Abstract
Background
BRAFV600E is the most common mutation in papillary thyroid carcinoma (PTC), and it is associated with high-risk prognostic factors. However, the significance of the BRAF mRNA level in PTC remains unknown. We evaluated the significance of BRAF mRNA expression level by analyzing PTC data from The Cancer Genome Atlas (TCGA) database.
Methods
Data from 499 patients were downloaded from the TCGA database. After excluding other PTC variants, we selected 353 cases of classic PTC, including 193 cases with BRAFV600E and 160 cases with the wild-type BRAF. mRNA abundances were measured using RNA-Seq with the Expectation Maximization algorithm.
Results
The mean BRAF mRNA level was significantly higher in BRAFV600E patients than in patients with wild-type BRAF (197.6 vs. 179.3, p = 0.031). In wild-type BRAF patients, the mean BRAF mRNA level was higher in cases with a tumor > 2 cm than those with a tumor ≤ 2.0 cm (189.4 vs. 163.8, p = 0.046), and was also higher in cases with lymph node metastasis than in those without lymph node metastasis (188.5 vs. 157.9, p = 0.040). Within BRAFV600E patients, higher BRAF mRNA expression was associated with extrathyroidal extension (186.4 vs. 216.4, p = 0.001) and higher T stage (188.1 vs. 210.2, p = 0.016).
Conclusions
A higher BRAF mRNA expression level was associated with tumor aggressiveness in classic PTC regardless of BRAF mutational status. Evaluation of BRAF mRNA level may be helpful in prognostic risk stratification of PTC.
Introduction
Thyroid carcinoma is the most common endocrine malignancy, and its incidence has increased rapidly over the past few decades. In 2014, an estimated 62,980 new patients were expected in the United States [1]. Fortunately, the prognosis is generally excellent, and the thyroid cancer mortality is as low as 0.5 cases per 100,000 people [2]. However, a subset of thyroid carcinomas has a poor prognosis that is not adequately explained by traditional staging systems. Recent advances in cancer genetics provide new opportunities for improved assessment, and the molecular markers now available represent an effective strategy for the diagnosis and prognostication of thyroid carcinoma.
BRAF somatic mutations, the most extensively investigated molecular markers, are the most common genetic alterations in papillary thyroid carcinoma (PTC). One somatic mutation, BRAFV600E, results in the substitution of a valine by a glutamate at reside 600 and is associated with adverse prognostic factors such as extrathyroidal extension (ETE), lymph node metastasis, advanced American Joint Committee on Cancer (AJCC) stage, recurrence, distant metastasis, and poor survival [3–6]. This is explained by the fact that BRAFV600E mutation, via activation of the MAP kinase pathway, causes loss of expression of thyroid genes and refractoriness to radioiodine, as well as up-regulation of angiogenic and tumor-promoting molecules [7, 8]. On the other hand, the incidence of the BRAFV600E mutation in classic PTC varies widely between studies (38–83%) [9], and its positive predictive value for cancer recurrence is only 28% [10]. Furthermore, the results of several studies have challenged the notion that the BRAFV600E mutation is a valuable prognostic marker [11, 12]. Consequently, the prognostic use of the BRAFV600E mutation may not be generalizable to all clinical settings.
Several studies have suggested that the behavior of PTC is influenced not only by BRAF mutation status, but also by the expression level of the Braf mutant protein or the percentage of BRAF mutant alleles. One study reported that higher expression of Braf mutant protein predicts aggressive tumor behavior in PTC [13]. Other studies assessed the correlation between the percentage of BRAF mutant alleles and clinical outcomes in PTC using pyrosequencing [14, 15]. The results showed that higher percentages of BRAF mutant alleles were associated with poor prognostic factors such as older age, larger tumor size, ETE, and higher recurrence rate.
In this study, we analyzed the BRAF mRNA expression level in cases of PTC from The Cancer Genome Atlas (TCGA) database to investigate the significance of the level of BRAF mRNA expression in PTC and evaluate its prognostic value.
Materials and Methods
Data acquisition
As of March 2015, TCGA group made available multiple types of genomic data regarding thyroid carcinoma, including somatic mutation, exome sequencing, methylation array, mRNA expression count, microRNA expression count and clinical information. We downloaded the data on somatic mutation, mRNA expression count, and clinical information from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). All patient information was anonymized and de-identified in this database. According to TCGA publication guidelines (http://cancergenome.nih.gov/publications/publicationguidelines), there are no restrictions on the publication of these somatic mutation and mRNA sequencing data and no specific permission is required for investigators to publish papers containing or referring to these data. Somatic mutation data were provided as a mutation call file by the Broad Institute and the Baylor College of Medicine. The Illumina Genome Analyzer was used as the platform for DNA sequencing (Illumina Inc., San Diego, CA, USA). mRNA sequencing data, obtained by Illumina HiSeq 2000 RNA Sequencing Version 2 analysis, were provided by the University of North Carolina. mRNA expression counts were obtained via the TCGA portal and are expressed as RNA-Seq by Expectation Maximization (RSEM) values. RSEM is an accurate software tool for quantifying transcript abundances from RNA-Seq data [16].
After excluding the patients with missing information, we downloaded data from a total of 499 patients. Two authors (YJC, JWY) independently reviewed every scanned original pathologic report file and revised incorrect or missing clinical information. In cases of multifocal PTC, the largest tumor was analyzed.
