Abstract
Uveal melanoma management is challenging due to its metastatic propensity. DecisionDx-UM is a prospectively validated molecular test that interrogates primary tumor biology to provide objective information about metastatic potential that can be used in determining appropriate patient care. To evaluate the continued clinical validity and utility of DecisionDx-UM, beginning March 2010, 70 patients were enrolled in a prospective, multicenter, IRB-approved study to document patient management differences and clinical outcomes associated with low-risk Class 1 and high-risk Class 2 results indicated by DecisionDx-UM testing. Thirty-seven patients in the prospective study were Class 1 and 33 were Class 2. Class 1 patients had 100% 3-year metastasis-free survival compared to 63% for Class 2 (log rank test p = 0.003) with 27.3 median follow-up months in this interim analysis. Class 2 patients received significantly higher-intensity monitoring and more oncology/clinical trial referrals compared to Class 1 patients (Fisher's exact test p = 2.1 × 10−13 and p = 0.04, resp.). The results of this study provide additional, prospective evidence in an independent cohort of patients that Class 1 and Class 2 patients are managed according to the differential metastatic risk indicated by DecisionDx-UM. The trial is registered with Clinical Application of DecisionDx-UM Gene Expression Assay Results (NCT02376920).
1. Introduction
Uveal melanoma (UM) is a rare intraocular cancer affecting the choroid, ciliary body, or iris. At diagnosis, almost all patients will present without evidence of metastatic disease. Despite high rates of primary tumor control, metastatic lesions, predominantly to the liver, occur in approximately 20–50% of UM patients [1]. Primary tumor gene expression profile (GEP) testing has shown that patients can be accurately and reliably classified into low-risk Class 1 and high-risk Class 2 with significantly different metastatic potentials [2–5]. These patient groups should be managed based upon their relative risk versus the overall population risk. For clinical care purposes, a prognostic tool should be highly accurate in its risk prediction (clinical validity), and its results should be implemented to inform patients' subsequent management plans (clinical utility) [6].
Clinical factors including age, extraocular extension, tumor size, and ciliary body involvement have been linked to higher risk for metastasis of UM tumors [7, 8]. However, none of these have offered the prognostic accuracy or reproducibility required for clinical implementation. Genetic analysis has led to the identification of metastasis-associated cytogenetic abnormalities on chromosomes 1, 3, 6, and 8 [9–14]. In particular, the presence of monosomy 3 in primary tumor cells is a significant factor for predicting metastatic risk [10, 15, 16], and fluorescence in situ hybridization (FISH), comparative genomic hybridization (CGH), multiplex ligation-dependent probe amplification (MLPA), and loss of heterozygosity (LOH) have been developed to identify chromosome abnormalities [17–20]. However, to our knowledge, these molecular tests to detect gains and losses of chromosomes 1, 3, 6, and 8, including the commercially available MLPA platform [17, 21], have not been clinically validated in prospective, multicenter studies and published clinical utility is limited to high-risk patients [22].
The DecisionDx-UM GEP test is an accurate prospectively validated molecular classifier, and its results are highly correlative to metastatic potential [2, 5], as reported by the Collaborative Ocular Oncology Group (COOG) [2]. Notably, because DecisionDx-UM has been extensively validated exclusively on pretreatment UM specimens and the training set, to which the machine-learning algorithm compares each patient sample to generate a Class 1 or 2 result, consists of only such samples, nonmelanoma samples are inappropriate for prognostication with the GEP panel, consistent with the exclusion criteria of the COOG study [2]. Similarly, given the lack of validation on posttreatment tumors and the potential for radiobiological effects on tumor's genomics, irradiated samples are also ineligible [23, 24]. DecisionDx-UM's clinical utility has also been reported, indicating that the majority of ophthalmologists who order the test use the results to guide risk-appropriate treatment and management strategies for UM patients [25].
Herein are the first results from an interim analysis of the ongoing, prospective, multicenter CLinical Application of DEcisionDx-UM Gene Expression Assay Results (CLEAR) registry. This study tracks decision impact (surveillance regimens and treatment referral patterns), as well as clinical outcomes for DecisionDx-UM patients [26]. These results add additional prospective, multicenter evidence underscoring DecisionDx-UM as a highly accurate and clinically actionable assay for determining risk associated with primary UM tumors.
2. Methods
2.1. Patient Enrollment
After IRB approval of the study (number NCT02376920; clinicaltrials.gov [26]) at participating centers (Mayo Clinic, Rochester MN; University of Virginia, Charlottesville, VA; Retinal Consultants, Sacramento, CA; Retina Specialists of Michigan, Grand Rapids, MI), patient consent was obtained. Data entry was performed at receipt of test results and semiannually (censor date: June 2015).
