A P. patens signaling pathway required for immunity triggered by PAMPs induces growth inhibition, a novel fluorescence burst, cell wall depositions, and accumulation of defense-related transcripts.
Abstract
MAP kinase (MPK) cascades in Arabidopsis thaliana and other vascular plants are activated by developmental cues, abiotic stress, and pathogen infection. Much less is known of MPK functions in nonvascular land plants such as the moss Physcomitrella patens. Here, we provide evidence for a signaling pathway in P. patens required for immunity triggered by pathogen associated molecular patterns (PAMPs). This pathway induces rapid growth inhibition, a novel fluorescence burst, cell wall depositions, and accumulation of defense-related transcripts. Two P. patens MPKs (MPK4a and MPK4b) are phosphorylated and activated in response to PAMPs. This activation in response to the fungal PAMP chitin requires a chitin receptor and one or more MAP kinase kinase kinases and MAP kinase kinases. Knockout lines of MPK4a appear wild type but have increased susceptibility to the pathogenic fungi Botrytis cinerea and Alternaria brassisicola. Both PAMPs and osmotic stress activate some of the same MPKs in Arabidopsis. In contrast, abscisic acid treatment or osmotic stress of P. patens does not activate MPK4a or any other MPK, but activates at least one SnRK2 kinase. Signaling via MPK4a may therefore be specific to immunity, and the moss relies on other pathways to respond to osmotic stress.
INTRODUCTION
Innate immune systems are required for pathogen detection and elimination. Plants and animals have evolved surface-localized, pattern recognition receptor kinases (PRRs) that detect conserved pathogen-associated molecular patterns (PAMPs; Zipfel, 2009). PAMP detection by PRRs induces PAMP-triggered immunity (PTI; Jones and Dangl, 2006). Examples of PAMP recognition by PRRs in flowering plants such as Arabidopsis thaliana include perception of bacterial flagellin by FLS2, elongation factor Tu by EFR1, and fungal chitin by CERK1 (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006; Miya et al., 2007).
Activated PRR signals are transmitted and amplified within minutes by MAP kinase cascades. These involve activation by phosphorylation of a MAP kinase kinase kinase (MEKK), a MAP kinase kinase (MKK), and a MAP kinase (MPK), which phosphorylates substrate proteins including transcription factors (Rodriguez et al., 2010). At least four Arabidopsis MPKs (MPK3/4/6 and 11) are activated by PRRs (Fiil et al., 2009; Bethke et al., 2012), and Arabidopsis MPK1 and 2 are also implicated in immunity to the fungal pathogen Botrytis cinerea (Weiberg et al., 2013). In addition to their roles in Arabidopsis immunity, MPK3/4/6 have been implicated in responses to abiotic stress including osmotic, salt, and cold stresses (Droillard et al., 2004; Teige et al., 2004; Rodriguez et al., 2010). However, the contributions of single MPKs in Arabidopsis immunity or stress responses remain unclear due to the pleiotropic phenotypes of some of their loss-of-function mutants and to their potential redundancies (Petersen et al., 2000; Ren et al., 2008; Zhang et al., 2012).
The Arabidopsis genome encodes 20 MPKs, including MPK4, which functions in a cascade(s) with MEKK1 and MKK1 and 2 (Ichimura et al., 1998; Qiu et al., 2008). In response to pathogens or PAMPs, this cascade permits the WRKY33 transcription factor to enhance the expression of defensive genes including PAD3 required for synthesis of antimicrobial camalexin (Suarez-Rodriguez et al., 2007; Qiu et al., 2008). The Arabidopsis mpk4 loss-of-function mutants exhibit dwarfism and ectopically express defense responses but are suppressed by mutation of SUMM2, which encodes a resistance (R) protein (Zhang et al., 2012). This indicates that the MPK4 pathway is a target of pathogen effectors and is monitored by SUMM2, in line with the guard hypothesis of effector-triggered immunity in plant-pathogen evolution (Jones and Dangl, 2006; Zhang et al., 2012).
The moss Physcomitrella patens is a bryophyte, an early branch of land plants (Zimmer et al., 2007). P. patens responds to pathogens, including fungi, oomycetes, and bacteria, by activating defenses including production of reactive oxygen species (ROS), cell wall fortifications, hormonal perturbations, and expression of defense-related genes (Lehtonen et al., 2012; Mittag et al., 2015; Oliver et al., 2009; Ponce de León et al., 2007, 2012). However, PTI components have not been identified in P. patens or other bryophytes.
The P. patens genome encodes only eight MPKs (Dóczi et al., 2012). They represent a basal set of pathways whose functions may be assessed by their targeted knockouts (KOs) or knockins (KIs) via homologous recombination, a genetic approach unavailable in vascular plants. Comparative analyses of such moss lines may elucidate aspects of the complexity of MPK activation in flowering plants and how plant immunity and stress responses have evolved.
To initiate such analyses, we examined if P. patens MPKs respond to PAMPs or abiotic stresses by assaying MPK phosphorylation and in-gel kinase activity. This showed that at least two MPKs are rapidly phosphorylated and activated in response to bacterial and fungal PAMPs. Targeted gene KOs and KIs identified components of this pathway, including a PRR, at least one MEKK and one MKK, and two MPKs required for responses to the fungal PAMP chitin and resistance to two fungal pathogens. In contrast, osmotic stress did not result in detectable MPK phosphorylation or activation in the moss but did result in rapid activation of at least one SnRK2 kinase of a canonical abscisic acid response pathway.
RESULTS
Phosphorylation and Activation of P. patens MPKs in Response to Chitin
We first assayed MPK phosphorylation in P. patens protein extracts using an anti-p44/42-ERK antibody (α-pTEpY) following treatment with bacterial and fungal PAMPs. MPK phosphorylation was undetectable following treatment with bacterial elongation factor Tu peptide (elf18; Roux et al., 2011), conserved flagellin peptides (Pseudomonas flg22; Gómez-Gómez and Boller, 2000; Salmonella flgII-28; Clarke et al., 2013) or full-length flagellins from Pseudomonas or Borrelia (Supplemental Figures 1A and 1B). This is in contrast to the elicitation patterns of at least three MPKs in Arabidopsis by these PAMPs, excluding Borrelia flagellin and Salmonella flgII-28 used as negative controls for Arabidopsis. The flagellin and elf18 insensitivity of P. patens is in line with the absence of close homologs of their respective Arabidopsis receptors FLS2 and EFR in P. patens (Boller and Felix, 2009). In contrast, treatment with the fungal PAMP chitin or its derivative chitosan induced phosphorylation and activation, assayed by phosphorylation of the general kinase substrate MBP, of two P. patens MPKs within 1 min (Supplemental Figures 1A and 1C). These findings are in line with those of Lehtonen et al. (2012, 2014) showing that chitosan triggers responses in P. patens including a ROS burst, defense transcript accumulation, and secretion of pathogen response proteins. In addition, these MPKs were rapidly phosphorylated in response to bacterial peptidyl glycans (PGNs; Supplemental Figure 1D), which have molecular patterns similar to chitin. This response is in line with studies showing that the Arabidopsis and rice (Oryza sativa) chitin receptors are also involved in PGN detection (Willmann et al., 2011; Ao et al., 2014). We conclude that at least two P. patens MPKs are activated in response to fungal and bacterial PAMPs.
The Moss Chitin Receptor CERK1
P. patens encodes four homologs of the Arabidopsis chitin receptor At CERK1 (Zhang et al., 2009), and the mRNA levels of two of these moss genes (Pp1s364 10V6.1 and Pp1s41 280V6.1) were enhanced by chitosan treatment (Supplemental Figures 2A and 2B and Supplemental Data Set 1). This may be relevant as stimuli that affect the activities of regulatory proteins may alter the levels of their corresponding mRNAs (Hruz et al., 2008). Targeted KO lines of these two P. patens CERK1 homologs were generated (Supplemental Figures 3A to 3C) and tested for PTI responses. Plants were transferred on cellophane sheets from normal media to media containing chitin to assess their responses. In contrast to the wild-type and KO lines of Pp1s41 280V6.1, chitin did not induce apparent cell wall modification readily stainable with Toluidine blue in Pp1s364 10V6.1 KO lines (hereafter designated Δcerk1; Figures 1A and 1B). This modification includes ethanol-insoluble, brownish red, cell wall-associated depositions in wild-type tissues, which become visible after 8 h of chitin treatment (Supplemental Figures 4A and 4B). Interestingly, chitin treatment of wild-type, but not Δcerk1 lines, led to complete growth inhibition within 1 min and to a strong fluorescence burst visible after 2 h, which peaked by 4 h and decayed by 8 h (Supplemental Figures 4A, 4C, and 4D, and Supplemental Movies 1 and 2). In addition, chitin, chitosan, chitin hexamer, or bacterial peptidyl glycan also did not induce phosphorylation of the two MPKs in Δcerk1 KO lines (Figures 1C and 1D). This effect was not due to a general impairment of PTI, as the MPKs were phosphorylated in these lines treated with bacterial cell-free culture filtrate from the harpin negative strain Pectobacterium wasabiae (ex Pectobacterium carotovorum strain SCC3193) (Figure 1E; Nykyri et al., 2012).
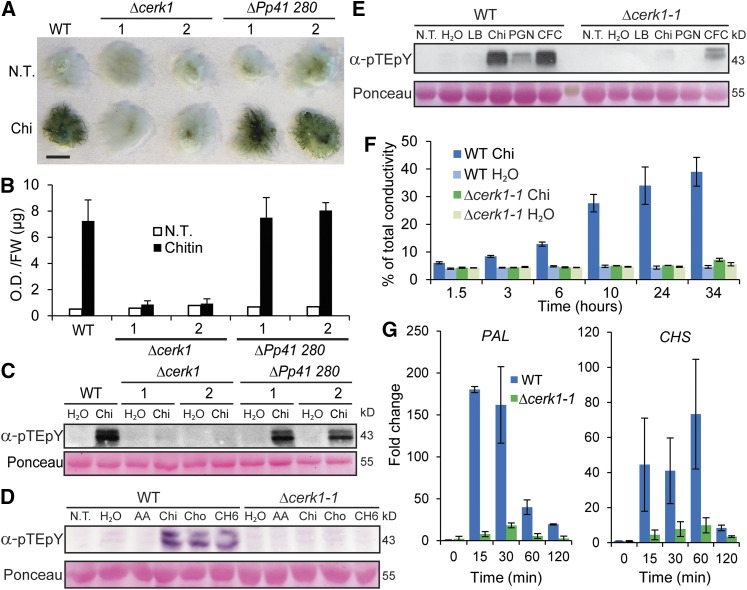
Figure 1.
