Abstract
Whole liver engineering holds the promise to create transplantable liver grafts that may serve as substitutes for donor organs, addressing the donor shortage in liver transplantation. While decellularization and recellularization of livers in animal models have been successfully achieved, scale up to human livers has been slow. There are a number of donor human livers that are discarded because they are not found suitable for transplantation, but are available for engineering liver grafts. These livers are rejected due to a variety of reasons, which in turn may affect the decellularization outcome. Hence, a one-size-fit-for all decellularization protocol may not result in scaffolds with consistent matrix quality, subsequently influencing downstream recellularization and transplantation outcomes. There is a need for a noninvasive monitoring method to evaluate the extent of cell removal, while ensuring preservation of matrix components during decellularization. In this study, we decellularized rat livers using a protocol previously established by our group, and we monitored decellularization through traditional destructive techniques, including evaluation of DNA, collagen, and glycosaminoglycan (GAG) content in decellularized scaffolds, as well as histology. In addition, we used computed tomography and perfusate analysis as alternative nondestructive decellularization monitoring methods. We found that DNA removal correlates well with the Hounsfield unit of the liver, and perfusate analysis revealed that significant amount of GAG is removed during perfusion with 0.1% sodium dodecyl sulfate. This allowed for optimization of our decellularization protocol leading to scaffolds that have significantly higher GAG content, while maintaining appropriate removal of cellular contents. The significance of this is the creation of a nondestructive monitoring strategy that can be used for optimization of decellularization protocols for individual human livers available for liver engineering.
Introduction
Orthotopic transplantation is the only definitive treatment for end-stage liver failure. While the procedure is successful in achieving high rates of survival, the treatment is limited by the significant shortage of donor organs and many patients die while waiting for a suitable liver. This significant gap has been the driving force behind tissue engineering, which aims to create tissue substitutes. Despite the efforts since its conception more than 20 years ago, construction of a large mass of liver tissue that is suitable for transplantation has been unsuccessful.
In the recent years, whole organ engineering has drawn attention due to its promise to fabricate whole organ substitutes for solid organ transplantation.1 In whole organ engineering, human cadaveric or animal organs are perfused with a detergent solution to remove the cellular components leaving behind the extracellular matrix (ECM) scaffold that still maintains the architecture and composition of the native organ. The resulting scaffold can subsequently be repopulated with healthy cells to create a transplantable functional organ substitute. We and others have shown the feasibility of this approach using rat,2,3 ferret,4 and pig5,6 livers. While these studies constitute the proof of concept of the whole liver engineering approach, there are several factors that need to be addressed for its clinical translation.
Some of the challenges include ensuring consistent matrix quality postdecellularization, recapturing native cellular architecture and attaining the physiological functions of the organ after recellularization, and maintaining graft viability and recipient survival upon transplantation. As the first step of the process, decellularization is critical because the downstream recellularization and transplantation outcomes are directly dependent on the quality of the scaffold. Ensuring consistent scaffold quality becomes more challenging when discarded donor livers are used for scaffold preparation because the donor liver quality varies greatly depending on donor characteristics. Therefore, it is essential to monitor progress of decellularization without damaging the organ and adjust its parameters to ensure complete removal of cellular components, while maintaining the ECM composition.
There are a number of approaches to organ decellularization with the most widely used methods involving chemical treatment of the organ with a combination of sodium dodecyl sulfate (SDS), Triton X-100, ammonium hydroxide, and phosphate-buffered saline (PBS) perfusion,7 with or without exertion of mechanical forces.8 Conventionally, the progress of decellularization is monitored macroscopically by change in gross appearance of the organ to pale or translucent color and the completeness of decellularization is confirmed when there is less than 50 nanograms of double-stranded DNA per milligram dry weight9 or less than 5% cell survival rate10 with no visible nuclear material in histologic analysis with DAPI or hematoxylin and eosin (H&E). While these criteria are widely accepted for preparation of decellularized organs, they are based on endpoint analysis of the scaffold by destructive assays and do not allow assessment of matrix integrity during cell removal progression. Subsequently, it becomes difficult to make adjustments to the decellularization protocol based on individual organ characteristics such as size and vascular anatomy. A recent study reported a method for monitoring the organ decellularization through rheological measurements of the perfusate and found correlations between overall protein, DNA and biomass content, and the viscosity of the perfusate.11 The technique prevents excessive exposure of individual organs to decellularization reagents that may potentially disrupt ECM integrity.
In this study, we present an alternative approach to evaluate the scaffold properties noninvasively and determine the extent of cell removal during rat liver decellularization. Computer tomography scanning was used to determine the Hounsfield units (HU) of the livers throughout decellularization, which correlated well with the DNA content of the scaffold. Combined with biochemical measurements of the perfusate for total protein, collagen, and glycosaminoglycan (GAG), a decellularization protocol previously established by our group was used to test this monitoring strategy and led to a modified protocol that was shorter and produced scaffolds that retained higher GAG content, while achieving the same amount of DNA removal. These results were validated using traditional characterization methods.