2.2. Tumor Sample Acquisition and Processing
Physician-obtained FNAB or FFPE UM tumor specimens were submitted to a centralized CAP-accredited, CLIA-certified laboratory for GEP. Frozen specimens, mostly FNAB, were dispersed into RNase-free stabilization buffer immediately following biopsy, and all samples were shipped to the recipient lab on dry ice. RNA isolation was performed with the PicoPure RNA Isolation Kit (Molecular Devices, Sunnyvale, CA). All FFPE samples were prepared from enucleated globes, and tumor sections on microscope slides were shipped at room temperature to the recipient lab. Tumor tissue was macrodissected from slides using a sterile, disposable scalpel, deparaffinized in xylene, and processed for RNA isolation with the Ambion RecoverAll Total Nucleic Acid Isolation Kit (Life Technologies Corporation, Grand Island, NY). All RNA was assessed for quantity and quality using the NanoDrop 1000 system (Life Technologies Corporation) and the Agilent Bioanalyzer 2100 and then converted to cDNA (Applied Biosystems High Capacity cDNA Reverse Transcription Kit; Life Technologies Corporation).
2.3. DecisionDx-UM Gene Expression Profile Assay
Each cDNA sample underwent a 14-cycle preamplification step and was then diluted 20-fold in Tris-EDTA buffer. Fifty microliters of each diluted sample was mixed with 50 μL of 2x TaqMan Gene Expression Master Mix (Life Technologies Corporation) and loaded onto a custom high-throughput microfluidics gene card containing primers specific for 12 class-discriminating genes and three endogenous control genes [5]. Each sample was run in triplicate on an Applied Biosystems HT7900 instrument (Life Technologies Corporation).
2.4. Expression Analysis Algorithm and Class Assignment
Delta Ct values were calculated by subtracting the mean Ct of each discriminating gene triplicate from the geometric mean of the three endogenous control genes' mean Ct values [5]. Molecular class assignments were determined by comparing the 12 discriminating gene ΔCt values from each sample to a well-characterized, proprietary UM training set of low-risk Class 1 and high-risk Class 2 GEPs using a support vector machine- (SVM-) learning algorithm [5]. The predicted confidence is indicated by the discriminant score, which is inversely proportional to the proximity of the sample to the hyperplane established between Class 1 and Class 2 training set samples. The summation of CDH1 and RAB31 expression identifies Class 1A and Class 1B subtypes [27].
2.5. Surveillance Categorization
Surveillance regimens were not prespecified but instead were independently decided upon by each participating physician utilizing the DecisionDx-UM result and documented as part of the registry data entry. The intensity of surveillance for each patient was categorized based upon a previously reported study [25] and determined by the frequency of imaging (ultrasound, PET/CT, or MRI) and liver function tests (LFTs) that a patient received in addition to their regular eye examination follow-up. A high-intensity schedule was characterized by imaging and/or LFTs every 3–6 months, whereas a low-intensity schedule was characterized by annual imaging and/or LFTs.
3. Results
3.1. Clinical Outcomes and Medical Management Associated with DecisionDx-UM
The prospective, multicenter CLEAR study was designed to assess (a) the management of patients according to their DecisionDx-UM results (clinical utility) and (b) their documented development of metastatic disease (clinical validity). Seventy patients have been enrolled from four centers across the US (Table 1). Thirty-seven (53%) were Class 1 and 33 (47%) were Class 2. None of the patients who consented to be in the registry study had technical failures with GEP testing. Of the Class 1 patients, 30 (81%) were Class 1A, while 7 (19%) were Class 1B. Consistent with their high-risk GEP, 12 (36%) Class 2 patients experienced a metastasis, whereas only 2 (5%) Class 1 patients experienced a metastasis (p = 0.002 by Fisher's exact test) with a median follow-up of 2.38 years. The median time to metastasis for Class 2 patients was 1.4 years (Table 2). Class 2 patients had a significantly worse 3-year metastasis-free survival (MFS) rate of 63% (95% confidence interval = 43%–83%) compared to 100% in Class 1 patients (log rank test p = 0.003) (Figure 1). The median rate of metastasis was 3.78 years for Class 2 patients, while the median was not reached for the Class 1 patients. The majority of the metastases were localized in the liver, but metastases were also found in the lungs, brain, and bone (Table 2). Of note, one of the Class 1 patients who experienced metastasis had a large tumor, while the other patient did not undergo treatment of their primary tumor. Nine out of the 12 Class 2 patients who ultimately had metastases were treated by enucleation, while 2 were treated with plaque radiotherapy, and one was treated by transpupillary thermotherapy (TTT) (Table 2).
Table 1.
