Abstract
Understanding the different molecular mechanisms responsible for gene expression has been a central interest of molecular biologists for several decades. Transcription, the initial step of gene expression, consists of converting the genetic code into a dynamic messenger RNA that will specify a required cellular function following translocation to the cytoplasm and translation. We now possess an in-depth understanding of the mechanism and regulations of transcription. By contrast, an understanding of the dynamics of an individual gene's expression in real time is just beginning to emerge following recent technological developments.
Introduction
Molecular events governing gene expression are among the best-studied and best-understood mechanisms in biology. In many cases, our understanding of these processes has reached the atomic level, especially with the recent crystallization of transcription complexes [1] and the unraveling of the atomic order within the nuclear pore complex [2]. This high-resolution picture of the transcription and export machineries contrasts with our lack of knowledge regarding the time scales of these reactions within live cells. While mechanistic approaches shed light on how these events are regulated, the consequences of this regulation for cell fate can only be understood by integrating these mechanisms into kinetic pathways, allowing an understanding of the inherent plasticity within living cells. In this review, we focus on the recent technological and conceptual advances that provide a better understanding of the kinetic rules that govern the molecular processes of mRNA transcription and export.
Dynamics of transcription initiation: few factors for many genes
Gene promoters can be viewed as static binding elements on which transcription factors assemble. It is the combinatorial variety of transcription factors in a cell that will presumably modulate the transcriptional activity of a specific gene. Recent approaches in which immunoprecipitation of chromatin using antibodies against transcription factors is followed by analysis of the bound sequences by DNA microarrays (‘ChIP-chip’ methods) have identified factor-specific yeast and mammalian promoter sequences [3, 4,5and 6••]. A complex network of interactions was revealed by a genome-wide ChIP-chip analysis of S. cerevisiae that identified 106 transcription factors bound to 2343 promoter sequences [ 6••]. In a recent study, 142 transcription factors interacting with 3420 target genes were linked to the expression of the genes they regulate, uncovering the underestimated dynamics of the ‘transcription factor network’ [7••]. One of the findings was that out of the 70 transcription factors that bind to transcription units during the cell cycle, 56 were also regulated by the cell cycle. Synthesis and binding of transcription factors could occur in the same phase of the cycle (as was the case for 13/56 genes) or also bind to genes expressed in subsequent phases (24/56 genes), providing a trigger to move to the next phase. The lag between transcription factor binding and gene expression, although highly reproducible, varied greatly from gene to gene and did not seem to depend on specific transcription factors but rather on the target gene [7••]. Although these findings correlating the binding of transcription factors to their expression can provide crucial information on the wiring of the transcriptional network, it is still unclear how the time lag separating transcription factor binding from gene expression is controlled at the molecular level. So far, the complexity of the mammalian genome has not allowed genome-wide ChIP-CHIP analysis [ 3 and 8]. However, new approaches where chromatin immunoprecipitation is analyzed by tandem cloning, known as serial analysis of chromosome occupancy (SACO), allow the genome-wide quantitative interrogation of the binding of specific transcription factors [9]. One way to extend this analysis would then be to investigate the correlation between transcription factor binding and polymerase loading of a gene. Technologies to address these questions now exist. For example, a time-course ChIP approach has shown that the binding of the estrogen receptor to its promoter is cyclical, defining permissive and non-permissive states for gene activation [ 10, 11 and 12]. These biochemical approaches can only address a population of cells at a fixed time determined by chemical fixation procedures, and often require cell synchronization [13]. Recent developments enable us to detect the transcriptional activity of individual genes in single cells by FISH (fluorescent in-situ hybridization) using color-coded probes for specific mRNAs at their transcription sites [14]. Ultimately, these experiments will have to be conducted in live cells and in real time, allowing the onset of gene activity to be correlated with upstream events of transcription factor binding and downstream (post-transcriptional) gene expression regulation events.
