ABSTRACT
The cyanobacterium Synechocystis sp. strain PCC 6803 encodes a single DEAD box RNA helicase, CrhR, whose expression is tightly autoregulated in response to cold stress. Subcellular localization and proteomic analysis results indicate that CrhR localizes to both the cytoplasmic and thylakoid membrane regions and cosediments with polysome and RNA degradosome components. Evidence is presented that either functional RNA helicase activity or a C-terminal localization signal was required for polysome but not thylakoid membrane localization. Polysome fractionation and runoff translation analysis results indicate that CrhR associates with actively translating polysomes. The data implicate a role for CrhR in translation or RNA degradation in the thylakoid region related to thylakoid biogenesis or stability, a role that is enhanced at low temperature. Furthermore, CrhR cosedimentation with polysome and RNA degradosome complexes links alteration of RNA secondary structure with a potential translation-RNA degradation complex in Synechocystis.
IMPORTANCE The interaction between mRNA translation and degradation is a major determinant controlling gene expression. Regulation of RNA function by alteration of secondary structure by RNA helicases performs crucial roles, not only in both of these processes but also in all aspects of RNA metabolism. Here, we provide evidence that the cyanobacterial RNA helicase CrhR localizes to both the cytoplasmic and thylakoid membrane regions and cosediments with actively translating polysomes and RNA degradosome components. These findings link RNA helicase alteration of RNA secondary structure with translation and RNA degradation in prokaryotic systems and contribute to the data supporting the idea of the existence of a macromolecular machine catalyzing these reactions in prokaryotic systems, an association hitherto recognized only in archaea and eukarya.
INTRODUCTION
Compartmentalization of metabolic processes is crucial for cells to avoid futile cycles. While this is efficiently achieved by lipid bilayers in eukaryotic cells, this solution does not generally apply in prokaryotes. Although prokaryotes have the ability to compartmentalize using invagination of the inner cell membrane, these compartments, for example, magnetosomes, tend to perform one specific function and are not widely used (1). Prokaryotes can also form proteinaceous microcompartments in which specific reactions occur, the primary cyanobacterial example being that of carboxysomes (2). Prokaryotes do not compartmentalize routine, everyday anabolic and catabolic processes such as transcription and translation or protein and RNA decay; however, there is a growing body of evidence indicating that prokaryotes spatially confine these cellular processes (3). A prime example is the localization of the RNA degradation complex, the degradosome, to the cytoplasmic membrane (CM) in Escherichia coli (4) and Bacillus subtilis (5).
The E. coli degradosome consists of RNase E, polynucleotide phosphorylase (PNPase), enolase, and the DEAD box RNA helicase RhlB (6). DEAD box RNA helicases are ubiquitous enzymes that participate in all aspects of RNA metabolism (7). While eukaryotes generally encode numerous DEAD box RNA helicases, 25 in yeast (8) and over 50 in Arabidopsis (9), prokaryotic genomes encode fewer members, 5 in E. coli (10) and 4 in B. subtilis (11). Structurally, RNA helicases consist of two tandem RecA-like helicase domains with N-terminal and/or C-terminal extensions that provide substrate and protein interaction domains (7). Functionally, in all organisms, RNA helicases are generally associated with ribonucleoprotein (RNP) complexes frequently involved in RNA degradation (12, 13, 14), translation initiation (15), and ribosome biogenesis (16, 17). While the majority of RNA helicases perform specific functions, some bacterial enzymes, including DeaD (CsdA) in E. coli (18) and CshA in Bacillus cereus (19), are associated with a variety of pathways. In some systems, the multifunctionality of these helicases originates from their association with different RNP complexes in response to environmental stress. For example, the degradosome-associated RNA helicase RhlB in E. coli can be functionally replaced by another DEAD box helicase, either DeaD or RhlE, during low-temperature stress, forming a cold-specific degradosome (20). In addition, three of the five DEAD box RNA helicases in B. cereus have divergent functions in response to a variety of stresses, including temperature (19). Thus, the reduction in the RNA helicase repertoire present in bacteria can be compensated by some helicases performing multiple functions.
Divergent RNA helicase functions are also associated with specific subcellular localization. For example, four of the five E. coli DEAD box RNA helicases localize with respect to their physiological function. RhlB is RNA degradosome associated at the cytoplasmic membrane, and SrmB and DbpA are solubility and ribosome associated, while the multifunctional helicase DeaD is associated with all three functions (18). Although RNA helicases do not contain canonical membrane-spanning domains, they are membrane associated in some bacteria (12, 21, 22) but not all (13). For example, RNA helicases associated with RNA degradosomes localize to the cytoplasmic membrane via RNase E in E. coli (4, 12) and RNase Y in B. subtilis (5) whereas CshA and CshB colocalize with CspB and ribosomes in areas surrounding the B. subtilis nucleoid, the localization being dependent on active transcription (23). Localization of these RNA helicase-containing complexes to specific cellular sites therefore confines the associated processes to restricted cellular regions. Thus, understanding RNA helicase localization provides insight into how the spatial separation of synthesis and degradation contributes to an integrated mechanism regulating cellular pathways in bacteria.
The Gram-negative, photosynthetic cyanobacteria also encode limited numbers of DEAD box RNA helicases, for example, one in Synechocystis sp. strain PCC 6803 (24, 25) and two in Anabaena sp. strain PCC 7120 (26–28). In Anabaena, crhB is expressed in response to a range of environmental conditions whereas crhC is exclusively expressed in response to temperature downshift (26, 27). In contrast, expression of the Synechocystis DEAD box RNA helicase encoded by crhR is regulated by abiotic stresses that alter the redox status of the electron transport chain in the thylakoid membrane (TM) (29), including temperature stress (24, 30) and salt stress (31). crhR expression is regulated at a number of CrhR-independent and CrhR-dependent checkpoints in response to temperature (24). The autoregulatory, CrhR-dependent checkpoints include temperature regulation of crhR transcript and protein half-life (24). CrhR protein half-life is controlled by conditional, temperature-upshift-induced proteolysis that generates the reduction in CrhR abundance observed at the optimal growth temperature, 30°C (24, 32). A truncated mutant of crhR, crhRTR, causes severe morphological and physiological aberrations in Synechocystis, including decreased photosynthetic electron transport, carbon fixation, and oxygen evolution, as well as significant disorganization of internal cell structures, including the thylakoid membrane (33). The results suggest that CrhR is associated with maintaining the photosynthetic capacity of the cells, possibly through a function associated with thylakoid assembly or function; however, the exact physiological function has not been determined.
Although subcellular localization is a crucial aspect required to decipher the physiological role performed by prokaryotic RNA helicases, RNA helicase localization in cyanobacteria has been reported only for CrhC in Anabaena sp. strain PCC 7120, where it localizes to the cytoplasmic membrane, primarily at the cell poles (21). In an attempt to identify the physiological pathway(s) associated with CrhR activity, we investigated the subcellular localization of this stress-induced RNA helicase and identified protein complexes with which it interacts. Here, we provide evidence that CrhR localizes to both the thylakoid membrane and cytoplasmic region, from which it cosediments with both polysome and RNA degradosome components. The results suggest that Synechocystis possesses a polysome-RNA degradosome complex to which CrhR localizes, linking alteration of RNA secondary structure with a translation-RNA turnover macromolecular complex in prokaryotic systems.
MATERIALS AND METHODS
Culture conditions and strains.
Wild-type Synechocystis sp. strain PCC 6803 (referred to here as Synechocystis) and a truncated crhR mutant strain, crhRTR, were cultivated photoautotrophically on BG-11 medium as described previously (24). The crhRTR mutant was created by insertion of a spectinomycin-streptomycin resistance cassette at the PmlI site halfway between motifs III (SAT) and IV (FVRTK), thereby removing the second RecA domain and C-terminal extension from crhR (33). CrhRTR, the 27-kDa truncated version of the CrhR polypeptide expressed in this strain (24, 32), is biochemically inactive (D. Chamot and G. W. Owttrim, unpublished data). The crhRTR mutant strain was grown on BG-11 medium containing sodium thiosulfate (0.3%) that was buffered with tricine (10 mM; pH 8.0) and supplemented with 50 μg/ml each of spectinomycin and streptomycin (34). For liquid cultures, cell mass was increased gradually, starting at 50 ml and progressing to 300 ml and finally to 4 liters at 30°C with continuous shaking (150 rpm) coupled with bubbling with humidified air at an illumination of 50 μmol photons m−2 s−1 (34). To induce CrhR expression, mid-log-phase cells were cold stressed for 3 h at 20°C. Representative data from a minimum of three biological replicates for each experiment are shown.
