ABSTRACT
Mycoviruses can have a marked effect on natural fungal communities and influence plant health and productivity. However, a comprehensive picture of mycoviral diversity is still lacking. To characterize the viromes of five widely dispersed plant-pathogenic fungi, Colletotrichum truncatum, Macrophomina phaseolina, Diaporthe longicolla, Rhizoctonia solani, and Sclerotinia sclerotiorum, a high-throughput sequencing-based metatranscriptomic approach was used to detect viral sequences. Total RNA and double-stranded RNA (dsRNA) from mycelia and RNA from samples enriched for virus particles were sequenced. Sequence data were assembled de novo, and contigs with predicted amino acid sequence similarities to viruses in the nonredundant protein database were selected. The analysis identified 72 partial or complete genome segments representing 66 previously undescribed mycoviruses. Using primers specific for each viral contig, at least one fungal isolate was identified that contained each virus. The novel mycoviruses showed affinity with 15 distinct lineages: Barnaviridae, Benyviridae, Chrysoviridae, Endornaviridae, Fusariviridae, Hypoviridae, Mononegavirales, Narnaviridae, Ophioviridae, Ourmiavirus, Partitiviridae, Tombusviridae, Totiviridae, Tymoviridae, and Virgaviridae. More than half of the viral sequences were predicted to be members of the Mitovirus genus in the family Narnaviridae, which replicate within mitochondria. Five viral sequences showed strong affinity with three families (Benyviridae, Ophioviridae, and Virgaviridae) that previously contained no mycovirus species. The genomic information provides insight into the diversity and taxonomy of mycoviruses and coevolution of mycoviruses and their fungal hosts.
IMPORTANCE Plant-pathogenic fungi reduce crop yields, which affects food security worldwide. Plant host resistance is considered a sustainable disease management option but may often be incomplete or lacking for some crops to certain fungal pathogens or strains. In addition, the rising issues of fungicide resistance demand alternative strategies to reduce the negative impacts of fungal pathogens. Those fungus-infecting viruses (mycoviruses) that attenuate fungal virulence may be welcome additions for mitigation of plant diseases. By high-throughput sequencing of the RNAs from 275 isolates of five fungal plant pathogens, 66 previously undescribed mycoviruses were identified. In addition to identifying new potential biological control agents, these results expand the grand view of the diversity of mycoviruses and provide possible insights into the importance of intracellular and extracellular transmission in fungus-virus coevolution.
INTRODUCTION
Recent metatranscriptomic and metagenomic studies of animals, fungi, insects, plants, and environmental samples have shown that mycoviruses are ubiquitous in nature (1–10). Analyses of viral metagenomes (i.e., viromes) of environmental samples suggest that the field of virology has discovered less than 1% of the existing viral diversity, and the rate of discovery by metagenomics is higher than those of all other methods combined (11). Yet, the biological and ecological roles that mycoviruses play in multitrophic interactions are largely unknown and complicated by the frequent infection of fungi by multiple mycoviruses (12–15). Better understanding of these interactions will enhance the utility of mycoviruses in improving animal and plant health.
As descriptions of fungal viromes expand, the picture of the origins and evolution of the genomes of fungal viruses become more complete (16). For example, the added information has reinforced the hypothesis that some plant viruses, such as endornaviruses, originated from mycoviruses (17) and added further support to the phylogenetic links between hypoviruses and potyviruses (18). The relationships between viruses infecting plants and fungal plant pathogens suggest the possibility that genetic exchanges between the two groups of viruses could lead to the emergence of novel deleterious plant-infecting viruses, as was reported for plant viruses of the family Amalgaviridae that appear to be the product of recombination between a double-stranded RNA (dsRNA) fungal virus and a negative-stranded RNA (nsRNA) virus (19). In addition, the discoveries of negative-stranded RNA viruses (20, 21) and single-stranded circular DNA viruses (22–24) have broadened the definition of what constitutes a mycovirus beyond dsRNA elements.
Many mycoviruses have been identified that reduce the virulence of fungal plant pathogens, but with the exception of Cryphonectria hypovirus 1 (CHV1), which infects and reduces the virulence of Cryphonectria parasitica, the causal agent of chestnut blight, few have been developed into effective biological control agents (25). Since capsidless mycoviruses are thought to be spread primarily through fungal anastomosis and within spores, differences in vegetative compatibility constrain the spread of mycovirus infections, which has been the primary limiting factor to the use of CHV1 for control of chestnut blight in North America (26). However, the first example of extracellular transmission of an encapsidated single-stranded circular DNA mycovirus of Sclerotinia sclerotiorum was recently reported (27), suggesting that other mycoviruses capable of extracellular transmission could exist. It is also possible that a virus capable of extracellular transmission could potentiate the transmission of a capsidless virus by transencapsidation, as has been reported for capsidless and encapsidated viruses coinfecting Rosellinia necatrix (15). In addition, coat protein (CP)-mediated transmission of plant viruses in the families Ophioviridae and Virgaviridae by internalization of virus particles within zoospores (28) suggests that mechanisms for the internalization of encapsidated viruses are not uncommon and may represent means to overcome the limitations on horizontal transmission that are characteristic of capsidless mycoviruses.
Given the numerous examples of mycoviruses capable of attenuating virulence in fungal pathogens, mycoviruses represent promising tools for the development of strategies to limit the impact of fungi on plant productivity. Mycoviruses also have potential application in human health for the control of fungicide-resistant human pathogens, which can produce mycoses in immunocompromised patients (29, 30). Analogous strategies are being investigated to control bacterial infections in humans using bacteriophages (31, 32). To overcome resistance to bacteriophage that frequently develops in bacteria, the use of antimicrobial proteins expressed by bacteriophages (e.g., endolysins) has been examined for control of human bacterial diseases (33–35). It may be possible to use individual proteins expressed by mycoviruses for control of fungal diseases, such as viral suppressors of RNA silencing that disrupt fungal small RNA metabolism and gene expression (14, 36).
Previously, we used a metatranscriptomics approach to characterize fungal viromes associated with field-grown soybean plants and identified genomes of at least 22 mycoviruses (37). In this study, we used a similar approach to sequence total RNA directly from 275 fungal isolates of five prevalent plant-pathogenic fungal species, Colletotrichum truncatum, Diaporthe longicolla, Macrophomina phaseolina, Rhizoctonia solani, and Sclerotinia sclerotiorum, which all have wide host ranges and the potential to significantly reduce crop yields. The genomic information provides insight into the diversity and taxonomy of mycoviruses and coevolution of mycoviruses and their fungal hosts.
MATERIALS AND METHODS
Fungal isolates, growth conditions and RNA extraction.
A collection of fungal isolates, including, 2 C. truncatum, 48 M. phaseolina, 3 D. longicolla, 84 R. solani, and 138 S. sclerotiorum isolates from various crop plants and locations in North America, maintained by the authors, was used in this research. Cultures were maintained and mycelia were pulverized as described previously (38). Total RNAs were extracted from fungal cultures using the RNeasy minikit (Qiagen, Valentia, CA) and depleted of rRNA with the Ribo-Zero plant kit (Epicentre, Madison, WI). Single-end or paired-end sequencing libraries were prepared from the rRNA-depleted samples using the ScriptSeq RNA-Seq library preparation kit (Illumina, San Diego, CA) and sequenced on an Illumina HiSeq2500 at the W. M. Keck Center, University of Illinois. Library 1 was prepared from total RNA of 2 C. truncatum, 2 M. phaseolina, 3 D. longicolla, 6 R. solani, and 12 S. sclerotiorum isolates. Library 2 was prepared from total RNA of 1 M. phaseolina isolate and 14 S. sclerotiorum isolates. Library 3 was prepared from total RNA of 64 S. sclerotiorum isolates. Library 4 was prepared from total RNA of 23 S. sclerotiorum isolates. Library 5 was prepared from total RNA of 45 M. phaseolina, 78 R. solani, and 16 S. sclerotiorum isolates. Library 6 was prepared from total RNA extracted from partially purified virus particles and double-stranded RNA (dsRNA). Virus particles and dsRNAs were extracted from a subset of M. phaseolina, R. solani, and S. sclerotiorum cultures that had been shown to be infected with mycoviruses from the first four libraries. Fractions enriched for virus particles were prepared by pulverizing 0.3 to 0.5 g mycelia in liquid nitrogen using a Mini Bead Beater (BioSpec, Bartlesville, OK). To each ground myceliuml sample, 1 ml of 0.5 M Na3PO4 (pH 7.0) buffer was added, and the mixture was allowed to incubate on ice for 30 min with occasional mixing. The sample then was centrifuged at 10,000 × g for 20 min. The supernatant was recovered and centrifuged at 50,000 rpm in a Beckman Ti71 rotor for 2 h. RNA was extracted from the pellet using the RNeasy minikit. Double-stranded RNAs were extracted from a subset of cultures as previously described (38). A sequencing library was prepared as described above, but without rRNA depletion.
Bioinformatic analyses.
Reads from each library were assembled de novo with Trinity (39) with kmers of 25 and 30. Assembled contigs were compared to a customized database that combined predicted proteomes from M. phaseolina (40), D. longicolla (41), R. solani, (http://rsolani.org), and S. sclerotiorum (42) and the NCBI viral reference amino acid sequence database (1 October 2015 release) using USEARCH (43). Hits annotated with “virus” or “viral” were retrieved and compared to the NCBI “non-redundant” (nr) database using BLASTX. The numbers of reads aligning to and depths of coverage for each sequence in each library were calculated using Bowtie (44). Sequences were aligned using MUSCLE (45), and neighbor-joining trees were constructed using MEGA 6.0 (46).
qRT-PCR and RACE.
Infection of individual fungal isolates with the new viruses was confirmed using virus-specific primers in quantitative reverse transcriptase PCR (qRT-PCR) as described previously (38). The origin and host information for fungal isolates shown to be infected with one or more of the identified viruses are presented in Table S2 in the supplemental material. For the tobamo-like virus from M. phaseolina that was similar to an endogenous virus of the fungus Eutypa lata UCREL1, PCR without a reverse transcription step using the same primer set as the qRT-PCR confirmed that it was not an endogenous virus. To complete the sequences of the tobamo-like and tombus-like virus genomes, the 5′- and 3′-terminal sequences were determined using the FirstChoice RLM-RACE kit (Thermo Fisher Scientific, Waltham, MA) and SMARTer RACE 5′/3′ kit (Clonetech Laboratories, Mountain View, CA), with and without the addition of poly(A) tails using Escherichia coli poly(A) polymerase (New England BioLabs, Waltham, MA). For the tobamo-like virus from M. phaseolina, primers 315R (5′-TCTTCTAGGGCACTACTCCACAGA-3′) and 264R (5′-GAAAATTCAGGACCATCACGCACA-3′) were used for 5′ random amplification of cDNA ends (RACE) as outer and inner primers, respectively. Primers 14391F (5′-ACATGTCAAGTCAAGAAGGGAGCA-3′) and 14604F (5′-GGTTACCTTCCATCCCCTCTCTTC-3′) were used for 3′ RACE as outer and inner primers, respectively.
Nucleotide sequence accession numbers.
Sequence data are available in the National Center for Biotechnology Information Short Read Archive (SRA) under accession number SRP074514 and in GenBank under the accession numbers given in Table 1.
TABLE 1.