CLEAR registry patient characteristics.
| Class 1 (n = 37) | Class 2 (n = 33) | p value | |
|---|---|---|---|
| Gender: | |||
| Female, n (%) | 13 (35%) | 24 (73%) | 0.002a |
| Male, n (%) | 24 (65%) | 9 (27%) | |
|
| |||
| Age at diagnosis (yrs) | |||
| Mean | 60.5 | 62.7 | 0.52b |
| Median | 62 | 63 | |
| Range | 29–84 | 31–87 | |
|
| |||
| Ciliary body involvement, n (%) | 15 (41%) | 12 (36%) | 0.81a |
|
| |||
| Tumor diameter (mm) | |||
| ≤10 (% of row) | 11 (73%) | 4 (27%) | |
| >10 to ≤15 (% of row) | 15 (58%) | 11 (42%) | |
| >15 (% of row) | 11 (38%) | 18 (62%) | |
| Mean (SD) | 12.8 (1.84) | 15.64 (4.95) | 0.02b |
|
| |||
| Tumor thickness (mm) | |||
| ≤3 (% of row) | 9 (69%) | 4 (31%) | |
| >3 to ≤8 (% of row) | 18 (54%) | 15 (46%) | |
| >8 (% of row) | 10 (42%) | 14 (58%) | |
| Mean (SD) | 5.89 (3.34) | 7.02 (3.39) | 0.16b |
|
| |||
| Treatment type | |||
| Enucleation | 9 (24%) | 17 (52%) | 0.026a |
| Plaque radiotherapy | 23 (62%) | 11 (33%) | 0.019a |
| Proton beam | 3 (8%) | 1 (3%) | 0.61a |
| TTT | 0 (0%) | 1 (3%) | 0.47a |
| None | 2 (5%) | 3 (9%) | 0.62a |
|
| |||
| Follow-up (years) | |||
| Mean | 2.7 | 2.5 | 0.62b |
| Median | 2.6 | 2.0 | |
|
| |||
| Metastatic events, n (%) | 2 (5%) | 12 (36%) | 0.002a |
|
| |||
| High-intensity management ∗ | 7 (19%) | 33 (100%) | 2.1 × 10−13 a |
|
| |||
| Referrals | 4 (11%) | 11 (33%) | 0.04a |
| Medical oncology | 4 (11%) | 6 (18%) | |
| Clinical trials | 0 (0%) | 8 (24%) | |
|
| |||
| Systemic adjuvant therapy | 0 (0%) | 4 (12%) | 0.04a |
TTT: transpupillary thermotherapy.
aFisher's exact test; bStudent's t-test.
∗Liver function tests and/or imaging every 3–6 months.
Table 2.
Clinical characteristics of CLEAR Class 1 and Class 2 UM patients who experienced metastasis.
| Patient | Class | Age | Ciliary body involvement | Largest basal diameter (mm) | Primary tumor treatment | Metastatic location (s) | Time to metastasis (yrs) | Time to death (yrs) | Follow-up (yrs) |
|---|---|---|---|---|---|---|---|---|---|
| CLASS 1 | |||||||||
| 1 | 1 | 75 | No | 9.4 | None | Liver/lungs | 3.24 | 3.27 | 3.27 |
| 2 | 1 | 53 | Yes | 18.5 | Enucleation | Liver | 4.37 | 5.86 | 6.18 |
|
| |||||||||
| CLASS 2 | |||||||||
| 1 | 2 | 31 | Yes | 18.2 | Enucleation | Liver | 1.37 | N/A | 1.37 |
| 2 | 2 | 84 | No | 9.1 | Enucleation | Liver | 6.77 | N/A | 7.83 |
| 3 | 2 | 48 | No | 23 | Enucleation | Bone | 1.11 | N/A | 1.71 |
| 4 | 2 | 74 | Yes | 14.8 | Enucleation | Bone | 1.31 | N/A | 1.36 |
| 5 | 2 | 77 | No | 19.9 | Enucleation | Liver/lungs | 0.85 | N/A | 1.32 |
| 6 | 2 | 54 | No | 20.8 | Enucleation | Liver | 1.47 | N/A | 1.95 |
| 7 | 2 | 61 | No | 11.9 | Plaque | Bone | 2.45 | N/A | 2.51 |
| 8 | 2 | 86 | No | 17.6 | Enucleation | Liver | 1.21 | 1.47 | 1.47 |
| 9 | 2 | 59 | No | 4.9 | TTT | Liver | 3.78 | 3.69 | 3.69 |
| 10 | 2 | 50 | No | 18 | Enucleation | Liver | 1.27 | 1.69 | 1.69 |
| 11 | 2 | 83 | No | 14.6 | Plaque | Liver | 3.19 | 3.5 | 3.5 |
| 12 | 2 | 54 | No | 12.6 | Enucleation | Lungs/brain | 2.57 | 2.66 | 2.66 |
|
| |||||||||
| Class 2 | |||||||||
| Median | 1.42 | 2.66 | 1.83 | ||||||
Figure 1.
