Abstract
In our aging society, diseases in the elderly come more and more into focus. An important issue in research is Mild Cognitive Impairment (MCI) and Alzheimer’s Disease (AD) with their causes, diagnosis, treatment, and disease prediction. We applied the Brain Age Gap Estimation (BrainAGE) method to examine the impact of the Apolipoprotein E (APOE) genotype on structural brain aging, utilizing longitudinal magnetic resonance image (MRI) data of 405 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. We tested for differences in neuroanatomical aging between carrier and non-carrier of APOE ε4 within the diagnostic groups and for longitudinal changes in individual brain aging during about three years follow-up. We further examined whether a combination of BrainAGE and APOE status could improve prediction accuracy of conversion to AD in MCI patients. The influence of the APOE status on conversion from MCI to AD was analyzed within all allelic subgroups as well as for ε4 carriers and non-carriers. The BrainAGE scores differed significantly between normal controls, stable MCI (sMCI) and progressive MCI (pMCI) as well as AD patients. Differences in BrainAGE changing rates over time were observed for APOE ε4 carrier status as well as in the pMCI and AD groups. At baseline and during follow-up, BrainAGE scores correlated significantly with neuropsychological test scores in APOE ε4 carriers and non-carriers, especially in pMCI and AD patients. Prediction of conversion was most accurate using the BrainAGE score as compared to neuropsychological test scores, even when the patient’s APOE status was unknown. For assessing the individual risk of coming down with AD as well as predicting conversion from MCI to AD, the BrainAGE method proves to be a useful and accurate tool even if the information of the patient’s APOE status is missing.
Introduction
During the last 20 years structural brain imaging was more and more integrated into research and diagnosis of neurological disorders [1]. It became part of the diagnostic workflow to assure clinical diagnosis, to clarify differential diagnoses [2] or to obtain longitudinal data for patient’s follow-up. Brain imaging is also increasingly used as diagnostic marker for abnormal brain atrophy processes such as in Alzheimer’s Disease (AD) [3–5]. AD is of great importance for research since it is the most common cause of dementia late in life, affecting approximately 1% of the population of 60–65 years, and 10–35% of 85 years and older [6].
Many AD patients suffer from Mild Cognitive Impairment (MCI) before fully developing all symptoms of AD. MCI is seen as prodromal state of AD [7] or transitional state between normal aging and AD [8]. In the case of cognitive impairment and dementia, the patterns and dimension of brain atrophy correlate strongly with the current and future extent of the disease [9–12]. Generally, whole brain atrophy rates are estimated to be about 1% per year in patients with very mild AD compared to about 0.5% in non-demented elderly [13], and approximately 2% per year for gray matter volume in AD patients [14].
During the last years, several methods have been developed to predict conversion from MCI to AD. Some of them are based on MR imaging, since it is easily applicable in clinics and widely available as well as non-invasive. MRI data can be also easily used for further analysis and calculations. Recently, a novel approach for estimating the individual neuroanatomical age based on structural MRI and a machine-learning pattern recognition method was presented, utilizing Relevance Vector Regression (RVR) to model brain aging in a large sample of healthy subjects [15]. Analyzing the local patterns of brain atrophy and matching them to the chronological age of the subject, a reliable biomarker based on the estimation of a person’s brain age gap estimation (BrainAGE) score was obtained. Applying the BrainAGE approach to clinical samples, this score discriminated those MCI subjects converting to AD within 36 months follow-up, i.e. progressive MCI (pMCI), from those remaining stable during 36 months follow-up, i.e. stable MCI (sMCI) [16]. Already at baseline MRI scan, pMCI and AD patients showed increased BrainAGE score of 6 to 7 years as compared to the control and sMCI groups. Additionally, brain aging was even accelerating by one year per follow-up year in the pMCI group and 1.5 years per follow-up year in the AD group during the follow-up period of about four years [17]. These findings are in line with other publications revealing dramatic shrinkage of brain tissue in MCI and AD [12, 13, 18, 19]. The scope of the present study was to investigate whether including individual apolipoprotein E (APOE) genotype status increases prediction accuracy for conversion from MCI to AD based on structural MRI.
The polymorphic APOE gene is located on chromosome 19q13.2 [20], with ε2, ε3, and ε4 being the three most common allelic isoforms [21, 22]. It is well known that APOE ε4 is a dose-dependent risk factor for developing late-onset AD [23–27]. Risk estimations vary between 3 times to a more than 4 times elevated risk per APOE ε4 allele [21, 23, 24]. Thus, AD affection would be around 8 to 15 times more likely in homozygous carriers of the APOE ε4 allele as compared to ε4 non-carriers [24, 26, 28]. APOE ε4 also influences the clinical course of AD [29–31], causing earlier onset of dementia [25–27, 32–35], higher degrees of brain atrophy [36], lower temporal [19, 37], hippocampal [2, 36–40], amygdala volumes [38, 41] and significant thinner cortices [42], and faster cognitive decline [30]. Most studies agree about the negative influence of APOE ε4 on disease severity of AD, manifesting in the deposit of neuritic plaques [43, 44] and neurofibrillary tangles [44]. In contrast, the APOE ε2-isoform is supposed to has a protective effect, e.g. manifesting in lower incidences of MCI and AD, older age of AD onset [21, 25, 27, 45, 46], and slower cognitive decline [30].
In the present study we analyzed the effects of the APOE status on individual deviations from normal brain aging trajectories, its longitudinal course as well as its relation to cognition and disease severity in healthy controls, MCI and AD patients. Additionally, we investigated whether a combination of BrainAGE and APOE status would increase prediction accuracy for conversion from MCI to AD.
Methods
Study samples
Longitudinal sample
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD).
To investigate the longitudinal pattern of BrainAGE changes as a function of the APOE ε4 carrier status, this sample included all subjects from the ADNI database, for whom the APOE ε4 status as well as a baseline MRI scan and at least one follow-up MRI scan (1.5T) were available, resulting in a sample size of 405 subjects (Table 1). For the exact procedures of data collection and up-to-date information, see www.adni-info.org. Subjects were grouped as (i) NO (normal control group), if subjects were diagnosed cognitively healthy at baseline and remained so during 3 years follow-up (n = 107); (ii) sMCI (stable MCI), if subjects were diagnosed with MCI at baseline and remained so during 3 years follow-up (n = 36), (iii) pMCI (progressive MCI), if subjects were diagnosed with MCI at baseline and classified AD at some point during follow-up, without reversion to MCI or NO (n = 112), (iv) AD, if subjects were diagnosed with AD at baseline and remained so at any follow-up (n = 150).
Table 1. Characteristics of the longitudinal test sample.
| NO (n = 107) | sMCI (n = 36) | pMCI (n = 112) | AD (n = 150) | ANOVA (p) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ε4 carriers (ε2/ε4; ε3/ε4; ε4/ε4) | ε4 non carriers (ε2/ε3; ε3/ε3) | ε4 carriers (ε2/ε4; ε3/ε4; ε4/ε4) | ε4 non-carriers (ε2/ε3; ε3/ε3) | ε4 carriers (ε2/ε4; ε3/ε4; ε4/ε4) | ε4 non-carriers (ε2/ε3; ε3/ε3) | ε4 carriers (ε2/ε4; ε3/ε4; ε4/ε4) | ε4 non-carriers (ε2/ε3; ε3/ε3) | Diagnostic group | ε4 status (carriers vs. non-carriers) | diagnostic group x ε4 status | |
| No. of subjects (by APOE genotypes) | 26 (1 / 21 / 4) | 81 (16 / 65) | 14 (0 / 12 / 2) | 22 (3 / 19) | 78 (5 / 52 / 21) | 34 (2 / 32) | 101 (4 / 66 / 31) | 49 (4 / 45) | - | - | - |
| Baseline | |||||||||||
| Age mean in years (SD) | 75.0 (5.1) | 75.9 (4.9) | 77.3 (5.6) | 76.8 (6.5) | 74.1 (6.5) | 75.5 (9.3) | 74.1 (6.8) | 75.7 (8.9) | 0.36 | 0.30 | 0.88 |
| MMSE mean (SD) | 29.3 (0.8) | 29.2 (0.9) | 27.7 (1.7) | 27.2 (2.0) | 26.7 (1.8) | 26.4 (1.7) | 23.4 (2.0) | 23.5 (1.9) | < 0.001 | 0.34 | 0.71 |
| CDR-SB mean (SD) | 0.0 (0.0) | 0.0 (0.1) | 1.3 (0.6) | 1.1 (0.6) | 1.9 (1.0) | 1.9 (1.1) | 4.2 (1.5) | 4.3 (1.7) | < 0.001 | 0.92 | 0.96 |
| ADAS mean (SD) | 8.3 (3.9) | 8.9 (3.8) | 17.3 (5.3) | 17.3 (6.3) | 21.8 (5.8) | 21.8 (5.4) | 28.7 (7.2) | 29.0 (9.1) | < 0.001 | 0.79 | 0.99 |
| BrainAGE score in years (SD) | -0.11 (6.79) | -1.35 (6.45) | -0.88 (6.13) | 0.09 (4.93) | 5.83 (6.44) | 5.54 (9.68) | 5.76 (7.68) | 6.20 (9.52) | < 0.001 | 0.97 | 0.85 |
| Last follow-up scan | |||||||||||
| Follow-up duration in days (SD) | 1171 (234) | 1197 (270) | 1121 (283) | 1110 (222) | 967 (381) | 974 (309) | 616 (223) | 595 (221) | < 0.001 | 0.99 | 0.94 |
| Age mean in years (SD) | 78.2 (5.1) | 79.1 (5.0) | 80.4 (5.4) | 79.9 (6.5) | 76.7 (6.7) | 78.1 (9.4) | 75.8 (6.9) | 77.4 (9.1) | < 0.05 | 0.31 | 0.89 |
| MMSE mean (SD) | 28.5 (1.6) | 29.2 (1.1) | 26.7 (2.8) | 27.4 (2.6) | 21.4 (4.1) | 21.9 (4.7) | 19.2 (5.8) | 19.2 (5.3) | < 0.001 | 0.31 | < 0.05 |
| CDR-SB mean (SD) | 0.2 (0.5) | 0.2 (0.5) | 1.9 (1.0) | 1.7 (1.2) | 5.5 (2.6) | 5.2 (2.5) | 7.6 (3.7) | 7.5 (3.7) | < 0.001 | 0.69 | 0.99 |
| ADAS mean (SD) | 10.0 (5.7) | 10.2 (5.4) | 18.0 (7.1) | 17.4 (6.2) | 32.1 (8.0) | 33.8 (12.2) | 38.9 (12.2) | 36.6 (12.1) | < 0.001 | 0.84 | 0.46 |
| BrainAGE score in years (SD) | -0.16 (7.94) | -1.40 (6.06) | -0.01 (6.05) | -0.64 (4.77) | 8.68 (7.24) | 7.34 (10.29) | 8.30 (8.03) | 7.67 (10.14) | < 0.001 | 0.32 | 0.98 |
| Changing rates (per follow-up year) | |||||||||||
| MMSE | -0.17 | -0.01 | -0.26 | 0.10 | -2.20 | -1.83 | -2.42 | -2.47 | < 0.001 | 0.38 | 0.87 |
| CDR-SB | 0.06 | 0.03 | 0.19 | 0.24 | 1.40 | 1.32 | 1.81 | 1.82 | < 0.001 | 0.92 | 0.99 |
| ADAS | 0.51 | -0.04 | -0.11 | -0.06 | 3.80 | 4.31 | 5.62 | 4.16 | < 0.001 | 0.42 | 0.26 |
| BrainAGE | -0.01 | 0.03 | 0.20 | -0.13 | 1.13 | 0.61 | 1.68 | 0.90 | < 0.001 | < 0.05 | 0.25 |
P-values are resulting from ANOVA. Bold type = significant test results.