To determine cell fate, transcription factors must be present at limiting concentrations so that changes in their levels can lead to differential gene expression. Using cell lines with tandem repeated genes has allowed observations of transcription in live cells. The recruitment of the glucocorticoid receptor to such gene arrays revealed that binding was transient, exhibiting half-residence of a few seconds [15, 16 and 17]. These findings were extended to other components of the transcription recruitment machinery [17 and 18] and suggest that a single transcription factor molecule can affect the transcription of several genes through successive interactions. Findings that gene expression is stochastic in single cells [19 and 20] would be explained by a model whereby transcription activation can be achieved by a small number of molecules. The probability of interaction defines the ‘transcriptional noise’ within the system. Differences in noise levels can arise from different transcription initiation activities [19 and 20] and it has been proposed that transcription processivity [21] or translation efficiency [22] could significantly influence the stochasticity of the system. Genes that are essential for cellular viability or proteins involved in macromolecular complexes are biased toward noise reduction mechanisms [22]. While transcriptional activation (‘promoter firing’) represents the input for gene expression, the many control points downstream in the transcriptional process will modulate the output.
Dynamics of post-initiation polymerases
Following assembly of factors on the promoter, the polymerase initiates transcription and then elongates the nascent pre-mRNA. The switch from the post-initiation stage to elongation (termed promoter clearance and promoter escape, respectively) is believed to be a limiting step leading to high levels of aborted short RNA molecules [23 and 24]. The instability of the polymerase on its DNA substrate results in aborted promoter clearance and it is unclear how different transcription factors can influence its processivity. Promoter usage could provide a positive regulation loop selecting for a conformation more favorable to polymerase progression. A more active control operates at the level of promoter escape, where the polymerase enters a paused state under the control of factors that inhibit elongation. Release from pausing is, at least partly, triggered by a combination of maturation events including capping of the nascent pre-mRNA and the action of transcription elongation factors. Genes controlled at the promoter escape stage, for example heat shock genes in Drosophila, can react rapidly to a stimulus since processive elongation-competent polymerases are available [ 25 and 26]. Paused polymerases have been detected in the proximal regions of genes including c-Myc [27]. Recently, the activity of TFIIH on c-Myc transcription was investigated using cell lines deficient in one of TFIIH's components, XPB. While c-Myc protein levels were only moderately affected in the population, cell-to-cell variance in protein level increased by a factor of three. Although the function of TFIIH is not completely understood, it plays an important role in the initial stages of transcription that occur before promoter clearance. Mathematical modeling showed that TFIIH might function as an integrator of promoter firing events, therefore establishing stable expression levels of c-Myc from a stochastic input [28•].
Dynamics of elongation
Single bacterial polymerases tethered to a fixed substrate and used to drag a DNA-conjugated bead enabled the determination of elongation speeds for single polymerases [29 and 30], and demonstrated that RNA polymerases have an intrinsic propensity to pause. The conservation between bacterial core polymerases and mammalian RNA polymerase II makes it likely that pausing is a property of the mammalian enzyme as well. It is difficult to address the synthesis speed of an mRNA as the kinetics will be determined by the limiting step in the transcriptional process (initiation, promoter escape or elongation). The human dystrophin gene (2.3 Mb) provides a model where initiation and promoter escape rates are minimal relative to elongation time. The measured transcription rates for this gene were 1.7–2.5 kb/min [31]. Other approaches using FISH to detect nascent pre-mRNAs at their locus of transcription following activation have found elongation speeds of 1.1–1.4 kb/min for the Drosophila UBX mRNA [ 32] and mammalian β-actin mRNA [33]. Similar rates were observed using nuclear run-on assays on the HSP70 gene following heat shock in insect cells [25].
The development of genetically encoded fluorescent markers such as GFP (green fluorescent protein), together with sophisticated imaging, have allowed kinetic measurements of many biochemical processes in live cells, including gene expression [34]. rRNA transcription is confined to the nucleolus, providing a system for studying a homogenous population of co-regulated genes. The kinetics of pol I transcription in live mammalian cells were calculated using FRAP (fluorescence recovery after photobleaching) and iFRAP (inverse FRAP) experiments. The kinetics of recruitment of this enzyme to the rDNA genes were correlated with the time it takes to transcribe one gene. This approach allowed the dissociation of the different steps of transcription (polymerase association with the promoter and elongation), and the experimental measurements were compared to simulations based on mathematical kinetic models and provided an in vivo kinetic model for Pol I transcription [ 35and 36••].
