Summary
Noncentrosomal microtubules play an important role in polarizing differentiated cells, but little is known about how these microtubules are organized. Here we identify the spectraplakin, Short stop (Shot), as the cortical anchor for noncentrosomal microtubule organizing centers (ncMTOCs) in the Drosophila oocyte. Shot interacts with the cortex through its actin-binding domain and recruits the microtubule minus-end-binding protein, Patronin, to form cortical ncMTOCs. Shot/Patronin foci do not co-localize with γ-tubulin, suggesting that they do not nucleate new microtubules. Instead, they capture and stabilize existing microtubule minus ends, which then template new microtubule growth. Shot/Patronin foci are excluded from the oocyte posterior by the Par-1 polarity kinase to generate the polarized microtubule network that localizes axis determinants. Both proteins also accumulate apically in epithelial cells, where they are required for the formation of apical-basal microtubule arrays. Thus, Shot/Patronin ncMTOCs may provide a general mechanism for organizing noncentrosomal microtubules in differentiated cells.
Graphical Abstract
Highlights
-
•
The Drosophila spectraplakin, Shot, recruits Patronin to form noncentrosomal MTOCs
-
•
The actin-binding domain of Shot anchors the ncMTOCs to the oocyte cortex
-
•
Par-1 excludes Shot from the posterior cortex to define the anterior-posterior axis
-
•
Shot/Patronin ncMTOCs lack γ-tubulin and grow MTs from stabilized minus-end stumps
Many differentiated cell types lack centrosomes but still form highly polarized microtubule networks. Nashchekin et al. describe how the spectraplakin Shot and the microtubule minus-end-binding protein Patronin form a cortical noncentrosomal microtubule organizing center that acts a source of growing microtubules independently of γ-tubulin.
Introduction
Many differentiated animal cells and all plant cells lack functional centrosomes, yet form highly organized microtubule (MT) arrays that play essential roles in cell polarity, organization, and function (Bartolini and Gundersen, 2006). For example, both Drosophila and rodent hippocampal neurons develop normally without active centrosomes, with the latter extending and even regenerating axons independently of centrosomal MT nucleation (Nguyen et al., 2011, Stiess et al., 2010). Most Drosophila tissues lack functional centrosomes or microtubule organizing centers (MTOCs) in interphase (Rogers et al., 2008).
Anterior-posterior axis formation in the Drosophila oocyte provides a well-studied example of the role of noncentrosomal MTs. Although the oocyte contains centrosomes, which cluster near the nucleus, oogenesis proceeds normally in their absence (Basto et al., 2006, Januschke et al., 2006, Stevens et al., 2007). Instead, the majority of MTs grow from the anterior/lateral cortex, but not from the posterior, where the plus ends concentrate (Clark et al., 1994, Clark et al., 1997, Parton et al., 2011, Theurkauf et al., 1992). This noncentrosomal MT array directs the localization of bicoid and oskar mRNAs to the anterior and posterior poles of the oocyte, respectively, to define the main body axis of the embryo (St Johnston, 2005, Zimyanin et al., 2008). 3D modeling of the oocyte MT cytoskeleton has shown that restricting MT minus ends to the anterior/lateral cortex is sufficient to generate an MT network that can direct the transport of oskar mRNA to the oocyte posterior by kinesin (Khuc Trong et al., 2015).
The formation of this polarized MT array is under the control of the PAR proteins, which localize in mutually antagonistic anterior and posterior cortical domains (Doerflinger et al., 2010, Shulman et al., 2000). The posterior crescent of the Par-1 kinase transmits this cortical polarity to the MT cytoskeleton by excluding minus ends from the oocyte posterior. It is not known, however, how PAR-1 activity is transduced into the asymmetric organization of MT minus ends, nor how the minus ends associate with the anterior/lateral cortex.
The recent discovery of the Patronin family of MT minus-end-binding proteins, consisting of Patronin in Drosophila, CAMSAP1, 2, and 3 in mammals, and PTRN-1 in worms, has begun to reveal how the minus ends of noncentrosomal MTs are organized and maintained (Akhmanova and Steinmetz, 2015, Baines et al., 2009, Goodwin and Vale, 2010, Marcette et al., 2014, Meng et al., 2008, Richardson et al., 2014). The Patronins recognize and stabilize free MT minus ends by protecting them from depolymerization (Goodwin and Vale, 2010, Hendershott and Vale, 2014, Jiang et al., 2014). Patronins appear to play a particularly important role in organizing MTs in differentiated cells. CAMSAP3 localizes to the apical domain in epithelial cells, where it is required for the formation of the apical-basal array of MTs (Tanaka et al., 2012, Toya et al., 2016, Zheng et al., 2013). CAMSAP2 stabilizes neuronal MTs in axon and dendrites, and its knockdown leads to defects in axon specification and dendritic branch formation (Yau et al., 2014). Similarly, Caenorhabditis elegans PTRN-1 is required for normal neurite morphology and axon regeneration (Chuang et al., 2014, Marcette et al., 2014, Richardson et al., 2014). The function of Drosophila Patronin has only been examined in cultured S2 cells, where its depletion leads to a decrease in MT number and an increase in free moving MTs (Goodwin and Vale, 2010).
Although it is now clear that the Patronins play an important role in organizing noncentrosomal MTs in differentiated cells, little is known about the regulation of the distribution and activity of the Patronins themselves. Here we show that Patronin is recruited to the anterior/lateral cortex of the Drosophila oocyte by the spectraplakin, Shot, under the control of Par-1. These Shot/Patronin complexes form the cortical noncentrosomal MTOCs that organize the polarized MT network in the oocyte, which specifies the anterior-posterior axis.
Results
Shot Is Required for the Polarized Organization of MTs in the Oocyte
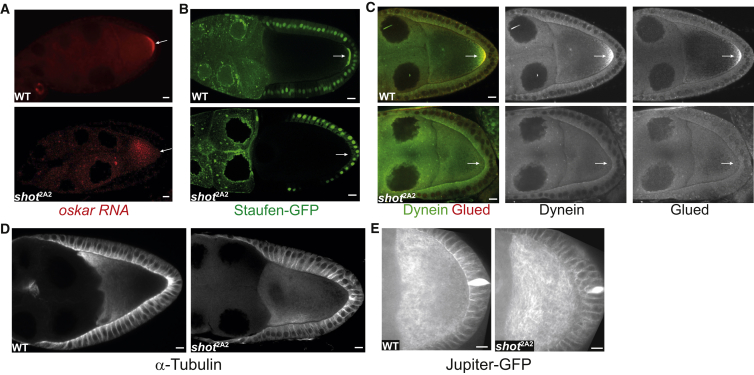
We previously isolated 11 new alleles of short stop (shot) in a screen for dominant suppressors of the bicaudal phenotype caused by mislocalizing oskar mRNA to the oocyte anterior (Chang et al., 2011). Shot is the single Drosophila spectraplakin, a giant cytoskeletal linker protein with an N-terminal actin-binding domain and two C-terminal domains that bind MT, the GAS2 domain, which binds to the MT lattice, and a more C-terminal domain that associates with MT plus ends through the +TIP, EB1 (Applewhite et al., 2010, Sun et al., 2001). Null alleles of shot block the specification of the oocyte, and this is also the case for 10 out of 11 of the new alleles (Roper and Brown, 2004). Some germline clones of shot2A2 are not blocked in oogenesis, however, and develop to later stages, occasionally laying fertilized eggs that develop into larvae that lack an abdomen. Since this is a typical posterior group phenotype, we examined whether the posterior determinant, oskar mRNA, is correctly localized in shot2A2 mutants. Both oskar RNA and Staufen-GFP (an RNA-binding protein associated with oskar) fail to localize to the oocyte posterior in shot2A2 germline clones (Figures 1A and 1B). To determine whether Shot is specifically required for oskar mRNA localization or plays a more general role in kinesin-dependent transport to the posterior, we also examined the localization of Dynein and the dynactin subunit, Glued, which are transported to the oocyte posterior by kinesin independently of oskar mRNA (Brendza et al., 2002, Palacios and St Johnston, 2002). Neither Dynein nor Glued are localized in shot2A2 oocytes, indicating that either kinesin activity is inhibited or the MT plus ends are not concentrated at the posterior pole (Figure 1C).
Figure 1.
