ABSTRACT
Cohesin is required for ES cell self-renewal and iPS-mediated reprogramming of somatic cells. This may indicate a special role for cohesin in the regulation of pluripotency genes, perhaps by mediating long-range chromosomal interactions between gene regulatory elements. However, cohesin is also essential for genome integrity, and its depletion from cycling cells induces DNA damage responses. Hence, the failure of cohesin-depleted cells to establish or maintain pluripotency gene expression could be explained by a loss of long-range interactions or by DNA damage responses that undermine pluripotency gene expression. In recent work we began to disentangle these possibilities by analyzing reprogramming in the absence of cell division. These experiments showed that cohesin was not specifically required for reprogramming, and that the expression of most pluripotency genes was maintained when ES cells were acutely depleted of cohesin. Here we take this analysis to its logical conclusion by demonstrating that deliberately inflicted DNA damage - and the DNA damage that results from proliferation in the absence of cohesin - can directly interfere with pluripotency and reprogramming. The role of cohesin in pluripotency and reprogramming may therefore be best explained by essential cohesin functions in the cell cycle.
KEYWORDS: cell cycle, cohesin, enhancer-promoter interactions, gene expression, pluripotency, reprogramming, stress
Introduction
Several studies reported an essential role for cohesin in ES cell self-renewal and in the iPS-mediated reprogramming of somatic cells to pluripotency.1-4 Given that in mammalian cells cohesin associates with CTCF, 5-8 NIPBL, Mediator and cell-type specific transcription factors1,9,10 at gene regulatory elements and can mediate long-range chromosomal interactions,1-4,11-16 these data suggested a special place for cohesin in the network of pluripotency where it enables the expression of pluripotency genes by forming connections between their regulatory elements.
However, cohesin has essential functions in preserving the integrity of the genome through the cell cycle. Cohesin consists of a heterodimer of SMC (structural maintenance of chromosomes) proteins - SMC1A and SMC3, and 2 non-SMC proteins - RAD21 and either STAG1 or STAG2 and forms a ring-like structure with a diameter of 40 nm. This is large enough to topologically entrap 2 strands of nucleosomal DNA.17,18 Cohesin's association with chromatin is carefully regulated during the cell cycle to facilitates its cell cycle-dependent and cell cycle-independent functions19 (Fig. 1A). In vertebrate cells, cohesin loading onto DNA is initiated in telophase20,21 and requires the activity of the cohesin loading factor NIPBL and its partner MAU2.22-24 During interphase, cohesin association with DNA is maintained in a dynamic equilibrium by the opposing unloading actions of the WAPL and PDS5 proteins.25 Current models suggest that cohesin acts as a transcriptional regulator and genome organizer by forming chromatin interactions between distant DNA regions.26,27 Locally, cohesin mediated enhancer-promoter interactions facilitate the rearrangement of the T cell receptor α chain locus Tcra in non-proliferating thymocytes.12 On a global scale, cohesin associated with CTCF at the boundaries of topologically associating domains (TADs) is important for the structural organization of the genome. Loss of cohesin allows increased inter-domain interactions across TAD boundaries28 and while architectural chromatin compartments are not affected, cohesin is required for specific interactions within the compartments.29 In S phase, cohesin facilitates DNA replication.30-32 The acetylation of SMC3 by ESCO1/2 establishes stable cohesin binding to DNA.33,34 Once stably bound, cohesin holds the sister chromatids together until they segregate during mitosis. The proximity of replicated DNA strands provided by cohesin also enables homology-based repair of post-replicative DNA lesions.35,36 After the onset of mitosis, most of the cohesin associated with chromosome arms is removed by the prophase pathway and the small fraction of cohesin retained at centromeres allows the continued alignment of chromosomes at the metaphase plate following spindle attachment. Cleavage of centromeric cohesin by separase at the onset of anaphase then facilitates the segregation of sister chromatids to daughter cells.37
Figure 1.
