Abstract
Background
The high mortality rate of hepatocellular carcinoma (HCC) is partly due to a lack of good diagnostic markers and treatment strategies. Recently, several microRNA (miRNA) profiling studies were conducted with HCC; however, their inconsistency means that their diagnostic or therapeutic value is debatable.
Aims
This study aims to systematically evaluate the consistency of miRNAs from multiple independent studies.
Methods
A systematic analysis of miRNAs from eligible publications was conducted, followed by real-time PCRs. The targets of highly consistent miRNAs were collected using online programs, followed by enrichment analyses for gene ontology terms and Kyoto encyclopedia of genes and genomes pathways.
Results
In total, 241 differentially expressed miRNAs were reported in 13 HCC profiling studies, of which 137 were upregulated and 104 downregulated. Among consistently upregulated miRNAs (cutoff > fourfold), miRNA-222, miRNA-21, miRNA-221, miRNA-210, and miRNA-224 were found increased in 8, 6, 6, 5, and 5 different studies, respectively. Among 137 downregulated miRNAs, miRNA-195, miRNA-199a, miRNA-125b, and miRNA-99a were reported in 8, 8, 5, and 5 studies, respectively. These results were confirmed by real-time PCR. Enrichment analyses demonstrated that programmed cell death and proliferation play important roles during the interplay of miRNA with HCC.
Conclusions
miRNAs most consistently related to HCC are oncomirs miRNA-221/222 and tumor suppressors miRNA-199a/195.
Electronic supplementary material
The online version of this article (doi:10.1007/s10620-016-4156-8) contains supplementary material, which is available to authorized users.
Keywords: Hepatocellular carcinoma, miRNA, Systematic study, Differential expression
Introduction
Hepatocellular carcinoma (HCC), cancer with an extremely poor prognosis, represents the fifth most commonly diagnosed cancer category and the third most common cause of cancer-related mortality worldwide. There are over 50 million cases of HCC every year in the world, and incidence of HCC in the USA has almost doubled or even tripled in recent decades [1–3]. Particularly because of the very poor prognosis and high fatality, the increasing incidence and high mortality rates of this debilitating cancer are almost equal worldwide. Indeed, HCC has become one of the major public health concerns in the world. Research on the etiology and pathogenesis of HCC has so far revealed that HCC usually occurs in the setting of underlying liver diseases; for example, hepatitis B and C are the major causes of HCC [3]. Additionally, genetic disorders like hemochromatosis and α1-anti-trypsin deficiency [4] and environmental factors such as alcohol abuse may increase the risk of HCC [5]. Despite some very promising advances in the understanding of HCC in recent years, the early diagnosis and treatment of HCC in the clinic is still quite challenging. The high mortality rate is largely due to its detection only at late stage with limited therapeutic options, which renders the disease a major challenge. MicroRNAs (miRNAs) regulate the expression of up to 30 % of protein-encoding genes by binding to specific mRNA targets and promoting their degradation and/or translational inhibition [6]. miRNAs play important roles in essential cellular processes including development, cell differentiation, inflammation, proliferation, and apoptosis [7]. miRNAs also play important roles in tumorigenesis [8], functioning as tumor suppressors or oncogenes (oncomirs), evidenced by the miRNA deregulation observed in various cancer types, including lung cancer [9]. Thus, miRNAs are potential biomarkers and intervention targets against various forms of cancer [10, 11]. Several studies have been conducted to search for biomarkers or therapeutic agents by identifying the differentially expressed miRNAs between hepatocellular carcinoma tissue samples and corresponding non-tumor liver tissue [12–24]. These studies generated hundreds of differentially expressed miRNAs; however, many of these are likely false positive. Additionally, only a small number of these may have clinical significance and act as diagnostic and prognostic biomarkers. Further, in miRNA-based therapeutics, miRNAs are tissue specific and their role depends on cellular context. For instance, overexpression of miRNA-143 in pancreatic xenografts can reduce tumor growth, but anti-miRNA-143 is required to suppress liver tumor formation [25]. It is therefore crucial to select both liver tissue-specific and consistently differentially expressed miRNAs to investigate their potential as biomarkers or therapeutic agents. Thus, it is very imperative to find the important and consistent miRNAs from a large number of candidate miRNAs in many individual and independent studies [26], and no HCC-related miRNAs have yet been identified as highly consistently and differentially expressed miRNAs in HCC from many independent miRNA profiling studies. Abnormal activation and inactivation of oncogenes and tumor suppressor genes are important factors leading to malignancy, including HCC. Clarifying the molecular mechanisms of HCC could provide a basis for HCC risk assessment, early diagnosis, effective treatment, and intervention.
In the present study, we conducted a systematic analysis to screen the most consistently and differentially expressed miRNAs from a number of miRNA profiling studies in HCC patients. Further, we confirmed the expression of most up- or downregulated miRNAs by quantitative PCR in 6 pairs of HCC and non-tumor liver tissue samples.
Materials and Methods
Search Strategy
Candidate profiling studies of miRNAs published in English were collected from PubMed, EMBASE, Web of Science, using a publication date range from April 2006 to July 2014 (last accessed on January 15, 2015) and the following medical subject heading terms: “miRNA” or “microRNA” or “miR,” “hepatocellular carcinoma” or “liver cancer” or “HCC,” “profiling” or “microarray.”
Inclusion Criteria of the Literature
The inclusion criteria of the literature were based on previously published studies, mainly including miRNA profiling studies (miRNA microarray methods) in HCC patients’ tissues and their corresponding adjacent non-tumor tissues for comparison, and the validation method set. Therefore, miRNA profiling studies that used serum samples of HCC patients or liver-relevant cell lines, those that compared HCC biopsies from tumors with different stages of disease, or those using different miRNA techniques were excluded. Review articles were also excluded. The final flowchart was made based on the guidance of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [27].
Data Extraction
Two investigators (L.Y. and Y.C.) independently evaluated and extracted the data using standard protocols, and all discrepancies were resolved by the corresponding investigator (Y.Y.). From the full text and corresponding supplementary information, the following eligibility items were collected and recorded for each study: author, journal and year of publication, location of study, methods and characteristics of animal modeling, platform of miRNA expression profiling, and author defined cutoff criteria of statistically significant differentially expressed miRNAs.
Ranking
All the included microarray investigations [12–24] reported lists of differentially expressed miRNAs related to HCC (Table S1). For example, Murakami et al. [12] carried out comprehensive analysis of HCC-related miRNAs based on a large number of miRNA profiling studies. The potential biomarkers for comparison groups were ranked by a method described previously [52], which has been used for profiling such as meta-analysis studies. Briefly, miRNAs were ranked to the criteria in the order of importance, such as the consistency, difference, and frequency of target miRNAs in included studies and their total sample size.
