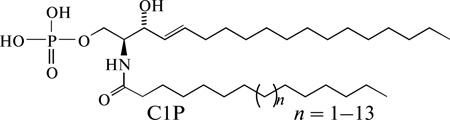
Abstract
A synthesis for fluorescent analogs of ceramide-1-phosphate bearing 9-anthrylvinyl or 4,4-difluoro-3a,4a-diaza-s-indacene-8-yl (Me4-BODIPY) fluorophore at co-position of fatty acid residue was carried out. The key stage of the synthesis is hydrolysis of corresponding sphingomyelins catalyzed by phospholipase D from Streptomyces chromofuscus; the enzymatic yield has been raised to 50–70% by appliance of organic solvent in the incubation medium.
Keywords: fluorescent lipid probes, ceramide-1-phosphate, BODIPY, anthrylvinyl, synthesis, phospholipase D
INTRODUCTION
Ceramide-1-phosphate (C1P) is a phospho-sphingolipid that is attracting increased interest as an important bioactive lipid molecule despite initial discovery over two decades ago in animal cells [1]. It is known now that C1P activates some enzymes, e.g. phospholipase A2, and blocks the activity of others (e.g. caspases), regulates eicosanoid biosynthesis, and participates in membrane fusion and in other essential vital functions—see reviews [2–4].
Continued elucidation of new C1P functional and regulatory roles in cells is expected to benefit from enhanced ways of obtaining different molecular C1P species (in animal cells it may contain fatty acyls ranging from C14 to C26 [4]) and different type analogs—radioactive, fluorescent etc. Currently, several enzymatic syntheses of C1P are known, including a number of chemical syntheses, which more precisely should be named semi-chemical syntheses because all start from natural compounds, sphingosine or ceramides.
Byun et al. [5] realized a ‘classical’ lipid rout that involved several stages and use of different protective groups and specific reagents. Szulc et al. developed a simplified synthesis that relied on phosphorylation of the primary HO group of ceramide or N-Boc-sphingosine in the presence of secondary one [6], but rather harsh conditions for dimethylphosphate group removal at the end of synthesis raise concerns about its effectiveness for obtaining C1P analogs with labile labels. Nussbaumer and co-authors described a simple way for obtaining C1Ps with different N-acyl species by direct acylation of sphingosine-1-phosphate [7]; the last is available but rather expensive. References to other C1P chemical syntheses are cited in the above papers [5–7].
In contrast to organic syntheses, C1P enzymatic syntheses currently involve two strategies: i—phosphorylation of ceramide with ATP and ceramide kinase, and; ii—hydrolytic splitting of sphingomyelin (SM) by phospholipase D (PLD). Schneider and Kennedy first obtained C1P in 1973 by phosphorylation of ceramide with ATP and ceramide kinase from E. coli [8]. Preiss et al. observed C1P formation when platelet lipids were treated with ATP and ceramide kinase [9]. On the basis of this reaction, Don and Rosen developed a method for C1P activity measurement [10]. Although this approach is convenient for preparation of 32P-labeled C1P [1], the complexity of the method limits its use for preparative purposes (see e.g. [12]).
Depending on the source of PLD, C1P production by hydrolysis of SM also can be inefficient. It is well known that PLD substrate specificity is limited to glycerophospholipids when this enzyme is obtained from various plant, animal or microbial sources [13]. However, Davidson and Long reported [14] that PLD from cabbage leaves splits SM, albeit at low rate. Analogous low activity was found in several microbial PLDs [15]. In contrast, moderate activity toward SM by PLD from the fungus Streptomyces chromofuscus has been reported [16–18]. Since we were looking for an efficient and direct way to obtain fluorescent-labeled C1Ps needed as probes for studies of lipid transfer mechanisms, our goal was to elaborate and improve the use of S. chromofuscus PLD to hydrolyze fluorescent anthrylvinyl (AV) and Me4BODIPY-labeled SM analogs [19, 20]. In this communication, the synthesis of fluorescent C1P analogs bearing residues of 12-(9-anthryl)-11E-dodecenic, 7-(Me4-BODIPY)heptanoic, and 15-(Me4-BODIPY)pentadecanoic acids, through PLD-mediated hydrolysis of the corresponding SMs is described.
