Abstract
In recent studies, strains of non-dysenteriae 1 Shigella (NDS) expressing Shiga toxin have been reported. In this study, we report a novel stx1a-converting bacteriophage of Shigella sonnei associated with travel to Mexico. Phylogenetic comparison between this and other stx-converting phages suggests that toxigenic NDS strains have arisen through separate horizontal transfer events from toxigenic Escherichia coli.
Keywords: bacteriophages, Shigella sonnei, Shiga toxin
Shiga toxin-producing Escherichia coli (STEC) has been associated with adverse clinical outcomes including hemolytic uremic syndrome (HUS) [1]. Within STEC strains, the stx gene operon is phage-encoded, and it exists as one of several stx subtypes (Stx1a, Stx1c, Stx1d, and Stx2a-f) [2]. These stx-converting phages vary considerably in genomic size, morphology, and integration site preference (Supplementary Table 1) [2]. The stx-converting phage lytic cycle and toxin expression is induced in response to deoxyribonucleic acid (DNA)-damaging agents including fluoroquinolone antibiotics [3].
Previously, the expression of Stx toxins in Shigella species was believed to be unique to Shigella dysenteriae type 1. More recently, stx-encoding isolates of Shigella sonnei [4–6], S dysenteriae type 4 [7], and Shigella flexneri [8] have been reported. These isolates were associated with travel to Eastern Europe [4, 5], Morocco [6], and Hispaniola [7, 8]. The details of stx acquisition by these non-dysenteriae 1 Shigella (NDS) isolates remain unclear, including the chronology of toxin acquisition (ie, recent vs remote), the origin of the converting phages, and their global distribution.
METHODS
Stool samples were screened for Stx by immunoassay according to the manufacturer's instructions (ImmunoSTAT! EHEC assay; Meridian Biosciences, Cincinnati, OH). Stool cultures were performed using Hektoen enteric and XLD selective agars, and automated biochemical speciation of suspicious colonies was performed (Vitek2, Durham, NC). A toxin-negative clinical isolate of S sonnei was isolated by similar means and used as a control. Identifications were confirmed by the San Diego County Public Health Laboratory.
Phage lysis was induced by treating Shigella cultures with 0.5 µg/mL mitomycin C (MMC) as previously described [8]. After MMC addition, culture aliquots were removed hourly for optical density (OD600) reading. Large-scale phage genomic DNA isolation was performed after the QIAGEN Lambda kit protocol (QIAGEN, Valencia, CA).
Restriction analysis was performed by digesting purified phage genomic DNA with EcoRI and HindIII according to the manufacturer's recommended conditions (New England BioLabs, Ipswich, MA), and restriction products were resolved on 0.7% agarose. Invitrogen 1 kb plus ladder was used as a size standard (Invitrogen, Carlsbad, CA).
Sequencing library preparation was performed from genomic bacterial DNA and purified phage genomic DNA. Deoxyribonucleic acid was fragmented to 350 base pairs (bp) using the Covaris M220 (Covaris, Woburn, MA). The Illumina TruSeq Nano Kit (Illumina, San Diego, CA) was used to create barcoded libraries according to the manufacturer's protocol. Equimolar amounts of each library were pooled, and bidirectional sequencing was performed using the Illumina MiSeq platform for 300 cycles, using v2 reagents. The phage genomic sequence was determined by de novo assembly using the SPAdes 3.5.0 algorithm. To identify the integration site, bacterial genomic sequencing reads containing the phage terminal sequences were extracted, aligned, and subjected to Basic Local Alignment Search Tool (BLAST) search. The complete phage genome sequence is available through GenBank under accession number KR781488.
For phylogenetic analysis, phage sequences were obtained from National Center for Biotechnology Information using accession numbers found in Supplementary Table 1. Multiple sequence alignment was performed using MAFFT version 7, and maximum likelihood trees were inferred using RAxML v7.0.3 with general time reversible + gamma nucleotide substitution model [9]. Branch support was performed using 100 bootstrap iterations; all branches had bootstrap scores of >90, except the HUN/2013 – Stx1phi branch, which had a score of 58.