The following neuropsychiatric scales, administered at baseline and follow-up examinations, were used to evaluate the degree of cognitive decline: Alzheimer’s Disease Assessment Scale (ADAS; ranging from 0 to 85, with higher test scores indicating worse cognitive functioning) [47], global Clinical Dementia Rating Scale Sum of Boxes (CDR; ranging from 0 to 3, with 0 indicating NO, 0.5 denoting MCI, 1 and more indicates stages of AD) [48], and Mini-Mental State Examination (MMSE; ranging from 0 to 30, with lower test scores indicating worse cognitive functioning) [49].
Sample for prediction of AD conversion
To explore the performance of the BrainAGE framework in predicting conversion from MCI to AD in APOE ε4 carriers and non-carriers, all MCI subjects were included for whom baseline MRI data (1.5T), at least moderately confident diagnoses (i.e. confidence >2), and test scores in certain cognitive scales (i.e., ADAS, CDR-SB, MMSE) were available. The MCI subjects (n = 193) were grouped as (i) sMCI (stable MCI), if diagnosis was MCI stable during follow-up, at least for 36 months (n = 62); (ii) pMCI_early (progressive MCI), if diagnosis was MCI at baseline measurement and conversion to AD occurred within the first 12 months after baseline, without reversion to MCI or cognitive normal (NO) at any follow-up (n = 57); (iii) pMCI_late, if diagnosis was MCI at baseline measurement and conversion to AD was diagnosed after the first 12 months (i.e. at 18, 24, or 36 months follow-up), without reversion to MCI or NO at any follow-up (n = 74). Hereby, time to conversion does refer to time from being enrolled in ADNI (i.e., individual baseline measurements) till first diagnose of AD. Details of the characteristics of the prediction sample are presented in Table 2. The participants were further grouped according to their APOE ε4 status, resulting in ε4 carrier groups (sMCIC, pMCIC_early, pMCIC_late) and non-carrier groups (sMCINC, pMCINC_early, pMCINC_late).
Table 2. Baseline characteristics of the MCI sample used for prediction of AD conversion.
| ε4 carriers (n = 117) | ε4 non-carriers (n = 76) | ANOVA (p) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| sMCIC | pMCIC_ early | pMCIC_ late | sMCINC | pMCINC_ early | pMCINC_ late | Diagnostic group | ε4 status | Group x ε4 status | |
| No. subjects | 26 | 33 | 58 | 36 | 24 | 16 | - | - | - |
| Males / Females | 23 / 3 | 20 / 13 | 36 / 22 | 26 / 10 | 13 / 11 | 11 / 5 | - | - | - |
| Age mean (SD) | 76.5 (5.2) | 72.9 (6.0) | 75.0 (6.4) | 76.2 (6.8) | 75.3 (8.3) | 76.4 (10.0) | 0.20 | 0.26 | 0.55 |
| Education years mean (SD) | 16.3 (2.7) | 15.7 (2.6) | 15.9 (3.0) | 16.6 (2.5) | 15.0 (3.4) | 16.1 (2.6) | 0.12 | 0.97 | 0.61 |
| MMSE mean (SD) | 28.0 (1.4) | 26.5 (2.0) | 26.8 (1.5) | 27.5 (2.0) | 26.4 (1.8) | 26.6 (1.7) | < 0.001 | 0.33 | 0.72 |
| CDR-SB mean (SD) | 1.4 (0.7) | 2.1 (0.9) | 1.7 (0.9) | 1.3 (0.6) | 1.9 (0.9) | 2.0 (1.1) | < 0.001 | 0.87 | 0.42 |
| ADAS mean (SD) | 17.1 (5.2) | 23.7 (6.6) | 20.6 (4.4) | 15.7 (6.1) | 23.1 (5.9) | 19.7 (4.2) | < 0.001 | 0.24 | 0.93 |
| BrainAGE mean (SD) | 0.0 (4.4) | 9.0 (6.3) | 5.7 (6.0) | 1.2 (4.0) | 8.0 (9.2) | 5.0 (7.7) | < 0.001 | 0.42 | 0.38 |
P-values are resulting from ANOVA. Bold type = significant test results.
MRI Data Preprocessing and Data Reduction
Preprocessing of the T1-weighted images was done using the SPM8 package (http://www.fil.ion.ucl.ac.uk/spm) and the VBM8 toolbox (http://dbm.neuro.uni-jena.de), running under Matlab. All T1-weighted images were corrected for bias-field inhomogeneities, then spatially normalized and segmented into gray matter, white matter, and cerebrospinal fluid within the same generative model [50]. The segmentation procedure was further extended by accounting for partial volume effects [51], by applying adaptive maximum a posteriori estimations [52], and by using a hidden Markov random field model [53]. Preprocessing the images further included affine registration and smoothing with 4-mm full-width-at-half-maximum (FWHM) smoothing kernels. Spatial resolution was set to 4 mm. Data reduction was performed by applying Principal Component Analysis (PCA) utilizing the “Matlab Toolbox for Dimensionality Reduction” (http://ict.ewi.tudelft.nl/~lvandermaaten/Home.html). PCA was performed on the training sample only. The estimated transformation parameters were subsequently applied to the test samples. No further data reduction or region pre-selection was accomplished.
Estimation of BrainAGE scores
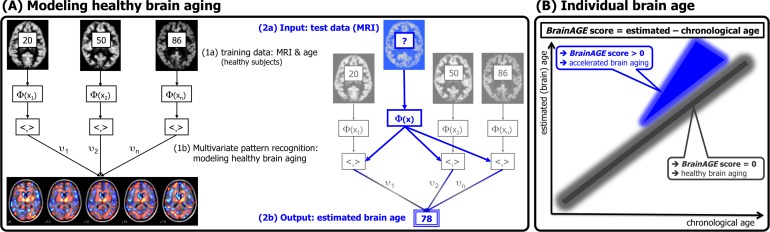
The BrainAGE framework utilizes a machine-learning pattern recognition method, namely relevance vector regression (RVR) [54], to estimate individual brain ages based on T1-weighted MR images [15]. The brain age of each test subject can be estimated using the individual tissue-classified MRI data, aggregating the complex, multidimensional aging pattern across the whole brain into one single value (Fig 1A). The difference between estimated and true chronological age will reveal the individual Brain Age Gap Estimation (BrainAGE) score. Consequently, the BrainAGE score directly quantifies the amount of acceleration or deceleration in brain aging. For example, if a 70 years old individual has a BrainAGE score of +5 years, this means that this individual shows the typical atrophy pattern of a 75 year old individual (Fig 1B). Recent work has demonstrated that this method provides reliable and stable estimates, with a correlation of r = 0.92 between the estimated and the chronological age and a mean absolute error of 5 years in healthy subjects aged 20–86 years [15]. Additionally, BrainAGE scores calculated from two shortly delayed scans resulted in an intraclass correlation coefficient (ICC) of 0.93 [55].
Fig 1. Depiction of the BrainAGE concept.
(A) The model of healthy brain aging is trained with the chronological age and preprocessed structural MRI data of a training sample (left, with an exemplary illustration of the most important voxel locations that were used by the age regression model). Subsequently, the individual brain ages of previously unseen test subjects are estimated, based on their MRI data (blue, picture modified from [56]). (B): The difference between the estimated and chronological age results in the BrainAGE score, indicating abnormal brain aging. [Image reproduced from [17], with permission from Hogrefe Publishing, Bern]
For the present study, the BrainAGE method was applied using the preprocessed gray matter images (as described above). To train the age estimation framework, we used T1-weighted MRI data of all subjects from the publicly accessible database “Information eXtraction from Images” (IXI; http://www.brain-development.org; data downloaded in September 2011) aged 20–86 years (mean age 48.6 ± 16.5 years; n = 560), which were collected on three different scanners (Philips 1.5T, General Electric 1.5T, Philips 3.0T). Additionally, MRI data of all healthy control subjects from the publicly accessible database “Open Access Series of Imaging Studies” (OASIS; http://www.oasis-brains.org; downloaded in June 2009) aged 51–94 years (mean age 71.3 ± 11.8 years; n = 126) were also included in the training sample. For training the model as well as for predicting individual brain ages, we used “The Spider” (http://www.kyb.mpg.de/bs/people/spider/main.html), a freely available toolbox running under Matlab. For an illustration of the most important features (i.e., the importance of voxel locations for regression with age) that were used by the RVR to model normal brain aging and more detailed information please refer to [15]. In both test samples, BrainAGE scores were calculated based on baseline MRI. In the longitudinal test sample, follow-up BrainAGE scores were calculated based on each available MRI data during follow-up.
Statistical Analysis
First, baseline BrainAGE scores, BrainAGE scores at last visit, and longitudinal changes in BrainAGE were compared among the 4 diagnostic groups and the APOE ε4 carrier status using analysis of variance (ANOVA). Post-hoc analyses (with Bonferroni correction to compensate for multiple comparisons) were conducted to further explore group differences. Additionally, the effects of the particular allelic isoforms (i.e., ε2/ε3, ε3/ε3, ε2/ε4, ε3/ε4, ε4/ε4) on BrainAGE were analyzed. Longitudinal changes in individual BrainAGE scores, i.e., the differences between follow-up and baseline BrainAGE scores were fitted against days from baseline with a multivariate linear regression model, including correction for age and gender. The relationships between BrainAGE scores and cognitive scales (i.e. MMSE, CDR-SB, ADAS) were explored using Pearson’s linear correlation coefficients.
In the second part of the study, prediction of conversion from MCI to AD in APOE ε4 carriers and non-carriers based on baseline BrainAGE scores was studied. Receiver operating characteristics (ROC) for discriminating MCI subjects who converted to AD from those who remained stable during follow-up were computed in early converting as well as all MCI subjects together, resulting in the area under the curve (AUC), also known as C-statistics or c-index. The AUC shows the quality of classification, with 1.0 indicating a perfect discrimination and 0.5 indicating a result obtained by chance only. In order to test whether the resulting AUC derived from ROC analysis based on BrainAGE scores is statistically greater than the AUCs of the cognitive scores, one-tailed z-tests were performed. Additionally, the McNemar test for paired data was performed in order to statistically test whether predictions of conversion based on baseline BrainAGE scores are significantly better than predictions based on cognitive scores. Furthermore, univariate Cox regression was used to estimate the hazard rate for conversion to AD, adjusted for age, gender, and education years. The time-to-event variable was time from baseline visit to first visit with AD diagnosis for pMCI subjects. For sMCI subjects, the duration of follow-up was truncated at 36 months. The main predictor was the baseline BrainAGE score as a continuous variable initially and with median split subsequently. Cox regression was also performed with baseline cognitive scores as main predictors. Furthermore, it was tested whether including the individual APOE status into the Cox regression model would significantly improve the model performance. As checked by log-minus-log-plots of survival, the assumption of proportional hazards was met for all Cox proportional hazard models. Cox regression was performed using SPSS. All other statistical testing was performed using Matlab.