Extraction of soluble and insoluble fractions of Synechocystis cells.
Cultures of wild-type Synechocystis and crhRTR cells were divided, with half of the culture volume maintained at 30°C and half of the culture volume maintained at 20°C for 3 h. Aliquots were collected by centrifugation at the indicated growth temperature. Cells were mechanically lysed by vortex mixing in the presence of glass beads in cyanobacterial protein extraction buffer (PEB; 20 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM MgCl2, 5 mM dithiothreitol [DTT], Roche cOmplete Mini EDTA-free protease inhibitor cocktail) as described previously (25, 34). An initial centrifugation at 1,000 × g for 1 min removed unlysed cells and glass beads and was followed by clarification of the lysate at 15,000 × g for 15 min. Following clarification, the pellet (membrane fraction) was washed with PEB and suspended in PEB at the same volume as the soluble, cytoplasmic fraction. For the solvent and RNase extraction of CrhR, aliquots of the membrane fraction corresponding to 60 μg cytoplasmic protein were utilized for each treatment. The STS buffer used for treatment contains 0.4% SDS, 0.5% Triton X-100, and 0.5% Sarkosyl. Solvent extractions were allowed to proceed on ice for 30 min, while RNase A (0.4 μg/μl) treatment was performed at 37°C for 30 min. Solubilized protein were separated from remaining insoluble material by centrifugation at 15,000 × g for 15 min. Supernatants were retained and pellets suspended in an equivalent volume of PEB. Protein concentrations of the cytoplasmic fractions were determined with the Bradford assay (Bio-Rad) using bovine serum albumin (BSA) as the standard. Cytoplasmic proteins (10 μg) or the equivalent volume of the insoluble fraction was resolved by 10% SDS-PAGE, electrophoretically transferred to Protran 0.45-μm-pore-size nitrocellulose membranes (Amersham), and subjected to Western blotting.
Polysome isolation.
Polysomes were isolated from cyanobacterial cytoplasmic extracts as described by Tyystjärvi et al. (35) with modifications. Two-liter mid-log-phase cell cultures were harvested by centrifugation at 7,500 × g, and the pellet was washed with 0.1× volume wash buffer (0.4 M sucrose; 50 mM Tris [pH 8.5], 10 mM MgCl2, 30 mM EDTA [pH 8.0], 500 μg/ml chloramphenicol) and harvested by centrifugation. For the polysome runoff experiment, cells were grown and the lysates processed as previously described except that chloramphenicol was omitted. The resulting pellet was washed with 0.1× volume polysome isolation buffer (0.4 M sucrose, 50 mM Tris, [pH 8.5], 10 mM MgCl2, 500 μg/ml chloramphenicol) and harvested, and the pellets were suspended in 0.0125× polysome isolation buffer containing protease inhibitor cocktail (Roche). Cells were lysed by passage through a continuous French pressure cell press (American Instrument Company) system at 20,000 lb/in2 three times. The cell lysate was cleared of intact cells by centrifugation at 1,000 × g and clarified twice at 18,000 × g, separating the cell debris from the cytoplasmic fraction. Aliquots of both pellet and supernatant fractions were retained for Western analysis. The supernatant was equilibrated with 0.5× volume polysome isolation buffer without sucrose and treated with polyoxyethylene (10) tridecyl ether (Sigma-Aldrich) (2% [vol/vol]) (35). Samples were incubated on ice for 10 min and unsolubilized components eliminated by two centrifugations at 18,000 × g for 20 min each time. Clarified supernatant was layered on a 1 M sucrose cushion (1.0 M sucrose, 50 mM Tris [pH 8.5], 10 mM MgCl2, 500 μg/ml chloramphenicol) and centrifuged at 4°C for 16 h at 243,500 × g in a SW40Ti rotor using a L8-60M ultracentrifuge (Beckman Coulter Inc., Pasadena, CA). Pigmented layers were individually harvested, and the polysome-containing pellet was suspended in polysome isolation buffer without sucrose. Samples of the supernatant layers (equivalent to 100 μg protein) and polysome pellet (A260 = 100 U) were analyzed by Western blotting. Blots were purposely overloaded to detect any CrhR in the fractions. Constituent proteins in the polysome pellet were identified by mass spectrometry.
Polysome fractionation.
The cytoplasmic fraction from a wild-type Synechocystis cell extract grown at 20°C for 3 h was overlaid on a continuous sucrose gradient (20% to 60% sucrose) prepared using a gradient former (DCode system; Bio-Rad) and centrifuged at 243,500 × g for 16 h at 4°C as described above. The absorbance of each fraction was measured at 254 nm using a NanoDrop spectrophotometer (ThermoScientific). Equal aliquots of each fraction were analyzed by Western blotting.
Membrane isolation.
The three membranes present in Synechocystis cells were separated by discontinuous flotation sucrose gradient fractionation as initially described by Murata and Omata (36) and modified according to the method of El-Fahmawi and Owttrim (21). Cultures grown at 30°C or cold shocked at 20°C for 3 h were harvested at 7,500 × g for 15 min at 4°C. The cell pellet was washed with 0.1× volume of potassium phosphate buffer (20 mM potassium phosphate; pH 7.8) and suspended in 0.1× volume potassium phosphate buffer supplemented with protease inhibitor cocktail (Roche). Cells were lysed by vortex mixing in the presence of glass beads (25, 34), and insoluble, membrane-containing material was pelleted by centrifugation at 111,000 × g for 30 min at 4°C in a SW40Ti rotor as described above. The crude membrane pellet was gently washed, centrifuged, and suspended in 20 mM potassium phosphate buffer and adjusted to a final sucrose concentration of 50%. A discontinuous sucrose gradient using 20 mM potassium phosphate and the indicated increasing sucrose concentrations (wt/vol) and volume ratios was assembled for the wild-type membrane isolation as follows: 10% (0.2×); 30% (0.09×); 39% (0.23×); and 50% (0.49×) (cell lysate adjusted with sucrose). For the crhRTR mutant membrane suspension, a finer discontinuous sucrose gradient was assembled, using 20 mM potassium phosphate buffer and the indicated increasing sucrose concentrations (wt/vol) and volume ratios as follows: 10% (0.03×); 30% (0.17×); 35% (0.17×); 38% (0.17×); 42% (0.19×) (cell lysate adjusted with sucrose); 50% (0.14×); and 60% (0.14×) (37). The crude membrane fraction was dissolved in 42% sucrose, and membrane separation was performed by floatation ultracentrifugation at 131,500 × g for 16 h at 4°C in a SW40Ti rotor as described above. A fraction from each membrane-containing layer was carefully harvested and diluted three times with 20 mM potassium phosphate. Individual membrane-containing fractions were pelleted at 188,000 × g for 45 min at 4°C as described above, and the pellets were suspended in 20 mM potassium phosphate buffer. The procedure also yielded a pellet that is referred to here as the cell wall pellet.
Western analysis.
Western analysis was performed on the fractionated samples as described previously (24, 34) with slight modifications. Proteins were separated on SDS–10% polyacrylamide gels and electroblotted onto nitrocellulose membrane using a semidry transfer apparatus (Tyler Research). Membranes were blocked with 5% skimmed milk–Tris-buffered saline (TBS; pH 7.4) for 3 h prior to the addition of polyclonal antisera. Anti-CrhR and anti-E. coli ribosomal protein S1 (S1) antibodies were used at a 1:5,000 dilution (24). For Synechocystis membrane-specific protein detection, anti-FtsH (raised against Arabidopsis thaliana) and anti-Vipp1 (immunoreactive to Synechocystis) were used at a 1:2,000 dilution as thylakoid and cytoplasmic membrane-specific markers, respectively. The Synechocystis thylakoid-specific protein PsbA (D1) antibody (1:5,000 dilution) was used to verify CrhR localization. All immunoreactive polypeptides were detected with anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Sigma) (1:20,000 dilution) using enhanced chemiluminescence (ECL; Amersham).
Mass spectrometry.
