Abstract
HIV is a major global epidemic that requires sophisticated clinical management. While there have been remarkable efforts to develop new strategies for detecting and treating HIV, it has been challenging to translate them into resource-limited settings. Significant research efforts have been recently devoted to developing point-of-care (POC) diagnostics that can monitor HIV viral load with high sensitivity by leveraging micro- and nano-scale technologies. These POC devices can be applied to monitoring antiretroviral therapy, early infant detection of HIV during mother-to-child transmission, and identification of latent HIV reservoirs. In this review, we discuss current challenges in HIV diagnosis and therapy in resource-limited settings and present emerging technologies that aim to solve these challenges using novel micro- and nanoscale solutions.
Keywords: HIV/AIDS, Point-of-Care, Emerging Technologies, Nanotechnology, Microfluidics
Graphical abstract
1. Introduction
HIV-1 remains a major epidemic despite significant efforts made with early diagnosis, treatment, and prevention of HIV-1. In 2014, there were 36.9 million people living with HIV, with 2 million new cases, and 1.2 million deaths worldwide [1]. The overall incidence of HIV-1 has decreased due to various factors including suppressive antiretroviral therapy and needle exchange programs in developed countries [2–4]. Prevalence of HIV-1 increases more slowly each year due to the expanding implementation of (ART), particularly in resource-limited settings [5,6]. As such, there is an increasing need to (i) reduce incidence of HIV-1 infection by strengthening current prevention programs, (ii) detect and diagnose HIV-1 infections as early as possible, and (iii) effectively monitor treatment efficacy, including management of secondary (non-AIDS) HIV-related illnesses such as cancer, cardiovascular, liver diseases, and co-infections [7–12]. However, solving these problems is expensive and challenging in resource-limited settings given the large number of HIV-1 infected individuals living in these settings.
In developed countries, nucleic acid testing (NAT)-based viral load testing and flow cytometry-based CD4 cell counting are routinely used to monitor ART therapy. However, these technologies are complex and costly making them unsuitable for resource-limited settings, where there is a shortage of laboratory infrastructure and financial support. Although first-line ART drugs may be available for free or inexpensively in resource-limited settings, the expansion and universal access of ART has been significantly thwarted by the lack of appropriate diagnostic tools for detecting HIV-1 infections and initiating and monitoring ART. Creating sensitive diagnostics would allow for early identification of acute HIV-1 infection (AHI), thus helping reduce the transmission rate among high-risk populations. Such tools could also be used to identify HIV-infection in pregnant women to reduce the risk of mother-to-child transmission (MTCT) during childbirth, currently a major problem in resource-limited settings [13].
To address this technological gap, researchers have been developing devices for HIV-infected individuals in resource-limited settings that are Affordable, Sensitive, Specific, User friendly, Robust and Rapid, Equipment-free, and Deliverable (ASSURED) [14]. Microfluidic and nanotechnologies have the potential to fulfill ASSURED criteria, because they require small sample volumes, have short assay turnaround times, and enable highly sensitive detection. We begin our discussion with recently developed diagnostic devices employing different micro- and nano-technologies for clinical management of HIV-1 in resource-limited settings. These devices utilize different sensing modalities including electrochemical, optical, and mechanical sensing. We then detail the latest advances in diagnostic tools currently being used for the detection of acute HIV-1 infection, CD4 cell count, viral load measurement, and latent HIV-1 reservoirs. Lastly, we provide future directions on the development of POC devices for improving HIV-1 management in resource-limited settings.
2. Recent advances in biosensors and chemical detection systems for HIV detection
Nanotechnology has had a large impact on the field of biosensing by allowing biomedical scientists and engineers to create tools that can directly monitor biological interactions for diagnosis of diseases, including HIV. In this section, we introduce emerging technologies, categorized by their transducing method, which are being used to both qualitatively and quantitatively detect HIV via direct (i.e., capture of intact virus), indirect (i.e., capture of host antibodies to the virus), or amplification methods (i.e., polymerase chain reaction).
2.1. Electrochemical-based assays
Electrical sensing technologies are used to report binding and recognition events occurring on a sensing surface, including protein-antibody, nucleic acid hybridization, and enzyme-cofactor coupling [15–17]. The broad class of sensors has several advantages, including short assay times and ease-of-use. However, there are technical challenges when used as point-of-care diagnostics in resource-limited settings, such as increasing the signal-to-noise ratio and eliminating electrical interference from highly ionic biological backgrounds. In the following section, we will provide a brief overview on different electrochemical sensing modalities and their applications in HIV detection.
2.1.1. Electrical sensing-based platforms
Electrical-based sensors monitor electrochemical reactions, such as enzymatic conversion or capture of biological targets, through an electrode interface by monitoring changes in electrical current, resistance, impedance, or voltage signals [18]. There are different strategies to measure electrical properties on a sensor surface, including amperometric, voltammetric, potentiometric, and impedance measurements.
Amperometric/voltammetric biosensors are mainly used to measure an electrochemical reaction, which is triggered by the generation or perturbation of a redox current. Thus, these biosensors simply record current generated by direct oxidation or reduction of target molecules that are immobilized on an electrode surface [18,19]. For affinity-based sensing strategies, antigen molecules are captured by specific antibodies/recognition elements (e.g., antibodies, aptamers) on an electrode surface and the resulting electrochemical reaction blocks electron transfer, thus reducing output current. The degree of output reduction provides a quantitative measurements of the captured target molecules [20]. For instance, an amperometric sandwich immunoassay was developed for HIV protein detection, where the electrode was modified with anti-p24 antibodies to capture HIV p24 protein, followed by labeling with a horseradish peroxidase secondary antibody [20]. Signal was produced by immersing the electrode in a solution containing hydrogen peroxide and hydroquinone, which are directly catalyzed by the HRP-secondary antibody complex. Results showed a linear dynamic range spanning from 0.01 ng/mL to 100 ng/mL of HIV p24 protein and a detection limit of 0.008 ng/mL was observed, which is two orders of magnitude better compared to conventional ELISA methods (~1 ng/mL). Amperometric sensors were also utilized to measure zidovudine (ZDV), a nucleoside analog reverse-transcriptase inhibitor that can be one of many drugs used for ART [21]. The concentrations of ZDV were measured using an amperometric sensor fabricated using silver nanofilm (Ag-NF) and multiwalled carbon nanotubes (MWCNTs) immobilized on glassy carbon electrode (GCE). This amperometric strategy under optimal conditions reported a linear detection range for ZDV concentrations spanning from 0.1 to 400 ppm (0.37 μM–1.5 mM) with a detection limit of 0.04 ppm (0.15 μM). Further examinations indicated a 98.6% of ZDV recovery from human serum samples. Results from this approach indicated that amperometric methods could be used to monitor HIV-drug levels.
Potentiometric biosensors mainly measure potential or charge accumulation on an electrode surface. These biosensors consist of (i) ion-selective electrodes, where the electric potential responds selectively to the concentration of a target ion, and (ii) a reference electrode, which records charge accumulation [18,22]. Using this signal transduction strategy, HIV-1 integrase activity was measured [23]. A series of metal complexes of two ligands (HL1 and HL2) were designed as clinical models to investigate the metal chelating mechanism of integrase inhibitors. To explain a metal speciation model, potentiometric analyses were performed using HL2 with Mg2+, Mn2+, Co2+, and Zn2+. Results indicated that metal ions played a pivotal role in HIV-1 integrase inhibition, suggesting that this strategy could be used for inhibitor design and development. Sensitive measures of active integration have the potential to explore potential sources of HIV-1 persistence by detecting the presence of low-level residual viral replication or de novo infection in various tissues in the setting of otherwise suppressive ART.
Impedance biosensors monitor electrical impedance of an interface in AC steady state with constant DC bias conditions [24]. This is performed by applying a minute sinusoidal voltage at a specific frequency, while monitoring the produced current response. The current-voltage ratio in this electrochemical impedance spectroscopy (EIS) provides the impedance signal and allows the precise monitoring of changes in conductivity/resistivity or charging capacity of an electrochemical reaction or interface where bio-chemical interactions occur. A micro-device was fabricated to record impedance changes for analysis of HIV viral nano-lysates [25]. In this study, multiple HIV-1 subtypes were captured using magnetic beads-coated with anti-gp120 antibodies, followed by viral lysis. The nano-lysate samples were then applied to microdevices consisting of a pyrex wafer with two gold microelectrodes. EIS was then recorded from 100 Hz to 1 MHz to evaluate impedance changes between multiple HIV-1 subtypes and control (sample without HIV-1). This device demonstrated that HIV-1 samples produced distinct impedance values compared to controls. HIV-1 samples mixed with Epstein-Barr Virus (EBV) were also tested and the results were not significantly different from that of HIV-1, demonstrating the specificity of HIV detection. To demonstrate portability of this device, the same strategy was applied to polyester-based flexible materials (Fig. 1A) [26]. Two silver microelectrodes were printed on a flexible polyester film and integrated into a microfluidic channel. Nano-lysate of HIV-1 samples from plasma and whole blood samples were then applied into the microchannels and the impedance was recorded from 100 Hz to 1 MHz. This flexible impedance biosensor showed its potential as an inexpensive (less than $2 in material cost) and disposable assay to selectively capture and detect HIV-1 from a clinical specimen. In another study, mass-producible and flexible impedance sensors were fabricated using conductive inks [27]. Multiple HIV-1 subtypes, EBV, and Kaposi’s Sarcoma-associated Herpes Virus (KSHV) were detected from a fingerprick volume (50 μL) of physiological buffer, plasma, and artificial saliva samples. Further developments in reducing the number of sample processing steps will significantly improve these sensors for POC applications.
Fig. 1. Electrochemical Assays for HIV-1 Detection.
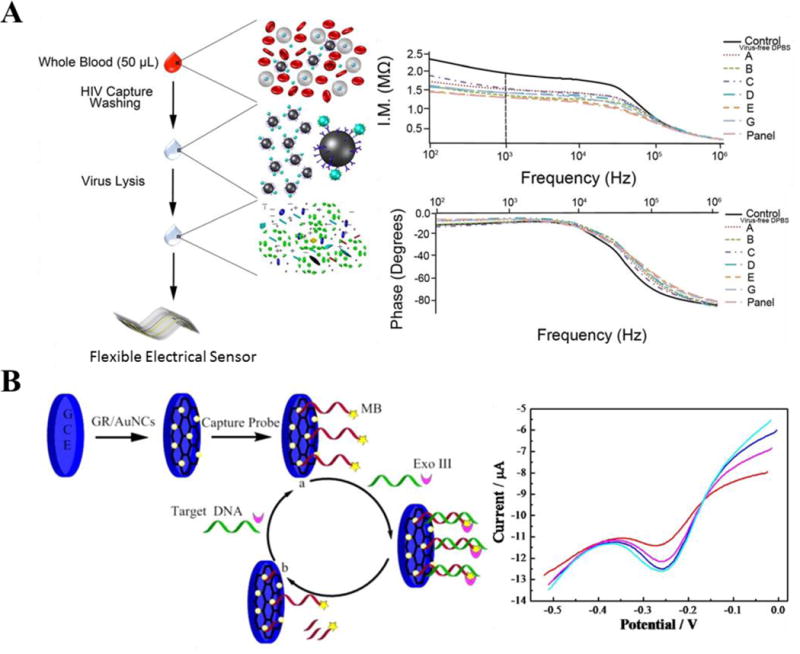
(A) Two-rail microelectrodes were printed on a flexible polyester-based material for detection of HIV nano-lysate. HIV was captured with magnetic particles coated with anti-gp120 antibodies (off-chip), followed signal measurements by electrochemical impedance spectroscopy [26]. (B) A gold nanocluster modified graphene electrode (GR/AuNCs) was fabricated to detect HIV target sequences using an exonuclease III (Exo III)-assisted target recycling amplification method [40]. In this method, the electrode surfaces were modified with capture probes (aptamers) and labeled with methylene blue (MB). During hybridization between target and capture probes, nucleic acids folded themselves into a duplex DNA structure, followed by the digestion of the capture probe by Exo III from its 3′-end, resulting in the release of MB molecules, which were recorded by differential pulse voltammetry technique (A) was adapted with permission from [26]. (B) was adapted with permission from [40]. Copyright (2015) American Chemical Society.