RESULTS AND DISCUSSION
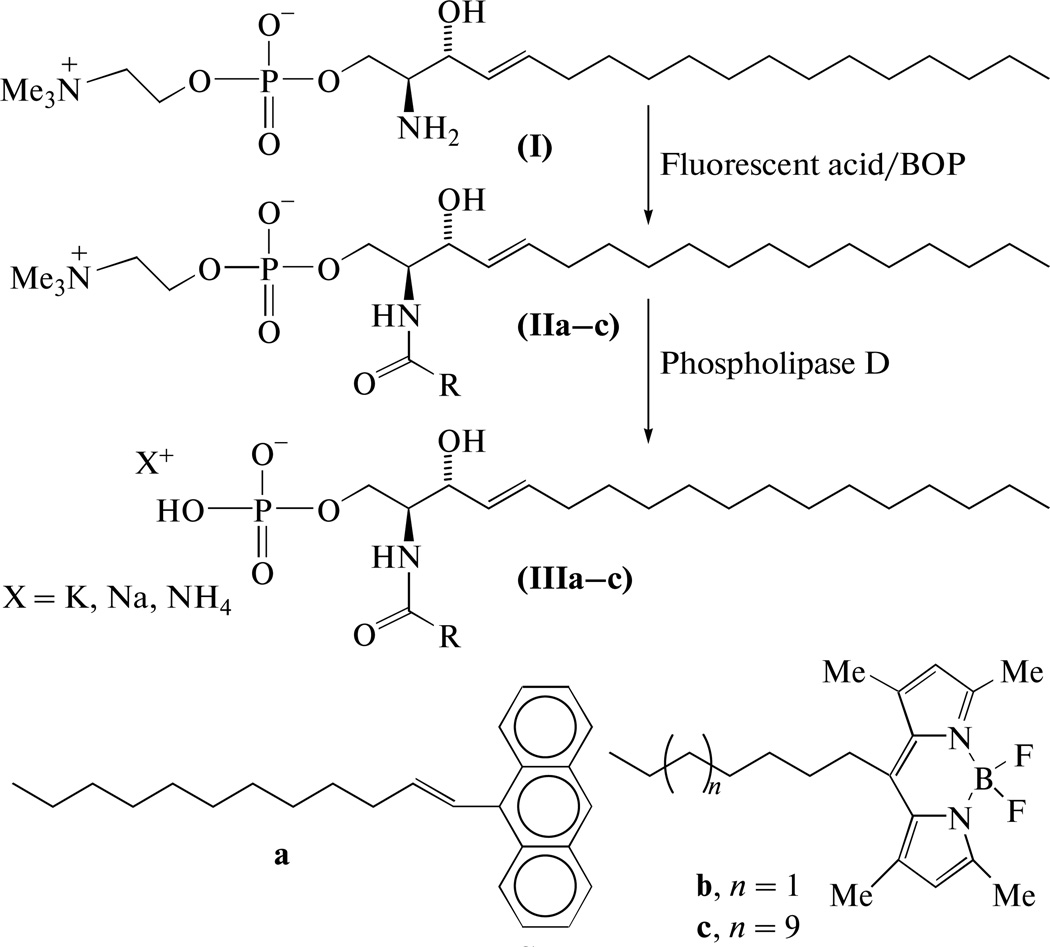
The starting compound for synthesis of fluorescent C1P analogs was sphingosine-1-phosphocholine (I) that then was acylated with fluorescent-labeled acid to yield SM (IIa–c; see Scheme). Removal of the choline moiety from SM by PLD isolated from S. chromofuscus yielded the desired probes (IIIa–c).
Scheme.
