Abstract
Previous studies have elevated the prognostic value of survivin in renal cell carcinoma (RCC). To increase statistical power and improve translation, we systematically searched PubMed, Web of Science, and Embase to identify relevant studies until December 2015 and conducted a standard meta-analysis. Based on the inclusion and exclusion criteria, a total of 12 studies, including 2051 patients, were eligible for further analysis. Results showed that high survivin expression in RCC was associated with poor OS (HR = 2.84, 95% CI 1.68–4.79), CSS (HR = 2.36, 95% CI 1.41–3.95), and PFS (HR = 2.20, 95% CI 1.58–3.08). Survivin expression was also correlated with TNM stage (RR = 2.75, 95% CI 2.21–3.44), pathological T stage (RR = 2.19, 95% CI 1.75–2.75), lymph node metastasis (RR = 2.28, 95% CI 1.61–3.25), distant metastasis (RR = 1.56, 95% CI 1.16–2.08), Fuhrman grade (RR = 2.81, 95% CI 2.29–3.45), tumor size (RR = 1.49, 95% CI 1.24–1.78). Our study suggested that survivin was a prognostic marker in RCC. High survivin expression was correlated with poor prognosis and more advanced clinicopathological features, and it could serve as a biomarker for disease management.
Renal cell carcinoma (RCC) accounts for 3% of all human malignancies and is the third most prevalent genitourinary cancers1. RCC is highly aggressive. Approximately 30% of patients have metastases at first diagnosis, and another 20% of RCC patients with clinically localized disease will develop metastasis even after curative nephrectomy2. Most metastatic RCCs still eventually cause death in spite of the application of targeted therapy3. In this regard, prediction models identifying patients with poor prognosis, who may benefit from early systematic therapy, are greatly needed. To date, the tumor, node, and metastasis (TNM) staging system is a widely used RCC prognostic predictor. However, such classic clinical and pathological factors fail to address the inherent biological heterogeneity of RCC4. Therefore, novel biomarkers that can stratify patients with poor prognosis of RCC are required to guide clinical decisions precisely.
Survivin is a member of the inhibitor of apoptosis protein family and is usually present in embryonic tissues5. Survivin plays a role in cell cycle regulation, inhibition of apoptosis, angiogenesis, and other biological effects6. Intriguingly, survivin is barely detectable in most normal adult tissues but overexpressed in many cancers, including RCC7. With further understanding of the molecular mechanisms of RCC, numerous studies focusing on survivin have been conducted in the fields of outcome prediction and potential therapeutic targets. To obtain a more precise evaluation of the prognostic and clinicopathological value of survivin expression in RCC, we conducted a systematic review and meta-analysis to evaluate the prognostic value of survivin quantitatively and explore the associations of survivin with the clinicopathological features of RCC.
Results
Search Results
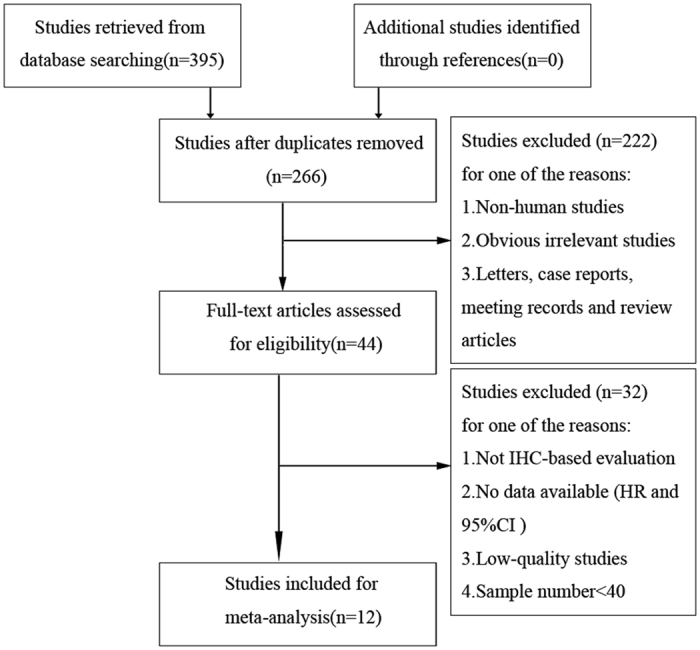
A total of 395 articles were retrieved from the primary literature search. A total of 129 duplicate reports were excluded. After screening the titles and abstracts, 222 articles were excluded for reasons such as non-human studies, letters, case reports, reviews, and other obvious irrelevant studies. The remaining articles were viewed in full text. To avoid the heterogeneity caused by the detection method, studies without IHC evaluation were excluded, and the remaining articles were further excluded for several reasons, such as no data available (HR and 95% CI), low-quality studies8, samples fewer than 40, and duplicate publication. Finally, only 12 articles with 2051 patients satisfied the criteria for meta-analysis9,10,11,12,13,14,15,16,17,18,19,20. A flowchart of the study selection process is shown in Fig. 1.
