Abstract
Specification of the germ cell lineage is required for sexual reproduction in all animals. However, the timing and mechanisms of germ cell specification is remarkably diverse in animal development. Echinoderms, such as sea urchins and sea stars, are excellent model systems to study the molecular and cellular mechanisms that contribute to germ cell specification. In several echinoderm embryos tested, the germ cell factor Vasa accumulates broadly during early development and is restricted after gastrulation to cells that contribute to the germ cell lineage. In the sea urchin, however, the germ cell factor Vasa is restricted to a specific lineage by the 32-cell stage. We therefore hypothesized that the germ cell specification program in the sea urchin/Euechinoid lineage has evolved to an earlier developmental time point. To test this hypothesis we determined the expression pattern of a second germ cell factor, Nanos, in four out of five extant echinoderm clades. Here we find that Nanos mRNA does not accumulate until the blastula stage or later during the development of all other echinoderm embryos except those that belong to the Echinoid lineage. Instead, Nanos is expressed in a restricted domain at the 32–128 cell stage in Echinoid embryos. Our results support the model that the germ cell specification program underwent a heterochronic shift in the Echinoid lineage. A comparison of Echinoid and non-Echinoid germ cell specification mechanisms will contribute to our understanding of how these mechanisms have changed during animal evolution.
Introduction
Eggs and sperm are essential for the reproduction of most animals. Thus, the germ cell lineage, any cell that retains the potential to give rise to an egg or sperm, is essential for animal development. Germ cell specification is the process when the germ cell lineage set aside from the rest of the somatic cells. Even though the germ cell lineage is a conserved requirement for sexual reproduction, there is not one common germ cell specification mechanism. Instead, studies of animal development reveal multiple mechanisms for germ cell specification that can belong to two major groups: inherited and inductive (also referred to as preformation and epigenesis) (Extavour and Akam, 2003).
An inherited mechanism of germ cell specification occurs relatively early in development. A defining characteristic of this mechanism is the early localization of maternally supplied germ cell determinant molecules (RNAs and proteins) in a restricted domain of the egg or early embryo. As a consequence of cellular division, whichever cells inherit these germ cell determinant molecules are instructed to take on a germ cell lineage fate. The other cells that did not receive these germ cell determinant molecules instead will become somatic lineages. Early development in animals such as fruit flies, nematode worms, frogs, and zebrafish is characteristic of the inherited mechanism of germ cell specification (Illmensee and Mahowald, 1974; Kawasaki et al., 1998; Kuznicki et al., 2000; Mello et al., 1992; Smith, 1966; Yoon et al., 1997). However, many other animals use a different cellular mechanism for germ cell specification.
An inductive mechanism of germ cell specification occurs relatively later in development. A defining characteristic of this mechanism is that maternally supplied germ cell determinant molecules (RNA’s and proteins) either do not accumulate during early development or only accumulate in large embryonic domains in early development. As a consequence, embryonic transcription, cell signaling, and cell interactions instruct which cells will become the germ cell lineage and which cells will become the somatic cell lineage. Early development in animals such as crickets, salamanders, and mice is more characteristic of the inductive mechanism of germ cell specification (Chatfield et al., 2014; Ewen-Campen et al., 2013; Tam and Zhou, 1996).
Despite the similarities that appear in germ cell specification mechanisms that allow categorization into two general mechanisms, it is clear that one if not both of these mechanisms have independently evolved many times throughout animal phylogeny. For example, within Chordates both mammals and salamanders use an inductive mechanism of germ cell specification whereas zebrafish and frogs use the inherited mechanism of germ cell specification (Supplement Figure 1). Similarly, within the protostomes both worms and fruit flies use the inherited mechanism of germ cell specification whereas crickets use the inductive mechanism of germ cell specification (Supplement Figure 1). Recent studies have aimed to understand how these different germ cell specification mechanisms have evolved so frequently in closely related organisms (Evans et al., 2014). However, in order to address this evolutionary question thoroughly, it is important to understand how germ cells are specified in diverse animal lineages.
Echinoderms are an informative group of animals to study the evolution of germ cell specification mechanisms for three reasons. First, echinoderm embryos are convenient models to study the evolution of early developmental processes. Embryos from each extant echinoderm clade are accessible and amenable for experimentation in early development. For example, they are large, optically clear, develop externally, and are mass-produced (on the order of hundreds of thousands). In addition, a variety of molecular tools are available for gene perturbation in the majority of echinoderm embryos. Second, previous studies suggest that different species within the echinoderm clade use different mechanisms for germ cell specification. Third, echinoderms occupy an evolutionarily important position within animal phylogeny because echinoderms and Chordates are sister groups that belong to the Deuterostome lineage (Supplemental Figure 1). Therefore, comparisons between Chordates and echinoderms are essential to understand ancestral Deuterostome mechanisms of germ cell specification. For these reasons the echinoderm clade is a great model to study the evolution of germ cell specification mechanisms.
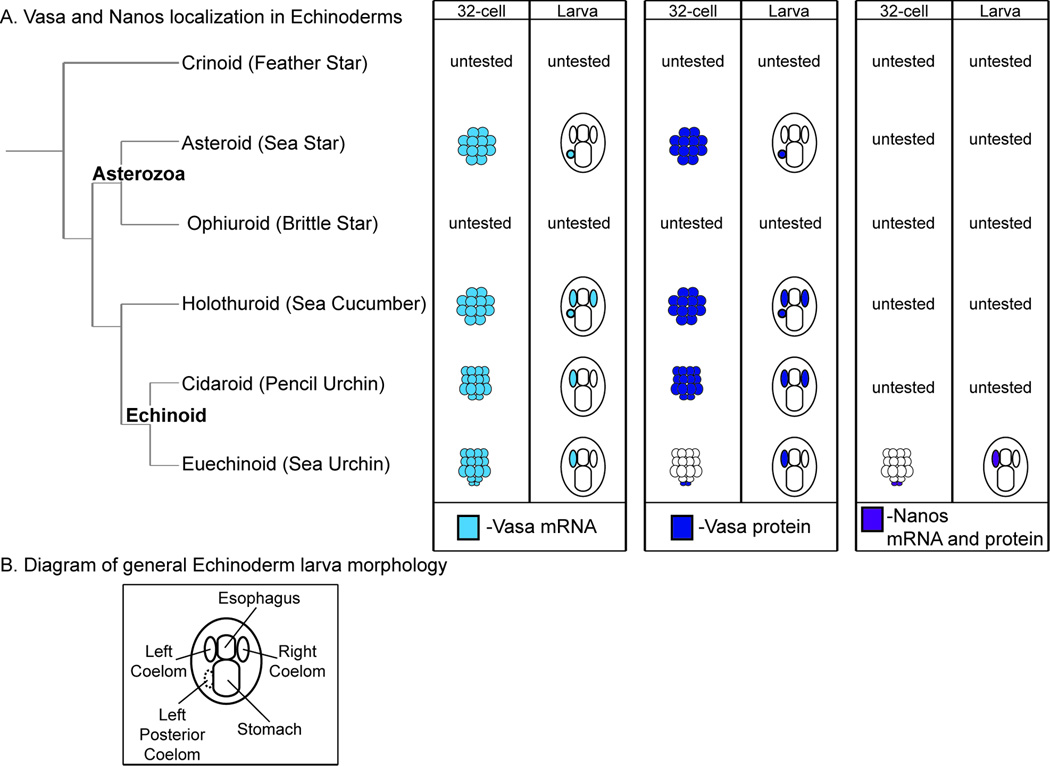
Previous studies of germ cell specification mechanisms in echinoderms have largely focused on the localization of the germ cell marker Vasa (Figure 1A) (Juliano and Wessel, 2009; Voronina et al., 2008; Yu et al., 2013). Vasa is an RNA helicase that is required to specify and/or maintain the germ cell lineage fate in many animals (Lasko, 2013). Vasa accumulates broadly during the early development of almost all echinoderm species tested (Figure 1A, light blue, dark blue, 32-cell stage) and is restricted to the coeloms by the larva stage (Figure 1A, light blue, dark blue, larva stage). These results have led previous authors to hypothesize that in most echinoderm species germ cell specification occurs later in development by inductive mechanisms (Wessel et al., 2014). By contrast, Vasa protein is restricted to a specific lineage early during sea urchin development, by the 32-cell stage. In fact, this specific lineage is an evolutionary derived characteristic that only occurs in the Euechinoid group of echinoderms. These data led to the hypothesis that during the evolution of the Euechinoid lineage there has been a shift for germ cell specification to occur earlier in development by inherited mechanisms (Figure 1A, dark blue, 32-cell stage, sea urchin) (Wessel et al., 2014). It is important to note that Vasa has other functions during the early development of some animals that are independent of germ cell specification (Schwager et al., 2015; Yajima and Wessel, 2011a, b, 2015). Therefore, in order to rigorously test these hypotheses, it is necessary to determine where other germ cell markers accumulate during the development of diverse echinoderm species.
