Abstract
Epigenetics provides the opportunity to revolutionize our understanding of the role of genetics and the environment in explaining human behavior, although the use of epigenetics to study human behavior is just beginning. In this introduction, the authors present the basics of epigenetics in a way that is designed to make this exciting field accessible to a wide readership. The authors describe the history of human behavioral epigenetic research in the context of other disciplines and graphically illustrate the burgeoning of research in the application of epigenetic methods and principles to the study of human behavior. The role of epigenetics in normal embryonic development and the influence of biological and environmental factors altering behavior through epigenetic mechanisms and developmental programming are discussed. Some basic approaches to the study of epigenetics are reviewed. The authors conclude with a discussion of challenges and opportunities, including intervention, as the field of human behavioral epigenetics continue to grow.
The emergence of the field of behavior epigenetics is revolutionizing biology and our understanding of the role of genetics in explaining human behavior. As a discipline, epigenetics is not new. The earliest hint of the term epigenetics can be traced to Aristotle who referred to “Epigenesis” to describe development based on a sequence of steps. Spemann and Mangold (1924) introduced the idea that cells can turn information on and off, and Conrad Waddington (1942) coined the term “Epigenetics” as the cross-talk between genetic information and the environment.
However, the application of epigenetic methods to the study of human behavior is just beginning (Lester et al., 2011); it is in its embryonic stage, if you will, and provides an unprecedented opportunity to identify molecular processes underlying child behavior and development. The articles in this special section represent early studies that advance the field in several ways: by demonstrating how epigenetic methodology can be applied to the study of child development, by showing relations between epigenetic phenomenon and child development describing developmental processes at the molecular level, and by generating new hypotheses that will lead to further discovery. But what exactly is epigenetics?
Conventional definitions of epigenetics can be daunting. These definitions include, “mitotically and meiotically heritable changes in gene expression that cannot be explained by changes in DNA sequence,” or “changes in phenotype or gene expression caused by mechanisms other than changes in the underlying DNA sequence.” It is also customary to mention the description of epigenetics by Waddington (1942) who coined the term as “the branch of biology which studies the causal interactions between genes and their products, which bring the phenotype into being.” The keys to understanding these and other definitions lies in the prefix “epi-” which literally means “on,” “upon,” or “over” and what is meant by “DNA sequence.” A less technical description of epigenetics is that it refers to processes and mechanisms that physically lie on top of the DNA that affect the activity of the DNA but do not change the DNA itself. But to understand the articles in this special section, a deeper understanding is required.
Recall that cell nuclei contain chromosomes composed of strands of DNA that contain genes. Within the genes, the DNA molecule contains genetic information or instructions stored as a code. The code is made up of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). These bases form units of base pairs. A pairs with T, and C pairs with G. Cytosine, as we shall see, is the most important base in epigenetics.
DNA is a double-stranded molecule twisted around to form a spiral structure (the double helix). A single strand of a double-stranded DNA molecule is a sequence of these bases held to together by chemical bonds. However, a base on one strand can only link up with its pair on the other strand. T must be connected to A, and C must be connected to G. A particular portion of the strand of a gene might read, for example, ACCCGCGGTATTTCGATC. These are the sequences or basic structure (code) of the gene that is often referred to as “unchanged” by epigenetic mechanisms.
The DNA code is executed by producing a mirrored copy of itself, transcribed in the form of RNA, which goes on to be processed and used to produce proteins. Gene expression is the process by which genes are transcribed into RNA, which, in turn, makes the specific proteins that determine the structure and function of the individual gene. Gene expression is initiated by transcription factors, and molecules that bind or attach to specific DNA sequences, thereby initiating and controlling the rate of transcription of genetic information from DNA to RNA. Epigenetic mechanisms regulate this transcriptional machinery, and in so doing control gene expression. Thus, epigenetics controls the activity of the gene or how the gene functions.
Epigenetic Mechanisms
DNA is wrapped around spools, or proteins known as histones, making up the fundamental unit of chromatin, the nucleosome that allows the DNA to be tightly packed enough to fit into the nucleus. Chromatin is the complex of DNA and the associated histone proteins. If the way in which DNA is wrapped around the histones changes, gene expression can change.
This chromatin remodeling is accomplished through two main mechanisms, histone modification and DNA methylation. Histone modifications are posttranslational, occurring after the protein is produced, often while it is in place at the nucleosome, and involve a number of ways in which molecules attach to the “tails” that protrude from the histones and alter the activity of the DNA wrapped around them. Although well studied in nonhuman models (Graff & Mansuy, 2008; LaPlant & Nestler, 2011), techniques to study histone modifications have not been effectively worked out for human studies and are not amenable to large-scale population studies.
