ABSTRACT
The increasing prevalence of extremely low frequency electromagnetic fields (ELF-EMFs) exposure has raised considerable public concern regarding the potential hazardous effects of ELF-EMFs on male reproductive function. Increasing evidence indicates that miRNAs are necessary for spermatogenesis and male fertility. However, the regulation of miRNA expression and the roles of miRNAs in response to ELF-EMFs remain unclear. In our study, mouse spermatocyte-derived GC-2 cells were intermittently exposed to a 50 Hz ELF-EMF for 72 h (5 min on/10 min off) at magnetic field intensities of 1 mT, 2 mT and 3 mT. MiR-26b-5p was differentially expressed in response to different magnetic field intensities of ELF-EMFs. The host gene CTDSP1 showed an unmethylation status in GC-2 cells at different magnetic field intensities of ELF-EMF exposure. MiR-26b-5p had no significant, obvious influence on the cell viability, apoptosis or cell cycle of GC-2 cells. However, the overexpression of miR-26b-5p significantly decreased the percentage of G0/G1 phase cells and slightly increased the percentage of S phase cells compared to the sham group that was exposed to a 50 Hz ELF-EMF. Computational algorithms identified Cyclin D2 (CCND2) as a direct target of miR-26b-5p. MiR-26b-5p and a 50 Hz ELF-EMF altered the expression of CCND2 at both the mRNA and protein levels. Overexpressed miR-26b-5p in GC-2 cells can change the mRNA expression of CCND2 following 50 Hz ELF-EMF at 3 mT. These findings demonstrate that miR-26b-5p could serve as a potential biomarker following 50 Hz ELF-EMF exposure, and miR-26b-5p-CCND2-mediated cell cycle regulation might play a pivotal role in the biological effects of ELF-EMFs.
Keywords: CCND2, cell cycle, extremely low frequency electromagnetic fields, miR-26b-5p, reproductive toxicity
Introduction
The prevalence of electric appliances from power supply lines and many household and commercial devices has increased the health risk of human beings who are progressively exposed to extremely low frequency electromagnetic fields (ELF-EMFs). This prevalence has also raised considerable public concern regarding the potential hazardous effects of ELF-EMFs.1,2 The male reproductive system is considered sensitive to electromagnetic radiation. Many studies have confirmed that ELF-EMFs can alter the reproductive endocrine hormones and decrease the semen quality of humans and animals, as well as gonadal fetal function.3-5 Despite numerous attempts, the biological mechanism facilitating the effects of ELF-EMFs remains unknown. Therefore, it is necessary to investigate and understand the potential effects of ELF-EMFs on the male reproductive system.
MiRNAs are a class of small endogenous non-coding RNAs that are 21–26 nucleotides in length.6,7 MiRNAs predominantly negatively regulate gene expression by binding to the 3′-untranslated region (3′-UTR) of the target genes.8 MiRNAs participate in the regulation of various cellular processes, including cell proliferation, cell cycle and apoptosis.9-11 Emerging evidence has demonstrated the critical role of miRNAs in the control of reproductive functions, especially in the processes of oocyte maturation, folliculogenesis, corpus luteum function, implantation and early embryonic development.12 In addition, increasing evidence indicates that miRNAs are necessary for spermatogenesis and male fertility.13 Therefore, we speculated that miRNA-mediated regulation might be linked to the adverse effects of ELF-EMFs on the male reproductive system.
MiR-26a and miR-26b, which are intrinsic miRNAs that are located in the intron of CTDSP1, are important for various types of cancer development.14,15 For example, the down-regulation of miR-26b in osteosarcoma increased the levels of CTGF and Smad1, facilitating osteosarcoma metastasis.16 The downregulation of miR-26b in carcinoma-associated fibroblasts from estrogen receptor-positive breast cancers can lead to enhanced cell migration and invasion.17 MiR-26b could modulate non-small cell lung cancer chemoresistance and migration through its association with PTEN.18 Recently, we found that a 50 Hz ELF-EMF could significantly change the expression of miR-26b-5p compared to a sham group in GC-2 cells. However, thus far, the role of miR-26b-5p in ELF-EMFs has never been investigated. In this study, we investigated the molecular regulation of miR-26b-5p in response to ELF-EMFs and examined whether miR-26b-5p could act as a biomarker of exposure to ELF-EMFs.
Materials and methods
Cell culture
Mouse spermatocyte-derived GC-2 cells (GC-2 cells) were purchased from the American Tissue Culture Collection (ATCC, Rockville, MD, USA). GC-2 cells were cultured in DMEM high-glucose medium (Hyclone, Logan, UT, USA) that was supplemented with 10% fetal bovine serum (Gibco BRL, Rockville, MD, USA) at 37 °C in a humidified atmosphere with 5% CO2. Germ cells of mouse were isolated as described previously.19
Exposure procedure and experimental design
The exposure system was designed and provided by the Foundation for Information Technologies in Society (IT′IS foundation, Zurich, Switzerland), as described previously.20, 21 Briefly, the exposure system consists of a power frequency generator, an arbitrary function generator, a narrow-band amplifier and 2 rectangular waveguides. The setup generated a vertical EMF that was composed of 2 4-coil systems (2 coils with 56 windings and 2 coils with 50 windings) and was placed inside a metal chamber. The system was composed of 2 identical exposure chambers. One of the chambers was sham-exposed, and the other chamber was exposed to radiation. Exposed and sham-exposed cell dishes were simultaneously placed in an incubator in which the environmental conditions were constant (37°C, 5% CO2). The exposure setup was controlled and monitored by a computer through specific sensors that can automatically control the exposure parameters, including exposure intensity and exposure time. The temperature difference between sham and ELF-EMF exposure never exceeded 0.3°C.
After overnight starvation, GC-2 cells were exposed to a 50 Hz ELF-EMF at magnetic intensities of 1 mT, 2 mT, and 3 mT for 72 h (5 min on /10 min off). The cultured cells were divided into 6 groups as follows: (i) sham group; (ii) ELF-EMF exposure group; (iii) GC-2 cells that were transfected with miR-26b-5p mimic or mimic negative control (100 nM); (iv) GC-2 cells that were transfected with miR-26b-5p inhibitor or inhibitor negative control (100 nM); (v) GC-2 cells that were transfected with miR-26b-5p mimic (100 nM) for 6 h and then exposed to a 50 Hz ELF-EMF; and (vi) GC-2 cells that were transfected with mimic negative control (100 nM) for 6 h and then exposed to a 50 Hz ELF-EMF.
Quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was extracted from the GC-2 cells using Trizol Reagent Kit (Invitrogen, Carlsbad, USA). cDNAs were obtained by the GoScript™ Reverse Transcription System Kit (Promega, WI, USA). For miR-26b-5p, total RNA (2 μg) was transcribed into cDNA using a microRNA First-Strand cDNA Synthesis Kit (Sangong Biotech, Shanghai, China) according to the manufacturer's protocol. The specific forward primer for miR-26b-5p is 5′-GGCTTCAAGTAATTCAGGATAGG-3′. To quantify the miR-26b-5p expression levels, the expression of small nuclear U6 was used as the internal control. MiRNA and mRNA expression was examined using a Bio-Rad IQ5 Detection System with SYBR Green PCR Master mix (Promega) as described previously.22 The primer sequences are listed in Table S1.
Methylation-specific PCR (MSP)
A DNA isolation kit (Omega Bio-Tek, USA) was used to extract DNA from the GC-2 cells following the manufacturer's instructions. DNA was bisulfite converted using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions. The primer sets were specific for amplifying either methylated or unmethylated sequences spanning the CpG island of the host gene of miR-26b-5p. The primer pairs that were used for MSP are listed in Table S1. The PCR conditions for MSP were standardized in our laboratory.22
Cell viability assay
Cell counting kit-8 (CCK-8, Dojindo, Japan) was used to detect cell viability. According to the manufacturer's instructions, 3000 cells per well were seeded into 96-well plates in 100 μl of cell culture medium and transfected with miR-26b-5p mimic, mimic negative control (mimic-nc), inhibitor, or inhibitor negative control (inhibitor-nc); some cells were transfected with miR-26b-5p mimic and mimic-nc. All cells were treated with 50 Hz ELF-EMF at a magnetic field intensity of 3 mT for 72 h. Then, the medium was mixed with 10 μl of CCK-8 reaction solution for 1 h at 37°C. The optical density was measured at 450 nm by a microplate reader (SpectraMax M2; Molecular Devices, Sunnyvale, CA, USA). Each measurement was repeated 3 times.
Detection of apoptosis and the cell cycle by flow cytometry
After transfection or ELF-EMF exposure, cellular apoptosis was analyzed by flow cytometry with Annexin V-FITC and propidium iodide (PI) double staining. For cell cycle analysis, GC-2 cells were harvested and fixed in 75% ice-cold ethanol overnight at 4°C. The fixed cells were washed twice with ice-cold PBS, stained with 50 mg/ml PI containing 50 mg/ml RNase A (DNase free) for 30 min at 37°C and then analyzed in a flow cytometer. The experiments were conducted 3 times.
Cell transfection
MiR-26b-5p mimic, inhibitor, and their negative controls were purchased from RiboBio (RiboBio, Guangzhou, China). GC-2 cells were cultured in a 96-well or 6-well plate 24 h before transfection. Prior to miRNA transfection, the cell culture medium of the GC-2 cells was replaced with serum-reduced medium (Opti-MEM I; Invitrogen, Carlsbad, CA, USA). The transfection was performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The GC-2 cells were incubated with the mimic or inhibitor/Lipofectamine mixture for 6 h. Then, the Opti-MEM I medium was switched to a growth medium. The miRNA mimic or inhibitor (100 nM) that was prepared with lipid carrier (1:2, v/v) was used for the transfection in Opti-MEM I medium. To determine the optimal transfection conditions, the transfection efficiency was confirmed by real-time PCR.
MiR-26b-5p target gene prediction
The target genes of miR-26b-5p were predicted by starBase v2.0 (http://starbase.sysu.edu.cn/targetSite.php), which included TargetScan, PicTar, RNA22, PITA, miRanda/mirSVR and other computational prediction methods. The predicted targets that were involved in proliferation, cell cycle and apoptosis were screened first.
Western blot analysis
The cells were lysed and centrifuged at 12,000 rpm for 30 min at 4 ºC, and the supernatants were collected. The protein concentrations were determined using the BCA protein assay kit (Pierce, Rockford, IL, USA). Western blotting was performed as previously described. Protein samples (60 μg) were separated by 10% SDS-PAGE and then transferred to an equilibrated PVDF membrane (Amersham Biosciences, Buckinghamshire, UK). After incubation at 4 ºC overnight with a primary antibody against CCND2 (Beyotime, Jiangsu, China) and then reacting with a secondary antibody, the proteins were detected by enhanced chemiluminescence (Amersham Corporation, Arlington Heights, IL, USA). The density of the bands was quantified using ImageJ software and normalized against β-actin.
Statistical analysis
All of the data were expressed as the mean ± SD from at least 3 independent experiments that were performed in duplicate. The differences between the sham group and the ELF-EMF group were measured by Student's t-test. Significant differences were established at P < 0.05.
Results
MiR-26b-5p is differentially expressed in GC-2 cells exposed to different magnetic field intensities of a 50 Hz ELF-EMF
Our previous microarray analysis showed that the degree of miR-26b-5p was higher than that of other miRNAs (Fig. 1A). The degree represents the number of target genes that are regulated by the miRNA in the miRNA-gene network. The results indicated that miR-26b-5p might play an important role in GC-2 cells following 50 Hz ELF-EMF. We further found that miR-26b-5p was differentially expressed at different magnetic field intensities of ELF-EMF exposure by real-time PCR (Fig. 1B). The expression level of miR-26b-5p was significantly up-regulated at a magnetic field intensity of 2 mT and was significantly down-regulated at 3 mT compared to the sham group. However, a 50 Hz ELF-EMF had no obvious influence on the expression of miR-26b-5p at 1 mT in GC-2 cells.
Figure 1.
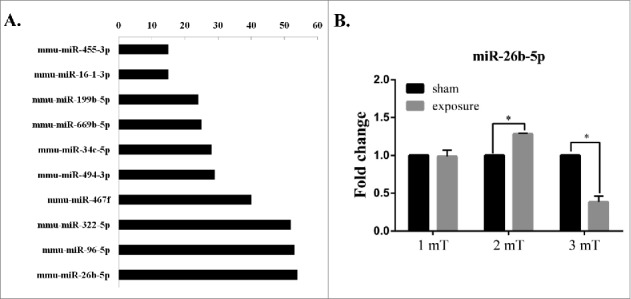
50 Hz ELF-EMF exposure alters the expression of miR-26b-5p. (A) MicroRNA-gene network in GC-2 cells between the sham and exposure groups at magnetic field intensity of 3 mT. The vertical axis corresponds to changed miRNAs, and the horizontal axis to the degree of miRNA, which represents the number of target genes. (B) The expression levels of miR-26b-5p were significantly higher in response to a magnetic field intensity of 2 mT but were significantly lower in response to a magnetic field intensity of 3 mT. However, 50 Hz ELF-EMF had no obvious influence on the expression of miR-26b-5p at 1 mT in GC-2 cells.
50 Hz ELF-EMF exposure does not alter the methylation status of the miR-26b-5p host gene CTDSP1
As DNA methylation is an important mechanism for miRNA deregulation and to investigate the potential mechanism of the down-regulation of miR-26b-5p, we further identified 3 typical CpG islands in the transcription start region of the CTDSP1 gene by MSP (Fig. 2A). Our MSP results showed that CTDSP1, the host gene of miR-26b-5p, had an unmethylated status in GC-2 cells at different magnetic intensities of ELF-EMF exposure (Fig. 2B). These findings indicated that the deregulation of miR-26b-5p is not related to the DNA methylation of the host gene.
