Abstract
Background
Prostatic Acid Phosphatase (PAP) is an enzyme that is produced primarily in the prostate and functions as a cell growth regulator and potential tumor suppressor. Understanding the genetic regulation of this enzyme is important because PAP plays an important role in prostate cancer and is expressed in other tissues such as the brain.
Methods
We tested association between 5.8 M SNPs and PAP levels in cerebrospinal fluid across 543 individuals in two datasets using linear regression. We then performed meta-analyses using METAL =with a significance threshold of p < 5 × 10−8 and removed SNPs where the direction of the effect was different between the two datasets, identifying 289 candidate SNPs that affect PAP cerebrospinal fluid levels. We analyzed each of these SNPs individually and prioritized SNPs that had biologically meaningful functional annotations in wANNOVAR (e.g. non-synonymous, stop gain, 3’ UTR, etc.) or had a RegulomeDB score less than 3.
Results
Thirteen SNPs met our criteria, suggesting they are candidate causal alleles that underlie ACPP regulation and expression.
Conclusions
Given PAP’s expression in the brain and its role as a cell-growth regulator and tumor suppressor, our results have important implications in brain health such as cancer and other brain diseases including neurodegenerative diseases (e.g., Alzheimer’s disease and Parkinson’s disease) and mental health (e.g., anxiety, depression, and schizophrenia).
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2787-y) contains supplementary material, which is available to authorized users.
Keywords: Brain, Cancer, CSF, PAP
Background
Prostatic Acid Phosphatase (PAP)—an enzyme expressed by the Acid Phosphatase, Prostate (ACPP) gene—is predominantly produced in the prostate, and is an important biomarker used to assess and monitor prostate cancer [1–3], but is also expressed in other tissues like the brain [4–7]. Recent research suggests PAP is a key cell growth regulator and potential tumor suppressor gene [8–10]. Additional studies demonstrate ACPP is expressed in the brain and suggest that PAP plays a critical role in preventing cell proliferation, cancer cell invasion, and neurite retraction [11, 12]. PAP’s function has critical implications in mental health diseases such as anxiety, depression, and schizophrenia [5], neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease [13], and possibly in brain cancer. Yet, little is known about ACPP’s function and regulation in the brain.
To date, research regarding ACPP function is almost exclusively limited to prostate cancer research, but ACPP is known to be expressed at lower levels in other tissues, such as the brain [4–7]. Specifically, ACPP is expressed in brain regions related to language, motor coordination, cognitive function, and self-awareness [4–7], suggesting it plays an important role in brain health. While PAP’s role within nervous tissue is not fully elucidated, it is known to localize in synaptic nerve endings [5] and co-localizes with SNAPIN [14], a protein that binds to SNAP-25 [15], which is associated with schizophrenia [16–19]. Recent studies further suggest PAP is a key cell growth regulator and potential tumor suppressor gene in the prostate [8–10], but PAP may have similar roles in the brain. A study by Tanaka et al. demonstrated that PAP degrades lysophosphatidic acid (LPA) in seminal plasma [11], and LPA is known to stimulate cell proliferation and prevent apoptosis [11] and has also been strongly implicated in cancer cell invasion [12]. Tanaka et al. further reported that lysophosphatidylcholine (LPC) and lysophospholipase D (lysoPLD) are found in CSF [11]. LPC is an LPA precursor and lysoPLD produces LPA from LPC, suggesting that LPA is likely present in the brain. These data suggest PAP may play a significant role protecting the brain from de novo brain tumors and metastatic tumors by inhibiting cell proliferation and cancer cell invasion, respectively.
LPA is also known to cause neurite retraction [11]. Neurites are a general term for axons and dendrites, generally used when it’s not possible to differentiate between the two (e.g., during development). A corresponding study by Sayas et al. demonstrated that inducing differentiated SY-SH5Y human neuroblastoma cells with LPA causes neurite retraction and site-specific Alzheimer's disease-like Tau phosphorylation [13]. These studies further suggest PAP plays a critical role in neuronal health, perhaps especially during development.
