Abstract
The Gene Expression Omnibus (GEO) database is an international public repository that archives and freely distributes high-throughput gene expression and other functional genomics data sets. Created in 2000 as a worldwide resource for gene expression studies, GEO has evolved with rapidly changing technologies and now accepts high-throughput data for many other data applications, including those that examine genome methylation, chromatin structure, and genome–protein interactions. GEO supports community-derived reporting standards that specify provision of several critical study elements including raw data, processed data, and descriptive metadata. The database not only provides access to data for tens of thousands of studies, but also offers various Web-based tools and strategies that enable users to locate data relevant to their specific interests, as well as to visualize and analyze the data. This chapter includes detailed descriptions of methods to query and download GEO data and use the analysis and visualization tools. The GEO homepage is at http://www.ncbi.nlm.nih.gov/geo/.
Keywords: microarray, high-throughput sequencing, gene expression, functional genomics, database, data mining
1. Introduction
Gene Expression Omnibus (GEO) is a database supported by the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM) that accepts raw and processed data with written descriptions of experimental design, sample attributes, and methodology for studies of high-throughput gene expression and genomics. The introduction of DNA microarrays and the Serial Analysis of Gene Expression (SAGE) protocol as methods of simultaneously assaying gene expression of multiple genes in 1995 enabled scientists to study gene expression of hundreds to thousands of genes, thereby vastly increasing the experimental scale and providing a far more complete understanding of biological processes compared to earlier single-gene studies [1,2]. Microarray technology quickly dominated the field of high-throughput gene expression studies and with the genome sequencing of humans [3] and many model organisms [4–7], genome-wide gene expression and other functional genomic studies became commonplace by the early 2000s. The accelerating pace of genomic-level data production and the bulky raw and processed data files they generated created a challenge for individual labs or journals to make the data available to the research community. In 2000, NCBI launched the GEO database as a repository for high-throughput gene expression data [8]. In 2002, major journals started to require deposit of microarray data into public repositories [9], and consequently, the content of GEO grew quickly. Furthermore, the nature of high-throughput genomic experiments expanded rapidly since the first microarrays used to analyze gene expression, and thus the GEO database similarly evolved to keep pace with the changing technologies and applications. Today, GEO accepts data from a wide variety of technologies, including DNA microarrays, protein or tissue arrays, high-throughput nucleic acid sequencing, SAGE, and RT-PCR. And while the majority, approximately 90%, of the data in GEO are indeed gene expression data, the applications have also expanded to include studies on genome methylation, genome binding/occupancy, protein profiling, chromosome conformation studies, and genome variation/copy number [10].
It is serendipitous that the word “geo” is a prefix meaning “earth” because not only does GEO primarily host global gene expression data, GEO itself is indeed a global resource; at the time of this writing GEO contains submissions from 72 nations. There are no fees to submit data to GEO, download data, or use GEO tools. Scientists submit to GEO in order to share their data with the research community and/or as a requirement of publication or grant directives. GEO supports the Minimum Information About a Microarray Experiment (MIAME) [11] and Minimum Information about a high-throughput SEQuencing Experiment (MINSEQE) guidelines set forth by the Functional Genomics Data Society (http://www.fged.org/) for standardization of information about microarray and sequencing experiments that enable the data to be interpreted and replicated by the research community. The GEO database handles the majority of direct submissions from the research community and at the time of this writing holds 54,640 public studies, comprising over 1.3 million samples, derived from 2889 different organisms. An up-to-date summary of GEO data types and content is provided at http://www.ncbi.nlm.nih.gov/geo/summary/.
While the chief role of GEO is to serve as a public data archive, the database is not simply an online warehouse of data. GEO strives to make the data it contains accessible to the research community. Due to the complex nature of the data generated by genomic experiments most studies are analyzed by bioinformaticians and statisticians, or researchers with specialized analysis software. Researchers who lack these skills or software face a substantial challenge if they wish to analyze genomics experiments themselves. In order to make such data analysis accessible to all researchers, GEO has developed several tools for data query, visualization, and analysis that can be performed directly on the GEO website and do not require the download or manipulation of the data files.
2. Methods
2.1 Retrieve a specific GEO record
If the GEO accession number is already known, for example, it has been cited in a manuscript describing the data, the user can type the accession number into the “GEO accession” query box. It recognizes Series (GSExxx), Sample (GSMxxx), Platform (GPLxxx) and DataSet (GDSxxx) accession numbers (see Note 1), and returns the queried record. The “GEO accession” query box is located on the upper right-hand corner of the GEO homepage at http://www.ncbi.nlm.nih.gov/geo/, and is also present at the top of all GEO records for easy movement between accessions. Alternatively, searching the Web directly with a major engine like Google will usually retrieve the correct record.
