ABSTRACT
Many bacteria utilize actin-like proteins to direct peptidoglycan (PG) synthesis. MreB and MreB-like proteins are thought to act as scaffolds, guiding the localization and activity of key PG-synthesizing proteins during cell elongation. Despite their critical role in viability and cell shape maintenance, very little is known about how the activity of MreB family proteins is regulated. Using a Bacillus subtilis misexpression screen, we identified two genes, yodL and yisK, that when misexpressed lead to loss of cell width control and cell lysis. Expression analysis suggested that yodL and yisK are previously uncharacterized Spo0A-regulated genes, and consistent with these observations, a ΔyodL ΔyisK mutant exhibited reduced sporulation efficiency. Suppressors resistant to YodL's killing activity occurred primarily in mreB mutants and resulted in amino acid substitutions at the interface between MreB and the highly conserved morphogenic protein RodZ, whereas suppressors resistant to YisK occurred primarily in mbl mutants and mapped to Mbl's predicted ATP-binding pocket. YodL's shape-altering activity appears to require MreB, as a ΔmreB mutant was resistant to the effects of YodL but not YisK. Similarly, YisK appears to require Mbl, as a Δmbl mutant was resistant to the cell-widening effects of YisK but not of YodL. Collectively, our results suggest that YodL and YisK likely modulate MreB and Mbl activity, possibly during the early stages of sporulation.
IMPORTANCE The peptidoglycan (PG) component of the cell envelope confers structural rigidity to bacteria and protects them from osmotic pressure. MreB and MreB-like proteins are thought to act as scaffolds for PG synthesis and are essential in bacteria exhibiting nonpolar growth. Despite the critical role of MreB-like proteins, we lack mechanistic insight into how their activities are regulated. Here, we describe the discovery of two B. subtilis proteins, YodL and YisK, which modulate MreB and Mbl activities. Our data suggest that YodL specifically targets MreB, whereas YisK targets Mbl. The apparent specificities with which YodL and YisK are able to differentially target MreB and Mbl make them potentially powerful tools for probing the mechanics of cytoskeletal function in bacteria.
INTRODUCTION
Bacterial cell growth requires that the machineries directing enlargement and division of the bacterial cell envelope be coordinated in both time and space (1). The cell envelope is comprised of membranes and a macromolecular mesh of peptidoglycan (PG) that possesses both rigid and elastic properties (2, 3). PG is highly cross-linked, allowing bacteria to maintain shapes and avoid lysis, even in the presence of several atmospheres of internal turgor pressure. PG rearrangements are required during the inward redirection of growth that occurs at the time of cell division, but they are also necessary when cells insert new PG and dynamically modify their morphologies in response to developmental or environmental signals (4, 5). To avoid lysis during PG rearrangements, bacteria must carefully regulate the making and breaking of glycan strands and peptide cross-links (3). In rod-shaped bacteria, PG enlargement during steady-state growth is constrained in one dimension along the cell's long axis and can either occur through polar growth, as is the case for Agrobacterium tumefaciens and Streptomyces coelicolor, or through incorporation of new cell wall material along the length of the cell cylinder, as observed with Escherichia coli, Bacillus subtilis, and Caulobacter crescentus (6).
To control cell diameter and create osmotically stable PG, bacteria that exhibit nonpolar growth require the activity of the highly conserved actin-like protein MreB. Biochemical, genetic, and cell biological data suggest that MreB likely directs PG synthesis during cell elongation, and in some bacteria MreB may also function during cell division (7–9). MreB possesses ATPase activity and polymerizes at sites along the cytoplasmic side of the inner membrane (10). ATP binding and hydrolysis are required for MreB polymerization and activity (11), and two S-benzylisothiourea derivatives, A22 and MP265, target the ATPase domain of MreB in Gram-negative organisms, possibly preventing nucleotide hydrolysis and/or release (12–15). Depletion or inactivation of MreB is lethal except in some conditional backgrounds (16, 17), so organisms sensitive to A22 and/or MP265 lose shape and eventually lyse (12–15).
MreB has been found to interact with several other proteins involved in PG synthesis, including the bitopic membrane protein RodZ (8, 16, 18–20). RodZ interacts directly with MreB through a cytoplasmic helix-turn-helix motif located at its N terminus (18). A cocrystal structure of RodZ and MreB shows the N terminus of RodZ extending into a conserved hydrophobic pocket located in subdomain IIA of MreB (18). Depletion of RodZ also leads to loss of cell shape and cell death (21–23). However, in various mutant backgrounds, rodZ can be deleted without loss of rod shape or viability, indicating that RodZ is not absolutely required for MreB's function in maintaining shape (24–26). Based on these observations and others, it has been proposed that MreB-RodZ interactions may regulate some aspect of MreB activity (10, 26).
Gram-positive bacteria often encode multiple paralogs (27). B. subtilis possesses three mreB family genes: mreB, mbl, and mreBH. mreB is distinguished from mbl and mreBH by its location within the highly conserved mreBCD operon. Although mreB, mbl, and mreBH are essential, it has been reported that each can be deleted under conditions in which cells are provided sufficient magnesium (28–30) or in strain backgrounds lacking ponA, the gene that encodes penicillin binding protein 1 (PBP1) (20). In addition, all three genes can be deleted in a single background with only minor effects on cell shape if any one of the paralogs is artificially overexpressed in trans from an inducible promoter (31). The ability of any one of the paralogs to compensate for the loss of the others, at least under some growth conditions, strongly suggests that MreB, Mbl, and MreBH share significant functional redundancy (31, 32).
At the same time, several lines of evidence suggest that the paralogs possess nonoverlapping functions. The genes themselves exhibit different patterns of transcriptional regulation, suggesting that each likely possesses specialized activities that are important in different growth contexts. For example, mreB and mbl are maximally expressed at the end of exponential growth but expression falls off sharply during stationary phase (33), whereas mreBH is part of the SigI heat shock regulon (34). There is also evidence suggesting that each protein may possess specialized activities. For example, MreBH interacts with the lytic transglycosylase LytE and is required for LytE localization (35), whereas the lytic transglycosylase CwlO depends on Mbl for wild-type function (35). More recently, MreB (but not Mbl or MreBH) was shown to aid in escape from the competent cell state (33).
Aside from RodZ (10, 26), only a few proteins targeting MreB activity in vivo have been identified. In E. coli, the YeeU-YeeV prophage toxin-antitoxin system is comprised of a negative regulator of MreB polymerization, CbtA (36), and a positive regulator of MreB bundling, CbeA (37). Another E. coli prophage toxin, CptA, is also reported to inhibit MreB polymerization (38). The MbiA protein of C. crescentus appears to regulate MreB in vivo; however, its physiological role is unknown (39). Given the importance of PG synthesis to cell viability and in cell shape control, it is likely that many undiscovered factors exist that modulate the activity of MreB and its paralogs.
In the present work, we describe the identification of YodL and YisK, modulators of MreB and Mbl activity that are expressed during early stages of B. subtilis sporulation. Misexpression of either yodL or yisK during vegetative growth results in loss of cell width control and cell death. Genetic evidence indicates that YodL targets and inhibits MreB activity, whereas YisK targets and inhibits Mbl. Our data also show that YisK activity affects cell length control through an Mbl- and MreBH-independent pathway.
MATERIALS AND METHODS
General methods.
All B. subtilis strains were derived from B. subtilis 168. E. coli and B. subtilis strains utilized in this study are listed in Table S2 in the supplemental material. Plasmids are listed in Table S3 in the supplemental material. Oligonucleotide primers are listed in Table S4 of the supplemental material. Details on plasmid and strain construction can be found in the supplemental material. Escherichia coli DH5α was used for cloning. All E. coli strains were grown in LB-Lennox medium supplemented with 100 μg/ml ampicillin. The following concentrations of antibiotics were used for generating B. subtilis strains: 100 μg/ml spectinomycin, 7.5 μg/ml chloramphenicol, 0.8 mg/ml phleomycin, 10 μg/ml tetracycline, 10 μg/ml kanamycin. To select for erythromycin resistance, plates were supplemented with 1 μg/ml erythromycin (ERM) and 25 μg/ml lincomycin. B. subtilis transformations were carried out as described previously (40). When indicated, the LB in the B. subtilis microscopy experiments was LB-Lennox broth. Sporulation by resuspension was carried out at 37°C according to the Sterlini-Mandelstam method (41). Penassay broth (PAB) is composed of 5 g peptone, 1.5 g beef extract, 1.5 g yeast extract, 1.0 g d-glucose (dextrose), 3.5 g NaCl, 3.68 g dipotassium phosphate, and 1.32 g monopotassium phosphate per liter of distilled water. To make solid media, the relevant medium was supplemented with 1.5% (wt/vol) Bacto agar.
Microscopy.
For microscopy experiments, all strains were grown in the indicated medium in volumes of 25 ml in 250-ml baffled flasks and placed in a shaking water bath set at 37°C and 280 rpm. Unless stated otherwise, misexpression was performed by inducing samples with 1.0 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) and imaging of samples 90 min postinduction. Fluorescence microscopy was performed with a Nikon Ti-E microscope equipped with a CFI Plan Apo lambda DM 100× objective, Prior Scientific Lumen 200 illumination system, C-FL UV-2E/C 4′,6-diamidino-2-phenylindole and C-FL green fluorescent protein (GFP) HC HISN Zero Shift filter cubes, and a CoolSNAP HQ2 monochrome camera. Membranes were stained with TMA-DPH [1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluene sulfonate; 0.02 mM] and imaged with exposure times of 1 s with a neutral density filter in place to reduce cytoplasmic background. All GFP images were captured with a 1-s exposure time. All images were captured and processed with the NIS Elements Advanced Research program (version 4.10) and ImageJ64 (42). Cells were mounted on glass slides with 1% agarose pads or polylysine-treated coverslips prior to imaging. To quantitate cell lengths for the strains imaged in Fig. 6, the cell lengths for 500 cells were determined for each population. The statistical significance of cell length differences between populations was determined using an unpaired Student's t test.