The second stage of the synthesis, enzymatic hydrolysis of SMs, was complicated with certain difficulties. The hydrolysis activity toward SM by PLD from S. chromofuscus, reportedly is only 20% of the rate for phosphatidylcholine [16, 17]. It also has been found that Triton X-100, BSA, fatty acids and diethyl ether activate hydrolysis, while SDS is inhibitory. The presence of Ca2+ ions is obligatory for S. chromofuscus PLD activity. Another feature distinguishing this enzyme from other PLDs is the hydrophobic site used by the protein to bind lipid substrate [16].
Our attempts to apply the procedure defined in the preceding works [16, 17] for hydrolysis of SMs (IIa–c), in a system containing Triton X-100 and CaCl2 (it is also recommended by enzyme supplier, Sigma-Aldrich—see http://www.sigmaaldrich.com) could be considered as only partially successful, at best. The desired reaction proceeded slowly to produce C1P analogs in yields <15%, together with unidentified products (detailed data are not shown). Substantial improvement of the reaction rates and yields was obtained when we substituted organic solvent for detergent in the enzymatic hydrolysis system. The best effect was obtained with methylene chloride: yields of C1P analogs (IIIa–c) increased to 54–71%. The spectral characteristics, 1H NMR, UV/visible and fluorescence, of the resulting probes were similar to those of other AV and Me4BODIPY probes (see Experimental).
The reason(s) for the dramatic improvement in the enzymatic yield is a matter of conjecture. Imamura and Horiuti suggested that diethyl ether accelerates the PLD activity due to the favorable solvent effect on the enzyme conformation [16]. But El Kirat et al., after studying the enzymolysis of a series of phospholipids by PLD from S. chromofuscus, came to the conclusion that the interfacial tension at a water–lipid interface is a key regulator of the hydrolytic process [21]. Hirche and Ulbrich-Hofman analyzed different organic solvents for their ability to stimulate phosphatidylcholine hydrolysis catalyzed by PLD from cabbage and from Streptomyces [22]. The worst was isooctane and the best, diethyl ether with activities differing 300-fold. By comparing the effects of the solvents on the critical concentration of lipid micelle formation, the size of the aggregates, the water content of the organic phase, and the interfacial tension, they also found that the enzymatic activity correlated with the interfacial tension on the water system/organic solvent interface and concluded that the packing density of the phosphatidylcholine aggregates was a key parameter controlling PLD activity.
It is quite probable that the preceding considerations can be applied to our data as well, but we also consider the solvent effect on lipid solubility to be another important parameter that should not be overlooked. Based on our observations, methylene chloride saturated with water is a rather good solvent for SMs, natural and fluorescent-labeled. It has been ascertained that in systems containing detergent (e.g. Triton X-100), PLD interacts with substrate included in micelles. However, in systems where detergent is replaced by organic solvent, a favorable effect is observed [18]. In such a case, enhanced solubility of the lipid substrate is a substantial factor that positively influences the enzymatic rate.
In conclusion, we find that the modified C1P synthesis described here offers a facile and convenient route for production of C1P probes.
EXPERIMENTAL
Mass spectra were recorded on a ESI spectrometer Agilent 6224 TOF LC/MS (USA) and on an ESI-TOF mass spectrometer MX-5311 (IAP RAS, St. Petersburg); 1H NMR spectra (δ, ppm, J, Hz) were recorded on a Bruker WM-700 spectrometer (United States). Electronic spectra of substances were measured on an SF-256 UVI spectrometer (LOMO Fotonika, St. Petersburg) in ethanol; fluorescence spectra (in ethanol) were recorded on a Hitachi F4000 spectrofluorimeter (Japan); the slit widths at excitation and emission were 3 nm.
DCC and DMSO were from Merck (Germany), DIPEA and S. chromofuscus phospholipase D were from Sigma-Aldrich (USA). Other reagents were from Reakhim (Russia). DIPEA was distilled over ninhydrin, then over powdery KOH; chloroform and methylene chloride were distilled over phosphorus pentoxide; other solvents were used after conventional purification. For column chromatography, silica gel Kieselgel 60 (Merck) was used, for TLC -Kieselgel 60 plates (Merck), detection with phosphomolybdic acid, ninhydrin, and UV irradiation. N-[15-(4,4-Difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene-8-yl)pentadecanoic acid [23], sphingosine-1-phosphocholine (I), N-[12-(9-anthryl)-11E-dodecenoyl]sphingosine-1-phosphocholine (IIa) [19], and N-[7-(4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4adiaza-s-indacene-8-yl)heptanoyl]sphingosine-1-phosphocholine (IIb) [20] were prepared as described. Evaporations in vacuo were performed at temperature <40°C.
