Abstract
Osmolyte transport is a pivotal part of bacterial life, particularly in high salt environments. Several low and high affinity osmolyte transport systems have been identified in various bacterial species. A lot of research has centered on characterizing the osmolyte transport systems of Gram‐negative bacteria, but less has been done to characterize the same transport systems in Gram‐positive bacteria. This review will focus on the previous work that has been done to understand the osmolyte transport systems in the species Staphylococcus aureus and how these transporters may serve dual functions in allowing the bacteria to survive and grow in a variety of environments, including on the surface or within humans or other animals.
Keywords: PutP, OpuD, Staphylococcus aureus, Proline transport, Osmolyte
INTRODUCTION
A well conserved, evolutionary strategy used by many organisms to adapt to high osmotic conditions is the transport of organic compounds, called compatible solutes[1]. These compatible solutes serve as cytoplasmic solutes that balance water relations, without interfering with normal cytoplasmic activities, within cells grown in high salt environments. Examination of the transport systems in Staphylococcus aureus (S. aureus) may provide insight into how proline and glycine betaine may be transported into Gram-positive bacteria.
GENERAL OSMOLYTE TRANSPORT FEATURES IN S. AUREUS
Although osmolyte transport is best described in E. coli[1–3], there are also compatible solute transport systems in S. aureus to adapt to high salt environments[4]. Studies have shown that S. aureus cells grown in very high salt environments had increased intracellular levels of proline and glycine betaine[5–11]. Other intracellular molecules that also increased in high NaCl environments were choline, proline betaine, taurine, and glutamic acid[6,7,12]. Of these accumulated solutes, proline and glycine betaine were the most effective osmoprotectants of S. aureus, since S. aureus growth was observed when these solutes were excluded from defined high osmotic media[6,8,12].
Identification of genes that encode transport proteins and their importance for the survival of S. aureus coincides with previous observations that S. aureus requires several amino acids as a source of carbon and nitrogen[4]. Of these essential amino acids, proline and other amino acids are not synthesized by S. aureus[4,13,14]. The accumulation of most of the proline in S. aureus occurs because of proline transport proteins.
Although prior research performed using other Gram-positive bacteria may not have specifically addressed proline transport, it does help in uncovering commonly conserved mechanisms of compatible solute transport in S. aureus. Several studies that have examined compatible solutes accumulation in S. aureus grown at high osmotic environments showed increased intracellular levels of proline, aminobutyric acid, glutamic acid, choline, taurine, and glycine betaine[5–7,15,16]. Of these compatible solutes, only glutamic acid is synthesized by S. aureus, whereas the other compatible solutes have to be imported from the external environment[5,7,8,17–19]. To substantiate the osmoprotective importance of these transported compatible solutes, the growth rates of S. aureus grown in defined high osmotic media was observed to increase when supplemented with either proline or glycine betaine[8]. Although S. aureus normally possess relatively large concentrations of glycine betaine and potassium ions, compatible solute transport is believed to aid in creating high intracellular pressure that enables S. aureus to survive in high osmotic environments[15].
SPECIFIC PROLINE TRANSPORT SYSTEMS IN S. AUREUS
Initial proline uptake research using whole cell assays on S. aureus has shown the presence of at least two proline transport systems[10,17,20]: Both a low- and high‐affinity system. These systems may be similar to the OpuE and OpuD transport systems found in B. subtilis[21,22] and they share properties with the PutP and ProP systems of E. coli[1]. They are both sodium-dependent transporters, since gramicidin D and monensin, which collapse Na+ gradients, inhibit proline transport in both systems[10]. Proline transport in either system showed low susceptibility to inhibition by glycolysis and ATP formation by a combination of NaF and sodium iodoacetate or sodium arsenate, respectively. Lastly, alterations of pH from 5.5 to 8.5 had little effect on the transport rates of proline[10].
