ABSTRACT
The lack of a peptide-swine leukocyte antigen class I (pSLA I) complex structure presents difficulties for the study of swine cytotoxic T lymphocyte (CTL) immunity and molecule vaccine development to eliminate important swine viral diseases, such as influenza A virus (IAV). Here, after cloning and comparing 28 SLA I allelic genes from Chinese Heishan pigs, pSLA-3*hs0202 was crystalized and solved. SLA-3*hs0202 binding with sβ2m and a KMNTQFTAV (hemagglutinin [HA]-KMN9) peptide from the 2009 pandemic swine H1N1 strain clearly displayed two distinct conformations with HA-KMN9 peptides in the structures, which are believed to be beneficial to stimulate a broad spectrum of CTL immune responses. Notably, we found that different HA-KMN9 conformations are caused, not only by the flexibility of the side chains of residues in the peptide-binding groove (PBG), but also by the skewing of α1 and α2 helixes forming the PBG. In addition, alanine scanning and circular-dichroism (CD) spectra confirmed that the B, D, and F pockets play critical biochemical roles in determining the peptide-binding motif of SLA-3*hs0202. Based on biochemical parameters and comparisons to similar pockets in other known major histocompatibility complex class I (MHC-I) structures, the fundamental motif for SLA-3*hs0202 was determined to be X-(M/A/R)-(N/Q/R/F)-X-X-X-X-X-(V/I) by refolding in vitro and multiple mutant peptides. Finally, 28 SLA-3*hs0202-restricted epitope candidates were identified from important IAV strains, and two of them have been found in humans as HLA-A*0201-specific IAV epitopes. Structural and biochemical illumination of pSLA-3*hs0202 can benefit vaccine development to control IAV in swine.
IMPORTANCE We crystalized and solved the first SLA-3 structure, SLA-3*hs0202, and found that it could present the same IAV peptide with two distinct conformations. Unlike previous findings showing that variable peptide conformations are caused only by the flexibility of the side chains in the groove, the skewing of the α1 and α2 helixes is important in the different peptide conformations in SLA-3*hs0202. We also determined the fundamental motif for SLA-3*hs0202 to be X-(M/A/R)-(N/Q/R/F)-X-X-X-X-X-(V/I) based on a series of structural and biochemical analyses, and 28 SLA-3*hs0202-restricted epitope candidates were identified from important IAV strains. We believe our structure and analyses of pSLA-3*hs0202 can benefit vaccine development to control IAV in swine.
INTRODUCTION
Influenza A virus (IAV) has always been a major cause of morbidity and mortality throughout history, and new emerging IAVs have posed an increasing threat to human health in recent years (1–3). IAV consists of eight single-stranded-RNA segments encoding 12 proteins: the nucleoprotein (NP), three polymerase proteins (PA, PB1, and PB2), two matrix proteins (M1 and M2), two nonstructural proteins (NS1 and NS2), two surface glycoproteins, hemagglutinin (HA), neuraminidase (NA), and two newly identified proteins (PB1-F2 and PB1-N40) (4–6). Reassortment of the eight gene segments from different IAV strains is a common cause of novel IV strains (7, 8). Pigs are considered a “mixing bowl” to produce new IAV strains and act as a primary host in cross-species transmission of IAV because they have receptors that bind to both avian and human IAV strains (9–11). This was highlighted by the emergence of a swine origin IAV in 2009, also called 2009 pandemic IAV (pH1N1), which caused a worldwide epidemic in the new century (2, 12). Elimination of IAV in swine is essential to control IAV in humans.
Currently, vaccination is the principal means to prevent IAV infection. The available vaccines are strain specific and are used to acquire neutralizing antibodies (13, 14). However, gene reassortment and rapid antigenic mutation make the vaccines ineffective against newly emerged IAV strains. In addition to the antibody-induced humoral immune response, cellular immunity is also critical in defending against IAV infection (15), which has been increasingly documented (16–19). Cytotoxic T lymphocytes (CTLs) have been shown to play a significant role in the control of primary IAV infection and to provide cross-protection against different IAV strains in mice and humans (20–24). New vaccine strategies are increasingly aimed at conserved CTL and B cell epitopes for IAV to overcome seasonal variations in influenza virus antigens (25–27).
The major histocompatibility complex class I (MHC-I) molecules can present viral peptides to specific T-cell receptors (TCRs), resulting in the proliferation of CTLs and eventually clearance of the virus from the host (28). Structural and biochemical studies have revealed that the MHC-I heavy chain, epitope peptide, and β2-microglobulin (β2m) could form a ternary complex (peptide–MHC-I complex [pMHC-I]) (29). Epitope peptides are fixed in the peptide-binding groove (PBG) of the MHC-I heavy chain by six pockets (A to F) (30). In human and mouse, MHC-I heavy chains are encoded by thousands of different alleles from several loci, which are highly polymorphic genes. Polymorphisms determine the distinct three-dimensional (3D) structure of the MHC-I PBG (31). Each classical MHC-I molecule is able to bind numerous CTL epitopes containing specific motifs, based on the compatibility of the pockets in the PBG. The crystal structure of MHC-I and its peptide-binding motif is the basis to identify CTL epitopes. Thus far, a number of the human and mouse pMHC-I structures have been solved, and these structures have greatly facilitated the identification of MHC-I-restricted CTL epitopes (http://www.rcsb.org/pdb/home/home.do). To date, more than 1,000 specific CTL epitopes for humans and mice have been identified and deposited in the Immune Epitope Database and Analysis Resource (IEDB).
The swine MHC is also called swine leukocyte antigen (SLA), and there are 3 expressed classical SLA I genes, SLA-1, SLA-2, and SLA-3. Currently, a total of 116 SLA I genes have been identified in the SLA-1, SLA-2, and SLA-3 loci and deposited in the Immune Polymorphism Database (IPD [http://www.ebi.ac.uk/ipd/index.html]). We solved the first SLA-1*0401 structure with a peptide from IAV and Ebola virus and found that SLA-1*0401 has its own characteristic peptide presentation; for example, Arg156 in the D pocket has a “one-ballot veto” function in peptide binding (32). Nevertheless, no pSLA I structures have been solved since then. The only SLA-1*0401 structure is limited in identifying CTL epitopes in swine IAV, especially in identifying the specific IAV epitopes from SLA-2 and SLA-3 allelic genes.
Chinese Heishan pigs are wild boars that are believed to be highly resistant to diseases (33, 34). In this study, 28 SLA I allelic genes were cloned from the swine, and their restricted IAV peptides were predicted in silico (http://www.cbs.dtu.dk/services/NetMHCpan/). The results suggested that the SLA-3*hs0202 allele may bind more IAV peptides than other cloned alleles, and six candidate IAV epitope peptides were then tested by in vitro refolding. To illuminate the structural basis, one peptide (KMNTQFTAV) derived from the HA protein, termed HA-KMN9, in complex with SLA-3*hs0202 and swine β2m (sβ2m) was crystalized. The crystal structure of SLA-3*hs0202-HA-KMN9-sβ2m (pSLA-3*hs0202) has a high resolution of 1.48 Å. Within one asymmetric unit, two pSLA-3*hs0202 molecules (referred to below as M1 and M2) with distinct overall structures were found. The differences between the PBGs of M1 and M2 lead to the different peptide conformations. Additionally, the peptide-binding motif of SLA-3*hs0202 was assessed using mutant peptides and circular dichroism (CD) spectral methods. Finally, 28 SLA-3*hs0202-restricted epitope candidates were identified from important IAV strains. The pSLA-3*hs0202 structures will contribute to new strategies in vaccine development to control IAV in swine.
MATERIALS AND METHODS
Prediction and synthesis of epitope peptides.
To identify the potential SLA I allele-specific peptides from pH1N1 IAV, two programs, accessed through the websites NetMHCpan2.8server (http://www.cbs.dtu.dk/services/NetMHCpan-2.8/) and SYFPEITHI (http://www.syfpeithi.de/bin/MHCServer.dll/EpitopePrediction.htm) (35, 36), were used. The results of in silico prediction indicated that SLA-3*hs0202 might have more IAV epitopes than other cloned SLA I alleles. Six peptides with high predicted scores for SLA-3*hs0202 were synthesized and purified by reverse-phase high-performance liquid chromatography (HPLC) (SciLight Biotechnology). The other 17 mutant peptides used to test the peptide-binding motif of SLA-3*hs0202 were also synthesized and purified in the same way.
Preparation of proteins.
A gene fragment corresponding to the full-length SLA-3*hs0202 (1,105 bp) was amplified by reverse transcription (RT)-PCR with allele group-specific PCR primer pairs designed for SLA-3 alleles using total RNA extracts isolated from the kidneys of Heishan pigs from Changbai Mountain. The gene fragment was cloned into the pMD-18T vector (TaKaRa) and named pMD-0202, and the extracellular region of the SLA-3*hs0202 heavy-chain gene was amplified from the pMD-0202 vector by PCR, cloned into the pET-21a vector (Novagen) with 5′ NdeI and 3′ XhoI restriction sites, and transformed into Escherichia coli strain BL21(DE3). The recombinant proteins were expressed as inclusion bodies and were purified as described previously (37). All the primer sequences will be made available on request.
Refolding of SLA-3*hs0202 with IAV peptides.
To form a complex with each peptide, SLA-3*hs0202 and sβ2m were refolded using the gradual-dilution method, as previously described (38, 39). As a negative control, SLA-3*hs0202 and sβ2m were also refolded without peptide. After 48 h of incubation at 277 K, the soluble fraction of the complexes was concentrated and then purified by size exclusion chromatography on a Superdex 200 16/60 column and by Resource-Q anion-exchange chromatography (GE Healthcare).
Crystallization and data collection.
One peptide with the sequence KMNTQFTAV, derived from the HA protein of the 2009 pandemic H1N1 (A/Beijing/01/2009) strain (GenBank accession no. ACR54994.1), could form a stable complex with SLA-3*hs0202 following in vitro refolding and was selected for crystallization with the SLA-3*hs0202 heavy chain and sβ2m. The pSLA-3*hs0202 complexes were ultimately concentrated to 10 mg/ml in a buffer containing 20 mM Tris (pH 8.0) and 50 mM NaCl for crystallization. The sample was mixed with reservoir buffer at a 1:1 ratio and crystallized by the hanging-drop vapor diffusion technique at 291 K. A PEG/Ion kit (Hampton Research, Riverside, CA) was used to screen for optimal crystal growth conditions. After several days, crystals of SLA-3*hs0202 complexed with the HA-KMN9 peptide and sβ2m were obtained with PEG/Ion Screen solution 3 (20% [wt/vol] polyethylene glycol 3350, 0.2 M ammonium fluoride). Diffraction data were collected at a resolution of 1.48 Å using an in-house X-ray source (Rigaku MicroMax007 desktop rotating anode X-ray generator with a Cu target operated at 40 kV and 30 mA) and an R-Axis IV++ imaging-plate detector at a wavelength of 1.5418 Å. The crystals were first soaked in reservoir solution containing 25% glycerol as a cryoprotectant and were then flash-cooled in a stream of gaseous nitrogen at 100 K (40). The collected intensities were indexed, integrated, corrected for absorption, scaled, and merged using the HKL2000 package (41).