Patient selection
Of the 499 cases of PTC, 101 cases of follicular variant, 35 cases of tall cell variant, and eight cases of other variants were excluded, leaving a total of 355 classic PTCs. Subtype classification of the PTC patients was based on a previously published paper by Cancer Genome Atlas Research Network [17]. Follicular variant or tall cell variant PTC was diagnosed when more than 99% of the tumor exhibited a follicular pattern or more than 50% of tall cell features, respectively [17]. Two patients with BRAF mutations other than V600E (N581T, V459V) were also excluded. Ultimately, 193 classic PTCs with BRAFV600E and 160 classic PTCs with wild-type BRAF were selected for the analysis.
Statistics
Data were analyzed using the R software version 3.0.2 (R Foundation for Statistical Computing, Vienna, Austria). Chi-square tests and Fisher’s exact test were used to compare categorical variables. Unpaired two sample t-test and linear regression analysis were performed to compare mRNA expression counts. Recursive Partitioning and Regression Trees were used to calculate the cut-off value of BRAFV600E mRNA expression above which the expression levels correlate with poor prognosis parameters. The Kaplan-Meier estimator was used for survival analysis. A p-value < 0.05 was considered significant.
Results
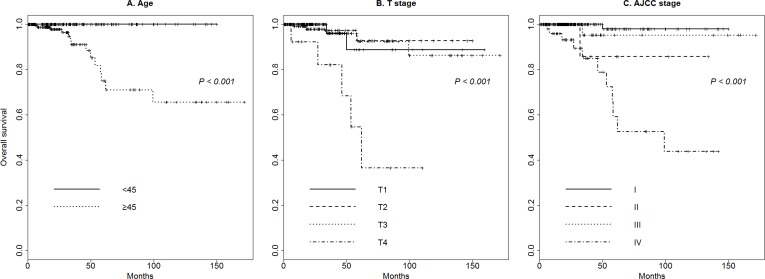
The median follow-up duration was 20.7 months (range, 0.03–171.70 months). Fig 1 shows Kaplan–Meier overall survival for the study population based on clinicopathological characteristics. Age ≥ 45 (p < 0.001), higher Tumor stage (T stage, p < 0.001), and higher AJCC stage (p < 0.001) were associated with poorer overall survival. Other clinicopathological characteristics such as gender (p = 0.293), size > 2.0 cm (p = 0.186), ETE (p = 0.14), presence of thyroiditis (p = 0.129), and lymph node metastasis (p = 0.225) were not significantly associated with overall survival.
Fig 1. Overall survival of classic papillary thyroid carcinoma patients.
(A) Age ≥ 45, (B) higher Tumor stage (T stage), and (C) higher American Joint Committee on Cancer (AJCC) stage were associated with poorer overall survival.
BRAF mutation status and clinicopathological characteristics
Clinicopathological characteristics of classic PTC patients classified according to BRAF mutation status are shown in Table 1. None of the factors, including age, gender, thyroiditis, ETE, tumor size, AJCC stage, recurrence, or vital status, differed significantly between the BRAFV600E and wild-type BRAF patients.
Table 1. Clinicopathological characteristics of classic papillary thyroid carcinoma patients with BRAFV600E or wild-type BRAF.
| Wild-type BRAF (n = 160) | BRAFV600E (n = 193) | p—value | |
|---|---|---|---|
| Age | |||
| < 45 years | 78 (48.8%) | 92 (49.7%) | 0.924 |
| ≥ 45 years | 82 (51.2%) | 101 (52.3%) | |
| Gender | |||
| Male | 41 (25.6%) | 55 (28.5%) | 0.629 |
| Female | 119 (74.4%) | 138 (71.5%) | |
| Thyroiditis | |||
| No | 47 (29.4%) | 65 (33.7%) | 1.000 |
| Yes | 49 (30.6%) | 67 (34.7%) | |
| n.a | 64 (40.0%) | 61 (31.6%) | |
| Extrathyroidal extension | |||
| No | 115 (71.9%) | 121 (62.7%) | 0.087 |
| Yes | 45 (28.1%) | 72 (37.3%) | |
| Tumor size | |||
| ≤ 2.0 cm | 63 (39.4%) | 83 (43.0%) | 0.561 |
| > 2.0 cm | 97 (60.6%) | 110 (57.0%) | |
| T stage | |||
| T1, T2 | 102 (63.8%) | 110 (57.0%) | 0.238 |
| T3, T4 | 58 (36.2%) | 83 (43.0%) | |
| N stage | |||
| N0 | 44 (27.5%) | 63 (32.6%) | 0.572 |
| N1 | 84 (52.5%) | 93 (48.2%) | |
| Nx | 32 (20.0%) | 37 (19.2%) | |
| AJCC stage | |||
| I, II | 104 (65.0%) | 129 (66.8%) | 0.802 |
| III, IV | 56 (35.0%) | 64 (33.2%) | |
| Recurrence | |||
| No | 149 (93.1%) | 176 (91.2%) | 0.637 |
| Yes | 11 (6.9%) | 17 (8.8%) | |
| Vital Status | |||
| Alive | 155 (96.9%) | 184 (95.3%) | 0.643 |
| Dead | 5 (3.1%) | 9 (4.7%) |
Abbreviations: AJCC = American Joint Committee on Cancer
BRAF mRNA expression levels in BRAFV600E and wild-type BRAF classic PTC
The BRAF mRNA expression counts for the study patients are shown in S1 Table. The count data for the wild-type BRAF and BRAFV600E patients can be summarized as follows: for wild-type BRAF patients, minimum 45.3, lower quartile 128.7, median 166.2, mean 179.3 and upper quartile 204.2; for BRAFV600E patients, minimum 56.1, lower quartile 154.7, median 194.1, mean 197.6, and upper quartile 243.5 (Fig 2). The mean BRAF mRNA expression count was significantly higher in BRAFV600E patients than in patients with wild-type BRAF (197.6 vs. 179.3, p = 0.031).