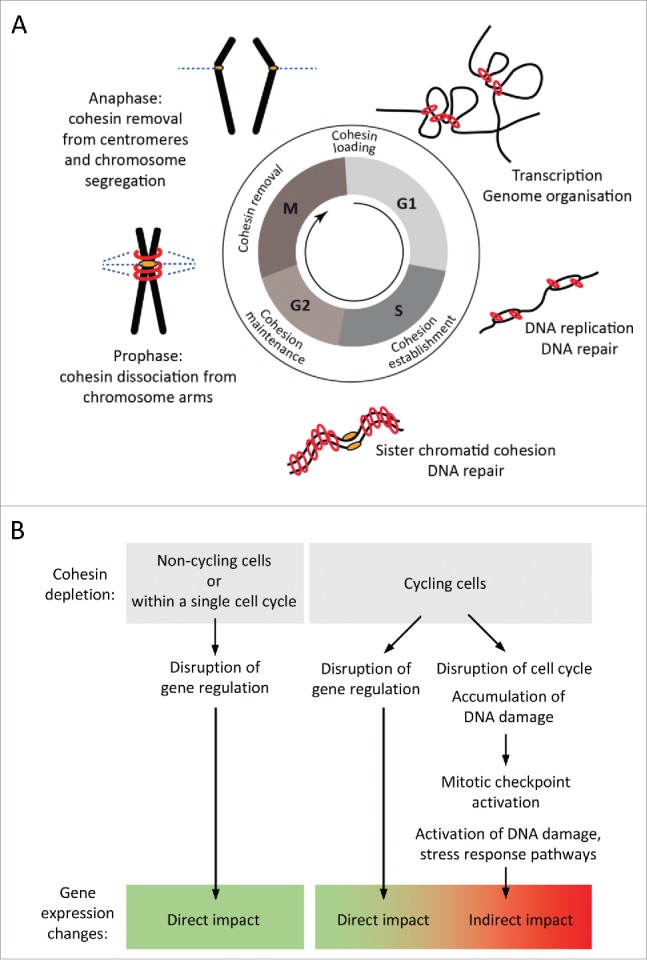
Cohesin functions during the cell cycle. (A) Cohesin dynamics during the cell cycle (see text for details). (B) Cohesin depletion in dividing cells can disrupt its cell cycle functions and indirectly impact gene expression due to the activation of mitotic checkpoints and cellular stress response pathways.
RNAi-mediated knockdown has been widely used to probe cohesin's role in gene regulation, and RNAi screens identified cohesin as a factor required for the self-renewal of pluripotent embryonic stem (ES) cells.1,38-40 However, gene expression analysis in ES cells 5 d after cohesin knockdown 1 revealed a preferential deregulation of genes related to cell cycle and DNA damage.41 Prolonged depletion of cohesin from rapidly dividing ES or iPS cells can therefore result in DNA damage, checkpoint activation, cell cycle arrest and the induction of p53 target gene expression (Fig. 1B). In turn, DNA damage responses abolish pluripotency gene expression42-44 and reprogramming.45-47 Hence, failure to establish or maintain pluripotency gene expression in cohesin-depleted cells does not necessarily implicate a loss of long-range interactions, but suggests the possibility that DNA damage responses could have interfered with pluripotency gene expression.
In a recent study we began to disentangle DNA damage responses from long-range interactions by conducting reprogramming experiments in the absence of cell division. Fusion of ES cells with somatic cells generates heterokaryons, which initiate reprogramming without cell division. In addition, nuclear transfer experiments eliminate the requirement for DNA replication. These experiments indicated that cohesin was not specifically required for reprogramming, and that ES cells maintained the expression of most pluripotency genes when analyzed after cohesin depletion but before the onset of DNA damage responses.41 Here we take this analysis to its logical conclusion by showing that deliberately inflicted DNA damage - or the DNA damage resulting from prolonged cohesin depletion in cycling ES cells - actively interferes with pluripotency and reprogramming. Our findings suggest that data concerning the role of cohesin in pluripotency and reprogramming derived from cells that cycle in the absence of cohesin should be re-interpreted in the context of essential cohesin functions in the cell cycle.
Results
A simple but restrictive approach to dissociate cell cycle-related and gene regulatory functions of cohesin is the genetic deletion of cohesin from non-cycling cells.12,28,48 Alternatively, cohesin can be acutely depleted from cycling cells at the gene level (by inducible deletion) or the protein level (by inducible cleavage or degradation), provided that depletion is sufficiently rapid to occur within a single cell cycle. Our experiments combined inducible ERt2Cre and conditional Rad21 alleles 41 to efficiently deplete mRNA (Fig. 2A) and protein (Fig. 2B) in ES cells within 24h of ERt2Cre induction by 4-hydroxy tamoxifen. This was achieved without significant induction of DNA damage (as indicated by phosphorylation of histone H2AX, γ-H2AX, Fig 2C) and in the absence of p53-dependent stress responses (such as Mdm2 induction, Fig. 2D) or cell cycle arrest (Fig. 2E).