Materials and Methods
Liver harvest
All procedures in regard to animal care, handling, and surgical procedures were in accordance to the Institutional Animal Care and Use Committee (IACUC) at Massachusetts General Hospital. A total of 35 Female Lewis rats (Charles River Laboratories, Wilmington, MA) weighing 150–200 g were used for liver harvest. After anesthetization, a transverse incision across the abdomen was made. To prevent coagulation of the blood, 20 U of heparin (Sagent Pharmaceuticals, Schaumburg, IL) was injected into the heart through the diaphragm. Ligaments connecting the liver lobes to the body cavity and to other lobes were carefully cut. The stomach and intestinal tract were removed to the side to expose the portal vein. An 18-gauge catheter was inserted into the portal vein and connected to a syringe filled with PBS (Sigma-Aldrich, St. Louis, MO). After cutting the inferior vena cava, 20 mL of PBS was slowly injected to clear the organ of blood. The catheter was secured with 5–0 silk sutures and the liver was carefully excised from the body cavity by freeing more ligaments and ligating the superior vena cava, bile duct, and portal vein inferior to the catheter. After excision, the liver was placed in a petri dish filled with PBS and the livers were kept at–80°C until decellularization.
Liver decellularization
Rat livers were thawed and decellularization was performed according to a previously established protocol. The perfusion setup consisted of a Masterflex peristaltic pump and a bubble trap. Briefly, livers were thawed, weighed, and cleared with PBS perfusion overnight. Next, livers were perfused with 0.01% SDS (Sigma-Aldrich, St. Louis, MO) for 5 min and with PBS for 1 h. These two steps were repeated thrice, with each repetition increasing the length of 0.01% SDS perfusion by an increment of 5 min. After the last hour of PBS perfusion, livers were perfused with 0.01% SDS overnight followed by 0.1% SDS for 4–18 h, 0.2% SDS for 1–3 h, and 0.5% SDS for 1–3 h. The livers were washed with distilled water for 15 min to remove SDS followed by 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) for 30 min to remove any residual cellular components in the scaffolds. Finally, the decellularized livers were washed with PBS for 2 h. The perfusion rate was kept at 1.2 mL/min throughout. During decellularization, the livers were imaged, their weights were measured, and biopsy samples were taken for later analysis. Perfusate samples were also collected during perfusion with 0.1% SDS and stored in–80°C for further analysis. The protocol timeline is outlined in Figure 1A.
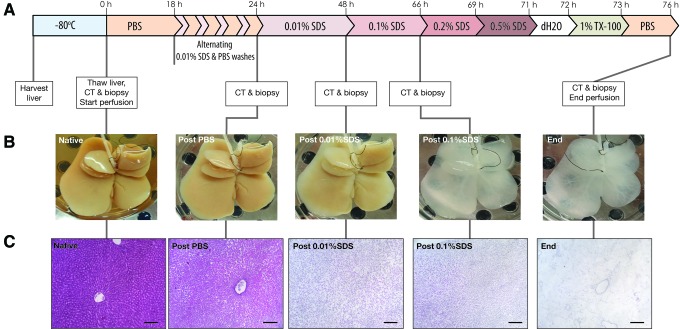
FIG. 1.
Rat liver decellularization. (A) Timeline of the decellularization protocol with noted biopsies and CT scans. (B) Macroscopic view of the whole liver matrix shows that a major color change occurs postperfusion with 0.1% SDS. (C) Histology of liver biopsies taken at each major change in solution shows that a major difference in cell density occurs after perfusion of PBS and after 0.01% SDS. Another change in cell density occurs after Triton X-100 perfusion removing any remnant cellular material (scale bar = 150 μm). CT, computed tomography; PBS, phosphate-buffered saline; SDS, sodium dodecyl sulfate. Color images available online at www.liebertpub.com/tec
Computed tomography scanning of decellularized livers
To evaluate the changes in tissue density during decellularization, rat livers were analyzed by computed tomography (CT) scanning at five different time points as indicated in Figure 1A. All livers were placed in a clear, sterile zip-lock bag and imaged using a CereTom® CT scanner (Neurologica, Danvers, MA) located in Shriners Hospital for Children in Boston. The machine was calibrated each time before use to ensure high-quality sections. After a measurement of the liver's dimensions was taken to ensure a full coverage scan, a noncontrast scan was executed. Dicom 3-compliant images were produced and reconstructed on the CereTom CT scanner software. The images were then compiled and HU of the liver was determined using AMIDE open source software (http://amide.sourceforge.net).