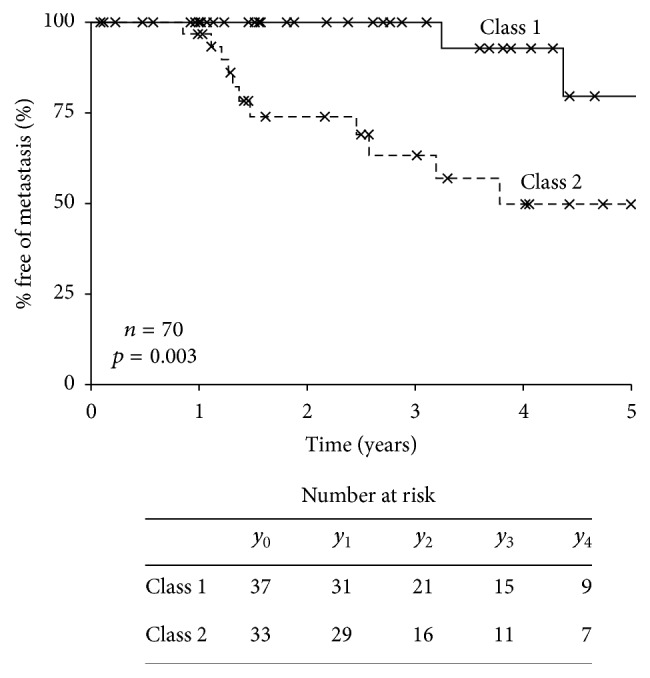
Metastasis-free survival (MFS) rates for CLEAR registry patients. Kaplan-Meier analysis of MFS for 70 patients enrolled in the CLEAR uveal melanoma study. Class 1 patients had a 3-year MFS rate of 100% versus 63% for Class 2 patients; the median rate of metastasis was 3.78 years for Class 2 patients but not reached for Class 1 patients.
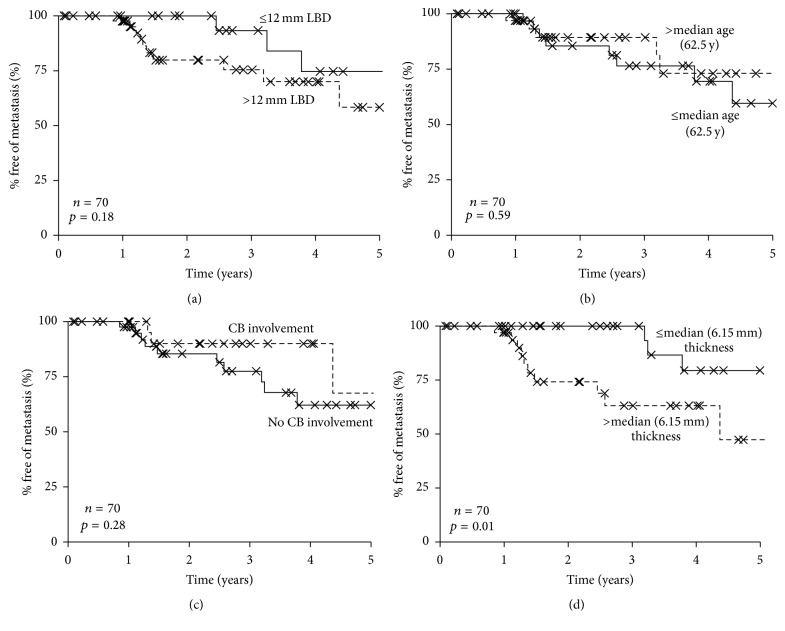
Tumor nodal metastasis (TNM) staging by AJCC includes tumor diameter, thickness, ciliary body involvement, and extraocular extension of the tumor. In this cohort, neither largest basal diameter nor ciliary body involvement performed as statistically significant prognostic markers (Figure 2). While tumor thickness did provide significant stratification of the patients, GEP showed stronger prognostic significance in Kaplan-Meier and multivariate analysis (Table 3), similar to previous studies [2, 28].
Figure 2.
Metastasis-free survival (MFS) rates according to clinicopathologic factors. The metastasis-free survival (MFS) rates for patients according to largest basal diameter (a), age (b), ciliary body (CB) involvement, (c) and tumor thickness (d). Only stratification according to tumor thickness was significant (p = 0.01).
Table 3.
Multivariate analysis of gene expression profiling (GEP) and tumor thickness.