2.1.2. Graphene-based sensors and assays
Graphene material consists of a two-dimensional (2D), single-layer sheet of sp2-hybridized carbon atoms, which form a hexagonal lattice structure [28–30]. From a structural and functional perspective, graphene is a semiconductor material and behaves as a semimetal due to its zero-bandgap [30–32]. This material has a remarkable ambipolar electric-field effect and exhibits a large theoretical surface area (2630 m2/g) with superior electrical conductance (64 mS/cm) [33,34]. In addition, graphene has a low charge-transfer resistance and a rapid electron transfer rate [30,35,36]. Due to these promising physical and electrochemical features, graphene has been broadly used as an electrode material for electrochemical sensing modalities, and been applied into a few clinical biosensing and diagnostic applications [17,37–39]. As a potential diagnostic tool, graphene-based materials have been modified with nucleic acids, aptamers, peptides, and antibodies to measure current and amperometric changes within biochemical reaction such as redox interactions. For instance, a gold nanocluster modified graphene electrode (GR/AuNCs) was developed to measure HIV-originated target sequences using an exonuclease III (Exo III)-assisted target recycling amplification strategy (Fig. 1B) [40]. GR/AuNCs were decorated with capture probes (aptamers), which were then labeled with methylene blue (MB) on 3′-end and Cytosine (C)-rich base on 5′-end. During hybridization between target and capture probes, nucleic acids folded themselves into a duplex DNA structure, followed by the digestion of the capture probe by Exo III from its 3′-end, resulting in the release of MB molecules. In this platform, the signal changes were monitored using differential pulse voltammetry technique and detected down to 30 aM of HIV target probe with a dynamic range spanning from 0.1 fM to 100 nM. Using this platform, serum samples were examined and results showed 99.8 % of a recovery rate when HIV target probe was tested at 10 fM.
Flexible and wearable materials could be used to perform tasks such as continuous HIV viral load monitoring, which may be useful for determining ART efficacy. Graphene-based sensors have also been decorated on flexible materials for HIV diagnostics [41]. In this work, a paper microchip modified with graphene-silver electrodes was designed to capture and detect HIV lysate on-chip. Geometry of electrodes and graphene notably affected the performance and sensitivity of microchips. First, HIV particles were captured on-chip using streptavidin-anti-gp 120 antibody complex. Then, captured viruses were lysed on-chip and the lysate was measured using impedance/capacitance spectroscopy between 1 Hz and 10 KHz. HIV-free controls had different capacitance values compared to HIV lysate samples.
2.2. Optical-based assays
Target binding-induced changes in optical signals, such as fluorescence, light absorbance and transmission, or refractive index change, have been widely used to identify or quantify biological interactions. Optical assays are highly sensitive and can potentially be used to detect captured intact HIV particles by eliminating background interference signals. In this section, we will discuss different optical methods developed for detection of HIV.
2.2.1. Fluorescence-based assays
Fluorescence is the most common optical method used for disease diagnostics. Antibodies conjugated with fluorescent markers provide specificity in tagging proteins or cellular components. The current gold standard for detecting and counting cells is a flow cytometer, which uses a laser beam focused on cells in stream to differentially separate and count cells based on their fluorescent marker. Early flow cytometers were only capable of functioning in centralized labs due to portability and electrical requirements. Traditional flow cytometry is used to characterize cells but can be implemented for viral nucleic acids. In one example, anti-Digoxigenin (anti-DIG) and anti-Dinitrophenyl (anti-DNP) coated microparticles were used to capture to PCR end-products. By performing flow cytometric analyses, they were able to detect different HIV-1 subtypes, including the subtypes O and N, with a detection range of 50 to 1 million copies [42].
Recently, new technologies have been developed to fluorescently label and count cells directly, thus eliminating the need for specialized flow cytometer equipment. For example, CD4 cells were labeled with quantum dots, captured on bioactivated PDMS nano-bio chips, and imaged using a portable single wavelength epi-fluorescent microscope (Fig. 2 A, B). The nano-bio chips were self-contained and had a waste reservoir and reagent storage on-chip (Fig. 2 C). CD4+ T cells were differentiated from other lymphocytes and monocytes (Fig. 2 E, F) [43]. CD4+ T cells can also be captured using controlled shear stress in a flow chamber and counted using fluorescently labeled antibodies [44]. This method was further improved by depleting monocytes to capture CD4+ T cell at lower concentrations [45]. Another novel approach to assess HIV infection and status was to count multinucleated giant cells based on induction on synctia – multi-nucleate enlarged cells formed by infected cells [46]. After infection of cell lines MT2 and SUPT1 with syncytium-inducing (SI) HIV-1 isolates, DNA staining was performed with propidium iodide and cell sizes were evaluated using phase contrast (Fig. 2 F) and fluorescent microscopy (Fig. 2 G–I). Results indicated that SI cells (Fig. 2 I) could be distinguished from non-SI (Fig. 2 G, H), suggesting that this method could be useful for assessment of HIV inhibition by different antiviral therapies. However, HIV-1 that uses CCR5 (Chemokine (C-C Motif) Receptor 5 (Gene/Pseudogene)) for entry tend not to produce syncycium in the above laboratory cell lines, which are the most common coreceptor-using strains isolated from patients during acute and early infection [47–49]. This assay is thus limited to HIV-1 that uses CXCR4 (C-X-C chemokine receptor type 4). Most recently, technology using HIV-specific transcription-mediated amplification (TMA) to cross link probes with fluorophores onto cells to flow, characterize, and sort RNA or DNA producing cells – as infected cells are as low as one in a million. While this technique is currently expensive and not yet high throughput, it represents the current state-of-the-art for flow based nucleic acid technologies [50].
Fig. 2. Integration of fluorescent microscopy into biochips for CD4 T cell enumeration.
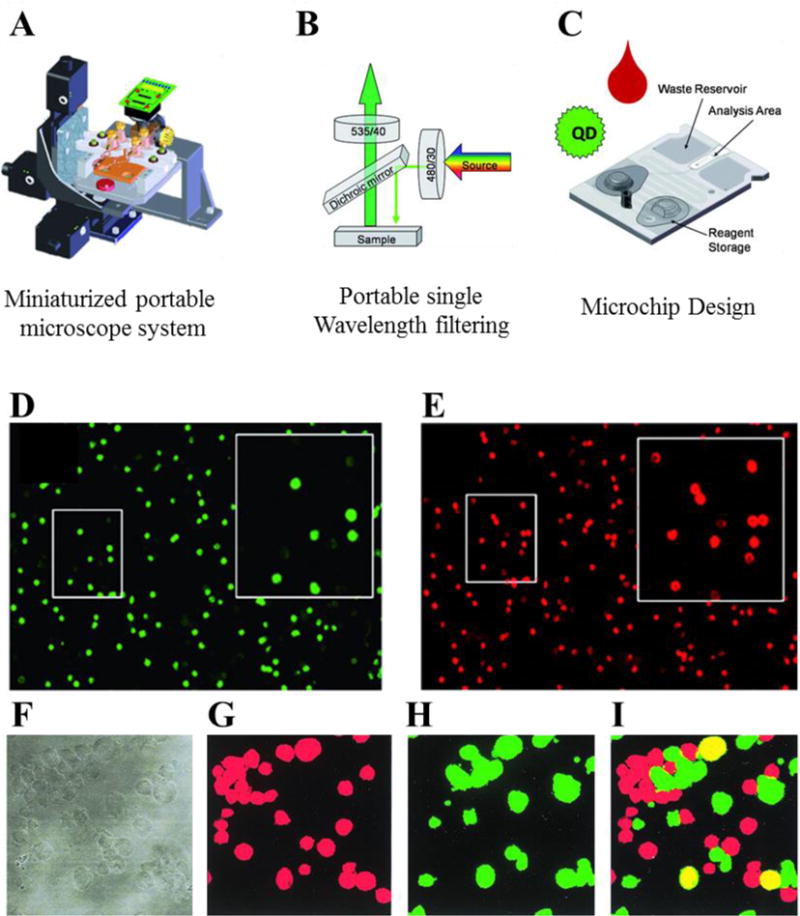
(A), (B) A benchtop single wavelength epi-fluorescence microscope was used with nano-bio chips to count CD4 T cells [43]. (C) The design of the nano-bio chips included on-chip waste reservoir and reagent storage. Fluorescent images were taken of CD specific antibody labeled whole blood using green and red quantum dots as secondary labels to identify monocytes and T lymphocytes (D) and CD4+ cells (E). A fluorescence-based method was developed to detect HIV that induce syncytia [46]. HIV-infected MT2 cells were labeled with red or green fluorescent dye. Panel (F) shows stained cells from phase contrast microscopy. HIV-1-induced fusion between cells could be detected as yellow-stained syncytia (I) from a background of cells labeled with red (G) and green (H) fluorescence. (A–E) Reproduced in part from [43] with permission of The Royal Society of Chemistry. (F–I) reproduced in part from [46] with permission from the American Society for Microbiology.
2.2.2. Lens-free imaging platform for HIV detection at the POC
Light microscopes are widely employed in the imaging of biomolecules and cells in clinical and research environments. However, they are bulky and costly, and thus have limited availability in resource-limited settings. Recently, a portable lens-free shadow imaging system was introduced, which incorporated a light source and an image sensor (such as CCD or CMOS) for the imaging of cells [44,51–56]. Briefly, LED light shining on a sample diffracts creating a pattern on the surface of a sensor located below the sample. A software program deconvolves these diffraction patterns and reconstructs a “shadow” image of the actual sample. Since there is no lens, a wide field-of-view can be imaged, which is two orders magnitude greater than that of a regular light microscope The entire platform is portable, rapid, and more practical than lens-based technologies and has also been integrated with a smartphone [57,58]. This device has been used to accurately enumerate CD4+ cells in patients recently infected with HIV in resource-limited settings [51]. CD4+ T cells were captured in a microfluidic channel using antibodies decorated on glass surfaces (Fig. 3A). Captured cells were then imaged with a lens-free shadow imaging system, and total cell number was calculated using computer-aided software in under one minute (Fig. 3B). The platform was demonstrated to work with HIV-positive blood samples collected from patients in Tanzania, and provided comparable results with conventional cell counting devices and microscopes (Fig. 3C).
Fig. 3. Lens-free shadow imaging system for HIV diagnosis.
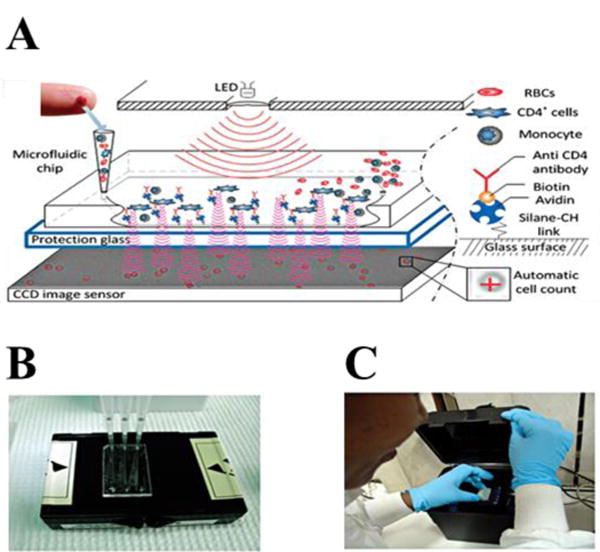
(A) General mechanism of lens-free shadow imaging system. Light from an LED passes through a pinhole and shines on a microfluidic channel in which CD4+ T-cells were captured. The light forms a diffraction pattern on a CCD surface. These holographic patterns reconstructed by software and cells were identified and counted. (B) Inset indicates the actual image of CCD sensor. (C) Filling the microfluidic channels with patient sample blood. (D) Portable system in actual trials in Tanzania for HIV patients. Images were reproduced with permission from [51].
2.2.3. Nanoplasmonic assays for HIV at the POC
Nanoplasmonic detection systems have been recently developed to quantify HIV and subtypes from unprocessed whole blood from patients. Nanoplasmonic systems measure the collective oscillation of electrons on metal nanoparticles by moniatoring optical resonance. In plasmonics, light is trapped in between metal structures inside nanometric volumes, which leads to enhancement in near-field signals. Evanescent waves bounded by metal dielectric propagate surface plasmons, which are caused by conduction electrons oscillating in metals. There are two commonly used optical biosensors that exploit plasmonic behavior: surface plasmon resonance (SPR) based on a planar, thin film gold surface and localized surface plasmon resonance (LSPR) based on confined gold nanoparticles. Both LSPR and SPR are sensitive to the local refractive index changes when target binds to the film or nanoparticles, and are capable of detecting single molecules [59]. Binding events of molecules on metallic nanoparticles cause changes in the absorbance spectrum, which can be monitored before and after sample incubation to determine the presence of target. LSPR provides broad spectral measurements with a higher sensitivity and signal-to-noise ratio than conventional SPR [60]. In a recent approach, plasmonic nanoprobes were used to detect the gag sequence HIV-1 based on hybridization between complementary target and probe DNA sequences [61]. Another recently developed platform detected clinically relevant concentrations of HIV-1 with a sensitivity as low as ~100 copies/ml across multiple HIV subtypes (A, B, C, D, E, G and subtype panel) [62]. In this method, surfaces were modified in several layers for immobilization of antibodies to capture HIV, consisting of a poly-l-lysine (PLL) layer, monolayer of gold, and antibody specific to the target. This platform has been validated with different infectious diseases, and has recently been developed as a portable device [63], showing its potential as a POC in resource-limited settings.