A study by Nousiainen et al. explored PAP’s function in the brain using PAP knockout mice [5]. Their results demonstrate that PAP knockout mice have enlarged lateral ventricles, a common phenotype in movement and neurodegenerative disorders such as Alzheimer’s disease, dementias, bipolar disorder, schizophrenia, Parkinson’s disease, and Huntington’s disease [5]. They also observed increased anxiety in the mice and decreased prepulsed inhibition. Molecular explanations for their observations may include increased GABAergic transmission and mislocalization of SNAPIN [5]. Increased GABAeric transmission inhibits neuronal excitability [20] while mislocalization of SNAPIN may affect neurotransmitter release [5], both of which may affect neuronal homeostasis and brain health.
Here we have conducted a genome-wide association study of PAP levels in cerebrospinal fluid (CSF) in 543 individuals from two datasets. Further characterization of the variants that we have identified may lead to a deeper understanding of PAP regulation and provide important insights into its effects on prostate cancer, brain cancer, mental health disorders, and neurodegenerative diseases.
Methods
-
A.
Subjects and data description
CSF samples were collected from 246 individuals in the Knight-Alzheimer’s Disease Research Center at Washington University School of Medicine (Knight ADRC) and from 297 individuals in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s Disease status was 93.5 % control and 6.5 % case in the Knight ADRC samples, and 86.5 % and 13.5 % in the ADNI samples. Levels for 190 biomarkers were measured for each sample using the Human DiscoveryMAP Panel v1.0 and a Luminex 100 platform [21] and samples were genotyped using the Illumina 610 or the Omniexpress chip. A description of the Knight ADRC samples and the associated CSF collection methods has been previously published [22, 23]. ADNI CSF samples were collected as part of the ADNI biomarker study [24], and were obtained from the ADNI database (adni.loni.usc.edu). All samples are of European descent. All individuals whose data were included in this study were explicitly consented, following appropriate Institutional Review Board policies.
-
B.
SNP imputation
SNPs were imputed as previously described. Briefly, data from the 1000 Genomes Project (June 2012 release) were used to impute SNPs using Beagle. Imputed SNPs with the following criteria were removed: (1) an r2 of 0.3 or lower, (2) a minor allele frequency (MAF) lower than 0.05 (3) out of Hardy-Weinberg equilibrium (p < 1 × 10 − 6), (4) a call rate lower than 95 %, or (5) a Gprobs score lower than 0.90. Exactly 5,815,690 SNPs passed the QC process.
-
C.
Data cleaning and analysis
All analyses were conducted using PLINK, a whole genome association analysis toolset [25]. We performed genotype quality control on the Knight ADRC and ADNI CSF datasets by first excluding SNPs that exceeded thresholds for Hardy-Weinberg Equilibrium [26, 27] (−−hwe 0.00001), missing genotype rate (−−geno 0.05), and minor allele frequency (−−maf 0.01). We then excluded individuals with a missing genotyping rate greater than 2 % (−−mind 0.02). Exactly 246 individuals from Knight ADRC and 282 samples from ADNI remained after data cleaning. Remaining Knight ADRC and ADNI samples consisted of 40 % and 61 % males, respectively. ADNI samples varied in age from 58 to 91 years, with an average age of 76 years, and Knight ADRC samples varied in age from 49 to 91 years, with an average age of 73 years.
After data cleaning, we conducted a linear regression for all remaining SNPs within each data set to test for an association with CSF PAP levels, adjusting for age, gender, and the first two principle components generated using EigenSoft [28, 29]. The full script for our PLINK analyses is found in the Appendix. We then performed a meta-analysis across both data sets, accounting for sample size, p-values, and direction of effect using the default METAL [30] settings. We have included our scripts in Additional files 1, 2, 3, 4, 5, 6 and 7 for convenience.
