;24:131. [Google Scholar]
- 196.Andersen RJ, Faulkner DJ. J. Am. Chem. Soc. 1975;94:936. [Google Scholar]
- 197.Schoenfeld RC, Ganem B. Tetrahedron Lett. 1998;39:4147. [Google Scholar]
- 198.Schoenfeld RC, Lumb JP, Fantini J, Ganem B. Bioorg. Med. Chem. Lett. 2000;10:2679. doi: 10.1016/s0960-894x(00)00543-6. [DOI] [PubMed] [Google Scholar]
- 199.Schoenfeld RC, Conova S, Rittschof D, Ganem B. Bioorg. Med. Chem. Lett. 2002;12:823. doi: 10.1016/s0960-894x(02)00022-7. [DOI] [PubMed] [Google Scholar]
- 200.Hoshino O, Murakata M, Yamada K. Bioorg. Med. Chem. Lett. 1992;2:1561. [Google Scholar]
- 201.Nicolaou KC, Hughes R, Pfefferkorn JA, Barluenga S, Roecker AJ. Chem. Eur. J. 2001;7:4280. doi: 10.1002/1521-3765(20011001)7:19<4280::aid-chem4280>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 202.Lehn J-M. Chem. Eur. J. 1999;5:2455. [Google Scholar]
- 203.Otto S, Furlan RLE, Sanders JKM. J. Am. Soc. Chem. 2001;122:12063. [Google Scholar]
- 204.Lees WJ, Whitesides GM. J. Org. Chem. 1993;58:642. [Google Scholar]
- 205.Boehlow TR, Harburn JJ, Spilling CD. J. Org. Chem. 2001;66:3111. doi: 10.1021/jo010015v. [DOI] [PubMed] [Google Scholar]
- 206.Forrester AR, Thomson RH, Woo S-O. J. Chem. Soc., Perkin Trans. 1975;1:2340. [Google Scholar]
- 207.Forrester AR, Thomson RH, Woo S-O. J. Chem. Soc., Chem. Comm. 1973:604. [Google Scholar]
- 208.Forrester AR, Thomson RH, Woo S-O. J. Chem. Soc., Perkin Trans. 1975;1:2348. [Google Scholar]
- 209.Boehlow TR, Spilling CD. Nat. Prod. Lett. 1995;7:1. [Google Scholar]
- 210.Conte V, Furi FD, Moro S. Tetrahedron Lett. 1994;35:7429. [Google Scholar]
- 211.Noda H, Niwa M, Yamamura S. Tetrahedron Lett. 1981;22:3247. [Google Scholar]
- 212.Kacan M, Koyuncu D, McKillop A. J. Chem. Soc., Perkin Trans. 1993;1:1771. [Google Scholar]
- 213.Nishiyama S, Yamamura S. Tetrahedron Lett. 1983;24:3351. [Google Scholar]
- 214.Murakata M, Yamada K, Hocino O. Tetrahedron. 1996;52:14713. [Google Scholar]
- 215.Murakata M, Tamura M, Hoshino O. J. Org. Chem. 1997;62:4428. doi: 10.1021/jo970082i. [DOI] [PubMed] [Google Scholar]
- 216.Forrester AR, Thomson RH, Woo S-O. Justus Liebigs Ann. Chem. 1978:66. [Google Scholar]
- 217.Nishiyama S, Yamamura S. Bull. Chem. Soc. Jap. 1985;58:3453. [Google Scholar]
- 218.Wasserman HH, Ennis DW, Power PL, Mitchell JR. J. Org. Chem. 1993;58:4785. [Google Scholar]
- 219.Wasserman HH, Ho WB. J. Org. Chem. 1994;59:4364. [Google Scholar]
- 220.Wasserman HH, Peterson AK. Tetrahedron Lett. 1997;38:953. [Google Scholar]
- 221.Wasserman HH, Peterson AK. J. Org. Chem. 1997;62:8972. [Google Scholar]
- 222.Wasserman HH, Wang J. J. Org. Chem. 1998;63:5581. [Google Scholar]
- 223.Murakata M, Yamada K, Hoshino O. Heterocycles. 1998;47:921. [Google Scholar]
- 224.Winterfeldt E. Chem. Rev. 1993;93:827. [Google Scholar]; b Borm C, Meibom D, Winterfeldt E. Chem. Commun. 1996:887. [Google Scholar]; c Winterfeldt E, Borm C, Nerenz F. Advances in Asymmetric Synthesis. 1997;2:1. [Google Scholar]