In S. aureus, proline transport kinetics is hard to interpret because of strain differences and the calculation setups used to determine the Km and Vmax values reported, one based on per mg protein and the other per mg dry weight. Reports have shown that the high-affinity proline transport system in S. aureus had a Km ranging from 1.7 to 7.0 mol/L, with a Vmax ranging from 1.1 nmol/min per milligram dry weight to 10 nmol/min per milligram protein[10,17]. Though these numbers are not directly comparative, they do give us a relative range of activity for this system, which correlates to a previously observed Km value of 3.5 mol/L for proline uptake with vesicles prepared from S. aureus grown in a low-osmolarity medium[23] and Km values of the PutP system in E. coli[1,17,24–26]. Moreover, like the PutP system of E. coli[1], the high-affinity proline transport system in S. aureus is specific for the transport of proline and it’s activity increases when proline deprivation is encountered, suggesting that this system may also be involved in scavenging low concentrations of proline from the environment[10]. Further proof of the relatedness of these systems can be seen from the complementation of a genetic defect in proline transport within E. coli by the high‐affinity proline transport system of S. aureus[27]. At the structural level, the PutP homolog of S. aureus shows a sodium-binding motif, the same ten conserved amino acids found in all other members of the sodium/solute symporters[28], and the predicted PutP protein of S. aureus[29] shares considerable similarity with the PutP protein of E. coli[1]. Although many similarities exist between the high-affinity proline transport systems in S. aureus and E. coli, major differences between these systems include: The concentration of NaCl appears to have no effect on proline transport in S. aureus[8,17]; the S. aureus putP gene is activated by high concentrations of osmolytes in the environment[30], whereas the E. coli putP gene is not[1,25,29]; and the S. aureus putP gene is regulated by SigB[30], which is similar to the regulation shown for opuE in B. subtilis[21]. Although PutP has a sodium binding motif and has homology with sodium/solute symporters, the concentration of NaCl does not affect proline transport[7,17], It is possible that when S. aureus is grown in an environment with a low sodium concentration that PutP behaves like other bacterial high affinity proline transporters that are driven by a sodium motive force. On the other hand, S. aureus grown in a high sodium environment may cause the PutP protein to use a proton motive force instead of a sodium motive force to bring proline into the cell.
The low-affinity proline transport system of S. aureus also has similarities to the low-affinity proline transport system (ProP) of E. coli. For proline transport, the Km value of S. aureus ATCC 12600 (Km of 420 mol/L and Vmax of 110 nmol/min per milligram protein) is similar to the Km value of ProP in E. coli (approximately 300 mol/L)[17]. For S. aureus (Km of 132 mol/L and Vmax of 22 nmol/min per milligram dry weight), a greater difference in the Km values for the low-affinity proline transport system can be seen between strains as compared to the difference in Km values for the high-affinity system. Again, the Km and Vmax values from the ProP system of E. coli fit within the overall range found for S. aureus[1,31–33], but strain variation along with calculation setup differences may again be the cause of these divergent numbers. Excluding the differences of the Km and Vmax values between strains, the low-affinity proline transport systems of different S. aureus strains possess identical characteristics[10,17]. Many of these characteristics are similar to the regulatory and functional properties of the ProP system of E. coli[34] (i.e., both of these systems transport proline and are stimulated by increasing osmolarity produced by either ionic or nonionic solutes)[17].
DIFFERENCES IN THE S. AUREUS OSMOLYTE TRANSPORT SYSTEMS COMPARED TO OTHER BACTERIA
Though these systems are similar, there are some major differences between the Gram-negative and Gram-positive low-affinity proline transport systems. One major difference is that the low‐affinity proline transport systems in S. aureus are optimally activated at NaCl concentrations ranging from 0.75 to 1.0 mol/L[17,35], whereas the low-affinity proline transport systems in E. coli are inhibited by NaCl concentrations greater than 0.2 to 0.3 mol/L[29,36]. Other major differences include glycine betaine transport activity by the low-affinity proline transport system has not been conclusively established and there conflicting opinions and data presented for the glycine betaine transport activity for the low-affinity system[9,17,18,20,37]. In part, the previous lack of any low-affinity system mutants in those studies complicated the examination of glycine betaine transport activities. Since glycine betaine accumulation has been linked to proline transporters in Gram-negative bacteria[1] and S. aureus has been shown to transport glycine betaine from the external environment[38], this suggests that an additional glycine betaine transporter that is osmotically stimulated may be present in S. aureus. Moreover, S. aureus cells shocked with 0.5 mol/L NaCl in the presence and absence of chloramphenicol (100 g/mL) showed identical levels of transported proline, suggesting that new protein synthesis is not necessary for rapid proline uptake and that osmotic shock activates a pre-existing proline transport system[10].
