Abstract
Understanding the structural basis of antibacterial resistance may enable rational design principles that avoid and subvert that resistance, thus leading to the discovery of more effective antibiotics. In this review, we explore the use of crystal structures to guide new discovery of antibiotics that are effective against resistant organisms. Structures of efflux pumps bound to substrates and inhibitors have aided the design of compounds with lower affinity for the pump or inhibitors that more effectively block the pump. Structures of β-lactamase enzymes have revealed the mechanisms of action toward key carbapenems and structures of gyrase have aided the design of compounds that are less susceptible to point mutations.
KEYWORDS : β-lactamase, crystal structures, efflux pump, gyrase
The global prevalence of organisms resistant to antibiotics continues to erode our ability to effectively treat infectious disease. Both Gram-positive (e.g., methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus or Streptococcus pneumoniae) and Gram-negative (e.g., carbapenem-resistant Enterobacteriaceae, Acinetobacter or Pseudomonas aeruginosa) bacteria exhibit resistance to multiple drugs and have been labeled as urgent or serious threats to human health by the Centers for Disease Control [1]. In some cases for Gram-negative Enterobacteriaceae, there are so few treatment options available that older drugs with known toxicity have become the preferred therapeutics. In response to these problems, it is critical to renew vigorous efforts to tackle the problem of drug resistance with new drug discovery.
Overcoming resistance to existing antibiotics often involves iteratively optimizing a first-generation drug to design second-generation compounds that are effective against the resistant organism. In this process, each generation of compounds undergoes expensive development and evaluation. An alternative approach focuses on incorporating and overcoming potential resistance early in the development cycle in order to produce a first-generation drug that is less susceptible to resistance. Ideally, a potential mechanism of resistance would be identified for the candidate and models of the mechanism would be employed to drive the design of improved candidates before introduction to clinical trials. This strategy of using structural models of resistance mechanisms has been particularly effective in other clinical indications with examples such as the development of HIV protease and reverse transcriptase inhibitors [2,3] as well as several tyrosine kinase inhibitors [4,5].
There are several major resistance mechanisms that affect antibiotics, namely efflux of the drug from the cell, enzymatic modification of the drug, target mutations that reduce affinity of the drug, upregulation of the target and permeability changes to the membrane. Of these types of resistance mechanisms, it is primarily efflux, enzymatic modification and target mutations that can be modeled at this time, mostly owing to the determination of critical crystal structures of efflux pumps, drug-modifying enzymes and modified targets. In the remainder of this review, we will discuss attempts to model each of these three resistance mechanisms and discuss the burgeoning efforts in drug discovery to use those models to understand resistance and aid in the discovery of new therapeutics.
Extrusion of the drug: efflux
Efflux is the active removal of antibiotics via a protein pump from the bacterial cell. While the direct decrease in intracellular drug concentration contributes to resistance in an obvious manner, efflux also contributes to resistance by providing bacteria time to accumulate mutations in other drug targets to increase resistance; this sequence is known to occur with fluoroquinolones [6]. Genes encoding efflux pumps are either located on the chromosome or acquired through plasmid transmission. It is often thought that overexpression of efflux pumps is among one of the first steps in the development of antibiotic resistance; however, native expression levels can confer resistance to both single agents as well as entire classes of antibiotics. Narrow spectrum efflux pumps have one specific substrate (e.g., tetracycline), whereas broad spectrum pumps have the ability to bind and export multiple substrates. To date, there have been five families of chromosomally encoded efflux pumps identified, four of which have been determined to be clinically relevant: the resistance nodulation division (RND) family, the major facilitator superfamily (MFS), the staphylococcal multiresistance (SMR) family and the multidrug and toxic compound extrusion families. Clinically relevant efflux pumps are found in both Gram-positive and Gram-negative species of bacteria.
