Abstract
Autosomal Dominant Polycystic Kidney Disease (ADPKD) is a common genetic disorder characterized by bilateral renal cyst formation1. Recent identification of signaling cascades de-regulated in ADPKD has led to the initiation of several clinical trials, but an approved therapy is still lacking2,3. Using a metabolomic approach here we identify a pathogenic pathway in ADPKD which can be safely targeted for therapy. We show that mutation in PKD1 results in enhanced glycolysis in cells, in a murine model of PKD, and in human-derived ADPKD kidneys. Glucose deprivation reduced proliferation and sensitized PKD1 mutant cells to apoptosis. Notably, treatment of two distinct PKD mouse models with 2-deoxyglucose (2DG) ameliorates kidney volume, cystic index and reduced proliferation rates. These metabolic alterations depend on the ERK pathway acting in a dual manner by inhibiting the LKB1-AMPK axis on the one hand while activating the mTORC1-glycolytic cascade on the other. Enhanced metabolic rates further inhibit AMPK. Forced activation of AMPK acts in a negative feedback loop restoring normal ERK activity. Taken together, these data indicate that defective glucose metabolism is intimately involved in the pathobiology of ADPKD. Our findings provide a strong rationale for a novel therapeutic paradigm using existing drugs, either individually or in combination.
ADPKD is a chronic progressive disease1. Cysts originate from any segment of the renal tubule in only 1-5% of the nephrons, a condition that should be compatible with a normal renal function1,4. However, the gradual expansion of cysts compresses and eventually replaces the normal tissue, causing end-stage renal disease in a majority of affected individuals1,4. Thus, therapeutic interventions targeting cyst expansion is currently being tested in multiple clinical trials to delay renal disease progression2,5,3.
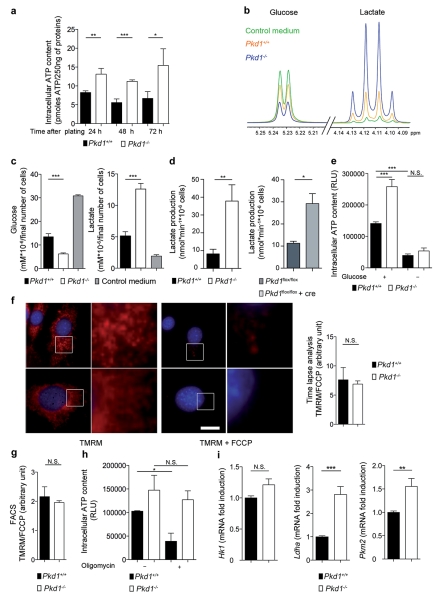
The disease is caused by loss-of-function mutations in either PKD1 or PKD21,2. To study alterations caused by defective PKD1 function we isolated Mouse embryonic fibroblasts (MEFs) from littermate Pkd1+/+ or Pkd1−/− embryos6, and using these cells we serendipitously identified a novel pathogenic process. During routine culture, we noticed that Pkd1−/− cells acidified the medium faster than the wild-type while the opposite was observed in cells over-expressing PKD1 (Supplementary Fig.1). This was also replicated in growth-arrested cells (100% density) suggesting an intrinsic, proliferation-independent metabolic increase in Pkd1−/− cells. Indeed, Pkd1−/− cells had much higher ATP content as compared to wild-type (Fig. 1a). To determine which metabolic pathways were altered in these cells, we performed a metabolomic profiling of the conditioned extracellular medium of wild-type and Pkd1−/− cells using NMR spectroscopy7,8 (Fig. 1b). An unsupervised statistical analysis revealed that the metabolomic profile of Pkd1−/− cells differed significantly from that of wild-type cells (Supplementary Fig.2 and Supplementary Table 1), the most prominent alteration being reduced glucose and increased lactate concentrations (Fig. 1c, d and Supplementary Fig. 2). These data suggest that Pkd1−/− cells use aerobic glycolysis as a source of energy. Indeed, glucose deprivation abrogated the increased ATP content of Pkd1−/− cells (Fig. 1e). Similar results were generated using a MEF cell line carrying conditional inactivation of a Pkd1 floxable allele upon treatment with a Cre (Fig. 1d and Supplementary Fig. 1). Since glucose metabolism is also the main source of energy through oxidative phosphorylation occurring in the mitochondria9, we analyzed the mitochondrial membrane potential in wild-type and Pkd1−/− cells using two independent assays and demonstrated no significant difference between the two cell lines (Fig. 1f, g). In line with this, treatment with oligomycin, an inhibitor of the mitochondrial ATP-synthase, decreased the ATP content in wild-type cells, but only had a minor effect in Pkd1−/− cells (Fig. 1h). These data suggest that an alternative metabolic pathway that is glucose-dependent is the source of the differential ATP content between the two cell lines. Previous studies demonstrated that transcriptional changes can regulate the glycolytic response in cells10,11. In line with this, real-time PCR analysis revealed that Pkd1−/− cells displayed a transcriptional signature of glycolytic enzymes10 (Fig. 1i). We conclude that Pkd1−/− cells preferentially use aerobic glycolysis, in a process similar to the Warburg’s effect observed in cancer12,9.