Fig 2. BRAF mRNA expression counts in classic papillary thyroid carcinoma patients.
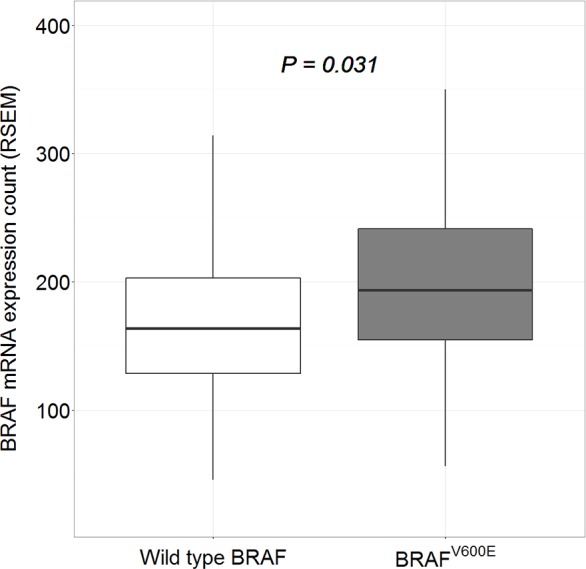
The mean BRAF mRNA expression count was significantly higher in BRAFV600E patients than in patients with wild-type BRAF.
Significance of the BRAF mRNA expression level
Table 2 shows the results of unpaired two sample t-test. BRAF mRNA expression was higher in patients < 45 years and in those without thyroiditis, with a tumor size > 2.0cm, and with N1 nodal stage. In wild-type BRAF patients, the BRAF mRNA expression count was higher in cases with a tumor size > 2 cm. In addition, the count was higher in cases with N1 nodal stage. Within BRAFV600E patients, higher BRAF mRNA expression was associated with the presence of ETE and higher T stage. Fig 3 summarizes the association between BRAF mRNA expression count and clinicopathological characteristics. Table 3 shows the results of linear regression analysis. The BRAF mRNA expression count was negatively correlated with age ≥ 45 years and thyroiditis, and was positively correlated with tumor size > 2 cm and N1 nodal stage. In wild-type BRAF patients, the BRAF mRNA expression count negatively correlated with age ≥ 45 years whereas it positively correlated with ETE and higher T stage in BRAFV600E patients.
Table 2. Unpaired t-test results of comparisons of each clinical variable, using BRAF mRNA expression counts as the response variable.
| All (n = 353) | Wild-type BRAF (n = 160) | BRAFV600E (n = 193) | ||||
|---|---|---|---|---|---|---|
| Mean (RSEM) | p-value | Mean (RSEM) | p-value | Mean (RSEM) | p-value | |
| Age | ||||||
| < 45 years | 199.046 | 0.023 | 193.7 | 0.051 | 203.5 | 0.207 |
| ≥ 45 years | 180.224 | 165.5 | 192.1 | |||
| Gender | ||||||
| Male | 183.224 | 0.310 | 168.3 | 0.271 | 194.4 | 0.647 |
| Female | 191.554 | 183.1 | 198.9 | |||
| Thyroiditis | ||||||
| No | 199.195 | 0.032 | 200.8 | 0.104 | 198.0 | 0.162 |
| Yes | 176.810 | 167.0 | 184.0 | |||
| Extrathyroidal extension | ||||||
| No | 196.560 | 0.178 | 185.0 | 0.111 | 186.4 | 0.001 |
| Yes | 185.683 | 164.8 | 216.4 | |||
| Tumor size | ||||||
| ≤ 2.0 cm | 179.442 | 0.028 | 163.8 | 0.046 | 191.4 | 0.209 |
| > 2.0 cm | 196.233 | 189.4 | 202.3 | |||
| T stage | ||||||
| T1, T2 | 183.554 | 0.079 | 178.7 | 0.908 | 188.1 | 0.016 |
| T3, T4 | 197.910 | 180.3 | 210.2 | |||
| N stage | ||||||
| N0 | 175.960 | 0.013 | 157.9 | 0.040 | 188.6 | 0.078 |
| N1 | 197.587 | 188.5 | 205.8 | |||
| AJCC stage | ||||||
| I, II | 193.133 | 0.157 | 187.3 | 0.073 | 197.9 | 0.934 |
| III, IV | 181.824 | 164.4 | 197.0 | |||
| Recurrence | ||||||
| No | 187.747 | 0.191 | 177.7 | 0.364 | 196.3 | 0.424 |
| Yes | 207.181 | 201.0 | 211.2 | |||
| Vital Status | ||||||
| Alive | 188.792 | 0.574 | 179.4 | 0.639 | 196.7 | 0.563 |
| Dead | 201.301 | 174.2 | 216.4 | |||
Abbreviations: RSEM = RNA-Seq by Expectation Maximization; AJCC = American Joint Committee on Cancer
Fig 3. BRAF mRNA expression counts according to clinicopathological features.