Shot Is Required for Oocyte Polarity and Microtubule Organization
(A–C) oskar mRNA (A), Staufen (B), Dynein, and Glued (C) localization in wild-type (WT; top) and shot2A2 mutant (bottom) oocytes. Arrows point to the oocyte posterior.
(D) MT organization detected by α-tubulin staining of WT (left) and shot2A2 mutant oocytes (right).
(E) Live imaging of Jupiter-GFP in WT (left) and shot2A2 mutant oocytes (right). The images are stills from Movies S1 (WT) and S2 (shot2A2).
Scale bars represent 10 μm.
We next examined the overall organization of the MTs in fixed and living oocytes. Staining of fixed oocytes with anti-tubulin and in vivo labeling of MTs in living oocytes with Jupiter-GFP (Karpova et al., 2006) reveals the anterior-posterior gradient of MTs in wild-type with the highest concentration of MTs at the anterior (Figures 1D and 1E; Movie S1). This anterior enrichment is lost in shot2A2 and the MT organization becomes somewhat variable, with a much more even distribution throughout the oocyte cytoplasm (Figures 1D and 1E, right panels; Movie S2).
Par-1 Regulates the Association of the Shot Actin-Binding Domain with the Cortex
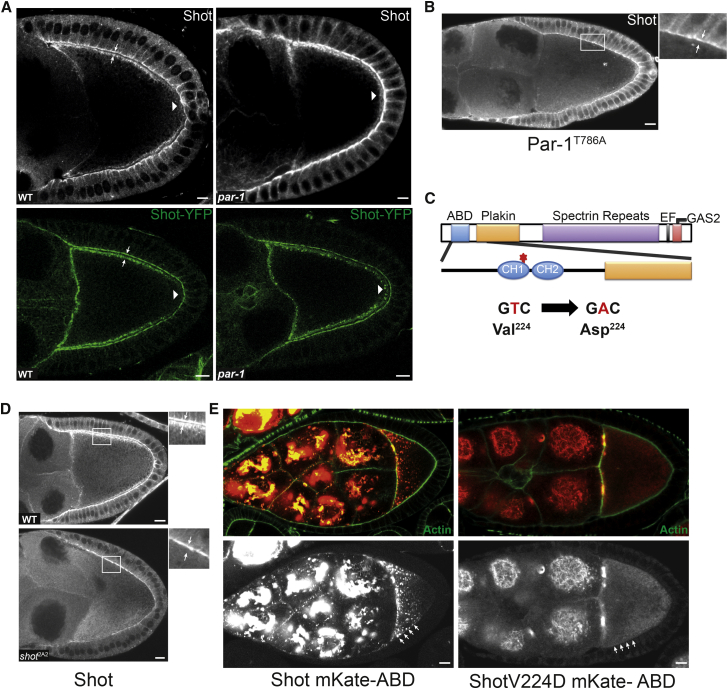
Shot localizes to the anterior and lateral cortex of the oocyte, but is absent from the posterior, following the predicted distribution of MT minus ends. Shot is also strongly enriched at the apical side of the epithelial follicle cells that surround the developing egg chamber (Figure 2A, left). YFP-tagged Shot expressed from a transgenic bacterial artificial chromosome (BAC) rescuing construct shows an identical distribution in both the follicle cells and oocyte. We therefore examined whether the interaction of Shot with the oocyte cortex is under the control of the cortical Par proteins that control the polarity of the MT cytoskeleton. In par-1 mutant oocytes, MTs grow from the posterior cortex as well as the anterior/lateral cortex, and the MT cytoskeleton loses its asymmetry, whereas Par-1T786A, which has a uniform cortical distribution, abolishes all MT growth from the cortex (Doerflinger et al., 2010, Parton et al., 2011). Shot responds to Par-1 activity in the same way as MTs: it extends around the posterior in the absence of Par-1, and is lost from the cortex in oocytes overexpressing Par-1T786A (Figures 2A and 2B). Thus, Shot is downstream of Par-1, consistent with it playing a role in MT minus-end localization.
Figure 2.
The Cortical Localization of Shot Depends on Its Actin-Binding Domain and Is Inhibited by Par-1
(A) Shot localizes to the anterior-lateral cortex and is excluded from the oocyte posterior (left). Shot spreads around the oocyte posterior in the par-16323/par-1W3 mutant (right). Top: Shot antibody. Bottom: Shot-YFP genomic BAC.
(B) Overexpression of Par-1T786A-GFP displaces Shot from the oocyte cortex.
(C) Diagram of the domain structure of Shot, indicating the position and the nature of the point mutation in shot2A2. CH, calponin homology domain. CH1 and CH2 form the actin-binding domain (ABD).
(D) shot2A2 disrupts the localization of Shot to the oocyte cortex. The small boxes on the right are higher-magnification views showing the localization of Shot to the lateral cortex of the wild-type (WT) oocyte and its absence in shot2A4. Shot also localizes to the apical cortex of the follicle cells.
(E) Wild-type Shot ABD (left) localizes to the anterior-lateral cortex, whereas the Shot ABD with a Val224 to Asp mutation (right) does not.
Arrows point to the cortical Shot signal in the oocyte and to the underlying apical signal in the epithelial follicle cells (A, B, D). Arrowheads in (A) point to posterior. Arrows in (E) indicate the cortical signal. Scale bars represent 10 μm.
Sequencing of shot2A2 reveals that it is a point mutation in the first calponin homology domain of the N-terminal actin-binding domain (ABD) of Shot, changing Val224 (isoform PE) to Asp (Figure 2C). Val224 is well conserved among ABD-containing proteins. Structural analysis of the interaction of fimbrin with F-actin showed that the equivalent to Val224 (Val212 in fimbrin) directly contacts F-actin (Hanein et al., 1998). In agreement with this, Shot loses its association with the actin-rich cortex in shot2A2 and is mainly cytoplasmic (Figure 2D). Like full-length Shot, the Shot ABD is enriched at the anterior-lateral cortex (Figure 2E, left). Introducing the Val224 to Asp mutation into the Shot ABD disrupts its cortical localization, although the protein still shows an enrichment at the ring canals, which is not observed with the full-length protein (Figure 2E, right). Thus, Shot is recruited to the cortex through its ABD, presumably by direct binding to cortical F-actin, and this interaction is inhibited at the posterior by Par-1.
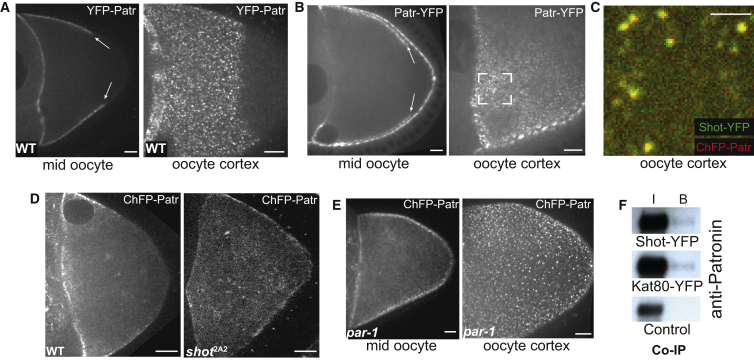
Shot Recruits Patronin Foci to the Oocyte Cortex
We took advantage of the recent identification of Patronin/CAMSAP as an MT minus-end-binding protein to analyze the relationship between cortical Shot and the distribution of MT minus ends in the oocyte (Goodwin and Vale, 2010, Jiang et al., 2014). Live imaging of both transgenic and endogenously tagged Patronin reveals that it localizes to anterior/lateral cortex in the expected distribution of MT minus ends (Figures 3A and 3B; Movie S3, left panel). Importantly, Patronin co-localizes with Shot in distinct cortical foci (Figure 3C). Patronin localization is Shot dependent, as it becomes largely cytoplasmic in shot2A2 (Figure 3D and Movie S3, right panel). Furthermore, the cortical Patronin foci extend around the posterior cortex in par-1 mutant oocytes, as Shot does, consistent with the two proteins being in the same complex (Figure 3E). In agreement with this, Patronin co-immunoprecipitates with Shot-YFP from ovary extracts (Figure 3F). The fact that both Patronin and Shot are no longer cortical in shot2A2 indicates that Shot anchors Patronin to the cortex, providing an explanation of how the asymmetric localization of Shot controls the polarized distribution of MT minus ends in the oocyte.
Figure 3.