Results
Longitudinal sample
BrainAGE scores and cognitive tests at baseline and follow-up were analyzed in all diagnostic groups (NO, sMCI, pMCI, AD) according to APOE ε4 carrier status (Table 1) and particular allelic isoform (Table 3). The allelic combination of ε2/ε2 was not represented in this sample. In line with other studies [22, 29], APOE ε2/ε4 was assigned to the ε4 carrier group.
Table 3. Mean BrainAGE scores at baseline and last follow-up for all particular allelic isoforms within the diagnostic groups of the longitudinal sample.
| NO | sMCI | pMCI | AD | |||||
|---|---|---|---|---|---|---|---|---|
| Baseline | Last follow-up | Baseline | Last follow-up | Baseline | Last follow-up | Baseline | Last follow-up | |
| APOE ε2 / ε3 | ||||||||
| No. subjects | 16 | 3 | 2 | 4 | ||||
| BrainAGE mean (SD) | -2.66 (5.32) | -3.01 (5.42) | +1.95 (6.92) | +2.71 (5.45) | +3.43 (5.29) | +9.24 (2.10) | +8.80 (4.86) | +11.31 (6.16) |
| APOE ε3 / ε3 | ||||||||
| No. subjects | 65 | 19 | 32 | 45 | ||||
| BrainAGE mean (SD) | -1.03 (6.69) | -1.01 (6.18) | -0.21 (4.73) | -1.16 (4.60) | +5.67 9.93) | +7.22 (10.60) | +5.97 (9.82) | +7.35 (10.40) |
| APOE ε2 / ε4 | ||||||||
| No. subjects | 1 | 0 | 5 | 4 | ||||
| BrainAGE mean (SD) | +13.28 (0.00) | +11.72 (0.00) | - | - | +3.39 (6.72) | +7.25 (6.05) | +2.10 (10.60) | +3.29 (7.97) |
| APOE ε3 / ε4 | ||||||||
| No. subjects | 21 | 12 | 52 | 66 | ||||
| BrainAGE mean (SD) | -1.42 (6.44) | -1.28 (8.07) | -0.15 (6.28) | +0.47 (6.45) | +5.38 (5.85) | +7.40 (7.03) | +6.19 (8.74) | +8.45 (8.60) |
| APOE ε4 / ε4 | ||||||||
| No. subjects | 4 | 2 | 21 | 31 | ||||
| BrainAGE mean (SD) | +3.44 (4.44) | +2.75 (4.94) | -5.29 (2.93) | -2.85 (0.59) | +7.52 (7.64) | +12.18 (7.12) | +5.33 (5.18) | +8.63 (6.69) |
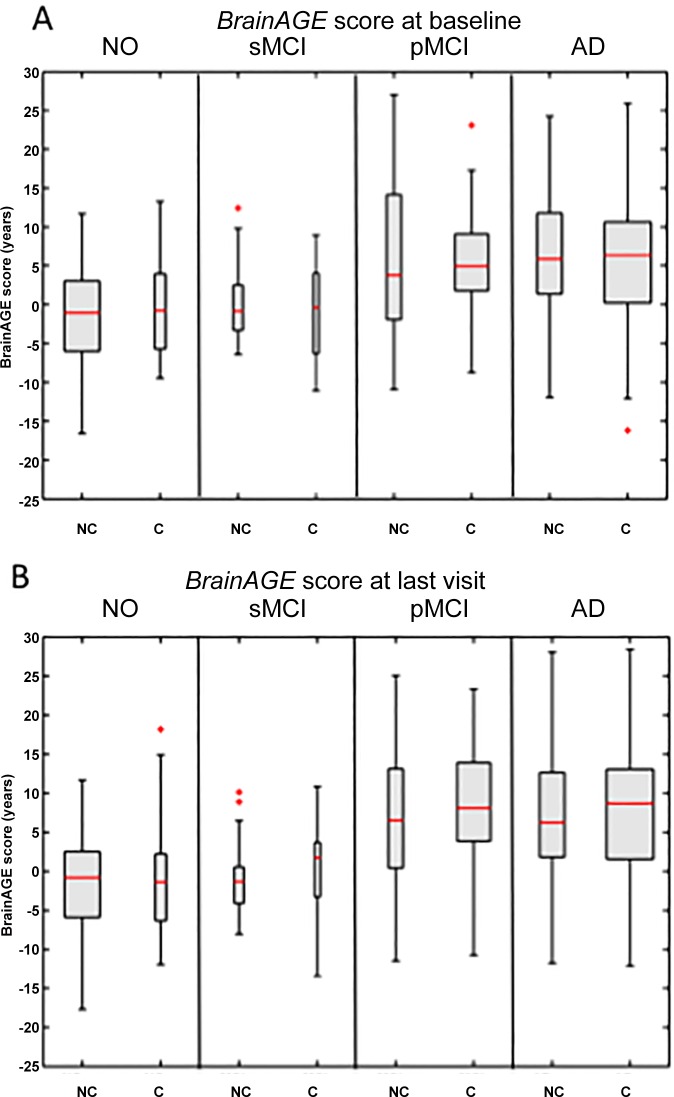
BrainAGE scores differed significantly among all 4 diagnostic groups at baseline (F = 18.86, p < 0.001; Table 1) and at last MRI scan (F = 30.56, p < 0.001; Table 1). As revealed by post-hoc t-tests, BrainAGE scores in NO as well as sMCI differed significantly from BrainAGE scores in pMCI as well as AD at baseline (p < 0.05; Fig 2A) and at last MRI scan (p < 0.05; Fig 2B), suggesting neuroanatomical changes that show patterns of advanced brain aging in pMCI and AD patients. At baseline as well as at last MRI scan, there was no significant effect regarding APOE ε4 status or interaction between diagnostic group and APOE ε4 status. Additionally, no significant effects were found for the particular allelic isoforms (Table 3), which may be due to the very small number of patients for some allelic isoforms.
Fig 2.
BrainAGE scores at (A) baseline and (B) the last visit for non-carriers and carriers of APOE ε4. Shown are boxplots, presenting the distribution of the BrainAGE scores for the 4 diagnostic groups NO, sMCI, pMCI and AD. BrainAGE scores differed significantly between diagnostic groups at baseline (F = 18.9, p < 0.001) and at follow-up scans (F = 30.6, p < 0.001). Post-hoc tests showed significant differences between BrainAGE scores in NO as well as sMCI from BrainAGE scores in pMCI as well as AD at baseline and last visit (p < 0.05). The boxes include values between the 25th and 75th percentiles and the median (red line). Lines extending the boxes below and above include data within 1.5 times the interquartile range. All outliers are symbolized with a red”+”. Width of the boxes symbolizes group size.
As mentioned above, patients with the allelic isoform ε2/ε4 were assorted to carriers. Since there were no representatives of this isoform in the sMCI cohort, we subsequently examined the possible effect of falsification by excluding all patients with a combination of ε2/ε4 from our longitudinal sample. However, test results did not change (F-statistics at baseline for diagnostic group: F = 9.22, p < 0.001; APOE ε4 status: F = 0.01, p = 0.99; interaction: F = 0.6, p = 0.79; follow-up scan for diagnostic group: F = 16.35, p < 0.001; APOE ε4 status: F = 0.62, p = 0.60; interaction: F = 0.74, p = 0.67).
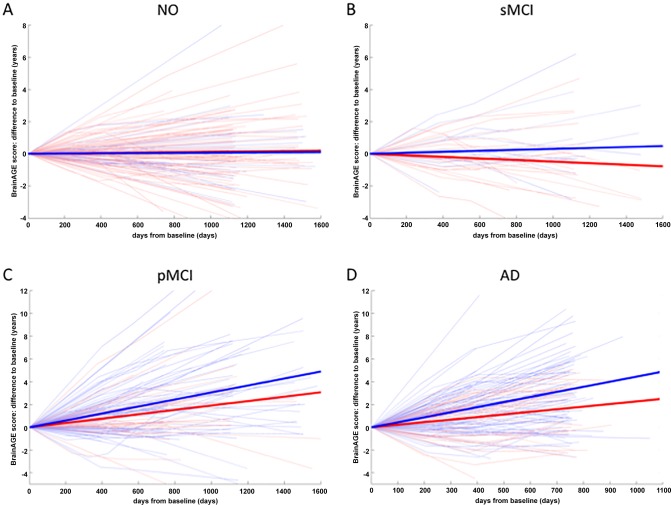
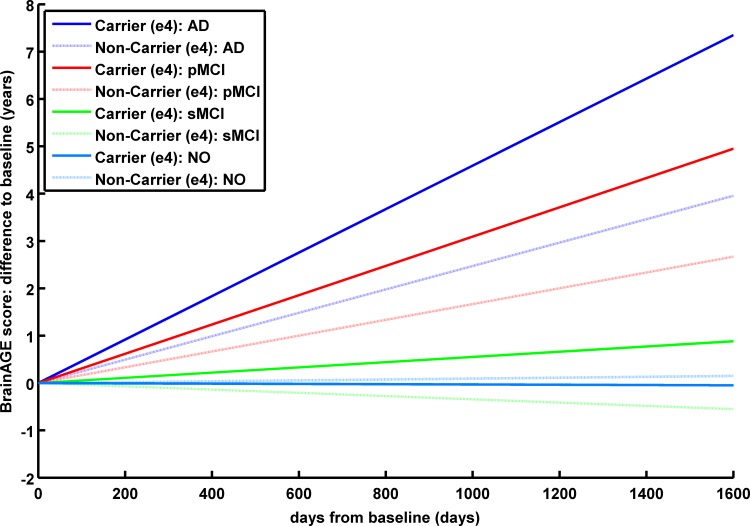
To further investigate individual trajectories of BrainAGE scores, longitudinal changes as compared to the baseline assessment were analyzed for each available time point during follow-up. BrainAGE scores remained stable in the NO and sMCI groups across the follow-up period of about three years, but increased in the pMCI and AD groups, suggesting additional acceleration in brain aging in the pMCI and AD groups (Fig 3). BrainAGE changing rates differed significantly between NO and sMCI subjects as compared to pMCI and AD subjects as well as between APOE ε4 carriers and non-carriers (p < 0.05; Fig 4), with ε4 carriers showing increased changing rates as compared to non-carriers (Table 1).
Fig 3.
Longitudinal changes in BrainAGE score for (A) NO, (B) sMCI, (C) pMCI and (D) AD patients. Thin lines represent individual trajectories of BrainAGE score over time of follow-up. Thick lines represent the estimated average regression lines for APOE ε4 non-carriers (red) and carriers (blue).
Fig 4.
Estimated longitudinal changes in BrainAGE scores for the 4 diagnostic groups: NO (light blue), sMCI (green), pMCI (red) and AD (blue), subdivided into APOE ε4 carriers and non-carriers. Post-hoc t-tests resulted in significant differences for ε4 carriers and non-carriers as well as for NO / sMCI vs. pMCI / AD (p < 0.05).