The second mechanism through which chromatin can be remodeled, and by far the most frequently studied epigenetic mechanism in human behavioral studies, and indeed, the only mechanism studied in the articles in this special section, is DNA methylation. DNA methylation is more stable than histone modifications and involves modification of the DNA itself in which a methyl group is added to a cytosine on the DNA. The methyl group is typically added to a C (cytosine) that is followed by a G (guanine) and is known as a cytosine–phosphate–guanine or CpG site. CpG methylation is an epigenetic mark added to (“on top of”) but does not change the underlying DNA sequence, meaning the methylated C still pairs with G and can still be read as a C by transcriptional or replication machinery.
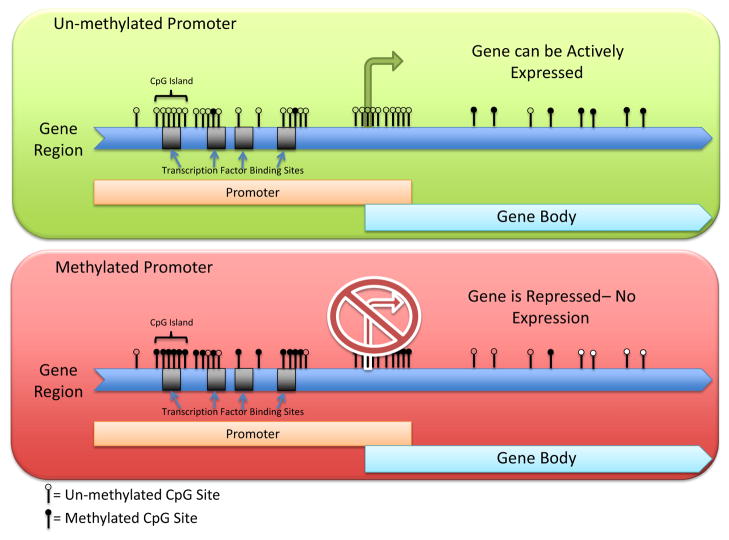
As shown in Figure 1, CpG pairs tend to be concentrated in the promoter, the region of the gene where the transcription of DNA to RNA begins. In most genes, the promoter is within regions where there is a high concentration of CpG sites known as CpG islands. A given C on one copy of the DNA in a single cell can only be methylated or not methylated. Thus, the percent of DNA methylation, which is typically reported in publications as amount or level, is the number of cells that is methylated at that site within the sample.
Figure 1.
Role of promoter region DNA methylation on gene expression. Upper panel depicts a gene promoter with unmethylated cytosine-phosphate-guanine (CpG) dinucleotides (open circles), allowing access to transcription binding sites and the opportunity for gene expression. Lower panel depicts methylated CpG dinucleotides (filled circles), which would block transcription factor binding and thus repress gene expression.
Levels of methylation are associated with how well the DNA is transcribed into RNA. As methylation levels increase there is less transcription until the level of DNA methylation reaches the point at which the gene is switched off (the double helix is closed). In the absence of DNA methylation, gene transcription is allowed to occur. Although DNA methylation is often described as an “on–off” switch, it is, in fact, more like a “dimmer” switch that gradually decreases gene expression as methylation increases, when it is thought of over a population of cells. In other words, if all of the cells associated with a particular gene are unmethylated, the population of cells can produce the amount of protein consistent with a fully active gene. Conversely, if the gene is fully methylated it will produce very little or none of the protein.
With some exceptions, the amount of DNA methylation related to human behavior is of the “dimmer switch” variety. For example, if a specific CpG site or region of the promoter is reported to have 50% methylation, which would indicate that within the sample examined, half of the cells within the population are methylated. If the RNA or protein product derived from that gene were measured in that same sample, we would expect to obtain an amount that is approximately half of what might be observed if the sample demonstrated 0% methylation.
This raises intriguing questions for human behavior: How much methylation of a given gene is necessary to affect human behavior? How does this amount vary by gene? As illustrated by the articles in the special section, the amount of methylation related to behavior varies by gene. Some behaviors may be affected by only slight changes in DNA methylation, while others may require a larger percent change in methylation; of course, the effects are also likely bidirectional, with behaviors impacting changes in methylation (Knopik, Maccani, Francazio, & McGeary, 2012). Could it be that the amount of methylation across many genes contributes to individual differences in behavior?