Patronin Is Recruited into Cortical Foci by Shot
(A and B) YFP-Patronin (expressed in the germline under the control of the maternal tubulin-α4 promoter) (A) and endogenously tagged Patronin-YFP (B) in living stage 9 oocytes. Patronin localizes to the anterior/lateral cortex of the oocyte, where it forms discrete foci. The right-hand panels are projections of several z sections spanning the oocyte cortex. The white rectangle in (B) marks a region where the oocyte cortex is in focus, showing the Patronin-YFP foci. The arrows point to the posterior boundary of the domain of Patronin foci in the oocyte. WT, wild-type.
(C) A close-up of a region of the lateral cortex of a living oocyte, showing the co-localization of Shot-YFP and Cherry-Patronin in cortical foci. UAS-Cherry-Patronin was expressed in the germline under the control of nanos-Gal4. Scale bar represents 2.5 μm.
(D) Cherry-Patronin localization in wild-type (WT; left) and shot2A2 mutant oocytes (right). UAS-Cherry-Patronin expression was driven by maternal α4tubulin-Gal4. These still images were taken from Movie S3.
(E) Cherry-Patronin foci extend around the oocyte posterior in par-1w3/par-16323 mutant oocytes (compare with A, B, and D). Images are projections of several z sections spanning the oocyte cortex.
(F) Co-immunoprecipitation (IP) of Patronin by Shot-YFP and Katanin 80-YFP. I, input. B, bound.
Scale bars represent 10 μm, except in (C). See also Figure S2.
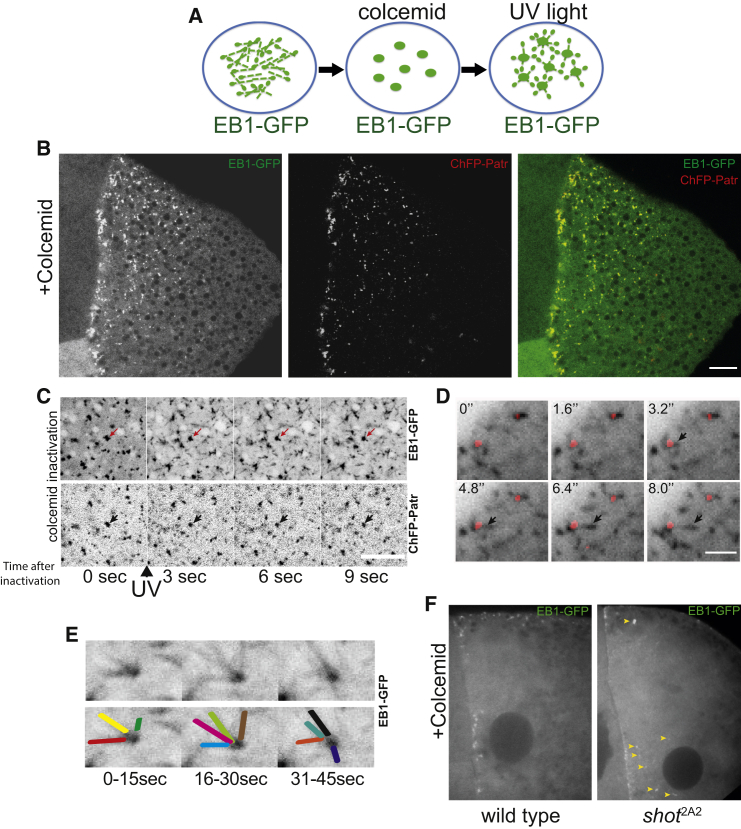
Patronin Cortical Foci Are Noncentrosomal MTOCs
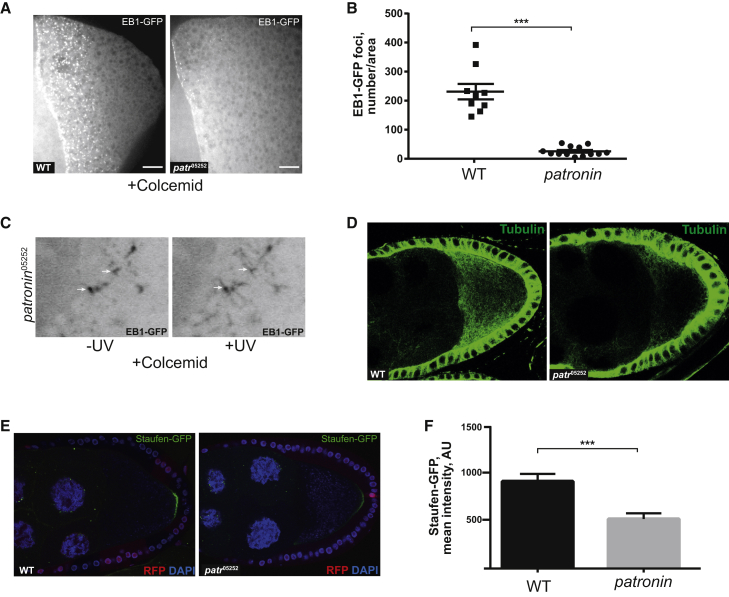
It has previously been shown, using Tau-GFP to label MTs and EB1-GFP to label the growing MT plus ends, that oocyte MTs grow out from noncentrosomal foci that can be visualized using an MT regrowth assay (Parton et al., 2011). Upon colcemid treatment, both proteins accumulate in cortical foci. Local inactivation of the colcemid with a pulse of UV light allows MTs to regrow from the cortex (Figure 4A). We therefore examined whether the MTs grow from the Patronin foci. Both EB1-GFP and Tau-GFP accumulate in the cortical Patronin foci upon colcemid treatment, indicating that these contain stable MT minus ends (Figures 4B and S1B). Furthermore, after colcemid inactivation with UV light, EB1-GFP and Tau-GFP label growing MTs that emerge from the Patronin foci (Figures 4C, S1C, and S1D; Movies S4 and S5). The Patronin foci also act as a source of growing MTs under steady-state conditions in the absence of colcemid (Figure 4D and Movie S6). After colcemid inactivation, each Patronin focus produces an average of 11.5 new MTs per minute (n = 15; SEM = 0.75), providing a source of MTs that grow in multiple directions (Figure 4E). Moreover, these foci are the only visible source of growing MTs at the oocyte cortex, strongly suggesting that they represent the noncentrosomal, cortical MT organizing centers (ncMTOCs) from which MTs grow to form the polarized cytoskeleton in the oocyte.
Figure 4.
Patronin Foci Are Cortical Noncentrosomal MTOCs
(A) Diagram of the MT regrowth assay.
(B) Patronin foci co-localize with the MT plus-end marker EB1-GFP in the presence of colcemid. Scale bar represents 10 μm.
(C) Still images from Movie S4 showing new EB1-GFP comets growing out from the Patronin foci a few seconds after colcemid inactivation. The arrows indicate a single active MTOC in successive frames. Scale bar represents 10 μm.
(D) Patronin foci are active MTOCs that produce new MTs in the absence of colcemid. Images taken from Movie S6. The arrows point to a new EB1-GFP comet that marks the plus end of a microtubule growing from a Patronin MTOC (red). Scale bar represents 2 μm.
(E) A single Patronin focus produces many MTs that grow in multiple directions. The images are projections of several time points over 15-s intervals. Each colored line represents a new EB1-GFP track (bottom panel). Images taken from Movie S4.
(F) Localization of EB1-GFP foci in wild-type (left) and shot2A2 (right) oocytes after colcemid treatment. Images taken from Movie S7. Arrowheads point to the cytoplasmic ncMTOCs in the shot2A2 mutant.
In shot2A2 mutant oocytes, many of the foci fail to be retained at the oocyte cortex and redistribute throughout the oocyte cytoplasm, consistent with the loss of most Shot and Patronin from the cortex in this mutant (Figure 4F and Movie S7). These cytoplasmic foci remain active, however, producing growing MTs after colcemid inactivation, explaining why the overall polarity of the MT network is disrupted (Movie S7).