BIOINFORMATIC TOOLS TO IDENTIFY OSMOLYTE TRANSPORT SYSTEMS IN S. AUREUS
Sequencing of several S. aureus genomes has provided a wealth of information on the existence of several putative osmolyte transport systems in S. aureus[14,39,40]. All of the strains appear to have a conserved putP gene for high affinity transport of proline, although there appears to be homologs for both a proP gene[1] and opuD gene[21,35] (Table 1). Additional analyses have shown that the opuD gene (encoding a low affinity proline transporter) is activated under osmotic stress conditions and OpuD transports proline under low affinity growth conditions[35]. Furthermore, a mutation in the S. aureus proP gene also causes lower proline transport in media with high concentrations of proline (Schwan WR unpublished data).
Table 1.
Distribution of proline and glycine betaine transport genes in some sequenced
| S. aureus strains | ||||
|
| ||||
| Gene | N315 | MW2 | COL | Mu50 |
| putP | SA1718 | MW1843 | SACOL1963 | SAV1902 |
| putP | SA0531 | MW0528 | SACOL0620 | SAV0573 |
| opuD | SA1183 | MW1236 | Yes (2)2 | SAV13494 |
| opuD1 | -1 | -1 | SACOL1384 | ND3 |
| opuD2 | -1 | -1 | SACOL2176 | ND3 |
| opuCA | SA2237 | MW2372 | ND3 | SAV2448 |
| opuCB | SA2236 | MW2371 | ND3 | SAV2447 |
| opuCC | SA2235 | MW2370 | ND3 | SAV2446 |
| opuCD | SA2234 | MW2369 | ND3 | SAV2445 |
Does not possess;
Multiple opuD genes in this species;
Not determined;
The gene appears to be fragmented into two pieces.
This is the first instance of both the ProP and OpuD low affinity proline/glycine betaine transport homologs being identified in one species and suggests the importance that proline transport must have in the survival of S. aureus cells in a variety of environments. Furthermore, the opuC system, which putatively transports glycine betaine/carnitine/choline, has also been observed. Together, the bioinformatic comparisons have uncovered some very interesting genomic features in S. aureus centered on osmolyte transport. A summary of the four osmolyte transport systems in S. aureus tied to proline transport and other known solutes is noted in Figure 1.
Figure 1.
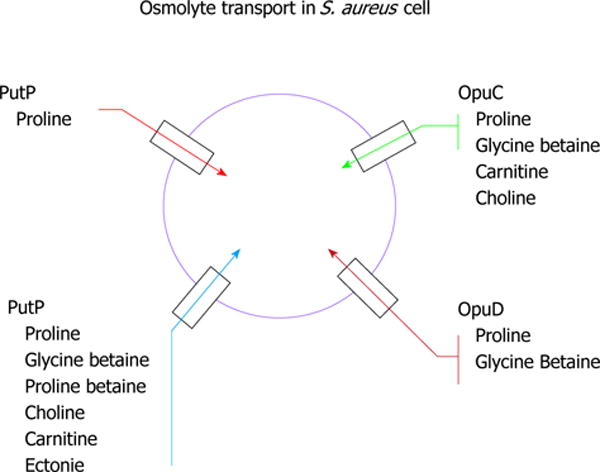
The four prominent osmolyte transport systems in Staphylococcus aureus tied to proline transport as well as other solutes.
OSMOLYTE TRANSPORT TIED TO S. AUREUS SURVIVAL IN HUMANS AND MICE
The rationale of investigating proline and glycine betaine transport in S. aureus is not purely academic. In planktonic S. aureus, the glycine betaine level is high, but lower in S. aureus found in biofilms[41]. Glycine betaine is the most effective osmoprotectant. To achieve the high glycine betaine level, an active glycine betaine transporter would need to be functioning in the planktonic S. aureus cells that are immersed in an environment of high osmotic stress, like the human skin.
