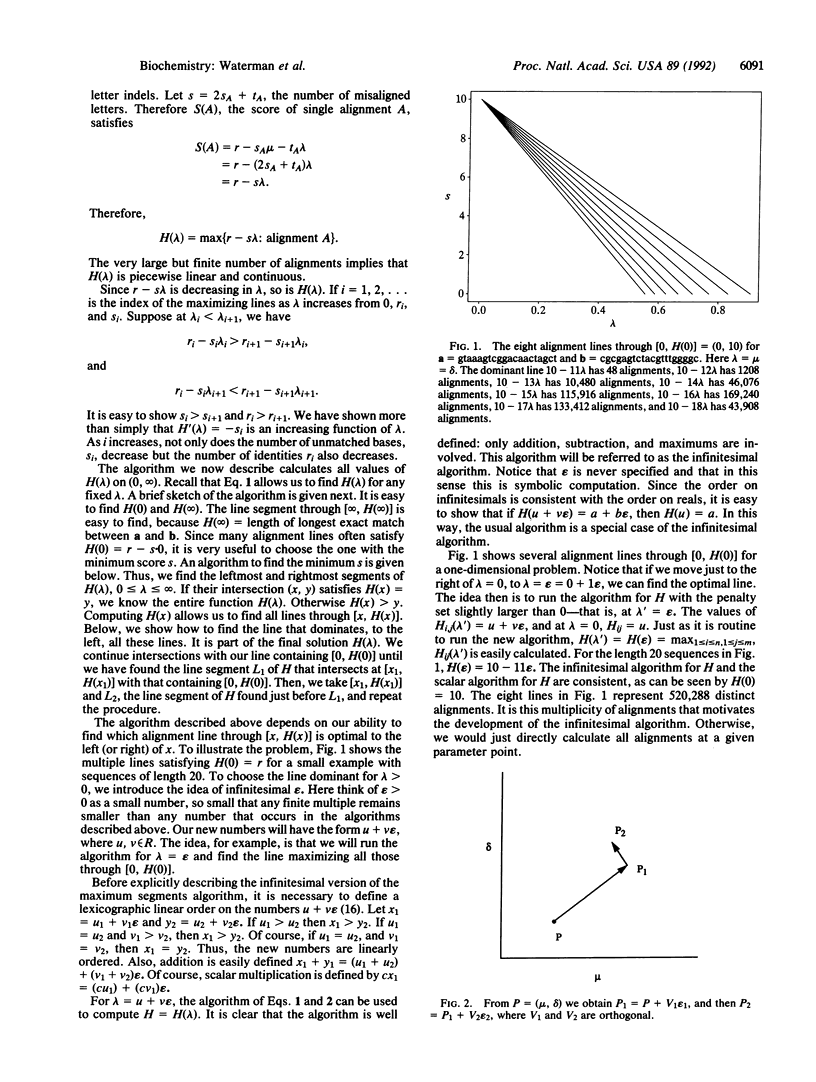
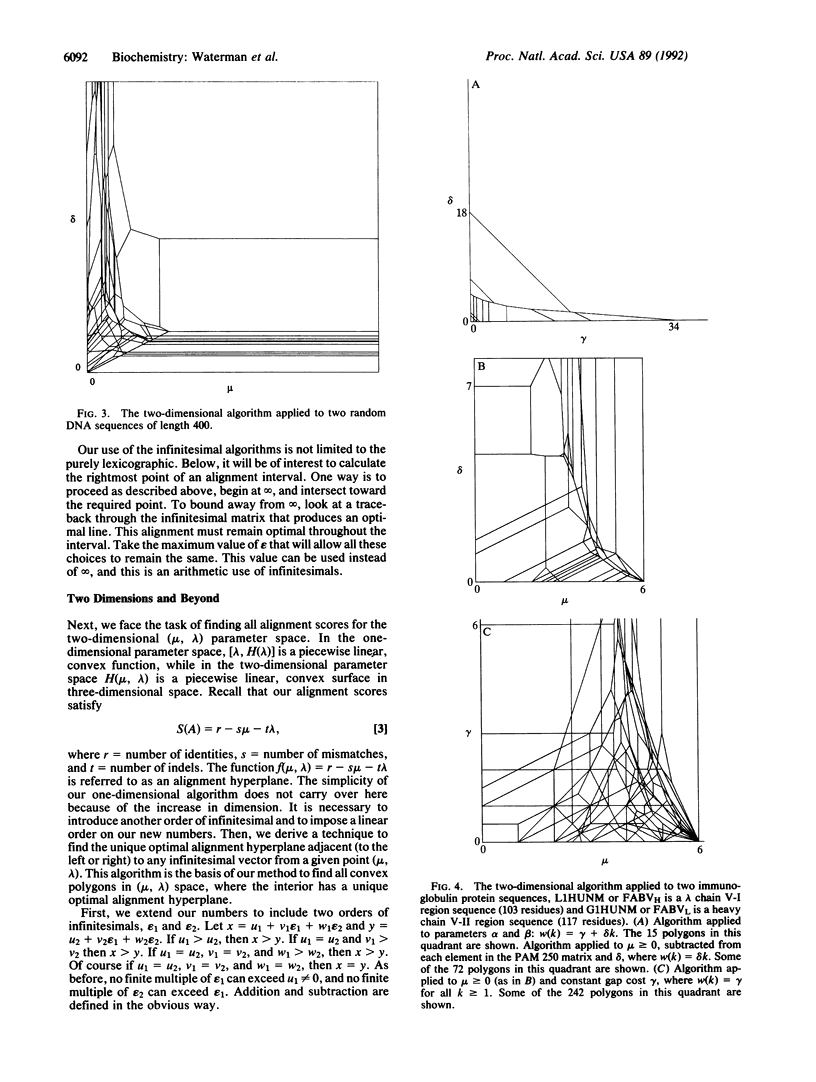
Abstract
Current algorithms can find optimal alignments of two nucleic acid or protein sequences, often by using dynamic programming. While the choice of algorithm penalty parameters greatly influences the quality of the resulting alignments, this choice has been done in an ad hoc manner. In this work, we present an algorithm to efficiently find the optimal alignments for all choices of the penalty parameters. It is then possible to systematically explore these alignments for those with the most biological or statistical interest. Several examples illustrate the method.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton G. J., Sternberg M. J. Evaluation and improvements in the automatic alignment of protein sequences. Protein Eng. 1987 Feb-Mar;1(2):89–94. doi: 10.1093/protein/1.2.89. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Hunkapiller M. W., Hood L. E., Devare S. G., Robbins K. C., Aaronson S. A., Antoniades H. N. Simian sarcoma virus onc gene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor. Science. 1983 Jul 15;221(4607):275–277. doi: 10.1126/science.6304883. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Smith T. F. Optimal sequence alignments. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1382–1386. doi: 10.1073/pnas.80.5.