Structure determination and refinement.
The structures of the pSLA-3*hs0202 complex were solved by molecular replacement using the MOLREP program with SLA-1*0401 (Protein Data Bank [PDB] code 3QQ3) as a search model. Extensive model building was performed by hand with COOT (42), and restrained refinement was performed using REFMAC5. Additional rounds of refinement were performed using the phenix refine program implemented in the PHENIX package (43) with isotropic atomic displacement parameter (ADP) refinement and bulk solvent modeling. The stereochemical quality of the final model was assessed with the PROCHECK program (44).
Circular-dichroism spectra.
CD experiments for the pSLA-3*hs0202 complexes with mutant HA-KMN9 peptides were performed on a CD instrument (Chirascan; Applied Photophysics Ltd.) with a thermal controller. The far-UV CD spectra (180 to 260 nm) were collected at a protein concentration of 0.2 mg/ml in 20 mM Tris (pH 8.0) buffer, using a 1-mm-path-length cuvette with a 0.1-nm spectral resolution. The ellipticity at 218 nm was continuously recorded during heating. The water jacket cell containing the sample was heated at a linear rate of 1°C/min. Thermal-denaturation curves were determined by monitoring the CD value at 218 nm as the temperature was raised from 25 to 94°C at a rate of 1°C/min. The fractions of unfolded protein were calculated from the mean residue ellipticity (θ) by the standard method. The unfolded fraction (%) is expressed as (θ − θN)/(θU − θN), where N and U are the mean residue ellipticity values in the fully folded and fully unfolded states. The midpoint transition temperature (Tm) was determined by fitting data to the denaturation curves using the Origin 8.0 program (Origin Lab) as described previously (45).
Biochemical assays and whole-genome screening of IAV strains.
A total of 17 mutant HA-KMN9 peptides with single-residue substitutions at position 2 (P2), 3 (P3), or 9 (P9) was synthesized. Their binding affinities to SLA-3*hs0202 were tested by in vitro refolding as described above. Finally, the candidate SLA-3*hs0202-restricted epitopes were screened throughout the whole genomes of four IAV strains, A/Beijing/01/2009 (H1N1), A/Texas/50/2012 (H3N2), A/chicken/Nigeria/SO494/2006 (H5N1), and A/Shanghai/4664T/2013 (H7N9), with the website NetMHCpan2.8server (35).
Mutation of pSLA-3*hs0202 to pSLA-3*hs0202-Ala156.
To investigate the function of Glu156 in SLA-3*hs0202, Glu156 was mutated to Ala156 by overlap PCR. The mutated gene, termed SLA-3*hs0202-Ala156, was inserted into the pET21a vector and expressed in BL21(DE3) cells. Recombinant SLA-3*hs0202-Ala156 was expressed in inclusion bodies and further purified, as described previously (38). SLA-3*hs0202-Ala156 was refolded with sβ2m and P3 mutant peptides of HA-KMN9 (P3 A, P3 Q, P3 R, P3 E, and P3 F). The complexes formed by refolding were further purified by anion-exchange chromatography as described above. In addition, the modeling structure of SLA-3*hs0202-Ala156 was built using SWISS-MODEL (http://swissmodel.expasy.org).
Accession numbers.
The crystal structures have been deposited in the Protein Data Bank (http://www.pdb.org/pdb/home/home.do) with accession number 5H94; the sequence of SLA-3*hs0202 is available at the National Center for Biotechnology Information (NCBI) database under the accession number KJ555032.
RESULTS
In silico prediction and in vitro refolding of SLA-3*hs0202-restricted IAV peptides.
Twenty-eight different SLA I genes were cloned from Heishan pigs, and the restricted peptides from pH1N1 HA and NP were predicted in silico (http://www.cbs.dtu.dk/services/NetMHCpan/). The results suggested that SLA-3*hs0202 could bind more IAV epitope peptides than other MHC molecules. Then, six predicted epitope peptides from HA and NP proteins were synthesized and tested for binding with SLA-3*hs0202 by in vitro refolding. One peptide, HA-KMN9 (KMNTQFTAV), could form a stable pMHC-I with SLA-3*hs0202 and sβ2m. Thus, the pSLA-3*hs0202 complex was further purified by gel filtration and anion-exchange chromatography and used for further experiments.
Distinct overall structures of two pSLA-3*hs0202 complexes.
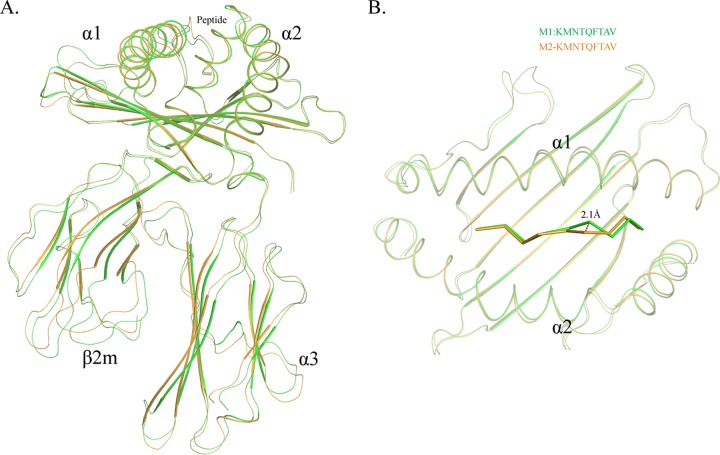
SLA-3*hs0202 complexed with HA-KMN9 was crystallized in the C121 space group with a high resolution of 1.48 Å (Table 1). pSLA-3*hs0202 displayed a canonical pMHC-I structure, which consisted of α1, α2, and α3 domains of the heavy-chain and the light-chain sβ2m, and the peptide was located in the PBG formed by the α1 and α2 domains (Fig. 1A). Within one asymmetric unit, there are two SLA-3*hs0202 molecules, referred to below as M1 and M2, with a root mean square difference (RMSD) of 0.873 Å, which is higher than the RMSD value of the two SLA-1*0401 (RMSD, 0.401; PDB code 3QQ3) and BoLA-N*01801 (RMSD, 0.295; PDB code 3PWV) molecules (32, 46). The high RMSD value indicated that the structures of M1 and M2 are not coincident, and there should be some differences between them (Fig. 1B).
TABLE 1.
X-ray diffraction data processing and refinement statistics
| Parameter | Value in SLA-3*hs0202a |
|---|---|
| Data processing | |
| Space group | C121 |
| Unit cell parameters (Å) | a = 206.46, b = 41.47, c = 106.74, α = 90.00, β = 117.46, γ = 90.00 |
| Resolution (Å) | 50.0–1.48 (1.518–1.48) |
| No. of reflections | 428,111 |
| No. of unique reflections | 127,299 |
| Completeness (%) | 99.8 (100) |
| Avg I/σ(I) | 19.482 (3.536) |
| Rmerge (%)b | 7.4 (38.4) |
| Redundancy | 3.7 (3.6) |
| Refinement | |
| Rwork (%)c | 14.975 |
| Rfree (%) | 19.025 |
| RMSD | |
| Bond lengths (Å) | 0.025 |
| Bond angles (°) | 2.171 |
| Avg B factor | 21.929 |
| Ramachandran plot quality | |
| Most favored region (%) | 98.4 |
| Allowed region (%) | 1.6 |
| Disallowed (%) | 0 |
The numbers in parentheses indicate the highest-resolution shell.
Rmerge = Σhkl Σi |Ii(hkl) − <I(hkl)>|/Σhkl Σi Ii(hkl), where Ii(hkl) is the observed intensity and áI(hkl)ñ is the average intensity from multiple measurements.
R = Σhkl ||Fobs| − k |Fcalc ||Σhkl |Fobs|, where Rfree is calculated for a randomly chosen 5% of reflections and Rwork is calculated for the remaining 95% of reflections used for structure refinement.
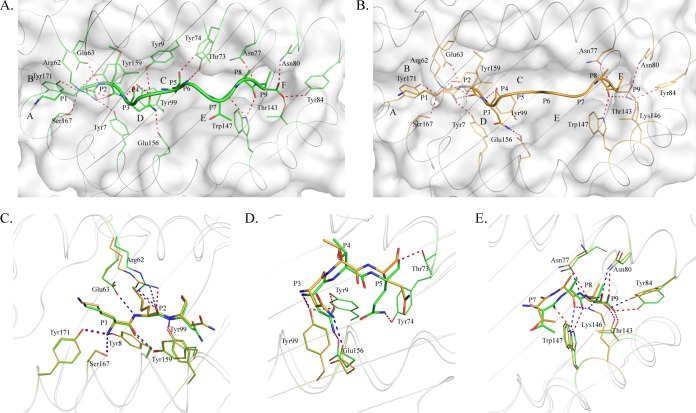
FIG 1.
Distinct structures of pSLA-3*hs0202 M1 and M2 in one asymmetric unit. (A) Superimposition of the overall structures of SLA-3*hs0202 M1 (green) and M2 (orange). They display a typical MHC-I structure but with obvious skewing in the α1, α2, and α3 domains. (B) Superimposition of the HA-KMN9 peptides presented by M1 and M2, respectively. The maximum distance between the same atoms in the two peptides, which could reach 2.1 Å, is represented by the dashed red line.
The overall structural alignment of M1 and M2 showed that their carbon backbones do not overlap. The skewing between M1 and M2 could be found in every domain of SLA-3*hs0202, and these discordances not only occur in the flexible loops, but also exist in the α1 and α2 helixes and the β-sheets of the α3 domain and sβ2m (Fig. 1A). The α1 and α2 helixes were not perfectly superimposed, even when only the peptide-binding domains were compared. The two peptides presented by M1 and M2 were not well aligned, and the distances between some atoms of the peptides could reach 2.1 Å (Fig. 1B).
Different exposed areas of HA-KMN9 peptides presented by pSLA-3*hs0202 M1 and M2.
