Gene regulatory networks (GRNs) integrate intrinsic and extrinsic signals to maintain cell identity, homeostasis and function. In stem cells, network models have been drafted to describe the molecular underpinnings of self-renewal and differentiation.1 However, previous studies in this area have proven to be transcription factor-centric, while molecular functions other than DNA binding and direct regulation of transcription have been largely overlooked.2 Recently, we described an integrative interaction network in mouse embryonic stem cells (mESCs) centered on a node that does not regulate transcription – a reported E3 ubiquitin ligase and newly characterized RNA-binding protein, Makorin 1 (MKRN1).3
We observed that MKRN1 was co-expressed with mESC pluripotency transcription factors in self-renewal and differentiation conditions and its promoter bound by OCT4,4 leading to our hypothesis that MKRN1 regulates pluripotency by its E3 ubiquitin ligase activity. However, MKRN1 inhibition did not alter self-renewal capacity. With our hypothesis disproved, we turned to an unbiasesd systems approach to elucidate the function of MKRN1 in mESCs (Fig. 1).
Figure 1.
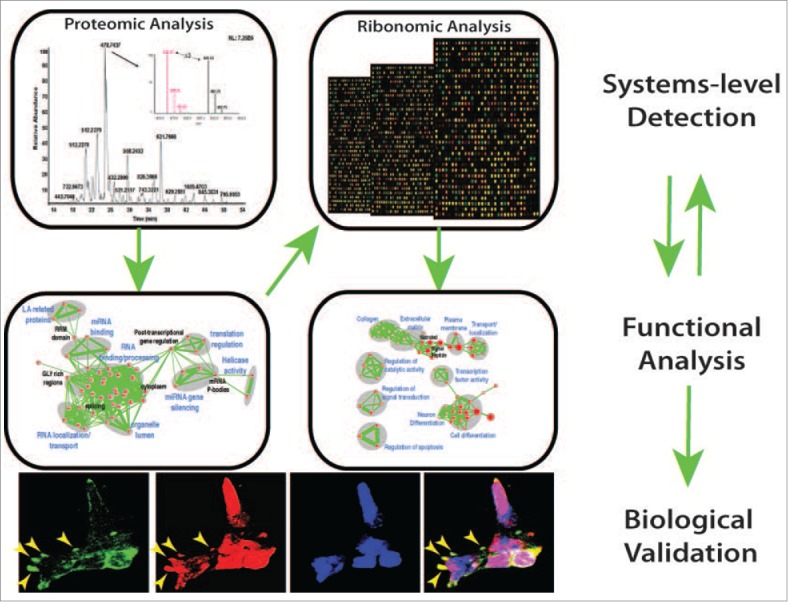
An unbiased systems approach was used to elucidate the role of MKRN1, a regulatory protein in mESCs. Proteomic analyses followed by functional annotation of interacting proteins uncovered a role in RNA-binding. Systems-level analyses of RNA-binding function revealed a role in RNA transport and cell death. Importantly, cellular and molecular assays were performed to confirm the functions inferred from systems-level datasets. We foresee broad applicability of this approach to identify the role of regulatory elements in a variety of cell types.
Initially, the protein interactome of MKRN1 was assessed by affinity purification-mass spectrometry. This analysis revealed that, despite the previous annotation of MKRN1 as an E3 ubiquitin ligase, MKRN1 associated proteins were not bound for proteasomal degradation, as treatment with a proteasome inhibitor did not affect the subset of proteins identified. Rather, MKRN1 was found associated with a variety of RNA-binding proteins, suggesting that MKRN1 is a component of the ribonucleoprotein complex. Because MKRN1 contains 4 C3H zinc finger domains, which are associated with RNA-binding capacity, the ability of MKRN1 to interact directly with RNA was assessed by UV crosslinking and immunoprecipitation (CLIP). This analysis indicated that MKRN1 indeed interacts directly with RNA, suggesting a previously uncharacterized RNA-binding function for MKRN1 in mESCs. The transcripts with which MKRN1 interacts were analyzed by RNA immunoprecipitation followed by microarray analysis (RIP-Chip). The transcripts pulled down with MKRN1 were generally expressed at low levels in undifferentiated mESCs. Functional analysis of the transcript interactome identified a strong enrichment for mRNAs encoding signal peptide-containing proteins, suggesting that MKRN1 may selectively interact with transcripts that are translated at the ER. Furthermore, enriched biological processes for MKRN1-bound transcripts included diverse terms such as signal transduction, differentiation, and regulation of apoptosis. We examined the apoptotic response of both MKRN1 knock-down and over-expression ESCs in unstressed conditions as well as in response to environmental and genotoxic stresses. Under basal conditions, manipulation of MKRN1 abundance did not affect ESC survival; however, following environmental stress, MKRN1 knock-down ESCs showed enhanced apoptosis, while genotoxic stress in overexpression ESCs resulted in a heightened apoptotic response. Collectively, this integrative analysis reveals that in mESCs, MKRN1 exhibits disparate function to its reported E3 ubiquitin ligase function. While MKRN1 may still possess E3 ubiquitin ligase activity in mESCs, its function as an RNA-binding protein suggests that MKRN1 may also contribute to a variety of gene regulatory processes, including regulation of apoptosis, transcript transport and control of translation.