Determination of DNA, total collagen, and GAGs
DNA extraction and purification from the liver biopsy samples weighing ∼20 mg were accomplished with the Purelink Genomic DNA kit (Life Technologies, Grand Island, NY) following manufacturer's recommendations. The concentration of extracted genomic DNA was determined using the Picogreen Assay kit (Life Technologies, Grand Island, NY).
To determine the GAG content of the liver scaffolds, a protocol modified from Farndale et al.12 was followed. Samples weighing ∼100 mg were lyophilized and hydrolyzed in 0.5 or 0.25 mL 6 M Hydrochloric acid (HCl) (Fisher, Waltham, MA) for 20 h at 95°C. The hydrolyzed samples were cooled to room temperature, 10 μL of sample was mixed with 250 μL dimethylene blue color reagent, and the absorbance was immediately measured at a wavelength of 525 nm. The results were compared to a calibration curve prepared using a serial dilution of 0.5 mg/mL chondroitin sulfate A (Sigma-Aldrich, St. Louis, MO) in PBS.
Collagen content was determined using the Quickzyme Total Collagen assay kit (Cedarlane Labs, Burlington, NC) following manufacturer's recommendations.
The results were normalized to wet tissue weight and corrected by the ratio of post- to predecellularization whole liver weights at each time point.
Western blot analysis
One hundred milligram whole liver tissue, decellularized liver matrix, or 1 million primary rat hepatocytes were crushed using a pestle and mortar and incubated in RIPA buffer containing protease inhibitors (Life Technologies, Grand Island, NY) at 4°C on a shaker for lysis. Next, total protein concentration was determined with the Pierce BCA Assay kit (Life Technologies, Grand Island, NY). Samples were prepared with equal volume of Laemmli Buffer (Bio-Rad, Hercules, CA) mixed with β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO). Postboiling, the samples were centrifuged at 14,000g for 1 min. Using precast gels (Bio-Rad), samples were loaded along with a protein ladder (Li-Cor, Lincoln, NE). After electrophoresis, samples were transferred on a nitrocellulose membrane (Bio-Rad) with a subsequent blocking step with the Odyssey blocking buffer (Li-Cor) for 1 h at room temperature. Primary antibody for β-actin (JLA-20; Developmental Studies Hybridoma Bank, Iowa City, IA) was added at a dilution of 0.2 μg/mL and membrane was incubated overnight at 4°C. Secondary antibody (Li-Cor) was added at a 1:10,000 dilution after membrane washes and left to incubate for 1 h at room temperature protected against light. After incubation, the membrane was washed and stored at 4°C until development. An Odyssey membrane scanner was used to develop the membrane and Image Studio Lite software was used for subsequent analysis.
Histology
Liver biopsies from each time point were fixed in 10% formalin for preparation of histological analysis. Samples were embedded in paraffin for tissue slicing, sliced at 10 μm, and stained with hematoxylin and eosin. Images of each biopsy were taken with a Nikon Eclipse 800 microscope and SPOT camera. Subsequent quantitative analysis was performed with Image J.
Statistical analysis
The results were reported as the average of four to eight independent experiments and error bars representing standard error of means. One-way ANOVA followed by Dunnett's multiple comparisons test with Bonferroni correction or unpaired two tailed t-test was performed using GraphPad Prism version 6.0 h for Mac OS × 10.11, GraphPad Software (La Jolla, CA).
Results
Morphological changes during decellularization
The rat liver decellularization protocol involved a gentle perfusion through the portal vein with increasing concentrations of SDS solution that continued for 76 h (Fig. 1A). After an initial perfusion with PBS, which allowed for clearance of any cellular debris that resulted from freeze and thaw cycle following storage, the liver was gently exposed to SDS solution between 18 and 24 h for short durations and alternating with PBS washes. This strategy was chosen to limit pressure buildup due to sudden lysis inside the organ and any damage to the Glisson's capsule. After the initial stepwise exposure, the SDS concentration was increased at time points where no change in the color was observed and the effluent was no longer cloudy. During decellularization, the gross shape of the organ remained the same with only changes in its color from brown to white as the decellularization progressed (Fig. 1B). The most drastic change in color occurred during perfusion with 0.1% SDS. The change in the gross liver morphology was evaluated microscopically by hematoxylin and eosin staining of histological sections, which showed that the cellular integrity was lost after freeze/thaw cycle and the cellular debris was gradually cleared afterward (Fig. 1C).