Assembled sequences with similarity to those of previously described viruses
| Name | Accession no. | Contig length (nt) | Best match | % aa identity | Genome type | Family or genus |
|---|---|---|---|---|---|---|
| Sclerotinia sclerotiorum negative-stranded RNA virus 2 | KP900931 | 9,608 | Sclerotinia sclerotiorum negative-stranded RNA virus 1 | 32 | ss(−)RNA | Mononegavirales |
| Sclerotinia sclerotiorum negative-stranded RNA virus 3 | KC601997 | 10,009 | Sclerotinia sclerotiorum negative-stranded RNA virus 1 | 77 | ss(−)RNA | Mononegavirales |
| Sclerotinia sclerotiorum negative-stranded RNA virus 4 | KP900930 | 9,707 | Sclerotinia sclerotiorum negative-stranded RNA virus 1 | 32 | ss(−)RNA | Mononegavirales |
| Sclerotinia sclerotiorum negative-stranded RNA virus 5 | KF913892 | 7,882 | Sandfly fever Naples virus | 25 | ss(−)RNA | Bunyaviridae |
| Rhizoctonia solani negative-stranded RNA virus 1 | KP900919 | 5,993 | Lettuce ring necrosis virus | 27 | ss(−)RNA | Ophioviridae |
| Rhizoctonia solani negative-stranded RNA virus 2 | KP900920 | 7,335 | Lettuce ring necrosis virus | 28 | ss(−)RNA | Ophioviridae |
| Rhizoctonia solani negative-stranded RNA virus 3 | KP900903 | 7,335 | Lettuce ring necrosis virus | 29 | ss(−)RNA | Ophioviridae |
| Rhizoctonia solani negative-stranded RNA virus 4 | KP900923 | 7,224 | Razdan virus | 23 | ss(−)RNA | Bunyaviridae |
| Macrophomina phaseolina negative-stranded RNA virus 1 | KP900899 | 6,081 | Watermelon silver mottle virus | 24 | ss(−)RNA | Bunyaviridae |
| Sclerotinia sclerotiorum hypovirus 2 Lactuca | KF898354 | 14,580 | Cryphonectria hypovirus 1 | 29 | ss(+)RNA | Hypoviridae |
| Macrophomina phaseolina hypovirus 1 | KP900893 | 12,468 | Cryphonectria hypovirus 1 | 39 | ss(+)RNA | Hypoviridae |
| Rhizoctonia solani endornavirus 2 | KT823701 | 15,849 | Rhizoctonia cerealis endornavirus 1 | 38 | ss(+)RNA | Endornaviridae |
| Sclerotinia sclerotiorum endornavirus 1 Lactuca | KM923990 | 10,619 | Sclerotinia sclerotiorum endornavirus 1 | 95 | ss(+)RNA | Endornaviridae |
| Sclerotinia sclerotiorum endornavirus 2-IL | KU299046 | 10,520 | Sclerotinia sclerotiorum endornavirus 2 | 90 | ss(+)RNA | Endornaviridae |
| Rhizoctonia solani ourmia-like virus 1 | KP900921 | 2,792 | Epirus cherry virus | 34 | ss(+)RNA | Ourmiavirus |
| Rhizoctonia solani ourmia-like virus 2 | KP900922 | 1,901 | Ourmia melon virus | 27 | ss(+)RNA | Ourmiavirus |
| Sclerotinia sclerotiorum ourmia-like virus 1 RNA1 | KP900928 | 3,180 | Epirus cherry virus | 27 | ss(+)RNA | Ourmiavirus |
| Sclerotinia sclerotiorum ourmia-like virus 2 RNA1 | KP900929 | 2,333 | Ourmia melon virus | 30 | ss(+)RNA | Ourmiavirus |
| Macrophomina phaseolina mitovirus 1 | KP900894 | 2,572 | Ophiostoma mitovirus 3a | 42 | ss(+)RNA | Narnaviridae |
| Macrophomina phaseolina mitovirus 2 | KP900895 | 2,445 | Cryphonectria parasitica mitovirus 1 | 32 | ss(+)RNA | Narnaviridae |
| Macrophomina phaseolina mitovirus 3 | KT823703 | 4,179 | Clitocybe odora virus | 41 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 1 | KP900905 | 2,462 | Botrytis cinerea debilitation-related virus | 35 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 2 | KP900911 | 2,865 | Cryphonectria parasitica mitovirus 1 | 39 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 3 | KP900912 | 2,924 | Tuber aestivum mitovirus | 34 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 4 | KP900913 | 2,605 | Botrytis cinerea debilitation-related virus | 36 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 5 | KP900914 | 2,357 | Tuber aestivum mitovirus | 37 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 6 | KP900915 | 2,615 | Ophiostoma mitovirus 6 | 37 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 7 | KP900916 | 3,085 | Cryphonectria parasitica mitovirus 1 | 35 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 8 | KP900917 | 3,225 | Cryphonectria parasitica mitovirus 1 | 33 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 9 | KP900918 | 2,273 | Clitocybe odora virus | 37 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 10 | KP900896 | 2,701 | Sclerotinia sclerotiorum mitovirus 1 | 33 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 11 | KP900906 | 3,283 | Cryphonectria parasitica mitovirus 1 | 35 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 12 | KP900907 | 3,378 | Clitocybe odora virus | 31 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 13 | KP900908 | 3,039 | Clitocybe odora virus | 31 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 14 | KP900909 | 3,219 | Clitocybe odora virus | 46 | ss(+)RNA | Narnaviridae |
| Rhizoctonia solani mitovirus 15 | KP900910 | 3,901 | Clitocybe odora virus | 38 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 5 | KF913880 | 2,871 | Ophiostoma mitovirus 6 | 31 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 6 | KF913881 | 2,535 | Ophiostoma mitovirus 4 | 34 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 7 | KF913882 | 2,648 | Cryphonectria parasitica mitovirus 1 | 41 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 7 IL-1 | KC601999 | 1,959 | Cryphonectria parasitica mitovirus 1 | 43 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 8 | KF913883 | 2,565 | Ophiostoma mitovirus 6 | 38 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 9 | KF913884 | 2,562 | Ophiostoma mitovirus 6 | 43 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 10 | KF913885 | 1,396 | Ophiostoma mitovirus 6 | 41 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 11 | KF913886 | 2,708 | Ophiostoma mitovirus 3a | 32 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 12 | KF913887 | 2,593 | Ophiostoma mitovirus 3a | 45 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 13 | KF913888 | 1,329 | Ophiostoma mitovirus 6 | 41 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 14 | KF913889 | 2,564 | Ophiostoma mitovirus 3a | 42 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 15 | KF913890 | 2,548 | Ophiostoma mitovirus 4 | 34 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 16 | KF913891 | 1,341 | Ophiostoma mitovirus 6 | 33 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 17 | KP900924 | 2,447 | Sclerotinia sclerotiorum mitovirus 1 | 42 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 18 | KP900925 | 2,509 | Sclerotinia sclerotiorum mitovirus 1 | 81 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 22 | KC602000 | 1,659 | Ophiostoma mitovirus 3a | 42 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum mitovirus 23 | KC601998 | 1,554 | Ophiostoma mitovirus 6 | 34 | ss(+)RNA | Narnaviridae |
| Sclerotinia sclerotiorum umbra-like virus 1 | KC601995 | 4,492 | Magnaporthe oryzae RNA virus | 42 | ss(+)RNA | Unclassified |
| Macrophomina phaseolina tobamo-like virus 1 | NC_025674 | 9,673 | Obuda pepper virus | 29 | ss(+)RNA | Virgaviridae |
| Macrophomina phaseolina tobamo-like virus 1a | KP900897 | 7,499 | Thunberg fritillary virus | 27 | ss(+)RNA | Virgaviridae |
| Rhizoctonia solani positive-stranded RNA virus 1 | KT823702 | 3,492 | Bee Macula-like virus | 30 | ss(+)RNA | Tymoviridae |
| Macrophomina phaseolina single-stranded RNA virus 1 | KP900890 | 6,356 | Penicillium roqueforti ssRNA mycovirus 1 | 40 | ss(+)RNA | unclassified |
| Macrophomina phaseolina single-stranded RNA virus 3 | KP900898 | 1,894 | Magnaporthe oryzae RNA virus | 50 | ss(+)RNA | Tombusviridae |
| Rhizoctonia solani barnavirus 1 | KP900904 | 3,915 | Mushroom bacilliform virus | 44 | ss(+)RNA | Barnaviridae |
| Rhizoctonia solani beny-like virus 1 | KP900902 | 1,306 | Beet soilborne mosaic virus | 41 | ss(+)RNA | Benyviridae |
| Macrophomina phaseolina double-stranded RNA virus 2 | KP900891 | 9,188 | Fusarium graminearum dsRNA mycovirus 3 | 35 | dsRNA | unclassified |
| Sclerotinia sclerotiorum double-stranded RNA virus 3 | KU299047 | 5,928 | Ustilago maydis virus H1 | 29 | dsRNA | Totiviridae |
| Diaporthe longicolla totivirus 1 | KP900901 | 4,005 | Coniothyrium minitans RNA virus | 62 | dsRNA | Totiviridae |
| Colletotrichum partitivirus 1 RNA1 | KR074421 | 1,820 | Verticillium dahliae partitivirus 1 | 87 | dsRNA | Partitiviridae |
| Colletotrichum partitivirus 1 RNA2 | KP900885 | 1,535 | Discula destructiva virus 1 | 63 | dsRNA | Partitiviridae |
| Rhizoctonia solani partitivirus 1 RNA1 | KU299048 | 1,810 | Sclerotinia sclerotiorum partitivirus S | 45 | dsRNA | Partitiviridae |
| Rhizoctonia solani partitivirus 1 RNA2 | KP900892 | 1,845 | Rosellinia necatrix partitivirus 7 | 26 | dsRNA | Partitiviridae |
| Macrophomina phaseolina chrysovirus 1 RNA1 | KP900886 | 3,712 | Penicillium chrysogenum virus | 55 | dsRNA | Chrysoviridae |
| Macrophomina phaseolina chrysovirus 1 RNA2 | KP900887 | 2,774 | Penicillium chrysogenum virus | 38 | dsRNA | Chrysoviridae |
| Macrophomina phaseolina chrysovirus 1 RNA3 | KP900888 | 2,927 | Penicillium chrysogenum virus | 24 | dsRNA | Chrysoviridae |
| Macrophomina phaseolina chrysovirus 1 RNA4 | KP900889 | 2,735 | Penicillium chrysogenum virus | 38 | dsRNA | Chrysoviridae |
RESULTS
Metatranscriptomic identification of mycoviruses infecting the selected fungal cultures.
