ABSTRACT
During the early steps of infection, retroviruses must direct the movement of the viral genome into the nucleus to complete their replication cycle. This process is mediated by cellular proteins that interact first with the reverse transcription complex and later with the preintegration complex (PIC), allowing it to reach and enter the nucleus. For simple retroviruses, such as murine leukemia virus (MLV), the identities of the cellular proteins involved in trafficking of the PIC in infection are unknown. To identify cellular proteins that interact with the MLV PIC, we developed a replication-competent MLV in which the integrase protein was tagged with a FLAG epitope. Using a combination of immunoprecipitation and mass spectrometry, we established that the microtubule motor dynein regulator DCTN2/p50/dynamitin interacts with the MLV preintegration complex early in infection, suggesting a direct interaction between the incoming viral particles and the dynein complex regulators. Further experiments showed that RNA interference (RNAi)-mediated silencing of either DCTN2/p50/dynamitin or another dynein regulator, NudEL, profoundly reduced the efficiency of infection by ecotropic, but not amphotropic, MLV reporters. We propose that the cytoplasmic dynein regulators are a critical component of the host machinery needed for infection by the retroviruses entering the cell via the ecotropic envelope pathway.
IMPORTANCE Retroviruses must access the chromatin of host cells to integrate the viral DNA, but before this crucial event, they must reach the nucleus. The movement through the cytoplasm—a crowded environment where diffusion is slow—is thought to utilize retrograde transport along the microtubule network by the dynein complex. Different viruses use different components of this multisubunit complex. We found that the preintegration complex of murine leukemia virus (MLV) interacts with the dynein complex and that regulators of this complex are essential for infection. Our study provides the first insight into the requirements for retrograde transport of the MLV preintegration complex.
INTRODUCTION
During early steps of infection, retroviruses must reach the nucleus to cross the nuclear membrane and access the host cell genome. The cytoplasm is extremely crowded, and simple diffusion is predicted to be too slow to explain the kinetics of virus infection (1). Many retroviruses are actively transported along microtubules (2–4) to move through the cytoplasm and reach the nucleus. Identifying the host proteins required for retrovirus infection has been a major challenge. Many screens have been performed for this purpose, including yeast two-hybrid screens of cDNA expression libraries with viral proteins as bait (5–8), biochemical screens of host proteins copurifying with viral proteins (9), and functional screens of cDNA overexpression libraries (10, 11) or knockdown libraries based on small interfering RNAs for those members that affect infection (12–14). In the case of the murine leukemia viruses (MLVs), most such screens have been performed with isolated viral proteins or expressed viral genes and not in the context of replicating virus or the complete preintegration complex (PIC).
The gammaretroviruses are a large group of simple retroviruses that include murine leukemia viruses, feline leukemia viruses, gibbon ape leukemia virus. The MLVs can be further subdivided by their species tropisms, as determined by their various envelope proteins and the receptors they utilize, into ecotropic, amphotropic, and xenotropic viruses. The ecotropic viruses are thought to enter the cell by receptor-mediated endocytosis and to gain access to the cytoplasm through fusion in the endosomes in a pH-dependent manner (15). The xenotropic viruses and the recombinant xenotropic murine leukemia virus-related virus (XMRV), using the distinctive XPR1 receptor, can infect cells in a pH-independent manner (16). The route of entry of the amphotropic viruses is controversial but is now thought to involve macropinocytosis (17) and not internalization by the caveolin or endosome route, as suggested previously (15). In spite of these profound differences in their receptor utilization and routes of entry, once in the cytoplasm, the viruses are thought to use similar mechanisms to achieve nuclear entry. All the gammaretroviruses are strongly dependent on cell division to gain entry into the nucleus. These viruses do not have the ability to cross the nuclear membrane in nondividing cells but, rather, must wait for nuclear-membrane breakdown to occur during mitosis to access the host chromatin.
Several screens have revealed that cytoskeleton-associated proteins are important in different stages of retrovirus replication (10, 13). Microtubules are particularly important during the early stages of viral infection. It has been shown that treating cells with drugs that disrupt microtubules before or at the moment of infection significantly reduces the infectivity of several viruses (reviewed in reference 18), indicating that virus transport in the early stages of infection relies critically on microtubules and/or the microtubule-associated motors (19). For MLV, it has been shown that nocodazole treatment, which impairs microtubule polymerization, reduces viral infection (20). In early stages of infection, as the incoming retroviral particles move toward the nucleus, the drivers of movement are expected to be associated with the microtubule retrograde transport, which is mediated by the cytoplasmic dynein. Some retroviruses, such as bovine immunodeficiency virus (BIV), directly interact with the cytoplasmic dynein complex for their retrograde transport (21). It has also been shown that dynein function is required for human immunodeficiency virus type 1 (HIV-1) transport along microtubules (3, 22), but direct interaction between HIV-1 and dynein has not been demonstrated in the context of infection. For MLV, it has not been shown that dynein function is required for retrograde transport, dynein complex, or adaptor proteins that would mediate interaction with the dynein complex.
Dynein, the motor associated with microtubule minus-end transport, is a multisubunit complex composed of two copies of heavy chains (DYNC1H1), two intermediate chains (DYNC1I1/2), two light-intermediate chains (DYNC1LI1/2), and two copies each of three different light chains (DYNLT1/3, DYNLRB1/2, and DYNLL1/2). The heavy chains are the ATPase motors that bind to microtubules and generate the mechanical force for movement, while the other subunits act as regulators and adaptors to different cargoes (reviewed in reference 23). Dynein function is regulated by two protein complexes, the dynactin complex and the NudE/NudEL-LIS1 complex; these complexes modulate cytoplasmic dynein processivity and mediate cargo binding. Dynein, along with its regulators, drives a wide range of functions in eukaryotic cells, including vesicular transport, spindle morphogenesis, spindle orientation, and the inactivation of the spindle assembly checkpoint. Dynein has also been shown to serve as the adaptor for microtubule transport of viruses from several different families (18, 19, 24–26).
To identify host proteins that associate with the incoming MLV PIC, we developed a screen using live virus. Using a combination of immunoprecipitation (IP) and mass spectrometry (MS), we show that the microtubule motor dynein regulator protein DCTN2/p50/dynamitin interacts with the MLV preintegration complex at early times of infection, suggesting a direct interaction between the incoming viral particles and the dynein complex regulators. Using RNA silencing, we demonstrate that DCTN2/p50/dynamitin and the dynein regulator NudEL are essential for ecotropic, but not amphotropic, MLV reporter infection. We propose that the cytoplasmic dynein regulators are a critical piece of the host machinery needed for infection by the retroviruses entering the cell via the ecotropic enveloped pathway.
