Abstract
We previously reported on cationic, pH-responsive p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) block copolymer micelles with high affinity for dental and biofilm surfaces and efficient anti-bacterial drug release in response to acidic pH, characteristic of cariogenic (tooth-decay causing) biofilm microenvironments. Here, we show that micelle pH-responsive behaviors can be enhanced through alterations in corona:core molecular weight ratios (CCR). Although similarly stable at physiological pH, upon exposure to acidic pH, micelles with CCR of 4.1 were less stable than other CCR examined. Specifically, a ~1.5-fold increase in critical micelle concentration (CMC) and ~50% decrease in micelle diameters were observed for micelles with CCR of 4.1, compared to no changes in micelles with CCR of 0.8. While high CCR was shown to enhance pH-responsive drug release, it did not alter drug loading and dental surface binding of micelles. Diblocks were shown to encapsulate the antibacterial drug, farnesol, at maximal loading capacities of up to ~27 wt% and at >94% efficiencies, independent of CCR or core size, resulting in micelle diameter increases due to contributions of drug volume. Additionally, micelles with small diameters (~17 nm) show high binding capacity to hydroxyapatite and dental pellicle emulating surfaces based on Langmuir fit analyses of binding data. Finally, micelles with high CCR that have enhanced pH-responsive drug release and binding were shown to exhibit greater antibiofilm efficacy in situ. Overall, these data demonstrate how factors essential for nanoparticle carrier (NPC)-mediated drug deliverycan be enhanced via modification of diblock characteristics, resulting in greater antibiofilm efficacy in situ.
Introduction
The majority of human infectious diseases are caused by biofilm formation, including those occurring in the mouth1–3. Biofilms are highly structured communities of microbial cells that are firmly adherent to surfaces and enmeshed in a matrix of extracellular polymeric substances such as exopolysaccharides (EPS). A distinctive feature of biofilm-related illnesses is that the embedded microorganisms become recalcitrant to antibiotic therapies1–3.
The cost to treat or prevent oral biofilm-related infectious diseases exceeds $81 billion in the US alone4. Among them, dental caries is the most prevalent and costly oral infectious disease5. Caries-producing (cariogenic) biofilms form and exist on pellicle-coated tooth enamel surfaces, which are negatively charged3, 6–9. Inefficacy of common chemotherapeutics such as antimicrobials against cariogenic biofilms is due to poor retention of topically introduced agents on dental surfaces, reduced drug availability within biofilm matrix, and drug recalcitrance of biofilm bacteria1–3, 10. Moreover, cariogenic bacteria within biofilms produce acidic pH (~4.5) microenvironments that further induces biofilm accumulation and acid-dissolution of the adjacent tooth enamel10–13.
Polymeric micelles have great potential in treatment of pathogenic biofilms due to tunable physicochemical parameters to match specific applications14–17. Specifically, for the treatment of cariogenic biofilms, drug delivery systems that destabilize and release antibacterial/antibiofilm drugs at acidic pH have shown to amplify drug efficacy in vivo17. Various pH-responsive delivery systems have been developed to release drugs within acidic environments, such as in tumors or cellular endosomes and lysosomes18–20. The incorporation of charged residues, such as DMAEMA, within micelle cores to disrupt micelle via protonation/deprotonation and electrostatic repulsion is a straightforward means to develop pH-responsive behaviors of nanoparticle assemblies18, 20–23. In addition, pH-responsive behaviors of micelles formed of block copolymers can be controlled through varying block copolymer compositions (i.e. ratio of hydrophilic coronas to hydrophobic cores)21. Generally, micelles with high ratios polar/hydrophilic coronas to hydrophobic cores respond more robustly to acidic pH21, 24, resulting in rapid release of drug via micelle destabilization.
Drug delivery characteristics, such as pH-responsive behavior and drug loading capacity are interrelated. For example, pH-triggered drug release may be enhanced by cores size reduction, whereas small cores may compromise drug-loading capacity for certain drugs. Both core size and polymer chemical constituents within hydrophobic cores have been shown to affect drug loading capacity25. For example, poly(styrene-alt-maleic anhydride)-b-poly(styrene) (PSMA-b-PS) micelles encapsulate the hydrophobic small molecule parthenolide (PTL) at capacities from 1% to 11 wt%. PTL loading increased with decreasing molecular weight ratios of PSMA/PS from 12.1 to 0.7, and with increasing micelle size26.
We previously reported that cationic p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) nanoparticle carriers (NPC) exhibit high affinity and drug loading capacity, pH-triggered drug delivery to hydroxyapatite surfaces (HA), dental pellicle (sHA), and biofilm exopolysaccharides (EPS) (gsHA), resulting in effective disruption of biofilm virulence in vivo17. NPC electrostatically bind to dental surfaces through multivalent interactions of corona p(DMAEMA) residues with negatively charged sites of hydroxyapatite and dental pellicle. In addition, hydrophobic and pH-responsive NPC cores mediate pH-triggered drug release behaviors17. However, the parameters that can be tuned to improve pH-responsive micelle structure destabilization have not been characterized. Here, we sought to achieve a robust pH-responsive micelle destabilization through alteration in corona:core molecular weight ratios (CCR). Meanwhile, farnesol, a hydrophobic anti-bacterial drug, was loaded into micelle nanoparticles. Alterations in drug loading capacity due to varied CCR were characterized. In addition to loading capacity, electrostatic binding to HA and sHA was characterized. Overall, a variety of polymers, with varied core and corona block molecular weights were synthesized and screened to extract the inter-related tunable parameters that can be employed in antibacterial drug delivery to oral biofilms. These properties can be applied in other micelle-based drug delivery system designs and optimization approaches.
Results and discussion
Polymer and micelle characterization
Reversible addition-fragmentation chain transfer (RAFT) polymerizations provide precise control over polymer molecular weights and polydispersity indices (PDI), and therefore were used to study the influence of distinct corona and core molecular weights on micelle stability. The structure, composition, and physical properties of pH-responsive p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) polymers are detailed in Table 1, along with other polymers used to further investigate the factors that enhance NPC mediated pH-responsive drug delivery. p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) diblocks were synthesized using a two-step RAFT polymerization with either constant (11 kDa) coronas and variable cores (2.7, 4.8, 7.4, 13.0 kDa) or constant cores (~7 kDa) and variable coronas (11.1, 16.7, 24.2 kDa) (Table 1). Amphiphilic p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) diblocks self-assemble into ~14–40 nm and monodisperse micelle-based nanoparticle carriers (NPC) (PDI<0.2) (Table 1). Micelle assembly occurs due to hydrophobic interactions among hydrophobic butyl methacrylate (BMA) residues, which represent 50% of p(DMAEMA-co-BMA-co-PAA) core blocks17. The p(DMAEMA) corona is 50% protonated at physiologic pH owing to tertiary amine residues (pKa ~7.5)27, contributing to cationic micelle surface potentials (ζ=+16–+19 mV) at physiologic pH (Table 1). Controls of p(DMAEMA) blocks (Table 1, DM1–4) were synthesized as well to assess how corona block molecular weight impacts binding to hydroxyapatite, used to emulate tooth mineral or dental enamel.
Table 1. Characteristics of amphiphilic diblock copolymers synthesized by RAFT polymerization.