Figure 2.
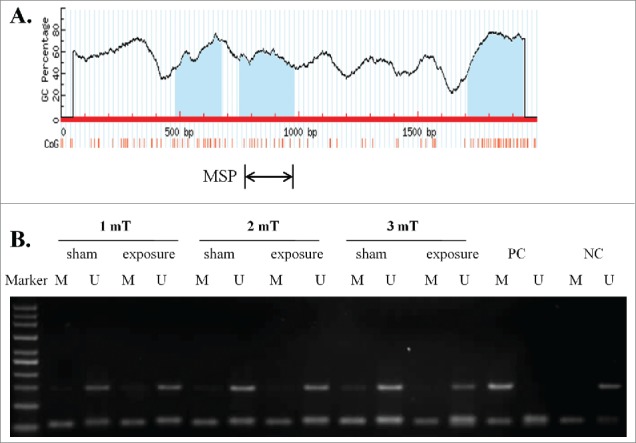
50 Hz ELF-EMF exposure does not alter the unmethylated status of the miR-26b-5p host gene CTDSP1. (A) Schematic of the CpG islands in the transcriptional region of CTDSP1. CpG sites are indicated by vertical orange lines. The regions of MSP are indicated. (B) CTDSP1 was unmethylated in GC-2 cells at different magnetic field intensities of ELF-EMF exposure. Each lane contains products that were amplified from separate PCRs using primers that were specific for methylated (M) or unmethylated (U) DNA templates. PC: positive control. NC: negative control.
MiR-26b-5p has no obvious influence on GC-2 cell growth
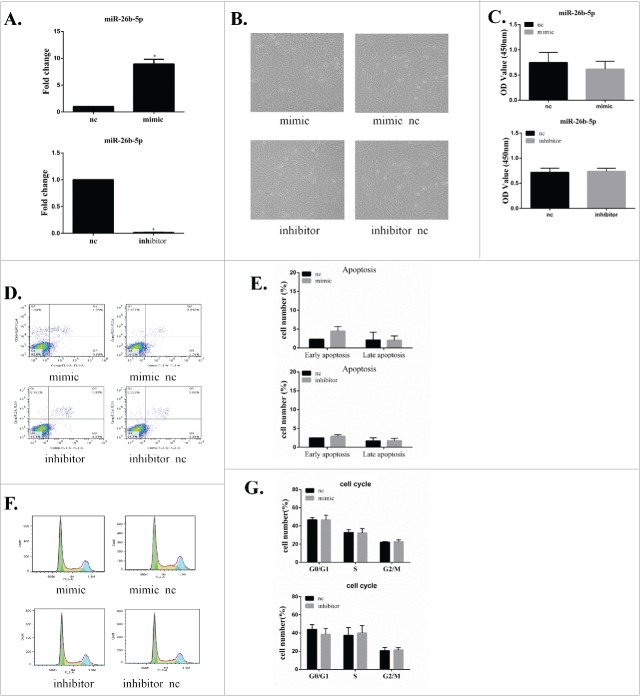
GC-2 cells were transfected for 6 h with a miR-26b-5p mimic, a mimic negative control (mimic-nc), a miR-26b-5p inhibitor or an inhibitor negative control (inhibitor-nc). The level of miR-26b-5p expression was used to evaluate the efficiency of cell transfection by qRT-PCR. The relative miR-26b-5p expression level in GC-2 cells was normalized to the mRNA level of the endogenous control gene U6. There was a significant increase in miR-26b-5p expression in GC-2 cells that were transfected with miR-26b-5p mimic (overexpressed group) compared to the mimic-nc group. The expression of miR-26b-5p in GC-2 cells that were transfected with miR-26b-5p inhibitor (knockdown group) was reduced significantly compared to that of the inhibitor-nc group. These results suggest that our protocol for the lipid-mediated miR-26b-5p transfection of GC-2 cells has high efficiency with undetectable cytotoxicity. Therefore, this protocol was suitable for use in the experiments described herein (Fig. 3A).
Figure 3.
MiR-26b-5p has no obvious influence on the cell growth, apoptosis or cell cycle of GC-2 cells. GC-2 cells were transfected for 6 h with miR-26b-5p mimic, mimic-nc, miR-26b-5p inhibitor or inhibitor-nc. (A) Evaluation of miR-26b-5p efficiency by real-time RT-PCR. (B) Representative images of the morphology of GC-2 cells transfected with miR-26b-5p. (C) MiR-26b-5p had no obvious influence on the cell viability of GC-2 cells at 24 and 48 h. Cell viability was examined by the CCK-8 assay. (D) Representative dot plot results of the apoptosis analysis by flow cytometry assays with Annexin V-FITC and PI double staining in GC-2 cells. (E) Quantitative analysis of GC-2 cell apoptosis. (F) Representative results of the cell cycle analysis by flow cytometry with PtdIns staining in GC-2 cells. (G) The percentages of cell cycle are represented by bar graphs. The data are expressed as the mean ± SD of 3 independent experiments.
GC-2 cells were transfected with miR-26b-5p mimics or mimic-nc, inhibitor or inhibitor-nc and then subjected to a cell viability analysis at 24 h and 48 h. MiR-26b-5p had no obvious influence on the morphology of GC-2 cells (Fig. 3B). The results of the cell proliferation assay showed that the overexpression or knockdown of miR-26b-5p had no obvious influence on the cell viability of GC-2 cells between the mimic, inhibitor and nc groups (Fig. 3C).
MiR-26b-5p does not induce apoptosis or cell cycle arrest in GC-2 cells
Apoptosis and the cell cycle are important characteristics that are associated with the process of proliferation. To further explore the effects of miR-26b-5p on GC-2 cells, apoptosis and the cell cycle of GC-2 cells after transfection were analyzed by flow cytometry. MiR-26b-5p did not induce apoptosis in GC-2 cells compared to the nc group (Fig. 3D-E). No significant difference was found in the percentages of G1, S, and G2 phase cells between GC-2 cells after transfection (Fig. 3F-G). These findings suggest that miR-26b-5p exerts no obvious effect on the proliferation of GC-2 cells.