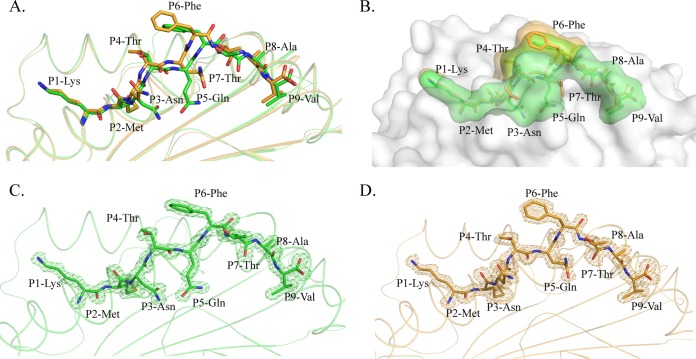
The comparison showed that the carbon backbones of the HA-KMN9 peptide in M1 and M2 are not coincident (Fig. 1B). Further analysis showed that the divergences occurred after P3 of the peptide (Fig. 2A). The P1 and P2 residues at the N termini of the two peptides are superimposed perfectly, but the two P3 Asn residues show an obvious shift, especially in their side chains. The mismatch reached a peak at the P6 residue, where the two Phe residues in M1 and M2 are the maximum distance apart. After P5 Phe, the conformational differences between the two peptides are diminished but still exist. The P7 to P9 residues in M1 are located deeper in the PBG than their counterparts in M2. Overall, the arch of the M2 peptide is higher than that of the M1 peptide, but the length is shorter. The side chains of P1, P4, P6, and P8 protruded from the PBG, especially P4 and P6, which are most likely recognized by specific TCRs. The most striking divergence between the two peptides is in the exposed region (Fig. 2B). The higher arch made the P4 and P6 residues of M2 more exposed than the two residues in the M1 peptide. The exposed area (solvent-accessible surface area [solvent ASA]) and buried area (buried ASA) of the M1 and M2 peptides are 1,447.5 and 1,114.8 Å, and 1,417.0 and 1,031.1 Å, respectively, and detailed information about each residue is provided in Table 2. The clear and complete electron density maps of the two peptides indicate that these differences are credible (Fig. 2C and D).
FIG 2.
Distinct conformations and electronic densities of HA-KMN9 peptides found in pSLA-3*hs0202 M1 and M2. (A) Detailed comparison of HA-KMN9 peptides shown as a stick model (green, M1; orange, M2). The mismatch of the two peptides is obvious, especially at P6 Phe. (B) Different exposed areas of HA-KMN9 peptides. The exposed P4 Thr and P6 Phe of the M2-HA-KMN9 peptide are substantially higher than the two residues of the M1-HA-KMN9 peptide. (C and D) Electron density at the 1σ contour level of HA-KMN9 peptides shown in the pSLA-3*hs0202 M1 and M2 structures. The electron density maps are clear and indicate that the two distinct peptide conformations are reliable.
TABLE 2.
Accessible surface area (ASA) and buried surface area (BSA) calculations for HA-KMN9 peptides in M1 and M2 of pSLA-3*hs0202
| Residue | Solvent ASA (Å2) | Buried ASA (Å2) |
|---|---|---|
| M1 | ||
| P1 Lys | 259.54 | 213.43 |
| P2 Met | 190.63 | 190.13 |
| P3 Asn | 115.88 | 114.70 |
| P4 Thr | 113.86 | 47.20 |
| P5 Gln | 143.83 | 137.95 |
| P6 Phe | 181.98 | 43.48 |
| P7 Thr | 134.68 | 92.37 |
| P8 Ala | 96.06 | 66.64 |
| P9 Val | 211.04 | 208.85 |
| M2 | ||
| P1 Lys | 259.14 | 213.06 |
| P2 Met | 190.27 | 189.93 |
| P3 Asn | 146.08 | 125.21 |
| P4 Thr | 89.14 | 30.46 |
| P5 Gln | 142.68 | 109.06 |
| P6 Phe | 191.15 | 9.74 |
| P7 Thr | 97.68 | 71.26 |
| P8 Ala | 90.94 | 72.73 |
| P9 Val | 209.99 | 209.66 |
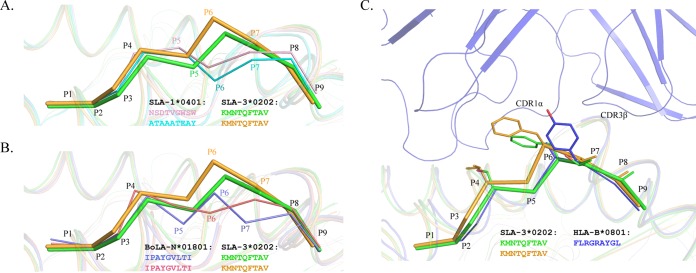
The phenomenon where peptides presented by the same MHC-I molecule show different conformations has been found in other known pMHC-I structures (46). Two nonapeptides presented by SLA-1*0401 showed similar M-like conformations, but their main chains obviously diverge after the P3 residue, similar to what we found in SLA-3*hs0202 (Fig. 3A). Using the same peptide, BoLA-N*01801 also showed results similar to those for SLA-3*hs0202. There are two BoLA-N*01801 molecules in one asymmetric unit, and the peptide conformations are very different (Fig. 3B) (46). By comparing all the known pMHC-I structures, the M1 peptide conformation was found to be almost identical to that of HLA-B*0801 (PDB code 1M15) (Fig. 3C). The complex structure of HLA-B*0801-TCR shows that the large side chain of P7 Tyr interacts with the TCRs CDR1α and CDR3β (47). In HA-KMN9 peptides of pSLA-3*hs0202, P6 Phe has a location and aromatic ring similar to those of P7 Tyr of HLA-B*0801.
FIG 3.
Peptide presentation characteristics of pSlA-3*hs0202 compared to those of other pMHC molecules. (A and B) HA-KMN9 peptide conformations (thick sticks) compared to those of other peptides (thin sticks) with distinct conformations presented by the same MHC-I allele. (A) Comparison with peptides presented by SLA-1*0401 (PDB 3QQ3, light pink; PDB 3QQ4, cyan). (B) Comparison with the same peptide presented by two BoLA-A11 (N*01801) molecules in one asymmetric unit (PDB 3PWU, pink and slate). (C) Peptide conformation similar to that of a peptide presented by HLA-B*0801 (PDB 1M15, blue) to a specific TCR. Phe6 of the M1-HA-KMN9 peptide has a location and an aromatic ring similar to those of Tyr7 of the peptide presented by HLA-B*0801.
Different PBG conformations found in M1 and M2 and various interactions with the HA-KMN9 peptide.
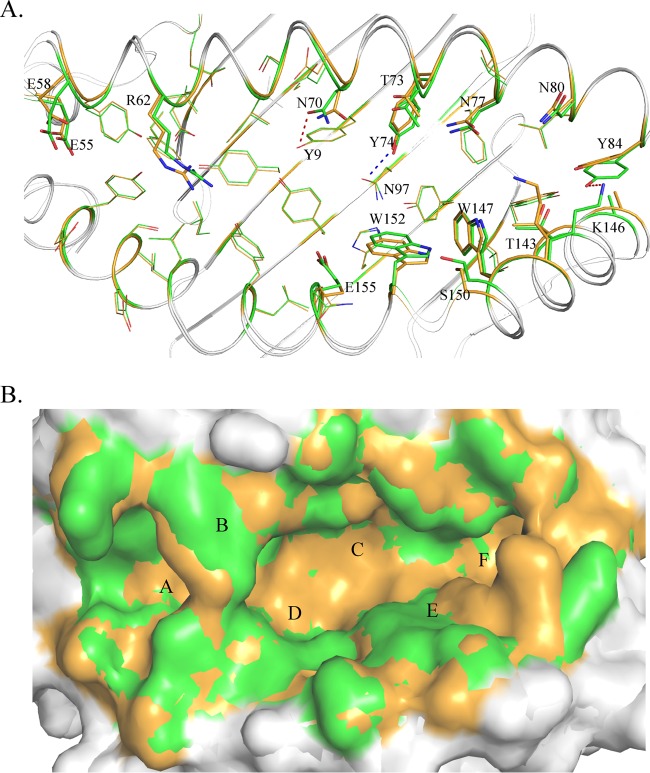
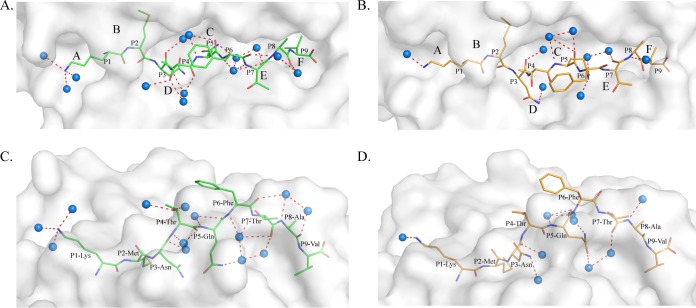
The PBG conformations of M1 and M2 are shown in Fig. 4. The PBG of pSLA-3*hs0202 consists of 6 pockets, similar to other known pMHC-I structures (48). However, superimposition of the M1 and M2 PBGs showed that they have different conformations. The distinct residues that directly interact with the HA-KMN9 peptide were selected and are shown as thick lines in the figure. The differences in these residues are caused not only by shifts in their side chains, but also by the mismatch of the α1 and α2 helixes. These distinct residues are mainly located in the middle region and C-terminal region of the PBG, where the helixes are highly divergent. Among these residues, N70 and Y74 could form different hydrogen bonds in M1 and M2. N70 forms a hydrogen bond only with Y9 in M1, and Y74 could interact only with N97 in M2 (Fig. 4A). Superimposition in a surface model (Fig. 4B) shows the side wall in the middle of the overlapped PBGs (mainly in green) and the bottom of the PBG (mainly in orange). This indicates that the PBG of M1 is narrower but deeper than the M2 PBG, which is consistent with the finding that the α1 and α2 helixes in M1 are closer than in M2 (Fig. 4B).
FIG 4.
Different conformations of HA-KMN9 peptides in the binding grooves of M1 and M2. (A) Superimposition of M1 and M2 PBGs in a stick model. The distinct residues that directly interact with the HA-KMN9 peptide are labeled and shown as thick sticks. They are mainly located in the middle and at the C terminus of the PBG, where the α1 and α2 helixes are mismatched. Two different hydrogen bonds formed by these residues are shown as dashed lines (M1, red; M2, blue). (B) Structural superimposition of M1 and M2 PBGs in a surface model. The six pockets (A, B, C, D, E, and F) are labeled in the PBGs. The M2 PBG is deeper and narrower than the M1 PBG, based on the fact that most of the bottom is yellow and the side wall is green.