Correlations between BrainAGE and cognitive scores were analyzed for baseline and last follow-up visit. In the whole sample, BrainAGE scores correlated significantly with each of the cognitive scores independent of the APOE ε4 carrier status (Table 4). Analyzing the diagnostic groups separately, correlations between BrainAGE and cognitive scores were only found in pMCI and AD patients, for both ε4 carriers and non-carriers. Probably, disease related neuroanatomical alterations might be reflected in the cognitive scores if they exceed a certain degree. Distinct differences between ε4 carriers and non-carriers were not found (Table 4).
Table 4. Correlation coefficients between BrainAGE scores and cognitive functioning (ADAS scores) as well as disease severity (MMSE & CDR-SB scores) for each diagnostic group and the whole test sample, separately for APOE ε4 carriers and non-carriers.
| NO | sMCI | pMCI | AD | Whole sample | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ε4 carriers | ε4 non-carriers | ε4 carriers | ε4 non-carriers | ε4 carriers | ε4 non-carriers | ε4 carriers | ε4 non-carriers | ε4 carriers | ε4 non-carriers | |
| No. of subjects | 26 | 81 | 14 | 22 | 78 | 34 | 101 | 49 | 219 | 186 |
| Correlation with BrainAGE score at baseline | ||||||||||
| MMSE score | 0.04 | -0.21 | 0.45 | -0.08 | -0.08 | -0.29 | -0.28** | -0.62*** | -0.34*** | -0.52*** |
| CDR-SB score | -0.04 | -0.15 | -0.12 | 0.02 | 0.27* | 0.13 | 0.10 | 0.60*** | 0.29*** | 0.50*** |
| ADAS score | -0.06 | 0.04 | -0.45 | -0.07 | 0.27* | 0.32 | 0.19 | 0.52*** | 0.33*** | 0.50*** |
| Correlation with BrainAGE score at last scan | ||||||||||
| MMSE score | 0.00 | -0.17 | 0.30 | -0.12 | -0.21 | -0.33 | -0.38*** | -0.66*** | -0.44*** | -0.59*** |
| CDR-SB score | -0.07 | 0.00 | -0.07 | 0.09 | 0.38** | 0.23 | 0.21* | 0.59*** | 0.40*** | 0.57*** |
| ADAS score | -0.06 | 0.04 | -0.34 | 0.09 | 0.38** | 0.38* | 0.25* | 0.56*** | 0.43*** | 0.58*** |
***p<0.001
**p<0.01
*p<0.05.
Prediction of conversion to AD
The subsample used to predict conversion to AD consisted of 193 MCI patients, including 117 APOE ε4 carriers and 76 non-carriers. Chronological age and education years did not differ between stable, early converting, and late converting MCI patients. Baseline BrainAGE scores differed significantly between groups (F = 8.96, p < 0.001; Table 2), as did the cognitive scores. There weren’t any effects for the APOE ε4 status or for interactions between diagnostic group and APOE ε4 status (Table 2).
A total number of 91 ε4 carriers and 40 non-carriers converted to AD during the 36 months of follow-up. That corresponds to a pre-test probability of 78% in carriers and 53% in non-carriers. In ε4 carriers, 28% of the MCI subjects converted to AD within the first 12 months after baseline examination, whereas 50% converted to AD after the first year of follow-up. In non-carriers, 32% of the MCI subjects converted to AD within the first 12 months after baseline examination, whereas 21% converted to AD after the first year of follow-up.
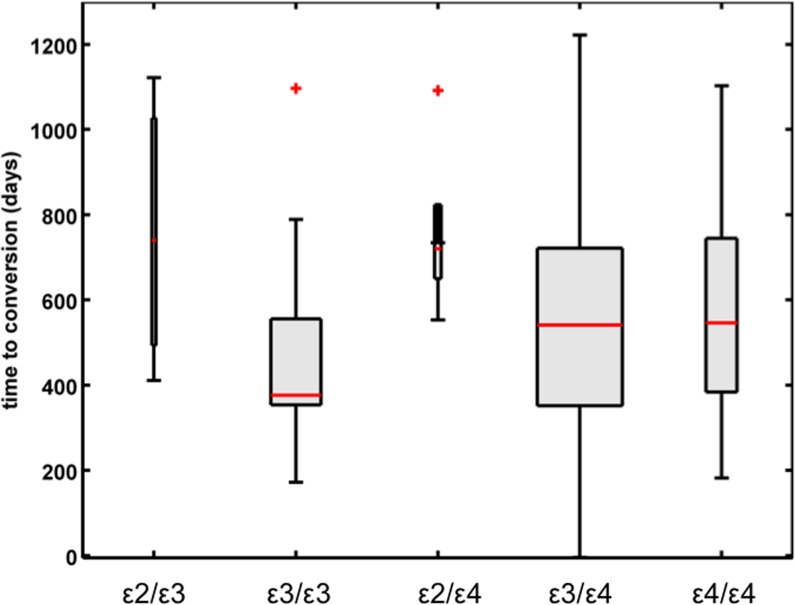
Regarding time to conversion from MCI to AD diagnosis, APOE ε4 carriers showed the tendency to take about 3 months longer to convert to AD (560 ± 280 days) as compared to non-carriers (471 ± 233 days; F = 3.14; p = 0.08; Fig 5). Interestingly, time to conversion was longer in homozygous ε4 carriers (591 days) as compared to homozygous ε3 carriers (448 days). Longest time to conversion was shown in heterozygous carriers of a protective ε2 allele with either ε3 (758 days) or ε4 (756 days). Time to conversion did not cover the whole time suffering from MCI, but rather corresponded to the individual time being enrolled in ADNI while suffering from MCI until AD was diagnosed for the first time.
Fig 5. Mean days to conversion from MCI to AD subdivided into all allelic combinations of APOE.
Presented are the mean ± SD days to conversion within the given allelic combinations of APOE (F = 3.14; p = 0.08): ε2/ε3 (n = 2): 758 ± 356, ε3/ε3 (n = 32): 448 ± 210, ε2/ε4 (n = 5): 756 ± 201, ε3/ε4 (n = 52): 534 ± 292, ε4/ε4 (n = 21): 591 ± 246. The boxes include values between the 25th and 75th percentiles and the median (red line). Lines extending the boxes below and above include data within 1.5 times the interquartile range. All outliers are symbolized with a red”+”. Width of the boxes symbolizes group size.
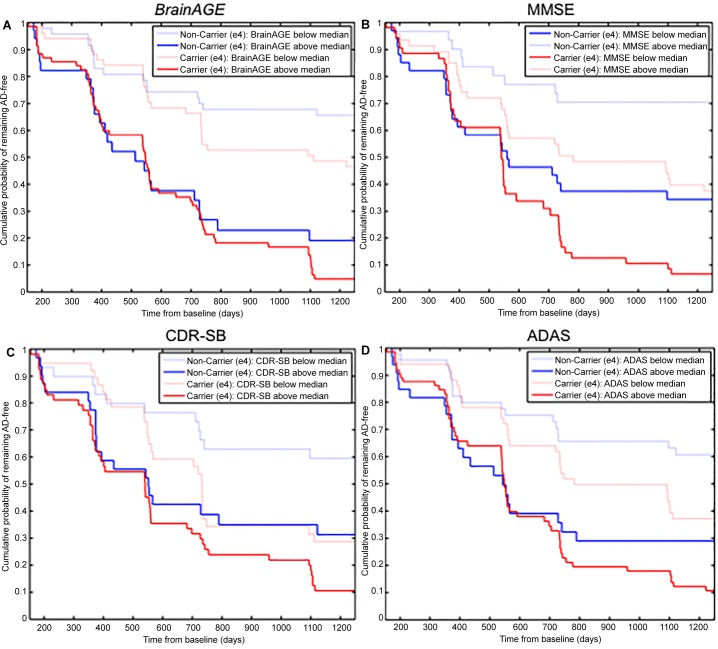
Cox regression for prediction of conversion to AD based on baseline BrainAGE scores resulted in higher baseline BrainAGE scores being associated with a higher risk of converting to AD independent of APOE status (χ2 = 53.88, p < 0.001; Table 5). Subjects with a BrainAGE score above median of 4.5 years had a nearly 4 times greater risk of converting to AD as compared to subjects with BrainAGE scores below the median (hazard ratio; HR: 3.76, p < 0.001; Table 5). Including the APOE status into the Cox regression model, the quality of the prediction model tended to improve (χ2 = 3.23, p = 0.07). The Cox regression model based on baseline BrainAGE scores outperformed all models based on baseline MMSE, CDR-SB, and ADAS scores, even when including the APOE ε4 status into the models (Table 5, Fig 6).
Table 5. Cox Regression values for cumulative AD incidence in APOE ε4 carriers and non-carriers in the BrainAGE, MMSE, CDR-SB, and ADAS scores alone and in combination with the APOE ε4 carrier status, based on a median split.
| Test of model | Change from model without APOE | Hazard ratio (HR) | Confidence interval (CI) | p | |||
|---|---|---|---|---|---|---|---|
| χ2 | p | χ2 | p | ||||
| BrainAGE | |||||||
| BrainAGE (only)* | 53.88 | < 0.001 | 3.76 | 2.58–5.48 | < 0.001 | ||
| BrainAGE & APOE | 56.79 | < 0.001 | 3.23 | 0.07 | |||
| ° BrainAGE | 3.58 | 2.44–5.24 | < 0.001 | ||||
| ° APOE | 1.41 | 0.96–2.05 | 0.08 | ||||
| MMSE | |||||||
| MMSE (only) | 18.46 | < 0.001 | 2.37 | 1.58–3.55 | < 0.001 | ||
| MMSE & APOE | 27.68 | < 0.001 | 9.62 | < 0.01 | |||
| ° MMSE | 2.42 | 1.61–3.63 | < 0.001 | ||||
| ° APOE | 1.91 | 1.25–2.93 | < 0.01 | ||||
| CDR-SB | |||||||
| CDR-SB (only) | 12.91 | < 0.001 | 2.05 | 1.37–3.05 | < 0.001 | ||
| CDR-SB & APOE | 19.61 | < 0.001 | 6.95 | < 0.01 | |||
| ° CDR-SB | 1.97 | 1.32–2.93 | < 0.001 | ||||
| ° APOE | 1.72 | 1.14–2.60 | < 0.01 | ||||
| ADAS | |||||||
| ADAS (only) | 22.57 | < 0.001 | 2.35 | 1.63–3.38 | < 0.001 | ||
| ADAS & APOE | 26.62 | < 0.001 | 4.27 | < 0.05 | |||
| ° ADAS | 2.22 | 1.54–3.20 | < 0.001 | ||||
| ° APOE | 1.48 | 1.01–2.18 | < 0.05 | ||||
Bold type = significant; asterisk = best performance of all models.
Fig 6.
Cumulative probability for MCI patients of remaining AD-free, divided into patients with the score of interest below the median (light lines) and above it (dark lines). Non-carriers of the APOE ε4 gene are painted in blue, carriers in red. Shown are Kaplan-Meier survival curves based on Cox regression, comparing the cumulative incidence of AD in ε4 carriers and non-carriers in (A) BrainAGE, (B) MMSE, (C) CDR-SB, and (D) ADAS scores. The follow-up duration is truncated at 1250 days.