Figure 1. Metabonomics revealed increased aerobic glycolysis in Pkd1−/− MEFs.
a. ATP content in Pkd1−/− and Pkd1+/+ cells at the indicated times after plating. b. Overlay of 1H-NMR spectra corresponding to the glucose or lactate regions in the extracellular medium alone (control) or incubated in the presence of Pkd1+/+ or Pkd1−/− cells. c. Quantitative analysis of NMR spectra depict glucose and lactate concentrations in the medium derived from Pkd1−/− as compared to Pkd1+/+ cells. d. Quantification of lactate production using a commercial assay in Pkd1−/− cells as compared to Pkd1+/+ or in Cre-treated Pkd1flox/flox as compared to Pkd1flox/flox cells. e. Quantification of ATP content in Pkd1+/+ cells compared to Pkd1−/− cells after 48 h of glucose starvation. f-g. Mitochondrial membrane potential in Pkd1+/+ and Pkd1−/− labelled with the fluorescent dye TMRM (red) before and after treatment with FCCP (uncoupling the membrane potential) was measured by time-lapse microscopy (f) or FACS (g). h. Measurement of ATP content in Pkd1+/+ and Pkd1−/− MEFs treated in the presence of oligomycin for 5 h. i. Real time analysis of the genes coding Hk1, Pkm2 and Ldha was performed in Pkd1+/+ and Pkd1−/−MEFs. N.S.: P ≥0.05; *: P < 0.05; **: P < 0.01; ***: P < 0.001; Means +/− SD, except for i means +/− SEM. Data are representative of three independent experiments performed in triplicate. In f and i the average value of all three experiments is provided. T-test in a, c, d, f, g and i; ANOVA followed by Bonferroni’s test in e and h. Bar = 10 μm.
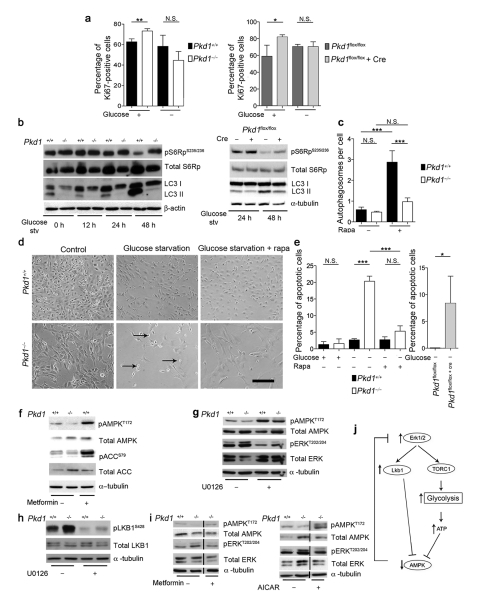
A defective balance between proliferation and apoptosis has been observed in ADPKD tissues and PKD1 mutant cells4,13,14. We thus tested if increased glucose metabolism contributes to de-regulation of this balance. Indeed, glucose deprivation restored the proliferation index of Pkd1−/− and Pkd1flox/flox:Cre cells similar to that of Pkd1+/+ or Pkd1flox/flox cells, respectively (Fig. 2a). In addition, while the Pkd1+/+ cells deprived of glucose activated cell autophagy to survive, Pkd1−/− cells failed to activate this response (Fig. 2b, c and Supplementary Fig. 3) but instead displayed increased apoptotic rates (Fig. 2d, e). Similar results were observed in the Pkd1flox/flox:Cre cells (Fig. 2b, c and Supplementary Fig. 1). Consistent with previous studies we also found that this effect is in part dependent on mTORC115,10. Treatment of Pkd1−/− cells with rapamycin partially restored autophagy (Fig. 2c), cell survival under glucose deprivation (Fig. 2e), and downregulated metabolic rates (Supplementary Fig. 4). Furthermore, we observed a mTORC1-dependent upregulation of HIF1-α in Pkd1−/− cells as compared to wild-type, likely responsible for the transcriptional changes observed (Supplementary Fig. 4)10,11.
Figure 2. Glucose-dependence, defective autophagy and altered AMPK and ERKs in Pkd1−/− cells.
a. Percentage of cells positive for Ki67 over total cells with or without 12 h glucose-starvation in Pkd1−/− (left) or Cre-treated Pkd1flox/flox cells (right). b. LC3-II western-blot upon glucose starvation for 12, 24 and 48 h in Pkd1+/+ and Pkd1−/− cells (left) or 48 h in Cre-treated Pkd1flox/flox cells (right). c. Quantification of the number of autophagosomes per cells evaluated by EM (Supplementary Fig. 3) in the presence or absence of rapamycin (50 nM). d. Cells were glucose-starved for 48 h in the presence or absence of rapamycin (20 nM) and bright field images captured. Arrows indicate dying cells. e. Quantification of apoptosis using the TUNEL assay after glucose starvation in Pkd1−/− and Pkd1+/+ cells (left) or Cre-treated Pkd1flox/flox MEFs (right). f. P-AMPK in Pkd1−/− cells. g. P-AMPK, P-ERK and P-S6RP in Pkd1−/− MEFs after 12 h in UO126 (30 μM). h. P-LKB1 in Pkd1−/− cells was decreased with 30 μM of UO126 (12 h). i-j. P-AMPK, P-ERK and P-S6RP in MEFs after treatment with 2 mM metformin (enhancing AMPK activity, left) or 2 mM AICAR (mimicking ADP, right) for 4 h showing ERK down-regulation in Pkd1−/− cells. k. Model of the molecular mechanism leading to increased aerobic glycolysis in ADPKD. N.S.: non significant; *: P < 0.05; **: P < 0.01; ***: P < 0.001; Means +/− SD are shown. T-test in a and e graph on the right; ANOVA followed by Bonferroni’s in a, c and e on the left. Graphs are representative of at least three independent experiments performed in triplicate. Bar = 200 μm.
