Abstract
Background. Human immunodeficiency virus (HIV)-1 drug resistance mutations (DRMs) often accompany treatment failure. Although subtype differences are widely studied, DRM comparisons between subtypes either focus on specific geographic regions or include populations with heterogeneous treatments.
Methods. We characterized DRM patterns following first-line failure and their impact on future treatment in a global, multi-subtype reverse-transcriptase sequence dataset. We developed a hierarchical modeling approach to address the high-dimensional challenge of modeling and comparing frequencies of multiple DRMs in varying first-line regimens, durations, and subtypes. Drug resistance mutation co-occurrence was characterized using a novel application of a statistical network model.
Results. In 1425 sequences, 202 subtype B, 696 C, 44 G, 351 circulating recombinant forms (CRF)01_AE, 58 CRF02_AG, and 74 from other subtypes mutation frequencies were higher in subtypes C and CRF01_AE compared with B overall. Mutation frequency increased by 9%–20% at reverse transcriptase positions 41, 67, 70, 184, 215, and 219 in subtype C and CRF01_AE vs B. Subtype C and CRF01_AE exhibited higher predicted cross-resistance (+12%–18%) to future therapy options compared with subtype B. Topologies of subtype mutation networks were mostly similar.
Conclusions. We find clear differences in DRM outcomes following first-line failure, suggesting subtype-specific ecological or biological factors that determine DRM patterns.
Keywords: drug resistance, evolution, first-line, HIV, subtype
Effective treatment of human immunodeficiency virus (HIV)-1 requires the lifelong administration of combination antiretroviral therapy (ART) [1]. In recent years, there has been rapid expansion of ART access in resource-limited settings. In these settings, the World Health Organization (WHO) recommends 2 nucleoside reverse-transcriptase inhibitors (NRTIs) with 1 nonnucleoside RTI (NNRTI) as first-line treatment followed by a second-line protease inhibitor-based (PI) regimen [2]. Although most data driving ART recommendations have been accrued in HIV-1 subtype B in the United States, Oceania, and Europe, treatment has expanded to regions with diverse group M subtypes and distinct circulating recombinant forms (CRFs) [3]. The influence of genetic background variation across subtypes on disease progression and resistance evolution is currently an active research area [4].
Subtype designations are based on phylogenic clades [5] and are widely used to represent genetic background variation in HIV sequences. Human immunodeficiency virus-1 subtypes are geographically distributed, with subtype C being the most prevalent worldwide and predominantly found in South Africa and India. Other common subtypes include A in East Africa and Asia, D in East Africa, F in South America and Eastern Europe, G in Africa and South-Western Europe, CRF01_AE in Southeast Asia, and CRF02_AG in West Africa. Subtype B is the most prevalent in the United States and Western Europe [6].
Although high viral genetic diversity and variable ART adherence are known to contribute to drug resistance and ART failure, subtype contributions to treatment outcomes remains actively researched through in vitro [7–9] and surveillance [10–13] methods. Subtype-specific codon use results in more frequent mutations at codons 65 and 106 in the HIV-1 subtype C RT gene under treatment selection [14, 15]. Other studies report lower genetic barriers to RT mutations at codons 151 and 210 in subtype F1 [16] and overall differences in polymorphisms and treatment-related mutation frequencies across subtypes [17, 18]. Subtype backgrounds have also been associated with differences in evolutionary responses to protease inhibitors [19, 20]. However, there are few comparative studies of drug resistance that integrate data from multiple subtypes in multiple cohorts [17, 21–23].
Combinations of first-line treatment regimens are the foundation of treatment administration and the prevention of transmission [24]. Yet information on drug resistance outcomes to first-line ART in resource-limited settings has been primarily from isolated regional studies [10, 11, 21, 22, 25], and a consensus view of the impact of drug resistance upon first-line failure in resource-limited settings has been lacking [26]. The WHO recently convened a conference to synthesize several consensus recommendations [26]. Although those data provided important insights, there has not been a comprehensive multicohort analysis comparing resistance mutation outcomes between subtypes after first-line triple-therapy regimens.
In collaboration with the non-subtype B working group, we previously examined differences in resistance mutation occurrence across subtypes from a global dataset of sequences [17]. A limitation of that study was that the ART-failure populations merged heterogeneous treatment histories and included both first-line/nonfirst-line ART exposures. In this study, we utilize a multicohort, multi-subtype sequence dataset from patients with known exposure to only first-line, triple-ART regimens. With these data, we determine differences in resistance mutation frequencies between ART regimens and common subtypes/CRFs following first-line failure. We develop a multilevel modeling approach to account for heterogeneities between combinations of treatments and subtypes. We perform a novel application of a network model to study combinatorial patterns of mutations and describe the implications for differences in second-line predicted susceptibility. Finally, we discuss the significance of our findings in the context of the underlying mechanistic determinants of HIV drug resistance evolution outcomes.