Effect of miR-26b-5p on cell viability in GC-2 cells following 50 Hz ELF-EMF exposure
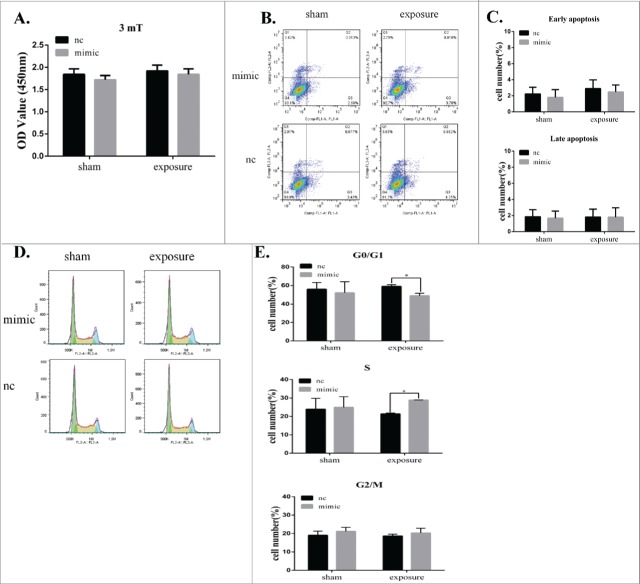
Given that miR-26b-5p was downregulated in GC-2 cells at a magnetic field intensity of 3 mT, we transiently transfected miR-26b-5p mimic into GC-2 cells to investigate the effects of miR-26b-5p following 50 Hz ELF-EMF. GC-2 cells were transfected for 6 h with a miR-26b-5p mimic and mimic-nc and then exposed to a 50 Hz ELF-EMF at 3 mT. After ELF-EMF radiation, no significant differences in cell viability were detected between the sham and exposure groups.(Fig. 4A).
Figure 4.
MiR-26b-5p regulates the G1/S phase transition in GC-2 cells following 50 Hz ELF-EMF exposure. GC-2 cells were transfected for 6 h with a miR-26b-5p mimic and a mimic negative control and then exposed to a 50 Hz ELF-EMF at a magnetic field intensity of 3 mT. (A) The cell viability showed no significant differences between the sham and exposure groups. (B) Representative dot plots of the flow cytometry assays in GC-2 cells. (C) Quantitative analysis of apoptosis in GC-2 cells. (D) Representative results of the cell cycle analysis in GC-2 cells. (E) The percentages of the cell cycle are represented with bar graphs. The data were expressed as the mean ± SD of 3 independent experiments.
MiR-26b-5p can regulate the G1/S phase transition in GC-2 cells following 50 Hz ELF-EMF
To further confirm the effect of miR-26b-5p on apoptosis and the cell cycle in GC-2 cells following 50 Hz ELF-EMF, apoptosis and the cell cycle were analyzed by flow cytometry. GC-2 cells overexpressing miR-26b-5p prior to 50 Hz ELF-EMF exposure showed no obvious change in apoptosis (Fig. 4B-C). However, the overexpression of endogenous miR-26b-5p significantly decreased the percentage of G0/G1 phase cells and slightly increased the percentage of S phase cells compared to the control group after 50 Hz ELF-EMF exposure (Fig. 4D-E). We also confirmed the expression of miR-26b-5p in GC-2 cell line and germ cells in the testes of mouse by real-time PCR (Supplementary Fig. 1A). The expression level of miR-26b-5p in germ cells of mouse was lower than that of in GC-2 cell line. According to the role of miR-26b-5p in GC-2 cells exposed to ELF-EMFs, lower expression of miR-26b-5p in germ cells suggested it was more sensitive to the EMFs. These results clearly indicate that miR-26b-5p plays an important role in modulating the cell cycle in GC-2 cells following 50 Hz ELF-EMF exposure.
CCND2 is a direct target of miR-26b-5p
To further elucidate the mechanism of miR-26b-5p participation in the regulation of ELF-EMFs, we identified the potential target genes of miR-26b-5p. A total of 85 genes were putative target genes of miR-26b-5p (Fig. 5A) as determined by computational prediction using 3 algorithms (TargetScan, PicTar and PITA). Among the target genes that were predicted from TargetScan, PicTar and PITA, we chose some target genes, such as USP25, PRKCD, FNIP1, EZH2 and CCND2, that are involved in the cell cycle and apoptosis. We found a negative correlation between miR-26b-5p and its potential target genes. Given that miR-26b-5p can regulate the G1/S phase transition in GC-2 cells following 50 Hz ELF-EMF, we focused on CCND2, which is a crucial cell cycle regulatory gene, as a direct target of miR-26b-5p. The effect of miR-26b-5p transfection on CCND2 mRNA and protein expression was subsequently measured in GC-2 cells by qRT-PCR and Western blotting. As shown in Figure 5B and 5C, the expression of CCND2 mRNA and protein decreased in GC-2 cells that were transfected with 100 nM miR-26b-5p mimics and increased in GC-2 cells that were transfected with 100 nM miR-26b-5p inhibitor compared to their negative controls, respectively. To investigate whether miR-26b-5p is involved in the regulation of CCND2 expression, we analyzed miR-26b-5p binding sites in the 3′-UTR of CCND2 and its cognate miRNAs using a bioinformatics approach. Using the TargetScan Release 6.2 database, we found a binding site for miR-26b in the 3′-UTR of CCND2 mRNA (Fig. 5D). These results suggest that miR-26b-5p can regulate the expression of CCND2 by directly targeting its 3′-UTR.
Figure 5.
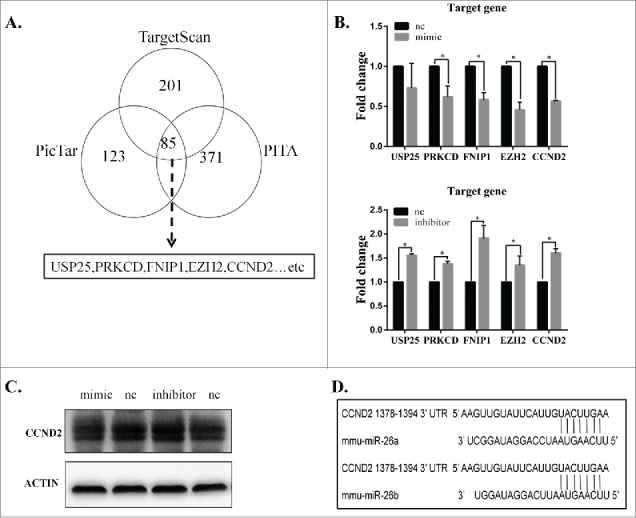
CCND2 is a direct target of miR-26b-5p. (A) Prediction results using 3 bioinformatics algorithms (TargetScan, PicTar and PITA). We identified USP25, PRKCD, FNIP1, EZH2 and CCND2 as potential target genes for further investigation. (B) The mRNA expression of potential target genes was measured by qRT-PCR. The mRNA expression showed a negative correlation between miR-26b-5p and the potential target genes. (C) CCND2 protein expression was analyzed using Western blotting. MiR-26b-5p can regulate the protein levels of CCND2. (D) The schematic representation of the binding sites for miR-26b-5p in the 3’-UTR of CCND2 using TargetScan software.