We retained all SNPs that had a meta-analysis p-value less than 5 × 10−8 and that had the same direction of effect in both the Knight ADRC and ADNI datasets. We searched for all SNPs in the NHGRI catalog of published genome-wide association studies [31] (downloaded July 16th, 2015) for known disease associations. We then used RegulomeDB [32] and functional annotations from wANNOVAR [33, 34] to identify SNPs that are biologically likely to modify gene function or expression. Specifically, we retained the 10 most significant SNPs and all significant SNPs with a RegulomeDB score less than 3 or that were non-synonymous, stop-gain, splice-site modifying, etc., or are in untranslated regions (UTRs). UTR SNPs have been shown to modify gene transcription and translation [35, 36]. RegulomeDB scores range from “1a” to “6”. Lower scores indicate stronger evidence that the SNP affects gene regulation based on both empirical data, such as ChIP-seq, and whether the SNP is within a known transcription factor binding motif. Any RegulomeDB score between “1a” and “1f” indicates the SNP is within a known expression quantitative trait locus (eQTL), is known to have transcription factors bind, and may have additional evidence. While these scores indicate functional effects, the associated SNP is not necessarily the causal variant.
All SNPs that met our inclusion criteria and are in or near the given region were also reanalyzed using conditional analyses to test whether there is one or multiple independent effects in the region [37]. Conditional analysis is a follow-up method used to test if there are secondary association signals within a region by retesting each SNP while including the top SNP as a covariate. We chose the most significant SNP in the region to use as a covariate in the conditional analyses.
Results
Our meta-analysis yielded 289 SNPs significantly associated with PAP CSF levels (Additional file 8). Of the 289 SNPs, 276 are located on chromosome 3, in or near the ACPP gene, which is the gene that codes for PAP. We generated plots in R failed to detect evidence of inflation (genomic inflation factor = 1; Additional files 9 and 10).
We explored potential causal SNPs and identified 23 that met our inclusion criteria, including the top 10 SNPs with the most significant p-values, and the 13 most biologically significant SNPs based on their RegulomeDB scores and functional annotations (Table 1). None of the 289 SNPs were associated with human disease in the NHGRI GWAS catalog. Two SNPs, rs16839055 and rs17182812, both received the score of “1f” from RegulomeDB. In this case, both SNPs are in LD with SNPs known to be associated with ACPP expression [31]. SNP rs3844501 has the strongest association with PAP CSF levels (p = 1.743e-20) and is approximately 2000 nucleotides upstream from the ACPP transcription start site (Fig. 1). Numerous SNPs proximal to rs3844501 are also highly associated and in strong LD. SNP rs3889987 has a RegulomeDB score of “2a”, implying it affects transcription factor binding because transcriptions factors have been observed binding to the SNP, the SNP is in a transcription factor motif, and matches a DNase footprint. There are 7 SNPs (rs11714139, rs56226080, rs73211958, rs56030168, rs4257547, rs11928839, and rs11917521) that received scores of 2b, showing they are also known to affect transcription factor binding and could affect PAP levels. The score of 2a is ranked higher than 2b, indicating rs3889987 is more likely to affect gene regulation. While 2a and 2b are similar, 2a is known to specifically match transcription factor motifs. SNPs, rs14192, rs1804136, and rs1042330 are all found in the 3’UTR, an important gene region for regulation.
Table 1.