Quantitative markers for monitoring decellularization
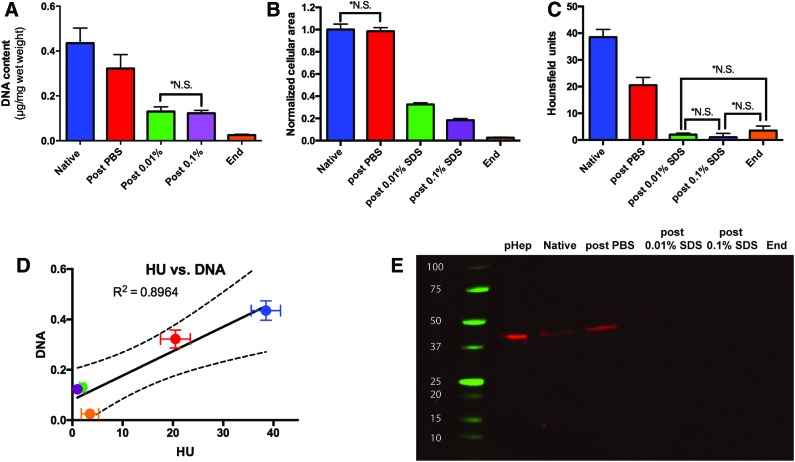
A widely accepted endpoint for completeness of decellularization is significant removal of DNA from the ECM scaffold. We found that DNA content of the liver during decellularization decreased with the most drastic drop during 0.01% SDS perfusion (Fig. 2A). While 0.1% SDS perfusion did not affect the DNA content significantly (p = 0.774), the second-largest decline in DNA content was observed in the last stage of the protocol where the liver was perfused with 0.2% and 0.5% SDS followed by 1% Triton X-100. Overall, the final decellularized liver matrix contained DNA that is less than 5% of the native liver (0.44 ± 0.07 vs. 0.02 ± 0.01 μg/mg tissue). Another marker for evaluation of decellularization is the absence of H&E staining in histological sections. We quantified the area that is positively stained with H&E by image analysis and found that perfusion with 0.01% SDS removes cellular components in the liver sections significantly. Further exposure to higher concentrations of SDS slightly reduced the cellular staining with almost no positive staining present at the end of decellularization (Fig. 2B).
FIG. 2.
Quantitative analysis of cellular removal during decellularization. (A) DNA analysis, (B) Quantitative analysis of the hematoxylin and eosin staining, and (C) HU determined using CT images show that cellular material is mostly removed postperfusion of 0.01% SDS. (D) A correlation is formed between DNA and HU demonstrating that CT scanning is an indicator of cellular removal during decellularization. (E) Western blot of β-actin confirms removal of cellular material after 0.01% SDS as β-actin is no longer present post 0.01% SDS (pHep: primary rat hepatocytes). All error bars represent SEM (n = 6, *NS differences are not significant at p = 0.05). HU, Hounsfield unit; SEM, standard error of means. Color images available online at www.liebertpub.com/tec
In an effort to evaluate the progression of decellularization noninvasively, we utilized CT imaging to determine the HU. Native liver tissue has an HU of 40–60 and water has an HU of zero, and we expect to see a decrease in HU as cellular components of the organ are removed during decellularization. As expected, HU of the liver decreased steadily from 38.5 ± 2.9 to 3.5 ± 1.7 until the end of 0.01% SDS perfusion. However, there was no further significant change in HU of the matrix as the decellularization progressed (Fig. 2C). This result agrees with the DNA content measurement (R2 = 0.8965, Pearson coefficient = 0.9441 and p = 0.016) suggesting that HU may be used to predict DNA removal during decellularization (Fig. 2D).
To further ensure removal of cellular components, we determined the presence of the most abundant cellular protein, β-actin, in the liver during decellularization. Western blot analysis demonstrated that perfusion of livers with 0.01% SDS for 24 h removed β-actin to undetectable levels at later stages of decellularization (Fig. 2E).