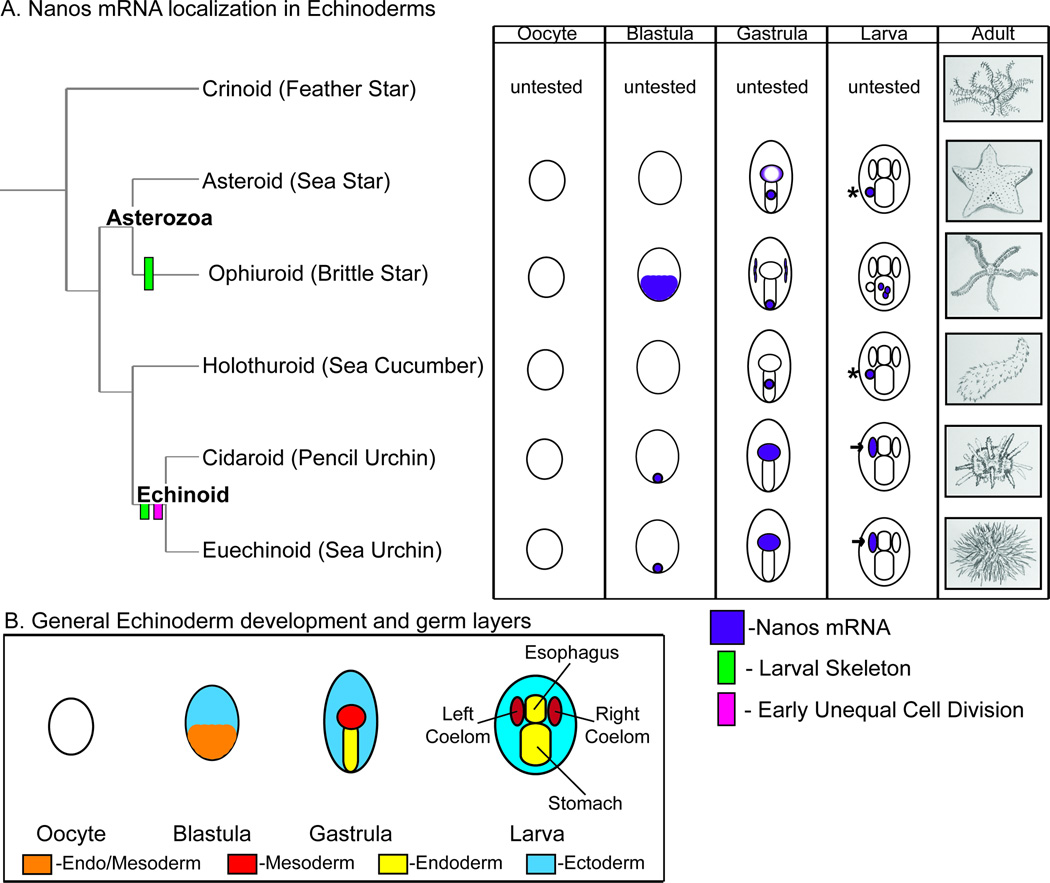
Figure 1. Vasa and Nanos localization in Echinoderms.
A. A summary of known Vasa and Nanos mRNA and protein localization in Echinoderm embryos before this study. Light blue= Vasa mRNA, Dark blue= Vasa protein, Purple= Nanos mRNA and protein. In Asteroids, Holothuroids and Cidaroids Vasa mRNA and protein localizes broadly at the 32-cell stage and both are restricted to the coeloms by the larva stage. In Euechinoids, Vasa mRNA localizes broadly at the 32-cell stage and accumulates in the left coelom at the larva stage. However, Vasa protein in Euechinoids is restricted to a subset of embryonic blastomeres at the 32-cell stage and accumulates in the left coelom at the larva stage. Nanos mRNA and protein expression has only been reported in Euechinoids and is restricted to a subset of embryonic blastomeres at the 32-cell stage. By the larva stage Nanos mRNA and protein accumulates in the left coelom. B. A diagram of general echinoderm larva morphology. The dotted left posterior coelom is not present in all Echinoderm clades. All larval views are dorsal images with the anterior side up as a standard for this paper.
Here, we test the localization of the germ cell marker Nanos during the embryonic development of several echinoderm species. Nanos is an RNA binding Zinc finger protein that is required for the specification and/or maintenance of the germ cell lineage in many animals (Ephrussi et al., 1991; Irish et al., 1989; Lai and King, 2013; Wharton and Struhl, 1989). In the sea urchin, Nanos is a more selective marker of the germ cell lineage as both the mRNA and protein is restricted to a small number of embryonic cells at the 32-cell stage (Figure 1A, purple, 32-cell stage, sea urchin) (Fujii et al., 2006; Juliano et al., 2006; Juliano et al., 2010). This is at a developmental time when Vasa mRNA accumulates in every cell of the sea urchin embryo (Figure 1A, light blue, 32-cell stage, sea urchin). Because Nanos is a more selective marker for the germ cell lineage, we use it here to further test how the germ cell lineage is specified in diverse echinoderm embryos.
Materials and Methods
Embryo Culture
Asteroid (sea star)
The species, Patiria miniata, were collected from the Southern coast of California, USA from either Pete Halmay (PeterHalmaygmail.com) or Josh Ross (infoscbiomarine.com) and cultured as described (Wessel et al., 2010). Briefly, oocytes and sperm were collected by dissection. Oocytes were isolated by passing diced ovary tissue through a cheesecloth and collecting the oocytes in a beaker. Oocytes were washed 2 times with filtered sea water and incubated 1 hour with 1-methyl adenine (3.0 µM, Acros Organics) to induce maturation. Eggs were fertilized with diluted sperm and excess sperm was washed out with filtered sea water. Embryos were cultured with continuous stirring at 16° C until the larval stage.
Ophiuroid (brittle star)
The species, Amphipholis kochii, were collected from Himi (Toyama Prefecture, Japan) and cultured as described (Koga et al., 2010). Briefly, animals were induced to spawn by a 1–2 hour cold shock at 4° C. Eggs were fertilized with diluted sperm and cultured in artificial sea water at 23° C.