Consistent with the high ATP content we also found reduced levels of AMPK phosphorylation in Pkd1−/− cells as compared to wild-type (Fig. 2f). In a previous study we have shown that the enhanced mTORC1 activity in Pkd1−/− cells is mostly driven by ERKs upregulation6. In line with this ERKs inhibitors restored the phosphoAMPK levels (Fig. 2g). Furthermore, a recent study reported that B-raf regulates AMPK activity via ERK-dependent phosphorylation of LKB116. We therefore tested if a similar mechanism might be involved here. Indeed, LKB1 was strongly phosphorylated at ERK-specific sites in Pkd1−/− cells compared to Pkd1+/+ and ERKs inhibitors reverted this (Fig. 2h). Thus, we propose a dual role for ERKs here: on the one hand they regulate LKB1 causing inhibition of AMPK16, on the other, they affect mTORC1 activity which in turn switches on aerobic glycolysis, increases ATP and further inhibits AMPK. Of great interest but unexpectedly, treatment of Pkd1−/− cells with metformin or AICAR (5-aminoimidazole-4-carboxamide ribonucleoside), both of which increased AMPK activity, also inhibited ERKs (Fig. 2i). These data suggest the existence of a negative feed-back loop whereby AMPK can down-regulate ERKs activity towards the basal conditions (Fig. 2j).
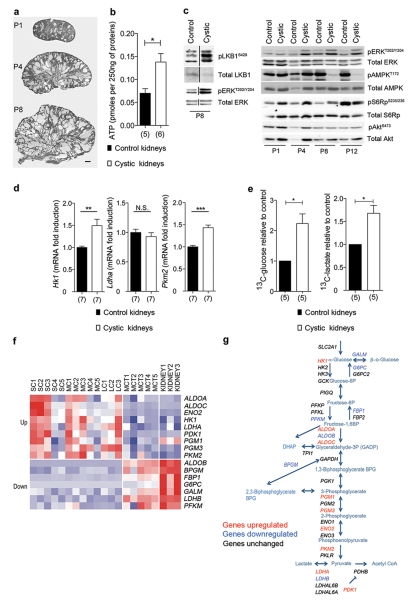
We next asked if the glycolytic switch observed in cells occurs in vivo, upon inactivation of the Pkd1 gene in the kidney. To test this, we used Ksp-Cre:Pkd1flox/− mice which develop early and severe PKD17 (Fig. 3a). Compared to non-cystic controls, we found that the cystic kidneys displayed higher ATP levels (Fig. 3b), biochemical alterations similar to those observed in vitro (Fig. 3c) and a transcriptional de-regulation of key glycolytic enzymes (Fig. 3d) consistent with a switch to glycolysis in vivo10. To experimentally test if enhanced glycolysis could be observed in these kidneys we injected subcutaneously uniformly-labelled 13C-glucose in Ksp-Cre:Pkd1flox/− or littermate controls (Ksp-Cre:Pkd1flox/+) and followed the levels of 13C-glucose or 13C-lactate using 13C-NMR spectroscopy (Fig. 3e). 40 minutes post-injection the cystic kidneys displayed a significantly higher uptake of 13C-glucose, which was more efficiently converted to 13C-lactate as compared to non-cystic control kidneys (Fig. 3e, n = 5). These data demonstrate that the cystic kidneys are characterized by aerobic glycolysis in vivo.
Figure 3. Defective Glycoslysis and ERKs/AMPK axis in vivo.
a. Representative images of Ksp-Cre:Pkd1−/flox kidneys at P1, P4 and P8. b. Measurement of ATP content in Ksp-Cre:Pkd1−/flox kidneys compared to control kidneys at P4. c. Right. Western blot analysis of pERK, pS6Rp and pAMPK in P1, P4, P8 and P12 Ksp-Cre:Pkd1flox/− kidneys. pAkt levels (S473) do not appear to change. Left. pLKB1 levels P8 in the cystic kidneys compared to control. d. Real time analysis of the genes coding for key glycolytic enzymes Hk1, Pkm2 and Ldha performed in KspCre:Pkd−/flox kidneys as compared to control kidneys (Ksp-Cre:Pkd1+/flox) at P4. e. The content of 13C-glucose in cystic kidneys (Ksp-Cre:Pkd1−/flox) was significantly higher compared to control non-cystic kidneys (Ksp-cre:Pkd1+/flox) (left, n = 5). Significantly increased levels of 13C-lactate (right, n = 5) could be detected in the cystic kidneys at P8. f. Panels showing genes coding for glycolytic and glucogenesis enzymes differentially expressed between the cysts and MCT samples. Up-regulated genes are shown in red, and down-regulated genes, in blue. SC, small cysts; MC, medium cysts; LC, large cysts; MCT, minimally cystic tissue; KIDNEY, normal renal cortical tissue. g. The scheme shows the glycolytic cascade, in red are indicated the genes up-regulated, in blue the ones down-regulated and in black the ones unchanged in cystic kidneys from ADPKD individuals compared to the normal kidneys. N.S.: P ≥0.05; *: P < 0.05; **: P < 0.01; ***: P < 0.001; Mean +/− SEM; the number of kidneys of three litters is indicated under the columns. Bar = 500 μm
We next asked if this is a general feature of the cystic kidneys from subjects with ADPKD. Thus, we examined the gene expression profile of both gluconeogenesis and glycolytic pathways using a previously established microarray database derived from PKD1 human renal cysts (compared to minimally cystic cortical tissues from the same kidneys as control)18. We found that many enzymes involved in gluconeogenesis and glycolysis were differentially expressed in the renal cysts. Detailed analysis revealed that most of the genes encoding enzymes involved in gluconeogenesis were down-regulated while several genes encoding enzymes involved in glycolysis were up-regulated in renal cysts as compared to minimally cystic or normal tissue (Fig. 3f, g and Supplementary Table 2). Overall, these data suggest that increased glucose consumption and enhanced glycolysis are likely features of human PKD1, as further supported by the presence of lactate in the ADPKD cyst fluid19.