Preservation of ECM components
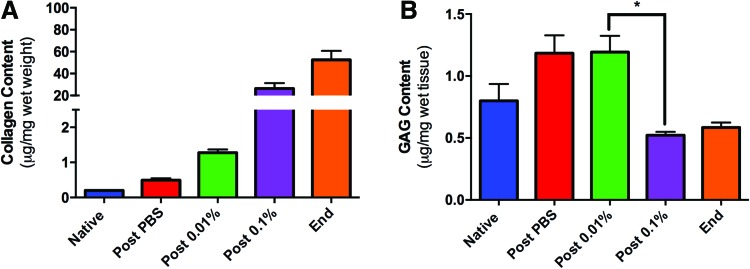
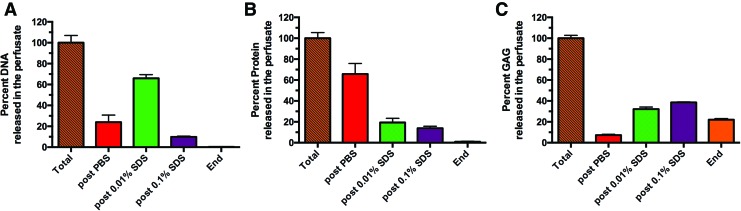
The results thus far show that decellularization of rat livers with increasing concentrations of SDS is effective and majority of the cellular components are removed during the second stage decellularization, that is, perfusion with 0.01% SDS, and further exposure to higher concentrations of SDS may be excessive. While the removal of cellular components from the organ is an essential aspect of decellularization, it is equally critical to preserve the ECM components in the final scaffold. Therefore, we measured two major ECM components, collagen and GAGs, in the liver matrix during decellularization. Total collagen content normalized to total liver weight was found to increase during the decellularization with statistically significant differences among means (p < 0.0001, one way ANOVA; Fig. 3A), which may be due to the significant loss of cellular protein mass (data not shown). It was found that GAG content of the matrix showed a slight increase after PBS wash, with no change until after the 0.01% SDS perfusion, but significantly decreased during the 0.1% SDS perfusion (Fig. 3B). The most significant change was observed after 0.1% SDS perfusion (p < 0.0001). Combined with the DNA, H&E, and β-actin data, these results suggest that the matrix may be excessively washed with 0.1% SDS removing ECM-related components, while not significantly altering its cell associated content. This observation was investigated further by analyzing the cellular and ECM-related components in the perfusate collected at the end of each stage of decellularization (Fig. 4). Interestingly, much of the DNA from the liver was released after PBS (23.9% ± 6.8%) and 0.01% SDS perfusion (66.0% ± 3.5%), while DNA measured after 0.1% SDS was significantly lower (9.8% ± 0.7%; Fig. 4A), possibly contributing to the fact that the DNA content of the scaffold remains unchanged after 0.1% SDS perfusion (Fig. 2A). Total protein content of the perfusate also declined as the decellularization progressed, more than 85% of the removal of proteins occurred after PBS (65.8% ± 5.0%) and 0.01% SDS (19.4% ± 2.0%) perfusion (Fig. 4B). We also confirmed that collagen was not present in the effluent at any stage of decellularization (data not shown). However, more than 60% of the total GAGs were removed during perfusion with 0.1% SDS (38.5% ± 0.6%) and higher concentrations of SDS (22.1% ± 1.1%; Fig. 4C), once again suggesting that decellularization beyond 0.01% SDS may be causing damage to matrix-associated components, while not affecting the cellular contents much.
FIG. 3.
Biochemical analysis of ECM components during decellularization. (A) Analysis of collagen concentration in the ECM confirms that collagen is retained in the ECM throughout decellularization (p < 0.0001 by one-way ANOVA). (B) Glycosaminoglycan (GAG) content remains constant until perfusion with 0.1% SDS after which it decreases (*p < 0.0001 by unpaired two-tailed t-test). All error bars represent SEM (n = 8). ECM, extracellular matrix. Color images available online at www.liebertpub.com/tec
FIG. 4.
Analysis of decellularization perfusate. (A) Cumulative DNA content of the perfusate after each stage shows the greatest amount of DNA is perfused out after 0.01% SDS perfusion. (B) Total protein analysis of the perfusate demonstrates that most proteins are expelled after the perfusion with PBS and slowly decreased after higher SDS concentrations. (C) Cumulative GAG shows that a lot of GAGs are removed from the ECM throughout the decellularization process with the most prominent removal happening during 0.1% SDS perfusion. All error bars represent SEM (n = 4). Color images available online at www.liebertpub.com/tec
Modified protocol for decellularization of rat livers
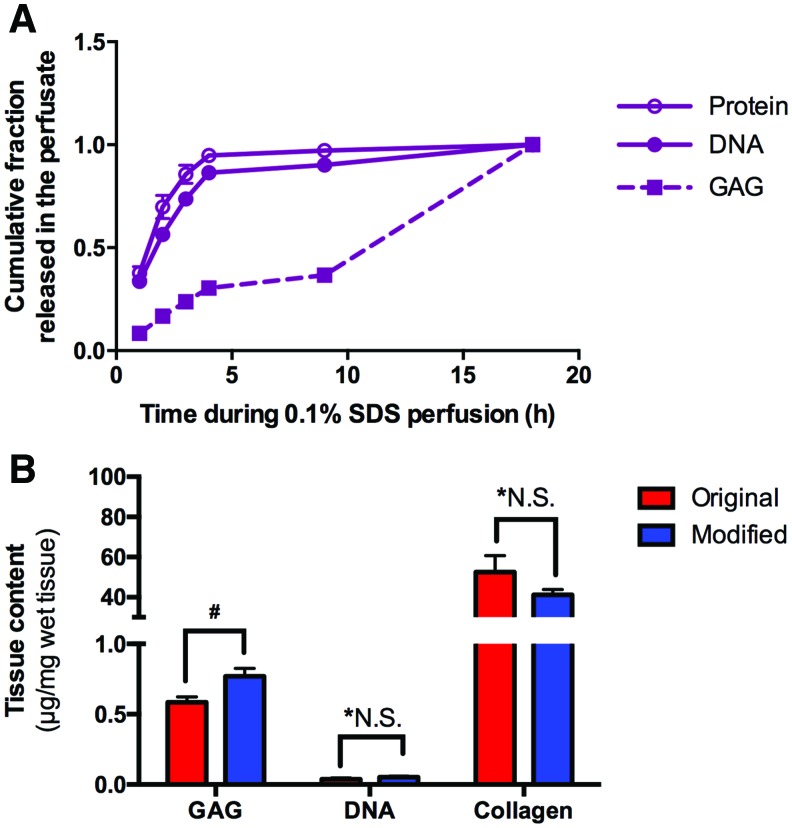