METHODS
Sequence Dataset and Drug Resistance Definitions
A multicohort, multi-subtype dataset of RT isolates was constructed and filtered to include only sequences from patients failing WHO-recommended first-line ART. This dataset builds on our previous study [17], adding new published sequences within the Stanford Database [27] as well as sequences from our studies in India, Thailand, China, and Kenya. Patients unexposed to ART or for whom the complete treatment history was unknown were excluded. Patients with heterogeneities in their history—prior histories of nonfirst-line therapies, prior usage of more than 1 first-line combination, or mono/duo therapy—were also excluded. Sequences were further excluded based on previously defined quality control criteria [28].
First-line ART was defined as standard combinations of NRTI/NNRTI triple therapy [29]: zidovudine (AZT)/stavudine (D4T)/tenofovir (TDF) + lamivudine (3TC) + efavirenz (EFV)/nevirapine (NVP). Treatment history was described using the total treatment duration and the composition of the specific ART regimens. Treatment start and stop dates were converted into an aggregate indicator of the total treatment duration. Treatment duration was represented as either failure after less than or equal to 1 year of first-line treatment or failure after more than 1 year of treatment.
Resistance mutations were based on definitions in the International Antiviral Society (IAS)-USA mutation list [30]. Because we were interested in the occurrence of all amino acids mutations associated with resistance, sequences were represented as a binary string with each position labeled as “mutation” or “no mutation”. A position has a mutation if its amino acid is listed in the IAS list. Sequences were subtyped using the Stanford database tools [27].
Hierarchical Model of Mutation Frequencies and Treatment Susceptibility
We constructed a hierarchical model to characterize resistance mutation variation with respect to treatment histories and subtype genetic backgrounds. Frequencies of each mutation were modeled as a beta binomial. The logit of the beta prior mean was parameterized by the position (1 of 31 resistance positions described in [30]), subtype (A, B, C, D, F, G, CRF01_AE, and CRF02_AG), ART regimen at the time of failure (1 of the triple-therapy combinations), and treatment duration. Additional details are discussed in the Supplementary Data.
Ising Model of Resistance Position Dependencies
We used an Ising/Boltzmann model to examine association between pairs of resistance positions as an undirected graph. For each subtype, we fitted the Ising model/Boltzmann distribution to the mutation statuses of the resistance positions, adjusted for the treatment type and duration of each subject. Details of the Ising model are in the Supplementary Data.
Predicted Susceptibility to Future Treatment
To examine the impact of first-line resistance outcomes, we determine the predicted susceptibility to future ART options. The susceptibility of each sequence was determined using the Stanford HIV database tools [27]. The distribution of susceptibility scores was determined for each subtype, treatment history composition, and treatment history duration.
RESULTS
From an initial aggregated, multicohort dataset of 59 002 sequences, most sequences (57 017) were excluded because patients were ART-naive at the time of genotyping or not failing a triple-therapy first-line ART regimen, leaving 1985 sequences. After applying quality control criteria [28] and removing sequences with missing treatment history information, 1425 sequences remained (Figure 1). Of these sequences, there were 202 of subtype B and 1223 of non-B subtypes including 696 of subtype C, 44 of subtype G, 351 of CRF01_AE, 58 of CRF02_AG, and 74 of other subtypes (A/D/F/H/K; each with <30 available sequences). Although all subtype data were used in estimating the model, the results focus on subtypes for which the largest number of sequences are available (B/C/G/CRF01_AE/CRF02_AG).
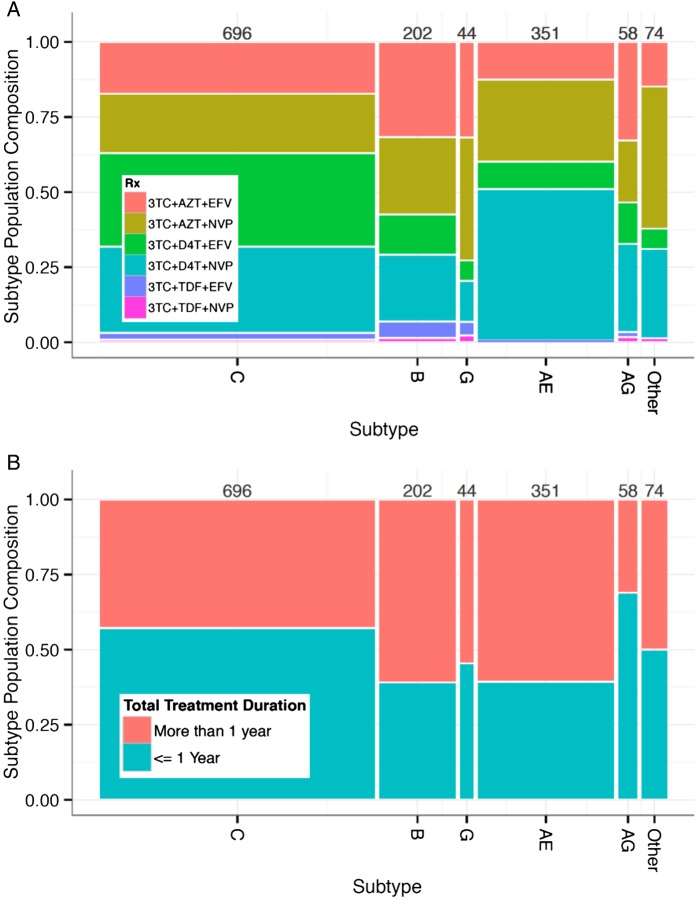
Figure 1.