2.2.4. Photonic-crystal based assays for HIV at POC
Photonic crystals consist of periodic arrangement of dielectric materials that selectively reflect a tunable wavelength (resonance wavelength) when light is incident on a surface [64]. A biomolecular interaction on the PC surface produces a shift in resonance wavelength that correlates with the concentration of a target analyte [65]. PC-based biosensors have been employed for the detection of a wide array of biomolecules including proteins, cells, nucleic materials, and pathogens [66–70], including rotavirus (Fig. 4E, F), human influenza virus (H1N1) (Fig. 4 C, D), and L. pneumophila bacteria [67,71–74]. Recently, a TiO2 coated 1D PC biosensor was utilized for capturing and quantifying HIV-1 viral load from serum, suggesting that such a device could be used to detect the virus at low concentrations [75]. In this study, a PC surface was functionalized with anti-gp120 antibodies, and used to specifically capture HIV-1 with a limit of detection in the range of 104 copies/mL to 108 copies/mL in serum (Fig. 4 A, B). PCs have also been shown to detect a single nanoparticle [76]. Although PC biosensors hold great promise as reliable and sensitive diagnostic tools, they require expensive optical read-out elements and have yet to be made portable. Smartphone incorporated PC systems have been recently developed in an effort to create a POC device aimed for application in resource-limited settings [77,78]. Recently, there has also been efforts to reduce the size of a PC system, such as using PCs integrated within a microfluidic channel to detect biotargets [79]. These recent works suggest that both smartphone integration and microfluidics with PCs may make possible the creation of a portable optical biosensor at the POC for detection of HIV and other infectious diseases in developing world.
Fig. 4. Photonic crystal (PC) biosensors for detection of HIV and other viruses.
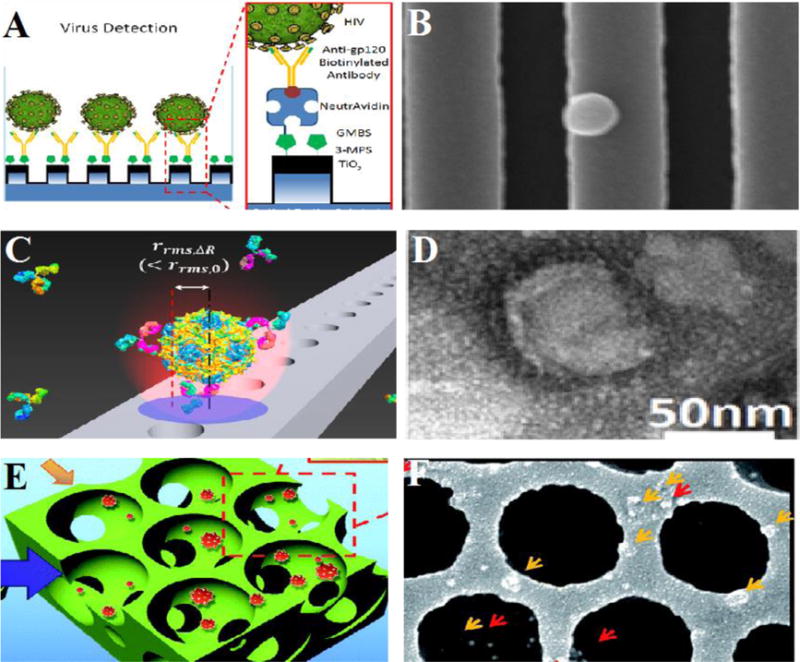
(A) General overview of 1D PC structure and the surface functionalization for HIV capturing using anti-gp120 antibody. (B) SEM image of the HIV on the PC surface. (C) 2D holes fabricated on a beam for capturing influenza virus. (D) TEM image of the influenza virus. (E) Schematic of inverse opals for capturing virus. (F) SEM image of captured rotaviruses. (A),(B) were adapted from [75] with permission from Macmillan Publishers Ltd. Copyright (2014). (C), (D) are reprinted from [80] with permission from Nature Publishing. Copyright (2015). (E),(F) are reproduced from [74] with permission of The Royal Society of Chemistry.
2.3. Mechanical assays
Measuring mechanical forces generated on the cellular scale, such as displacement and mass changes, provides fundamental information about biological systems at the micro- and nanoscale including, adhesion, transport, and binding affinity parameters of biochemical reactions and cellular processes [81,82]. Mechanical sensors have been broadly used in binding and biosensing studies, including measuring antigen-antibody interactions, monitoring drug binding, and measuring virus-cell surface and virus-antibody interactions. In the following section we present various mechanical sensing strategies using cantilever-based and quartz crystal microbalance (QCM) based sensors with applications for detecting HIV and related diseases.
2.3.1. Cantilever-based tests
A micro-cantilever beam is an element that mechanically senses the presence of target molecules by measuring its deflection or dynamic frequency resonance on a micrometer sized suspended beam. When a biomolecule binds to a cantilever via electrostatic repulsion/attraction, hydration effects, entropic effects, or steric interactions, a deflection of the cantilever is measured by using either a reflecting laser beam or electrical readout [83–87]. A dynamic cantilever mode can also be used to determine the frequency response of the cantilever [88,89]. When a biomolecule binds to the surface, the resonant frequency is altered. One of the key benefits to using a driven cantilever is they can be used in various environments (e.g., humid, fluid, and vacuum environments) and in different modes (e.g., suspended microchannel resonator) [81,90].
In HIV detection strategies, cantilever-based arrays and sensors are mainly decorated with specific antibodies that capture HIV antigens. For instance, a cantilever-based modality was designed to detect HIV-1 envelope glycoprotein (Env) gp120 from physiological buffers [91]. In this work, a cantilever surface was decorated with monoclonal anti-A32 antibodies (primary antibody) via self-assembled monolayers to capture gp120 proteins, followed by capture of a secondary antibody (monoclonal anti-17b antibodies) (Fig. 5 A). In each stage, cumulative deflections were observed. In this study, the limit of detection was observed to be greater than 400 ng/mL. Although this detection limit is the range for in vitro cultures, it is still unsatisfactory for monitoring gp120 in vivo validation (0.2–100 ng/mL) [92,93]. Such a strategy would work best for capturing free circulating virus, as cells express very low amounts of gp120 on their surface. In another study, membrane receptors were immobilized onto nanomechanical cantilevers without requiring passivation of the underlying surface [94]. By using equilibrium theory, quantitative mechanical responses were recorded for vancomycin, HIV-1 antigens, and coagulation factor VIII captured on the sensor surface. A cantilever was then used for HIV experiments, where an N-terminal fragment (VHH) of llama single chain antibodies (~15 kDa) against HIV-1 trimeric envelope glycoprotein (gp140) was immobilized on the cantilever surface, which had been previous shown to have high sensitivity and specificity against gp140 protein [95]. A response was observed after 80–90% of the surface was coated with gp140. Results demonstrated a dependence between molecular footprint of the target and the induced stress. Further experiments were performed to detect gp140 antigens from HIV-1 B/C subtypes, with a limit of detection down to 500 fM, which is orders of magnitude lower than the previous study [96]. Cantilever sensors can also be used to differentiate between fusion and binding events of viral particles and living cells. For example, a single-molecule force spectroscope (SMFS) was employed to monitor the strength and lifetime of molecular bonds with a single-molecule resolution [97]. In this study, viruses were first immobilized on a cantilever surface of an atomic force microscope (AFM) and then operated in contact mode with a living cell (Fig. 5B) Force-time curves were collected to distinguish between adhesion and fusion when viruses interacted with cell surfaces. Results indicated that cantilever-based sensors could be potentially used for monitoring molecular interactions with small deflections (pN/s levels).
Fig. 5. Emerging technologies incorporating mechanical assays.
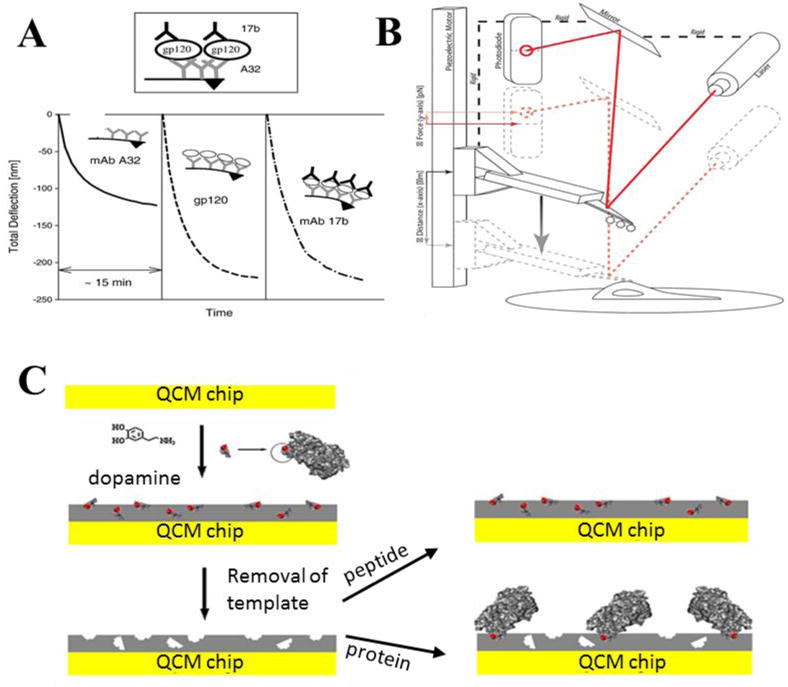
(A) A microcantilever was demonstrated to detect HIV-1 gp120 protein using a sandwich assay with anti-A32 and anti-17b antibodies. In this study, deflection versus-time curves were collected for each step [91]. (B) A single-molecule force spectroscope (SMFS) capable of single-molecule resolution was developed to determine viral adhesion and fusion events to a cell surface. The cantilever surface was functionalized with whole viruses to contact with cell surfaces expressing viral surface receptors. Displacement values of the cantilever were recorded to monitor molecular interactions [97]. (C) A quartz crystal microbalance surface was decorated with molecularly imprinting polymer to mimic biomolecular confirmation of gp41 epitope of HIV-1. In this study, dopamine was used as functional monomer, having analog residues with HIV-1 transmembrane protein (gp41). Then, gp41 proteins were applied to these sensor surfaces, and captured on the grafted surfaces [104]. (A) was reprinted from [91], Copyright (2006), with permission from Elsevier. (B) was adapted from [97] with permission from Macmillan Publishers Ltd. Copyright (2015). (C) was reprinted from [104], Copyright (2012), with permission from Elsevier.
Overall, cantilever sensor-based approaches can provide new insights of cellular mechanisms by monitoring mechanical responses to biochemical interactions and also be potentially used to detect ultralow concentrations of biomarkers, making these sensors and assays an ideal candidate for use in the POC diagnostic settings [97,98].
2.3.2. QCM-based tests
Quartz crystal microbalance (QCM)-based sensors are centimeter-sized mechanical sensors that contain a piezoelectric material coated with a metal film, such as gold or silver. QCM sensors are used to measure masses of small analytes in fluid, vacuum, or gas environments by real-time monitoring the changes in oscillation frequency [99–102]. These sensors are commercially available and can be purchased with temperature and oscillation modules that enable detection of sub-nanogram mass loads [103]. QCM surfaces are often modified through self-assembled monolayer, polymeric materials, or molecular imprinting approaches. A molecularly imprinting polymer was applied QCM sensing platforms to mimic biomolecular confirmation of gp41 epitope of HIV-1 [104] (Fig. 5 C). In this study, dopamine was utilized as functional monomer with 35 amino acid residues, which were analogues to residues 579–613 of HIV-1 transmembrane protein (gp41) due to its high abundance in AIDS patients (98%). Thus, a grafted MIP surface was designed for capture gp41 proteins. Kinematic parameters for this system indicated a binding strength of 3.17 nM, which is similar to using monoclonal antibodies. A detection limit was observed to be 2 ng/mL, which was comparable to ELISA. The platform was further validated with gp41 in human urine samples, and between 86.5–94.1% of recovery rate was observed.