50 Hz ELF-EMF can alter the mRNA expression and protein expression of CCND2
To further confirm the effect of 50 Hz ELF-EMF, the mRNA expression and protein expression of CCND2 was measured by real-time PCR and Western blotting, respectively. As shown in Figure 6A, the expression of CCND2 was significantly lower in the group at a magnetic intensity of 2 mT than that in the sham group and was significantly higher than that of the sham group at 3 mT compared to that of the sham exposure group; however, there was no obvious changes in CCND2 expression between the exposure and sham groups at 1 mT. The protein expression of CCND2 was consistent with the mRNA expression (Fig. 6B). Most importantly, the expression of CCND2 was negatively correlated with the expression miR-26b-5p. These results suggest that 50 Hz ELF-EMF can alter the expression of CCND2 and that miR-26b-5p might play an important role in this process.
Figure 6.
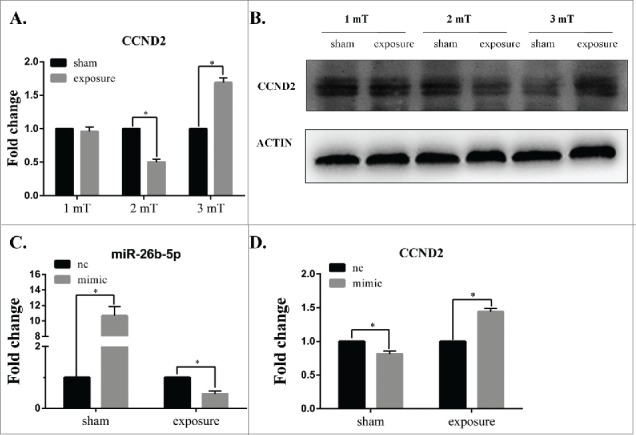
Overexpressed miR-26b-5p inhibits the expression of CCND2 following 50 Hz ELF-EMF exposure at 3 mT in GC-2 cells. (A) The expression of CCND2 was significantly lower than that of the sham group at a magnetic intensity of 2 mT and was significantly higher than that of the sham group at 3 mT compared to that of the sham-exposure group, while there were no obvious changes in CCND2 expression between the exposure and sham groups at 1 mT. (B) The protein expression of CCND2 was consistent with the results of mRNA expression detection. (C) The expression of miR-26b-5p was significantly higher than the negative control following transfection, and 50 Hz ELF-EMF exposure decreased the expression of miR-26b-5p. (D) The overexpression of miR-26b-5p in GC-2 cells can lower the expression of CCND2. A 50 Hz ELF-EMF can increase the expression of CCND2 by down-regulating the expression of miR-26b-5p compared to that of the sham group.
Overexpressed miR-26b-5p in GC-2 cells can change the expression of CCND2 following 50 Hz ELF-EMF at 3 mT
Following transfection with miR-26b-5p mimic and mimic-nc for 6 h, GC-2 cells were exposed to 50 Hz ELF-EMF at 3 mT. We then determined the levels of miR-26b-5p and CCND2 by qRT-PCR. As shown in Figure 6C, the expression of miR-26b-5p was significantly higher than the negative control following transfection. After 50 Hz ELF-EMF, the expression of miR-26b-5p was lower than the negative control. These results further confirmed the effect of 50 Hz ELF-EMF on the expression of miR-26b-5p. Overexpressed miR-26b-5p in GC-2 cells can inhibit the expression of CCND2. However, the expression of CCND2 increased when exposed to 50 Hz ELF-EMF compared to the sham group (Fig. 6D). Taken together, 50 Hz ELF-EMF and miR-26b-5p overexpression were strongly attributed to the mRNA expression of CCND2. These data further confirm that miR-26b-5p deregulation may play an important role in the regulation of the cell cycle in GC-2 cells that were exposed to 50 Hz ELF-EMF.
Discussion
The major findings of this study show that intermittent exposure to a 50 Hz ELF-EMF altered the expression of miR-26b-5p in GC-2 cells and that the overexpression of miR-26b-5p could block the G1/S phase transition following 50 Hz ELF-EMF exposure by directly targeting the 3′-UTR of CCND2 and simultaneously degrading the mRNA expression of CCND2. Thus, we showed for the first time that the regulation of miR-26b-5p might play an important role in the biological effects of a 50 Hz ELF-EMF.
Given the wide use of power supply lines, as well as many household and commercial devices in everyday life, whether ELF-EMF exposure can affect the male reproductive system is a topic of concern. A large body of evidence has confirmed that exposure to ELF-EMFs can induce spermatogenic cell apoptosis, decrease the numbers of spermatogenic cells and significantly increase abnormalities in cells.5 However, the underlying molecular mechanism of this influence remains unknown.
Recently, miRNAs have been shown to play an important role in the control of reproductive functions, especially in the processes of oocyte maturation, implantation and early embryonic development.23,24 The aberrant expression of specific miRNAs is also linked to certain male reproductive disorders.25,26 The miR-26 family comprises miR-26a and miR-26b. MiR-26a and miR-26b are important for lung development and human breast cancer.14,15 Furthermore, miR-26a also suppresses cell proliferation and activates tumor-specific apoptosis in nasopharyngeal carcinoma and liver cancer.27,28 In our study, we found that miR-26b-5p was significantly upregulated at a magnetic field intensity of 2 mT and significantly down-regulated at 3 mT compared to the sham group. Thus, miR-26b-5p may serve as a candidate biomarker of early-stage ELF-EMF exposure and may participate in the biological effects of ELF-EMFs.
Many studies have shown that the expression of miRNAs is linked to the DNA methylation of their host gene.29-31 CTDSP1 is the host gene of miR-26b-5p. Therefore, we identified the methylation status of CTDSP1 via MSP. The MSP results showed an unmethylated status in GC-2 cells at different electric field intensities of ELF-EMF exposure, suggesting that the deregulation of miR-26b-5p might have no relationship with the DNA methylation of the host gene. The deregulation of miR-26b-5p in response to different magnetic field intensities of 50 Hz ELF-EMF may have different biological effects and requires further investigation.
Previous studies have demonstrated that miR-26b overexpression inhibited cellular growth by targeting PTGS2.32 Liu et al. found that miR-26b could promote the apoptosis of MCF7 cells and directly regulate the expression of SLC7A11.15 In our study, miR-26b-5p had no obvious influence on the GC-2 cell growth. These results are not consistent with previous studies, possibly due to the different cell model. Spermatocyte-derived GC-2 cells are normal mouse cells and may be have the ability to balance the expression of some key regulatory molecules.20,33
To the best of our knowledge, previous studies have not addressed the biological effects of ELF-EMF-induced changes in miR-26b-5p expression in the GC-2 cell line. The overexpression of miR-26b-5p significantly decreased the percentage of G0/G1 phase cells and slightly increased percentage of S phase cells following 50 Hz ELF-EMF exposure. Although there were no detectable changes in the viability of GC-2 cells, we speculate that long-term ELF-EMF exposure might regulate some key signaling pathways, thereby inducing alterations in cell growth. ELF-EMFs can induce a significant reduction in miR-30a expression and affect autophagy via Beclin1 expression in human neuroblastoma cells.34 Dasdag et al. reported that 2.4 GHz radiofrequency radiation emitted from Wi-Fi equipment can alter the expression of some miRNAs, such as miR-106b-5p and miR-107, in brain tissue, which may have adverse effects, such as neurodegenerative diseases.35 In addition, the long-term and excessive use of 900 MHz radiofrequency radiation can alter miRNA expression in the brain.36 These findings support our results that a 50 Hz ELF-EMF can alter the expression of miR-26b-5p, which may account for the biological effects of ELF-EMF.