2.2 Quick search using keywords
NCBI uses a search and retrieval system that can be used to search the content of its entire network of integrated databases including PubMed, GenBank, Genomes, Taxonomy, and many others [12] (see http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi). GEO data are available in two separate NCBI databases:
GEO DataSets: Users should use this database to search for studies of interest. The database stores all original submitter-supplied Platform, Sample and Series records, as well as curated gene expression DataSet records. Retrievals include the title, summary, organism, and accession for each record, as well as links to related data (Fig. 1). The GEO DataSets search interface is available at http://www.ncbi.nlm.nih.gov/gds/ or can be selected from the dropdown databases menu from the main search box on the NCBI home page at http://www.ncbi.nlm.nih.gov/.
GEO Profiles: Users should use this database to search for expression profiles of genes. The database stores gene expression profiles derived from curated DataSet records. Retrievals include the gene name, DataSet title, and a thumbnail image that depicts the expression values of that gene across each Sample in that DataSet. Experimental context is provided in the blocks at the foot of the charts making it possible to see immediately whether that gene is differentially expressed across experimental conditions (Fig. 2). Clicking on the thumbnail image enlarges the chart to reveal the full profile details, expression values, and the DataSet subsets that reflect experimental design, see Section 2.7 for more details). The GEO Profiles search interface is available at http://www.ncbi.nlm.nih.gov/geoprofiles/ or can be selected from the dropdown databases menu from the main search box on the NCBI home page at http://www.ncbi.nlm.nih.gov/.
Simple keyword searches work very well in these databases. For example, if a user wants to find studies that examine hepatocellular carcinoma, it is only necessary to type “hepatocellular carcinoma” into the GEO DataSets search box to retrieve all the DataSet, Series, and Sample records that mention that term. Similarly, if a user is studying the gene CREB5, it is only necessary to type “CREB5” into the GEO Profiles search box to retrieve all gene expression profile records for that gene across all DataSets.
Fig. 1.
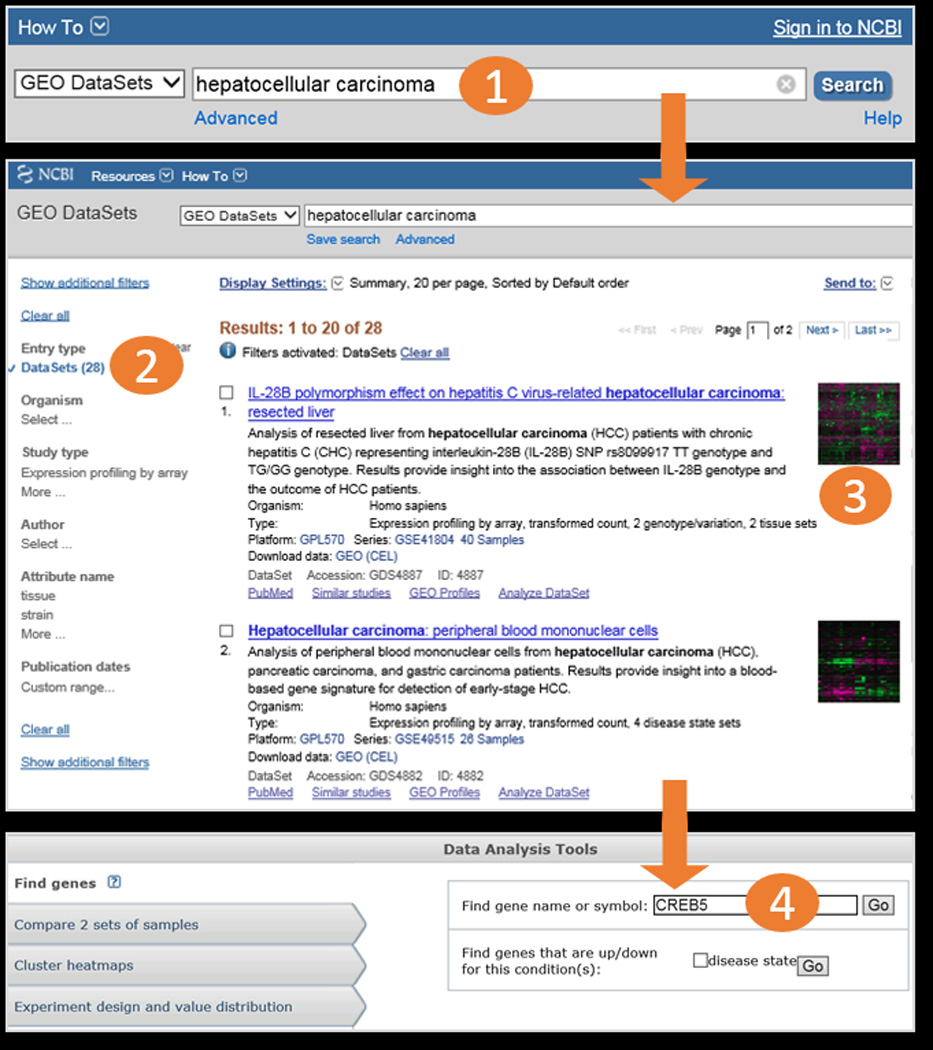
Workflow screenshots. After typing a search term into the GEO DataSets search box (1), and using the filter feature to restrict to DataSet entries (2), the user retrieves 28 relevant records (3). The user selects the second DataSet, GDS4882, and uses the “Find genes” feature in the DataSet Analysis Tools to search for gene CREB5 in that DataSet (4). Workflow continues in Fig 2.