The graphic illustrates the sequence dataset composition. The spine plot shows dataset subtype composition (horizontal axes) as a function of (A) first-line antiretroviral therapy (ART)-specific regimens and (B) first-line ART duration. Each column corresponds to a subtype, and the width of the column represents the proportion of sequences available from each subtype. The height of each tile represents the proportion of each regimen within that subtype. Thus, the area of each tile is proportional to the fraction of the dataset composed of a particular subtype and treatment composition/duration. Colors correspond to the (A) first-line regimen composition and (B) treatment duration presented in the legends. Numbers above each subtype column correspond to the total number of sequences in each subtype.
The population composition of treatment regimens and durations is displayed in Figure 1, which shows substantial variability across subtypes. The most common ART regimens in this dataset were nevirapine-containing zidovudine/stavudine regimens (57%) followed by efavirenz-containing zidovudine/stavudine regimens (40%). The remaining 3% of sequences were tenofovir-containing efavirenz and nevirapine regimens, which were incorporated more recently into WHO recommendations [2]. Although tenofovir was added to treatment guidelines more recently, prior alternatives remain in widespread use. Treatment duration prior to failure was approximately evenly divided between within 1 year of treatment (with a range of 31%–61%) and more than 1 year of treatment (with a range of 39%–69%).
Estimates of DRM frequencies across subtypes are shown in Figure 2. Differences in mutation frequencies between subtypes are shown in Figure 3. We also examined variation in DRM frequency across the different first-line treatment regimens (Supplementary Figures 1 and 2). Together, these figures illustrate how resistance mutation frequencies after first-line ART failure differ between subtypes.
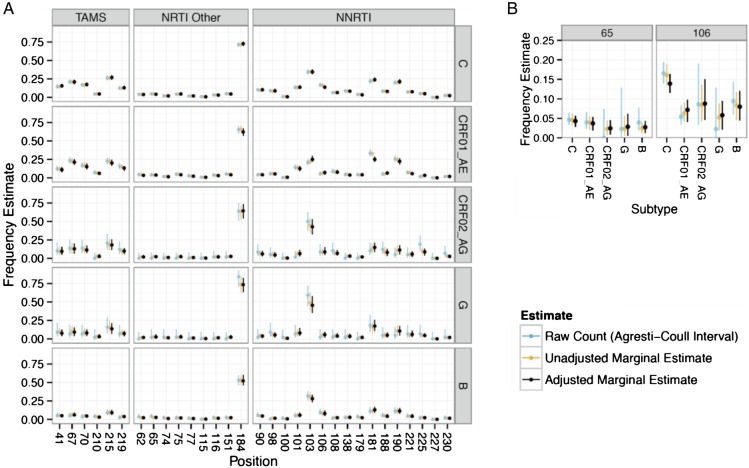
Figure 2.
The graphic illustrates frequency estimates of mutations at resistance positions across subtypes. Frequencies of mutations are shown for all (A) reverse-transcriptase resistance positions and specifically for (B) K65R and 106 M/I/A. In both (A) and (B), 3 estimates of mutation frequency (y axis) are shown: (1) the raw counting estimate with an Agresti-Coull binomial 95% confidence interval in blue, (2) the posterior estimate for each position within each treatment composition without poststratification with a 95% credible interval in green, and (3) the posterior estimate within each treatment composition with poststratification with a 95% credible interval in black. Abbreviations: NRTI, nucleoside reverse-transcriptase inhibitor; NNRTI, nonnucleoside RTI; TAMS, thymidine analog mutations.
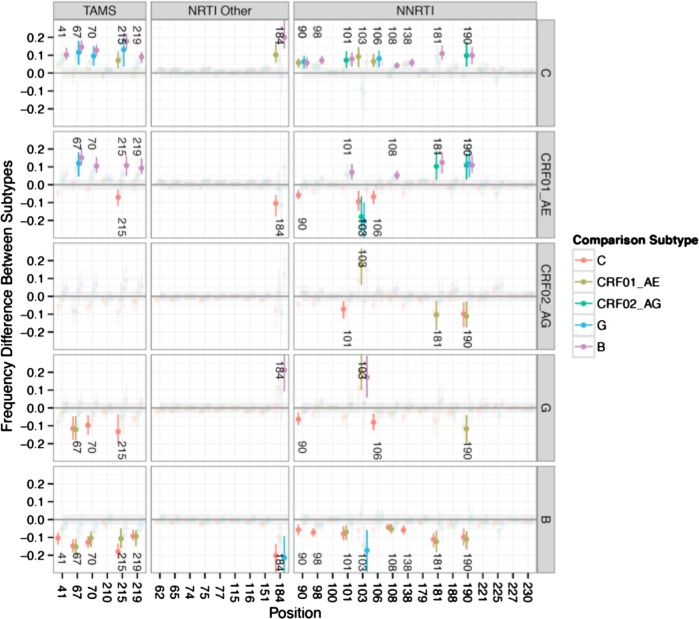
Figure 3.