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol. 1982 Dec 15;162(3):705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Optimal sequence alignment allowing for long gaps. Bull Math Biol. 1990;52(3):359–373. doi: 10.1007/BF02458577. [DOI] [PubMed] [Google Scholar]
- Graves P. V., Bégu D., Velours J., Neau E., Belloc F., Litvak S., Araya A. Direct protein sequencing of wheat mitochondrial ATP synthase subunit 9 confirms RNA editing in plants. J Mol Biol. 1990 Jul 5;214(1):1–6. doi: 10.1016/0022-2836(90)90138-c. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Naharro G., Robbins K. C., Reddy E. P. Gene product of v-fgr onc: hybrid protein containing a portion of actin and a tyrosine-specific protein kinase. Science. 1984 Jan 6;223(4631):63–66. doi: 10.1126/science.6318314. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Schöniger M., Waterman M. S. A local algorithm for DNA sequence alignment with inversions. Bull Math Biol. 1992 Jul;54(4):521–536. doi: 10.1007/BF02459633. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Waterman M. S., Eggert M. A new algorithm for best subsequence alignments with application to tRNA-rRNA comparisons. J Mol Biol. 1987 Oct 20;197(4):723–728. doi: 10.1016/0022-2836(87)90478-5. [DOI] [PubMed] [Google Scholar]
- Weiss R. Cancer. Oncogenes and growth factors. Nature. 1983 Jul 7;304(5921):12–12. doi: 10.1038/304012a0. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



