The dynamics of histones H3.1 and H3.3 characterize cells with different division potential in the growing root and a longer G2 phase in cells undergoing the last cell cycle before differentiation.
Abstract
A coordinated transition from cell proliferation to differentiation is crucial for organogenesis. We found that extensive chromatin reorganization, shown here for histone H3 proteins, characterizes cell population dynamics in the root developmental compartments. The canonical H3.1 protein, incorporated during S-phase, is maintained at high levels in cells dividing at a high rate but is massively evicted in cells undergoing their last cell cycle before exit to differentiation. A similar pattern was observed in the quadruple mutant for the H3.1-encoding genes HTR1, HTR2, HTR3, and HTR9 (htr1,2,3,9), in which H3.1 is expressed only from the HTR13 gene. H3 eviction is a fast process occurring within the G2 phase of the last cell cycle, which is longer than G2 in earlier cell cycles. This longer G2 likely contributes to lower the H3.1/H3.3 ratio in cells leaving the root meristem. The high H3.1/H3.3 ratio and H3.1 eviction process also occurs in endocycling cells before differentiation, revealing a common principle of H3 eviction in the proliferating and endocycling domains of the root apex. Mutants in the H3.1 chaperone CAF-1 (fas1-4) maintain a pattern similar to that of wild-type roots. Our studies reveal that H3 incorporation and eviction dynamics identify cells with different cell division potential during organ patterning.
INTRODUCTION
Developing organs consist of very strictly balanced populations of proliferating and differentiating cells. In plants, organogenesis is entirely postembryonic and occurs in a continuous manner throughout the life of the organism. Plant roots are made up of three major regions: the root apical meristem (RAM), where new cells are produced; the elongation zone, where cells arrest proliferation and initiate endoreplication; and the differentiated or mature zone (Petricka et al., 2012). In the apical part of the RAM, the stem cell niche is organized by a group of approximately four cells, the quiescent center (QC), that rarely divide and the surrounding stem cells in contact with the QC (Heyman et al., 2014). Stem cells undergo asymmetrical divisions and their derivatives maintain proliferation potential for a limited number of cell cycles within the transient amplifying compartment of the RAM, which is the proliferation domain (the most rootward half; Baskin et al., 2010). Afterwards, the division potential decreases in cells within the transition domain of the RAM, where mitoses are infrequent (Ivanov and Dubrovsky, 2013). Thus, root development depends on the activity of stem cells, the proliferation of their derivatives, the decrease in cell division competence, and the initiation of endoreplication and differentiation (Petricka et al., 2012). However, while the size of all these cell populations depends on the proliferative potential, the mechanisms determining the lifespan of stem cell derivatives is not fully understood (Scheres, 2007). Likewise, the defining features of cells with different proliferation potential are also poorly understood.
Specific gene expression patterns guided by master regulatory genes determine the boundaries within growing organs (Scheres, 2007; Heidstra and Sabatini, 2014). In the case of Arabidopsis thaliana roots, detailed transcriptomic analyses have provided information about genes expressed in 12 arbitrarily defined regions along the root as well as in the different cell types (Birnbaum et al., 2003; Brady et al., 2007). One key feature contributing to the spatiotemporal regulation of gene expression patterns, in both plants and animals, is the appropriate organization of chromatin, including DNA sequence, nucleosome positioning, and histone composition and posttranslational modifications at specific loci. There are numerous chromatin changes that occur in association with cell cycle progression as well as in the transition to cell differentiation (Desvoyes et al., 2014). Together they generate particular chromatin states that direct gene activity (Pfluger and Wagner, 2007; Maze et al., 2014; Sequeira-Mendes et al., 2014; Kundaje et al., 2015). H3 is one of the histone family members that contains variants and can be posttranslationally modified in many different residues (Kouzarides, 2007). While there are two canonical histones in vertebrates, the only canonical histone in plants is H3.1, differing from the variant H3.3 in only four amino acids that confer them different properties regarding genome distribution, posttranslational modifications, and binding to chaperones (Loyola and Almouzni, 2007; Goldberg et al., 2010; Ingouff and Berger, 2010; Shi et al., 2011; Stroud et al., 2012; Jacob et al., 2014; Otero et al., 2014). Remarkably, H3.1 is incorporated only during DNA replication in the S-phase, while H3.3 is exchanged in a cell cycle-independent manner (Loyola and Almouzni, 2007).
We reasoned that the dynamics of the H3 isoforms might be in a strict relationship with the cell population dynamics within the RAM. In this study, we tested this hypothesis, and we demonstrate that changes in the H3.1/H3.3 balance occur along the root in association with changes in the proliferative potential of root meristem cells. In short, we found that (1) the H3.1/H3.3 ratio in individual nuclei is an indicator of the proliferative potential of the cell; (2) a large and fast H3.1 replacement in the last G2, which is longer than in previous cell cycles, identifies cells undergoing their last cell cycle; (3) the same pattern of high H3.1/H3.3 ratio and H3.1 eviction is reproduced in the root elongation zone after endoreplication; and (4) the H3 eviction pattern seems to be a general feature since it also occurs in other cell types and developmental conditions, e.g., stomatal lineage and hypocotyl cells. Together, our results serve to define functionally different cell populations in the root domains and reveal that H3 replacement, cell division potential, and differentiation are intimately coordinated.