Validation of miRNAs Using Real-Time PCR
To confirm the profiling results, 31 fresh HCC tissue samples and their paired non-tumor liver samples were obtained from the Second Hospital of Hangzhou City, affiliated to Hangzhou Normal University School of Medicine. These patients developed HCC as a consequence of infection by hepatitis B virus (HBV) and were at early (A) or intermediate stages (B) as categorized by the Barcelona Clinic Liver Cancer (BCLC) staging system.
The diagnosis of HCC was based on clinical and histological criteria. Clinical data for HCC patients were obtained by medical record review. Collection of samples from patients was approved by the Institutional Review Board of Hangzhou Normal University and performed after written informed consent was obtained. Total RNA was extracted from 6 pairs of matched human HCC specimens (including cancer and adjacent non-cancerous tissues) using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The differentially expressed amount of the miRNAs was validated in triplicate by real-time PCR. Briefly, reverse transcription from 3 µg RNA was carried out using SuperScript III First-Strand Synthesis System (Life Technologies) according to the manufacturer’s protocol. Real-time PCR was performed using iQ SYBR Green SuperMix Kit (Bio-Rad, Hercules, CA, USA) with the iCycler sequence detection system (Bio-Rad) with specific primers (Table 1). Small nucleolar RNA U6 was used as an internal standard for normalization. The cycle threshold (CT) value was calculated. The (ΔCT = CTmiR − CTU6 RNA) method was used to quantify the relative amount of miRNAs.
Table 1.
Characteristics of miRNA datasets in hepatocellular carcinoma
| Author | Year | Region | Platform | No. of tissues | Differently expressed miRNAs | Criteria | Upregulated miRNAs in HCC | Downregulated miRNAs in HCC |
|---|---|---|---|---|---|---|---|---|
| Han | 2014 | China | GPL10122 Platform | 19 (9/10) | 32 | FDR < 0.05 Log FC > 1 |
14 | 18 |
| Peveling-Oberhag | 2014 | Germany | Affymetrix GeneChip | 12 (6/6) | 8 | p < 0.05 | 8 | |
| He | 2014 | China | Agilent-021 | 8 (6/2) | 40 | FDR < 0.05 Log FC > 1 |
4 | 36 |
| Li | 2014 | China | Microcirculation pump | 6 (3/3) | 10 | ? | 4 | 6 |
| Karakatsanis | 2013 | Greece | Exiqon | 158 (60/98) | 9 | p < 0.05 | 5 | 4 |
| Alpini | 2011 | USA | SBI | 38 (19/19) | 6 | p < 0.05 | 4 | 2 |
| Yang | 2010 | China | miRCURYTM LNA Array | 13 (8/5) | 14 | p < 0.01 | 6 | 8 |
| Chung | 2009 | Korea | ? | 50 (25/25) | 24 | p < 0.05 | 17 | 7 |
| Huang | 2009 | China | mirMASA technique | 40 (20/20) | 31 | p < 0.05 | 12 | 19 |
| Pineau | 2009 | USA | Exigon | 40 (20/20) | 12 | p < 0.01 | 11 | 1 |
| Su | 2009 | China | Agilent | 8 (5/3) | 29 | p < 0.05 | 14 | 15 |
| Meng | 2007 | USA | GenePix 4000A array | 6 (3/3) | 15 | p < 0.05 | 9 | 6 |
| Murakami | 2006 | USA | Agilent | 46 (24/22) | 8 | p < 0.05 | 3 | 5 |
Identify the Experimentally Validated miRNA Target Genes
miRTarBase (mirtarbase.mbc.nctu.edu.tw), microRNA.org, and TargetScanHuman 6.2 were used to explore all the target genes of miRNAs and highlighted the genes that are experimentally validated by luciferase reporter assay, Western blot, or microarray experiments with overexpression or knockdown of miRNAs.
Enrichment Analysis
Enrichment analyses for gene ontology (GO) terms and KEGG pathways were carried out with Database for Annotation, Visualization, and Integrated Discovery (DAVID) [53]. For this, we listed all the target genes of all the differentially expressed miRNAs, including upregulated and downregulated miRNAs. The top 17 or 18 GO terms and KEGG pathways showing association with target genes were listed with GO terms, KEGG pathway, and number of genes in the GO term, number of genes in the KEGG pathways.
Statistical Analysis
Student’s t test was used to compare values between two independent groups.
Results
Included Independent Studies
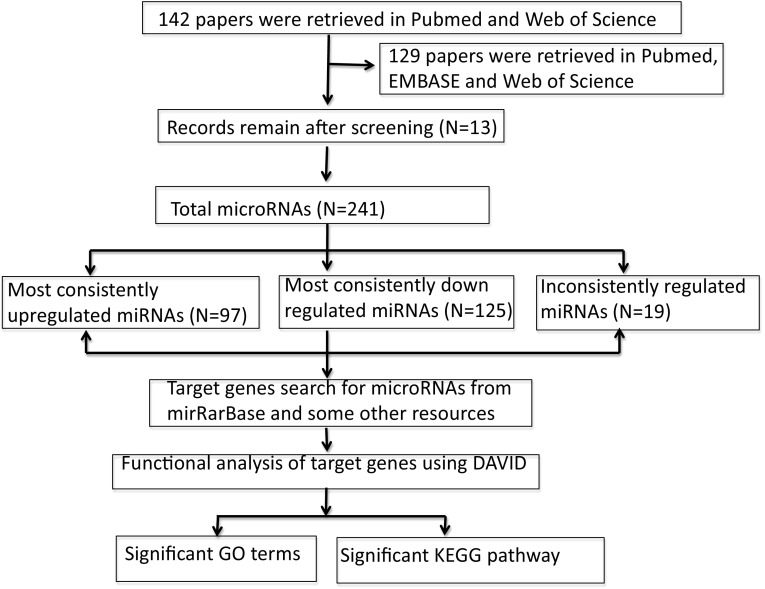
A total of 142 studies were recorded using PubMed, EMBASE, and Web of Science, 78 of which were excluded after screening the titles and abstracts and further 51 studies were excluded after reading the full text based on the inclusion and exclusion criteria. Only 13 independent studies (only the downregulated miRNAs were shown in study conducted by Peveling-Oberhag group [24]) were included in this systematic analysis. The detailed workflow used in our analysis is shown in Fig. 1, which is adapted and modified from PRISMA guidance [27]. The detailed characteristics of each study are given in Table 1 in descending order of year of publication.
Fig. 1.