Six sequencing libraries were prepared from rRNA-depleted total RNA, dsRNA, and partially purified virus particles and generated 0.7 × 108 to 3.2 × 108 100-nucleotide (nt) paired-end sequence reads (with four libraries sequenced as paired-end reads and two as single-end reads), with 0.5% to 15.1% of the reads predicted to be of viral origin. From the assembled sequence reads, 2 sequences were identified from C. truncatum (2 isolates), 14 sequences from M. phaseolina (48 isolates), 1 sequence from D. longicolla (3 isolates), 27 sequences from R. solani (84 isolates), and 28 sequences from S. sclerotiorum (138 isolates), whose predicted amino acid sequences showed significant identity to previously described viruses. The depth of coverage of the assembled sequences ranged from 15- to 247,742-fold with an average of 20,687-fold coverage for all sequences and an average of 7,216-fold coverage for nonmitovirus sequences. Of the 72 sequences, 62 had at least 50-fold coverage (see Table S1 in the supplemental material). The numbers of reads that aligned with each of the assembled contigs represented an unbiased estimate of abundance of the prevalence of each virus in the pooled RNA sample. The putative viral genomes showed affinity with 15 distinct lineages, including Barnaviridae, Benyviridae, Chrysoviridae, Endornaviridae, Fusariviridae, Hypoviridae, Mononegavirales, Narnaviridae, Ophioviridae, Ourmiavirus, Partitiviridae, Tombusviridae, Totiviridae, Tymoviridae, and Virgaviridae (Table 1). Among the 72 putative mycovirus sequences identified, 5 showed strong affinity with three families (Benyviridae, Ophioviridae, and Virgaviridae) that previously contained no mycovirus species. Some viruses were detected in single fungal isolates, while others infected multiple isolates (see Table S1). The majority (73%) of the putative virus sequences were predicted to represent single-stranded positive-sense RNA [ss(+)RNA] genomes: 49% were related to virus that replicate within mitochondria, and 25% were related to cytoplasmic viruses. Of the remaining sequences, 15% were predicted to represent dsRNA viruses and 12% were predicted to have negative-sense RNA (nsRNA) genomes, a class of fungal viruses that was only recently described (20). In one library (BN64), 30% of the viral sequences were derived from an ourmia-like virus. As previously reported (47–49), many of the mycovirus-infected isolates did not display abnormal growth phenotypes in culture. For example, greater than 90% of the S. sclerotiorum isolates analyzed produced sclerotia, survival structures needed for overwintering (data not shown).
Sequences related to members of the Mononegavirales.
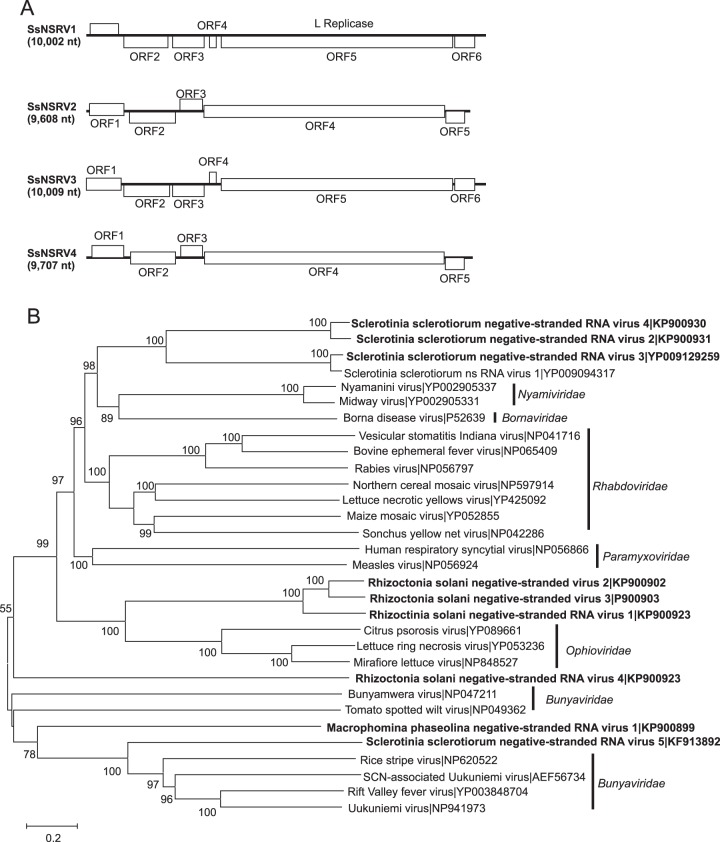
Nine sequences showed similarity to nsRNA viruses in the order Mononegavirales, which contains eight families of viruses with nsRNA genomes of 8.9 to 19 kb (50). One sequence was identified from M. phaseolina, five from R. solani, and four from S. sclerotiorum (Fig. 1). The assembled sequences ranged in length from 2,422 to 10,009 nt and contained from one to six open reading frames (ORFs). Sequences of Macrophomina phaseolina negative-stranded RNA virus 1 (MpNSRV1; KP900899 [6,081 nt]), Rhizoctonia solani negative-stranded RNA virus 1 (RsNSRV1; KP900919 [5,993 nt]), Rhizoctonia solani negative-stranded RNA virus 2 (RsNSRV2; KP900920 [7,335 nt]), Rhizoctonia solani negative-stranded RNA virus 3 (RsNSRV3; KP900903 [7,335 nt]), Rhizoctonia solani negative-stranded RNA virus 4 (RsNSRV4; KP900923 [7,224 nt]), and Sclerotinia sclerotiorum negative-stranded RNA virus 5 (SsNSRV5; KF913892 [7,882 nt]) each contained a single large ORF and grouped phylogenetically with families within the Mononegavirales with segmented genomes. The predicted amino acid sequences of RsNSRV1, RsNSRV2, and RsNSRV3 were most similar to those of the L proteins encoded by RNA1 of Lettuce ring necrosis virus (NC_006051 [7,651 nt]) and other members of the family Ophioviridae (Fig. 1A). The sizes of RNA1 segments of members of the Ophioviridae ranged from 7,651 to 8,186 nt, suggesting the assembled sequences represented at least 73.2% coverage of the corresponding genome segments. Because the three sequences were 5′ incomplete (relative to the coding strand), the sequences did not contain the small ORF upstream of the L protein-encoding ORF observed in members of the Ophioviridae.
FIG 1.
Genome organizations and phylogenetic relationships of putative negative-stranded RNA virus genomes detected from Macrophomina phaseolina, Rhizoctonia solani, and Sclerotinia sclerotiorum. (A) Comparison of the organizations of putative monopartite negative-stranded RNA viruses Sclerotinia sclerotiorum negative-stranded RNA virus 2 (SsNSRV2), Sclerotinia sclerotiorum negative-stranded RNA virus 3 (SsNSRV3), and Sclerotinia sclerotiorum negative-stranded RNA virus 4 (SsNSRV4) to Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV1). Open reading frames (ORFs) are shown as boxes and staggered to indicate the reading frame. (B) Maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of SlaMyfV1 and other confirmed and proposed members of the order Mononegavirales. Predicted RdRp amino acid sequences were aligned with MUSCLE, and trees were inferred using MEGA6. Branch lengths are scaled to the expected underlying number of amino acid substitutions per site. Bootstrap percentages greater than 50% are shown.
The sequences designated Sclerotinia sclerotiorum negative-stranded RNA virus 2 (SsNSRV2; KP900931 [9,608 nt]), Sclerotinia sclerotiorum negative-stranded RNA virus 3 (SsNSRV3; KC601997 [10,009 nt]), and Sclerotinia sclerotiorum negative-stranded RNA virus 4 (SsNSRV4; KP900930 [9,707 nt]) contained five or six ORFs (Fig. 1A). The products of the largest ORFs were related to L proteins of families within the Mononegavirales that all contained nonsegmented genomes. The predicted amino acid sequences of the RNA-dependent RNA polymerases (RdRps; L proteins) of the three viruses were most similar to that of Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV1; NC_025383 [10,002 nt]), the type member in the recently established family Mymonaviridae (20, 50). Upstream sequence elements required for transcription termination were identified for each viral genome. The genome of SsNSRV2 contained five ORFs with the sequence 5′-AGATTTAAGAAAAACC-3′ an average of 44 nt upstream of ORFs 2, 3, 4 (which encodes L protein), and 5. In the case of SsNSRV3, six ORFs were identified, each of which had the sequence 5′-TTATTTAATAAAACTTAGGA-3′ an average of 32 nt upstream of ORFs 2, 3, 4, 5 (which encodes L protein), and 6. SsNSRV4 contained five ORFs with the sequence 5′-ATTTAAGAAAAACC-3′ an average of 40 nt upstream of ORFs 2, 3, 4 (which encodes L protein), and 5. Similarly, conserved sequences were reported in the genomes of other nonsegmented members of the Mononegavirales: these sequences serve as polyadenylation signals terminating each cistron, signaling the start of the next mRNA species, and templates for capping complexes (51). Unlike the other nonsegmented negative-stranded RNA viruses, the sequences of SsNSRV1, SsNSRV2, SsNSRV3, and SsNSRV4 contained an ORF downstream of the ORF encoding the L protein. The presence of a transcription termination initiation sequence upstream of the ORF suggests that it is expressed from a separate mRNA and not as a fusion with the product of the L protein-encoding ORF. None of the products of the ORFs upstream or downstream of the L protein-encoding ORF showed significant similarity to previously described proteins. The sequences putatively derived from segmented nsRNA viruses grouped with members of the Bunyaviridae but did not show close affinity with any established genera. Similarly, the sequences putatively derived from nonsegmented nsRNA viruses were most closely related to members of the Bornaviridae and Nyamiviridae, but differences in genome organizations and amino acid sequences suggest that the viruses represent a distinct family within the Mononegavirales.
Fusariviridae- and Hypoviridae-related sequences.
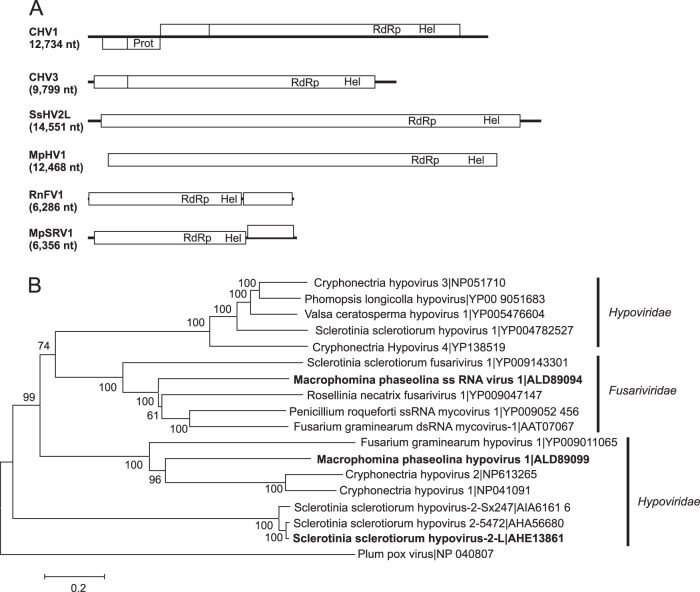
Two large (>12-kb) viral genome sequences showed similarity to those of members of the family Hypoviridae, one from M. phaseolina (Macrophomina phaseolina hypovirus 1 [MpHV1]; KP900893 [12,468 nt]) and one from S. sclerotiorum (Sclerotinia sclerotiorum hypovirus 2 Lactuca [SsHV2L]; KF898354 [14,580 nt]) (Fig. 2). Viruses in this family typically have ss(+)RNA genomes of 9 to 13 kb with one or two ORFs (52). The hypovirus from M. phaseolina was initially identified in two contigs of approximately 7,000 nt each, but multi-kmer and multilibrary assemblies showed that they were in fact part of the same genome, resulting in a final contig length of 12,468 nt. Both genomes contained one large ORF encoding putative polyproteins with RdRp and helicase domains similar to those of members of the family Hypoviridae. However, the sequence of the large ORF of MpHV1 was 5′ and 3′ incomplete. In a separate study, we further characterized SsHV2L, a naturally occurring recombinant strain of Sclerotinia sclerotiorum hypovirus 2 from North America, by determining its complete nucleotide sequence and constructing an infectious clone of the virus (38). Alignments of the predicted amino acid sequences showed that they share from 9 to 29% amino acid sequence identity with previously reported members of the Hypoviridae.