MATERIALS AND METHODS
Cell lines.
Human embryonic fibroblasts (293T), mouse fibroblasts (NIH 3T3), and human medulloblastoma cells (TE671) were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 mg/ml streptomycin. All the cells were cultured at 37°C in 5% CO2.
Plasmids and mutagenesis.
Plasmid DNAs were as follows. pCMVI expresses gag and pol from Moloney MLV (MoMLV). pHit123 expresses the ecotropic envelope of MLV. pHit456 expresses the amphotropic envelope of MLV. pFBLuc (Stratagene) is a reporter plasmid containing the firefly luciferase coding sequence flanked by MLV-based long terminal repeats (LTRs). p8.91 carries gag and pol of HIV-1. pMD.G expresses the vesicular stomatitis virus envelope glycoprotein. pNCS (27) contains the full-length Moloney murine leukemia virus. pNCS IN3×FLAG was generated by overlapping PCR using primers containing the FLAG epitope in frame with the integrase coding sequence and the pNCS vector as a template. IN is located at positions 5063 to 6283 of pNCS; the primers used for the overlapping PCR fragments were as follows: for the 5′ fragment, pNCS-4154-SalI (5′-CTTTGTCGACGAGAAGCAGGGCTACGCC-3′)/3FLAGINaa10rev (5′-CTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCCATATGTTCTGAGGTGTA-3′), and for the 3′fragment, 3FLAGINaa10for (5′-ATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGTTTCATTACACAGTG-3′)/pNCS5043SacIIrev (5′-ATGCCCGCGGACCCTAGTTCCCTG-3′). After amplification of the full-length product using the external primers, the PCR product was digested with the restriction enzymes SalI (position 4155 of pNCS) and SacII (position 5402 of pNCS) and cloned into pNCS digested with the same enzymes.
Identification of MLV preintegration complex-associated proteins.
NIH 3T3 cells transfected with pNCS or pNCS IN3×FLAG were maintained as producer cells and were expanded to 10 100-mm plates. Once the cells were confluent, wild-type (WT) MoMLV or the tagged version was collected from the supernatants, filtered (0.45 μm), and used to infect 10 150-mm plates of semiconfluent NIH 3T3 cells in the presence of 8 μg/ml Polybrene. After 30 min at 4°C, the cells were transferred to 37°C for 6 h. Cytosolic extracts were prepared by incubating the cells in buffer A (10 mM Tris-HCl, pH 7.4, 225 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], and Complete protease inhibitors). The cells were disrupted by Dounce homogenization with 20 strokes, and the lysates were clarified by centrifugation at 1,000 × g for 3 min and again by centrifugation at 8,000 × g for 20 min. The extracts were fractionated using a Sephacryl S300 column (GE) in buffer B (10 mM Tris-HCl, pH 7.4, 120 mM KCl, 5 mM MgCl2), and 4-ml fractions were collected. The presence of DNA and proteins was analyzed by measuring the A260 and A280. Fractions 6 to 15 were kept. DNA was extracted from 50 μl of each fraction using a PCR product purification kit (Qiagen) after proteinase K treatment. The presence of viral DNA on the fraction with a high A260 was confirmed by PCR using DNA primers specific for linear viral DNA (8). Proteins were precipitated from 200 μl of each fraction with trichloroacetic acid, and the presence of CA and the tagged IN was analyzed by Western blotting. The fractions with viral DNA were pooled and concentrated using an Amicon centrifuge filter unit with a cutoff of 3,000 Da, 20% sucrose was added, and the samples were stored at −80°C until use.
For immunoprecipitations, 400 μg of each extracted protein and 5 μg of anti-FLAG (M2; Sigma) conjugated to M270 epoxy (Invitrogen) were incubated for 6 h at room temperature in IP/wash buffer (20 mM HEPES, pH 7.5, 5 mM MgCl2, 120 mM KCl, 6 mM EDTA, 0.1% IGEPAL CA-630, and Complete protease inhibitor). After 3 washes, the beads were isolated, and the bound material was eluted in buffer containing 3×FLAG peptide (Sigma). Samples were concentrated in a speed vacuum, and sodium dodecyl sulfate (SDS) sample buffer was added. Proteins were separated in SDS-10% polyacrylamide gel electrophoresis (PAGE), bands that were unique to the IN3×FLAG extract were excised, and the proteins were subjected to tryptic digestion followed by peptide identification by liquid chromatography-tandem MS (LC–MS-MS) using a hybrid high-resolution quadrupole time of flight electrospray mass spectrometer at the Protein Core Facility at Columbia University. The results were analyzed using the MASCOT database search tool (Matrix Science).
Generation of stable cell lines.
Lentiviruses for transduction were produced by transfection of 293T cells with a mixture of plasmid DNAs: 1 μg pMD.G, 1 μg p8.91, and 1.5 μg pGIPz or pGIPzDync1i2 (containing short hairpin RNAs [shRNAs] 1 to 3), pGIPzDCTN1 (containing shRNAs 1 and 2), pGIPzDCTN2 (containing shRNAs 1 and 2), pGIPzNudE (containing shRNAs 1 and 2), pGIPzLIS1 (containing shRNAs 1 and 2), or pGIPzNudEL (containing shRNAs 1 and 2) (Open Biosystems). Viruses were harvested 48 h after infection, filtered (0.45 μm), and used to infect 5 × 104 NIH 3T3 cells in 35-mm dishes in the presence of 8 μg/ml Polybrene. Cells were selected in 1 μg/ml puromycin. For knockdown (KD) in human cells, TE671 cells were infected with the human version of pGIPzDCTN2 (containing shRNAs 1 and 2) and pGIPzNudEL (containing shRNAs 1, 2, and 3) and selected with 1 μg/ml puromycin.
Western blotting.