2.4. Miniaturizing conventional assays
The need for diagnostic devices to detect and monitor HIV viral load, CD4+ T cell number, and therapeutic monitoring before and during antiretroviral therapy (ART) in resource-limited settings has not been yet been satisfied. Although many testing platforms exist in the developed world, which have a low unit price per test for a system, they still require centralized laboratories and specialized devices, which makes them inadequate as ASSURED POC tools in resource-limited settings. The focus of this section is to discuss the recent research efforts toward miniaturizing conventional assays by incorporating microfluidics and nanotechnologies to reduce the size and complexity of many commercially successful assays and adapting them in resource-limited settings applications.
2.4.1. ELISA incorporating microfluidics
Enzyme-linked immunosorbent assays (ELISA) are colorimetric and fluorometric based tests that are composed of an antibody/antigen interaction detected via a secondary reporter conjugated to an antibody (i.e., horseradish peroxidase, fluorescent molecules, quantum dots). ELISA tests have had remarkable success with detecting p24 antigens as shown by a recent publication, which used an inexpensive approach to achieve an attomolar limit of detection [105]. ELISA based tests have also been used to detect T cell CD4 proteins collected from a fingerpick blood sample, and have a 97% sensitivity when the CD4+ T lymphocyte count is as low as 350 cells/μL, suggesting that it could be used to quantify response to ART in resource-limited settings. Visitect® CD4 is a commercially available semi-quantitative POC test for the enumeration of CD4+ T lymphocytes in whole blood within 40 minutes using an automated reader that eliminates visual, potentially biased interpretations [106]. Another recent technology is an automated cell phone based imaging system called a micro-a-fluidic ELISA (m-ELISA) [107]. This was developed to detect and quantify CD4+ T lymphocytes from unprocessed whole blood using magnetic beads to capture analytes from multi-channel reservoirs. The method has an accuracy of 97% for capturing and quantifying CD4+ T lymphocyte from 35 blood samples with cut off 350 cells/μL. A platform, named MyT4, was also developed for detection of CD4+ cells, where micro capillary columns were used to collect microparticles conjugated with CD4+ anti-bodies and CD4+ T lymphocytes cells. After centrifuging the particles and cells the height of the pellet was measured for the quantification of CD4+ T cells. This technique had a sensitivity of 95% and specificity of 87% at the threshold of 350 cells/μL [108]. There is also a microfluidics-based mChip technology (mobile microfluidic chip for immunoassay on protein markers) that has been used to detect HIV-1 and only requires 1 microliter of unprocessed whole blood [109]. Another ELISA based assay has been recently developed which combines microfluidics with a smartphone to multiplex immunoassays in a single test obtained within 15 min with a sensitivity of 92% and specificity of 79% [110].
2.4.2. Microfluidic-PCR assays
Reverse-transcription polymerase chain reaction (RT-PCR) is a commonly employed viral load assay. Several research groups have attempted to lower the cost of RT-PCR, so that it can be made portable for resource-limited settings [111,112]. One of these technologies is a Liat™ analyzer, which extracts and amplifies HIV RNA from whole blood with a detection limit of 57 copies/mL. The system is user-friendly, automated, and requires minimum operator involvement, which makes this technology ideal for resource-limited settings [113]. Nucleic acid based tests are sensitive, but require multiple reactions during sample processing, thereby lowering its throughput. Portable cartridges have been developed with the use of a dipstick-based nucleic acid detector for HIV-1 RNA detection with a sensitivity of 75% and a detection limit of 50 copies of HIV-1 RNA [114]. Another recent portable PCR approach has been developed to integrate microfluidics with a battery-powered quantitative PCR platform. Results using this integrated device have shown a limit of detection of 5 copies/μL, which is superior to the current gold standard RT-qPCR method, with a detection limit of 20 copies/μL [115,116]. In an effort to lower the equipment cost and size for resource-limited settings, it may be beneficial to use super helicases, which are faster, more efficient and can replace the automated heating and cooling steps [117].
2.4.3. Digital droplet-based PCR
Instead of relying on large volumes for PCR, microfluidic channels can be used to create small droplets that encapsulate cells or genetic material to quickly analyze nucleic acids in less than a minute, referred to as digital droplet based PCR (ddPCR) [118]. Due to high sensitivity, ddPCR has been used to measure the number of CD4+ T lymphocytes or peripheral blood mononuclear cell (PBMC)-associated genomic HIV-1 DNA or PBMCs and episomal HIV-1 DNA in these cells. With this technology, cellular DNA extracts are portioned into picoliters in a single copy level in thousands of droplets, and these droplets are then individually amplified. If a droplet contains HIV-1 DNA, it will generate fluorescent signals and be deemed positive. The number of positive droplets is enumerated to determine the copies of HIV-1 DNA. HIV-1 DNA and 2-LTR was successfully detected from patients with suppressive ART from PBMCs using a ddPCR system from Bio-Rad [118]. Another study subsequently showed that ddPCR had improved precision for quantification of HIV-1 pol gene and 2-LTR by 5-fold and 20-fold, respectively [119]. Other potential advantages of ddPCR methods include the minimization of the detrimental effects of primer or probe-sequence mismatches and efficiency on quantitation, and the potential to reduce PCR inhibitors by partitioning assays into micro or nano-scale reaction droplets.
2.4.4. Paper microfluidics
Paper-based detection systems are one of the earliest point-of-care devices developed for resource-limited settings. Because paper is abundant, it can be mass-produced and incinerated when disposed reducing the cost per test. Applications for paper-based technologies were first developed as cheap pregnancy tests, but later developed into more complex resource-limited settings devices such as anti-HIV antibody [120], and HIV p24 antigen tests [121]. Paper has also been used for a broad range of biofluids in both home and clinical settings. Recent studies have focused on lateral flow assays that give quantitative, rather than qualitative ‘yes/no’ results [120]. Microfluidic paper-based analytical devices (μPads) use different patterning technologies depending on their application [122,123]. They are compatible with optical, electrochemical, chemiluminescence, and microelectromechanical methods, and use small sample volumes and are easier to read compared to lateral flow immunoassays [124]. A magnetic immunochromatographic test (MICT) method was implemented for detection of HIV-1 p24 within a 40 minute window. This system had a limit of detection of 30 pg/mL for spiked HIV-1 p24 antigen in buffer and spiked into 50% plasma [121]. Lateral flow assay materials and lyophilized enzymes were employed on paper that could amplify and detect 10 template copies of HIV DNA in 15 min [120]. The device performed polymerase recombinase amplification using a paper based plastic layer, acetate layer, two small pads for soaking “master mix” and magnesium acetate buffer, and a sample wicking pad. (Fig. 6 A). A rectangular window was prepared on the paper that is covering plastic base for enzyme pellet (Fig. 6B). A master mix pad placed on to enzyme pellet and a sample wicking strip were placed on the right side of the device (Fig. 6 C). The protective paper was peeled from the all surfaces (Fig. 6D). Acetate layer was placed on the right hand side of the device covering sample wicking strip and master mix pad. (Fig. 6E). Magnesium acetate pad was placed on the window prepared on acetate layer (Fig. 6F). Master mix and magnesium acetate were soaked to the membranes (Fig. 6G–H). Sample was introduced to the sample wicking strip (Fig 6I). Sample wicking strip was folded to match the strip and master mix pad (Fig. 6J). The left hand side of the device were fold from the midline to match magnesium acetate pad to the other components as enzyme pellet, master mix and sample (Fig. 6K). Results of the amplification reaction were generated on paper substrates, and had a limit of detection of 10 template copies of HIV DNA (Fig. 6 L). Paper-based assays are one of the best-studied detection platforms for resource-limited settings because of their low cost components and well-established production platforms.
Fig. 6. Paper-based viral load detection system to perform recombinase polymerase amplification of HIV DNA, system assembly and operation.
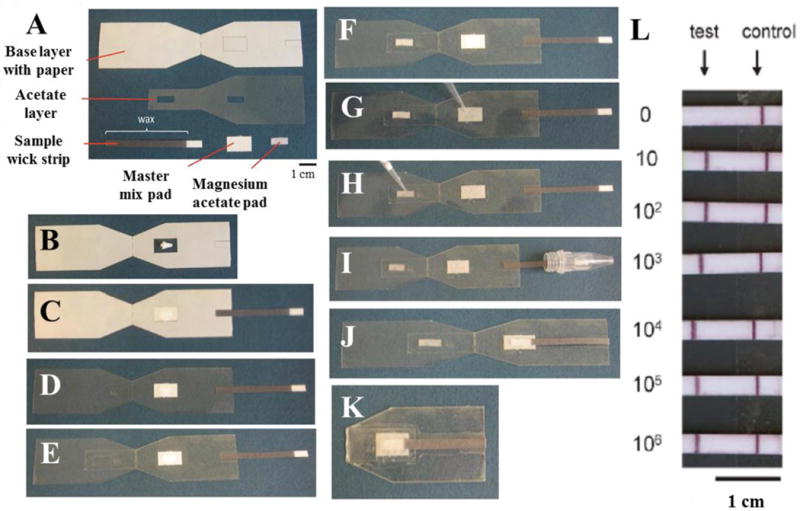
(A) Components of the device. (B) An enzyme pellet is placed on prepared adhesive window. (C) Pad for master mix and sample strip is placed. (D) Remaining paper is peeled to uncover all adhesive surfaces. (E) Acetate layer is fixed to the right side of the system. (F) Magnesium acetate pad is placed on the window of acetate layer. (G–H) Master mix and magnesium acetate are soaked to pads. (I) Sample is applied through sample wick strip. (J) Sample wick is folded on to the master mix pad. (K) Device is folded through the midline allowing interacting master mix, sample and magnesium acetate. (L) Number of HIV DNA copies introduced to system before recombinase polymerase amplification. This figure was reproduced from [125] with permission of The Royal Society of Chemistry.
2.5. Technology comparison
Below, Table 1 compares a select set of technologies and platforms discussed in the previous subsections. A comparison was made between detection modality (type), relevant HIV-1 detection target, target matrix, and our estimated cost of the method including disposables instrument where applicable. We have also listed some advantages and disadvantages to each technology to give the reader an idea of the current state of the art. Much of the information that is included was taken directly from the references. However, estimated cost was determined based on our own evaluation of the methods sections from within each reference to obtain an order of magnitude cost for the chips and instruments (where applicable).
Table 1.
Comparison of technologies and platforms discussed in Section 2. Some words were abbreviated to simplify the table – LSPR (Localized Surface Plasmon Resonance), ELISA (Enzyme Linked Immunosorbent Assay), PCR (Polymerase Chain Reaction), RT-PCR (real-time Polymerase Chain Reaction), MICT (Magnetic immunochromatography).
| Type | Target | Matrix | Est. Cost | Adv. | Disadv. | Ref. |
|---|---|---|---|---|---|---|
| Amperometric | p24 protein | Serum, physiological buffers | >$1000 per instrument | High sensitivity | Bulky, requires multiple wash steps. | 20 |
| Potentiometric | HIV-1 integrase activity | Reaction buffer | >$1000 per instrument | High sensitivity | Sensitive to changes in temperature and ionic buffer strength | 23 |
| Impedance | Intact HIV-1 | 1% Triton-X 100 in water. | <$10 per chip, >$1000 per instrument | Highly specific measurements and can be multiplexed. | Bulky, requires multiple wash steps. | 25 |
| Impedance | Intact HIV-1 | Physiological buffers, plasma, artificial saliva | <$2 per chip, >$1000 per instrument | Highly specific measurements and can be multiplexed. | Requires multiple wash steps. | 26,27 |
| Voltametric | HIV-1 complementary target DNA | Human serum | >$1000 per instrument | High sensitivity, good selectivity. | Bulky and expensive. | 40 |
| Impedance | Intact HIV-1 | Serum | <$2 per chip, >$1000 per instrument | Flexible substrate. | High LoD. | 41 |
| Fluorescence | CD4+ T cells | Blood | >$1000 per instrument | Highly specific cell capture. | Bulky and expensive. | 45 |
| Lens-free Imaging | CD4+ T cells | Blood | <$2 per chip, <$1000 per instrument | Portable and disposable | Chips require storage. | 51 |
| LSPR | Intact HIV-1 | Blood | >$1000 per instrument | Highly sensitive | Bulky, expensive. | 62,63 |
| Photonic Crystal | HIV-1 | Serum | >$1000 per instrument | Highly specific direct virus capture. | Bulky and expensive. | 75 |
| Mechanical | gp120 protein | Physiological buffers | >$100 per chip, >$1000 per instrument | Highly sensitive to changes on virus surface. | Bulky, high LoD. | 91 |
| Mechanical | gp120 protein | Physiological buffers | > $100 per chip, >$1000 per instrument | Highly sensitive to changes on virus surface. | Bulky, expensive, and requires trained staff. | 94 |
| Mechanical | gp41 protein | Urine | > $100 per chip, >$1000 per instrument | Antibody-free. | Bulky, expensive, and requires trained staff. | 104 |
| ELISA | CD4+ T cells | Blood | <$2 per chip, <$1000 per instrument | Highly sensitive and specific. | Bulky, expensive, and requires trained staff. | 108 |
| RT-PCR | HIV-RNA | Blood | <$1000 per plate, >$1000 per instrument | Highly sensitive. | Multiple reaction steps. | 113 |
| PCR | HIV-1 RNA | Blood | <$100 per plate | Highly specific. | Expensive and requires trained staff. | 115,116 |
| MICT (magnetic) | HIV-1 p24 | Plasma | <$10 per chip | Short assay time. | Low sensitivity | 121 |
| Lateral flow assay | HIV-1 DNA | Buffer | <$10 per strip | Short assay time, no refrigeration required. | Electric heater required. | 125 |
3. HIV management using emerging technologies
One of the gold-standard technologies for HIV detection is nucleic acid amplification. This method has been implemented in many different ways to help solve important and challenging problems with HIV management including detection of acute HIV-1 infection, mother-to-child transmission and HIV-1 reservoir quantification. Fig. 7 below illustrates the many ways nucleic acid amplification is currently being used.