Holothuroid (sea cucumber)
Adults of the species Apostichopus japonicus, were collected near Misaki Marine Biological Station, the University of Tokyo in Kanagawa Prefecture, Japan and cultured as described (Kikuchi et al., 2015). Briefly, spawning was induced by injecting animals with 10 µM cubifrin. Eggs were fertilized with sperm that had been obtained by mincing of the testis. Embryos and larvae were cultured at 20° C in filtered sea water containing 75 mg/L of penicillin and streptomycin.
Cidaroid (pencil urchin)
The species, Eucidaris tribuloides, were collected from Florida, USA by KP Aquatics LLC (www.sealifeinc.net). Animals were induced to spawn by injection with 0.5 M KCl. Eggs were fertilized with diluted sperm and excess sperm was washed off 1 time with filtered sea water. Cultures were kept in 10 cm Petri dishes at 15° C.
Euechinoid (sea urchin)
The species, Lytechinus variegatus, were collected from Florida, USA by Pelagic Corporation. Animals were induced to spawn by injection with 0.5 M KCl. Eggs were fertilized with diluted sperm and excess sperm was washed off 1 time with filtered sea water. Cultures were kept stirring at 25° C until the larval stage.
Fixing Embryos
Ophiuroid (brittle star)
Embryos were fixed overnight at 4° C in a solution containing 4% PFA, 0.1 M MOPS (pH 7.0), and 0.5 M NaCl. Embryos were first washed with 50% Ethanol in PBS, then washed in 70% Ethanol, and stored at −20° C in 70% Ethanol.
All other embryos were fixed essentially as described with a few exceptions (Arenas-Mena et al., 2000). Briefly, embryos were resuspended in MOPS buffered PFA (5% PFA, 162.5 mM NaCl, 32.5 mM MOPS pH 7.0, 32.5% filtered sea water) and incubated overnight at 4° C. Embryos were washed 5 times with MOPS Buffer (0.1 M MOPS pH 7.0, 0.5 M NaCl, 0.1% Tween 20) and stored at −20° C in 70% Ethanol.
Whole Mount RNA in situ Hybridization
General
RNA in situ analyses were done essentially as described (Arenas-Mena et al., 2000). Briefly, embryos were hybridized with 0.1–0.5 ng/ul of DIG labeled RNA probe for 1 week at 50° C. Embryos were incubated with anti-DIG Alkaline Phosphatase FAB fragments overnight at room temperature (1:1500, Roche Diagnostics). After washing, embryos were stained with NBT/BCIP for 6–24 hours. The reaction was stopped with 5 mM EDTA in MOPS Buffer. Embryos were stored up to 1 day at 4° C and imaged on a Zeiss Axiovert 200M Microscope with an AxioCam MRc5 color camera. In all cases embryos were also incubated with the same concentration of Neomycin probe as a negative control.
Asteroid (sea star)
0.2 ng/ul of probe was used for hybridization. Alkaline Phosphatase staining took approximately 6 hours.
Ophiuroid (brittle star)
0.2 ng/ul of probe was used for hybridization of eggs through the mid-gastrula stage embryos. 0.5 ng/ul of probe was used for hybridization of late-gastrula and larva stage embryos. Alkaline Phosphatase staining took approximately 13.5 hours and 10 hours, respectively.
Holothuroid (sea cucumber)
0.5 ng/ul of probe was used for hybridization. Alkaline Phosphatase staining took approximately 7 hours.
Cidaroid (pencil urchin)
0.5 ng/ul of probe was used for hybridization. Alkaline Phosphatase staining took approximately 6 hours.
Euechinoid (sea urchin)
0.1 ng/ul of probe was used for hybridization. Alkaline Phosphatase staining took approximately 24 hours.
Sequence Analysis for Probe Design
Asteroid (sea star) and Ophiuroid (brittle star)
Sequences were obtained using de novo development transcriptome databases. The sea urchin, Stronglyocentrotus purpuratus, Nanos2 protein sequence (NP_001073023.1) was used as a query to find Nanos gene sequences by performing a tblastn analysis. A reciprocal blastn was then performed using the non-redundant nucleotide database on NCBI to test authenticity.
Holothuroid (sea cucumber) and Cidaroid (pencil urchin)
Sequences were obtained using de novo ovary transcriptome databases (Reich et al., 2015) using the same method as described above for Asteroids.
Euechinoid (sea urchin)
Sequences were obtained by degenerate PCR followed by 5’ and 3’ RACE. Degenerate PCR primers are as follows: Forward-TTYTGYAARMAYAAYGGIGAR, Reverse- RTTIAVIGGRCARTAYTTIA. 5’ RACE primers are as follows: Outer- ACTCTAGTCATTTGCACACC, Inner- GGGCTATTGTCCAACTGCAA. 3’ RACE primers are as follows: Outer- CAATTCTAAGGGCGTACACC, Inner- GCTCTGTGGGACGAATGG.
Probe Design and Synthesis
General
Nanos sequences were amplified by PCR and ligated into PGEM-T Easy Vector (Promega) and transformed into competent XL-1 Blue Bacteria. Primers used for PCR amplification are listed in Supplement Table 1. Following sequence verification, plasmids were digested and used for antisense transcription using the DIG RNA Labeling Kit (Roche).
Holothuroid (sea cucumber) and Cidaroid (pencil urchin)
Nanos sequences were amplified by PCR. However, the reverse primer contained a T7 RNA polymerase recruitment sequence at its 5’ end. Following sequence verification, PCR products were used for antisense transcription as described above.
Phylogenomic Analyses
Sequences used for alignments were found as described above for Asteroid, Ophiuroid, Holothuroid, Cidaroid, and Euechinoid Nanos. Crinoid Nanos sequences were obtained using the species Oxycomanthus japonicus Ovary transcriptome using the same method described above for Asteroids, accession number: GAZO01037160 (Reich et al., 2015).
The Nanos protein alignment was made using BioEdit software that used a ClustalW analysis (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Nanos protein phylogram was made using phylogeny.fr (Dereeper et al., 2010; Dereeper et al., 2008).
Vasa Immunofluorescence
Embryos were fixed with 90% ethanol for 1 hour. After a PBS wash Vasa primary antibody (Voronina et al., 2008) was added to embryos at a 1:200 dilution for 3 hours at 25° C or overnight at 4° C Embryos were washed with PBS, stained with a fluorescent secondary antibody, and counterstained with Hoechst.