Figure 2.
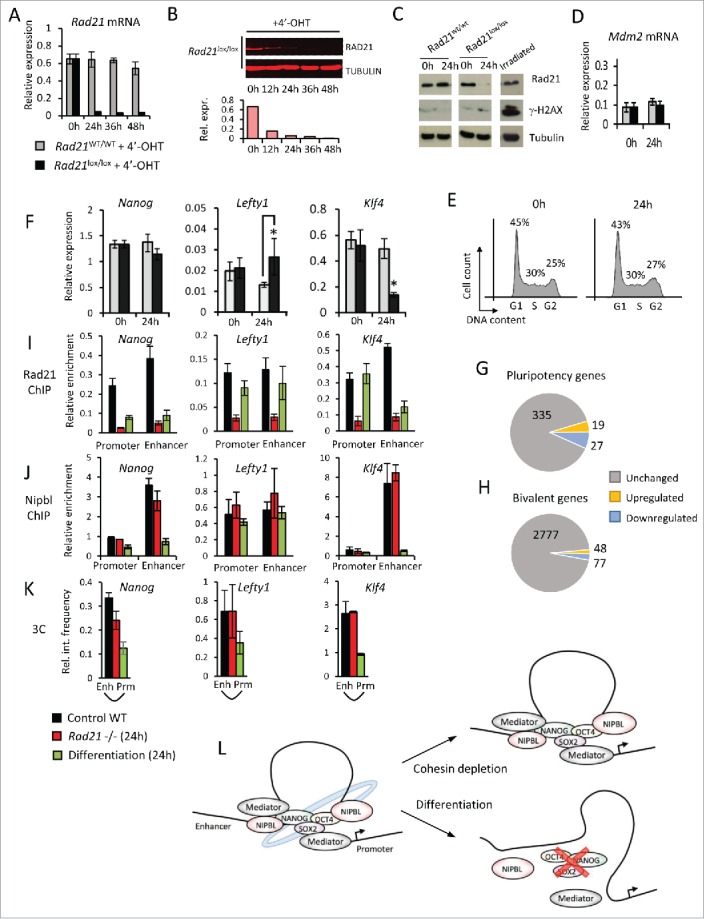
Acute cohesin depletion is compatible with pluripotency gene expression and enhancer-promoter interactions. (A-E) Time course of Rad21 mRNA (A) and RAD21 protein depletion (B) induced by 4′-OHT-mediated activation of ERt2Cre in ERT2Cre-Rad21lox/lox ES cells (100nM 4′-OHT). Acute cohesin depletion did not result in significant DNA damage as indicated by phosphorylation of H2AX (γ-H2AX), irradiated ES cells were used as positive control (C); upregulation of the p53 target gene Mdm2 (D) or cell cycle arrest (E). (F) Quantitative RT-PCR analysis of selected pluripotency genes in ES cells before (0h) and after acute cohesin depletion (24h). (G, H) Genome-wide expression analysis of pluripotency genes (G) and bivalent genes (H) in acutely cohesin-depleted ES cells at 24 hours. (I,J) ChIP for RAD21 (I) and NIPBL (J) at the promoters and enhancers of Nanog, Lefty1 and Klf4 in control ES cells (black), 24h Rad21-deleted ES cells (red) and differentiating ES cells (green). (K) Chromosome conformation capture (3C) assays for promoter-enhancer interactions at Nanog, Lefty1 and Klf4 in control ES cells (black), 24h Rad21-deleted ES cells (red) and differentiating ES cells (green) (L) Enhancer-promoter interactions at pluripotency loci in ES cells (left) are maintained after acute cohesin depletion (right, top) but lost during ES cell differentiation (right, bottom).