The different PBG conformations of M1 and M2 lead to various peptide interactions (Fig. 5). In M1, there are 19 hydrogen bonds between the PBG and the HA-KMN9 peptide (Fig. 5A), but in M2, the number is 16 hydrogen bonds (Fig. 5B). In the A and B pockets of M1 and M2, the interactions with P1 Lys and P2 Met are the same (Fig. 5C). The differences are mainly found in the middle of the groove and in the C-terminal region. In M1, P3 Asn formed two hydrogen bonds, with Glu156 and Tyr9, but in M2, P3 Asn formed a hydrogen bond only with Glu156 (Fig. 5D). P5 Gln formed two hydrogen bonds, with Thr73 and Tyr74, in M1 but did not form any hydrogen bonds in M2 (Fig. 5D and Table 3). In the F pockets, Trp147 could bind to P7 and P8 in M1 but could interact only with P8 in M2. Lys146 formed only two hydrogen bonds with P8 and P9 in M2 (Fig. 5E). In addition to the hydrogen bonds, the interactions formed by van der Waals forces (VDWs) are also different in the two SLA-3*hs0202 H chains. The total numbers of VDWs were 373 and 299 in M1 and M2, respectively. Complete interaction information is provided in Table 3. Moreover, the water molecules involved in the hydrogen bond network between the PBG and the HA-KMN9 peptide are different in M1 and M2. In M1, 13 water molecules form 17 hydrogen bonds with the HA-KMN9 peptide (Fig. 6A and C). In M2, only 9 water molecules and 10 direct bonds are found (Fig. 6B and D). The diverse water molecules could fit into the different PBG conformations of M1 and M2 to stabilize the distinct peptide conformations (49).
FIG 5.
Different peptide interactions found in pSLA-3*hs0202. (A and B) Hydrogen bonds found in pSLA-3*hs0202 M1 (A) and M2 (B) (both shown as dashed red lines). (C) Comparison of the interactions of P1 Lys and P2 Met in the A and B pockets (the N terminus of PBG), respectively. The numbers and positions of hydrogen bonds are the same in M1 and M2. (D) Comparison of the interactions of P3 Asn and P5 Gln in the D and C pockets (the middle of the PBG), respectively. P3 Glu forms two hydrogen bonds with Tyr99 in M1 (dashed red lines) but forms only one hydrogen bond with Tyr99 in M2 (dashed blue line). P5 Gln forms two hydrogen bonds with Thr73 and Tyr74 in M1 but does not form any hydrogen bonds in M2. (E) Comparison of the interactions of P7 Thr, P8 Ala, and P9 Val in the E and F pockets (the C terminus of PBG). Trp147 could form two hydrogen bonds with P7 and P8 in M1 (dashed red lines) but only one hydrogen bond with P8. Lys146 could form two hydrogen bonds in M2 (dashed blue lines) but could not form any hydrogen bonds in M1.
TABLE 3.
Hydrogen bonds and van der Waals interactions between HA-KMN9 peptides and peptide-binding domains of M1 and M2 molecules
| Complex | Peptide |
Hydrogen bond partner |
Van der Waals contact residuesa | ||
|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | ||
| M1 | P1 Lys | N | Tyr171 | OH | Leu5, Tyr7, Tyr59, Arg62, Glu63, Tyr59, Leu163, Ser167, Glu170, Tyr171 (59) |
| N | Ser167 | OG | |||
| N | Tyr7 | OH | |||
| O | Tyr159 | OH | |||
| P2 Met | N | Glu63 | OE1 | Tyr7, Tyr9, Glu45, Arg62, Glu63, Ile66, Asn70, Ser67, Tyr99, Tyr59, Leu163 (74) | |
| O | Arg62 | NH1 | |||
| O | Arg62 | NH2 | |||
| P3 Asn | N | Tyr99 | OH | Tyr9, Ile66, Tyr99, His114, Glu156, Tyr159 (65) | |
| ND2 | Tyr99 | OE1 | |||
| ND2 | Glu156 | OE2 | |||
| OD1 | Tyr9 | OH | |||
| P4 Thr | Arg62, Ile66, Trp152 (5) | ||||
| P5 Gln | NE2 | Tyr74 | OH | Tyr9, Asn70, Thr73, Tyr74, Asn97, Tyr99, His114, Tyr116, Trp152 (37) | |
| O | Thr73 | OG1 | |||
| P6 Phe | Thr73, Asp69 (13) | ||||
| P7 Thr | O | Trp147 | NE1 | Thr73, Trp147, Ser150, Trp152 (27) | |
| P8 Ala | O | Trp147 | NE1 | Thr73, Val76, Asn77, As80, Trp147 (15) | |
| P9 Val | N | Asn77 | OD1 | Asn77, Asn80, Leu81, Tyr84, Phe95, Tyr123, Thr143, Lys146, Trp147 (59) | |
| O | Asn80 | ND2 | |||
| OXT | Thr143 | OG1 | |||
| OXT | Tyr84 | OH | |||
| M2 | P1 Lys | N | Tyr7 | OH | Leu5, Tyr7, Tyr59, Arg62, Glu63, Tyr159, Leu163, Ser167, Glu170, Tyr171 (53) |
| N | Tyr171 | OH | |||
| N | Ser167 | OG | |||
| O | Tyr159 | OH | |||
| P2 Met | N | Glu63 | OE2 | Tyr7, Tyr9, Ala24, Glu45, Arg62, Glu63, Ile66, Ser67, Tyr99, Tyr159, Leu163 (63) | |
| O | Arg62 | NH1 | |||
| O | Arg62 | NH2 | |||
| P3 Asn | N | Tyr99 | OH | Tyr9, Ile66, Tyr99, Trp152, Gln155, Glu156, Tyr159 (48) | |
| ND2 | Glu156 | OE2 | |||
| P4 Thr | Arg62, Ile66 (3) | ||||
| P5 Gln | Asn70, Thr73, Tyr116, Trp152 (18) | ||||
| P6 Phe | |||||
| P7 Thr | Thr73, Trp147, Trp152 (8) | ||||
| P8 Ala | O | Lys146 | NZ | Thr73, Val76, Asn77, As80, Lys146, Trp147 (24) | |
| O | Trp147 | NE1 | |||
| P9 Val | N | Asn77 | OD1 | Asn77, Asn80, Leu81, Tyr84, Phe95, Tyr123, Thr143, Lys146, Trp147 (67) | |
| O | Asn80 | ND2 | |||
| OXT | Tyr84 | OH | |||
| OXT | Thr143 | OG1 | |||
| O | Lys146 | NZ | |||
The numbers in parentheses are the amounts of van der Waals force.
FIG 6.
Different water molecules found in pSLA-3*hs0202 M1 and M2 structures. (A and C) Water molecules of pSLA-3*hs0202 M1 in a top view (A) and a side view (C); 13 water molecules form 17 hydrogen bonds with the HA-KMN9 peptide. (B and D) Water molecules of SLA-3*hs0202 M2 in a top view (B) and a side view (D); 9 water molecules in M2 form 10 direct bonds. The blue balls represent water molecules.
Anchor residues determined by alanine scanning and CD spectra.
Although the conformations of HA-KMN9 peptides in M1 and M2 are different, the accommodation of residues by the 6 pockets is roughly the same. The major proportion of P1, P2, P3, P5, and P9 in the HA-KMN9 peptide is accommodated by the A, B, D, C, and F pockets, respectively (Fig. 5). To determine the primary anchor residues, these five residues were tested by alanine scanning (Fig. 7). The P1 Ala mutant peptide could efficiently form a complex with SLA-3*hs0202, similar to the wild-type HA-KMN9 peptide. As in the known pMHC-I structures, P1 Lys binds the A pocket with its main chain (Fig. 5C) (45). Therefore, although the binding between P1 and the A pocket is strong, the substitution of the P1 residue cannot affect the peptide binding. For the other four residues, interactions with the pockets largely depend on their side chains. The variation of these residues may affect the binding between HA-KMN9 and the PBG of SLA-3*hs0202. The in vitro refolding results for these mutant peptides showed that mutation of P2, P3, and P9 decreased the refolding efficiency and stability of the complex (Fig. 7A and B). Interestingly, although the M1 structure showed P5 Gln could form two hydrogen bonds with pocket C, the refolding efficiency of P5 Ala was not affected.
FIG 7.