Fig. 2.
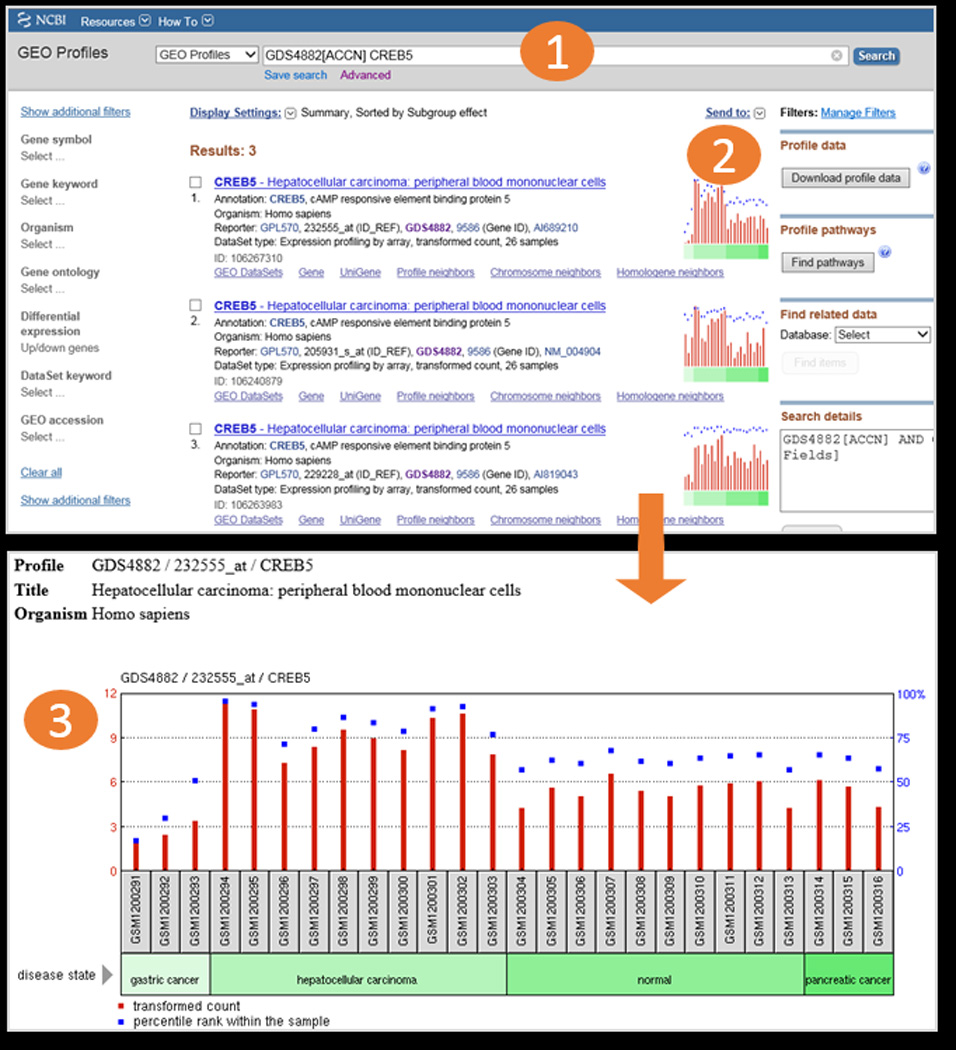
Workflow screenshots (continued). The “Find genes” feature in the DataSet Analysis Tools (in Fig. 1) creates a search for gene CREB5 in DataSet GDS4882 (1). The user is presented with 3 results in GEO Profiles (2), meaning that the CREB5 gene is represented by 3 separate probesets on the Platform in GDS4882. Looking at the chart images, the user can immediately see that all 3 CREB5 probesets exhibit a similar expression pattern. Clicking on the top chart reveals a detailed graphic (3), where the user can see that CREB5 is more highly expressed in the hepatocellular Samples, compared to the other Samples examined in that DataSet.
Typical workflows within and between these databases depend on the aims of the user. Generally, if users want to identify particular studies of interest, they should search the GEO DataSets database first, and then they have the option to use either GEO2R (Section 2.8), GEO DataSets analysis tools (Section 2.6) or GEO Profiles analysis tools (Section 2.7) to identify specific genes or interesting gene expression patterns within those studies (Fig. 1 and Fig. 2). On the other hand, users want to see expression patterns of a favorite gene across any study, they can search the GEO Profiles database directly to see how that gene behaves across all DataSets.