| Multivariate | |||
|---|---|---|---|
| Hazard ratio | 95% CI | p value | |
| Thickness (>median) | 4.09 | 1.1–15.3 | 0.037 |
| GEP class 2 | 6.43 | 1.4–29.4 | 0.016 |
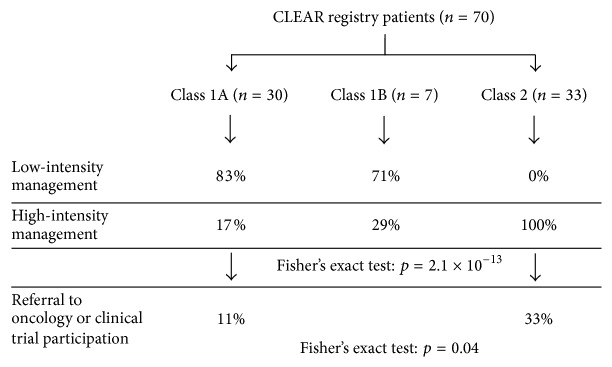
One primary objective of CLEAR was to document clinical management differences that are implemented for Class 1 compared to Class 2 patients. Of the 37 Class 1 patients, the majority (n = 30) were treated with low-intensity follow-up (imaging and/or LFTs every year), while all 33 Class 2 patients were treated with high-intensity follow-up (imaging and/or LFTs every 3–6 months) (Figure 3). Two of the Class 1 patients who received high-intensity surveillance had intermediate risk Class 1B results. Only 4 out of the 37 (11%) Class 1 patients were referred to medical oncology. Of note, two of these referred Class 1 patients had ciliary body involvement, one of whom also had a large (22 mm diameter) tumor and displayed loss of chromosome 3. Therefore, it is possible that the presence of these clinical features contributed to their medical oncology referrals. Due to their high-risk disease, 6 out of 33 (18%) of Class 2 patients were referred to medical oncology and 8 Class 2 patients (24%) were referred to adjuvant clinical trials. Importantly, four Class 2 patients went on to receive systemic adjuvant therapy; three patients received combinatorial chemotherapy (tamoxifen, sunitinib, and cisplatin) within a clinical trial, while one received IVIG immunotherapy. Three of the four patients remained metastasis-free at last follow-up (mean follow-up: 4.48 years; median follow-up: 4.02 years). No Class 1 patients were referred to clinical trials or had systemic adjuvant therapy. Taken together, these results indicate that Class 2 patients that have imaging and LFTs are managed by medical oncology and are offered clinical trial participation significantly more often than Class 1 patients (Fisher's exact test for intensity of surveillance p < 0.0001; for medical oncology/clinical trial referral p = 0.04; Table 1; Figure 3).
Figure 3.
Clinical management decisions associated with DecisionDx-UM classification results for CLEAR registry patients. Monitoring plans associated with Class 1 and Class 2 test results. Low-intensity management is defined as liver function tests (LFTs) and/or imaging studies annually. High-intensity management is defined as LFTs and/or imaging studies every 3–6 months.
4. Discussion
The National Comprehensive Cancer Network (NCCN) has emphasized the critical importance that all participants in the medical management of cancer patients, including the patients themselves, are provided with timely, reliable, and actionable education regarding molecular testing for diagnosis and treatment [6]. As molecular biomarker tests are increasingly available for diagnostic, prognostic, and therapeutic purposes, there is a growing need for transparency in reporting clinical validity and utility to ensure confidence in the accuracy and clinical impact of their results [6, 29]. Previous studies have reported the validity and utility of DecisionDx-UM to provide highly accurate prognostic information to guide individualized management of UM patients [2, 5, 30]. Herein, we report the initial results of the CLEAR prospective registry study that confirms the clinical utility of DecisionDx-UM, since it became clinically available. As this trial is ongoing, a final analysis of 5-year survival rates will be performed at the end of the study.
The NCCN recommends that the clinical utility of a biomarker test should be determined in a prospective clinical trial [6]. The CLEAR results represent a second independent, prospective, multicenter study making the test unique, not only in UM, but in comparison to the majority of high-complexity advanced diagnostic tests. The multicenter COOG study validated DecisionDx-UM's ability to accurately predict patient outcomes, reporting that metastatic events were observed in only 1% of Class 1 cases versus 26% of Class 2 cases after 50 months of follow-up time (median follow-up was 17.4 months) [2]. Similarly, interim results from our prospective registry study to track DecisionDx-UM patients indicated a low-risk of metastasis for Class 1 patients compared to Class 2 patients (5% versus 36%, resp.; median follow-up of 27.3 months). Taken together, the results from the CLEAR registry add to the compelling evidence-based clinical validity of DecisionDx-UM.
As has been demonstrated in other cancers for which prognostic and predictive testing are employed, tailoring a treatment strategy according to patient's individual tumor biology offers the potential for improved quality-of-life and more efficient utilization of healthcare resources [31–34]. The clinical utility of DecisionDx-UM was initially reported following a review of Medicare medical records that indicated the test significantly impacted patient management plans, resulting in less aggressive management of Class 1 versus Class 2 patients, and similar management impact results were observed from blinded surveys of ocular oncologists [25]. The CLEAR results also demonstrate that the clinical management of Class 2 patients is associated with significantly higher surveillance intensity, including more frequent imaging, LFTs, and referral to medical oncology compared to Class 1 patients. The higher-intensity surveillance for Class 2 patients is consistent with the goal of potentially identifying metastases earlier, thus permitting intervention, while the patient is asymptomatic and likely more amenable to treatment(s). Conversely, unnecessary surveillance can potentially be avoided for patients in whom extraocular recurrence of disease is unlikely. It should be noted that it is possible, based on a Class 1 result, for a patient who ultimately experiences metastasis to receive lower frequency management, thus potentially delaying the identification of metastatic disease by a few months (i.e., at 12 months versus 3–6 months with annual versus more frequent surveillance, resp.). However, given the consistency of the MFS rates between the COOG study and our data presented here (Table 4), we expect metastasis would occur in a small minority of Class 1 patients [2]. Furthermore, the small percentage of Class 1 patients who experienced metastasis (5%) that we report here is in line with previously reported metastasis rates reported for UM patients who are identified as low-risk [21, 35, 36] and for breast and colon cancer patients classified as low-risk by other tests [37, 38].