Characterization of Chitin Receptor Knockout Lines.
(A) Toluidine blue staining of untreated (N.T.) and 48 h chitin (Chi) treated wild type and targeted (KOs) Pp CERK1 (Δcerk1-1 and 2) and another homolog (ΔPp41 280-1 and 2) of Arabidopsis CERK1. Bar = 5 mm.
(B) Quantitation of stainings in (A) (see Methods).
(C) Chitin (Chi) induced MAP kinase phosphorylation in the wild type and ΔPp41 280 but not in Δcerk1 KO lines assayed by immunoblotting with anti-phospho-p44/42 MPK antibody (α-pTEpY). Loading control is Ponceau-stained total protein.
(D) Immunoblot assay for MAP kinase phosphorylation of wild type and Δcerk1-1 KO line in response to no treatment (N.T.), water, 0.01% acetic acid (AA), 100 µg/mL chitin (Chi), 100 µg/mL chitosan (Cho), or 100 µg/mL hexa-N-acetylchitohexaose (CH6) for 5 min.
(E) Immunoblot assay for MAP kinase phosphorylation of wild type and Δcerk1-1 KO line in response to no treatment (N.T.), water, Luria broth (LB), 100 µg/mL chitin (Chi), 200 µg/mL peptidyl glycan from Staphylococcus aureus (PGN), or cell-free culture filtrate (CFC) from the harpin negative strain P. wasabiae SCC3193.
(F) Tissue ion leakage assay of the wild type and Δcerk1-1 treated with water or chitin for times specified. Data are presented as percent of total conductivity as an average of four replicates each with five plants. Error bars represent sd.
(G) RT-qPCR of PAL4 and CHS transcript levels in the wild type and Δcerk1-1 as fold change relative to the wild type untreated at times specified after 100 µg/mL chitin treatment. Data points represent an average of three independent experiments (three biological replicates) each with three technical replicates for each PCR reaction. Error bars represent se of the means.
Δcerk1 KO lines were also reduced in two other responses to chitin compared with the wild type. First, Δcerk1 tissues failed to exhibit enhanced conductivity (Figure 1F) using a standard assay for immunity-associated electrolyte leakage in Arabidopsis (Mackey et al., 2003). Second, Δcerk1 lines were compromised in the accumulation of mRNAs of the defense genes Phenylalanine Ammonia Lyase4 (PAL4) and Chalcone Synthase (CHS) (Figure 1G; Oliver et al., 2009). Finally, expression of Pp CERK1 complemented the Arabidopsis cerk1 mutant (Miya et al., 2007), as measured by chitin-induced MPK phosphorylation and accumulation of WRKY22 transcripts (Supplemental Figures 5A and 5B). We conclude that Pp CERK1 is required for moss responses to chitin and bacterial peptidyl glycans and is therefore an ortholog of the Arabidopsis chitin receptor At CERK1.
Two P. patens MEKKs Function Redundantly in Responses to Chitin
Arabidopsis PRR signaling via MAP kinase cascades includes MEKK1-MKK1/MKK2-MPK4 as well as MEKK1-MKK4/MKK5-MPK3/MPK6 (Asai et al., 2002; Qiu et al., 2008). The P. patens genome encodes 22 MEKK homologs (Sun et al., 2014). From the phylogenetic relationships with 21 MEKKs from Arabidopsis (Supplemental Figure 6A and Supplemental Data Set 2), two P. patens homologs of At MEKK1 were chosen as candidates and are here referred to as Pp MEKK1a (Pp1s136 5V6.3) and Pp MEKK1b (Pp1s31 252V6.1). The mRNA levels of these two genes increase in response to chitosan, although Pp MEKK1a displayed a stronger response (Supplemental Figure 6B). Targeted KO lines of these two MEKK genes (Δ mekk1a and Δ mekk1b; Supplemental Figures 3D and 3E) were generated and tested for responses to chitin.
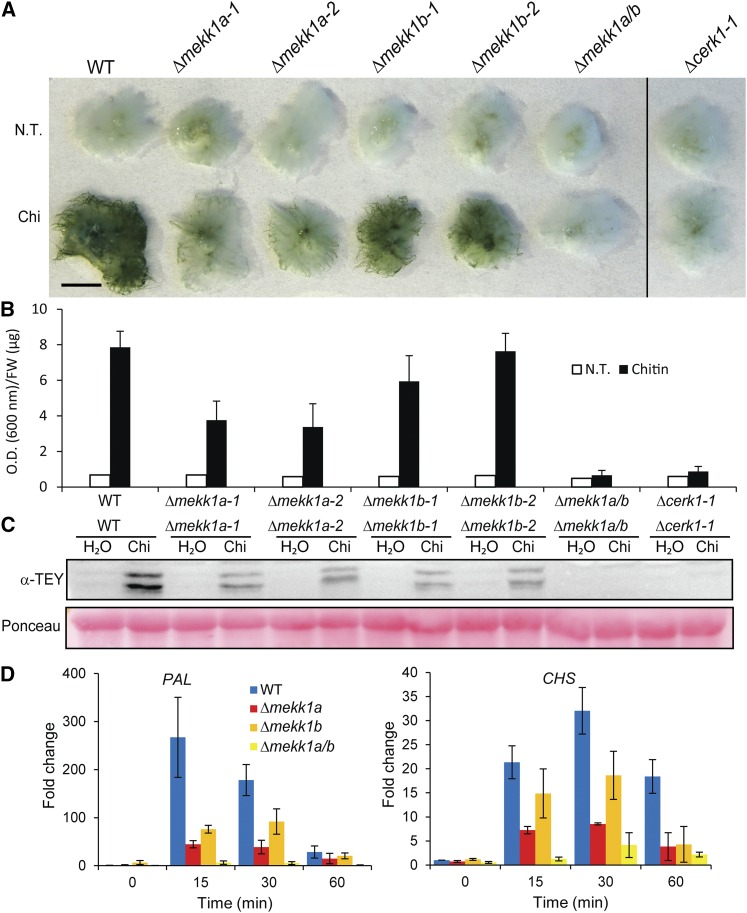
Δmekk1a exhibited strongly reduced chitin induced cell wall associated depositions compared with the wild type and Δmekk1b (Figures 2A and 2B). To assess potential redundancy of MEKK1a and MEKK1b, we generated the double KO Δmekk1a/b (Supplemental Figure 3F). This line, like the Δcerk1 KO line, was unable to induce cell wall depositions in response to chitin (Figures 2A and 2B). Chitin-induced MPK phosphorylation was also reduced in both Δmekk1b and Δmekk1a and absent in Δmekk1a/b (Figure 2C). The same trend was seen in these lines on the levels of mRNAs of the defense-related PAL4 and CHS genes (Figure 2D). In addition, Δmekk1a/b failed to induce the fluorescent burst when exposed to chitin, similarly to Δcerk1 (Supplemental Figure 7A and Supplemental Movie 3). We conclude that Pp MEKK1a and b have redundant functions in these chitin responses. Interestingly, the growth of Δmekk1a/b was inhibited within 1 min after chitin exposure as seen in wild-type moss (Supplemental Figure 7B and Supplemental Movie 4). This chitin response is therefore independent of MEKK1a and b.
Figure 2.
Characterization of MEKK KO Lines.
(A) Toluidine blue staining of untreated and 48 h chitin-treated wild type, Δcerk1-1, and single KOs of two MEKK homologs (Δmekk1a and b) and their double KO (Δmekk1a/b). Bar = 5 mm.
(B) Quantitation of stainings in (A).
(C) MAP kinase phosphorylation in the same moss lines treated with 100 µg/mL chitin and assayed by immunoblotting with anti-phospho-p44/42 MPK antibody (α-pTEpY). Loading control is Ponceau staining of total protein.
(D) Quantitative RT-PCR of PAL4 and CHS transcript levels in the wild type and Δmekk1a-1, Δmekk1b-1, and Δmekk1a/b as fold change relative to the wild type untreated at times specified after 100 µg/mL chitin treatment. Data points represent an average of three independent experiments, each with three technical replicates. Error bars represent se of the means.
Redundancy among MKKs in Response to Chitin
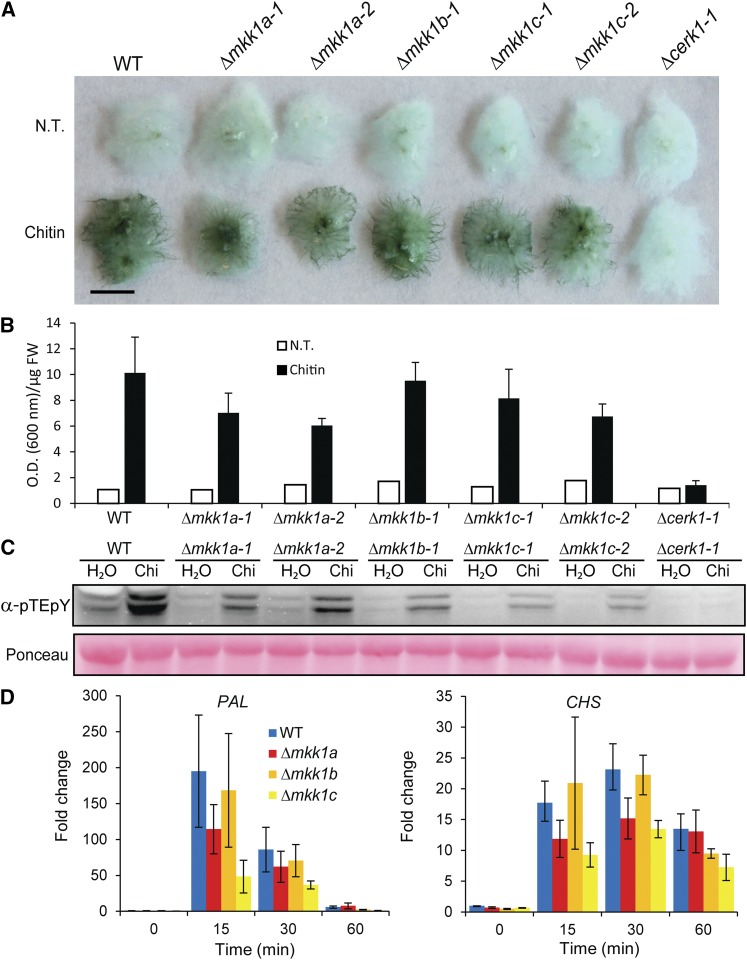
The P. patens genome encodes seven MKK homologs, three of which are most closely related to Arabidopsis MKK1, 2, and 6. Since At MKK1 and 2 are strongly implicated in immunity, we designated the three P. patens homologs Pp MKK1a (Pp1s32 219v6.1), Pp MKK1b (Pp1s106 83v6.1), and Pp MKK1c (Pp1s50 83v6.1) (Supplemental Figure 8A and Supplemental Data Set 3). The mRNAs of Pp MKK1a and b were slightly induced in response to chitosan, while Pp MKK1c mRNA was not (Supplemental Figure 8B). Targeted KO lines of these MKK genes were generated (Supplemental Figures 3G to 3I) and tested for responses to chitin. Toluidine blue staining revealed that chitin induced cell wall modification was slightly reduced in the two Δmkk1a and c KO lines compared with the wild type, but not in the Δmkk1b line (Figures 3A and 3B). Chitin-induced MPK phosphorylation also appeared slightly reduced in Δmkk1a and b lines and further reduced in the Δmkk1c lines (Figure 3C). In addition, the levels of mRNAs of the defense-related PAL4 and CHS showed a trend of being less induced following chitin treatment in Δmkk1a and c compared with the wild type and Δmkk1b (Figure 3D). Although these results do not pinpoint the involvement of a single moss MKK in chitin responses, they indicate that KOs of individual MKKs, in particular mkk1a and c, reduce responses to chitin. Combinatorial, multiple KOs of MKK1a, b, and c would be required to fully delineate their involvement in PAMP signal transduction.