Flowchart for this systematic analysis
Differentially Expressed miRNAs
These 13 studies from 13 different groups employed different platforms, different numbers of samples, and various statistical analyses for miRNA profiling analysis to compare HCC tissue with corresponding non-cancerous liver tissue. The number of differentially expressed miRNAs ranged from 6 to 38. A total of 239 differentially expressed miRNAs were reported in the 13 profiling studies (differentially expressed miRNAs in each study are detailed in supplemental data S1, S2, and S3). In total, 135 miRNAs are downregulated in HCC, and 104 miRNAs are upregulated. Among the 135 downregulated miRNAs, two were reported in 8 microarray studies (miRNA-195 and miRNA-199a), miRNA-125b and miRNA-99a were reported in five studies, and four miRNAs were reported in four studies (miRNA-126, miRNA-145, miRNA-378, and miRNA-497). Among the 104 upregulated miRNAs, we identified 7 (36.5 %) that are reported to be affected in at least four times (miRNA-21, miRNA-25, miRNA-93, miRNA-210, miRNA-221, miRNA-222, miRNA-224), miRNA-222 was found increased in 8 different studies, two miRNAs were reported in 6 studies (miRNA-21 and miRNA-221), and miRNA-210 and miRNA-224 were reported to be upregulated in 5 studies. Interestingly, miRNA-222 and its paralog miRNA-221 were represented 14 times in our selected studies. Consistently expressed miRNAs, the corresponding microarray study, and the total number of tissue samples are shown in Table 2. We also noticed that the differential expression of these selected miRNAs was very inconsistent in our selected profiling studies. For example, in the groups of inconsistently reported miRNAs, miRNA-122 was reported downregulated in two studies but upregulated in one study; miRNA-130a was increased in one study but downregulated in 3 studies (Table 3), respectively.
Table 2.
Consistently reported up-/downregulated miRNAs in profiling studies (HCC tissue vs. normal)
| Upregulated miRNA | Frequency [references] | No of tissue samples (HCC patients/healthy) | Downregulated miRNA | Frequency [references] | No of tissue samples (HCC patients/healthy) |
|---|---|---|---|---|---|
| miRNA-222 | 8 [26–28, 32–37] | 407 (228/179) | miRNA-199a | 8 [25–31, 33, 38, 40] | 183 (97/86) |
| miRNA-221 | 6 [26, 28, 32, 34–36, 39] | 373 (209/164 | miRNA-195 | 8 [25, 27–31, 33, 37, 38, 40] | 184 (97/87) |
| miRNA-21 | 6 [26, 32, 34–37, 39] | 352 (198/154) | miRNA-125b | 5 [26–28, 34, 40] | 104 (53/51 |
| miRNA-210 | 5 [26, 27, 29–33] | 202 (145/57) | miRNA-99a | 5 [27, 33, 37, 39, 40] | 58 (31/27) |
| miRNA-224 | 5 [25, 27, 28, 32, 37] | 225 (121/104) |
Table 3.
Inconsistency of differentially expressed miRNAs
| miRNA | No. of in upregulated miRNA | No. of in downregulated miRNA |
|---|---|---|
| miRNA-122 | 1 | 2 |
| miRNA-130a | 1 | 3 |
| miRNA-146 | 1 | 2 |
| miRNA-27a | 1 | 2 |
| miRNA-376 | 1 | 2 |
| miRNA-520c | 1 | 1 |
| Let-7a | 1 | 1 |
Validation of the Selected miRNAs in HCC Patients
To validate the expression of the nine most consistently reported miRNAs (miRNA-199a, miRNA-195, miRNA-125b, miRNA-99a, miRNA-222, miRNA-221, miRNA-21, miRNA-210, and miRNA-224), the expression of these miRNAs in HCC biopsies and adjacent non-cancerous tissues was compared in 31 HCC patients using real-time PCR with specific primers (Table 4). The results showed that the expression of miRNA-199a, miRNA-195, miRNA-125b, and miRNA-99a was downregulated in HCC tissues, whereas the other five miRNAs (miRNA-222, miRNA-221, miRNA-21, miRNA-210, and miRNA-224) were upregulated significantly in HCC patients (Fig. 2), which were consistent with those of the corresponding original profiling studies.
Table 4.
Primers used for real-time PCR
| Primers | Sequence |
|---|---|
| miRNA-222 | For 5′-CGCAGCTACATCTGGCTACTG-3′ Rev 5′-GTGCAGGGTCCGAGGT-3′ |
| miRNA-224 | For 5′-CACTAGTGGTTCCGTTTAGTAG-3′ Rev 5′-TTGTAGTCACTAGGGCACC-3′ |
| miRNA-21 | For 5′-GGGCTTTCAAGTCACTAGTG-3′ Rev 5′-GGGCTTTGTAGTCACTAGGGC-3′ |
| miRNA-125b | For 5′-TGCGCTCCTCTCAGTCCCTGAG-3′ Rev 5′-AGCACGACTCGCAGCTCCCA3′ |
| miRNA-22 | For 5′-TGAACATCCAGGTCTGGGGC-3′ Rev 5′GAGAACATGTTTCCAGGTAGCC-3′ |
| miRNA-99a | For 5′-CCCATTGGCATAAACCCG-3′ Rev 5′-CGGGTTTATGCCAATGGG-3′ |
| miRNA-199a | For 5′-AGGAAGCTTCTGGAGATCCTGC-3′ Rev 5′-TGCTCTCCCTTGCCCAGTCTAAC-3′ |
| miRNA-195 | For 5′-AGCTTCCCTGGCTCTAGCAG-3′ Rev 5′-CACCACCCTGCCTGGAGCAG-3′ |
| miRNA-210 | For 5′-ACCCGGCAGTGCCTCCAGGCG-3′ Rev 5′-GGGTCGCGCTGCCCAGGCAC-3′ |
| U6 | For 5′-GCGCGTCGTGAAGCGTTC-3′ Rev 5′-GTGCAGGGTCCGAGGT-3′ |
Fig. 2.