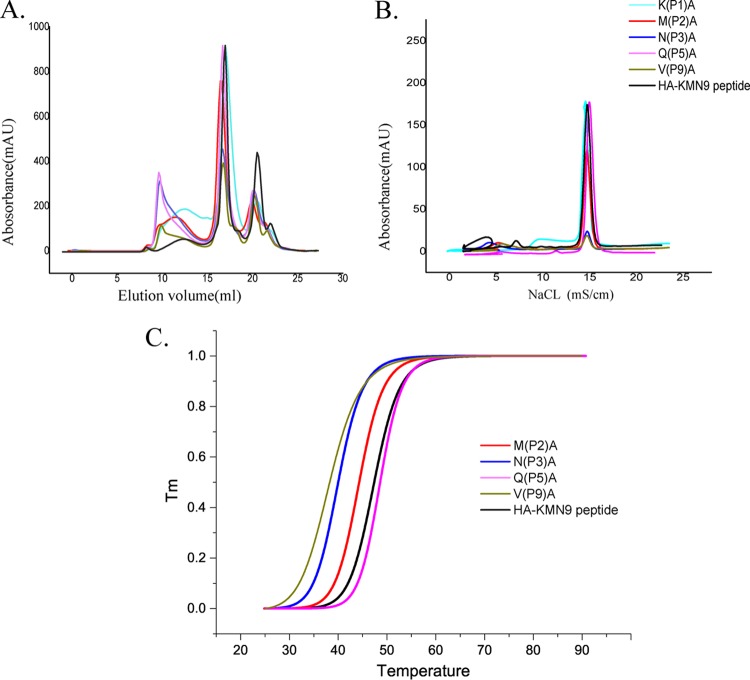
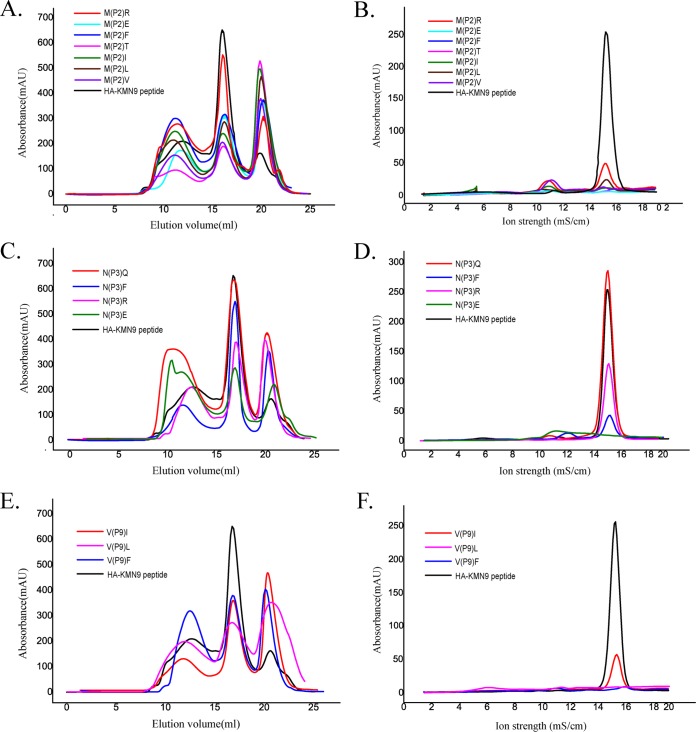
Anchor residues of the HA-KMN9 peptide verified by alanine scanning and CD spectra. (A) Gel filtration chromatograms of the refolded products of the HA-KMN9 peptide and its Ala mutants. Peak 1, peak 2, and peak 3 represent the aggregated H chain, the correctly refolded SLA-3*hs0202 complex (44 kDa), and the redundant sβ2m, respectively. The refolding efficiencies are represented by the relevant concentration ratios and heights of peak 2 formed by complexes with mutant peptides. A high peak 2 indicates high efficiency of the peptide to help the MHC renature. However, a low or no peak 2 indicates that the peptide cannot stabilize the complex. (B) The stabilization of refolded complexes with mutant peptides was further tested by anion exchange. Under the anion-exchange conditions, complexes with P1 A and P5 A mutant peptides can be eluted at NaCl concentrations of 14 to 16% as well as the complex with the wild-type peptide. The peak of the complex with the P2 A mutant peptide is lower than the three high peaks and indicates less stability. With peptides P3 A and P9 A, the refolded complex proteins dissociated at NaCl concentrations of 14 to 16%, implying very poor stability. (C) The thermostabilities of SLA-3*hs0202 with five peptides (wild-type HA-KMN9 peptide, P2 A, P3 A, P5 A, and P9 A) were tested by CD spectroscopy. The temperature was increased by 1°C/min. The curves for the unfolded fractions were determined by monitoring the CD value at 218 nm (45).
CD spectra were further used to test the stabilities of pSLA-3*hs0202 complexes with the mutant peptides (Fig. 7C). The Tm value of the wild-type HA-KMN9 peptide is 47.4°C, similar to the Tm value of pSLA-1*0401, which we tested previously (47.1°C). The results indicated that the binding between the HA-KMN9 peptide and SLA-3*hs0202 is quite stable. The Tm value of the SLA-3*hs0202 and P2 Ala mutant peptide complex (pSLA-3*hs0202-P2A) is 44.3°C, which is slightly lower than that of the wild-type HA-KMN9 and SLA-3*hs0202, demonstrating that the side chains of P2 residues contributed to the peptide binding. The Tm values of pSLA-3*hs0202-P3A and pSLA-3*hs0202-P9A are 40.0°C and 38.3°C, respectively, which are significantly decreased compared to that of the wild-type HA-KMN9. The results showed that the side chains of P3 and P9 play key roles in HA-KMN9 peptide binding and that these two residues are the primary anchor residues. The Tm value of pSLA-3*hs0202-P5A is 48.6°C and is higher than that of the wild-type complex. This suggested that the interactions formed by the side chains of P5 residues are not essential for the stability of pSLA-3*hs0202. Based on these results, we determined that the P2, P3, and P9 residues are the primary anchor residues of epitope peptides presented by SLA-3*hs0202.
The peptide-binding motif of SLA-3*hs0202.
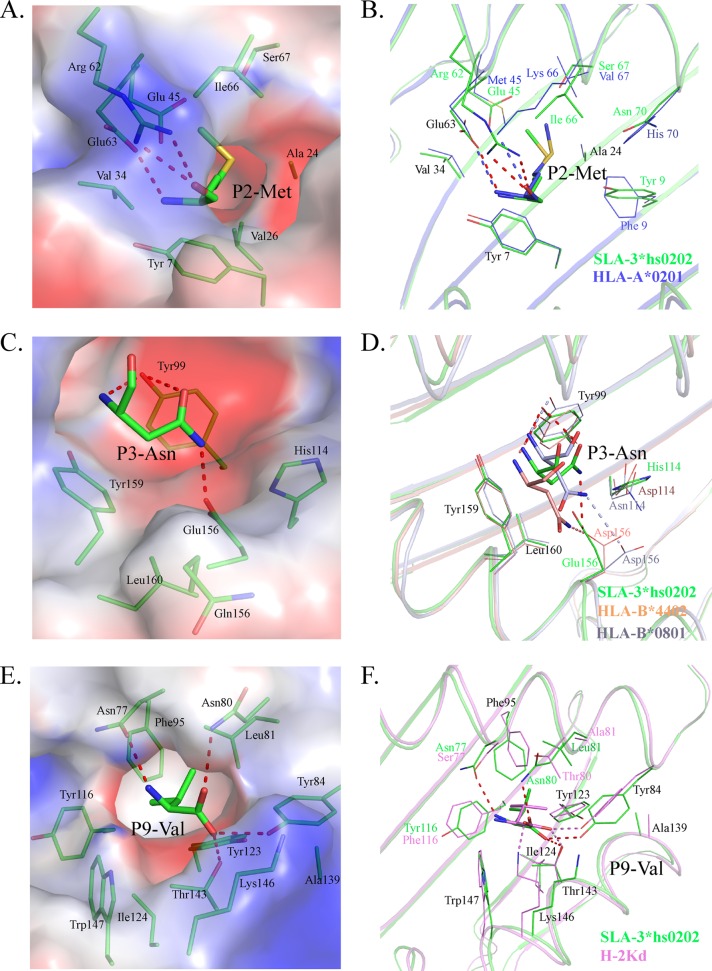
The peptide-binding motif of SLA-3*hs0202 is the set of acceptable combinations of P2, P3, and P9 residues, because they are the primary anchor residues for the binding peptides. The B, D, and F pockets accommodated the side chains of P2, P3, and P9, respectively, so they are very critical to determine the peptide-binding motif of SLA-3*hs0202. The B pocket consists of Tyr7, Ala24, Val25, Gly26, Val34, Glu45, Arg62, Glu63, Ile66, and Ser67, and its polarity is shown in Fig. 8A. The long side chain of Arg62 stretches to the α2 helix and seals the top of the B pocket. There are three charged residues (Glu45, Arg62, and Glu63) in the B pocket, and two of them form hydrogen bonds with the main-chain atoms of P2 M. The side chain of P2 M stretches to the α1 helix and is inserted into the B pocket, and it is surrounded by uncharged amino acids, except for Glu45. The mutant HA-KMN9 peptides with charged P2 E or P2 R were used to test the influence of Glu45, and the refolding results showed that the binding of P2 R is better than that of P2 E, but both of them are not as efficient as the wild-type HA-KMN9 peptide (Fig. 9A). P2 M as a primary anchor residue is rare in the other known pMHC-I structures, and only HLA-A*0201 has been found to present peptides with P2 M, similar to SLA-3*hs0202 (50). Although the residues in the B pockets of SLA-3*hs0202 and HLA-A*0201 are not same, they bind P2 M in very similar manners (Fig. 8B). The B pocket of HLA-A*0201 could accommodate uncharged residues, such as M, L, V, I, T, and F. We used the P2 V/L/I/T/F mutant HA-KMN9 peptides to verify whether the binding motif of the SLA-3*hs0202 B pocket is the same as that of the HLA-A*0201 B pocket. The refolding results showed that SLA-3*hs0202 could not bind these mutant peptides as efficiently as the wild-type KMN peptide (Fig. 9A and B). Only the P2 A/R mutant peptides could form stable complexes that survived the strong ionic buffer during anion exchange (Fig. 7B and 9B). Our results confirmed that P2 M/A/R is suitable for binding the B pocket of SLA-3*0202 and that P2 M is the better choice.
FIG 8.
Compositions of the B, D, and F pockets of SLA-3*hs0202 with other solved pMHC-I structures. The residues comprising the pockets and the residues of bound peptides accommodated by the pockets are shown as stick models. The hydrogen bonds between the peptides and the pockets are shown as dashed red lines. (A) The electrostatic potential of pocket B with the P2 residue (red, negative; blue, positive; gray, neutral) is shown, with the HA-KMN9 peptide in green. (B) B pocket alignment between SLA-3*hs0202 and HLA-A*0201 (PDB 3HPJ), which both accommodate P2 Met. (C) Electrostatic potential of pocket D with the P3 residue. (D) D pocket alignment between SLA-3*hs0202 with HLA-B*4402 (PDB 3DX6) and HLA-B*0801 (PDB 4QRT). They all bind P3 Asn in similar manners. (E) Electrostatic potential of pocket F with the P9 residue. (F) F pocket alignment between SLA-3*hs0202 and H-2Kd (PDB 2FWO), which both accommodate P9 Val or P9 Ile.
FIG 9.
Determination of the peptide-binding motif of SLA-3*hs0202 by in vitro refolding with mutant peptides. (A) Gel filtration chromatograms of the wild-type HA-KMN9 peptide and its P2 mutants (P2 R, P2 E, P2 F, P2 T, P2 I, P2 L, and P2 V). The P2 R (magenta) peptide is more efficient than other mutant peptides in helping MHC to renature, but its efficiency is lower than that of the wild-type HA-KMN9 peptide. (B) Results from further stabilization assays of the refolded complexes with P2 mutants by anion exchange. Consistent with the results shown in Fig. 8A, only partial complexes of P2 R can be eluted at normal NaCl concentrations of 14 to 16%, implying less stability. (C) Gel filtration chromatograms of the HA-KMN9 peptide and its P3 mutants (P3 Q, P3 F, P3 R, and P3 E). P3 Q (red) efficiently helps the MHC to renature as well as the wild-type peptide, and the P3 F (blue) peptide has the second highest peak. (D) Results from further stabilization assays of the refolded complexes with P3 mutant peptides by anion exchange. The complex with P3 Q could be eluted normally at NaCl concentrations of 14 to 16%, but very little of the complex with P3 F survived. (E) Gel filtration chromatograms of the wild-type peptide and its P9 mutants (P9 I, P9 L, and P9 F). The P9 I (magenta) and P9 F (blue) peptides can help the MHC renature. (F) Results from further stabilization assays of the P9 mutants by anion exchange. Only a small amount of P9 I could be eluted normally at NaCl concentrations of 14 to 16%.