Figure 3.
Characterization of MKK KO Lines.
(A) Toluidine blue staining of untreated and 48 h chitin-treated wild type, Δcerk1, and KOs of three MKK homologs (Δmkk1a, b, and c). Bar = 5 mm.
(B) Quantitation of stainings in (A).
(C) MAP kinase phosphorylation in the same moss lines treated with 100 µg/mL chitin and assayed by immunoblotting with anti-phospho-p44/42 MPK antibody (α-pTEpY). Loading control is Ponceau staining of total protein.
(D) Quantitative RT-PCR of PAL4 and CHS transcript levels in the wild type and Δmkk1a-2, Δmkk1b-1, and Δmkk1c-2 as fold change relative to the wild type untreated at times specified after 100 µg/mL chitin treatment. Data points represent an average of three independent experiments each with three technical replicates. Error bars represent se of the means.
MPK4a and b Activation in Response to Chitin and B. cinerea Inoculation
A phylogeny of MPKs from 10 plant species (Dóczi et al., 2012) identified eight putative MPKs in P. patens. Two of these homologs group with the Arabidopsis group B MPKs MPK4/5/11/12/13 (Supplemental Figure 9A and Supplemental Data Set 4; Ichimura et al., 2002). Arabidopsis MPK3/4/6/11 have all been shown to be phosphorylated and activated upon PAMP treatment (Gómez-Gómez and Boller, 2000; Bethke et al., 2012). Since the two P. patens MPKs are most similar to At MPK4 (Supplemental Figure 9A), we refer to them as Pp MPK4a (Pp1s149 39V6.1) and Pp MPK4b (Pp1s59 325V6.1). Pp MPK4a mRNA accumulated in response to chitosan within 15 min, peaked 8-fold at 2 h, and returned to basal levels by 8 h (Supplemental Figure 9B). In contrast, Pp MPK4b transcripts were only marginally affected by chitosan. Since the sizes of the two P. patens MPKs phosphorylated and activated in response to chitin (Figures 1C to 1E; Supplemental Figures 1C and 1D) match those predicted for MPK4a (42.8 kD) and MPK4b (43.5 kD), we created stable KI lines of these two MPK genes containing C-terminal GFP reporter fusions. We also created KO lines of MPK4a (Δmpk4a) but were unable to obtain stable knockout lines of MPK4b despite several attempts using different constructs. The recombined loci were verified in the three lines (Δmpk4a, MPK4a-GFP, and MPK4b-GFP; Supplemental Figures 3K to 3N).
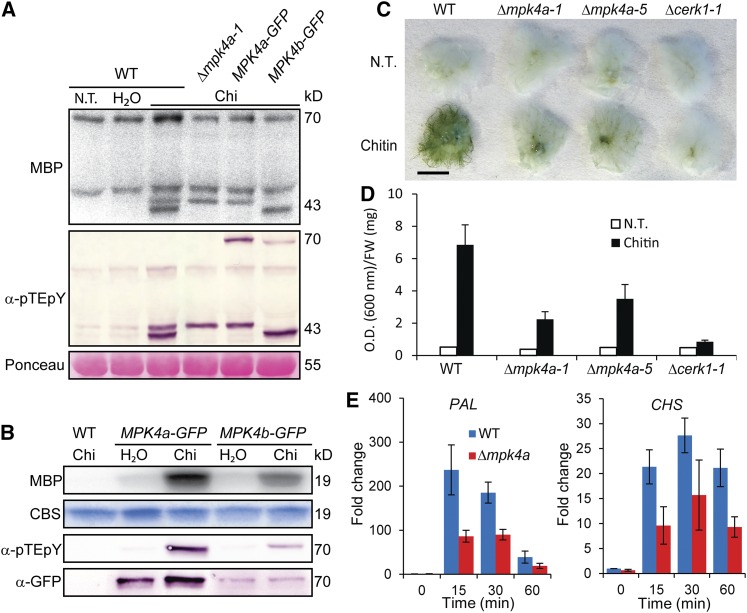
Wild-type and selected Δmpk4a, MPK4a-GFP, and MPK4b-GFP lines were treated with chitin and protein extracts subjected to immunoblot analysis of MPK activation loop phosphorylation with α-pTEpY and to in-gel kinase assays with MBP as substrate. Both the 42.8- and 43.5-kD MPKs became phosphorylated and activated in the wild type, but the 42.8-kD band could not be detected in the Δmpk4a or MPK4a-GFP lines (Figure 4A). In contrast, the 43.5-kD band was absent in MPK4b-GFP lines (Figure 4A). In addition, phosphorylated MPK4a-GFP and MPK4b-GFP fusion proteins with expected sizes of ∼70 kD (∼43-kD MPK with ∼27-kD GFP fusion) appeared after chitin treatment, although these bands were masked in the kinase assay by an unspecific band in all samples (Figure 4A). However, kinase assays of the ∼70-kD MPK-GFP fusions immunoprecipitated with an anti-GFP antibody confirmed that MPK4a-GFP and MPK4b-GFP could be activated by chitin treatment (Figure 4B). Thus, MPK4a and MPK4b represent the 42.8- and 43.5-kD bands, respectively, and are the two MPKs phosphorylated and activated in response to chitin and other PAMPs tested (Figures 1C to 1E).
Figure 4.
MPK Activation, Phosphorylation, and Expression upon Chitin Treatment.
(A) Total protein extracts assayed for kinase activity in gels with myelin basic protein (MBP) as substrate and for MPK phosphorylation by immunoblotting with anti-phospho-p44/42 MPK antibody (α-pTEpY). Loading control is Ponceau staining of total protein. The wild type subjected to no treatment (N.T.), water treatment, or 100 µg/mL chitin (chi), and Δmpk4a knockout and Pp MPK4a-GFP and Pp MPK4b-GFP knockin lines treated with 100 μg/mL chitin (Chi) for 5 min.
(B) Pp MPK4a-GFP and Pp MPK4b-GFP are activated and phosphorylated in response to chitin. The wild type and MPK4a-GFP and MPK4b-GFP lines subjected to water treatment or 100 μg/mL chitin (Chi) for 5 min. MPK4a-GFP and MPK4b-GFP were isolated with GFP-trap beads and assayed for kinase activity in solution with MBP as substrate, for MPK phosphorylation with α-pTEpY, and for MPK-GFP fusion levels with α-GFP. Loading control is Coomassie blue staining (CBS) of MBP.
(C) Toluidine blue staining of untreated and 48 h chitin-treated wild type, Δcerk1, and KO lines of Pp MPK4a (Δmpk4a-1 and 5). Bar = 5 mm.
(D) Quantitation of stainings in (C).
(E) Quantitative RT-PCR of PAL4 and CHS transcript levels in the wild type and Δmpk4a-5 as fold change relative to the wild type untreated at times specified after 100 µg/mL chitin treatment. Data points represent an average of three independent experiments, each with three technical replicates. Error bars represent se of the means.
We examined the localization of the MPK4a-GFP and MPK4b-GFP fusions as their chitin-inducible activities indicated that they were functional. Moss plants have three cell types that extend by tip growth: (1) chloronema with many large chloroplasts and transverse cell walls perpendicular to the growth axis; (2) caulonema with fewer, smaller chloroplasts and oblique transverse cell walls; and (3) rhizoids lacking chloroplasts that develop from the mature gametophyte. The MPK4a-GFP signal was strong in the apical cells of caulonemal air filaments and rhizoids in fully developed plants and less strong but readily detectable in filamentous protonemal tissue at the edge of the plant consisting of both chloronema and caulonema (Supplemental Figure 10A). When grown on media overlayed with cellophane, which promotes filamentous growth of protonema, the MPK4a-GFP signal was strongest in newly formed apical tip cells of protonemal tissue, whereas a 35S promoter-driven GFP control showed uniform expression throughout the plant (Supplemental Figure 10B). MPK4a-GFP localized to the cytoplasm and nucleus, and this pattern did not appear to change significantly following chitin treatment (Supplemental Figure 10C). This pattern of expression is similar to that of At MPK4-GFP in Arabidopsis (Andreasson et al., 2005). In contrast to MPK4a-GFP, MPK4b-GFP was undetectable. This may reflect the ∼20-fold lower transcript level of MPK4b compared with MPK4a in untreated plants (Supplemental Figure 9C).