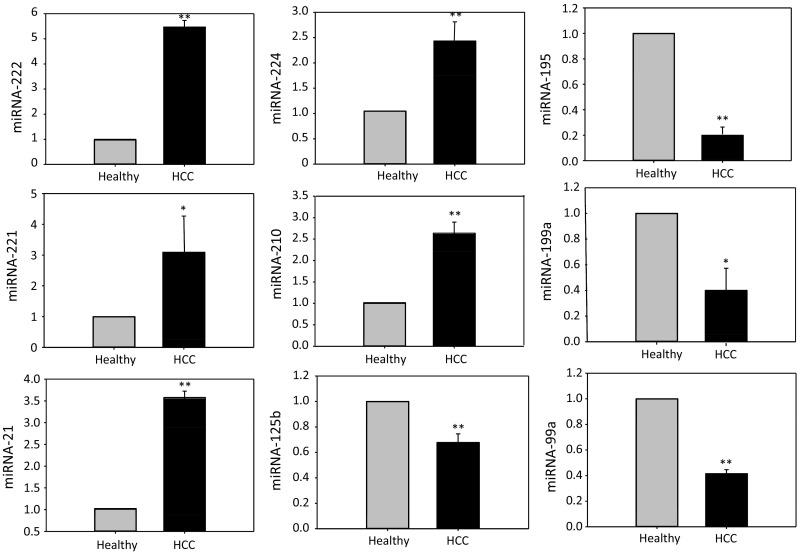
Real-time PCR analysis of the most consistent differentially expressed miRNAs. Expression levels of miR-222, miR-221, miR-21, miR-224, miR-210, miR-199a, miR-99a miRNA-125b, and miR-195 in HCC and adjacent non-cancerous tissue samples were analyzed by real-time PCR, and U6 here functions as an internal control. The expression of miR-222, miR-221, miR-21, miR-224, and miR-210 was significantly higher in HCC tissues, while the expression of miR-199a, miR-195, miR-99a, and miR-125b was significantly reduced. Here, values represent mean ± SE of triplicate of experiments. *p < 0.05; **p < 0.01
Primary miRNA Analysis
The identified and selected miRNAs all play specific roles in hepatocellular homeostasis, liver development, or pathogenesis of HCC. Particularly, all these miRNAs are either oncogenic miRNAs or tumor suppressors. Specifically, miRNA-222, miRNA-221, miRNA-21, miRNA-210, and miRNA-224 are oncomirs, and miRNA-199a, miRNA-195, miRNA-125b, miRNA-99a are tumor suppressors. These selected oncomirs are overexpressed consistently in the selected profiling studies; however, all the tumor suppressors are consistently inhibited in all the corresponding studies.
Target Genes of Differentially Expressed miRNAs
After we identified the most consistently downregulated or upregulated miRNAs in HCC, we screened their potential target genes with programs such as miRTarBase (mirtarbase.mbc.nctu.edu.tw), microRNA.org, and TargetScanHuman 6.2, focusing on the genes which were confirmed by either real-time PCR, Western blot, microarray, or luciferase assay. As a result, we identified 142 and 267 target genes (409 in total) corresponding to those downregulated and upregulated miRNAs in HCC (Table 5, supplemental data S1, S2, and S3), respectively.
Table 5.
Selected miRNAs and their predicated target genes
| MicroRNAs | Predicted targets |
|---|---|
| Upregulated miRNAs | |
| miRNA-222 | TRPS1, CERS2, RECK, DICER1, ETS1, DIRAS3, TCEAL1, CORO1A, TP53, SELE, PTEN, BBC3, ESR1, Kit, ICAM1, FOS Bcl2l11, Cdkn1b, TNFSF10, TIMP3, PPP2R2A, TMED7, Cdkn1c, FOXO3, MMP1, SOD2, STAT5A |
| miRNA-221 | CDKN1B, BCL2L11, BMF, FOXO3, Cdkn1c, kit, Psmd9, HOXB5, TMED7, DDIT4, BNIP3L, TBK1, MYBL1, DKK2, CREBZF, BRAP, USP18, ARIH2, BBC3, HMGXB4, TIMP3, TNFSF10, ICAM1, FOS, DDIT4, BNIP3, NAIP, ESR1, TICAM1, MDM2, PTEN, SELE, TP53, CORO1A, TCEAL1, DIRAS3, ETS1, DICER1, TRPS1, CERS2, SOX9, FN1 |
| miRNA-21 | BTG2, CDKAP1, PDCD4, LRRFIP1, CDC25A, PELI1, PDCD4, TP63, PTEN, HNRPK, TGFBR2, TGFBR2, SMARCA4, SPRY1/2, PRARA, TIMP3, RHOB, MSH2/6, ANP32A, BCL2, ANP32A, BMPR2, MARCK5, ANKRD46, RECK |
| miRNA-210 | TCF7L2, HOXA9, TP53I11, PIM1, HOXA1, FGFRL1, RAD52, NPTX1, EFNA3, BDNF, PTPN1, P4HB, UBQLN1, SERTAD2, SEH1L, NCAM1, MID1IP1, MDGA1, KIAA1161, ISCU, HOXA3, GPD1L, DENND6A, CPEB2, CDK10, ABCB9, CBX1, XIST, TNPO1, SMCHD1, PTAR1, NIPBL, MIB1, HECTD1, ELK3, DDAH1, CLASP2, CHD9, ATP11C, APC, E2F3, ACVR1B, MRE11A, XPA, UCP2, MNT, AIFM3, CASP8AP2, VMP1, TFRC, NFKB1, BCL2 |
| miRNA-224 | KLK10, CXCR4, CDC4, AP2M1, NIT1, FOSB, NCOA6, API5, EYA4, EDNRA, DIO1, SMAD4, PEBP1 |
| Downregulated miRNAs | |
| miRNA-99a | RAVER2, FGFR3, SERPINE1, IGF1R, mTOR, SNF2H |
| miRNA-199a | SIRT1, HIF1A, MET, KRT7, CD44, EZH2, IKBKB, CCNL1, MAPK9, MAPK1, AKT1, MAPK8, MAPK14, LIF, DDR1, EDN1, MAP3K11, HIF1A, SOX9, FUT4, UBE2G1, UBE2I, SMARCA2, SMAD1, RUNX1, COX2, CAV2, TMEM54, SMAD4, SULT1E1, GPR78, ERBB2, UNG, CAV1, ET1 |
| miRNA-195 | WEE1, E2F3, CDK6, CCND1, ARL2, CCND1, VEGFA, CCL4, KRT7, BCL2, RAF1, RUNX2, SLC2A3, TBCCD1, CCND3, CDK4, SIRT1, BACE1, CDC42, CAB39, CHUK, TAB 3, MBD1, CCNE1, BCL2L2, ARL2 |
| miRNA-125b | BMPR1B, EIF4EBP1, HMGA2, HMGA1, GLI1, NKIRAS2, TP53, SMO, VDR, SGPL1, BAK1, ERBB3, ERBB2, BMF, KLF13, NTRK3, LIN28A, CBFB, AKT1, CYP24A1, RAF1, SMO, PRDM1, IRF4, GRIN2A, CDKN2A, LIN28A, MAP2K7, JUB, KRT7, TNF, TP53INP1, E2F3, TRIM71, IGF2, LIN28B, BAK1, BBC3, BMF, KLF13, TEF, STAT3, BAK1, JUN, JUND, PPP1CA, PPKRA, PRKRA, PPP1CAB, SRF, NKX2-5, PRPF8, BCL2, ETS1, RPS6KA1, TNFAIP3, PIGF, BCL3, TBC1D1, DGAT1, FGFR2, SUV39H1, ARID3B, SMAD4, MCL1, IL6R, STARD13, ABTB1, CBFB, HK2, MMP13, SNAI1, MAPK14, MUC1, NES |
Enrichment Analysis Result
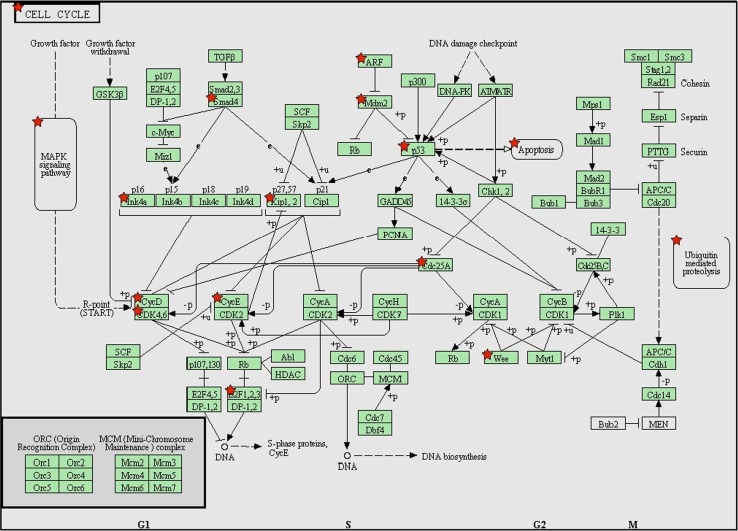