Genome-wide transcriptional profiling showed that ∼600 genes were deregulated. These genes were enriched for developmental functions but not for cell cycle or DNA damage responses. Deregulated expression was highly correlated with cohesin binding by ChIP-seq, indicating that many deregulated genes were direct targets of cohesin. 41 Quantitative reverse transcription PCR (RT-PCR) of selected pluripotency markers confirmed our array data indicating that the expression of Nanog remained unaffected, Klf4 was downregulated and Lefty1 was upregulated (Fig. 2F). Overall, 8% of deregulated genes were pluripotency-associated (Fig. 2G) and 12% of pluripotency genes were affected by cohesin depletion. Hence, acute cohesin depletion in ES cells did not cause a global collapse in pluripotency gene expression but had a selective and gene-specific impact where most pluripotency genes remained unaffected, whereas a minority were either up- or downregulated, most to a moderate extent. Many developmental genes in ES cells are marked by bivalent chromatin marks49,50 and can be rapidly activated upon differentiation. Of 2902 bivalent genes, 125 were deregulated within 24 hours of Rad21 deletion. Of these, only a minority (48) were upregulated, while 77 were downregulated (Fig. 2H). These data indicate that cohesin-depleted cells do not undergo wholesale differentiation and corroborate the conclusion that cohesin depletion does not result in a collapse of pluripotency gene expression.
As cohesin is thought to promote the expression of pluripotency genes by mediating enhancer-promoter interactions1-4 we carefully assessed how acute cohesin depletion affected the binding of cohesin to gene regulatory elements and interaction between enhancers and promoters in ES cells. ChIP-PCR showed that RAD21 was indeed efficiently depleted from the promoters and enhancers of Nanog, Lefty1, and Klf4 (red bars, Fig. 2i). RAD21 association in differentiating cells is shown for comparison (green bars, Fig. 2i). In contrast to RAD21, the cohesin loading protein NIPBL remained associated with the promoters and enhancers of Nanog, Lefty1 and Klf4 in Rad21-deleted ES cells (red bars, Fig. 2J). Unexpectedly, Nanog, Lefty1 and Klf4 enhancer-promoter interactions remained strong 24 hours after Rad21 deletion as detected by chromatin conformation capture (3C) (red bars, Fig. 2K), despite reduced cohesin occupancy (red bars, Fig. 2i). As a control for the ability of our 3C assays to detect change, reduced enhancer-promoter interactions were readily detected in differentiating ES cells (green bars, Fig. 2K). Hence, in contrast to expectations based on ES cells suffering DNA damage,1 enhancer-promoter interactions can be maintained at least at some pluripotency loci even after cohesin depletion. These interactions may be mediated by transcription factors, mediator or Nipbl (Fig. 2L).
When cohesin-depleted ES cells were allowed to proliferate, significant DNA damage occurred within 36 hours as indicated by γ-H2AX (Fig. 3A) and upregulation of the p53 target gene Mdm2 to levels similar to those induced by causing deliberate DNA damage by exposure of ES cells to doxorubicin (Fig. 3B). After 48 hours of cohesin depletion ES cells were arrested in G2/M phase of the cell cycle (Fig. 3C). With the exception of Lefty1 the expression of the pluripotency genes tested was downregulated (Fig. 3D, right) to levels that were comparable to those after deliberate DNA damage by exposure to Doxorubicin (Fig. 3D, center) or ES cell differentiation induced by withdrawal of 2i (Fig. 3D, right). These experiments show that (i) proliferation in the absence of cohesin causes DNA damage, and that (ii) deliberate DNA damage is sufficient to trigger a collapse of pluripotency gene expression in ES cells, reminiscent of what was reported in RNAi screens after prolonged depletion of cohesin in proliferating cells. These results are important because they question models where cohesin has a universal role in maintaining enhancer-promoter interactions.
Figure 3.