Mn is number average molecular weight, PDI is diblock copolymer molecular weight polydispersity index, ζ are micelle surface zeta-potentials, CCR is corona;core molecular weight ratio, Size is diameter in monometer (nm). NPC is nanoparticle. DMAEMA is dimethylaminoethyl methacrylate. BMA is butyl methacrylate. PAA is propylacrylic acid. DM is p(DMAEMA) polymer. NA: not appicable (no micelle structure), and ND: not detectable.
| Polymers | Micelles | |||||||
|---|---|---|---|---|---|---|---|---|
| Corona Blocks | Core Blocks | Diblock Copolymers | ||||||
| Abbreviation | Composition | Mn | Composition | Mn | PDI | Size (nm) | ζ (mV) | CCR |
| NPC9/13 | p(DMAEMA) | 9.3 kDa | p(DMAEMA-co-BMA-co-PAA) | 21.9 kDa | 1.10 | 21±0.4 | +16±1.3 | 0.7 |
| NPC11/3 | p(DMAEMA) | 11.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 13.9 kDa | 1.03 | 14±0.3 | +15±2.1 | 4.1 |
| NPC11/5 | p(DMAEMA) | 11.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 16.0 kDa | 1.12 | 18±1.4 | +16±1.4 | 2.3 |
| NPC11/7 | p(DMAEMA) | 11.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 18.6 kDa | 1.10 | 22±2.4 | +16±1.1 | 1.5 |
| NPC11/13 | p(DMAEMA) | 11.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 24.2 kDa | 1.07 | 26±1.1 | +18±0.7 | 0.8 |
| NPC16/7 | p(DMAEMA) | 16.7 kDa | p(DMAEMA-co-BMA-co-PAA) | 24.7 kDa | 1.09 | 24±0.1 | +17±0.7 | 2.1 |
| NPC24/7 | p(DMAEMA) | 24.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 30.9 kDa | 1.12 | 29±1.7 | +19±1.3 | 3.6 |
| NPC24/13 | p(DMAEMA) | 24.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 37.8 kDa | 1.10 | 28±2.1 | +17±1.2 | 1.8 |
| NPC24/20 | p(DMAEMA) | 24.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 43.8 kDa | 1.12 | 26±0.8 | +16±0.9 | 1.2 |
| NPC24/22 | p(DMAEMA) | 24.2 kDa | p(DMAEMA-co-BMA-co-PAA) | 46.1 kDa | 1.14 | 28±1.5 | +15±1.5 | 1.1 |
| NPC50/5 | p(DMAEMA) | 50.5 kDa | p(DMAEMA-co-BMA-co-PAA) | 55.4 kDa | 1.21 | 41±3.4 | +18±1.8 | 10.3 |
| DM1 | p(DMAEMA) | 15.6 kDa | NA | NA | NA | ND (soluble) | ND | ND |
| DM2 | p(DMAEMA) | 25.9 kDa | NA | NA | NA | ND (soluble) | ND | ND |
| DM3 | p(DMAEMA) | 28.4 kDa | NA | NA | NA | ND (soluble) | ND | ND |
| DM4 | p(DMAEMA) | 32.1 kDa | NA | NA | NA | ND (soluble) | ND | ND |
Impact of NPC corona:core molecular weight ratios (CCR) on micelle pH-responsive behaviors
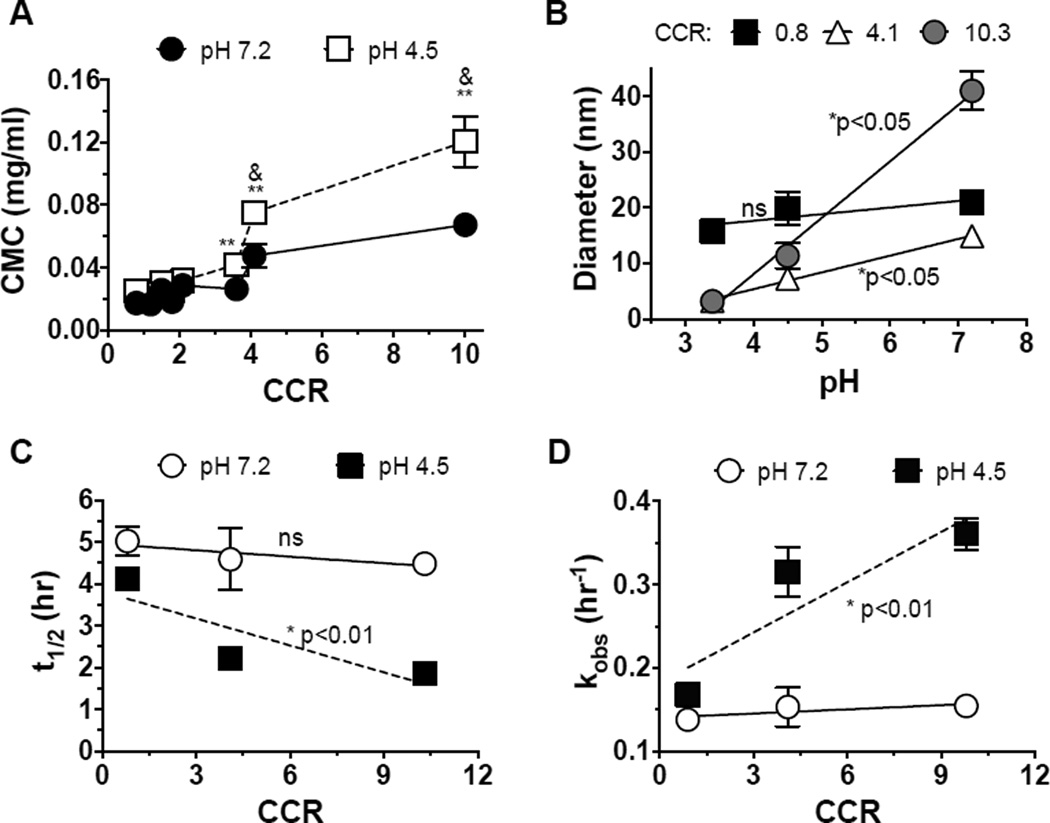
NPC stability is dependent on a balance between hydrophobic and electrostatic interactions within micelle cores and electrostatic interactions within micelle coronas. At physiological pH (pH 7.2), 50% protonated DMAEMA and 50% deprotonated PAA results in amphiphilic cores as they are present at equivalent ratios. At acidic pH (pH 4.5), DMAEMA is ~100% protonated, while PAA becomes neutralized. Thus, electrostatic repulsion due to protonation of micelle coronas and cores at acidic pH may lead to disruption of micelle structures. Typically, micelles with higher CCR yield less stable micelles25. To explore how micelle CCR impacts micelle pH-responsive behaviors, critical micelle concentrations (CMC) of seven NPC forming polymers with different CCR were characterized in the absence of farnesol. More robust pH-responsive behaviors of NPC, which result in high CMC at acidic pH (~4.5), were achieved via increases in CCR. As shown in Figure 1A, micelle CMC at pH 7.2 increased from ~0.02 mg/ml to ~0.69 mg/ml with increased CCR of micelles from 0.9 to 10. Similar trends of increasing CMC with increased CCR were observed at pH 4.5 (Figure 1A). In addition, significant increase in CMC was observed at acidic pH. At pH 4.5, NPC with CCR of 4.1 and 10.3 exhibited a robust increase in CMC, from ~0.05 mg/ml to ~0.08 and from ~0.07 mg/ml to ~0.12 mg/ml, respectively (Figure 1A). This is presumably due to strong electrostatic repulsion of DMAEMA within NPC coronas and cores, and relatively weak hydrophobic interactions within small NPC cores. Therefore, micelles with larger CCR have greater pH-responsive behavior, which may increase drug release kinetics from micelle cores at acidic pH18, 28–30. This destabilization is similar in principle to previously described pH-mediated membrane disruption of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) micelles for siRNA delivery20–23.
Figure 1. Impact of CCR on NPC pH-responsiveness and farnesol release.
A. Change in critical micelle concentration (CMC) at acidic pH as a function of pH. ** denote a significant difference when comparing pH 4.5 and pH 7.2 (p<0.001). & denotes a significant difference when comparing different CCR (p<0.01). The error bars represent standard deviation (n=3). B. NPC diameters as a function of pH. NPC with larger CCR (smaller cores) exhibit smaller diameters at acidic pH. C–D. Farnesol release half-life and release constants of nanoparticles with different CCR. Raw release data is shown in Supplementary Figure 1. * denotes a significant difference of the slope versus 0. ns = no significant difference. Note that A–B is NPC alone while C–D is farnesol-loaded NPC.
As less stable micelles result in smaller diameters at acidic pH21, 22, NPC diameters were analyzed for various CCR at a range of pH in the absence of farnesol (Figure 1B). No significant changes in NPC diameters as a function of pH were observed for nanoparticles with CCR of 0.8. However, NPC with CCR of 4.1 and 10.3 exhibited smaller diameters at acidic pH. For example, at pH 4.5, diameters of NPC50/5 micelles drop to the limits of detection (~10 nm), likely as repulsion of protonated DMAEMA residues renders the large micelle assemblies energetically unfavorable, resulting in transition to diblock unimers21. Similarly, at each pH, NPC with larger CCR exhibited smaller diameters, phenomena that is particularly apparent at acidic versus physiologic pH, as hydrophobic, core-stabilizing interactions are reduced due to protonation of both DMAEMA and PAA groups. The robust pH-responsive behavior of NPC with CCR of 4.1 and 10.3 is likely a result of weak hydrophobic interactions within relatively small molecular weight cores. This finding is in line with reports on block copolymer micelle structure destabilization at acidic pH for micelles with relatively low percentages of hydrophobic BMA residues within cores21. Thus, NPC with larger CCR are favorable for robust micelle destabilization at acidic pH.