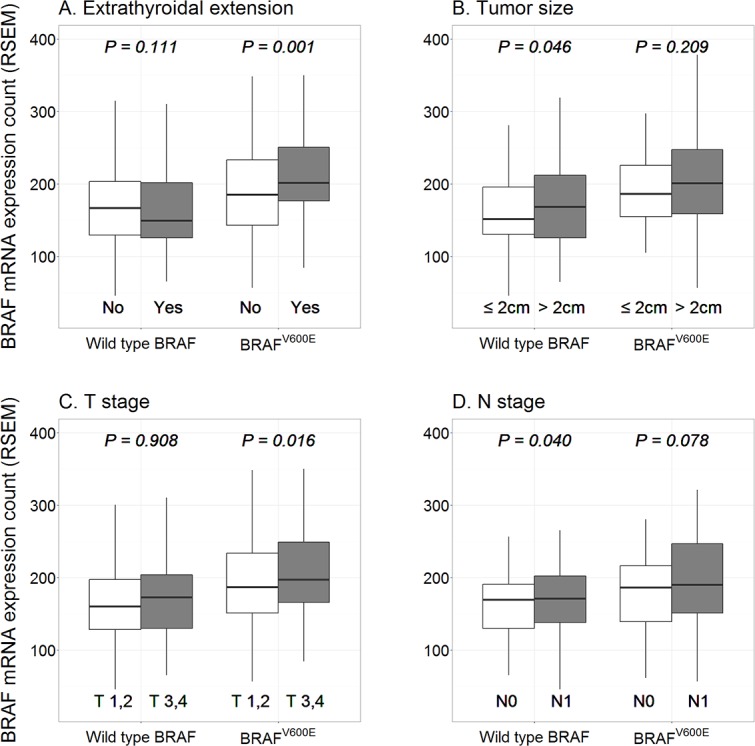
In BRAFV600E patients, higher BRAF mRNA expression was associated with (A) the presence of extrathyroidal extension and (B) higher Tumor stage (T stage). In wild-type BRAF patients, higher BRAF mRNA expression was associated with (C) tumor size > 2 cm and (D) N1 stage.
Table 3. Linear regression analysis of comparisons of each clinical variable, using BRAF mRNA expression counts as the response variable.
| All (n = 353) | Wild-type BRAF (n = 160) | BRAFV600E (n = 193) | ||||
|---|---|---|---|---|---|---|
| t-value | p-value | t-value | p-value | t-value | p-value | |
| Age ≥ 45 years | -2.318 | 0.021 | -2.001 | 0.047 | -1.263 | 0.208 |
| Male gender | 0.908 | 0.365 | 0.909 | 0.365 | 0.448 | 0.655 |
| Thyroiditis | -2.213 | 0.028 | -1.855 | 0.066 | -1.299 | 0.196 |
| Extrathyroidal extension | 1.255 | 0.210 | -1.281 | 0.202 | 3.305 | 0.001 |
| Tumor size > 2.0 cm | 2.035 | 0.043 | 1.772 | 0.078 | 1.201 | 0.231 |
| T stage (T3, T4) | 1.727 | 0.085 | 0.108 | 0.914 | 2.462 | 0.015 |
| N stage (N1) | 2.314 | 0.021 | 1.839 | 0.068 | 1.685 | 0.094 |
| AJCC stage (III, IV) | -1.314 | 0.190 | -1.538 | 0.126 | -0.086 | 0.932 |
| Recurrence | 1.288 | 0.199 | 0.83 | 0.408 | 0.935 | 0.351 |
| Mortality | 0.598 | 0.551 | -0.129 | 0.897 | 0.921 | 0.358 |
Abbreviations: RSEM = RNA-Seq by Expectation Maximization; AJCC = American Joint Committee on Cancer
Discussion
Since the start of the TCGA project in 2006, multiplatform genomic data from more than 20 types of carcinomas have been provided through the TCGA data portal, and multiple studies have been performed using this powerful open resource. Several studies of thyroid carcinoma using this data resource have been reported recently [14, 17, 18]. The TCGA portal provides an enormous amount of information, including data regarding microsatellite instability, DNA sequencing, miRNA sequencing, protein expression, mRNA sequencing, DNA methylation, copy number variation, clinical information, and clinical images. Although TCGA genomic data are archived under standardized and strictly controlled condition, some concerns remain regarding the correlation of genomic data with clinical information due to the potential inaccuracy of the clinical information. Such potential discrepancies exist because the surgical procedures and pathologic reporting systems differed among the institutes that collected the data. Therefore, to ensure the reliability of the TCGA data, we examined each scanned original pathologic file and revised the clinical information. Moreover, to examine the reliability of the TCGA data, we performed survival analysis; the results showed that overall survival was associated with older patient age, absence of thyroiditis, higher T stage, and higher AJCC stage, consistent with the known relationships between these parameters and overall survival in PTC.