This table includes the 10 most significant SNPs and 13 other significant SNPs that have a RegulomeDB score less than 3, are non-synonymous, a splicing variant, or are located in untranslated regions (UTRs). All 23 SNPs are located in or near the ACPP gene on chromosome 3, which codes for PAP. There were no significant non-synonymous SNPs. The table includes SNP ids, reference and alternate alleles, minor allele frequency, predicted SNP function, and its directions in the Knight ADRC and ADNI data sets. Also included are the p-values from ADNI and Knight ADRC alone, their meta-analysis p-value and their RegulomeDB score, where available
| SNP | Minor Allele | Major Allele | MAF | Predicted Function | Direction | ADNI p-value | Knight ADRC p-value | Meta-analysis p-value | RegulomeDB score |
|---|---|---|---|---|---|---|---|---|---|
| rs3844501 | T | G | 0.1458 | Intergenic | ++ | 1.972e-13 | 7.974e-09 | 1.743e-20 | 6 |
| rs3762671 | T | C | 0.1060 | Intergenic | ++ | 3.815e-13 | 3.245e-08 | 1.515e-19 | 6 |
| rs11716607 | G | A | 0.0923 | Intronic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | No Data |
| rs56158166 | T | G | 0.0921 | Intronic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | No Data |
| rs73213842 | A | T | 0.0923 | Intergenic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | No Data |
| rs11706024 | A | G | 0.0921 | Intergenic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | 5 |
| rs113143077 | A | G | 0.0923 | Intergenic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | 6 |
| rs17344445 | A | G | 0.0923 | Intergenic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | 6 |
| rs2887519 | A | G | 0.0923 | Intergenic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | 6 |
| rs56073503 | A | T | 0.0921 | Intronic | ++ | 1.708e-12 | 3.615e-08 | 6.397e-19 | 6 |
| rs3889987 | T | G | 0.0913 | Intergenic | ++ | 2.15E-13 | 4.08E-07 | 1.77E-18 | 2a |
| rs56030168 | G | A | 0.1769 | 5upstream | ++ | 2.94E-11 | 1.34E-06 | 4.65E-16 | 2b |
| rs11714139 | A | G | 0.1232 | Intergenic | ++ | 1.25E-07 | 4.81E-04 | 5.41E-10 | 2b |
| rs56226080 | C | G | 0.1228 | Intergenic | ++ | 1.25E-07 | 4.81E-04 | 5.41E-10 | 2b |
| rs73211958 | A | C | 0.1218 | Intergenic | ++ | 1.25E-07 | 5.11E-4 | 5.82E-10 | 2b |
| rs16839055 | T | C | 0.3081 | Intergenic | ++ | 2.49E-05 | 2.52E-04 | 2.55E-08 | 1f |
| rs1804136 | T | G | 0.3962 | UTR3 | ++ | 4.80E-05 | 1.39E-04 | 2.57E-08 | No Data |
| rs14192 | C | T | 0.3964 | UTR3 | ++ | 4.80E-05 | 1.39E-04 | 2.57E-08 | 5 |
| rs1042330 | A | G | 0.3962 | UTR3 | ++ | 6.49E-05 | 1.39E-04 | 3.43E-08 | No Data |
| rs17182812 | T | C | 0.3962 | Intronic | ++ | 6.49E-05 | 1.39E-04 | 3.43E-08 | 1f |
| rs4257547 | G | C | 0.4113 | Intronic | ++ | 5.19E-05 | 1.94E-04 | 3.87E-08 | 2b |
| rs11928839 | A | C | 0.3954 | Intronic | ++ | 6.49E-05 | 1.60E-04 | 3.93E-08 | 2b |
| rs11917521 | C | T | 0.3954 | Intronic | ++ | 6.49E-05 | 1.60E-04 | 3.93E-08 | 2b |
Fig. 1.