FIG 2.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of assembled sequences from Macrophomina phaseolina and Sclerotinia sclerotiorum with similarities to members of the Fusariviridae or Hypoviridae. (A) Comparisons of the organizations of Cryphonectria hypovirus 1 (CHV1), Cryphonectria hypovirus 3 (CHV3), Macrophomina phaseolina hypovirus 1 (MpHV1), Macrophomina phaseolina single-stranded RNA virus 1 (MpSRV1), Rosellinia necatrix fusarivirus 1 (RnFV1), and Sclerotinia sclerotiorum hypovirus 2-L (SsHV2L). Open reading frames (ORFs) are shown as boxes and staggered to indicate the reading frame. (B) For the maximum likelihood tree, predicted RdRp amino acid sequences were aligned, and phylogenetic trees were constructed as described in the legend to Fig. 1. The Plum pox virus RdRp amino acid sequence was used as an outgroup to root the tree.
The sequence designated Macrophomina phaseolina single-stranded RNA virus 1 (MpSRV1; KP900890 [6,356 nt]) was most similar to that of Penicillium roqueforti ssRNA mycovirus 1 (NC_024699 [6,002 nt]) and other viruses in the recently proposed Fusariviridae family (53). The genomes of most viruses in the Fusariviridae family contain two ORFs, the first of which expresses replicases that are similar to those of members of the Hypoviridae and have genome sizes that range from 6,002 to 6,621 nt (53), suggesting that at 6.3 kb, the MpSRV1 sequence is nearly complete.
Endornaviridae-related sequences.
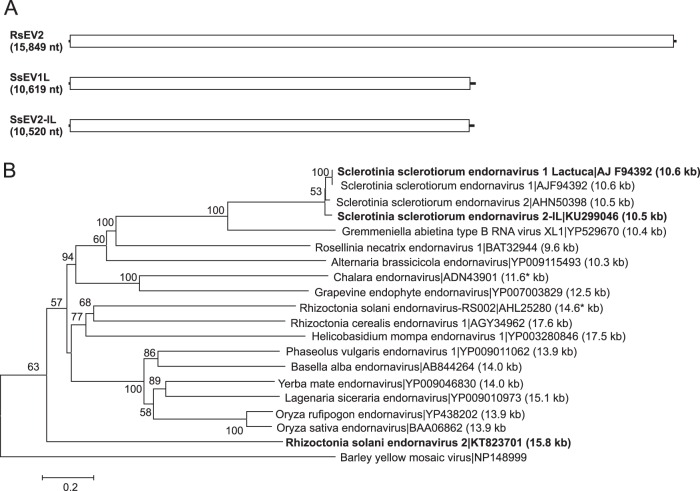
The predicted amino acid sequences of three putative viral genomes were similar to those of viruses in the family Endornaviridae: Rhizoctonia solani endornavirus 2 (RsEV2; KT823701 [15,849 nt]), Sclerotinia sclerotiorum endornavirus 1 Lactuca (SsEV1L; KM923990 [10,619 nt]), and Sclerotinia sclerotiorum endornavirus 2 (SsEV2-IL; KU299046 [10,520 nt]) (Fig. 3). The family Endornaviridae contains a single genus, Endornavirus, members of which have linear ssRNA genomes that range in length from about 10 kb to more than 17 kb and contain a single large ORF (54). The three sequences each contained a single large ORF predicted to encode a polyprotein that included domains for a viral RNA helicase and RdRp. The predicted amino acid sequence of the polyprotein of RsEV2 was most similar to that of Rhizoctonia cerealis endornavirus 1 (KF311065) at 38% identity. The nucleotide sequence of SsEV1L represented greater than 98% genome coverage and was 92% identical to that of Sclerotinia sclerotiorum endornavirus 1 (KC852908) and 81% identical to that of Sclerotinia sclerotiorum endornavirus 2 (KJ123645) (38, 55). The predicted amino acid sequence of the polyprotein of SsEV2-IL was 90% identical to the polyprotein of Sclerotinia sclerotiorum endornavirus 2 (KJ123645). Hence, SsEV1L and SsEV2-IL represents strains of S. sclerotiorum-infecting endornaviruses originally reported from New Zealand (55).
FIG 3.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of contigs assembled from Rhizoctonia solani and Sclerotinia sclerotiorum and confirmed and proposed members of the Endornaviridae. (A) Comparisons of the organizations of Rhizoctonia solani endornavirus 2 (RsEV2), Sclerotinia sclerotiorum endornavirus 1 Lactuca (SsEV1L), and Sclerotinia sclerotiorum endornavirus 2 (SsEV2-IL). (B) Predicted RdRp amino acid sequences were aligned and phylogenetic trees were constructed as described in the legend to Fig. 1. The Barley yellow mosaic virus RdRp amino acid sequence was used as an outgroup to root the tree. Asterisks indicate incomplete sequences.
Narnaviridae and Ourmiavirus-related sequences.
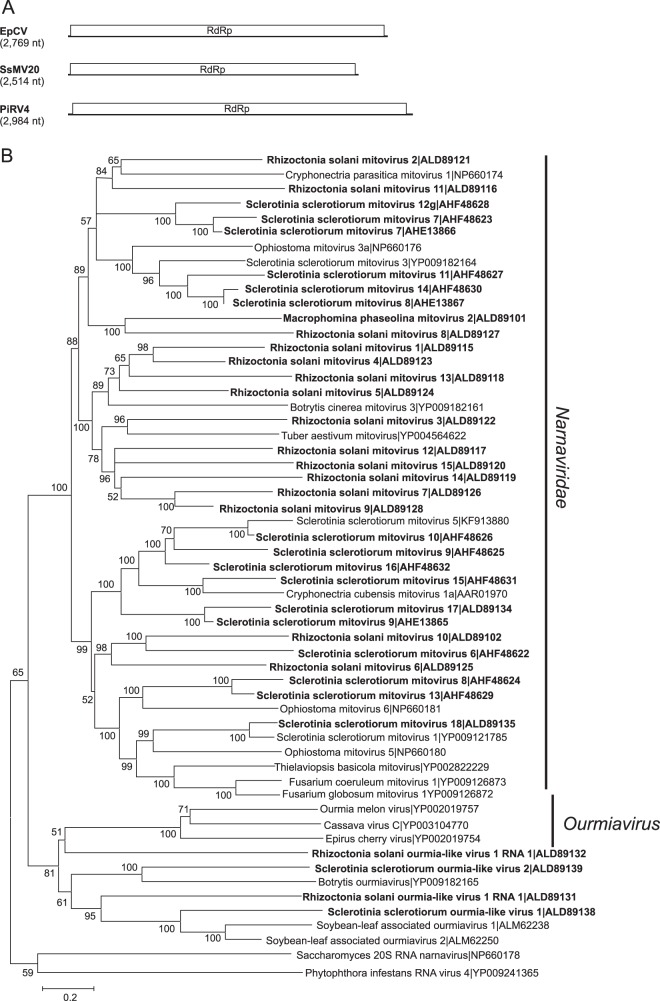
A total of 35 sequences (17 from S. sclerotiorum, 15 from R. solani, and 3 from M. phaseolina) showed similarity to mitoviruses in the family Narnaviridae (Fig. 4). Mitoviruses typically have genome sizes of about 2.3 to 2.7 kb containing a single ORF encoding only the RdRp. Eleven of the putative mitovirus sequences from S. sclerotiorum were at least 2.5 kb and therefore represented nearly complete sequences. In contrast to previously reported mycoviruses, six of the putative mitoviruses from R. solani and M. phaseolina were larger than 3.2 kb. Phylogenetic analysis grouped the sequences into two well-supported clades where sequences were most similar to previously identified mitoviruses from the same fungal host (Fig. 4).
FIG 4.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of contigs assembled from Macrophomina phaseolina, Rhizoctonia solani, and Sclerotinia sclerotiorum with confirmed and proposed members of the family Narnaviridae and genus Ourmiavirus. (A) Comparisons of the organizations of Epirus cherry virus (EpCV), Phytophthora infestans RNA virus 4 (PiRV4), and Sclerotinia sclerotiorum mitovirus 20 (SsMV20). (B) Predicted RdRp amino acid sequences were aligned and phylogenetic trees were constructed as described in the legend to Fig. 1.
Four viral genomes showed similarity to viruses in the genus Ourmiavirus, two from R. solani and two from S. sclerotiorum. Ourmiaviruses have tripartite ssRNA genomes with RNAs of 2.8, 1.1, and 1.0 kb. The largest RNA encodes the viral replicase, which is phylogenetically related to those encoded by the Narnaviridae (56). However, mitoviruses replicate in mitochondria, while ourmiaviruses are thought to replicate in the cytoplasm (100). Sclerotinia sclerotiorum ourmia virus 1 (KP900928 [3,180 nt]) had a nearly complete genome sequence and had the most reads of all the viral contigs (see Table S1 in the supplemental material). The sequences from Sclerotinia sclerotiorum ourmia-like virus 2 (KP900929 [2,333 nt]), Rhizoctonia solani ourmia-like virus 1 (KP900921 [2,792 nt]), and Rhizoctonia solani ourmia-like virus 2 (KP900922 [1,901 nt]) were less complete and represent 59 to 87% of the genomes. Even though the genomes of plant ourmiaviruses typically contain three segments, with the smallest segment encoding the coat protein (CP) (56), contigs encoding proteins similar to ourmiavirus CPs were not identified. All three sequences shared about 30% amino acid sequence identity with cassava virus C, Epirus cherry virus, and Ourmia melon virus.
Putative positive-sense single-stranded extramitochondrial RNA virus genomes.
The predicted amino acid of one contig, Sclerotinia sclerotiorum umbra-like virus 1 (SsULV1; KC601995 [4,492 nt]), showed similarity to members of the family Tombusviridae. The initial contig produced by multi-kmer assembly was 4,277 nt in length, which we extended to 4,347 nt by RACE, indicating that the initial assembly was 98.4% complete. Members of the family Tombusviridae have monopartite genomes composed of one or two 3.7- to 4.8-kb ssRNA segments with up to six ORFs (57). Most commonly, the two 5′-proximal ORFs express the viral replicase by read-through of a termination codon at the end of the first ORF. The ORF encoding the CP is 3′ proximal and expressed from a subgenomic RNA (57). The genome sequence of SsULV1 contained two large ORFs. The second ORF was in the same reading frame as the first ORF and could be translated by suppression of an amber (UAG) termination codon. The predicted amino acid sequence of the second ORF (527 amino acids) was 42% identical to the product of ORF2 of Magnaporthe oryzae RNA virus (NC_026137 [3,246 nt]) and the next hit to Diaporthe ambiqua RNA virus 1 (NC_001278 [4,113 nt]) (Fig. 5). The product of the first ORF was less conserved and did not contain identifiable conserved domains. The genomes of the three fungal viruses related to members of the Tombusviridae had long 5′ or 3′ noncoding regions that contained one or more small reading frames that also failed to show similarity to other sequences in GenBank. ORFs potentially encoding CPs were not identified in the genomes of the three fungal tombus-like viruses. Because the three sequences show similarity to members of the Tombusviridae, they are expected to have a monopartite ss(+)RNA genome that lacks a cap structure and a poly(A) tail. The absence of a poly(A) tail was confirmed in the RACE analysis.