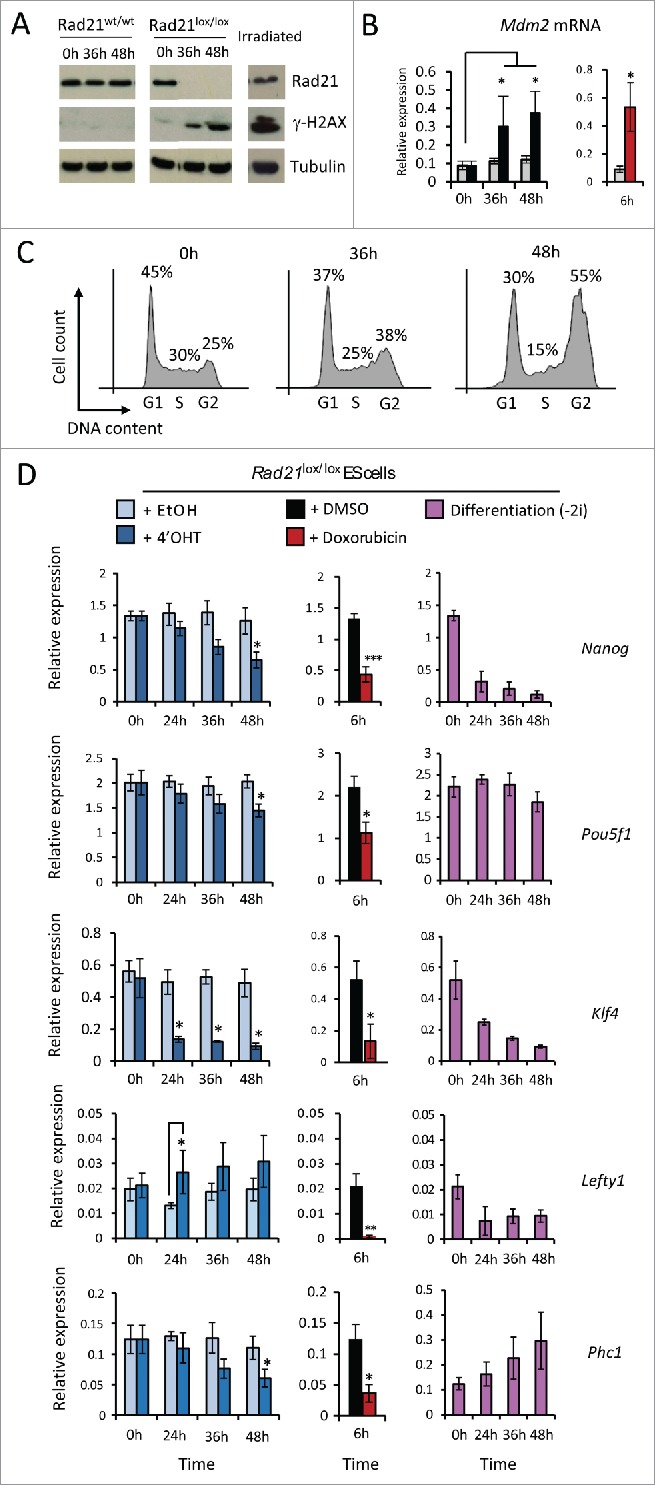
Incidental or deliberate DNA damage abolishes pluripotency gene expression. (A-C) Time course of DNA damage accumulation after cohesin depletion in proliferating ES cell indicated by phosphorylation of γ-H2AX (A), expression of the p53 target gene Mdm2 (B) and cell cycle arrest (C). (D) Quantitative RT-PCR analysis of selected pluripotency genes in ES cells undergoing incidental DNA damage as a result of prolonged cohesin depletion (left), deliberate DNA damage inflicted by doxorubicin treatment (6h, middle) or induced differentiation (right).
The idea that cohesin has special functions in promoting the expression of pluripotency genes is not restricted to the maintenance of pluripotency gene expression in ES cells, but extends to the induction of pluripotency gene expression during the reprogramming of somatic cells to pluripotency by iPS. 2-4 Given that iPS reprogramming also requires multiple rounds of cell division and is sensitive to activation of stress responses 45-47 we wondered to what extent the requirement for cohesin in reprogramming reflects essential cohesin functions in the cell cycle. We addressed this question by examining early reprogramming events that occur when ES cells and somatic cells are fused to form heterokaryons because reprogramming in heterokaryons is initiated in the absence of proliferation.51 Interestingly, acute cohesin depletion did not impair the ability of ES cells to initiate the expression of pluripotency genes in somatic nuclei.41 On the contrary, acutely cohesin-depleted ES cells reprogrammed better than control ES cells (Table 1, top). This was explained by the expression of Myc, which was increased in cohesin-depleted ES cells in 2i conditions.41 Increased Myc expression drove increased DNA replication in somatic nuclei, which is known to promote reprogramming in ES cell heterokaryons.52 Conversely, cohesin-depleted somatic cells showed reduced DNA replication and impaired reprogramming in ES cell heterokaryons, but reprogramming was rescued by nuclear transfer experiments in Xenopus oocytes, where reprogramming occurs in the absence of DNA replication.53-55 Taken together, these experiments demonstrated that cohesin was not required for the re-expression of pluripotency genes in somatic nuclei.