- 225.Jones PG, Weinmann H, Winterfelt E. Angew. Chem. 1995;107:489. [Google Scholar]; Angew. Chem. Int. Ed. 1995;34:448. [Google Scholar]
- 226.Beil W, Jones PG, Nerenz F, Winterfeldt E. Tetrahedron. 1998;54:7273. [Google Scholar]
- 227.Goldenstein K, Fendert T, Proksch P, Winterfeldt E. Tetrahedron. 2000;56:4173. [Google Scholar]
- 228.Noda H, Niwa M, Yamamura S. Tetrahedron Lett. 1981;22:3247. [Google Scholar]
- 229.Nishiyama S, Suzuki T, Yamamura S. Chem. Lett. 1851. 1982 [Google Scholar]
- 230.Nishiyama S, Yamamura S. Tetrahedron Lett. 1982;23:1281. [Google Scholar]
- 231.Nishiyama S, Yamamura S. Tetrahedron Lett. 1982;23:3699. [Google Scholar]
- 232.Guo ZW, Machiya K, Salamonczyk GM, Sih CJ. J. Org. Chem. 1998;63:4269. [Google Scholar]
- 233.Guo ZW, Salamonczyk GM, Han K, Machiya K, Sih CJ. J. Org. Chem. 1997;62:6700. [Google Scholar]
- 234.Couladouros EA, Moutsos VI, Pitsinos EN. ARKIVOC. Gainesville; FL, United States: 2003. p. 92. http://www.arkat-usa.org/ark/journal/2003/General_Part(xv)/AV-910A/910A.pdf. [Google Scholar]
- 235.Couladouros EA, Moutsos VI. Tetrahedron Lett. 1999;40:7027. [Google Scholar]
- 236.Couladouros EA, Moutsos VI. Tetrahedron Lett. 1999;40:7023. [Google Scholar]
- 237.Carreno MC, Ruano JG, Sanz G, Toledo MA, Urbano A. J. Org. Chem. 1995;60:5328. [Google Scholar]
- 238.Bailey KL, Molinski TF. J. Org. Chem. 1999;64:2500. [Google Scholar]
- 239.Bailey KL, Molinski TF. Tetrahedron Lett. 2002;43:9657. [Google Scholar]
- 240.Masuno MN, Molinski TF. J. Nat. Prod. 2003;66:112. doi: 10.1021/np020382h. [DOI] [PubMed] [Google Scholar]
- 241.Leone-Stumpf D, Lindel T. Chem. Eur. J. 2001;7:3961. doi: 10.1002/1521-3765(20010917)7:18<3961::aid-chem3961>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 242.Leone-Stumpf D, Lindel T. Eur. J. Org. Chem. 2003:1853. [Google Scholar]
- 243.Andreas KT, Caroline R, Wolfgang S. Org. Lett. 2002;4:3287. [Google Scholar]
- 244.Grieco PA, Hon YS, Perez-Medrano A. J. Am. Chem. Soc. 1988;110:1630. [Google Scholar]
- 245.Chan WR, Tinto WF, Manchand PS, Todaro LJ. J. Org. Chem. 1987;52:3091. [Google Scholar]
- 246.Grieco PA, Perez-Medrano AA. Tetrahedron Lett. 1988;29:4225. [Google Scholar]
- 247.Irreland CM, Molinski TF, Roll DM, Zabriskie TM, Mckee TC, Swersey JC, Foster MP. In: Bioorganic Marine Chemistry. Scheuer PJ, editor. Vol. 3. Springer-Verlag; New York: 1989. p. 1. [Google Scholar]
- 248.Shield LS, Rinehart KL., Jr. Antibiotics, “Isolation, Separation and Purification”. Elsevier; New York: 1978. p. 309. [Google Scholar]
- 249.Hashimoto Y. Marine Toxins and Other Bioactive Marine Metabolites. Japan Scientific Societies Press; Tokyo: 1976. p. 245. [Google Scholar]
- 250.Teeyapant R, Woerdenbag HJ, Kreis P, Hacker J, Wray V, Witte L, Proksch P. Z. Naturforsch. 1993;48c:939. doi: 10.1515/znc-1993-11-1218. [DOI] [PubMed] [Google Scholar]
- 251.Peng J, Shen X, El Sayed KA, Dunbar DC, Perry TL, Wilkins SP, Hamann MT. J. Food Agr. Chem. 2003;51:2246. doi: 10.1021/jf0207880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Encarnacion-Dimayuga R, Ramirez MR, Luna-Herrera J. Pharm. Bio. 2003;41:384. [Google Scholar]