There are also epigenetically related mechanisms, including those that modify histones, that occur in conjunction with DNA methylation, and act to regulate transcription. Beyond DNA and histones, microRNAs (miRNAs) are molecules that act as posttranscriptional regulators of gene expression and have been related to child behavior in a few studies (Maccani, Padbury, Lester, Knopik, & Marsit, 2013; Mundalil Vasu et al., 2014). In the same way in which DNA methylation regulates transcription of DNA to RNA, miRNAs regulate the steps of translation, from RNA to protein, either through accelerating the destruction of the RNA or blocking the RNA’s ability to be translated into a protein.
In short, epigenetic mechanisms change gene expression, the action or activity of the gene, thereby altering cell function. The gene is the blueprint; it carries the code, the instructions. But how those instructions are carried out can be modified through epigenetic mechanisms. There are many metaphors that are used to describe the differences between genetics and epigenetics. The gene is the hardware of the computer; epigenetics is the software. Genes load the gun, epigenetics pulls the trigger. The prose of Macbeth is the genes, but the many interpretations, the variations in performance, is epigenetics. Dare we borrow from Hamlet, “the play’s the thing. ” Epigenetic mechanisms control how the gene is expressed. This is the fundamental importance of epigenetics.
A non-Shakespearian example: NR3C1 is a well-studied gene, arguably the most studied gene in child developmental studies (Bick et al., 2012; Romens, McDonald, Svaren, & Pollak, 2015) including studies in this special section. NR3C1 encodes, that is, specifies the genetic code, for the glucocorticoid receptor (GR) and is involved in the regulation of cortisol. GR is the receptor to which cortisol binds. Methylation of NR3C1 results in reduced expression, a smaller number of GR proteins available, and so fewer binding sites resulting in higher levels of circulating cortisol.
It is well known that too much or too little cortisol is related to changes in behavior including behavior disorders and psychopathology (Doom & Gunnar, 2013). There are numerous prenatal and postnatal biological, social, and environmental factors that have been related to DNA methylation of NR3C1 altering its expression resulting in overproduction or underproduction of cortisol levels (Glover, O’onnor, & O’Donnell, 2010; Lester, Conradt, & Marsit, 2013; Romens et al., 2015; Tyrka et al., 2015).
Recent Human Epigenetics Research
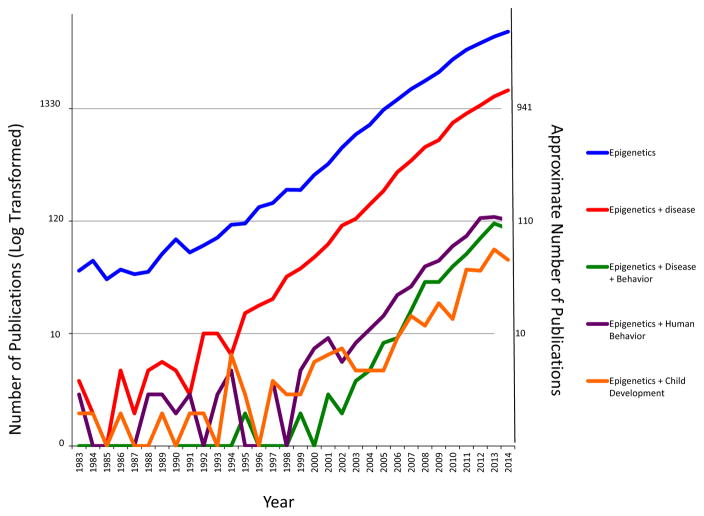
There are thousands of studies of epigenetics that have been conducted over the last 40 years. However, as mentioned earlier, the application of epigenetics to the study of human behavior is just beginning. Figure 2 is a citation search for epigenetic articles by year of publication from 1983 to 2014 using PubMed and shows the recent emergence of the field of behavioral epigenetics. The figure shows the number of published articles using the search terms “epigenetics,” “epigenetics and disease,” “epigenetics, disease, and behavior” “epigenetics and human behavior,” and “epigenetics and child development.” The search includes filters for the search terms with the number of publications plotted on a logarithmic scale because of the low number of publications involving epigenetics and behavior relative to the other epigenetic categories (see Figure 2). In 1983, altered DNA methylation was found in some cancers and, as shown in the Figure 2 (blue line), the field of epigenetics virtually exploded and was (and still is) dominated by cancer research. There were 42 “epigenetic” publications in 1983 and almost 7,000 by 2014.
Figure 2.