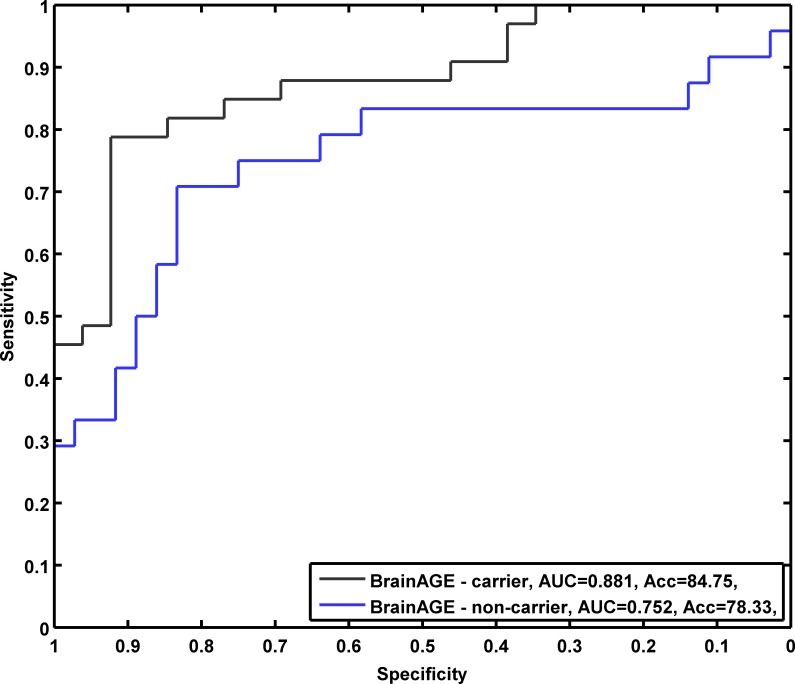
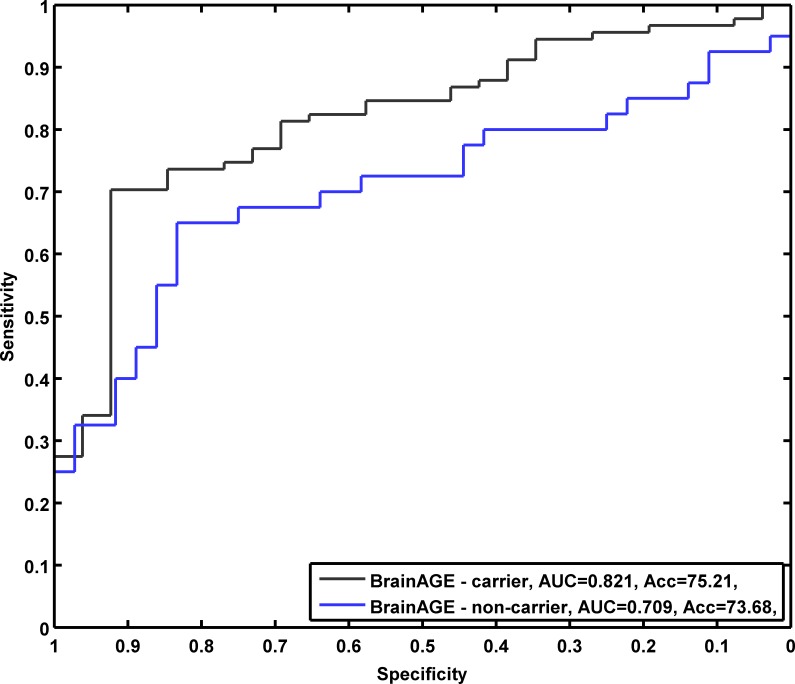
The effect of APOE ε4 status on prediction accuracy was further examined with ROC analyses. By varying the threshold applied to the BrainAGE score, ROC curves were constructed for a binary discrimination between MCI subjects who remained stable during 3 years follow-up from those who converted to AD. For the discrimination of pMCI_early from sMCI, ROC analyses at baseline BrainAGE scores resulted in an AUC (or c-index) of 0.88 with an accuracy rate of 85% in APOE ε4 carriers. In APOE ε4 non-carriers, prediction performances were slightly lower with an AUC of 0.75 and an accuracy rate of 78% (Fig 7). For discriminating early and late converting MCI together from sMCI patients, the AUC resulted in 0.82 for APOE ε4 carriers and 0.71 for non-carriers. Achieved accuracies for APOE ε4 carriers were 75%, for APOE ε4 non-carriers 74% (Fig 8).
Fig 7.
ROC curves for pMCI_early subjects, analyzing individual subject classification based on baseline BrainAGE score in either remaining MCI or converting to AD, subdivided into APOE ε4 carriers and non-carriers.
Achieved accuracies (sensitivity / specificity) for predicting conversion from MCI to AD for APOE ε4 carriers: 85% (0.79 / 0.92), non-carriers: 78% (0.71 / 0.83).
Fig 8. ROC curves of APOE ε4 carriers vs. non-carriers based on baseline BrainAGE scores in all MCI patients.
Achieved accuracies (sensitivity / specificity) for predicting conversion from MCI to AD during follow-up for APOE ε4 carriers: 75% (0.70 / 0.92) and for non-carriers: 74% (0.65 / 0.83).
Furthermore, the McNemar test was applied to explore whether predictions of future conversion to AD in MCI patients based on BrainAGE are significantly better than predictions based on chronological age or cognitive test scores. Predicting conversion based on baseline BrainAGE scores showed significantly better results as compared to chronological age and cognitive scores, especially in APOE ε4 carriers (Tables 6 and 7).
Table 6. Results for predicting conversion to AD in MCI subjects (APOE ε4 carriers).
| sMCI vs. pMCIC_early | sMCI vs. pMCIC (all) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (CI) | Sensitivity (CI) | Specificity (CI) | McNemar test | Accuracy (CI) | Sensitivity (CI) | Specificity (CI) | McNemar test | |||
| Error rate (CI) | χ2 | Error rate (CI) | χ2 | |||||||
| BrainAGE score* | 0.85 (0.76–0.94) | 0.79 (0.68–0.89) | 0.92 (0.85–0.99] | 0.15 (0.06–0.22) | - | 0.75 (0.67–0.83] | 0.70 (0.62–0.79) | 0.92 (0.87–0.97) | 0.25 (0.17–0.33) | - |
| Chronological age | 0.39 (0.27–0.51) | 0.39 (0.27–0.52) | 0.92 (0.85–0.99] | 0.61 (0.49–0.73) | 17.79 (p < 0.001) | 0.54 (0.45–0.63] | 0.35 (0.26–0.44) | 0.88 (0.83–0.94) | 0.46 (0.37–0.55) | 7.78 (p < 0.01) |
| MMSE score | 0.46 (0.33–0.58) | 0.52 (0.39–0.64) | 0.85 (0.75–0.94] | 0.54 (0.42–0.67) | 8.53 (p < 0.01) | 0.23 (0.15–0.31] | 0.68 (0.60–0.77) | 0.65 (0.57–0.74) | 0.77 (0.69–0.85) | 40.01 (p < 0.001) |
| CDR-SB score | 0.49 (0.36–0.62) | 0.70 (0.58–0.81) | 0.81 (0.71–0.91] | 0.51 (0.38–0.64) | 14.28 (p < 0.001) | 0.26 (0.18–0.34] | 0.52 (0.43–0.61) | 0.81 (0.74–0.88) | 0.74 (0.65–0.81) | 46.12 (p < 0.001) |
| ADAS score | 0.69 (0.58–0.81) | 0.79 (0.68–0.89) | 0.69 (0.57–0.81] | 0.31 (0.19–0.42) | 3.20 (n.s.) | 0.43 (0.34–0.52] | 0.71 (0.63–0.80) | 0.69 (0.61–0.78) | 0.57 (0.48–0.66) | 22.44 (p < 0.001) |
Bold type = significant; asterisk = best performance of all models; n.s. = not significant
Table 7. Results for predicting conversion to AD in MCI subjects (APOE ε4 non-carriers).
| sMCI vs. pMCINC_early | sMCI vs. pMCINC (all) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (CI) | Sensitivity (CI) | Specificity (CI) | McNemar test | Accuracy (CI) | Sensitivity (CI) | Specificity (CI) | McNemar test | |||
| Error rate (CI) | χ2 | Error rate (CI) | χ2 | |||||||
| BrainAGE score | 0.78 (0.68–0.89) | 0.71 (0.59–0.82) | 0.83 (0.74–0.93) | 0.22 (0.11–0.32) | - | 0.74 (0.64–0.84) | 0.65 (0.54–0.76) | 0.83 (0.75–0.92) | 0.26 (0.16–0.36) | - |
| Chronological age | 0.50 (0.37–0.63) | 0.83 (0.74–0.93) | 0.31 (0.19–0.42) | 0.50 (0.37–0.63) | 8.53 (p < 0.01) | 0.47 (0.36–0.59) | 0.15 (0.07–0.23) | 0.97 (0.93–1.00) | 0.53 (0.41–0.64) | 6.81 (p < 0.01) |
| MMSE score | 0.60 (0.48–0.72) | 0.79 (0.69–0.89) | 0.58 (0.46–0.71) | 0.40 (0.28–0.52) | 4.17 (p < 0.05) | 0.47 (0.36–0.59) | 0.78 (0.68–0.87) | 0.58 (0.47–0.69) | 0.53 (0.41–0.64) | 9.00 (p < 0.01) |
| CDR-SB score | 0.67 (0.55–0.79) | 0.58 (0.46–0.71) | 0.75 (0.64–0.86) | 0.33 (0.21–0.45) | 1.64 (n.s.) | 0.51 (0.40–0.63) | 0.53 (0.41–0.64) | 0.75 (0.65–0.85) | 0.49 (0.37–0.60) | 8.53 (p < 0.01) |
| ADAS score | 0.68 (0.57–0.80) | 0.92 (0.85–0.99) | 0.58 (0.46–0.71) | 0.32 (0.20–0.43) | 0.93 (n.s.) | 0.64 (0.54–0.75) | 0.90 (0.83–0.97) | 0.58 (0.47–0.69) | 0.36 (0.25–0.46) | 1.20 (n.s.) |
Bold type = significant; n.s. = not significant
Discussion
This study explored the effects of individual APOE ε4 status on the performance of a novel MRI-based biomarker based on the recently presented BrainAGE framework [15, 55] in (i) recognizing advanced brain aging in a longitudinal design and (ii) predicting prospective conversion to AD on an individual subject level.
Longitudinal sample
AD patients as well as MCI subjects, who cognitively declined and thus converted to AD within 3 years of follow-up (pMCI), exhibited significantly larger baseline BrainAGE scores compared to control subjects and those with MCI, who remained cognitively stable (sMCI), but did not differ between APOE ε4 carriers and non-carriers. In contrast, brain aging accelerates more in APOE ε4 carriers during follow-up as compared to non-carriers in the pMCI and AD groups, i.e. already starting with a higher baseline BrainAGE score of about 6 years, brain aging accelerates during follow-up with the speed of 1.1 additional year in brain atrophy per follow-up year in pMCI ε4 carriers, but only about 0.6 years in pMCI ε4 non-carriers, and 1.7 additional years in brain atrophy per follow-up year in AD ε4 carriers and 0.9 years in AD ε4 non-carriers. This accumulated to mean BrainAGE scores between 7 to 9 years at the last scan, with mean follow-up durations of 2.7 years for pMCI and 1.7 years for AD. Compared to that, healthy control as well as sMCI subjects did not show any deviations from normal brain aging trajectories at baseline and follow-up.