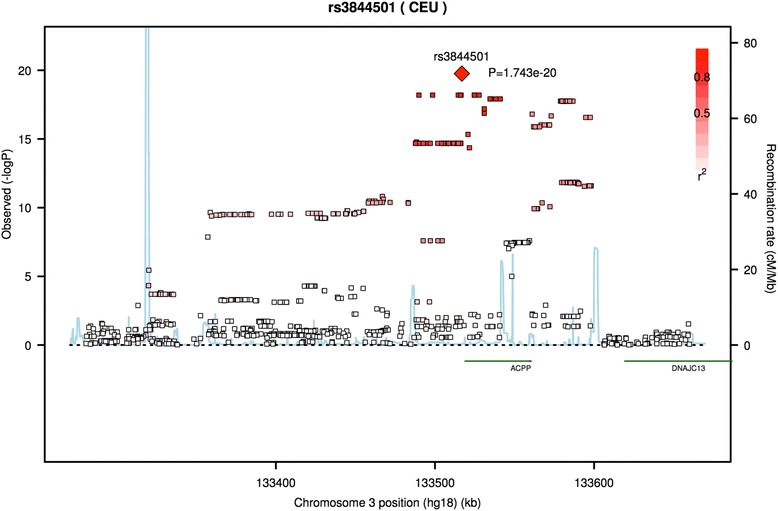
Regional association plot showing rs3844501 has the strongest association with PAP CSF levels. We identified several SNPs associated with PAP levels in CSF that are in or near the ACPP gene, which codes for PAP, and plotted association p-values in the region. rs3844501 has the strongest association with PAP CSF levels and is located approximately 2000 nucleotides upstream of the ACPP transcription start site. LD between rs3844501 and other SNPs in the region is strong
We performed conditional analyses including rs3844501 as a covariate in the linear regression. No SNPs in or near ACPP were significant, suggesting there is one association signal within the ACPP region.
Discussion
We identified 289 SNPs that show replicable association with CSF PAP levels, the majority of which are found in or close to ACPP, the gene that codes for PAP. Among these, 23 SNPs in or near ACPP are highly significant and demonstrate evidence of functional effects, making them top candidates for being the causal allele for this association.
Conditional analyses strongly suggest there is a single signal within the region. As is the case with any association study, LD facilitates the discovery of associations, but makes identifying the causal allele difficult. The SNP most strongly associated with PAP CSF levels, rs3844501, is approximately 2000 nucleotides upstream of the ACPP transcription start site and has no known functional impact. Data from RegulomeDB identifies rs16839055 and rs17182812 as the two most likely candidate variants, however RegulomeDB is not exhaustive, and there are several other plausible candidates.
Based on current evidence, there are numerous mechanisms that can affect PAP protein levels in CSF including transcription, translation, feedback loops, and damaged proteins. Transcription is often most suspect in expression analyses and one of the top SNPs we identified, rs56030168, is most likely to affect transcription because it is less than 300 nucleotides upstream from the ACPP transcription start site and, according to the RegulomeDB score of “2b”, is known to have transcription factors bound at that location and is a known DNase peak. Given rs56030168’s location and other evidence, it may affect transcription factor binding. Rs16839055 is also upstream of ACPP and may affect transcription, but is more than 26,000 nucleotides upstream from the transcription start site.
The 3’UTR SNPs rs14192, rs1804136, and rs1042330 could affect PAP levels by affecting both transcription and translation. UTR regions are known to affect mRNA stability, transport, and translation processes [38–41]. Any of these three SNPs may play an important role in ACPP or PAP regulation.
None of the top 23 SNPs we identified were in ACPP exons, but several are intronic. Intronic variants can affect protein structure through splice modifications and have even been experimentally shown to affect transcription [42]. While intronic variants are typically less suspect, they may play an important role in PAP levels.
While these SNPs are the most significant and biologically likely eQTLs based on our criteria, there may be other candidate eQTLs in the list of 289. They all met the genome-wide significance p-value threshold and have matching effect directions, but more biological data will be necessary to support or refute their direct involvement. Additionally, our data are not whole exome or genome and there may be causal variants in LD with our top hits that were not genotyped. Full sequencing data within the region may reveal other candidate causal variations. Further research will be necessary to know which SNPs affect PAP CSF levels, and particularly whether they contribute to prostate cancer and other PAP-related functions and diseases.
Conclusions
A plethora of studies suggest PAP plays an important role in prostate cancer, but recent studies suggest that PAP may play a critical role in brain health ranging from cancers to various brain disorders. Based on current research, we hypothesize that PAP’s role in brain health includes protecting against cancer development and metastasis, protecting against neuronal death by regulating LPA levels, and generally protecting brain health by contributing to neuronal homeostasis. In summary, this study has identified a clear and replicable QTL in ACPP for CSF PAP levels. Additional investigation of this locus may lead to a better understanding of ACPP regulation in the brain and additional insights into PAP’s role in the brain.