Figure 7. Nucleic acid amplification strategies for HIV management.
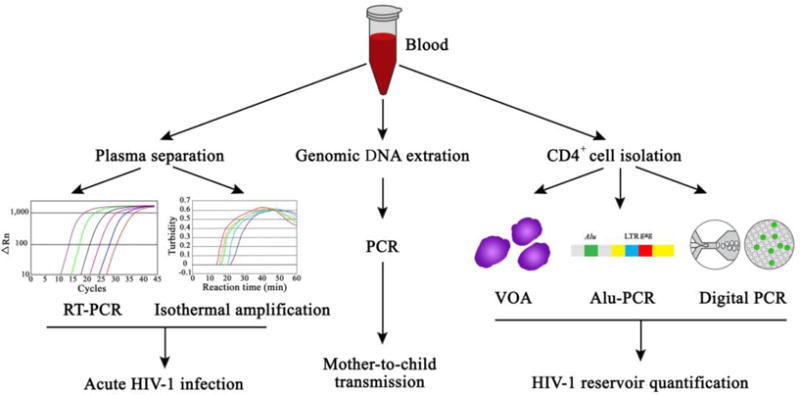
Nucleic acid amplification strategies are currently being used for challenging HIV management problems, such as acute HIV-1 infection (via reverse transcription PCR and isothermal amplification), mother-to-child transmission, and HIV-1 reservoir quantification (via viral outgrowth assay (VOA), Alu-PCR, and digital PCR).
Due to the large success with implementing these strategies in developed countries, significant efforts have been made to further develop these technologies to meet ASSURED criteria so that these advancements can be implemented in resource-limited settings. The following section describes current challenges with HIV clinical management in resource-limited-settings and discusses emerging technologies that may be useful in solving them.
3.1. Detection and diagnosis of acute HIV infection
Early or acute HIV infection (AHI) has become more important in HIV management for both prevention of HIV transmission and improved ART efficacy [126–128]. As acute retroviral syndromes present non-specifically, individuals with AHI may not be aware of their HIV status and may continue high-risk behaviors such as unprotected sex, intravenous drug use, or even blood donation. Moreover, AHI is associated with a high level of viremia, which renders them highly contagious and leads to further HIV-1 transmission. Thus, it is essential to detect AHI as early as possible and to provide appropriate clinical management. However, AHI cannot be detected using traditional immunoassays due to having a long window period, during which HIV-1 specific antibodies are not yet generated in AHI patients. The window period varies from a few weeks to 6 months among individuals following AHI, posing a great challenge for the prevention and control of HIV [127,128]. Acute HIV infection (AHI) is also of importance for clinical treatment [126,129]. During the early stage of HIV-1 infection, HIV-1 rapidly establishes its reservoir in the infected individuals, preventing complete eradication or functional cure with current ART regimens [130,131]. Early diagnosis of AHI followed by timely treatment can reduce the magnitude of HIV-1 reservoir and may improve the prognosis. Thus, early diagnosis of AHI followed by prompt ART initiation provides an invaluable opportunity for both HIV prevention and treatment.
Diagnosis of AHI is generally made using nucleic acid amplification technology to detect high level of HIV-1 RNA in plasma, in combination with traditional immunoassays (rapid lateral flow assays, ELISA and Western Blot) to confirm the absence of HIV-1 specific antibodies. [132]. Although the multi-testing algorithm can be established in developed countries, it presents a significant challenge for POC testing in resource-limited settings due to cost and differences in infrastructure. Additionally, in some resource-limited settings, clinical management is based completely on CD4 counts rather than viral load, which may be particularly troublesome since current medical practice is to treat immediately upon diagnosis [127]. Thus, rapid, inexpensive, and simple-to-use nucleic acid assays have been developed. For example, a miniaturized SAMBA device with pre-loaded freeze-drying reagents can amplify HIV-1 RNA with a detection of limit down to 78 copies/mL [133]. In comparison with other laboratory-based NAT assays such as Abbott Realtime, bioMerieux NucliSENS, and Roche COBAS, SAMBA obtained comparable sensitivity but also eliminated the need for refrigerated -transportation and storage, increased the feasibility for POC testing in resource-limited settings. In another study, the Ismagilov group developed a “SlipChip” that can perform digital reverse-transcription loop-temperature mediated amplification (RT-LAMP) for robust detection of HIV-1 RNA using a mobile phone [134]. As demonstrated, this SlipChip can differentiate 1×105 and 2×105 copies/mL with a temperature variation of 6 °C. Taken together, these two technologies represent advances in rapid detection of HIV-1 RNA in a miniaturized microfluidic device for identifying individuals with AHI in resource-limited settings.
3.2. Mother-to-child transmission
MTCT is another major area for HIV-1 prevention and control. As reported, the rate of mother-to-child transmission can be reduced to 0.7% with a combined measure including ART in HIV-1-infected mothers, C-cession, and formula feeding [13]. Thus, two aspects need to be focused on; one is the viral load (HIV-1 RNA in plasma) in HIV-1 -infected mothers, and the other is early detection of HIV-1 DNA in infants for timely care. Viral load in mothers can be measured using RNA amplification technologies with either a bulky instrument or a miniature device. For detection of HIV in infants, traditional immunoassays cannot be used since passive transfer of specific antibodies from HIV-1 infected mothers interferes with the diagnosis of infected infants up to 18 months. Instead, amplification of HIV-1 DNA from peripheral blood mononuclear cells (PBMCs) is used achieve early diagnosis of HIV-1 infected infants. PCR [115], loop-mediated isothermal amplification (LAMP) [135], and recombinase polymerase amplification (RPA) [125] have also been employed to detect HIV-1 DNA. Based on the LAMP technology, a microfluidic biomolecular amplification reader (μBAR) was developed, which integrated molecular biology, microfluidics, optics and electronics. However, this system could only detect the HIV-1 integrase gene at a level of 106 copies/mL [135]. In addition, this system did not solve one of the most common bottlenecks with molecular diagnostics: many sample preparation steps at the POC. RPA is another isothermal amplification technology developed by TwistDX. The RPA technology was miniaturized into a paper and plastic device [125]. As demonstrated, this light-weight device amplified HIV-DNA by RPA and detected the amplicons down to 10 copies by a lateral flow strip in 15 minutes. However, this system was designed to amplify HIV-1 DNA after nucleic acid extraction from dried-blood spots. Recently, a fully integrated HIV-1 DNA PCR system was developed to detect HIV-1-infected infants in resource-limited settings, though this method could only detect HIV-1 DNA when using more than 5000 cells per reaction. Thus, a fully integrated nucleic acid amplification system is preferred to facilitate the diagnosis of HIV-1 infected infants in resource-limited settings.
3.3. HIV eradication
Either achieving HIV eradication or a functional cure is the next big challenge [129,136]. Curing HIV can occur either by: (1) a sterilizing cure, in which HIV-infected cells that harbor replication competent virus are eliminated from infected individuals, or, (2) a functional cure such as ART-free remission, in which ART can be stopped with zero or low detectable viral loads without viremia rebound [137]. Strategies for achieving an HIV cure include reducing HIV reservoirs with simultaneously enhancing antiviral immunity or inducing cellular resistance to de novo HIV-1 infection [138–140]. The persistent nature of HIV arises from its ability to integrate into the host genomes of cells in a variety of tissues, creating a long-lived viral reservoir that is relatively untouched by long-term ART [141,142]. As a result, the main challenge in achieving a “cure” for HIV-1 is the persistence of these viral reservoirs.
Reservoir studies of HIV-1 infection in the setting of ART intensification show that HIV-1 can persist indefinitely [142,143]. HIV-1 can infect both activated and resting cells, followed by integration of viral genomes into host cell chromosomes. Latently infected, memory CD4+ T-cells are the best characterized reservoir for HIV-1, which can exist in very low numbers (e.g. as low as one integrated and replication competent proviral infected cell per million resting CD4+ T cells) [142–144]. Interestingly, a majority of integrated HIV DNA does not code for replication competent virus, and does not contribute to long-term HIV persistence despite being detected by quantitative PCR-based assays [145]. Furthermore, latently infected cell are able to evade normal innate and HIV-specific immune responses regardless if integrated proviruses are capable of producing replication competent HIV-1. Proliferation of these immune quiescent cells likely plays an important role in the maintenance of the long-lived viral reservoir [146,147]. After treatment initiation, the number of infected cells decreases initially, but reaches a low but stable level after several months. In the setting of ART, the resting latent reservoir decays very slowly with an estimated half-life of over 40 months [142].
Some studies suggest that HIV replication may continue in certain tissues despite the use of otherwise fully suppressive ART [148–150]. Historical data suggest, however, that significantly impairing viral replication with ART is not sufficient to significantly reduce viral reservoirs [142,151]. Very early initiation of ART or the prompt initiation of ART in infants born to infected mothers has been shown to lead to smaller HIV reservoirs, which may be due, in part, to the limited reservoir seeding of long-lived memory CD4+ T cells [152]. However, allogeneic stem cell transplantation of HV-uninfected donor cells under the protection of ART or genetic modifications that prohibit HIV entry is one of the few strategies that has shown to lead to significant reductions in the viral reservoir size in individuals with established reservoirs who started ART during chronic infection [152–154]. Unfortunately, mathematical modeling studies have predicted and treatment interruptions studies have shown that HIV may rebound months to years after stopping ART, even after several total body log reductions in infected cells and no detectable infected cells in peripheral blood [155].
A majority of the HIV reservoir exists in tissues, such as gut-associated lymphoid tissue (GALT) and the lymphoreticular systems (lymph nodes, spleen, etc.) that is more difficult to access than peripheral blood, and hence, less well understood [156]. Tissue-derived cells may act quite differently than those collected from the peripheral blood. For example, B cell follicles appear to be an important sanctuary for SIV-infected CD4+ T lymphocytes [157]. Follicular helper (TFH) cells located in these follicles are particularly enriched in cell-associated HIV DNA and RNA and support high levels of replication in vitro [157,158]. Other infected cell types, such as macrophages or neuroresident glial cells, may also play a role in the persistence of HIV on therapy, although their precise contribution to the long-term reservoir is still under investigation [156,159,160].
Quantitative co-culture assays (qVOAs) have been developed to quantify the functional component of the reservoir. However, these platforms rely on large numbers of resting CD4+ T cells (hundreds of millions) cultured in static culture-plate formats over a period of weeks [144,161,162]. As a result, they are time-consuming and labor-intensive, and have not been adapted for use in various infected tissues outside of PBMCs. Although there have been recent important improvements in the workflow of such assays [163], miniaturization and automation may allow for increased assay sensitivity and throughput. Integrated HIV-1 DNA has also been used to quantify HIV-1 reservoir by Alu-PCR [164]. However, this method cannot indicate whether infected cells with integrated HIV-1 DNA can produce infectious HIV-1 virions. Recently, digital PCR or RT-PCR has been increasingly used to characterize HIV-1 reservoir. Since there is much interest in studying HIV reservoirs in resource-limited settings where the burden of persistent disease is greatest, development of platforms involving microfluidic chip-based or other technologies that may be deployed efficiently and cost-effectively in RLS are urgently needed.