Number of PubMed citations from 1983 to 2014 using the following search terms: (1) “epigenetics,” including articles on cancer and nonhuman studies; (2) “epigenetics and disease”; (3) “epigenetics, disease, and behavior,” filtering out cancer articles, but including nonhuman studies; (4) “epigenetics and human behavior,” filtering out the nonhuman studies; and (5) “epigenetics and child development.” We used a logarithmic scale to display publications by year because there were a large number of studies published using search terms 1–3, and fewer for search terms 4–5, that is, to adjust for the negative skewness in the data. The actual number of studies that correspond to the log-transformed values is shown on the z-axis.
The human genome project was published in 2000. Mapping the human genome led to the need to explain how genes were regulated. Thus, the European Epigenome Project began in 2004 followed by the NIH Roadmap Epigenome Project in 2009. The study of epigenetics expanded beyond cancer to the study of other diseases including diabetes, and metabolic disorders, nutrition, and conditions such as undernutrition with almost 2,000 “epigenetic and disease” studies (red line) as of 2014.
Before 2006 there were fewer than 10 behavioral studies in nonhuman models (green line) and 103 by 2014. Severe mental and physical disorders in children due to the failure of normal epigenetic mechanisms were identified since 1992 with the discovery that the developmental disorders Prader–Willi and Angelman syndromes were due to epigenetic processes (Flint, 1992; Hall, 1992). As of 2014, there were 124 studies of “epigenetics and human behavior” (purple line). There were fewer than 10 studies of “epigenetics and child development” (orange line) before 2006 and 53 publications by 2014. We can see from this figure that the trajectory of the number of epigenetic studies reflects the explosion of research across all fields of study including our, albeit, nascent discipline, further reinforcing why this is a “special” section.
Epigenetics and Development
Epigenetics is part of, in fact necessary, for normal development especially during sensitive developmental periods and plays a critical role in maintaining genomic stability. In fact the term was initially used as part of the description of normal embryonic development. The best example of this is the process of cellular differentiation. Following fertilization, cells divide and undifferentiated stem cells become differentiated (specialized) in specific regions. Stem cells are generally unmethylated. Through DNA methylation, some genes are activated while others are inhibited or shut down. By controlling which genes are expressed or repressed, the cell adopts its specialized phenotype, giving rise to all types of cells of the body, including neurons, muscle cells, epithelium, endothelium of blood vessels, and so on, with different methylation patterns specific to each cell type.
In addition to its role in embryonic development, epigenetics is also involved in many other aspects of both normal and abnormal development throughout the life span. The concept has expanded to include epigenetic changes related to pre- and postnatal environmental and biological factors. Certainly there is a rich and extensive literature on how these environmental and biological factors affect behavior and development through adolescence, some which are represented in this special section (see Beach et al., 2016; Naumova et al., 2016).
The role of epigenetics is most likely to mediate relations between these environmental and biological factors and child outcome. Epigenetic changes have been related to prenatal factors including poor nutrition, maternal depression, stress, posttraumatic stress syndrome smoking, prematurity, and neurodevelopmental disorders (Conradt, Lester, Appleton, Armstrong, & Marsit, 2013; Essex et al., 2013; Heijmans et al., 2008; Romens et al., 2015; Uddin et al., 2010). Postnatally, epigenetic alterations have been related to factors including environmental adversity, child maltreatment, and abuse, stress, parenting, psychopathology, and behavior disorders (Lester, Marsit, Conradt, Bromer, & Padbury, 2012). With respect to child development, epigenetics typically refers to how signals from the environment (prenatal or postnatal) trigger molecular changes, presumably in brain cells, but potentially in various cells that alter behavior. Neural plasticity enables the organism to change (i.e., reprogram) structure and function in response to environmental cues. The adaptive significance is that plasticity enables a range of phenotypes to develop from a single genotype depending on environment factors.