Pol II transcription was addressed in live cells by complementing a non-functional endogenous copy of the large subunit with a GFP-tagged functional version [37]. Pol II recruitment to nuclear transcription units comprised two detectable components, a fast component presenting a half life in the order of several seconds that was attributed to Pol II diffusion and binding to the promoter, and a slow component showing a half life of 14 to 20 min, resulting from engaged polymerases. The average transcription unit length in mammalian cells (~14 kb) and the previously reported polymerase speeds (1.1–2.5 kb/min) would predict an average elongation time in the order of 6–13 min, significantly shorter than that observed [38••]. This discrepancy could be explained if the polymerases were engaged on the DNA longer than the elongation process, raising the possibility that promoter escape, termination or pausing could add a rate-limiting step within the transcription unit. Ultimately, kinetic analyses at the single gene level will resolve these issues.
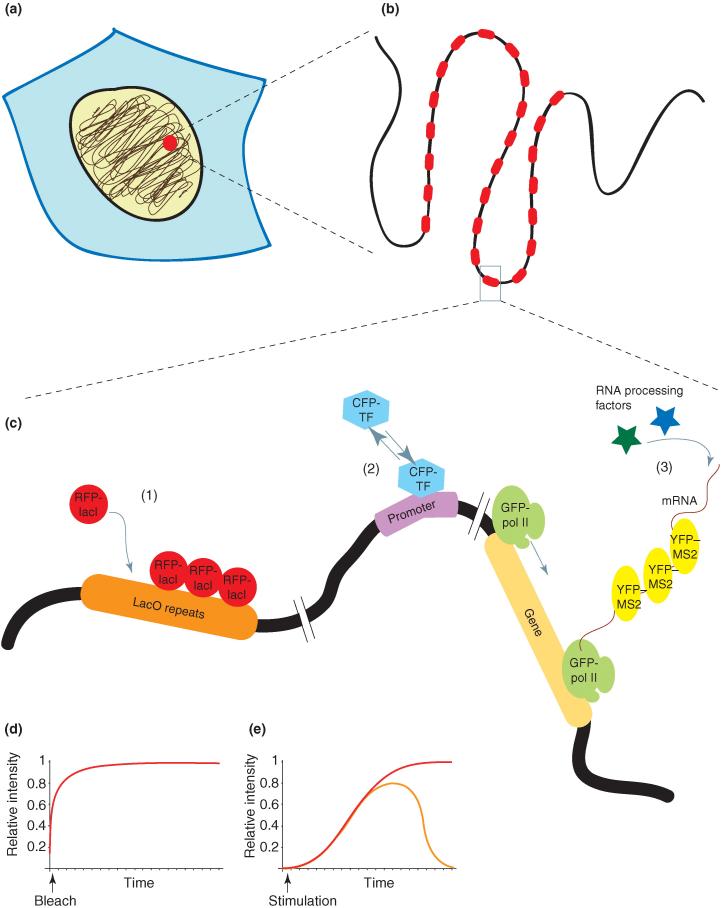
To follow the activity of specific genes in single living cells, cell lines have been generated where a particular gene is integrated as multiple tandem repeats in the genome (Figure 1) and can be visualized either using the lac operator repeat system (lacO/LacI) [39,40 and 41] or by the recruitment of fluorescent chimeric transcription factors to higher levels than the nucleoplasmic background [15, 16, 17 and 18]. Using this technology, a GFP–RNA-pol-II subunit was used to monitor the transcriptional activity of an MMTV tandem gene array: it was found that its activation was transient even in the constant presence of dexamethasone, showing a maximal activity 30 min post-activation [17]. Observing the loading of the RNA polymerases on the tandem gene array over time revealed variability among individual cells. The lacO/LacI system combined with a live-cell approach to RNA visualization [42 and 43] has provided information on the dynamics of chromatin at the locus of transcriptional activity and on the temporal resolution of gene activation (Figure 2) [44••]. Applying such techniques to the study of transcription kinetics within a gene will undoubtedly lead to a more precise understanding of this process.
Figure 1.