The MAPK pathway [Ras Raf MEK MAPK/ERK] is a fundamental intracellular signaling pathway that plays a central role in cellular functions such as proliferation, differentiation, apoptosis, and survival [19]. Several mutations in the MAPK pathway are involved in the tumorigenesis of PTC. Of these, the BRAFV600E mutation is the most potent activator of the MAPK pathway [20]. The BRAFV600E kinase is more active than the wild-type BRAF, and its transformation efficacy is 138- to 500-fold higher [21, 22]. The clinical impact of the BRAFV600E mutation is well known, but the significance of the BRAF mRNA expression level has not been previously studied well.
Although there are concerns that the mRNA expression level may not provide a true reflection of the protein expression level, RNA sequencing has the advantage in that the results obtained using this method can be objectively measured and there are studies reporting positive correlation between the mRNA expression level and protein expression level. One study showed that differentially expressed mRNA correlated well with their protein levels [23] and another study showed that more than 85% of the variation in steady-state protein levels could be explained by changes in mRNA levels [24]. In this regard, we used the BRAF mRNA expression level as a surrogate marker for Braf function in thyroid carcinoma, although a substantial proportion of variation in the protein level cannot be explained wholly by changes in the mRNA level alone [25].
The significance of BRAF mRNA expression in thyroid tumors has been reported previously [26], and recurrence or distant metastasis in PTC patients has been shown to be related to higher BRAF mRNA expression [26]. In the present study, we found an association between the BRAF mRNA expression level and clinicopathological characteristics in a large number of cases. We found that higher BRAF mRNA expression levels were associated with aggressive clinical features in both wild-type BRAF and BRAFV600E patients except patient age < 45 years which is controversial as a cutoff for favorable prognosis [27]. On the other hand, we observed no differences in the clinicopathological characteristics of wild-type BRAF and BRAFV600E patients, which is in contrast to the results of other studies showing that the BRAFV600E mutation is associated with poor prognostic factors [28]. This might be because a relatively small number of patients with short term follow-up weakened the statistical power. On the other hand, it can be also postulated that the expression level of BRAF mRNA contributes to aggressiveness of PTC, and PTC with BRAFV600E may not exhibit aggressive features unless mRNA expression of the BRAFV600E reaches a certain level. Additionally, the present dataset might have included relatively low numbers of PTCs with high BRAFV600E mRNA expression counts, which would have prevented observing a statistical difference in clinicopathological features between wild-type BRAF and BRAFV600E patients. In fact, a number of studies with small sample size could not demonstrate that BRAF mutation is a poor prognostic factor [11, 12, 29, 30], whereas studies with larger sample sizes could [8, 31].
The significance of the Braf protein expression level has been demonstrated in previous studies using immunohistochemistry. Immunohistochemical studies suggest that higher Braf protein expression is associated with poor prognostic factors in PTC and melanoma. Zagzag et al. [13] performed an immunohistochemical study using a mutation-specific antibody (VE1; Springer-Bio, Maisons-Alfort Cedex, France) and reported that higher BrafV600E mutant protein expression is associated with the presence of ETE in PTC. Although immunohistochemical studies are difficult to evaluate objectively or quantitatively, and the results of such studies are sometimes not reproducible [32], detection of BrafV600E mutant protein using a mutation-specific antibody has been validated in several studies [33–35]. An association between high Braf expression and adverse prognostic outcomes was also demonstrated in melanoma [36].
The possible heterogeneous distribution of the BRAF mutation in PTC tumors is an important consideration in interpreting BRAF mRNA expression. Whether the BRAF mutation in PTC is clonal or subclonal has been the subject of much debate. Up until recently, the general perception was that BRAF mutations in PTC are strong driver mutations and PTCs with the BRAF mutation are clonal. This viewpoint is supported by the results of an immunohistochemical study using a BrafV600E mutation-specific antibody [37] as well by a recent publication from the TCGA Research Network [17]. Conversely, a number of recent studies suggest that tumors are heterogeneous for the BRAF mutation and that not every cell in a BRAF mutation-positive tumor has a heterozygous mutation [14, 15, 38–40]. These studies also demonstrated that the percentage of BRAF mutant alleles in PTC with the BRAFV600E mutation were significantly below 50%, the theoretical percentage for a clonal heterozygous mutation [14, 38–40]. Additionally, Guerra et al. subsequently demonstrated that a higher percentage of BRAF mutant alleles are associated with older age, larger tumor sizes, and higher recurrence rates in PTC [15]. Other studies demonstrated that a higher proportion of BRAF mutant cells in PTC is associated with larger tumor size [40, 41] and lymph node metastasis [40]. Likewise, the present study showed that high BRAF mRNA expression correlates with aggressive clinical features of PTC, probably because the tumors with high BRAF mRNA expression had a greater number of BRAFV600E mutant cells. Further molecular investigations, including single cell sequencing, might help resolve the heterogeneity issue. If the BRAF mutation proves to be heterogeneous in PTC, it may turn out that PTCs cannot be categorized merely on whether they are BRAF mutated or not.
Regarding the role of BRAF mRNA expression in wild-type PTC, the issue of tumor heterogeneity may have little impact on the aggressive clinicopathological characteristics. There may be increased activities of the Braf kinase in association with presumptive increased Braf protein levels as a result of the overexpression of wild-type BRAF mRNA in aggressive PTC compared with indolent PTC. Consequently, the downstream signaling activities of the MAP kinase pathway may be more active in aggressive PTC than in indolent PTC; this would be consistent with the fact that overexpression of wild-type BRAF also causes increased activation of downstream signaling of the MAP kinase pathway [42–44]. This may explain the finding in the present study that BRAF mRNA overexpression was associated with aggressive features even in PTC with the wild-type BRAF. The clinical significance of this finding needs further studies to define. It is also interesting that in this study the mean BRAF mRNA expression level was significantly higher in PTC with BRAFV600E than in PTC with the wild-type BRAF although there was no significant difference in the tumor aggressiveness between the two groups. The roles of the wild-type BRAF and BRAFV600E in tumor progression are apparently complex. Further mechanistic studies are needed to fully understand these findings.