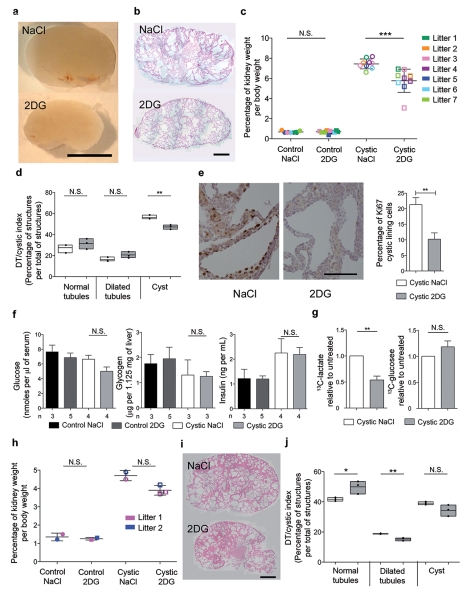
Based on the above data we hypothesized that interfering with glucose metabolism might present a novel strategy to retard cyst expansion. To test this, we treated the Ksp-Cre:Pkd1flox/− mice with a glucose analogue that cannot be metabolized (2-deoxyglucose, 2DG) and compared the results with littermate Ksp-Cre:Pkd1flox/− treated with vehicle only (NaCl) and with littermate controls (Ksp-Cre:Pkd1flox/+ or Pkd1flox/+ interchangeably) treated with NaCl or 2DG. We found that 2DG reduced the kidney/body weight (Fig. 4a-c) only in mutants and had no effect on the weight of other organs or on the total body weight (Supplementary Fig. 5). Histological evaluation of the kidneys revealed that 2DG reduced the cystic index in these kidneys (Fig. 4d), likely by interfering with the cellular proliferation rates of the cystic kidneys (Fig. 4e). Analysis of key metabolic parameters (insulin and glucose blood concentrations as well as glycogen levels in the liver) showed that the effect of 2DG is not secondary to a general effect on metabolism (Fig. 4f). Importantly, using the uniformly-labelled 13C-glucose strategy described above, we demonstrated that 2DG acts indeed by reducing glycolysis in Ksp-Cre:Pkd1flox/− kidneys (Fig. 4g, n = 3). Finally, to determine the efficacy of 2DG treatment in a second, less aggressive model of PKD, we used the previously described Pkd1v/v model20 and found that 2DG reduced the kidney/body weight and ameliorated the histology and cystic index of Pkd1v/v kidneys as well (Fig. 4h-j).
Figure 4. Treatment with 2DG ameliorates cystic kidney disease in two ADPKD orthologous models.
a. Representative example of P8 Ksp-Cre:Pkd−/flox kidneys treated daily (from P6 to P8) with 500 mg/kg 2DG or with vehicle (NaCl). b. Representative examples of the histology of kidneys treated as in a. c. Ratio of kidney-over-body weight in controls or mutant littermates (each litter in a different color) after treatment with 2DG or NaCl. d. Cystic index in 2DG vs NaCl-treated KspCre:Pkd1−/flox kidneys shows a significant reduction in the number of cysts in 2DG-treated animals. e. Ki67 assay shows the proliferation index in the Ksp-Cre:Pkd1−/flox mice treated with 2DG vs vehicle (NaCl). Right: quantification of Ki67 in the cyst-lining epithelium in 2DG vs NaCl-treated kidneys. f. Serum concentrations of glucose and insulin and liver levels of glycogen in littermate controls and mutant Ksp-Cre:Pkd1flox/− kidneys treated with 2DG or with vehicle (NaCl). g. 13C-lactate levels are significantly reduced in the Ksp-Cre:Pkd1flox/− cystic kidneys treated with 2DG as compared to vehicle-only treated, while the levels of 13C-glucose remain higher in the 2DG treated kidneys. h. Kidney-over-body weight in Pkd1v/v mice treated with 500 mg/kg 2DG from P5 until P7. i. Cystic kidneys from Pkd1v/v mice treated with 2DG as in h. N.S.: P ≥ 0.05; *: P < 0.05; **: P < 0.01; ***:P < 0.001. e, f and g Mean +/− SEM; c and h Mean +/− SD. Statistical analysis by ANOVA followed by Bonferroni’s test in d, f and j; t-test in e and g; Mann-Whitney test in c and h. Bar = 5mm in a, 1 mm in b, 100 μm in e and 1 mm in i.
Thus, our study shows that the use of glucose analogues, such as 2DG, may be a promising therapeutic approach. The kidney is an organ with a great functional redundancy and loss of a small percentage of nephrons is not sufficient to cause its functional loss. As outlined above, renal failure in ADPKD is caused by the progressive renal cyst expansion affecting only a minority of nephrons1,4. Therefore, the use of a molecule able to selectively reduce the viability or proliferation of cells lining the cysts would most likely be an effective therapy. 2DG might serve this purpose as it is a well tolerated molecule already in use in humans for some forms of cancer21,12.