The graphic illustrates estimates of differences in mutation frequency at resistance-associated positions between sequences from patients failing first-line antiretroviral therapy across different subtypes. Mutations are categorized into thymidine analog mutations (TAMS), nonnucleoside reverse-transcriptase inhibitors (NRTI Other), and nucleoside reverse-transcriptase inhibitors (NNRTI) mutations. Contrasts are performed using posterior estimates of each subtype with poststratification to account for different treatment composition within each subtype population. The y axis indicates the differences in mutation frequency. Every position in each subtype is compared with that position in all other subtypes, and position number in each row indicates significantly different results. The colors indicate the comparison subtype.
The lamivudine-associated mutations 184 V/I were the most common in all subtypes (53%–84% of sequences), followed by the NNRTI-associated mutations K103N/S (21%–59%) (Figure 2). Other common mutations were thymidine analog mutations (TAMs) 41L, 67N, 70R/E, 215F/Y, and 219Q/E and NNRTI-associated mutations 90I, 101I/P/E/H, 106N/S, 181C/I/V, and 190S/A. The 151 complex mutations (62 V/75I/77L/116Y/151 M), other non-TAM NRTI, and NNRTI-associated mutations were relatively rare across subtypes (all <5%). Our frequency estimates of 2 known subtype-related mutations, 65R and 106 M/I/A [14, 31], were consistent with prior reports suggesting that these mutations are more frequent in subtype C, although the frequency of K65R in subtype C was not significantly higher than other subtypes in this dataset (Figure 2B).
Overall, subtype B exhibited lower resistance mutation frequencies, and subtype C exhibited higher resistance mutation frequencies over both NRTI and NNRTI resistance-associated positions (Figure 3). Every subtype C position comparison that resulted in a significant difference (15 of 31 positions, indicated in Figure 3) exhibited a higher frequency compared with other subtypes. Every subtype B position comparison that exhibited a significant difference (14 of 31 positions) exhibited a lower frequency compared with other subtypes. Differences were observed in 5 of 6 TAMs: 41L (+10% with a high posterior density interval of [7–14] in subtype C vs B; relative risk [RR], 1.9–5.2), 67N (+15% [11–18]; RR, 2.2–5.3), 70R/E (+13% [10–16]; RR, 2.4–5.8) 215F/Y (+18% [13–22]; RR, 1.9–4.3), and 219Q/E (+9% [6–12]; RR, 1.8–5.6). Thymidine analog mutation 210W was relatively rare across all subtypes compared with other TAMs. The largest absolute difference in mutation frequency was observed for 184 V/I (+20% [14–30]; RR, 1.2–1.6), whereas the largest average RR was observed for 98G (+7% [5–10], RR 2.6–10.7). The general trend of higher resistance in C and lower resistance in B was also observed in earlier analyses that excluded newer samples from India, China, and Thailand as well as analyses of only these newer samples, which excluded the United States, suggesting the trend was not driven by a single subtype/country cohort such as C/India or B/United States.
Mutation frequencies in CRF01_AE were also higher than in subtype B overall and comparable to subtype C but with lower frequencies than C for mutations 90I (−6% [−8 to −3]; RR, .43−.60), 103N/S (−9% [−14 to −3]; RR, .73−.88), 106 M/I/A (−7% [−11 to −3]; RR, .53−.71), 184 V/I (−10% [−18 to −6]; RR, .86−.92), and 215F/Y (−7% [−12 to −3]; RR, .74−.88). The CRF02_AG and subtype G had fewer samples, and thus their estimates exhibited higher uncertainty compared with other subtypes. Estimated mutation frequencies of these other subtype populations were between the low and high extremes of subtypes B and C.