The crude polysome pellet from the first ultracentrifugation was treated with 2% polyoxyethylene (10) tridecyl ether (35), layered over a 9-ml 1 M sucrose cushion, and centrifuged as described above. The resulting pellet was suspended in polysome isolation buffer without sucrose. A260 (100 U) fractions of this polysome pellet were resolved on an SDS–4 to 15% polyacrylamide gel (Bio-Rad) and stained with colloidal Coomassie (0.08% Coomassie brilliant blue G250, 1.6% orthophosphoric acid, 8% ammonium sulfate, 20% methanol). Mass spectrometry analysis was performed at the Alberta Proteomics and Mass Spectrometry Facility (APM), University of Alberta. In-gel trypsin digestion was performed on the samples. Each excised lane was cut into 10 equal gel sections, destained twice in 100 mM ammonium bicarbonate/acetonitrile (ACN) (50:50), reduced (10 mM beta-mercaptoethanol [BME]–100 mM bicarbonate), and alkylated (55 mM iodoacetamide–100 mM bicarbonate). After dehydration, trypsin digestion (6 ng/μl) was allowed to proceed overnight at room temperature. Tryptic peptides were extracted from the gel using 97% water–2% acetonitrile–1% formic acid followed by a second extraction using 50% of the initial extraction buffer and 50% acetonitrile. Fractions containing tryptic peptides were resolved and ionized using nanoflow high-performance liquid chromatography (HPLC) (Easy-nLC II; Thermo Scientific) coupled to an LTQ XL-Orbitrap hybrid mass spectrometer (Thermo Scientific). Nanoflow chromatography and electrospray ionization were accomplished by using a PicoFrit fused silica capillary column (ProteoPepII; C18) with a 100-μm inner diameter (New Objective) (300 Å, 5 μm pore size). Peptide mixtures were injected onto the column at a flow rate of 3,000 nl/min and resolved at 500 nl/min using 70-min linear gradients of 4% to 45% (vol/vol) aqueous ACN with 0.2% (vol/vol) formic acid. The mass spectrometer was operated in data-dependent acquisition mode, recording high-accuracy and high-resolution Orbitrap survey spectra using external mass calibration, with a resolution of 60,000 and m/z range of 400 to 2,000. The 10 most intensely multiply charged ions were sequentially fragmented by using collision-induced dissociation, and spectra of their fragments were recorded in the linear ion trap; after two fragmentations, all precursors selected for dissociation were dynamically excluded for 60 s. Data were processed using Proteome Discoverer 1.4 (Thermo Scientific), and the Uniprot cyanobacteria database was searched using SEQUEST (Thermo Scientific). Search parameters included a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.8 Da. Peptides were searched with carbamidomethyl cysteine as a static modification and oxidized methionine and deamidated glutamine and asparagine as dynamic modifications.
Immunogold electron microscopy (IEM).
Ultrathin sections were prepared as described by El-Fahmawi and Owttrim (21) with modifications. Cells were fixed with 4% paraformaldehyde–0.8% glutaraldehyde–0.1 M phosphate buffer (pH 7.2) for 60 min with shaking and washed three times with 0.1 M phosphate buffer for 15 min each time followed by ascending ethanol dehydration (1× [30%]; 1× [50%]; 1× [70%]; 2× [90%]) for 15 min each time. Dehydrated cell pellets were infiltrated with an LR White resin-ethanol solution (2:1) for 3 h and finally with 100% LR White resin overnight. Cell pellets were washed with fresh LR White resin, transferred to gelatin capsules, and polymerized at 50°C for 24 h. Ultrathin sections (80 nm) were prepared on a Reichert-Jung Ultra Cut E microtome, mounted on nickel-coated grids, and blocked with phosphate buffer-gelatin solution (PBG; 10 mM sodium phosphate [pH 7.2], 600 mM NaCl, 0.5% fish gelatin). Primary antibodies (1:100 dilution) were included as indicated for 1 h, washed extensively with phosphate-buffered saline (PBS), and labeled with goat anti-rabbit IgG coupled with 5-nm-diameter gold particles (Sigma-Aldrich) (1:20 dilution) for 1 h. Grids were washed extensively with PBST (0.1% Tween 20–PBS), stained with 4% uranyl acetate for 10 min and with 2.5% lead citrate for 3 min, and fixed with 2% glutaraldehyde for 1 min and washed with distilled water for 1 min. Sections were viewed using a Morgagni 268 transmission electron microscope (TEM) (Philips) and Morgagni 268 3.0 software (33).
RESULTS
CrhR cytoplasmic and membrane localization.
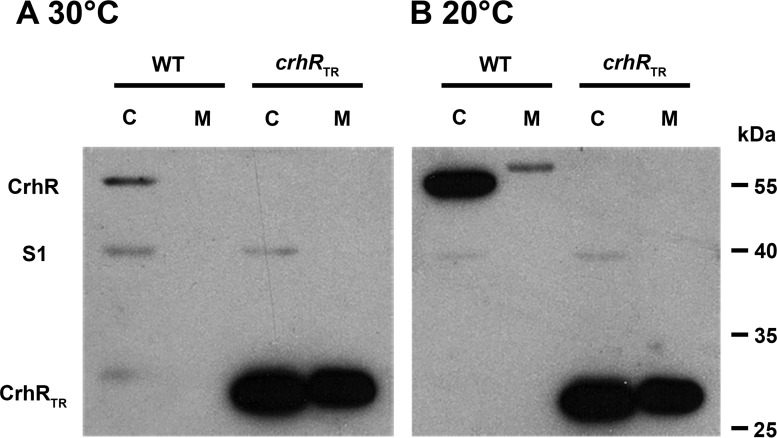
Differential extraction revealed that CrhR was detected in both the soluble and insoluble fractions of cell lysates, suggesting association with the cytoplasm and membrane fractions, respectively (Fig. 1). Previously, CrhR had been detected only in soluble lysate, where it was present at background levels in cells grown at 30°C and increased in abundance by ∼10-fold at 20°C (24). The same change in absolute protein abundance was observed here, with CrhR abundance increasing at 20°C in both the cytoplasmic and membrane fractions. Unexpectedly, and consistently, CrhR migrated slower in the membrane fraction (Fig. 1B), a result of excess lipid present in these samples, as only a single peptide was detected when the cytoplasmic and membrane fractions were mixed (data not shown). Although the majority of the wild-type CrhR was detected in the cytoplasmic fraction, the results of extraction into this fraction were variable in our hands, likely due to the intensity of the mechanical extraction (data not shown). In contrast, CrhRTR, a truncated form of the CrhR protein, did not change with respect to either the abundance or the segregation of the protein (Fig. 1), consistent with the previously observed lack of expression regulation of the truncated protein (24). This suggests that CrhRTR is overexpressed in the mutant at 30°C and that the truncation alters CrhRTR localization. Localization of the control, primarily soluble ribosome-associated protein S1, was detected in supernatant fractions only under the conditions tested (Fig. 1). These observations led us to further analyze CrhR association with both the cytoplasmic and membrane fractions.
FIG 1.
CrhR abundance in cytoplasmic and membrane fractions. Wild-type (WT) and crhRTR mutant strains were grown at 30°C (A), and half of the culture was incubated at 20°C for 3 h (B). Cells were mechanically lysed and clarified by centrifugation at 13,000 × g to generate crude soluble cytoplasmic (lanes C) and membrane (lanes M) fractions. Soluble protein (10 μg) or an equivalent volume of the membrane fraction was subjected to Western analysis. CrhR (55 kDa), S1 (40 kDa), and CrhRTR (27 kDa) were detected using anti-CrhR and anti-S1 antibodies and ECL.
The degree to which CrhR associates with the membrane fraction was investigated by determining CrhR release in response to classic membrane-disrupting agents. As shown in Fig. S1A in the supplemental material, CrhR was relatively weakly associated with this fraction, as high-salt and/or high-pH treatments are sufficient to remove a portion of CrhR from the membrane pellet. Among the gentle, nonionic detergents tested, only polyoxyethylene (10) tridecyl ether was able to remove a portion of the CrhR from the membrane fraction. The treatment using STS buffer, a mixture of ionic and nonionic detergents, was the only treatment that completely removed CrhR from the membrane fraction. RNase A treatment also did not remove CrhR from the membrane fraction (see Fig. S1B). The extraction results indicate that a portion of the cellular CrhR in Synechocystis consists of a peripheral membrane-associated protein.
CrhR polysome cosedimentation.