These results are in line with recent studies that showed AD-like MRI-based indices in pMCI subjects [4, 57], increased GM atrophy of approximately 2% per year in AD [58], accelerated changes in whole brain volume in MCI [18], acceleration in atrophy rates as subjects progress from MCI to AD [59], and greater GM loss in certain regions in pMCI subjects [60, 61]. Furthermore, our results also support the assumption of AD being a form of or at least being associated with accelerated aging [18, 62, 63]. Taking into account the patients APOE genotype revealed significant differences within the MCI groups at baseline and follow-up measurements in a recent study using a MRI-based index for diagnosis and prediction of conversion [57]. In the present study, we did not find significant differences between ε4 carriers and non-carriers in baseline or follow-up BrainAGE scores. In contrast, ε4 carriers showed increased acceleration of individual brain aging as compared to non-carriers in pMCI and AD patients. This is in line with recent studies suggesting that APOE ε4 carriers are suffering from faster pathologic processes than non-carriers [64] and therefore have higher atrophy rates [59].
Additionally, individual BrainAGE scores were profoundly related to measures of clinical disease severity, most pronounced in APOE ε4 carriers and non-carriers already diagnosed with AD at baseline, as well as to measures of cognitive functioning, most pronounced in APOE ε4 carriers diagnosed with MCI at baseline and converting to AD within the next three years. Cognitive decline was recently found to progressively accelerate years before being diagnosed with AD [65], and to be correlated with the atrophy rates in specified brain regions [60]. Our results support the suggested relationship between progressive acceleration in brain aging and rate of change in cognitive functioning as well as clinical severity in pMCI and AD during follow-up, especially in APOE ε4 carriers. Furthermore, we could even show that accelerated brain aging is more closely related to the worsening of higher cognitive functions, but slightly less with disease severity in pMCI subjects, whereas in AD patients accelerated brain aging was more closely related to disease severity and slightly less with worsening of higher cognitive functions. Regarding NO and sMCI subjects a ceiling effect as well as a slightly lower variance within the cognitive scores was observed. This may be mainly due to the fact, that the scales analyzed in this study were used specifically to identify clinical disease severity as well as deterioration in cognitive functioning in the ADNI sample.
Cross-sectional sample
Analysing the effect of APOE on the risk of conversion to AD, we compared ε4 carriers and non-carriers within the whole MCI sample. A total of 78% of ε4 carriers converted to AD within 3 years of follow-up, compared to only 53% of non-carriers, underlining a higher risk for carriers to convert to AD. Within this sample, ε4 carriers tend to convert slower than non-carriers. Several studies suggested the APOE ε4 genotype to be associated with a faster cognitive decline [66] and clinical progression of AD [30], whereas other studies doubt accelerated deterioration in APOE ε4 carriers [29, 30, 67–70]. APOE ε4 homozygosity was even suggested to slow down disease progession, since biological processes involved in AD onset and disease progression are of a different nature [29].
In our sample, having at least one ε4 allele increases time of conversion to AD by about 3 to 4 months as compared to homozygous ε3/ε3 carriers. Heterozygous ε2 carriers showed slowest conversion times. Since our sample did not include subjects with the allelic combination of ε2/ε2, we can not make any statements about average conversion times of the assumed protective allelic combination. Although ε4 carriers did not show the shortest conversion time in our study, they show the fastest brain aging, fastest cognitive decline, and highest mortality rate once converted to AD. In line with that, some studies suggest survival rates in AD patients mainly depending on their age at disease onset rather than on their APOE genotype [21, 67].
For APOE ε4 carriers as well as for non-carriers, prediction of conversion from MCI to AD was most accurate when based on baseline BrainAGE scores as compared to chronological age and cognitive test scores, even after inclusion of the APOE ε4 carrier-status, although prediction accuracy did not significantly improve in the BrainAGE prediction model. Nevertheless, prediction accuracy based on BrainAGE scores was higher for APOE ε4 carriers than for non-carriers, being in line with another study relating disease progression to decreases in hippocampal volume [71]. These results strongly suggest the usage of the BrainAGE method for screening MCI patients, aiming to find those, who are in a special high risk of conversion to AD in opposition to patients, who remain at a stable cognitive level. Identifying the quickly progressing subjects as early and secure as possible could help to prepare them best for their probable illness progression and supply them early with potential disease programs, cognitive training and medical treatments [72, 73]. Although a genetical test for APOE cannot replace any part of the clinical diagnostic procedure [35, 74, 75], including the patient’s APOE carrier status improves prediction accuracy of diagnostic tests such as memory tests or MR imaging in individuums who meet clinical MCI or AD criteria [8, 24, 76–78].
Limitations
The present study focused on the influence of APOE status on individual brain aging trajectories in healthy subjects as well as MCI and AD patients. Therefore we divided our cohort in different APOE carrier types, based on the three allele haplotypes of the Apolipoprotein E gene, composed of ε2, ε3 and ε4. The distribution was 4% for ε2, 62% for ε3 and 34% for ε4. In caucasians, frequencies of the 3 allelic types were previously estimated 11% for ε2, 72% for ε3 and 17% for ε4 [79], respectively 8% for ε2, 77% for ε3 and 15% for ε4 [80, 81]. The underrepresentation of ε2 and ε3 and the overrepresentation of ε4 in our sample could be due to a sort of preselection in the ADNI database. Homozygous APOE ε4/ε4 carriers form about 1% to 2% of the general population [25, 34], whereas our sample included 14%. Besides, we found a relative overrepresentation of ε4 within the group of AD patients in our sample (ε4/ε4 in ADs: 21%, compared to 4% in NOs), whereas the frequency of ε2 was lower (ε2/ε3 in ADs: 3%, compared to 15% in NOs), which was also reported in several other studies [20, 21, 35]. The ADNI cohort, which was used for this study, may differ from the general population, since it only includes individuals from memory clinics, patient registries, and people recruited in public media campaigns or other forms of public advertisements. There could also exist differences in the population of North American as compared to Central Europe. This might also explain the differences in the frequencies of APOE isoforms within our sample from the estimations for the general Caucasian population.
The clinical follow-up for our ADNI cohort was done in average 1.7 years in ADs, 2.9 years in MCIs and 3.2 years in NOs. We cannot make any statement if some sMCI patients would have converted later on. Besides, group sizes of some allelic subgroups were limited to a very small number (e.g., 14 carriers and 22 non-carriers in sMCIs) due to low prevalence of some APOE isoforms or limiting selection criterions for our study. Additionally, misdiagnoses of prodromal states of AD as well as AD itself may have occured due to the possibility of mixed dementia forms or overlaying physical illness [82]. Besides, cognitive decline is a continuous process; therefore it is not always easy to securely classify the disease stage [7]. It would be of interest to repeat the study in some years in order to include longer follow-up periods and more secure diagnoses.
Furthermore, it would be interesting to examine age-specific effects of the APOE genotype on BrainAGE and cognitive scores, respective disease burden, as well as to verify, whether the APOE ε4 genotype is associated with an earlier age of onset, or the risk of coming down with AD, or time to conversion [26, 33, 83]. Examining age specific effects also has the potential to test, whether the correlation of APOE ε4 with the AD risk and gross brain morphology diminishes in very old age [2, 21, 84]. Aside from APOE, there are also other genetical risk factors, which probably influence MCI and AD pathogenesis. However, the inheritence of AD predisposition is very complex, with gene polymorphism and mutations interacting with each other as well as with non-genetic factors. So far, only four genes could be identifyied to influence the AD pathogenesis [7, 23, 27].
In addition, when predicting individual progression from MCI to AD, it’s impossible to take into account all possible risk factors and influencing variables like comorbidities or cognitive reserve [85]. The variability of disease progression is als reflected in the strong variations of slopes for the longitudinal BrainAGE changes. Generaly, abnormal brain atrophy were also found in asymptomatic subjects, whose sufficient cognitive reserve or well adapted coping methods prolongated appearance of dementia [42, 57, 85–87]. This, in turn, provokes a strong divergency between anatomical and clinical findings. But since we’ve examined flexible biological systems, we will only be able to provide estimations, but not certainty in AD diagnosis and prediction.
Conclusions
In summary, the present study showed the potential of the BrainAGE method to provide more accurate results in prediciting conversion from MCI to AD than the already well-established cognitive tests, like MMSE, CDR-SB and ADAS. The knowledge of patients’ APOE genotype additionally tended to even improve prediction performance. Compared to a wide range of existing classification approaches that require disease-specific data for training, the BrainAGE framework uses an independent database of healthy, non-demented subjects to model the normal brain-aging pattern and consequently recognizing subtle deviations from age-related brain atrophy in new test samples. As the BrainAGE approach utilizes only a single T1-weighted image per subject and already has proven to work fast and fully automated with multi-centre data, it can be easily implemented in clinical routine to encourage the identification of subtly abnormal atrophy patterns as well as for monitoring treatment options.
Acknowledgments
We would like to thank Robert Dahnke for his ongoing support with our work. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of Southern California.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found in the Acknowledgments and at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgment_List.pdf
Data Availability
Data for the training sample were taken from the IXI cohort (http://www.brain-development.org; downloaded in September 2011) and OASIS (http://www.oasis-brains.org; downloaded in June 2009). Information on these data can be found the “Estimation of BrainAGE scores” section of the paper.