To test how CCR increase impacts pH-triggered drug release, farnesol release from three micelles with different CCR (NPC11/3, NPC11/13 and NPC50/5) was characterized as a function of pH (Figure 1C–D). Farnesol release profiles were determined under sink conditions to emulate the in vivo environment where continuous drug losses after topical application would occur due to saliva clearance. For NPC11/13 with CCR 0.8, farnesol release half-life was t1/2 = 4.1 h and t1/2 = 5.0 h for release at pH 4.5 and pH 7.2, according to first-order release fits. The release half-life of micelles at pH 7.2 decreased to 4.5 h when increasing CCR to ~10, while the release half-life at pH 4.5 decreased to 1.9 h with CCR of ~10 (Figure 1C). As a result, the release rate constant increased ~2-fold from 0.17 hr−1 to 0.36 hr−1 at pH 4.5 versus pH 7.2 for NPC with CCR 10 (Figure 1D). Within a CCR range of 0.8 to 10, micelles are stable at pH 7.2. However, due to full protonation of DMAEMA and neutralization of PAA at pH 4.5, the overall cationic charge of the core and corona results in electrostatic repulsion at acidic pH, which destabilizes micelle structures thereby triggering farnesol release. Thus, NPC with higher CCR showed greater repulsion and disruption, resulting in significant increases in farnesol release rate. CCR correlation with drug release was consistent with reduction in micelle stability as measured by increased CMC and reduced micelle diameters (Figure 1A&B). Therefore, the high CCR micelle-based drug delivery system more favorably releases drug in response to acidic pH, such as of acidic pH microenvironments within cariogenic biofilms.
Characterization of farnesol-loaded NPC
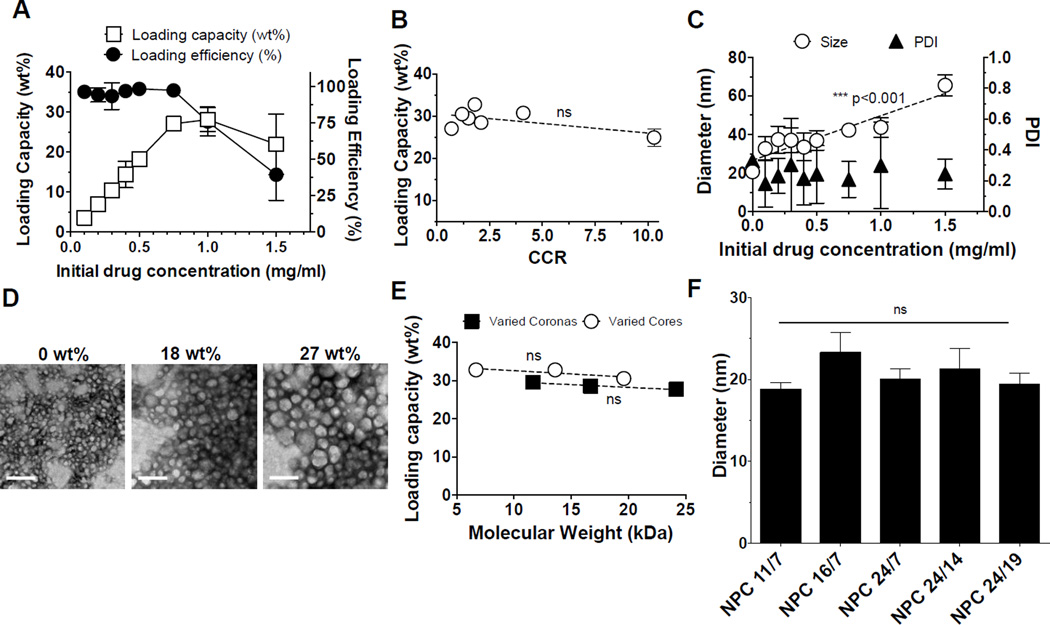
Optimally, NPC characteristics that enhance pH-responsive drug release would not alter drug loading. Therefore, we explored farnesol, a hydrophobic antibacterial agent, to identify how loading is impacted by NPC characteristics. NPC9/13, a formulation previously identified to have promising antibiofilm characteristics via local, pH-responsive farnesol release17, were loaded with farnesol at 27 wt% capacity (Figure 2A). NPC loading capacities for farnesol, quantified as the weight of drug loaded per weight of nanoparticle, were at the upper level of loading capacities for hydrophobic drugs reported for polymeric micelles (~20–37 wt%)14, 25. For example, upon loading 0.75 mg/ml farnesol per 2.7 mg/ml NPC at 97% loading efficiency (27 wt%) (Figure 2A), farnesol can be delivered at 27-fold higher amounts than its minimum inhibitory concentration (MIC) of ~0.028 mg/ml for Streptococcus mutans, a common cariogenic bacteria31–33. Additionally, the concentration of farnesol within nanoparticles is 440 times higher than its estimated aqueous solubility limit (~1.7*10−3 mg/ml, based on US EPA; Estimation Program Interface (EPI) Suite V.3.12) in the absence of a carrier. Below the maximal loading capacity of around 27wt%, farnesol loading efficiencies were above 94% (Figure 2A). However, at farnesol concentrations above 0.75 mg/ml, loading efficiencies decrease from 97% to 39% (Figure 2A) as hydrophobic NPC cores are saturated with encapsulated drug.
Figure 2. Characterization of farnesol-loaded NPC.
A. NPC9/13 loading capacity (le6 axis) and loading efficiency (right axis) was examined as a function of farnesol concentration. B. Loading capacities of micelles as a function of CCR. C. NPC9/13 diameters increase (circles) upon loading farnesol, and NPC PDI (triangles) were <0.3 upon loading with farnesol. D. Morphological characteristics of NPC loaded with farnesol by transmission electron micrographs (scale bar = 100 nm). 18 and 27 wt% of farnesol was loaded into NPC, respectively. E. Maximal drug loading capacity was measured as a function of corona and core molecular weights. Maximal drug loading capacity was independent of corona and core sizes within the tested NPC range. F. Increased diameters of micelles tested in Figure 2E a6er loading farnesol at maximum loading capacities compared to micelles alone. *** denote a significant difference of a trend from 0 as determined by two-tailed T-test (C). ns = no significant difference (B). Error bars represent standard deviation (n=4).
As discussed, a micelle with the greatest pH-responsive behavior is a priority to achieve the greatest antibiofilm drug efficacy. Thus, micelles with different CCR ranging from 0.8 to 10.3 were loaded with farnesol at maximum loading capacities(Figure 2B). No significant trends in drug loading were found as a function of CCR. All explored micelles showed similar farnesol loading capacity of ~28 wt%. This finding is counter to previously-reported trends of drug loading within micelles. Typically, the greatest drug loading is observed in micelles with the largest hydrophobic blocks and micelle sizes26, 34, 35, as large hydrophobic blocks allow for more favorable drug interactions. Thus, no alteration of drug loading among the examined micelles supports an alternative loading phenomena for farnesol.
For NPC9/13, when loaded at 27 wt%, NPC size increased from 21 nm to 61 nm (Figure 2C). NPC imaging via transmission electron microscopy (TEM) before and after loading confirms the observed size increases upon drug encapsulation at 18 wt% and 27 wt% (Figure 2D). Similar effects on size upon drug loading have been reported due to the contribution of encapsulated drug to micelle volume. For example, poly(ethylene glycol)-b-poly(lactic-co-glycolic acid) (PEG-b-PLGA) and poly(ethylene glycol)-b-poly(styrene) (PEG-b-PS) micelle diameters increased by 15 nm upon loading with β-carotene, a drug similar in structure to farnesol36. In addition, p(DMAEMA)-b-p(BMA) micelle sizes increased by 8–15 nm upon loading with doxorubicin18. Drug-loaded NPC diameter increases, as calculated from specific volumes of farnesol and NPC loaded at 18 wt%, were ~17 nm, which is similar to measured increases of ~16.5 nm. Therefore, nanoparticle size increases may be due to formation of a liquid drug phase within NPC cores that volumetrically contributes to increases in nanoparticle volumes. To test this hypothesis, two groups of nanoparticles with either the same corona sizes (~11 kDa) or core sizes (~7 kDa) were loaded with farnesol. As a result, NPC loading capacities did not significantly change as a function of corona or core size (Figure 2E). Also, encapsulation of farnesol volumetrically increased micelle diameter equally across all micelles (Figure 2F). These data imply that farnesol, reported herein to load extremely well into NPC structures, resulting in substantial increases in nanoparticle diameters (Figure 2C), may form an oily (hydrophobic) phase that can be exploited as an integral “lipid drop” within NPC cores for delivery of other drugs with similar properties. Enhancement of aqueous solubility of bioactive agents is critical for development of formulations for therapeutic use clinically.