Cells were lysed in 20 mM Tris-HCl (pH 8.0), 137 mM KCl, 10% (vol/vol) glycerol, 1% (vol/vol) NP-40, and Complete protease inhibitor (Roche) or Reporter lysis buffer (Promega). Samples were then boiled in 5× SDS loading buffer, and the proteins were resolved by SDS-PAGE. After transfer to nitrocellulose membranes, the blots were probed with goat anti-MLV CA, rabbit monoclonal anti-dynamitin (EPR5094; Abcam), mouse anti-tubulin (DM1A; Sigma), or mouse anti p150Glued (catalog no. 612709; BD). Secondary antibodies conjugated to horseradish peroxidase (HRP) and the enhanced chemiluminescence (ECL) reagents were used for developing.
Single-cycle infectivity assay.
Moloney MLV-luciferase reporters were produced by transfection of 293T cells with 10 μg pCMVI, 10 μg pHIT123 or pHIT456, and 20 μg pFBluc (per 100-mm plate) using calcium phosphate. HIV-1–luciferase reporter was produced by transfection of 293T cells with 15 μg pNL43-env luciferase and 5 μg pHIT123 or pHIT456. Reporter virus stocks were harvested 48 h after transfection, filtered (0.45 μm), and stored at −80°C. NIH 3T3 cells (2.5 × 104 per well) were seeded in 24-well plates and infected with MLV luciferase reporters. Forty-eight hours postinfection, the cells were collected and assayed for firefly luciferase activity (Promega) in a luminometer.
Analysis of viral DNA synthesized in vivo.
NIH 3T3 cells (1 × 105) plated in 35-mm dishes or nonsilenced control and p50 or NudEL KD cells (1 × 106) plated in 10-cm dishes were infected with MoMLV or the tagged version for 16 h. Low-molecular-weight DNA was isolated using the modified Hirt method (28). The linear DNA (linearDNAF [5′-CCCCTGCGCGCAGGAGGAAAC-3′]/linearDNAR [5′-CCCAGTCTGGGCGCTCGAGGGG-3′]) and 2-LTR circles (LTR-LTR junctions) (MR5784 [5′-AGTCCTCCGATTGACTGAG-3′]/MR4091 [5′-CTCTTTTATTGAGCTCGGG-3′]) were detected by PCR, as previously described (8). The mitochondrial DNA was used as a control (MmmithoconF [5′-CGTATGACCGCGGTGGCTGG-3′]/MmmitochonR [5′-CTAGCCACACCCCCACGGGA-3′]).
Immunostaining.
Control and knockdown cells were transfected with TransIT-LT1 transfection reagent (Mirus) for 16 h to express mCAT-hemagglutinin (HA) and processed after 48 h for indirect immunofluorescence (IFI) assay. The cells were fixed with 4% paraformaldehyde on phosphate-buffered saline (PBS)-CM (1 mM MgCl2, 0.1 mM CaCl2, pH 7.4) at room temperature for 15 min and then rinsed 3 times with PBS-CM and permeabilized with PBS-CM-0.2% Triton X-100. Then, the cells were incubated with monoclonal anti-HA antibody (clone 16B12; Covance) on PBS-CM-0.2% bovine serum albumin (BSA) for 1 h at room temperature, rinsed with PBS-CM, and incubated with goat-anti-mouse–Alexa 555 secondary antibody for 1 h. The cells were then mounted for IFI with Fluoromount-G (Electron Microscopy Sciences), and fluorescence images were acquired at ×40 magnification with a DFC3000G Leica charge-coupled-device (CCD) camera mounted on a Leica DMIL microscope running Leica LASX software. Digital images were processed with Photoshop CS3 (Adobe) and are shown with inverted contrast.
Analysis of knockdown.
NIH 3T3 cells were harvested, and total RNA was extracted using TRIzol reagent (Invitrogen). Two micrograms of total RNA per cell line was used for reverse transcription reactions to produce cDNA using random hexamers and the Affinity Script qPCR cDNA Synthesis kit (Agilent Technologies); 2 μl of each cDNA was used for quantitative PCR (qPCR) analysis of Dync1i2 (Dync1i2f [5′-GTGGTTGTCCAGCTTTTGCC-3′]/Dync1i2r [5′-CCGACATGGTTGTGACCTTTTG-3′]), p50 (p50f [5′-ATGGCGGACCCTAAATACGC-3′]/p50r [5′-GAACTTGTCGTAGGCAGCGT-3′]), p150Glued (p150f [5′-CAAGAGAGAGGGAGCCGATG-3′]/p150r [5′-GGGCTCACTGCTGCTTAGTT-3′]), NudE (NudEf [5′-GGGCGATAAAGGGTCAGGAA-3′]/NudEr [5′-GCGCAGTTCTCATTGGTCCT-3′]), LIS1 (LIS1f [5′-GAATGGATTCCCCGTCCACC-3′]/LIS1r [5′-GTGACCGTGCATGGTTCTGA-3′]), NudEL (NudELf [5′-CTTGCGGAAAGTAGGGGCTT-3′]/NudELr [5′-GATTTGCGGGATGCTTGGTC-3′]), and CypA (CypAf [5′-GAGCTCTGAGCACTGGAGAGA-3′]/CypAr [5′-CCACCCTGGCACATGAAT-3′]) transcript levels. In human cells, we proceeded in the same manner except that the primers we used detected the human versions of p50 (p50f [5′-TAAATACGCCGACCTTCCCG-3′]/p50r [5′-GCTGTCGGTGCCTTCTTAGG-3′]), NudEL (NudELf [5′-CGGAGGTACGCTGAGTGGA-3′]/NudELr [5′-CCATGATCAAGAAAGCCTCCAA-3′]), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (GAPDHf [5′-CCTTCATTGACCTCAACTACATGG-3′]/GAPDHr [5′-CATGGTTCACACCCATGACGAACAT-3′]). The fold change was calculated using the relative standard curve method.
Statistical analysis.
All statistical analyses were performed in GraphPad Prism using one-way analysis of variance (ANOVA) with 95% confidence.
RESULTS
Identification of MLV PIC-associated proteins.