Figure 1.
Characteristics of Studies
The detailed data of the 12 studies are summarized in Table 1. All of the included studies were published recently (2005–2015). Patients in these studies were all diagnosed with RCC with different tumor types and received radical or partial nephrectomy. Five studies originated from the United States, four from China, one from Germany, one from Italy, and one from Korea. Among the studies, four studies were carried out to analyze OS, seven studies were conducted to investigate CSS, and four studies reported PFS. Various clinicopathological data were reported in seven studies (TNM stage in five studies, pathological T stage in five studies, lymph node metastasis in six studies, distant metastasis in four studies, Fuhrman grade in six studies, tumor size in four studies). All studies applied immunohistochemical staining to investigate survivin expression. The cutoff values of positive survivin expression varied among different studies, so we classified all the cases according to their original studies (negative or positive staining).
Table 1. Characteristics of eligible studies in the meta-analysis.
| Study | Country | Study design | Extent of tumora | Histopathological subtype | Case number | Age (years) | Expression Location | Positive staining | follow-up (months) | Survival analysis | Quality score* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kosari12 | USA | Cohort study | all-stage | ccRCC | 183 | NA | N | IHC scorec ≥2 | >28.8 | CSS | 7 |
| Parker11 | USA | Cohort study | all-stage | ccRCC | 312 | 157/155(≥65 y/<65 y)b | N | ≥2% | >26.4 | CSS, PFS | 8 |
| Byun10 | Korea | Cohort study | all-stage | ccRCC + non-ccRCC | 85 | Mean 53.2 | C | ≥10% | 45 | OS, PFS | 7 |
| Krambeck13 | USA | Cohort study | all-stage | ccRCC | 228 | 113/115(≥65 y/<65 y) | C | ≥2% | >25.2 | CSS, PFS | 8 |
| Zamparese14 | Italy | Cohort study | all-stage | ccRCC | 49 | Mean 62 | C | ≥25% | 47.1 | CSS | 6 |
| Parker15 | USA | Cohort study | all-stage | ccRCC | 634 | 312/322(≥65 y/<65 y) | N | ≥15 positive cells/mm2 | >25.2 | CSS | 7 |
| Eckel-Passow16 | USA | Cohort study | all-stage | ccRCC | 100 | NA | N | NA | >19.2 | CSS | 6 |
| Lei9 | China | Cohort study | all-stage | ccRCC + non-ccRCC | 75 | 31/34(≥50 y/<50 y) | C | ≥25% | NA | OS | 8 |
| Weber17 | Germany | Cohort study | localized | ccRCC | 132 | Median 63.5 | C+N | ≥10% | 122.4 | CSS | 8 |
| Liu18 | China | Cohort study | all-stage | ccRCC | 90 | Mean 52.2 | C | >0 | 48.7 | PFS | 8 |
| Lu19 | China | Cohort study | all-stage | ccRCC | 98 | Mean 55.2 | C | ≥10% | NA | OS | 6 |
| Shi20 | China | Cohort study | all-stage | ccRCC | 65 | Mean 59.8 | C | ≥5% | 19 | OS | 7 |
C: cytoplasm; N: nucleus; OS: overall survival; CSS: cancer-specific survival; PFS: progression-free survival; NA: not available.
aReported at time of dianosis
b157 patients ≥65 years, and other 155 patients <65 years.
cIHC score was measured by computer assisted analysis with the IHCScore software.
*The quality of the included studies was evaluated using the Newcastle–Ottawa scale.