Micelle electrostatic binding to hydroxyapatite (HA) or saliva coated surfaces (sHA)
We previously reported that the family of micelles explored herein exhibits strong affinities to dental pellicle and biofilm EPS matrix surfaces17. Thus, characterization of factors, including CCR, that impact NPC affinity to these surfaces was explored. Adsorption to surfaces depends on collective contributions of several parameters among which are surface potential, pH, micelle diameters and polymer molecular weight37–39. Therefore, micelle corona block molecular weights, and micelle diameters were altered to study NPC binding affinity to dental surfaces. Although teeth are covered by proteinaceous dental pellicle in vivo, for simplicity, hydroxyapatite surfaces (HA) were also used herein, as hydroxyapatite is the prominent mineral in teeth with overall negative ζ-potentials. Additionally, similar trends in electrostatic binding of NPC to HA, dental pellicle (sHA), and EPS surfaces (gsHA) were reported17.
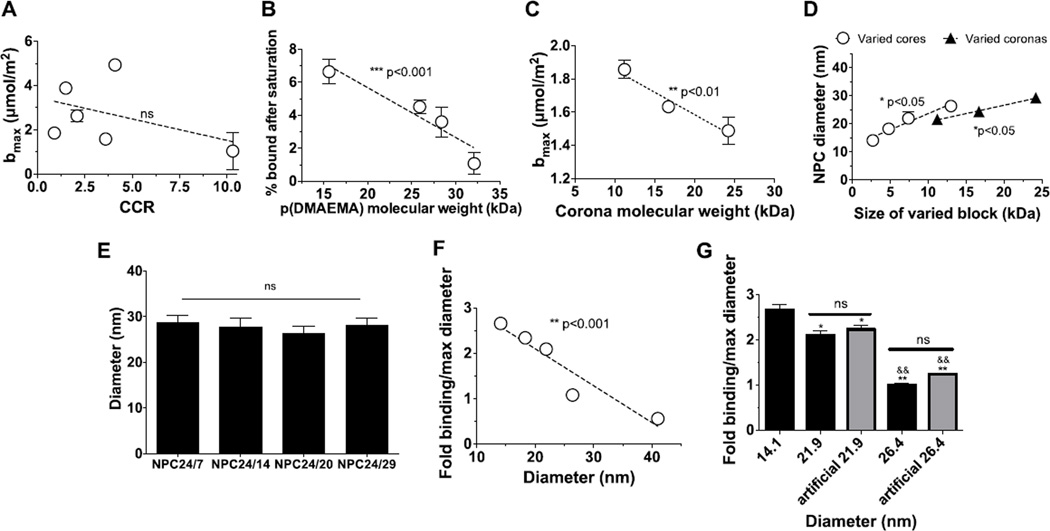
Binding of nanoparticles was first examined as a function of CCR, as robust pH-responsive drug release mediated by large CCR was the priority for biofilm treatment. Similar to micelle loading capacity, no significant alterations of micelle binding affinity to hydroxyapatite were found when increasing the CCR (Figure 3A). This phenomenon inspired us to explore the micelle characteristics that do affect binding to HA. Binding of p(DMAEMA) alone, in the absence of hydrophobic core-forming blocks, was first examined as a function of molecular weight. As shown in Figure 3B, p(DMAEMA) binding decreases slightly as molecular weight increases. Decreased binding with increased polymer chain length is attributed to entropic losses upon binding of long versus short polymer chains39–42. Therefore, smaller micelle coronas may be preferable for binding. However, polymers and NPC may interact differently with HA surfaces, due to micelle multivalent corona charge densities or micelle curvature. Thus, further examination was focused on binding of NPC with various corona sizes and diameters.
Figure 3. Electrostatic binding of micelles to HA surfaces.
A. Maximal binding capacities of micelles as a function of CCR. B. Decreases in p(DMAEMA) binding as a function of molecular weight. C. Confirmation of enhanced binding of NPC with small p(DMAEMA) molecular weights with uniform diameters. D. Increases in diameters as a function of corona and core sizes. E. Similar sizes were observed with micelles with greater overall molecular weights. F. Increases in HA binding observed as a function of diameter. G. Confirmation of enhanced binding of NPC with small diameters. Fold binding relative to micelles with maximum diameters (26.4 nm). ***p<0.001, **p<0.01 and *p<0.05 denote a significant difference of a trend from 0 as determined by two-tailed T-test (A, B, C, and D). ns = no significant difference. Specifically in G, * denotes a significant difference among 14.1 nm group and rest of the groups. (*p<0.05, **p<0.01); & denotes a significant different among 21.9 nm group and the rest of the groups (&&p<0.01). The error bars represent standard deviation (n=3).
To examine how corona sizes impact NPC binding affinity, nanoparticles with various (11.2 kDa, 16.7 kDa, and 24.2 kDa) corona sizes but similar diameters (~26 nm) were used. No change in NPC surface potentials was observed over these NPC (Table 1). Similar to the binding of p(DMAEMA) (Figure 3B), electrostatic binding of NPC to hydroxyapatite increased from 1.4 to 1.9 µmol/mm2 for nanoparticles with smaller, 11.2 kDa, corona sizes relative to NPC with 24.2 kDa corona sizes (Figure 3C). Therefore, small coronas are preferable for multivalent, electrostatic targeting of dental surfaces, which, similar to polymers, is likely due to lower entropic losses upon binding of small versus large molecular weight chains.
Other than corona sizes, the effect of micelle diameters on HA affinity was examined as variation of micelle diameters could alter the charge density on micelle surface. Micelle sizes were analysed to be between 17 and 30 nm via DLS (Table 1). For lower molecular weight polymers (micelles with 11 kDa corona or 7 kDa core), micelle size was dependent on diblock molecular weight and relative chain length of corona and core block. As shown in Figure 3D, size increases with either corona or core molecular weights within the range of explored block sizes. Accordingly, sizes of micelles with small core molecular weights would increase proportionally with overall diblock molecular weights. However, as shown in Figure 3E, micelles with greater 24 kDa corona molecular weights and varied cores sizes exhibited similar diameters. Therefore, it is possible that introduction of large charged p(DMAEMA) blocks results in water infiltration into micelle coronas and micelle diameter increases independent of core size, unlike in densely packed small corona micelles with robust core interactions. Accordingly, large corona NPC50/5 micelles (Table 1) possessed the largest diameters and exhibit greatest hydrophilicity (CCR=10.3).
To focus more on the impact of corona sizes on micelle binding, binding of NPC with 11.2 kDa corona size but different (14 nm, 18 nm, 22 nm, and 26 nm) diameters due to varied core molecular weights was examined. Binding of NPC to hydroxyapatite increased by ~2.7-fold for nanoparticles with the smallest diameter versus the largest17(Figure 3F). Micelle binding capacities increased with decreasing micelle sizes (1.9, 3.6, 4.3, and 5.0 µmol/m2 for diameter of 26, 22, 18, and 14 nm, respectively), suggesting that micelles with smaller diameters are likely to interact more with hydroxyapatite surfaces. Though not controlled by corona size, NPC50/5 binding capacity was also included in this data for comparison with data on small corona micelles. As large corona sizes and large micelle diameters are unfavorable for micelle binding, NPC50/5 exhibited reduced binding capacity to HA of ~1.0 µmol/m2. Therefore, NPC with similar coronas, but small diameters are preferable for multivalent, electrostatic targeting of dental surfaces.