Results
Diversity in Echinoderm Nanos sequences
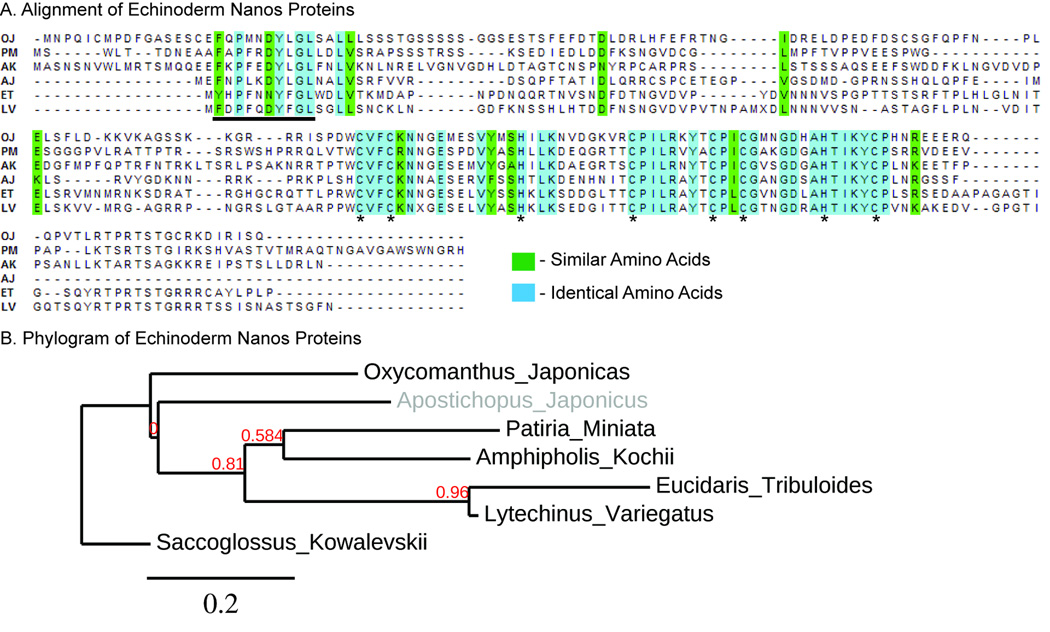
Nanos protein sequences were found in one species from each extant echinoderm clade. An alignment of the echinoderm Nanos protein sequences reveal each protein has 2 conserved CCHC zinc fingers that are characteristic of the Nanos protein family and are necessary for function (Figure 2A, asterisks) (Arrizabalaga and Lehmann, 1999; Hashimoto et al., 2010). In addition, echinoderm Nanos proteins contain a conserved upstream domain that is found in non-protostome Nanos sequences and is also required for function (Figure 2A, underline) (Torras and Gonzalez-Crespo, 2005; Torras et al., 2004). This domain has been called the NIM domain in vertebrates and is required to recruit the deadenylase protein CNOT1 to Nanos (Bhandari et al., 2014). Sequence analyses reveal there is a Nanos ortholog in every echinoderm database that has been examined.
Figure 2. Nanos sequence analysis in Echinoderms.
A. An alignment of Nanos proteins found in Echinoderms from each major clade using ClustalW2 and BioEdit. All similar amino acids are highlighted in green and all identical amino acids are highlighted in blue. Asterisks identify the conserved Nanos CCHC zinc fingers. Underlined amino acids identify the NIM domain, a non-protostome conserved protein interaction domain. OJ= Oxycomanthus japonicus, Crinoid; PM= Patiria miniata, Asteroid; AK = Amphipholis kochii, Ophiuroid; AJ= Apostichopus japonicus, Holothuroid; ET = Eucidaris tribuloides, Cidaroid; LV = Lytechinus variegatus, Euechinoid. B. A phylogram of Echinoderm Nanos protein sequences using phylogeny.fr. Clades in grey do not share the same relationships as the species phylogenetic tree.
A phylogram of echinoderm Nanos protein sequences reveals that all Nanos sequences do not follow the species tree (Figure 1A; Figure 2B, grey). We note that the Apostichopus japonicus Nanos protein, a Holothuroid sequence, is not placed as a sister group to Echinoids (Figure 1A and Figure 2B, grey). This disparity could be explained by the relatively low amount of conserved amino acids between each species (Figure 2A, non-highlighted amino acids). Nanos appears to be a protein where many amino acid changes have occurred throughout echinoderm evolution with the exception of the zinc finger and the NIM domains, both regions essential for its function.
Nanos expression in Asterozoa
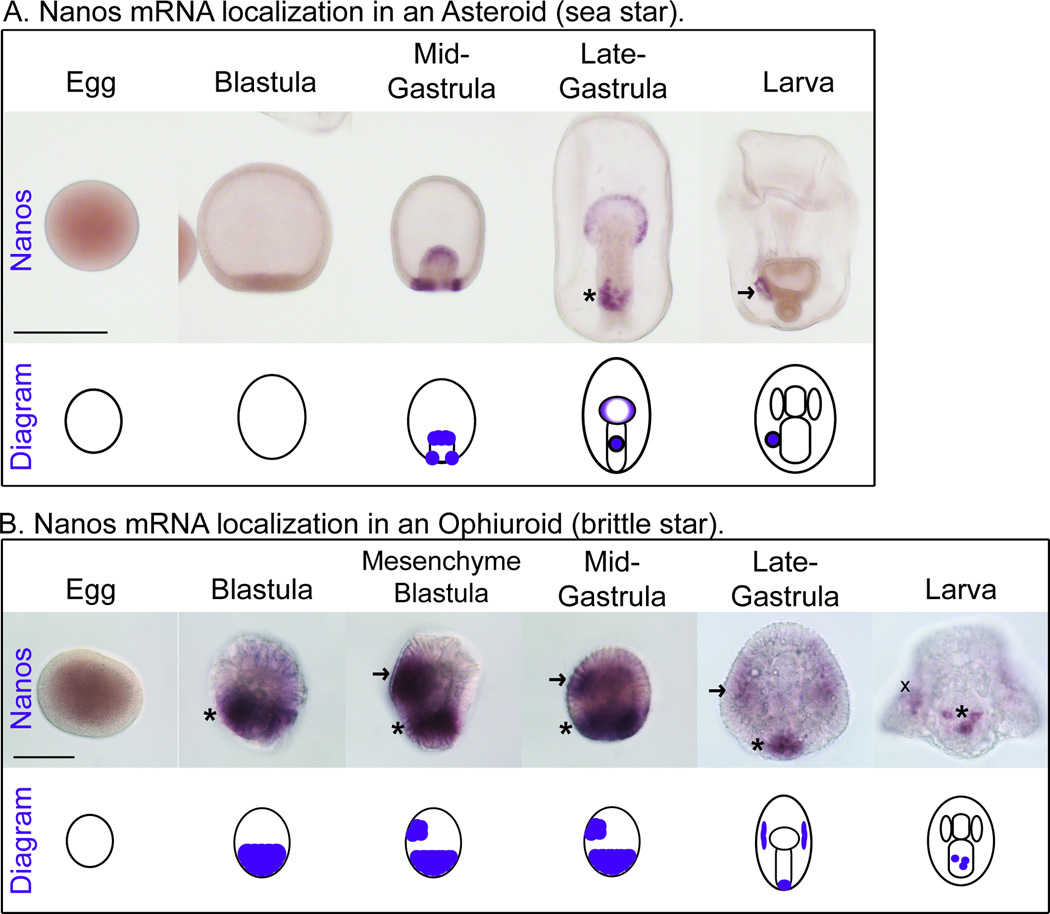
The sister groups Asteroid and Ophiuroid comprise the Asterozoa clade (Cannon et al., 2014; Reich et al., 2015; Telford et al., 2014). In the Asteroid, Patiria miniata (a sea star), Nanos is not detectable by in situ RNA hybridization until the mid-gastrula stage when it accumulates in the endomesoderm (Figure 3A). By the late-gastrula stage, Nanos is further restricted to the precursor cells that give rise to the left posterior coelom, a dorsal patch in the middle of the developing gut (Figure 3A, asterisk). In the larva stage, Nanos accumulates in the left posterior coelom (Figure 3A, arrow). This expression pattern is consistent with Vasa expression in sea stars and further supports the idea that the left posterior coelom is the site of germ cell formation (Figure 1A, light blue and dark blue, larva stage, sea star) (Juliano and Wessel, 2009). Unlike the initial widespread early expression of Vasa, Nanos is first expressed at the mid-gastrula stage in a restricted number of cells. Because Nanos expression is based upon embryonic events, Asteroids likely use an inductive mechanism for germ cell specification.
Figure 3. Nanos mRNA localization in an Asteroid and an Ophiuroid.