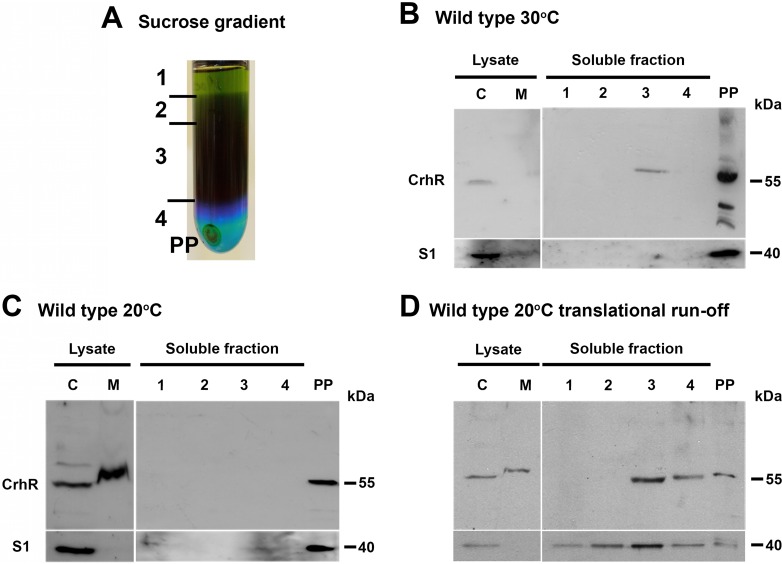
CrhR association with both cytoplasmic and membrane fractions prompted us to investigate the localization of CrhR within the Synechocystis cytoplasm. Sucrose ultracentrifugation separated the soluble Synechocystis extract into colored layers, as well into as a pellet that contained proteins capable of sedimentation through a 1 M sucrose cushion (Fig. 2A). Western analysis detected the slower-migrating, 58-kDa form of CrhR in the dark green (third) layer obtained from the sucrose gradient ultracentrifugation (Fig. 2B). This layer contained residual amounts of chlorophyll and thus most likely contained thylakoid membranes, consistent with the membrane-associated CrhR observed (Fig. 1). CrhR and S1 were enriched in the pellet fraction, indicating cosedimentation of CrhR with polysome-containing material and not soluble translation components. CrhR cosedimented with this material from cells grown at both 30 and 20°C, suggesting that temperature does not influence CrhR association with this fraction (Fig. 2B and C). RNase A treatment extracted CrhR from the polysome pellet (data not shown).
FIG 2.
CrhR localization in soluble Synechocystis lysates. (A) Lysate fractionation. A soluble cell lysate was overlaid on a 1 M sucrose cushion, resulting in a crude polysome-containing pellet (PP) and colored soluble layers (layers 1 to 4) by ultracentrifugation. (B and C) CrhR localization. Wild-type Synechocystis cells were treated with chloramphenicol to stabilize polysome complexes before lysis. CrhR and S1 were detected in the gradient fractions obtained from wild-type cells grown at 30°C (B) and cold shocked at 20°C for 3 h (C). (D) CrhR localization under translational runoff conditions. CrhR and S1 were localized in sucrose gradient fractions from wild-type cells grown at 20°C that were not treated with chloramphenicol. For reference, CrhR was also detected in the cytoplasmic (lanes C) and membrane (lanes M) fractions obtained after clarification of French press lysates. CrhR and S1 were detected as described in the Fig. 1 legend.
We also localized CrhR in wild-type cells that were not treated with chloramphenicol. The polysome runoff analysis indicated that CrhR distribution was dramatically altered, with CrhR being predominately detected in colored supernatant fractions 3 and 4 and less abundant in the polysome pellet obtained from cells grown at 20°C (Fig. 2D). S1 distribution was dramatically altered, shifting from the polysome pellet to the colored supernatant fractions (Fig. 2D). Similar results were obtained from cells grown at 30°C (data not shown). These results suggest that soluble S1 and CrhR are associated with actively translating polysomes; the absence of chloramphenicol allowed polysomes to finish translation during isolation, releasing CrhR and S1 into the supernatant fraction. These results also indicate that CrhR localization to the pellet fractions was not a result of aggregation.
In order to separate CrhR association with ribosome biogenesis from active translation, a linear sucrose gradient was used to fractionate soluble wild-type Synechocystis extracts to isolate ribosome subunits from 70S ribosomes and polysomes. Absorbance (A254) and Western analysis of the sucrose gradient fractions from cells grown at 20°C further confirmed the cosedimentation of CrhR only with the polysome fraction and not significantly with the 30S or 50S subunits or free 70S ribosomes (see Fig. S2 in the supplemental material). The slower-migrating (58-kDa) CrhR band was detected at a low level in lanes 15 to 18, at an approximate sucrose concentration of 44% to 49%. The molecular mass and sucrose banding data suggest that the detected CrhR corresponded to the thylakoid membrane-localized CrhR, as total Synechocystis extract was utilized in the analysis. As a control, S1 was detected in all ribosome-containing fractions (see Fig. S2).
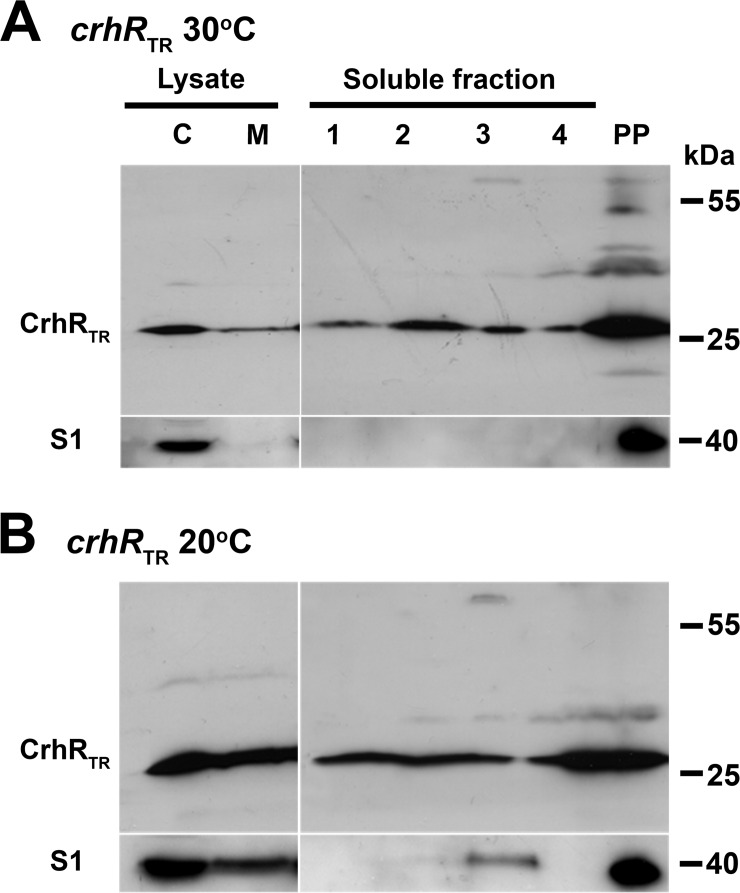
crhR truncation results in improper polysome association.
A similar polysome analysis was performed using the crhRTR mutant to determine the effect of CrhR truncation on localization. In striking contrast to the polysome-specific localization of CrhR in wild-type cells, the CrhRTR polypeptide was detected in all sucrose layers and in the pellet, irrespective of growth temperature (Fig. 3). In comparison, S1 localization was not altered in the crhRTR mutant (Fig. 3). Similarly to the observations in wild-type cells (Fig. 2B), S1 was detected only in the pellet fraction and also in chlorophyll-containing layer 3 of the sucrose gradient of soluble lysate obtained from cells grown at 20°C (Fig. 3B). This suggests that full-length CrhR RNA helicase is not required for S1 localization to polysomes but is required for CrhR association with polysomes.
FIG 3.
CrhRTR localization in soluble Synechocystis lysates. CrhRTR mutant Synechocystis cells were treated with chloramphenicol to stabilize polysome complexes before lysis. Soluble cell lysate from crhRTR mutant cells was overlaid on a 1 M sucrose cushion and a polysome-containing pellet (PP) obtained by ultracentrifugation. CrhRTR and S1 were detected in the gradient fractions obtained from crhRTR cells grown at 30°C (A) and cold shocked at 20°C for 3 h (B). For reference, CrhRTR was also detected in the cytoplasmic (lane C) and membrane (lane M) fractions obtained after clarification of French press lysates. CrhRTR and S1 were detected as described in the Fig. 1 legend.
CrhR cosediments with polysome- and degradosome-associated proteins.
Proteins cosedimenting with polysomes from soluble wild-type and crhRTR cells grown at 30 and 20°C were identified by mass spectrometry following a second purification through a 1 M sucrose cushion. The complete data set was organized into functional categories and is presented in Table S1 in the supplemental material. The major categories include subunits of the small and large ribosomal complexes, translation initiation and elongation factors, ribosome binding factors, and transcription factors. Furthermore, RNA modification enzymes involved in rRNA processing and RNA binding and the RNase E and PNPase degradosome subunits were also detected. Photosynthesis-associated proteins, including phycobilisome, ATP synthase subunits, and photosystem I (PSI) and II components, were primarily thylakoid membrane localized. Other proteins associated with energy metabolism, primarily glycolytic enzymes, proteins involved in regulation of transcription, including LexA and RNA polymerase components, protein degradation subunits, bacterioferritin, and a number of unknown and hypothetical proteins, were also detected.