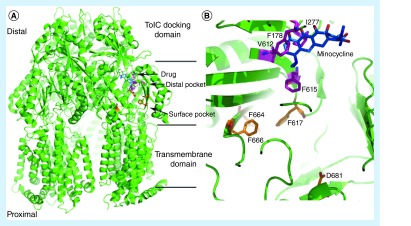
For the majority of efflux pump families, the pump transports antibiotics from the cytoplasm to the periplasmic space using a transmembrane proton gradient. Efflux pumps of the RND family have been of particular interest because of their ability to transport drugs directly to the external medium from the cytoplasm. The RND pumps are comprised of three components: a transporter protein, a periplasmic accessory protein, and an outer membrane protein channel. One of the more well-studied RND pumps is the AcrAB-TolC complex found in E. coli. Crystal structures reveal that AcrA, the periplasmic protein, coordinates the linkage of AcrB, the transporter protein, with TolC, the outer membrane channel. Structures suggest that substrates bind a pocket at the surface of AcrB and are then transported to a deeper (distal) pocket near the periplasmic domain before being funneled to the TolC unit (Figure 1A). Lipophilic substrates can diffuse toward the pockets where they bind to the wall of AcrB and are pumped out with the proton gradient.
Figure 1. . Structure of efflux pumps reveal substrate and inhibitor pockets.
(A) AcrB shown with the transmembrane, transporter and TolC docking domains shown (PDB ID 2DRD [11]). Minocycline is shown in blue, the residues observed from [9] in the distal pocket that interact with substrates are shown in magenta and the residues observed to interact with inhibitors shown in orange. The residues of the surface pocket overlap with the residues that interact with inhibitors. (B) An expanded view of the residues that interact with substrates (magenta) and inhibitors (orange). For clarity, the view in (B) is rotated 180 from that shown in (A).
There has been strong motivation to overcome or avoid resistance by efflux during the antibiotic development phase. These efforts have primarily focused on two strategies: one, the design of antibiotics that are not substrates of efflux pumps and two, the development of efflux pump inhibitors (EPIs) for coadministration with antibiotic substrates. Toward the first goal, cocrystallization of AcrB with various ligands has led to the hypothesis that the cavity of AcrB is filled with a lipid bilayer that is confluent with the inner membrane; therefore, substrate binding involves both protein residues as well as interactions with phospholipids. This organization allows pumps to bind compounds based on charge and lipophilicity. Positively charged ligands are bound using the head groups of negatively charged phospholipids. For example, fluoroquinolones behave as amphiphilic cations that bind the phospholipid and then diffuse to the central cavity for transport. Likewise, β-lactam antibiotics with lipophilic side chains are known to be good substrates of AcrB as they are expected to partition partially in the bilayer with the carboxylate groups exposed to the solvent [6]. The flexibility of the AcrB complex explains the wide range of substrates but does not necessarily provide a solution to avoiding the potential for novel antibiotics to be substrates of the pump.
Comparisons of AcrB with homologues within the RND family such as AcrD of E. coli and MexB of P. aeruginosa show there is some variation in the pocket residues that may explain pump/substrate selectivity. For example, the drug uptake channel of AcrD is lined with more acidic residues, which may potentially explain why AcrD transports aminoglycosides and AcrB does not. Likewise, where AcrB contains an acidic residue at Asp 301 in the ‘vestibule’ site outside the surface pocket, MexB of Pseudomonas contains more basic residues. As such, AcrB is better equipped to efflux cationic compounds [7].
Overall, there has been only preliminary advancement in the determination of structure-efflux activity relationships mostly owing to the vast diversity of substrates and lack of detailed understanding of substrate binding. Site-directed mutagenesis of residues near ligands observed in crystal structures has identified residues essential to AcrB activity [8]. Recently, molecular dynamics studies investigating the binding of several known substrates and two inhibitors have shown that known substrates bind the distal pocket, making specific interactions with hydrophobic residues Phe 178, Ile 277, Val 612 and Phe 615 [9]. Interestingly, simulations of two known inhibitors show that the compounds dock outside the distal pocket, possibly preventing the movement of substrates from the surface to the distal pocket. Although these studies are just getting underway, these detailed investigations of substrate and inhibitor-binding sites should aid the development of new antibiotics that avoid efflux as well as novel EPIs.