RESULTS
The stx1a-converting phage described here was isolated from S sonnei that infected 2 epidemiologically unrelated subjects who presented to our hospital in 2014. Both subjects had recently returned from Baja California Norte, Mexico, and presented with diarrhea and abdominal discomfort. Both were treated with intravenous fluids and oral fluoroquinolone antibiotics, and both made uneventful recoveries. Stool samples from both individuals were positive for Stx by immunoassay, and, unexpectedly, stool culture on selective agar grew S sonnei from both subjects. Shigella sonnei species was confirmed by automated biochemical species determination and confirmed by polymerase chain reaction targeting a methylase gene sequence that is unique to S sonnei [10] (data not shown). The Stx-producing isolates were named Ss-VASD01 and Ss-VASD02.
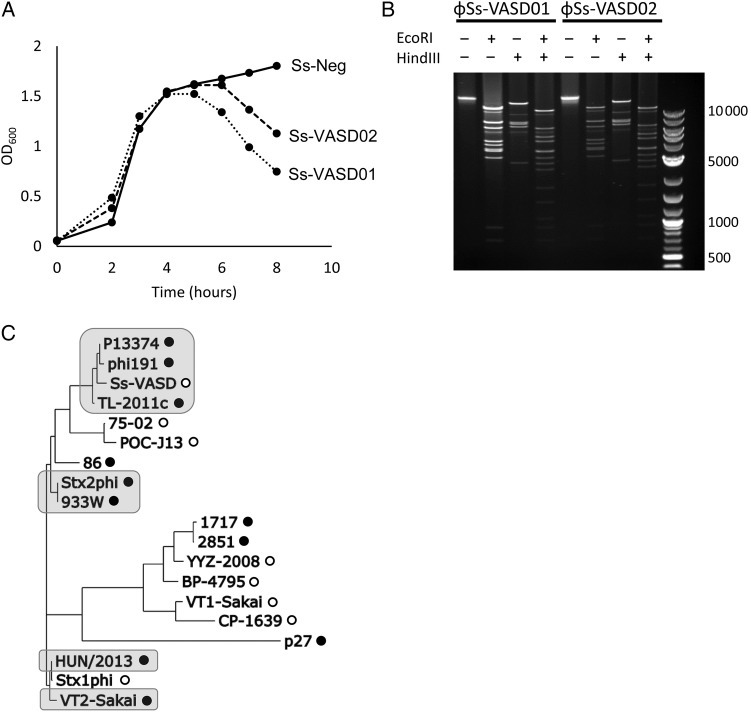
Treatment of S sonnei cultures with the DNA-alkylating agent MMC induced lysis of both Stx-expressing isolates but not of an Stx-negative S sonnei isolate (Figure 1A), suggesting the presence of an inducible bacteriophage. Phage particles were purified from MMC-induced cultures by polyethylene glycol precipitation, and restriction analysis of purified phage genomic DNA revealed a large molecular weight genome from both isolates, which shared identical restriction patterns (Figure 1B).
Figure 1.
(A) Induction of bacterial lysis by mitomycin C (MMC). Culture optical density (OD600) of Ss-VASD01, Ss-VASD02, and a nontoxigenic Shigella sonnei strain (Ss-Neg) was measured over time after addition of 0.5 µg/mL MMC (added at the 2-hour time point). (B) Restriction analysis of phage genomic deoxyribonucleic acid (DNA) obtained from purified phage particles after MMC induction. The DNA was digested with EcoRI, HindIII, or both. Invitrogen 1 kb plus ladder was run in the rightmost lane. (C) Whole genome phylogenetic analysis of ɸSs-VASD and other stx-converting bacteriophages. Phylogenetic tree of the complete ɸSs-VASD genome and other stx-converting phage genomes was inferred using the maximum likelihood method. Phages encoding stx1 are denoted by open circles, and those encoding stx2 are denoted by closed circles. Phages known to integrate into the wrbA gene are shaded in gray.