A Venn diagram depicting the polypeptides detected in the 1 M cushion pellet obtained from soluble extracts of wild-type and crhRTR mutants at 20 and 30°C is presented in Fig. S3 in the supplemental material. A total of 38% of the detected polypeptides that pelleted through the 1 M sucrose cushion under all conditions were shared among all conditions (see Fig. S3A). Differential detection of the remainder could have resulted from either temperature effects or the absence of functional CrhR RNA helicase activity. The extensive numbers of polypeptides detected in the 1 M sucrose cushion that are associated with translation and RNA metabolism, including degradation, are depicted in Fig. S3B and C, respectively. The majority of these polysome- and degradosome-associated polypeptides were detected under all four conditions tested. The polysome proteomic data indicating that CrhR cosediments with peptides associated with translation and RNA turnover are supported by the results of protein association network analysis. STRING analysis predicted that helicases related to the DeaD subfamily of DEAD box RNA helicases colocalize with translation, RNA degradation, and RNA binding proteins (see Fig. S4).
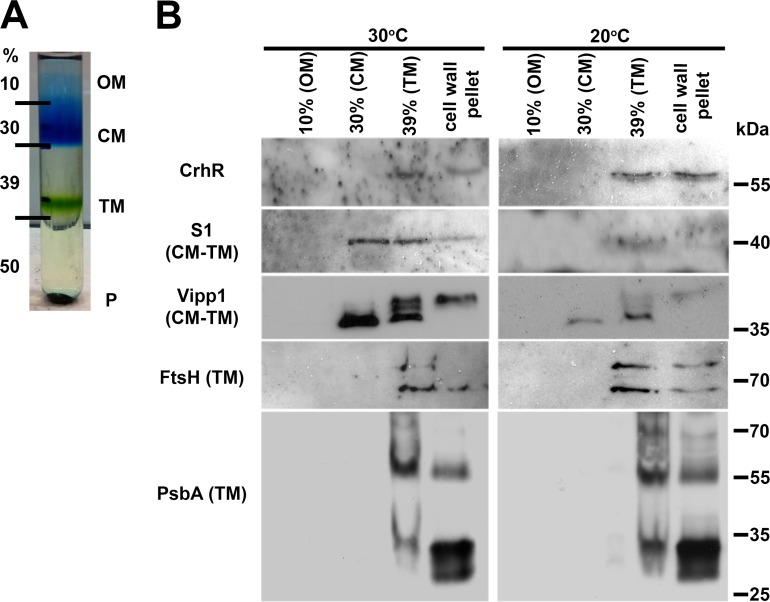
Thylakoid membrane localization.
The presence of the ∼58-kDa polypeptide cross-reacting with the CrhR antibody from the membrane-containing pellet fractions necessitated analysis of the three distinct membranes found in Synechocystis. We utilized the classic flotation ultracentrifugation method using a discontinuous sucrose gradient described by Murata and Omata (36) to separate the Synechocystis membrane systems from total cell lysates. The fractions corresponded to the outer membrane (OM; 10% sucrose), the cytoplasmic membrane (CM; 30% sucrose), the thylakoid membrane (TM; 39% sucrose), and cell wall and other material that pelleted through the 50% sucrose cushion (cell wall pellet). CrhR was detected only in the thylakoid membrane-containing 39% sucrose layer and the pellet (Fig. 4). To verify the purity of the thylakoid membrane fraction, the thylakoid-associated integral FtsH protease and the photosystem-associated PsbA (D1) protein were used as thylakoid membrane-specific markers. As expected, the anti-FtsH and anti-PsbA antibodies detected polypeptides in the thylakoid membrane-containing 39% sucrose layer and not in the cytoplasmic membrane (Fig. 4). Similarly, Vipp1 and S1 were detected in the cytoplasmic and thylakoid membrane fractions in which they are known to function (Fig. 4). Vipp1 and S1 were also detected in the cell wall pellet, indicating that this fraction contained thylakoid membrane and associated ribosomes, as this experiment was performed in the absence of chloramphenicol. Detection of the tested polypeptides in the pellet fraction indicates that this fraction was contaminated with thylakoid membrane and/or membrane-associated polysomes that would pellet through the 50% sucrose cushion. Overall, the parallel detection of CrhR with FtsH and PsbA suggests colocalization of these peptides within the thylakoid membrane fraction in Synechocystis.
FIG 4.
CrhR localization in wild-type Synechocystis membranes. Total membrane pelleted from cells grown at 30°C and cold shocked at 20°C for 3 h was adjusted to 50% sucrose, and individual membranes were isolated by flotation ultracentrifugation. (A) Sucrose gradient showing the relative positions of the membrane fractions. OM, outer membrane; CM, cytoplasmic membrane; TM, thylakoid membrane; P, cell wall pellet. (B) The indicated peptides were detected in the resolved membrane fractions using anti-CrhR (Synechocystis), anti-FtsH (Arabidopsis thaliana), anti-S1 (E. coli), anti-Vipp1 (Synechocystis), and anti-PsbA (Synechocystis) antibodies, as described in the Fig. 1 legend.
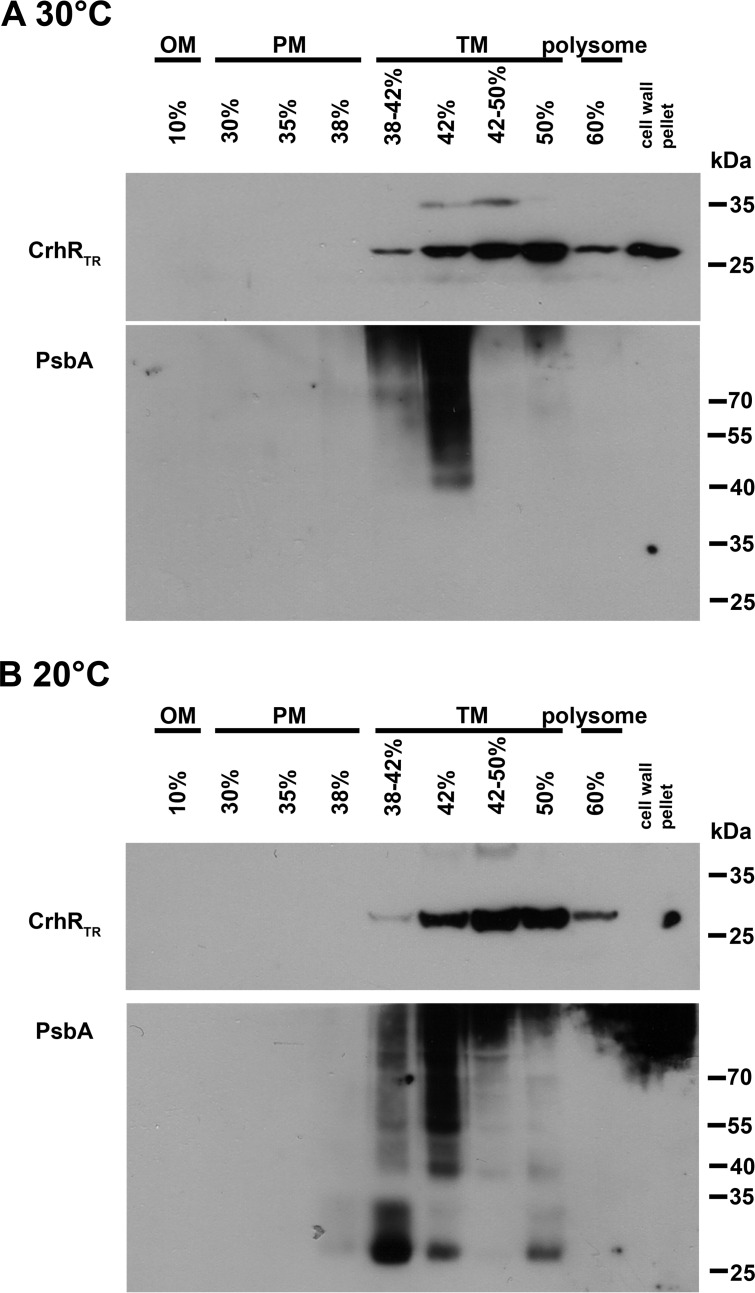
Since the truncated CrhR peptide was observed in all fractions of the polysome preparation, a finer discontinuous sucrose gradient (37) was used to isolate the three membrane types from crhRTR mutant cells. In contrast to the disrupted polysome localization shown in Fig. 3, CrhRTR was primarily detected only in the thylakoid-enriched fractions at both 30 and 20°C even though it was overexpressed at the higher temperature (Fig. 5). Similarly to the observations presented in Fig. 4, PsbA was detected only in the sucrose layers corresponding to thylakoid membrane-containing fractions. The results suggest that the second helicase core RecA domain and the C-terminal domain are not explicitly required for CrhR or PsbA localization to the thylakoid membrane region.