2.3 Advanced search using structured queries and filters
While simple keyword searches work well, the ever-growing volume of data in GEO means it is increasingly necessary to use structured and filtered queries to find the most relevant data. The GEO DataSets and GEO Profiles databases enable both simple and sophisticated queries to identify data of interest. Basic keyword searches can be performed alone or in combination with Boolean operators (AND, OR, NOT) to refine the search. Keyword searches with multiple parameters are structured with the following general format:
term[field] OPERATOR term[field]
where term is the search term, field is the search field (can be omitted to search for the term across all fields), and OPERATOR is the Boolean operator (“AND,” “OR,” “NOT” must be capitalized). For example, a simple search term “hepatocellular carcinoma” can be refined to “hepatocellular carcinoma AND human[organism]” if a researcher is only interested in studies of hepatocellular carcinoma in humans. Searchable fields include but are not limited to: organism, e.g., macaca mulatta[organism]; study type, e.g., expression profiling by array[DataSet type]; number of samples, e.g., 100:300[number of samples]; author, e.g., smith, a[author]; supplementary file type, e.g., cel[Supplementary Files]. The “How to construct a query’ link from the GEO home page provides detailed directions and examples for building queries (http://www.ncbi.nlm.nih.gov/geo/info/qqtutorial.html) (see Note 2).
The results presented in either GEO DataSets or GEO Profiles can be further filtered or refined in several ways. Clicking on the word “Advanced” under the search box displaying the original query takes the user to the “Advanced Search Builder” page where searches can be built from drop-down menus. Here queries can be expanded to include multiple fields and operators. The clickable text “show index list” provides the options for available terms to use. For example, the search “hepatocellular carcinoma AND human[organism]” can be further refined to identify only those studies assaying gene expression by array by adding AND DataSet Type and choosing “expression profiling by array” in the Builder.
Search results can also be refined on the same page as the returned results by using the filters available on the left sidebar (Fig. 1). Filters available on GEO DataSets and GEO Profiles search results are specific to the study- or gene-level type of data contained within each database. For the results in GEO DataSets, filters exist for entry type (DataSets, Series, Samples, Platforms), organism, study type, publication date, and author, among others. Filters can be applied by clicking on the text beneath each header (entry type, organism, etc.). For some filters, choosing “select” will open a dialog box to enter the text to be used in the filtering. Once a filter is applied, only the results matching the filter requirements will be available in the results page. Each filter that is applied can be removed by clicking the word “clear” available in small, gray text adjacent to the filter header. At the bottom of the left sidebar is the text “Clear all” which will remove all applied filters. The filters on GEO Profiles function in a similar manner but are specific to gene-level queries. Filters on GEO Profiles include gene symbol, gene keyword, gene ontology and a filter called “Differential expression” to identify genes that have been flagged as being differentially expressed based on the effect of treatment or condition in a DataSet. The filters provide a flexible way to restrict searches to drill down to relevant data.
2.4 Search programmatically
Data in both GEO DataSets and GEO Profiles can be searched programmatically. A suite of NCBI programs called Entrez Programming Utilities (E-utils) are used to conduct queries. E-utils enable sophisticated queries to be performed similar to the nature of the keyword searches or filtering as described above. The utilities are designed to be called from within a computer program that can process their output, which is in XML format.
A typical E-utils workflow might have the following steps:
Use the qualifier fields in the GEO DataSets database to locate data of interest and construct the appropriate eSearch query in your script or program
Run the query, retrieve the results in the form of unique identifiers or history parameters as needed
Run eSummary or eFetch and/or eLink depending on specific needs to retrieve the final metadata or accessions.
If full data tables or supplementary files need to be downloaded, use the accession information to construct an FTP URL and download the data.
More information for constructing programmatic queries with E-utils can be found at http://www.ncbi.nlm.nih.gov/geo/info/geo_paccess.html.
2.5 Query with a nucleotide sequence
The GEO BLAST database contains all GenBank sequences represented on microarray Platforms that participate in DataSets. The GEO BLAST search function provides the opportunity to perform a sequence-based search against GEO using either a nucleotide sequence or a GenBank accession number. The output of GEO BLAST is standard BLAST format where each alignment contains a list of “Related Information” at the right side of the page. Therein is a link to GEO Profiles where gene expression on the BLASTed-sequence can be explored. A link to the GEO BLAST utility is available on the GEO home page, http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch
2.6 DataSet analysis tools
The first analysis features developed at GEO were based on curated DataSet records which are created at periodic intervals by GEO staff from selected Series; over 3800 DataSets exist at the time of this writing. DataSet records are designed to provide both visualization and data analysis tools for normalized, array-based gene expression studies stored in GEO [13].