Experimental treatment with rapamycin reduces renal cyst expansion in murine ADPKD22,23. However, the results in human beings are more controversial for reasons possibly related to the side-effects of this drug24,25,26. Our data (Fig. 3f) highlight the possibility of combination therapy with several drugs to exploit their synergistic effects while reducing potential side-effects5. For example, metformin is a well-tolerated drug used for treatment of type 2 diabetes mellitus12 and was shown to effectively reduce cyst expansion in a PKD mouse model27,28. Our current study provides the mechanistic explanation for this finding and further suggests that the use of metformin or AICAR (enhancing AMPK activity) in combination with 2DG might provide an effective combination therapy, as suggested for cancer29.
In summary, our finding of a metabolic switch to glycolysis in the absence of functional polycystin-1 signaling provides the rationale for a novel therapeutic approach in ADPKD. We also speculate that this might represent a paradigm whereby several drugs that are expected to work synergistically can be exploited to enhance both efficacy and tolerance in the treatment of ADPKD.
ONLINE METHODS
Antibodies, reagents, and inhibitors
We used antibodies to pAkt (Ser473) #9271, Akt #9272, pAMPK (Thr172) 40H9 #2535, AMPK #2532, pS6RP (Ser235 and Ser236) #2211, S6RP #2217, pERK (Thr202 and Tyr204) #9101, ERK #9102, p4EBP1 (Ser65) #9451, 4EBP1 #9452, pLKB1 (Ser428) #3051 and pP70S6K (Thr389) #9205 from Cell Signaling Technology at 1:1,000; antibodies to LKB1 (N-19) #sc-8185 from Santa Cruz at 1:1,000; to pACC (Ser 79) #07-303 and ACC #05-1098 from Millipore at 1:500 and 1:300, respectively; antibodies to LC3 NB100 #2331 from Novus Biologicals at 1:300; antibodies to actin #A5441 and tubulin #T5168 from Sigma Aldrich at 1:5,000; and antibodies to Ki67 #NCL-L-Ki67-MM1 from Novocastra at 1:400.
We employed: U0126 and rapamycin (Cell Signaling Technologies) at final concentrations of 30 μM and 20 or 50 nM, respectively; AICAR and Metfomin (Sigma Aldrich) at the final concentration of 2 mM; Oligomycin (Sigma Aldrich) at the final concentration of 30 μg/ml.
For experiments of glucose starvation the composition of the medium is: basic DMEM (Gibco) 2,3 g/L, Sodium Bicarbonate (Gibco) 3,7 g/L, L-glutamine 0,584 g/L (Gibco), serum (Euroclone), 10%, pen–strep 1% (Gibco) with high glucose (Sigma) 4,5 g/L (equivalent to 25 mM) or with 1 mM glucose for starvation.
TatCre treatment in vitro
For conditional inactivation of the Pkd1flox/flox lines6, we incubated cells twice in the presence of a recombinant TAT-Cre protein at 1 μM (Excellgen) for 2 h with 100 μM of chloroquine (Sigma Aldrich).
NMR and Metabolomic profiling
For NMR analysis of the extracellular medium we mixed 530 μl of cell culture medium with 60 μl of deuterated PBS solution containing DSS as chemical shift reference for both proton and carbon dimensions, and 10 μl of 1.2% NaN3 water solution. Final sample volume was 600 μl, containing 50 mM PBS, 0.02% NaN3 and 90 μM DSS. Further details on NMR sample preparation, spectra acquisition, metabolites recognition and statistical analysis are described in Supplementary Methods.
13C-Glucose experiments
We injected litters of Ksp-cre:Pkd1flox/− and Pkd1flox/+ at P8 intracutaneously with 1000 mg/Kg of 13C- glucose (Cortecnet). After 40 min, we sacrificed the mice, washed the kidneys in PBS and snap-frozed them in liquid nitrogen. For 2DG treatment, we injected litters intracutaneously with 2DG at 500 mg/Kg or NaCl at P7 and P8 and the second injection was followed by an injection with 1000 mg/Kg of 13C- glucose and we collected kidneys as above. Sample preparation and detection of 13C-glucose and 13C-lactate by 13C-NMR spectroscopy are described in Supplementary Methods.
Glucose, glycogen and insulin quantification
We determined serum glucose levels using the BioVision Glucose Assay Kit, liver glycogen levels with the BioVision Glycogen Assay Kit and serum levels of insulin using the Millipore Rat/Mouse insulin ELISA kit following the manufacturer’s recommendations.
TD/Cystic index
To quantify the rate of cystic kidneys we applied a grid of squares 13.625 μm large to sections of kidneys stained with Hematoxylin-Eosin. We marked each cross with a dot and we counted the number of dots inside the lumen on three litters (2.99 mm2 for each). We determined the degree of dilatation according to criteria previously established in Bastos et al30. Briefly:1 dot: normal tubules; 2 dots: dilatated tubules; >3 dots: Cysts.
ATP and lactate quantification
For ATP content evaluation, we prepared whole-cell extracts of control and treated MEFs by suspending pellets in lysis buffer as described6. We measured intracellular ATP quantification of lysates on 250 ng of protein by luciferase activity as showed in the standard protocol present in the ATP Determination kit (Invitrogen). We used medium of control and treated MEFs after 24 h at 100% cell density. We determined the concentration of lactate using EnzyChromTM L-lactate Assay Kit (BioAssay Systems) and quantified on the final number of cells.