To identify cellular proteins that associate with the incoming MLV virion and PIC by coimmunoprecipitation, we developed a replication-competent MLV genome in which the integrase protein was modified by the insertion of a 3×FLAG epitope tag after amino acid 10 in the context of the complete provirus genome (Fig. 1A). To verify that this new construct, dubbed pNCS IN3×FLAG, was able to produce replication-competent viruses, we transfected 293T cells with the wild-type parent pNCS or pNCS IN3×FLAG DNA and analyzed cell lysates and culture supernatants for viral proteins after 48 h. We observed that similar amounts of Gag and CA were present and that proteins with the epitope tag were detected only in the supernatant of pNCS IN3×FLAG-transfected cells (Fig. 1B). To verify that the 3×FLAG insertion did not affect viral entry, reverse transcription, or nuclear entry, we analyzed the levels of linear viral DNA and the LTR-LTR junction sequence, a marker of nuclear entry, in cells infected with WT or IN3×FLAG virus. We observed that the two viruses were able to accomplish reverse transcription and nuclear entry at similar levels (Fig. 1C). To analyze the capability of the IN3×FLAG virus to complete all steps of the life cycle, we performed a virus-spreading assay of the IN3×FLAG virus in permissive NIH 3T3 cells in parallel with WT virus. We observed that the two viruses were able to spread through the cell cultures with similar efficiencies and kinetics, as shown by the accumulation of reverse transcriptase (RT) activity in culture supernatants (Fig. 1D). Furthermore, the IN3×FLAG viruses collected after 6 days of infection still retained the 3×FLAG tag (Fig. 1E), which indicated that the insertion did not affect the stability of the genome and that the virus did not revert.
FIG 1.
Characteristics of MLV IN3×FLAG. (A) The coding sequence for a 3×FLAG epitope was inserted after amino acid 10 of IN in the proviral MLV vector pNCS. A schematic representation of the construct is shown. The amino terminus of the IN3×FLAG protein is presented, and the 3×FLAG tag is shown in boldface. (B) The plasmid pNCS and the mutant version, pNCS- IN3×FLAG, were transfected in parallel into 293T cells, and the presence of the tagged IN was detected in the pelleted supernatant (PS) and cell lysates (CL) by Western blotting using an anti-FLAG antibody (bottom). The presence of the Gag and CA proteins is shown as a control (top). (C) NIH 3T3 cells were infected with the IN3×FLAG virus and the WT MLV; 18 h postinfection, low-molecular-weight DNA was recovered, and the presence of linear DNA and LTR-LTR circles was detected by PCR. (D and E) NIH 3T3 cells were infected with preparations of WT virus, IN3×FLAG virus, or mock virus at two dilutions. (D) RT activity in the supernatants was assayed on different days postinfection. (E) On day 6, the supernatants were collected, and virions were isolated by ultracentrifugation. The viral proteins Gag, CA, and FLAG-tagged IN were detected by Western blotting. (F) NIH 3T3 cells were infected with WT or IN3×FLAG virus for 6 h. Then, they were lysed, and proteins in the cytosolic extracts were immunoprecipitated using anti-FLAG antibody. Viral DNA in the immunoprecipitated material was detected by PCR (top), and the presence of the IN3×FLAG protein was detected by Western blotting using FLAG antibody (bottom).
To test that the 3×FLAG tag present in the integrase was accessible to antibodies for IP, we infected cells with the WT and IN3×FLAG viruses and prepared lysates that were subjected to IP using an anti-FLAG antibody. We observed that viral proteins in the IN3×FLAG PICs were indeed immunoprecipitated from the extracts and that the viral DNA was associated with it (Fig. 1F). Importantly, we did not observe immunoprecipitation of the viral DNA in extracts derived from the WT MLV-infected cells (Fig. 1F), indicating that the immunoprecipitation was specific and that the viral DNA detected was part of the PIC.
To identify cellular proteins specifically associated with the IN3×FLAG virus, we infected cells with WT or tagged virus, and 6 h postinfection, the cells were lysed and the lysates were subjected to gel filtration to eliminate any IN proteins that were not associated with the PIC (Fig. 2A). The eluted fractions that contained viral DNA and IN were pooled and subjected to immunoprecipitation using anti-FLAG antibody. The immunoprecipitated material was then separated by SDS-PAGE, and proteins in various size classes were analyzed using mass spectrometry. The proteins that were present only in the IN3×FLAG IP are shown in Fig. 2B. From that list, our attention was drawn to the presence of two microtubule-associated proteins, the cytoplasmic dynein intermediate chain and dynactin subunit 2. The dynein intermediate chain (Dync1i2) is part of the dynein complex, the motor for retrograde transport of different cargos along the microtubule network. Dynactin subunit 2 (dynamitin/p50/DCTN2, referred to below as p50) is part of the dynactin complex, which regulates processivity of and cargo association with the dynein complex. It has been previously shown that microtubules (20) and microtubule-associated proteins (6) are important for MLV infection and that retroviruses move from the cell periphery in a retrograde manner using the dynein motors (9, 21, 29, 30), but no association with the dynein complex or the dynein regulator has been shown. Therefore, we decided to investigate whether these proteins are important for MLV infection.
FIG 2.

Proteins specifically associated with MLV PIC. (A) NIH 3T3 cells were infected with IN3×FLAG MoMLV for 6 h, and cytosolic extracts were prepared and gel filtered through a Sephacryl S300 column. The presence of viral DNA and viral proteins IN and CA in fractions 6 to 15 is shown by PCR and Western blotting, respectively. (B) Proteins identified by mass spectrometry as specifically associated with IN3×FLAG MoMLV.
Dynein and dynactin proteins are required for MLV infection.
To explore the potential importance of Dync1i2 and p50 for infection, we generated stable KD cells using at least two different shRNAs and then challenged the cells with MLV reporters. Many of the dynein chains and dynactin subunits are essential for cell survival, since they have an important role in the maintenance of the cytoskeleton, organelle positioning, and cell division (reviewed in references 31 and 32). For this reason, we expected to select for cells that produced sufficient quantities of these proteins to survive and function in the long term but had reduced mRNA or protein levels compared to nonsilenced, stable cell lines.
Cells engineered for KD of Dyncli2 exhibited at most a 70% reduction in the levels of RNA. When these Dync1i2 KD cells (Fig. 3A) were challenged with ecotropic enveloped MLV-luciferase, we observed that infection was modestly (Fig. 3B) but significantly (Fig. 3C) reduced to ∼50% compared to control cells. We conclude that Dyncli2 is required but not essential for infection or that the level of KD in our cell lines was not sufficient to impact infection in a more severe manner.
FIG 3.