Patronin Is Required for ncMTOC Formation
A patronin null mutant blocks oogenesis at an early stage. To test whether Patronin is required for the activity of the cortical ncMTOCs in the oocyte, we therefore used a hypomorphic allele, patronin05252, which strongly reduces Patronin levels (Bellen et al., 2004). patronin05252 homozygous oocytes contain 90% fewer cortical EB1-GFP foci after colcemid treatment than wild-type, and the remaining foci also generally recruit less EB1-GFP (Figures 5A and 5B). Nevertheless, the Patronin foci that form are still active, acting as a source of growing MTs after colcemid inactivation (Figure 5C and Movie S8). The density of MTs is also significantly reduced in patronin05252 clones, as expected from the reduced number of cortical ncMTOCs (Figure 5D). Despite the dramatic reduction in MT number, there are still sufficient MTs to direct the localization of Staufen/oskar mRNA complexes to the oocyte posterior, although the levels of localization are reduced by >40% (Figures 5E and 5F).
Figure 5.
Patronin Is Required for the Formation of Cortical MTOCs
(A and B) The number of cortical MTOCs marked by EB1-GFP is reduced in patronin05252 mutant oocytes. (A) Images of wild-type (WT; left) and patronin05252 mutant (right) oocytes expressing nanos>UAS-EB1-GFP after colcemid treatment. The images are projections of the several z sections spanning the oocyte cortex. (B) Quantification of the number of cortical EB1-GFP foci after colcemid treatment in WT and patronin05252 oocytes. ∗∗∗p < 0.0001. Error bars indicate the SEM.
(C) EB1-GFP foci before (left) and after (right) colcemid inactivation in a patronin05252 mutant oocyte. Close-up still images from Movie S8. The arrows indicate two of the activated MTOCs.
(D) MT density is strongly reduced in patronin05252 mutant oocytes. WT (left) and patronin05252 mutant (right) oocytes stained with anti-tubulin.
(E and F) Localization of Stau-GFP to the oocyte posterior is reduced in patronin05252 mutant oocytes. (E) Localization of Stau-GFP in WT (left) and patronin05252 (right) oocytes. patronin05252 germline clones were marked by the absence of nlsRFP. (F) Quantification of the mean fluorescence intensity of posteriorly localized Stau-GFP in patronin05252 and WT oocytes. ∗∗∗p = 0.0005. Error bars indicate the SEM.
Scale bars represent 10 μm.
Patronin ncMTOCs Do Not Co-localize with γ-Tubulin
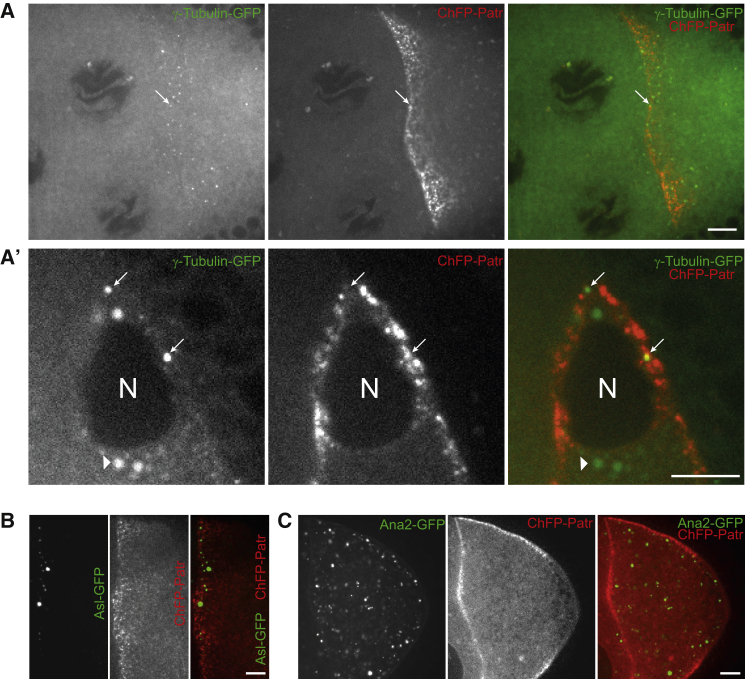
To further investigate the nature of the Shot/Patronin noncentrosomal MTOCs, we asked whether they contain γ-tubulin as the source of new MTs. Antibody staining of oocytes for γ-tubulin label only the centrosomes adjacent to the oocyte nucleus, but overexpressed γ-tubulin 37C-GFP is also seen in weak foci along the anterior/lateral cortex (Januschke et al., 2006, Parton et al., 2011). We therefore co-expressed γ-tubulin-GFP and Cherry-Patronin to determine whether the two proteins co-localize (Figures 6A and 6A′). Patronin labels some of the nuclear-associated, γ-tubulin foci, which probably correspond to the active centrosomes. The cortical Patronin foci do not co-localize with the γ-tubulin-GFP foci, however, and Shot/Patronin ncMTOCs contain no detectable γ-tubulin. Since MTs start to grow out from Patronin foci within 1 s of colcemid inactivation, and these foci are the only visible source of cortical MTs, it seems most likely that the MTs are seeded from Patronin-stabilized MT minus-end stumps and not from de novo nucleation by the γ-tubulin ring complex.
Figure 6.
Patronin MTOCs and Centrosomal Components
(A and A′) Ectopically expressed γTub37C-GFP accumulates in cortical foci (A) and in the centrosomes around the nucleus (A′), but does not localize to the Patronin foci. Arrows point to γ-Tub-GFP-positive centrosomes. Arrowheads point to autofluorescent yolk particles. N, nucleus.
(B) Asl-GFP ectopically expressed under the control of nanos>Gal4 forms foci at oocyte cortex, but does not co-localize with Cherry-Patronin MTOCs.
(C) Ana2-GFP ectopically expressed under the control of nanos>Gal4 forms foci in the oocyte cytoplasm, but does not co-localize with the Cherry-Patronin MTOCs.
Scale bars represent 10 μm.
Overexpression of the centriolar duplication factors dSAS6, dSas4, Sak/PLK4, and Ana2/STIL can promote the formation of acentriolar MTOCs in the oocyte (Dzhindzhev et al., 2010, Peel et al., 2007, Stevens et al., 2010). Moreover, expression of membrane-tethered Cep152/Asl and PLK4 is sufficient to induce formation of ectopic acentriolar MTOCs in mouse oocytes (Coelho et al., 2013). To test whether any of these acentriolar MTOC components are involved in the formation of the Shot/Patronin ncMTOCs, we co-expressed Cherry-Patronin with Asl-GFP (Figure 6B), Ana2-GFP (Figure 6C), dSas6-GFP, dSas4-GFP, and Sak-GFP (data not shown). None of these proteins co-localize with the Patronin foci, however, indicating that they are not components of the ncMTOCs (Figures 6B and 6C, and data not shown).
An alternative mechanism that can contribute to the formation of new MTs is the severing of existing MTs to generate minus ends that act as seeds for new microtubule growth (Baas and Ahmad, 1992, Lindeboom et al., 2013, Roll-Mecak and Vale, 2006). The mammalian Patronin orthologs, CAMSAP2 and CAMSAP3, associate with the microtubule severing protein, Katanin (Jiang et al., 2014). This association is conserved in Drosophila, as a protein trap insertion that labels endogenous Katanin 80 co-localizes with Patronin in the cortical foci in the oocyte and at the apical side of the follicle cells (Lowe et al., 2014) (Figure S2). Furthermore, Katanin 80-YFP co-immunoprecipitates with Patronin from ovary extracts, confirming that it is a component of the cortical Patronin complex (Figure 3F). Thus, MT severing by Katanin may contribute to the generation of new MTs in the Patronin ncMTOCs.