Importantly, our findings underscore the importance of using unbiased approaches to interrogate gene function. We initially hypothesized that MKRN1 would play a role in mESC self-renewal given co-expression with and regulation by pluripotency factors. When this hypothesis was refuted through loss-of-function assays, we used an unbiased mass spectrometry assay to identify protein-protein interactions. Again, our hypothesis that MKRN1 targets would be destined for proteasome degradation based on previously reported ubiquitin ligase activity of MKRN1 was not supported by this experiment, but it instead suggested that MKRN1 functioned in ribonucleoprotein complexes with RNA-binding proteins with established roles in regulating RNA turnover, transport, and/or translational control. Finally, transcriptomic analysis of MKRN1-bound RNA suggested diverse roles for the novel RNA-binding protein, including transcript transport and regulation of programmed cell death.
Additionally, we centered our integrative genomics analysis around a node that is unrelated to gene transcription. Network-level analyses in stem cells have largely focused on transcriptional regulation by transcription factors,1 and recently, on epigenetic regulation.5,6 While post-transcriptional regulation has been recognized as an important element of GRNs, many efforts to include post-transcriptional control are still focused on species that control transcript abundance (i.e. microRNA).7 Here, we identify a component of the mESC GRN that does not regulate transcript abundance of its targets. Instead, MKRN1 appears to regulate the transport of target transcripts and contribute to stress responses. How MKRN1 binding affects the translation of its targets remains unknown, though the enrichment of low abundance transcripts in the MKRN1 interactome suggests that it may play a repressive role in translation. Furthermore, the interaction of MKRN1 with polyA binding protein (PABP) may implicate a role for MKRN1 in cap-dependent translation. The function of MKRN1 in stress response may be related to its potential role in translation repression, as cap-mediated translation repression may be circumvented by cap-independent translation, which occurs during genotoxic stress.
In conclusion, we have used an unbiased systems approach to identify MKRN1 function in mESCs. We found that contrary to expectations, MKRN1 is not required for self-renewal and does not target proteins for degradation. Instead, we uncovered a novel role for MKRN1 as an RNA-binding protein that does not regulate target transcript abundance, but instead plays a role in transcript transport and stress response. Thus, the integration of systems-level detection and functional analyses to elucidate the role of regulatory elements is a powerful approach that is broadly applicable in a variety of cell types.
References
- 1.Chen X, et al.. Cell 2008; 133:1106-17. 18555785; PMID:18555785; http://dx.doi.org/ 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- 2.Cassar PA, et al.. J Cell Physiol 2012; 227:439-49. 21503874; PMID:21503874; http://dx.doi.org/ 10.1002/jcp.22787 [DOI] [PubMed] [Google Scholar]
- 3.Cassar PA, et al.. EMBO Rep 2015; 16:1334-57. 26265008; PMID:26265008; http://dx.doi.org/ 10.15252/embr.201540974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker E, et al.. Cell Stem Cell 2007; 1:71-86. 18371337; PMID:18371337; http://dx.doi.org/ 10.1016/j.stem.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 5.Ang YS, et al.. Cell 2011; 145:183-97. 21477851; PMID:21477851; http://dx.doi.org/ 10.1016/j.cell.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker E, et al.. Cell Stem Cell 2010; 6:153-66. 20144788; PMID:20144788; http://dx.doi.org/ 10.1016/j.stem.2009.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez NJ, et al.. Cell Stem Cell 2010; 7:31-5. 20621047; PMID:20621047; http://dx.doi.org/ 10.1016/j.stem.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]