A. Nanos mRNA localization in an Asteroid (sea star, Patiria miniata). Nanos mRNA first accumulates in mid-gastrula embryos in the endomesoderm. Subsequently, Nanos accumulates in the tip of the developing gut and in a dorsal patch in the middle of the developing gut, asterisk. By the larva stage Nanos is restricted to the left posterior coelom, arrow. Scale bar is 200 µM. B. Nanos mRNA localization in an Ophiuroid (brittle star, Amphipholis kochii). Nanos mRNA first accumulates in blastula embryos in half of the cells of the embryo, asterisk. Subsequently, Nanos accumulates in a second expression domain at the mid-gastrula stage, arrow. At the late gastrula stage, Nanos is restricted to a few cells at the blastopore, asterisk, as well as to some mesenchymal cells within the blastocoel (arrow). By the larva stage, Nanos is further restricted to a few cells within the epithelium of the stomach, asterisk. Note: The larval stage is viewed dorsally, pigment cells are stained non-selectively throughout the blastocoel, X. Scale bar is 50 µM.
In the Ophiuroid, Amphipholis kochii (a brittle star), Nanos first accumulates at the blastula stage in a domain that contains roughly half of the cells in the embryo (Figure 3B, asterisk). We hypothesize this first expression domain gives rise to the endomesoderm by homology to the sea star. Later in development, Nanos is expressed in a second domain (Figure 3B, arrow). We suggest the second expression domain appears dorsally as Nanos is restricted dorsally in the sea star. However, further endomesoderm and dorsal markers will be necessary to test the identity of these early Nanos-positive expression domains. In the late gastrula stage the first Nanos expression domain is restricted to a few cells in half of the blastopore (Figure 3B, asterisk) and the second Nanos expression domain is restricted to a few cells in the blastocoel (Figure 3B, arrow). By the 3-day old larval stage the first Nanos expression domain becomes restricted further to a few cells in half of the stomach epithelium. This is the first report of germ cell marker expression in Ophiuroid embryos, to the best of our knowledge. Furthermore, using Vasa protein labeling we have identified a left posterior coelom in an Ophiuroid embryo for the first time (Supplemental Figure 2B, asterisk). It remains to be tested if the Nanos enriched cells at the 3-day larva stage will migrate to the left posterior coelom at a later time point in larval development. However, we hypothesize that this will occur because Vasa protein accumulates in the left posterior coelom at the 1-week old larval stage (Supplemental Figure 2B, asterisk). Together, these results suggest germ cell markers accumulate in the left posterior coelom of an Ophiuroid larva. In addition, the relatively later initial expression of Nanos (blastula stage) and the dependence on embryonic events to restrict the Nanos positive domains suggests Ophiuroids also use inductive mechanisms for germ cell specification.
Nanos expression in Holothuroids
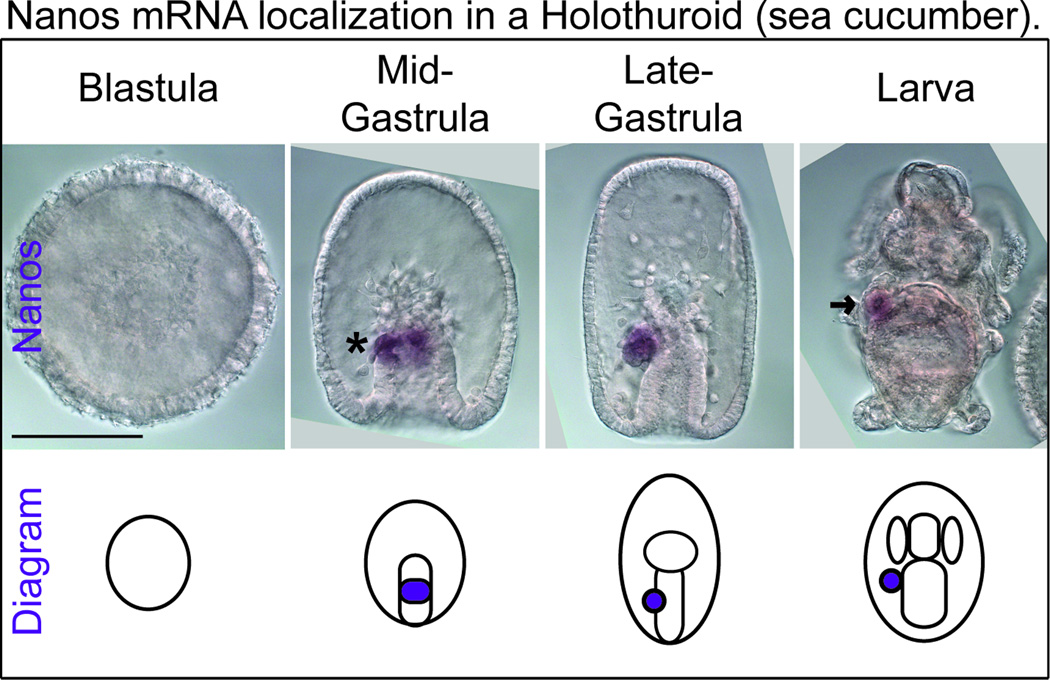
In the Holothuroid, Apostichopus japonicus (a sea cucumber), Nanos is first expressed in the mid-gastrula stage in a dorsal patch biased to the left side of the developing gut (Figure 4, asterisk). These Nanos positive cells likely develop into the left posterior coelom visible in the larva stage (Figure 4, arrow). This expression pattern is similar to Vasa expression in the sea cucumber because Vasa also accumulates in the left posterior coelom (Figure 1A, light blue and dark blue, larva stage, sea cucumber) (Supplemental Figure 2A, arrow) (Yu et al., 2013). However, Nanos expression is more restricted than Vasa because Vasa accumulates in all of the other coeloms at the early-larva stage. These results demonstrate that germ cell markers accumulate in the left posterior coelom of a Holothuroid embryo. In addition, the initial expression of Nanos in Holothuroid embryos is relatively late in development (mid-gastrula stage) and suggests they use inductive mechanisms for germ cell specification.
Figure 4. Nanos mRNA localization in a Holothuroid (sea cucumber).
In the sea cucumber, Apostichopus japonicus, Nanos mRNA first accumulates in mid-gastrula embryos in a dorsal domain in the middle of the developing gut, asterisk. Nanos is restricted to the left posterior coelom during the gastrula and larva stages (viewed dorsally), arrow. Scale bar is 100 µM.