Tandem gene arrays for the analysis of nuclear factor dynamics in living cells. The establishment of integrated tandem gene arrays in the genome provides a platform on which a signal from a fluorescently labeled DNA- or RNA-binding protein can be amplified, detected and analyzed kinetically. (a) The locus of integration occurs randomly in the genome and (b) many copies of the gene are inserted in tandem at this locus. (c) Different types of gene arrays have been used. (1) lac operator repeats (lacO) are bound by the lac repressor protein (LacI), which can either be tagged with a fluorescent protein to mark the locus of integration or can be fused to a protein of interest, thus tethering it to the chromatin. (2) Tandem arrays of promoters can serve for the analysis of transcription factor (TF) binding dynamics. (3) The production of mRNA can be followed using gene arrays that transcribe mRNAs containing MS2 repeats, which are bound by the YFP–MS2 protein. The kinetics of RNA pol II or RNA processing factors can also be analyzed on such arrays. (d) Photobleaching methods such as FRAP are used to measure transcription-factor binding to gene arrays. (e) The onset of transcription can be measured upon cellular stimulation (red line shows a classical activation with saturation; orange line shows activation with a negative feedback).
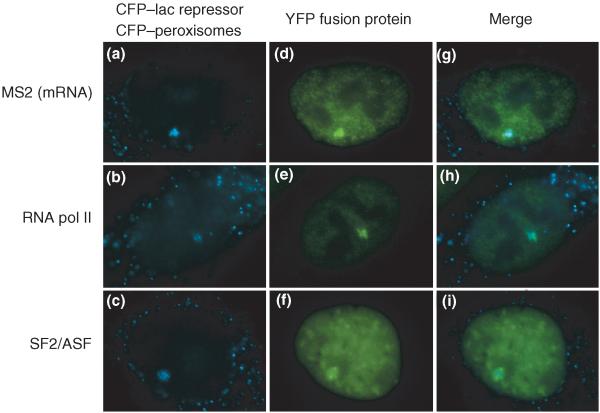
Figure 2.
Visualizing the binding of nuclear factors to tandem gene arrays. Human cells containing a tandem gene array (200 copies) were used for real-time detection of the recruitment of different factors to the site of active transcription. The tetracycline-inducible gene module gives rise to an intron-containing mRNA coding for a peroxisome-targeted CFP (cyan fluorescent protein) and containing 24 MS2 repeats in the 3′UTR. Each gene module is flanked by 256 repeats of the lacO binding site. (a,b,c) After induction of transcription, the gene locus is detected by the binding of the CFP–LacI protein to the lacO repeats and the peroxisome-targeted CFP protein product is observed in the cytoplasm. (d) The production of the mRNA at the gene locus is visualized by the binding of the yellow fluorescent protein (YFP)–MS2 protein to the MS2 stem-loops in the mRNA. (e) The recruitment of the transcription machinery to this active site of transcription is seen by the enrichment of YFP–RNA pol II at the locus. (f) The pre-mRNA splicing factor YFP–SF2/ASF is also enriched at the locus. (g,h,i) Merge. (Adapted from Janicki et al. [ 44••]).
Dynamics of mRNA within the nuclear environment
The mechanisms that regulate RNA flow from the transcription site are not well understood. It is now clear that mRNAs traverse the nucleoplasm by a diffusion-based mechanism. Measurements of nuclear RNAs labeled either with fluorescent probes [45,46 and 47] or with specific GFP-fused protein tags [48••and 49] have yielded a range of diffusion coefficients. Single-particle tracking of messenger ribonucleoprotein particles (mRNPs) in the nuclei of living cells allowed the detection of random movements [48••]. This motion was energy-independent and was not directed, although corralled motion implied the presence of intra-nuclear structures hindering the movements; mRNPs could be seen ‘bouncing off’ nucleoli, suggesting the presence of chromatin domains inaccessible to mRNPs [50 and 51]. Saturation of the export machinery by RNA overexpression revealed a reticular network throughout the nucleus space [52], corroborating previous studies showing mRNA movement within inter-chromatin channels [46]. A simplistic view of chromatin domains separated from inter-chromatin channels has been complicated by the fact that large macromolecule complexes can move through condensed chromatin regions [53•]. Chromatin regions were completely accessible to fluorescent dextrans with sizes of 3–10 kDa, but 70-kDa dextrans could not penetrate highly condensed areas. The radius of gyration of these inert molecules was calculated and found to be 4, 6 or 10 nm, values which correspond to either spherical proteins with molecular weights of approximately 400, 1400 and 6600 kDa or to ellipsoid proteins of 55, 180 and 850 kDa, respectively. Therefore, large macromolecule complexes such as RNA polymerases could gain access to most chromatin areas, whereas larger complexes in the megadalton range, such as RNP complexes (~50 nm diameter [54]), would appear to be excluded.