FIG 5.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of an assembled sequence from Sclerotinia sclerotiorum with members of the Tombusviridae. (A) Comparisons of the organizations of carnation mottle virus (CarMV), Diaporthe ambiqua RNA virus 1 (DaRV1), Magnaporthe oryzae RNA virus (MoRV), and Sclerotinia sclerotiorum umbra-like virus 1 (SsULV1). Open reading frames (ORFs) are shown as boxes and staggered to indicate the reading frame. (B) For the maximum likelihood tree, predicted RdRp amino acid sequences were aligned and phylogenetic trees were constructed as described in the legend to Fig. 1.
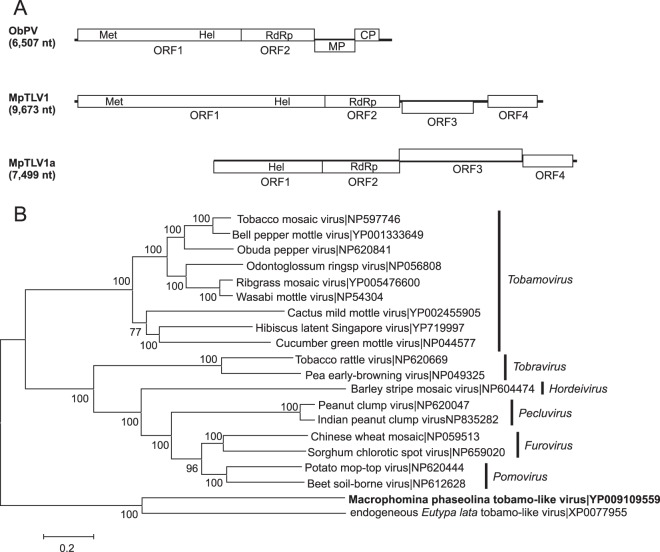
Two sequences were recovered from M. phaseolina with similarity to viruses in the family Virgaviridae. The sequence of one, Macrophomina phaseolina tobamo-like virus 1 (MpTLV1), was nearly complete at 9,497 nt from high-throughput sequencing (HTS) (Fig. 6). The ends of the genome sequence were analyzed by RACE, which extended the sequence to 9,673 nt (NC_025674). However, the MpTLV1 genomic sequence contained a short 3′ poly(A) tract that was detected by four different RACE methods. The sequence likely represents an internal poly(A) tract similar to that in hibiscus latent Singapore virus (58). MpTLV1 was recovered from a highly debilitated M. phaseolina isolate, which was coinfected by a hypovirus (KP900893) and a negative-sense RNA virus (KP900899), described above. The MpTLV1 genome contained four ORFs encoding a methyltransferase/helicase, RdRp, putative movement protein, and coat protein with a gene order and genome organization typical of those of members of the genus Tobamovirus (Fig. 6). Like plant tobamoviruses, MpTLV1 has a short (71-nt) 5′ noncoding region that contains AAC repeats, but at 9.5 kb, the genome is much larger than those of plant tobamoviruses that have ss(+)RNA genomes of 6.3 to 6.6 kb. It was not possible to determine if the genome contained a 3′ tRNA-like structure because of the putative internal poly(A) tract. The predicted masses of the proteins encoded by MpTLV1 were larger than those for plant tobamoviruses: 189 kDa for ORF1 compared to 124 to 132 kDa, 258 kDa for the ORF1-2 read-through protein compared to 181 to 189 kDa, 54 kDda for the putative movement protein compared to 28 to 31 kDa, and 38 kDa for the putative CP compared to 17 to 18 kDa. The predicted amino acid sequence of MpTLV1 was most closely related to a tobamovirus-like genome integrated into the genome of the grapevine dieback fungus Eutypa lata (59). Among nonendogenous tobamoviruses, the predicted amino acid sequence of the MpTLV1 RdRp domain was 29% identical to the corresponding region of Obuda pepper virus (NC_003852 [6,507 nt]). A less complete tobamovirus-related sequence, Macrophomina phaseolina tobamo-like virus 1a (MpTLV1a; KP900897 [7,499 nt]) was recovered from a second M. phaseolina isolate that was nearly identical to MpTLV1, except for the size of the putative movement protein (MP) ORF (Fig. 6). The predicted mass of the protein encoded by MpTLV1 was 54 kDa compared to 80 kDa for MpTLV1a. Comparison of the nucleotide sequences of MpTLV1 and MpTLV1a suggested that the MpTLV1 sequence contained a deletion in the putative MP ORF that also produced a longer noncoding intergenic region in MpTLV1 than in MpTLVa1.
FIG 6.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of sequences assembled from two isolates of Macrophomina phaseolina with members of the Virgaviridae. (A) Comparisons of the genome organizations of Obuda pepper virus (ObPV), Macrophomina phaseolina tobamo-like virus (MpTLV1), and Macrophomina phaseolina tobamo-like virus a (MpTLV1a). Open reading frames (ORFs) are shown as boxes and staggered to indicate the reading frame. (B) For the maximum likelihood tree, predicted RdRp amino acid sequences were aligned and phylogenetic trees were constructed as described in the legend to Fig. 1. Regions encoding the coat protein (CP), helicase (Hel), methyltransferase (Met), movement protein (MP), and RdRp are indicated in the corresponding ORFs.
The predicted amino acid sequence of one contig named Rhizoctonia solani positive-stranded RNA virus 1 (RsPSRV1; KT823702 [3,492 nt]) was 30% identical to the replicase of Bee Macula-like virus (NC_027631 [6,258 nt]) in the order Tymovirales (Fig. 7). Viruses in the family Tymoviridae contain a single ssRNA genome of 6.0 to 7.5 kb (60). The organization of the predicted RsPSRV1 ORFs (one large 5′-proximal ORF and multiple smaller 3′-proximal ORFs) was similar to those of viruses in the Alphaflexiviridae and Betaflexiviridae families (61, 62).
FIG 7.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) of sequences assembled from Rhizoctonia solani with members of the order Tymovirales. (A) Comparisons of the organizations of grapevine fleck virus (GFkV), mushroom bacilliform virus (MBV), Rhizoctonia solani barnavirus 1 (RsBV1), and Rhizoctonia solani positive-stranded RNA virus 1 (RsPSV1). Open reading frames (ORFs) are shown as boxes and staggered to indicate the reading frame. (B) For the maximum likelihood tree, predicted RdRp amino acid sequences were aligned, and phylogenetic trees were constructed as described in the legend to Fig. 1. Regions encoding the coat protein (CP), helicase (Hel), methyltransferase (Met), movement protein (MP), and RdRp are indicated in the corresponding ORFs.
One sequence, Rhizoctonia solani barnavirus 1 (RsBarV1; KP900904 [3,915 nt]), was highly enriched in crude virus preparations and contained three ORFs. The predicted amino acid sequence of the third ORF showed 47% identity to the RdRp of Mushroom bacilliform virus (MBV; NC_001633 [4,009 nt]), the sole member of the family Barnaviridae and genus Barnavirus. The MBV genome is monopartite ss(+)RNA with four major and three minor ORFs (63). The assembled sequence contained ORFs predicted to encode the ORF2 polyprotein, ORF3 RdRp, and ORF4 CP with ORF1 missing, suggesting an incomplete 5′ end for the contig.
The sequence designated Rhizoctonia solani beny-like virus 1 (RsBenV1; KP000902 [1,306 nt]) was most closely related to Beet soilborne mosaic virus (NC_003506 [6,683 nt]) in the genus Benyvirus, in the recently approved Benyviridae family (64). Viruses in the Benyvirus genus have four to five linear positive-sense ssRNAs of 6.7, 4.6, 1.8, 1.4, and 1.3 kb (65), where the first RNA encodes the RdRp. Only RNA1 encoding RdRp was identified for RsBenV1.
Putative double-stranded RNA virus genomes.
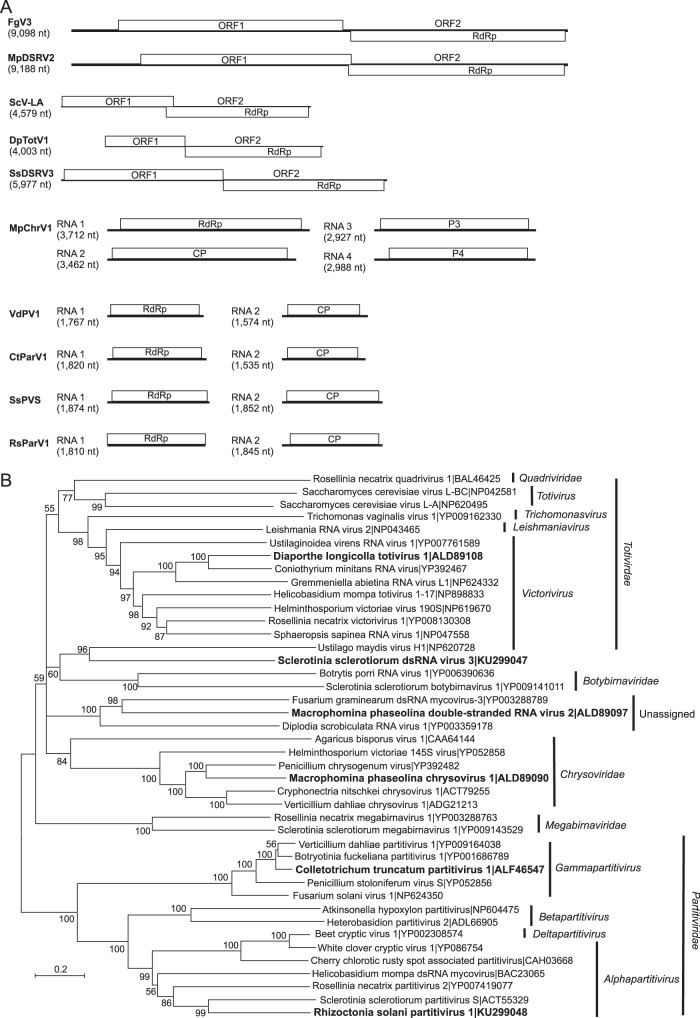
One sequence, Macrophomina phaseolina double-stranded RNA virus 2 (MpDSRV2; KP000891 [9,188 nt]), was similar to Fusarium graminearum dsRNA mycovirus 3 (FgV3; NC_013469 [9,098 nt]) (Fig. 8), an unclassified dsRNA virus. Like FgV3, the MpDSRV2 sequence was about 9.1 kb and contained two large ORFs, suggesting that the assembled sequence represents a nearly complete virus genome. The predicted amino acid sequence of ORF1 was similar to the products of the first ORFs of other unclassified bicistroinc dsRNA mycoviruses with genomes of 8.9 to 9.5 kb (66–69). Unlike the genomes of the other large bicistronic dsRNA mycoviruses, the two ORFs in MpDSRV2 overlapped by 28 nt, and ORF2 could be translated by a −1 frameshift from ORF1. The predicted amino acid sequence of MpDSRV2 ORF2 contained an RdRp domain and was 35% identical to the product of ORF2 of FgV3.
FIG 8.