- 253.El Sayed KA, Bartyzel P, Shen X, Perry TL, Zjawiony JK, Hamann MT. Tetrahedron. 2000;56:949. [Google Scholar]
- 254.Nicolaou KC, Hughes R, Pfefferkorn JA, Barluenga S, Roecker AJ. Chem. Eur. J. 2001;7:4280. doi: 10.1002/1521-3765(20011001)7:19<4280::aid-chem4280>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 255.Nicolaou KC, Hughes R, Pfefferkorn JA, Barluenga S. Chem. Eur. J. 2001;7:4280. doi: 10.1002/1521-3765(20011001)7:19<4280::aid-chem4280>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 256.Kim D, II, Lee S, Jung JH, Yang S-II. Arch. Pharm. Res. 1999;22:25. doi: 10.1007/BF02976431. [DOI] [PubMed] [Google Scholar]
- 257.Burkholder PR. In: Biology and Geology of Coral Reefs. Jones OA, Endean R, editors. Academic Press; New York: 1973. p. 117. [Google Scholar]
- 258.Breakman JC, Daloze D. Pure Appl. Chem. 1986;58:357. [Google Scholar]
- 259.MaCaffrey EJ, Endean R. Mar. Biol. 1985;89:1. [Google Scholar]
- 260.Weiss B, Ebel R, Elbraechter M, Kirchner M, Proksch P. Biochem. System. Ecol. 1996;24:1. [Google Scholar]
- 261.Bulich AA, Ker-Kong T, Scheibner G. J. Bioluminesc. Chemoluminesc. 1990;5:71. doi: 10.1002/bio.1170050202. [DOI] [PubMed] [Google Scholar]
- 262.Lawton LA, Campbell DL, Beattie KA. Lett. Appl. Microbiol. 1990;11:205. doi: 10.1111/j.1472-765x.1990.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 263.Teeyapant R, Kreis P, Wray V, Witte L, Proksch P. Z. Naturforsch. 1993;48c:640. doi: 10.1515/znc-1993-11-1218. [DOI] [PubMed] [Google Scholar]
- 264.Proksch P. Dtsch. Apoth. Ztg. 1994;134:5069. [Google Scholar]
- 265.Kreuter MH, Bernd A, Holzmann H, Maidhof WA, Wei mann N, Kljajic Z, Batel R. Z. Naturforsch. 1989;44c:680. doi: 10.1515/znc-1989-7-822. [DOI] [PubMed] [Google Scholar]
- 266.Müller WEG, Kreuter MH. Ger. Offen. 1989:91780. CODEN: GWXXBX DE 3737108 A1 19890511 CAN 112. [Google Scholar]
- 267.Müller WEG, Rohde HJ, Beyer R, Maidhof A, Bachmann M, Taschner H, Zahn RK. Cancer Res. 1975;35:2160. [PubMed] [Google Scholar]
- 268.Müller WEG, Zahn RK, Maidhof A, Beyer R, Arendes J. Chem.-Biol. Interact. 1978;23:141. doi: 10.1016/0009-2797(78)90001-7. [DOI] [PubMed] [Google Scholar]
- 269.Müller WEG, Maidhof A, Arendes J, Geurtsen W, Zahn RK, Schmidseder R. Cancer Res. 1979;39:3768. [PubMed] [Google Scholar]
- 270.Müller WEG, Obermeier J, Maidhof A, Zahn RK. Chem.-Biol. Interact. 1974;8:183. doi: 10.1016/0009-2797(74)90040-4. [DOI] [PubMed] [Google Scholar]
- 271.Oda Y, Nakamura S, Oki I, Kato T, Shinagawa H. Mut. Res. 1985;147:219. doi: 10.1016/0165-1161(85)90062-7. [DOI] [PubMed] [Google Scholar]
- 272.Corbett TH, Leopold WR, Dykes DJ, Roberts BJ, Griswold DP, Schabel FW. Cancer Res. 1982;42:1707. [PubMed] [Google Scholar]
- 273.Kreuter MH, Leake RE, Rinaldi F, Mueller-Klieser W, Maidhof A, Mueller WEG, Schroeder HC. Comp. Biochem. Physiol. B Comp. Biochem. 1990;97:151. doi: 10.1016/0305-0491(90)90194-x. [DOI] [PubMed] [Google Scholar]
- 274.Pepinsky RB, Sinclair LK. Nature, Lond. 1986;321:81. doi: 10.1038/321081a0. [DOI] [PubMed] [Google Scholar]