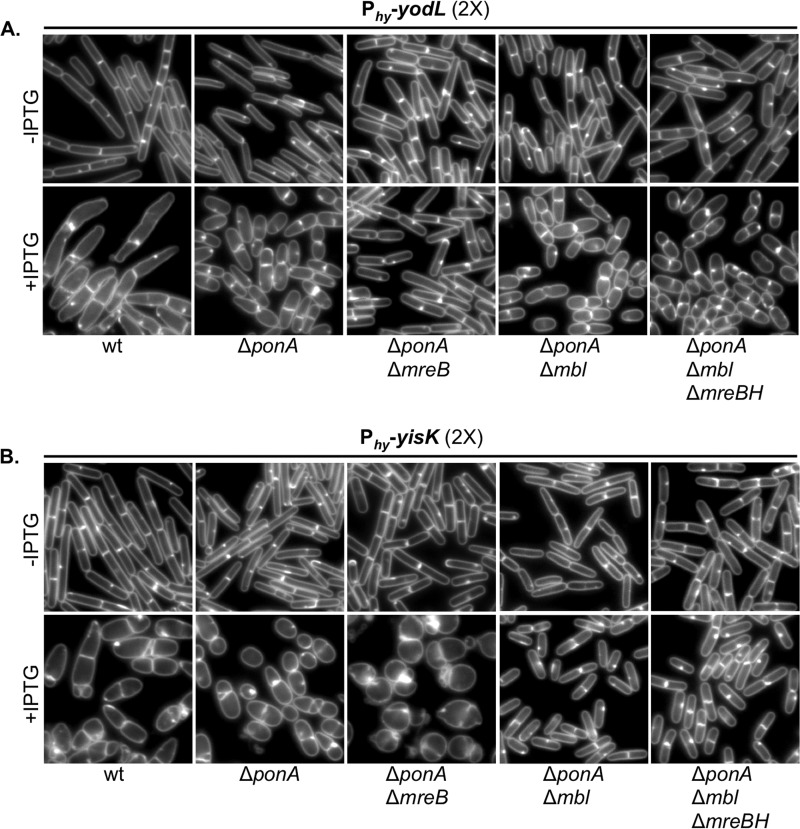
FIG 6.
YodL and YisK cell-widening activities require MreB and Mbl, respectively. (A) Cells harboring two copies of Phy-yodL in the wild-type (BAS191), ΔponA (BYD176), ΔponA ΔmreB (BYD263), ΔponA Δmbl (BYD259), or ΔponA Δmbl ΔmreBH (BAS249) background were grown at 37°C in LB supplemented with 10 mM MgCl2 to mid-exponential phase. To induce yodL expression, cells were back-diluted to an OD600 of ∼0.02 in LB with 10 mM MgCl2 and IPTG (1 mM) was added. Cells were grown for 1.5 h at 37°C before image capture. Membranes were stained with TMA-DPH. All images are shown at the same magnification. (B) Cells harboring two copies of Phy-yisK in the wild-type (BYD074), ΔponA (BYD175), ΔponA ΔmreB (BYD262), ΔponA Δmbl (BYD258), or ΔponA Δmbl ΔmreBH (BAS248) background were grown at 37°C in LB supplemented with 10 mM MgCl2 to mid-exponential phase. To induce yisK expression, cells were back-diluted to an OD600 of ∼0.02 in LB with 10 mM MgCl2, and IPTG (1 mM) was added. Cells were grown for 1.5 h at 37°C before image capture. Membranes were stained with TMA-DPH. All images are shown at the same magnification.
Plate growth assays.
B. subtilis strains were streaked on LB-Lennox plates containing 100 μg/ml spectinomycin and 1 mM IPTG. The plates were supplemented with the indicated concentrations of MgCl2 when necessary. Plates were incubated at 37°C overnight, and images were captured on a ScanJet G4050 flatbed scanner (Hewlett Packard).
Heat kill.
Spore formation was quantified by growing cells in Difco sporulation medium (DSM) (43). A freshly grown single colony of each strain was inoculated into 2 ml of DSM and placed in a roller drum at 37°C, 60 rpm, for 36 h. To determine the number of CFU per milliliter, an aliquot of each culture was serially diluted and plated on DSM agar plates. To enumerate the heat-resistant spores per milliliter, the serially diluted cultures were subjected to a 20-min heat treatment at 80°C and plated on DSM agar plates. The plates were incubated at 37°C overnight, and the next day colony counts were determined. The relative sporulation frequency compared to the wild type was determined by calculating the number of spores per CFU of each experimental and dividing it by the spores per CFU of the wild type. The reported statistical significance was determined using an unpaired Student's t test.
Transcriptional fusions.
Transcriptional fusions were constructed by fusing an ∼200-bp region up to the start codon of either yodL or yisK to gfp or lacZ and integrating the fusions into the B. subtilis chromosome at the amyE locus (for more details, see the descriptions of strain constructions in the supplemental material). Microscopy was conducted on each strain over a time course in sporulation by resuspension media (see the supplemental material) or in a nutrient exhaustion time course experiment in CH (41). Beta-galactosidase assays were performed as described previously (44), except all samples were frozen at −80°C before processing. All experiments were performed on at least three independent biological replicates.
Suppressor selections.
Single colonies of BYD048 (harboring three copies of Phy-yodL [3× Phy-yodL] and one copy of Phy-lacZ) or BYD076 (3× Phy-yisK Phy-lacZ) were used to inoculate independent 5-ml LB-Lennox cultures. Six independent cultures were grown for each strain. The cultures were grown for 6 h at 37°C, and 0.3 μl of each culture was diluted in 100 μl LB and plated on an LB-Lennox agar plate containing 100 μg/ml spectinomycin and 1 mM IPTG. After overnight growth, suppressors that arose were patched on both LB-Lennox agar plates supplemented with 100 μg/ml spectinomycin and LB-Lennox agar plates supplemented with 100 μg/ml spectinomycin, 1.0 mM IPTG, and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and grown at 37°C overnight. Only blue colonies were selected for further analysis; this screen eliminated mutants unable to derepress Phy in the presence of IPTG. In addition, each Phy-yodL or Phy-yisK construct was transformed into a wild-type background to ensure that the construct remained fully functional with respect to preventing cell growth on LB-Lennox agar plates supplemented with the relevant antibiotic and 1 mM IPTG.
Whole-genome sequencing and analysis.
Genomic DNA was isolated from six YodL-resistant suppressors obtained from independent cultures as well as the parent strain (BYD048) by inoculating a single colony in 6 ml LB-Lennox medium and growing at 37°C for 4 h in a roller drum. Cells were collected by spinning at 21,130 × g for 2 min at room temperature, resuspending the pellets in lysis buffer (20 mM Tris-HCl [pH 7.5], 50 mM EDTA [pH 8], 100 mM NaCl, and 2 mg/ml lysozyme) and incubating at 37°C for 30 min. Sarkosyl was added to a final concentration of 1% (wt/vol). Protein was removed by extracting with 600 μl phenol, centrifuging at 21,130 × g for 5 min at room temperature, and transferring the top (aqueous) layer to a new microcentrifuge tube. This was followed by an extraction with 600 μl phenol-saturated chloroform and centrifugation at 21,130 × g for 5 min at room temperature. After transferring the aqueous layer to a new microcentrifuge tube, a final extraction was performed with 100% chloroform, followed by centrifugation at 21,130 × g for 5 min at room temperature. The aqueous layer was transferred to a new microcentrifuge tube, being careful to avoid the interphase material. To precipitate the genomic DNA, a 1/10 volume of 3.0 M Na-acetate and 1 ml of 100% ethanol were added, and the tube was inverted multiple times. The sample was centrifuged at 21,130 × g for 1 min at room temperature in a microcentrifuge. The pellet was washed with 150 μl 70% ethanol and resuspended in 500 μl TE (10 mM Tris [pH 7.5], 1 mM EDTA [pH 8.0]). To eliminate potential RNA contamination, RNase was added to a final concentration of 200 μg/ml and the sample was incubated at 55°C for 1 h. To remove the RNase, the genomic DNA was repurified by phenol-chloroform extraction and ethanol precipitation as described above. The final pellet was resuspended in 100 μl TE. Bar-coded libraries were prepared from each genomic DNA sample by using a TruSeq DNA kit (Illumina) according to the manufacturer's specifications, and the samples were subjected to Illumina-based whole-genome sequencing using a MiSeq 250 paired-end run. CLC Genomics Workbench (Qiagen) was used to map the sequence reads against the Bs168 reference genome and to identify single nucleotide polymorphisms, insertions, and deletions. Mutations associated with the Phy integration constructs and those in which less than 40% of the reads differed from the reference genome were excluded as candidate changes responsible for suppression in our initial analysis (see Table S1 in the supplemental material). The remaining suppressor mutations were identified by PCR amplification of mreB (using primer set OAS044 and OAS045) and mbl (using primer set OAS046 and OAS047) and sequencing with the same primers. To determine if the candidate suppressor alleles identified were sufficient to confer resistance to the original selective pressure, each was linked to a kanamycin resistance cassette and moved by transformation into a clean genetic background (see the supplemental material for a further description of strain construction).
RESULTS
YodL and YisK affect cell width.