The majority of EPIs that have been explored immobilize the G-loop of AcrAB that is responsible for ushering substrates from the surface pocket to the distal pocket. EPIs that have this mechanism of action are generally structurally similar to the peptidomimetic, PAβN, but have not been used clinically owing to some toxicity issues. MBX2319, a novel pyranopyridine, is thought to inhibit AcrB via binding at the interface between the surface pocket and distal pocket [24]. Molecular dynamic simulations suggest that a cluster of phenylalanine residues plays an important role in inhibitor binding [24], which is supported by recently published crystal structures of AcrAB bound with pyridopyrimidine EPI derivatives [25]. MBX2319 shows specificity toward AcrB but has activity over a range of organisms [10] and demonstrates the ability to potentiate the activity of fluoroquinolones in bacterial strains overexpressing efflux pumps by up to eightfold [26].
Modification of the drug: β-lactamase enzymes
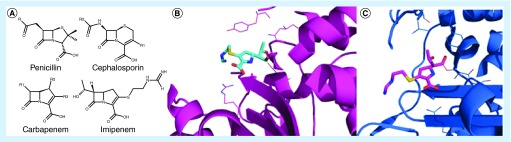
β-lactam antibiotics, including penicillins, cephalosporins and carbapenems (Figure 2A), have a broad spectrum of activity and are often used as a staple of therapy in clinically relevant infections caused by both Gram-positive and Gram-negative bacteria. Resistance to this class of antibiotics, primarily through drug modification via β-lactamases, has become a major barrier to effective treatment.
Figure 2 . Structures of β-lactam antibiotics bound to β-lactamase enzymes reveal mechanisms of cleavage and inhibition.
(A) β-lactam antibiotic structures and imipenem, the first clinically available carbapenem antibiotic [13]. Crystal structures of imipenem bound to (B) TEM-1 (PDB: 1BT5, magenta) and (C) AmpC (PDB: 1LL5, blue) β-lactamases [14,15].
β-lactam antibiotics target the transpeptidase domain of penicillin-binding proteins (PBPs) involved in bacterial cell wall synthesis. PBPs are essential to bacterial survival and the maintenance of cell wall integrity by acting as transpeptidases, cross-linking adjacent glycan strands in the cell membrane. The β-lactam ring is the defining characteristic of this class of antibiotics. It mimics the geometry of the D-alanine-D-alanine linkages of the glycan strands and is the basis for affinity of these antibiotics toward the PBPs [12]. Chemically, the ring strain drives a rapid reaction leading to irreversible inhibition. Inhibition, via acylation of the PBP to the β-lactam ring, halts cell wall synthesis while peptidoglycan autolysis continues, creating a compromised cell wall. The cells succumb to osmotic pressure, resulting in cell lysis [12].
The prevalence of chromosomal and plasmid-acquired β-lactamases has diminished the efficacy of most β-lactam antibiotics [12]. There are four different classes of β-lactamases based on structural homology. Classes A, C and D β-lactamases employ a nucleophilic serine to acylate the β-lactam bond while Class B uses a Zn2+ ion in the active site [13]. As β-lactamases have a high degree of structural homology to the PBPs, β-lactam antibiotics exhibit high affinity for these enzymes. β-lactams bind Class A β-lactamases and the nucleophilic active site serine acylates the carboxylate on the β-lactam ring via a ring opening reaction [15]. Once acylated, an active site water molecule hydrolyzes the acyl bond and the cleaved, deactivated β-lactam antibiotic is released from the active site [15]. Extended-spectrum β-lactamases (ESBLs), mainly from Class A, arose from early β-lactam use and are active in certain Gram-negative bacteria. ESBLs inactivate cephalosporin antibiotics that are typically resistant to acylation via β-lactamases [12].