RESULTS AND DISCUSSION
Cell Fate Acquisition in the Embryo Associates with Massive H3.1 Replacement
In Arabidopsis, five genes code for identical H3.1 proteins and three genes code for identical H3.3 histones. We designed an approach to monitor the in vivo dynamics of H3.1 and H3.3 using plants that express tagged H3.1 (HTR3 or HTR13 genes) and H3.3 (HTR4 or HTR5 genes), under their own promoters (Supplemental Figures 1A and 1B; see Methods). The expression levels of endogenous genes and the transgenes differed by less than 2- to 4-fold (Supplemental Figure 1C). The H3-GFP fusion proteins showed the expected subnuclear localization and immunoblot analysis revealed their integrity (Supplemental Figures 1D and 1E). It has been demonstrated previously that the use of different tags on H3 proteins render similar incorporation results (Goldberg et al., 2010). Since the tagged proteins recapitulated the expected pattern for endogenous proteins using cellular and genomic approaches (Ingouff et al., 2010; Stroud et al., 2012; Wollmann et al., 2012; Otero et al., 2014), we considered them bona fide markers of H3.1 and H3.3.
We first focused on the H3.1/H3.3 balance during Arabidopsis embryo development. At the globular stage, the embryo contains cells with high division potential. We found that most nuclei contained a relatively high H3.1 level (Figures 1A, 1B, 1E, and 1F). In fact, the patchy pattern that is typical of cell cycle-regulated proteins was not very apparent at this early stage. Since H3.1 is incorporated only during S-phase, this is consistent with the occurrence of fast cell cycle (perhaps due to a shorter G1). The patchy pattern was recovered at later embryo stages (Figures 1C, 1D, 1G, and 1H). Interestingly, the QC cells showed high levels of H3.1 at early stages (Figures 1A, 1B, 1E, 1F, and insets A1, B1, E1, and F1), but a drastic reduction of H3.1 was observed in QC cells at heart and torpedo embryos (Figures 1C, 1D, 1G, 1H, and insets C1, D1, G1, and H1). Since it is after the 16-cell stage that the QC cells acquired their fate and start to express QC markers, e.g., WOX5 (Scheres et al., 2002; Haecker et al., 2004), these H3.1 dynamics suggest an association between cell division potential and H3.1 content. This is also clearly observed in plants expressing both H3.1-GFP and H3.3-mRFP, in which QC cells at late embryo stages appeared almost exclusively labeled with H3.3-RFP (Supplemental Figures 2C and 2D), whereas they contain high levels of H3.1-GFP at early stages (Supplemental Figures 2A and 2B), except in the rare cases where they divide (Heyman et al., 2013). We found the same pattern using two different H3.1 genes (HTR3 and HTR13; Figures 1A to 1D and 1E to 1H, respectively), suggesting that our observations are not exclusive to one H3 gene. These observations revealed an extensive H3.1 replacement in late embryo stages and suggest a correlation between low H3.1 content and a reduction of proliferative potential.
Figure 1.
Histone H3.1 Labeling Pattern during Arabidopsis Embryo Development.
Embryos of plants expressing H3.1(HTR3)-GFP ([A] to [D]) or H3.1(HTR13)-GFP ([E] to [H]) are shown at different stages: globular ([A] and [E]), early ([B] and [F]) and late ([C] and [G]) heart, and torpedo ([D] and [H]). Cell membranes were stained with FM4-64 (red). Insets focus on the region containing the QC (white arrowheads) in each case.
A Low H3.1/H3.3 Balance Identifies Cells Undergoing Their Last Cell Cycle in the Transient Amplifying Compartment of the RAM
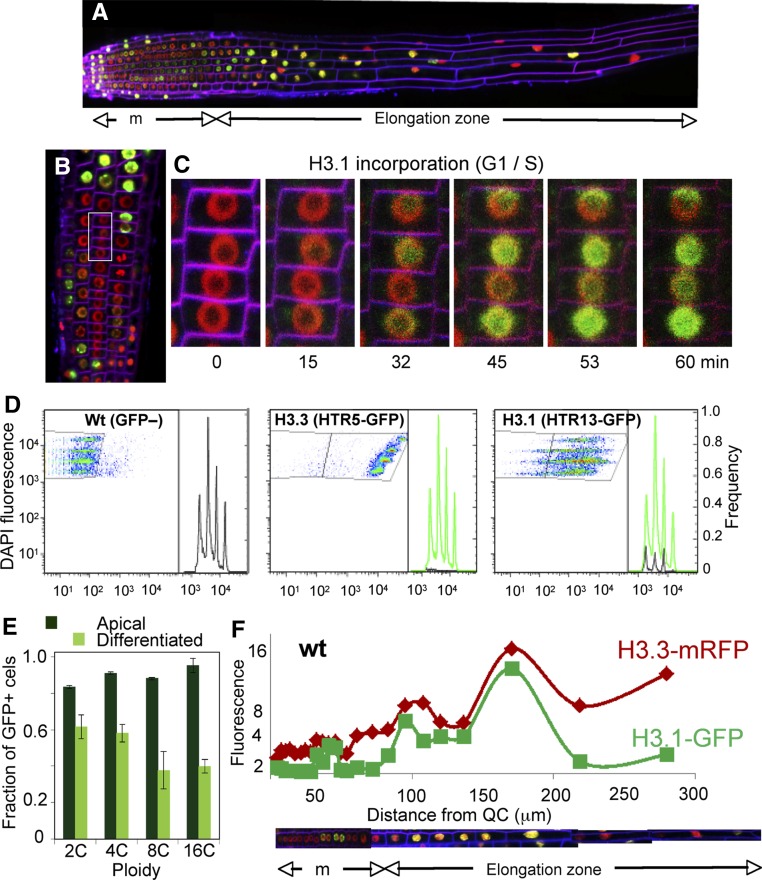
Next, we investigated the H3.1/H3.3 balance within the RAM. Here, H3.3 was expressed constitutively while the patchy pattern of H3.1 was obvious (Figures 2A and 2B), confirming previous observations (Ingouff et al., 2010). Interestingly, the QC cells were not labeled with H3.1-GFP (Figures 2C and 2D), consistent with the observation in late embryos and with the arrest in G1 of QC cells (Forzani et al., 2014). A similar pattern was observed in other root cell layers, e.g., epidermis and cortex, and with other H3 genes (Supplemental Figures 2E to 2P).