Silencing of the dynein intermediate chain Dync1i2 partially blocks MLV infection. (A) NIH 3T3 cells in which the intermediate chain of cytoplasmic dynein was silenced in a stable manner were generated. Dync1i2 mRNA levels were determined by quantitative PCR. The values were normalized to CypA mRNA and expressed relative to the nonsilenced (non-sil) control (**, P = 0.086). (B) Control and Dync1i2 KD cells were challenged with increasing amounts of ecotropic MLV-luciferase reporter. Forty-eight hours after infection, luciferase activity was measured. One representative of three independent experiments is shown. The error bars indicate standard deviations between triplicates in the same experiment. (C) Relative infection of ecotropic MLV-luciferase reporter in Dync1i2 KD cells compared to the nonsilenced control. The error bars represent the standard deviations among the 3 independent experiments (***, P < 0.001; **, P = 0.0064).
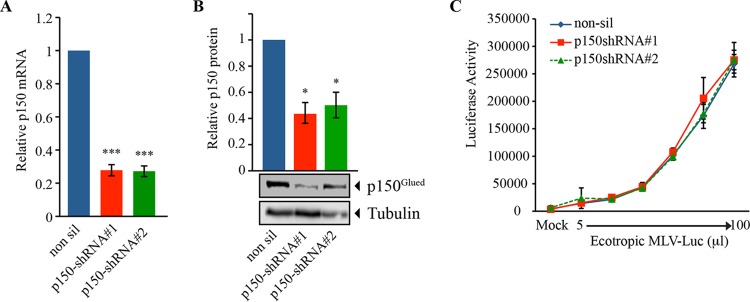
The cell lines with p50 KD vectors exhibited an approximately ∼80% reduction in p50 mRNA (Fig. 4A) and an ∼50% reduction of p50 protein levels (Fig. 4B). This reduction in the protein level had no effect on the timing of cell division (data not shown). Strikingly, despite the limited extent of the KD, infection of the p50 KD cells with the ecotropic MLV-luciferase was profoundly (Fig. 4C) and significantly (Fig. 4E) reduced. We then tested whether a similar effect was seen with amphotropic MLV, which uses a different entry mechanism. The ecotropic enveloped MLV in NIH 3T3 cells is endocytosed and the virion is released to the cytoplasm after acidification of the endosome (20), whereas the amphotropic virus does not require acidification for release to the cytoplasm (17, 33). We challenged the p50 KD cells with an amphotropic MLV-luciferase and observed that the infection was moderately reduced compared to the nonsilenced cell line (Fig. 4D), and although it was statistically significant (Fig. 4E), it was not as striking as for the ecotropic enveloped MLV-luciferase. If the ecotropic envelope fusion releases the viral cores closer to the membrane than the amphotropic envelope, then the PIC from ecotropic MLV would be expected to exhibit a greater requirement for the retrograde transport machinery, consistent with the more severe effects observed for this particular virus.
FIG 4.
The dynein regulator p50 is required for ecotropic MLV infection in NIH 3T3 cells. (A) NIH 3T3 cells in which the dynactin subunit p50 was silenced in a stable manner were generated. p50 mRNA levels were determined by quantitative PCR. The values were normalized to CypA mRNA and expressed as fold reduction over the nonsilenced control (***, P < 0.0001). (B) The protein levels of p50 on the KD cells were determined by Western blotting. α-Tubulin was used as a normalization control. A representative Western blot is shown. The quantifications were derived from the results of three independent assays (***, P < 0.0001). (C and D) Control and p50 KD cells were challenged with increasing amounts of ecotropic (C) and amphotropic (D) MLV-luciferase reporter. Forty-eight hours after infection, luciferase activity was measured. One representative of three independent experiments is shown. The error bars indicate standard deviations between triplicates in the same experiment. (E) Relative infection by ecotropic (Eco) and amphotropic (Ampho) MLV-luciferase reporter in p50 KD cells compared to the nonsilenced control. The error bars represent the standard deviations among 3 independent experiments (***, P < 0.0001).
The above-mentioned results indicate that p50—which is a fundamental component of the dynactin complex and a fundamental regulator of the dynein complex—is important for ecotropic MLV infection. We then asked whether another component of the dynactin complex, p150Glued, and other regulators of the dynein complex, such as NudEL-NudE and LIS1, were relevant for MLV infectivity. The dynactin complex binds to dynein and microtubules via p150Glued, and the overexpression of a dominant-negative version of p150Glued has been shown to interrupt the trafficking of several viruses (3, 34). We generated stable cell lines in which p150Glued was knocked down. The cells showed efficient reduction of the p150Glued mRNA (Fig. 5A) and protein (Fig. 5B), but this KD of p150Glued had no effect on the infectivity of ecotropic MLV-luciferase (Fig. 5C), suggesting that p150Glued, unlike p50, is not likely required for MLV infection.
FIG 5.
Silencing of the dynactin complex protein p150Glued does not affect MLV infection. (A) NIH 3T3 cells in which the dynactin subunit p150Glued was silenced in a stable manner were generated. mRNA levels of p150Glued in cells engineered for KD of p150Glued were determined by quantitative PCR. The values were normalized to CypA mRNA and expressed as fold reduction over the nonsilenced control (***, P < 0.0001). (B) The protein levels of p150Glued on KD cells were determined by Western blotting using α-tubulin as a normalization control. A representative Western blot is shown. The quantification was derived from the results of three independent assays (*, P = 0.0248). (C) p150Glued KD and control cells were challenged with increasing amounts of ecotropic MLV-luciferase reporter. Forty-eight hours after infection, luciferase activity was measured. One representative of five independent experiments is shown. The error bars indicate standard deviations between triplicates in the same experiment.
The dynein regulator NudEL is essential for ecotropic MLV infection.
We next investigated whether the NudEL-NudE-LIS1 complex, which regulates many of the key functions of the dynein complex, such as recruitment to kinetochores, early endosome recruitment, and dynactin association (35–37), was required for MLV infection. We generated stable KD cell lines for NudEL, NudE, or LIS1. When the NudE (Fig. 6A) and LIS1 (Fig. 6C) KD cells were challenged with ecotropic MLV-luciferase, no difference was observed compared to the nonsilenced control cells (Fig. 6B and D, respectively). Strikingly, the NudEL KD cells (Fig. 7A) almost completely abolished the infectivity of the ecotropic MLV-luciferase reporter (Fig. 7B) but had no significant effect on the amphotropic MLV-luciferase reporter (Fig. 7C and D). Similar to the p50 KD cells, the NudEL KD cells had a normal duplication rate (data not shown) and the Golgi apparatus was not fragmented (data not shown), as has been shown before in NudEL silenced cells (38).