Given that miR-26b-5p plays a pivotal role in GC-2 cells following 50 Hz ELF-EMF exposure, it is necessary to clarify the precise mechanism of how miR-26b contributes to ELF-EMF effects. Computational prediction was used to predict the putative target genes of miR-26b-5p. Among the target genes that were predicted from miRBase, PicTar and PITA, we chose some target genes, including USP25, PRKCD, FNIP1, EZH2 and CCND2, to test and verify by qRT-PCR. We found a negative correlation between miR-26b-5p and its potential target genes. USP25 (ubiquitin-specific protease 25) can regulate TLR4-dependent innate immune responses through the deubiquitination of the adaptor protein TRAF3 and negatively regulate IL-17-triggered signaling.37,38 PRKCD is involved in the regulation of a variety of cellular functions, including apoptosis and cell growth. The overexpression of PRKCD can induce phenotypic changes indicative of apoptosis in several cell types.39,40 The folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn.41 Nicholas et al. reported that Fnip1 can also regulate skeletal muscle fiber-type specification and susceptibility to muscular dystrophy.42 EZH2 is a histone lysine methyltransferase that is part of the PRC2 polycomb repressive complex. EZH2 overexpression occurs in multiple cancers, including breast cancer, lung cancer and prostate cancer.14,43 CCND2 is a crucial cell cycle regulatory gene, and its aberrant expression can lead to abnormal cell proliferation.44,45 The aberrant expression of CCND2 has been observed for testicular germ cell tumor cell lines,46 gastric cancer,47 and colorectal cancer,48 and its overexpression has been reported in prostate cancer tissues and cell lines.49
According to our results, the overexpression of miR-26b-5p can decrease the percentage of G0/G1 phase cells and can slightly increase the percentage of S phase cells of the total GC-2 cells following 50 Hz ELF-EMF exposure. Therefore, we focused on CCND2 as a direct target of miR-26b-5p. Further computational algorithms revealed that the 3′-UTR of CCND2 contains a binding site for miR-26b. Recently, consistent findings have also reported that miR-26a and miR-26b can block the G1/S phase transition by directly targeting the 3′-UTR of CCND2.50-52 MiR-26a and miR-26b belong to the miR-26 family, whose conserved region has the same seed sequence ‘AUGAACU′. Our findings suggest that miR-26b-5p could regulate the expression of CCND2 by directly targeting its 3′-UTR. Cell cycle progression is precisely regulated by a series of cell cycle regulators, including cyclins, CDKs and CDK inhibitors (CDKIs).53,54 CCND2 can be regulated by miR-26a, miR-154, miR-206 and miR-29a.50,55-57 Herein, CCND2 was negatively regulated by miR-26b-5p in GC-2 cells at 2 mT and 3 mT. However, there was no obvious change in CCND2 or miR-26b-5p expression between the exposure and sham groups at 1 mT. Therefore, further research is required to illustrate this regulation mechanism.
Taken together, we demonstrate that intermittent exposure to a 50 Hz ELF-EMF can alter the expression of miR-26b-5p. MiR-26b-5p can directly regulate the expression of CCND2 and induce the G1/S phase transition by directly targeting the 3′-UTR of CCND2. Our findings suggest that miR-26b-5p can be used as a biomarker of early-stage ELF-EMF exposure, and the miR-26b-5p-CCND2-mediated regulation of the cell cycle might play a key role in the biological effects of ELF-EMFs. These claims require further investigation.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Basic Research Program of China (National 973 Program) (Grant No. 2011CB503700).
References
- 1.Pedersen C, Raaschou-Nielsen O, Rod NH, Frei P, Poulsen AH, Johansen C, Schuz J. Distance from residence to power line and risk of childhood leukemia: a population-based case-control study in Denmark. Cancer Causes Control 2014; 25:171–7; PMID:24197706; http://dx.doi.org/ 10.1007/s10552-013-0319-5 [DOI] [PubMed] [Google Scholar]
- 2.Grellier J, Ravazzani P, Cardis E. Potential health impacts of residential exposures to extremely low frequency magnetic fields in Europe. Environ Int 2014; 62:55–63; PMID:24161447; http://dx.doi.org/ 10.1016/j.envint.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 3.Lee JS, Ahn SS, Jung KC, Kim YW, Lee SK. Effects of 60 Hz electromagnetic field exposure on testicular germ cell apoptosis in mice. Asian J Androl 2004; 6:29–34; PMID:15064831 [PubMed] [Google Scholar]
- 4.Kim YW, Kim HS, Lee JS, Kim YJ, Lee SK, Seo JN, Jung KC, Kim N, Gimm YM. Effects of 60 Hz 14 microT magnetic field on the apoptosis of testicular germ cell in mice. Bioelectromagnetics 2009; 30:66–72; PMID:18839413; http://dx.doi.org/ 10.1002/bem.20448 [DOI] [PubMed] [Google Scholar]
- 5.Roychoudhury S, Jedlicka J, Parkanyi V, Rafay J, Ondruska L, Massanyi P, Bulla J. Influence of a 50 hz extra low frequency electromagnetic field on spermatozoa motility and fertilization rates in rabbits. J Environ Sci Health A Tox Hazard Subst Environ Eng 2009; 44:1041–7; PMID:19827497; http://dx.doi.org/ 10.1080/10934520902997029 [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 7.Moreno-Moya JM, Vilella F, Simon C. MicroRNA: key gene expression regulators. Fertil Steril 2014; 101:1516–23; PMID:24314918; http://dx.doi.org/ 10.1016/j.fertnstert.2013.10.042 [DOI] [PubMed] [Google Scholar]
- 8.Lai EC. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet 2002; 30:363–4; PMID:11896390; http://dx.doi.org/ 10.1038/ng865 [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. microRNAs: tiny regulators with great potential. Cell 2001; 107:823–6; PMID:11779458; http://dx.doi.org/ 10.1016/S0092-8674(01)00616-X [DOI] [PubMed] [Google Scholar]
- 10.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett 2011; 585:2087–99; PMID:20708002; http://dx.doi.org/ 10.1016/j.febslet.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 11.Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Curr Opin Oncol 2010; 22:35–45; PMID:19907325; http://dx.doi.org/ 10.1097/CCO.0b013e328333dcbb [DOI] [PubMed] [Google Scholar]