The mechanism for this is thought to be developmental programming or the resetting of physiological parameters due to environmental events. Thus, although epigenetic changes are generally stable, they are also dynamic and can be altered by environmental events. Research on epigenetics offers the opportunity to study the molecular underpinnings of these effects as seen in the eight studies in this special section. Of particular interest to developmentalists might be epigenetic marks that regulate transcriptional activity involved in brain development including DNA differentiation and synthesis, neuronal plasticity, cell proliferation, cell cycle regulation, and apoptosis (cell death). The example above of the role of DNA methylation of NR3C1 regulating cortisol levels is instructive for several reasons. It is thought to be due to DNA methylation of NR3C1 in the brain (hippocampus) in the region of the promoter that is controlled by the transcription nerve growth factor (NGFI-A) that affects brain development. It is also related to cortisol, the stress hormone that has been implicated in the development of behavior problems and psychopathology in children as well as long-term medical problems in adults (Miller, Chen, & Zhou, 2007; Reynolds, 2013). The manuscripts in this special section by Stroud et al. (2016) and Parade et al. (2016) specifically focus on DNA methylation of the NR3C1 gene, examining how factors such as maternal smoking and early adversity, respectively, impact the status of methylation of this gene. Additional genes of interest in this special section include SLC6A4, which regulates serotonin exposure and is sensitive to variations in maternal care (Champagne & Curley, 2009). Montirosso et al. (2016) found that methylation of SLC6A4 is predictive of infant temperament at 3 months. A comparatively understudied gene in human behavioral epigenetic research is OXTR, the oxytocin receptor. This is surprising given oxytocin’s association with social affiliation, bonding, and stress (Feldman, 2012). Smearman et al. (2016) found that methylation of OXTR was associated with abuse in childhood, and moderated the effect of this abuse on symptoms of depression and anxiety in adulthood.
Approaches to the Study of DNA Methylation
Our discussion so far has focused on epigenetic mechanisms that affect specific or individual genes. However, it would be naïve to think that human behavior is affected by epigenetic alterations of single genes. The candidate or target gene approach provides detailed information on small regions and enables us to study genes and their pathways as in the above example of DNA methylation of NR3C1 and the regulation of cortisol levels.
A step beyond examining single genes is to examine a set of genes that are involved in a common pathway of signaling. For example, the hypothalamic–pituitary–adrenal (HPA) axis is a consistent topic of studies of development and programming as it plays a central role in stress reactivity and can be programmed by various environmental factors. (Glover et al., 2010; Lester et al., 2015; Meaney, 2010). Although the GR (encoded by NR3C1) is a key gene in this pathway, a number of other genes also are involved. 11-Beta-hydroxysteroid dehydrogenase Type II (HSD11B2) is a gene that encodes an enzyme that controls the levels of active cortisol by inactivating cortisol to cortisone, thus inhibiting its ability to bind to active GRs. The study by Conradt et al. (2016) examines how methylation of these two genes (NR3C1 and HSD11B2) are impacted by maternal sensitivity and depression, and in turn, how their epigenetic state is related to cortisol levels. The HPA axis has additional levels of control, from the release of cortisol following stress regulated by corticotropin-releasing hormone (CRH), the inactivation of CRH by the CRH binding protein, and within cells where GR activity can be controlled by FK506-binding protein 5 (FKBP5). The study by Kertes et al. (2016) examines all of these members of the pathway and the way that traumatic stress, including the stress of war, impacts the DNA methylation status of these genes.
It is also possible to interrogate thousands of genes throughout the genome. Microarray technology is a technique for the simultaneous measurement of many genes at once and was used in a study of the DNA methylation status of 28,000 CpG sites in relation to childhood stress (Essex et al., 2011). The entire genome can also be scanned. A genome-wide scan was used, for example, in a study to profile the methylation of > 485,000 CpG loci relating prenatal mercury exposure to newborn neurobehavior (Maccani et al., 2015). Genome-wide approaches can identify patterns of epigenetic alterations throughout the genome and point to new critical genes or pathways and inform overall processes. Thus, candidate gene and genome-wide approaches should be seen as complementary and can generate new hypotheses. In this special section, the studies by Beach et al. (2016) and Naumova et al. (2016) utilize these genome-wide approaches to generate novel hypotheses regarding how parenting can impact the epigenome, and in the case of Naumova et al., how that epigenomic variation is associated with psychosocial adjustment.
Challenges and Opportunities for Epigenetics Research
Any new field of science raises a multitude of issues and epigenetics is no exception. Due to space limitations, we can only briefly describe some of the major challenges being faced. At the same time we want to highlight the optimism in the epigenetics research community in addressing these challenges and limitations.
Tissue
Clearly we cannot conduct experimental studies and examine brain tissue as can be done with other species (other than studies of postmortem brain, which has its own caveats). In human studies, other accessible tissues are used, the most common being placenta, blood, and saliva/cheek swabs. Thus, we must take care to consider the tissue specificity of epigenetic mechanisms and the interpretation of findings from studies of accessible tissues in studies of human development.