We used the DAVID program to build up the molecular networks corresponding to the target genes of downregulated and upregulated miRNAs. The top 10 of 428 GO terms for upregulated miRNAs are shown in Table 6 (p < 10E−9, Benjamini < 1.0E−6 false discovery rate (FDR) < 1.0E−5). Predicted target genes are mainly involved in cancer-related cellular processes including programmed cell death and proliferation probably through regulating cell cycle and cell differentiation. The top 10 of 670 GO terms for downregulated miRNAs are listed in Table 7 (p < 8E−15, Benjamini < 1.0E−11, FDR < 1.0E−10); predicted target genes are also involved in cancer-related cellular process including apoptosis, proliferation, cellular biosynthetic process, and metabolic process. Table 8 (p < 1.0E−16, Benjamini < 1.0E−13, FDR < 1.0E−13) shows the top 10 of 818 GO terms for combined target genes of all upregulated or downregulated differentially expressed miRNAs; similarly, predicted target genes are mainly involved in cancer-related cellular processes including apoptosis, proliferation, cellular biosynthesis, and metabolism. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways combined with different functional and signaling transduction pathways with number of target genes were analyzed, as shown in Table 9 (25 KEGG pathways are related to target genes of upregulated miRNAs), Table 10 (44 KEGG pathways for downregulated miRNAs), and Table 11 (46 KEGG pathways for all selected miRNAs). These KEGG pathways are very similar to, and largely overlap with, relevant signaling pathways and cellular functions, even though the target genes themselves may function quite differently. Among these, the cell cycle pathways seem unique and important for both cell proliferation and apoptosis as listed as example in Figs. 3, 4, and 5. Importantly, the KEGG pathways in cancer from total target genes, similar to the ones from upregulated or downregulated genes, were combined with various types of pathways related to tumorigenesis of cancers, like cell cycle (such as ARF, SMAD4, p53; Fig. 5), adherens junction (such as SMAD4, CDC42, and PTPN1; Supplemental data S4), cytokine–cytokine receptor interaction (such as ERBB2, IGFR, and Met; Supplemental data S5), and some signaling pathways like mTOR (such as mTOR, IKK, and Bcl-2; Supplemental data S6), Toll-like receptor (AP-1, Erk, NF-κB; Supplemental data S7), MAPK (such as Raf, Erk, and c-Fos; Supplemental data S8), p53 (such as p53, p14, and MDM2; Supplemental data S9), apoptosis (Trail, TNF, BCL-2; Supplemental data S10), VEGF signaling pathways (such as VEGF, TGF, and HIF; Supplemental data S11), TGF-β (such as TGF-βR1, TGF-βR11, and SMAD4; Supplemental data S12), and WNT (APC, JNK, p53; Supplemental data S13). All these pathways work synergistically to evade apoptosis and increase proliferation.
Table 6.
Top 10 of 428 GO terms of target genes related to upregulated miRNAs
| Term | Gene count | p value | Benjamini | FDR |
|---|---|---|---|---|
| Regulation of programmed cell death | 33 | 6.1E−14 | 1.0E−10 | 1.0E−10 |
| Regulation of cell death | 33 | 6.8E−14 | 5.8E−11 | 1.1E−10 |
| Regulation of apoptosis | 32 | 3.0E−13 | 1.7E−10 | 5.1E−10 |
| Regulation of cell proliferation | 28 | 2.0E−10 | 8.6E−8 | 3.4E−7 |
| Programmed cell death | 24 | 9.7E−10 | 3.3E−7 | 1.2E−5 |
| Regulation of cell cycle | 18 | 2.0E−9 | 5.7E−7 | 1.2E−5 |
| Negative regulation of programmed cell death | 18 | 6.9E−9 | 1.7E−6 | 1.6E−5 |
| Negative regulation of cell death | 18 | 7.1E−9 | 1.5E−6 | 1.6E−5 |
| Induction of apoptosis | 17 | 9.2E−9 | 1.7E−6 | 2.7E−5 |
| Induction of programmed cell death | 17 | 9.6E−9 | 1.6E−6 | 3.0E−5 |
Table 7.
Top 10 of 670 GO terms of target genes related to downregulated miRNAs
| Term | Gene count | p value | Benjamini | FDR |
|---|---|---|---|---|
| Regulation of cell proliferation | 38 | 4.9E−19 | 1.0E−15 | 8.4E−16 |
| Positive regulation of macromolecule metabolic process | 38 | 8.1E−18 | 8.4E−15 | 1.4E−14 |
| Positive regulation of cell differentiation | 21 | 2.2E−15 | 1.5E−12 | 3.8E−12 |
| Positive regulation of cellular biosynthetic process | 32 | 2.3E−15 | 1.2E−12 | 4.0E−12 |
| Positive regulation of biosynthetic process | 32 | 3.4E−15 | 1.4E−12 | 5.9E−12 |
| Regulation of apoptosis | 34 | 3.9E−15 | 1.3E−12 | 6.7E−12 |
| Positive regulation of macromolecule biosynthetic process | 31 | 5.0E−15 | 1.6E−12 | 8.6E−12 |
| Regulation of programmed cell death | 34 | 5.2E−15 | 4.8E−12 | 9.0E−12 |
| Regulation of cell death | 34 | 5.7E−15 | 1.9E−11 | 1.0E−11 |
| Positive regulation of developmental process | 22 | 7.8E−15 | 3.6E−11 | 1.3E−11 |
Table 8.