Meta-Analysis
The results showed that high survivin expression in RCC was associated with poor OS (a fixed-effect model, HR = 2.84; 95% CI: 1.68–4.79; p < 0.001; I2 = 6.9%, p = 0.359; Fig. 2A), CSS (a random-effect model, HR = 2.36; 95% CI: 1.41–3.95; p < 0.001; I2 = 90.8%, p < 0.001; Fig. 2B), and PFS (a fixed-effect model, HR = 2.20; 95% CI: 1.58–3.08; p < 0.001; I2 = 0.0%, p = 0.857; Fig. 2C). Furthermore, subgroup analysis stratified by ethnicity, expression location, extent of tumor at time of diagnosis, histopathological subtype and cutoff of staining were also performed. With regard to ethnicity, high survivin expression was associated with poor OS (HR, 2.84; 95% CI: 1.68–4.79; p < 0.001) and PFS (HR, 2.64; 95% CI: 1.54–4.53; p < 0.001) in Asian patients; with poor CSS (HR, 2.36; 95% CI: 1.41–3.95; p = 0.001) and PFS (HR, 1.96; 95% CI: 1.28–3.01; p = 0.002) in Caucasian patients (Table 2). For expression location, high cytoplasmic expression of survivin was associated with poor OS (HR, 2.84; 95% CI: 1.68–4.79; p < 0.001), CSS (HR, 2.33; 95% CI: 1.40–3.88; p = 0.001), and PFS (HR, 2.41; 95% CI: 1.56–3.72; p < 0.001). High nuclear expression of survivin was associated with poor CSS (HR, 2.28; 95% CI: 1.20–4.35; p = 0.012) and PFS (HR, 1.93; 95% CI: 1.14–3.27; p = 0.015). Regarding extent of tumor, high survivin expression was correlated with poor OS (HR, 2.84; 95% CI: 1.68–4.79; p < 0.001), CSS (HR, 2.31; 95% CI: 1.30–4.10; p = 0.004), and PFS (HR, 2.20; 95% CI: 1.58–3.08; p < 0.001) for all stages of RCC; with poor CSS (HR, 2.75; 95% CI: 1.23–6.15; p = 0.014) for localized RCC. For histopathological subtype, high survivin expression was correlated with poor OS (HR, 7.37; 95% CI: 2.21–24.58; p = 0.001), CSS (HR, 2.36; 95% CI: 1.41–3.95; p = 0.001), and PFS (HR, 2.14; 95% CI: 1.51–3.04; p < 0.001) for ccRCC. In the cutoff of staining subgroup analysis, high survivin expression was correlated with poor OS (HR, 5.98; 95% CI: 1.13–31.67; p = 0.036), CSS (HR, 1.95; 95% CI: 1.35–2.83; p < 0.001), and PFS (HR, 2.14; 95% CI: 1.51–3.04; p < 0.001) when the cutoff value was less than 10%. Studies with cutoff value over 10% showed that high survivin expression was related to poor OS (HR, 2.61; 95% CI: 1.51–4.54; p = 0.001) and poor CSS (HR, 3.03; 95% CI: 1.54–5.98; p = 0.001) but not to PFS (HR, 2.92; 95% CI: 0.97–8.80; p = 0.057).
Figure 2. Forest plots of studies evaluating the association of survivin expression and prognostic outcomes of RCC patients and subgroup analysis in terms of different subcellular localization of survivin expression.
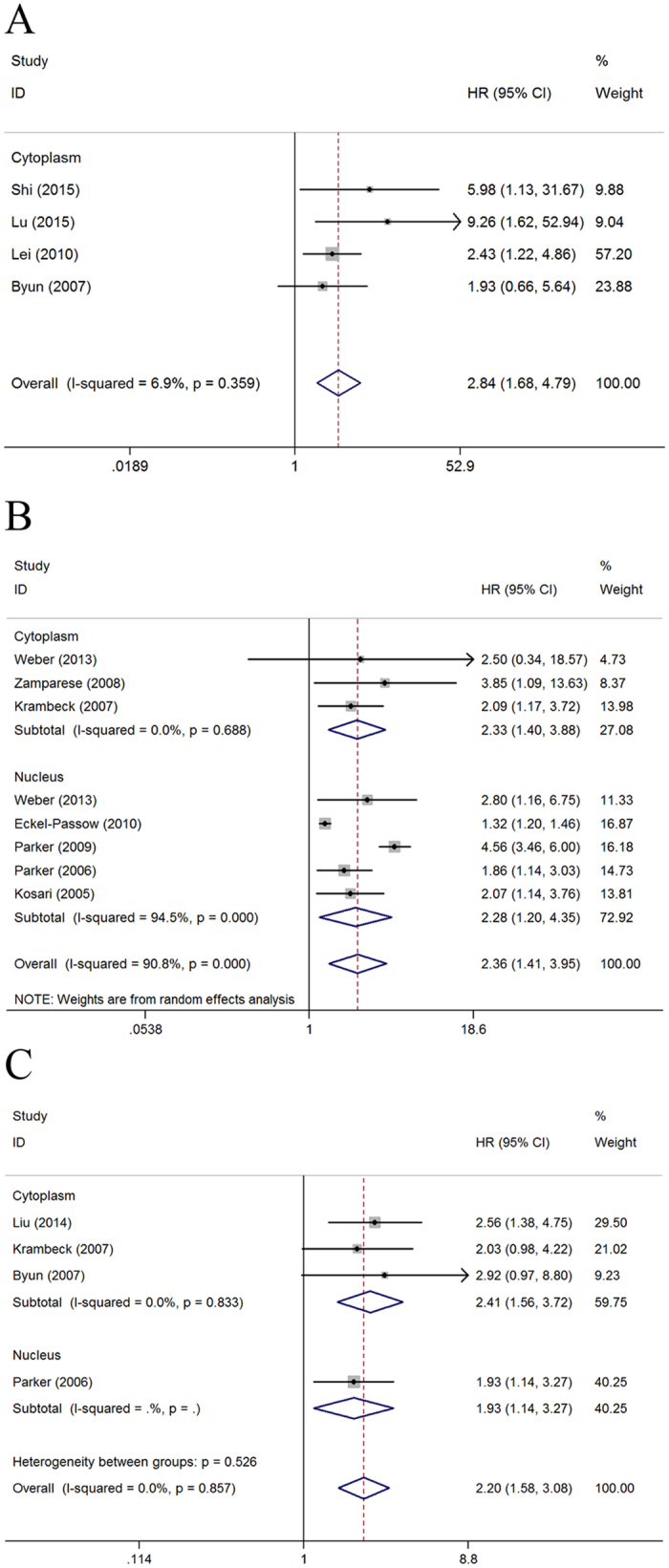
(A) effect of survivin overexpression on OS, (B) CSS, and (C) PFS. HR: hazard ratio; CI: confidence interval; OS: overall survival; CSS: cancer-specific survival; PFS: progression-free survival; RCC: renal cell carcinoma. HR > 1 implies unfavorable prognosis for patients with high survivin expression.