Table 1.
Incidental or deliberate DNA damage abrogates ES cell reprogramming potential.
| ES cells: DNA damage: | Control No | Rad21 ko (24h) No | |
|---|---|---|---|
| ES cell # | 100×106 | 100×106 | |
| Somatic cell # | 100×106 | 100×106 | |
| Heterokaryon # | ∼6×106* | ∼6×106* | |
| Reprogramming |
++ |
+++ |
|
| ES cells: DNA damage: |
Control No |
Rad 21 ko (48h) Incidental |
Doxorubicin Deliberate |
| ES cell # | 25×106 | 25×106 | 25×106 |
| Somatic cell # | 25×106 | 25×106 | 25×106 |
| Heterokaryon # | ∼1.5×106* | None | None |
| Reprogramming | ++ | N/A | N/A |
Estimate based on 3% fusion efficiency determined by flow cytometry
Acutely cohesin-depleted ES cells not only retained the ability to reprogram somatic cells in heterokaryons but in addition showed an unexpected increase in their reprogramming potential (top, n = 3 biological replicates, 100 × 106 ES cells per fusion). 41 Fusion with Rad21 KO (48h) ES cells and doxorubicin-treated (6h) ES cells did not result in viable heterokaryon formation or reprogramming. Poor survival meant that lower ES cell numbers were available (25 × 106), and control ES cell numbers were reduced accordingly (bottom, n=2 biological replicates per treatment condition).
To explore how prolonged cohesin depletion and DNA damage affect reprogramming we carried out cell fusions between somatic cells and ES cells that were Rad21-deleted 48 hours earlier and showed DNA damage, cell cycle arrest and reduced pluripotency gene expression. These fusions did not result in viable heterokaryon formation or successful reprogramming (Table 1, bottom). To explore whether this failure could be ascribed to DNA damage we induced deliberate DNA damage by treating ES cells with doxorubicin for 6 hours prior to fusion with somatic cells. ES cells with DNA damage also failed to form viable heterokaryons and did not induce successful reprogramming (Table 1, bottom).
Discussion
We have addressed the role of cohesin in pluripotency and reprogramming. To this end we designed experimental systems with the power to separate the spectrum of cohesin functions in the cell cycle from cohesin functions in gene regulation. Unexpectedly, cohesin-depleted ES cells maintained pluripotency gene expression and the ability to reprogram somatic nuclei in heterokaryons, provided that ES cells did not incur DNA damage as a result of attempting cell division in the absence of cohesin. 41 Data presented in the current manuscript show that experimentally induced DNA damage was sufficient to erase pluripotency gene expression and to abolish heterokaryon formation and reprogramming. These findings affect the interpretation of data from previous studies that had linked cohesin with pluripotency and reprogramming where cohesin was depleted over the course of several cell divisions. We suggest that results obtained after protracted cohesin depletion should not be ascribed to long-range chromosomal interactions or other functions of cohesin in transcription. Rather, they should be re-interpreted in the context of essential cohesin functions in the maintenance of genome integrity during the cell cycle.
Abbreviations
- 2i
combination of MEK and GSK3 inhibitors
- ES
cells embryonic stem cells
- RT-PCR
reverse transcriptase polymerase chain reaction
- TAD
Topologically associating domain
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Medical Research Council, UK, Wellcome Trust Investigator Award 099276 (MM) and a Commonwealth Scholarship (PG).