The top section of a DataSet record provides information about the study including title, study summary, organism, citation, and Platform and Series accession numbers upon which the DataSet is based. The lower portion of the DataSet record has 4 tabs encompassing customizable data analysis tools to assist with identification of genes of interest within that DataSet (Fig. 1) (see Note 3):
Find genes: Provides a search box for looking up specific gene names or symbols in this DataSet, as well as an option to identify genes that have been flagged as being differentially expressed according to the specific experimental variables in this study. Both of these types of searches take the user to the resulting gene(s) in GEO Profiles.
Compare 2 sets of samples: Enables a user to perform a customized Student’s t-test of self-selected Samples in order to identify differentially expressed genes in this DataSet. In order to start the analysis, the user firsts select the test to perform and P-value significance level from the drop-down menus. Second, the Samples to be included in the analysis are selected. For example if a user is interested in genes with differential expression only in hepatocellular carcinoma compared to healthy controls from DataSet GDS4882, the user would select the hepatocellular carcinoma Samples for group A by clicking on the corresponding Sample GSM accession numbers under “Group A” and the “normal” Samples under group B. The analysis is initiated by clicking “Query Group A vs. B”. The calculation is performed and takes the user to the resulting genes in GEO Profiles (see Note 4).
Cluster heatmaps: Presents precalculated and interactive cluster heatmap images that help detect natural groups of coordinately regulated genes. Genes with high levels of expression are represented in pink while genes with low levels of expression are represented in green. This tool allows for choice of hierarchical and partitional (K-means/medians) clustering or clustering genes by chromosome position. The hierarchical and K-means/median clustering contain options so that users can specify the method for linkage and distance, number of clusters, and color display options using drop-down menus. After clicking on the heatmap, the user can zoom in to areas of the cluster, select and export underlying expression values, or view the genes in GEO Profiles.
Experiment design and value distribution: draws boxplots for the expression values for all Samples in a study with corresponding Sample identifiers and Sample subset labels (e.g., drug-treated or control). The boxplot provides a visual overview of the data distribution and Sample categories in this DataSet. Boxplot images that display the distribution of expression values together with experimental design are useful for quality control checks.
2.7 GEO Profiles analysis tools
Regardless of how a user arrives at GEO Profiles results, either through direct searches (Section 2.2) or as a result of performing DataSet analyses (Section 2.6), various features exist on GEO Profiles records to assist with further analysis and exploration.
Each entry in GEO Profiles displays the name of the gene and the title of the DataSet that the data are from, and additional annotation and information about the organism, Platform and probe identifier. Several links that enable further analysis are also presented on the page:
Profile Neighbors: Retrieves Profiles with similar patterns of expression within the same DataSet, as calculated by Pearson correlation coefficients between pairs of Profiles. The top 200 results are arbitrarily considered to be Profile Neighbors, which may help identify genes with coordinated regulation.
Chromosome Neighbors: Retrieves Profiles for up to 20 of the closest-found chromosome neighbors within the same DataSet, helping identify expression data for genes within the same chromosomal region.
Sequence Neighbors: Retrieves Profiles based on BLAST nucleotide sequence similarity across all DataSets, assisting in the identification of genes representing sequence homologs and orthologs.
Homologene neighbors: Retrieves Profiles that belong to the same HomoloGene group across all DataSets. HomoloGene is a NCBI resource for automated detection of homologs among the annotated genes of several completely sequenced eukaryotic genomes.
Download Profile Data button: Downloads the values, experimental factors and gene annotations for each Profile on the page. Download files are tab-delimited and suitable for opening in a spreadsheet application such as Excel (see Note 5).
Find Pathways button: Maps the Profiles to a frequency weighted list of pathways in NCBI’s BioSystems database. Knowing the pathways the set of Profiles participate in can help characterize that list of genes.
Each record also has a thumbnail image of a chart of gene expression across all Samples in that DataSet, and are useful for rapidly scanning and comparing multiple Profile retrievals (Fig. 2). Clicking this thumbnail image opens a new window with a large view of the same chart. The red bars represent expression values while the blue squares represent the percentile rank of that expression value within the Sample (see Note 6). The blocks at the bottom of the chart represent experimental variable subsets within the DataSet. Each subset has a type, e.g., 'disease state', and a description, e.g., “hepatocellular carcinoma” so users can see at a glance how a gene is behaving across experimental variables (Fig. 2) (see Note 7).
2.8 Analyze with GEO2R
While the curated DataSets and Profiles records described in Sections 2.6 and 2.7 provide analysis and visualization tools for many GEO Series records, the pace of GEO submissions now greatly exceeds the rate at which DataSets can be produced. In order to provide immediate, Web-based, and user-driven analysis for GEO data, GEO2R was developed. GEO2R is an interactive tool that enables the analysis of approximately 90% of GEO Series as soon as they are released. It uses a Web-based program that employs the Bioconductor [14] packages GEOQuery [15] and limma [16] in R, with the Benjamini-Hochberg false-discovery rate method [17] for multiple-testing correction as its default method. A typical GEO2R workflow would be:
Search GEO DataSets (see Section 2.3) to identify a suitable study of interest (see Note 8).