Abbreviations
ACPP, Acid Phosphatase, Prostate; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CSF, cerebrospinal fluid; eQTL, expression quantitative trait locus; Knight ADRC, Knight-Alzheimer’s Disease Research Center at Washington University School of Medicine; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; lysoPLD, lysophospholipase D; MAF, minor allele frequency; PAP, Prostatic Acid Phosphatase; SNP, single nucleotide polymorphism; UTRs, untranslated regions.
Acknowledgements
The authors acknowledge that many scientists contributed in developing the clinical and genetic resources necessary to collect these data and complete this project. The authors also gratefully acknowledge the efforts of hundreds of individuals who participated as subjects in these studies.
This work was supported by grants from NIH (R01 AG035053, R01 AG042611, P50 AG05681, P01 AG03991, P01 AG026276), the Alzheimer’s Association (MNIRG-11-205368), and the Brigham Young University Gerontology Program. We also acknowledge the Alzheimer’s Disease Genetics Consortium (ADGC) and Genetic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD) for providing genotype data used in this work.
GERAD was supported by the Medical Research Council (Grant nu 503480), Alzheimer’s Research UK (Grant nu 503176), the Wellcome Trust (Grant nu 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant nu 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; ElanPharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Some of the samples used in this study were genotyped by the ADGC and GERAD. ADGC is supported by grants from the NIH (#U01AG032984) and GERAD from the Wellcome Trust (GR082604MA) and the Medical Research Council (G0300429).
Some data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
The ADNI Executive Committee consists of: Michael Weiner, MD UC San Francisco; Paul Aisen, MD UC San Diego; Ronald Petersen, MD, PhD Mayo Clinic, Rochester; Clifford R. Jack, Jr., MD Mayo Clinic, Rochester; William Jagust, MD UC Berkeley; John Q. Trojanowki, MD, PhD U Pennsylvania; Arthur W. Toga, PhD USC; Laurel Beckett, PhD UC Davis; Robert C. Green, MD, MPH Brigham and Women’s Hospital/Harvard Medical School; Andrew J. Saykin, PsyD Indiana University; John Morris, MD Washington University St. Louis; Leslie M. Shaw University of Pennsylvania. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Declarations
Publication of this article was funded by Brigham Young University’s Department of Biology.
This article has been published as part of BMC Genomics Volume 17 Supplement 3, 2016: Selected articles from the 12th Annual Biotechnology and Bioinformatics Symposium: genomics. The full contents of the supplement are available online at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-17-supplement-3.
Availability of data and materials
Data are available to researchers by applying to the respective organizations, ADNI and ADGC consortia. Application is required to protect participant confidentiality. The ADNI data are available at (http://adni.loni.usc.edu/), the Knight ADRC data are available through dbGAP (http://www.ncbi.nlm.nih.gov/gap).
Authors’ contributions
LS, DB, MD, PGR and JSKK out the analyses. LS and ME annotated and analyzed the SNPs for significance and drafted the manuscript. All other authors participated in the conception of the project and obtaining data. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Study data and analyses were approved by the Brigham Young University Institutional Review Board.