A detailed analysis of perfusate contents during 0.1% SDS perfusion revealed that a significant fraction of the total DNA (86.4% ± 2.2%) and proteins (94.8% ± 2.3%) were washed during the first 4 h, while 69.6% ± 1.6% of GAGs were removed during the remainder of this stage (Fig. 5A). To limit the loss of matrix-associated components from the liver ECM during decellularization, the protocol was modified by reducing the duration of 0.1% SDS perfusion to 4 h and the resulting matrix scaffold was analyzed for DNA, collagen, and GAG content. As expected, it was found that the DNA and collagen content remained unchanged at the final stage of the decellularization protocol (p = 0.369 and 0.251, respectively). The GAG content increased from 0.584 ± 0.040 to 0.769 ± 0.056 μg/mg tissue when the duration of the 0.1% SDS perfusion stage was 4 h instead of 18 h, and the difference was found to be statistically significant (p = 0.01; Fig. 5B).
FIG. 5.
Cumulative DNA, GAG, and total protein during 0.1% SDS perfusion and scaffold analysis after modified protocol. (A) Most of the DNA and proteins are removed during the first 4 h of 0.1% SDS perfusion, while GAG is removed during the last 8 h of perfusion. This set up the platform for our modified decellularization to shorten the 0.1% SDS perfusion to 4 h to prevent significant removal of GAG. (B) Comparison between modified and original decellularization protocols in GAG, DNA, and collagen concentration analysis. Retention of glycosaminoglycan was higher in the modified protocol (#p = 0.01 by unpaired two-tailed t-test), while DNA removal was similar between both protocols (*p = 0.251). Retention of collagen was also similar between both protocols (*p = 0.369). All error bars represent SEM (n ≥ 6, *NS differences are not significant). Color images available online at www.liebertpub.com/tec
Discussion
Perfusion decellularization of whole organs, including the liver, has become an attractive method to create whole liver scaffolds for use in tissue engineering. Several characteristics such as the preserved vascular architecture and the ECM composition resembling the native liver have made decellularized whole liver scaffolds an ideal platform for regenerative medicine applications. While the decellularization protocols differ widely in the type of detergent and/or enzyme utilized to remove the cells or whether mechanical forces are used to aid cell removal, one common feature is that the progress of decellularization is monitored by observing the color change in the liver from its native color to a vaguely defined “white” color. The removal of cells is later confirmed by destructive techniques such as H&E staining in histological sections and DNA content of the resultant scaffold. While this monitoring technique works to a great extent for livers from animal models, its clinical scale-up to human livers may be problematic because variability in the vascular anatomy and liver quality will potentially lead to variable outcomes with a one-size fits-all decellularization protocol, and there is need for a reliable and noninvasive method for monitoring decellularization of whole livers.13
The aim of this study was to understand the changes a rat liver undergoes during decellularization using our previously developed protocol2,14 and identify a noninvasive technique to evaluate the extent of decellularization, which can be scaled up to human or large animal livers. The decellularization protocol involves gentle perfusion of a cadaveric rat liver through the portal vein with increasing concentrations of SDS solution. The process is slow and lasts for about 3 days, but is sufficiently gentle that the gross appearance of the liver is not altered and the Glisson's capsule is not damaged during the decellularization. The most significant color change takes place after perfusion with 0.1% SDS, but interestingly, the notable decrease in both DNA and cellular protein content in the liver occurs during perfusion with 0.01% SDS, which is before the color change. This observation was also confirmed by perfusate analysis, which suggests that majority of DNA and proteins, specifically β-actin, are removed during PBS and 0.01% SDS perfusion, and prolonged perfusion with 0.1% SDS does not affect the liver's cellular components beyond the first few hours. This result suggests that the color change of the liver during decellularization may be due to a component that was not measured in this study. With the ubiquity of heme and heme proteins along with their abundance in the liver, the color change could be explained by the removal of heme,15 but it requires further investigation.