The D pocket is composed of Tyr99, His114, Gln155, Glu156, Tyr159, and Leu160 and showed obvious negatively charged polarity (Fig. 8C). Glu156 could form a hydrogen bond with the side chain of P3 N, and this interaction is critical to anchor the peptide because the P3 A mutant attenuates the peptide-binding stability. Tyr99 could form two hydrogen bonds with the P3 residue in M1 and is important in stabilizing the peptide; however, it does not restrict the scope of the binding peptide because it contacts the main chain of the P3 residue. HLA-B*0801 and HLA-B*4402 have D pockets similar to those of SLA-3*hs0202 (51, 52). Their D pockets can accommodate P3 N and bind it in a manner similar to that of SLA-3*hs0202 (Fig. 8D). These three D pockets have similar compositions, including a negatively charged residue in the same position (156); therefore, they should have similar preferences for P3 residues. The D pockets of HLA-B*0801 and HLA-B*4402 could accommodate R, N, and F. The P3 Q/R/F/E mutant HA-KMN9 peptides were used to test the binding motif of the D pocket of SLA-3*hs0202 (Fig. 9C and D). The refolding results showed that the P3 Q mutant peptide could bind to SLA-3*hs0202 as well as the wild-type HA-KMN9 peptide. The P3 R mutant peptide could also form a stable complex with SLA-3*hs0202, but its refolding efficiency was lower than that of the wild-type and P3 Q mutant peptides. The refolding efficiency of the P3 F mutant peptide was better than that of the P3 R mutant peptide, but the complex formed by the P3 F peptide was not stable and was mostly dissociated during anion exchange. As expected, refolding of the P3 E mutant peptide was the least efficient among these four mutant peptides. These results suggested P3 N and Q are the best choices, and P3 R and F were also acceptable for the D pocket of SLA-3*hs0202.
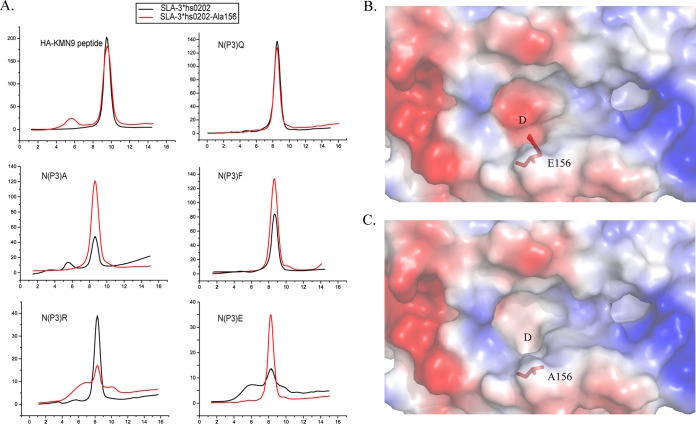
Our previous work proved that the charged Arg156 of SLA-1*0401 is vital to the peptide binding of the D pocket. Here, Glu156 is also a charged residue in the D pocket of SLA-3*hs0202. To clarify the role of Glu156, a mutated heavy chain, SLA-3*hs0202-Ala156 was co-refolded with the HA-KMN9 and the P3 mutant peptides. The refolding results showed that the binding properties of HA-KMN9 and P3 Q mutant peptides are not much affected by Ala156 mutation, and P3 A, F, and E peptides bind with SLA-3*hs0202-Ala156 better than with SLA-3*hs0202. Only the P3 R peptide could not bind with SLA-3*hs0202-Ala156 as well as the wild-type heavy chain (Fig. 10A). By comparison with SLA-3*hs0202, the modeling structure of SLA-3*hs0202-Ala156 was shown to have a bigger and more hydrophobic D pocket (Fig. 10B and C). This change made the D pocket of SLA-3*hs0202-Ala156 prefer the hydrophobic P3 residues, such as P3 A and P3 F, and have weaker resistance to P3 E. Therefore, these three peptides can bind with the mutant heavy chain better than with the wild-type chain. The mutant D pocket cannot form the salt bridge with P3 R, so it cannot bind well with the P3 R peptide. The results indicate that Glu156 is a vital residue for the peptide motif of SLA-3*hs0202.
FIG 10.
Comparative analysis of D pockets between SLA-3*hs0202 and SLA-3*hs0202-Ala156 mutants. (A) Anion-exchange chromatography results for HA-KMN9 and its P3 N mutant peptides co-refolding with SLA-3*hs0202 and SLA-3*hs0202-Ala156. (B) D pocket of pSLA-3*hs0202 in the electrostatic potential surface model (red, negative; blue, positive; gray, neutral). Glu156 is shown in red as a stick model. (C) 3D model of the pSLA-3*hs0202-Ala156 D pocket built by the SWISS-MODEL program. Ala156 is shown in red as a stick model. Compared with that of SLA-3*hs0202, the D pocket of SLA-3*hs0202-Ala156 became bigger and more hydrophobic without the negative side chain of Glu156.
The F pocket is composed of the conserved residues Tyr84, Tyr123, Thr143, Lys146, and Trp147, as well as the less conserved residues Asn77, Asn80, Leu81, Phe95, Ser97, Tyr116, Ile124, and Ala139. It showed strong hydrophobic properties (Fig. 8E). The nonpolar side chain of P9-V stretched into the F pocket and formed many VDWs with the surrounding residues on the bottom and wall of the pocket (Table 3). Four hydrogen bonds are formed by the main chain of P9 V and the residues at the top of the F pocket. The F pocket of H-2Kd could accommodate P9 V in a similar manner (Fig. 8F) and could also bind other P9 residues, such as I, L, and F (53). Therefore, mutant HA-KMN9 peptides with P9 I/L/F were used to test the binding of the SLA-3*hs0202 F pocket (Fig. 9E and F). The refolding efficiencies of P9 mutant peptides were lower the those of the wild-type HA-KMN9 peptide, and only P9 I could form a stable complex that could survive in the anion-exchange buffer. These results suggest P9 V is the most suitable anchor residue for the F pocket of SLA-3*hs0202, and P9 I is another possible choice. Therefore, the 9-mer peptide-binding motif of SLA-3*hs0202 was preliminarily determined to be X-(M/A/R)-(N/Q/R/F)-X-X-X-X-X-(V/I).
Biochemical data predicted in H1N1, H3N2, H7N9, and H5N1 strains.
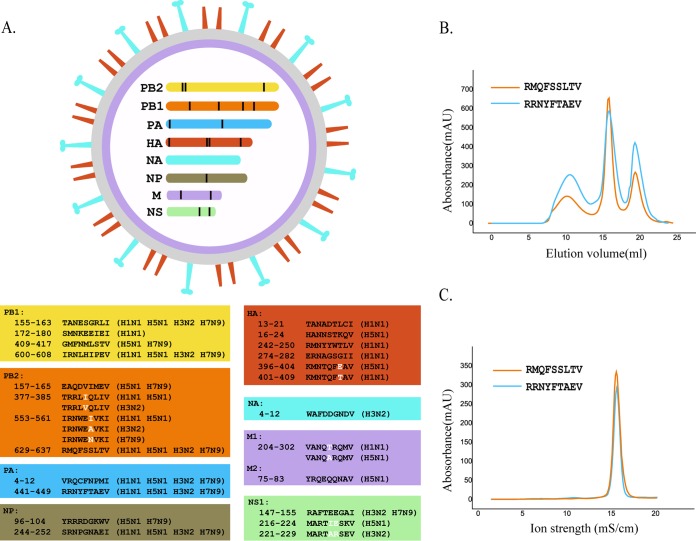
Based on the preliminary motif described above, we screened the whole protein sequences of four current epidemic IAV strains: A/Beijing/01/2009 (H1N1), A/Texas/50/2012 (H3N2), A/chicken/Nigeria/SO494/2006 (H5N1), and A/Shanghai/4664T/2013 (H7N9) (54–57). There were 28 total 9-mer epitope peptides that were predicted to fit the motif. Six of them were completely conserved in the H1N1, H3N2, H5N1, and H7N9 strains, and five have amino acid substitutions among different strains (Fig. 11A). Two conserved peptides, RMQFSSLTV (PB2-RMQ9) and RRNYFTAEV (PA-RRN), were selected to test their affinities to SLA-3*hs0202 by in vitro refolding, and the results confirmed that both of them could form stable complexes with SLA-3*hs0202 (Fig. 11B and C). Both HA-KMN9 and PB2-RMQ9 were considered HLA-A*0201-restricted epitopes, but only PB2-RMQ9 could stimulate an effective CTL response after injection (58). None of the other peptides we predicted here have been deposited in the IEDB.
FIG 11.
Distribution of SLA-3*hs0202-restricted epitope peptides in important IAV strains. (A) Genome-wide scanning for SLA-3*hs0202-restricted peptides in four IAV strains, A/Beijing/01/2009 (H1N1), A/Texas/50/2012 (H3N2), A/chicken/Nigeria/SO494/2006 (H5N1), and A/Shanghai/4664T/2013 (H7N9). Twenty-eight nonapeptides derived from the PB1, PB2, PA, HA, NA, NP, M1, and NS1 proteins were selected based on the motif X-(M/A/R)-(N/Q/R/F)-X-X-X-X-X-(V/I). (B) Gel filtration chromatograms of peptides RMQFSSLTV and RRNYFTAEV. (C) Further stabilization assays of the two peptides by anion exchange. The results confirmed that both could form stable complexes with SLA-3*hs0202.