Top 10 of 818 GO terms of target genes related to all selected differentially expressed miRNAs
| Term | Gene count | p value | Benjamini | FDR |
|---|---|---|---|---|
| Regulation of programmed cell death | 61 | 1.2E−24 | 3.1E−21 | 2.1E−21 |
| Regulation of cell death | 61 | 1.4E−24 | 1.9E−21 | 2.6E−21 |
| Regulation of apoptosis | 60 | 4.6E−24 | 3.9E−21 | 8.1E−21 |
| Regulation of cell proliferation | 59 | 9.9E−24 | 6.4E−21 | 1.7E−20 |
| Positive regulation of macromolecule metabolic process | 57 | 2.2E−20 | 1.1E−17 | 3.9E−17 |
| Positive regulation of developmental process | 33 | 9.4E−19 | 4.0E−16 | 1.7E−15 |
| Positive regulation of cell differentiation | 30 | 3.3E−18 | 1.2E−15 | 5.8E−15 |
| Positive regulation of cellular biosynthetic process | 46 | 1.9E−17 | 6.1E−15 | 3.3E−14 |
| Regulation of biosynthetic process | 47 | 6.3E−17 | 1.0E−14 | 6.4E−14 |
| Positive regulation of cell cycle | 33 | 1.6E−16 | 2.9E−14 | 2.0E−13 |
Table 9.
The 25 KEGG pathways of target genes related to upregulated miRNAs
| Term | Gene count | p value | Benjamini | FDR |
|---|---|---|---|---|
| Pathways in cancer | 19 | 2.9E−10 | 2.0E−8 | 3.0E−7 |
| Chronic myeloid leukemia | 9 | 3.6E−7 | 1.2E−5 | 3.7E−4 |
| Colorectal cancer | 8 | 1.1E−5 | 2.6E−4 | 1.1E−2 |
| Prostate cancer | 8 | 1.6E−5 | 2.8E−4 | 1.1E−2 |
Table 10.
The 44 KEGG pathways of target genes related to downregulated miRNAs
| Term | Gene count | p value | Benjamini | FDR |
|---|---|---|---|---|
| Pathways in cancer | 31 | 1.1E−17 | 1.1E−15 | 1.2E−14 |
| Focal adhesion | 17 | 1.4E−8 | 2.0E−7 | 1.6E−5 |
| Neurotrophin signaling pathway | 14 | 1.4E−8 | 1.8E−7 | 1.6E−5 |
| MAPK signaling pathway | 19 | 2.1E−8 | 2.1E−7 | 2.3E−5 |
| T cell receptor signaling pathway | 12 | 2.8E−7 | 2.0E−6 | 3.1E−4 |
| erbB signaling pathway | 11 | 3.3E−7 | 2.2E−6 | 3.6E−4 |
| mTOR signaling pathway | 9 | 5.5E−7 | 3.5E−6 | 6.2E−4 |
| Toll-like receptor signaling pathway | 11 | 1.3E−6 | 7.9E−6 | 1.5E−3 |
| NOD-like receptor signaling pathway | 9 | 2.2E−6 | 1.2E−5 | 2.5E−3 |
| p53 | 9 | 4.5E−6 | 2.2E−5 | 5.0E−3 |
Table 11.
The 46 KEGG pathways of target genes related to all selected differentially expressed miRNAs
| Term | Gene count | p value | Benjamini | FDR |
|---|---|---|---|---|
| Pathways in cancer | 44 | 5.1E−23 | 5.7E−21 | 5.7E−20 |
| Colorectal cancer | 17 | 2.9E−11 | 5.4E−10 | 3.3E−8 |
| Toll-like receptor signaling pathway | 16 | 4.8E−9 | 6.8E−8 | 5.5E−6 |
| MAPK signaling pathway | 24 | 1.7E−8 | 1.7E−7 | 1.9E−5 |
| Focal adhesion | 19 | 4.2E−7 | 3.6E−6 | 4.7E−4 |
| erbB signaling pathway | 13 | 4.3E−7 | 3.0E−6 | 4.9E−4 |
| T cell receptor signaling pathway | 14 | 6.9E−7 | 4.6E−6 | 7.9E−4 |
| NOD-like receptor signaling pathway | 11 | 9.5E−7 | 5.9E−6 | 1.1E−3 |
| mTOR signaling pathway | 10 | 1.8E−6 | 1.1E−5 | 2.1E−3 |
| Cell cycle | 14 | 3.7E−6 | 2.0E−5 | 4.2E−3 |
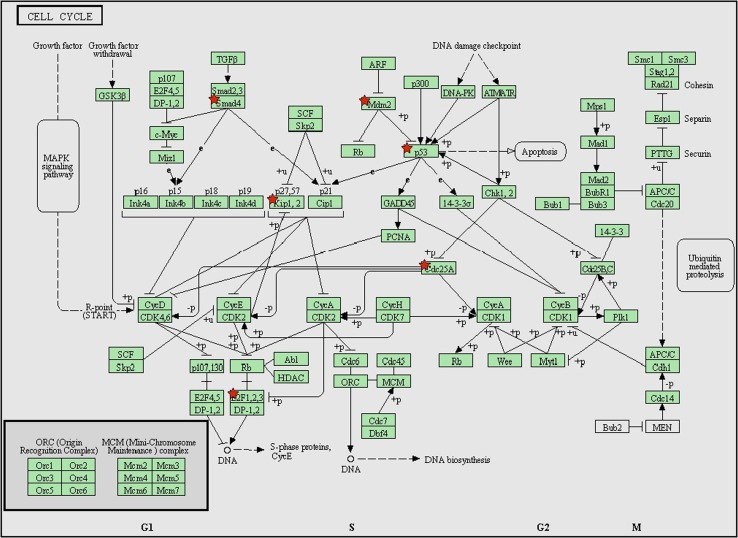
Fig. 3.
KEGG pathway (pathways in cancer) analysis with predicated target genes from upregulated miRNAs. These pathways in cancer are a combination of many different but relevant pathways, such as MAPK signaling pathway, mTOR signaling pathway, and apoptosis signaling pathway. Green boxes with red star represent the potential genes targeted by selected miRNAs
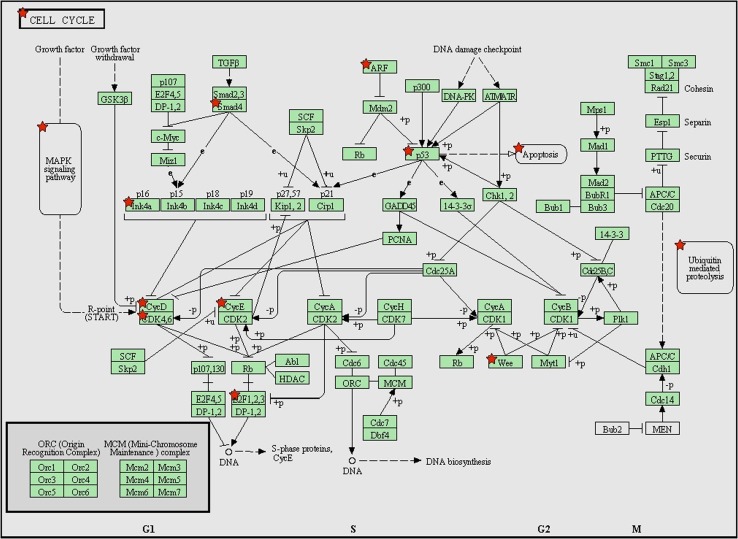
Fig. 4.