Since chitin is a component of fungal cell walls, we examined if MPK4a and MPK4b are activated in response to the fungal pathogen B. cinerea. To this end, we assayed the kinase activities and phosphorylation patterns of immunoprecipitated MPK4a-GFP and MPK4b-GFP following inoculation with B. cinerea spores (Supplemental Figure 11). MPK4a-GFP was clearly activated and phosphorylated in response to B. cinerea. In contrast, MPK4b-GFP activation and phosphorylation were less marked. The signals detected after inoculation with fungal spores were weaker than those detected after chitin treatment (Figure 4B). This is presumably because even high spore concentrations only trigger a fraction of the PRR receptors compared with those triggered by soluble chitin or other PAMPs.
Kinase Activation in Response to Osmotic Stress
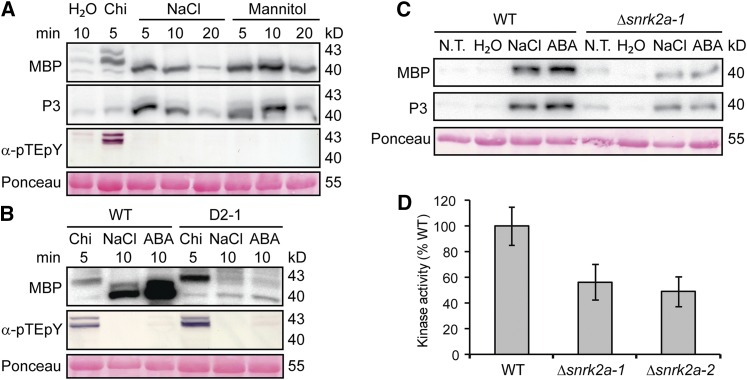
In addition to their PAMP responsiveness, At MPK3/4/6 are phosphorylated in response to abiotic stresses including hyperosmolarity and salt (Ichimura et al., 2000; Droillard et al., 2004; Teige et al., 2004). P. patens also accumulates specific transcripts and proteins in response to osmotic stress (Cuming et al., 2007; Lunde et al., 2007; Wang et al., 2008). To examine if Pp MPK4a or Pp MPK4b are activated in response to osmotic stress, we treated lines with 500 mM NaCl or 800 mM mannitol and assayed kinase activity. We did not detect activation or phosphorylation of Pp MPK4a or Pp MPK4b, or of any other MPKs, in response to these treatments (Figure 5A).
Figure 5.
Kinase Activation in Response to Abiotic Stress.
(A) In-gel kinase activity assay with MBP or synthetic peptide P3 as substrates, and immunoblot analysis of MPK phosphorylation with α-pTEpY of extracts from wild type treated with water, 100 μg/mL chitin (Chi), 500 mM NaCl, or 800 mM mannitol for indicated times. Loading control is Ponceau staining.
(B) In-gel kinase assay with MBP and immunoblotting of MPK phosphorylation with α-pTEpY of extracts of the wild type or the ABA-insensitive line (D2-1) treated with 100 μg/mL chitin (Chi), 500 mM NaCl, or 10 μM ABA. Loading control is Ponceau staining of total protein.
(C) In-gel kinase assay with MBP of extracts of the wild type and Δsnrk2a-1 untreated (N.T.) or treated with water, 500 mM NaCl, or 10 µM ABA for 10 min.
(D) Quantitation of kinase activities in extracts relative to the wild type of two Δsnrk2a lines treated with 500 mM NaCl for 10 min. Data points represent an average of three biological replicates with sd as error bars.
However, both treatments activated a kinase of ∼40 kD, which phosphorylated MBP. Since MBP is a substrate of many kinases, including stress-activated SnRKs (Boudsocq et al., 2004), we performed in-gel kinase assays with the SnRK-specific synthetic peptide P3 (Halfter et al., 2000). This demonstrated that the P3 phosphorylation patterns by the ∼40-kD kinase were the same as those of MBP in response to the NaCl and mannitol (Figure 5A). Importantly, P3 was not phosphorylated by the MPKs activated in response to chitin, and α-pTEpY immunoblotting only detected MPK phosphorylation in response to chitin and not to NaCl or mannitol (Figure 5A). It thus appears that one or more SnRK2s, but not MPKs, are involved in early responses to osmotic stress in P. patens.
P. patens encodes four SnRK2 group III kinases with sizes of 38.5 to 40 kD (Fujita et al., 2013). The osmoregulatory phytohormone abscisic acid (ABA) activates both MPKs and SnRK2s in Arabidopsis (Lu et al., 2002; Boudsocq et al., 2004; Ortiz-Masia et al., 2007; Xing et al., 2008). We therefore assayed kinase activity and found that ABA activated the same P. patens SnRK2 kinase(s) of ∼40 kD as did NaCl treatment, but not activities with sizes of MPKs (Figure 5B). This SnRK2 activation by NaCl is dependent on ABA since it was strongly reduced in the ABA-insensitive moss line D2-1 (Figure 5B; Tougane et al., 2010). Since the kinase assay of NaCl-treated moss indicated activation of SnRK2s, we measured the transcript levels of the four moss SnRK2 group III kinases in response to NaCl treatment. Transcript levels of all four SnRK2s responded similarly in the wild type and Δmpk4a (Supplemental Figure 12), indicating that MPK4a is not involved in signaling osmotic changes. Interestingly, SnRK2a transcripts accumulated to higher levels than those of the other three. We therefore generated a targeted KO of this kinase (Δsnrk2a; Supplemental Figure 3J). The levels of SnRK2 activated by NaCl or ABA were markedly less in Δsnrk2a lines compared with the wild type (Figures 5C and 5D).
Characterization of Moss MPK4a in Immunity
Analysis of Δmpk4 KO lines showed that they are markedly reduced in chitin-induced cell wall-associated depositions assayed by Toluidine blue staining over a time course up to 48 h (Figures 4C and 4D; Supplemental Figure 13). To assess whether the levels of pathogen defense-related transcripts were altered in Δmpk4a, we measured the response to chitosan of five transcripts involved in defense against pathogens and/or are induced in response to chitosan in both Arabidopsis and P. patens: PAL4, CHS, ERF2, α-DOX, and LOX7 (Oliver et al., 2009; Lehtonen et al., 2012; Machado et al., 2015; Overdijk et al., 2016). Although the response to chitosan (Supplemental Figures 14A and 14B) is slower than to chitin (Figure 4E), all these transcripts accumulated less in Δmpk4a compared with the wild type (Supplemental Figures 14A to 14E). These data implicate P. patens MPK4a in the transduction of signals from chitosan perception to the activation of defense genes.
In Arabidopsis, the contribution of PTI to resistance is often studied using pathogens that are unable to overcome PTI (Heese et al., 2007). While such host-pathovar systems have yet to be developed for P. patens, the broad host range necrotrophic fungus B. cinerea proliferates on P. patens (Ponce de León et al., 2007, 2012) and delivers effectors and secretes metabolites aimed at circumventing PTI in vascular plant hosts (Frías et al., 2011; Weiberg et al., 2013). We therefore examined the susceptibility of the Δmpk4a mutant to B. cinerea. After 2 d, symptoms of susceptibility characterized by browning protonema and stems became visible in both the wild type and Δmpk4a (Supplemental Figures 15A to 15H). We also stained tissues for the production of ROS and phenolic compounds that accumulate during B. cinerea infection (Ponce de León et al., 2007, 2012). While ROS production was detected in infected tissues, clear differences could not be seen between the wild type and Δmpk4a (Supplemental Figures 15K to 15N). Although phenolic compounds were incorporated in moss cell walls in contact with B. cinerea hyphae, especially around the point of hyphal penetration, no clear differences were noted between the wild type and Δmpk4a (Supplemental Figures 15I and 15J).
In contrast to such qualitative analyses, host cell death is a hallmark of B. cinerea infection and can be quantified by Evans blue staining (Ponce De León et al., 2012). Using this assay, significantly more staining was quantified 2 d after inoculation in two independent Δmpk4a lines than in the wild type (Figure 6A). To substantiate these observations, we assessed susceptibility to another necrotrophic fungus, Alternaria brassicicola. In this assay, 14-d-old wild-type and Δmpk4a lines were inoculated with a suspension of A. brassicicola spores and the moss plants harvested after 4 d for spore counting (van Wees et al., 2003). A. brassicicola produced significantly more spores on the independent Δmpk4a lines compared with the wild type (Figure 6B). Taken together, these results indicate that although the fungal pathogens B. cinerea and A. brassicicola establish infections on wild-type moss, the absence of MPK4a results in further loss of basal resistance.
Figure 6.
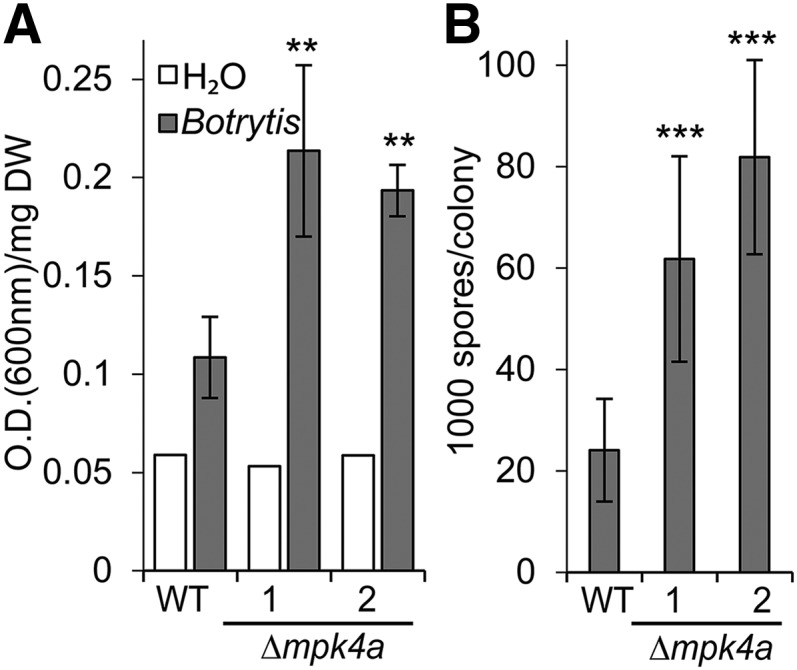
Δmpk4a Plants Are More Susceptible to Necrotrophic Fungi Than the Wild Type.
(A) Evans blue staining 2 d after spraying with water (white bars) or 2 × 105 B. cinerea spores per milliliter (gray). Staining measured as OD600 per mg dry weight (DW). Treated samples are presented as an average of four independent biological replicates with sd as error bars. Analysis of variance followed by Tukey's test determined statistical differences indicated by two asterisks (P < 0.01) between the wild type and the two independent knockout lines (Δmpk4a-1 and 2). The experiment was repeated twice with similar results.