Among the β-lactam antibiotics that are not susceptible to deactivation by serine β-lactamases (Classes A, C and D) is a class of antibiotics called carbapenems. Structurally, carbapenems have a similar 4–5 fused ring scaffold as penicillins differing by carbon substitution at the C1 position and a double bond at the C2-C3 position of the pyrroline ring (Figure 2). Of the clinically relevant β-lactams, carbapenems have the greatest potency and broadest spectrum of activity toward Gram-negative and Gram-positive bacteria [13]. Carbapenems act as β-lactamase inhibitors and are thus inherently resistant to deactivation by β-lactamases. Imipenem, the first clinically available carbapenem, binds the β-lactamase active site with high affinity and like other β-lactam antibiotics becomes acylated. However, owing to a slow rate of hydrolysis, imipenem acts more as an inhibitor than a substrate. A crystal structure of imipenem bound to TEM-1, a Class A β-lactamase (PDB: 1BT5) and AmpC, a Class C β-lactamase (PDB:1LL5) captures the acyl intermediate that is key to its action as a β-lactamase inhibitor (Figure 2). The structures show the 6α-1R-hydroxyethyl substitutions on the β-lactam ring cause the ligand to adopt an unexpected, strained conformation once bound to the enzyme. This strained conformation displaces the hydrolytic water in the active site, protecting it from deacylation [14,15].
Discovery of newer generation carbapenems is critical in order to overcome emerging resistance by Class B β-lactamases and the metabolic instability of imipenem. Imipenem is susceptible to elimination via dehydropeptidase-1 (DHP-1) found in the renal tubules, requiring the co-administration of a DHP-1 inhibitor to improve stability. Doripenem, a later generation carbapenem, corrects for imipenem's metabolic instability, increases its Gram-negative bacterial activity and has reduced β-lactamase susceptibility [16].
In addition to overcoming metabolism by DHP-1, attempts have been made to overcome resistance to Class B, metallo-β-lactamases. This has proven to be difficult, although strides have been made using the carbapenem transition state as a basis for novel β-lactamase inhibitors [17]. Additionally, crystal structures of metallo-β-lactamases, such as the recently published New Delhi metallo-β-lactamase [18], will aid the design of inhibitors. Other motivating factors for new carbapenem discovery include overcoming lowered PBP affinity due to point mutations, resistance due to efflux, and issues overcoming poor cellular penetration [13].
Modification of the target: binding site mutations
Strains with mutations in the target protein that reduce contacts, either directly or indirectly, between the drug and protein may arise by selection in the presence of the drug. The mutations may occur in the first shell of residues that contact the drug and eliminate key contacts or may arise in the outer shell and affect the conformation or dynamics of the drug-binding site [19]. If target mutation is a mechanism by which resistance arises for a given drug, it is possible to determine the structures of both the wild-type and mutated (resistant) proteins in order to understand the basis of resistance, thus enabling a modeling approach to predicting and overcoming the loss of interactions.
Bacterial topoisomerases, including DNA gyrase (topoisomerase II) and DNA topoisomerase IV, are essential for replication. The mechanism of the heterotetrameric topoisomerases (Gyr A/B or Par C/E) is characterized by the creation of a DNA strand break, the passage of another DNA duplex through the broken strand followed by the religation of the broken strand. Over time, the inhibition of this reaction leads to an accumulation of strand breaks and chromosomal fragmentation, eventually resulting in cell death. The topoisomerases have formed a successful platform for drug discovery for the past several decades, beginning with the discovery of the Gram-negative antibacterial activity of nalidixic acid. Over the years, there have been several generations of quinolone antibiotics [20], with notable examples including ciprofloxacin, levofloxacin and moxifloxacin. Each generation of quinolones has expanded the spectrum of coverage of these compounds such that they now inhibit Gram-negative and Gram-positive bacteria, Pseudomonas and Mycobacterium tuberculosis. The quinolones exert their inhibition by stabilizing the cleaved DNA/enzyme complex.