Figure 2.
Developmental Pattern of H3.1/H3.3 Dynamics.
(A) Confocal image of root epidermis expressing H3.1(HTR3)-GFP. Cell walls were labeled with propidium iodide (red).
(B) Same as in (A) but for H3.3(HTR5)-GFP.
(C) Detail of QC cells of a root expressing H3.1(HTR3)-GFP.
(D) Same as in (C) but for H3.3(HTR5)-GFP plants.
(E) Epidermal layer of a root expressing both H3.1(HTR3)-GFP and H3.3(HTR5)-RFP. Cell membranes stained with FM4-64 appear in magenta.
(E1) Detail showing two cells in metaphase, one containing both H3.1 and H3.3 and another with an undetectable H3.1 signal.
(F) Relative position of mitotic figures in the root epidermal layer labeled with H3.3(HTR5)-RFP only or with both H3.1(HTR3)-GFP and H3.3(HTR5)-RFP, as indicated, along the RAM. Position 0 indicates the end of the RAM and −1.0 the first epidermal cell that can be identified in the confocal plane (n = 53; P = 0.05; t test).
(G) Epidermal layer of a root expressing both H3.1(HTR13)-RFP and CYCB1;1-GFP. Cell membranes stained with FM4-64 appear in magenta.
(H) Relative position of mitotic figures labeled with H3.1(HTR13)-RFP or CYCB1;1-GFP, as indicated, along the RAM. Positions are as in (F). The two populations are statistically similar (n = 77; P = 0.05; t test).
(I) Epidermal layer of roots expressing both H3.1(HTR3)-GFP and H3.3(HTR5)-RFP control plants and treated with proteasome inhibitors (50 µM MG132 plus 0.5 µM epoxomycin) for 3 h, as indicated. Note that the overall H3.1-GFP labeling pattern is not affected by the treatment. Arrowheads point to mitotic cells that accumulate in the presence of proteasome inhibitors.
Root growth is the combination of continuous production of new cells (within the RAM) and the increases in cell length in the longitudinal root axis that occur in the elongation/differentiation zone (Beemster and Baskin, 1998). Thus, the stem cell derivatives undergo several cell divisions to produce cells that constitute the transient amplifying compartment (Scheres, 2007) and finally exit the cell cycle and initiate differentiation. This defines a rootward half of the RAM (the proliferation domain), where all cells divide, and a shootward half (the transition domain), where cells arrest in the cell cycle. Therefore, the proliferative fraction (i.e., the fraction of dividing cells within the meristematic population) decreases from 1.0 in the proliferation domain to ∼0.0 along the transition domain (Beemster and Baskin, 1998; Ivanov and Dubrovsky, 2013).
To determine H3 dynamics in relation to the proliferation potential, we assessed in detail the H3.1/H3.3 balance along the RAM by focusing first on mitotic cells. Visual inspection of root meristems of doubly labeled H3.1-GFP/H3.3-mRFP plants revealed that most mitoses occur in the rootward half of the RAM, as expected from the known reduction of the proliferative fraction in the shootward half of the RAM. Unexpectedly, we found that two types of fluorescently labeled mitoses were detected within the proliferation domain of the root (Figure 2E): one doubly labeled with GFP and mRFP and another labeled almost exclusively with mRFP. Moreover, the distribution of these two types of labeled mitoses appeared to be nonrandom. Thus, doubly GFP/mRFP-labeled mitoses were more frequent in the most rootward part of the proliferation domain of the RAM while its shootward part, next to the transition domain of the RAM, was enriched in mRFP-labeled mitoses, as exemplified here for the epidermal cells (Figures 2E, 2E1, and 2F). The epidermis contains two different cell types, trichoblasts and atrichoblasts, which differ in their proliferation rate (Berger et al., 1998). By scoring GFP and/or mRFP-labeled mitoses separately in these cell types, we found that the H3.3-mRFP-labeled cell distribution in trichoblasts was wider than in atrichoblasts (Supplemental Figure 3), likely accounting for the different division rate and potential between these cell types (Berger et al., 1998). To rule out that these observations were GFP specific, we generated plants expressing H3.1-mRFP and found that mRFP-labeled mitoses were also highly enriched in the most rootward part of the proliferation domain (Figures 2G and 2H). Moreover, crossing these plants with the CYCB1;1-GFP marker of cycling cells allowed us to show that the distribution of H3.1-mRFP-containing mitoses largely colocalizes with the CYCB1;1-GFP expression domain of the RAM (Figures 2G and 2H).
The finding that there are mitoses with high and low H3.1/H3.3 ratios and that they have a defined spatial localization pattern within the RAM is highly informative since H3.1 is exclusively incorporated during S-phase (Gurard-Levin et al., 2014). As the mitoses labeled only with H3.3-mRFP are enriched toward the end of the proliferation domain, we concluded that these mitotic cells have completed their last division (characterized by a low H3.1/H3.3 ratio, as a result of massive H3.1 eviction prior to mitosis). Furthermore, the presence of mitoses with a low H3.1/H3.3 ratio is indicative of a high H3.1 exchange that must have occurred during the G2 phase in cells undergoing their last cell division in the RAM (Supplemental Movie 1; see for example the 2nd nucleus from the bottom, labeled in red, in the leftmost atrichoblast cell file of the root that enters mitosis). Therefore, it is reasonable to conclude that we are visualizing the histone replacement event that occurs at the exit of cell proliferation before initiating the endocycle program in the elongation zone of the root.