References
- 1.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al.. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467:430-5; PMID:20720539; http://dx.doi.org/ 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei Z, Gao F, Kim S, Yang H, Lyu J, An W, Wang K, Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell 2013; 13:36-47; PMID:23747203; http://dx.doi.org/ 10.1016/j.stem.2013.05.010 [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Jiao W, Sun L, Fan J, Chen M, Wang H, Xu X, Shen A, Li T, Niu B, et al.. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell 2013; 13:30-5; PMID:23747202; http://dx.doi.org/ 10.1016/j.stem.2013.05.012 [DOI] [PubMed] [Google Scholar]
- 4.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al.. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 2013; 12:699-712; PMID:23665121; http://dx.doi.org/ 10.1016/j.stem.2013.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al.. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 2008; 132:422-33; PMID:18237772; http://dx.doi.org/ 10.1016/j.cell.2008.01.011 [DOI] [PubMed] [Google Scholar]
- 6.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish J a, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A 2008; 105:8309-14; PMID:18550811; http://dx.doi.org/ 10.1073/pnas.0801273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 2008; 27:654-66; PMID:18219272; http://dx.doi.org/ 10.1038/emboj.2008.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al.. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008; 451:796-801; PMID:18235444; http://dx.doi.org/ 10.1038/nature06634 [DOI] [PubMed] [Google Scholar]
- 9.Schmidt D, Schwalie PC, Ross-innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 2010; 20(5):578-88; PMID:20219941; http://dx.doi.org/ 10.1101/gr.100479.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 2012; 22:2163-75; PMID:22780989; http://dx.doi.org/ 10.1101/gr.136507.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 2009; 460:410-3; PMID:19458616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al.. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 2011; 476:467-71; PMID:21832993; http://dx.doi.org/ 10.1038/nature10312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters J-M, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 2009; 5:e1000739; PMID:19956766; http://dx.doi.org/ 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A 2010; 107:3651-6; PMID:20133600; http://dx.doi.org/ 10.1073/pnas.0912087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J 2009; 28:1234-45; PMID:19322193; http://dx.doi.org/ 10.1038/emboj.2009.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkenschlager M, Odom DT. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell 2013; 152:1285-97; PMID:23498937; http://dx.doi.org/ 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- 17.Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 2002; 9:773-88; PMID:11983169; http://dx.doi.org/ 10.1016/S1097-2765(02)00515-4 [DOI] [PubMed] [Google Scholar]
- 18.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell 2003; 112:765-77; PMID:12654244; http://dx.doi.org/ 10.1016/S0092-8674(03)00162-4 [DOI] [PubMed] [Google Scholar]
- 19.Remeseiro S, Losada A. Cohesin, a chromatin engagement ring. Curr Opin Cell Biol 2013; 25:63-71; PMID:23219370; http://dx.doi.org/ 10.1016/j.ceb.2012.10.013 [DOI] [PubMed] [Google Scholar]
- 20.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 1998; 12:1986-97; PMID:9649503; http://dx.doi.org/ 10.1101/gad.12.13.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters J-MM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 2000; 151:749-62; PMID:11076961; http://dx.doi.org/ 10.1083/jcb.151.4.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, et al.. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol 2006; 4:e242; PMID:16802858; http://dx.doi.org/ 10.1371/journal.pbio.0040242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters J-M. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 2006; 16:863-74; PMID:16682347; http://dx.doi.org/ 10.1016/j.cub.2006.03.049 [DOI] [PubMed] [Google Scholar]
- 24.Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 2014; 505:367-71; PMID:24291789; http://dx.doi.org/ 10.1038/nature12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shintomi K, Hirano T. Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma 2010; 119:459-67; PMID:20352243; http://dx.doi.org/ 10.1007/s00412-010-0271-z [DOI] [PubMed] [Google Scholar]
- 26.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell 2013; 152:1285-97; PMID:23498937; http://dx.doi.org/ 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- 27.Gorkin DU, Leung D, Ren B. The 3D Genome in Transcriptional Regulation and Pluripotency. Cell Stem Cell 2014; 14:762-75; PMID:24905166; http://dx.doi.org/ 10.1016/j.stem.2014.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sofueva S, Yaffe E, Chan W-C, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 2013; 32:1-11; PMID:23211745; http://dx.doi.org/ 10.1038/emboj.2013.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seitan VC, Faure AJ, Zhan Y, Mccord RP, Lajoie BR, Ing-simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al.. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 2013; 23:2066-77; PMID:24002784; http://dx.doi.org/ 10.1101/gr.161620.