- 12.Imbar T, Galliano D, Pellicer A, Laufer N. Introduction: MicroRNAs in human reproduction: small molecules with crucial regulatory roles. Fertil Steril 2014; 101:1514–5; PMID:24882615; http://dx.doi.org/ 10.1016/j.fertnstert.2014.04.030 [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Xu C. Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction 2015; 149:R127–37; PMID:25352684; http://dx.doi.org/ 10.1530/REP-14-0239 [DOI] [PubMed] [Google Scholar]
- 14.Wu T, Chen W, Liu S, Lu H, Wang H, Kong D, Huang X, Kong Q, Ning Y, Lu Z. Huaier suppresses proliferation and induces apoptosis in human pulmonary cancer cells via upregulation of miR-26b-5p. FEBS Lett 2014; 588:2107–14; PMID:24815696; http://dx.doi.org/ 10.1016/j.febslet.2014.04.044 [DOI] [PubMed] [Google Scholar]
- 15.Liu XX, Li XJ, Zhang B, Liang YJ, Zhou CX, Cao DX, He M, Chen GQ, He JR, Zhao Q. MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett 2011; 585:1363–7; PMID:21510944; http://dx.doi.org/ 10.1016/j.febslet.2011.04.018 [DOI] [PubMed] [Google Scholar]
- 16.Duan G, Ren C, Zhang Y, Feng S. MicroRNA-26b inhibits metastasis of osteosarcoma via targeting CTGF and Smad1. Tumour Biol 2015; 36(8):6201–9 [DOI] [PubMed] [Google Scholar]
- 17.Verghese ET, Drury R, Green CA, Holliday DL, Lu X, Nash C, Speirs V, Thorne JL, Thygesen HH, Zougman A, et al. MiR-26b is downregulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J Pathol 2013; 231:388–99; PMID:23939832; http://dx.doi.org/ 10.1002/path.4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang N, Zhou X, Zhao M, Zhao D, Zhu Z, Li S, Yang H. Downregulation of microRNA-26b modulates non-small cell lung cancer cells chemoresistance and migration through the association of PTEN. Acta Biochim Biophys Sin (Shanghai) 2015; 47:530–8; PMID:26068649; http://dx.doi.org/ 10.1093/abbs/gmv046 [DOI] [PubMed] [Google Scholar]
- 19.Guan K, Wolf F, Becker A, Engel W, Nayernia K, Hasenfuss G. Isolation and cultivation of stem cells from adult mouse testes. Nat Protoc 2009; 4:143–54; PMID:19180086; http://dx.doi.org/ 10.1038/nprot.2008.242 [DOI] [PubMed] [Google Scholar]
- 20.Ma Q, Deng P, Zhu G, Liu C, Zhang L, Zhou Z, Luo X, Li M, Zhong M, Yu Z, et al. Extremely low-frequency electromagnetic fields affect transcript levels of neuronal differentiation-related genes in embryonic neural stem cells. PLoS One 2014; 9:e90041; PMID:24595264; http://dx.doi.org/ 10.1371/journal.pone.0090041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuderer J, Oesch W, Felber N, Spat D, Kuster N. In vitro exposure apparatus for ELF magnetic fields. Bioelectromagnetics 2004; 25:582–91; PMID:15515036; http://dx.doi.org/ 10.1002/bem.20037 [DOI] [PubMed] [Google Scholar]
- 22.Han F, Liu W, Jiang X, Shi X, Yin L, Ao L, Cui Z, Li Y, Huang C, Cao J, et al. SOX30, a novel epigenetic silenced tumor suppressor, promotes tumor cell apoptosis by transcriptional activating p53 in lung cancer. Oncogene 2014; 34(33):4391-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development 2011; 138:1653–61; PMID:21486922; http://dx.doi.org/ 10.1242/dev.056234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev 2007; 21:644–8; PMID:17369397; http://dx.doi.org/ 10.1101/gad.418707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbar T, Eisenberg I. Regulatory role of microRNAs in ovarian function. Fertil Steril 2014; 101:1524–30; PMID:24882616; http://dx.doi.org/ 10.1016/j.fertnstert.2014.04.024 [DOI] [PubMed] [Google Scholar]
- 26.Galliano D, Pellicer A. MicroRNA and implantation. Fertil Steril 2014; 101:1531–44; PMID:24882617; http://dx.doi.org/ 10.1016/j.fertnstert.2014.04.023 [DOI] [PubMed] [Google Scholar]
- 27.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med 2009; 361:1437–47; PMID:19812400; http://dx.doi.org/ 10.1056/NEJMoa0901282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji Y, He Y, Liu L, Chong X. MiRNA-26b regulates the expression of cyclooxygenase-2 in desferrioxamine-treated CNE cells. FEBS Lett 2010; 584:961–7; PMID:20100477; http://dx.doi.org/ 10.1016/j.febslet.2010.01.036 [DOI] [PubMed] [Google Scholar]
- 29.Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev 2012; 26:25–30; PMID:22215807; http://dx.doi.org/ 10.1101/gad.177774.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia H, Zhang Z, Zou D, Wang B, Yan Y, Luo M, Dong L, Yin H, Gong B, Li Z, et al. MicroRNA-10a is down-regulated by DNA methylation and functions as a tumor suppressor in gastric cancer cells. PLoS One 2014; 9:e88057; PMID:24498243; http://dx.doi.org/ 10.1371/journal.pone.0088057 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang Q, Wang LQ, Wong KY, Li ZY, Chim CS. Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple myeloma. J Clin Pathol 2015; 68:557–61; PMID:25855800; http://dx.doi.org/ 10.1136/jclinpath-2014-202817 [DOI] [PubMed] [Google Scholar]
- 32.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int 2013; 13:7; PMID:23374284; http://dx.doi.org/ 10.1186/1475-2867-13-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Ma Q, Liu C, Deng P, Zhu G, Zhang L, He M, Lu Y, Duan W, Pei L, et al. Exposure to 1800 MHz radiofrequency radiation impairs neurite outgrowth of embryonic neural stem cells. Sci Rep 2014; 4:5103; PMID:24869783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchesi N, Osera C, Fassina L, Amadio M, Angeletti F, Morini M, Magenes G, Venturini L, Biggiogera M, Ricevuti G, et al. Autophagy is modulated in human neuroblastoma cells through direct exposition to low frequency electromagnetic fields. J Cell Physiol 2014; 229:1776–86; PMID:24676932; http://dx.doi.org/ 10.1002/jcp.24631 [DOI] [PubMed] [Google Scholar]
- 35.Dasdag S, Akdag MZ, Erdal ME, Erdal N, Ay OI, Ay ME, Yilmaz SG, Tasdelen B, Yegin K. Effects of 2.4 GHz radiofrequency radiation emitted from Wi-Fi equipment on microRNA expression in brain tissue. Int J Radiat Biol 2015; 91(7):555–61, 1-7 [DOI] [PubMed] [Google Scholar]