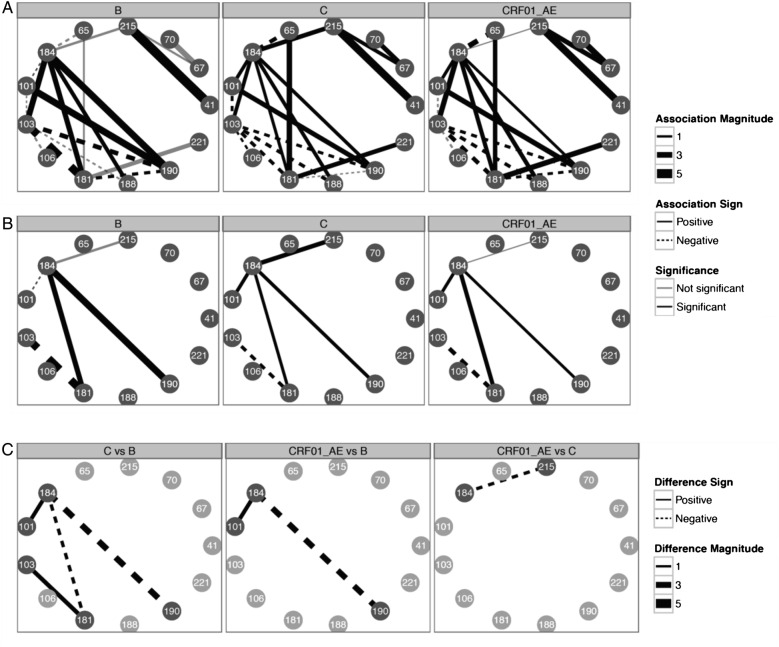
We examined relationships between resistance mutations using an Ising network model of dependence (see Methods) and compared these networks across subtypes. Network figures showing dependence between mutations at resistance positions for the subtypes B, C, and CRF01_AE (subtypes with the most sequences) are shown in Figure 4. Thirteen of the 31 RT resistance positions were found to be linked to at least 1 other resistance position in the 3 subtype datasets analyzed. These mutations were TAMs 41L, 67N, 70R/E, and 215F/Y; other NRTI mutations 65R and 184 V/I; and NNRTI mutations 101P/E/H, 103N/S, 106 M/I/A, 181C/I/V, 188L/C/H, 190S/A, and 221F/Y. We find that the directionality between almost every edge in the mutation network is preserved across subtypes. The 3TC-associated mutations 184 V/I connect the TAM network via their association with 215F/Y with NNRTI mutations 103N/S, 181C/I/V, 188L/C/H, and 190S/A. The 184 V/I are also negatively associated with 65R in all subtypes examined. The 103N/S is a hub that is (1) negatively associated with other mutations in the NNRTI network, (2) positively associated with only 184 V/I, and (3) negatively associated with 5 NNRTI mutations. The 215F/Y is positively associated with 41L and 67N. There is no association between 41L and 67N, consistent with the inclusion of patients who were likely sequenced early in the TAM pathway progression [32].
Figure 4.
The graphic illustrates the Boltzmann/Ising network model of correlation between resistance-associated positions. (A) illustrates mutation networks for subtypes B, C, and CRF01_AE. Solid edges indicate positive correlations, whereas dashed edges indicate negative correlations with varying magnitude according to line width. Black edges indicate significant correlations, whereas gray edges indicate correlations that are not significant. (B) shows resistance position networks only with edges for which correlations differ between subtypes. (C) illustrates differences in resistance position correlation between subtypes.
We also examined differences between subtype networks and the magnitude of those differences. The NNRTI mutations (P/E/H) at position 101 are slightly negatively associated with 184 V/I in subtype B and positively associated with 184 V/I in other subtypes. Other differences are due to weaker, but still positive, association between 184 V/I and NNRTI mutations 181C/I/V, 190S/A, and TAM 215F/Y. Finally, the mutual exclusivity of 103 and 181 is attenuated in subtype C compared with subtype B.
Susceptibility scores, which predict phenotypic effects of resistance mutations on ART, showed lower levels of resistance to future ART options in the subtype B population compared with CRF01_AE and C (Figure 5A). It is important to note that the Stanford Database scores do not account for subtype-dependent mutation effects on susceptibility. These scores should be interpreted as a qualitative summary of mutation status and allow for the possibility of subtype differences, not captured by current models of the relationship between mutations and susceptibility.
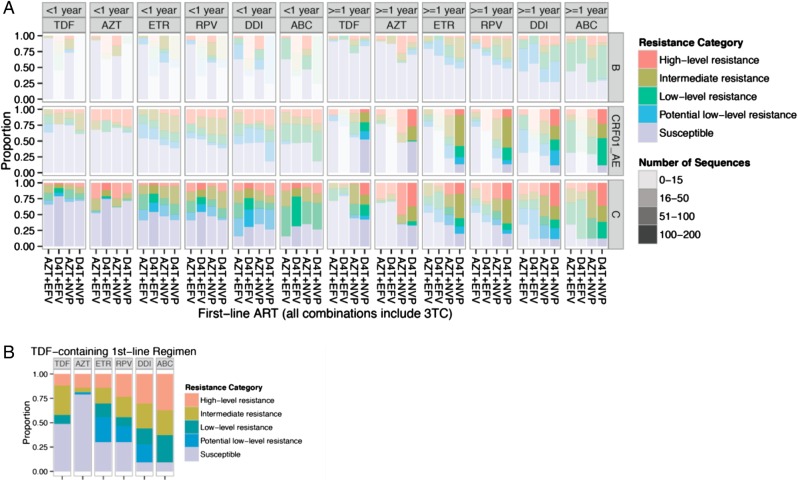
Figure 5.
The graphic illustrates predicted antiretroviral therapy (ART) susceptibility after first-line treatment failure. Proportions of sequences are shown for each of 6 drugs (columns), according to the specific ART regimen (x axis), subtype (rows: B, C, CRF01_AE), ART duration (<1 year, left side of figure; and ≥1 year, right side of figure), and 5 resistance categories (colors). Number of sequences are indicated by the color strength according to the legend. (A) Non-tenofovir (TDF)-based regimens, sorted from left to right by decreasing susceptibility within each ART duration category; (B) TDF-based regimens, pooled across subtypes and prior treatments due to the limited sample size. Abbreviations: ABC, abacavir; AZT, zidovudine; DDI, didanosine; D4T, stavudine; EFV, efavirenz; ETR, etravirine; NVP, nevirapine; RPV, rilpivirine; 3TC, lamivudine.