One possibility is that the proteasome is involved in H3.1 eviction and degradation, thus contributing to the observed H3.1 pattern in mitotic cells in different RAM domains. We performed experiments in which plants expressing both H3.1-GFP and H3.3-mRFP were incubated for 3 h with proteasome inhibitors (MG132 and epoxomycin). This treatment was physiologically informative since we observed a clear accumulation of mitotic figures, as expected (Figure 2I). The total amount of GFP or mRFP labeling did not change significantly after this treatment. We did not observe either accumulation of H3.1-GFP signal in the cytoplasm of mitotic cells (Figure 2I). Furthermore, the H3.1-GFP labeling pattern was maintained in the presence of proteasome inhibitors. Together, our results strongly suggest that the proteasome is not a major component responsible for the observed H3.1 incorporation/eviction dynamics.
We analyzed high-resolution microarray data along the root (Brady et al., 2007) and found that the high expression of many cell cycle and chromatin genes in the more rootward slices (Supplemental Table 1) declined drastically at the beginning of the transition domain. It is worth noting that this pattern precisely colocalizes with the H3.1 decrease and is consistent with the loss of proliferation capacity. These changes in gene expression patterns along the root may be highly dependent upon the presence or absence of specific transcription factors, likely including cell cycle regulators, developmentally regulated patterning genes, and cell type-specific factors, among others. Therefore, we conclude that a complex network of signals contributes to coordinate cell proliferation potential and gene expression pattern within the root meristem.
Cells Undergoing Their Last Cell Cycle in the RAM Possess a Longer G2
Our data suggest that H3 dynamics allows discrimination between cells undergoing cell division in the rootward and shootward halves of the RAM. Furthermore, a striking feature is that H3.1 replacement in the last cell cycle is very fast, occurring within a single G2 phase. We considered the possibility that these cells experience an extended G2 phase compared with that in the earlier divisions of stem cell derivatives, thus contributing to more efficient H3.1 replacement. Preliminary support for this idea was obtained by analyzing the labeling pattern of mitoses a few hours after a short pulse with EdU and the analysis of EdU-labeled mitotic figures at different time points afterward. For example, after a few hours chase (see also below), we consistently found that EdU-labeled mitotic figures appeared to be enriched in the most rootward half of the meristem, whereas in the other half most mitotic figures were not labeled with EdU (Figure 3A). This finding suggested that cells in their last cell cycle undergo a longer G2. Accordingly, we directly measured the G2 phase length along the RAM by determining the kinetics of appearance of EdU-labeled mitotic cells. Root meristems were labeled with EdU for 15 min and then chased for different times. EdU-labeled mitotic figures were scored in whole-mount 4′,6-diamidino-2-phenylindole (DAPI)-stained and chemically detected EdU-treated roots in individual epidermal cell files along the RAM (from the first epidermal cell up to the last in the meristem). At short chase times (3 h and 15 min) all mitotic cells were unlabeled with EdU (only DAPI positive). At a chase time of 3 h and 30 min, we observed the first EdU-labeled mitotic figures and they appeared largely located in the most rootward positions (Figure 3B). EdU-labeled mitotic figures in the more shootward locations were detected at later chase times (4 h and 15 min), consistent with the idea that the G2 is longer in these cells. Quantification of the rate of increase of EdU-labeled mitotic figures allowed us to estimate that cells in their last cell cycle undergo a G2 phase that is 40 to 45 min longer than the G2 of the first stem cell derivatives (Figure 3C).
Figure 3.
Estimation of the G2 Phase Length along the Root Meristem.
(A) Epidermal cell layer of a root labeled with EdU (15 min) and analyzed after a 4 h and 15 min chase. EdU was detected (red) and all nuclei stained with DAPI (blue). The inset contains an enlarged image of the box in the left panel to highlight the presence of EdU-positive (yellow arrowheads) and -negative (white arrowheads) metaphase nuclei.
(B) Distribution of EdU-labeled (red) and unlabeled (blue) mitotic figures along individual epidermal cell files at different chase times after a 15-min EdU pulse. The relative position within the RAM was determined as in Figure 2. Statistical analysis of the distributions was performed with the Wilcoxon-Mann-Whitney test, as indicated: **P < 0.01 and ***P < 0.005.
(C) Kinetics of appearance of EdU-labeled mitotic figures in two regions of the RAM at different chase times after a 15-min pulse with EdU: between positions −0.2 and −0.6 (cells undergoing their last cell cycle; circles) and between positions −0.6 and −1.0 (cells in previous cell cycles; squares).
(D) Distribution of EdU-labeled (red) and unlabeled (blue) mitotic figures in the wild type (filled circles) and htr1,2,3,9 quadruple mutant (circles). The analysis was performed as in (B). The EdU-labeled cell distributions (Col-0 versus htr1,2,3,9 mutant) were not significantly different based on the Wilcoxon-Mann-Whitney test (P < 0.05).
We next explored whether the observed dynamic pattern in the RAM is due to differences in histone H3 availability. To test this, we used a quadruple htr1,2,3,9 mutant that expresses H3.1 from its only wild-type HTR13 gene (Jacob et al., 2014). We performed an EdU pulse-chase experiment under the same conditions used for wild-type plants. Based on the results shown in Figure 3B, we chose to analyze in detail the time points when the amount of EdU-labeled nuclei starts to increase in the shootward part of the RAM. Importantly, we found that the mutant exhibited a pattern undistinguishable from that of wild-type control seedlings (Figure 3D). This suggests that the incorporation/eviction dynamics observed in the RAM may occur largely independently of H3 availability, although this point has not been experimentally addressed. Furthermore, the extended G2 phase during the last cell cycle might be a major determinant of H3.1 eviction.