Meanwhile, various BRAF inhibitors are currently being tested in clinical trials on patients with refractory thyroid carcinoma, with varying results. Subclonality, splice variants, or copy number gains of BRAF may be responsible for the lack of responses to BRAF inhibitors in many cases [45–47]. In selecting patients who may benefit from BRAF inhibitors, the BRAF mRNA expression level might serve as a potentially useful selection indicator if, in the future, it is proven to be specifically correlated with the response rate to BRAF inhibitors. As such, BRAF mRNA expression level might be helpful in predicting therapeutic effects. Similarly, it could also help exclude patients who would likely not respond to unnecessary treatments with BRAF inhibitors, and thereby avoid unnecessary exposure of such patients to the undesirable side effects of these drugs.
In conclusion, high BRAF mRNA expression levels were associated with aggressive clinical features in both wild-type BRAF and BRAF V600E patients with classic PTC, although the association with recurrence or survival could not be shown due to limited follow-up. Although heterogeneity of the BRAF mutation could weaken the correlation between the BRAF mRNA expression level and aggressive clinical features, the correlation was probably more robust in the PTC samples with higher percentages of BRAF mutant cells. Evaluation of the BRAF mRNA expression level in patients could provide a way to establish more accurate prognoses and risk stratification.
Supporting Information
(XLSX)
Acknowledgments
Authors acknowledge the contribution of specimen donors and The Cancer Genome Atlas Research Network.
Data Availability
All relevant data are available within the paper and its Supporting Information files.
Funding Statement
This study was conducted with research grants from the Korean Foundation for Cancer Research (Grant Number: CB-2011-03-01) and the Korean Health Technology R&D Project, Ministry of Health and Welfare (HI13C2164). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. Epub 2014/01/09. 10.3322/caac.21208 . [DOI] [PubMed] [Google Scholar]
- 2.Davies L, Welch HG. Increasing incidence of thyroid cancer in the United States, 1973–2002. JAMA: the journal of the American Medical Association. 2006;295(18):2164–7. Epub 2006/05/11. 10.1001/jama.295.18.2164 . [DOI] [PubMed] [Google Scholar]
- 3.Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. The Journal of clinical endocrinology and metabolism. 2005;90(12):6373–9. Epub 2005/09/22. 10.1210/jc.2005-0987 . [DOI] [PubMed] [Google Scholar]
- 4.Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, et al. Association Between BRAF V600E Mutation and Recurrence of Papillary Thyroid Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015;33(1):42–50. Epub 2014/10/22. 10.1200/jco.2014.56.8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abubaker J, Jehan Z, Bavi P, Sultana M, Al-Harbi S, Ibrahim M, et al. Clinicopathological analysis of papillary thyroid cancer with PIK3CA alterations in a Middle Eastern population. The Journal of clinical endocrinology and metabolism. 2008;93(2):611–8. Epub 2007/11/15. 10.1210/jc.2007-1717 . [DOI] [PubMed] [Google Scholar]
- 6.Lang BH, Chai YJ, Cowling BJ, Min HS, Lee KE, Youn YK. Is BRAFV600E mutation a marker for central nodal metastasis in small papillary thyroid carcinoma? Endocrine-related cancer. 2014;21(2):285–95. Epub 2014/01/10. 10.1530/erc-13-0291 . [DOI] [PubMed] [Google Scholar]
- 7.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocrine reviews. 2007;28(7):742–62. Epub 2007/10/18. 10.1210/er.2007-0007 . [DOI] [PubMed] [Google Scholar]
- 8.Kim TH, Park YJ, Lim JA, Ahn HY, Lee EK, Lee YJ, et al. The association of the BRAF(V600E) mutation with prognostic factors and poor clinical outcome in papillary thyroid cancer: a meta-analysis. Cancer. 2012;118(7):1764–73. Epub 2011/09/02. 10.1002/cncr.26500 . [DOI] [PubMed] [Google Scholar]
- 9.Xing M. BRAF mutation in thyroid cancer. Endocrine-related cancer. 2005;12(2):245–62. Epub 2005/06/11. 10.1677/erc.1.0978 . [DOI] [PubMed] [Google Scholar]
- 10.Xing M. Prognostic utility of BRAF mutation in papillary thyroid cancer. Molecular and cellular endocrinology. 2010;321(1):86–93. Epub 2009/11/04. 10.1016/j.mce.2009.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walczyk A, Kowalska A, Kowalik A, Sygut J, Wypiorkiewicz E, Chodurska R, et al. The BRAF(V600E) mutation in papillary thyroid microcarcinoma: does the mutation have an impact on clinical outcome? Clinical endocrinology. 2014;80(6):899–904. Epub 2013/12/21. 10.1111/cen.12386 . [DOI] [PubMed] [Google Scholar]