Indirect effects on S. aureus survival have been tied to osmolyte transport systems. Defects in the cell wall caused by a femAB mutation caused an upregulation of opuC (glycine betaine/carnitine/choline transporter) and downregulation of opuD to compensate for the defect[42]. YhcSR encodes a two-component signal transduction system that is required for S. aureus survival. This two-component regulatory system regulates transcription of the opuCABCD operons affecting proline and glycine betaine levels in S. aureus[43]. One study examining daptomycin resistance revealed an accumulation of glycine betaine within S. aureus cells that was coupled with upregulation of the cudT (choline transporter) gene, a beta choline dehydrogenase gene, a gbsA gene (glycine betaine aldehyde dehydrogenase), an opuD2 gene, and the proP gene[44]. Uptake of choline is needed to produce glycine betaine internally, the best osmoprotectant[19].
More directly, a transposon mutation in the gene for the high affinity (PutP) proline transport system of S. aureus rendered the bacteria less able to survive in several animal infection models[45–47]. Within cardiac vegetations, the viable S. aureus count was 1–3 logs lower than the wild-type parent strain[45]. Transcription of putP was shown to increase 105-fold shortly after S. aureus infection of murine kidneys[30]. In S. aureus infected murine bladders, spleen and livers, putP transcription was also elevated very quickly and then dropped markedly as the infection progressed. Proline levels in livers and spleens are very low[47] and the levels are likely low in the other organs (e.g., bladder and kidney), but through tissue damage by staphylococcal toxins, the concentration of proline may increase substantially and in turn shut off transcription of the high affinity proline transport gene.
Conversely, transcription of the low affinity proline transport gene opuD was shown to be the highest after 4 h post-infection in murine bladders and 18 h post-infection in murine thigh abscesses[35]. Within murine bladders and kidneys, high osmotic conditions prevail. Initial observations demonstrated that at least one of the low‐affinity proline transport systems of S. aureus was activated under moderate to high osmotic conditions[17], which has been subsequently confirmed[35].
Our model is that PutP is important in the early stages of an infection when proline concentrations are low, but OpuD expression is not as important (Figure 2). As the infection proceeds, tissue damage occurs, which releases free proline. By 18 h post-infection, the level of free proline is higher and OpuD becomes important at this stage of the infection.
Figure 2.
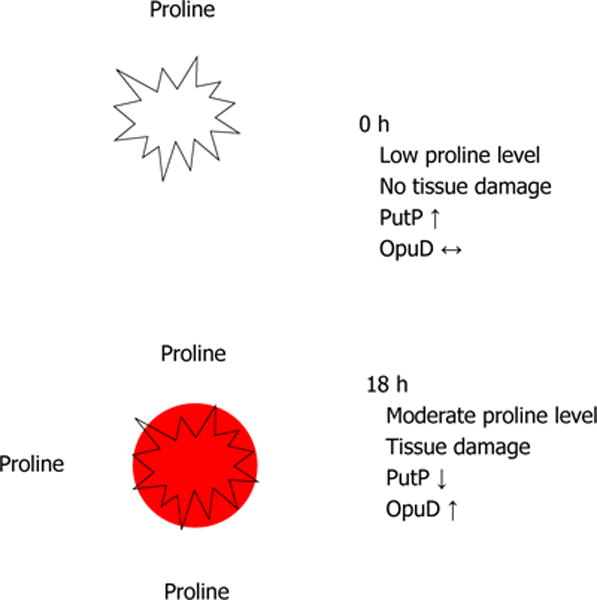
Model for the roles of proline transporters in Staphylococcus aureus pathogenesis within a murine abscess.
These studies suggest that osmolyte transport systems may play essential roles in survival of S. aureus within humans or mice. Characterization of the proline and glycine betaine transport systems will provide us with experimental proof of the importance of these systems during growth in high osmotic conditions, how these systems are regulated, and will further our understanding of the significance of the proline/glycine betaine transport to the survival of S. aureus in vivo.
Core tip.
Staphylococcus aureus (S. aureus) is the number one cause of skin and soft tissue infections. In the United States, S. aureus is usually the number one hospital-acquired pathogen. The skin and urinary tract organs are high osmotic stress environments. Osmolyte transport is essential for S. aureus survival in different environmental niches, such as within human skin abscesses or the human urinary tract.