(B) A. brassicicola spore counts 4 d after inoculation with ∼2500 spores per P. patens plant. Each data point is an average of four pools of 16 plants per genotype. Analysis of variance followed by Tukey's test determined statistical differences indicated by three asterisks (P < 0.001) between the wild type and the two knockout lines. The experiment was repeated twice with similar results.
DISCUSSION
Many studies have shown that MAP kinase cascades contribute to PTI in vascular plants (Rasmussen et al., 2012). Much less is known about PTI in nonvascular plants including the bryophyte moss model P. patens. We provide a primary example of how the activation of a specific MPK (Pp MPK4a) in response to the fungal PAMP chitin is reduced in targeted gene knockout lines of a chitin receptor or an upstream MEKK or MPKK. In addition, KO of moss MPK4a results in reduced accumulation of defense-related transcripts and increased susceptibility to pathogens. MPK4a is not activated by ABA or osmotic stress and therefore appears to function primarily in PTI. Apart from their disease susceptibility phenotype, Δmpk4a KO lines appear indistinguishable from wild-type moss. Loss of Pp MPK4a activity is therefore not associated with the kinds of pleiotropic effects detected in MPK KO lines seen in Arabidopsis.
More specifically, our experiments showed that two P. patens MPKs are rapidly activated within 1 min in response to PAMPs, including chitin, chitin derivatives, bacterial peptidyl glycan, and bacterial culture filtrates. These moss MPKs were not activated in response to the bacterial PAMP elf18 peptide, flg22 peptides from P. syringae or Salmonella, or full-length flagellin proteins from P. syringae or Borrelia. This is in line with the lack of close homologs of the Arabidopsis EF2 and FLS2 receptors in P. patens (Boller and Felix, 2009).
The rapid and robust MPK activation by chitin and derivatives prompted us to try to identify components of this signaling pathway using targeted gene KOs and KIs by homologous recombination. Of the two moss homologs of the Arabidopsis chitin receptor At CERK1, one KO mutant (Δcerk1) failed to activate the MPKs in response to chitin. In addition, expression of Pp CERK1 restored chitin sensitivity in the Arabidopsis cerk1 mutant, indicating that Pp CERK1 is an ortholog of At CERK1. Furthermore, Δcerk1 loss of function in the moss reduced ion leakage and the induced accumulation of defense related mRNAs compared with the wild type. Loss of chitin sensitivity in Δcerk1 lines versus the wild type was also easily quantified by staining of brownish-red cell wall-associated depositions with Toluidine blue. Time-lapse photography revealed that chitin treatment of wild-type, but not Δcerk1 lines, led to growth inhibition within minutes. Molecular components involved in PAMP-triggered growth inhibition have been identified in Arabidopsis (Li et al., 2009), and studies of their P. patens homologs may provide tools to study tip growth in the moss (Vidali and Bezanilla, 2012). In addition, chitin treatment of wild-type moss, but not Δcerk1, led to a strong fluorescence burst by 4 h, which decayed by 8 h. The identities of these fluorescent molecules and the brownish-red depositions remain to be determined. We note that the chitin-induced depositions resemble those seen around hyphal penetrations during B. cinerea infection.
We used the same targeted KO approach to identify components of kinase cascades functioning downstream of CERK1 in moss. This revealed that KO of the MAP kinase kinase kinase MEKK1a had markedly reduced chitin sensitivity, while the chitin sensitivity of the KO of MEKK1b was slightly reduced. However, as chitin responsiveness was reduced in a double KO Δmekk1a/b line to the level of the chitin receptor KO Δcerk1, MEKK1a and b may have partially redundant functions in responses to chitin. However, it is also possible that the absence of MEKK1a in Δmekk1a lines leads to ectopic recruitment of MEKK1b to the pathway. Such effects have been documented in other systems (Madhani and Fink, 1998). Interestingly, chitin induced the same rapid growth inhibition in Δmekk1a/b as in the wild type. Since chitin failed to inhibit the growth of Δcerk1, this inhibitory response to chitin is independent of the kinase cascade in which MEKK1a and b function.
Targeted KOs of three MKK genes revealed that KO of MKK1a and c caused a modest reduction in chitin sensitivity. This indicates that MKK1a and c may be partially redundant with other MKK genes in responses to chitin, with the above caveat of potential ectopic recruitment of other MKK s in the Δmkk1a KO line. However, in Arabidopsis, At MPKK1 and 2 act redundantly in signal transduction downstream of PRR activation; therefore, redundancy at this level of the MPK signaling cascade is expected (Gao et al., 2008).
A recent phylogenic analysis could not establish clear orthologous relationships between MPKs from Arabidopsis and basal land plants including P. patens (Dóczi et al., 2012). Since we and others have shown that the Arabidopsis group B MAP kinase MPK4 is involved in immunity, we created KO and KI lines of the two P. patens group B MPKs MPK4a and MPKb. Analysis of KI lines expressing Pp MPK4a or b GFP fusions demonstrated that these two kinases are the two enzymes activated in response to chitin and other PAMPs. While we were unsuccessful in generating Δmpk4b KO lines, Δmpk4a lines were strongly compromised in (1) accumulation of chitin-induced cell wall-associated depositions, (2) expression of defense-related genes, and, most significantly, (3) basal resistance against necrotrophic fungi.
Interestingly, we did not detect phosphorylation or activation of P. patens MPKs in response to osmotic stress treatments or ABA. This is in contrast to recent reports of MPK functions in ABA and osmotic stress responses in Arabidopsis (Montillet et al., 2013; Danquah et al., 2015). We also found no difference between the wild type and Δmpk4a in the accumulation of abiotic stress responsive transcripts upon NaCl treatment or in the survival of the wild type and Δmpk4a on high salt media (S. Bressendorff, unpublished data). These results indicate that Pp MPK4a primarily functions in PTI signaling and thus differs from its close homologs in vascular plants, which are also activated by abiotic stress (Ichimura et al., 2000; Droillard et al., 2004; Teige et al., 2004; González Besteiro et al., 2011). We conclude that MPK signaling in response to the abiotic stresses examined here could either have been lost in the P. patens lineage or have evolved subsequently in vascular lineages.
While ABA and osmotic stress treatments did not induce the activity of kinases of similar size of moss MPKs using MBP as substrate (≥42 kD), they did induce the activity of one or more kinases of ∼40 kD within 5 min. This activity was presumably not that of an MPK, as it was not detected with α-pTEpY antibodies. We showed that this activity corresponds to that of Pp SnRK2a, and likely of other redundant SnRK2s, as seen in Arabidopsis (Fujii et al., 2007). This activity was strongly reduced in an ABA insensitive moss line expressing a constitutively active PP2C phosphatase (Tougane et al., 2010). Such ectopic PP2C activity deactivates SnRK2s (Yoshida et al., 2006; Boudsocq et al., 2007; Vlad et al., 2010). This is in accordance with the findings of Komatsu et al. (2013) on the activation of one or more SnRK2s of <40 kD by ABA in P. patens and of D’Souza and Johri (2002) on an ∼38-kD kinase activated by NaCl and ABA in the closely related moss Funaria hygrometrica. We showed that this kinase activity phosphorylates the SnRK-specific substrate peptide P3 (Halfter et al., 2000) using a novel in-gel kinase assay with acrylamide cross-linked acryloylated P3. In addition, this osmotic stress and ABA-inducible SnRK2 activity was strongly reduced in the targeted knockout lines of Δsnrk2a. These results indicate that SnRK2a, but not MPKs, functions in a canonical ABA signaling pathway in P. patens. Saruhashi et al. (2015) recently reported similar findings of SnRK2 and not MPK activity induced in P. patens in response to ABA and hyperosmotic stress. They further found that this SnRK2 activity was dependent on a Raf-like kinase.
The apparent phosphorylation of moss MPK4b in response to chitin and PGN indicates that it is also involved in PAMP signaling, perhaps redundantly with MPK4a. While our inability to produce Δmpk4b lines currently obscures such a function, it suggests that MPK4b is also required for metabolism and/or development in the moss. Future studies using kinase chemical genetics in P. patens could elucidate MPK4b functions (Brodersen et al., 2006).
Pp MPK4a is a close homolog of At MPK4 but, in apparent contrast to the dwarfed Arabidopsis mpk4 mutant, the moss Δmpk4a mutant appears phenotypically normal under all conditions tested. While this suggests that Pp MPK4a and At MPK4 have different functions, both are responsive to pathogens and PAMPs and At MPK4 is implicated in the regulation of PTI (Qiu et al., 2008). One explanation for their different mutant phenotypes is that At MPK4 and orthologs in vascular plants have evolved additional functions. A second explanation relates to the evolution of host-pathogen interactions. Successful pathogens deliver effectors to modify host proteins and suppress immune responses (Bent and Mackey, 2007). Plants have developed mechanisms to detect pathogen effectors via cytoplasmic immune receptors (termed resistance or R proteins) that typically trigger a rapid, localized host cell death known as the hypersensitive response (DeYoung and Innes, 2006). R proteins may thus guard effector host targets or guardees (Jones and Dangl, 2006). Zhang et al. (2012) recently reported that the absence of At MPK4 in the mpk4 mutant triggers immunity via an R protein. Thus, Arabidopsis MPK4 or its pathway may be targeted by pathogen effectors to block PTI and thereby prevent defense gene activation. If so, it is possible that moss MPK4a remains unguarded in the moss. This difference may have enabled us to assess the phenotype and function of Pp MPK4 more readily than has been the case for At MPK4. In a similar vein, our failure to isolate Δmpk4b mutants may be because Pp MPK4b performs essential functions or because it is in fact guarded. Although some 18 R genes are found in P. patens (Xue et al., 2012), nothing is currently known of their functions.