Table 2. Subgroup analysis of pooled HR for RCC patients with survivin overexpression.
| Outcome | Subgroup | Studies | Pooled HR | 95% CI | P Value | Model | HeterogeneityI2 (%) | P Value |
|---|---|---|---|---|---|---|---|---|
| OS | Ethnicity | |||||||
| Caucasian | 0 | – | – | – | – | – | – | |
| Asian | 4 | 2.84 | 1.68–4.79 | <0.001 | fixed | 6.9 | 0.359 | |
| Subcellular location | ||||||||
| nucleus | 0 | – | – | – | – | – | – | |
| cytoplasm | 4 | 2.84 | 1.68–4.79 | <0.001 | fixed | 6.9 | 0.359 | |
| Extent of tumor | ||||||||
| all-stage | 4 | 2.84 | 1.68–4.79 | <0.001 | fixed | 6.9 | 0.359 | |
| localized | 0 | – | – | – | – | – | – | |
| metastatic | 0 | – | – | – | – | – | – | |
| Histopathological subtype | ||||||||
| ccRCC | 2 | 7.37 | 2.21–24.58 | 0.001 | fixed | 0 | 0.722 | |
| Cutoff of staining | ||||||||
| < 10% | 1 | 5.98 | 1.13–31.67 | 0.036 | – | – | – | |
| ≥ 10% | 3 | 2.61 | 1.51–4.54 | 0.001 | fixed | 15.7 | 0.306 | |
| CSS | Ethnicity | |||||||
| Caucasian | 8 | 2.36 | 1.41–3.95 | 0.001 | random | 90.8 | <0.001 | |
| Asian | 0 | – | – | – | – | – | – | |
| Subcellular location | ||||||||
| nucleus | 5 | 2.28 | 1.20–4.35 | 0.012 | random | 94.5 | <0.001 | |
| cytoplasm | 3 | 2.33 | 1.40–3.88 | 0.001 | fixed | 0 | 0.688 | |
| Extent of tumor | ||||||||
| all-stage | 6 | 2.31 | 1.30–4.10 | 0.004 | random | 93.2 | <0.001 | |
| localized | 2 | 2.75 | 1.23–6.15 | 0.014 | fixed | 0 | 0.919 | |
| metastatic | 0 | – | – | – | – | – | – | |
| Histopathological subtype | ||||||||
| ccRCC | 8 | 2.36 | 1.41–3.95 | 0.001 | random | 90.8 | <0.001 | |
| Cutoff of staining | ||||||||
| < 10% | 2 | 1.95 | 1.35–2.83 | <0.001 | fixed | 0 | 0.762 | |
| ≥ 10% | 3 | 3.03 | 1.54–5.98 | 0.001 | fixed | 0 | 0.903 | |
| PFS | Ethnicity | |||||||
| Caucasian | 2 | 1.96 | 1.28–3.01 | 0.002 | fixed | 0 | 0.913 | |
| Asian | 2 | 2.64 | 1.54–4.53 | <0.001 | fixed | 0 | 0.838 | |
| Subcellular location | ||||||||
| nucleus | 1 | 1.93 | 1.14–3.27 | 0.015 | – | – | – | |
| cytoplasm | 3 | 2.41 | 1.56–3.72 | <0.001 | fixed | 0 | 0.833 | |
| Extent of tumor | ||||||||
| all-stage | 4 | 2.20 | 1.58–3.08 | <0.001 | fixed | 0 | 0.857 | |
| localized | 0 | – | – | – | – | – | – | |
| metastatic | 0 | – | – | – | – | – | – | |
| Histopathological subtype | ||||||||
| ccRCC | 3 | 2.14 | 1.51–3.04 | <0.001 | fixed | 0 | 0.782 | |
| Cutoff of staining | ||||||||
| < 10% | 3 | 2.14 | 1.51–3.04 | <0.001 | fixed | 0 | <0.001 | |
| ≥ 10% | 1 | 2.92 | 0.97–8.80 | 0.057 | – | – | – | |
OS: overall survival; CSS: cancer-specific survival; PFS: progression-free survival; HR: hazard ratio; CI: confidence interval; RCC: renal cell carcinoma; ccRCC: clear cell renal cell carcinoma.