Access the GEO2R tool by clicking the text “Analyze with GEO2R” on the Series record. This brings the user to the GEO2R page which is organized with a table of all Samples in the Series complete with Sample accession numbers, titles and attributes such as cell type and tissue. Below the Sample table is a set of tabs (“GEO2R,” “Value Distribution,” “Options,” “Profile Graph,” and “R Script”) where analysis by GEO2R is initiated and data visualization and analysis options are available.
Check the distribution of the Sample values using the “Value Distribution” tab. Before performing an analysis with GEO2R it is important to check the distribution of the values that are to be analyzed. The quantitative data available for use with GEO2R comes directly from the user-provided Sample data tables with no GEO curation. These data may not be median-centered indicating that they have not been normalized and thus may not be suitable for cross-comparison. The value distributions can be viewed as graphically as a boxplot, or exported and saved as a number summary table.
Choose the groups of Samples to be analyzed. For example, GSE18388 is a study of gene expression in the thymuses of mice flown in space compared to those of earth-bound control mice. In order to identify upregulated and downregulated genes in the mouse thymus after space flight, the Samples are placed into two groups. The groups are created and named by clicking “Define groups” at the top of the list of Samples and entering a group name, such as “space flown” and “control”. To assign Samples to groups, either click or drag the cursor on the Samples and once highlighted, click the group to which they belong (“space flown” or “control”). The Samples will be highlighted in the group color. More than two groups can be defined for testing across multiple factors.
Perform the analysis with default parameters by clicking “Top 250” on the “GEO2R” tab. The top 250 differentially expressed genes are presented in a new window, ordered by P-value. The expression pattern of each gene in the table can be visualized by clicking the row to depict expression profile graphs. The complete set of ordered results can be downloaded as a table by clicking “Save all results” (see Note 9).
If desired, default parameters GEO2R can be customized by choosing an alternative method for multiple-testing correction of p-values in the “Options” tab. Options are also available to skip or force a log transformation of the input data. If options are changed, GEO2R must be run again, performed by returning to the “GEO2R” tab and clicking “Recalculate”.
If desired, the “R Script” tab provides all of the R commands used in that analysis, and this information can be saved and used as a reference for how a set of results were calculated.
Alternatively, if a user is not interested in performing a comparison to identify differentially expressed genes, but rather only wants to view the expression profile of a specific gene in the Series, the “Profile Graph” tab can be used to draw the gene expression profile graph for that gene. A YouTube video demonstrating GEO2R features is available at http://www.youtube.com/watch?v=EUPmGWS8ik0.
2.9 Visualize data as a genome track
Data visualization is an important aspect of the analysis of many high-throughput sequencing experiments such as chromatin immunoprecipitation (ChIP-seq) and DNA methylation profiling (bisulfite-seq). Files with raw or normalized genome-wide signal are produced which can be loaded into online genome browsers. Once the data are entered into the genome browser, it is very easy to move to a gene of interest or zoom out to see signal across an entire chromosome. Since GEO receives files types appropriate for genome browser visualization (.WIG, .bedGraph, .BED, etc.) as processed data, GEO is in the process of making these files viewable as tracks in NCBI’s Genome Data Viewer. GEO records with tracks include a button with the text “See the Data on the Genome Data Viewer”. Clicking this button takes the user directly to the Genome Data Viewer where the track has been preloaded (Fig. 3). On the left side of the Genome Data Viewer page are several boxes with search options to view the desired genomic region: 1) An ideogram view of chromosomes to view signal over an entire chromosome 2) A search box for entering chromosome coordinates or gene name or accession, 3) A box called “Your Data” where additional files can be uploaded for side-by-side viewing with the preloaded tracks from GEO. At the time of this writing there are almost 10,000 GEO Samples with links to tracks preloaded in the Genome Data Viewer. Track creation is an ongoing project and the number of GEO records with tracks is expected to increase in the near future.
Fig. 3.