METHODS
Growth Conditions
Physcomitrella patens (Gransden 2004 strain) was grown on BCDAT media (Ashton and Cove, 1977) containing (250 mg/L MgSO4·7H2O, 250 mg/L KH2PO4, 1010 mg/L KNO3, 920 mg/L ammonium tartrate, 12.5 mg/L FeSO4·7H2O, 147 mg/L CaCl2·2H2O, trace elements [614 μg/L H3BO3, 389 μg/L MnCl2·4H2O, 110 μg/L AlK(SO4)2·12H2O, 55 μg/L CoCl2·6H2O, 55 μg/L CuSO4·5H2O, 55 μg/L ZnSO4·7H2O, 28 μg/L KBr, 28 μg/L KI, 28 μg/L LiCl, 28 μg/L SnCl2·2H2O, 25 μg/L Na2MoO4·2H2O, 59 μg/μl NiCl2·6H2O], pH 6.5 adjusted with KOH, solidified with agar 8 g/L, and overlaid with cellophane discs (AA Packaging) when needed. For growth inhibition and fluorescent burst assays, moss was grown on minimal media, BCD, which is BCDAT without ammonium tartrate. Moss plants were grown at 22°C for 2 or 3 weeks in 55 µE m−2 s−1 from fluorescent light bulbs (Philips TLD36W/33) in 16 h light/8 h dark. Arabidopsis thaliana Columbia and transgenic lines were grown in 150 µE m−2 s−1 at 21°C and short-day cycle (8 h light/16 h dark) in a growth chamber with controlled temperature and humidity. Botrytis cinerea and Alternaria brassicicola were cultivated on 24 g/L potato dextrose agar (Difco) at 22°C for 2 weeks until sporulation was dense. Spores were collected in H2O and filtered through Miracloth, and their concentration was determined with a hemocytometer. Pectobacterium wasabiae SCC3193 was grown on LB medium at 28°C.
Generation of P. patens Mutants
Cloning primers are listed in Supplemental Table 1. The Δmpk4a transformation vector was constructed by cloning a PCR product of the 5′ upstream region of MPK4a into the NotI and BamHI sites of P. patens transformation vector pMBL10a. The 3′ downstream region of MPK4a was then cloned in the EcoRV site of this vector. The KO transformation vectors for P. patens CERK1, 1s41 280V6.1, MEKK1a, MEKK1b, MKK1a, MKK1b, MKK1c, and SnRK2a were initially constructed by adding USER cloning sites on both sides of the NptII selection cassette in vector pMBL6 (Nour-Eldin et al., 2006). The respective genes were cloned into the resulting USER cloning vector pMBLU by PCR amplification of their 5′ upstream and 3′ downstream regions with USER compatible primers. These two PCR products were combined in a single, four-fragment USER cloning step into pMBLU cut with PacI and AsiSI and nicked with Nt.BbvCI (Jacobsen et al., 2011). GFP and a nos terminator were cloned in the pMBLU KpnI site to make the USER cloning vector pMBLU-GFP for tagging proteins with a C-terminal GFP. P. patens MPK4a-GFP and MPK4b-GFP were cloned by PCR amplification of their genomic regions and of 3′ regions downstream of the genes with USER compatible primers. To make the double KO line Δmekk1a/b, pMBLU was modified to contain an HtpII selection cassette (hygromycin resistance) by cutting pMBLU with SalI and KpnI and inserting HtpII amplified from pUNI33. P. patens MEKK1b was cloned into the resulting vector pMBLU-hyg and transformed into the single KO line Δmekk1a-1 to create Δmekk1a/b. The pro35S:GFP line was made for random integration by cloning the 35S promoter from pEarly103 into the pMBLU-GFP by cutting with PacI and nicked with Nt.BbvCI. P. patens protoplasts were transformed with 30 μg of linearized vectors using polyethylene glycol as previously described (Liu and Vidali, 2011). Stable transformants were selected by transferring the protoplasts on cellophane to media containing 50 µg/mL G418 or 30 µg/mL hygromycin B for 2 weeks, followed by transfer to media without selection for 2 weeks, and finally by transfer onto media with selection for a further 2 weeks (Schaefer et al., 1991).
Complementation of Arabidopsis cerk1 with P. patens CERK1
P. patens CERK1 was cloned into pENTR/D-TOPO, modified with InFusion Overhangs, to be fused with a Gateway destination vector. First, the cDNA corresponding to CERK1 was isolated by RT-PCR using RevertAid reverse transcriptase (Thermo Scientific) from 0.5 µg of total P. patens RNA. The open reading frame of CERK1 was obtained by PCR using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and gene-specific primers. pENTR was digested with AscI and NotI and the CERK1 cDNA cloned into the vector using In-Fusion HD Cloning Plus (Clontech). The plasmid obtained was then transferred to Gateway destination vector pGWB514 with a C-terminal HA tag using Gateway LR Clonase II Enzyme mix (Life Technologies) according with the manufacturer’s specifications. Escherichia coli XL blue cells were transformed with these plasmid constructs that were then transformed into Agrobacterium tumefaciens Agl1 electro-competent cells. Arabidopsis cerk1 sil1-2 (Gabi-kat line) was transformed using Agrobacterium floral dipping as previously described (Clough and Bent, 1998).
Treatments for Kinase Assays
Unless otherwise stated, all treatments were applied for 5 min by spraying 3 mL of the solution used onto 16 moss plants grown for 14 d on a Petri dish (9 cm) with BCDAT media overlaid with cellophane. Chitin oligosaccharide (Yaizu Suisankagaku Industry) was diluted to 100 μg/mL in water. Hexa-N-acetylchitohexaose (Sigma-Aldrich) was diluted to 100 µg/mL in water. Chitosan (Sigma-Aldrich), 75 to 85% deacetylated and low molecular weight (50 to 190 kD), was solubilized at 100 µg/mL in 0.010% acetic acid followed by adjusting to pH 5.5 with NaOH. Acetic acid (0.010%) adjusted to pH 5.5 was used as a control. Full-length flagellins from Pseudomonas aeruginosa (Invitrogen) and from Borrelia (Alpha Diagnostic), and flg22 and flgII-28 purified peptides (Schafer-N) were sprayed on P. patens and applied to Arabidopsis in liquid culture at the indicated concentrations. Then, 1 × 107 B. cinerea spores per milliliter were sprayed onto plants. Abscisic acid (Sigma-Aldrich) was diluted to a concentration of 10 μM in 0.01% (w/w) ethanol. Peptidyl glycan, PGN-Staphylococcus aureus (Sigma-Aldrich), was applied by gentle shaking in a suspension of 200 µg/mL and Xanthomonas campestris PGN, obtained as previously described (Erbs et al., 2008), was similarly applied at 100 µg/mL. NaCl was applied at 500 mM and mannitol at 800 mM. Cell-free culture filtrates of P. wasabiae SCC3193 were made by growing bacteria in LB broth overnight, removing bacterial cells by centrifugation (10 min at 4000g), and filter sterilizing the supernatant. Detached leaves from three independent 14-d-old T1 complementing lines expressing Pp CERK1 in the Arabidopsis cerk1 mutant background were preincubated for 2 h in 10 mM MgCl2. Chitin oligosaccharide (Yaizu Suisankagaku Industry) was added to a final concentration of 200 µg/mL for 10 min to the treated samples.
Toluidine Blue Staining
Wild-type and mutant moss were grown for 14 d on BCDAT overlaid with cellophane and then transferred on the cellophane onto BCDAT plates supplemented with 300 µg/mL chitin. After 48 h the five plants per sample were weighed in microfuge tubes and plants stained in 0.05% Toluidine blue (Sigma-Aldrich) in citrate-citric acid buffer (50 mM, pH 3.5) with 50% ethanol for 10 min. The plants were quickly washed twice in water and then washed twice in 96% ethanol and left in 96% ethanol overnight. Representative plants were then photographed. The Toluidine blue staining was quantified by destaining in 10% acetic acid, 86% ethanol at 60°C for 4 h, and the OD of the supernatant was measured at 600 nm as previously described (Beasley et al., 1974) and data presented as OD600/µg fresh weight. Treated samples represent an average of three samples of five plants each with sd as error bars. Nontreated controls were treated equally except plants were not exposed to chitin.
Electrolyte Leakage Assay
Wild-type and Δcerk1-1 moss were grown for 14 d on BCDAT overlaid with cellophane. Five plants were then briefly washed in milli-Q water and transferred to culture tubes containing either 5 mL water or 5 mL 1 mg/mL chitin. The conductivity was measured at indicated times, and at the end of the time series (34 h) total conductivity of each sample was established by freezing the samples in liquid nitrogen, thawing them, and measuring conductivity. Electrolyte leakage is presented as the percentage of total conductivity for each sample as an average of four samples with each five plants. Error bars represent sd.
Evans Blue Staining for Detection of Cell Death
Cell death quantitation by Evans blue staining was similar to the assay of Oliver et al. (2009). Four plates of each line, each with 16 plants, were sprayed with 3 mL of 2 × 105 B. cinerea spores per milliliter and one plate sprayed with water. Two days later, plants were incubated in 0.1% Evans blue (Bie and Berntsen) in 0.5× PBS for 2 h, washed four times in water, then destained in 50% (v/v) methanol, 1% SDS at 60°C for 30 min, and the OD600 measured. Plants were dried over night at 70°C and weighed. Each data point is presented as OD600/mg dry weight as an average of four samples of 16 Plants each. Error bars represent sd. An analysis of variance followed by a Tukey's test was applied to assess significant differences. The experiment was repeated twice with similar results.
A. brassicicola Spore Count Assay
The assay was adapted from van Wees et al. (2003). A. brassicicola spores were harvested in water from 14-d-old plates and counted in a hemocytometer. Moss plants were inoculated by placing 5 μL of 2 × 105 A. brassicicola spores/mL on top of each plant. Four plates of each line, each with 16 plants, were inoculated, and 4 d later the plants were transferred to a tube containing 8 mL 0.01% (v/v) Tween 20 (Sigma-Aldrich) and shaken vigorously. Spores in the suspension were counted in a hemocytometer. Each data point is an average of four pools of 16 plants per genotype. Analysis of variance followed by a Tukey's test assessed differences between the wild-type and two Δmpk4a lines. The experiment was repeated twice with similar results.