4. Future directions
Rapid, inexpensive and simple-to-use HIV diagnostics are urgently needed for early diagnosis and ART monitoring in resource-limited settings. To address these challenges, researchers have been developing systems to detect HIV using portable and inexpensive nanotechnologies integrated with microfluidics. Such devices may one day be capable of routinely monitoring ART or identifying latent HIV reservoirs. Smartphones utilizing mobile applications have been integrated with microfluidics and paper-based portable platforms for the quantification of target biomolecules [77,110,165–168], and we envision that future POC technologies will likely incorporate mobile technologies since they are portable, inexpensive, and have high processing capabilities. Another technology that is likely to become important in resource-limited settings is the creation of wearable/implantable sensors, which can be used for patient health status monitoring including blood parameters, body temperature, pressure, and pH of the bio-environment [169–174]. Integration of flexible materials with mobile systems will also allow for the transmission of information. In addition, wearable biosensors can be manufactured by printing technologies on flexible materials to be potentially utilized as skin patches [175–177]. Although wearable/implantable sensors are at the early stage of development, they represent one of the promising areas for developing POC diagnostic tools for rapid HIV diagnoses and potentially monitoring ART in resource-limited settings. Since finding a cure for HIV infection is the ultimate goal in the clinic, significant research efforts have been dedicated to identifying and eradicating HIV-1 reservoirs. A portable or wearable device that can perform this task would be ideal for informing physicians when to stop ART. We believe that convergence of emerging technologies targeting unmet clinical challenges in HIV-1 management will bring hope to eventually curing the HIV-1 pandemic.
Acknowledgments
U Demirci would like to acknowledge R01 AI093282, R01 DE02497101, and U54 EB015408. S Wang acknowledges the support of the Fundamental Research Funds for the Central Universities (2015QNA7026) from China. T Henrich receives support from the Foundation for AIDS Research (amfAR), R21AI110277 and R21AI113117.
Abbreviations
- ART
antiretroviral therapy
- RLS
resource-limited settings
- POC
point of care
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
U Demirci is a founder of, and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions, and (ii) Koek Biotech, a company that is developing microfluidic IVF technologies for clinical solutions. U Demirci’s interests were viewed and managed in accordance with the conflict of interest policies. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.AIDS by the numbers, 2015.
- 2.Mathers BM, Degenhardt L, Ali H, Wiessing L, Hickman M, Mattick RP, Myers B, Ambekar A, Strathdee SA. HIV prevention, treatment, and care services for people who inject drugs: a systematic review of global, regional, and national coverage. Lancet. 2010;375:1014–1028. doi: 10.1016/S0140-6736(10)60232-2. [DOI] [PubMed] [Google Scholar]
- 3.Abdul-Quader AS, Feelemyer J, Modi S, Stein ES, Briceno A, Semaan S, Horvath T, Kennedy GE, Des Jarlais DC. Effectiveness of structural-level needle/syringe programs to reduce HCV and HIV infection among people who inject drugs: A systematic review. AIDS Behav. 2013;17:2878–2892. doi: 10.1007/s10461-013-0593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan MO, Lutalo T, Gray RH. Viral Load and Heterosexual Transmission of Human Immunodeficiency Virus Type 1. N Engl J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 5.Tanser F, Bärnighausen T, Grapsa E, Zaidi J, Newell ML. High Coverage of ART Associated with Decline in Risk of HIV Acquisition in Rural KwaZulu-Natal, South Africa. Science (80-) 2013;339:966–972. doi: 10.1126/science.1228160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bor J, Herbst AJ, Newell ML, Bärnighausen T. Increases in Adult Life Expectancy in Rural South Africa: Valuing the Scale-Up of HIV Treatment. Science (80-) 2011;339:961–965. doi: 10.1126/science.1230413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigel K, Dubrow R, Silverberg M, Crothers K, Braithwaite S, Justice A. Cancer screening in patients infected with HIV. Curr HIV/AIDS Rep. 2011;8:142–152. doi: 10.1007/s11904-011-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science (80-) 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockstroh JK, Spengler U. HIV and hepatitis C virus co-infection. Lancet Infect Dis. 2004;4:437–444. doi: 10.1016/S1473-3099(04)01059-X. [DOI] [PubMed] [Google Scholar]
- 10.Pawlowski A, Jansson M, Sköld M, Rottenberg ME, Källenius G. Tuberculosis and HIV co-infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemkens LG, Bucher HC. HIV infection and cardiovascular disease. Eur Heart J. 2014;35:1373–1381. doi: 10.1093/eurheartj/eht528. [DOI] [PubMed] [Google Scholar]
- 12.Grinspoon S, Carr A. Cardiovascular Risk and Body-Fat Abnormalities in HIV-Infected Adults. N Engl J Med. 2005;352:48–62. doi: 10.1056/NEJMra041811. [DOI] [PubMed] [Google Scholar]
- 13.Townsend CL, Cortina-Borja M, Peckham CS, de Ruiter A, Lyall H, Tookey PA. Low rates of mother-to-child transmission of HIV following effective pregnancy interventions in the United Kingdom and Ireland, 2000–2006. AIDS. 2008;22:973–981. doi: 10.1097/QAD.0b013e3282f9b67a. [DOI] [PubMed] [Google Scholar]
- 14.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Tropical infectious diseases: Diagnostics for the developing world. Nat Rev Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 15.Ren K, Wu J, Yan F, Ju H. Ratiometric electrochemical proximity assay for sensitive one-step protein detection. Sci Rep. 2014;4:4360. doi: 10.1038/srep04360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–9. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Wang J, Wu H, Liu J, Aksay IA, Lin Y. Graphene based electrochemical sensors and biosensors: a review. Electroanalysis. 2010;22:1027–1036. [Google Scholar]
- 18.Luo X, Davis JJ. Electrical biosensors and the label free detection of protein disease biomarkers. Chem Soc Rev. 2013;42:5944–62. doi: 10.1039/c3cs60077g. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Mullens C, Gorski W. Insulin oxidation and determination at carbon electrodes. Anal Chem. 2005;77:6396–6401. doi: 10.1021/ac0508752. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Jia L, Li B, Situ B, Liu Q, Wang Q, Gan N. A sandwich HIV p24 amperometric immunosensor based on a direct gold electroplating-modified electrode. Molecules. 2012;17:5988–6000. doi: 10.3390/molecules17055988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafati AA, Afraz A. Amperometric sensing of anti-HIV drug zidovudine on Ag nanofilm-multiwalled carbon nanotubes modified glassy carbon electrode. Mater Sci Eng C. 2014;39:105–112. doi: 10.1016/j.msec.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 22.Dimeski G, Badrick T, John AS. Ion Selective Electrodes (ISEs) and interferences - A review. Clin Chim Acta. 2010;411:309–317. doi: 10.1016/j.cca.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Bacchi A, Carcelli M, Compari C, Fisicaro E, Pala N, Rispoli G, Rogolino D, Sanchez TW, Sechi M, Sinisi V, Neamati N. Investigating the role of metal chelation in HIV-1 integrase strand transfer inhibitors. J Med Chem. 2011;54:8407–8420. doi: 10.1021/jm200851g. [DOI] [PubMed] [Google Scholar]
- 24.Daniels JS, Pourmand N. Label-free impedance biosensors: Opportunities and challenges. Electroanalysis. 2007;19:1239–1257. doi: 10.1002/elan.200603855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafiee H, Jahangir M, Inci F, Wang S, Willenbrecht RBM, Giguel FF, Tsibris AMN, Kuritzkes DR, Demirci U. Acute on-chip HIV detection through label-free electrical sensing of viral nano-lysate. Small. 2013;9:2553–2563. doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shafiee H, Asghar W, Inci F, Yuksekkaya M, Jahangir M, Zhang MH, Durmus NG, Gurkan UA, Kuritzkes DR, Demirci U. Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Sci Rep. 2015;5:8719. doi: 10.1038/srep08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafiee H, Kanakasabapathy MK, Juillard F, Keser M, Sadasivam M, Yuksekkaya M, Hanhauser E, Henrich TJ, Kuritzkes DR, Kaye KM, Demirci U. Printed Flexible Plastic Microchip for Viral Load Measurement through Quantitative Detection of Viruses in Plasma and Saliva. Sci Rep. 2015;5:9919. doi: 10.1038/srep09919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss NO, Zhou H, Liao L, Liu Y, Jiang S, Huang Y, Duan X. Graphene: an emerging electronic material. Adv Mater. 2012;24:5782–5825. doi: 10.1002/adma.201201482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X, Qi X, Boey F, Zhang H. Graphene-based composites. Chem Soc Rev. 2012;41:666–686. doi: 10.1039/c1cs15078b. [DOI] [PubMed] [Google Scholar]
- 30.Wu S, He Q, Tan C, Wang Y, Zhang H. Graphene-Based Electrochemical Sensors. Small. 2013;9:1160–1172. doi: 10.1002/smll.201202896. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, Yin Z, Wu S, Qi X, He Q, Zhang Q, Yan Q, Boey F, Zhang H. Graphene-based aterials: synthesis, characterization, properties, and applications. Small. 2011;7:1876–1902. doi: 10.1002/smll.201002009. [DOI] [PubMed] [Google Scholar]
- 32.Tian JF, Jauregui LA, Lopez G, Cao H, Chen YP. Ambipolar graphene field effect transistors by local metal side gates. Appl Phys Lett. 2010;96:263110. [Google Scholar]
- 33.Stoller MD, Park S, Zhu Y, An J, Ruoff RS. Graphene-based ultracapacitors. Nano Lett. 2008;8:3498–3502. doi: 10.1021/nl802558y. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Alwarappan S, Chen Z, Kong X, Li CZ. Membraneless enzymatic biofuel cells based on graphene nanosheets. Biosens Bioelectron. 2010;25:1829–1833. doi: 10.1016/j.bios.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Pumera M, Ambrosi A, Bonanni A, Chng ELK, Poh HL. Graphene for electrochemical sensing and biosensing. TrAC Trends Anal Chem. 2010;29:954–965. [Google Scholar]
- 36.Brownson DAC, Banks CE. Graphene electrochemistry: an overview of potential applications. Analyst. 2010;135:2768–2778. doi: 10.1039/c0an00590h. [DOI] [PubMed] [Google Scholar]
- 37.Feng L, Wu L, Qu X. New horizons for diagnostics and therapeutic applications of graphene and graphene oxide. Adv Mater. 2013;25:168–186. doi: 10.1002/adma.201203229. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, Ratinac KR, Ringer SP, Thordarson P, Gooding JJ, Braet F. Carbon nanomaterials in biosensors: should you use nanotubes or graphene? Angew Chemie Int Ed. 2010;49:2114–2138. doi: 10.1002/anie.200903463. [DOI] [PubMed] [Google Scholar]
- 39.Viswanathan S, Narayanan TN, Aran K, Fink KD, Paredes J, Ajayan PM, Filipek S, Miszta P, Tekin HC, Inci F, Demirci U, Li P, Bolotin KI, Liepmann D, Renugopalakrishanan V. Graphene-protein field effect biosensors: Glucose sensing. Mater Today. 2015;18:513–522. [Google Scholar]
- 40.Wang Y, Bai X, Wen W, Zhang X, Wang S. Ultrasensitive electrochemical biosensor for HIV gene detection based on graphene stabilized gold nanoclusters with exonuclease amplification. ACS Appl Mater Interfaces. 2015;7:18872–18879. doi: 10.1021/acsami.5b05857. [DOI] [PubMed] [Google Scholar]
- 41.Safavieh M, Khetani S, Kaul V, Kuritzkes DR, Shafiee H. A graphene-modified cellulose paper microchip for HIV detection. SPIE Sens Technol Appl, International Society for Optics and Photonics. 2015:94900G–94900G–7. [Google Scholar]
- 42.Greve B, Weidner J, Cassens U, Odaibo G, Olaleye D, Sibrowski W, Reichelt D, Nasdala I, Göhde W. A new affordable flow cytometry based method to measure HIV-1 viral load. Cytometry A. 2009;75:199–206. doi: 10.1002/cyto.a.20676. [DOI] [PubMed] [Google Scholar]
- 43.Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, McDevitt JT. Integration of semiconductor quantum dots into nano-bio-chip systems for enumeration of CD4+ T cell counts at the point-of-need. Lab Chip. 2008;8:2079–90. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng X, Irimia D, Dixon M, Ziperstein JC, Demirci U, Zamir L, Tompkins RG, Toner M, Rodriguez WR. A microchip approach for practical label-free CD4+ T-cell counting of HIV-infected subjects in resource-poor settings. J Acquir Immune Defic Syndr. 2007;45:257–61. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- 45.Cheng X, Gupta A, Chen C, Tompkins RG, Rodriguez W, Toner M. Enhancing the performance of a point-of-care CD4+ T-cell counting microchip through monocyte depletion for HIV/AIDS diagnostics. Lab Chip. 2009;9:1357–64. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wünschmann S, Stapleton JT. Fluorescence-based quantitative methods for detecting human immunodeficiency virus type 1-induced syncytia. J Clin Microbiol. 2000;38:3055–60. doi: 10.1128/jcm.38.8.3055-3060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J Exp Med. 1997;185:621–8. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribeiro RM, Hazenberg MD, Perelson AS, Davenport MP. Naïve and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J Virol. 2006;80:802–9. doi: 10.1128/JVI.80.2.802-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwa D, Vingerhoed J, Boeser B, Schuitemaker H. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J Infect Dis. 2003;187:1397–403. doi: 10.1086/374650. [DOI] [PubMed] [Google Scholar]