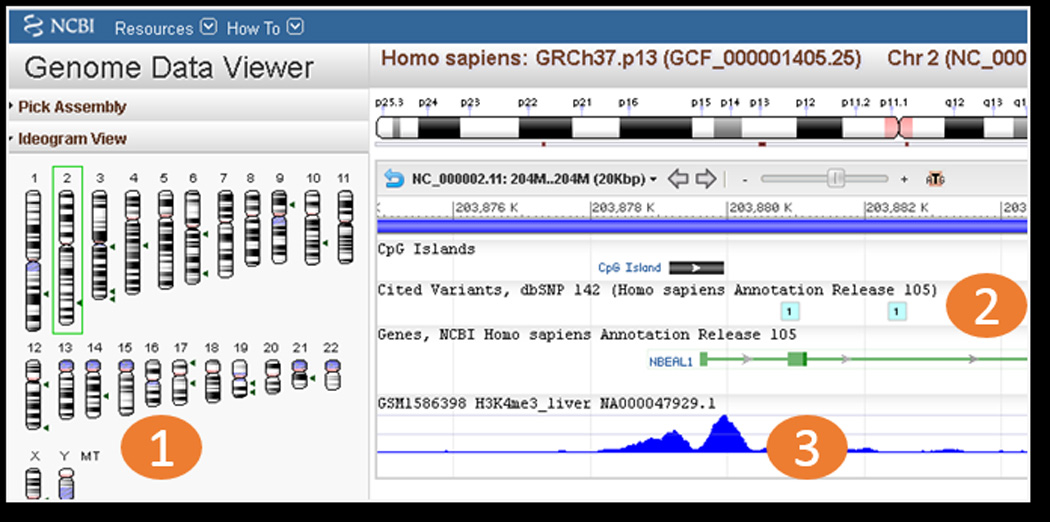
Screenshot of NCBI Genome Data Viewer. The left side of the viewer has tools for locating specific regions of the genome (1). The tracks area depicts RefSeq gene, CpG island, and SNP tracks which are set as default for context (2), and a track for GEO Sample GSM1586398 which is a H4K3me3 histone ChIP-seq experiment performed on liver tissue (3). This track shows a typical H3K4me3 double peak with depletion at the transcriptional start site of gene NBEAL1.
2.10 Download GEO data
All GEO data can be downloaded in various formats using a variety of mechanisms. A popular method for downloading data for specific studies is to download directly from Series pages. At the bottom of each Series page, there is a banner with the text “Download family” under which there are links for downloading the data for that Series in 3 different formats:
SOFT formatted family file(s) is a link for downloading all of the Series, Sample and Platform data in a single SOFT formatted file. SOFT is an acronym that stands for “Simple Omnibus Format in Text” and formats the data as line-based, plain text.
MINiML formatted family file(s) is a link for downloading all of the Series, Sample, and Platform data in MiNIML formatted files. MiNIML is an acronym that stands for MIAME Notation in Markup Language, and formats the data as XML with separate data tables. MINiML is essentially an XML rendering of SOFT format.
Series Matrix File(s) is a link for downloading a tab-delimited value-matrix table generated from the “VALUE” column of each Sample record, headed by Sample and Series metadata. This format is convenient for uploading into data programs such as Microsoft Excel or R.
The Series page also contains links to any supplementary files associated with the Series and a link to a tar archive of all supplementary files provided with the Samples, typically raw data files (see Note 10). If only a subset of the supplementary files are required there is an option to customize the set of files in the tar archive by clicking the word “custom” on same line as “GSExxx_RAW.tar”. Clicking the “custom” button expands the page to include a list of all Sample supplementary files in the Series with check boxes to select the desired files. Once the boxes next to the needed files have been selected, pressing “Download” initiates the download of a tar archive containing only the selected files.
Additional options for downloading data, including downloading specific portions of records, or programmatic approaches are described at http://www.ncbi.nlm.nih.gov/geo/info/download.html.
3. Conclusion
The GEO database is now 15 years old, and continues to serve as the leading public repository for direct deposits of high-throughput gene expression and other functional genomics data sets. GEO offers fast and efficient accessioning of raw and processed data with experimental descriptions for the worldwide research community at no cost to the submitter or user. GEO archives these data and makes the data available through flexible querying and download capabilities, and offers several Web-based tools and graphical renderings that facilitate data interpretation and exploration. These tools enable researchers to analyze GEO data with no prerequisite computational skills or software, and without time-consuming download or processing, thereby greatly increasing the utility of the data. GEO continues to evolve to accommodate new data types and increase and improve access to data.
The availability of the high-throughput data in GEO is driving new research. Several thousand publications exist where GEO data have been reused and reanalyzed to develop and test new hypotheses (http://www.ncbi.nlm.nih.gov/geo/info/citations.html). It is evident that the community is using GEO data to address matters far beyond those the initial studies were intended to tackle. Examples include using GEO data to test new or improved algorithms [18], create new subject-specific databases [19], identify disease biomarkers [20], and further characterize gene function [21]. These types of reuse of large data increase the pace and efficiency of scientific discovery and demonstrate the power of the GEO database as a resource for all scientists.