Averaged across categories of prior ART, treatment duration, and regimens, 60% of sequences from subtype B-infected patients were predicted to be susceptible (score <10) to treatment options to which they were not exposed, compared with 48% of CRF01_AE and 42% subtype C. Seventeen percent of subtype B genotypes were predicted to have intermediate or high level resistance to any treatment option, compared with 36% in CRF01_AE and 34% in subtype C. Thus, observed differences in resistance mutation outcomes after first-line therapy between subtypes have consequences for cross-resistance to nonfirst-line options.
Consistent with recent reports [33], patients failing D4T-containing regimens are more susceptible to TDF than to AZT (Figure 5, D4T+ EFV, D4T+ NVP, columns 1–2, 7–8). This holds regardless of subtype or ART duration. However, in patients failing TDF-containing first-line regimens, predicted cross-resistance was substantial (19%, 56%, and 63% intermediate or high level resistance to AZT, didanosine (DDI), and abacavir (ABC), respectively (Figure 5B).
Overall, there were lower levels of resistance to TDF (72% of patients susceptible, averaged across prior histories) and higher levels of resistance to ABC (32% susceptible), except in patients with prior exposure to TDF. Between the extremes of TDF and ABC was resistance to AZT (67% susceptible), etravirine (49%), rilpivirine (49%), and DDI (32%). Within each drug susceptibility column in Figure 5, NVP-exposed patients with durations of treatment exceeding 1 year tended to exhibit higher levels of resistance than EFV-exposed patients. These susceptibility trends are robust across subtypes and are clearly observed in subtypes B, C, and CRF01_AE.
DISCUSSION
We characterized HIV-1 drug resistance mutation patterns in a multicohort, multi-subtype dataset of patients failing first-line triple ART. Overall, we find lower resistance mutation frequencies in subtype B and higher resistance mutation frequencies in subtype C and CRF01_AE. This suggests that much of these differences likely reflect biological or ecological factors that simultaneously influence multiple DRMs. The trend is less evident in prior studies [17], likely due to greater treatment heterogeneity among the sequences included.
Subtype mutation pattern differences affect the predicted cross-resistance to subsequent regimens, with CRF01_AE and subtype C exhibiting high levels of cross-resistance (Figure 5). For D4T-failing patients, there is evidence of more AZT than TDF cross-resistance across subtypes, consistent with current knowledge [30, 33]. More broadly, resistance to TDF and AZT was lower and resistance to second-generation NNRTIs, DDI, and ABC was higher, regardless of subtype and prior first-line ART history. Our findings extend a report on second-line susceptibility following first-line therapy in sub-Saharan Africa, which also suggested high cross-resistance to second-generation NNRTIs in patients failing first-line therapy [22]. However, it should be noted that patients who fail TDF-based first-line ART exhibit relatively high levels of cross-resistance. Although the number of TDF-failing patients in our dataset is limited, resistance to TDF should be closely monitored as it becomes more widely used.
Resistance mutation outcomes can be explained by the underlying determinants of the observed genotype upon treatment failure. During viral population expansion before treatment, subtype differences in genetic barriers and tolerance to mutations determine the baseline spectrum of mutations [15]. Fitness considerations determine relative frequencies of resistant genotypes within the viral quasispecies and introduce negative correlations between high fitness-cost mutations such as K65R [34]. Differential tolerance to resistance mutations is also consistent with measurements of the effect of genetic background on fitness tolerance to TAMs. Competition assays have found that mutants with TAMs 67N and 70R are less fit in subtype B compared with subtype C [8], and susceptibility assays have found that TAMs contribute more to resistance in a CRF01_AE background [35, 36]. It is not yet possible to synthesize generalizable principles from such individual studies. Nevertheless, such experiments provide the most direct measurement available of the impact of subtype on phenotype.
Ecological factors such as the frequency of monitoring [21] also determine the level of resistance upon treatment failure (see model in the Supplementary Figure 3). Given the higher levels of resistance observed in subtype C and CRF01_AE populations, interventions that minimize ecological contributions to resistance such as increased monitoring, viral load testing, or use of boosted PI regimens may warrant consideration. If these subtype differences reflect ecological factors, then they suggest a need for programmatic interventions to optimize ART and mitigate the accumulation of resistance among subtypes C and CRF01_AE. An additional ecological explanation incorporates temporal differences among subtypes in time of sample collection, reflecting changes in clinical practice and treatment monitoring. Although the collection periods for the major variants (subtypes B, C, and CRF01_AE) overlap (all start between 1994 and 1996 and end in 2010 and 2011), subtype B sequences are enriched in earlier collection dates (mean = 2002.2, standard deviation [SD] = 3.8), whereas subtype C and CRF01_AE are enriched in later collection dates (subtype C mean = 2005.9, SD = 2.4; subtype CRF01_AE mean = 2005.1, SD = 3.2).