In the comprehensive analyses of the role of survivin expression in RCC as a biomarker, we investigated the association of high survivin expression and clinicopathological characteristics. As reported in Table 3, high survivin expression was significantly associated with TNM stage (III/IV vs. I/II: RR, 2.75; 95% CI: 2.21–3.44; p < 0.001), pathological T stage (T3/T4 vs. T1/T2: RR, 2.19; 95% CI: 1.75–2.75; p < 0.001), lymph node metastasis (yes vs. no: RR, 2.28; 95% CI: 1.61–3.25; p < 0.001), distant metastasis (yes vs. no: RR, 1.56; 95% CI: 1.16–2.08; p = 0.003), Fuhrman grade (III/IV vs. I/II: RR, 2.81; 95% CI: 2.29–3.45; p < 0.001), and tumor size (>7 vs. ≤7: RR, 1.49; 95% CI: 1.24–1.78; p < 0.001). Some interstudy significant heterogeneity was observed in distant metastasis and tumor size, but analysis on other parameters did not exhibit significant heterogeneity.
Table 3. Meta-analysis of the association between high survivin expression and clinicopathological features of RCC.
| Variables | Studies | Pooled RR | 95% CI | P Value | Model | HeterogeneityI2 (%) | P Value |
|---|---|---|---|---|---|---|---|
| TNM stage | 5 | 2.75 | 2.21–3.44 | <0.001 | fixed | 0 | 0.520 |
| pT stage | 5 | 2.19 | 1.75–2.75 | <0.001 | fixed | 12.2 | 0.336 |
| Lymph node metastasis | 6 | 2.28 | 1.61–3.25 | <0.001 | fixed | 0 | 0.514 |
| Distant metastasis | 4 | 1.56 | 1.16–2.08 | 0.003 | random | 85.7 | <0.001 |
| Fuhrman grade | 6 | 2.81 | 2.29–3.45 | <0.001 | fixed | 0 | 0.618 |
| Tumor size | 4 | 1.49 | 1.24–1.78 | <0.001 | random | 91.9 | <0.001 |
RR: relative ratio; CI: confidence interval; RCC: renal cell carcinoma.
Sensitivity Analyses
To validate the reliability of our results and investigate the source of significant heterogeneity, sensitivity analysis was performed. Sensitivity analyses showed that the pooled HR was not significantly influenced after omitting any single study for the effect of survivin expression on CSS and revealed that the study15 was the source of statistical heterogeneity (Table 4). When this study was deleted, no significant heterogeneity was observed in the remaining studies (I2 = 47%, p = 0.079) and pooled HR was 1.39 (a fixed-effect model, 95% CI: 1.27–1.53; p < 0.001) or 1.84 (a random-effect model, 95% CI: 1.37–2.47; p < 0.001). No significant heterogeneity was detected in either OS or PFS; hence, we did not conduct sensitivity analysis.
Table 4. Pooled HR (95% CI) of sensitivity analysis for the effect of survivin expression on CSS.
| Study Omitted | Pooled HR | 95% CI | P Value | Model | Heterogeneity I2 (%) | P Value |
|---|---|---|---|---|---|---|
| Kosari12 | 2.42 | 1.36–4.32 | 0.003 | random | 92 | <0.001 |
| Parker11 | 2.47 | 1.36–4.50 | 0.003 | random | 92 | <0.001 |
| Krambeck13 | 2.42 | 1.35–4.33 | 0.003 | random | 92 | <0.001 |
| Zamparese14 | 2.26 | 1.32–3.88 | 0.003 | random | 91.9 | <0.001 |
| Parker15 | 1.39 | 1.27–1.53 | <0.001 | fixed | 47 | 0.079 |
| Eckel-Passow16 | 2.68 | 1.81–3.97 | <0.001 | random | 61.3 | 0.017 |
| Weber(Nucleus)17 | 2.32 | 1.33–4.03 | 0.003 | random | 91.9 | <0.001 |
| Weber(Cytoplasm)17 | 2.36 | 1.39–4.01 | 0.002 | random | 92.1 | <0.001 |
| Combined | 2.36 | 1.41–3.95 | 0.001 | random | 90.8 | <0.001 |
HR: hazard ratio; CI: confidence interval; CSS: cancer-specific survival.