Additional files
File contains the PLINK script used to clean data and find associations between SNPs and prolactin levels in the samples. (DOCX 101 kb)
File contains the forge_metal.py program, which is used to run METAL on the samples form each dataset to combine data. (DOCX 85 kb)
File includes the command for running forge_metal.py on ADNI dataset. (DOCX 36 kb)
File includes the command for running forge_metal.py on WU dataset. (DOCX 35 kb)
File include the contents of METAL meta analysis script. (DOCX 44 kb)
File include the command to run METAL software. (DOCX 29 kb)
File contains the command to sort combined METAL results. (DOCX 40 kb)
File contains a table of 289 Significant SNPs. (DOCX 124 kb)
File contains a Q-Q plot of the CSF in the ADNI samples. (DOCX 54 kb)
File contains a Q-Q plot of the CSF in the Knight ADRC samples. (DOCX 52 kb)
References
- 1.Azumi N, Traweek ST, Battifora H. Prostatic acid phosphatase in carcinoid tumors. Immunohistochemical and immunoblot studies. Am J. Surg. Pathol. 1991;15:785–90. doi: 10.1097/00000478-199108000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Gunia S, Koch S, May M, Dietel M, Erbersdobler A. Expression of prostatic acid phosphatase (PSAP) in transurethral resection specimens of the prostate is predictive of histopathologic tumor stage in subsequent radical prostatectomies. Virchows Arch. Int. J. Pathol. 2009;454:573–9. doi: 10.1007/s00428-009-0759-1. [DOI] [PubMed] [Google Scholar]
- 3.Yi Wang MH. Prostatic acid phosphatase as a target molecule in specific immunotherapy for patients with nonprostate adenocarcinoma. J. Immunother. Hagerstown Md 1997. 2005;28:535–41. doi: 10.1097/01.cji.0000175490.26937.22. [DOI] [PubMed] [Google Scholar]
- 4.Quintero IB, Araujo CL, Pulkka AE, Wirkkala RS, Herrala AM, Eskelinen E-L, et al. Prostatic Acid Phosphatase Is Not a Prostate Specific Target. Cancer Res. 2007;67:6549–54. doi: 10.1158/0008-5472.CAN-07-1651. [DOI] [PubMed] [Google Scholar]
- 5.Nousiainen HO, Quintero IB, Myöhänen TT, Voikar V, Mijatovic J, Segerstråle M, et al. Mice Deficient in Transmembrane Prostatic Acid Phosphatase Display Increased GABAergic Transmission and Neurological Alterations. PLoS ONE. 2014;9:e97851. doi: 10.1371/journal.pone.0097851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen Brain Atlas Resources [Internet]. Allen institute for brain science; Available from: www.brain-map.org.
- 7.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong HY, Byun J. Emerging Roles of Human Prostatic Acid Phosphatase. Biomol. Ther. 2013;21:10–20. doi: 10.4062/biomolther.2012.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakthivel Muniyan MAI. Cellular prostatic acid phosphatase, a PTEN-functional homologue in prostate epithelia, functions as a prostate-specific tumor suppressor. Biochim. Biophys. Acta. 2014;1846 [DOI] [PMC free article] [PubMed]
- 10.Veeramani S, Yuan T-C, Chen S-J, Lin F-F, Petersen JE, Shaheduzzaman S, et al. Cellular prostatic acid phosphatase: a protein tyrosine phosphatase involved in androgen-independent proliferation of prostate cancer. Endocr. Relat. Cancer. 2005;12:805–22. doi: 10.1677/erc.1.00950. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka M, Kishi Y, Takanezawa Y, Kakehi Y, Aoki J, Arai H. Prostatic acid phosphatase degrades lysophosphatidic acid in seminal plasma. FEBS Lett. 2004;571:197–204. doi: 10.1016/j.febslet.2004.06.083. [DOI] [PubMed] [Google Scholar]
- 12.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer. 2003;3:582–91. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 13.Sayas CL, Moreno-Flores MT, Avila J, Wandosell F. The Neurite Retraction Induced by Lysophosphatidic Acid Increases Alzheimer’s Disease-like Tau Phosphorylation. J. Biol. Chem. 1999;274:37046–52. doi: 10.1074/jbc.274.52.37046. [DOI] [PubMed] [Google Scholar]