FIG 5.
CrhRTR localization in crhRTR mutant Synechocystis membranes. Total membrane pelleted from crhRTR cells grown at 30°C (A) and 20°C (B) was adjusted to 42% sucrose, and individual membranes were purified by flotation ultracentrifugation in a discontinuous sucrose gradient. CrhRTR and PsbA distributions were detected in equal volumes of each of the gradient fractions using anti-CrhR and anti-PsbA antibodies as described in the Fig. 1 legend. OM, outer membrane; CM, cytoplasmic membrane; TM, thylakoid membrane.
CrhR association with the thylakoid membrane region in vivo.
Since discontinuous sucrose gradient purification of Synechocystis membranes revealed a strong association with the thylakoid membrane-enriched fraction, in vivo analysis of CrhR localization using immunogold electron microscopy was performed on both wild-type and crhRTR mutant cells. CrhR was detected in both the cytoplasm and thylakoid membrane regions of both warm-grown and cold-shocked cells, with approximately 50% more CrhR localized to the thylakoid membrane region (Table 1). CrhR partitioning was not temperature dependent, as similar ratios (1.64 and 1.54) were observed at 30 and 20°C, suggesting that the increases in CrhR levels at 20°C segregate equally to the two subcellular regions. A similar partitioning ratio was observed for the CrhRTR protein at 20°C; however, the truncated protein did not partition to the thylakoid membrane properly at 30°C. The results suggest that full-length CrhR is required for partitioning between the cytoplasm and thylakoid membrane regions. As expected, PsbA was predominately thylakoid membrane localized, with crhR mutation not affecting the distribution (Table 1). Together, these results corroborate the findings that CrhR localizes to both the cytoplasm and thylakoid membrane regions in vivo and that full-length CrhR is required for the localization.
TABLE 1.
CrhR and PsbA ratio between thylakoid membrane region and cytoplasm of immunogold-labeled wild-type and crhRTR Synechocystis cellsa
| Protein and cell type | Result for: |
|||
|---|---|---|---|---|
| 20°C |
30°C |
|||
| No. of gold particles | TM/C ratio ± SD | No. of gold particles | TM/C ratio ± SD | |
| CrhR | ||||
| Wild type | 442 | 1.64 ± 0.22 | 264 | 1.54 ± 0.25 |
| crhRTR | 1,367 | 1.44 ± 0.24 | 200 | 0.70 ± 0.19 |
| PsbA (D1) | ||||
| Wild type | 372 | 16.36 ± 11.7 | 107 | 10.18 ± 1.14 |
| crhRTR | 126 | 21.46 ± 9.65 | 250 | 15.34 ± 6.67 |
Warm-grown (30°C) and cold-shocked (20°C for 3 h) cells were viewed using immunoelectron microscopy. Gold particles were enumerated manually from the digital images obtained from ultrathin sections. Normalization was performed by accounting for the area of the thylakoid membrane region (TM), the cytoplasm (C), and the surrounding resin background.
DISCUSSION
Prokaryotes generally compartmentalize biosynthetic pathways by sequestering processes to confined regions within the cell. A prime example is the differential localization of DEAD box RNA helicases within RNP complexes that are associated with ribosome biogenesis, translation initiation, or RNA degradation (18). The primary objective of this investigation was to identify the pathway in which CrhR functions, utilizing a variety of cellular localization techniques. The results establish that CrhR, the single DEAD box RNA helicase encoded in the model cyanobacterium Synechocystis, localizes to the cytoplasmic and thylakoid membrane regions and cosediments with translating polysome complexes. CrhR localization was temperature independent and differentially required CrhR RNA helicase activity, as thylakoid association and polysome association were unaffected and defective, respectively.
Although many prokaryotic DEAD box RNA helicases have roles in ribosome biogenesis (16–18, 38), our polysome fractionation results suggest that this function is unlikely in Synechocystis, as CrhR was detected only in the polysome-containing fraction. In addition, our runoff polysome analysis indicated that CrhR is associated with translating polysomes. The data therefore suggest that CrhR is associated with translation, consistent with the proteomic analysis of the polysome pellet. However, the mass spectrometric analysis of the polysome pellet also detected PNPase and RNase E degradosome components that correspond to the recently identified minimal degradosome in cyanobacteria (39). Interaction with an RNA helicase such as CrhR was not tested when it was shown previously that the Synechocystis RNase E and PNPase interact as a complex (39). Thus, while our data indicate that CrhR cosediments with polysome and degradosome components, we cannot determine if it directly interacts with the degradosome complex. The association of CrhR with translating polysomes does not preclude association with the RNA degradosome, however. The mass spectrometry-generated peptide spectrum match (PSM) values indicate that the presence of CrhR is substoichiometric with respect to ribosomal proteins and is more closely related to the levels detected for translation factors and degradosome components. Thus, since CrhR is encoded by the only DEAD box RNA helicase in the Synechocystis genome, given the multitasking of other bacterial RNA helicases (18–20), CrhR may be performing divergent roles and thus may associate with a range of cellular complexes under different growth conditions.
CrhR association with the translation and/or RNA degradation machinery is not unexpected, as RNA helicase association with the bacterial degradosome (4–6, 12–14) and with ribosome complexes (15–17) has been well documented. How CrhR is recruited to and interacts with the thylakoid membrane and translating polysome and/or degradosome components remains unknown. The mechanisms of localization to these compartments appear to differ, as CrhR was removed from the polysome pellet but not the membrane fraction by RNase treatment. This suggests that protein-protein and protein-RNA associations dictate CrhR localization to the thylakoid membrane and polysome, respectively. RNA- and protein-protein-dependent association of RNA helicases has been observed in other systems (14, 40, 41). The ease with which CrhR was removed from the membrane fraction also indicates that protein-protein interactions and not direct membrane interactions are involved. While RNA helicases, including CrhR, do not contain traditional membrane-spanning domains (12, 21, 22), the N-terminal and/or C-terminal amino acid extensions outside the helicase core are typically involved in specific RNA substrate recognition or protein-protein interactions (7, 8, 14). Indeed, we observed aberrant localization of CrhRTR, which lacks the second RecA domain and C-terminal extension to the polysome pellet. Altered localization of CrhRTR to the membrane region was also observed. These localization defects could have partially resulted from the CrhRTR overexpression observed at 30°C in the truncation mutant. Overall, the results indicate that intact CrhR is required for proper cellular localization. This conclusion is similar to those from studies revealing similar responses to C-terminal deletion of a cold-induced RNA helicase, Lmo1722, which resulted in disassociation from the 50S ribosome in Listeria monocytogenes (38) and removal of CshA from the Staphylococcus aureus degradosome (14). While thylakoid membrane-associated polysomes have been reported in cyanobacteria (42), the subcellular localization of the cyanobacterial degradosome has not been determined (39). It is predicted that the Synechocystis degradosome is soluble since the RNaseE does not contain the C-terminal scaffolding domain required for membrane attachment (39), as observed in some bacteria (4, 5, 12–14). Although other studies investigating Synechocystis subcellular localization failed to detect CrhR, the lack of CrhR detection in these studies most likely resulted from growth conditions that did not induce abundant CrhR expression, as the cultures were grown at 30 and 34°C, temperatures at which CrhR is expressed at a basal level (24). Results of those studies included the lack of detection of a DEAD box RNA helicase in the Synechocystis minimal degradosome (39) and membrane (37, 43–45) or cytoplasmic (46) extracts.
CrhR localization to the thylakoid membrane coincides with the significant morphological and physiological effects observed in response to crhR mutation (24). As was particularly evident when Synechocystis was stressed at 20°C, crhRTR cells exhibited a reduction in photosynthesis due to defects in photosynthetic carbon fixation which are associated with decreased pigmentation and alterations in electron transport chain function and thylakoid membrane structure (33). Defects in transitory adaptation of the photosynthetic apparatus to low temperature have also been observed in a crhR mutant which showed decreased PSII activity, a loss of PSI, and an oxidized plastoquinone (PQ) pool (47). These effects are directly related to the conditions required for regulation of crhR expression, namely, the redox potential of the electron transport chain (29). Although CrhR function is not known, the small RNA (sRNA) PsrR1 regulates expression of many photosynthesis-related genes (48). CrhR mutation-mediated disruption of PsrR1 function would be expected to generate similar phenotypic alterations.