Publication Bias
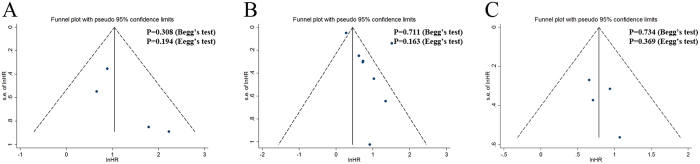
In the present meta-analysis, we introduced Begg’s and Egger’s tests, as well as funnel plots, to assess publication bias. As presented in Fig. 3, the funnel plots revealed that the included studies had no evident asymmetry. Further, the results from Begg’s test (P value) and Egger’s test (intercept with corresponding 95% CI, P value) for the included studies evaluating the survival outcomes were PBegg’s = 0.308, intercept 1.97 with 95% CI −2.43 to 6.36, PEgger’s = 0.194 (OS); PBegg’s = 0.711, intercept 2.27 with 95% CI −1.22 to 5.76, PEgger’s = 0.163 (CSS); PBegg’s = 0.734, intercept 1.13 with 95% CI −3.11 to 5.38, PEgger’s = 0.369 (PFS), respectively. Therefore, the aforementioned evidences suggested significant publication bias did not exist in our meta-analysis.
Figure 3. Funnel plots and Begg’s and Egger’s tests for the evaluation of potential publication bias.
(A) Overall survival; (B) Cancer-specific survival; (C) Progression-free survival.
Discussion
High expression of survivin is observed in many malignancies, but it is barely detectable in most normal adult tissues; thus, survivin is an attractive prognostic prediction marker and potential therapeutic target for several cancer types21,22,23,24. The new paper by Gulati et al. validated survivin gene expression is adverse implications in the prognosis of RCC25. To increase statistical power and improve translation, we conducted a meta-analysis to determine a pooled conclusion and provide evidence on the correlation.
In the current study, we focused exclusively on validating survivin IHC expression and evaluated the prognostic values of survivin IHC expression in RCC. We concluded that high survivin expression predicted poor prognosis in RCC patients. In particular, RCC patients with high survivin expression had shorter OS, CSS, and PFS. Subgroup analysis revealed that high survivin expression was significantly associated with poor OS and CSS, regardless of ethnicity, subcellular localizations, extent of tumor, histopathological subtype, and staining cutoff. Among patients with staining cutoff ≥10%, no significant association was identified between high survivin expression and poor PFS, even though patients with high survivin expression presented a relatively unfavorable PFS. The absence of a significant association was possibly due to the relatively limited studies in the subgroups. The reasons why high survivin expression may be of prognostic relevance in patients suffering from RCC remain speculative. Survivin is an anti-apoptotic protein that has been associated with cellular apoptosis inhibition function through preferential blocking of mitochondrial-dependent apoptosis by targeting caspase 9 and second mitochondria-derived activator of caspases/direct inhibitory apoptotic protein-binding protein with a low isoelectric point. Moreover, survivin presents a mitosis-regulated pattern of expression during the G2/M phase of the cell cycle. In addition to the widely accepted apoptosis inhibition function, survivin also plays a critical role in mitosis and microtubule stability26,27. Abnormal inhibition of apoptosis and cell division during cellular homeostasis is a critical process for the development and progression of RCC.
Consistent with previous reports, our results also suggested that RCC patients with high survivin expression were likely to have a higher TNM stage, pathological T stage, positive lymph nodes, distant metastasis, a higher Fuhrman grade, and a larger tumor size. Currently, survivin is believed to be related to angiogenesis by interacting with vascular endothelial growth factor, angiopoietin, and basic fibroblast growth factor. In addition, survivin is associated with metastatic behavior by activating various signaling pathways28. These mechanisms can explain the association between high survivin expression and clinicopathological characteristics in RCC patients.
This study is comprehensive analysis on the effect of survivin on prognostic and clinicopathological significance in patients with RCC, but several limitations should be pointed out. First, although all included studies measured survivin expression via IHC, the criteria to determine the positive or negative expression of survivin were inconsistent in different studies, which may generate heterogeneity of the overall results. Thus, a more standard cutoff value should be defined in the future. Second, marked heterogeneity of studies was observed in CSS analysis. The heterogeneity of CSS analysis was probably caused by differences in factors such as the patients’ characteristics (ethnicity, nationality, gender, age, and tumor stage and grade), variation of cut-off values for survivin expression, and different duration of follow-up. Third, in evaluating associations between survivin expression and clinicopathological characteristics of RCC, some studies lacked complete data, which may cause heterogeneity and contribute to the low reliability of the results. Furthermore, positive results were more likely to be published in most studies, whereas studies with negative results were often rejected or less assessable, which could lead to publication bias29, although this bias was not detected in the current analysis.
In conclusion, despite the abovementioned limitations, our meta-analysis suggested the prognostic and clinicopathological values of survivin expression in RCC. High survivin expression was correlated with poor prognosis and more advanced clinicopathological features, which may potentially serve as risk stratification markers and even therapeutic targets in RCC. However, more multicenter prospective studies with standardized methods and long-term follow up are needed to verify our results.