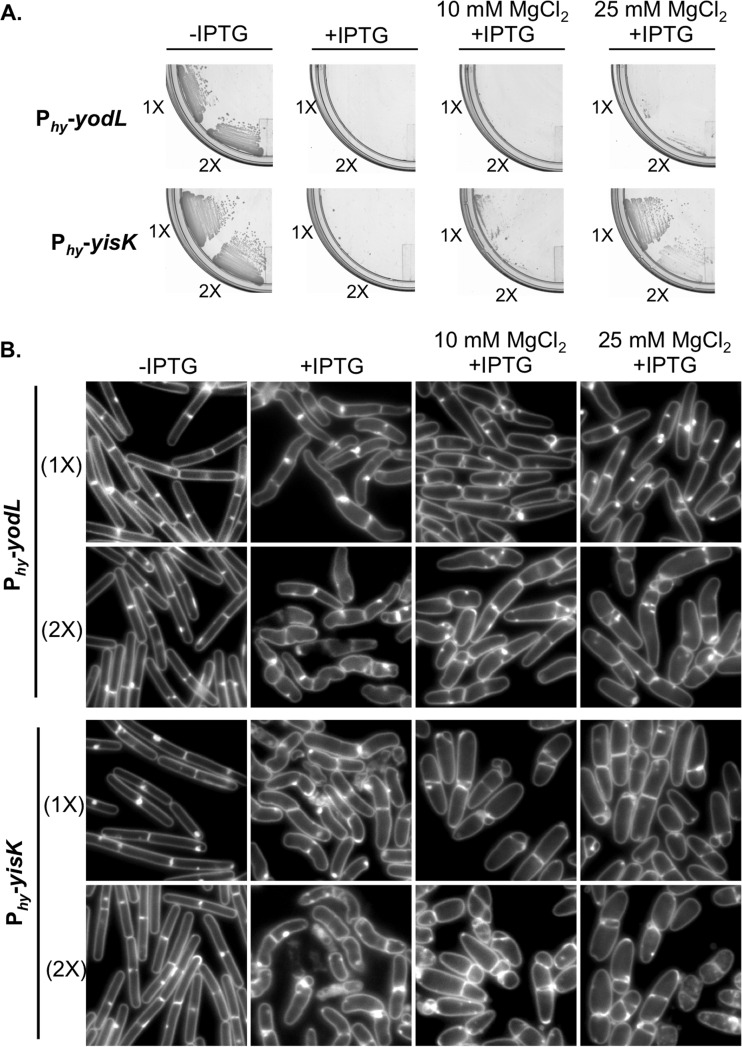
To identify novel factors involved in cellular morphogenesis, we created an ordered gene misexpression library comprising over 800 previously uncharacterized genes from B. subtilis. Each gene was placed under the control of an IPTG-inducible promoter (Phy) and integrated in single copy (1×) at amyE, a nonessential locus in the B. subtilis chromosome. The library (called the BEIGEL, for Bacillus ectopic inducible gene expression library) was screened for misexpression phenotypes that perturbed growth on solid media and also resulted in obvious defects in nucleoid morphology, changes in cell division frequency, and/or perturbations in overall cell shape in liquid cultures. Two strains, one harboring Phy-yodL and one harboring Phy-yisK, were unable to form colonies on plates containing inducer (Fig. 1A) and also produced wide, irregular cells with slightly tapered poles following misexpression in LB liquid medium (Fig. 1B). Cell lysis and aberrant cell divisions were also observed. Introducing a second copy (2×) of each Phy misexpression construct into the chromosome did not appreciably enhance cell widening at the 90-min postinduction time point, although cell lysis was more readily observed (Fig. 1B). Phy-yisK (2×) misexpression also led to a drop in optical density over time (see Fig. S1A in the supplemental material), consistent with the cell lysis observed microscopically. We concluded that the activities of yodL and yisK target one or more processes integral to width control during cell elongation.
FIG 1.
Misexpression of YodL and YisK prevents cell growth on solid medium and causes loss of cell shape in liquid medium. (A) Cells harboring one (1×) or two (2×) copies of Phy-yodL (BAS040 and BAS191) or Phy-yisK (BAS041 and BYD074) were streaked on an LB plate supplemented with 100 μg/ml spectinomycin and, where indicated, 1 mM IPTG or 1 mM IPTG and the denoted concentration of MgCl2. Plates were incubated for ∼16 h at 37°C before image capture. (B) The strains shown in panel A were grown in LB-Lennox medium at 37°C to mid-exponential phase and back-diluted to an OD600 of ∼0.02. Where indicated, 1 mM IPTG or 1 mM IPTG and the denoted concentration of MgCl2 were added. Cells were grown for 1.5 h at 37°C before image capture. Membranes were stained with TMA-DPH. All images were scaled identically.
The yodL and yisK misexpression phenotypes are similar to those observed when proteins involved in cell elongation are perturbed in B. subtilis (20, 31, 45). Since the addition of magnesium was previously reported to suppress the lethality and/or morphological phenotypes associated with depletion or deletion of some proteins important for cell elongation in B. subtilis (16, 20, 29, 31, 46), we assessed if the Phy-yodL and Phy-yisK misexpression phenotypes could be rescued by growing cells with media supplemented with two different concentrations of MgCl2. The YodL-producing cells failed to grow on any LB media containing inducer, regardless of MgCl2 concentration (Fig. 1A). In contrast, LB supplemented with 25 mM MgCl2 restored viability to the strain producing YisK (Fig. 1A). Interestingly, even 25 mM MgCl2 was not sufficient to suppress the cell-widening effect associated with YodL and YisK misexpression (Fig. 1B), although these cells did not lyse (see Fig. S1C in the supplemental material). Since PAB medium was often used in the prior studies that showed that MgCl2 supplementation rescued cell shape (16, 20, 29, 31, 46), we also assayed for growth on PAB following YodL and YisK expression. PAB supplemented with 25 mM MgCl2 rescued growth on plates (see Fig. S2A in the supplemental material), but it still did not rescue morphology in liquid culture (see Fig. S2B).
yodL and yisK expression.
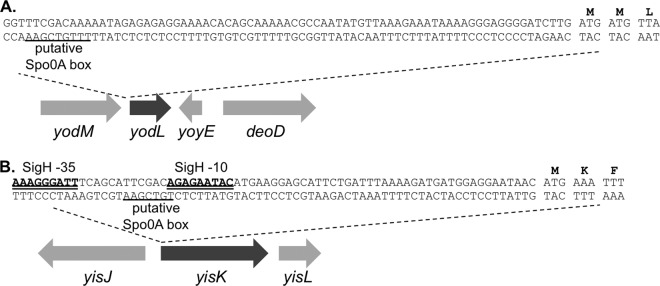
To better understand the possible physiological functions of the yodL and yisK gene products, we analyzed the genes and their genetic contexts bioinformatically. yodL is predicted to encode a 12.5-kDa hypothetical protein which, based on amino acid similarity, is conserved in the Bacillus genus. In data from a global microarray study analyzing conditional gene expression in B. subtilis, yodL was expressed as a monocistronic mRNA, exhibiting peak expression ∼2 h after entry into sporulation (47). yodL expression is most strongly correlated with expression of racA and refZ (yttP) (47), genes directly regulated by Spo0A (48). yodL was not previously identified as a member of the Spo0A regulon controlling early sporulation gene expression (48, 49); however, a more recent study found that yodL expression during sporulation was reduced in a Δspo0A mutant (50). Consistent with this observation, we identified a putative Spo0A box approximately ∼75 bp upstream of the annotated yodL start codon (Fig. 2A). yisK is predicted to encode a 33-kDa protein and is annotated as a putative catabolic enzyme, based on its similarity to proteins involved in the degradation of aromatic amino acids (51). yisK was previously identified as a member of the SigH regulon and possesses a SigH −35/−10 motif (49) (Fig. 2B). Expression of yisK peaks ∼2 h after entry into sporulation (39) and is most strongly correlated with expression of kinA (47), a gene regulated by both SigH (the stationary-phase sigma factor) (49, 52–54) and Spo0A (48, 55). As with yodL, we identified a putative Spo0A box in the regulatory region upstream of the yisK start codon (Fig. 2B).
FIG 2.
DNA sequence upstream of yodL and yisK. (A) Putative Spo0A box (underlined) upstream of the yodL start codon. (B) SigH binding motifs (double underline) and putative Spo0A box (underlined) upstream of the yisK start codon.