Gene positioning and mRNA export
An efficient and rapid path of mRNA export could result if active genes were associated with nuclear pores. Although studies have indicated that the nuclear periphery is actually involved in gene silencing, a screen operated on yeast has shown that classes of highly active genes, for instance genes involved in glycolysis or protein biosynthesis, are preferentially associated with nucleoporins, karyopherins and nuclear-pore-associated proteins [55••]. Moreover, transcriptional induction of the GAL genes caused their relocation towards the nuclear envelope, suggesting a mechanism for ‘express shipping’ of highly required transcripts. In another study, an active gene region was tethered to the nuclear pore by export proteins, including the transportin Cse1 [ 56]. Live imaging of GFP-tagged lac repressor protein (LacI) fused to the C-terminus of Cse1 showed not only the preferential relocation of a lacO locus to the nuclear periphery, but the occasional penetration of these chromosomal domains into the cytoplasm. The proximity of genes to the pore might be different in mammalian cells where the nucleus is much larger and chromosome organization more complex. Inactive genes in mammalian cells have been found to be associated with heterochromatin and the nuclear periphery, whereas active genes were sometimes interiorly positioned [57]. Yet the nuclear distribution of a specific mRNP was not influenced by the relative position of the transcription site in comparison to the nuclear envelope, implying that transcriptional proximity to nuclear pores might have more subtle or indirect effects [48••]. Genes distally separated by large DNA regions on the same chromosome had a high probability of sharing the same nuclear transcription space, demonstrating a high degree of spatial linkage within chromosome territories [58•], and other studies have shown that genes adopt preferential locations during differentiation [ 59, 60 and 61]. An analysis of the travels of endogenous mRNAs with respect to their distance from the nuclear envelope may reveal whether gene positioning can influence mRNA export in mammalian cells.
Nucleo-cytoplasmic transit of mRNA
Distinct RNA transport mechanisms are responsible for the export of different species of nuclear RNAs (mRNAs, spliceosomal U snRNAs and tRNAs). This implies the existence of mechanisms that are able to distinguish between RNA species while in transit. For example, mRNAs and spliceosomal U snRNAs are both produced by RNA polymerase II transcription, yet are preferentially exported by different pathways. Interestingly, RNA length was found to be a parameter measured by the export machinery [62 and 63]. If shortened to <120 nt, mRNAs were exported by the U snRNA pathway, whereas U1 snRNA with a 300 nt insertion behaved like an mRNA. It remains to be defined whether the commitment to a certain pathway occurs during the formation of an mRNP at the transcription site or during its travels to the nuclear pore [64]. If the mRNP is indeed remodeled during transit, it would be of interest to see whether this occurs in a specific nuclear compartment.
The time necessary for transport of an mRNA molecule from its transcription site to the cytoplasm is still an open question due to the lack of suitable living cell systems for measuring the rates of transport of endogenous RNAs. The study of mRNA export in purified nuclei from Xenopus oocytes has yielded export times ranging from 10 min to 1 h. These biochemical studies depend on viral RNA systems or on microinjection of mRNAs that have been transcribed in vitro. The rabbit β-globin gene expressed under the control of a doxycycline-controlled inducible promoter allowed the kinetic analysis of mRNP assembly and transport following induction [ 65]. This approach yielded nuclear residency half-lives of 2.5–4.4 min, which obeyed first-order kinetics. Kinetic modeling of these data showed that the nuclear pool of mRNA built up slowly, whereas cytoplasmic accumulation was exponential. Differences in the nuclear dwell-times of the mRNAs were not observed if actin polymerization was inhibited or if cells were plated at low or high densities. The availability of new methods to label endogenous mRNAs [66] and the development of live cell systems for the study of mRNA transport, combined with kinetic modeling approaches, have been successfully applied to nuclear protein transport [ 67,68 and 69], and will provide new insights into the process of mRNA export.