Shot and Patronin Play a Role in the Formation of Apical-Basal MT Arrays in Follicle Epithelial Cells
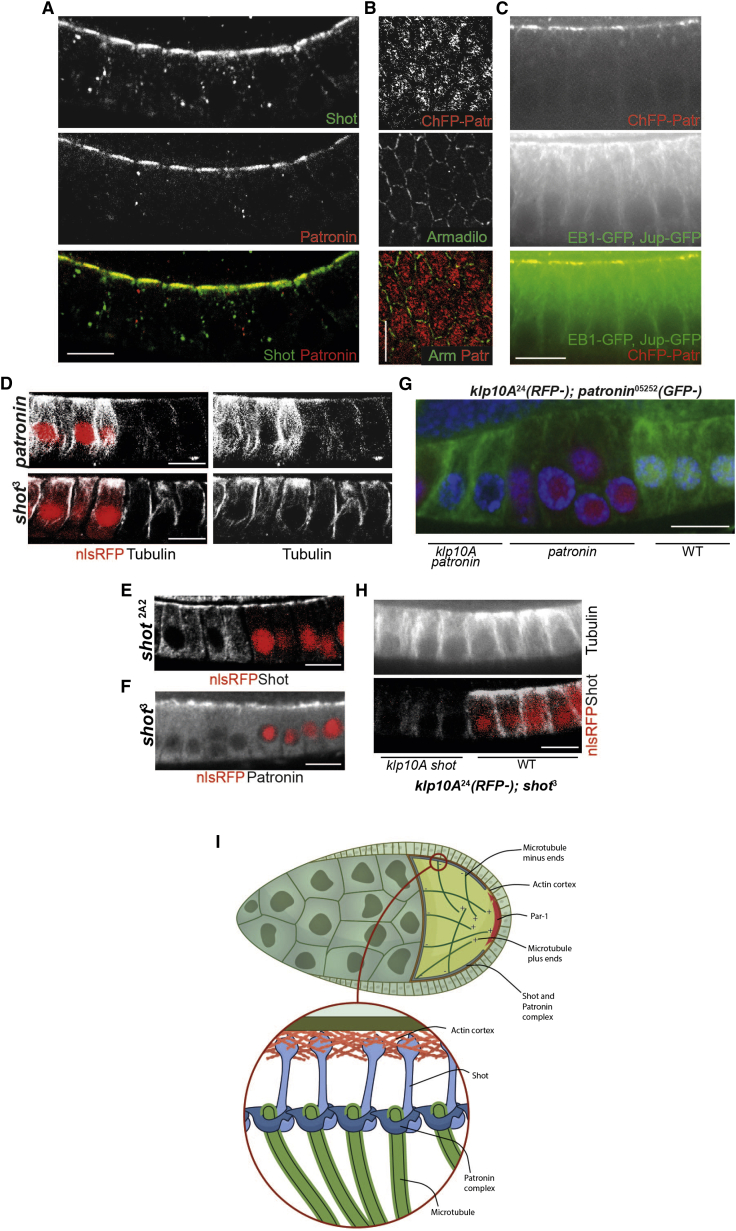
In epithelial cells, noncentrosomal MTs form apical-basal arrays with their MT minus ends concentrated at the apical cortex (Bacallao et al., 1989, Jankovics and Brunner, 2006). The mammalian Patronin homolog, CAMSAP3, localizes to the apical cortex of mouse intestinal cells and human Caco2 cells, and mutation of camsap3 leads to a random orientation of MTs (Toya et al., 2016). To test whether Patronin ncMTOCs play a similar role in the formation of the apical-basal array of MTs in Drosophila epithelia, we analyzed the localization of Patronin in the follicle cells, larval salivary glands, and male ejaculatory duct (Figures 3B, 7A, and S3). Patronin localizes apically in all three epithelia, forming multiple apical foci in the follicle cells, but is excluded from the adherens junctions (Figure 7B). Live imaging of EB1-GFP and Jupiter-GFP reveals that most MTs grow from the region of apical Patronin foci (Figure 7C and Movie S9). Although capsap3 null cells contain relatively normal numbers of MTs, patronin05252 mutant cells have very few MTs (Figure 7D), presumably because it is the only copy of this gene in Drosophila. In addition, larger patronin05252 mutant clones often lead to tissue disorganization and multi-layering (Figures 7G and S4A). This suggests that Patronin apical foci act as ncMTOCs in epithelial cells and that they are crucial for tissue integrity.
Figure 7.
Shot and Patronin Are Required for MT Organization in the Epithelial Follicle Cells
(A) Shot and Patronin co-localize at the apical cortex of the follicle cells. An optical section through the epithelia monolayer with apical at the top and basal at the bottom. See also Figure S3.
(B) Follicle cells contain multiple apical Patronin foci. Top: view of the apical region of follicle cells expressing ubi>Cherry-Patronin. Patronin does not localize to the adherens junctions marked by Armadillo (green) staining.
(C) Apical Patronin foci co-localize with MTs. MTs were marked by ubi>EB1-GFP and Jupiter-GFP. The image is a temporal merge of several frames from Movie S9.
(D) MT organization in patronin05252 and shot3 mutant follicle cell clones marked by the loss of nuclear RFP (red). Top: patronin05252 mutant cells contain many fewer microtubules than their heterozygous neighbors. Bottom: shot null mutant cells lose the apical enrichment of MTs, but retain lateral MTs. See also Figure S4.
(E) Shot protein is still enriched apically in shot2A2 mutant follicle cells, but the protein is also diffusely distributed throughout the cytoplasm. shot2A2 mutant cells were marked by the absence of nlsRFP.
(F) Shot is not required for the apical recruitment of Patronin in the follicle cells. shot3 mutant cells were marked by the absence of nlsRFP.
(G) Patronin protects microtubule minus ends from the depolymerizing kinesin KLP10A in the follicle cells. The removal of KLP10A from patronin05252 mutant cells partially rescues the loss of MTs caused by the patronin mutant alone. Mutant cells were marked by the absence of nlsGFP (patronin) and nlsRFP (klp10A). Double-mutant cells lack both GFP and RFP.
(H) Mutation of klp10A does not rescue the MT phenotype of shot3 mutant clones. Double-mutant cells were marked by the absence of nlsRFP (klp10A) and by the loss of Shot staining (bottom).
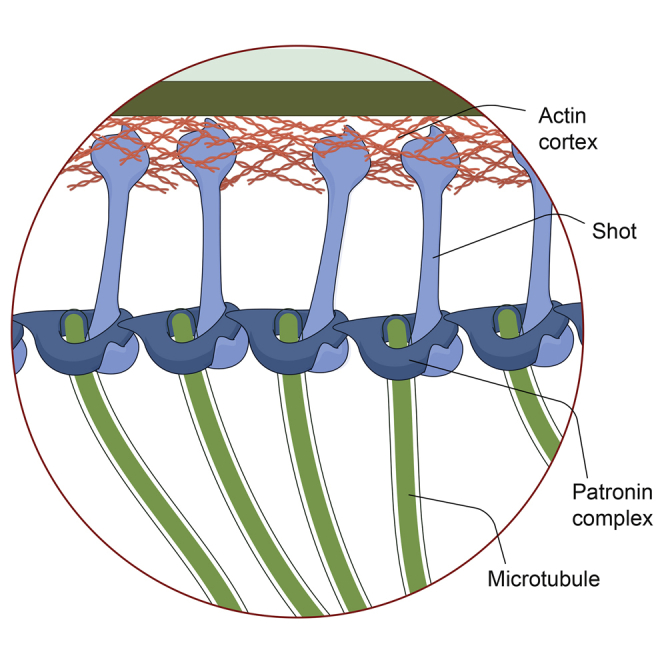
(I) A model showing how Shot exclusion by Par-1 generates the polarized MT cytoskeleton in the oocyte. Par-1 is localized to the posterior of the oocyte, where it inhibits the association of Shot with the actin-rich cortex. Shot recruits Patronin to the anterior and lateral cortex to stabilize free MT minus ends and induce the formation of ncMTOCs that are the source of the MTs that localize oskar mRNA.
Scale bars represent 10 μm.
Shot also localizes apically in the follicle cells and the embryonic salivary gland epithelium, and has been proposed to link acentrosomal MT minus ends to medial actomyosin, although this does not appear to require its ABD (Booth et al., 2014, Roper and Brown, 2003). This suggests that Shot may have similar role as an anchor of Patronin ncMTOCs in epithelial cells. In agreement with previous studies, we observed that Shot is strongly enriched at the apical side of the follicle cells, where it co-localizes with Patronin (Figures 2A and 7A). In homozygous clones of the ABD mutant, shot2A2, Shot protein at the apical cortex is slightly reduced and the protein is found throughout the cytoplasm, indicating that the ABD contributes to efficient apical recruitment (Figure 7E).
To examine the role of Shot in MT organization, we generated clones of shot3, a null mutation (Lee et al., 2000, Roper and Brown, 2003). Mutant clones lose the pronounced apical enrichment of MTs seen in wild-type cells and have fewer MTs than normal, with the remaining MTs mainly along the lateral cortex (Figures 7D and S4B). shot3 mutant cells contain more MTs than patronin mutant cells, however, and the absence of Shot does not disrupt the apical localization of Patronin (Figure 7F).