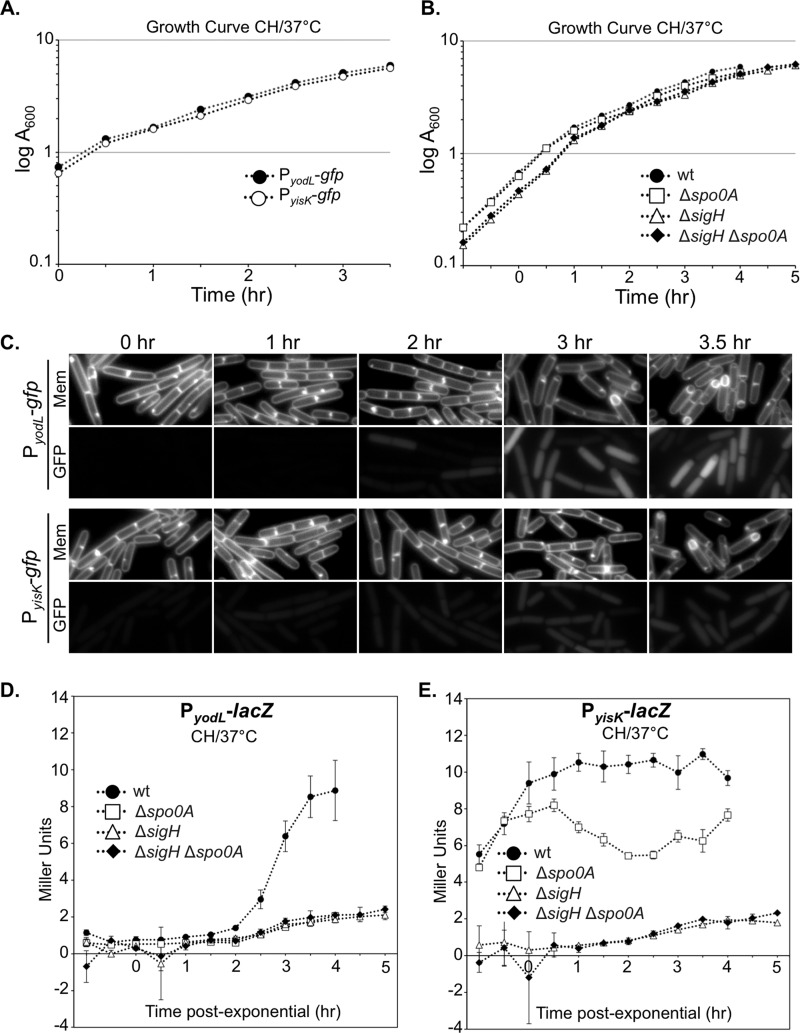
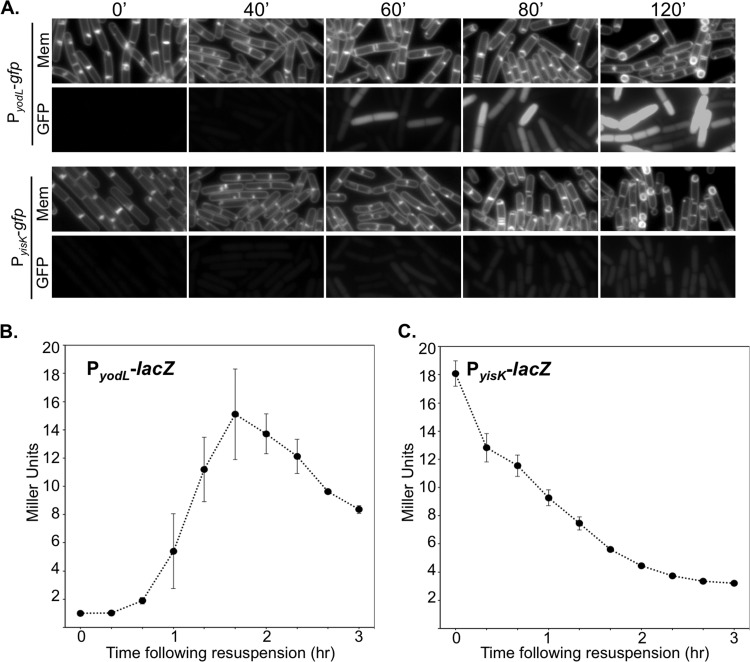
To independently test if yodL and yisK expression are consistent with Spo0A-dependent regulation, we fused the putative regulatory regions upstream of each gene to a gfp reporter gene and integrated the fusions into the amyE locus. We then followed expression from the promoter fusions over a time course in CH liquid broth, a rich medium in which the cells first grow exponentially, transition to stationary phase, and finally gradually enter sporulation (Fig. 3A to C). In this time course, the GFP signal from PyisK-gfp increased dramatically from time zero (optical density at 600 nm [OD600], ∼0.6) to time 1 h (OD600, ∼1.6) (Fig. 3C), consistent with yisK's prior characterization as a SigH-regulated gene (49). In contrast, GFP fluorescence from PyodL-gfp became evident at a later time point (120 min) and was more heterogeneous (Fig. 3C), consistent with expression patterns previously observed for other Spo0A-P regulated genes (56, 57).
FIG 3.
Expression levels of yodL and yisK promoters during a CH time course experiment. Expression from the putative yodL and yisK promoter regions was monitored in CH medium at 37°C over the time course. The OD600 (A and B) and production of either GFP (C) or beta-galactosidase (D and E) were monitored at 30-min intervals. Membranes were stained with TMA-DPH. All GFP channel images were captured with 1-s exposures and scaled identically to allow for direct comparisons. In this assay, time zero represents the last exponential time point, not the initiation of sporulation. Vertical bars in the graphs represent standard deviations.
To quantitate expression from the promoters, we generated PyodL-lacZ and PyisK-lacZ reporter strains and collected samples over a CH time course beginning with early exponential phase (OD600, 0.2). Expression from PyodL-lacZ rose steadily, beginning about 2 h after exit from exponential growth, and continued to rise at least until the final time point (Fig. 3D). In contrast, expression from PyisK-lacZ rose as cells transitioned from early to late exponential growth, reached peak levels shortly after exit from exponential growth, and remained steady for the remainder of the time points (Fig. 3E). Wild-type expression from both PyodL-lacZ and PyisK-lacZ required both SigH and Spo0A and was largely eliminated in the absence of both regulators (Fig. 3D and E). We did not attempt to draw further conclusions from these data, since Spo0A and SigH each require the other for wild-type levels of expression (see Discussion).
We then followed expression from the promoter fusions over a time course following the sporulation by using the resuspension method, which generates a more synchronous entry into sporulation (58). At time zero, neither the strain harboring PyodL-gfp nor the strain harboring PyisK-gfp showed appreciable levels of fluorescence (Fig. 4A) and appeared similar to a negative control harboring gfp without a promoter (see Fig. S3 in the supplemental material). Between 0 and 40 min, both strains showed detectable increases in fluorescence. At 60 min, when the first polar divisions characteristic of sporulation typically begin to manifest, both strains were more strongly fluorescent (Fig. 4A). GFP fluorescence from PyodL was qualitatively more intense than fluorescence produced from PyisK (all images were captured and scaled with identical parameters to allow for direct comparisons). Moreover, the GFP signal continued to accumulate in the strain harboring PyodL-gfp for at least 2 h (Fig. 4A) and was heterogenous, consistent with activation by Spo0A. In contrast, the fluorescence signal produced from PyisK-gfp was similar across the population and appeared similar at the 60- and 120-min time points (Fig. 4A), consistent with SigH regulation.
FIG 4.
Expression from yodL and yisK promoters following sporulation by resuspension. Expression from the putative yodL and yisK promoter regions was monitored in resuspension medium. The production of either GFP (A) or beta-galactosidase (B and C) was monitored at 20-min intervals. Membranes were stained with TMA-DPH. All GFP channel images were captured with 1-s exposures and scaled identically to allow for direct comparisons. Vertical bars in the graphs represent standard deviations.
To quantitate expression from the promoters during sporulation via a resuspension time course, we collected data from time points for strains harboring either the PyodL-lacZ or PyisK-lacZ reporter constructs and performed beta-galactosidase assays. Expression from PyodL-lacZ rose rapidly between the 40-min and 100-min time points and steadily declined thereafter (Fig. 4B). The decline in signal was not observed for the GFP reporter, likely because GFP is stable once synthesized (59). In contrast, expression from PyisK-lacZ was highest at the time of resuspension (T0) and declined until the final time point (Fig. 4C).
Collectively, the patterns of expression we observed for yodL are consistent with those observed for genes activated by high-threshold levels of Spo0A during sporulation, including racA, spoIIG, and spoIIA (60). In contrast, yisK's expression pattern is similar to that observed for kinA (47, 53, 61), with expression increasing in late exponential and stationary phases and early sporulation in a SigH-dependent manner (Fig. 3) but decreasing during sporulation in the resuspension time course experiment (Fig. 4). We do not exclude the possibility that YodL and YisK also function in other growth contexts.
A ΔyodL ΔyisK mutant is defective in sporulation.
Since yodL and yisK expression levels correlate with those for other early sporulation genes, we next investigated if the gene products influenced the production of heat-resistant spores. To determine the number of heat-resistant spores in a sporulation culture, we quantified the number of CFU present in cultures before (total CFU) and after (heat-resistant CFU) a heat treatment that kills vegetative cells. These values were normalized to display the sporulation efficiency of the mutants relative to wild type. Single mutants in which either yodL or yisK was deleted displayed only mild (97% and 94%, respectively) reductions in relative sporulation efficiency (Table 1). Although the single mutants always sporulated less efficiently than wild type in each experimental replicate, the differences were not statistically significant with only six experimental replicates. In contrast, the ΔyodL ΔyisK double mutant produced ∼20% fewer heat-resistant spores than the wild type (P < 0.0006) (Table 1). No decrease in total CFU was observed for any of the mutants compared to the wild type, indicating that the reduction in heat-resistant spores in the ΔyodL ΔyisK mutant was not due to reduced cell viability before heat treatment (Table 1). The gene downstream of yisK, yisL, is transcribed in the same direction as yisK. To determine if the reduction in sporulation we observed might be partially attributable to polar effects of the yisK deletion on yisL expression, we introduced PyisK-yisK at an ectopic locus (amyE) in the ΔyodL ΔyisK mutant and repeated the heat-kill assay. The ectopic copy of PyisK-yisK restored sporulation in the ΔyodL ΔyisK double mutant to levels statistically indistinguishable from the those of the ΔyodL single mutant (Table 1). These results lend support to the idea that YodL and YisK function during early sporulation and possess activities that, directly or indirectly, affect the production of viable spores. We do not exclude the possibility that YodL and YisK also function outside the context of sporulation.
TABLE 1.