For example, studies in blood must consider how the variability in DNA methylation could be affecting pathways involved in immune responses, which has, of course, been linked to mental health outcomes including stress. Buccal cells from saliva/cheek swabs have been posited to be potentially reflective of the epigenetic status of the brain as these cells derive from the same primitive germ layer (the ectoderm) and so early programming effects during development may be coincident in these tissues. The extent to which epigenetic changes in these (or other) tissues can be used as a proxy for brain tissue is not known. Another possibility is that different tissues provide different epigenetic information, that the meaning of DNA methylation could be tissue specific and provide complementary information. What they have in common is that they provide evidence of an epigenetic “footprint,” that is, an epigenetic event or processes was involved, even if the specificity of the event is uncertain.
All tissue samples, including blood, placenta, or even saliva samples are heterogeneous collections of cells, and the DNA methylation or other epigenetic features examined represent the averaged state across these heterogeneous cells. Methods have been developed, particularly for the genome-wide studies, to address these issues of cell mixture, allowing researchers an opportunity to control for this potential confounder and identify signals beyond those that can be explained by this heterogeneity.
Associational Studies
As a result of our inability to conduct the kind of experimental work in humans that is done in nonhuman models, human behavioral epigenetic studies are, to date, associational. Of course, this could be said about most developmental studies but behavioral experimental designs have been used in developmental research and could be developed for human behavioral epigenetic work. The approach from the field of epidemiology offers another perspective in which even if we do not know (and we may never know) the specific mechanisms by which epigenetics affects behavior, the fact that epigenetics predicts behavior may be sufficient. For example, as discussed next, evidence-based use of epigenetics for diagnostic purposes and intervention would be warranted, even perhaps ethically mandated, if they relieved human suffering. We know full well that in medicine as well as other disciplines diagnosis and treatment are not necessarily based on mechanistic understanding.
Normative Studies
Little is known about what are normal epigenetic marks, such as levels of methylation in candidate genes, in genome-wide analysis, in different tissues, and the longitudinal stability (or lack thereof) of epigenetic changes across the life span including the identification of sensitive periods.
Intervention and Diagnosis
There are epigenetically based pharmacological treatments approved by the Food and Drug Administration for some cancers. In patients with the glioblastoma brain tumor, epigenetic markers are used to predict longer survival and to determine response to pharmacological treatment. Dietary changes and supplements such as folates and methyl donors are being used, sometimes in the name of epigenetics, albeit with little supporting research. Meaney and colleagues (Liu et al., 1997) showed that the quality of maternal care in rodents resulted in epigenetic changes in offspring that reduced cortisol stress reactivity. This opens the door for the consideration of behavioral interventions to reduce stress during pregnancy or, in a more general sense, to develop interventions based on epigenetic effects. In a translational study with humans, Murgatroyd, Quinn, Sharp, Pickles, and Hill (2015) found greater methylation of NR3C1 among infants whose mothers had low prenatal depression, but high postnatal depression, and that this effect was reversed by maternal self-reported stroking of infants during the first 5 weeks of life. These results pave the way for intervention studies, including randomized clinical trials, to test whether maternal touch may dampen the infant stress response via DNA methylation of NR3C1. Behavioral interventions could be developed to improve parenting and reduce or prevent behavioral problems in infants and children. Epigenetically based pharmacological treatments for psychiatric disorders and developmental disabilities could also be developed and there may be epigenetic signatures or biomarkers for the later development of these impairments. The ability to understand the molecular basis for why some children develop mental disorders and others do not could have far-reaching implications for personalized medicine. Especially in cases in which a specific risk factor has been identified, such as mood disorders, epigenetic biomarkers may lead to the early identification of those most predisposed to these disorders and key to developing early and personalized preventive interventions. Although it may seem premature to even think about using epigenetics as a basis for treatment and diagnosis, such consideration might be warranted given the fact that there is precedence for it in other domains.
Summary
The burgeoning field of behavioral epigenetics has “reprogrammed” preexisting notions of how genes and environment interact to influence human behavior across development. Now that we know that environmental differences can change gene expression through epigenetic mechanisms at the cellular level, epigenetics is, arguably, the quintessential gene–environment interaction. It has given us a new perspective on the importance of the prenatal and early caregiving environment, and has provided us with more agencies in considering other avenues of shaping our own development and health outcomes, given that we now know our DNA per se is not our “destiny.” In the first sentence of this introduction we used the word “revolutionize.” While not exactly a Copernican revolution, epigenetics will revolutionize developmental theory for the simple reason that any developmental theory will have to consider the role of epigenetics in at least two ways: by incorporating how our understating of the molecular underpinning of behavior informs developmental theory, and by the dynamic changes in behavior and development inherent from the epigenetic perspective. Epigenetics joins the ranks of the most fundamental basics of developmental science including “genetics,” “environment,” and “gene–environment interaction” precisely because epigenetics is nature and nurture. We hope the research in this special section stimulates new ideas for future research and has made the epigenetic field more accessible to developmental psychologists who wish to explore age-old questions using novel epigenetic methods.