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terret M-E, Sherwood R, Rahman S, Qin J, Jallepalli P V. Cohesin acetylation speeds the replication fork. Nature 2009; 462:231-4; PMID:19907496; http://dx.doi.org/ 10.1038/nature08550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tittel-Elmer M, Lengronne A, Davidson MB, Bacal J, François P, Hohl M, Petrini JHJ, Pasero P, Cobb JA. Cohesin association to replication sites depends on rad50 and promotes fork restart. Mol Cell 2012; 48:98-108; PMID:22885006; http://dx.doi.org/ 10.1016/j.molcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Méndez J. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 2010; 24:2812-22; PMID:21159821; http://dx.doi.org/ 10.1101/gad.608210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman A a, Mechtler K, et al.. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 2010; 143:737-49; PMID:21111234; http://dx.doi.org/ 10.1016/j.cell.2010.10.031 [DOI] [PubMed] [Google Scholar]
- 34.Gerlich D, Koch B, Dupeux F, Peters J-M, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 2006; 16:1571-8; PMID:16890534; http://dx.doi.org/ 10.1016/j.cub.2006.06.068 [DOI] [PubMed] [Google Scholar]
- 35.Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol 2001; 11:991-5; PMID:11448778; http://dx.doi.org/ 10.1016/S0960-9822(01)00271-8 [DOI] [PubMed] [Google Scholar]
- 36.Watrin E, Peters J-M. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells. EMBO J 2009; 28:2625-35; PMID:19629043; http://dx.doi.org/ 10.1038/emboj.2009.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutiérrez-Caballero C, Cebollero LR, Pendás AM. Shugoshins: from protectors of cohesion to versatile adaptors at the centromere. Trends Genet 2012; 28:351-60; PMID:Can't; http://dx.doi.org/ 10.1016/j.tig.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 38.Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger A-K, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al.. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 2009; 4:403-15; PMID:19345177; http://dx.doi.org/ 10.1016/j.stem.2009.03.009 [DOI] [PubMed] [Google Scholar]
- 39.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 2009; 23(7):837-48; PMID:19339689; http://dx.doi.org/ 10.1101/gad.1769609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al.. RAD21 Cooperates with Pluripotency Transcription Factors in the Maintenance of Embryonic Stem Cell Identity. PLoS One 2011; 6:e19470; PMID:21589869; http://dx.doi.org/ 10.1371/journal.pone.0019470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavagnolli T, Gupta P, Hörmanseder E, Mira-Bontenbal H, Dharmalingam G, Carroll T, Gurdon JB, Fisher AG, Merkenschlager M. Initiation and maintenance of pluripotency gene expression in the absence of cohesin. Genes Dev 2015; 29:23-38; PMID:25561493; http://dx.doi.org/ 10.1101/gad.251835.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 2005; 7:165-71; PMID:15619621; http://dx.doi.org/ 10.1038/ncb1211 [DOI] [PubMed] [Google Scholar]
- 43.Maimets T, Neganova I, Armstrong L, Lako M. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene 2008; 27:5277-87; PMID:18521083; http://dx.doi.org/ 10.1038/onc.2008.166 [DOI] [PubMed] [Google Scholar]
- 44.Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 2012; 46:30-42; PMID:22387025; http://dx.doi.org/ 10.1016/j.molcel.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al.. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 2009; 23:2134-9; PMID:19696146; http://dx.doi.org/ 10.1101/gad.1811609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009; 460:1145-8; PMID:19668190; http://dx.doi.org/ 10.1038/nature08285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009; 460:1136-9; PMID:19668188; http://dx.doi.org/ 10.1038/nature08290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al.. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 2013; 501:564-8; PMID:23975099; http://dx.doi.org/ 10.1038/nature12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al.. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006; 8:532-8; PMID:16570078; http://dx.doi.org/ 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- 50.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al.. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125:315-26; PMID:16630819; http://dx.doi.org/ 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 51.Pereira CF, Terranova R, Ryan NK, Santos J, Morris KJ, Cui W, Merkenschlager M, Fisher AG. Heterokaryon-based reprogramming of human B lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet 2008; 4:e1000170; PMID:18773085; http://dx.doi.org/ 10.1371/journal.pgen.1000170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsubouchi T, Soza-Ried J, Brown K, Piccolo FMM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AGG. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell 2013; 152:873-83; PMID:23415233; http://dx.doi.org/ 10.1016/j.cell.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin 2012; 5:17; PMID:23102146; http://dx.doi.org/ 10.1186/1756-8935-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gurdon JB, Partington GA, De Robertis EM. Injected nuclei in frog oocytes:RNA synthesis and protein exchange. J Embryol Exp Morphol 1976; 36:541-53; PMID:1010978 [PubMed] [Google Scholar]
- 55.Gurdon JB. Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J Embryol Exp Morphol 1976; 36:523-40; PMID:1010977 [PubMed] [Google Scholar]