Conclusions
mRNA transcription and export constitute a chain of molecular events offering many points of control. Although there are many combinations of promoter–transcription-factor associations, the downstream cellular responses cannot be explained by promoter firing alone. To understand the subtleties of these transcriptional pathways, single cell approaches are necessary. Recent developments in our ability to probe single cells in real time have yielded new information on the dynamics of gene expression. These studies will ultimately take us to the complex task of unraveling the dynamics of transcription within the live organism.
Acknowledgements
We thank Olivier Bensaude and Claire Dugast-Darzacq for critical reading of the manuscript. This work was supported by grants from the NIH EB 2060, DOE ER63056 and P20 EB4930 to RHS.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Cramer P. RNA polymerase II structure: from core to functional complexes. Curr Opin Genet Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 3.Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 5.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 6••.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. This work reveals the complexity of the transcriptional regulatory network and provides a genome-wide view of gene regulation in yeast. [DOI] [PubMed] [Google Scholar]
- 7••.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. The transcription network [4] is integrated with available expression data. The transcription factor expression patterns associated with different cellular treatments or with different phases of the cell cycle are used to define ‘active transcription networks’. This work shows the high dynamism and plasticity of this network. [DOI] [PubMed] [Google Scholar]
- 8.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 9.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 10.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen-receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 11.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 12.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 13.Levsky JM, Singer RH. Gene expression and the myth of the average cell. Trends Cell Biol. 2003;13:4–6. doi: 10.1016/s0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 14.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 15.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 17.Becker M, Baumann C, John S, Walker DA, Vigneron M, McNally JG, Hager GL. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenoien DL, Nye AC, Mancini MG, Patel K, Dutertre M, O’Malley BW, Smith CL, Belmont AS, Mancini MA. Ligand-mediated assembly and real-time cellular dynamics of estrogen-receptor-α-coactivator complexes in living cells. Mol Cell Biol. 2001;21:4404–4412. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 20.Blake WJ, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. M KA. [DOI] [PubMed] [Google Scholar]
- 21.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 24.Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 27.Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 28•.Weber A, Liu J, Collins I, Levens D. TFIIH operates through an expanded proximal promoter to fine-tune c-myc expression. Mol Cell Biol. 2005;25:147–161. doi: 10.1128/MCB.25.1.147-161.2005. Transcriptional variation observed at the single-cell level does not lead to protein variations in cell cultures. It has been proposed that the stochastic initiation events of transcription onset are balanced by the regulations that apply to post-initiation events in the chain of gene expression [21 and 22]. This paper illustrates this theory by giving an example where post-initiation transcriptional regulation of the c-Myc maintains constant c-Myc protein content. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 30.Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci USA. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 32.Shermoen AW, O’Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 34.Shav-Tal Y, Singer RH, Darzacq X. Imaging gene expression in single living cells. Nat Rev Mol Cell Biol. 2004;5:855–861. doi: 10.1038/nrm1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phair RD, Misteli T. Kinetic modelling approaches to in vivo imaging. Nat Rev Mol Cell Biol. 2001;2:898–907. doi: 10.1038/35103000. [DOI] [PubMed] [Google Scholar]
- 36••.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. This study breaks new ground in its approaches to study transcription in live cells. The recruitment of the enzyme to the gene is correlated with its activity using mathematical kinetic modeling (see also [35]) [DOI] [PubMed] [Google Scholar]