Biography
Elisabeth Conradt is the recipient of the Society for Research in Child Development’s 2015 Victoria S. Levin Award for Early Career Success in Young Children’s Mental Health Research.
Contributor Information
Barry M. Lester, Alpert Medical School of Brown University and Women and Infants Hospital of Rhode Island
Elisabeth Conradt, University of Utah.
Carmen Marsit, Geisel School of Medicine at Dartmouth.
References
- Beach SRH, Lei MK, Brody GH, Kim S, Barton AW, Dogan MV, Philibert RA. Parenting, SES risk, and later young adult health: Exploration of opposing indirect effects via DNA methylation. Child Development. 2016;87:111–121. doi: 10.1111/cdev.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick J, Naumova O, Hunter S, Barbot B, Lee M, Luthar SS, … Grigorenko EL. Childhood adversity and DNA methylation of genes involved in the hypothalamus-pituitary-adrenal axis and immune system: whole-genome and candidate-gene associations. Development and Psychopathology. 2012;24:1417–1425. doi: 10.1017/S0954579412000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Curley J. Epigenetic mechanisms mediating the long-term effects of maternal care on development. Neuroscience and Biobehavioral Reviews. 2009;33:593–600. doi: 10.1016/j.neubiorev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Conradt E, Hawes K, Guerin D, Armstrong DA, Marsit CJ, Tronick E, Lester BM. NR3C1: The contributions of maternal sensitivity and maternal depressive symptoms to epigenetic processes and neuroendocrine functioning. Child Development. 2016;87:73–85. doi: 10.1111/cdev.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt E, Lester B, Appleton A, Armstrong D, Marsit C. The roles of DNA methylation of NR3C1 and 11β-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8:1321–1329. doi: 10.4161/epi.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doom JR, Gunnar MR. Stress physiology and developmental psychopathology: past, present, and future. Development and Psychopathology. 2013;25(4, Pt 2):1359–1373. doi: 10.1017/S0954579413000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex M, Boyce W, Hertzman C, Lam L, Armstrong J, Neumann S, Kobor M. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Development. 2013;84:58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex M, Shirtcliff E, Burk L, Ruttle P, Klein M, Slattery M, … Armstrong J. Influence of early life stress on later hypothalamic-pituitary-adrenal axis functioning and its covariation with mental health symptoms: a study of the allostatic process from childhood into adolescence. Development and Psychopathology. 2011;23:1039–1058. doi: 10.1017/S0954579411000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R. Oxytocin and social affiliation in humans. Hormones and Behavior. 2012;61:380–391. doi: 10.1016/j.yhbeh.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Flint J. Implications of genomic imprinting for psychiatric genetics. Psychological Medicine. 1992;22:5–10. doi: 10.1017/s0033291700032669. [DOI] [PubMed] [Google Scholar]
- Glover V, O’Connor T, O’Donnell K. Prenatal stress and the programming of the HPA axis. Neuroscience and Biobehavioral Reviews. 2010;35:17–22. doi: 10.1016/j.neubiorev.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behavioural Brain Research. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Hall JG. Genomic imprinting and its clinical implications. New England Journal of Medicine. 1992;326:827–829. doi: 10.1056/NEJM199203193261210. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, … Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertes DA, Kamin HS, Hughes DA, Rodney NC, Bhatt S, Mulligan CJ. Prenatal maternal stress predicts methylation of genes regulating the hypothalamic-pituitary-adrenocortical system in mothers and newborns in the Democratic Republic of Congo. Child Development. 2016;87:61–72. doi: 10.1111/cdev.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopik VS, Maccani MA, Francazio S, McGeary JE. The epigenetics of maternal cigarette smoking during pregnancy and effects on child development. Development and Psychopathology. 2012;24:1377–1390. doi: 10.1017/S0954579412000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Nestler EJ. CRACKing the histone code: cocaine’s effects on chromatin structure and function. Hormones and Behavior. 2011;59:321–330. doi: 10.1016/j.yhbeh.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester B, Conradt E, Marsit C. Epigenetic basis for the development of depression in children. Clinical Obstetrics and Gynecology. 2013;56:556–565. doi: 10.1097/GRF.0b013e318299d2a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester B, Marsit C, Conradt E, Bromer C, Padbury J. Behavioral epigenetics and the developmental origins of child mental health disorders. Journal of Developmental Origins of Health and Disease. 2012;3:395–408. doi: 10.1017/S2040174412000426. [DOI] [PubMed] [Google Scholar]