Methods
Search Strategy
This meta-analysis followed the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)30.
A systematic literature search was performed in the electronic databases PubMed, Web of Science, and Embase on 31 December 2015 using the following search strategy: (“survivin” or “BIRC5” or “baculoviral inhibitor of apoptosis repeat-containing 5”) and (“carcinoma” or “neoplasm” or “tumor” or “cancer” or “malignancy”) and (“kidney” or “renal”) and (“prognosis” or “prognostic” or “survival” or “outcome” or “mortality”). Furthermore, we manually searched the reference lists of relevant literature.
Selection Criteria
Studies were included based on the following criteria: (1) the association of survivin with prognosis significance in RCC should be described; (2) studies detected survivin protein expression by immunohistochemistry(IHC); and (3) studies reported survival outcomes [overall survival (OS), cancer-specific survival (CSS), or progression-free survival (PFS)] with hazard ratio (HR) and 95% confidence interval (CI). Exclusion criteria were as follows: (1) non-English papers; (2) case reports, letters, commentaries, meeting records, or review articles; (3) sample number fewer than 40 patients; (4) the study focused on animal models or cancer cells; (5) the study did not analyze survivin protein expression, clinical features, and survival outcome; (6) the study lacked sufficient data for obtaining HR and 95% CI. All evaluations were independently performed by three individual researchers to ensure the accurate inclusion of studies. For duplicate studies, we only retrieved the most informative and recently studied one for further analyses.
Because this study was based on published literature, ethical approval from ethics committees was not needed.
Data Extraction
Three investigators independently extracted data from eligible studies using a predefined form. Discrepancies in data extraction were resolved by discussion. The following data were extracted: surname of the first author, publication year, origin of the studied population, study design, extent of tumor, histopathological subtype, sample size, patient’s age, location of survivin expression, cutoff value, follow-up time, and effect estimates, namely, HR of survivin expression for OS, CSS, or PFS, as well as their 95% CI (Table 1). If the HR and 95% CI were not directly available, we calculated HRs and their 95% CI based on the methods reported by Tierney et al.31.
Quality Assessment
The quality of the included studies was evaluated using the Newcastle–Ottawa scale, which was recommended by the Cochrane Non-Randomized Studies Methods Working Group8. Each study can be assessed by eight methodology items with a score ranging from 0 to 9. High scores indicated high quality, and we considered studies with scores of 6 or more as high quality for the meta-analysis.
Statistical Analysis
Pooled HR and risk ratio (RR) with 95% CI were used to evaluate the association of survivin expression with RCC prognosis and clinicopathological characteristics, respectively. An observed HR > 1 implied worse prognosis for patients with high survivin expression. An observed RR > 1 implied more advanced clinicopathological characteristics for the group of high survivin expression. A heterogeneity test of pooled HR and RR was conducted using Cochran’s Q test and Higgins I-squared statistic. I2 values >50% indicated heterogeneity among studies32. When heterogeneity was observed (I2 > 50%), a random-effect model was used; otherwise, a fixed-effect model was used. For additional analyses, meta-analyses were subgrouped based on ethnicity, subcellular localizations, extent of tumor, histopathological subtype, and staining cutoff. For the investigation of heterogeneous studies, we also conducted sensitivity analysis to evaluate the influence of individual studies on the robustness of pooled results. Publication bias was assessed by funnel plot visual inspection and statistically evaluated by Begg’s33 and Egger’s tests34. All statistical analyses were performed using Stata 12.0 software (StatCorp, College Station, TX, USA) and p < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Xie, Y. et al. Prognostic and Clinicopathological Significance of Survivin Expression in Renal Cell Carcinoma: A Systematic Review and Meta-Analysis. Sci. Rep. 6, 29794; doi: 10.1038/srep29794 (2016).
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (2014AA020607)
Footnotes
Author Contributions Conceived and designed the experiments: Y.X., X.M. and X.Z. Performed the experiments: Y.X. and L.G. Analyzed the data: H.L., L.C., X.L., Y.G. and Y.F. Contributed analysis tools: Y.Z. and Y.Y. Wrote the paper: Y.X. and X.Z. Revised the manuscript: H.L. and L.G. All authors have reviewed the manuscript.