It has previously been shown that Patronin functions during spindle elongation in the embryo and in interphase S2 cells to protect MT minus ends from the depolymerizing kinesin, Klp10A (kinesin-13), as simultaneous knockdown of Klp10A and Patronin rescues the MT phenotype of Patronin knockdown alone (Goodwin and Vale, 2010, Wang et al., 2013). To ask whether Patronin also antagonizes KLP10a in epithelial cells, we examined the MT phenotype of klp10a patronin double-mutant clones. Loss of KLP10a partially rescues MT abundance in patronin mutant cells, but does not rescue the apical enrichment of MTs, resulting in an MT phenotype that is similar to that seen in shot3 (Figure 7G). By contrast, klp10a has no effect on MT density or organization in shot3 cells (Figure 7H). Thus, Patronin is required both to position MT minus ends apically and to protect them from depolymerization by Klp10A. Shot is not required for Patronin's activity in protecting MT minus ends, but the fact that shot and klp10a patronin mutants produce very similar defects in MT organization suggests that Shot and Patronin act in the same pathway to anchor MTs apically. We also tested whether Patronin functions in the oocyte to protect MT minus ends from depolymerization by Klp10A. However, klp10a patronin double-mutant germline clones show the same reduction in MT density as the patronin single mutant, suggesting that Klp10A plays little role in the germline (Figure S5).
Discussion
The polarized arrangement of the MTs in the Drosophila oocyte depends on the posterior crescent of the Par-1 kinase, which excludes MT minus ends from the posterior cortex (Doerflinger et al., 2010, Parton et al., 2011). Here we show that Par-1 acts by preventing the association of Shot with the posterior actin cortex, thereby restricting the formation of noncentrosomal MTOCs to the anterior and lateral cortex. Computer modeling has shown that this asymmetric localization of MT minus ends is sufficient to explain the formation of the weakly polarized MT network that directs the transport of oskar mRNA to the posterior pole (Khuc Trong et al., 2015). Thus, the regulation of the interaction of Shot with the cortex by Par-1 transmits cortical PAR polarity into the polarization of the MT cytoskeleton that localizes the axis determinants (Figure 7I).
The mechanism by which Par-1 excludes Shot is unknown. The interaction of Shot with the cortex depends on its N-terminal calponin homology domains, which bind to F-actin (Lee and Kolodziej, 2002, Leung et al., 1999). Thus, Par-1 could phosphorylate Shot to inhibit its binding to the cortex. If this is the case, Par-1 would have to modify the activity or accessibility of the N-terminal ABD of Shot, as this domain recapitulates the posterior exclusion and cortical recruitment of the full-length protein. We have not detected any phosphorylation of the ABD by Par-1 in vitro, however, and it seems more likely that Par-1 acts by modifying the cortex to block the binding of Shot.
Shot and its vertebrate ortholog, MACF1, have previously been shown to interact with the MT plus-end tracking protein EB1 through their C-terminal SxIP motifs and with the MT lattice through their Gas2 and C-terminal domains (Alves-Silva et al., 2012, Applewhite et al., 2010, Honnappa et al., 2009, Kodama et al., 2003, Sun et al., 2001). Our results indicate that in addition to binding to MT plus ends and to the MT lattice, Shot also interacts with MT minus ends through its association with the Patronin/Katanin complex. The exact nature of the interaction between Shot and the Patronin complex is unclear, but Shot was found to interact with Katanin 60 in a high-throughput yeast two-hybrid screen (Giot et al., 2003). Thus, one possibility is that Katanin acts as a link between Shot and Patronin. Since Shot is exclusively cortical in the oocyte, the protein does not appear to bind to MT plus ends or along the body of MTs in this system. It will therefore be interesting to investigate whether the different modes of MT binding by Shot are mutually exclusive and how this is regulated.
Several models have been proposed to explain the formation of noncentrosomal MTs. Upon centrosome inactivation in postmitotic Drosophila tracheal cells and C. elegans intestinal cells, γ-TuRC complexes and other pericentriolar material (PCM) components are released from the centrosome and transported toward the apical membrane, where they nucleate MT (Brodu et al., 2010, Feldman and Priess, 2012). Whole MTs released from the centrosome can also be delivered and anchored to the apical domain or cell junctions by Ninein (Lechler and Fuchs, 2007, Mogensen et al., 2000). Alternatively, new MTs can grow from MT ends generated by severing enzymes, a mechanism that is thought to be important in plant cells and neurons (Baas and Ahmad, 1992, Lindeboom et al., 2013, Roll-Mecak and Vale, 2006). Here, we present evidence that this latter mechanism is responsible for the formation of the MT array that directs Drosophila axis formation. Firstly, Shot/Patronin ncMTOCs contain stable minus ends even after treatment with the MT-depolymerizing drug, colcemid, as shown by the persistent recruitment of Tau-GFP and EB1-GFP to these foci. This is consistent with the ability of Patronin and CAMPSAPs to capture and stabilize minus ends of single MTs in vitro and in cells (Goodwin and Vale, 2010, Hendershott and Vale, 2014, Jiang et al., 2014, Meng et al., 2008). Secondly, MTs start to grow out in all directions from the Shot/Patronin foci immediately after colcemid inactivation. Indeed all visible growing MTs emanate from Patronin foci, indicating that they are the principal source of MTs in the oocyte. Thirdly, the foci contain no detectable γ-tubulin and do not co-localize with PCM proteins. This is consistent with observations in Caco-2 cells, which showed that CAMSAP2 and CAMSAP3 do not co-localize with γ-tubulin and in the C. elegans epidermis, where PTRN-1 and γ-tubulin function in parallel pathways to assemble circumferential MTs (Tanaka et al., 2012, Wang et al., 2015).
Taken together, these results suggest a model in which the Shot/Patronin foci act as ncMTOCs by capturing and stabilizing MT minus-end stumps that then act as templates for new MT growth. One attractive feature of this model is that it uncouples MT organization from MT nucleation in both space and time. The Shot/Patronin complex bypasses the need to continually nucleate new MTs by preventing existing microtubules from completely depolymerizing. Thus, once a cell has nucleated sufficient MTs, it can maintain and reorganize its MT cytoskeleton by stabilizing MT minus-end stumps in appropriate locations and using these, rather than the γ-tubulin ring complex, to provide the seeds from which new MTs grow. The number of MTs can even increase in the absence of new MT nucleation if MT-severing proteins chop up existing MTs to produce new minus ends that can then be captured and stabilized. The presence of the severing protein, Katanin, in the Shot/Patronin foci is intriguing in this context, as it raises the possibility that it severs existing MTs to provide a local source of minus ends for Patronin to capture.
Shot and Patronin also co-localize at the apical cortex of the epithelial follicle cells, where they are required for apical-basal MT organization. This consistent with the recent observation that CAMSAP3 is required for the recruitment of MT minus ends to the apical cortex of mammalian intestinal epithelial cells (Toya et al., 2016). Thus, this function of Patronin has been evolutionarily conserved. Furthermore, the similarities between roles of Shot and Patronin in the oocyte and the follicle cells suggest that the complex may provide a general mechanism for organizing noncentrosomal MTs. The relationship between Shot and Patronin is different in the follicle cells compared with the oocyte, however, as Shot is not required for the apical recruitment of Patronin. Nevertheless, loss of either protein produces a very similar loss of apical MT and a reduction in overall MT density. Although we cannot rule out the possibility that they act in parallel pathways, this observation suggests that they collaborate to anchor MTs to the apical cortex. The combination of Patronin binding to the MT minus ends and Shot binding to the MT lattice may therefore provide a robust anchor to retain MTs at the apical cortex.
Experimental Procedures
Colcemid Treatment
The protocol was modified from Parton et al. (2011). Flies were starved for 3 hr and then fed colcemid (Sigma) in yeast paste (66 μg/ml) for 2–3 hr. Ovaries were dissected and imaged as described below. Colcemid was inactivated with a brief UV pulse (3–5 s).
Imaging
For live imaging, ovaries were dissected and imaged in Voltalef oil 10S (VWR International) on an Olympus IX81 inverted microscope with a Yokogawa CSU22 spinning disk confocal imaging system (40× 1.35 NA Oil UPlanSApo, 60× 1.35 NA Oil UPlanSApo, and 100× 1.3 NA Oil UPlanSApo). Fixed preparations were imaged using Olympus IX81 (40× 1.35 NA Oil UPlanSApo, 60× 1.35 NA Oil UPlanSApo) and Zeiss LSM510 (40× NA 1.3 Oil Plan-NeoFluor) confocal microscopes. Images were collected with Olympus Fluoview, LSM510 AIM software, or MetaMorph software and processed using ImageJ. The oocyte cortex was imaged by collecting 10–15 z sections spaced 0.5 μm apart and then merging them.