As mentioned, farnesol is likely to form a continuous lipid phase within NPC cores upon drug loading, increasing NPC diameters proportionally to farnesol loading amount. Thus, specific amounts of farnesol can be loaded into micelles to alter NPC size independent of micelle composition and block sizes. To strengthen the conclusion that smaller micelle diameter are preferable for micelle binding, farnesol was used herein to change diameter of NPC11/3. NPC11/3 was loaded with ~4 and 16 wt% for loading capacity ~0.11 mg/ml and ~0.42 mg/ml farnesol per 2.7 mg/ml NPC to achieve diameters of ~22 nm and ~26 nm, comparable to the diameters of NPC11/7 and NPC11/13, respectively (Table 1). Binding of NPC to hydroxyapatite was significantly reduced in micelles with larger diameters or higher farnesol loading. Moreover, nanoparticles with farnesol-mediated “artificial” diameters have the same binding capacity as nanoparticles alone (no farnesol) with equivalent diameters (e.g., farnesol-loaded NPC11/3 versus NPC11/7, both exhibiting 22 nm diameters) (Figure 3G). Therefore, with uniform corona sizes, diameter is the only parameter identified here that influences NPC binding.
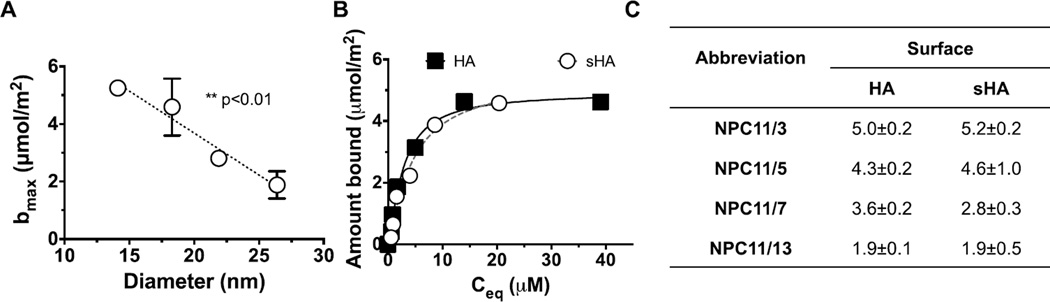
Uncoated HA surfaces rarely occur in the oral cavity, as in vivo, teeth are covered by a proteinaceous salivary film known as pellicle43. Thus, small micelles with high HA binding capacities were assessed for binding to more clinically-relevant dental pellicle surfaces of saliva coated HA (sHA) (Figure 4)7, 33, 43–45. Comparisons of NPC binding properties to HA and sHA versus micelle size were performed using the Langmuir equation. Specifically, micelles with similar coronas (11 kDa) and different core weights (3, 5, 7, 13 kDa) that exhibit different diameters, were characterized for binding of sHA surfaces (Figure 4A). Similar to targeting of HA surfaces, small micelles were favorable for binding to saliva-coated HA (sHA) (Figure 4A). Moreover, according to Langmuir fits to adsorption data, average maximal binding capacities of nanoparticles to HA versus sHA did not differ statistically (Figure 4B–C).
Figure 4. Electrostatic binding of micelles to saliva-coated dental surfaces.
A. Increased binding to sHA as a function of micelle diameter (n=4). B. Equilibrium binding profile of NPC11/3 at increasing polymer concentrations. Other micelles’ binding profiles are not shown here for simplicity. The solid and dotted lines represent Langmuir fits to the adsorption data (n=4 independent experiments). C. Langmuir fit parameters that define binding capacity (bmax). The equation parameters were calculated based on data presented in B (r2<0.98). HA, uncoated hydroxyapatite; sHA, saliva- coated HA. The error bars represent standard error (n=4).
Antibiofilm effects of farnesol delivery via optimized nanoparticles in situ
We demonstrated that at acidic pH, NPC with high CCR exhibit increased micelle destabilization and pH-triggered farnesol release. Additionally, no compromise in either maximal drug loading capacities for farnesol or binding to dental surfaces was observed for high CCR NPC. Moreover, small corona and diameter micelles exhibited better binding to HA and sHA dental surfaces. Therefore, high CCR NPC with small diameters (i.e., relative to NPC50/5), namely NPC11/3, was chosen as the optimized candidate for testing of antibiofilm NPC effects. NPC11/13, which has similar corona and core molecular weights to our previous work was used here for biofilm treatment comparison17.
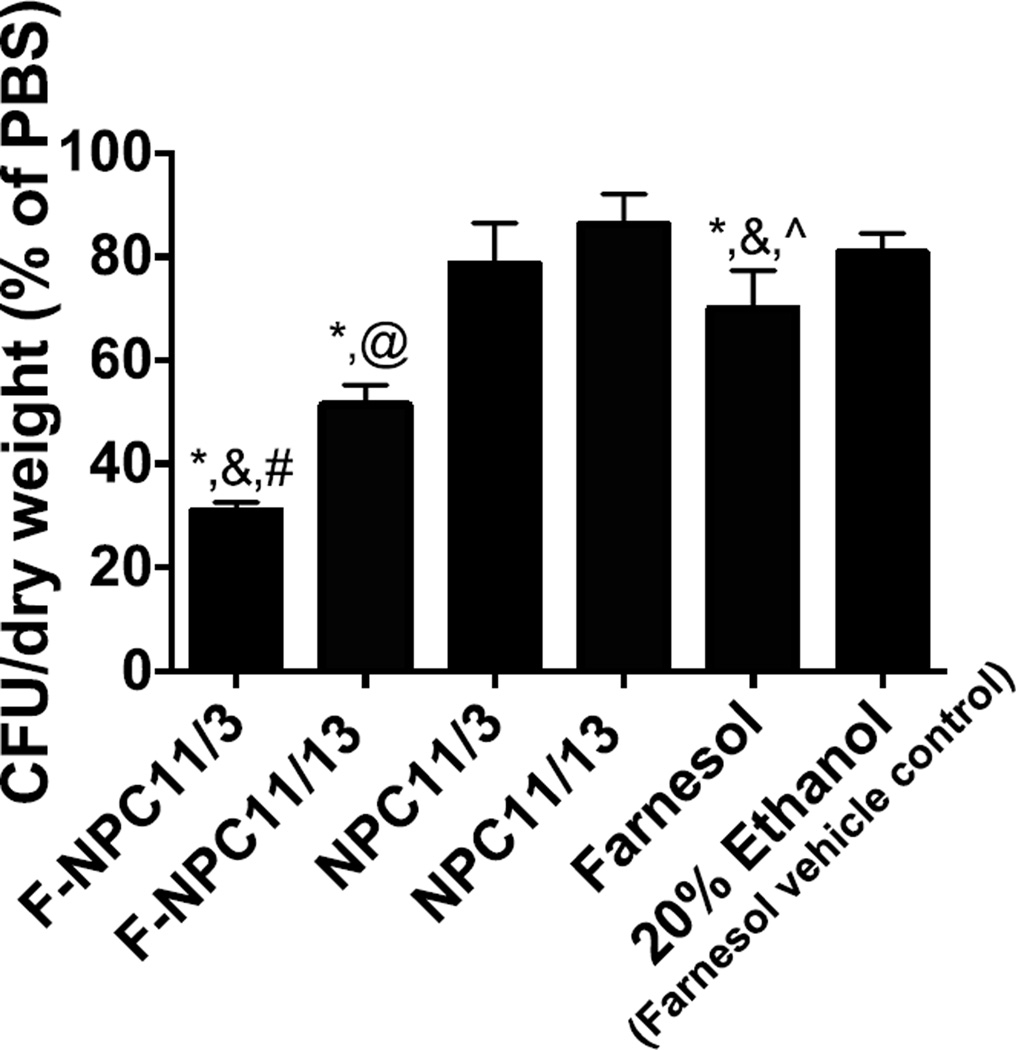
The impact of NPC-delivered farnesol against S. mutans biofilms was explored using an in vitro saliva-coated hydroxyapatite biofilm model. Farnesol was encapsulated within nanoparticles at maximal loading capacities. Equivalent farnesol concentrations were used for free-drug treatments, which were dissolved in 20% ethanol (vehicle) due to poor aqueous solubility. In addition, free nanoparticles (NPC11/3 and NPC11/13) or 20% ethanol were used as controls for various treatments. Treatments were applied topically 2 or 3 times per day (a total of 7 applications over 54 h), emulating clinically relevant dental treatment regimen17. We observed significant decreases in colony forming units (CFUs) per biofilm dry weight in both farnesol-loaded nanoparticle treated groups (Figure 5). In contrast, only a modest reduction in S. mutans viability within biofilms was observed when treated with free farnesol, consistent with previous observations46, 47. Most importantly, F-NPC11/3 showed more robust antibiofilm efficacy was observed as compared to F-NPC11/13 (~2-fold enhancement compared with free farnesol). The enhanced antibiofilm activity resulting from NPC11/3 is likely due to optimized characteristics including robust pH-responsiveness and higher maximal binding capacity, which drastically enhances farnesol retention at the pellicle-biofilm interface.