- 50.Hanley MB, Lomas W, Mittar D, Maino V, Park E. Detection of low abundance RNA molecules in individual cells by flow cytometry. PLoS One. 2013;8:e57002. doi: 10.1371/journal.pone.0057002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moon S, Gurkan UA, Blander J, Fawzi WW, Aboud S, Mugusi F, Kuritzkes DR, Demirci U. Enumeration of CD4+ T-cells using a portable microchip count platform in tanzanian HIV-infected patients. PLoS One. 2011;6:e21409. doi: 10.1371/journal.pone.0021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobieranski AC, Inci F, Tekin HC, Yuksekkaya M, Comunello E, Cobra D, von Wangenheim A, Demirci U. Portable lensless wide-field microscopy imaging platform based on digital inline holography and multi-frame pixel super-resolution. Light Sci Appl. 2015;4:e346. doi: 10.1038/lsa.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozcan A, Demirci U. Ultra wide-field lens-free monitoring of cells on-chip. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 54.Moon S, Keles HO, Khademhosseini A, Kuritzkes D, Demirci U. Integrating microfluidics and lensless imaging for point-of-care testing. 2009 IEEE 35th Annu Northeast Bioeng Conf, IEEE. 2009:1–2. doi: 10.1016/j.bios.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. A microfluidic device for practical label-free CD4+ T cell counting of HIV-infected subjects. Lab Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim YG, Moon S, Kuritzkes DR, Demirci U. Quantum dot-based HIV capture and imaging in a microfluidic channel. Biosens Bioelectron. 2009;25:253–8. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gurkan UA, Moon S, Geckil H, Xu F, Wang S, Lu TJ, Demirci U. Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnol J. 2011;6:138–149. doi: 10.1002/biot.201000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alyassin MA, Moon S, Keles HO, Manzur F, Lin RL, Hæggstrom E, Kuritzkes DR, Demirci U. Rapid automated cell quantification on HIV microfluidic devices. Lab Chip. 2009;9:3364–9. doi: 10.1039/b911882a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer KM, Hao F, Lee S, Nordlander P, Hafner JH. A single molecule immunoassay by localized surface plasmon resonance. Nanotechnology. 2010;21:255503. doi: 10.1088/0957-4484/21/25/255503. [DOI] [PubMed] [Google Scholar]
- 60.Willets KA, Van Duyne RP. Localized surface plasmon resonance spectroscopy and sensing. Annu Rev Phys Chem. 2007;58:267–97. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 61.Wabuyele MB, Yan F, Vo-Dinh T. Plasmonics nanoprobes: detection of single-nucleotide polymorphisms in the breast cancer BRCA1 gene. Anal Bioanal Chem. 2010;398:729–36. doi: 10.1007/s00216-010-3992-1. [DOI] [PubMed] [Google Scholar]
- 62.Inci F, Tokel O, Wang S, Gurkan UA, Tasoglu S, Kuritzkes DR, Demirci U. Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano. 2013;7:4733–4745. doi: 10.1021/nn3036232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inci F, Filippini C, Baday M, Ozen MO, Calamak S, Durmus NG, et al. Multitarget, quantitative nanoplasmonic electrical field-enhanced resonating device (NE2RD) for diagnostics. Proc Natl Acad Sci U S A. 2015;112:E4354–4363. doi: 10.1073/pnas.1510824112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yablonovitch E. Photonic band-gap structures. J Opt Soc Am B. 1993;10:283. [Google Scholar]
- 65.Fenzl C, Hirsch T, Wolfbeis OS. Photonic crystals for chemical sensing and biosensing. Angew Chemie - Int Ed. 2014;53:3318–3335. doi: 10.1002/anie.201307828. [DOI] [PubMed] [Google Scholar]
- 66.Macconaghy KI, Geary CI, Kaar JL, Stoykovich MP. Photonic crystal kinase biosensor. J Am Chem Soc. 2014;136:6896–6899. doi: 10.1021/ja5031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pal S, Yadav AR, Lifson MA, Baker JE, Fauchet PM, Miller BL. Selective virus detection in complex sample matrices with photonic crystal optical cavities. Biosens Bioelectron. 2013;44:229–234. doi: 10.1016/j.bios.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan LL, Gosangari SL, Watkin KL, Cunningham BT. A label-free photonic crystal biosensor imaging method for detection of cancer cell cytotoxicity and proliferation. Apoptosis. 2007;12:1061–8. doi: 10.1007/s10495-006-0031-y. [DOI] [PubMed] [Google Scholar]
- 69.Coscelli E, Sozzi M, Poli F, Passaro D, Cucinotta A, Selleri S, Corradini R, Marchelli R. Toward A Highly Specific DNA Biosensor: PNA-Modified Suspended-Core Photonic Crystal Fibers. IEEE J Sel Top Quantum Electron. 2010;16:967–972. [Google Scholar]
- 70.Lin B, Li P, Cunningham BT. A label-free biosensor-based cell attachment assay for characterization of cell surface molecules. Sensors Actuators, B Chem. 2006;114:559–564. [Google Scholar]
- 71.Pineda MF, Chan LL, Kuhlenschmidt T, Choi CJ, Kuhlenschmidt M, Cunningham BT. Rapid Specific and Label-Free Detection of Porcine Rotavirus Using Photonic Crystal Biosensors. IEEE Sens J. 2009;9:470–477. [Google Scholar]
- 72.Endo T, Ozawa S, Okuda N, Yanagida Y, Tanaka S, Hatsuzawa T. Reflectometric detection of influenza virus in human saliva using nanoimprint lithography-based flexible two-dimensional photonic crystal biosensor. Sensors Actuators, B Chem. 2010;148:269–276. [Google Scholar]
- 73.Kang P, Schein P, Serey X, O’Dell D, Erickson D. Nanophotonic detection of freely interacting molecules on a single influenza virus. Sci Rep. 2015;5:12087. doi: 10.1038/srep12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maeng B, Park Y, Park J. Direct label-free detection of Rotavirus using a hydrogel based nanoporous photonic crystal. RSC Adv. 2016;6:7384–7390. [Google Scholar]
- 75.Shafiee H, Lidstone E, Jahangir M, Inci F, Hanhauser E, Henrich TJ, Kuritzkes DR, Cunningham BT, Demirci U. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci Rep. 2014;4:4116. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhuo Y, Hu H, Chen W, Lu M, Tian L, Yu H, Long KD, Chow E, King WP, Singamaneni S, Cunningham BT. Single nanoparticle detection using photonic crystal enhanced microscopy. Analyst. 2014;139:1007–15. doi: 10.1039/c3an02295a. [DOI] [PubMed] [Google Scholar]
- 77.Gallegos D, Long KD, Yu H, Clark PP, Lin Y, George S, Nath P, Cunningham BT. Label-free biodetection using a smartphone. Lab Chip. 2013;13:2124–32. doi: 10.1039/c3lc40991k. [DOI] [PubMed] [Google Scholar]
- 78.Yu H, Tan Y, Cunningham BT. Smartphone fluorescence spectroscopy. Anal Chem. 2014;86:8805–8813. doi: 10.1021/ac502080t. [DOI] [PubMed] [Google Scholar]
- 79.Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442:381–6. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 80.Kang P, Schein P, Serey X, O’Dell D, Erickson D. Nanophotonic detection of freely interacting molecules on a single influenza virus. Sci Rep. 2015;5:12087. doi: 10.1038/srep12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arlett JL, Myers EB, Roukes ML. Comparative advantages of mechanical biosensors. Nat Nanotechnol. 2011;6:203–215. doi: 10.1038/nnano.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kang D, Pikhitsa PV, Choi YW, Lee C, Shin SS, Piao L, Park B, Suh KY, Kim T, Choi M. Ultrasensitive mechanical crack-based sensor inspired by the spider sensory system. Nature. 2014;516:222–226. doi: 10.1038/nature14002. [DOI] [PubMed] [Google Scholar]
- 83.Wee KW, Kang GY, Park J, Kang JY, Yoon DS, Park JH, Kim TS. Novel electrical detection of label-free disease marker proteins using piezoresistive self-sensing micro-cantilevers. Biosens Bioelectron. 2005;20:1932–1938. doi: 10.1016/j.bios.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 84.Rasmussen PA, Thaysen J, Hansen O, Eriksen SC, Boisen A. Optimised cantilever biosensor with piezoresistive read-out. Ultramicroscopy. 2003;97:371–376. doi: 10.1016/S0304-3991(03)00063-9. [DOI] [PubMed] [Google Scholar]
- 85.Ibach H. The role of surface stress in reconstruction, epitaxial growth and stabilization of mesoscopic structures. Surf Sci Rep. 1997;29:195–263. [Google Scholar]
- 86.Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature. 2007;446:1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 87.Squires TM, Messinger RJ, Manalis SR. Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 88.Johnson BN, Mutharasan R. Biosensing using dynamic-mode cantilever sensors: a review. Biosens Bioelectron. 2012;32:1–18. doi: 10.1016/j.bios.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 89.Olcum S, Cermak N, Wasserman SC, Manalis SR. High-speed multiple-mode mass-sensing resolves dynamic nanoscale mass distributions. Nat Commun. 2015;6:7070. doi: 10.1038/ncomms8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savran CA, Knudsen SM, Ellington AD, Manalis SR. Micromechanical detection of proteins using aptamer-based receptor molecules. Anal Chem. 2004;76:3194–3198. doi: 10.1021/ac049859f. [DOI] [PubMed] [Google Scholar]
- 91.Lam Y, Abu-Lail NI, Alam MS, Zauscher S. Using microcantilever deflection to detect HIV-1 envelope glycoprotein gp120. Nanomedicine Nanotechnology, Biol Med. 2006;2:222–229. doi: 10.1016/j.nano.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 92.Gilbert M, Kirihara J, Mills J. Enzyme-linked immunoassay for human immunodeficiency virus type 1 envelope glycoprotein 120. J Clin Microbiol. 1991;29:142–147. doi: 10.1128/jcm.29.1.142-147.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh S, Cruikshank WW, Raina J, Blanchard GC, Adler WH, Walker J, Kornfeld H. Identification of HIV-1 envelope glycoprotein in the serum of AIDS and ARC patients. JAIDS J Acquir Immune Defic Syndr. 1992;5:251–256. [PubMed] [Google Scholar]
- 94.Patil SB, Vögtli M, Webb B, Mazza G, Pinzani M, Soh YA, McKendry RA, Ndieyira JW. Decoupling competing surface binding kinetics and reconfiguration of receptor footprint for ultrasensitive stress assays. Nat Nanotechnol. 2015;10:899–907. doi: 10.1038/nnano.2015.174. [DOI] [PubMed] [Google Scholar]
- 95.McCoy LE, Rutten L, Frampton D, Anderson I, Granger L, Bashford-Rogers R, Dekkers G, Strokappe NM, Seaman MS, Koh W, Grippo V, Kliche A, Verrips T, Kellam P, Fassati A, Weiss RA. Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1. PLoS Pathog. 2014;10:e1004552. doi: 10.1371/journal.ppat.1004552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huber F, Lang HP, Backmann N, Rimoldi D, Gerber C. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. Nat Nanotechnol. 2013;8:125–129. doi: 10.1038/nnano.2012.263. [DOI] [PubMed] [Google Scholar]
- 97.Dobrowsky TM, Rabi SA, Nedellec R, Daniels BR, Mullins JI, Mosier DE, Siliciano RF, Wirtz D. Adhesion and fusion efficiencies of human immunodeficiency virus type 1 (HIV-1) surface proteins. Sci Rep. 2013;3:3014. doi: 10.1038/srep03014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lang HP, Hegner M, Gerber C. Cantilever array sensors. Mater Today. 2005;8:30–36. [Google Scholar]
- 99.Bruckenstein S, Shay M. Experimental aspects of use of the quartz crystal microbalance in solution. Electrochim Acta. 1985;30:1295–1300. [Google Scholar]
- 100.Rodahl M, Höök F, Krozer A, Brzezinski P, Kasemo B. Quartz crystal microbalance setup for frequency and Q-factor measurements in gaseous and liquid environments. Rev Sci Instrum. 1995;66:3924–3930. [Google Scholar]
- 101.Fredriksson C, Kihlman S, Rodahl M, Kasemo B. The piezoelectric quartz crystal mass and dissipation sensor: a means of studying cell adhesion. Langmuir. 1998;14:248–251. [Google Scholar]
- 102.Steinem C, Janshoff A. Piezoelectric sensors. Springer Science & Business Media; 2007. [Google Scholar]
- 103.Hong SR, Jeong HD, Hong S. QCM DNA biosensor for the diagnosis of a fish pathogenic virus VHSV. Talanta. 2010;82:899–903. doi: 10.1016/j.talanta.2010.04.065. [DOI] [PubMed] [Google Scholar]