Genome organizations and maximum likelihood tree depicting the relationships of the predicted amino acid sequences of RNA-dependent RNA polymerases (RdRps) from contigs assembled from Colletotrichum truncatum, Macrophomina phaseolina, Diaporthe longicolla, Rhizoctonia solani, and Sclerotinia sclerotiorum and mycoviruses with double-stranded RNA genomes. (A) Comparisons of the genome organizations of Colletotrichum truncatum partitivirus 1 (CtParV1), Diaporthe longicolla totivirus 1 (DpTotV1), Fusarium graminearum double-stranded RNA mycovirus 3 (FgV3), Macrophomina phaseolina double-stranded RNA virus 2 (MpDSRV2), Macrophomina phaseolina chrysovirus 1 (MpChrV1), Rhizoctonia solani partitivirus 1 (RsParV1), Saccharomyces cerevisiae virus L-A (ScV-LA), Sclerotinia sclerotiorum double-stranded RNA virus 3 (SsDSRV3), Sclerotinia sclerotiorum partitivirus S (SsPVS), and Verticillium dahliae partitivirus 1 (VdPV1). Open reading frames are shown as boxes. (B) Predicted RdRp amino acid sequences were aligned and phylogenetic trees were constructed as described in the legend to Fig. 1. Regions encoding the coat protein (CP) and RdRp are indicated in the corresponding ORFs.
Two sequences showed similarity to dsRNA viruses in the family Totiviridae, Sclerotinia sclerotiorum double-stranded RNA virus 3 (SsDSRV3; KU299047 [5,928 nt]) and Diaporthe longicolla totivirus 1 (DpTotV1; KP900901 [4,005 nt]). The family Totiviridae contains four genera, Giardiavirus, Leishmaniavirus, Totivirus, and Victorivirus, all with dsRNA genomes of 4.6 to 7.0 kb. Most contain two large overlapping ORFs (70). The sequence of SsDSRV3 contained two large ORFs. The predicted amino acid sequence of SsDSRV3 ORF2 was related to RdRps encoded by viruses in the genus Totivirus and was most similar to Ustilago maydis virus H1 (NC_003823 [6,099 nt]) at 29% identity. In SsDSRV3, a sequence (5′-AAATTTA-3′) similar to a slippery heptanucleotide was present near the end of ORF1 that could serve as a part of a frameshift signal. However, ORFs 1 and 2 were separated by 5 nt and did not overlap. Hence, SsDSRV3 may represent a new species within the genus Totivirus. The DpTotV1sequence also contained two ORFs, but the first was 5′ incomplete. The predicted amino acid sequence of DpTotV1 ORF2 contained an RdRp domain and was 62% identical to the ORF2-encoded peptide of Coniothyrium minitans RNA virus (NC_007523 [4,975 nt]), a member of the genus Victorivirus. Like viruses in the genus Victorivirus, the stop codon of ORF1 overlapped the start codon of ORF2 in the sequence AUGA in DpTotV1, which suggests that DpTotV1 is a new virus in the genus Victorivirus.
Four sequences, Colletotrichum truncatum partitivirus 1 (CtParV1) RNA1 (KR074421 [1,820 nt]), CtParV1 RNA2 (KP900885 [1,535 nt]), Rhizoctonia solani partitivirus 1 (RsParV1) RNA1 (KU299048 [1,810 nt]), and RsParV1 RNA2 (KP900892 [1,845 nt]), were each predicted to encode a single protein related to the nonstructural or structural proteins of viruses in the family Partitiviridae (Fig. 8). Viruses in the family Partitiviridae have genomes that consist of two dsRNA segments of 1.3 to 2.4 kb each, where the larger of the two RNAs encodes the replicase and the smaller the structural proteins (71). The predicted amino acid sequence from CtParV1 RNA1 was 87% identical to the product of RNA1 of Verticillium dahliae partitivirus 1 (NC_002801 [1,767 nt]), a member of the Gammapartitivirus genus. The sizes of the CtParV1 RNA1 and RNA2 were similar to the average sizes for RNA1 (1,715 nt) and RNA2 (1,537 nt) of members of the Gammapartitivirus genus. Similarly, the sizes of the polypeptides predicted to be encoded by RNA1 (539 amino acids [aa]) and RNA2 (430 aa) were very similar to the average sizes of products from RNA1 (537 aa) and RNA2 (423 aa) for gammapartitiviruses. The predicted amino acid sequence from RsParV1 RNA1 was 45% identical to product of RNA1 of Sclerotinia sclerotiorum partitivirus S (NC_028494 [1,874 nt]), a member of the Alphapartitivirus genus. The size of RsParV1 RNA1 (1,810 nt) was smaller than the average for alphapartitiviruses (1,964 nt), but it was predicted to encode a peptide of 602 amino acids, which is very close to the 606-aa average for alphapartitiviruses. This suggests that the 5′ and 3′ noncoding regions are incomplete for the RNA. The RsParV1 RNA2 (1,845 nt) was longer than the average (1,793 nt) for alphapartitiviruses and was predicted to encode a protein of 525 aa, which also was larger than the average (494 aa) for alphapartitiviruses. Hence, CtParV1 and RsParV1 likely represent new virus species within the Gammapartitivirus and Alphapartitivirus genera, respectively.
Four sequences from M. phaseolina (Macrophomina phaseolina chrysovirus 1 [MpChrV1]: RNA1, KP900886 [3,712 nt]; RNA2, KP900887 [3,462 nt]; RNA3, KP900888 [2,774 nt]; and RNA4, KP900889 [2,988 nt]) were predicted to encode proteins similar to those encoded by RNAs 1 to 4, respectively, of viruses in the family Chrysoviridae. The family Chrysoviridae contains one genus, Chrysovirus, with genomes that consist of four dsRNA segments that typically range from 2.4 to 3.6 kb (72). RNAs 1 and 2 encode the replicase and major CP. RNAs 3 and 4 encode proteins of unknown functions. The predicted amino acid sequence of MpChrV1 RNA1 was 55% identical to the RdRp encoded by Penicillium chrysogenum virus (YP_392482) and hence likely represents the genome of a new virus species in the genus Chrysovirus.
DISCUSSION
In this study, we identified sequences that putatively represented 72 fungal virus genome segments, many of which were nearly full length. The analysis also expanded the types of viruses associated with plant-pathogenic fungi with the identification of a virus from M. phaseolina with a genome architecture very similar to that of tobamoviruses in the family Virgavirdae and viruses related to members of the families Benyviridae and Ophioviridae from R. solani. Even though an increasing number of single-stranded circular DNA viruses have been reported from fungi and environmental samples (22–24, 73, 74), none were detected in the cultures analyzed. However, we detected two contigs representing the two transcripts of a putative ssDNA mycovirus in rRNA-depleted RNA samples from field-grown soybean plants using similar methods and used the information to amplify and sequence the complete circular genome (37). Hence, it is likely that if single-stranded circular DNA viruses had been present, they would have been detected by the methods used.
Viral metagenomics in the last decade has discovered many novel viruses (11), and our method of using rRNA-depleted total RNA extracts directly from host tissue (i.e., fungal mycelia) in combination with HTS for metagenomic analysis further accelerated the rate of virus discovery. Some metagenomics studies have enriched samples for viruses by biological, chemical, or physical methods prior to HTS (74–79). However, enrichment strategies based on the properties of virus particles or host range may bias results toward only the viruses that form particles and may further bias toward viral particles with specific properties based on the methods selected, including charge, density, and size. Although useful in finding large DNA viruses, the detection of few RNA viruses from marine samples may result from bias during the enrichment process (80). Our method permitted detection of viruses regardless of genome type and showed that enrichment was not necessary. By foregoing the enrichment step, the possibility of biased detection was greatly reduced, as is illustrated by the detection of diverse lineages of novel viruses. A limitation of our approach is that only viral sequences containing the conserved domains previously associated with virus genomes can be retrieved, which may explain why we did not identify RNAs encoding structural proteins from some putative multipartite nsRNA and ss(+)RNA viruses.
Members of the Furovirus, Pecluvirus, and Pomovirus genera in the Virgaviridae are transmitted horizontally by fungi in the genus Olpidium and two genera of plasmodiophorid protists, Polymyxa and Spongospora (81). Similarly, viruses in the family Ophioviridae and members of several genera in the family Tombusviridae are transmitted by soilborne fungi in the genus Olpidium, and some members of the Tombusviridae can be transmitted through soil in the absence of their fungal vectors (57, 82). Transmission of viruses by Olpidium, Polymyxa, and Spongospora involves coat protein-mediated interactions that result in the internalization of virus particles into zoospores or the adherence of virus particles to the external surfaces of zoospores, possibly in receptor-mediated processes (81). Because some of the viruses are acquired and borne by their vectors intracellularly, opportunities for horizontal virus transfer between taxa exist. It is not uncommon for deletions to occur in the CP coding regions of viruses that are normally vectored by Olpidium, Polymyxa, or Spongospora when they are repeatedly transmitted mechanically from plant to plant that result in loss of transmission by fungi or protists (83, 84). In plant benyviruses and ourmiaviruses, entire CP-encoding genome segments can be lost as a result of maintenance in plants for extended periods of time (85–87). The retention of CP coding regions in genomes similar to those of ophioviruses and tobamoviruses suggests that these sequences are required for virus function. The apparent loss of CP coding sequences from the three mycoviruses that show similarity to tombusviruses may represent large deletions in CP coding regions that resulted from repeated cytoplasmic transmission of the viruses. Alternatively, structural proteins could be expressed by an RNA segment that was not detected in these studies. In addition, loss of CP coding region could also be compensated for by the CP of an encapsidated coinfecting virus (15).
Consistent with their cytoplasmic mode of horizontal transmission, our study showed that mitoviruses, which replicate within mitochondria, were the most prevalent capsidless viruses in the viromes of S. sclerotiorum and R. solani. It is likely that capsidless viruses exist in other eukaryotic or prokaryotic systems where cell fusion is the predominant means for exchange of genetic material. However, even though sequences related to members of the capsidless Hypoviridae and Endornaviridae were detected, most of the extramitochondrial monopartite virus genomes recovered from M. phaseolina, R. solani, and S. sclerotiorum were predicted to express structural proteins. Among the viruses predicted to have segmented genomes, genome segments encoding the structural proteins were detected for a putative chrysovirus from M. phaseolina and for putative partitiviruses infecting M. phaseolina and R. solani. As mentioned above, it is possible that RNAs encoding structural proteins associated with other viruses with segmented genomes had been lost by repeated cytoplasmic transmission. Additional analysis will be required to determine whether this is an outcome of horizontal cytoplasmic virus transfer or the bioinformatic methods used to detect structural protein genes, which are typically much less conserved than replication-related protein genes (88, 89).
In this study, we identified viral genomes with affinity to viruses in the families Benyviridae, Ophioviridae, and Virgaviridae, which previously contained no mycovirus species. The discovered tobamo-like virus infecting M. phaseolina was most similar to an endogenized virus in the genome of the plant-pathogenic fungus Eutypa late. Failure to amplify the MpTLV1 sequence in the absence of reverse transcriptase confirmed that the M. phaseolina tobamo-like virus was not integrated into the host genome. Multiple studies in the 1970s suggested that tobacco mosaic virus (TMV) could replicate in and debilitate oomycetes in the genus Pythium (90–92), which provided early support to the infection of fungus-like organisms by tobamoviruses. More recently, Mascia et al. (93) infected Colletotrichum acutatum with a TMV-based vector for virus-induced gene silencing using Agrobacterium-mediated transformation. In addition, tobamo-like viruses were observed naturally infecting cultures of Agaricus bisporus (Basidiomycota) and Plicaria spp. (Ascomycota) (94). Analysis of a tobamo-like virus from the ascomycete fungus Peziza ostracoderma by X-ray defraction showed that the fungal virus had a helical pitch of about 2.7 nm compared to 2.3 nm for TMV (95), which could result from the predicted larger mass of the CPs of fungus-associated tobamoviruses. These data suggest the existence of a group of large-genome tobamo-like viruses that infect plant-pathogenic fungi. Furthermore, members of the Hordeivirus and Tobamovirus genera in the Virgaviridae are transmitted exclusively without the aid of biological vectors and produce highly stable virus particles. Hence, it is possible that MpTLV1 virions could be relatively stable in nature and be acquired extracellularly by M. phaseolina.