Figure 5. Antibiofilm effects of farnesol delivery via different nanoparticles.
Increased efficacy of antibiofilm effects was achieved in biofilms treated with farnesol loaded NPC11/3 as compared to farnesol loaded NPC11/13. F-NPC = farnesol loaded nanoparticles. *p<0.05 versus PBS, &p<0.05 versus F-NPC11/13, #p<0.05 versus NPC11/3, @p<0.05 versus NPC11/13, ^p<0.05 versus F-NPC11/3 as assessed using Student’s t-tests for relevant pairwise comparisons (α= 0.05, n=6 independent replicates). The error bars represent standard error.
Conclusions
We characterized the primary parameters of pH-responsive nanoparticle carriers (NPC), formed of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA), that enhance micelle destabilization and drug release at acidic pH, and impact micelle drug loading and targeting to negatively-charged dental surfaces. First, pH-triggered and robust micelle destabilization behaviors resulted in micelles with CCR=4.1 or greater. At acidic pH, micelles with high CCR likely exhibit greater pH response due to repulsion and hydration of protonated DMAEMA residues within micelle coronas and cores relative to weak hydrophobic interactions within small micelle cores. Neither CCR nor overall molecular weight affected farnesol loading capacity. Similarly, binding capacity of NPC did not vary as a function of CCR. However, decreases in micelle diameters resulted in stronger micelle binding. Finally, optimized micelles with enhanced pH-responsive drug release and binding were shown to exhibit greater antibiofilm efficacy in situ. Thus, micelles with high CCR and small coronas and diameters are preferable to achieve enhanced drug bioavailability on surface and within acidic biofilm microenvironments.
Materials and methods
Materials
Chemicals and materials were supplied by Sigma-Aldrich unless otherwise specified. 4-Cyano-4-[(ethylsulfanylthiocarbonyl)-sulfanyl]pentanoic acid (ECT) and propylacrylic acid (PAA) were synthesized as described previously. 2,2-azobisisobutyronitrile (AIBN) was recrystallized from methanol. Dimethylaminoethyl methacrylate (DMAEMA) and butyl methacrylate (BMA) were distilled prior to use, and poly(ethylene glycol) monomethylether methacrylate was filtered over basic alumina to remove inhibitor.
Methods
Polymer synthesis
Polymers were synthesized by reversible-addition fragmentation chain transfer (RAFT) polymerizations that provide precise control over polymer molecular weights and polydispersity indices (Mw/Mn, PDI<1.3). Specifically the following polymers were synthesized: p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA), p(DMAEMA). RAFT polymerizations were performed in the presence of monomers, 2,2-azobisisobutyronitrile (AIBN) as the initiator, and ECT as chain transfer agent (CTA). The specific reaction conditions for each polymer are detailed below.
Synthesis of poly(dimethylaminoethyl methacrylate), p(DMAEMA)
3 g of dimethylformamide (DMF) (40 wt% monomer) and 2 g of distilled DMAEMA was introduced into reaction vessels. The molecular weights, PDI and initial monomer to CTA ratio (DP, [M]0: [CTA]0) for p(DMAEMA) that was used for binding experiments or for p(DMAEMA) that was used as macroCTA (1st block) for synthesis of block copolymers with p(DMAEMA-co-BMA-co-PAA) are reported in Table 1. CTA to initiator ratios ([CTA]0: [I]0) of 10:1 were used. Reactions were purged with nitrogen using for 40 min using a Schlenk line prior to transfer to an oil bath at 60 °C for polymerization (t = 6 h). The resulting polymers p(DMAEMA) were isolated by precipitation in 30:70 diethyl ether: pentane and centrifugation. p(DMAEMA) polymers were redissolved in acetone and precipitated in pentane three times and dried overnight in vacuo.
Synthesis of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) block copolymers
Diblock copolymers of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) were synthesized using p(DMAEMA) macroCTA (Table 1). The desired stoichiometric quantities of DMAEMA, PAA, and BMA (25:25:50 %, respectively) were added to the p(DMAEMA) macroCTA dissolved in DMF (25 wt% monomers, DP=[M]0:[CTA]0 (Table 1)). CTA to initiator ratios ([CTA]0: [I]0) were 5:1 for NPCs except for 10:1 for NPC9/13, with AIBN as the initiator. Following the addition of AIBN, the solutions were purged with nitrogen for 40 minutes and allowed to react at 60 °C for 24 hr. The resulting diblock copolymers were isolated by precipitation in 30:70 diethyl ether/pentane and centrifugation. The polymers were then redissolved in acetone and precipitated in pentane three times and dried overnight in vacuo.
Polymer labeling with Fluorescein
Polymers used for confirmation of binding to HA and sHA surfaces were labeled with Fluorescein cadaverine (Biotium, Hayward, CA). Labeling was performed at 1:10:10 molar ratios of NPC polymer: fluorescein: ethyldimethylaminopropyl carbodiimide (EDC) for 3 hours in PBS at 5 mM N-hydroxysulfosuccinimide (sulfo-NHS). Labeled polymers were purified using dialysis against distilled, deionized water (ddH2O) using 3500 kDa MWCO membranes (Spectra/Por®, Spectrum Labs, Rancho Dominquez, CA). Dialysis water was changed twice a day for 10 days and polymers were collected via lyophilization.
Characterization of polymers
Molecular weight determination and confirmation of polymer compositions
Absolute molecular weights and polydispersities (Mw/Mn, PDI) of all polymers were determined by gel permeation chromatography (GPC, 1200 Series (Shimadzu Technologies, Santa Clara, CA) equipped with a miniDAWN TREOS, multi-angle light scattering (MALS) instrument (Wyatt Technologies, Santa Barbara, CA) and a refractive index detector (Shimadzu Technologies, Santa Clara, CA) [columns: TSK Gel Super H-H guard; TSK Gel HM-N for gel separation, Tosoh Bioscience, Montgomeryville, PA]. HPLC-grade DMF containing 0.05 M LiBr at 60 °C was used as the mobile phase at a flow rate of 0.35 mL/min. Absolute molecular weights were determined using reported dn/dc values for p(DMAEMA) (0.06 ml/g)48, 49. Block copolymers that included pH-responsive blocks ((p(DMAEMA-co-BMA-co-PAA)) were analyzedvia 1H NMR spectroscopy (Bruker Avance 400) to confirm second block composition, as previously described20.
Formation and characterization of NPC
Size, polydispersity indices (PDI), and zeta (ζ) potentials of nanoparticles of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) were measured using a Zetasizer (Malvern Instruments, UK). The measurements were performed at 0.2 mg/ml and 2.7 mg/ml for size measurements. Data is reported as number average. ζ-potentials were measured at 0.2 mg/ml and pH 7.2 for all micelles.
Critical micelle concentrations (CMC) of NPC
CMC of micelle-based nanoparticle carriers (NPC) composed of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) was approximated using solvatochromic shifts in fluorescence emission of PRODAN® (Molecular Probes, Eugene, Oregon)50, 51. Briefly, PRODAN® dissolved in methanol was aliquoted into black 96-well plates. After drying overnight, micelle solutions at a range of concentrations (0–2 mg/ml) were added and incubated overnight to achieve final PRODAN® concentrations of 5.45*10−4 mg/ml. PRODAN® emission was measured at two wavelengths (Ex/Em1: 360 nm/436 nm and Ex/Em2: 360 nm/518 nm) that corresponds to emission of PRODAN® in hydrophobic and hydrophilic phases, respectively. The ratio of emissions (hydrophobic phase/hydrophilic phase, Em1/Em2) was plotted versus of log(micelle concentration), and CMC was determined as a concentration at which the emission ratio begins to increase with polymer concentration.