Quite interestingly, there are precedents for cell cycle time differences in the amplification stage of cell populations in other systems. For example, the last cell cycle during synchronized divisions of the Drosophila melanogaster embryo can be distinguished from the previous ones by several features, including increased cell cycle duration (with an extended G2) (Yuan et al., 2014; Blythe and Wieschaus, 2015; Zaballos et al., 2015). We believe that an analogous scenario may apply to the RAM during the last cell cycle just before cell proliferation arrest and initiation of differentiation, associated with changes in the gene expression program. The H3.1/H3.3 dynamics that we have observed likely reflect intrinsic aspects of proliferation dynamics within the RAM that were not noticed previously. Interestingly, histone H2B has been reported to decrease in mobility in differentiated cells in the Arabidopsis root (Rosa et al., 2014). Even though this and the H3 replacement studied here are likely unrelated phenomena, they highlight the relevance of histone dynamics at different levels in proliferating and differentiating cells.
A Massive Eviction of H3.1 Also Defines the End of the Endocycle Program
Histone H3.1 is the isoform incorporated during the S-phase by the CAF-1 chaperone (Gurard-Levin et al., 2014), and the H3.1-GFP signal was undetectable in G2 nuclei near the end of RAM boundary (Figure 4A). First, we confirmed using live imaging that H3.1-GFP was incorporated upon entering the S-phase of the first endoreplication round (Figures 3B and 3C; Supplemental Movie 1). We found that nuclei leaving the endoreplication zone and entering the differentiated part of the root showed a drastic reduction in H3.1-GFP labeling (Figure 4A). This pattern is reminiscent of that at the exit of the proliferative domain of the RAM, suggesting that a massive H3.1 exchange was also concomitant with the entry into full differentiation. To study this in detail, we used flow cytometry of nuclei isolated from the apical ∼5 mm. We found that nuclei in all ploidy levels were labeled with H3.3-GFP, with an increase proportional to the DNA content (Figure 4D). By contrast, the distribution ranges of H3.1-GFP-labeled cells were very wide, with all ploidy levels displaying different amounts of GFP (Figure 4D). Remarkably, a similar pattern was observed in nuclei isolated from the differentiated part of the root (Supplemental Figure 4A), indicating that events occurring early in the elongation zone are maintained in the mature root. Strikingly, the fraction of H3.1-GFP-positive nuclei decreased in differentiated tissue, which was more evident in the highest ploidy levels. This may indicate that cells negative or with a small amount of H3.1 could have reached the last endocycle round (Figure 4E). To confirm these results, we repeated the study with other H3.1 and H3.3 genes and obtained similar results (Supplemental Figure 4B).
Figure 4.
H3 Dynamics during the Developmentally Controlled Endocycle.
(A) Composite image of a root expressing H3.1(HTR3)-GFP and H3.3(HTR5)-mRFP to visualize the labeling pattern along the root developmental zones. Cell membranes stained with FM4-64 appear in magenta.
(B) Detail of the epidermal layer at the meristem/elongation boundary. Inset: nuclei followed in the live-imaging experiments summarized in (C).
(C) Time-lapse confocal microscopy series at the indicated times of nuclei highlighted in (B) to show the onset of the first endocycle as revealed by H3.1(HTR3)-GFP incorporation. See full experiment in Supplemental Movie 1.
(D) Analysis of ploidy level of wild-type (GFP–), H3.3(HTR5)-GFP, and H3.1(HTR13)-GFP nuclei of the 5-mm apical part of the root by two-channel flow cytometry. DAPI was used for nuclear staining and determination of DNA content (2C to 16C). Wild-type Col-0 and H3.3(HTR5)-GFP nuclei were used to define the GFP– (left panel) and GFP+ (middle panel) gates.
(E) Fraction of H3.1(HTR13)-GFP-positive nuclei in the 5-mm apical (dark green) and differentiated (light green) parts of the root. Data are mean values ± sd (n = 3).
(F) The H3.1/H3.3 balance along the root developmental zones. The relative fluorescence intensity of H3.1(HTR3)-GFP and H3.3(HTR5)-RFP of each nucleus in an entire cell file was plotted against its position in the RAM (µm). The cell file is reconstructed at the bottom, indicating the meristem (m) and the elongation and differentiation zones. The relative amount of H3.3 intensity was used to estimate the ploidy level (y axis). A representative example of an epidermal cell file expressing H3.1(HTR3)-GFP and H3.3(HTR5)-RFP in wild-type Col-0 is shown.
To further validate these observations, we measured the amount of H3.1-GFP and H3.3-mRFP in each nucleus combined with its position along individual cell files. Importantly, while the amount of H3.3 relates to the ploidy level, we observed that H3.1 incorporation paralleled that of H3.3 in each endocycle round, ruling out the possibility that the H3.1 decrease results from a progressive lack of incorporation (Figure 4F). The decrease in the GFP signal at the onset of full differentiation confirms that H3.1-GFP is efficiently evicted from chromatin, most likely by replacement with H3.3. Together, these data reveal the extensive chromatin reorganization that relies on a large H3.1/H3.3 exchange at the end of the endoreplication program and the onset of terminal differentiation.