Sporulation efficiencies of yodL and yisK mutantsa
| Genotype | Strain | Total CFU | Heat-resistant CFU | % sporulation efficiency | % relative sporulation efficiency |
|---|---|---|---|---|---|
| Wild type | B. subtilis 168 | 2.8 × 108 (4.7 × 107) | 1.9 × 108 (4.5 × 107) | 66.9 (5) | 100 |
| ΔyodL | BYD276 | 2.6 × 108 (3.9 × 107) | 1.7 × 108 (2.8 × 107) | 65.2 (7) | 97 |
| ΔyisK | BYD278 | 2.7 × 108 (4.6 × 107) | 2.4 × 108 (2.7 × 107) | 63.1 (6) | 94 |
| ΔyodL ΔyisK | BYD279 | 3.1 × 108 (6.5 × 107) | 1.7 × 108 (4.1 × 107) | 54.1 (4) | 81 |
| ΔyodL ΔyisK PyisK-yisK | BYD510 | 3.4 × 108 (3.3 × 107) | 2.3 × 108 (4.1 × 107) | 66.2 (7) | 99 |
Sporulation efficiency was calculated as the number of spores per milliliter, divided by the total CFU per milliliter, times 100. Relative sporulation efficiency is the sporulation efficiency normalized to that of the wild type (times 100). Values are means, with standard deviations in parentheses. The data shown are the average results for six independent biological replicates. The difference in sporulation efficiency between the wild type and the ΔyodL ΔyisK double mutant was statistically significant (P < 0.0006).
Given that yisK and yodL expression during vegetative growth leads to cell widening, we hypothesized that yisK and yodL mutants might produce thinner cells or spores during sporulation. However, no qualitative differences in cell or spore width were observed for the ΔyodL, ΔyisK, or ΔyodL ΔyisK mutant populations compared to the wild type during a sporulation time course assay (see Fig. S4 in the supplemental material). We also observed no qualitative differences in the shapes of germinating cells (data not shown). Thus, although YodL and YisK contribute to the production of heat-resistant spores, they do not appear to be required to generate any of the major morphological changes required for spore production.
MreB and Mbl are targets of YodL and YisK activity.
To identify genetic targets associated with YodL and YisK activity, we took advantage of the fact that misexpression of the proteins during vegetative growth prevents colony formation on plates, and we performed suppressor selection analysis. Strains harboring three copies of each misexpression cassette were utilized to reduce the chances of obtaining trivial suppressors in the misexpression cassette itself. In addition, Phy-lacZ was used as a reporter to eliminate suppressors unable to release LacI repression following addition of inducer. In total, we obtained 14 suppressors resistant to YodL expression and 13 suppressors resistant to YisK expression. Six of the suppressors resistant to YodL were subjected to whole-genome sequencing. The results of the sequencing are shown in Table S1 in the supplemental material. All of the suppressors possessed mutations in either mreB or mbl, genes previously shown to be important in regulating cell width (see Table S1). Using targeted sequencing, we determined that the remaining suppressor strains resistant to YodL also harbored mutations in mreB or mbl. Since the phenotypes of YodL and YisK expression were similar, we also performed targeted sequencing of the mreB and mbl chromosomal regions in the YisK-resistant suppressors. All but one of the YisK-resistant suppressors possessed mutations in mbl; the remaining suppressor harbored a mutation in mreB.
To determine if the point mutations we identified were sufficient to confer resistance to YodL or YisK misexpression, we generated the mutant alleles in clean genetic backgrounds (see the supplemental material) and assayed for resistance to three copies (3×) of each misexpression construct (Table 2). In all cases but one, the engineered strains were resistant to the same selective pressure applied in the original selections (either 3× yodL or 3× yisK) (Table 2), indicating that the mreB or mbl mutations identified through sequencing were sufficient to confer resistance. When we attempted to engineer a strain harboring only MreBS154R, all but one of the strains also possessed a second substitution, MreBR230C. Although the remaining strain possessed only the MreBS154R substitution in MreB, unlike the original suppressor identified by whole-genome sequencing (see Table S1 in the supplemental material), the MreBS154R-producing strain was also sensitive to YodL expression. Based on these data, we suspect that the strain harboring the gene for MreBS154R might be unstable and possibly predisposed to the accumulation of second-site mutations.
TABLE 2.
Analysis of suppressor strains resistant to YodL and/or YisKa
| Variant(s) | Gene mutation | Amino acid change | Phenotype with misexpression of: |
|
|---|---|---|---|---|
| YodL | YisK | |||
| Variants obtained via YodL misexpression | ||||
| mreB mutants | CGC → GGC | R117Gb | R | R |
| GGA → GCA | G143A | R | R | |
| AAT → GAT* | N145Dc | R | S | |
| CCA → CGA* | P147Rb,c | R | S | |
| AGC → AGA | S154Rb,c | NA | NA | |
| AGC → AGA | S154Rb | R | R | |
| CGC → TGC | R230C | |||
| AGA → AGT* | R282Sb,c | R | S | |
| GGG → GAG | G323Eb | R | R | |
| mbl mutants | ACG → ATG | T158M | R | R |
| GAA → AAA* | E250Kb | R | R | |
| ACA → ATA | T317I | R | R | |
| Variants obtained via YisK misexpression | ||||
| mreB mutant | CGC → TGC | R117G | R | R |
| mbl mutants | ATG → ATA | M51Id | R | R |
| CGC → TGC | R63Cd | R | S | |
| GAC → AAC* | D153Nd | R | R | |
| GGC → GAC | G156Dd | R | R | |
| ACG → GCG* | T158Ad | R | R | |
| GAG → GGG | E204Gd | R | R | |
| GAA → AAA | E250K | R | R | |
| TCT → ΔΔΔ | ΔS251 | R | S | |
| CCT → CTT | P309Ld | R | S | |
| GCC → ACC | A314Td | R | R | |
Details for the suppressor selections are described in Materials and Methods. Candidate mutations were introduced into clean genetic backgrounds harboring three copies of Phy-yodL or three copies of Phy-yisK, and the resultant strains were assessed for resistance (R) or sensitivity (S) to either yodL or yisK expression, as judged by the ability to grow on LB plates supplemented with 1 mM IPTG and 100 μg/ml spectinomycin. An asterisk indicates that two suppressors possessing the same nucleotide change were obtained in the original selection. The underlined residues displayed specificity in resistance to YodL over YisK (after YodL misexpression) or YisK over YodL (after YisK misexpression). NA, not assessed.
Originally identified using whole-genome sequencing (see Table S1 in the supplemental material).
Residues previously implicated in the RodZ-MreB interaction (18).
The YodL-resistant strains generally possessed mutations resulting in amino acid substitutions with charge changes (Table 2). When mapped to the Thermotoga maritima MreB structure, 5/7 of the unique suppressor strains possessed amino acid substitutions in a region important for mediating the interaction between MreB and the bitopic membrane protein RodZ (MreBG143A, MreBN145D, MreBP147R, MreBS154R, and MreBR282S) (Table 2; see also Fig. S5 in the supplemental material) (18, 62); three of these substitutions occurred in residues that make up the RodZ-MreB binding surface (MreBN140, MreBP142, and MreBR279 in T. maritima) (18).
A majority of the mutations in YisK-resistant Mbl variants clustered in regions of Mbl that are predicted to make up the ATP binding pocket (Table 2; see also Fig. S6 in the supplemental material). Moreover, seven of the substitutions occurred in amino acids previously associated with resistance to the MreB inhibitor A22 in C. crescentus and Vibrio cholerae (see Fig. S6) (12, 63, 64).
MreBR117G and MblE250K were independently isolated in both the YodL and YisK suppressor selections, raising the possibility that at least some of the other MreB and Mbl variants exhibit cross-resistance to YodL and YisK misexpression. To test for cross-resistance, we generated the mutant alleles in clean genetic backgrounds and then introduced 3 copies of Phy-yisK into the YodL-resistant suppressors and 3 copies Phy-yodL into the YisK-resistant suppressors. We then assayed for the ability of the misexpression strains to grow on medium in the presence of inducer. The results, summarized in Table 2, showed that several of the variants exhibited resistance to both YodL and YisK. Three MreB variants, MreBN145D, MreBP147R, and MreBR282S, exhibited specificity in their resistance to YodL compared to YisK. Three Mbl variants, MblR63C, MblΔS251, and MblP309L, showed specificity in their resistance to YisK over YodL. These results suggest that the alleles exhibiting cross-resistance to both YisK and YodL are likely to be general, possibly conferring gain of function to either MreB or Mbl activity.
YodL and YisK cell-widening activities require MreB and Mbl, respectively.
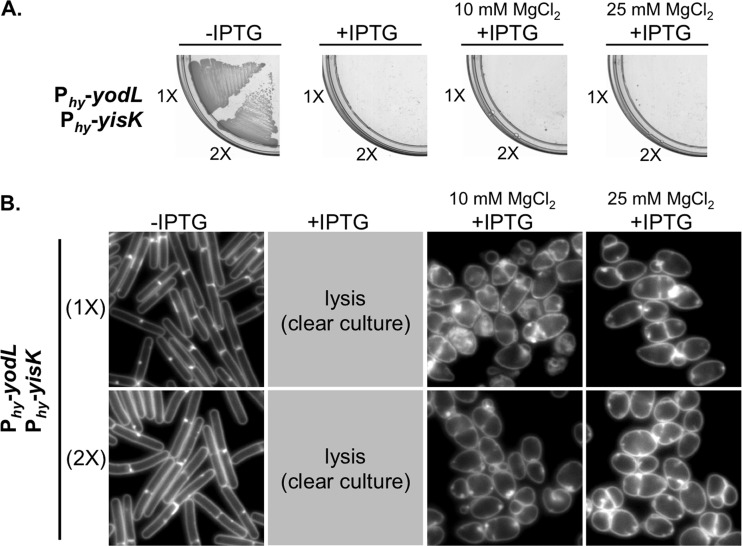
The phenotypic consequences of YodL and YisK misexpression are similar but not identical (Fig. 1B), suggesting that YodL and YisK might have distinct targets. Consistent with this idea, YodL and YisK coexpression resulted in phenotypes distinct from misexpression of either YodL or YisK alone. More specifically, cells coexpressing YodL and YisK did not grow on plates, regardless of the medium or MgCl2 concentration (Fig. 5A; see also Fig. S2A in the supplemental material), and growth without lysis in liquid media required the presence of MgCl2 (Fig. 5B; see also Fig. S1 and S2B in the supplemental material). Importantly, the coexpressing cells displayed a round morphology that strongly contrasted with strains expressing either YodL or YisK alone (Fig. 5B; seee also Fig. S2B). The round morphology was unlikely due to higher expression of gene products (1× Phy-yodL plus 1× Phy-yisK), since cells harboring two copies (2×) of either Phy-yodL or Phy-yisK did not become round (Fig. 1B; see also Fig. S2B).