Phylogenetic analyses of viruses infecting lower eukaryotes often show clusters of viral genomes that are more closely related to viruses that infect the same host species than to viruses that infect other species. For example, in the family Totiviridae, members of the genera Leishmaniavirus and Trichomonasvirus infect protozoans in the species Leishmania braziliensis and Trichomonas vaginalis, respectively. Here, clades of putative nsRNA viruses were identified from R. solani and S. sclerotiorum with segmented and monopartite genomes, respectively. This may be a function of sample size: for example, all hypoviruses were at one time thought to be derived from C. parasitica. Alternatively, they could result from adaptation to a specific host and/or radiation of viral species within a particular host from a common ancestral sequence. The diversity of hosts for viruses related to nsRNA mycoviruses suggests that horizontal virus transfer among taxa has occurred, as has been proposed for viruses in the family Partitiviridae (16).
In addition to their potential for use in managing and understanding fungal disease etiology, identification of such a diverse set of mycoviruses from curated fungal cultures has implications in other areas of research as well. For example, because of the importance of pathogenic fungi to human and plant health, fungal genomes are being characterized to identify virulence effectors and other sequences associated with pathogenesis (96–99). However, infection of pathogenically diverse fungal isolates with virulence-altering mycoviruses could reduce the abilities of such studies to identify chromosomal regions linked to disease severity. This underscores the importance of understanding the virus loads of fungal isolates in association studies and the mechanisms by which mycoviruses alter the interactions between fungi and their hosts.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the National Sclerotinia Initiative and the United States Department of Agriculture/Agricultural Research Service.
Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the United States Department of Agriculture or the University of Illinois and does not imply its approval to the exclusion of other products or vendors that may also be suitable.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00357-16.
REFERENCES
- 1.Adams IP, Glover RH, Monger WA, Mumford R, Jackeviciene E, Navalinskiene M, Samuitiene M, Boonham N. 2009. Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol Plant Pathol 10:537–545. doi: 10.1111/j.1364-3703.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiu CY. 2013. Viral pathogen discovery. Curr Opin Microbiol 16:468–478. doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. 2012. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu ZQ, Ren XW, Yang L, Hu YF, Yang J, He GM, Zhang JP, Dong J, Sun LL, Du J, Liu LG, Xue Y, Wang JM, Yang F, Zhang SY, Jin Q. 2012. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol 86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook S, Chung BYW, Bass D, Moureau G, Tang SY, McAlister E, Culverwell CL, Glucksman E, Wang H, Brown TDK, Gould EA, Harbach RE, de Lamballerie X, Firth AE. 2013. Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in Dipteran hosts. PLoS One 8:e80720. doi: 10.1371/journal.pone.0080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraberger S, Stainton D, Dayaram A, Zawar-Reza P, Gomez C, Harding JS, Varsani A. 2013. Discovery of Sclerotinia sclerotiorum hypovirulence-associated virus-1 in urban river sediments of Heathcote and Styx Rivers in Christchurch City, New Zealand. Genome Announc 1:e00559-13. doi: 10.1128/genomeA.00559-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al Rwahnih M, Daubert S, Urbez-Torres JR, Cordero F, Rowhani A. 2011. Deep sequencing evidence from single grapevine plants reveals a virome dominated by mycoviruses. Arch Virol 156:397–403. doi: 10.1007/s00705-010-0869-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coetzee B, Freeborough M-J, Maree HJ, Celton J-M, Rees DJG, Burger JT. 2010. Deep sequencing analysis of viruses infecting grapevines: virome of a vineyard. Virology 400:157–163. doi: 10.1016/j.virol.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 9.Bruenn JA, Warner BE, Yerramsetty P. 2015. Widespread mitovirus sequences in plant genomes. PeerJ 3:e876. doi: 10.7717/peerj.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nerva L, Ciuffo M, Vallino M, Margaria P, Varese GC, Gnavi G, Turina M. 2015. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res doi: 10.1016/j.virusres.2015.10.028. [DOI] [PubMed] [Google Scholar]
- 11.Mokili JL, Rohwer F, Dutilh BE. 2012. Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khalifa ME, Pearson MN. 2013. Molecular characterization of three mitoviruses co-infecting a hypovirulent isolate of Sclerotinia sclerotiorum fungus. Virology 441:22–30. doi: 10.1016/j.virol.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Jiang D, Fu Y, Guoqing L, Ghabrial SA. 2013. Viruses of the plant pathogenic fungus Sclerotinia sclerotiorum. Adv Virus Res 86:215–248. doi: 10.1016/B978-0-12-394315-6.00008-8. [DOI] [PubMed] [Google Scholar]
- 14.Chiba S, Suzuki N. 2015. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc Natl Acad Sci U S A 112:E4911–E4918. doi: 10.1073/pnas.1509151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Hisano S, Tani A, Kondo H, Kanematsu S, Suzuki N. 2016. A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat Microbiol 1:15001. doi: 10.1038/nmicrobiol.2015.1. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roossinck MJ, Sabanadzovic S, Okada R, Valverde RA. 2011. The remarkable evolutionary history of endornaviruses. J Gen Virol 92:2674–2678. doi: 10.1099/vir.0.034702-0. [DOI] [PubMed] [Google Scholar]
- 18.Linder-Basso D, Dynek JN, Hillman BI. 2005. Genome analysis of Cryphonectria hypovirus 4, the most common hypovirus species in North America. Virology 337:192–203. doi: 10.1016/j.virol.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 19.Krupovic M, Dolja VV, Koonin EV. 2015. Plant viruses of the Amalgaviridae family evolved via recombination between viruses with double-stranded and negative-strand RNA genomes. Biol Direct 10:12. doi: 10.1186/s13062-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D. 2014. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A 111:12205–12210. doi: 10.1073/pnas.1401786111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo H, Chiba S, Toyoda K, Suzuki N. 2013. Evidence for negative-strand RNA virus infection in fungi. Virology 435:201–209. doi: 10.1016/j.virol.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Du Z, Tang Y, Zhang S, She X, Lan G, Varsani A, He Z. 2014. Identification and molecular characterization of a single-stranded circular DNA virus with similarities to Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1. Arch Virol 159:1527–1531. doi: 10.1007/s00705-013-1890-5. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Li B, Fu YP, Jiang DH, Ghabrial SA, Li GQ, Peng YL, Xie JT, Cheng JS, Huang JB, Yi XH. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci U S A 107:8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikorski A, Massaro M, Kraberger S, Young LM, Smalley D, Martin DP, Varsani A. 2013. Novel myco-like DNA viruses discovered in the faecal matter of various animals. Virus Res 177:209–216. doi: 10.1016/j.virusres.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N. 2015. 50-plus years of fungal viruses. Virology 479-480:356–368. doi: 10.1016/j.virol.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 26.Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, Nuss DL. 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190:113–127. doi: 10.1534/genetics.111.133983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, Li B, Fu YP, Xie JT, Cheng JS, Ghabrial SA, Li GQ, Yi XH, Jiang DH. 2013. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci U S A 110:1452–1457. doi: 10.1073/pnas.1213755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rochon D, Kakani K, Robbins M, Reade R. 2004. Molecular aspects of plant virus transmission by olpidium and plasmodiophorid vectors. Annu Rev Phytopathol 42:211–241. doi: 10.1146/annurev.phyto.42.040803.140317. [DOI] [PubMed] [Google Scholar]
- 29.Refos JM, Vonk AG, Eadie K, Lo-Ten-Foe JR, Verbrugh HA, van Diepeningen AD, van de Sande WWJ. 2013. Double-stranded RNA mycovirus infection of Aspergillus fumigatus is not dependent on the genetic make-up of the host. PLoS One 8:e77381. doi: 10.1371/journal.pone.0077381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Sande WWJ, Lo-Ten-Foe JR, van Belkum A, Netea MG, Kullberg BJ, Vonk AG. 2010. Mycoviruses: future therapeutic agents of invasive fungal infections in humans? Eur J Clin Microbiol 29:755–763. doi: 10.1007/s10096-010-0946-7. [DOI] [PubMed] [Google Scholar]
- 31.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semler DD, Goudie AD, Finlay WH, Dennis JJ. 2014. Aerosol phage therapy efficacy in Burkholderia cepacia complex respiratory infections. Antimicrob Agents Chemother 58:4005–4013. doi: 10.1128/AAC.02388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gervasi T, Lo Curto R, Minniti E, Narbad A, Mayer MJ. 2014. Application of Lactobacillus johnsonii expressing phage endolysin for control of Clostridium perfringens. Lett Appl Microbiol 59:355–361. doi: 10.1111/lam.12298. [DOI] [PubMed] [Google Scholar]
- 34.Tillman GE, Simmons M, Garrish JK, Seal BS. 2013. Expression of a Clostridium perfringens genome-encoded putative N-acetylmuramoyl-l-alanine amidase as a potential antimicrobial to control the bacterium. Arch Microbiol 195:675–681. doi: 10.1007/s00203-013-0916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer M, Vukov N, Scherer S, Loessner MJ. 2002. The murein hydrolase of the bacteriophage phi 3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl Environ Microbiol 68:5311–5317. doi: 10.1128/AEM.68.11.5311-5317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Nuss DL, Suzuki N. 2006. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J Gen Virol 87:3703–3714. doi: 10.1099/vir.0.82213-0. [DOI] [PubMed] [Google Scholar]
- 37.Marzano SL, Domier LL. 2016. Novel mycoviruses discovered from metatranscriptomics survey of soybean phyllosphere phytobiomes. Virus Res 213:332–342. doi: 10.1016/j.virusres.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Marzano SYL, Hobbs HA, Nelson BD, Hartman GL, Eastburn DM, McCoppin NK, Domier LL. 2015. Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus. J Virol 89:5060–5071. doi: 10.1128/JVI.03199-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam MS, Haque MS, Islam MM, Emdad EM, Halim A, Hossen QMM, Hossain MZ, Ahmed B, Rahim S, Rahman MS, Alam MM, Hou SB, Wan XH, Saito JA, Alam M. 2012. Tools to kill: genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics 13:493. doi: 10.1186/1471-2164-13-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Darwish O, Alkharouf N, Matthews B, Ji P, Domier L, Zhang N, Bluhm BH. 2015. Draft genome sequence of Phomopsis longicolla isolate MSPL 10-6. Genomics Data 3:55–56. doi: 10.1016/j.gdata.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier J-M, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun M-H, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7:e1002230. doi: 10.1371/journal.pgen.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuss DL. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol 3:632–642. doi: 10.1038/nrmicro1206. [DOI] [PubMed] [Google Scholar]