To further characterize the phage (named ɸSs-VASD), deep sequencing of purified phage and the whole bacterial genome was performed. De novo assembly of sequencing reads resulted in a 62 851-bp genome with 712.5× mean coverage. A total of 189 sequencing reads were recovered that spanned the host-phage junction, and BLAST alignment of the non-phage portions revealed 100% identity to the wrbA gene of S sonnei reference strain Ss046 (host-phage junction at Ss046 position 1088102).
To assess the relationship of ɸSs-VASD to other stx-converting phages, we performed phylogenetic comparisons between the genomic sequences of ɸSs-VASD and 18 previously reported stx-converting phages (Supplementary Table 1) [2, 5, 8]. The phage genome sequences were noted to cluster by integration site preference, with distinct clusters containing wrbA-integrating isolates (Figure 1C). The closest genetic relationships were found between ɸSs-VASD and stx2-converting phages of E coli.
Analysis of the stx coding sequence of ɸSs-VASD revealed 100% amino acid identity to 7/8 stx1-converting phages, with only POC-J13 varying from the consensus sequence at 2 positions (P284Q in stxA and A11T in stxB). ɸSs-VASD was found to contain the stx operon promoter PR’, which drives fluoroquinolone-mediated toxin production from stx2-encoding phage and contributes to toxin expression in stx1-encoding phages [3, 11, 12].
DISCUSSION
In this study, we report a novel stx1a-converting phage of S sonnei associated with travel to Baja California, Mexico. Prior reports of Stx-expressing NDS have been associated with travel (Hispaniola for S dysenteriae serotype 4 and S flexneri, and to the Eastern Europe and Morocco for S sonnei). To our knowledge, these are the first such isolates originating from North America. Furthermore, it appears that the subjects in this study may be part of a larger outbreak of Stx-toxigenic S sonnei, presently under investigation by the California Department of Public Health [13]. These findings suggest an expanding global emergence of Stx-toxigenic NDS. Alternatively, the presence of Stx-toxigenic phage in NDS may have been overlooked because, until recently, it was not common practice to screen diarrheal stools for Shiga-like toxins.
One concern surrounding Shiga toxin-producing bacterial strains is the potential for toxin induction by antibiotics. Fluoroquinolone antibiotics are of particular concern, because they can induce toxin production by inducing bacterial DNA damage responses. Even though ɸSs-VASD is inducible by DNA damage and encodes a fully intact stx operon, treatment with fluoroquinolone antibiotics in both subjects reported here did not appear to trigger any clinical worsening or complications. In fact, to our knowledge, there have been no cases of complications associated with Stx-NDS infection, an observation that could be explained by several theories including the predominance of the less-pathogenic Stx1 variant [2] or variability in toxin induction by antibiotics. Alternatively, there may simply be an insufficient numbers of cases detected thus far (even in outbreaks involving high-risk STEC isolates, HUS occurs in the minority of cases).
It is interesting to note that phylogenetic analysis of ɸSs-VASD reveals a relationship to other wrbA-integrating phages. This relationship seems to be irrespective of stx type, because the closest relationship was to ɸ191 and p13374, 2 stx2-converting phages of STEC associated with outbreaks of enteroaggregative hemorrhagic E coli. Given the overall similarity of these phages apart from the stx operon, the findings implicate a recombination event leading to similar phages with differing toxin operons, a phenomenon that has been observed in STEC phages [14]. A weaker phylogenetic relationship was observed between ɸSs-VASD and the S flexneri phage POC-J13, and the S sonnei-converting phage 75-02.