Acknowledgments
The authors acknowledge the expertise and efforts of the whole GEO curation and development team - Steve Wilhite, Pierre Ledoux, Carlos Evangelista, Irene Kim, Kimberly Marshall, Katherine Phillippy, Patti Sherman, Cynthia Robertson, Hyeseung Lee, Maxim Tomashevsky, Andrey Yefanov, Nadezhda Serova, Naigong Zhang, and Alexandra Soboleva.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. This chapter is an official contribution of the National Institutes of Health; not subject to copyright in the USA
Footnotes
Record types, accession codes, and their relationships to each other are described in detail at http://www.ncbi.nlm.nih.gov/geo/info/overview.html. Three primary record types, referred to as Platform (GPLxxx), Sample (GSMxxx), and Series (GSExxx), are supplied by submitters. A Platform record is composed of a summary description of the array or sequencer and, for array-based Platforms, a data table defining the array template. A Sample record describes the conditions under which an individual Sample was handled, the manipulations it underwent, and the measurements derived from it. A Series record links together a group of related Samples and provides a focal point and description of the whole study. A fourth record type, referred to as DataSets (GDSxxx), are assembled by the GEO curation staff from the three primary records. A DataSet represents a curated collection of biologically and statistically comparable GEO Samples and forms the basis of the GEO DataSets and GEO Profiles analysis tools. Only array-based expression data are currently considered for DataSet creation, and not all expression data qualify (for instance, due to having experimental designs incompatible with GEO tools). Furthermore, many expression studies have not yet been reviewed by the curation staff for DataSet creation.
It is advisable to be aware of full search capabilities including: using asterisks as a wild cards; the fact that some fields accept ranges of data; putting quotes around text to retrieve specific phrases; and proper placement of parentheses.
Analyses with GEO DataSet tools and GEO2R cannot be performed across multiple Series. Each data normalization is performed only for the Samples within a Series thus the normalized signals may be quite different across Samples from different Series, making direct cross-Series analysis without re-normalization invalid. However, by using GEO DataSets or GEO2R to identify differentially expressed genes in two independent Series in the same subject area and with similar experimental designs can be a powerful way to identify genes that are consistently identified in specific diseases, cell types, treatments, etc.
Not all the Samples within the DataSet need be included in the comparison.
The download file only includes Profiles shown on the current page; to get the maximum number of Profiles, go to the “Display Settings” link and set the “Items per page” to 500.
It is important to note that the values (red columns) and ranks (blue squares) are charted on different scales - the blue ranks are always on a scale of 1–100% (right Y axis of the chart) while the red value scale slides to fit the values of a particular profile (left Y axis of the chart). This sliding value scale allows subtle differences in values to be more clearly visualized.
Clicking the subset type names resorts the chart according to a particular experimental variable - this can assist in clearer visualization of an expression trend in DataSets with multiple variables.
Data for use in GEO2R are provided directly by submitters. Data for all Samples in a Series may not meant to be comparable due to a loop design, non-standard or no normalization. Users should thoroughly read through the Series and Sample descriptions to make sure the planned analysis with GEO2R is appropriate for the Samples.
The GEO2R results table contains various additional categories of gene annotation that are not immediately visible in default settings, including Gene Ontology (GO) terms and chromosome locations. Use the “Select columns” link to amend your table with this information.
The Series tar archive of supplementary files is a convenient way to download all supplementary files at one time, instead of individually downloading the files from each Sample record.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270(5235):484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, International Human Genome Sequencing C. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274(5287):546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 5.Mouse Genome Sequencing C. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 6.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 7.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 8.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Microarray standards at last (2002) Nature. 419(6905):323. doi: 10.1038/419323a. [DOI] [PubMed] [Google Scholar]
- 10.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 12.Gibney G, Baxevanis AD. Searching NCBI Databases Using Entrez. Current protocols in human genetics / editorial board, Jonathan L Haines [et al] 2011;Chapter 6(Unit6):10. doi: 10.1002/0471142905.hg0610s71. [DOI] [PubMed] [Google Scholar]
- 13.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33(Database issue):D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 16.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 18.Tejera E, Bernardes J, Rebelo I. Co-expression network analysis and genetic algorithms for gene prioritization in preeclampsia. BMC medical genomics. 2013;6:51. doi: 10.1186/1755-8794-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni M, Ye F, Zhu J, Li Z, Yang S, Yang B, Han L, Wu Y, Chen Y, Li F, Wang S, Bo X. ExpTreeDB: web-based query and visualization of manually annotated gene expression profiling experiments of human and mouse from GEO. Bioinformatics. 2014;30(23):3379–3386. doi: 10.1093/bioinformatics/btu560. [DOI] [PubMed] [Google Scholar]
- 20.Meng J, Li P, Zhang Q, Yang Z, Fu S. A radiosensitivity gene signature in predicting glioma prognostic via EMT pathway. Oncotarget. 2014;5(13):4683–4693. doi: 10.18632/oncotarget.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Zhang Y, Gao Y, Cui Y, Liu H, Li M, Tian Y. Downregulation of HNF1 homeobox B is associated with drug resistance in ovarian cancer. Oncology reports. 2014;32(3):979–988. doi: 10.3892/or.2014.3297. [DOI] [PubMed] [Google Scholar]