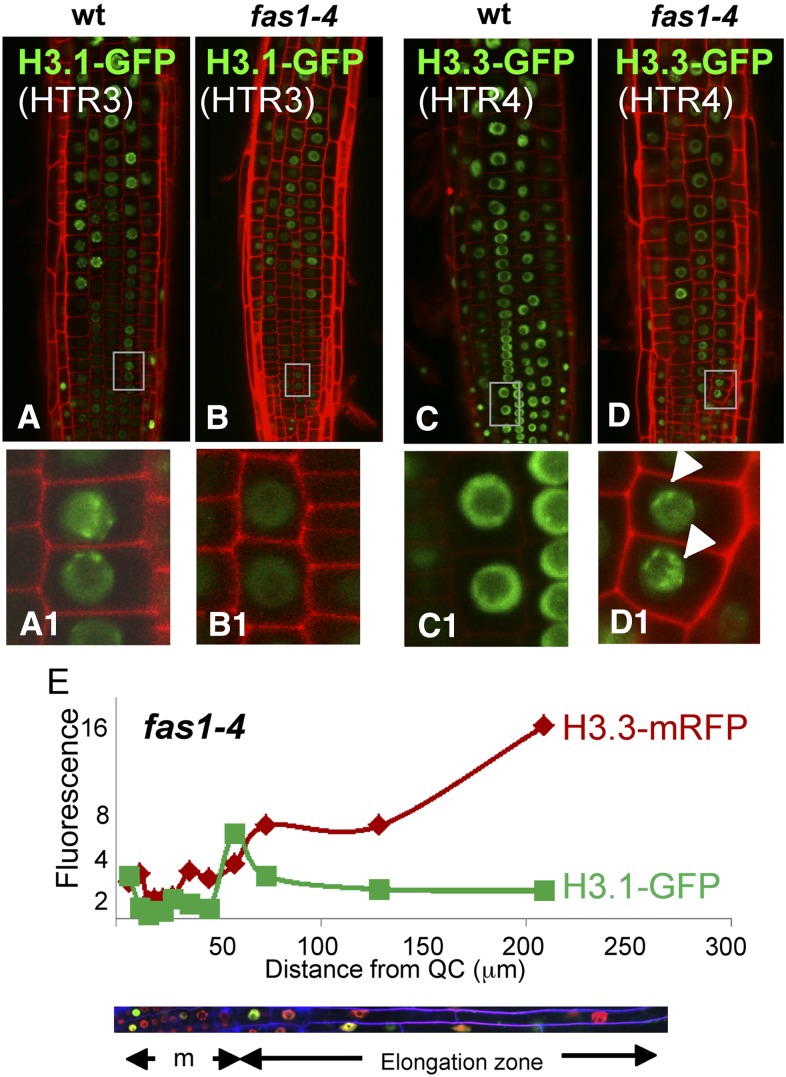
To answer whether the developmental domains defined by the cell division potential and the H3.1/H3.3 ratio are affected when H3.1 incorporation is impaired, we used fas1 plants, carrying a mutation in the large subunit of the DNA replication-dependent H3.1 chaperone CAF-1 (Exner et al., 2006; Ramirez-Parra and Gutierrez, 2007b). Among other phenotypic features, fas1 cells exhibit a strong G2 cell cycle arrest and a premature onset of the endocycle program that leads to a systemic increase in nuclear ploidy (Ramirez-Parra and Gutierrez, 2007a) and reduced root and shoot apical meristems (Kaya et al., 2001). Still, some cells escape the G2 arrest and undergo mitosis, although with various degrees of chromosomal aberrations, allowing growth of viable plants. There was an overall decrease in H3.1 incorporation (Figures 5A and 5B) and an abnormal increase of H3.3 in chromocenters (Figures 5C and 5D). In spite of the protracted RAM of fas1-4 plants as a consequence of a highly reduced cell division potential, the longitudinal zonation pattern was maintained. Thus, cells undergoing their last cell cycle and cells at the end of the endocycle program were also characterized by a change in the H3.1/H3.3 ratio (Figure 5E). This result in the fas1 mutant, together with the data using the htr1,2,3,9 histone mutant, reveals the robustness of the root developmental domains identified herein based on the H3.1/H3.3 ratio since the cell population and H3 dynamics are maintained even when H3.1 and its chaperone are altered. Furthermore, given the reduced proliferation and increased differentiation phenotype of fas1 mutant cells, our results reinforce the idea of a correlation between high levels of H3.3 (or reduced levels of H3.1) concomitant with cell differentiation.
Figure 5.
H3 Dynamics in Wild-Type and fas1-4 Mutant Roots.
(A) to (D) Images of the root epidermal cell layer of H3.1(HTR3)-GFP ([A] and [B]) and H3.3(HTR4)-GFP ([C] and [D]) expressed in wild-type ([A] and [C]) and fas1-4 mutant background ([B] and [D]). Insets show a magnification of nuclear labeling patterns. Note the decrease of the HTR3-GFP signal (B) and the enrichment of H3.3(HTR4)-GFP in the chromocenters (D) in fas1-4 plants.
(E) The H3.1/H3.3 balance along the root developmental zones in fas1-4 mutant plants. Measurements were carried as described for Figure 4F.
H3.1 Replacement at Proliferation to Differentiation Transitions throughout the Plant Body
The results described so far show that a high H3.1/H3.3 ratio is indicative of proliferative potential of cells in a developing organ, while a fast reduction in the H3.1/H3.3 ratio identifies cells undergoing their last cell cycle. Our results in the root also reveal that the massive H3.1 replacement is a characteristic associated with exit to differentiation after endoreplication. We examined whether this is also the case in two extreme situations outside the root: the stomatal lineage, cells that differentiate in the absence of ploidy increase, and hypocotyl cells that undergo extra endoreplication cycles in response to darkness.