FIG 5.
YodL and YisK comisexpression causes cell lysis. (A) BYD361 (Phy-yodL Phy-yisK) and BYD281 (2× Phy-yodL 2× Phy-yisK) were streaked on an LB plate with 100 μg/ml spectinomycin and, where indicated, 1 mM IPTG or 1 mM IPTG and the denoted concentration of MgCl2. (B) Cells were grown in LB-Lennox medium at 37°C to mid-exponential phase and back-diluted to an OD600 of ∼0.02. Where indicated, 1 mM IPTG or 1 mM IPTG and the denoted concentration of MgCl2 were added. Cells were then grown for 1.5 h at 37°C before image capture. Membranes were stained with TMA-DPH. All images are shown at the same magnification.
Based on the observation that YodL and YisK coexpression yields distinct phenotypes, and the fact that all of the YodL-specific suppressor mutations occurred in mreB (yielding MreBN145D, MreBP147R, and MreBR282S), while all of the YisK-specific suppressor mutations occurred in mbl (yielding MblR63C, MblΔS251, and MblP309L), we hypothesized that YodL targets MreB whereas YisK targets Mbl. To test these hypotheses, we assessed if MreB and Mbl were specifically required for YodL and YisK function by taking advantage of the fact that mreB and mbl can be deleted in a ΔponA background with only minor changes in cell shape (20, 31). The ΔponA strain, which does not make PBP1, produces slightly longer and thinner cells than the parent strain and requires MgCl2 supplementation for normal growth (65, 66). We generated ΔponA ΔmreB and ΔponA Δmbl strains and then introduced either two copies of Phy-yodL or two copies of Phy-yisK into each background. We reasoned that 2× expression would provide a more stringent test for specificity than 1× expression, as off-target effects (if any) would be easier to detect. To assess the requirement of either mreB or mbl for YodL or YisK activity, cells were grown to exponential phase in LB medium supplemented with 10 mM MgCl2, back-diluted to a low optical density, and induced for 90 min before images were captured for microscopy. Uninduced controls all appeared as regular rods, although ΔponA deletion strains were noticeably thinner than wild-type parents (Fig. 6). The ΔponA cells became wider following YodL expression, indicating that PBP1 is not required for YodL activity. We also observed that the poles of the ΔponA mutant were less elongated and tapered than the wild-type control following YodL expression, suggesting that this particular effect of YodL expression is PBP1 dependent (Fig. 6A). A ΔponA Δmbl mutant phenocopied the ΔponA parent following YodL expression (Fig. 6A), indicating that Mbl is not required for YodL's activity. In contrast, the ΔponA ΔmreB strain did not show morphological changes following YodL expression and instead appeared similar to the uninduced control. We conclude that YodL requires MreB for its cell-widening activity.
We performed a similar series of experiments for YisK misexpression. The ΔponA mutant was sensitive to YisK expression, indicating that PBP1 is not required for YisK-dependent cell widening. Similarly, expression of YisK in a ΔponA ΔmreB mutant also resulted in loss of cell width control (Fig. 6B), indicating that MreB is not required for YisK activity; however, unlike YisK expression in a wild-type or ΔponA background, the cells became round (Fig. 6B), more similar to the YodL- and YisK-coexpressing cells (Fig. 5; see also Fig. S2B in the supplemental material). In contrast, a ΔponA Δmbl mutant did not lose control over cell width following YisK expression (Fig. 6B), indicating that YisK activity requires Mbl for its cell-widening activity. We conclude that YodL requires MreB but not Mbl for its cell-widening activity, whereas YisK requires Mbl but not MreB.
YisK possesses at least one additional target.
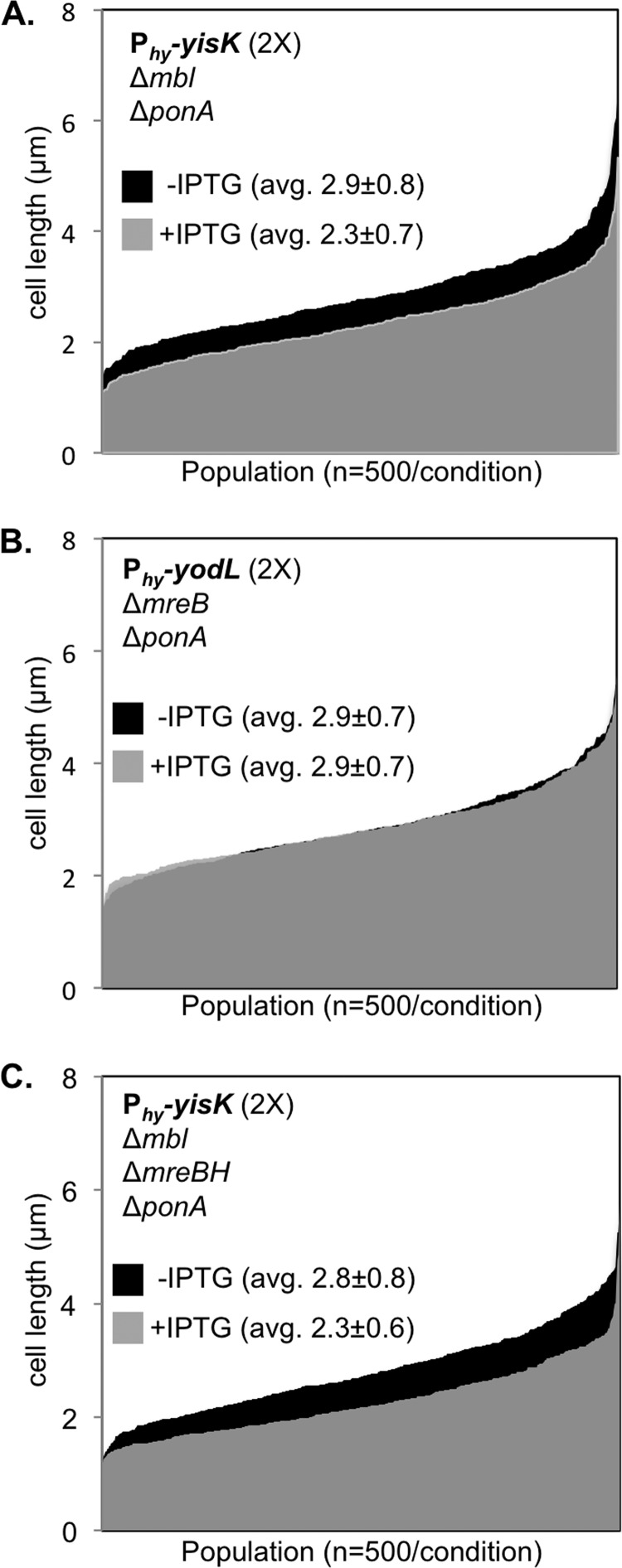
Although YisK expression in a ΔponA Δmbl mutant did not result in cell widening, we observed that the induced cells appeared qualitatively shorter than the uninduced controls, suggesting that YisK might possess a second activity (Fig. 6B). Quantitation of cell length in a ΔponA Δmbl mutant following YisK expression revealed that the YisK-induced cells were ∼20% shorter than the uninduced cells (Fig. 7A). In contrast, YodL expression did not result in a change in cell length in a ΔponA ΔmreB mutant (Fig. 7B), suggesting that the cell shortening effect is specific to YisK. We hypothesized that MreBH, the third and final B. subtilis MreB family member, might be YisK's additional target. We hypothesized that if MreBH is the additional target, then the cell shortening observed upon YisK expression in a ΔponA Δmbl mutant strain should be lost in a ΔponA Δmbl ΔmreBH mutant background. However, we found that even when mreBH was additionally deleted, YisK expression still resulted in cell shortening (Fig. 7C). We conclude that YisK likely has at least one additional target that is not MreB or Mbl dependent and that this additional target regulates some aspect of cell length.
FIG 7.
YisK expression results in cell shortening. (A) Cells harboring two copies of Phy-yisK in a ΔponA Δmbl background (BYD262) were grown at 37°C in LB supplemented with 10 mM MgCl2 to mid-exponential phase. To induce yisK expression, cells were back-diluted to an OD600 of ∼0.02 in LB with 10 mM MgCl2, and IPTG (1 mM) was added. Cells were grown for 1.5 h at 37°C before image capture. Membranes were stained with TMA-DPH. Cell lengths (n = 500 cells/condition) were measured before and after yisK expression and rank-ordered from smallest to largest along the x axis so that the entire population could be visualized without binning. The uninduced population (black) is juxtaposed behind the induced population (semitransparent, gray). The differences in average cell length before and after Phy-yisK induction were statistically significant (P < 0.0001). (B) Cells harboring two copies of Phy-yodL in a ΔponA ΔmreB background (BYD263) were grown, quantitated, and plotted as described above. The differences in average cell length before and after Phy-yodL induction were not statistically significant. (C) Cells harboring two copies of Phy-yisK in a ΔponA Δmbl ΔmreBH background (BAS248) were grown, quantitated, and plotted as described above. The differences in average cell length before and after Phy-yisK induction were statistically significant (P < 0.0001).