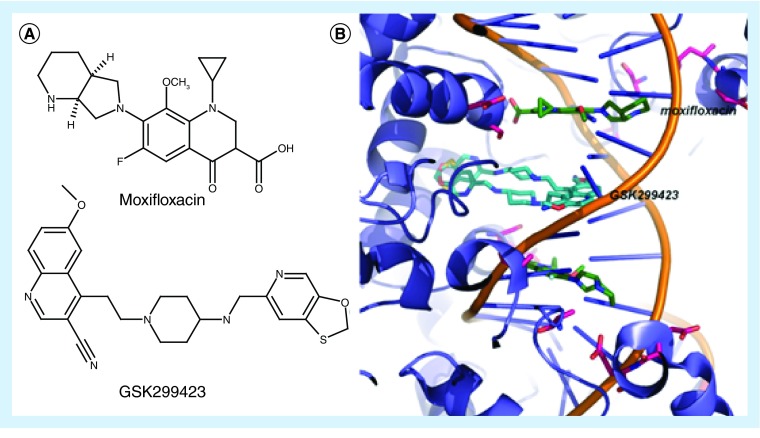
The primary mechanism of resistance to fluroquinolones is a stepwise accumulation of mutations in TopoIV and gyrase, many of which are in the ‘quinolone resistance determining region’ (QRDR) in the GyrA subunit. High levels of resistance are often achieved through mutations in the target combined with upregulation of efflux pump activity, for example norA in S. aureus. Crystal structures of Acinetobacter baumanii [21] or Streptococcus pneumoniae [22] ParC/E bound to DNA and moxifloxacin (Figure 3) have revealed many of the details of the mechanisms of topoisomerases, the mode of inhibition of the quinolone antibiotics and the structural basis of quinolone resistance.
Figure 3. . Point mutations in gyrase confer resistance to fluoroquinolones.
(A) Chemical structures of moxifloxacin and GSK299423 (B) the crystal structure of gyrase (purple) bound to DNA (backbone shown in orange) and two copies of GSK299423 (cyan) from PDB ID: 2XCS [23]. The figure shows a superposition of moxifloxacin (green) from PDB ID: 3FOF [22]. Typical resistance mutations are shown in magenta.
In response to the observed resistance to quinolones, several groups have worked to design more effective molecules that are active against the resistant strains. For example, scientists at GlaxoSmithKline designed compound GSK299423 [23] to act via a mechanism distinct from the quinolones based on stabilization of a precleavage enzyme-DNA complex. Structures of GSK299423 show that the compound occupies a novel noncatalytic-binding pocket between the two GyrA subunits (Figure 3), which is adjacent to but not overlapping with the ciprofloxacin site. As such, the affinity of the compound is not affected by the existing resistance mutations. In fact, GSK299423 shows potent activity against several bacterial strains, both wild-type and fluoroquinolone-resistant.
Conclusion
As resistance to current antibiotics spreads world-wide, it is critical to design and develop new antibiotics that overcome these existing mechanisms while possessing traits that extend their own effective lifetimes. High-resolution crystal structures of targets or resistance-conferring enzymes are critical in aiding this development of new antibiotics. Structures have assisted our understanding of the mechanisms of efflux pumps and β-lactamases, providing a rational basis for the design of new compounds that will avoid acting as substrates or will act as inhibitors. Structures of the mutated gyrase have supported the design and development of inhibitors unaffected by existing mutations.
Future perspective
As more high resolution crystal structures of efflux pumps, β-lactamase enzymes or drug targets bound to various antibiotics or inhibitors become available, it is increasingly possible to rationally design next-generation antibiotics to overcome existing mechanisms. Examples of the new generation of compounds such as some efflux pump or β-lactamase inhibitors may be administered as adjuvants, bringing a revised perspective into antibiotic treatment. Additional structures of enzymes that undergo specific point mutations will also aid our understanding of the structural basis of resistance and the greater impact on fitness, the role of compensatory mutations and the discovery of next-generation antibiotics that oppose resistance.