Table 4.
Comparison of the Cooperative Ocular Oncology Group (COOG) [2] and CLEAR studies.
| COOG [2] | CLEARa | |||
|---|---|---|---|---|
| Class 1 | Class 2 | Class 1 | Class 2 | |
| n (%) | 276 (61.9%) | 170 (38.1%) | 37 (52.8%) | 33 (47.2%) |
| Age (yrs) (median, mean) | 59, 59.6 | 65.9, 65.3 | 62, 60.5 | 63, 62.7 |
| Tumor diameter (mm) (median, mean) |
12.0, 12.1 | 14.0, 13.9 | 12.7, 12.8 | 16, 15.64 |
| Tumor thickness (mm) (median, mean) |
5.0, 5.7 | 6.2, 7.0 | 5, 5.89 | 7.1, 7.02 |
| Ciliary body involvement | 61 (22.7%) | 72 (43.6%) | 15 (41%) | 12 (36%) |
| 3-year metastasis-free survival | 97% | 50% | 100% | 63% |
aThe CLEAR study is a registry and therefore may not be entirely reflective of the general population of UM patients.
The data reported in this study are important for UM because numerous clinical, pathological, and genetic characteristics of the primary tumor have been proposed as being significantly prognostic for UM metastasis, yet these methods have achieved neither adequate NCCN level of evidence for clinical utility nor clinical validity. Based upon multiple prospective and retrospective studies published to date, DecisionDx-UM has achieved Level IA evidence, according to NCCN biomarker guidelines [6]. Given its robust ability to identify high-risk patients, rational intervention trials to identify effective adjuvant therapies have been initiated [39–42]. Four Class 2 patients within this cohort pursued adjuvant treatment for their high-risk disease; three of whom were metastasis-free at last follow-up, underscoring the importance of making more clinical trials accessible for high-risk UM patients. The decision to enroll Class 2 patients in clinical trials is directly related to the level of evidence for metastatic propensity that has been reported for the test. The continued clinical performance and utility demonstrated in this study contributes to the high level of evidence regarding the clinical validity and utility of the DecisionDx-UM test.
5. Conclusions
This study demonstrates that the 15-gene expression assay DecisionDx-UM continues to accurately predict metastatic risk for UM patients, thus enhancing the molecular test's established clinical validity. Furthermore, this is the first prospective analysis of clinical utility of the assay, and this study demonstrates that the test results are being used to guide decision-making for physicians and patients in the clinic.
Acknowledgments
The authors would like to acknowledge Dr. Jose Pulido, MD (Mayo Clinic, Rochester, MN) for enrolling patients and contributing data to this study.
Disclosure
Drs. Pulido, Tsai, Shildkrot, and Aaberg did not receive any compensation for this study. Robert W. Cook had full access to all of the data in the study and takes responsibility for the integrity of the data and accuracy of the data analysis.
Competing Interests
Financial support/sponsorship for this research was provided by Castle Biosciences, Inc. The following authors are employees of study's sponsor, Castle Biosciences: Kristen Meldi Plasseraud, Robert W. Cook, Brooke Middlebrook, Jeff Wilkinson, John Stone, Kristen Oelschlager, and Derek Maetzold. The following authors have stock in the sponsoring company: Kristen Meldi Plasseraud, Robert W. Cook, Brooke Middlebrook, Jeff Wilkinson, John Stone, Kristen Oelschlager, and Derek Maetzold. The following author has a board position in Castle: Derek Maetzold.
Authors' Contributions
Kristen Meldi Plasseraud, Robert W. Cook, Brooke Middlebrook, Kristen Oelschlager, Derek Maetzold, and John Stone wrote the paper; Tony Tsai, Yevgeniy Shildkrot, and Thomas M. Aaberg enrolled patients and contributed outcomes/management data; Jeff Wilkinson and John Stone performed GEP and generated risk classifications; Kristen Oelschlager and Clare Johnson collected data; Kristen Meldi Plasseraud, Brooke Middlebrook, Robert W. Cook, Kristen Oelschlager, and Clare Johnson analyzed the data. Kristen Meldi Plasseraud, Robert W. Cook, Brooke Middlebrook, Kristen Oelschlager, Derek Maetzold, Tony Tsai, Yevgeniy Shildkrot, and Thomas M. Aaberg edited the paper. All authors approved submission of the final paper.