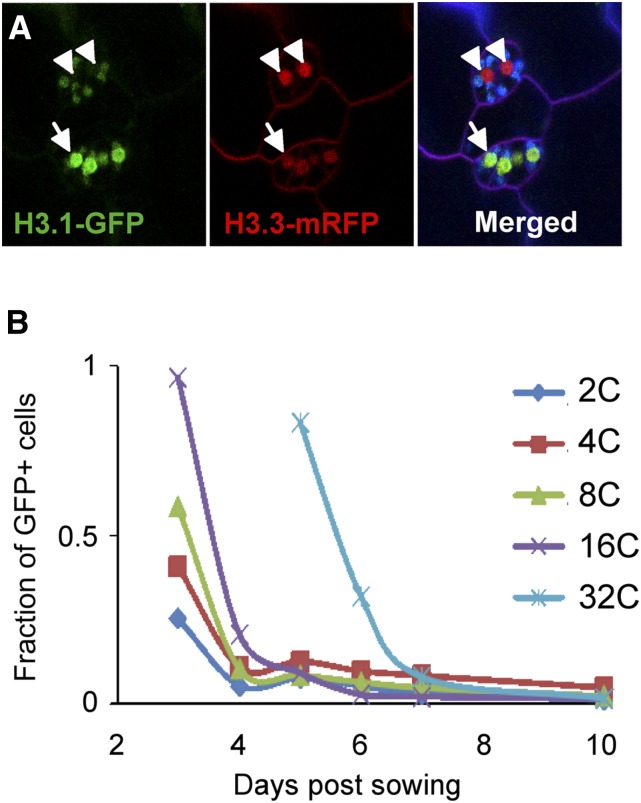
Meristemoid mother cells divide asymmetrically to generate meristemoids, which in turn undergo a few rounds of asymmetric divisions to produce guard mother cells. These commit to generate a mature guard cell that will divide to produce the two young stomatal guard cells that later differentiate into the mature guard cell (Dow and Bergmann, 2014; Adrian et al., 2015). We found that meristemoids, which still maintain a limited proliferation potential, exhibit a high H3.1 content until the last division of the guard mother cell in the lineage that gives rise to the two young guard cells (Figure 6A). Then, a drastic reduction in the H3.1 content is observed and their nuclei appear labeled with H3.3-mRFP, with undetectable or very low amounts of H3.1-GFP (Figure 6A).
Figure 6.
H3 Dynamics during Stomatal and Hypocotyl Cell Differentiation.
(A) Confocal image of the cotyledon epidermis of plants expressing both H3.1-GFP and H3.3-mRFP. Cell membranes were stained with FM4-64 (purple). Chloroplasts are visible due to autofluorescence (green or blue). Arrowheads point to the nuclei of the two mature guard cells that are largely devoid of H3.1-GFP. Arrows point to nuclei of undifferentiated cells that still contain both H3.1-GFP and H3.3-mRFP.
(B) Time-course flow cytometry measurements of the fraction of H3-1-GFP-positive nuclei in each ploidy level during hypocotyl development in the dark.
To establish whether the H3.1 eviction observed at the exit of the endoreplication program in root cells can be modulated depending on the number of endocycles, we chose the hypocotyl, an organ that is exclusively made by cell expansion associated with endoreplication. During skotomorphogenic development, the length of epidermal hypocotyl cells can expand ∼100 times (Gendreau et al., 1997) after undergoing one extra endoreplication round. We analyzed the H3.1-GFP content in the different ploidy levels by flow cytometry, isolating nuclei from the aerial part of etiolated seedlings grown in the dark at various time points. We observed that the number of cells positive for H3.1-GFP protein was higher at the beginning of hypocotyl development (3 to 4 d after sowing) than in nuclei isolated 7 d after sowing (Figure 6B). The first 32C cells appear 5 d after sowing. Interestingly, at this time when the H3.1 content (HTR3) is already low or undetectable in all other ploidy levels, most 32C cells are positive for H3.1, and, later in development, the number of H3.1-GFP positive cells also decreases in the 32C nuclei. These results indicate that massive H3.1 replacement coincides with the last endocycle round but is independent of the number of endocycles occurring during the differentiation process.
One related question is whether H3.1 is linked to active cycling cells by being deposited again when differentiated and/or arrested cells reenter the cell cycle. To answer this question, we analyzed H3 dynamics during reentry into the cell cycle in a physiological condition, as occurs during initiation of lateral root development. We found that at very early stages of lateral root primordia a few nuclei are labeled with H3.1-GFP, revealing that lateral root primordia founder cells have reentered S-phase (Supplemental Figure 5).
Conclusion
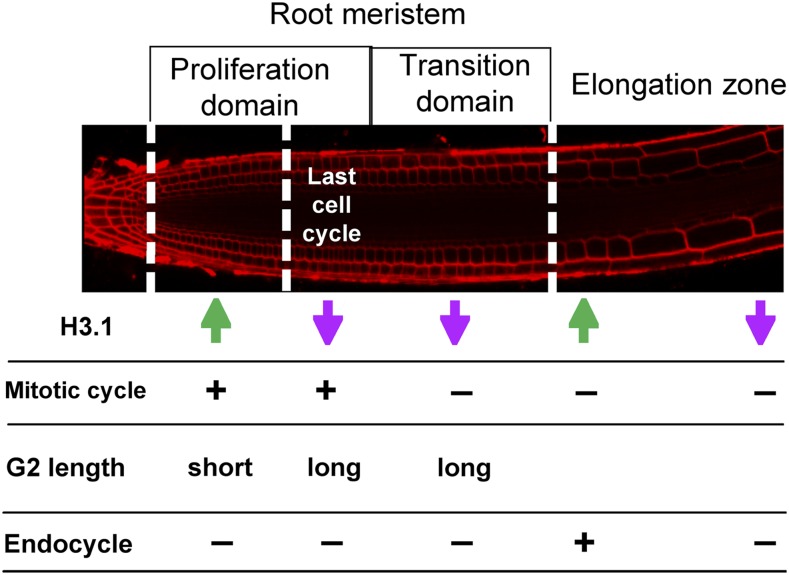
Our studies of the H3.1/H3.3 dynamics within the root served to identify distinct cell populations, in particular, cells undergoing their last cell cycle within the transient amplifying compartment, which is the proliferation half of the RAM. Likewise, changes in the H3.1/H3.3 ratio allow the identification of cells undergoing their last endocycle before initiating full differentiation. A similar H3.1 replacement process occurs in other cell types (stomatal and hypocotyl cells) with very different differentiation programs. We conclude that the H3.1/H3.3 dynamics reported herein are a feature of cells undergoing their last cell cycle or endocycle in their differentiation program (Figure 7). It is remarkable that the same principle applies to the end of cell proliferation and the end of the endocycle program, which are very different processes (Edgar et al., 2014). Interestingly, distinct degrees of chromatin dynamics have been reported during reprogramming at very early stages of mouse development (Bošković et al., 2014; Burton and Torres-Padilla, 2014) and along the root (Rosa et al., 2014).