RNA Extraction and RT-qPCR
Three 14-d-old moss plants grown on media overlaid with cellophane were sampled at each time point after spraying with 500 mM NaCl, 100 µg/mL chitosan (in 0.010% acetic acid followed by adjusting to pH 5.5 with NaOH), or 100 µg/mL chitin and flash-frozen in liquid nitrogen. Arabidopsis plants were grown for 14 d on soil, and detached leaves were preincubated for 2 h in 10 mM MgCl2 before chitin was added to a final concentration of 200 µg/mL for 30 and 60 min and the samples flash-frozen in liquid nitrogen. RNA was extracted with NucleoSpin RNA plant (Macherey-Nagel) according to the manufacturer’s protocol. RNA concentrations were measured in a Nano Drop 1000 (Thermo Scientific) and adjusted to 5 ng/μL. Quantitative PCR used Brilliant II SYBR green one-step kit (Agilent Technologies), with 10 pmol of each primer and 12.5 ng total RNA in 10 µL. Reactions were run on a CFX 96 thermocycler (Bio-Rad) with three technical replicates for each PCR reaction, and means of normalized expression relative to untreated wild type calculated with β-TUBULIN as internal normalization control for the P. patens samples (Zhang et al., 2015) and UBQ10 for the Arabidopsis samples. Primers used for qPCR are in Supplemental Table 1.
Protein Extraction and Immunoblotting
Three 14-d-old moss plants grown on media overlaid with cellophane and Arabidopsis plants grown for 14 d in soil or liquid cultures were flash-frozen at the times given. Protein was extracted in Lacus buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 15 mM EGTA, 100 mM NaCl, 2 mM DTT, 30 mM β-glycero-phosphate, and 0.1% Nonidet P-40) plus phosphatase inhibitor (PhosSTOP; Roche) and protease inhibitor cocktails (Complete; Roche). Samples were cleared by centrifugation, boiled for 5 min in SDS-loading buffer, and subjected to 12% SDS-PAGE and electroblotting. Immunoblots were blocked for 1 h in a standard solution of TBS-Tween (0.1% v/v) and 5% milk (Sigma-Aldrich). Phosphorylated MAP kinases were detected by incubation with primary antibody anti-p42/p44-erk (1/2000; Cell Signaling Technology), followed by incubation with anti-rabbit-IgG-HRP secondary antibody (1/10,000; Promega) or anti-rabbit-IgG-AP (1/5000; Promega). GFP was detected with anti-GFP (1/1000; Promega) followed by incubation with anti-mouse-IgG-AP secondary antibody (1/5000; Promega). HA was detected using anti-HA SC 7392 (1/1000; Santa Cruz Biotechnology) followed by anti-mouse-IgG-HRP (1/5000; Promega). Horseradish peroxidase-conjugated antibodies were visualized with Pierce ECL Plus Western Blotting Substrate (Thermo Scientific) and pictures taken with a Sony A7S camera. Alkaline phosphatase-conjugated antibodies were detected with NBT/BCIP substrate (Roche).
Immunoprecipitation Kinase Assay
Four 14-d-old moss plants grown on media overlaid with cellophane were flash-frozen at given times. Protein was extracted in 500 μL extraction buffer (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM EDTA, 5 mM DTT, 0.5% [w/v] PVPP, and 1% Nonidet P-40) plus phosphatase inhibitor (PhosSTOP; Roche) and protease inhibitor cocktails (Complete; Roche). Protein concentrations were determined by Bradford assay (Bio-Rad) and samples adjusted to 1 mg/mL. Twenty-five microliters of GFP-trap beads (Chromotec) were added and incubated at 4°C for 2 h. Beads were washed three times in extraction buffer without PVPP. Immunoprecipitates were incubated in 40 μL kinase buffer (12.5 µM ATP, 20 mM Tris HCl, pH 7.5, 30 mM NaCl, 30 mM MgCl2, 2 mM EGTA, and 1 mM DTT) with 5 µg MBP (Millipore), 10 µCi of [γ-32P]ATP plus phosphatase inhibitor (PhosSTOP; Roche), and protease inhibitor cocktail (Complete; Roche) at 30°C for 30 min. Reactions were stopped with SDS loading buffer and products resolved by 12% SDS-PAGE. Gels were stained with 0.04% Coomassie Brilliant Blue in 3.5% perchloric acid, dried, and MBP phosphorylation determined by phosphor imaging.
In-Gel Kinase Assay
Three 14-d-old plants grown on media overlaid with cellophane were flash-frozen at given times after treatment. Protein was extracted in Lacus buffer. Forty micrograms of protein was resolved on 13% SDS-PAGE with 14 µM MBP (Millipore) or 14 µM P3 (N-terminal acryolated by 6-amino hexanoic acid peptide sequence, ALARAASAAALARRRALARAASAAALARRR; Schafer-N) polymerized in separating gels as substrate (Zhang and Klessig, 1997). Gels were washed six times for 3 h in 25 mM Tris-HCl (pH 7.5), 0.5 mM DTT, 5 mM NaF, 0.1 mM Na3VO4, 0.5 mg/mL BSA, and 0.1% Triton X-100. Proteins were denatured by rinsing gels twice for 30 min at room temperature in 25 mM Tris-HCl (pH 7.5), 6 M guanidine hydrochloride, and 5 mM DTT, followed by incubation overnight with five changes of 25 mM Tris-HCl (pH 7.5), 1 mM DTT, 5 mM NaF, and 0.1 mM Na3VO4. After gel equilibration for 1 h in kinase activity buffer (25 mM Tris-HCl, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, and 0.1 mM Na3VO4), the assay was performed for 1 h at room temperature in that buffer with 100 nM ATP plus 100 µCi [γ-32P]ATP. The reaction was stopped with 5% TCA and 1% sodium pyrophosphate, and unincorporated [γ-32P]ATP was removed by washing five times for 15 min in the same buffer. Gels were dried and phosphorylation determined by phosphor imaging.
Time-Lapse Movies of Growth Inhibition and Fluorescent Burst
Wild-type and Δcerk1-1 or wild-type and Δmekk1a/b moss plants were grown in close proximity on BCD media overlaid with cellophane for 10 d and then transferred on cellophane onto BCD plates supplemented with 300 µg/mL chitin. Pictures were taken every 2nd or 4th minute using a Sony A7S camera. Growth inhibition (Supplemental Movies 1 and 4) was recorded in white light, while the fluorescent burst (Supplemental Movies 2 and 3) was recorded using GFP settings (excitation <488 nm and detection >510 nm).
Confocal Microscopy
Confocal images were acquired using a Zeiss LSM 700 and analyzed with ZEN imaging software (version 2011) with GFP settings (excitation 488 nm and detection 300 to 610 nm). The 14-d-old MPK4a-GFP plants were mounted on a glass slide embedded in BCDAT with 1% agar and supplemented with 300 µg/mL chitin.
Staining of B. cinerea Infection Symptoms
The 14-d-old moss plants of wild type and Δmpk4a-1 were sprayed with 2 × 105 B. cinerea spores per milliliter. Toluidine blue staining of cell wall modifications was conducted 1 d after infection by staining tissue with 0.05% Toluidine blue in citrate-citric acid buffer (50 mM, pH 3.5). Cell death was visualized by staining for 2 h with 0.1% Evans blue (Sigma-Aldrich) in 0.5× PBS and washed four times in water to remove excess unbound dye. Intracellular ROS production was analyzed by incubating moss tissues with 10 mM 2′,7′-dichlorodihydrofluorescein diacetate for 15 min in 0.1 M phosphate buffer (pH 7.5) in the dark.
Accession Numbers
A list of accession numbers is provided in Supplemental Table 1.
Supplemental Data
Supplemental Figure 1. MAP kinase activities in response to PAMP treatments.
Supplemental Figure 2. Phylogeny of CERK1 homologs and mRNA levels of P. patens CERK1 homologs.
Supplemental Figure 3. Genotyping of knockout and GFP-knockin lines.
Supplemental Figure 4. Phenotypes, growth inhibition, fluorescence burst, and cell wall depositions in wild type and Δcerk1.
Supplemental Figure 5. Complementation of the Arabidopsis cerk1 mutant with P. patens CERK1.
Supplemental Figure 6. Phylogeny of MEKK homologs and mRNA levels of P. patens MEKK1 homologs.
Supplemental Figure 7. Growth inhibition and fluorescent burst of wild type and Δmekk1a/b.
Supplemental Figure 8. Phylogeny of MKK homologs and mRNA levels of P. patens MKK1 homologs.
Supplemental Figure 9. Phylogeny of MPK homologs and mRNA levels of P. patens MPK4 homologs.
Supplemental Figure 10. Localization of P. patens MPK4a-GFP.
Supplemental Figure 11. Activation and phosphorylation of P. patens MPK4a-GFP and MPK4b-GFP following B. cinerea inoculation.
Supplemental Figure 12. NaCl-induced SnRK2 mRNA levels in wild type and Δmpk4a.
Supplemental Figure 13. Time series of chitin-induced Toluidine blue staining in wild type and Δmpk4a.
Supplemental Figure 14. mRNA levels of defense genes after chitosan treatment.
Supplemental Figure 15. Symptoms of B. cinerea infection on wild type and Δmpk4a.
Supplemental Table 1. Oligos used for cloning, genotyping, RT-qPCR, and accession numbers.
Supplemental Data Set1. Protein sequences used to generate the CERK1 phylogeny presented in Supplemental Figure 2.
Supplemental Data Set 2. Protein sequences used to generate the MEKK phylogeny presented in Supplemental Figure 6.
Supplemental Data Set 3. Protein sequences used to generate the MKK phylogeny presented in Supplemental Figure 7.
Supplemental Data Set 4. Protein sequences used to generate the MPK phylogeny presented in Supplemental Figure 8.
Supplemental Movie 1. Growth inhibition in response to chitin in wild type but not in Δcerk1.
Supplemental Movie 2. Fluorescence burst in response to chitin in wild type but not in Δcerk1.
Supplemental Movie 3. Fluorescence burst in response to chitin in wild type but not in Δmekk1a/b.
Supplemental Movie 4. Growth inhibition in response to chitin in wild type and Δmekk1a/b.
Supplementary Material
Acknowledgments
We thank Andrew Cuming for the pMBL10a and pMBL6 transformation vectors, Christine Lunde for wild-type P. patens (Gransden 2004 strain), Yaizu Suisankagaku Industry for chitin, and Daisuke Takezawa for the ABA insensitive line (D2-1). This work was supported by grants to R.A. from the Portuguese Foundation for Science and Technology (SFRH/BD/75955/2011) and to J.M. and M.P. from the Danish Basic Research Foundation (Comparative Genomics) and Natural Science Council (1323-00267A).
AUTHOR CONTRIBUTIONS
S.B. designed and performed research, analyzed data, and wrote the article. I.P.d.L., R.A., C.S.K., J.V.O., and M.W.R. performed research and analyzed data. G.E. and M.-A.N. provided a new analytic tool (Xanthomonas campestris PGN). M.P. and J.M. designed research and wrote the article.