References
- 1.Collaborative Ocular Melanoma Study Group. The COMS randomized trial of iodine 125 brachytherapy for choroidal melanoma: V. twelve-year mortality rates and prognostic factors: COMS report no. 28. Archives of Ophthalmology. 2006;124(12):1684–1693. doi: 10.1001/archopht.124.12.1684. [DOI] [PubMed] [Google Scholar]
- 2.Onken M. D., Worley L. A., Char D. H., et al. Collaborative ocular oncology group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119(8):1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onken M. D., Worley L. A., Dávilla R. M., Char D. H., Harbour J. W. Prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. Journal of Molecular Diagnostics. 2006;8(5):567–573. doi: 10.2353/jmoldx.2006.060077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onken M. D., Worley L. A., Ehlers J. P., Harbour J. W. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Research. 2004;64(20):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onken M. D., Worley L. A., Tuscan M. D., Harbour J. W. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. Journal of Molecular Diagnostics. 2010;12(4):461–468. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Febbo P. G., Ladanyi M., Aldape K. D., et al. NCCN Task Force report: evaluating the clinical utility of tumor markers in oncology. Journal of the National Comprehensive Cancer Network. 2011;9(supplement 5):S1–S33. doi: 10.6004/jnccn.2011.0137. [DOI] [PubMed] [Google Scholar]
- 7.Augsburger J. J., Gamel J. W. Clinical prognostic factors in patients with posterior uveal malignant melanoma. Cancer. 1990;66(7):1596–1600. doi: 10.1002/1097-0142(19901001)66:7<1596::AID-CNCR2820660726>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Shammas H. F., Blodi F. C. Prognostic factors in choroidal and ciliary body melanomas. Archives of Ophthalmology. 1977;95(1):63–69. doi: 10.1001/archopht.1977.04450010065005. [DOI] [PubMed] [Google Scholar]
- 9.Harbour J. W., Onken M. D., Roberson E. D. O., et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330(6009):1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilic E., Van Gils W., Lodder E., et al. Clinical and cytogenetic analyses in uveal melanoma. Investigative Ophthalmology and Visual Science. 2006;47(9):3703–3707. doi: 10.1167/iovs.06-0101. [DOI] [PubMed] [Google Scholar]
- 11.Tschentscher F., Prescher G., Zeschnigk M., Horsthemke B., Lohmann D. R. Identification of chromosomes 3, 6, and 8 aberrations in uveal melanoma by microsatellite analysis in comparison to comparative genomic hybridization. Cancer Genetics and Cytogenetics. 2000;122(1):13–17. doi: 10.1016/S0165-4608(00)00266-1. [DOI] [PubMed] [Google Scholar]
- 12.Prescher G., Bornfeld N., Becher R. Nonrandom chromosomal abnormalities in primary uveal melanoma. Journal of the National Cancer Institute. 1990;82(22):1765–1769. doi: 10.1093/jnci/82.22.1765. [DOI] [PubMed] [Google Scholar]
- 13.Sisley K., Rennie I. G., Parsons M. A., et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes, Chromosomes and Cancer. 1997;19(1):22–28. doi: 10.1002/(SICI)1098-2264(199705)19:1<22::AID-GCC4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Aalto Y., Eriksson L., Seregard S., Larsson O., Knuutila S. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Investigative Ophthalmology and Visual Science. 2001;42(2):313–317. [PubMed] [Google Scholar]
- 15.Worley L. A., Onken M. D., Person E., et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clinical Cancer Research. 2007;13(5):1466–1471. doi: 10.1158/1078-0432.CCR-06-2401. [DOI] [PubMed] [Google Scholar]
- 16.Prescher G., Bornfeld N., Hirche H., Horsthemke B., Jöckel K.-H., Becher R. Prognostic implications of monosomy 3 in uveal melanoma. The Lancet. 1996;347(9010):1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 17.Damato B., Dopierala J. A., Coupland S. E. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clinical Cancer Research. 2010;16(24):6083–6092. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- 18.Naus N. C., Verhoeven A. C. A., van Drunen E., et al. Detection of genetic prognostic markers in uveal melanoma biopsies using fluorescence in situ hybridization. Clinical Cancer Research. 2002;8(2):534–539. [PubMed] [Google Scholar]
- 19.Sandinha M. T., Farquharson M. A., Roberts F. Identification of monosomy 3 in choroidal melanoma by chromosome in situ hybridisation. The British Journal of Ophthalmology. 2004;88(12):1527–1532. doi: 10.1136/bjo.2004.044768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onken M. D., Worley L. A., Person E., Char D. H., Bowcock A. M., Harbour J. W. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clinical Cancer Research. 2007;13(10):2923–2927. doi: 10.1158/1078-0432.CCR-06-2383. [DOI] [PubMed] [Google Scholar]
- 21.Damato B., Dopierala J., Klaasen A., van Dijk M., Sibbring J., Coupland S. E. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Investigative Ophthalmology and Visual Science. 2009;50(7):3048–3055. doi: 10.1167/iovs.08-3165. [DOI] [PubMed] [Google Scholar]
- 22.Marshall E., Romaniuk C., Ghaneh P., et al. MRI in the detection of hepatic metastases from high-risk uveal melanoma: a prospective study in 188 patients. The British Journal of Ophthalmology. 2013;97(2):159–163. doi: 10.1136/bjophthalmol-2012-302323. [DOI] [PubMed] [Google Scholar]
- 23.Mooy C., Vissers K., Luyten G., et al. DNA flow cytometry in uveal melanoma: the effect of pre-enucleation irradiation. British Journal of Ophthalmology. 1995;79(2):174–177. doi: 10.1136/bjo.79.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dogrusöz M., Kroes W. G. M., van Duinen S. G., et al. Radiation treatment affects chromosome testing in uveal melanoma. Investigative Ophthalmology & Visual Science. 2015;56(10):5956–5964. doi: 10.1167/iovs.15-17092. [DOI] [PubMed] [Google Scholar]
- 25.Aaberg T. M., Cook R. W., Oelschlager K., Maetzold D., Rao P. K., Mason J. O. Current clinical practice: differential management of uveal melanoma in the era of molecular tumor analyses. Clinical Ophthalmology. 2014;8:2449–2460. doi: 10.2147/opth.s70839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook R. W. CLEAR registry study: an open, 5-year registry study to track clinical application of decisionDx-UM multigene assay results and associated patient outcomes. Clinical Trials, 2015, https://clinicaltrials.gov/ct2/show/NCT02376920.