FIG 6.

Silencing of the dynein regulators NudE and LIS1 does not affect MLV infection. (A and C) Cells KD for NudE (A) and LIS1 (C) were generated. RNA was extracted, and the mRNA levels of NudE and LIS1 were determined by quantitative PCR. The values were normalized to CypA mRNA and expressed as fold reduction over the nonsilenced control (***, P < 0.0001). (B and D) Control and NudE (B) and LIS1 (D) KD cells were challenged with increasing amounts of ecotropic MLV-luciferase reporter. Forty-eight hours after infection, luciferase activity was measured. One representative of four independent experiments is shown. The error bars indicate standard deviations between triplicates in the same experiment.
FIG 7.

The dynein regulator NudEL is important for ecotropic MLV infection in NIH 3T3 cells. (A) Cells KD for the dynein regulator NudEL were generated. NudEL mRNA levels in cells engineered by KD of NudEL were determined by quantitative PCR. The values were normalized to CypA mRNA and expressed as fold reduction over the nonsilenced control (*, P = 0.0119). (B and C) Control and NudEL KD cells were challenged with increasing amounts of ecotropic (B) or amphotropic (C) MLV-luciferase reporter. Forty-eight hours after infection, luciferase activity was measured. One representative of three independent experiments is shown. The error bars indicate standard deviations between triplicates in the same experiment. (D) Relative infection by ecotropic and amphotropic MLV-luciferase reporters in NudEL KD cells compared to the nonsilenced control. The error bars represent the standard deviations among 3 independent experiments (***, P < 0.0001; n.s., nonsignificant).
To determine the stage of early MLV infection during which p50 and NudEL are required and to discard defects in fusion of MLV due to the KD of these proteins, we challenged nonsilenced control and KD cells with MoMLV and determined the levels of total viral DNA produced during the process of reverse transcription and the levels of the LTR-LTR circles, a nuclear entry marker. Challenge viruses were able to complete reverse transcription in p50 and NudEL KD cells to form viral DNA at normal levels, but the levels of circular viral DNA containing the LTR-LTR junction, the hallmark of nuclear entry, were profoundly reduced compared to the nonsilenced control cell line (Fig. 8A). These experiments indicate that the major block to infection in cells with KD of p50 or NudEL occurs at the time of nuclear entry and that neither fusion nor reverse transcription was affected.
FIG 8.
The dynein regulators p50 and NudEL are required for a post-reverse transcription step in MLV infection. (A) Control, p50, or NudEL KD cells were infected with Mo-MLV. Low-molecular-weight DNA was isolated 20 h after infection, and the viral DNA synthesized in the infected cells was detected by PCR using primer pairs specific for the different forms of the linear DNA (primers directed against the CA region) or the LTR-LTR junction nuclear entry marker. The amplification of mitochondrial DNA was used as a control. (B) The correct localization of the ecotropic receptor mCAT1 in p50 and NudEL KD cells was determined. Control (nonsilenced), p50, and NudEL KD cells were transfected with a plasmid encoding an HA-tagged version of mCAT1. The subcellular localization of the overexpressed receptor was detected by indirect immunofluorescence using an α-HA antibody. The arrowheads indicate membrane-localized receptors. (C) Stable TE671 cells in which p50 and NudEL were silenced were generated and challenged with amphotropic HIV-1. (Bottom) NudEL and p50 mRNA levels in cells engineered by KD of NudEL or p50 were determined by quantitative PCR. The values were normalized to GAPDH mRNA and expressed as fold reduction over the nonsilenced control (**, P < 0.001). (Top) Relative infection by amphotropic HIV-1–luciferase reporter in the KD cell compared to the nonsilenced control. The error bars represent the standard deviations among the 3 independent experiments (n.s., nonsignificant). (D) Relative infection of ecotropic HIV-1–luciferase reporter in NIH 3T3 p50 and NudEL KD cells compared to the nonsilenced control. The error bars represent the standard deviations among the 3 independent experiments (*, P < 0.05).
To corroborate that the KD of p50 and NudEL does not have an effect on the membrane localization of the ecotropic MLV receptor (mCAT-1), we analyzed the subcellular localization of transiently expressed mCAT1-HA protein. We did not observe any significant difference in the localization of mCAT1-HA when expressed in nonsilenced cells and p50 or NudEL KD cells (Fig. 8B). We also verified that the endogenous mCAT-1 was working properly as a viral receptor in the p50 and NudEL cells by infecting them with HIV-1 pseudotyped with the ecotropic envelope of MLV. First, we verified that in human cells where p50 and NudEL were knocked down (Fig. 8C, bottom), the infectivity of an amphotropic HIV-1–luciferase was not significantly affected (Fig. 8C, top); this allowed us to use HIV-1 to avoid problems in fusion or receptor-trafficking defects on the KD cells. We observed a moderate reduction in HIV-1 infection in all the cell lines (Fig. 8D), and although it was statistically significant in one of the two lines for each set of KD cells, it was not as striking as for MLV infection. Thus, since HIV-1 with the MLV envelope infection is not affected in the mouse cells where p50 and NudEL are knocked down, the function of the receptor is intact, and localization assays show that the receptor is at the membrane on those cells.
DISCUSSION
Here, we identified cellular proteins that associate with the incoming MLV preintegration complex using a modified replication-competent Moloney MLV with an epitope-tagged integrase protein. After several attempts to tag the integrase at different locations and with different tags, we found a modification that did not affect the capacity of the virus to replicate and thus allowed us to IP the incoming PIC and its associated proteins after virus infection. We found specifically associated proteins that suggested a direct interaction between incoming viral particles and the microtubule retrograde transport machinery. We screened for a requirement for those proteins and others that participate in the retrograde transport along microtubules and observed that p50 and NudEL were essential for MLV infection. By infecting cell lines in which the p50 or NudEL protein was depleted and analyzing for the presence of different viral DNA intermediates, we demonstrated that these cellular proteins are required in a replication step that follows reverse transcription and precedes nuclear entry, corresponding to the step of retrograde transport of the PIC and its subsequent association with mitotic chromosomes.