Funding Statement
This work was supported by the European Community [FB7 HEALTH Project 279281 (Brain Age) to K.F.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC. Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. NeuroImage. 2009;48(1):138–49. Epub 2009/06/02. 10.1016/j.neuroimage.2009.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigler ED, Lowry CM, Anderson CV, Johnson SC, Terry J, Steed M. Dementia, quantitative neuroimaging, and apolipoprotein E genotype. AJNR American journal of neuroradiology. 2000;21(10):1857–68. Epub 2000/12/08. . [PMC free article] [PubMed] [Google Scholar]
- 3.Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ Jr., et al. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol. 2010;31(2):347–54. Epub 2010/01/16. ajnr.A1809 [pii] 10.3174/ajnr.A1809 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiology of aging. 2011;32(12):2322 e19-27. Epub 2010/07/03. 10.1016/j.neurobiolaging.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jack CR Jr., Wiste HJ, Vemuri P, Weigand SD, Senjem ML, Zeng G, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer's disease. Brain: a journal of neurology. 2010;133(11):3336–48. Epub 2010/10/12. 10.1093/brain/awq277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kester MI, Scheltens P. Dementia: the bare essentials. Pract Neurol. 2009;9(4):241–51. 10.1136/jnnp.2009.182477 . [DOI] [PubMed] [Google Scholar]
- 7.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–9. 10.1016/j.jalz.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen RC. Mild cognitive impairment as a diagnostic entity. Journal of internal medicine. 2004;256(3):183–94. Epub 2004/08/25. 10.1111/j.1365-2796.2004.01388.x . [DOI] [PubMed] [Google Scholar]
- 9.Davatzikos C, Xu F, An Y, Fan Y, Resnick SM. Longitudinal progression of Alzheimer's-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain. 2009;132(Pt 8):2026–35. Epub 2009/05/07. awp091 [pii] 10.1093/brain/awp091 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack CR Jr., Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62(4):591–600. Epub 2004/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spulber G, Niskanen E, MacDonald S, Smilovici O, Chen K, Reiman EM, et al. Whole brain atrophy rate predicts progression from MCI to Alzheimer's disease. Neurobiology of aging. 2010;31(9):1601–5. Epub 2008/10/03. 10.1016/j.neurobiolaging.2008.08.018 . [DOI] [PubMed] [Google Scholar]
- 12.Karas GB, Scheltens P, Rombouts SA, Visser PJ, van Schijndel RA, Fox NC, et al. Global and local gray matter loss in mild cognitive impairment and Alzheimer's disease. NeuroImage. 2004;23(2):708–16. Epub 2004/10/19. 10.1016/j.neuroimage.2004.07.006 . [DOI] [PubMed] [Google Scholar]
- 13.Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology. 2005;64(6):1032–9. Epub 2005/03/23. 10.1212/01.WNL.0000154530.72969.11 . [DOI] [PubMed] [Google Scholar]
- 14.Anderson VM, Schott JM, Bartlett JW, Leung KK, Miller DH, Fox NC. Gray matter atrophy rate as a marker of disease progression in AD. Neurobiology of aging. 2012;33(7):1194–202. Epub 2010/12/18. 10.1016/j.neurobiolaging.2010.11.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke K, Ziegler G, Klöppel S, Gaser C, the Alzheimer's Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. NeuroImage. 2010;50(3):883–92. Epub 2010/01/15. S1053-8119(10)00010-8 [pii] 10.1016/j.neuroimage.2010.01.005 . [DOI] [PubMed] [Google Scholar]
- 16.Gaser C, Franke K, Kloppel S, Koutsouleris N, Sauer H. in Mild Cognitive Impaired Patients: Predicting the Conversion to Alzheimer's Disease. PloS one. 2013;8(6):e67346 Epub 2013/07/05. 10.1371/journal.pone.0067346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke K, Gaser C. Longitudinal Changes in Individual BrainAGE in Healthy Aging, Mild Cognitive Impairment, and Alzheimer's Disease. GeroPsych. 2012;25(4):235–45. 10.1024/1662-9647/a000074 [DOI] [Google Scholar]
- 18.Driscoll I, Davatzikos C, An Y, Wu X, Shen D, Kraut M, et al. Longitudinal pattern of regional brain volume change differentiates normal aging from MCI. Neurology. 2009;72(22):1906–13. Epub 2009/06/03. 72/22/1906 [pii] 10.1212/WNL.0b013e3181a82634 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sluimer JD, van der Flier WM, Karas GB, van Schijndel R, Barnes J, Boyes RG, et al. Accelerating regional atrophy rates in the progression from normal aging to Alzheimer's disease. European radiology. 2009;19(12):2826–33. Epub 2009/07/21. 10.1007/s00330-009-1512-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(5):1977–81. Epub 1993/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr., et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nature genetics. 1994;7(2):180–4. Epub 1994/06/01. 10.1038/ng0694-180 . [DOI] [PubMed] [Google Scholar]
- 22.Nalbantoglu J, Gilfix BM, Bertrand P, Robitaille Y, Gauthier S, Rosenblatt DS, et al. Predictive value of apolipoprotein E genotyping in Alzheimer's disease: results of an autopsy series and an analysis of several combined studies. Annals of neurology. 1994;36(6):889–95. Epub 1994/12/01. 10.1002/ana.410360614 . [DOI] [PubMed] [Google Scholar]
- 23.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68(2):270–81. Epub 2010/10/20. 10.1016/j.neuron.2010.10.013 . [DOI] [PubMed] [Google Scholar]
- 24.Bertram L, Tanzi RE. The current status of Alzheimer's disease genetics: what do we tell the patients? Pharmacological research: the official journal of the Italian Pharmacological Society. 2004;50(4):385–96. Epub 2004/08/12. 10.1016/j.phrs.2003.11.018 . [DOI] [PubMed] [Google Scholar]
- 25.Saunders AM. Apolipoprotein E and Alzheimer disease: an update on genetic and functional analyses. Journal of neuropathology and experimental neurology. 2000;59(9):751–8. Epub 2000/09/27. . [DOI] [PubMed] [Google Scholar]
- 26.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–3. Epub 1993/08/13. . [DOI] [PubMed] [Google Scholar]
- 27.Brouwers N, Sleegers K, Van Broeckhoven C. Molecular genetics of Alzheimer's disease: an update. Annals of medicine. 2008;40(8):562–83. Epub 2008/07/09. 10.1080/07853890802186905 . [DOI] [PubMed] [Google Scholar]
- 28.Deuschl G, Maier W. S3- Leitlinie Demenzen.: Deutsche Gesellschaft f√°r Psychiatrie, Psychotherapie und Nervenheilkunde (DGPPN), Deutsche Gesellschaft f√°r Neurologie (DGN); 2009.
- 29.Hoyt BD, Massman PJ, Schatschneider C, Cooke N, Doody RS. Individual growth curve analysis of APOE epsilon 4-associated cognitive decline in Alzheimer disease. Archives of neurology. 2005;62(3):454–9. Epub 2005/03/16. 10.1001/archneur.62.3.454 . [DOI] [PubMed] [Google Scholar]
- 30.Martins CA, Oulhaj A, de Jager CA, Williams JH. APOE alleles predict the rate of cognitive decline in Alzheimer disease: a nonlinear model. Neurology. 2005;65(12):1888–93. Epub 2005/12/29. 10.1212/01.wnl.0000188871.74093.12 . [DOI] [PubMed] [Google Scholar]
- 31.Cosentino S, Scarmeas N, Helzner E, Glymour MM, Brandt J, Albert M, et al. APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology. 2008;70(19 Pt 2):1842–9. Epub 2008/04/11. 01.wnl.0000304038.37421.cc [pii] 10.1212/01.wnl.0000304038.37421.cc . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jack CR Jr., Petersen RC, Xu YC, O'Brien PC, Waring SC, Tangalos EG, et al. Hippocampal atrophy and apolipoprotein E genotype are independently associated with Alzheimer's disease. Annals of neurology. 1998;43(3):303–10. Epub 1998/03/20. 10.1002/ana.410430307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liddell MB, Lovestone S, Owen MJ. Genetic risk of Alzheimer's disease: advising relatives. The British journal of psychiatry: the journal of mental science. 2001;178(1):7–11. Epub 2001/01/03. . [DOI] [PubMed] [Google Scholar]
- 34.Roses AD, Strittmatter WJ, Pericak-Vance MA, Corder EH, Saunders AM, Schmechel DE. Clinical application of apolipoprotein E genotyping to Alzheimer's disease. Lancet. 1994;343(8912):1564–5. Epub 1994/06/18. . [DOI] [PubMed] [Google Scholar]
- 35.Kurz A, Altland K, Lautenschlager N, Zimmer R, Busch R, Gerundt I, et al. Apolipoprotein E type 4 allele and Alzheimer's disease: effect on age at onset and relative risk in different age groups. Journal of neurology. 1996;243(6):452–6. Epub 1996/06/01. . [DOI] [PubMed] [Google Scholar]
- 36.Honea RA, Vidoni E, Harsha A, Burns JM. Impact of APOE on the healthy aging brain: a voxel-based MRI and DTI study. Journal of Alzheimer's disease: JAD. 2009;18(3):553–64. Epub 2009/07/09. 10.3233/JAD-2009-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hua X, Lee S, Yanovsky I, Leow AD, Chou YY, Ho AJ, et al. Optimizing power to track brain degeneration in Alzheimer's disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. NeuroImage. 2009;48(4):668–81. Epub 2009/07/21. 10.1016/j.neuroimage.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehtovirta M, Soininen H, Laakso MP, Partanen K, Helisalmi S, Mannermaa A, et al. SPECT and MRI analysis in Alzheimer's disease: relation to apolipoprotein E epsilon 4 allele. Journal of neurology, neurosurgery, and psychiatry. 1996;60(6):644–9. Epub 1996/06/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, et al. Accelerated hippocampal atrophy in Alzheimer's disease with apolipoprotein E epsilon4 allele. Annals of neurology. 2002;51(2):209–14. Epub 2002/02/09. . [DOI] [PubMed] [Google Scholar]
- 40.Manning EN, Barnes J, Cash DM, Bartlett JW, Leung KK, Ourselin S, et al. APOE epsilon4 is associated with disproportionate progressive hippocampal atrophy in AD. PloS one. 2014;9(5):e97608 Epub 2014/06/01. 10.1371/journal.pone.0097608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basso M, Gelernter J, Yang J, MacAvoy MG, Varma P, Bronen RA, et al. Apolipoprotein E epsilon4 is associated with atrophy of the amygdala in Alzheimer's disease. Neurobiology of aging. 2006;27(10):1416–24. Epub 2005/09/27. 10.1016/j.neurobiolaging.2005.08.002 . [DOI] [PubMed] [Google Scholar]
- 42.Querbes O, Aubry F, Pariente J, Lotterie JA, Demonet JF, Duret V, et al. Early diagnosis of Alzheimer's disease using cortical thickness: impact of cognitive reserve. Brain. 2009;132(Pt 8):2036–47. Epub 2009/05/15. awp105 [pii] 10.1093/brain/awp105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olichney JM, Hansen LA, Galasko D, Saitoh T, Hofstetter CR, Katzman R, et al. The apolipoprotein E epsilon 4 allele is associated with increased neuritic plaques and cerebral amyloid angiopathy in Alzheimer's disease and Lewy body variant. Neurology. 1996;47(1):190–6. Epub 1996/07/01. . [DOI] [PubMed] [Google Scholar]
- 44.Mortimer JA, Snowdon DA, Markesbery WR. The effect of APOE-epsilon4 on dementia is mediated by Alzheimer neuropathology. Alzheimer disease and associated disorders. 2009;23(2):152–7. Epub 2009/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berlau DJ, Corrada MM, Head E, Kawas CH. APOE epsilon2 is associated with intact cognition but increased Alzheimer pathology in the oldest old. Neurology. 2009;72(9):829–34. Epub 2009/03/04. 10.1212/01.wnl.0000343853.00346.a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genin E, Hannequin D, Wallon D, Sleegers K, Hiltunen M, Combarros O, et al. APOE and Alzheimer disease: a major gene with semi-dominant inheritance. Molecular psychiatry. 2011;16(9):903–7. Epub 2011/05/11. 10.1038/mp.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohs RC, Cohen L. Alzheimer's Disease Assessment Scale (ADAS). Psychopharmacology bulletin. 1988;24(4):627–8. Epub 1988/01/01. . [PubMed] [Google Scholar]