- 36.Dasdag S, Akdag MZ, Erdal ME, Erdal N, Ay OI, Ay ME, Yilmaz SG, Tasdelen B, Yegin K. Long term and excessive use of 900 MHz radiofrequency radiation alter microRNA expression in brain. Int J Radiat Biol 2015; 91:306–11; PMID:25529971; http://dx.doi.org/ 10.3109/09553002.2015.997896 [DOI] [PubMed] [Google Scholar]
- 37.Zhong B, Liu X, Wang X, Chang SH, Wang A, Reynolds JM, Dong C. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat Immunol 2012; 13:1110–7; PMID:23042150; http://dx.doi.org/ 10.1038/ni.2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong B, Liu X, Wang X, Li H, Darnay BG, Lin X, Sun SC, Dong C. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci Signal 2013; 6:ra35; PMID:23674823; http://dx.doi.org/ 10.1126/scisignal.2003708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Okamoto M, Panzhinskiy E, Zawada WM, Das M. PKCdelta/midkine pathway drives hypoxia-induced proliferation and differentiation of human lung epithelial cells. Am J Physiol Cell Physiol 2014; 306:C648–58; PMID:24500281; http://dx.doi.org/ 10.1152/ajpcell.00351.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia L, Wang TD, Shen SM, Zhao M, Sun H, He Y, Xie L, Wu ZX, Han SF, Wang LS, et al. Phosphoproteomics study on the activated PKCdelta-induced cell death. J Proteome Res 2013; 12:4280–301; PMID:23879269; http://dx.doi.org/ 10.1021/pr400089v [DOI] [PubMed] [Google Scholar]
- 41.Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, Matsuo M, Merino MJ, Yao M, Ito Y, et al. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc Natl Acad Sci U S A 2015; 112:E1624–31; PMID:25775561; http://dx.doi.org/ 10.1073/pnas.1419502112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes NL, Banks GB, Tsang M, Margineantu D, Gu H, Djukovic D, Chan J, Torres M, Liggitt HD, Hirenallur SD, et al. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc Natl Acad Sci U S A 2015; 112:424–9; PMID:25548157; http://dx.doi.org/ 10.1073/pnas.1413021112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget 2011; 2:669–83; PMID:21941025; http://dx.doi.org/ 10.18632/oncotarget.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malumbres M, Pevarello P, Barbacid M, Bischoff JR. CDK inhibitors in cancer therapy: what is next? Trends Pharmacol Sci 2008; 29:16–21; PMID:18054800; http://dx.doi.org/ 10.1016/j.tips.2007.10.012 [DOI] [PubMed] [Google Scholar]
- 45.Ando K, Ajchenbaum-Cymbalista F, Griffin JD. Regulation of G1/S transition by cyclins D2 and D3 in hematopoietic cells. Proc Natl Acad Sci U S A 1993; 90:9571–5; PMID:8415743; http://dx.doi.org/ 10.1073/pnas.90.20.9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skotheim RI, Abeler VM, Nesland JM, Fossa SD, Holm R, Wagner U, Florenes VA, Aass N, Kallioniemi OP, Lothe RA. Candidate genes for testicular cancer evaluated by in situ protein expression analyses on tissue microarrays. Neoplasia 2003; 5:397–404; PMID:14670177; http://dx.doi.org/ 10.1016/S1476-5586(03)80042-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, Luo M, Zhao H, Dong L, Song W, et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis 2014; 35:497–506; PMID:24130168; http://dx.doi.org/ 10.1093/carcin/bgt337 [DOI] [PubMed] [Google Scholar]
- 48.Bartkova J, Thullberg M, Slezak P, Jaramillo E, Rubio C, Thomassen LH, Bartek J. Aberrant expression of G1-phase cell cycle regulators in flat and exophytic adenomas of the human colon. Gastroenterology 2001; 120:1680–8; PMID:11375949; http://dx.doi.org/ 10.1053/gast.2001.24880 [DOI] [PubMed] [Google Scholar]
- 49.Dong Q, Meng P, Wang T, Qin W, Wang F, Yuan J, Chen Z, Yang A, Wang H. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One 2010; 5:e10147; PMID:20418948; http://dx.doi.org/ 10.1371/journal.pone.0010147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Liu L, Shen Y, Wang T, Chen L, Xu D, Wen F. MicroRNA-26a modulates transforming growth factor β-1-induced proliferation in human fetal lung fibroblasts. Biochem Biophys Res Commun 2014; 454:512–7; PMID:25451270; http://dx.doi.org/ 10.1016/j.bbrc.2014.10.106 [DOI] [PubMed] [Google Scholar]
- 51.Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, Li MF, Chen GQ, Zhao Q. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 2011; 32:2–9; PMID:20952513; http://dx.doi.org/ 10.1093/carcin/bgq209 [DOI] [PubMed] [Google Scholar]
- 52.Xu G, Ji C, Song G, Shi C, Shen Y, Chen L, Yang L, Zhao Y, Guo X. Obesityassociated microRNA26b regulates the proliferation of human preadipocytes via arrest of the G1/S transition. Mol Med Rep 2015; 12:3648–54; PMID:26016996 [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Chen J, Wang L, Huang Y, Leng Y, Wang G. Fangchinoline induces G0/G1 arrest by modulating the expression of CDKN1A and CCND2 in K562 human chronic myelogenous leukemia cells. Exp Ther Med 2013; 5:1105–12; PMID:23596478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks G. Cyclins, cyclin-dependent kinases, and cyclin-dependent kinase inhibitors: detection methods and activity measurements. Methods Mol Biol 2005; 296:291–8; PMID:15576940 [DOI] [PubMed] [Google Scholar]
- 55.Zhu C, Shao P, Bao M, Li P, Zhou H, Cai H, Cao Q, Tao L, Meng X, Ju X, et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol 2014; 32:31 e9-16; PMID:23428540; http://dx.doi.org/ 10.1016/j.urolonc.2012.11.013 [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Liu X, Jin H, Guo X, Xia L, Chen Z, Bai M, Liu J, Shang X, Wu K, et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett 2013; 332:94–101; PMID:23348698; http://dx.doi.org/ 10.1016/j.canlet.2013.01.023 [DOI] [PubMed] [Google Scholar]
- 57.Cao X, Wang J, Wang Z, Du J, Yuan X, Huang W, Meng J, Gu H, Nie Y, Ji B, et al. MicroRNA profiling during rat ventricular maturation: A role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS Lett 2013; 587:1548–55; PMID:23587482; http://dx.doi.org/ 10.1016/j.febslet.2013.01.075 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.