- 275.Kreuter MH, Robitzki A, Chang S, Steffen R, Michaelis M, Kljajic Z, Bachmann M, Schroder HC, Muller WEG. Comp. Biochem. Physiol. C Comp. Pharmacol. 1992;101:183. doi: 10.1016/0742-8413(92)90217-u. [DOI] [PubMed] [Google Scholar]
- 276.Koulman A, Proksch P, Beekman RACE, Uden WV, Konings AWT, Pedersen JA, Pras N, Woerdenbag HJ. J. Nat. Prod. 1996;59:591. doi: 10.1021/np960167z. [DOI] [PubMed] [Google Scholar]
- 277.Meister A. Science. 1982;220:472. doi: 10.1126/science.6836290. [DOI] [PubMed] [Google Scholar]
- 278.Arrick BA, Nathan CF. Cancer Res. 1984;44:4224. [PubMed] [Google Scholar]
- 279.Workman P. Oncology Res. 1994;6:461. [PubMed] [Google Scholar]
- 280.Rodriguez-Nieto S, Gonzalez-Iriarte M, Carmona R, Munoz-Chapuli R, Medina MA, Quesada AR. FASEB J. 2002;16:261. doi: 10.1096/fj.01-0427fje. [DOI] [PubMed] [Google Scholar]
- 281.Quesada AR, Torres MAM, Chapuli RM, Puentes JLF. Aeroplysinin-1 and related compounds for the treatment of angiogenic diseases. PCT Int. Appl. 2002:14. CODEN: PIXXD2 WO 2002043718 A2 20020606.
- 282.Folkman J. N. Engl. J. Med. 1971;285:1182. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 283.Harris AL. Recent Results Cancer Res. 1998;152:341. doi: 10.1007/978-3-642-45769-2_33. [DOI] [PubMed] [Google Scholar]
- 284.Paleolog EM, Miotla JM. Angiogenesis. 1998;2:295. doi: 10.1023/a:1009229508096. [DOI] [PubMed] [Google Scholar]
- 285.Perez LB, Bair KW, Karl D, Green MA, Lamberson C, Remiszewski SW, Sambucetti LC. Proc. 92nd AACR. 2001;42:927. [Google Scholar]
- 286.Shim SS, Lee H-S, Shin J, Kwon HJ. Cancer Lett. 2004;203:163. doi: 10.1016/j.canlet.2003.08.036. [DOI] [PubMed] [Google Scholar]
- 287.Layman PL. Chem. Eng. News, May. 1995;1:23. [Google Scholar]
- 288.Pawlik JR. Chem. Rev. 1993;93:191. [Google Scholar]
- 289.Clare AS, Rittschof D, Gerhart DJ, Maki JS. Invert. Reprod. Dev. 1992;22:67. [Google Scholar]
- 290.Thompson JE. Mar. Biol. 1985;88:23. [Google Scholar]
- 291.Teeyapan R, Proksch P. Naturwissenschaften. 1993;80:369. [Google Scholar]
- 292.Yamada A, Kitamura H, Yamaguchi K, Fukuzawa S, Kamijima C, Yazawa K, Kuramoto M, Wang G-Y-S, Fujitani Y, Uemura D. Bull. Chem. Soc. Jap. 1997;70:3061. [Google Scholar]
- 293.Diers JA, Pennaka HK, Peng J, Bowling JJ, Duke SO, Hamann MT. J. Nat. Prod. 2004;67:2117. doi: 10.1021/np040097t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Takito J, Nakamura H, Kobayashi J, Ohizumi Y, Ebisawa K, Nonomura Y. J. Biol. Chem. 1986;261:13861. [PubMed] [Google Scholar]
- 295.Fang Y-II, Yokota E, Mabuchi I, Nakamura H, Ohizumi Y. Biochemistry. 1997;36:15561. doi: 10.1021/bi971363n. [DOI] [PubMed] [Google Scholar]
- 296.Nakamura Y, Kobayashi M, Nakamura H, Wu H, Kobayashi J, Ohizumi Y. Eur. J. Biochem. 1987;167:1. doi: 10.1111/j.1432-1033.1987.tb13296.x. [DOI] [PubMed] [Google Scholar]
- 297.Nakamura E, Kobayashi J, Nakamura Y, Oizumi Y. Tyrosine derivatives and salts. Jpn. Kokai Tokkyo Koho. 1986:111799. CODEN: JKXXAF JP 61130274 A2 19860618 CAN 107.