The origin of stx-converting phages in NDS stains is uncertain, as is the chronology of their existence. Possible theories include (1) recent horizontal transfer of phage from STEC and (2) longstanding presence of stx-converting phages in NDS that was previously undetected. The high degree of genetic similarity between ɸSs-VASD and previously reported stx-converting phages of STEC supports the theory that Ss-VASD arose from horizontal transfer between STEC and NDS strains, rather than by parallel evolution within NDS. We are not able to perform precise molecular clock analysis; however, the absence of major sequence divergence between ɸSs-VASD and STEC phages is suggestive of a recent transfer event. The occurrence of horizontal transfer events such as this seems plausible, given the discovery of large amounts of stx-converting phage found in human and agricultural wastewater [15].
CONCLUSIONS
Important future directions will include characterization of additional Stx-NDS phages, which will create better understanding of their diversity and may allow more precise inference of their origins. These findings suggest the need for more careful clinical and epidemiological monitoring for Shiga toxigenic S sonnei, because these strains seem to be broadly distributed globally, and infections therewith have the potential to cause complications not previously associated with S sonnei.
Supplementary Material
Acknowledgments
Financial support. This work was funded by the National Institutes of Health (grant AI093163).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Karmali MA, Petric M, Lim C et al. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli . J Infect Dis 1985; 151:775–82. [DOI] [PubMed] [Google Scholar]
- 2.Krüger A, Lucchesi PM. Shiga toxins and stx-phages: highly diverse entities. Microbiology 2014; 161:451–62. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, McDaniel AD, Wolf LE et al. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis 2000; 181:664–70. [DOI] [PubMed] [Google Scholar]
- 4.Strauch E, Lurz R, Beutin L. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect Immun 2001; 69:7588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tóth I, Sváb D, Bálint B et al. Comparative analysis of the Shiga toxin converting bacteriophage first detected in Shigella sonnei. Infect Genet Evol 2016; 37:150–7. [DOI] [PubMed] [Google Scholar]
- 6.Nyholm O, Lienemann T, Halkilahti J et al. Characterization of Shigella sonnei isolate carrying Shiga toxin 2-producing gene. Emerg Infect Dis 2015; 21:891–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta SK, Strockbine N, Omondi M et al. Emergence of Shiga toxin 1 genes within Shigella dysenteriae type 4 isolates from travelers returning from the island of Hispañola. Am J Trop Med Hyg 2007; 76:1163–5. [PubMed] [Google Scholar]
- 8.Gray MD, Lampel KA, Strockbine NA et al. Clinical isolates of Shiga toxin 1a-producing Shigella flexneri with an epidemiological link to recent travel to Hispañiola. Emerg Infect Dis 2014; 20:1669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006; 22:2688–90. [DOI] [PubMed] [Google Scholar]
- 10.Cho MS, Ahn TY, Joh K et al. A novel marker for the species-specific detection and quantitation of Shigella sonnei by targeting a methylase gene. J Microbiol Biotechnol 2012; 22:1113–7. [DOI] [PubMed] [Google Scholar]
- 11.Wagner PL, Neely MN, Zhang X et al. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J Bacteriol 2001; 183:2081–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner PL, Livny J, Neely MN et al. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol Microbiol 2002; 44:957–70. [DOI] [PubMed] [Google Scholar]
- 13.Shiga toxin-producing Shigella sonnei cases in San Diego County, 2015. Available at: http://www.sdcms.org/Portals/18/Assets/pdf/germ/02-24-2015%5B1%5D.pdf Accessed 13 April 2015. [Google Scholar]
- 14.Sváb D, Bálint B, Maróti G, Tóth I. A novel transducible chimeric phage from Escherichia coli O157:H7 Sakai strain encoding Stx1 production. Infect Genet Evol 2015; 29:42–7. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Castillo A, Muniesa M. Implications of free Shiga toxin-converting bacteriophages occurring outside bacteria for the evolution and the detection of Shiga toxin-producing Escherichia coli. Front Cell Infect Microbiol 2014; 4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.