Nanos expression in Echinoids
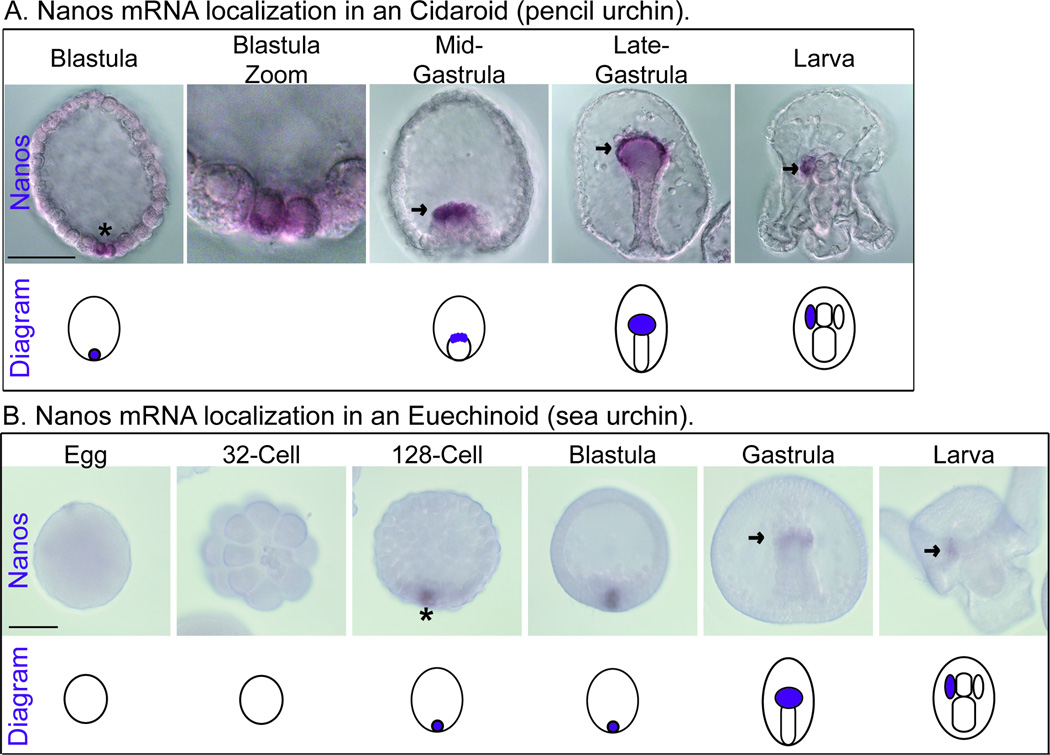
The sister groups Cidaroid and Euechinoid comprise the Echinoid clade. In the Cidaroid, Eucidaris tribuloides (a pencil urchin), Nanos is first detected at the early blastula stage in 2–4 vegetal cells (Figure 5A, asterisk). These cells are likely the micromeres, the smaller cells that arise from a variable number of unequal cell divisions at the 16-cell stage (Schroeder, 1981). Subsequently, these Nanos positive cells accumulate in the left coelom (Figure 5A, arrows). Although Vasa is not restricted to the micromere lineage in this animal, it does end up accumulating in the left coelom (Figure 1A, light blue and dark blue, 32-cell stage and larva stage, pencil urchin) (Juliano and Wessel, 2009). Together, these results indicate that the left coelom accumulates germ cell markers in a Cidaroid embryo. Furthermore, Nanos is initially expressed early in development (during the first few cleavages), which is likely more dependent upon the localization of maternal molecules. This is more consistent with an inherited mechanism of germ cell specification.
Figure 5. Nanos mRNA localization in a Cidaroid and an Euechinoid.
A. Nanos mRNA localization in a Cidaroid (pencil urchin, Eucidaris tribuloides). Nanos mRNA first accumulates in blastula embryos in a restricted number of cells at the vegetal plate, asterisk. Subsequently, Nanos accumulates in the tip of the developing gut during the gastrula stage, arrow. By the larva stage (viewed dorsally) Nanos is restricted to the left coelom, arrow. Scale bar is 100 µM. B. Nanos mRNA localization in an Euechinoid (sea urchin, Lytechinus variegatus). Nanos mRNA first accumulates in 128-cell embryos in a restricted population of cells at the vegetal plate, asterisk. Subsequently, Nanos accumulates at the tip of the developing gut during the gastrula stage, arrow. By the larva stage (viewed dorsally) Nanos is restricted to the left coelom, arrow. Scale bar is 60 µM.
In the Euechinoid, Lytechinus variegatus (a sea urchin), Nanos is first detected at the 128-cell stage in a restricted number of vegetal cells (Figure 5B, asterisk). These are likely the small micromeres, the smaller cells that arise from two series of unequal cell divisions at both the 16 and 32-cell stages. Subsequently, these cells migrate to the tip of the developing gut and end up in the left coelom (Figure 5B, arrows). This expression is very similar to both Nanos and Vasa protein expression in the related sea urchin, Stronglyocentrotus purpuratus (Figure 1A, dark blue, 32-cell stage and larva stage, sea urchin) (Juliano et al., 2010; Voronina et al., 2008). In summary, these results demonstrate that the left coelom accumulates germ cell markers and that the small micromeres have a conserved germ cell fate in the Euechinoid lineage. In addition, Euechinoids also likely use an inherited mechanism of germ cell specification because Nanos is initially expressed very early in development (at the 32-cell stage).
Discussion
Three major developmental shifts in Nanos expression
We notice three major developmental stages in which Nanos mRNA first begins to accumulate.
Nanos expression is associated with cells that arise as a result of unequal cell divisions in Echinoids. The study of Nanos expression in Echinoid embryos is particularly important to understand evolutionary transitions in germ cell specification because the Echinoid clade has a derived feature in their early development related to germ cell specification. They are the only echinoderm embryos that undergo unequal cell divisions during early cleavage (Figure 6A, pink). It was already known that Nanos is first expressed in the small micromere lineage of sea urchins, cells that arise at the 32-cell stage as a result of two unequal cell divisions (Figure 1A, purple, 32-cell stage, sea urchin) (Fujii et al., 2006; Juliano et al., 2010). Here, we test if Nanos expression is associated with cells that arise from unequal cell divisions in a different Echinoid lineage, the pencil urchin. The pencil urchin undergoes a variable number of unequal cell divisions at the 16-cell stage to give rise to the micromere lineage (Figure 1A, 32-cell stage, pencil urchin). This study suggests Nanos is expressed in the micromere lineage of the pencil urchin (Figure 5A, asterisk). We hypothesize that unequal cell divisions are linked to an earlier restricted expression of Nanos. Furthermore, the sea urchin Nanos-positive small micromere cells have previous been shown to directly contribute to the germ cell lineage (Yajima and Wessel, 2011c). Because Nanos is expressed early in development in a restricted number of cells and these cells have been shown to contribute to the germ cell lineage (at least in the sea urchin) we hypothesize that Echinoids display inherited mechanisms of germ cell specification.
In contrast to the Echinoid lineage, both Asteroids and Holothuroids do not start expressing Nanos until later in development (Figure 3A; Figure 4, asterisk). Animals in both of these clades contain a left posterior coelom and Nanos is first expressed at the mid-gastrula stage in left posterior coelom precursor cells (Figure 3A; Figure 4, asterisk). In addition, removal studies in the sea star show that the left posterior coelom contributes to morphologically distinct germ cells (Inoue et al., 1992). Because Nanos is expressed later in development and in cells that contribute to germ cells (in the sea star) we hypothesize that these animals use inductive mechanisms to specify the germ cell lineage.
Nanos expression in the Ophiuroid lineage is the most perplexing. It is expressed at the blastula stage in a large embryonic domain equal to roughly half of the cells of the embryo (Figure 3B, asterisk). Because Nanos expression is not restricted to a small number of cells early in development we postulate that Ophiuroids use inductive mechanisms for germ cell specification. Ultimately though, it will be important to examine more species from each echinoderm clade to better refine these models.
Figure 6. A Summary of Nanos mRNA localization in Echinoderms.
A. In both Asteroids and Holothuroids, Nanos is enriched in the left posterior coelom by the larva stage, asterisks. In both Cidaroids and Euechinoids Nanos accumulates in the left coelom by the larva stage, arrows. In contrast, Nanos accumulates in the stomach at the larva stage in Ophiuroids. Purple= Nanos mRNA, Green= larval skeleton, Pink= early unequal cell divisions. B. A diagram of general Echinoderm development and the embryonic germ layers.