Acknowledgments
I would like to thank the University of Wisconsin-La Crosse for grant support for my laboratory and also thank all of the undergraduate and graduate students whom I have mentored.
Supported by NIH grant, No. 1R15AI47801-01A.
Footnotes
Author contributions: All the authors contribute to the manuscript.
Conflict-of-interest statement: Authors declare no conflicts of interest.
P- Reviewer: García-Elorriaga G, Jung H, Krishnan T
S- Editor: Qiu S L- Editor: A E- Editor: Jiao XK
References
- 1.Wood JM. Proline porters effect the utilization of proline as nutrient or osmoprotectant for bacteria. J Membr Biol. 1988;106:183–202. doi: 10.1007/BF01872157. [DOI] [PubMed] [Google Scholar]
- 2.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fichman Y, Gerdes SY, Kovács H, Szabados L, Zilberstein A, Csonka LN. Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev Camb Philos Soc. 2015;90:1065–1099. doi: 10.1111/brv.12146. [DOI] [PubMed] [Google Scholar]
- 4.Ruoff KL. Algorithm for identification of aerobic Gram-positive cocci, p. 262–282. In: Murray PR, Baron EJ, Pflaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. 7th. American Society for Microbiology; Washington, D.C: 1999. [Google Scholar]
- 5.Anderson CB, Witter LD. Glutamine and proline accumulation by Staphylococcus aureus with reduction in water activity. Appl Environ Microbiol. 1982;43:1501–1503. doi: 10.1128/aem.43.6.1501-1503.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham JE, Wilkinson BJ. Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J Bacteriol. 1992;174:2711–2716. doi: 10.1128/jb.174.8.2711-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koujima I, Hayashi H, Tomochika K, Okabe A, Kanemasa Y. Adaptational change in proline and water content of Staphylococcus aureus after alteration of environmental salt concentration. Appl Environ Microbiol. 1978;35:467–470. doi: 10.1128/aem.35.3.467-470.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller KJ, Zelt SC, Bae J-H. Glycine betaine and proline are the principle compatible solutes of Staphylococcus aureus. Curr Micorobiol. 1991;23:131–137. doi: 10.1007/BF02091971. [DOI] [Google Scholar]
- 9.Pourkomailian B, Booth IR. Glycine betaine transport by Staphylococcus aureus: evidence for two transport systems and for their possible roles in osmoregulation. J Gen Microbiol. 1992;138:2515–2518. doi: 10.1099/00221287-138-12-2515. [DOI] [PubMed] [Google Scholar]
- 10.Townsend DE, Wilkinson BJ. Proline transport in Staphylococcus aureus: a high‐affinity system and a low‐affinity system involved in osmoregulation. J Bacteriol. 1992;174:2702–2710. doi: 10.1128/jb.174.8.2702-2710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijaranakul U, Nadakavukaren MJ, Bayles DO, Wilkinson BJ, Jayaswal RK. Characterization of an NaCl-sensitive Staphylococcus aureus mutant and rescue of the NaCl-sensitive phenotype by glycine betaine but not by other compatible solutes. Appl Environ Microbiol. 1997;63:1889–1897. doi: 10.1128/aem.63.5.1889-1897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amin US, Lash TD, Wilkinson BJ. Proline betaine is a highly effective osmoprotectant for Staphylococcus aureus. Arch Microbiol. 1995;163:138–142. doi: 10.1007/BF00381788. [DOI] [PubMed] [Google Scholar]
- 13.Iandolo JJ, Worrell V, Groicher KH, Qian Y, Tian R, Kenton S, Dorman A, Ji H, Lin S, Loh P, Qi S, Zhu H, Roe BA. Comparative analysis of the genomes of the temperate bacteriophages phi 11, phi 12 and phi 13 of Staphylococcus aureus 8325. Gene. 2002;289:109–118. doi: 10.1016/S0378-1119(02)00481-X. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 15.Kunin CM, Rudy J. Effect of NaCl-induced osmotic stress on intracellular concentrations of glycine betaine and potassium in Escherichia coli, Enterococcus faecalis, and staphylococci. J Lab Clin Med. 1991;118:217–224. [PubMed] [Google Scholar]