DISCUSSION
Distinct conformations of peptides presented by the same MHC class I molecule, and even the same peptide, have been found in mammalian pMHC-I structures (32, 46, 59). The pMHC-I structures of swine, cattle, and rhesus macaques show that the flexible PBG adopts various conformations to accommodate the peptides, leading to the distinct peptide conformations (32, 46, 59). It is believed that the various peptide conformations presented by the same MHC-I could elicit a broad range of CTL responses (60). The distinct peptide conformations are mainly caused by the flexible side chains of the pockets and do not involve the α-helixes and β-sheets that form the PBG. Our data showed that the HA-KMN9 peptide displays different conformations in M1 and M2. However, the distinct HA-KMN9 peptide conformations are not only caused by the side chains in the PBG, but also by the skewing of the α1 and α2 helixes. SLA-3*hs0202 M1 and M2 are in one asymmetric unit, but the differences in their overall structures are obvious. The RMSD between M1 and M2 is 0.873 Å, which is higher than the RMSD of the SLA-1*0401 (0.401 Å) and BoLA-N*01801 (0.267 Å) molecules, which are also within one asymmetric unit (32, 46). As described in detail in Results above, these discordances not only occur in the flexible loops but are also present in the α1 and α2 helixes and the β-sheets of the α3 domain and sβ2m (Fig. 1A), which has not been described in the structures of SLA-1*0401 and other molecules. The major differences in the HA-KMN9 peptides occur at the mismatches of the α-helixes of M1 and M2. This finding indicates that, besides the side chains of the PBG, the flexibility of the swine MHC class I carbon backbone might expand the peptide conformation and facilitate the activation of an increased TCR repertoire.
Pockets B, D, and F accommodate the side chains of the P2, P3, and P9 residues, respectively, and determine the peptide-binding motif of SLA-3*hs0202. In addition to the canonical B and F pockets of HLA molecules, the D pocket of SLA-3*hs0202 also plays a vital role in binding peptides. This is similar to SLA-1*0401, in which the D pocket is the major limitation to peptide binding (32). In the D pocket, residue 156 interacting with the P3 peptide residue is critical to determine the motif in both SLA-3*hs0202 and SLA-1*0401. The peptide-binding motif determined by the B, D, and F pockets may be common among SLA I alleles.
The conformations of the SLA-3*hs0202 pockets are unique, but similar pockets were found in other solved pMHC-I structures, such as the B pocket of HLA-A*0201, the D pockets of HLA-B*4401 and HLA-B*0801, and the F pocket of H2-Kd (50–53). These pockets could accommodate the same residues of the HA-KMN9 peptide in similar manners. Therefore, we tested the peptide-binding motif of SLA-3*hs0202 based on the known binding residues. Indeed, the strategy we used to determine the motif of SLA-3*hs0202 is same one used to classify the HLA I supertypes and to predict allele-specific epitopes in silico. The in vitro refolding results showed that some candidate residues are acceptable to SLA-3*hs0202, at least in mutant HA-KMN9 peptides, and the 9-mer peptide-binding motif of SLA-3*hs0202 was determined to be X-(M/A/R)-(N/Q/R/F)-X-X-X-X-X-(V/I). The structural pocket alignment showed accuracy in determining the peptide-binding motif of a new MHC-I allele.
Based on our analysis and previous studies, the peptide-binding motif of SLA-3*hs0202 partly overlaps that of the most popular HLA allele, HLA-A*0201, which prefers L or M in position 2 and I or V at the C terminus (50). Previous studies demonstrated that HA-KMN9 and PB2-RMQ9 peptides derived from other subtypes of IAV are HLA-A*0201-restricted epitope peptides (58). The sequences of the HA-KMN9 peptide derived from the PR8 and Qinghai IAV strains are KMNIQFTAV and KMNTQFEAV, with a substitution of P4/P8 residues in the HA-KMN9 peptide from H1N1, respectively (19, 58). However, the anchor residues are conserved in all the HA-KMN9 peptides. Both HA-KMN9 peptides from the PR8 and Qinghai strains cannot elicit significant CTL responses in HLA-A*0201 transgenic mice (19, 58). This may be caused by the intermediate affinity between the HA-KMN9 peptide and HLA-A*0201, which was tested by both quantitative inhibition assays and in vitro refolding, because the low affinity could result in impaired pMHC stability and loss of immunogenicity. However, in our experiment, the HA-KMN9 peptide showed high binding affinity for SLA-3*hs0202, and its immunogenicity to SLA-3*hs0202 pigs may be different from that of HLA-A*0201 individuals. The sequence of the PB2-RMQ9 peptide from the PR8 strain is RMQFSSFTV, with a substitution at the P7 residue (F/L) compared to the pH1N1 peptide we used. The PR8 PB2-RMQ9 peptide has a high affinity for HLA-A*0201 and could elicit CTL responses after the injection of the peptide (58). Notably, the HA-KMN9 peptide is part of a B cell epitope located at HA391 to HA410 and has been documented in previous studies (61–63). The PB2-RMQ9 peptide we used has the same anchor residues and high affinity for SLA-3*hs0202. The substitution located in the region recognized by TCR may affect its immunogenicity, but L also has a long side chain that forms the “featured” conformation. Other peptides we identified here have not been described previously, and they are conserved in different subtypes of IAV, especially the peptides from the PB1, PB2, PA, NP, and M1 proteins.
In brief, the first structure of the SLA-3 allele determined here should help increase our understanding of antigen presentation in swine, and the “epitope map” for SLA-3*hs0202 can benefit the development of CTL vaccines against IAV diseases.
ACKNOWLEDGMENTS
This work was supported by the 863 Project of the China Ministry of Science and Technology (grant no. 2013AA102503), the State Key Program of the National Natural Science Foundation of China (grant no. 31201887), the 973 Project of the China Ministry of Science and Technology (grant no. 2013CB835302), and the Youth Teacher Scientific Research and Innovation Program of Zhoukou Normal University (grant no. ZKNUB115208).
We thank the Shanghai Synchrotron Radiation Facility (SSRF), Shanghai, People's Republic of China, for diffraction data on the crystals.
We declare no conflict of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M, Webby RJ. 2011. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 17:1624–1629. doi: 10.3201/1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naffakh N, van der Werf S. 2009. April 2009: an outbreak of swine-origin influenza A(H1N1) virus with evidence for human-to-human transmission. Microbes Infect 11:725–728. doi: 10.1016/j.micinf.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. 2015. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol 89:6218–6226. doi: 10.1128/JVI.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 5.Kingsford C, Nagarajan N, Salzberg SL. 2009. 2009 Swine-origin influenza A (H1N1) resembles previous influenza isolates. PLoS One 4:e6402. doi: 10.1371/journal.pone.0006402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol 83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stincarelli M, Arvia R, De Marco MA, Clausi V, Corcioli F, Cotti C, Delogu M, Donatelli I, Azzi A, Giannecchini S. 2013. Reassortment ability of the 2009 pandemic H1N1 influenza virus with circulating human and avian influenza viruses: public health risk implications. Virus Res 175:151–154. doi: 10.1016/j.virusres.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Tao H, Steel J, Lowen AC. 2014. Intrahost dynamics of influenza virus reassortment. J Virol 88:7485–7492. doi: 10.1128/JVI.00715-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claas EC, Kawaoka Y, de Jong JC, Masurel N, Webster RG. 1994. Infection of children with avian-human reassortant influenza virus from pigs in Europe. Virology 204:453–457. doi: 10.1006/viro.1994.1553. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kundin WD. 1970. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature 228:857. [DOI] [PubMed] [Google Scholar]
- 12.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 13.Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, Garcia-Sastre A, Moran TM, Palese P. 2010. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A 107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yusuf M, Konc J, Sy Bing C, Trykowska Konc J, Ahmad Khairudin NB, Janezic D, Wahab HA. 2013. Structurally conserved binding sites of hemagglutinin as targets for influenza drug and vaccine development. J Chem Infect Model 53:2423–2436. doi: 10.1021/ci400421e. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Shi Y, Zhang W, Li Q, Liu D, Vavricka C, Yan J, Gao GF. 2010. In silico characterization of the functional and structural modules of the hemagglutinin protein from the swine-origin influenza virus A (H1N1)-2009. Sci China Life Sci 53:633–642. doi: 10.1007/s11427-010-4010-8. [DOI] [PubMed] [Google Scholar]
- 16.Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. 2007. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A 104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMichael AJ, Gotch FM, Noble GR, Beare PA. 1983. Cytotoxic T-cell immunity to influenza. N Engl J Med 309:13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 18.Rimmelzwaan GF, Fouchier RA, Osterhaus AD. 2007. Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development. Curr Opin Biotechnol 18:529–536. doi: 10.1016/j.copbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Liu J, Yang M, Gao F, Zhou J, Kitamura Y, Gao B, Tien P, Shu Y, Iwamoto A, Chen Z, Gao GF. 2010. Identification and structural definition of H5-specific CTL epitopes restricted by HLA-A*0201 derived from the H5N1 subtype of influenza A viruses. J Gen Virol 91:919–930. doi: 10.1099/vir.0.016766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender BS, Croghan T, Zhang L, Small PA Jr. 1992. Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J Exp Med 175:1143–1145. doi: 10.1084/jem.175.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langley WA, Bradley KC, Li ZN, Talekar GR, Galloway SE, Steinhauer DA. 2010. The effects of preexisting immunity to influenza on responses to influenza vectors in mice. Vaccine 28:6305–6313. doi: 10.1016/j.vaccine.2010.06.112. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Wu B, Zhang S, Tan S, Sun Y, Chen Z, Qin Y, Sun M, Shi G, Wu Y, Liu N, Ning K, Ma Y, Gao B, Yan J, Zhu F, Wang H, Gao GF. 2013. Conserved epitopes dominate cross-CD8+ T-cell responses against influenza A H1N1 virus among Asian populations. Eur J Immunol 43:2055–2069. doi: 10.1002/eji.201343417. [DOI] [PubMed] [Google Scholar]