- 27.Field M. G., Harbour J. W. Recent developments in prognostic and predictive testing in uveal melanoma. Current Opinion in Ophthalmology. 2014;25(3):234–239. doi: 10.1097/ICU.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corrêa Z. M., Augsburger J. J. Independent prognostic significance of gene expression profile class and largest basal diameter of posterior uveal melanomas. American Journal of Ophthalmology. 2016;162:20–27.e1. doi: 10.1016/j.ajo.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Collins G. S., Reitsma J. B., Altman D. G., Moons K. G. M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. The British Medical Journal. 2015;350 doi: 10.1136/bmj.g7594.g7594 [DOI] [Google Scholar]
- 30.Correa Z. M., Augsburger J. J. Sufficiency of FNAB aspirates of posterior uveal melanoma for cytologic versus GEP classification in 159 patients, and relative prognostic significance of these classifications. Graefe's Archive for Clinical and Experimental Ophthalmology. 2014;252(1):131–135. doi: 10.1007/s00417-013-2515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blohmer J. U., Rezai M., Kümmel S., et al. Using the 21-gene assay to guide adjuvant chemotherapy decision-making in early-stage breast cancer: a cost-effectiveness evaluation in the German setting. Journal of Medical Economics. 2013;16(1):30–40. doi: 10.3111/13696998.2012.722572. [DOI] [PubMed] [Google Scholar]
- 32.Sanft T., Aktas B., Schroeder B., et al. Prospective assessment of the decision-making impact of the Breast Cancer Index in recommending extended adjuvant endocrine therapy for patients with early-stage ER-positive breast cancer. Breast Cancer Research and Treatment. 2015;154(3):533–541. doi: 10.1007/s10549-015-3631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed S. D., Dinan M. A., Schulman K. A., Lyman G. H. Cost-effectiveness of the 21-gene recurrence score assay in the context of multifactorial decision making to guide chemotherapy for early-stage breast cancer. Genetics in Medicine. 2013;15(3):203–211. doi: 10.1038/gim.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vataire A.-L., Laas E., Aballéa S., Gligorov J., Rouzier R., Chéreau É. Cost-effectiveness of a chemotherapy predictive test. Bulletin du Cancer. 2012;99(10):907–914. doi: 10.1684/bdc.2012.1652. [DOI] [PubMed] [Google Scholar]
- 35.Thomas S., Pütter C., Weber S., Bornfeld N., Lohmann D. R., Zeschnigk M. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study. British Journal of Cancer. 2012;106(6):1171–1176. doi: 10.1038/bjc.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCannel T. A., Chang M. Y., Burgess B. L. Multi-year follow-up of fine-needle aspiration biopsy in choroidal melanoma. Ophthalmology. 2012;119(3):606–610. doi: 10.1016/j.ophtha.2011.08.046. [DOI] [PubMed] [Google Scholar]
- 37.Gray R. G., Quirke P., Handley K., et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. Journal of Clinical Oncology. 2011;29(35):4611–4619. doi: 10.1200/jco.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 38.Paik S., Shak S., Tang G., et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England Journal of Medicine. 2004;351(27):2817–2826. doi: 10.1056/nejmoa041588. [DOI] [PubMed] [Google Scholar]
- 39.Kamrava M. Prophylactic Hepatic Irradiation for Uveal Melanoma. Clinical Trials 2015, https://clinicaltrials.gov/ct2/show/NCT02336763.
- 40.Sato T. A Randomized Phase ll Study of Adjuvant Sunitinib or Valproic Acid in High-Risk Patients with Uveal Melanoma. Clinical Trials 2014, https://clinicaltrials.gov/ct2/show/NCT02068586?term=NCT02068586.
- 41.Chapman P. Phase II Trial of Adjuvant Crizotinib in High-Risk Uveal Melanoma Following Definitive Therapy, 2014, https://clinicaltrials.gov/ct2/show/NCT02223819. [DOI] [PMC free article] [PubMed]
- 42.Sunitinib M. E. Tamoxifen, and Cisplatin in Treating Patients with High-Risk Ocular Melanoma, 2007, https://clinicaltrials.gov/ct2/show/NCT00489944.