References
- DeSantis C. E. et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 64, 252–271 (2014). [DOI] [PubMed] [Google Scholar]
- Athar U. & Gentile T. C. Treatment options for metastatic renal cell carcinoma: a review. Can J Urol 15, 3954–3966 (2008). [PubMed] [Google Scholar]
- Oudard S. & Vano Y. The role of rechallenge with targeted therapies in metastatic renal-cell carcinoma. Curr Opin Urol 25, 402–410 (2015). [DOI] [PubMed] [Google Scholar]
- Frank I. et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol 168, 2395–2400 (2002). [DOI] [PubMed] [Google Scholar]
- Altieri D. C. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer 8, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- Sah N. K., Khan Z., Khan G. J. & Bisen P. S. Structural, functional and therapeutic biology of survivin. Cancer Lett 244, 164–171 (2006). [DOI] [PubMed] [Google Scholar]
- Mahotka C. et al. Distinct in vivo expression patterns of survivin splice variants in renal cell carcinomas. Int J Cancer 100, 30–36 (2002). [DOI] [PubMed] [Google Scholar]
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25, 603–605 (2010). [DOI] [PubMed] [Google Scholar]
- Lei Y., Geng Z., Guo-Jun W., He W. & Jian-Lin Y. Prognostic significance of survivin expression in renal cell cancer and its correlation with radioresistance. Mol Cell Biochem 344, 23–31 (2010). [DOI] [PubMed] [Google Scholar]
- Byun S. S., Yeo W. G., Lee S. E. & Lee E. Expression of survivin in renal cell carcinomas: association with pathologic features and clinical outcome. Urology 69, 34–37 (2007). [DOI] [PubMed] [Google Scholar]
- Parker A. S. et al. High expression levels of survivin protein independently predict a poor outcome for patients who undergo surgery for clear cell renal cell carcinoma. Cancer 107, 37–45 (2006). [DOI] [PubMed] [Google Scholar]
- Kosari F. et al. Clear cell renal cell carcinoma: Gene expression analyses identify a potential signature for tumor aggressiveness. Clinical Cancer Research 11, 5128–5139 (2005). [DOI] [PubMed] [Google Scholar]
- Krambeck A. E. et al. Survivin and b7-h1 are collaborative predictors of survival and represent potential therapeutic targets for patients with renal cell carcinoma. Clin Cancer Res 13, 1749–1756 (2007). [DOI] [PubMed] [Google Scholar]
- Zamparese R. et al. Survivin expression in renal cell carcinoma. Cancer Investigation 26, 929–935 (2008). [DOI] [PubMed] [Google Scholar]
- Parker A. S. et al. Development and evaluation of BioScore: a biomarker panel to enhance prognostic algorithms for clear cell renal cell carcinoma. Cancer 115, 2092–2103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Passow J. E. et al. Tissue microarrays: one size does not fit all. Diagn Pathol 5, 48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T. et al. Immunohistochemical analysis of prognostic protein markers for primary localized clear cell renal cell carcinoma. Cancer Invest 31, 51–59 (2013). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. Survivin and HLA-I expression predicts survival of patients with clear cell renal cell carcinoma. Tumour Biol 35, 8281–8288 (2014). [DOI] [PubMed] [Google Scholar]
- Lu H. et al. The expression and role of MEKK3 in renal clear cell carcinoma. Anat Rec (Hoboken) 298, 727–734 (2015). [DOI] [PubMed] [Google Scholar]
- Shi Z. G. et al. Expression of vimentin and survivin in clear cell renal cell carcinoma and correlation with p53. Clin Transl Oncol 17, 65–73 (2015). [DOI] [PubMed] [Google Scholar]
- Lv S. et al. Prognostic and clinicopathological significance of survivin expression in bladder cancer patients: a meta-analysis. Tumour Biol 35, 1565–1574 (2014). [DOI] [PubMed] [Google Scholar]
- Liu Y., Teng Z., Wang Y., Gao P. & Chen J. Prognostic Significance of Survivin Expression in Osteosarcoma Patients: A Meta-Analysis. Med Sci Monit 21, 2877–2885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S. et al. The impact of survivin on prognosis and clinicopathology of glioma patients: a systematic meta-analysis. Mol Neurobiol 51, 1462–1467 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Prognostic and Clinicopathological Value of Survivin in Diffuse Large B-cell Lymphoma: A Meta-Analysis. Medicine (Baltimore) 94, e1432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S. et al. Systematic evaluation of the prognostic impact and intratumour heterogeneity of clear cell renal cell carcinoma biomarkers. Eur Urol 66, 936–948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer 92, 212–216 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H. & Mak T. W. Pathways of apoptotic and non-apoptotic death in tumour cells. Nat Rev Cancer 4, 592–603 (2004). [DOI] [PubMed] [Google Scholar]
- Semba S. et al. Fhit modulation of the Akt-survivin pathway in lung cancer cells: Fhit-tyrosine 114 (Y114) is essential. Oncogene 25, 2860–2872 (2006). [DOI] [PubMed] [Google Scholar]
- Sutton A. J., Song F., Gilbody S. M. & Abrams K. R. Modelling publication bias in meta-analysis: a review. Stat Methods Med Res 9, 421–445 (2000). [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. & Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6, e1000097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney J. F., Stewart L. A., Ghersi D., Burdett S. & Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8, 16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J. & Altman D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994). [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]