- 12.Ito Y, Yoshida H, Maruo R, Morita S, Takano T, Hirokawa M, et al. BRAF mutation in papillary thyroid carcinoma in a Japanese population: its lack of correlation with high-risk clinicopathological features and disease-free survival of patients. Endocrine journal. 2009;56(1):89–97. Epub 2008/10/09. . [DOI] [PubMed] [Google Scholar]
- 13.Zagzag J, Pollack A, Dultz L, Dhar S, Ogilvie JB, Heller KS, et al. Clinical utility of immunohistochemistry for the detection of the BRAF v600e mutation in papillary thyroid carcinoma. Surgery. 2013;154(6):1199–204; discussion 204–5. Epub 2013/08/13. 10.1016/j.surg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MH, Bae JS, Lim DJ, Lee H, Jeon SR, Park GS, et al. Quantification of BRAF V600E alleles predicts papillary thyroid cancer progression. Endocrine-related cancer. 2014;21(6):891–902. Epub 2014/10/01. 10.1530/erc-14-0147 . [DOI] [PubMed] [Google Scholar]
- 15.Guerra A, Fugazzola L, Marotta V, Cirillo M, Rossi S, Cirello V, et al. A high percentage of BRAFV600E alleles in papillary thyroid carcinoma predicts a poorer outcome. The Journal of clinical endocrinology and metabolism. 2012;97(7):2333–40. Epub 2012/04/18. 10.1210/jc.2011-3106 . [DOI] [PubMed] [Google Scholar]
- 16.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323 Epub 2011/08/06. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–90. Epub 2014/11/25. 10.1016/j.cell.2014.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chai YJ, Kim YA, Jee HG, Yi JW, Jang BG, Lee KE, et al. Expression of the embryonic morphogen Nodal in differentiated thyroid carcinomas: Immunohistochemistry assay in tissue microarray and The Cancer Genome Atlas data analysis. Surgery. 2014;156(6):1559–68. Epub 2014/12/03. 10.1016/j.surg.2014.08.050 . [DOI] [PubMed] [Google Scholar]
- 19.Steelman LS, Abrams SL, Shelton JG, Chappell WH, Basecke J, Stivala F, et al. Dominant roles of the Raf/MEK/ERK pathway in cell cycle progression, prevention of apoptosis and sensitivity to chemotherapeutic drugs. Cell Cycle. 2010;9(8):1629–38. Epub 2010/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer research. 2003;63(7):1454–7. Epub 2003/04/03. . [PubMed] [Google Scholar]
- 21.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. Epub 2002/06/18. 10.1038/nature00766 . [DOI] [PubMed] [Google Scholar]
- 22.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–67. Epub 2004/03/24. . [DOI] [PubMed] [Google Scholar]
- 23.Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep. 2015;5:10775 Epub 2015/06/09. 10.1038/srep10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csardi G, Franks A, Choi DS, Airoldi EM, Drummond DA. Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast. PLoS Genet. 2015;11(5):e1005206 Epub 2015/05/08. 10.1371/journal.pgen.1005206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227–32. Epub 2012/03/14. 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araujo PP, Marcello MA, Tincani AJ, Guilhen AC, Morari EC, Ward LS. mRNA BRAF expression helps to identify papillary thyroid carcinomas in thyroid nodules independently of the presence of BRAFV600E mutation. Pathology, research and practice. 2012;208(8):489–92. Epub 2012/07/10. 10.1016/j.prp.2012.05.013 . [DOI] [PubMed] [Google Scholar]
- 27.Nixon IJ, Wang LY, Migliacci JC, Eskander A, Campbell MJ, Aniss A, et al. An International Multi-Institutional Validation of Age 55 Years as a Cutoff for Risk Stratification in the AJCC/UICC Staging System for Well-Differentiated Thyroid Cancer. Thyroid. 2016;26(3):373–80. Epub 2016/02/26. 10.1089/thy.2015.0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing M, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA: the journal of the American Medical Association. 2013;309(14):1493–501. Epub 2013/04/11. 10.1001/jama.2013.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam JK, Jung CK, Song BJ, Lim DJ, Chae BJ, Lee NS, et al. Is the BRAF(V600E) mutation useful as a predictor of preoperative risk in papillary thyroid cancer? Am J Surg. 2012;203(4):436–41. Epub 2011/08/02. 10.1016/j.amjsurg.2011.02.013 . [DOI] [PubMed] [Google Scholar]
- 30.Ahn D, Park JS, Sohn JH, Kim JH, Park SK, Seo AN, et al. BRAFV600E mutation does not serve as a prognostic factor in Korean patients with papillary thyroid carcinoma. Auris, nasus, larynx. 2012;39(2):198–203. Epub 2011/08/25. 10.1016/j.anl.2011.07.011 . [DOI] [PubMed] [Google Scholar]