KEGG pathway (pathways in cancer) analysis with predicated target genes from downregulated miRNAs. These pathways in cancer are a combination of many different but relevant pathways, such as MAPK signaling pathway, mTOR signaling pathway, and apoptosis signaling pathway. Green boxes with red star represent the potential genes targeted by selected miRNAs
Fig. 5.
KEGG pathway (pathways in cancer) analysis with predicated target genes from all selected differentially expressed miRNAs. These pathways in cancer are a combination of many different but relevant pathways, such as MAPK signaling pathway, mTOR signaling pathway, and apoptosis signaling pathway. Green boxes with red star represent the potential genes targeted by selected miRNAs
Discussion
In the current study, we conducted a systematic analysis to establish the concept that only a few consistently differentially expressed miRNAs may be of significance for tumorigenesis and development of HCC. The potential targets of these selected miRNAs conduct the signaling pathways critical for programmed cell death and proliferation. Together, this research sheds some light on the potential of certain miRNAs as biomarkers or therapeutic target for HCC.
The usage of many conventional tumor biomarkers, such as ovarian cancer biomarker CA 125 and HCC biomarker alpha-fetoprotein (AFP), is limited due to their poor sensitivity and specificity. For example, only 50 % of patients with early-stage ovarian cancer have elevated levels of CA125, AFP with variety of sensitivity (20–90 %) for HCC [28]. Thus, there is a critical need for reliable biomarkers that allow high-sensitivity diagnosis and precise monitoring of the HCC pathogenic process and novel therapeutic alternatives to treat HCC. Many miRNAs are deregulated significantly even in early stages of tumorigenesis and metastasis of many cancer categories, exhibiting their diagnostic and therapeutic potential. However, as miRNA profiling studies have been performed on various platforms using diverse processing methods by investigators, consistency is lacking between these studies. This may result in inconsistent miRNA expression profiles [12–24].
miRNAs can act as both tumor suppressors and oncogenes and are important in the tumorigenesis and development of HCC. By systematic analysis, we found that five of the screened miRNAs (miRNA-221/222, miRNA-21, miRNA-224, miRNA-210) exhibit enhanced expression and act as oncomirs, and four of the screened miRNAs in 13 investigations (miRNA-199a, miRNA-99a, miRNA-195, miRNA-125b) decreased markedly, functioning as tumor suppressors. Oncomirs miRNA-221 and its paralog miRNA-222 are the most consistently expressed miRNAs in 13 profiling studies (14 times) and can aggravate the tumorigenesis of HCC by regulating cell growth, cell cycle, or apoptosis by directly targeting p27, p57, PUMA, Ki67, NF-κB, STAT3, CDKN1B, and CDKN1C [31]. Oncogenic miRNA-21 has been reported in a wide range of tumor types with mechanisms underlying the inhibition of apoptosis and the promotion of cell proliferation by targeting at PTEN, PDCD4, and TIMP3 [13, 32]. Particularly, miRNA-21 is upregulated during HCC migration and invasion [33], implying that it may be a novel marker for HCC progression and treatment. Besides being upregulated in HCC, hypoxia-inducible miRNA-210 can increase the metastatic potential of HCC cells by promoting entry into the cell cycle and inhibiting apoptosis [34] by acting on the cell cycle regulator E2F3, homeobox proteins (HOXA1, HOXA9), and the myc antagonist MNT [35, 36]. miRNA-224, another important oncomir, is upregulated and persists throughout HCC development, acting to inhibit apoptosis [37]. In hepatitis B virus (HBV)-related HCC, accumulation of miRNA-224 has a significant role in promoting cell proliferation, migration, and invasion by targeting SMAD4 [38].
miRNAs can also function as tumor suppressors in different cancer types by inhibiting the oncogenic process and preventing tumor development. In our systematic study, both miRNA-199a and 99a consistently decreased in 8 out of 13 studies. As a tumor suppressor, miRNA-199a negatively regulates oncogene c-Met, an important oncogene involved in HCC invasion and metastasis [39]. Overexpression of miRNA-199a decreases invasion and proliferation of HCC cell lines [40]. Additionally, miRNA-199a regulates the tumor suppressor mitogen-activated protein kinase kinase kinase 11 in gastric cancer [41]. miRNA-99a functions as another tumor suppressor, inhibits HCC growth, and correlates with prognosis of HCC patients [42]. Furthermore, miRNA-99a can inhibit proliferation of c-Src-transformed cells and prostate cancer cells by targeting mTOR [43]. miRNA-195 is downregulated frequently in multiple cancer types, including HCC, indicating its growth-suppressive function [44]. miRNA-195 has been shown to block the G1/S transition of the cell cycle by targeting CCND1/3, CDK4/6, and E2F3 [44, 45] and to promote apoptosis by suppressing the expression of BCL2 and BCL-w [46, 47]. miRNA-125b performs tumor suppressor functions in several cancers including hepatocellular carcinoma, by suppressing HCC cell proliferation and metastasis [48]. In our analysis, there are several other miRNAs that may contribute to a lesser extent to HCC development and progression, such as upregulated miRNA-93 and miRNA-95 or downregulated miRNA-145 and 497. Taking miRNA-145 as an example, the tumor suppressor miRNA-145, normally transcribed together with miRNA-143, regulates renewal and differentiation of cancer-related stem cells (CSCs) by negatively regulating the expression of certain key genes in stem cells such as CD44, Oct4, Sox2, and Klf4 [49].
miRNAs function as oncogenes or tumor suppressors by involving multiple molecular mechanisms. By enrichment analysis, we found these oncomirs and tumor suppressors can target various substrate genes. It is of great significance that these target genes cluster with cancer-related cellular process, including apoptosis and proliferation. This is in agreement with the fact that apoptosis and proliferation play checkpoint roles in tumorigenesis of most, if not all, cancers [50, 51], which further highlight the importance of these miRNAs as potential biomarkers or therapeutic alternatives. KEGG pathway analysis using DAVID demonstrated that the target genes of these miRNAs build up very meaningful networks, combined with different pathways and molecule–molecule interactions. For example, the cell cycle and p53 signaling pathways contribute to apoptosis inhibition and stimulate proliferation; the Wnt signaling pathway is vital for the integrity of endothelial cells and for HCC invasion and metastasis; the inhibition of mTOR signaling pathway can reduce hepatocyte apoptosis; and the MAPK signaling pathway is crucial for hepatocytes proliferation. All these pathways work together to evade apoptosis and increase proliferation. Several other pathways that related to the block of differentiation, resistance to chemotherapy, failed repair of genes, insensitivity to anti-growth signals are also implicated. The pathways related to these miRNAs and target genes exactly mimic the pathways involved in HCC tumorigenesis. This makes these miRNAs even more important as potential biomarkers and therapeutic candidates for clinical application. Extensive studies should focus these miRNAs to establish them as biomarkers for cancer detection and progression.