Due to differences in data availability across cohorts, potentially relevant metadata at the subject level was not always available. Viral load and CD4 counts measure an important effect of potential treatment monitoring differences, and a limitation of this study is the lack of their measurements across these multiple resource-limited cohorts. In a mathematical model presented in the Supplementary Data, we specifically discuss how both treatment monitoring frequency and genetic background effects interact with viral population expansion before treatment and can influence the observed pattern of resistance upon treatment failure. Patterns of transmitted resistance and mutations before treatment may also exhibit population-specific heterogeneities not apparent in this cross-sectional dataset. In addition, sequences in any observational monitoring database do not constitute a random sample of the treatment population. Interpretation of observed subtype differences must allow for potential sampling biases favoring patient subpopulations with a higher propensity to be sequenced.
We have discussed both ecological and biological contributions to subtype differences, but the correlation between the subtype of a patient and their geographical location inherently limits the determination of causal attribution from observational data. Dividing a patient population by subtype inevitably results in ecologically distinct comparisons. Even comparisons within locales including multiple subtypes, subtype populations segregate along transmission networks and reflect distinct ecological circumstances [37]. Our central limitation remains the causal identification of the genetic background effect regardless of sample size. Secondary limitations include the limited availability of certain subtypes and aggregation of discretization of treatment durations.
Observational data from clinical genotypes should be used in conjunction with evidence from in vitro phenotypic characterizations, for which direct effects are causally identifiable. Furthermore, observational comparisons between subtypes should incorporate as much background information regarding ecological heterogeneity as possible. The multilevel modeling approach we developed here enables modeling of population heterogeneity while minimizing analytical problems arising from the large number of parameters inherent to mutational analyses [38]. Focusing on estimation of subtype differences has interpretational advantages over the standard approach of multiple hypothesis testing [38, 39].
CONCLUSIONS
Previous characterizations of subtype differences have focused on mechanisms related to position-specific associations among subtype backgrounds, such as codon usage in subtype C, which impact the frequency of K65R and V106 M [14, 15, 19, 31]. Although such position-specific mechanisms are clearly important, variation in resistance between subtypes appears dominated by shared influences affecting multiple mutations. Future studies should further disentangle the role of ecological and biological contributors for higher levels of resistance in subtype C and CRF01_AE. Identifying the correct causal explanations for the patterns of resistance characterized here is crucial to optimizing ART among populations infected with non-B HIV-1 subtypes.
Supplementary Data
Supplementary material is available online at Open Forum Infectious Diseases (http://OpenForumInfectiousDiseases.oxfordjournals.org/).
Acknowledgments
Disclaimer. This work was not funded or sponsored by Pfizer nor do the contents of this manuscript reflect the views of Pfizer.
Financial support. This work was supported by National Institutes of Health grants R01AI66922, P30AI042853, and T32DA13911–10. The Stanford HIV Sequence Database is maintained by Dr. Robert Shafer and Soo-Yon Rhee.
Potential conflicts of interest. A. H. is currently employed as a Principal Scientist at Pfizer, Inc. His contributions to this manuscript were performed while he was working at Brown University and prior to his employment at Pfizer. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Palella FJ Jr, Delaney KM, Moorman AC et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med 1998; 338:853–60. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Consolidated guidelines on general HIV care and the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. Available at: http://apps.who.int//iris/handle/10665/85321. Accessed 1 February 2014.
- 3.Piot P, Quinn TC. Response to the AIDS pandemic—a global health model. N Engl J Med 2013; 368:2210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med 2008; 358:1590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson DL, Anderson JP, Bradac JA et al. HIV-1 nomenclature proposal. Science 2000; 288:55–6. [DOI] [PubMed] [Google Scholar]
- 6.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 2011; 25:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong KL, Lee TH, Essex M. Replicative capacity differences of thymidine analog resistance mutations in subtype B and C human immunodeficiency virus type 1. J Virol 2009; 83:4051–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong KL, Lee TH, Essex M. Replicative fitness costs of nonnucleoside reverse transcriptase inhibitor drug resistance mutations on HIV subtype C. Antimicrob Agents Chemother 2011; 55:2146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunha RD, Abreu CM, Gonzalez LM et al. Differential in vitro kinetics of drug resistance mutation acquisition in HIV-1 RT of subtypes B and C. PLoS One 2012; 7:e46622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaplin B, Eisen G, Idoko J et al. Impact of HIV type 1 subtype on drug resistance mutations in Nigerian patients failing first-line therapy. AIDS Res Hum Retroviruses 2011; 27:71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallis CL, Mellors JW, Venter WD et al. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. J Acquir Immune Defic Syndr 2010; 53:480–4. [DOI] [PubMed] [Google Scholar]