Mitochondrial transmembrane potential (Ψm) assay
We assessed the mitochondrial transmembrane potential (Ψm) using the tetramethylrhodamine (TMRM, Invitrogen) and analyzed cells by time-lapse imaging and cytofluorimetrically. For FACS analysis, 24 h after plating, we resuspended the cells in phenol-red free HBSS with 10 mM HEPES with 20 nM TMRM in the presence of multidrug resistance pump inhibitor cyclosporine-H 2 μm and incubated for 30 min at 37 °C. In parallel, we incubated the cells with an uncoupling agent FCCP 4 μm to measure the specific mitochondria staining. We measured the TMRM fluorescence by FACS analysis6. For quantitative real-time analysis of mitochondrial transmembrane potential, we incubated the cells for 30 min at 37 °C in phenol-red free HBSS (Gibco) with 10 mM HEPES (Gibco), 20 nM TMRM, cyclosporine-H 2 μM and 2 μg/ml Hoechst 33342. We acquired images with IN CELL Analyser 1000 (LKT laboratories) before and after FCCP 4 μm was injected in a motorized way and sequential images were taken for TMRM and Hoechst in different regions of interest every 3 min. Images were automatically analysed with IN CELL INVESTIGATOR ANALYSIS software (GE Healthcare) to define the TMRM intensity.
Proliferation and apoptosis assays
For proliferation assays we carried out immunostaining using an antibody directed against Ki67 followed by analysis. For cells, we measured fluorescence on triplicates of 300 cells using the microscope Axioplot (Zeiss). For kidney sections, we took images at 20x using the camera Axio MRc5 (Zeiss) with the microscope Axioplan 2 (Zeiss) and we counted positive cells on 6 sections for each group for each experiment using the ImageJ software.
For apoptosis assays we analyzed the cells using the DeadEnd Flurometric transferase-mediated dUTP nick-end labeling (TUNEL) system kit (Promega) following the manifacturer’s instructions.
Generation and 2DG treatment of Pkd1flox/−: Ksp-Cre mice and Pkd1v/v mice
We previously described the generation of the Ksp-Cre:Pkd1flox/− mice31. Briefly, we crossed Pkd1flox/flox,32 and Pkd1+/−:Ksp-Cre mice33 in pure C57/Bl6 genetic background. For treatments, we injected subcutaneously 2DG (Sigma-Aldrich) or vehicle (NaCl) daily from P6 until P8 at 500 mg/kg. We previously described the generation of Pkd1v/v mice20. For treatments, we injected subcutaneously 2DG (Sigma-Aldrich) or vehicle (NaCl) daily from P5 until P7 at 500 mg/kg. For all animal work the female to male ratio is 1:1. All animal care and experimental protocols were conducted upon approval of a specific protocol (IACUC-401) by the institutional care and use ethical committee (I.A.C.U.C.) at the San Raffaele Scientific Institute. Pkd1V/V animal care and experimental protocols were conducted upon approval of a specific protocol (MO10M387) by the institutional care and use ethical committee (I.A.C.U.C.) at the Johns Hopkins University School of Medicine.
Histology, Immunohistochemistryand Immunofluorescence
After sacrifice, we removed the kidneys, washed them in phosphate-buffered saline (PBS), weighed and fixed in 4% Paraformaldehyde (PFA). After incubation in a sucrose in PBS gradient scale from 10% to 30% we incubated the samples in 10% glycerol (Sigma) in a mixture of OCT (BIO-OPTICA) and sucrose 30%, finally we embedded them in OCT. We air-dried criostat sections for 1h, rehydrated in PBS, incubated in Harris Hematoxylin 1:10 (Sigma Aldrich) for 2 min, washed, incubated in Eosin G (BIO-OPTICA) for 7 min, washed deshydrated and mounted in DEPEX (Sigma).
For Immunohistochemistry we washed 10 μm cryosections in Tween-0.1% in PBS (like all washes), we fixed them in 4%PFA, permeabilized in 0.1% TritonX100 in PBS, incubated in 0.3% hydrogen peroxide for 30 min, blocked for 1h at room temperature with 5% Normal Goat Serum (Sigma Aldrich) 3% bovine serum albumin (BSA, Sigma) in phosphate-buffered saline and incubated them O/N at 4 °C with the antibody (ab) in blocking buffer diluted at 1:500. We finally incubated sections with the Dako EnVision+ System-HRP (Dako). We completed the staining by an incubation with 3, 3′-diaminobenzidine tetrahydrochloride (DAB) DAB+ Substrate Chromogen System (DakoCytomation) and counterstained with Harris Hematoxylin (Sigma Aldrich). Diluted 1 to 10 for 3 min.
For immunofluorescence: we fixed cells in 4% PFA, washed them in PBS, permeabilized in PBS-TritonX100 (0,2%), blocked with 3% BSA in PBS, incubated 45 min at 37 °C with the ab directed against ki67 described above at 1:1000 in blocking solution, washed and incubated with the secondary Ab diluted at 1:1000 for 1 h.