Limitations
Several limitations of this research should be considered when interpreting the results due to some unsolvable reasons. Firstly, our literature searching was based on English databases only, and as a result, language bias may present. Secondly, our study only included Chinese, Korean, American, German, Greece, and Japanese populations, so the result may not be applicable to other populations such as Latin American and African.
Conclusions
We performed a comprehensive literature search in multiple databases by limiting publication language, date, and key words. By systematic analysis, we identified the most consistently expressed upregulated (5) and downregulated (4) miRNAs, and further confirmed by real-time PCR, specially, highlight will be given to miRNA-221/222, miRNA-195, and miRNA-199a since they consistently exhibited differential expression (14 and 8 studies, respectively). By DAVID analysis, we showed that the target genes are mainly involved in the regulation of apoptosis and proliferation mediated by the combined action of several signaling pathways.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by Grants from the Natural Science Foundation of China (81570479) and the Climbing Program in Hangzhou Normal University-Phase II (PF14002004021).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest (financial or otherwise).
Footnotes
Yanhu Li and Chunhong Di have contributed equally to this work.
Contributor Information
Guoqiang Lou, Phone: (86) 571-88312277, Email: Louguoqiang2012@126.com.
Yutao Yan, Phone: (86) 571-28867302, Email: yutaoyan@yahoo.com.
References
- 1.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 2.Siegel AB, Olsen SK, Magun A, Brown RS., Jr Sorafenib: where do we go from here? Hepatology. 2010;52:360–369. doi: 10.1002/hep.23633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 5.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42:S206–S214. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Xie B, Chen S, Jiang F, Meng W. Association study of two inflammation-related polymorphisms with susceptibility to hepatocellular carcinoma: a meta-analysis. BMC Med Genet. 2014;15:92. doi: 10.1186/s12881-014-0092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothschild SI, Tschan MP, Federzoni EA, et al. MicroRNA-29b is involved in the Src-ID1 signaling pathway and is dysregulated in human lung adenocarcinoma. Oncogene. 2012;31:4221–4232. doi: 10.1038/onc.2011.578. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 13.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su H, Yang JR, Xu T, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 15.Huang XH, Wang Q, Chen JS, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 16.Chung GE, Yoon JH, Myung SJ, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23:113–119. doi: 10.3892/or_00000682. [DOI] [PubMed] [Google Scholar]
- 17.Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803–808. doi: 10.4161/cbt.9.10.11440. [DOI] [PubMed] [Google Scholar]
- 19.Alpini G, Glaser SS, Zhang JP, et al. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J Hepatol. 2011;55:1339–1345. doi: 10.1016/j.jhep.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog. 2013;52:297–303. doi: 10.1002/mc.21864. [DOI] [PubMed] [Google Scholar]
- 21.Han K, Li J, Zhao H, et al. Identification of the typical miRNAs and target genes in hepatocellular carcinoma. Mol Med Rep. 2014;10:229–235. doi: 10.3892/mmr.2014.2194. [DOI] [PubMed] [Google Scholar]
- 22.He TL, Zheng KL, Li G, Song B, Zhang YJ. Identification of typical miRNAs and target genes in hepatocellular carcinoma by DNA microarray technique. Eur Rev Med Pharmacol Sci. 2014;18:108–116. [PubMed] [Google Scholar]
- 23.Li T, Yin J, Yuan L, et al. Downregulation of microRNA-139 is associated with hepatocellular carcinoma risk and short-term survival. Oncol Rep. 2014;31:1699–1706. doi: 10.3892/or.2014.3032. [DOI] [PubMed] [Google Scholar]
- 24.Peveling-Oberhag J, Seiz A, Doring C, et al. MicroRNA profiling of laser-microdissected hepatocellular carcinoma reveals an oncogenic phenotype of the tumor capsule. Transl Oncol. 2014;7:672–680. doi: 10.1016/j.tranon.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 26.Wang JL, Hu Y, Kong X, et al. Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PloS One. 2013;8:e73683. doi: 10.1371/journal.pone.0073683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witjes CD, van Aalten SM, Steyerberg EW, et al. Recently introduced biomarkers for screening of hepatocellular carcinoma: a systematic review and meta-analysis. Hepatol Int. 2013;7:59–64. doi: 10.1007/s12072-012-9374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 31.Santhekadur PK, Das SK, Gredler R, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J Biol Chem. 2012;287:13952–13958. doi: 10.1074/jbc.M111.321646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 33.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ying Q, Liang L, Guo W, et al. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54:2064–2075. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]
- 35.Huang X, Ding L, Bennewith KL, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Sun H, Dai H, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 37.Chen PJ, Yeh SH, Liu WH, et al. Androgen pathway stimulates microRNA-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology. 2012;56:632–643. doi: 10.1002/hep.25695. [DOI] [PubMed] [Google Scholar]
- 38.Lan SH, Wu SY, Zuchini R, et al. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy. 2014;10:1687–1689. doi: 10.4161/auto.29959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornari F, Milazzo M, Chieco P, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 40.Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453–460. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Song G, Zeng H, Li J, et al. miR-199a regulates the tumor suppressor mitogen-activated protein kinase kinase kinase 11 in gastric cancer. Biol Pharm Bull. 2010;33:1822–1827. doi: 10.1248/bpb.33.1822. [DOI] [PubMed] [Google Scholar]
- 42.Li D, Liu X, Lin L, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. 2011;286:36677–36685. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oneyama C, Ikeda J, Okuzaki D, et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene. 2011;30:3489–3501. doi: 10.1038/onc.2011.63. [DOI] [PubMed] [Google Scholar]
- 44.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 45.Lin Y, Wu J, Chen H, et al. Cyclin-dependent kinase 4 is a novel target in microRNA-195-mediated cell cycle arrest in bladder cancer cells. FEBS Lett. 2012;586:442–447. doi: 10.1016/j.febslet.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 46.Liu L, Chen L, Xu Y, Li R, Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem Biophys Res Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 47.Yang X, Yin J, Yu J, et al. miRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol Rep. 2012;27:250–257. doi: 10.3892/or.2011.1472. [DOI] [PubMed] [Google Scholar]
- 48.Liang L, Wong CM, Ying Q, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52:1731–1740. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 49.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 50.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 51.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffith OL, Melck A, Jones SJ, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol. 2006;24:5043–5051. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- 53.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation. Vis Integr Discov Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.