The liver was monitored using noncontrast CT scan during decellularization and the captured images were used to determine the HU of the tissue. HU is an x-ray attenuation unit used in CT scan interpretation and it characterizes the relative density of a substance. Each pixel in a CT scan image is assigned a value between −1000 (air) and +1000 (bone). Each tissue has a specific attenuation depending on its density, for example liver is 40–60 HU, whereas fat is −50 HU.16 As the liver gets decellularized, the attenuation of the tissue is expected to decline reaching that of water, that is, 0 HU. As expected, the liver attenuation decreased from 39 HU as the liver was perfused with PBS and 0.01% SDS, reaching a plateau at an average value of 2.2 HU after 0.01% SDS and beyond. This trend correlated well with that of DNA content and intensity of H&E staining of the matrix during decellularization, strongly suggesting that the liver attenuation obtained from noncontrast CT scans may be used to predict DNA and cellular protein removal. As a noninvasive tool to monitor decellularization of whole livers, this will allow customization of the SDS perfusion duration for human livers, which are expected to display donor to donor variability in their quality and vascular architecture, which will subsequently affect their perfusion characteristics. However, it is important to point out that at this point, we have not established the sensitivity of this method, hence, we suggest its use in conjunction with other assessment techniques such as perfusate analysis.
While the removal of DNA and cellular proteins is essential for a successful decellularization, preservation of the ECM components during the process is equally important because it will affect the behavior of the cells once the scaffold is recellularized. Our decellularization protocol employs SDS that is an anionic detergent, which is very effective in solubilizing phospholipids and proteins but also known to remove matrix-associated components and may be altering the native ECM composition.17 Previously, we reported by proteomic analysis that a large number of cellular proteins are removed and a significant number of ECM proteins are retained with decellularization.18 In this study, it was shown that the total collagen in the liver is retained after decellularization, whereas the 50% of native liver's GAGs are removed in the final scaffold. This was largely attributed to the 0.1% SDS perfusion step as confirmed by the perfusate analysis. GAGs include heparin and hyaluronan, which are found to promote maintenance of hepatocyte function in vitro,19–22 therefore, we modified the decellularization protocol by shortening the duration of 0.1% SDS perfusion to limit the loss of GAGs from the matrix and found that GAG content of the scaffold increases by more than 30%. Although we initially attempted to eliminate 0.2% and 0.5% SDS perfusion steps to further improve the GAG content, those steps were necessary to remove the residual DNA from scaffolds such that the resulting DNA content is less than 5% (results not shown). H&E staining of the modified decellularization tissue with the shortened 0.1% SDS perfusion and the incorporated 0.2% and 0.5% SDS perfusion shows similar intensity of staining as the original decellularization. Suggestively, it is possible to perfuse the liver with a lower concentration of SDS for a longer perfusion time to potentially prevent GAG removal and still have sufficient DNA removal to produce a completely decellularized liver tissue with a structurally strong and viable ECM.
Alternative decellularization protocols exist to effectively remove cellular components from whole organ scaffolds. We used our previously established protocol for the purpose of this study, which was mainly the development of nondestructive monitoring strategies of decellularization. By using this strategy, we were able to optimize the decellularization protocol, which highlights the utility of this approach, and we believe that CT scanning in combination with biochemical analysis of the perfusate can be used to evaluate different decellularization methods to determine the best decellularization approach.
Future investigations hold for determining what causes the color change when the liver is perfused with 0.1% SDS solution. In addition, finding the exact perfusion time and SDS concentration that retain important ECM components as well as sufficient DNA removal is necessary. CT scan monitoring during decellularization may prove to be useful to evaluate DNA removal, but currently it does not provide the required sensitivity to achieve low levels of DNA content. However, analysis of perfusate for DNA and cellular proteins by online rheological measurements11 combined with CT scan monitoring may hold the promise for a sensitive and noninvasive monitoring technique for decellularization. It is also essential that the quality of the matrix scaffolds should be tested in subsequent recellularization and transplantation experiments. The studies presented in this study were performed on rat livers as proof of concept studies; however, the natural progression is testing this system in more clinically relevant models such as donor human livers found not suitable for transplantation or livers from larger animals. In addition, this system can be used to create a decellularization assessment method, including establishing a quantitative definition of decellularization success, using these and other nondestructive evaluation techniques.
Conclusion
Liver tissue engineering using decellularized whole organ scaffolds has drawn much attention due to its promise in developing functional and transplantable liver grafts as substitutes for donor livers. While progress has been made in decellularization and recellularization of livers, there is no method to evaluate scaffold contents during decellularization noninvasively and customize decellularization duration based on initial donor liver characteristics. In this study, we present for the first time that CT scanning and HU measurements can be used to predict DNA removal from rat livers during decellularization. Although HU is not sensitive to low levels of DNA content in the scaffolds, it can be combined with biochemical analysis of the perfusate to yield an enhanced decellularization protocol to produce a well-preserved ECM scaffold. This approach is applicable to decellularization of discarded human donor livers to achieve consistent scaffold quality for preparation of engineered liver grafts for transplantation.