Establishing which host proteins are required for retrovirus infection has been a longstanding challenge in retrovirology, and many screens have been performed using yeast two-hybrid procedures with viral proteins as bait (6–8), by biochemical assays for copurifying cellular proteins (9), and by functional tests of cDNA expression libraries (10, 11) or knockdown libraries (12–14) for effects on infection. The screens have helped to reveal the roles of many cellular proteins that are required for retroviral infection, as well as proteins that block or restrict infection. Our screen uncovered two particular factors, the dynein intermediate chain and dynamitin. These two proteins are part of the microtubule retrograde transport system regulated by the cytoplasmic dynein complex. Because retroviruses move toward the nucleus in the early phase of infection and MLV requires microtubules for this retrograde transport (20), it is logical to propose that the process involves the dynein complex. The requirement for dynein function in MLV infection has not been previously demonstrated, and cellular proteins that can act as adaptors to couple the MLV PIC to the dynein complex have not been identified. Our findings are consistent with studies of other retroviruses. It has been shown that retrograde transport of other retroviruses, such as BIV and prototype foamy virus (PFV), is mediated by direct binding to a specific dynein light chain (21, 39). Other work has shown that transient KD of the dynein heavy chain or the use of dynein ATPase activity inhibitors reduced trafficking and uncoating of HIV-1 (22, 40), indicating that a requirement for dynein is common to many retroviruses.
We also demonstrated that the dynein intermediate chain is partially required for MLV infection. Considering that dynactin interacts with the dynein complex via p150 and Dync1i2 and that NudEL also directly interacts with the dynein intermediate chain (41, 42), we can attribute the 50% reduction of MLV infection in Dync1i2 KD cells to disruption of this interaction. It is possible that in Dync1i2 KD cells the quantities of dynein-dynactin and dynein-NudEL are reduced and that this affects the probability of virus attachment to the motor complex. It is also possible that the KD of Dync1i2 affects the binding of light chains to the dynein complex.
To address the possibility of off-target effects in KD experiments, it would be desirable to express an shRNA-resistant form of the KD protein and to reverse the KD phenotype. We attempted to complement the KD cells with an shRNA-resistant version of p50, but without success. Others have found that the overexpression of p50 disassembles the dynactin complex and eliminates dynein activity, which ultimately results in cell death (43). It is likely that overexpression of p50 in the KD cells similarly acted in a dominant-negative manner. Similar results were obtained with NudEL.
Two major reasons can be proposed for why two different dynein regulators are required for MLV infection. First, it is possible that MLV PICs coexist in several forms containing different components, that they dynamically change during retrograde transport, and that they need to interact with the dynein regulators in succession. Second, MLV PICs may also associate with different cellular proteins according to the phase of the cell cycle: for example, p50 could act during interphase as the cargo adaptor for association of the PIC with microtubules, whereas p50 and NudEL may be required during mitosis to associate the PIC with the mitotic chromosome and to position the dynein complex at kinetochores (35, 36, 44). Since MLV requires the breakdown of the nuclear envelope for nuclear entry, the protein could help first to bring the PIC into close proximity with the nucleus and subsequently to associate it with mitotic chromosomes via p12 (45). After attachment to the chromosomes, MLV presumably can remain inside the new nucleus and proceed to integrate its genome into the host DNA.
A particularly striking observation is that NudEL and p50 silencing drastically affect ecotropic MLV reporters but have only a partial effect (p50) or no effect (NudEL) on amphotropic MLV-luciferase. The ecotropic enveloped MLV is endocytosed, and the virion is released to the cytoplasm after acidification of the endosome (20); the amphotropic enveloped MLV is endocytosed via caveola-mediated coated vesicles and macropinocytosis and does not require acidification for its release to the cytoplasm (17, 33). One possible explanation for this observation is that the dynein regulators do not actually interact with the virus but rather with the vesicles that transport it. Since the endocytic pathway is different for viruses entering the cell with different envelopes, the machinery used for their transport is different and is therefore differentially affected. Another possible explanation is that viral particles are released at different locations in the cytoplasm, from which they require further transport along microtubules to arrive at their final destination. A third possibility is that the acidification of the endosome is somehow sensed by the viral core, affecting the conformation of the viral proteins and allowing them to interact differently with cellular proteins than the cores derived from the amphotropic enveloped viruses, which do not undergo acidification. The requirement for acidification for a viral core to interact with the dynein complex has been shown for nonenveloped viruses (46), but whether this phenomenon occurs in enveloped viruses remains to be determined. A final possibility is that the knocked down cells that were generated had reduced expression of the ecotropic receptor (mCAT1) at the cell surface. We observed that in p50 and NudEL KD cells, MoMLV was able to carry on reverse transcription (Fig. 8) at levels similar to those in the nonsilenced control cell line, indicating that entry was normal in the KD cells. We have also tested the localization of an overexpressed receptor in the nonsilenced and KD cells; we observed the receptor at the plasma membrane, and no obvious difference between the control and KD cells was detected (Fig. 8B). Furthermore, we tested the infection of an ecotropic HIV-1–luciferase in the KD cells and observed only a small reduction in infection, which does not account for the huge deficit in MLV infection. These observations suggest that there is no defect in the receptor levels or endocytosis. Moreover, they reveal that p50 and NudEL are essential for MLV infection and not for HIV-1 infection.
ACKNOWLEDGMENTS
We thank Andrea Garces, Tomas Schwenke, and Felipe Barraza for technical help.
R.V.-T., S.P.G., and G.A. designed the research; R.V.-T., T.O., J.C., and G.A. performed the research; R.V.-T., T.O., J.C., S.P.G., and G.A. analyzed the data; and G.A. wrote the paper. We all reviewed and edited the manuscript.