- 14.Quintero IB, Herrala AM, Araujo CL, Pulkka AE, Hautaniemi S, Ovaska K, et al. Transmembrane Prostatic Acid Phosphatase (TMPAP) Interacts with Snapin and Deficient Mice Develop Prostate Adenocarcinoma. PLoS ONE. 2013;8:e73072. doi: 10.1371/journal.pone.0073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilardi JM, Mochida S, Sheng Z-H. Snapin: a SNARE–associated protein implicated in synaptic transmission. Nat. Neurosci. 1999;2:119–24. doi: 10.1038/5673. [DOI] [PubMed] [Google Scholar]
- 16.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am. J. Hum. Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatemi SH, Earle JA, Stary JM, Lee S, Sedgewick J. Altered levels of the synaptosomal associated protein SNAP-25 in hippocampus of subjects with mood disorders and schizophrenia. Neuroreport. 2001;12:3257–62. doi: 10.1097/00001756-200110290-00023. [DOI] [PubMed] [Google Scholar]
- 18.Thompson PM, Egbufoama S, Vawter MP. SNAP-25 reduction in the hippocampus of patients with schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2003;27:411–7. doi: 10.1016/S0278-5846(03)00027-7. [DOI] [PubMed] [Google Scholar]
- 19.Thompson PM, Kelley M, Yao J, Tsai G, van Kammen DP. Elevated cerebrospinal fluid SNAP-25 in schizophrenia. Biol. Psychiatry. 2003;53:1132–7. doi: 10.1016/S0006-3223(02)01599-8. [DOI] [PubMed] [Google Scholar]
- 20.Dichter MA, Ayala GF. Cellular mechanisms of epilepsy: a status report. Science. 1987;237:157–64. doi: 10.1126/science.3037700. [DOI] [PubMed] [Google Scholar]
- 21.Kauwe JSK, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, et al. Genome-Wide Association Study of CSF Levels of 59 Alzheimer’s Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation. PLoS Genet. 2014;10:e1004758. doi: 10.1371/journal.pgen.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–68. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anne M, Fagan MAM. Inverse relation between in vivo amyloid imaging load and CSF Aβ42 in humans. Ann. Neurol. 2006;59:512–9. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 24.Trojanowski JQ, Vandeerstichele H, Korecka M, Clark CM, Aisen PS, Petersen RC, et al. Update on the biomarker core of the Alzheimer’s Disease Neuroimaging Initiative subjects. Alzheimers Dement. J. Alzheimers Assoc. 2010;6:230–8. doi: 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wigginton JE, Cutler DJ, Abecasis GR. A Note on Exact Tests of Hardy-Weinberg Equilibrium. Am. J. Hum. Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jan Graffelman VM. The mid p-value in exact tests for Hardy-Weinberg equilibrium. Stat. Appl. Genet. Mol. Biol. 2013;12:433–48. doi: 10.1515/sagmb-2012-0039. [DOI] [PubMed] [Google Scholar]
- 28.Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinforma. Oxf. Engl. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Med. Genet. 2012;49:433–6. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delay C, Calon F, Mathews P, Hébert SS. Alzheimer-specific variants in the 3’UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 2011;6:70. doi: 10.1186/1750-1326-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006;38:813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Ferreira T, Morris AP, Medland SE, Consortium GI of AnT (GIANT) Consortium DiaGRAM (DIAGRAM) et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to Tango: Regulation of Mammalian Iron Metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012;69:3613–34. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, et al. RNA binding protein/RNA element interactions and the control of translation. Curr. Protein Pept. Sci. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mignone F, Pesole G. mRNA Untranslated Regions (UTRs). eLS [Internet]. John Wiley & Sons, Ltd; 2001 [cited 2015 Sep 4]. Available from: http://onlinelibrary.wiley.com/doi/10.1002/9780470015902.a0005009.pub2/abstract
- 42.Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–20. doi: 10.1016/S0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available to researchers by applying to the respective organizations, ADNI and ADGC consortia. Application is required to protect participant confidentiality. The ADNI data are available at (http://adni.loni.usc.edu/), the Knight ADRC data are available through dbGAP (http://www.ncbi.nlm.nih.gov/gap).