- 12.Yu XF, Chen J, Shao Y et al. Two subtypes of HIV-1 among injection-drug users in southern China. Lancet 1998; 351:1250. [DOI] [PubMed] [Google Scholar]
- 13.De Oliveira T, Pillay D, Gifford RJ. The HIV-1 subtype C epidemic in South America is linked to the United Kingdom. PloS One 2010; 5:e9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner BG, Oliveira M, Doualla-Bell F et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 2006; 20:F9–13. [DOI] [PubMed] [Google Scholar]
- 15.Brenner BG, Coutsinos D. The K65R mutation in HIV-1 reverse transcriptase: genetic barriers, resistance profile and clinical implications. HIV Ther 2009; 3:583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumans AT, Soares MA, Machado ES et al. Synonymous genetic polymorphisms within Brazilian human immunodeficiency virus type 1 subtypes may influence mutational routes to drug resistance. J Infect Dis 2004; 189:1232–8. [DOI] [PubMed] [Google Scholar]
- 17.Kantor R, Katzenstein DA, Efron B et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med 2005; 2:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang A, Hogan JW, Istrail S et al. Global analysis of sequence diversity within HIV-1 subtypes across geographic regions. Future Virol 2012; 7:505–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman Z, Paxinos EE, Averbuch D et al. Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob Agents Chemother 2004; 48:2159–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neogi U, Sahoo PN, Kumar R et al. Characterization of HIV type 1 subtype C protease gene: selection of L63P mutation in protease inhibitor-naive Indian patients. AIDS Res Hum Retroviruses 2011; 27:1249–53. [DOI] [PubMed] [Google Scholar]
- 21.Gupta RK, Hill A, Sawyer AW et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis 2009; 9:409–17. [DOI] [PubMed] [Google Scholar]
- 22.Hamers RL, Sigaloff KC, Wensing AM et al. Patterns of HIV-1 drug resistance after first-line antiretroviral therapy (ART) failure in 6 sub-Saharan African countries: implications for second-line ART strategies. Clin Infect Dis 2012; 54:1660–9. [DOI] [PubMed] [Google Scholar]
- 23.Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis 2006; 19:594–606. [DOI] [PubMed] [Google Scholar]
- 24.Cohen J. Breakthrough of the year. HIV Treatment as Prevention. Science 2011; 334:1628. [DOI] [PubMed] [Google Scholar]
- 25.Manosuthi W, Butler DM, Chantratita W et al. Patients infected with HIV type 1 subtype CRF01_AE and failing first-line nevirapine- and efavirenz-based regimens demonstrate considerable cross-resistance to etravirine. AIDS Res Hum Retroviruses 2010; 26:609–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertagnolio S, Perno CF, Vella S, Pillay D. The impact of HIV drug resistance on the selection of first- and second-line ART in resource-limited settings. J Infect Dis 2013; 207 (Suppl 2):S45–8. [DOI] [PubMed] [Google Scholar]
- 27.Rhee SY, Gonzales MJ, Kantor R et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 2003; 31:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kantor R, Delong A, Wu M et al. SQUAT (Sequence Quality Assessment Tool): a standalone computational tool for HIV sequence quality evaluation. AIDS Res Hum Retroviruses 2012; 28:894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. Rapid Advice: Antiretroviral Therapy for HIV Infection in Adults and Adolescents. Geneva: World Health Organization; 2009; 10. [Google Scholar]
- 30.Johnson VA, Calvez V, Gunthard HF et al. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 2013; 21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner BG, Turner D, Oliveira M et al. A V106 M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2003; 17:F1–5. [DOI] [PubMed] [Google Scholar]
- 32.Novitsky V, Wester CW, DeGruttola V et al. The reverse transcriptase 67N 70R 215Y genotype is the predominant TAM pathway associated with virologic failure among HIV type 1C-infected adults treated with ZDV/ddI-containing HAART in southern Africa. AIDS Res Hum Retroviruses 2007; 23:868–78. [DOI] [PubMed] [Google Scholar]
- 33.Tang MW, Rhee SY, Bertagnolio S et al. Nucleoside reverse transcriptase inhibitor resistance mutations associated with first-line stavudine-containing antiretroviral therapy: programmatic implications for countries phasing out stavudine. J Infect Dis 2013; 207 (Suppl 2):S70–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cong M, Heneine W, García-Lerma JG. The fitness cost of mutations associated with human immunodeficiency virus type 1 drug resistance is modulated by mutational interactions. J Virol 2007; 81:3037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delviks-Frankenberry KA, Nikolenko GN, Maldarelli F et al. Subtype-specific differences in the human immunodeficiency virus type 1 reverse transcriptase connection subdomain of CRF01_AE are associated with higher levels of resistance to 3′-azido-3′-deoxythymidine. J Virol 2009; 83:8502–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanuma J, Hachiya A, Ishigaki K et al. Impact of CRF01_AE-specific polymorphic mutations G335D and A371 V in the connection subdomain of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on susceptibility to nucleoside RT inhibitors. Microbes Infect 2010; 12:1170–7. [DOI] [PubMed] [Google Scholar]
- 37.Mastro TD, Kunanusont C, Dondero TJ, Wasi C. Why do HIV-1 subtypes segregate among persons with different risk behaviors in South Africa and Thailand? AIDS 1997; 11:113–6. [DOI] [PubMed] [Google Scholar]
- 38.Chanock SJ, Manolio T, Boehnke M et al. Replicating genotype-phenotype associations. Nature 2007; 447:655–60. [DOI] [PubMed] [Google Scholar]
- 39.Gelman A, Hill J, Yajima M. Why we (usually) don't have to worry about multiple comparisons. J Res Educ Eff 2012; 5:189–211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.