- 48.Pearson MN, Beever RE, Boine B, Arthur K. 2009. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384. doi: 10.1146/annurev-phyto-080508-081932. [DOI] [PubMed] [Google Scholar]
- 50.Afonso CL, Amarasinghe GK, Bányai K, Baò Y, Basler CF, Bavari S, Bejerman N, Blasdell K, Briand F, Briese T, Bukreyev A, Calisher CH, Chandran K, Chéng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astúa J, Formenty P, Fouchier AM, Fù Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li C, Lin X, Liú L, Longdon B, Marton S, Maisner A, Mühlberger E, Netesov SV, Nowotny N, Patterson JL, Payne SL, Paweska JT, Randall RE, Rima BK, Rota P, Rubbenstroth D, Schwemmle M, Shi M, Smither SJ, Stenglein MD, Stone DM, Takada A, Terregino C, Tesh RB, Tian J, Tomonaga K, Tordo N, Towner JS, Vasilakis N, Verbeek M, Volchkov VE, Wahl-Jensen V, Walsh JA, Walker PJ, Wang D, Wang L, Wetzel T, Whitfield AE, Xiè J, Yuen K, Zhang Y, Kuhn JH. 23 May 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Easton AJ, Pringle CR. 2012. Order Mononegavirales, p 654–657. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 52.Nuss DL, Hillman BI. 2012. Family Hypoviridae, p 1029–1033. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 53.Zhang R, Liu S, Chiba S, Kondo H, Kanematsu S, Suzuki N. 2014. A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front Microbiol 5:360. doi: 10.3389/fmicb.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukuhara T, Gibbs MJ. 2012. Family Endornaviridae, p 519–521. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 55.Khalifa ME, Pearson MN. 2014. Molecular characterisation of an endornavirus infecting the phytopathogen Sclerotinia sclerotiorum. Virus Res 189:303–309. doi: 10.1016/j.virusres.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 56.Rastgou M, Turina M, Milne RG. 2012. Genus Ourmiavirus, p 654–657. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 57.Rochon D, Lommel S, Martelli GP, Rubino L, Russo M. 2012. Family Tombusviridae, p 1069–1089. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 58.Srinivasan KG, Min BE, Ryu KH, Adkins S, Wong SM. 2005. Determination of complete nucleotide sequence of Hibiscus latent Singapore virus: evidence for the presence of an internal poly(A) tract. Arch Virol 150:153–166. doi: 10.1007/s00705-004-0404-x. [DOI] [PubMed] [Google Scholar]
- 59.Blanco-Ulate B, Rolshausen PE, Cantu D. 2013. Draft genome sequence of the grapevine dieback fungus Eutypa lata UCR-EL1. Genome Announc 1:e00228-13. doi: 10.1128/genomeA.00228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreher TW, Edwards MC, Gibbs AJ, Haenni A-L, Hammond RW, Jupin I, Koenig R, Sabanadzovic S, Martelli GP. 2012. Family Tymoviridae, p 944–952. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 61.Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba S, Pearson MN, Ryu KH, Saldarelli P, Yoshikawa N. 2012. Family Betaflexiviridae, p 920–941. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 62.Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba S, Pearson MN, Ryu KH, Vaira AM. 2012. Family Alphaflexiviridae, p 904–919. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 63.Revill PA. 2012. Family Barnaviridae, p 961–964. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 64.Adams MJ, Lefkowitz EJ, King AMQ, Carstens EB. 2014. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014). Arch Virol 159:2831–2841. doi: 10.1007/s00705-014-2114-3. [DOI] [PubMed] [Google Scholar]
- 65.Gilmer D, Ratti C. 2012. Genus Benyvirus, p 1163–1168. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 66.Yu L, Sang W, Wu MD, Zhang J, Yang L, Zhou YJ, Chen WD, Li GQ. 2015. Novel hypovirulence-associated RNA mycovirus in the plant-pathogenic fungus Botrytis cinerea: molecular and biological characterization. Appl Environ Microbiol 81:2299–2310. doi: 10.1128/AEM.03992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu HQ, Fu YP, Xie JT, Cheng JS, Ghabrial SA, Li GQ, Peng YL, Yi XH, Jiang DH. 2012. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol Biol 14:241. doi: 10.1186/s12862-014-0241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marvelli RA, Hobbs HA, Li S, McCoppin NK, Domier LL, Hartman GL, Eastburn DM. 2014. Identification of novel double-stranded RNA mycoviruses of Fusarium virguliforme and evidence of their effects on virulence. Arch Virol 159:349–352. doi: 10.1007/s00705-013-1760-1. [DOI] [PubMed] [Google Scholar]
- 69.Yu J, Kwon SJ, Lee KM, Son M, Kim KH. 2009. Complete nucleotide sequence of double-stranded RNA viruses from Fusarium graminearum strain DK3. Arch Virol 154:1855–1858. doi: 10.1007/s00705-009-0507-5. [DOI] [PubMed] [Google Scholar]
- 70.Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC. 2012. Family Totiviridae, p 639–650. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 71.Nibert ML, Ghabrial SA, Maiss E, Lesker T, Vainio EJ, Jiang DH, Suzuki N. 2014. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res 188:128–141. doi: 10.1016/j.virusres.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 72.Ghabrial SA, Castón JR. 2012. Family Chrysoviridae, p 509–513. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 73.Conceição-Neto N, Zeller M, Heylen E, Lefrère H, Mesquita JR, Matthijnssens J. 2015. Fecal virome analysis of three carnivores reveals a novel nodavirus and multiple gemycircularviruses. Virol J 12:79. doi: 10.1186/s12985-015-0305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosario K, Dayaram A, Marinov M, Ware J, Kraberger S, Stainton D, Breitbart M, Varsani A. 2012. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J Gen Virol 93:2668–2681. doi: 10.1099/vir.0.045948-0. [DOI] [PubMed] [Google Scholar]
- 75.Aw TG, Howe A, Rose JB. 2014. Metagenomic approaches for direct and cell culture evaluation of the virological quality of wastewater. J Virol Methods 210:15–21. doi: 10.1016/j.jviromet.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 76.Van Twest R, Kropinski A. 2009. Bacteriophage enrichment from water and soil. Methods Mol Biol 501:15–21. doi: 10.1007/978-1-60327-164-6_2. [DOI] [PubMed] [Google Scholar]
- 77.Lysholm F, Wetterbom A, Lindau C, Darban H, Bjerkner A, Fahlander K, Lindberg AM, Persson B, Allander T, Andersson B. 2012. Characterization of the viral microbiome in patients with severe lower respiratory tract infections, using metagenomic sequencing. PLoS One 7:e30875. doi: 10.1371/journal.pone.0030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bibby K, Peccia J. 2013. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ Sci Technol 47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edwards RA, Rohwer F. 2005. Viral metagenomics. Nat Rev Microbiol 3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 80.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol 18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams MJ, Heinze C, Jackson AO, Kreuze JF, Macfarlane SA, Torrance L. 2012. Family Virgaviridae, p 1139–1162. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 82.Vaira AM, Gago-Zachert S, Garcia ML, Guerri J, Hammond J, Milne RG, Moreno P, Morikawa T, Natsuaki T, Navarro JA, Pallas V, Torok V, Verbeek M, Vetten HJ. 2012. Family Ophioviridae, p 743–748. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 83.Kakani K, Sgro JY, Rochon D. 2001. Identification of specific cucumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J Virol 75:5576–5583. doi: 10.1128/JVI.75.12.5576-5583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reavy B, Arif M, Cowan GH, Torrance L. 1998. Association of sequences in the coat protein/readthrough domain of potato mop-top virus with transmission by Spongospora subterranea. J Gen Virol 79:2343–2347. doi: 10.1099/0022-1317-79-10-2343. [DOI] [PubMed] [Google Scholar]
- 85.Tamada T, Shirako Y, Abe H, Saito M, Kiguchi T, Harada T. 1989. Production and pathogenicity of isolates of beet necrotic yellow vein virus with different numbers of RNA components. J Gen Virol 70:3399–3409. doi: 10.1099/0022-1317-70-12-3399. [DOI] [Google Scholar]
- 86.Lemaire O, Merdinoglu D, Valentin P, Putz C, Zieglergraff V, Guilley H, Jonard G, Richards K. 1988. Effect of beet necrotic yellow vein virus RNA composition on transmission by Polymyxa betae. Virology 162:232–235. doi: 10.1016/0042-6822(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 87.Rossi M, Vallino M, Abba S, Ciuffo M, Balestrini R, Genre A, Turina M. 2015. The importance of the KR-rich region of the coat protein of Ourmia melon virus for host specificity, tissue tropism, and interference with antiviral defense. Mol Plant Microbe Interact 28:30–41. doi: 10.1094/MPMI-07-14-0197-R. [DOI] [PubMed] [Google Scholar]
- 88.Koonin EV, Dolja VV. 2014. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev 78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nasir A, Caetano-Anollés G. 2015. A phylogenomic data-driven exploration of viral origins and evolution. Sci Adv 1:e1500527. doi: 10.1126/sciadv.1500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pahlow G. 1976. Tobacco mosaic-virus translocated by mycelium of artificially infected Pythium spp. J Phytopathol 86:361–364. doi: 10.1111/j.1439-0434.1976.tb01711.x. [DOI] [Google Scholar]
- 91.Brants DH. 1971. Infection of Pythium sylvaticum in vitro with tobacco mosaic virus. Neth J Plant Pathol 77:175–177. doi: 10.1007/BF02000009. [DOI] [Google Scholar]
- 92.Nienhaus F, Mack C. 1974. Infection of Pythium arrhenomanes in vitro with tobacco mosaic virus and tobacco necrosis virus. Z Pflanzenkr Pflanzenschutz 81:728–731. [Google Scholar]
- 93.Mascia T, Nigro F, Abdallah A, Ferrara M, De Stradis A, Faedda R, Palukaitis P, Gallitelli D. 2014. Gene silencing and gene expression in phytopathogenic fungi using a plant virus vector. Proc Natl Acad Sci U S A 111:4291–4296. doi: 10.1073/pnas.1315668111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dieleman-Van Zaayen A. 1967. Virus-like particles in a weed mould growing on mushroom trays. Nature 216:595–596. [Google Scholar]
- 95.Dieleman-Van Zaayen A, Igesz O, Finch JT. 1970. Intracellular appearance and some mrophological features of viruslike particles in an ascomycete fungus. Virology 42:534–537. doi: 10.1016/0042-6822(70)90296-5. [DOI] [PubMed] [Google Scholar]
- 96.Persoons A, Morin E, Delaruelle C, Payen T, Halkett F, Frey P, De Mita S, Duplessis S. 2014. Patterns of genomic variation in the poplar rust fungus Melampsora larici-populina identify pathogenesis-related factors. Front Plant Sci 5:16. doi: 10.3389/fpls.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muller LAH, Lucas JE, Georgianna DR, McCusker JH. 2011. Genome-wide association analysis of clinical vs. nonclinical origin provides insights into Saccharomyces cerevisiae pathogenesis. Mol Ecol 20:4085–4097. doi: 10.1111/j.1365-294X.2011.05225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sperschneider J, Gardiner DM, Thatcher LF, Lyons R, Singh KB, Manners JM, Taylor JM. 2015. Genome-wide analysis in three Fusarium pathogens identifies rapidly evolving chromosomes and genes associated with pathogenicity. Genome Biol Evol 7:1613–1627. doi: 10.1093/gbe/evv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dalman K, Himmelstrand K, Olson A, Lind M, Brandstrom-Durling M, Stenlid J. 2013. A genome-wide association study identifies genomic regions for virulence in the non-model organism Heterobasidion annosum s.s. PLoS One 8:e53525. doi: 10.1371/journal.pone.0053525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hillman BI, Cai G. 2013. The family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176. doi: 10.1016/B978-0-12-394315-6.00006-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.