DISCUSSION
Functional targets of YodL and YisK.
Misexpression of YodL during vegetative growth results in cell widening and lysis, and spontaneous suppressor mutations conferring resistance to YodL occur primarily in mreB. MreB is also required for YodL's cell-widening activity, whereas Mbl is not. By comparison, expression of YisK during vegetative growth also results in cell widening and lysis; however, spontaneous suppressor mutations conferring resistance to YisK occur primarily in mbl. YisK's cell-widening activity requires Mbl but not MreB. The simplest interpretation of these results is that YodL targets MreB function while YisK targets Mbl function. Alternatively, YodL and YisK could target other factors that affect cell shape and simply require MreB and Mbl for their respective functions.
MreB variants specifically resistant to YodL activity, MreBN145D, MreBP147R, and MreBR282S, all result in charge change substitutions in residues previously shown to constitute the RodZ-MreB interaction surface (equivalent T. maritima residues: MreBN140, MreBN142, and MreBR279) (18). MreBG143A, which exhibits cross-resistance to YisK, also maps near the RodZ-MreB interaction interface. The two remaining YodL-resistant MreB variants occur in (MreBR117G) or near (MreBG323E) residues previously associated with bypass of RodZ essentiality in E. coli (see Fig. S5 in the supplemental material) (25). A simple model explaining both the nature of the MreB variants we obtained in the suppressor selections and YodL's MreB-dependent effect on cell shape is based on YodL acting by disrupting the interaction between RodZ and MreB. In this model, MreB's RodZ-independent activities would remain functional, and several observations are consistent with this idea. If YodL were to completely inactivate MreB function, then we would expect that expressing YodL in a ΔponA Δmbl ΔmreBH background would generate round cells, similar to the phenotype observed when MreB is depleted in a Δmbl ΔmreBH mutant background (31), or when mreB, mbl, and mreBH are deleted in backgrounds with upregulated sigI expression (the triple mutant is otherwise lethal) (30). However, we observed that cells expressing YodL in a ΔponA Δmbl ΔmreBH mutant instead formed wide rods (Fig. 6A). If YodL does specifically target MreB activity, then these results suggest that MreB likely retains at least some of its width maintenance function. Morgenstein et al. recently found that the interaction between RodZ and MreB in E. coli is required for MreB rotation but that MreB rotation was not required for rod shape or cell viability under standard laboratory conditions (26). The findings of that study are consistent with prior findings indicating that RodZ is not absolutely required for maintenance of rod shape (25).
We hypothesize that the substitutions obtained in residues near the RodZ-MreB interface either enhance the RodZ-MreB interaction or decrease the ability of YodL to disrupt the RodZ-MreB interface. Although we did not identify YodL-resistant suppressor mutations in rodZ, it is possible that the requisite rodZ mutations are rare or lethal for the cell; thus, we cannot rule out the possibility that YodL targets RodZ function. Similarly, although we found that MreB is required for YodL activity, we can envision a scenario in which a YodL-MreB interaction may be necessary to localize YodL to a cellular location where it can be effective against RodZ or some other cellular component. We think this possibility is less likely, as cells expressing YodL have a distinct phenotype from RodZ depleted B. subtilis cells. More specifically, YodL expression results in cell widening and tapered poles (Fig. 1B), whereas RodZ-depleted cells generate wide rods (22), similar to the phenotype we observed following YodL expression in a ΔponA Δmbl ΔmreBH mutant (Fig. 6A). These results argue against the idea that YodL could work by inactivating RodZ function completely. Future work aimed at characterizing the nature of the YodL-resistant suppressors and the effect of YodL on MreB function will shed light on the mechanism underlying YodL's observed activity.
Only three Mbl variants MblR63C, MblΔS251, and MblP309L showed specificity in resistance to YisK over YodL. MblR63C, MblD153N, MblG156D, MblT158A, MblE204G, MreBP309L, and MblA314T occur in residues that form Mbl's predicted ATP binding pocket (see Fig. S6 in the supplemental material), and substitutions in all seven of these residues have been previously implicated in A22 resistance (see Fig. S6) (12, 63, 64). We speculate that most, if not all, of the substitutions in Mbl's ATP binding pocket result in a gain of function with respect to Mbl polymerization, a hypothesis that can ultimately be tested in vitro. Similarly, we hypothesize that the MblM51I substitution, located at the MreB head-tail polymerization interface (62), may overcome YisK activity by promoting Mbl polymerization. MreBE262 of C. crescentus, equivalent to B. subtilis MblE250 (see Fig. S6), is located at the interaction interface of antiparallel MreB protofilament bundles (67). If B. subtilis MblE250 is also present at this interface (this has not been tested to our knowledge), then MblE250K could promote resistance to YodL and YisK by enhancing Mbl bundling. How might YisK exert its activity? One idea is that YisK disrupts Mbl bundling, possibly by competing for sites required for protofilament formation. An alternative possibility is that YisK somehow prevents Mbl from effectively binding or hydrolyzing ATP. It is also possible that Mbl is simply required for YisK to target one or more other factors involved in cell width control.
In addition to Mbl-dependent cell widening, YisK expression resulted in cell shortening, an effect that only became apparent in a ΔponA Δmbl mutant background (Fig. 6B and 7A). Given the similarities of MreB, Mbl, and MreBH to each other, we initially hypothesized that YisK-dependent effects on MreB and/or MreBH might be responsible for the decrease in cell length we observed; however, we found that mreBH was not required for cell shortening (Fig. 6B and 7C). Since YisK expression results in a dramatic loss of cell shape in ΔmreB mutant backgrounds (Fig. 6A), we were unable to confidently assess cell length changes to determine if there is a requirement for MreB in the cell-shortening phenotype. It is unlikely that YisK's additional activity affects MreB's role in maintaining cell width (at least not without Mbl), as YisK-expressing cells retain a rod shape when mbl and mreBH are both deleted (Fig. 6B). An exciting alternative possibility is that YisK activity affects another factor involved in cell length control. One attractive candidate is the cell wall hydrolase CwlO, a known modulator of cell length in B. subtilis (68), which recent genetic data also suggest depends at least in part on Mbl (35). Future experiments aimed at determining the identity and function of YisK's additional target should shed light on how cells regulate both cell length and cell width.
Identification of genes involved in cellular organization through a novel gene discovery pipeline.
To systematically identify and characterize novel genes involved in cellular organization, we developed a gene discovery pipeline that combines known regulatory information (47), phenotypes obtained from misexpression screening, and suppressor selection analysis. The ability to identify genetic targets associated with the unknown genes provides a key parameter beyond phenotype from which to formulate testable hypotheses regarding each gene's possible function. The misexpression constructs we generated are inducible and present in single copy on the chromosome. We have found that to obtain phenotypes, our strategy works best when the unknown genes are expressed outside their native regulatory context. Thus far, we have restricted our gene function discovery pipeline to B. subtilis; however, the general approach should be broadly applicable to other organisms and diverse screening strategies.
In this work, we have described the use of the pipeline to identify and characterize two B. subtilis genes, yodL and yisK, that produce proteins capable of targeting activities intrinsic to cell width control. Although yodL and yisK were not previously recognized as members of the Spo0A regulon, both genes have putative Spo0A boxes and possess promoters that exhibit expression patterns consistent with other Spo0A-regulated genes (Fig. 2 to 4). YisK is also a member of the SigH regulon (49), and our expression analysis results are also consistent with expression of yisK during stationary phase (Fig. 3). If the putative Spo0A box we identified is utilized in vivo, then we would predict based on our expression profiling that yisK is transcribed during exponential and early stationary phases via SigH and then repressed as Spo0A-P accumulates during early sporulation. Such a pattern is similar to the regulation that has been proposed for kinA (55, 60). We also observed that expression levels of PyodL and PyisK are reduced in the absence of Spo0A and SigH (Fig. 3D and E). The specific contributions of these global regulators to yodL and yisK regulation cannot be determined by analyzing the expression profiles of the sigH and spo0A deletion strains alone, since spo0A depends on SigH for upregulation during the early stages of sporulation (52, 55). Moreover, since Spo0A inhibits expression of the sigH repressor AbrB (69–72), a spo0A mutant also has reduced sigH expression.
A ΔyodL ΔyisK double mutant reproducibly produces ∼20% fewer heat-stable spores than the wild type, suggesting that the YodL and YisK have functions that affect spore development (either directly or indirectly). Most studies on sporulation genes are biased toward factors that reduce sporulation efficiency by an order of magnitude or more in a standard heat-kill assay. However, even small differences in fitness (if reproducible) can contribute significantly to the ability of an organism to persist, especially in competitive environments (73). The 20% reduction in heat-resistant spores we observed in cells lacking YisK and YodL would likely result in a substantial fitness disadvantage to cells in the environment. We do not currently understand how YodL and YisK might function in spore development, but the identification of MreB and Mbl as genetic targets suggests that the proteins likely regulate some aspect of PG synthesis. Future studies will be aimed at understanding the molecular and biochemical bases of the activities of YodL and YisK.