The data presented here indicate that CrhR localizes to both the cytoplasmic and thylakoid membrane regions. CrhR in the cytoplasm is associated with translation, as it cosediments with actively translating polysomes. CrhR association with and disruption of the thylakoid membrane in a crhR mutant suggests that CrhR performs a role in thylakoid biogenesis and/or stability. In this capacity, CrhR RNA helicase activity in the thylakoid membrane could aid translation initiation and thus transertion of proteins into the thylakoid membrane, similarly to the role proposed for the RNA helicase, CrhC, in cytoplasmic membrane protein transertion in Anabaena sp. strain PCC 7120 (21). Adjustment of thylakoid membrane polysome profiles in response to temperature and light conditions indicates that abiotic stresses influence thylakoid function by altering translation (35, 45, 49). In this context, it would be of interest to determine if CrhR was involved in synthesis of thylakoid membrane proteins such as the D1 photosystem II protein. D1 translation initiates on soluble ribosomes and halts in the dark (49). The stalled ribosomal complex is targeted to the thylakoid membrane in the light, D1 synthesis is completed, and D1 is cotranslationally inserted into the membrane (35).
Cosedimentation of degradosome components with the polysome indicates the possibility that a functional association between the two complexes exists in Synechocystis. This association is known to occur in archaea and eukarya (50–53), and polysome fractionation on sucrose gradients has recently provided evidence for polysome-RNA degradosome association in E. coli (54) and Helicobacter pylori (55). Indeed, other than the results from photosynthesis-associated peptides, our proteomic data set closely resembles that obtained in the E. coli (54) and H. pylori (55) studies. Similarly to the results presented here, the respective RNA helicase peptides were associated with the identified polysome-RNA degradosome complexes (54, 55). In E. coli, the degradosome-associated RNA helicase RhlB contributes to ribosome binding and thus to formation of the polysome-RNA degradosome complex (54). Although Zhang et al. (39) did not detect CrhR in the minimal degradosome isolated from Synechocystis at 30°C, CrhR association was seen at lower temperatures. Temperature-dependent alteration of the degradosome-associated RNA helicase has been reported in a variety of bacteria, for example, E. coli (20) and Psychrobacter arcticus 273-4 (56). Similarly, Redko et al. (55) showed that a minimal degradosome consisting of RNase J and RhpA, the only DEAD box RNA helicase in the genome, associated with translating ribosomes and not 30S or 50S subunits in H. pylori. Although it is different from other Gram-negative organisms in this respect, Synechocystis encodes an RNase J homologue that appeared in our polysome data set. The potential for CrhR to be associated with a minimal degradosome and with RNase J at low temperature deserves further investigation. These observations are similar to those we report here for CrhR, suggesting that CrhR may also contribute to the formation of a polysome-RNA degradosome complex in Synechocystis.
Potential roles for RNA helicase alteration of RNA secondary structure in a polysome-RNA degradosome complex include unwinding of an inhibitory RNA secondary structure that blocks RNase access and sRNA annealing to create a RNase site. CrhR could participate in either of these scenarios, as it catalyzes both double-stranded RNA (dsRNA) unwinding and annealing reactions in vitro (25). In prokaryotes, related systems include termination of translation by A-site cleavage in translating ribosomes by RNase toxins in toxin-antitoxin pairs (57), and binding of the sRNA RhyB to the 5′ untranscribed region (UTR) of the sodB transcript activates RNaseE cleavage within the open reading frame (ORF) during translation (58). CrhR RNA helicase activity could be involved in similar regulatory mechanisms, as Tsai et al. (54) also detected Hfq in their polysome-degradosome preparations and speculated that Hfq contributed to RhyB binding and thus to cleavage of sodB. Cyanobacteria do not encode an Hfq homologue that functions in sRNA metabolism (59), raising the possibility that CrhR replaces Hfq in a Synechocystis degradosome-polysome complex. In eukaryotes, RNA helicases are associated with RNA degradation of actively translating transcripts. For example, a variety of ribonucleases are targeted to the ribosome by UPF1 during nonsense-mediated decay (NMD) (52), and Dhh1 slows translation elongation and promotes decapping of polysome-associated transcripts, creating substrates for Dhh1-mediated RNA decay (53). Whether CrhR is associated with similar regulatory pathways remains to be elucidated.
Here we provide evidence that CrhR, the single DEAD box RNA helicase encoded in the Synechocystis genome, localizes to both the cytoplasmic and thylakoid membrane regions and cosediments with polysome and RNA degradosome components. The results suggest that the RNA degradation machinery is coupled with translation in Synechocystis, contributing to the emerging picture showing that these processes are intimately linked in bacterial systems.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to P. Baumann (University of California, Davis) for the E. coli S1 antisera, W. Hess (University of Freiburg) for the Synechocystis Vipp1 antisera, Z. Adam (Hebrew University of Jerusalem) for the Arabidopsis FtsH antisera, and D. Campbell (Mount Allison University) for the PsbA (D1) antisera. We appreciate receiving data in advance of publication from P. Linder. We also thank Arlene Oatway of the Advanced Microscopy Facility and Jack Moore of the Alberta Proteomics and Mass Spectrometry Facility for their assistance.
A.R.R.R. and D.S.W. equally contributed to designing the research, performing experiments, analyzing data, and writing the manuscript; R.P.F. performed the protein mass spectrometry analysis, analyzed data, and cowrote the manuscript; G.W.O. designed the research, analyzed data, and cowrote the manuscript.
This work was supported by National Sciences and Engineering Research Council of Canada Discovery Grants 171319 to G.W.O. and 341453 to R.P.F.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00267-16.
REFERENCES
- 1.Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 2.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 3.Campos M, Jacobs-Wagner C. 2013. Cellular organization of the transfer of genetic information. Curr Opin Microbiol 16:171–176. doi: 10.1016/j.mib.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khemici V, Poljak L, Luisi BF, Carpousis AJ. 2008. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol 70:799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stülke J. 2011. RNase Y in Bacillus subtilis: a natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol 193:5431–5441. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Py B, Higgins CF, Krisch HM, Carpousis AJ. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 7.Linder P, Jankowsky E. 2011. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 8.Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mingam A, Toffano-Nioche C, Brunaud V, Boudet N, Kreis M, Lecharny A. 2004. DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J 2:401–415. doi: 10.1111/j.1467-7652.2004.00084.x. [DOI] [PubMed] [Google Scholar]
- 10.Kalman M, Murphy H, Cashel M. 1991. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol 3:886–895. [PubMed] [Google Scholar]
- 11.Lehnik-Habrink M, Rempeters L, Kovács Á Wrede TC, Baierlein C, Krebber H, Kuipers OP, Stülke J. 2013. DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J Bacteriol 195:534–544. doi: 10.1128/JB.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liou GG, Jane WN, Cohen SN, Lin NS, Lin-Chao S. 2001. RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc Natl Acad Sci U S A 98:63–68. doi: 10.1073/pnas.98.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardwick SW, Chan VS, Broadhurst RW, Luisi BF. 2011. An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res 39:1449–1459. doi: 10.1093/nar/gkq928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraud C, Hausmann S, Lemeille S, Prados J, Redder P, Linder P. 2015. The C-terminal region of the RNA helicase CshA is required for the interaction with the degradosome and turnover of bulk RNA in the opportunistic pathogen Staphylococcus aureus. RNA Biol 12:658–673. doi: 10.1080/15476286.2015.1035505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäreclev C, Vaitkevicius K, Netterling S, Johansson J. 2014. DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression. RNA Biol 11:1457–1466. doi: 10.1080/15476286.2014.996099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol 48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 17.Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res 32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iost I, Bizebard T, Dreyfus M. 2013. Functions of DEAD-box proteins in bacteria: current knowledge and pending questions. Biochim Biophys Acta 1829:866–877. doi: 10.1016/j.bbagrm.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Pandiani F, Brillard J, Bornard I, Michaud C, Chamot S, Nguyen-the C, Broussolle V. 2010. Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures. Appl Environ Microbiol 76:6692–6697. doi: 10.1128/AEM.00782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prud'homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. 2004. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome'. Mol Microbiol 54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 21.El-Fahmawi B, Owttrim GW. 2003. Polar biased localization of the cold stress-induced RNA helicase, CrhC, in the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol 50:1439–1448. doi: 10.1046/j.1365-2958.2003.03783.x. [DOI] [PubMed] [Google Scholar]