- 31.Li F, Chen G, Sheng C, Aaron GM, Huang Y, Lv Z, et al. BRAFV600E mutation in papillary thyroid microcarcinoma: a meta-analysis. Endocrine-related cancer. 2015. Epub 2015/01/17. 10.1530/erc-14-0531 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koperek O, Kornauth C, Capper D, Berghoff AS, Asari R, Niederle B, et al. Immunohistochemical detection of the BRAF V600E-mutated protein in papillary thyroid carcinoma. The American journal of surgical pathology. 2012;36(6):844–50. Epub 2012/05/18. 10.1097/PAS.0b013e318246b527 . [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann AK, Camenisch U, Rechsteiner MP, Bode-Lesniewska B, Rossle M. Value of immunohistochemistry in the detection of BRAF(V600E) mutations in fine-needle aspiration biopsies of papillary thyroid carcinoma. Cancer cytopathology. 2014;122(1):48–58. Epub 2013/09/17. 10.1002/cncy.21352 . [DOI] [PubMed] [Google Scholar]
- 34.Crescenzi A, Guidobaldi L, Nasrollah N, Taccogna S, Cicciarella Modica DD, Turrini L, et al. Immunohistochemistry for BRAF(V600E) antibody VE1 performed in core needle biopsy samples identifies mutated papillary thyroid cancers. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 2014;46(5):370–4. Epub 2014/02/27. 10.1055/s-0034-1368700 . [DOI] [PubMed] [Google Scholar]
- 35.Ilie MI, Lassalle S, Long-Mira E, Bonnetaud C, Bordone O, Lespinet V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAF(V600E) mutation in papillary thyroid carcinoma: comparative analysis with three DNA-based assays. Thyroid. 2014;24(5):858–66. Epub 2014/01/15. 10.1089/thy.2013.0302 . [DOI] [PubMed] [Google Scholar]
- 36.Safaee Ardekani G, Jafarnejad SM, Khosravi S, Martinka M, Ho V, Li G. Disease progression and patient survival are significantly influenced by BRAF protein expression in primary melanoma. The British journal of dermatology. 2013;169(2):320–8. Epub 2013/04/05. 10.1111/bjd.12351 . [DOI] [PubMed] [Google Scholar]
- 37.Ghossein RA, Katabi N, Fagin JA. Immunohistochemical detection of mutated BRAF V600E supports the clonal origin of BRAF-induced thyroid cancers along the spectrum of disease progression. The Journal of clinical endocrinology and metabolism. 2013;98(8):E1414–21. Epub 2013/06/19. 10.1210/jc.2013-1408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerra A, Sapio MR, Marotta V, Campanile E, Rossi S, Forno I, et al. The primary occurrence of BRAF(V600E) is a rare clonal event in papillary thyroid carcinoma. The Journal of clinical endocrinology and metabolism. 2012;97(2):517–24. Epub 2011/12/16. 10.1210/jc.2011-0618 . [DOI] [PubMed] [Google Scholar]
- 39.Cheng SP, Hsu YC, Liu CL, Liu TP, Chien MN, Wang TY, et al. Significance of allelic percentage of BRAF c.1799T > A (V600E) mutation in papillary thyroid carcinoma. Annals of surgical oncology. 2014;21 Suppl 4:S619–26. Epub 2014/04/22. 10.1245/s10434-014-3723-5 . [DOI] [PubMed] [Google Scholar]
- 40.Finkel A, Liba L, Simon E, Bick T, Prinz E, Sabo E, et al. Subclonality for BRAF Mutation in Papillary Thyroid Carcinoma Is Associated With Earlier Disease Stage. The Journal of clinical endocrinology and metabolism. 2016;101(4):1407–13. Epub 2016/02/03. 10.1210/jc.2015-4031 . [DOI] [PubMed] [Google Scholar]
- 41.de Biase D, Cesari V, Visani M, Casadei GP, Cremonini N, Gandolfi G, et al. High-sensitivity BRAF mutation analysis: BRAF V600E is acquired early during tumor development but is heterogeneously distributed in a subset of papillary thyroid carcinomas. The Journal of clinical endocrinology and metabolism. 2014;99(8):E1530–8. Epub 2014/05/02. 10.1210/jc.2013-4389 . [DOI] [PubMed] [Google Scholar]
- 42.Kondo T, Nakazawa T, Murata S, Kurebayashi J, Ezzat S, Asa SL, et al. Enhanced B-Raf protein expression is independent of V600E mutant status in thyroid carcinomas. Human pathology. 2007;38(12):1810–8. Epub 2007/08/24. 10.1016/j.humpath.2007.04.014 . [DOI] [PubMed] [Google Scholar]
- 43.Mitsutake N, Miyagishi M, Mitsutake S, Akeno N, Mesa C Jr, Knauf JA, et al. BRAF mediates RET/PTC-induced mitogen-activated protein kinase activation in thyroid cells: functional support for requirement of the RET/PTC-RAS-BRAF pathway in papillary thyroid carcinogenesis. Endocrinology. 2006;147(2):1014–9. Epub 2005/10/29. 10.1210/en.2005-0280 . [DOI] [PubMed] [Google Scholar]
- 44.Ouyang B, Knauf JA, Smith EP, Zhang L, Ramsey T, Yusuff N, et al. Inhibitors of Raf kinase activity block growth of thyroid cancer cells with RET/PTC or BRAF mutations in vitro and in vivo. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12(6):1785–93. Epub 2006/03/23. 10.1158/1078-0432.ccr-05-1729 . [DOI] [PubMed] [Google Scholar]
- 45.Xing M. BRAFV600E mutation and papillary thyroid cancer: chicken or egg? The Journal of clinical endocrinology and metabolism. 2012;97(7):2295–8. Epub 2012/07/10. 10.1210/jc.2012-2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480(7377):387–90. Epub 2011/11/25. 10.1038/nature10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724 Epub 2012/03/08. 10.1038/ncomms1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are available within the paper and its Supporting Information files.