- 37.Sugaya K, Vigneron M, Cook PR. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J Cell Sci. 2000;113:2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- 38••.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. The nuclear association of the RNA pol II with chromatin is addressed by FRAP and iFRAP and the results are discussed in light of the transcriptional activity of this enzyme. This work lays the groundwork for the kinetic analysis of pol II in live cells. (See also [37] for the establishment of the cellular system.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 40.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mais C, Wright JE, Prieto JL, Raggett SL, McStay B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005;19:50–64. doi: 10.1101/gad.310705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 43.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. Tandem gene arrays [17, 39, 40 and 41] are combined with technologies enabling live cell detection of mRNA [42 and 43] to reveal the early events leading to transcriptional activation at a single locus. The recruitment of chromatin remodeling factors, chromatin decondensation factors, transcription factors and mRNA processing factors is analyzed in real time following gene activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Politz JC, Browne ES, Wolf DE, Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc Natl Acad Sci USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr Biol. 1999;9:285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 47.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. Single mRNPs transcribed from a specific gene are tracked in the nucleoplasm of living mammalian cells and are found to move by a diffusion-based mechanism. Diffusion coefficients for these mRNPs are then measured by different techniques. This study shows that energy is not required for mRNP movement per se, but is essential for the integrity of the chromatin structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calapez A, Pereira HM, Calado A, Braga J, Rino J, Carvalho C, Tavanez JP, Wahle E, Rosa AC, Carmo-Fonseca M. The intranuclear mobility of messenger RNA binding proteins is ATP-dependent and temperature-sensitive. J Cell Biol. 2002;159:795–805. doi: 10.1083/jcb.200203046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zachar Z, Kramer J, Mims IP, Bingham PM. Evidence for channeled diffusion of pre-mRNAs during nuclear RNA transport in metazoans. J Cell Biol. 1993;121:729–742. doi: 10.1083/jcb.121.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 52.Bridger JM, Kalla C, Wodrich H, Weitz S, King JA, Khazaie K, Krausslich HG, Lichter P. Nuclear RNAs confined to a reticular compartment between chromosome territories. Exp Cell Res. 2005;302:180–193. doi: 10.1016/j.yexcr.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 53•.Verschure PJ, van der Kraan I, Manders EM, Hoogstraten D, Houtsmuller AB, van Driel R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003;4:861–866. doi: 10.1038/sj.embor.embor922. Using fluorescent dextrans of various molecular sizes, this work shows that condensed chromatin domains are accessible to large macromolecules, implying that the nuclear machinery can freely enter highly ordered regions of the genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spann P, Feinerman M, Sperling J, Sperling R. Isolation and visualization of large compact ribonucleoprotein particles of specific nuclear RNAs. Proc Natl Acad Sci USA. 1989;86:466–470. doi: 10.1073/pnas.86.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55••.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. Using genome location analysis, all genes associated with nuclear pore proteins are analyzed, revealing preferential binding of transcriptionally active genes. Some of the genes tested can reposition next to the nuclear periphery upon transcriptional activation. [DOI] [PubMed] [Google Scholar]
- 56.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 57.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. Several active genes distributed along one chromosome (in a 40-Mb-long region) are found by 3D FISH to have a high frequency of colocalization in the same transcription factories, indicating that genes are recruited to active domains of transcription. [DOI] [PubMed] [Google Scholar]
- 59.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 60.Kim SH, McQueen PG, Lichtman MK, Shevach EM, Parada LA, Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet Genome Res. 2004;105:292–301. doi: 10.1159/000078201. [DOI] [PubMed] [Google Scholar]
- 61.Delaire S, Huang YH, Chan SW, Robey EA. Dynamic repositioning of CD4 and CD8 genes during T cell development. J Exp Med. 2004;200:1427–1435. doi: 10.1084/jem.20041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno M, Segref A, Kuersten S, Mattaj IW. Identity elements used in export of mRNAs. Mol Cell. 2002;9:659–671. doi: 10.1016/s1097-2765(02)00454-9. [DOI] [PubMed] [Google Scholar]
- 63.Masuyama K, Taniguchi I, Kataoka N, Ohno M. RNA length defines RNA export pathway. Genes Dev. 2004;18:2074–2085. doi: 10.1101/gad.1216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 65.Audibert A, Weil D, Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol Cell Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kehlenbach RH. In vitro analysis of nuclear mRNA export using molecular beacons for target detection. Nucleic Acids Res. 2003;31:e64. doi: 10.1093/nar/gng063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG. Systems analysis of Ran transport. Science. 2002;295:488–491. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- 68.Gorlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 2003;22:1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kustanovich T, Rabin Y. Metastable network model of protein transport through nuclear pores. Biophys J. 2004;86:2008–2016. doi: 10.1016/S0006-3495(04)74262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]