Renal cysts and microarray data analysis
We obtained renal cysts of different sizes from 5 polycystic kidneys. We obtained a minimally cystic tissue (MCT), which might have contained a few microscopic cysts from the renal cortex, as PKD control tissue from the same kidneys. We used non-cancerous renal cortical tissue from 3 nephrectomized kidneys with isolated renal cell carcinoma as normal control tissue. We previously described the surgical technique, RNA extraction, purification, quality control, microarray hybridization, profiling and quality assessments18. In brief, after extraction with Absolutely RNA RT–PCR Miniprep Kit (Stratagene), we labelled 50 to 100 ng total RNA and hybridized onto GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix) according to the manufacturer’s protocol. We processed scanned raw data images with GeneChip Operating Software (GCOS) 1.4. We extracted probe set signal intensities and normalized by the robust multi-array average algorithm, which can be found in the R package affy that can be downloaded from the Bioconductor project website (http://www.bioconductor.org). Microarray data are available at GEO website (accession number: GSE7869).
Western blot analysis
For western blot analysis, we lysed the cells in lysis buffer [150 mM NaCl, 20 mM Na4HPO4/NaH2PO4, 10% Glycerol, 1% Triton-X 100, pH 7.2, Complete protease inhibitors (Roche) and phosphatase inhibitors (1 mM final of glycerophosphate, sodium orthovanadate and sodium fluoride)]. We quantified total lysates and added Laemmli buffer. We resolved proteins in a SDS-PAGE gel and transferred onto PVDF membranes. Next, we used 5% milk in TBS-T for blocking and for secondary antibody incubations, while 3% BSA in TBS-T for incubations with primary antibodies. We visualized HRP-conjugated secondary antibodies (from Amersham) using the ECL System (Amersham), after mixing with Signal West Femto Maximum sensitivity substrate from Thermo Scientific when necessary.
Realtime PCR Analysis
We isolated total RNA from cells or whole kidneys using the RNAspin kit (GE Healthcare) and obtained complementary DNA using oligo(dT) primers (Invitrogen) and Superscript II Reverse Transcriptase (Invitrogen). We performed quantitative real-time PCR in duplicates using LightCycler480 (Roche Molecular Diagnostics) using SYBR Green I master mix. The complete sequence of primers that we used is provided in Supplementary Methods.
Statistical analysis
For statistical analysis of the NMR data we performed PCA analysis using R-statistical open source software (http://www.r-project.org/) using in-house statistical package called MUMA (free available upon request). For further details see Supplementary Methods.
For other in vitro and in vivo sudies we performed statistical analysis by applying either an unpaired T-test or a one-way analysis of variance (ANOVA) followed by Bonferroni’s test. The statistical analysis tool that we employed is in the legends. NS: > 0.05; *: P ≤ 0.05; **: P ≤ 0.01; ***: P ≤ 0.001.
For the microarrays studies, we provide details in the Supplementary Methods. In brief, we defined differentially expressed pathways by a NOM P-value ≤ 0.05 with a false discovery rate (FDR) ≤ 0.25, and used FDR ≤ 0.5% for the individual gene comparisons.
Supplementary Material
Acknowledgements
The authors are grateful to other members of the lab Boletta and to G. Di Grigoli for helpful discussion, to R.M. Moresco for helpful suggestions and for critically reading the manuscript, to G. Casari and L. Cassina for help with the experiments on mitochondria, to Giorgio M. for analysis of oxygen consumption and to the San Raffaele microscopy facility (Alembic) for the electron microscopy studies (M.C. Panzeri) and TMRM analysis (M. Ascagni). VM is a student in the PhD Program of Biochemical, Nutritional and Metabolic Sciences, University of Milan. This work was supported by Telethon-Italy (TCR05007 to AB and TCP99035 to GM), by the US National Institutes of Health Grants DK62199 (to F.Q.) and DK090868 (Johns Hopkins Polycystic Kidney Disease Research and Clinical Core Center, P30) and by the Canadian Institutes for Health Research Grant MOP123429 (to YP). AB and GM are Associate Telethon Scientists.
The lab Boletta is especially indebted to Bramani S. for her continuous, intelligent and motivating support.
Footnotes
Authors’ Contributions
IR designed and performed the experiments, interpreted them, wrote the manuscript. MC designed and performed the in vivo data with 2DG treatment and 13-C glucose injections, interpreted the results. VU performed experiments in vitro on autophgy and signaling. MP. Generated the KspCre:Pkd1 mice, analyzed the kidneys biochemically and performed qRT-PCR. AB designed the studies, supervised the work and collaborations, wrote the manuscript.
VM prepared samples for metabolomic analysis, analyzed NMR spectra and performed statistical analysis. GQ acquired NMR spectra and performed statistical analysis. SM prepared samples for metabolomic analysis, acquired and analyzed NMR spectra, performed statistical analysis. GM supervised metabolomic analysis, discussed results.
XWS and YP designed, performed, and interpreted the human PKD1 renal cyst microarray experiment.
HS and FQ. designed and carried out the 2DG treatment experiment of Pkd1V/V mice and analyzed and interpret its data.