N-[15-(4,4-Difluoro-1,3,5,7-tetramethyl-4-bora3a,4a-diaza-s-indacene-8-yl)pentadecanoyl]sphingosine-1-phosphocholine (IIc)
A solution of 15-(Me4-BODIPY)-pentadecanoic acid (16 mg, 33 µmol) and 12 µL DIDEA (68 µmol) in 1 mL dry CH2Cl2 was added to sphingosine-1-phosphocholine (15 mg, 31 µmol) dissolved in 0.5 mL dry DMSO, followed by 30 µL 50% DCC (~70 µmol) solution in chloroform and the mixture was stirred for 12 h. Column chromatography in gradient system chloroform against methanol with 5% 7 N NH4OH (10 → 25%) afforded 23 mg (76%) sphingomyelin (IIc) as a dark orange gum, Rf 0.48 in chloroform–methanol–7 N NH4OH, 65 : 35 : 8, system. UV, λmax, nm (ε, M−1 cm−1): 495 (7.8 × 104). MS, m/z: calculated for C51H91 F2N4O6P [M +H]+ 935.6737, found 935.672. 1H NMR (CD3OD–CDCl3): 6.06 (2H, s, ar), 5.68 (1 H, dt, J1 14.8, J2 7.2, =CHCH2), 5.42 (1H, dt, J1 14.8, J2 6.6, CH(OH)CH=), 3.59 (2H m, CH2N), 3.20 (9H, s, (9H, s, NMe3), 2.95 (2H, t, J 7.7, arCH2), 2.47 and 2.42 (12H, two s, arCH3), 2.15 (2H, t, J 7.4, CH2CO), 1.99 (2H, m, =CHCH2), 1.62 (2H, m, COCH2CH2), 1.30–1.20 (40H, br m, CH2), 0.86 (3H, t, J 7.0, CH2CH3). Fluorescence: λex 496 nm (λem 530 nm), λem 504 nm (λex 480 nm).
Enzymatic hydrolysis of sphingomyelin → ceramide-1-phosphate (general procedure)
The original preparation of PLD from S. chromofuscus (solution in a water–glycerol mixture, 53435 units/mL) was diluted by bidistillate to the ~500 units/mL concentration. Initially, sphingomyelin (II) (10–12 mg, 10–15 µmol) was added to a medium composed of 2 mL CH2Cl2, 1.5 mL 50 mM Na-borate buffer, pH 8.0, containing 0.02% NaN3, 1.5 mL bidistillate, and 40 µL 0.1 M CaCl2; the mixture was dispersed by ultrasonic water-bath treatment (1–2 min). Then 40 µL PLD solution (~20 units) was added to the mixture, the free volume of the vessel was filled with argon gas, and the vessel was stoppered and vortexed at 32°C. The hydrolysis course was checked by thin layer chromatography using a chloroform–methanol–7 N NH4OH, 65 : 35 : 8, solvent system (SM: Rf ~ 0.5; C1P: Rf ~ 0.2). When the initial SM content in the mixture appeared negligible (usually, 2–4 h) it was treated with 0.5 mL acetic acid, evaporated and dried in vacuo (up to 20 Pa). The residue was extracted with chloroform–methanol, 2 : 1, mixture (3 × 15 mL; phase separation by centrifugation), and the extract was filtered through filter cel and evaporated. C1P analogs (III) were separated by column chromatography with stepwise gradient system chloroform–methanol–7 N NH4OH, 90 : 10 : 0.5 → 70 : 27 : 3, followed by gel filtration on Sephadex LH-20 in a chloroform–methanol, 1 : 1, mixture.