Taken together, the results are consistent with the hypothesis that echinoderms ancestrally specified their germ cell lineage following gastrulation via inductive mechanisms. This is because the majority of extant echinoderm clades (Asteroids, Ophiuroids, and Holothuroids) use inductive mechanisms for germ cell specification. In sea stars and Holothuroids (brittle stars are less clear at this point), germ cell markers accumulate in a restricted embryonic domain late in embryogenesis (during the late gastrula stage/early larva stage). Furthermore, our results are also consistent with the hypothesis that an evolutionary transition has occurred in the Echinoid lineage for the germ cell lineage to be specified earlier by inherited mechanisms. In these animals, germ cell markers accumulate in a restricted number of cells very early in development, during the first few cleavages. This is consistent with a previous comprehensive study in animals which proposes that the inductive germ cell specification mechanism is ancestral, while there have been several independent acquisitions of the inherited germ cell specification mechanism (Extavour and Akam, 2003).
Cells within left coeloms contribute to the germ cell lineage in echinoderms
In most of the animals studied here, Nanos and Vasa accumulate together in either the left coelom or the left posterior coelom (Figure 1A; Figure 6A, purple, larva stage). We notice that germ cell markers accumulate in the left posterior coelom in almost every echinoderm lineage (Figure 6A, asterisk). However, the Echinoid lineage seems to have lost this structure and germ cell markers accumulate instead in the left coelom (Figure 6A, arrows).
In addition to accumulating germ cell markers we argue that the posterior left coelom also contributes to the germ cell lineage in Asteroids, Ophiuroids, and Holothuroids. In support of this hypothesis, the left posterior coelom has been removed from sea star larva and results in juveniles with about half of the normal number of morphologically distinct germ cells (Inoue et al., 1992). We also argue that the left coelom contributes to the germ cell lineage in all Echinoids. The small micromere cells have been removed from sea urchins before they reach the left coelom and results in an adult that develops normally, but is lacking any gametes (Yajima and Wessel, 2011c). From these results, we propose that left-sided coeloms contribute to the germ cell lineage in all echinoderm species.
It is important to note that the left coelom is a mesodermal lineage (Figure 6B, red). In addition, the left posterior coelom also accumulates many mesodermal markers in sea stars and sea cucumbers (McCauley et al., 2012) (unpublished results). This is significant because in all animals that have been studied that use an inductive mechanism for germ cell specification their germ cell lineage was induced from a mesodermal lineage (Chatfield et al., 2014; Ewen-Campen et al., 2013; Saitou et al., 2002). These results further support the hypothesis that the most recent common ancestor of Bilaterian animals had a germ cell lineage induced from a mesodermal lineage.
Nanos expression correlates with the expression of larval skeletogenic genes
We notice that the germ cell lineage is linked to genes that are required for specification of the larval skeleton. Both the Echinoid and Ophiuroid lineages likely acquired larval skeletons independently because larval skeletons are absent from other clades (Figure 6A, green). In the sea urchin (Echinoid), it has already been shown that the cell lineage that gives rise to the germ cells at the 16-cell stage also gives rise to the larval skeleton (Ettensohn, 1997; Gustafson and Wolpert, 1967; Okazaki, 1975; Yajima and Wessel, 2012). In addition, the cell lineage that gives rise to the larval skeleton in the pencil urchin (Echinoid) also accumulates Nanos (Wray and McClay, 1988) (Figure 5A, asterisk). This shows that both the germ cell lineage and the larval skeletal cell lineage share a common precursor cell lineage in Echinoids. In addition, this study found that Nanos is expressed in half of the cells during the blastula stage of an Ophiuroid embryo (Figure 3B, asterisk). This expression pattern resembles the vegetal expression pattern domain where five larval skeletal genes accumulate in Ophiuroids, although the skeletal genes appear to localize in a more restricted domain (Dylus et al., 2016; Koga et al., 2010; Morino et al., 2012). We hypothesize that the gene regulatory networks that control the germ cell lineage and the larval skeleton lineage are co-dependent in both Echinoids and Ophiuroids and that this may be an evolutionary restriction placed on the evolution of both cell lineages. In other words, whenever the gene regulatory networks that control the skeletal cell lineage or germ cell lineage experience a heterochronic shift in development the other lineage may follow.
In support of this hypothesis, the expression of germ cell markers also correlates with the expression of genes associated with larval skeletal specification even in animals that do not have larval skeletons. Asteroids do not have larval skeletons, yet genes that are involved in specification of the larval skeleton in Euechinoids (e.g. Alx and Ets) are also co-expressed with Nanos in the left posterior coelom of the Asteroid larva (Figure 3A, arrow) (Koga et al., 2010; McCauley et al., 2012). Together, these results support the hypothesis that the gene regulatory network that specifies the echinoderm germ cell lineage is in some way intertwined with the gene regulatory network that specifies the Echinoid and Ophiuroid larval skeletons.
Mechanisms that may contribute to evolutionary transitions in germ cell specification
We notice three developmental changes that are involved in the evolution of an inherited germ cell specification mechanism in the Echinoid clade. First, we propose that the evolution of early unequal cell divisions within the Echinoid clade was important for the evolution of earlier accumulation of germ cell markers. Therefore, early unequal cell divisions may be a broader strategy for the evolution of an inherited mechanism of germ cell specification. To explore this model further it will be important to understand the cellular and molecular tools that are required for an early unequal cell division in the Echinoid lineage. In addition, it will be important to understand the upstream regulatory molecules that are required for the earlier accumulation of germ cell markers. Second, we propose that the earlier specification of the germ cell lineage in the Echinoid lineage is accompanied by an earlier specification of the larval skeletal lineage because these two cell fates overlap. This suggests there may be an upstream factor that is required for the specification of both cell lineages, such as a general determinant of the endo/mesoderm cell fate (ie. Wnt). This may be an evolutionary constraint placed upon the evolution of an earlier germ cell specification in Echinoids. To explore this model further it will be important to define the upstream regulators of both of these cell lineages. Third, there is a loss of the posterior left coelom in the Echinoid lineage. This further emphasizes the importance of the posterior left coelom in echinoderms that use the ancestral inductive mechanism of germ cell specification, while this structure is not required for germ cell specification in Echinoids and has been lost. To explore how the posterior left coelom cell lineage has been lost in the Echinoid lineage it will be necessary to understand the molecules that are required to induce the left posterior coelom cell lineage in non-Echinoid embryos. The future study of these three developmental changes in diverse echinoderm embryos will significantly impact the broader field examining the evolution of germ cell mechanisms within all animals.
Supplementary Material
Acknowledgments
Funding for this research was provided by the NIH: (2RO1HD028152 to GMW) (NIGMS 1F31GM112479-01, Award No. 004102 to TMF) (Training grant T32GM007601 to Brown University) and by the NSF: (EPSCoR-RI Graduate student fellowship, 1004057 to TMF). Also, thank you to the members of PRIMO for a rich working environment.