- 104.Lu CH, Zhang Y, Tang SF, Fang ZB, Yang HH, Chen X, Chen GN. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens Bioelectron. 2012;31:439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 105.Nakatsuma A, Kaneda M, Kodama H, Morikawa M, Watabe S, Nakaishi K, Yamashita M, Yoshimura T, Miura T, Ninomiya M, Ito E. Detection of HIV-1 p24 at Attomole Level by Ultrasensitive ELISA with Thio-NAD Cycling. PLoS One. 2015;10:e0131319. doi: 10.1371/journal.pone.0131319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Arora DR, Maheshwari M, Arora B. Rapid Point-of-Care Testing for Detection of HIV and Clinical Monitoring. ISRN AIDS. 2013;2013:287269. doi: 10.1155/2013/287269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang S, Inci F, De Libero G, Singhal A, Demirci U. Point-of-care assays for tuberculosis: Role of nanotechnology / micro fl uidics. Biotechnol Adv. 2013;31:438–449. doi: 10.1016/j.biotechadv.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mwau M, Kadima S, Mwende J, Adhiambo M, Akinyi C, Prescott M, Lusike J, Hungu J, Vojnov L. Technical performance evaluation of the MyT4 point of care technology for CD4+ T cell enumeration. PLoS One. 2014;9:e107410. doi: 10.1371/journal.pone.0107410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, Wang J, Moore H, Rouse R, Umviligihozo G, Karita E, Mwambarangwe L, Braunstein SL, van de Wijgert J, Sahabo R, Justman JE, El-Sadr W, Sia SK. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17:1015–9. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 110.Laksanasopin T, Guo TW, Nayak S, Sridhara Aa, Xie S, Olowookere OO, et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med. 2015;7:273re1–273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 111.Drosten C, Panning M, Drexler JF, Hänsel F, Pedroso C, Yeats J, de Souza Luna LK, Samuel M, Liedigk B, Lippert U, Stürmer M, Doerr HW, Brites C, Preiser W. Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5′ long terminal repeat domain. Clin Chem. 2006;52:1258–66. doi: 10.1373/clinchem.2006.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rouet F, Ekouevi DK, Chaix ML, Burgard M, Inwoley A, Tony TD, Danel C, Anglaret X, Leroy V, Msellati P, Dabis F, Rouzioux C. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J Clin Microbiol. 2005;43:2709–17. doi: 10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tanriverdi S, Chen L, Chen S. A rapid and automated sample-to-result HIV load test for near-patient application. J Infect Dis. 2010;201(Suppl):S52–8. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- 114.Tang W, Chow WHA, Li Y, Kong H, Tang YW, Lemieux B. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J Infect Dis. 2010;201(Suppl):S46–51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jangam SR, Agarwal AK, Sur K, Kelso DM. A point-of-care PCR test for HIV-1 detection in resource-limited settings. Biosens Bioelectron. 2013;42:69–75. doi: 10.1016/j.bios.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 116.Zhang C, Xing D. Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends. Nucleic Acids Res. 2007;35:4223–37. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Arslan S, Khafizov R, Thomas CD, Chemla YR, Ha T. Protein structure. Engineering of a superhelicase through conformational control. Science. 2015;348:344–7. doi: 10.1126/science.aaa0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods. 2012;186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly Precise Measurement of HIV DNA by Droplet Digital PCR. PLoS One. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mansfield M. Lateral Flow Immunoassay. Humana Press; Totowa, NJ: 2009. [Google Scholar]
- 121.Workman S, Wells SK, Pau CP, Owen SM, Dong XF, LaBorde R, Granade TC. Rapid detection of HIV-1 p24 antigen using magnetic immuno-chromatography (MICT) J Virol Methods. 2009;160:14–21. doi: 10.1016/j.jviromet.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 122.Martinez AW, Phillips ST, Wiley BJ, Gupta M, Whitesides GM. FLASH: A rapid method for prototyping paper based microfluidic devices. Lab Chip. 2008;8:2146. doi: 10.1039/b811135a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: Microfluidic paper-based analytical devices. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 124.Parolo C, Merkoçi A. Paper-based nanobiosensors for diagnostics. Chem Soc Rev. 2013;42:450–457. doi: 10.1039/c2cs35255a. [DOI] [PubMed] [Google Scholar]
- 125.Rohrman BA, Richards-Kortum RR. A paper and plastic device for performing recombinase polymerase amplification of HIV DNA. Lab Chip. 2012;12:3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shafiee H, Wang S, Inci F, Toy M, Henrich TJ, Kuritzkes DR, Demirci U. Emerging Technologies for Point-of-Care Management of HIV Infection. Annu Rev Med. 2015;66:387–405. doi: 10.1146/annurev-med-092112-143017. [DOI] [PubMed] [Google Scholar]
- 128.Wang S, Xu F, Demirci U. Advances in developing HIV-1 viral load assays for resource-limited settings. Biotechnol Adv. 2010;28:770–781. doi: 10.1016/j.biotechadv.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ananworanich J, Dubé K, Chomont N. How does the timing of antiretroviral therapy initiation in acute infection affect HIV reservoirs? Curr Opin HIV AIDS. 2015;10:18–28. doi: 10.1097/COH.0000000000000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ananworanich J, Chomont N, Fletcher JL, Pinyakorn S, Schuetz A, Sereti I, Rerknimitr R, Dewar R, Kroon E, Vandergeeten C, Trichavaroj R, Chomchey N, Chalermchai T, Michael NL, Kim JH, Phanuphak P, Phanuphak N. Markers of HIV reservoir size and immune activation after treatment in acute HIV infection with and without raltegravir and maraviroc intensification. J Virus Erad. 1:116–122. doi: 10.1016/S2055-6640(20)30482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cohen MS, Gay CL, Busch MP, Hecht FM. The detection of acute HIV infection. J Infect Dis. 2010;202(Suppl):S270–S277. doi: 10.1086/655651. [DOI] [PubMed] [Google Scholar]
- 133.Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA. Simple amplification-based assay: a nucleic acid-based point-of-care platform for HIV-1 testing. J Infect Dis. 2010;201(Suppl):S65–72. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- 134.Selck DA, Karymov MA, Sun B, Ismagilov RF. Increased robustness of single-molecule counting with microfluidics, digital isothermal amplification, and a mobile phone versus real-time kinetic measurements. Anal Chem. 2013;85:11129–11136. doi: 10.1021/ac4030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Myers FB, Henrikson RH, Bone J, Lee LP. A Handheld Point-of-Care Genomic Diagnostic System. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol. 2016;14:55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 137.Lewin SR, Rouzioux C. HIV cure and eradication: how will we get from the laboratory to effective clinical trials? AIDS. 2011;25:885–897. doi: 10.1097/QAD.0b013e3283467041. [DOI] [PubMed] [Google Scholar]
- 138.Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, Schneider T, Hofmann J, Kücherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–8. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 139.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–47. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12:607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–7. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 142.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 143.Coiras M, López-Huertas MR, Pérez-Olmeda M, Alcamí J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol. 2009;7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 144.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 145.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–51. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–3. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science (80-) 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Günthard HF, Frost SD, Leigh-Brown AJ, Ignacio CC, Kee K, Perelson AS, Spina CA, Havlir DV, Hezareh M, Looney DJ, Richman DD, Wong JK. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J Virol. 1999;73:9404–12. doi: 10.1128/jvi.73.11.9404-9412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Buzón MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, Palmer S, Stevenson M, Clotet B, Blanco J, Martinez-Picado J. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. 2010;16:460–5. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- 150.Sharkey M, Babic DZ, Greenough T, Gulick R, Kuritzkes DR, Stevenson M. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog. 2011;7:e1001303. doi: 10.1371/journal.ppat.1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gandhi RT, Bosch RJ, Aga E, Albrecht M, Demeter LM, Dykes C, Bastow B, Para M, Lai J, Siliciano RF, Siliciano JD, Eron JJ. No evidence for decay of the latent reservoir in HIV-1-infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. J Infect Dis. 2010;201:293–6. doi: 10.1086/649569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M, Chun TW, Strain M, Richman D, Luzuriaga K. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med. 2013;369:1828–35. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, Gallien S, Lin NH, Giguel FF, Lavoie L, Ho VT, Armand P, Soiffer RJ, Sagar M, Lacasce AS, Kuritzkes DR. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013;207:1694–702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161:319–27. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hill AL, Rosenbloom DIS, Fu F, Nowak MA, Siliciano RF. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci U S A. 2014;111:13475–80. doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schrager LK, D’Souza MP. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA. 1998;280:67–71. doi: 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- 157.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–9. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–56. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in Human Immunodeficiency Virus Type I infection. Retrovirology. 2012;9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Abbas W, Tariq M, Iqbal M, Kumar A, Herbein G. Eradication of HIV-1 from the macrophage reservoir: an uncertain goal? Viruses. 2015;7:1578–98. doi: 10.3390/v7041578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bergren CT, Chan FN, Bodzin JH. Intravenous pyelogram results in association with renal pathology and therapy in trauma patients. J Trauma. 1987;27:515–8. doi: 10.1097/00005373-198705000-00010. [DOI] [PubMed] [Google Scholar]
- 162.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 163.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liszewski MK, Yu JJ, O’Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chhablani J, Kaja S, Shah VA. Smartphones in ophthalmology. Indian J Ophthalmol. 2012;60:127–131. doi: 10.4103/0301-4738.94054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Phillippi J, Wyatt T. Smartphones in Nursing Education. Comput Informatics Nurs. 2011;29:449–454. doi: 10.1097/NCN.0b013e3181fc411f. [DOI] [PubMed] [Google Scholar]
- 167.Suki NM. Students’ demand for smartphones. Campus - Wide Inf Syst. 2013;30:236–248. [Google Scholar]
- 168.Cheng J, Wong SHY, Yang H, Lu S. Proc 5th Int Conf Mob Syst Appl Serv - MobiSys ’07. ACM Press; New York, New York, USA: 2007. SmartSiren; p. 258. [Google Scholar]
- 169.Honda W, Harada S, Arie T, Akita S, Takei K. Wearable, human-interactive, health-monitoring, wireless devices fabricated by macroscale printing techniques. Adv Funct Mater. 2014;24:3299–3304. [Google Scholar]
- 170.Kim D-H, Ghaffari R, Lu N, Rogers Ja. Flexible and Stretchable Electronics for Biointegrated Devices. Annu Rev Biomed Eng. 2012;14:113–128. doi: 10.1146/annurev-bioeng-071811-150018. [DOI] [PubMed] [Google Scholar]
- 171.Liao C, Zhang M, Yao MY, Hua T, Li L, Yan F. Flexible Organic Electronics in Biology: Materials and Devices. Adv Mater. 2015;27:7493–7527. doi: 10.1002/adma.201402625. [DOI] [PubMed] [Google Scholar]
- 172.Baday M, Calamak S, Durmus NG, Davis RW, Steinmetz LM, Demirci U. Integrating Cell Phone Imaging with Magnetic Levitation (i-LEV) for Label-Free Blood Analysis at the Point-of-Living. Small. 2016;12:1222–9. doi: 10.1002/smll.201501845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Durmus NG, Tekin HC, Guven S, Sridhar K, Arslan Yildiz A, Calibasi G, Ghiran I, Davis RW, Steinmetz LM, Demirci U. Magnetic levitation of single cells. Proc Natl Acad Sci. 2015;112:E3661–E3668. doi: 10.1073/pnas.1509250112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tasoglu S, Khoory JA, Tekin HC, Thomas C, Karnoub AE, Ghiran IC, Demirci U. Levitational Image Cytometry with Temporal Resolution. Adv Mater. 2015;27:3901–3908. doi: 10.1002/adma.201405660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pohlenz P, Adler W, Li L, Schmelzle R, Klatt J. Medial orbital wall reconstruction with flexible Ethisorb® patches. Clin Oral Investig. 2013;17:511–516. doi: 10.1007/s00784-012-0716-2. [DOI] [PubMed] [Google Scholar]
- 176.Boland JJ. Flexible electronics: Within touch of artificial skin. Nat Mater. 2010;9:790–792. doi: 10.1038/nmat2861. [DOI] [PubMed] [Google Scholar]
- 177.Xu Y, Tai YC, Huang A, Ho CM. IC-integrated flexible shear-stress sensor skin. J Microelectromechanical Syst. 2003;12:740–747. [Google Scholar]