- 23.Sridhar S, Begom S, Bermingham A, Ziegler T, Roberts KL, Barclay WS, Openshaw P, Lalvani A. 2012. Predominance of heterosubtypic IFN-gamma-only-secreting effector memory T cells in pandemic H1N1 naive adults. Eur J Immunol 42:2913–2924. doi: 10.1002/eji.201242504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yap KL, Ada GL. 1978. Cytotoxic T cells in the lungs of mice infected with an influenza A virus. Scand J Immunol 7:73–80. doi: 10.1111/j.1365-3083.1978.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 25.Choo JA, Liu J, Toh X, Grotenbreg GM, Ren EC. 2014. The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans. J Virol 88:10613–10623. doi: 10.1128/JVI.00855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan PT, Khan AM, August JT. 2011. August. Highly conserved influenza A sequences as T cell epitopes-based vaccine targets to address the viral variability. Hum Vaccin 7:402–409. [DOI] [PubMed] [Google Scholar]
- 27.Wu KW, Chien CY, Li SW, King CC, Chang CH. 2012. Highly conserved influenza A virus epitope sequences as candidates of H3N2 flu vaccine targets. Genomics 100:102–109. doi: 10.1016/j.ygeno.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 28.MacDonald IK, Harkiolaki M, Hunt L, Connelley T, Carroll AV, MacHugh ND, Graham SP, Jones EY, Morrison WI, Flower DR, Ellis SA. 2010. MHC class I bound to an immunodominant Theileria parva epitope demonstrates unconventional presentation to T cell receptors. PLoS Pathog 6:e1001149. doi: 10.1371/journal.ppat.1001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. 1987. Structure of the human class I histocompatibility antigen, HLA-A2 Nature 329:506–512. [DOI] [PubMed] [Google Scholar]
- 30.Madden DR. 1995. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol 13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 31.Reimann J, Miller RG. 1983. Polymorphism and MHC gene function. Dev Comp Immunol 7:403–412. doi: 10.1016/0145-305X(83)90025-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang N, Qi J, Feng S, Gao F, Liu J, Pan X, Chen R, Li Q, Chen Z, Li X, Xia C, Gao GF. 2011. Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides. J Virol 85:11709–11724. doi: 10.1128/JVI.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, Li S, Larkin DM, Kim H, Frantz LA, Caccamo M, Ahn H, Aken BL, Anselmo A, Anthon C, Auvil L, Badaoui B, Beattie CW, Bendixen C, Berman D, Blecha F, Blomberg J, Bolund L, Bosse M, Botti S, Bujie Z, Bystrom M, Capitanu B, Carvalho-Silva D, Chardon P, Chen C, Cheng R, Choi SH, Chow W, Clark RC, Clee C, Crooijmans RP, Dawson HD, Dehais P, De Sapio F, Dibbits B, Drou N, Du ZQ, Eversole K, Fadista J, Fairley S, Faraut T, Faulkner GJ, Fowler KE, Fredholm M, Fritz E, Gilbert JG, Giuffra E, Gorodkin J, Griffin DK, Harrow JL, Hayward A, Howe K, Hu ZL, Humphray SJ, Hunt T, Hornshoj H, Jeon JT, Jern P, Jones M, Jurka J, Kanamori H, Kapetanovic R, Kim J, Kim JH, Kim KW, Kim TH, Larson G, Lee K, Lee KT, Leggett R, Lewin HA, Li Y, Liu W, Loveland JE, Lu Y, Lunney JK, Ma J, Madsen O, Mann K, Matthews L, McLaren S, Morozumi T, Murtaugh MP, Narayan J, Nguyen DT, Ni P, Oh SJ, Onteru S, Panitz F, Park EW, Park HS, Pascal G, Paudel Y, Perez-Enciso M, Ramirez-Gonzalez R, Reecy JM, Rodriguez-Zas S, Rohrer GA, Rund L, Sang Y, Schachtschneider K, Schraiber JG, Schwartz J, Scobie L, Scott C, Searle S, Servin B, Southey BR, Sperber G, Stadler P, Sweedler JV, Tafer H, Thomsen B, Wali R, Wang J, Wang J, White S, Xu X, Yerle M, Zhang G, Zhang J, Zhang J, Zhao S, Rogers J, Churcher C, Schook LB. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491:393–398. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, Wang T, Yeung CK, Chen L, Ma J, Zhang J, Jiang A, Li J, Zhou C, Zhang J, Liu Y, Sun X, Zhao H, Niu Z, Lou P, Xian L, Shen X, Liu S, Zhang S, Zhang M, Zhu L, Shuai S, Bai L, Tang G, Liu H, Jiang Y, Mai M, Xiao J, Wang X, Zhou Q, Wang Z, Stothard P, Xue M, Gao X, Luo Z, Gu Y, Zhu H, Hu X, Zhao Y, Plastow GS, Wang J, Jiang Z, Li K, Li N, Li X, Li R. 2013. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet 45:1431–1438. doi: 10.1038/ng.2811. [DOI] [PubMed] [Google Scholar]
- 35.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. 2009. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Gao F, Chu F, Zhang J, Gao GF, Xia C. 2010. Crystal structure of a bony fish beta2-microglobulin: insights into the evolutionary origin of immunoglobulin superfamily constant molecules. J Biol Chem 285:22505–22512. doi: 10.1074/jbc.M109.095000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu F, Lou Z, Gao B, Bell JI, Rao Z, Gao GF. 2005. Complex assembly, crystallization and preliminary X-ray crystallographic studies of rhesus macaque MHC Mamu-A*01 complexed with an immunodominant SIV-Gag nonapeptide. Acta Crystallogr Sect F Struct Biol Cryst Commun 61:614–616. doi: 10.1107/S1744309105016453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou M, Xu Y, Lou Z, Cole DK, Li X, Liu Y, Tien P, Rao Z, Gao GF. 2004. Complex assembly, crystallization and preliminary X-ray crystallographic studies of MHC H-2Kd complexed with an HBV-core nonapeptide. Acta Crystallogr D Biol Crystallogr 60:1473–1475. doi: 10.1107/S0907444904013587. [DOI] [PubMed] [Google Scholar]
- 40.Harp JM, Timm DE, Bunick GJ. 1998. Macromolecular crystal annealing: overcoming increased mosaicity associated with cryocrystallography. Acta Crystallogr D Biol Crystallogr 54:622–628. doi: 10.1107/S0907444997019008. [DOI] [PubMed] [Google Scholar]
- 41.Jensen LH. 1997. Refinement and reliability of macromolecular models based on X-ray diffraction data. Methods Enzymol 277:353–366. doi: 10.1016/S0076-6879(97)77020-4. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 45.Tobita T, Oda M, Morii H, Kuroda M, Yoshino A, Azuma T, Kozono H. 2003. A role for the P1 anchor residue in the thermal stability of MHC class II molecule I-Ab. Immunol Lett 85:47–52. doi: 10.1016/S0165-2478(02)00206-7. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Liu J, Qi J, Gao F, Li Q, Zhang N, Xia C, Gao GF. 2011. Two distinct conformations of a rinderpest virus epitope presented by bovine major histocompatibility complex class I N*01801: a host strategy to present featured peptides. J Virol 85:6038–6048. doi: 10.1128/JVI.00030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kjer-Nielsen L, Clements CS, Purcell AW, Brooks AG, Whisstock JC, Burrows SR, McCluskey J, Rossjohn J. 2003. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity 18:53–64. doi: 10.1016/S1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 48.Saper MA, Bjorkman PJ, Wiley DC. 1991. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol 219:277–319. doi: 10.1016/0022-2836(91)90567-P. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Wu P, Gao F, Qi J, Kawana-Tachikawa A, Xie J, Vavricka CJ, Iwamoto A, Li T, Gao GF. 2010. Novel immunodominant peptide presentation strategy: a featured HLA-A*2402-restricted cytotoxic T-lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J Virol 84:11849–11857. doi: 10.1128/JVI.01464-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borbulevych OY, Do P, Baker BM. 2010. Structures of native and affinity-enhanced WT1 epitopes bound to HLA-A*0201: implications for WT1-based cancer therapeutics. Mol Immunol 47:2519–2524. doi: 10.1016/j.molimm.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Archbold JK, Macdonald WA, Gras S, Ely LK, Miles JJ, Bell MJ, Brennan RM, Beddoe T, Wilce MC, Clements CS, Purcell AW, McCluskey J, Burrows SR, Rossjohn J. 2009. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med 206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C, Gras S, Brennan RM, Bird NL, Valkenburg SA, Twist KA, Burrows JM, Miles JJ, Chambers D, Bell S, Campbell S, Kedzierska K, Burrows SR, Rossjohn J, Khanna R. 2014. Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire. Sci Rep 4:3993. doi: 10.1038/srep03993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitaksov V, Fremont DH. 2006. Structural definition of the H-2Kd peptide-binding motif. J Biol Chem 281:10618–10625. doi: 10.1074/jbc.M510511200. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, Wen Y. 2015. Global dynamic analysis of a H7N9 avian-human influenza model in an outbreak region. J Theor Biol 367:180–188. doi: 10.1016/j.jtbi.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Grontvedt CA, Er C, Gjerset B, Hauge AG, Brun E, Jorgensen A, Lium B, Framstad T. 2013. Influenza A(H1N1)pdm09 virus infection in Norwegian swine herds 2009/10: the risk of human to swine transmission. Prev Vet Med 110:429–434. doi: 10.1016/j.prevetmed.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hillaire ML, Vogelzang-van Trierum SE, Kreijtz JH, de Mutsert G, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2013. Human T-cells directed to seasonal influenza A virus cross-react with 2009 pandemic influenza A (H1N1) and swine-origin triple-reassortant H3N2 influenza viruses. J Gen Virol 94:583–592. doi: 10.1099/vir.0.048652-0. [DOI] [PubMed] [Google Scholar]
- 57.Yu X, Jin T, Cui Y, Pu X, Li J, Xu J, Liu G, Jia H, Liu D, Song S, Yu Y, Xie L, Huang R, Ding H, Kou Y, Zhou Y, Wang Y, Xu X, Yin Y, Wang J, Guo C, Yang X, Hu L, Wu X, Wang H, Liu J, Zhao G, Zhou J, Pan J, Gao GF, Yang R. 2014. Influenza H7N9 and H9N2 viruses: coexistence in poultry linked to human H7N9 infection and genome characteristics. J Virol 88:3423–3431. doi: 10.1128/JVI.02059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gianfrani C, Oseroff C, Sidney J, Chesnut RW, Sette A. 2000. Human memory CTL response specific for influenza A virus is broad and multispecific. Hum Immunol 61:438–452. doi: 10.1016/S0198-8859(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 59.Wu Y, Gao F, Liu J, Qi J, Gostick E, Price DA, Gao GF. 2011. Structural basis of diverse peptide accommodation by the rhesus macaque MHC class I molecule Mamu-B*17: insights into immune protection from simian immunodeficiency virus. J Immunol 187:6382–6392. doi: 10.4049/jimmunol.1101726. [DOI] [PubMed] [Google Scholar]
- 60.Tynan FE, Elhassen D, Purcell AW, Burrows JM, Borg NA, Miles JJ, Williamson NA, Green KJ, Tellam J, Kjer-Nielsen L, McCluskey J, Rossjohn J, Burrows SR. 2005. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J Exp Med 202:1249–1260. doi: 10.1084/jem.20050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, Vincent AL, Golding H. 2013. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci Transl Med 5:200ra114. doi: 10.1126/scitranslmed.3006366. [DOI] [PubMed] [Google Scholar]
- 62.Lee JS, Chowdhury MY, Moon HJ, Choi YK, Talactac MR, Kim JH, Park ME, Son HY, Shin KS, Kim CJ. 2013. The highly conserved HA2 protein of the influenza A virus induces a cross protective immune response. J Virol Methods 194:280–288. doi: 10.1016/j.jviromet.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 63.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. 2013. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity 38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]