In this study, the morphological phenotypes associated with YodL and YisK occurred when the genes were expressed during vegetative growth. Consequently, it is formally possible (although we think unlikely) that the targeting of MreB and Mbl is simply a coincidence that is unrelated to the potential functions of the proteins during stationary phase or sporulation. Regardless of what YodL's and YisK's physiological roles turn out to be, we have already been able to utilize misexpression of the proteins to obtain interesting variants of both MreB and Mbl that can now be used to generate testable predictions regarding how MreB and Mbl function in B. subtilis. Moreover, the apparent specificities with which YodL and YisK appear to target MreB and Mbl, respectively, make them potentially powerful tools to differentially target the activities of these two highly similar paralogs in vivo. Of course, more studies will be required to determine if YodL and YisK interact directly or indirectly to modulate MreB and Mbl activities. In the meantime, it is exciting to speculate that many undiscovered modulators of MreB and MreB-like proteins exist and that we have only just begun to scratch the surface regarding regulation of this important class of proteins. The identification and characterization of such modulators could go a long way toward addressing the significant gaps in our knowledge that exist regarding the regulation of PG synthesis in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Herman Lab for critical reading of the manuscript, Benjamin Mercado for help with the whole-genome sequencing analysis, and Allyssa Miller, Benjamin Mercado, Jared McAnulty, and Simon Rousseau for their efforts toward generating and screening the BEIGEL.
Funding Statement
This work was funded by a National Science Foundation grant (1514629) to J.K.H. and by start-up funds from the Center for Phage Technology and the Department of Biochemistry and Biophysics at Texas A&M University.
Footnotes
This work is dedicated to the memory of Arthur L. Koch.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00183-16.
REFERENCES
- 1.Young KD. 2010. Bacterial shape: two-dimensional questions and possibilities. Annu Rev Microbiol 64:223–240. doi: 10.1146/annurev.micro.112408.134102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holtje JV. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62:181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young KD. 2007. Bacterial morphology: why have different shapes? Curr Opin Microbiol 10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young KD. 2006. The selective value of bacterial shape. Microbiol Mol Biol Rev 70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randich AM, Brun YV. 2015. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol 6:580. doi: 10.3389/fmicb.2015.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenton AK, Gerdes K. 2013. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J 32:1953–1965. doi: 10.1038/emboj.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figge RM, Divakaruni AV, Gober JW. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 9.Ouellette SP, Karimova G, Subtil A, Ladant D. 2012. Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85:164–178. doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- 10.Salje J, van den Ent F, de Boer P, Lowe J. 2011. Direct membrane binding by bacterial actin MreB. Mol Cell 43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colavin A, Hsin J, Huang KC. 2014. Effects of polymerization and nucleotide identity on the conformational dynamics of the bacterial actin homolog MreB. Proc Natl Acad Sci U S A 111:3585–3590. doi: 10.1073/pnas.1317061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Iwai N, Nagai K, Wachi M. 2002. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem 66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 14.Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. 2009. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takacs CN, Poggio S, Charbon G, Pucheault M, Vollmer W, Jacobs-Wagner C. 2010. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol 192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55:78–89. doi: 10.1111/j.1365-2598.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 17.Bendezu FO, de Boer PA. 2008. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Ent F, Johnson CM, Persons L, de Boer P, Lowe J. 2010. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J 29:1081–1090. doi: 10.1038/emboj.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varma A, Young KD. 2009. In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J Bacteriol 191:3526–3533. doi: 10.1128/JB.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai Y, Daniel RA, Errington J. 2009. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol 71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- 21.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muchova K, Chromikova Z, Barak I. 2013. Control of Bacillus subtilis cell shape by RodZ. Environ Microbiol 15:3259–3271. doi: 10.1111/1462-2920.12200. [DOI] [PubMed] [Google Scholar]
- 23.Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci U S A 106:1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niba ET, Li G, Aoki K, Kitakawa M. 2010. Characterization of rodZ mutants: RodZ is not absolutely required for the cell shape and motility. FEMS Microbiol Lett 309:35–42. doi: 10.1111/j.1574-6968.2010.02014.x. [DOI] [PubMed] [Google Scholar]
- 25.Shiomi D, Toyoda A, Aizu T, Ejima F, Fujiyama A, Shini T, Kohara Y, Niki H. 2013. Mutations in cell elongation genes mreB, mrdA, and mrdB suppress the shape defect of RodZ-deficient cells. Mol Microbiol 87:1029–1044. doi: 10.1111/mmi.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgenstein RM, Bratton BP, Nguyen JP, Ouzounov N, Shaevitz JW, Gitai Z. 2015. RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc Natl Acad Sci U S A 112:12510–12515. doi: 10.1073/pnas.1509610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabeen MT, Jacobs-Wagner C. 2010. The bacterial cytoskeleton. Annu Rev Genet 44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 28.Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. 2006. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell 11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Formstone A, Errington J. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 30.Schirner K, Errington J. 2009. The cell wall regulator σI specifically suppresses the lethal phenotype of mbl mutants in Bacillus subtilis. J Bacteriol 191:1404–1413. doi: 10.1128/JB.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai Y, Asai K, Errington J. 2009. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol Microbiol 73:719–731. doi: 10.1111/j.1365-2958.2009.06805.x. [DOI] [PubMed] [Google Scholar]
- 32.Defeu Soufo HJ, Graumann PL. 2006. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Mol Microbiol 62:1340–1356. doi: 10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- 33.Mirouze N, Ferret C, Yao Z, Chastanet A, Carballido-Lopez R. 2015. MreB-dependent inhibition of cell elongation during the escape from competence in Bacillus subtilis. PLoS Genet 11:e1005299. doi: 10.1371/journal.pgen.1005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tseng CL, Shaw GC. 2008. Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis. J Bacteriol 190:1561–1567. doi: 10.1128/JB.01497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez-Cuevas P, Porcelli I, Daniel RA, Errington J. 2013. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol 89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda H, Tan Q, Awano N, Wu KP, Inouye M. 2012. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol 84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- 37.Tan Q, Awano N, Inouye M. 2011. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol 79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda H, Tan Q, Awano N, Yamaguchi Y, Inouye M. 2012. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol Lett 328:174–181. doi: 10.1111/j.1574-6968.2012.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakhnina AA, Gitai Z. 2012. The small protein MbiA interacts with MreB and modulates cell shape in Caulobacter crescentus. Mol Microbiol 85:1090–1104. doi: 10.1111/j.1365-2958.2012.08159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ababneh QO, Herman JK. 2015. CodY regulates SigD levels and activity by binding to three sites in the fla/che operon. J Bacteriol 197:2999–3006. doi: 10.1128/JB.00288-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley, New York, NY. [Google Scholar]
- 42.Rasband W. 2015. ImageJ. U.S. National Institutes of Health, Bethesda, MD. [Google Scholar]
- 43.Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A 54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ababneh QO, Herman JK. 2015. RelA inhibits Bacillus subtilis motility and chaining. J Bacteriol 197:128–137. doi: 10.1128/JB.02063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abhayawardhane Y, Stewart GC. 1995. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J Bacteriol 177:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leaver M, Errington J. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol 57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 47.Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Volker U, Bessieres P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 48.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 49.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol 184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arrieta-Ortiz ML, Hafemeister C, Bate AR, Chu T, Greenfield A, Shuster B, Barry SN, Gallitto M, Liu B, Kacmarczyk T, Santoriello F, Chen J, Rodrigues CD, Sato T, Rudner DZ, Driks A, Bonneau R, Eichenberger P. 2015. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol Syst Biol 11:839. doi: 10.15252/msb.20156236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Predich M, Nair G, Smith I. 1992. Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H. J Bacteriol 174:2771–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antoniewski C, Savelli B, Stragier P. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol 172:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujita M, Sadaie Y. 1998. Promoter selectivity of the Bacillus subtilis RNA polymerase sigmaA and sigmaH holoenzymes. J Biochem 124:89–97. doi: 10.1093/oxfordjournals.jbchem.a022102. [DOI] [PubMed] [Google Scholar]
- 55.Fujita M, Sadaie Y. 1998. Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis. J Biochem 124:98–104. doi: 10.1093/oxfordjournals.jbchem.a022103. [DOI] [PubMed] [Google Scholar]
- 56.Chastanet A, Vitkup D, Yuan GC, Norman TM, Liu JS, Losick RM. 2010. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 107:8486–8491. doi: 10.1073/pnas.1002499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Jong IG, Veening JW, Kuipers OP. 2010. Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J Bacteriol 192:2053–2067. doi: 10.1128/JB.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan Q, Losick R. 2003. Unique degradation signal for ClpCP in Bacillus subtilis. J Bacteriol 185:5275–5278. doi: 10.1128/JB.185.17.5275-5278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fujita M, Gonzalez-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol 187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang M, Shao W, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 62.van den Ent F, Amos LA, Lowe J. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 63.Dye NA, Pincus Z, Fisher IC, Shapiro L, Theriot JA. 2011. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Mol Microbiol 81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Srivastava P, Demarre G, Karpova TS, McNally J, Chattoraj DK. 2007. Changes in nucleoid morphology and origin localization upon inhibition or alteration of the actin homolog, MreB, of Vibrio cholerae. J Bacteriol 189:7450–7463. doi: 10.1128/JB.00362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murray T, Popham DL, Setlow P. 1998. Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. J Bacteriol 180:4555–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popham DL, Setlow P. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J Bacteriol 178:2079–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Ent F, Izore T, Bharat TA, Johnson CM, Lowe J. 2014. Bacterial actin MreB forms antiparallel double filaments. eLife 3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubnau EJ, Cabane K, Smith I. 1987. Regulation of spo0H, an early sporulation gene in bacilli. J Bacteriol 169:1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weir J, Predich M, Dubnau E, Nair G, Smith I. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J Bacteriol 173:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perego M, Spiegelman GB, Hoch JA. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol 2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 72.Strauch M, Webb V, Spiegelman G, Hoch JA. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci U S A 87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiser MJ, Lenski RE. 2015. A comparison of methods to measure fitness in Escherichia coli. PLoS One 10:e0126210. doi: 10.1371/journal.pone.0126210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A 101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.