- 48.Morris JC. The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology. 1993;43(11):2412–4. Epub 1993/11/01. . [DOI] [PubMed] [Google Scholar]
- 49.Cockrell JR, Folstein MF. Mini-Mental State Examination (MMSE). Psychopharmacology bulletin. 1988;24(4):689–92. Epub 1988/01/01. . [PubMed] [Google Scholar]
- 50.Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26(3):839–51. Epub 2005/06/16. S1053-8119(05)00110-2 [pii] 10.1016/j.neuroimage.2005.02.018 . [DOI] [PubMed] [Google Scholar]
- 51.Tohka J, Zijdenbos A, Evans A. Fast and robust parameter estimation for statistical partial volume models in brain MRI. NeuroImage. 2004;23(1):84–97. Epub 2004/08/25. 10.1016/j.neuroimage.2004.05.007 S1053811904002745 [pii]. . [DOI] [PubMed] [Google Scholar]
- 52.Rajapakse JC, Giedd JN, Rapoport JL. Statistical approach to segmentation of single-channel cerebral MR images. IEEE Transactions on Medical Imaging. 1997;16(2):176–86. Epub 1997/04/01. 10.1109/42.563663 . [DOI] [PubMed] [Google Scholar]
- 53.Cuadra MB, Cammoun L, Butz T, Cuisenaire O, Thiran JP. Comparison and validation of tissue modelization and statistical classification methods in T1-weighted MR brain images. IEEE Transactions on Medical Imaging. 2005;24(12):1548–65. Epub 2005/12/15. 10.1109/TMI.2005.857652 . [DOI] [PubMed] [Google Scholar]
- 54.Tipping ME. The Relevance Vector Machine In: Solla SA, Leen TK, Müller K-R, editors. Advances in Neural Information Processing Systems 12 Cambridge, MA: MIT Press; 2000. p. 652–8. [Google Scholar]
- 55.Franke K, Gaser C, for the Alzheimer's Disease Neuroimaging Initiative. Longitudinal changes in individual BrainAGE in healthy aging, mild cognitive impairment, and Alzheimer’s disease. GeroPsych: The Journal of Gerontopsychology and Geriatric Psychiatry. 2012;25(4):235–45. [Google Scholar]
- 56.Schölkopf B, Smola A. Learning with Kernels: Support vector machines, regularization, optimization, and beyond Cambridge: MA: MIT; 2002. [Google Scholar]
- 57.Aguilar C, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, et al. Application of a MRI based index to longitudinal atrophy change in Alzheimer disease, mild cognitive impairment and healthy older individuals in the AddNeuroMed cohort. Frontiers in aging neuroscience. 2014;6:145 Epub 2014/07/30. 10.3389/fnagi.2014.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson VM, Schott JM, Bartlett JW, Leung KK, Miller DH, Fox NC. Gray matter atrophy rate as a marker of disease progression in AD. Neurobiology of Aging. 2012;33(7):1194–202. Epub 2010/12/18. S0197-4580(10)00475-6 [pii] 10.1016/j.neurobiolaging.2010.11.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jack CR Jr., Weigand SD, Shiung MM, Przybelski SA, O'Brien PC, Gunter JL, et al. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008;70(19):1740–52. Epub 2007/11/23. 01.wnl.0000281688.77598.35 [pii] 10.1212/01.wnl.0000281688.77598.35 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desikan RS, Fischl B, Cabral HJ, Kemper TL, Guttmann CR, Blacker D, et al. MRI measures of temporoparietal regions show differential rates of atrophy during prodromal AD. Neurology. 2008;71(11):819–25. Epub 2008/08/02. 01.wnl.0000320055.57329.34 [pii] 10.1212/01.wnl.0000320055.57329.34 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sluimer JD, van der Flier WM, Karas GB, van Schijndel R, Barnes J, Boyes RG, et al. Accelerating regional atrophy rates in the progression from normal aging to Alzheimer's disease. European Radiology. 2009;19(12):2826–33. Epub 2009/07/21. 10.1007/s00330-009-1512-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dukart J, Schroeter ML, Mueller K. Age correction in dementia—matching to a healthy brain. PLoS One. 2011;6(7):e22193 Epub 2011/08/11. 10.1371/journal.pone.0022193 PONE-D-11-03484 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones DT, Machulda MM, Vemuri P, McDade EM, Zeng G, Senjem ML, et al. Age-related changes in the default mode network are more advanced in Alzheimer disease. Neurology. 2011;77(16):1524–31. Epub 2011/10/07. WNL.0b013e318233b33d [pii] 10.1212/WNL.0b013e318233b33d . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu L, Boyle P, Schneider JA, Segawa E, Wilson RS, Leurgans S, et al. APOE epsilon4, Alzheimer's disease pathology, cerebrovascular disease, and cognitive change over the years prior to death. Psychol Aging. 2013;28(4):1015–23. 10.1037/a0031642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson RS, Leurgans SE, Boyle PA, Bennett DA. Cognitive decline in prodromal Alzheimer disease and mild cognitive impairment. Archives of Neurology. 2011;68(3):351–6. Epub 2011/03/16. 68/3/351 [pii] 10.1001/archneurol.2011.31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirono N, Hashimoto M, Yasuda M, Kazui H, Mori E. Accelerated memory decline in Alzheimer's disease with apolipoprotein epsilon4 allele. The Journal of neuropsychiatry and clinical neurosciences. 2003;15(3):354–8. Epub 2003/08/21. . [DOI] [PubMed] [Google Scholar]
- 67.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC Jr., Rimmler JB, et al. Apolipoprotein E, survival in Alzheimer's disease patients, and the competing risks of death and Alzheimer's disease. Neurology. 1995;45(7):1323–8. Epub 1995/07/01. . [DOI] [PubMed] [Google Scholar]
- 68.Growdon JH, Locascio JJ, Corkin S, Gomez-Isla T, Hyman BT. Apolipoprotein E genotype does not influence rates of cognitive decline in Alzheimer's disease. Neurology. 1996;47(2):444–8. Epub 1996/08/01. . [DOI] [PubMed] [Google Scholar]
- 69.Holmes C, Levy R, McLoughlin DM, Powell JF, Lovestone S. Apolipoprotein E: non-cognitive symptoms and cognitive decline in late onset Alzheimer's disease. Journal of neurology, neurosurgery, and psychiatry. 1996;61(6):580–3. Epub 1996/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stern Y, Brandt J, Albert M, Jacobs DM, Liu X, Bell K, et al. The absence of an apolipoprotein epsilon4 allele is associated with a more aggressive form of Alzheimer's disease. Annals of neurology. 1997;41(5):615–20. Epub 1997/05/01. 10.1002/ana.410410510 . [DOI] [PubMed] [Google Scholar]
- 71.Apostolova LG, Hwang KS, Kohannim O, Avila D, Elashoff D, Jack CR Jr., et al. ApoE4 effects on automated diagnostic classifiers for mild cognitive impairment and Alzheimer's disease. NeuroImage Clinical. 2014;4:461–72. Epub 2014/03/19. 10.1016/j.nicl.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franko E, Joly O. Evaluating Alzheimer's disease progression using rate of regional hippocampal atrophy. PloS one. 2013;8(8):e71354 Epub 2013/08/21. 10.1371/journal.pone.0071354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's & dementia: the journal of the Alzheimer's Association. 2011;7(3):280–92. Epub 2011/04/26. 10.1016/j.jalz.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alzheimer'sAssociationWorkingGroup. Apolipoprotein E genotyping in Alzheimer's disease. National Institute on Aging/Alzheimer's Association Working Group. Lancet. 1996;347(9008):1091–5. Epub 1996/04/20. . [PubMed] [Google Scholar]
- 75.Tsuang D, Larson EB, Bowen J, McCormick W, Teri L, Nochlin D, et al. The utility of apolipoprotein E genotyping in the diagnosis of Alzheimer disease in a community-based case series. Archives of neurology. 1999;56(12):1489–95. Epub 1999/12/11. . [DOI] [PubMed] [Google Scholar]
- 76.Mayeux R, Saunders AM, Shea S, Mirra S, Evans D, Roses AD, et al. Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer's disease. Alzheimer's Disease Centers Consortium on Apolipoprotein E and Alzheimer's Disease. The New England journal of medicine. 1998;338(8):506–11. Epub 1998/02/19. 10.1056/NEJM199802193380804 . [DOI] [PubMed] [Google Scholar]
- 77.Boissonneault GA. MCI and dementia: diagnosis and treatment. JAAPA: official journal of the American Academy of Physician Assistants. 2010;23(1):18, 21–2. Epub 2010/02/09. . [DOI] [PubMed] [Google Scholar]
- 78.Susanto TA, Pua EP, Zhou J. Cognition, brain atrophy, and cerebrospinal fluid biomarkers changes from preclinical to dementia stage of Alzheimer's disease and the influence of apolipoprotein e. Journal of Alzheimer's disease: JAD. 2015;45(1):253–68. Epub 2014/12/20. 10.3233/JAD-142451 . [DOI] [PubMed] [Google Scholar]
- 79.Zannis VI, Just PW, Breslow JL. Human apolipoprotein E isoprotein subclasses are genetically determined. American journal of human genetics. 1981;33(1):11–24. Epub 1981/01/01. [PMC free article] [PubMed] [Google Scholar]
- 80.Thakkinstian A, Bowe S, McEvoy M, Smith W, Attia J. Association between apolipoprotein E polymorphisms and age-related macular degeneration: A HuGE review and meta-analysis. American journal of epidemiology. 2006;164(9):813–22. Epub 2006/08/19. 10.1093/aje/kwj279 . [DOI] [PubMed] [Google Scholar]
- 81.Utermann G, Langenbeck U, Beisiegel U, Weber W. Genetics of the apolipoprotein E system in man. American journal of human genetics. 1980;32(3):339–47. Epub 1980/05/01. [PMC free article] [PubMed] [Google Scholar]
- 82.Jicha GA, Parisi JE, Dickson DW, Johnson K, Cha R, Ivnik RJ, et al. Neuropathologic outcome of mild cognitive impairment following progression to clinical dementia. Archives of neurology. 2006;63(5):674–81. Epub 2006/05/10. 10.1001/archneur.63.5.674 . [DOI] [PubMed] [Google Scholar]
- 83.Meyer MR, Tschanz JT, Norton MC, Welsh-Bohmer KA, Steffens DC, Wyse BW, et al. APOE genotype predicts when—not whether—one is predisposed to develop Alzheimer disease. Nature genetics. 1998;19(4):321–2. Epub 1998/08/11. 10.1038/1206 . [DOI] [PubMed] [Google Scholar]
- 84.Sobel E, Louhija J, Sulkava R, Davanipour Z, Kontula K, Miettinen H, et al. Lack of association of apolipoprotein E allele epsilon 4 with late-onset Alzheimer's disease among Finnish centenarians. Neurology. 1995;45(5):903–7. Epub 1995/05/01. . [DOI] [PubMed] [Google Scholar]
- 85.Stern Y. What is cognitive reserve? Theory and research application of the reserve concept. Journal of the International Neuropsychological Society: JINS. 2002;8(3):448–60. Epub 2002/04/10. . [PubMed] [Google Scholar]
- 86.Stern Y, Zarahn E, Hilton HJ, Flynn J, DeLaPaz R, Rakitin B. Exploring the neural basis of cognitive reserve. Journal of clinical and experimental neuropsychology. 2003;25(5):691–701. Epub 2003/06/20. 10.1076/jcen.25.5.691.14573 . [DOI] [PubMed] [Google Scholar]
- 87.Winblad B, Palmer K, Kivipelto M, Jelic V, Fratiglioni L, Wahlund LO, et al. Mild cognitive impairment—beyond controversies, towards a consensus: report of the International Working Group on Mild Cognitive Impairment. Journal of internal medicine. 2004;256(3):240–6. Epub 2004/08/25. 10.1111/j.1365-2796.2004.01380.x . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data for the training sample were taken from the IXI cohort (http://www.brain-development.org; downloaded in September 2011) and OASIS (http://www.oasis-brains.org; downloaded in June 2009). Information on these data can be found the “Estimation of BrainAGE scores” section of the paper.