- 22.Strahl H, Turlan C, Khalid S, Bond PJ, Kebalo JM, Peyron P, Poljak L, Bouvier M, Hamoen L, Luisi BF, Carpousis AJ. 2015. Membrane recognition and dynamics of the RNA degradosome. PLoS Genet 11:e1004961. doi: 10.1371/journal.pgen.1004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunger K, Beckering CL, Wiegeshoff F, Graumann PL, Marahiel MA. 2006. Cold-induced putative DEAD box RNA helicases CshA and CshB are essential for cold adaptation and interact with cold shock protein B in Bacillus subtilis. J Bacteriol 188:240–248. doi: 10.1128/JB.188.1.240-248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosana AR, Chamot D, Owttrim GW. 2012. Autoregulation of RNA helicase expression in response to temperature stress in Synechocystis sp. PCC 6803. PLoS One 7:e48683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamot D, Colvin KR, Kujat-Choy SL, Owttrim GW. 2005. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J Biol Chem 280:2036–2044. [DOI] [PubMed] [Google Scholar]
- 26.Chamot D, Magee WC, Yu E, Owttrim GW. 1999. A cold shock-induced cyanobacterial RNA helicase. J Bacteriol 181:1728–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chamot D, Owttrim GW. 2000. Regulation of cold shock-induced RNA helicase gene expression in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 182:1251–1256. doi: 10.1128/JB.182.5.1251-1256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu E, Owttrim GW. 2000. Characterization of the cold stress-induced cyanobacterial under DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res 28:3926–3934. doi: 10.1093/nar/28.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kujat SL, Owttrim GW. 2000. Redox-regulated RNA helicase expression. Plant Physiol 124:703–714. doi: 10.1104/pp.124.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol 40:235–244. doi: 10.1046/j.1365-2958.2001.02379.x. [DOI] [PubMed] [Google Scholar]
- 31.Vinnemeier J, Hagemann M. 1999. Identification of salt-regulated genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 by subtractive RNA hybridization. Arch Microbiol 172:377–386. doi: 10.1007/s002030050774. [DOI] [PubMed] [Google Scholar]
- 32.Tarassova OS, Chamot D, Owttrim GW. 2014. Conditional, temperature-induced proteolytic regulation of cyanobacterial RNA helicase expression. J Bacteriol 196:1560–1568. doi: 10.1128/JB.01362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosana AR, Ventakesh M, Chamot D, Patterson-Fortin LM, Tarassova O, Espie GS, Owttrim GW. 2012. Inactivation of a low temperature-induced RNA helicase in Synechocystis sp. PCC 6803: physiological and morphological consequences. Plant Cell Physiol 53:646–658. [DOI] [PubMed] [Google Scholar]
- 34.Owttrim GW. 2012. RNA helicases in cyanobacteria: biochemical and molecular approaches. Methods Enzymol 511:385–403. doi: 10.1016/B978-0-12-396546-2.00018-8. [DOI] [PubMed] [Google Scholar]
- 35.Tyystjärvi T, Herranen M, Aro EM. 2001. Regulation of translation elongation in cyanobacteria: membrane targeting of the ribosome nascent-chain complexes controls the synthesis of D1 protein. Mol Microbiol 40:476–484. doi: 10.1046/j.1365-2958.2001.02402.x. [DOI] [PubMed] [Google Scholar]
- 36.Murata N, Omata T. 1988. Isolation of cyanobacterial plasma-membranes. Methods Enzymol 167:245–251. doi: 10.1016/0076-6879(88)67026-1. [DOI] [Google Scholar]
- 37.Huang F, Parmryd I, Nilsson F, Persson AL, Pakrasi HB, Andersson B, Norling B. 2002. Proteomics of Synechocystis sp. strain PCC 6803 identification of plasma membrane proteins. Mol Cell Proteomics 1:956–966. doi: 10.1074/mcp.M200043-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Netterling S, Vaitkevicius K, Nord S, Johansson J. 2012. A Listeria monocytogenes RNA helicase essential for growth and ribosomal maturation at low temperatures uses its C terminus for appropriate interaction with the ribosome. J Bacteriol 194:4377–4385. doi: 10.1128/JB.00348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JY, Deng XM, Li FP, Wang L, Huang QY, Zhang CC, Chen WL. 2014. RNase E forms a complex with polynucleotide phosphorylase in cyanobacteria via a cyanobacterial-specific nonapeptide in the noncatalytic region. RNA 20:568–579. doi: 10.1261/rna.043513.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Resch A, Većerek B, Palavra K, Bläsi U. 2010. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol 7:796–802. doi: 10.4161/rna.7.6.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senissar M, Le Saux A, Belgareh-Touzé N, Adam C, Banroques J, Tanner NK. 2014. The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res 42:10005–10022. doi: 10.1093/nar/gku584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Meene AM, Hohmann-Marriott MF, Vermaas WF, Roberson RW. 2006. The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol 184:259–270. [DOI] [PubMed] [Google Scholar]
- 43.Norling B, Zak E, Andersson B, Pakrasi H. 1998. 2D-isolation of pure plasma and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 436:189–192. [DOI] [PubMed] [Google Scholar]
- 44.Huang F, Hedman E, Funk C, Kieselbach T, Schroder WP, Norling B. 2004. Isolation of outer membrane of Synechocystis sp. PCC 6803 and its proteomic characterization. Mol Cell Proteomics 3:586–595. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava R, Pisareva T, Norling B. 2005. Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5:4905–4916. [DOI] [PubMed] [Google Scholar]
- 46.Simon WJ, Hall JJ, Suzuki I, Murata N, Slabas AR. 2002. Proteomic study of the soluble proteins from the unicellular cyanobacterium Synechocystis sp. PCC6803 using automated matrix-assisted laser desorption/ionization time of flight peptide mass fingerprinting. Proteomics 2:1735–1742. [DOI] [PubMed] [Google Scholar]
- 47.Sireesha K, Radharani B, Krishna PK, Sreedhar N, Subramanyam R, Mohanty P, Prakash JSS. 2012. RNA helicase, CrhR is indispensable for the energy redistribution and the regulation of photosystem stoichiometry at low temperature in Synechocystis sp. PCC6803. Biochim Biophys Acta 1817:1525–1536. doi: 10.1016/j.bbabio.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Georg J, Dienst D, Schürgers N, Wallner T, Kopp D, Stazic D, Kuchmina E, Klähn S, Lokstein H, Hess WR, Wilde A. 2014. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 26:3661–3679. doi: 10.1105/tpc.114.129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyystjärvi T, Sirpio S, Aro EM. 2004. Post-transcriptional regulation of the psbA gene family in the cyanobacterium Synechococcus sp. PCC 7942. FEBS Lett 576:211–215. [DOI] [PubMed] [Google Scholar]
- 50.Koonin EV, Wolf YI, Aravind L. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res 11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiorini F, Bonneau F, Le Hir H. 2012. Biochemical characterization of the RNA helicase UPF1 involved in nonsense-mediated mRNA decay. Methods Enzymol 511:255–274. doi: 10.1016/B978-0-12-396546-2.00012-7. [DOI] [PubMed] [Google Scholar]
- 53.Sweet T, Kovalak C, Coller J. 2012. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol 10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai YC, Du D, Domínguez-Malfavón L, Dimastrogiovanni D, Cross J, Callaghan AJ, García-Mena J, Luisi BF. 2012. Recognition of the 70S ribosome and polysome by the RNA degradosome in Escherichia coli. Nucleic Acids Res 40:10417–10431. doi: 10.1093/nar/gks739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Redko Y, Aubert S, Stachowicz A, Lenormand P, Namane A, Darfeuille F, Thibonnier M, Reuse HD. 2013. A minimal bacterial RNase J-based degradosome is associated with translating ribosomes. Nucleic Acids Res 41:288–301. doi: 10.1093/nar/gks945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bergholz PW, Bakermans C, Tiedje JM. 2009. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol 191:2340–2352. doi: 10.1128/JB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schureck MA, Dunkle JA, Maehigashi T, Miles SJ, Dunham CM. 2015. Defining the mRNA recognition signature of a bacterial toxin protein. Proc Natl Acad Sci U S A 112:13862–13867. doi: 10.1073/pnas.1512959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. 2011. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. 2009. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J 276:3904–3915. doi: 10.1111/j.1742-4658.2009.07104.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.