Immunohistochemistry
Ovaries were fixed for 10 min in 10% paraformaldehyde and 2% Tween in PBS. Ovaries were then blocked with 10% BSA in PBS for 1 hr at room temperature. Ovaries were incubated with the primary antibody for 16 hr in PBS with 0.2% Tween and for 4 hr with the secondary antibody. In situ hybridizations were performed as previously described (Doerflinger et al., 2010). We used the following primary antibodies: mouse anti-α-tubulin fluorescein isothiocyanate at 1:250 (Sigma); mouse anti-Dynein heavy chain at 1:50 (DSHB); rabbit anti-Glued antibody raised against amino acid residues 1–400 of Glued and used at 1:100; mouse anti-DIG Cy3 at 1:200 (Jackson Immunoresearch), rabbit anti-Patronin (Goodwin and Vale, 2010) at 1:300 (gift from R. Vale, HHMI and UCSF, USA); mouse anti-Armadillo at 1:100 (DSHB); and guinea pig anti-Shot antibody raised against amino acid residues 2,602–3,640 (isoform PE) and used at 1:500. Conjugated secondary antibodies (Jackson Immunoresearch) were used at 1:100.
Molecular Biology
To generate a rescuing genomic shot transgene with C-terminal YFP tag, we used the PACMAN CH321-44M3 BAC clone (Venken et al., 2009) covering the entire shot locus. The BAC was modified using the galK positive/counter-selection cassette and recombineering (Warming et al., 2005). Transgenic flies were created by Genetivision.
The Patronin C-terminal YFP knockin was made by injecting nos>Cas9 embryos (Port et al., 2014) with a single guide RNA targeting the region of the stop codon in patronin (5′-GGCGCTTGTAATCTAAGCGG-3′, the stop codon is in bold) and a donor plasmid with 4-kb homology arms surrounding the Venus sequence.
pUASP-mKate-ABD was constructed by amplifying Shot ABD and mKate2 with the following primers: 5′-ATGTAGCGGCCGCCCGCGATGCCATTCAGAAGA-3′ and 5′-ATGTATCTAGATCAAATGTACGTGATGAGGGACT-3′; 5′ACGTGGTACCATGGTGAGCGAGCTGATT-3′ and 5′ATGTAGCGGCCGCGGAAGAGGAAGATCTGTGCCCCAGTTTGCT-3′. The amplified fragments were cloned into the pUASP vector (Rørth, 1998). The mutated Shot ABD was amplified with 5′-GATCAAACTGGACAACATACG-3′ and 5′-CGTATGTTGTCCAGTTTGATC-3′. Shot RE cDNA was obtained from A. Prokop (University of Manchester, UK).
For generation of pUASP-mCherry-Patronin and pUMAT-mCherry-Patronin, patronin RI and mCherry were amplified with 5′-ATGTAGGTACCATGGTGAGCAAGGGCGAGGAGGATAACA-3′ and 5′-GCATTCTAGATTAGATTACAAGCGCCATGTCTTTT-3′ from the pMT-mCherry-Patronin plasmid (Goodwin and Vale, 2010) (Addgene) and cloned into the pUASP vector (Rørth, 1998) and the pUMAT vector (Irion et al., 2006).
For generation of pUMAT-YFP-Patronin, patronin RI and YFP were amplified with 5′-ATGGACGAGCTGTACAAGCACCGGTATACAAGT-3′ and 5′-GCATTCTAGATTAGATTACAAGCGCCATGTCTTTT-3′, and 5′-TAGTAGGTACCCATGAGCAAGGGCGAGG-3′ and 5′-ACTTGTATACCGGTGCTTGTACAGCTCGTCCAT-3′, respectively and cloned into the pUMAT vector (Irion et al., 2006).
shot2A2 genomic DNA was isolated from homozygous embryos and larvae using the Gentra Puregene Cell Kit (Qiagen), and exonic regions were amplified by PCR and sequenced. Primer sequences are available on request.
Author Contributions
D.N. performed most of the experiments and data analysis. A.R.F. performed immunoprecipitations. D.N. and D.St J. planned the experiments. D.N. and D.St J. conceived the project and wrote the manuscript.
Acknowledgments
We are grateful to R. Vale, A. Prokop, N. Brown, J. Raff, S. Endow, K. McKim, the Kyoto Stock Center and the Bloomington Stock Center for flies and reagents. We thank N. Dawney for technical assistance and C. Flandoli for technical art work. This work was supported by a Wellcome Trust PRF to D.S.J. (080007) and by core support from the Wellcome Trust (092096) and Cancer Research UK (A14492). D.N. was supported by a postdoctoral fellowship from the Swedish Research Council. A.R.F. is funded by a University of Cambridge PhD studentship.
Published: July 11, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and nine movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2016.06.010.
Supplemental Information
References
- Akhmanova A., Steinmetz M.O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Alves-Silva J., Sánchez-Soriano N., Beaven R., Klein M., Parkin J., Millard T.H., Bellen H.J., Venken K.J.T., Ballestrem C., Kammerer R.A. Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins) J. Neurosci. 2012;32:9143–9158. doi: 10.1523/JNEUROSCI.0416-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite D.A., Grode K.D., Keller D., Zadeh A., Slep K.C., Rogers S.L. The spectraplakin Short stop is an actin-microtubule cross-linker that contributes to organization of the microtubule network. Mol. Biol. Cell. 2010;21:1714–1724. doi: 10.1091/mbc.E10-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P.W., Ahmad F.J. The plus ends of stable microtubules are the exclusive nucleating structures for microtubules in the axon. J. Cell Biol. 1992;116:1231–1241. doi: 10.1083/jcb.116.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacallao R., Antony C., Dotti C., Karsenti E., Stelzer E.H., Simons K. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J. Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines A.J., Bignone P.A., King M.D.A., Maggs A.M., Bennett P.M., Pinder J.C., Phillips G.W. The CKK Domain (DUF1781) binds microtubules and defines the CAMSAP/ssp4 family of animal proteins. Mol. Biol. Evol. 2009;26:2005–2014. doi: 10.1093/molbev/msp115. [DOI] [PubMed] [Google Scholar]
- Bartolini F., Gundersen G.G. Generation of noncentrosomal microtubule arrays. J. Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth A.J.R., Blanchard G.B., Adams R.J., Roper K. A dynamic microtubule cytoskeleton directs medial actomyosin function during tube formation. Dev. Cell. 2014;29:562–576. doi: 10.1016/j.devcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza R.P., Serbus L.R., Saxton W.M., Duffy J.B. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr. Biol. 2002;12:1541–1545. doi: 10.1016/s0960-9822(02)01108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodu V., Baffet A.D., Le Droguen P.-M., Casanova J., Guichet A. A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Dev. Cell. 2010;18:790–801. doi: 10.1016/j.devcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Chang C.-W., Nashchekin D., Wheatley L., Irion U., Dahlgaard K., Montague T.G., Hall J., St Johnston D. Anterior-posterior axis specification in Drosophila oocytes:identification of novel bicoid and oskar mRNA localization factors. Genetics. 2011;188:883–896. doi: 10.1534/genetics.111.129312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang M., Goncharov A., Wang S., Oegema K., Jin Y., Chisholm A.D. The microtubule minus-end-binding protein Patronin/PTRN-1 is required for axon regeneration in C. elegans. Cell Rep. 2014;9:874–883. doi: 10.1016/j.celrep.2014.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I., Giniger E., Ruohola-Baker H., Jan L.Y., Jan Y.-N. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 1994;4:289–300. doi: 10.1016/s0960-9822(00)00068-3. [DOI] [PubMed] [Google Scholar]
- Clark I.E., Jan L.Y., Jan Y.N. Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development. 1997;124:461–470. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- Coelho P.A., Bury L., Sharif B., Riparbelli M.G., Fu J., Callaini G., Glover D.M., Zernicka-Goetz M. Spindle formation in the mouse embryo requires Plk4 in the absence of centrioles. Dev. Cell. 2013;27:586–597. doi: 10.1016/j.devcel.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H., Vogt N., Torres I.L., Mirouse V., Koch I., Nüsslein-Volhard C., St Johnston D. Bazooka is required for polarisation of the Drosophila anterior-posterior axis. Development. 2010;137:1765–1773. doi: 10.1242/dev.045807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D.M. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- Feldman J.L., Priess J.R. A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr. Biol. 2012;22:575–582. doi: 10.1016/j.cub.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goodwin S.S., Vale R.D. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D., Volkmann N., Goldsmith S., Michon A.M., Lehman W., Craig R., DeRosier D., Almo S., Matsudaira P. An atomic model of fimbrin binding to F-actin and its implications for filament crosslinking and regulation. Nat. Struct. Biol. 1998;5:787–792. doi: 10.1038/1828. [DOI] [PubMed] [Google Scholar]