- Lester BM, Marsit CJ, Giarraputo J, Hawes K, LaGasse LL, Padbury JF. Neurobehavior related to epigenetic differences in preterm infants. Epigenomics. 2015 doi: 10.2217/epi.15.63.26585459. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester B, Tronick E, Nestler E, Abel T, Kosofsky B, Kuzawa C, … Wood MA. Behavioral epigenetics. Annals of the New York Academy of Sciences. 2011;1226:14–33. doi: 10.1111/j.1749-6632.2011.06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, … Meaney M. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- Maccani J, Koestler D, Lester B, Houseman E, Armstrong D, Kelsey K, Marsit CJ. Placental DNA Methylation Related to Both Infant Toenail Mercury and Adverse Neurobehavioral Outcomes. Environmental Health Perspectives. 2015;123:723–729. doi: 10.1289/ehp.1408561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani M, Padbury J, Lester B, Knopik V, Marsit C. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatric Research. 2013;74:272–278. doi: 10.1038/pr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of Gene × Environment interactions. Child Development. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Miller G, Chen E, Zhou E. If it goes up, must it come down? Chronic stress and the hypothalamic-pituitary-adrenocortical axis in humans. Psychological bulletin. 2007;133:25–45. doi: 10.1037/0033-2909.133.1.25. [DOI] [PubMed] [Google Scholar]
- Montirosso R, Provenzi L, Fumagalli M, Sirgiovanni I, Giorda R, Pozzoli U, … Borgatti R. Serotonin transporter gene (SLC6A4) methylation associates with NICU stay and 3-month-old temperament in pre-term infants. Child Development. 2016;87:38–48. doi: 10.1111/cdev.12492. [DOI] [PubMed] [Google Scholar]
- Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, … Mori N. Serum microRNA profiles in children with autism. Molecular Autism. 2014;5:40. doi: 10.1186/2040-2392-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Quinn J, Sharp H, Pickles A, Hill J. Effects of prenatal and postnatal depression, and maternal stroking, at the glucocorticoid receptor gene. Translational Psychiatry. 2015;5:e560. doi: 10.1038/tp.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova OY, Hein S, Suderman M, Barbot B, Lee M, Raefski A, Grigorenko EL. Epigenetic patterns modulate the connection between developmental dynamics of parenting and offspring psychosocial adjustment. Child Development. 2016;87:98–110. doi: 10.1111/cdev.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parade SH, Ridout KK, Seifer R, Armstrong DA, Marsit CJ, McWilliams MA, Tyrka AR. Methylation of the glucocorticoid receptor gene promoter in preschoolers: Links with internalizing behavior problems. Child Development. 2016;87:86–97. doi: 10.1111/cdev.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RM. Glucocorticoid excess and the developmental origins of disease: two decades of testing the hypothesis--2012 Curt Richter Award Winner. Psychoneuroendocrinology. 2013;38:1–11. doi: 10.1016/j.psy-neuen.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Romens SE, McDonald J, Svaren J, Pollak SD. Associations between early life stress and gene methylation in children. Child Development. 2015;86:303–309. doi: 10.1111/cdev.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smearman EL, Almli LM, Conneely KN, Brody GH, Sales JM, Bradley B, Ressler KJ, Smith AK. Oxytocin receptor genetic and epigenetic variation: Association with child abuse and adult psychiatric symptoms. Child Development. 2016;87:122–134. doi: 10.1111/cdev.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spemann H, Mangold H. Induction of embryonic primordia by implantation of organizers from a different species. International Journal of Developmental Biology. 2001;45:13–38. [PubMed] [Google Scholar]
- Stroud LR, Papandonatos GD, Salisbury AL, Phipps MG, Huestis MA, Niaura R, Lester B. Epigenetic regulation of placental NR3C1: Mechanism underlying prenatal programming of infant neurobehavior by maternal smoking? Child Development. 2016;87:49–60. doi: 10.1111/cdev.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Eslinger NM, Marsit CJ, Lesseur C, Armstrong DA, … Seifer R. Methylation of exons 1D, 1F, and 1H of the glucocorticoid receptor gene promoter and exposure to adversity in preschool-aged children. Dev Psychopathol. 2015;27:577–585. doi: 10.1017/S0954579415000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, … Galea S. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]