Encapsulation of Antibiofilm Agent, Farnesol, in Polymer Micelles
Micelles were loaded with farnesol by sonication similar to Tang et al52. Briefly, farnesol emulsions at a range of concentrations (0.2–1.5 mg/ml) were prepared by sonication (Sonic Raptor 250, Omni International, Kennesaw, GA) in ddH2O at 40 Watts. Emulsions were then mixed with 2.7 mg/ml of p(DMAEMA)-b-p(DMAEMA-co-BMA-co-PAA) micelles in glass scintillation vials. These solutions were placed in a bath sonicator (50 HT, VWR) for 15 minutes. Based on calibration curves, the change in absorption of farnesol emulsions at 700 nm (as a measure of turbidity) was correlated to the amount of drug loaded. According to the amount of farnesol loaded, loading capacities () and efficiencies () were calculated. Where Wtloaded is the amount of loaded drug, Wtmicelle is the amount of micelle, and Wt0 is the initial amount of farnesol in emulsion. Nanoparticle sizes both before and after farnesol loading were examined using Zetasizer (Malvern Instruments, UK) as described above, and confirmed by transmission electron microscopy (TEM). Briefly, micelles were loaded with farnesol at loading capacities of 0 wt%, 18 wt%, and 28 wt%, transferred to carbon coated nickel grids, and dried for 2–5 minutes in the presence of 2% (w/v) phosphotungstic acid as a contrast agent. The images of free and loaded NPC were taken at magnifications of 200,000x using a Hitachi 7650 transmission electron microscope (Hitachi, Schaumburg, IL), attached to 11 megapixel Erlangshen digital camera system (Gatan, Pleasanton, CA).
Adsorption of polymers onto dental mimetic surfaces
Preparation of mimetic dental surfaces
Two materials that emulate dental surfaces were used to assess polymer binding: uncoated hydroxyapatite (HA) that emulates tooth mineral (enamel) and hydroxyapatite coated with saliva (sHA) which mimics salivary pellicle formed on tooth surface. Hydroxyapatite (CHTTM, BioRad) beads were used for HA and sHA surfaces. For formation of pellicle surfaces, HA beads were washed twice with buffer (50 mM KCl, 1 mM KPO4, 1 mM CaCl2, 1 mM MgCl2, 0.1 mM PMSF and 0.02% NaN3, in ddH2O, pH 6.5). Washed HA beads were incubated with human whole saliva, process known to result in formation of saliva coated-hydroxyapatite (sHA), similar to pellicle layer formed on teeth, as detailed elsewhere45.
Assessment of polymer binding
Quantitative assessments of NPC binding to dental surfaces as a function of corona and core sizes and with increase in polymer concentrations were performed in triplicate by incubation of polymers with HA surfaces for 40 minutes at 37°C. The amount of bound polymer was analyzed based on the difference in absorption/emission of Fluorescein fluorescence (Ex/Em: 480 nm /530 nm) before and after incubation as NPC exhibit linear, concentration dependent increase in emission at 530 nm. The binding was measured by an Infinite N200 PRO microplate reader (Tecan, Switzerland) at optimal gain. Binding of p(DMAEMA) to HA versus molecular weight (DM1–4) was similarly.
Adsorption equilibrium curves
Adsorption of NPC to HA and sHA was further analyzed at concentrations of 0–25 µM, and Langmuir () equation was fit to adsorption equilibrium data by GraphPad Prism software (v.6.03), where Cbound and Ceq are the bound and equilibrium NPC concentrations respectively, and n is the cooperativity constant. From the fits, binding capacity of NPC to HA surfaces (bmax [µmol/m2]) was calculated. NPC adsorption was expressed relative to a surface area of hydroxyapatite beads, which was calculated according to the average diameters and density of the beads as provided by the manufacturer (80µm and 0.63 g/ml, respectively).
Antibiofilm activity of nanoparticle-mediated farnesol delivery
Streptococcus mutans UA159, a well-characterized EPS matrix producing and cariogenic pathogen53, was used to assess the effect of nanoparticle-mediated delivery of farnesol on biofilm formation using a topical treatment regimen17, 46. The treatment solutions were: free nanoparticles (1.9 mg/mL in PBS, pH 7.0), farnesol-loaded nanoparticles (1.9 mg/mL loaded with 0.5 mg/mL farnesol, in PBS, pH 7.0), free farnesol (0.5 mg/mL farnesol, in PBS, pH 7.0, 20 % ethanol (EtOH)), vehicle control for free farnesol (PBS, pH 7.0, 20 % EtOH), and PBS (pH 7.0). A 20 % v/v ethanol solution was used as a vehicle to solubilize free farnesol, which is otherwise insoluble in aqueous media46. Biofilms of S. mutans UA159 were formed on saliva-coated hydroxyapatite surfaces (12.7 mm in diameter, 1 mm in thickness, Clarkson Chromatography Products Inc., South Williamsport, PA, USA) as detailed elsewhere54. The HA discs were placed vertically using a custom-made holder and grown in UFTYE (pH 7.0) with 1% sucrose at 37°C and 5% CO2.
The sHA discs and biofilms were treated for 10 min, washed twice with sterile saline (0.89% NaCl), and transferred to culture media. The first treatment was applied directly after salivary pellicle formation (sHA), and then treated disks were transferred to culture media containing S. mutans (105 CFU/mL). Biofilms were allowed to form on the discs without interruption for 6 h, at which point a second treatment was applied. The next day (20 h), the biofilms were treated three times with 6 h intervals. The following day the biofilms were treated at 44 h and 50 h, and then collected to assess the amount of colony forming units per dry weight of biofilm. For CFU and dry weight assessment, biofilms were removed from sHA discs via sonication46, 47; our sonication procedure does not kill bacterial cells, while providing optimum dispersal and maximum recoverable counts. Aliquots of biofilm suspension were serially diluted and plated onto blood agar plates, and after 48 h incubation (37°Cand 5% CO2), the colonies were visually counted. The remaining biofilm suspension was washed twice with ddH2O, oven-dried (into preweighed foil boats) for 2 h, and weighed.
Statistical analyses
Significance among groups was assessed by One or Two-Way AVOVA followed by Tukey’s tests for multiple comparisons at p<0.01. Alternatively, a significant difference of regression trend slopes from 0, were assessed by Two-Tailed t-tests and p-values were reported. Goodness of fits to Langmuir equation for binding equilibrium was assessed by extra sum-of-squares F test on compatibility of each data set to binding data (p<0.05), and by Shapiro-Wilk W normality tests on residuals (p<0.05). A significant difference among fit parameters was assessed by extra sum-of-squares F test on fit parameters (p<0.05). In situ biofilm treatment data was analyzed using unpaired t-tests of relevant pairwise comparisons, as indicated (p<0.05).
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the NIH (K12ES019852, career development award to DB; R01DE018023, PI: HK), NSF (BMAT-DMR1206219, PI: DB), and IADR/GlaxoSmithKline Innovation in Oral Care Awards (PI: MK) for funding this work. The authors would like to thank James L. McGrath lab for access to the Malvern Zetasizer and TECAN microplate reader, Karen Bentley of URMC Electron Microscope Research Core for assistance with TEM imaging, and Dr. Linda M. Callahan and Dr.Paivi M. Jordan of URMC Confocal and Conventional Microscopy Core for assistance with laser scanning microscopy.