N-[12-(9-Anthryl)-11E-dodecenoyl]sphingosine-1-phosphate (IIIa)
Yellowish gum, Rf 0.25, UV, λmax, nm (ε, M−1 cm−1): 255 (1.2 × 105), 349 (5 900), 367 (7800) and 385 (7000). MS, m/z: calculated for C44H67NO6P [M + H]+ 736.4706, found 736.4712. 1H NMR: 8.35 (1 H, s, ar H10), 8.27–8.31 (2 H, m, ar), 8.13 (2H, m, ar), 7.96 (2H, m, ar), 7.41 (2 H, m, ar), 6.92 (1H, d, J 11.3, arCH=CH), 6.20 (1H, dd, J1 11.3, J2 7.3, arCH=CH), 5.65 and 5.41 (2H, two m, CH=CH), 4.11 (1H, m, CHOH), 3.85 (1H, m, CHNH), 2.28 (2H, t, J 7.3, CH2CO), 2.25 (2H, dt, J1 7.3, J2 6.9, arCH=CHCH2), 2.09 (2H, t, J 7.2, CH2CO) 1.96 (2H, m, =CHCH2) 1.62 (2H, m, CH2CH2CO), 1.33–1.15 (34H, br. m, CH2), 0.84 (3H, t, J 6.5, CH3). Fluorescence: λex 256, 350, 368 and 387 nm (λem 430 nm), λem 420 nm (λex 370 nm).
N-[7-(4,4-Difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene-8-yl)heptanoyl]-sphingosine-1-phosphate (IIIb)
Red gum, Rf 0.22. UV/visible λmax, nm (ε, M−1cm−1): 495 (7.8 × 104). MS, m/z: calculated for C38H63BFN3O6P [M – F]+ 718.4532, found 718.4525. 1H NMR: 6.04 (2H, s, ar), 5.68 (1H, dt, J1 13.5, J2 6.5 =CHCH2), 5.42 (1H, dd, J1 13.5, J2 6.2, CH(OH)CH=), 2.93 (2H, t, J 7.8, arCH2), 2.46 and 2.39 (12H, two c, arCH3), 2.15 (2H, t, J 7.3, CH2CO) 1.97 (2H, m, =CHCH2), 1.60 (2H, m, COCH2CH2), 1.28–1.17 (24H, br m, CH2), 0.85 (3H, t, J 7.1, CH3). Fluorescence: λex 496 nm (λem 530 nm), λem 503 nm (λex 480 nm).
N-[15-(4,4-Difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene-8-yl)pentadecanoyl]-sphingosine-1-phosphate (IIIc)
Red gum, Rf 0.24. UV/visible λmax, nm (ε, M−1 cm−1): 495 (7.7 × 104). MS, m/z: calculated for C46H79BFN3O6P [M – F]+ 830.5784, found 830.5804. 1H NMR: 6.01 (2H, s, ar), 5.67 (1H, dt, J1 13.9, J2 6.6 =CHCH2), 5.43 (1H, dd, J1 13.9, J2 6.6, CH(OH)CH=), 2.89 (2H, m, arCH2), 2.45 and 2.36 (12H, two s, arCH3), 2.13 (2H, t, J 7.4, CH2CO), 1.98 (2H, m, =CHCH2), 1.58 (2H, m, COCH2CH2), 1.30–1.19 (40H, br m, CH2), 0.85 (3H, t, J 7.3, CH3). Fluorescence: λex 496 nm (λem 530 nm), λem 505 nm (λex 480 nm).
Acknowledgments
The authors are thankful to K.V. Antonov and I.D. Konstantinova (IBCh RAS) for MS spectra registration. This work was supported by the Russian Foundation for Basic Research (project 12-04-00168), also USPHS NIH-GM45928, and the Hormel Foundation (USA).
Abbreviations
- AV
9-anthrylvinyl
- C1P
ceramide-1-phosphate
- DIPEA
diisopropylethylamine
- Me4BODIPY
4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacen-8-yl
- PLD
phospholipase D
- SM
sphingomyelin
Footnotes
The article was translated by the authors.