EXECUTIVE SUMMARY.
Understanding the mechanisms of potential resistance to antibacterial drugs and ideally attempting to overcome that resistance early in the development process will enable a more efficient path for new antibiotic discovery.
Models of resistance mechanisms based on crystal structures have begun to aid in the design of drugs that are less susceptible to resistance.
This review highlighted the design of compounds to overcome three mechanisms of antibacterial drug resistance.
Structures of efflux pumps have been used to elucidate critical interactions between amino acids and drugs that are substrates for these pumps as well as to understand the mechanism by which inhibitors function.
Crystal structures of serine β-lactamase enzymes have revealed the mechanisms by which β-lactam substrates are cleaved and the structural basis of inhibition by carbapenems. New structures of metallo-β-lactamases will aid the design of inhibitors for these enzymes.
Structures of wild-type and fluoroquinolone-resistant gyrase have been instrumental in guiding the design of inhibitors active against the resistant enzyme.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Centers for Disease Control. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013–508.pdf
- 2.Ghosh A, Dawson Z, Mitsuya H. Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV. Bioorg. Med. Chem. 2007;15:7576–7580. doi: 10.1016/j.bmc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das K, Clark AD, Lewi PJ, et al. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (Etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 2004;47(10):2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 4.Huang W-S, Metcalf CA, Sundaramoorthi R, et al. Discovery of 3-[2-(Imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J. Med. Chem. 2012;53(12):4701–4719. doi: 10.1021/jm100395q. [DOI] [PubMed] [Google Scholar]
- 5.Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4701–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piddock L. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu E, Aires J, Nikaido H. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 2003;185:5657–5664. doi: 10.1128/JB.185.19.5657-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vargiu A, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics. Proc. Natl Acad. Sci. USA. 2012;109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Opperman T, Kwasny S, Kim H-S, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli . Antimicrob. Agents Chemother. 2014;58(2):722–733. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 12.Drawz S, Bonomo R. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papp-Wallace K, Endimiani A, Taracila M, Bonomo R. Carbapenems: past, present and future. Antimicrob. Agents Chemother. 2011;55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beadle B, Shoichet B. Structural basis for imipenem inhibition of class C β-lactamases. Antimicrob. Agents Chemother. 2002;46:3978–3980. doi: 10.1128/AAC.46.12.3978-3980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maveyraud L, Mourey L, Korta L, et al. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. J. Amer. Chem. Soc. 1988;120:9748–9752. [Google Scholar]
- 16.Bonfidlio G, Russo G, Nicoletti G. Recent developments in carbapenems. Expert Opin. Investig. Drugs. 2002;11:529–544. doi: 10.1517/13543784.11.4.529. [DOI] [PubMed] [Google Scholar]
- 17.Yang K, Feng L, Yang S, et al. New β-phospholactam as a carbapenem transition state analog: synthesis of a broad-spectrum inhibitor of metallo-β-lactamases. Bioorg. Med. Chem. Lett. 2013;23:5855–5859. doi: 10.1016/j.bmcl.2013.08.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King D, Worrall L, Gruninger R, Strynadka N. New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J. Am. Chem. Soc. 2012;134:11362–11365. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- 19.Anderson A. Winning the arms race by improving drug discovery against mutating targets. ACS Chem. Biol. 2012;7:278–288. doi: 10.1021/cb200394t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohlkonig A, Chan P, Fosberry A, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struc. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 22.Laponogov I, Sohi M, Veselkov D, et al. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struc. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 23.Bax B, Chan P, Eggleston D, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 24.Vargiu A, Ruggerone P, Opperman T, et al. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014;58:6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakashima R, Sakurai K, Yamasaki S, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 26.Opperman T, Kwasny S, Kim H, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli . Antimicrob. Agents Chemother. 2014;58:722–733. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]