- 298.Takito J, Nakamura H, Kobayashi J, Ohizumi Y. Biochim. Biophys. Acta. 1987;912:404. doi: 10.1016/0167-4838(87)90045-8. [DOI] [PubMed] [Google Scholar]
- 299.Takito J, Ohizumi Y, Nakamura H, Kobayashi J, Ebisawa K, Nonomura Y. Euro. J. Pharmacol. 1987;142:189. doi: 10.1016/0014-2999(87)90107-5. [DOI] [PubMed] [Google Scholar]
- 300.Yoshida M, Kijima M, Akita M, Beppu T. J. Biol. Chem. 1990;265:17174. [PubMed] [Google Scholar]
- 301.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. J. Biol. Chem. 1993;268:22429. [PubMed] [Google Scholar]
- 302.Remiszewski SW, Sambucetti LC, Atadja P, Bair KW, Cornell WD, Green MA, Howell KL, Jung M, Kwon P, Trogani N, Walker H. J. Med. Chem. 2002;45:753. doi: 10.1021/jm015568c. [DOI] [PubMed] [Google Scholar]
- 303.Remiszewski SW, Sambucetti LC, Atadja P, Bair KW, Cesarz D, Chandramouli N, Cheung M, Cornell W, Dean K, Green MA, Howell KL, Lassota P, Sorensen E, Sheng T, Taplin F, Trogani N, Versace R, Walker H, Wood A. Proc. 92nd AACR. 2002;43:740. [Google Scholar]
- 304.Barre-Sinoussl F, Chermann JC, Rey F, Nugeyre MT, Chamarel S, Gruest J, Daguet C, Axler-Blin C, Vezlnet-Brun F, Rozenbaum C, Monlagnler L. Science. 1983;220:868. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 305.Loya S, Rudi A, Kashman Y, Hizi A. Biochem. J. 1999;344:85. [PMC free article] [PubMed] [Google Scholar]
- 306.Chen L, Molinski TF, Pessah IN. J. Biol.Chem. 1999;274:32603. doi: 10.1074/jbc.274.46.32603. [DOI] [PubMed] [Google Scholar]
- 307.Gonzalez A, Kirsch WG, Shirokova N, Pizarro G, Brum G, Pessah IN, Stern MD, Cheng H, Rios E. Proc. Natl. Acad. Sci. 2000;97:4380. doi: 10.1073/pnas.070056497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.DiJulio DH, Watson EL, Pessah IN, Jacobson KL, Ott SM, Buck ED, Singh JC. J. Biol. Chem. 1997;272:15687. doi: 10.1074/jbc.272.25.15687. [DOI] [PubMed] [Google Scholar]
- 309.Kotte A, Myers J, Shih N-Y, Egan RW, Clark MA. Biochem. Biophys. Res. Commun. 1990;168:979. doi: 10.1016/0006-291x(90)91125-c. [DOI] [PubMed] [Google Scholar]
- 310.Trzeciakowski JP. J. Pharmacol. Exp. Ther. 1987;243:874. [PubMed] [Google Scholar]
- 311.Gochfeld DJ, El Sayed KA, Yousaf M, Hu J, Bartyzel P, Dunbar DC, Wilkins SP, Zjawiony JK, Schinazi RF, Wirtz S, Tharnish PM, Hamann MT. Mini Rev. Med. Chem. 2003;3:451. doi: 10.2174/1389557033487962. [DOI] [PubMed] [Google Scholar]