- Hendershott M.C., Vale R.D. Regulation of microtubule minus-end dynamics by CAMSAPs and Patronin. Proc. Natl. Acad. Sci. USA. 2014;111:5860–5865. doi: 10.1073/pnas.1404133111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J.A., Buey R.M., Lawera A. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Irion U., Adams J., Chang C.-W., St Johnston D. Miranda couples oskar mRNA/Staufen complexes to the bicoid mRNA localization pathway. Dev. Biol. 2006;297:522–533. doi: 10.1016/j.ydbio.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Jankovics F., Brunner D. Transiently reorganized microtubules are essential for zippering during dorsal closure in Drosophila melanogaster. Dev. Cell. 2006;11:375–385. doi: 10.1016/j.devcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Gillet L., Keryer G., Bornens M., Guichet A. The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development. 2006;133:129–139. doi: 10.1242/dev.02179. [DOI] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F.M., Heck A.J.R., Hoogenraad C.C. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 2014;28:295–309. doi: 10.1016/j.devcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., Debec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- Khuc Trong P., Doerflinger H., Dunkel J., St Johnston D., Goldstein R.E. Cortical microtubule nucleation can organise the cytoskeleton of Drosophila oocytes to define the anteroposterior axis. Elife. 2015;4:e06088. doi: 10.7554/eLife.06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A., Karakesisoglou I., Wong E., Vaezi A. ACF7 an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Lechler T., Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J. Cell Biol. 2007;176:147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kolodziej P. Short Stop provides an essential link between F-actin and microtubules during axon extension. Development. 2002;129:1195–1204. doi: 10.1242/dev.129.5.1195. [DOI] [PubMed] [Google Scholar]
- Lee S., Harris K.L., Whitington P.M., Kolodziej P.A. Short stop is allelic to kakapo, and encodes rod-like cytoskeletal-associated proteins required for axon extension. J. Neurosci. 2000;20:1096–1108. doi: 10.1523/JNEUROSCI.20-03-01096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C., Sun D., Zheng M., Knowles D., Liem R. Microtubule actin cross-linking factor (MACF): a hybrid of Dystonin and Dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 1999;147:1275–1286. doi: 10.1083/jcb.147.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom J.J., Nakamura M., Hibbel A., Shundyak K., Gutierrez R., Ketelaar T., Emons A.M.C., Mulder B.M., Kirik V., Ehrhardt D.W. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science. 2013;342:1245533. doi: 10.1126/science.1245533. [DOI] [PubMed] [Google Scholar]
- Lowe N., Rees J.S., Roote J., Ryder E., Armean I.M., Johnson G., Drummond E., Spriggs H., Drummond J., Magbanua J.P. Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development. 2014;141:3994–4005. doi: 10.1242/dev.111054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcette J.D., Chen J.J., Nonet M.L., Hobert O. The Caenorhabditis elegans microtubule minus-end binding homolog PTRN-1 stabilizes synapses and neurites. Elife. 2014;3:e01637. doi: 10.7554/eLife.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W., Mushika Y., Ichii T., Takeichi M. Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–959. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of Ninein. J. Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Nguyen M.M., Stone M.C., Rolls M.M. Microtubules are organized independently of the centrosome in Drosophila neurons. Neural Dev. 2011;6:38. doi: 10.1186/1749-8104-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I.M., St Johnston D. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 2002;129:5473–5485. doi: 10.1242/dev.00119. [DOI] [PubMed] [Google Scholar]
- Parton R.M., Hamilton R.S., Ball G., Yang L., Cullen C.F., Lu W., Ohkura H., Davis I. A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 2011;194:121–135. doi: 10.1083/jcb.201103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N., Stevens N.R., Basto R., Raff J.W. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., Bullock S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA. 2014;111:E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C.E., Spilker K.A., Cueva J.G., Perrino J., Goodman M.B., Shen K., Hobert O. PTRN-1, a microtubule minus end-binding CAMSAP homolog, promotes microtubule function in Caenorhabditis elegans neurons. Elife. 2014;3:e01498. doi: 10.7554/eLife.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Peifer M., Rogers S.L. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., Vale R.D. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 2006;175:849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper K., Brown N.H. Maintaining epithelial integrity: a function for gigantic spectraplakin isoforms in adherens junctions. J. Cell Biol. 2003;162:1305–1315. doi: 10.1083/jcb.200307089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper K., Brown N.H. A spectraplakin is enriched on the fusome and organizes microtubules during oocyte specification in Drosophila. Curr. Biol. 2004;14:99–110. [PubMed] [Google Scholar]
- Rørth P. Gal4 in the Drosophila female germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Shulman J.M., Benton R., St Johnston D. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Stevens N.R., Raposo A.A.S.F., Basto R., St Johnston D., Raff J.W. From stem cell to embryo without centrioles. Curr. Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N.R., Dobbelaere J., Brunk K., Franz A., Raff J.W. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiess M., Maghelli N., Kapitein L.C., Gomis-Rüth S., Wilsch-Bräuninger M., Hoogenraad C.C., Tolić-Nørrelykke I.M., Bradke F. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 2010;327:704–707. doi: 10.1126/science.1182179. [DOI] [PubMed] [Google Scholar]
- Sun D., Leung C.L., Liem R.K. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): identification of a novel group of microtubule associated proteins. J. Cell Sci. 2001;114:161–172. doi: 10.1242/jcs.114.1.161. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Meng W., Nagae S., Takeichi M. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc. Natl. Acad. Sci. USA. 2012;109:20029–20034. doi: 10.1073/pnas.1218017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E., Smiley S., Wong M.L., Alberts B.M. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 1992;115:923–936. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- Toya M., Kobayashi S., Kawasaki M., Shioi G., Kaneko M., Ishiuchi T., Misaki K., Meng W., Takeichi M. CAMSAP3 orients the apical-to-basal polarity of microtubule arrays in epithelial cells. Proc. Natl. Acad. Sci. USA. 2016;113:332–337. doi: 10.1073/pnas.1520638113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J.T., Carlson J.W., Schulze K.L., Pan H., He Y., Spokony R., Wan K.H., Koriabine M., de Jong P.J., White K.P. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Brust-Mascher I., Civelekoglu-Scholey G., Scholey J.M. Patronin mediates a switch from kinesin-13-dependent poleward flux to anaphase B spindle elongation. J. Cell Biol. 2013;4:1343. doi: 10.1083/jcb.201306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wu D., Quintin S., Green R.A., Cheerambathur D.K., Ochoa S.D., Desai A., Oegema K. NOCA-1 functions with γ-tubulin and in parallel to Patronin to assemble non-centrosomal microtubule arrays in C. elegans. Elife. 2015;4:e08649. doi: 10.7554/eLife.08649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. Simple and highly efficient BAC recombineering using galK selection. Nucl. Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau K.W., van Beuningen S.F.B., Cunha-Ferreira I., Cloin B.M.C., van Battum E.Y., Will L., Schätzle P., Tas R.P., van Krugten J., Katrukha E.A. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron. 2014;82:1058–1073. doi: 10.1016/j.neuron.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Zheng J., Furness D., Duan C., Miller K.K., Edge R.M., Chen J., Homma K., Hackney C.M., Dallos P., Cheatham M.A. Marshalin, a microtubule minus-end binding protein, regulates cytoskeletal structure in the organ of Corti. Biol. Open. 2013;2:1192–1202. doi: 10.1242/bio.20135603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin V.L., Belaya K., Pecreaux J., Gilchrist M.J., Clark A., Davis I., St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.