Glossary
- PRR
pattern recognition receptor kinase
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- ROS
reactive oxygen species
- KO
knockout
- KI
knockin
- PGN
peptidyl glycan
- ABA
abscisic acid
Footnotes
Articles can be viewed without a subscription.
References
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Y., Li Z., Feng D., Xiong F., Liu J., Li J.-F., Wang M., Wang J., Liu B., Wang H.-B. (2014). OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80: 1072–1084. [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Ashton N.W., Cove D.J. (1977). The isolation and preliminary characterisation of auxotrophic and analogue resistant mutants of the moss Physcomitrella patens. Mol. Gen. Genet. 154: 87–95. [Google Scholar]
- Beasley C.A., Birnbaum E.H., Dugger W.M., Ting I.P. (1974). A quantitative procedure for estimating cotton fiber growth. Stain Technol. 49: 85–92. [DOI] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436. [DOI] [PubMed] [Google Scholar]
- Bethke G., Pecher P., Eschen-Lippold L., Tsuda K., Katagiri F., Glazebrook J., Scheel D., Lee J. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25: 471–480. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279: 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Droillard M.-J., Barbier-Brygoo H., Laurière C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63: 491–503. [DOI] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M.-A., Shokat K.M., Rietz S., Parker J., Mundy J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47: 532–546. [DOI] [PubMed] [Google Scholar]
- Clarke C.R., Chinchilla D., Hind S.R., Taguchi F., Miki R., Ichinose Y., Martin G.B., Leman S., Felix G., Vinatzer B.A. (2013). Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol. 200: 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cuming A.C., Cho S.H., Kamisugi Y., Graham H., Quatrano R.S. (2007). Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 176: 275–287. [DOI] [PubMed] [Google Scholar]
- Danquah A., et al. (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82: 232–244. [DOI] [PubMed] [Google Scholar]
- DeYoung B.J., Innes R.W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7: 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóczi R., Okrész L., Romero A.E., Paccanaro A., Bögre L. (2012). Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 17: 518–525. [DOI] [PubMed] [Google Scholar]
- Droillard M.-J., Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett. 574: 42–48. [DOI] [PubMed] [Google Scholar]
- D’Souza J.S., Johri M.M. (2002). ABA and NaCl activate myelin basic protein kinase in the chloronema cells of the moss Funaria hygrometrica. Plant Physiol. Biochem. 40: 17–24. [Google Scholar]
- Erbs G., Silipo A., Aslam S., De Castro C., Liparoti V., Flagiello A., Pucci P., Lanzetta R., Parrilli M., Molinaro A., Newman M.A., Cooper R.M. (2008). Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chem. Biol. 15: 438–448. [DOI] [PubMed] [Google Scholar]
- Fiil B.K., Petersen K., Petersen M., Mundy J. (2009). Gene regulation by MAP kinase cascades. Curr. Opin. Plant Biol. 12: 615–621. [DOI] [PubMed] [Google Scholar]
- Frías M., González C., Brito N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192: 483–495. [DOI] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.-K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 147: 15–27. [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- González Besteiro M.A., Bartels S., Albert A., Ulm R. (2011). Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 68: 727–737. [DOI] [PubMed] [Google Scholar]
- Halfter U., Ishitani M., Zhu J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97: 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Irie K., Morris P., Giraudat J., Matsumoto K., Shinozaki K. (1998). Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 253: 532–543. [DOI] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Yoshida R., Yuasa T., Shinozaki K. (2000). Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24: 655–665. [DOI] [PubMed] [Google Scholar]
- Ichimura K., et al. ; MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7: 301–308. [DOI] [PubMed] [Google Scholar]
- Jacobsen J.H., Rosgaard L., Sakuragi Y., Frigaard N.-U. (2011). One-step plasmid construction for generation of knock-out mutants in cyanobacteria: studies of glycogen metabolism in Synechococcus sp. PCC 7002. Photosynth. Res. 107: 215–221. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Komatsu K., Suzuki N., Kuwamura M., Nishikawa Y., Nakatani M., Ohtawa H., Takezawa D., Seki M., Tanaka M., Taji T., Hayashi T., Sakata Y. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4: 2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen M.T., Akita M., Frank W., Reski R., Valkonen J.P.T. (2012). Involvement of a class III peroxidase and the mitochondrial protein TSPO in oxidative burst upon treatment of moss plants with a fungal elicitor. Mol. Plant Microbe Interact. 25: 363–371. [DOI] [PubMed] [Google Scholar]
- Lehtonen M.T., Takikawa Y., Rönnholm G., Akita M., Kalkkinen N., Ahola-Iivarinen E., Somervuo P., Varjosalo M., Valkonen J.P.T. (2014). Protein secretome of moss plants (Physcomitrella patens) with emphasis on changes induced by a fungal elicitor. J. Proteome Res. 13: 447–459. [DOI] [PubMed] [Google Scholar]
- Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J.D.G. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. USA 106: 15973–15978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-C., Vidali L. (2011). Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens. J. Vis. Exp. 50: 2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Han M.-H., Guevara-Garcia A., Fedoroff N.V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 99: 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde C., Drew D.P., Jacobs A.K., Tester M. (2007). Exclusion of Na+ via sodium ATPase (PpENA1) ensures normal growth of Physcomitrella patens under moderate salt stress. Plant Physiol. 144: 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado L., Castro A., Hamberg M., Bannenberg G., Gaggero C., Castresana C., de León I.P. (2015). The Physcomitrella patens unique alpha-dioxygenase participates in both developmental processes and defense responses. BMC Plant Biol. 15: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Madhani H.D., Fink G.R. (1998). The riddle of MAP kinase signaling specificity. Trends Genet. 14: 151–155. [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag J., Šola I., Rusak G., Ludwig-Müller J. (2015). Physcomitrella patens auxin conjugate synthetase (GH3) double knockout mutants are more resistant to Pythium infection than wild type. J. Plant Physiol. 183: 75–83. [DOI] [PubMed] [Google Scholar]
- Montillet J.-L., et al. (2013). An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Hansen B.G., Nørholm M.H.H., Jensen J.K., Halkier B.A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykyri J., Niemi O., Koskinen P., Nokso-Koivisto J., Pasanen M., Broberg M., Plyusnin I., Törönen P., Holm L., Pirhonen M., Palva E.T. (2012). Revised phylogeny and novel horizontally acquired virulence determinants of the model soft rot phytopathogen Pectobacterium wasabiae SCC3193. PLoS Pathog. 8: e1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.P., Castro A., Gaggero C., Cascón T., Schmelz E.A., Castresana C., Ponce de León I. (2009). Pythium infection activates conserved plant defense responses in mosses. Planta 230: 569–579. [DOI] [PubMed] [Google Scholar]
- Ortiz-Masia D., Perez-Amador M.A., Carbonell J., Marcote M.J. (2007). Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 581: 1834–1840. [DOI] [PubMed] [Google Scholar]
- Overdijk E.J.R., DE Keijzer J., DE Groot D., Schoina C., Bouwmeester K., Ketelaar T., Govers F. (2016). Interaction between the moss Physcomitrella patens and Phytophthora: a novel pathosystem for live-cell imaging of subcellular defence. J. Microsc. 00: 1–10. [DOI] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120. [DOI] [PubMed] [Google Scholar]
- Ponce de León I., Oliver J.P., Castro A., Gaggero C., Bentancor M., Vidal S. (2007). Erwinia carotovora elicitors and Botrytis cinerea activate defense responses in Physcomitrella patens. BMC Plant Biol. 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce De León I., Schmelz E.A., Gaggero C., Castro A., Álvarez A., Montesano M. (2012). Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 13: 960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.-L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M.W., Roux M., Petersen M., Mundy J. (2012). MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.-Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saruhashi M., Kumar Ghosh T., Arai K., Ishizaki Y., Hagiwara K., Komatsu K., Shiwa Y., Izumikawa K., Yoshikawa H., Umezawa T., Sakata Y., Takezawa D. (2015). Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. USA 112: E6388–E6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D., Zryd J.-P., Knight C.D., Cove D.J. (1991). Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 226: 418–424. [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.-H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang C., Yang B., Wu F., Hao X., Liang W., Niu F., Yan J., Zhang H., Wang B., Deyholos M.K., Jiang Y.Q. (2014). Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J. Exp. Bot. 65: 2171–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152. [DOI] [PubMed] [Google Scholar]
- Tougane K., Komatsu K., Bhyan S.B., Sakata Y., Ishizaki K., Yamato K.T., Kohchi T., Takezawa D. (2010). Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol. 152: 1529–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees S.C., Chang H.S., Zhu T., Glazebrook J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Bezanilla M. (2012). Physcomitrella patens: a model for tip cell growth and differentiation. Curr. Opin. Plant Biol. 15: 625–631. [DOI] [PubMed] [Google Scholar]
- Vlad F., Droillard M.-J., Valot B., Khafif M., Rodrigues A., Brault M., Zivy M., Rodriguez P.L., Merlot S., Laurière C. (2010). Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 63: 778–790. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang P., Gao Q., Liu X., Kuang T., Shen S., He Y. (2008). Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 228: 167–177. [DOI] [PubMed] [Google Scholar]
- Weiberg A., Wang M., Lin F.-M., Zhao H., Zhang Z., Kaloshian I., Huang H.-D., Jin H. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342: 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann R., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 108: 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54: 440–451. [DOI] [PubMed] [Google Scholar]
- Xue J.-Y., Wang Y., Wu P., Wang Q., Yang L.-T., Pan X.-H., Wang B., Chen J.-Q. (2012). A primary survey on bryophyte species reveals two novel classes of nucleotide-binding site (NBS) genes. PLoS One 7: e36700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281: 5310–5318. [DOI] [PubMed] [Google Scholar]
- Zhang F., Han M., Lv Q., Bao F., He Y. (2015). Identification and expression profile analysis of NUCLEAR FACTOR-Y families in Physcomitrella patens. Front. Plant Sci. 6: 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-C., Cannon S.B., Stacey G. (2009). Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263. [DOI] [PubMed] [Google Scholar]
- Zimmer A., Lang D., Richardt S., Frank W., Reski R., Rensing S.A. (2007). Dating the early evolution of plants: detection and molecular clock analyses of orthologs. Mol. Genet. Genomics 278: 393–402. [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12: 414–420. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.