REFERENCES
- 1.Bajjalieh SM, Martin TFJ, Floor E. J. Biol. Chem. 1989;264:14354–14360. [PubMed] [Google Scholar]
- 2.Gómez-Muñoz A. FEBS Letters. 2004;562:5–10. doi: 10.1016/s0014-5793(04)00211-x. [DOI] [PubMed] [Google Scholar]
- 3.Fyrst H, Saba JD. Nat. Chem. Biol. 2010;6:489–497. doi: 10.1038/nchembio.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornancin F. Cell. Signal. 2011;23:999–1008. doi: 10.1016/j.cellsig.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Byun H-S, Erukulla RK, Bittman R. J. Org. Chem. 1994;59:6495–6498. [Google Scholar]
- 6.Szulc ZM, Hannun YA, Bielawska A. Tetrahedron Lett. 2000;41:7821–7824. [Google Scholar]
- 7.Nussbaumer P, Hornillos V, Ghobrial M, Ullrich T. Chem. Phys. Lipids. 2008;151:125–128. doi: 10.1016/j.chemphyslip.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Schneider EG, Kennedy EP. J. Biol. Chem. 1973;248:3739–3741. [PubMed] [Google Scholar]
- 9.Preiss J, Loomis CR, Bishop WR, Stein R, Niedel JE, Bell RM. J. Biol. Chem. 1986;261:8597–8600. [PubMed] [Google Scholar]
- 10.Don AS, Rosen H. Anal. Biochem. 2008;375:265–271. doi: 10.1016/j.ab.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez-Muñoz A, Frago LM, Alvarez L, Varela-Nieto I. Biochem. J. 1997;325:435–440. doi: 10.1042/bj3250435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauzin L, Graf C, Sun M, Rovina P, Bouveyron N, Jaritz M, Winiski A, Hartmann N, Staedtler F, Billich A, Baumruker T, Zhang M, Bornancin F. J. Lipid Res. 2007;48:66–76. doi: 10.1194/jlr.M600399-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Heller M. Adv. Lipid Res. 1978;16:267–326. doi: 10.1016/b978-0-12-024916-9.50011-1. [DOI] [PubMed] [Google Scholar]
- 14.Davidson FM, Long C. Biochem. J. 1958;69:458–466. doi: 10.1042/bj0690458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Chem. Rev. 2011;111:6064–6119. doi: 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura S, Horiuti Y. J. Biochem. 1979;85:79–95. doi: 10.1093/oxfordjournals.jbchem.a132334. [DOI] [PubMed] [Google Scholar]
- 17.Estrada R, Stolowich N, Yappert MC. Anal. Biochem. 2008;380:41–50. doi: 10.1016/j.ab.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Uesugi Y, Hatanaka T. Biochim. Biophys. Acta. 2009;1791:962–969. doi: 10.1016/j.bbalip.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Molotkovsky JG, Manevich YM, Babak VI, Bergelson LD. Biochim. Biophys. Acta. 1984;778:281–288. [Google Scholar]
- 20.Boldyrev IA, Molotkovsky JG. Russ. J. Bioorg. Chem. 2006;32:78–83. doi: 10.1134/S106816201305004X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Kirat K, Prigent AF, Chauvet JP, Roux B, Besson F. Eur. J. Biochem. 2003;270:4523–4530. doi: 10.1046/j.1432-1033.2003.03841.x. [DOI] [PubMed] [Google Scholar]
- 22.Hirche F, Ulbrich-Hofmann R. Biochim. Biophys. Acta. 1999;1436:383–389. doi: 10.1016/s0005-2760(98)00143-x. [DOI] [PubMed] [Google Scholar]
- 23.Sachl R, Boldyrev I, Johansson L. Phys. Chem. Chem. Phys. 2010;12:6027–6034. doi: 10.1039/b926953c. [DOI] [PubMed] [Google Scholar]
- 24.Bittman R, Verbicky CA. J. Lipid Res. 2000;41:2089–2093. [PubMed] [Google Scholar]