REFERENCES
- 1.Luby-Phelps K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol 192:189–221. [DOI] [PubMed] [Google Scholar]
- 2.Campbell EM, Hope TJ. 2008. Live cell imaging of the HIV-1 life cycle. Trends Microbiol 16:580–587. doi: 10.1016/j.tim.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. 2002. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desfarges S, Salin B, Calmels C, Andreola ML, Parissi V, Fournier M. 2009. HIV-1 integrase trafficking in S. cerevisiae: a useful model to dissect the microtubule network involvement of viral protein nuclear import. Yeast 26:39–54. doi: 10.1002/yea.1651. [DOI] [PubMed] [Google Scholar]
- 6.Leung J, Yueh A, Appah FS Jr, Yuan B, de los Santos K, Goff SP. 2006. Interaction of Moloney murine leukemia virus matrix protein with IQGAP. EMBO J 25:2155–2166. doi: 10.1038/sj.emboj.7601097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Studamire B, Goff SP. 2008. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology 5:48. doi: 10.1186/1742-4690-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yueh A, Leung J, Bhattacharyya S, Perrone LA, de los Santos K, Pu SY, Goff SP. 2006. Interaction of Moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J Virol 80:342–352. doi: 10.1128/JVI.80.1.342-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghavendra NK, Shkriabai N, Graham R, Hess S, Kvaratskhelia M, Wu L. 2010. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology 7:66. doi: 10.1186/1742-4690-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. 2007. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. EMBO J 26:41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valente ST, Gilmartin GM, Mott C, Falkard B, Goff SP. 2009. Inhibition of HIV-1 replication by eIF3f. Proc Natl Acad Sci U S A 106:4071–4078. doi: 10.1073/pnas.0900557106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 13.Gallo DE, Hope TJ. 2012. Knockdown of MAP4 and DNAL1 produces a post-fusion and pre-nuclear translocation impairment in HIV-1 replication. Virology 422:13–21. doi: 10.1016/j.virol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katen LJ, Januszeski MM, Anderson WF, Hasenkrug KJ, Evans LH. 2001. Infectious entry by amphotropic as well as ecotropic murine leukemia viruses occurs through an endocytic pathway. J Virol 75:5018–5026. doi: 10.1128/JVI.75.11.5018-5026.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cote M, Zheng YM, Liu SL. 2012. Membrane fusion and cell entry of XMRV are pH-independent and modulated by the envelope glycoprotein's cytoplasmic tail. PLoS One 7:e33734. doi: 10.1371/journal.pone.0033734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen I, Vilhardt F. 2015. Macropinocytosis is the entry mechanism of amphotropic murine leukemia virus. J Virol 89:1851–1866. doi: 10.1128/JVI.02343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merino-Gracia J, Garcia-Mayoral MF, Rodriguez-Crespo I. 2011. The association of viral proteins with host cell dynein components during virus infection. FEBS J 278:2997–3011. doi: 10.1111/j.1742-4658.2011.08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopold PL, Pfister KK. 2006. Viral strategies for intracellular trafficking: motors and microtubules. Traffic 7:516–523. doi: 10.1111/j.1600-0854.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- 20.Kizhatil K, Albritton LM. 1997. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J Virol 71:7145–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y, Qiao W, Guo T, Tan J, Li Z, Chen Y, Li X, Li Y, Zhou J, Chen Q. 2010. Microtubule-dependent retrograde transport of bovine immunodeficiency virus. Cell Microbiol 12:1098–1107. doi: 10.1111/j.1462-5822.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 22.Pawlica P, Berthoux L. 2014. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses 6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kardon JR, Vale RD. 2009. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greber UF, Way M. 2006. A superhighway to virus infection. Cell 124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 26.Kelkar S, De BP, Gao G, Wilson JM, Crystal RG, Leopold PL. 2006. A common mechanism for cytoplasmic dynein-dependent microtubule binding shared among adeno-associated virus and adenovirus serotypes. J Virol 80:7781–7785. doi: 10.1128/JVI.00481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yueh A, Goff SP. 2003. Phosphorylated serine residues and an arginine-rich domain of the Moloney murine leukemia virus p12 protein are required for early events of viral infection. J Virol 77:1820–1829. doi: 10.1128/JVI.77.3.1820-1829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arad U. 1998. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques 24:760–762. [DOI] [PubMed] [Google Scholar]
- 29.Fontenot DR, den Hollander P, Vela EM, Newman R, Sastry JK, Kumar R. 2007. Dynein light chain 1 peptide inhibits human immunodeficiency virus infection in eukaryotic cells. Biochem Biophys Res Commun 363:901–907. doi: 10.1016/j.bbrc.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 30.Petit C, Giron ML, Tobaly-Tapiero J, Bittoun P, Real E, Jacob Y, Tordo N, De The H, Saib A. 2003. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J Cell Sci 116:3433–3442. doi: 10.1242/jcs.00613. [DOI] [PubMed] [Google Scholar]
- 31.Allan VJ. 2011. Cytoplasmic dynein. Biochem Soc Trans 39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 32.Bader JR, Vaughan KT. 2010. Dynein at the kinetochore: timing, interactions and functions. Semin Cell Dev Biol 21:269–275. doi: 10.1016/j.semcdb.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beer C, Andersen DS, Rojek A, Pedersen L. 2005. Caveola-dependent endocytic entry of amphotropic murine leukemia virus. J Virol 79:10776–10787. doi: 10.1128/JVI.79.16.10776-10787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engelke MF, Burckhardt CJ, Morf MK, Greber UF. 2011. The dynactin complex enhances the speed of microtubule-dependent motions of adenovirus both towards and away from the nucleus. Viruses 3:233–253. doi: 10.3390/v3030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Y, Yu W, Li Y, Yu L, Zhang Q, Wang F, Yang Z, Du J, Huang Q, Yao X, Zhu X. 2007. NudEl modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol Biol Cell 18:2656–2666. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stehman SA, Chen Y, McKenney RJ, Vallee RB. 2007. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol 178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Ketcham SA, Schön A, Goodman B, Wang Y, Yates J, Freire E, Schroer TA, Zheng Y. 2013. NudEl/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell 24:3522–3533. doi: 10.1091/mbc.E13-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y, Yu W, Li Y, Yang Z, Yan X, Huang Q, Zhu X. 2004. NudEl functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J Cell Biol 164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saib A, Puvion-Dutilleul F, Schmid M, Peries J, de The H. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J Virol 71:1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. 2014. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol 88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenney RJ, Weil SJ, Scherer J, Vallee RB. 2011. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem 286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaughan K, Vallee R. 1995. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150 glued. J Cell Biol 131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. 1997. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol 139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. 1990. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- 45.Prizan-Ravid A, Elis E, Laham-Karam N, Selig S, Ehrlich M, Bacharach E. 2010. The Gag cleavage product, p12, is a functional constituent of the murine leukemia virus pre-integration complex. PLoS Pathog 6:e1001183. doi: 10.1371/journal.ppat.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scherer J, Vallee RB. 2011. Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to pH-primed hexon. Viruses 3:1417–1431. doi: 10.3390/v3081417. [DOI] [PMC free article] [PubMed] [Google Scholar]