- 16.Measures JC. Role of amino acids in osmoregulation of non-halophilic bacteria. Nature. 1975;257:398–400. doi: 10.1038/257398a0. [DOI] [PubMed] [Google Scholar]
- 17.Bae JH, Miller KJ. Identification of two proline transport systems in Staphylococcus aureus and their possible roles in osmoregulation. Appl Environ Microbiol. 1992;58:471–475. doi: 10.1128/aem.58.2.471-475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae JH, Anderson SH, Miller KJ. Identification of a high-affinity glycine betaine transport system in Staphylococcus aureus. Appl Environ Microbiol. 1993;59:2734–2736. doi: 10.1128/aem.59.8.2734-2736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaenjak A, Graham JE, Wilkinson BJ. Choline transport activity in Staphylococcus aureus induced by osmotic stress and low phosphate concentrations. J Bacteriol. 1993;175:2400–2406. doi: 10.1128/jb.175.8.2400-2406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pourkomailian B, Booth IR. Glycine betaine transport by Staphylococcus aureus: evidence for feedback regulation of the activity of the two transport systems. Microbiology. 1994;140(Pt 11):3131–3138. doi: 10.1099/13500872-140-11-3131. [DOI] [PubMed] [Google Scholar]
- 21.Kappes RM, Kempf B, Bremer E. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J Bacteriol. 1996;178:5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Blohn C, Kempf B, Kappes RM, Bremer E. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol Microbiol. 1997;25:175–187. doi: 10.1046/j.1365-2958.1997.4441809.x. [DOI] [PubMed] [Google Scholar]
- 23.Short SA, Kaback HR. Amino acid transport and staphylococcal membrane vesicles. Ann N Y Acad Sci. 1974;236:124–143. doi: 10.1111/j.1749-6632.1974.tb41487.x. [DOI] [PubMed] [Google Scholar]
- 24.Bracher S, Guérin K, Polyhach Y, Jeschke G, Dittmer S, Frey S, Böhm M, Jung H. Glu-311 in External Loop 4 of the Sodium/Proline Transporter PutP Is Crucial for External Gate Closure. J Biol Chem. 2016;291:4998–5008. doi: 10.1074/jbc.M115.675306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CC, Wilson TH. Solubilization and functional reconstitution of the proline transport system of Escherichia coli. J Biol Chem. 1986;261:2599–2604. [PubMed] [Google Scholar]
- 26.Myers RS, Townsend D, Maloy S. Dissecting the molecular mechanism of ion-solute cotransport: substrate specificity mutations in the putP gene affect the kinetics of proline transport. J Membr Biol. 1991;121:201–214. doi: 10.1007/BF01951554. [DOI] [PubMed] [Google Scholar]
- 27.Wengender PA, Miller KJ. Identification of a PutP proline permease gene homolog from Staphylococcus aureus by expression cloning of the high‐affinity proline transport system in Escherichia coli. Appl Environ Microbiol. 1995;61:252–259. doi: 10.1128/aem.61.1.252-259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reizer J, Reizer A, Saier MH. A functional superfamily of sodium/solute symporters. Biochim Biophys Acta. 1994;1197:133–166. doi: 10.1016/0304-4157(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 29.Grothe S, Krogsrud RL, McClellan DJ, Milner JL, Wood JM. Proline transport and osmotic stress response in Escherichia coli K-12. J Bacteriol. 1986;166:253–259. doi: 10.1128/jb.166.1.253-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwan WR, Lehmann L, McCormick J. Transcriptional activation of the Staphylococcus aureus putP gene by low-proline-high osmotic conditions and during infection of murine and human tissues. Infect Immun. 2006;74:399–409. doi: 10.1128/IAI.74.1.399-409.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Culham DE, Lasby B, Marangoni AG, Milner JL, Steer BA, van Nues RW, Wood JM. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- 32.MacMillan SV, Alexander DA, Culham DE, Kunte HJ, Marshall EV, Rochon D, Wood JM. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim Biophys Acta. 1999;1420:30–44. doi: 10.1016/S0005-2736(99)00085-1. [DOI] [PubMed] [Google Scholar]