References
- 1.Davies D. Nature reviews. Drug discovery. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 2.del Pozo JL, Patel R. Clin Pharmacol Ther. 2007;82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 3.Flemming HC, Wingender J. Nature reviews. Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 4.Flemmig TF, Beikler T. Periodontology 2000. 2011;55:9–15. doi: 10.1111/j.1600-0757.2010.00383.x. [DOI] [PubMed] [Google Scholar]
- 5.Dye BA, Tan S, Smith V, Lewis BG, Barker LK, Thornton-Evans G, Eke PI, Beltran-Aguilar ED, Horowitz AM, Li CH. Vital and health statistics. Series 11, Data from the national health survey. 2007:1–92. [PubMed] [Google Scholar]
- 6.Young A, Smistad G, Karlsen J, Rolla G, Rykke M. Advances in dental research. 1997;11:560–565. doi: 10.1177/08959374970110042501. [DOI] [PubMed] [Google Scholar]
- 7.Weerkamp AH, Uyen HM, Busscher HJ. Journal of dental research. 1988;67:1483–1487. doi: 10.1177/00220345880670120801. [DOI] [PubMed] [Google Scholar]
- 8.Miyake N, Sato T, Maki Y. The Bulletin of Tokyo Dental College. 2013;54:97–101. doi: 10.2209/tdcpublication.54.97. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds EC, Wong A. Infection and immunity. 1983;39:1285–1290. doi: 10.1128/iai.39.3.1285-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koo H, Falsetta ML, Klein MI. Journal of dental research. 2013;92:1065–1073. doi: 10.1177/0022034513504218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, 3rd, Heydorn A, Koo H. PLoS pathogens. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlafer S, Raarup MK, Meyer RL, Sutherland DS, Dige I, Nyengaard JR, Nyvad B. PloS one. 2011;6:e25299. doi: 10.1371/journal.pone.0025299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vroom JM, De Grauw KJ, Gerritsen HC, Bradshaw DJ, Marsh PD, Watson GK, Birmingham JJ, Allison C. Applied and environmental microbiology. 1999;65:3502–3511. doi: 10.1128/aem.65.8.3502-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SC, Huh KM, Lee J, Cho YW, Galinsky RE, Park K. Biomacromolecules. 2007;8:202–208. doi: 10.1021/bm060307b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F, Liu XM, Rice KC, Li X, Yu F, Reinhardt RA, Bayles KW, Wang D. Antimicrobial agents and chemotherapy. 2009;53:4898–4902. doi: 10.1128/AAC.00387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F, Rice KC, Liu XM, Reinhardt RA, Bayles KW, Wang D. Pharmaceutical research. 2010;27:2356–2364. doi: 10.1007/s11095-010-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horev B, Klein MI, Hwang G, Li Y, Kim D, Koo H, Benoit DS. ACS nano. 2015;9:2390–2404. doi: 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benoit DS, Henry SM, Shubin AD, Hoffman AS, Stayton PS. Molecular pharmaceutics. 2010;7:442–455. doi: 10.1021/mp9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee ES, Oh KT, Kim D, Youn YS, Bae YH. Journal of controlled release : official journal of the Controlled Release Society. 2007;123:19–26. doi: 10.1016/j.jconrel.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benoit DS, Convertine AJ, Duvall CL, Hoffman AS, Stayton PS. Journal of controlled release : official journal of the Controlled Release Society. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manganiello MJ, Cheng C, Convertine AJ, Bryers JD, Stayton PS. Biomaterials. 2012;33:2301–2309. doi: 10.1016/j.biomaterials.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Convertine AJ, Diab C, Prieve M, Paschal A, Hoffman AS, Johnson PH, Stayton PS. Biomacromolecules. 2010;11:2904–2911. doi: 10.1021/bm100652w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benoit DS, Srinivasan S, Shubin AD, Stayton PS. Biomacromolecules. 2011;12:2708–2714. doi: 10.1021/bm200485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. ACS nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douroumis D, Fahr A. Drug delivery strategies for poorly water-soluble drugs. West Sussex: John Wiley & Sons, Chichester; 2012. [Google Scholar]
- 26.Baranello MP, Bauer L, Benoit DS. Biomacromolecules. 2014;15:2629–2641. doi: 10.1021/bm500468d. [DOI] [PubMed] [Google Scholar]
- 27.van de Wetering P, Moret EE, Schuurmans-Nieuwenbroek NM, van Steenbergen MJ, Hennink WE. Bioconjugate chemistry. 1999;10:589–597. doi: 10.1021/bc980148w. [DOI] [PubMed] [Google Scholar]
- 28.Fan JQ, Zeng F, Wu SZ, Wang XD. Biomacromolecules. 2012;13:4126–4137. doi: 10.1021/bm301424r. [DOI] [PubMed] [Google Scholar]
- 29.Cottenye N, Cui ZK, Wilkinson KJ, Barbeau J, Lafleur M. Biofouling. 2013;29:817–827. doi: 10.1080/08927014.2013.807505. [DOI] [PubMed] [Google Scholar]
- 30.Wei T, Liu J, Ma H, Cheng Q, Huang Y, Zhao J, Huo S, Xue X, Liang Z, Liang XJ. Nano letters. 2013;13:2528–2534. doi: 10.1021/nl400586t. [DOI] [PubMed] [Google Scholar]
- 31.Jabra-Rizk MA, Shirtliff M, James C, Meiller T. FEMS yeast research. 2006;6:1063–1073. doi: 10.1111/j.1567-1364.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- 32.Cordeiro RA, Teixeira CE, Brilhante RS, Castelo-Branco DS, Paiva MA, Giffoni Leite JJ, Lima DT, Monteiro AJ, Sidrim JJ, Rocha MF. Medical mycology. 2013;51:53–59. doi: 10.3109/13693786.2012.692489. [DOI] [PubMed] [Google Scholar]
- 33.Koo H, Rosalen PL, Cury JA, Park YK, Bowen WH. Antimicrobial agents and chemotherapy. 2002;46:1302–1309. doi: 10.1128/AAC.46.5.1302-1309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S, Shi Y, Kim JY, Park K, Cheng JX. Expert opinion on drug delivery. 2010;7:49–62. doi: 10.1517/17425240903380446. [DOI] [PubMed] [Google Scholar]
- 35.Lin LY, Lee NS, Zhu J, Nystrom AM, Pochan DJ, Dorshow RB, Wooley KL. Journal of controlled release : official journal of the Controlled Release Society. 2011;152:37–48. doi: 10.1016/j.jconrel.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z. Biomaterials. 2013;34:10238–10248. doi: 10.1016/j.biomaterials.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorbunoff MJ. Analytical biochemistry. 1984;136:425–432. doi: 10.1016/0003-2697(84)90239-2. [DOI] [PubMed] [Google Scholar]
- 38.Sakai H, Ebana H, Sakai K, Tsuchiya K, Ohkubo T, Abe M. Journal of colloid and interface science. 2007;316:1027–1030. doi: 10.1016/j.jcis.2007.08.042. [DOI] [PubMed] [Google Scholar]
- 39.Bhat RR, Tomlinson MR, Genzer J. Macromol Rapid Comm. 2004;25:270–274. [Google Scholar]
- 40.Suzuki M. Fundamentals of adsorption : proceedings of the Fourth International Conference on Fundamentals of Adsorption, Kyoto; May 17–22, 1992; Kodansha ; Elsevier, Tokyo Amsterdam ; New York. 1993. [Google Scholar]
- 41.Klushin LI, Polotsky AA, Hsu HP, Markelov DA, Binder K, Skvortsov AM. Physical review. E, Statistical, nonlinear, and soft matter physics. 2013;87:022604. doi: 10.1103/PhysRevE.87.022604. [DOI] [PubMed] [Google Scholar]
- 42.Mahltig B, Gohy JF, Jerome R, Stamm M. J Polym Sci Pol Phys. 2001;39:709–718. [Google Scholar]
- 43.Lendenmann U, Grogan J, Oppenheim FG. Advances in dental research. 2000;14:22–28. doi: 10.1177/08959374000140010301. [DOI] [PubMed] [Google Scholar]
- 44.Ambatipudi KS, Hagen FK, Delahunty CM, Han X, Shafi R, Hryhorenko J, Gregoire S, Marquis RE, Melvin JE, Koo H, Yates JR., 3rd Journal of proteome research. 2010;9:6605–6614. doi: 10.1021/pr100786y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koo H, Vacca Smith AM, Bowen WH, Rosalen PL, Cury JA, Park YK. Caries Res. 2000;34:418–426. doi: 10.1159/000016617. [DOI] [PubMed] [Google Scholar]
- 46.Koo H, Hayacibara MF, Schobel BD, Cury JA, Rosalen PL, Park YK, Vacca-Smith AM, Bowen WH. The Journal of antimicrobial chemotherapy. 2003;52:782–789. doi: 10.1093/jac/dkg449. [DOI] [PubMed] [Google Scholar]
- 47.Koo H, Schobel B, Scott-Anne K, Watson G, Bowen WH, Cury JA, Rosalen PL, Park YK. Journal of dental research. 2005;84:1016–1020. doi: 10.1177/154405910508401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallow KC, Jhon YK, Genzer J, Loo YL. Polymer. 2012;53:1131–1137. [Google Scholar]
- 49.Kryuchkov MA, Detrembleur C, Jerome R, Prud'homme RE, Bazuin CG. Macromolecules. 2011;44:5209–5217. [Google Scholar]
- 50.Adhikary R, Mukherjee P, Kee TW, Petrich JW. The journal of physical chemistry. B. 2009;113:5255–5261. doi: 10.1021/jp901234z. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez ME, Kim J, Delos Santos GB, Azizuddin K, Berlin J, Anderson VE, Kenney ME, Oleinick NL. Journal of biomedical optics. 2010;15:051604. doi: 10.1117/1.3484256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Y, Liu SY, Armes SP, Billingham NC. Biomacromolecules. 2003;4:1636–1645. doi: 10.1021/bm030026t. [DOI] [PubMed] [Google Scholar]
- 53.Bowen WH, Koo H. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koo H, Xiao J, Klein MI, Jeon JG. Journal of bacteriology. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.