References
- Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–4643. doi: 10.1242/dev.127.21.4631. [DOI] [PubMed] [Google Scholar]
- Arrizabalaga G, Lehmann R. A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics. 1999;153:1825–1838. doi: 10.1093/genetics/153.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. Structural basis for the Nanos-mediated recruitment of the CCR4-NOT complex and translational repression. Genes Dev. 2014;28:888–901. doi: 10.1101/gad.237289.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JT, Kocot KM, Waits DS, Weese DA, Swalla BJ, Santos SR, Halanych KM. Phylogenomic resolution of the hemichordate and echinoderm clade. Curr Biol. 2014;24:2827–2832. doi: 10.1016/j.cub.2014.10.016. [DOI] [PubMed] [Google Scholar]
- Chatfield J, O'Reilly MA, Bachvarova RF, Ferjentsik Z, Redwood C, Walmsley M, Patient R, Loose M, Johnson AD. Stochastic specification of primordial germ cells from mesoderm precursors in axolotl embryos. Development. 2014;141:2429–2440. doi: 10.1242/dev.105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal Phylogeny and Its Evolutionary Implications. Annu Rev Ecol Evol S. 2014;45:371-+. [Google Scholar]
- Dylus DV, Czarkwiani A, Stangberg J, Ortega-Martinez O, Dupont S, Oliveri P. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo. 2016;7:2. doi: 10.1186/s13227-015-0039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Guss KA, Hodor PG, Malinda KM. The morphogenesis of the skeletal system of the sea urchin embryo. New York: Wiley and Sons; 1997. [Google Scholar]
- Evans T, Wade CM, Chapman FA, Johnson AD, Loose M. Acquisition of germ plasm accelerates vertebrate evolution. Science. 2014;344:200–203. doi: 10.1126/science.1249325. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Donoughe S, Clarke DN, Extavour CG. Germ cell specification requires zygotic mechanisms rather than germ plasm in a basally branching insect. Curr Biol. 2013;23:835–842. doi: 10.1016/j.cub.2013.03.063. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Fujii T, Mitsunaga-Nakatsubo K, Saito I, Iida H, Sakamoto N, Akasaka K, Yamamoto T. Developmental expression of HpNanos, the Hemicentrotus pulcherrimus homologue of nanos. Gene Expr Patterns. 2006;6:572–577. doi: 10.1016/j.modgep.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Gustafson T, Wolpert L. Cellular movement and contact in sea urchin morphogenesis. Biol Rev Camb Philos Soc. 1967;42:442–498. doi: 10.1111/j.1469-185x.1967.tb01482.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Hara K, Hishiki A, Kawaguchi S, Shichijo N, Nakamura K, Unzai S, Tamaru Y, Shimizu T, Sato M. Crystal structure of zinc-finger domain of Nanos and its functional implications. EMBO Rep. 2010;11:848–853. doi: 10.1038/embor.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci U S A. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue C, Kiyomoto M, Shirai H. Germ-Cell Differentiation in Starfish - the Posterior Enterocoel as the Origin of Germ-Cells in Asterina-Pectinifera. Dev Growth Differ. 1992;34:413–418. doi: 10.1111/j.1440-169X.1992.00413.x. [DOI] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Wessel GM. An evolutionary transition of Vasa regulation in echinoderms. Evol Dev. 2009;11:560–573. doi: 10.1111/j.1525-142X.2009.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Yajima M, Wessel GM. Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Dev Biol. 2010;337:220–232. doi: 10.1016/j.ydbio.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kikuchi M, Omori A, Kurokawa D, Akasaka K. Patterning of anteroposterior body axis displayed in the expression of Hox genes in sea cucumber Apostichopus japonicus. Dev Genes Evol. 2015;225:275–286. doi: 10.1007/s00427-015-0510-7. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsubara M, Fujitani H, Miyamoto N, Komatsu M, Kiyomoto M, Akasaka K, Wada H. Functional evolution of Ets in echinoderms with focus on the evolution of echinoderm larval skeletons. Dev Genes Evol. 2010;220:107–115. doi: 10.1007/s00427-010-0333-5. [DOI] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 2000;127:2907–2916. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translational control in germ cells. Mol Reprod Dev. 2013;80:665–676. doi: 10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- Lasko P. The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim Biophys Acta. 2013;1829:810–816. doi: 10.1016/j.bbagrm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- McCauley BS, Wright EP, Exner C, Kitazawa C, Hinman VF. Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. Evodevo. 2012;3:17. doi: 10.1186/2041-9139-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70:163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Morino Y, Koga H, Tachibana K, Shoguchi E, Kiyomoto M, Wada H. Heterochronic activation of VEGF signaling and the evolution of the skeleton in echinoderm pluteus larvae. Evol Dev. 2012;14:428–436. doi: 10.1111/j.1525-142X.2012.00563.x. [DOI] [PubMed] [Google Scholar]
- Okazaki K. Normal development to metamorphosis. New York: Springer-Verlag; 1975. [Google Scholar]
- Reich A, Dunn C, Akasaka K, Wessel G. Phylogenomic analyses of Echinodermata support the sister groups of Asterozoa and Echinozoa. PLoS One. 2015;10:e0119627. doi: 10.1371/journal.pone.0119627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. Development of a Primitive Sea-Urchin (Eucidaris-Tribuloides) - Irregularities in the Hyaline Layer, Micromeres, and Primary Mesenchyme. Biol Bull. 1981;161:141–151. [Google Scholar]
- Schwager EE, Meng Y, Extavour CG. vasa and piwi are required for mitotic integrity in early embryogenesis in the spider Parasteatoda tepidariorum. Dev Biol. 2015;402:276–290. doi: 10.1016/j.ydbio.2014.08.032. [DOI] [PubMed] [Google Scholar]
- Smith LD. The role of a "germinal plasm" in the formation of primordial germ cells in Rana pipiens. Dev Biol. 1966;14:330–347. doi: 10.1016/0012-1606(66)90019-4. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Lowe CJ, Cameron CB, Ortega-Martinez O, Aronowicz J, Oliveri P, Copley RR. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proc Biol Sci. 2014;281 doi: 10.1098/rspb.2014.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torras R, Gonzalez-Crespo S. Posterior expression of nanos orthologs during embryonic and larval development of the anthozoan Nematostella vectensis. Int J Dev Biol. 2005;49:895–899. doi: 10.1387/ijdb.051980rt. [DOI] [PubMed] [Google Scholar]
- Torras R, Yanze N, Schmid V, Gonzalez-Crespo S. nanos expression at the embryonic posterior pole and the medusa phase in the hydrozoan Podocoryne carnea. Evol Dev. 2004;6:362–371. doi: 10.1111/j.1525-142X.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–286. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Brayboy L, Fresques T, Gustafson EA, Oulhen N, Ramos I, Reich A, Swartz SZ, Yajima M, Zazueta V. The biology of the germ line in echinoderms. Mol Reprod Dev. 2014;81:679–711. doi: 10.1002/mrd.22223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Reich AM, Klatsky PC. Use of sea stars to study basic reproductive processes. Syst Biol Reprod Med. 2010;56:236–245. doi: 10.3109/19396361003674879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. Structure of the Drosophila BicaudalD protein and its role in localizing the the posterior determinant nanos. Cell. 1989;59:881–892. doi: 10.1016/0092-8674(89)90611-9. [DOI] [PubMed] [Google Scholar]
- Wray GA, McClay DR. The origin of spicule-forming cells in a 'primitive' sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103:305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development. 2011a;138:2217–2222. doi: 10.1242/dev.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. The multiple hats of Vasa: its functions in the germline and in cell cycle progression. Mol Reprod Dev. 2011b;78:861–867. doi: 10.1002/mrd.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Small micromeres contribute to the germline in the sea urchin. Development. 2011c;138:237–243. doi: 10.1242/dev.054940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Autonomy in specification of primordial germ cells and their passive translocation in the sea urchin. Development. 2012;139:3786–3794. doi: 10.1242/dev.082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Essential elements for translation: the germline factor Vasa functions broadly in somatic cells. Development. 2015;142:1960–1970. doi: 10.1242/dev.118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- Yu L, Yan M, Sui J, Sheng WQ, Zhang ZF. Gonadogenesis and expression pattern of the vasa gene in the sea cucumber Apostichopus japonicus during early development. Mol Reprod Dev. 2013;80:744–752. doi: 10.1002/mrd.22207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.