- 33.Milner JL, Grothe S, Wood JM. Proline porter II is activated by a hyperosmotic shift in both whole cells and membrane vesicles of Escherichia coli K12. J Biol Chem. 1988;263:14900–14905. [PubMed] [Google Scholar]
- 34.Racher KI, Culham DE, Wood JM. Requirements for osmosensing and osmotic activation of transporter ProP from Escherichia coli. Biochemistry. 2001;40:7324–7333. doi: 10.1021/bi002331u. [DOI] [PubMed] [Google Scholar]
- 35.Wetzel KJ, Bjorge D, Schwan WR. Mutational and transcriptional analyses of the Staphylococcus aureus low-affinity proline transporter OpuD during in vitro growth and infection of murine tissues. FEMS Immunol Med Microbiol. 2011;61:346–355. doi: 10.1111/j.1574-695X.2011.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faatz E, Middendorf A, Bremer E. Cloned structural genes for the osmotically regulated binding-protein-dependent glycine betaine transport system (ProU) of Escherichia coli K-12. Mol Microbiol. 1988;2:265–279. doi: 10.1111/j.1365-2958.1988.tb00028.x. [DOI] [PubMed] [Google Scholar]
- 37.Stimeling KW, Graham JE, Kaenjak A, Wilkinson BJ. Evidence for feedback (trans) regulation of, and two systems for, glycine betaine transport by Staphylococcus aureus. Microbiology. 1994;140(Pt 11):3139–3144. doi: 10.1099/13500872-140-11-3139. [DOI] [PubMed] [Google Scholar]
- 38.Peddie BA, Wong-She J, Randall K, Lever M, Chambers ST. Osmoprotective properties and accumulation of betaine analogues by Staphylococcus aureus. FEMS Microbiol Lett. 1998;160:25–30. doi: 10.1111/j.1574-6968.1998.tb12885.x. [DOI] [PubMed] [Google Scholar]
- 39.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 40.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Junka AF, Deja S, Smutnicka D, Szymczyk P, Ziółkowski G, Bartoszewicz M, Młynarz P. Differences in metabolic profiles of planktonic and biofilm cells in Staphylococcus aureus - (1)H Nuclear Magnetic Resonance search for candidate biomarkers. Acta Biochim Pol. 2013;60:701–706. [PubMed] [Google Scholar]
- 42.Hübscher J, Jansen A, Kotte O, Schäfer J, Majcherczyk PA, Harris LG, Bierbaum G, Heinemann M, Berger-Bächi B. Living with an imperfect cell wall: compensation of femAB inactivation in Staphylococcus aureus. BMC Genomics. 2007;8:307. doi: 10.1186/1471-2164-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan M, Hall JW, Yang J, Ji Y. The essential yhcSR two-component signal transduction system directly regulates the lac and opuCABCD operons of Staphylococcus aureus. PLoS One. 2012;7:e50608. doi: 10.1371/journal.pone.0050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y, Rubio A, Jayaswal RK, Silverman JA, Wilkinson BJ. Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. PLoS One. 2013;8:e58469. doi: 10.1371/journal.pone.0058469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer AS, Coulter SN, Stover CK, Schwan WR. Impact of the high-affinity proline permease gene (putP) on the virulence of Staphylococcus aureus in experimental endocarditis. Infect Immun. 1999;67:740–744. doi: 10.1128/iai.67.2.740-744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwan WR, Coulter SN, Ng EY, Langhorne MH, Ritchie HD, Brody LL, Westbrock-Wadman S, Bayer AS, Folger KR, Stover CK. Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models. Infect Immun. 1998;66:567–572. doi: 10.1128/iai.66.2.567-572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwan WR, Wetzel KJ, Gomez TS, Stiles MA, Beitlich BD, Grunwald S. Low-proline environments impair growth, proline transport and in vivo survival of Staphylococcus aureus strain-specific putP mutants. Microbiology. 2004;150:1055–1061. doi: 10.1099/mic.0.26710-0. [DOI] [PubMed] [Google Scholar]