Acknowledgments
Funding was received from the National Institutes of Health (R01DK084053 M.L.Y., R00DK088962 B.E.U.), Shriners Hospitals for Children (M.L.Y.).
Disclosure Statement
B.E.U. has a financial interest in Organ Solutions, LLC, which is reviewed and arranged by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. The rest of the authors of this article have no conflicts of interest to disclose.
References
- 1.Badylak S.F., Taylor D., and Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng 13, 27, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uygun B.E., Soto-Gutierrez A., Yagi H., Izamis M.-L., Guzzardi M.A., Shulman C., et al. . Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med 16, 814, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao J., Shi Y., Sun H., Yin X., Yang R., Li L., et al. . Construction of a portal implantable functional tissue-engineered liver using perfusion-decellularized matrix and hepatocytes in rats. Cell Transplant 20, 753, 2011 [DOI] [PubMed] [Google Scholar]
- 4.Baptista P.M., Siddiqui M.M., Lozier G., Rodriguez S.R., Atala A., and Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 53, 604, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Barakat O., Abbasi S., Rodriguez G., Rios J., Wood R.P., Ozaki C., et al. . Use of decellularized porcine liver for engineering humanized liver organ. J Surg Res 173, e11, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Bao J., Wu Q., Sun J., Zhou Y., Wang Y., Jiang X., et al. . Hemocompatibility improvement of perfusion-decellularized clinical-scale liver scaffold through heparin immobilization. Sci Rep 5, 10756, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faulk D.M., Wildemann J.D., and Badylak S.F. Decellularization and cell seeding of whole liver biologic scaffolds composed of extracellular matrix. J Clin Exp Hepatol 5, 69, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bühler N.E.M., Schulze-Osthoff K., Königsrainer A., and Schenk M. Controlled processing of a full-sized porcine liver to a decellularized matrix in 24 h. J Biosci Bioeng 119, 609, 2015 [DOI] [PubMed] [Google Scholar]
- 9.Gilbert T.W., Sellaro T.L., and Badylak S.F. Decellularization of tissues and organs. Biomaterials 27, 3675, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Sawa Y., Taketani S., Iwai S., Matsuda H., Hara M., Uchimura E., et al. , inventors; Cardio, Inc., Tissue Engineering Research Center, assignee. Decellularized tissue. United States patent US 20050256588. 2005, November 17 [Google Scholar]
- 11.Hülsmann J., Aubin H., Bandesha S.T., Kranz A., Stoldt V.R., Lichtenberg A., et al. . Rheology of perfusates and fluid dynamical effects during whole organ decellularization: a perspective to individualize decellularization protocols for single organs. Biofabrication 7, 035008, 2015 [DOI] [PubMed] [Google Scholar]
- 12.Farndale R.W., Buttle D.J., and Barrett A.J. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta 883, 173, 1986 [DOI] [PubMed] [Google Scholar]
- 13.Uygun B.E., Yarmush M.L., and Uygun K. Application of whole-organ tissue engineering in hepatology. Nat Rev Gastroenterol Hepatol 9, 738, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uygun B.E., Price G., Saeidi N., Izamis M.-L., Berendsen T., Yarmush M., et al. Decellularization and recellularization of whole livers. J Vis Exp pii: , 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan A.A., and Quigley J.G. Control of intracellular heme levels: heme transporters and heme oxygenases. Biochim Biophys Acta 1813, 668, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moses S. Hounsfield Unit [Internet]. FP Notebook. 2016. [cited 2016 Apr 2]. Available at: www.fpnotebook.comradCTHnsfldUnt.htm
- 17.Keane T.J., Swinehart I.T., and Badylak S.F. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods 84, 25, 2015 [DOI] [PubMed] [Google Scholar]
- 18.Li Q., Uygun B.E., Geerts S., Ozer S., Scalf M., Gilpin S.E., et al. . Proteomic analysis of naturally-sourced biological scaffolds. Biomaterials 75, 37, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J., Shang Y., Yuan Y., and Yang J. Preparation and characterization of chitosan/galactosylated hyaluronic acid scaffolds for primary hepatocytes culture. J Mater Sci Mater Med 21, 319, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Skardal A., Smith L., Bharadwaj S., Atala A., Soker S., and Zhang Y. Tissue specific synthetic ECM hydrogels for 3-D in vitro maintenance of hepatocyte function. Biomaterials 33, 4565, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y., Larkin A.L., Davis R.M., and Rajagopalan P. The design of in vitro liver sinusoid mimics using Chitosan–Hyaluronic acid polyelectrolyte multilayers. Tissue Eng 16, 2731, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M., Lee J.Y., Jones C.N., Revzin A., and Tae G. Heparin-based hydrogel as a matrix for encapsulation and cultivation of primary hepatocytes. Biomaterials 31, 3596, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]