References
- 1.Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 2.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–37. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takiar V, Caplan MJ. Polycystic kidney disease: pathogenesis and potential therapies. Biochim Biophys Acta. 2011;1812:1337–43. doi: 10.1016/j.bbadis.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grantham JJ, Geiser JL, Evan AP. Cyst formation and growth in autosomal dominant polycystic kidney disease. Kidney Int. 1987;31:1145–52. doi: 10.1038/ki.1987.121. [DOI] [PubMed] [Google Scholar]
- 5.Torres VE, et al. Prospects for mTOR inhibitor use in patients with polycystic kidney disease and hamartomatous diseases. Clin J Am Soc Nephrol. 2010;5:1312–29. doi: 10.2215/CJN.01360210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Distefano G, et al. Polycystin-1 regulates extracellular signal-regulated kinase-dependent phosphorylation of tuberin to control cell size through mTOR and its downstream effectors S6K and 4EBP1. Mol Cell Biol. 2009;29:2359–71. doi: 10.1128/MCB.01259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forseth RR, Schroeder FC. NMR-spectroscopic analysis of mixtures: from structure to function. Curr Opin Chem Biol. 2011;15:38–47. doi: 10.1016/j.cbpa.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Manteiga JM, et al. Metabolomics of B to plasma cell differentiation. J Proteome Res. 2011;10:4165–76. doi: 10.1021/pr200328f. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res. 2011;71:2815–20. doi: 10.1158/0008-5472.CAN-10-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiaradonna F, et al. From cancer metabolism to new biomarkers and drug targets. Biotechnol Adv. 2012;30:30–51. doi: 10.1016/j.biotechadv.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Woo D. Apoptosis and loss of renal tissue in polycystic kidney diseases. N Engl J Med. 1995;333:18–25. doi: 10.1056/NEJM199507063330104. [DOI] [PubMed] [Google Scholar]
- 14.Merrick D, et al. The gamma-Secretase Cleavage Product of Polycystin-1 Regulates TCF and CHOP-Mediated Transcriptional Activation through a p300-Dependent Mechanism. Dev Cell. 2012;22:197–210. doi: 10.1016/j.devcel.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choo AY, et al. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38:487–99. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng B, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009;33:237–47. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibazaki S, et al. Cyst formation and activation of the extracellular regulated kinase pathway after kidney specific inactivation of Pkd1. Hum Mol Genet. 2008;17:1505–16. doi: 10.1093/hmg/ddn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song X, et al. Systems biology of autosomal dominant polycystic kidney disease (ADPKD): computational identification of gene expression pathways and integrated regulatory networks. Hum Mol Genet. 2009;18:2328–43. doi: 10.1093/hmg/ddp165. [DOI] [PubMed] [Google Scholar]
- 19.Foxall PJ, et al. High resolution proton magnetic resonance spectroscopy of cyst fluids from patients with polycystic kidney disease. Biochim Biophys Acta. 1992;1138:305–14. doi: 10.1016/0925-4439(92)90008-b. [DOI] [PubMed] [Google Scholar]
- 20.Yu S, et al. Essential role of cleavage of Polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc Natl Acad Sci U S A. 2007;104:18688–93. doi: 10.1073/pnas.0708217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csibi A, Blenis J. Appetite for destruction: the inhibition of glycolysis as a therapy for tuberous sclerosis complex-related tumors. BMC Biol. 2011;9:69. doi: 10.1186/1741-7007-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shillingford JM, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci U S A. 2006;103:5466–71. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shillingford JM, Piontek KB, Germino GG, Weimbs T. Rapamycin ameliorates PKD resulting from conditional inactivation of Pkd1. J Am Soc Nephrol. 2010;21:489–97. doi: 10.1681/ASN.2009040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serra AL, et al. Clinical proof-of-concept trial to assess the therapeutic effect of sirolimus in patients with autosomal dominant polycystic kidney disease: SUISSE ADPKD study. BMC Nephrol. 2007;8:13. doi: 10.1186/1471-2369-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walz G, et al. Everolimus in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2010;363:830–40. doi: 10.1056/NEJMoa1003491. [DOI] [PubMed] [Google Scholar]
- 26.Serra AL, et al. Sirolimus and kidney growth in autosomal dominant polycystic kidney disease. N Engl J Med. 2010;363:820–9. doi: 10.1056/NEJMoa0907419. [DOI] [PubMed] [Google Scholar]
- 27.McCarty MF, Barroso-Aranda J, Contreras F. Activation of AMP-activated kinase as a strategy for managing autosomal dominant polycystic kidney disease. Med Hypotheses. 2009;73:1008–10. doi: 10.1016/j.mehy.2009.05.043. [DOI] [PubMed] [Google Scholar]
- 28.Takiar V, et al. Activating AMP-activated protein kinase (AMPK) slows renal cystogenesis. Proc Natl Acad Sci U S A. 2011;108:2462–7. doi: 10.1073/pnas.1011498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheong JH, et al. Dual inhibition of tumor energy pathway by 2-deoxyglucose and metformin is effective against a broad spectrum of preclinical cancer models. Mol Cancer Ther. 2011;10:2350–62. doi: 10.1158/1535-7163.MCT-11-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastos AP, et al. Pkd1 haploinsufficiency increases renal damage and induces microcyst formation following ischemia/reperfusion. J Am Soc Nephrol. 2009;20:2389–402. doi: 10.1681/ASN.2008040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wodarczyk C, et al. Nephrocystin-1 forms a complex with polycystin-1 via a polyproline motif/SH3 domain interaction and regulates the apoptotic response in mammals. PLoS One. 2010;5:e12719. doi: 10.1371/journal.pone.0012719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wodarczyk C, et al. A novel mouse model reveals that polycystin-1 deficiency in ependyma and choroid plexus results in dysfunctional cilia and hydrocephalus. PLoS One. 2009;4:e7137. doi: 10.1371/journal.pone.0007137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao X, Somlo S, Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol. 2002;13:1837–46. doi: 10.1097/01.asn.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.