Figure 7.
Cell Proliferation Dynamics in the RAM.
Schematic representation of the root cell populations according to their anatomical positions. The H3.1/H3.3 ratio is indicated with arrows (green, high H3.1; magenta, low H3.1). Other characteristics, such as the G2 length and the mitotic and endocycling potential, are also indicated.
In addition to revealing that the fast H3.1 eviction is a feature of differentiating cells, most likely in all plant locations, the approaches used in our study show their usefulness to identify five functionally relevant domains associated with Arabidopsis root development (Figure 7): (1) the transit amplifying compartment with cells proliferating at a high rate, (2) the domain occupied by cells undergoing their last cell cycle, (3) the meristem domain where the cells arrest cell division, (4) cells in active endocycle, and (5) the cells undergoing their last endocycle before initiating full differentiation. Strikingly, the functional proliferation domains uncovered by H3 dynamics are not altered in fas1 and htr1,2,3,9 mutants, indicating that the longitudinal zonation patterning is not disturbed by the H3 unbalance. Therefore, the H3.1/H3.3 ratio reveals specific domains in the root and chromatin reorganization events, but it is not the causal agent, pointing to a higher order regulation of the H3 and related genes. Further experiments using these tools should facilitate functional studies of distinct cell populations within a developing organ to expand this concept and confirm if it is a general mechanism of cell proliferation and developmental dynamics in all organisms under normal and pathological conditions.
METHODS
Plant Materials and Constructs
A genomic fragment containing the promoter and coding region, except the termination codon, of different histone H3.1 (HTR3 and HTR13) and H3.3 (HTR4 and HTR5) genes was amplified by PCR using Pfx (Life Technologies). Primers used are listed in Supplemental Table 2. The PCR products were cloned into pDONR221 (Life Technologies). Clones without mutations were transferred to the corresponding Gateway destination vectors (pGWB4 and pGWB453; Nakagawa et al., 2007). Expression clones were generated with the C-terminal region of the different histones fused to fluorescent tags (GFP or mRFP). The constructs were confirmed by sequencing and introduced into Agrobacterium tumefaciens (C58C1 strain). Arabidopsis thaliana plants (Col-0 ecotype) were stably transformed by the floral dip method (Clough and Bent, 1998), and transformed seeds were selected on Murashige and Skoog agar plates containing kanamycin (50 µg/mL). Plants with only one insertion were selected and homozygous lines were used in the experiments. pHTR3:HTR3-GFP × pHTR5:HTR5-mRFP was used to follow H3 dynamics along the root. fas1-4 mutants (Exner et al., 2006) were crossed to the histone-GFP tagged lines and fas1-4−/− × pHTR3:HTR3-GFP+/+, fas1-4−/− × pHTR4:HTR4-GFP+/−, and fas1-4−/− × pHTR3:HTR3-GFP+/− pHTR5:HTR5-mRFP+/− were used in this study. Heterozygous lines for the tagged histones were used when a seedling-lethal phenotype was observed in the crosses.
Plants were grown in an incubator at 22°C (or 20°C) and 60% humidity under standard long-day conditions (16-h light/8-h darkness; cool white fluorescent bulbs, 37 W, 4200K, at 100 to 110 µmol/m2/s) in MSS 1% agar plates (1% sucrose, 0.5× MS, and 0.5 g/L MES, pH 5.7).
RNA Extraction, cDNA Synthesis, and qPCR
Roots from seedlings grown for 7 d after sowing were used to extract RNA. Tissue was ground in Silamat S5 (Ivoclar Vivadent) using glass beads, and RNA was extracted with Trizol (Life Technologies). To obtain cDNA, 500 ng total RNA was used. The reverse transcriptase reaction was performed using oligo(dT) as a primer and the SuperScript III kit from Life Technologies, following kit instructions. qPCR was performed with GoTaqPCR master mix (Promega) in an ABI Prism 7900HT (see Supplemental Table 2 for the primers used). To measure the expression of the different histones, the absolute copy number of each cDNA was calculated with the help of standards where the copy number is known. For this purpose, amplicons for the different primers were amplified by PCR and purified directly from a low-melting agarose gel. The concentration of each amplicon was measured using NanoDrop, and the number of copies of double-stranded DNA was calculated using the algorithm described in http://cels.uri.edu/gsc/cndna.html. Several dilutions of these standards were run in the ABI Prism, and the crossing points of each dilution and the number of copies were plotted in standard curves where the unknown samples were interpolated to obtain the absolute copy number.
Immunoblot Analysis
Nuclear extracts were prepared from 6-d-old seedlings as described previously (Stroud et al., 2012). Proteins were fractionated in 13.5% SDS-PAGE gels and proteins identified by immunoblots using anti-GFP (Abcam; catalog no. 5450, lot GR136040).
Confocal Microscopy
Roots were stained with either propidium iodide (50 µg/mL) or 10 µM FM4-64 (Life Technologies) and observed using a confocal LSM510 (Zeiss) microscope. Embryos were extracted from the siliques and transferred to a slide with a 25-μL drop of embryo extraction buffer (4% paraformaldehyde and 5% glycerol in PBS) and dissected using two syringes. Then, 25 μL of 10 µM FM4-64 was added and the embryos were infiltrated for a few seconds to allow staining. Embryos were then observed using the LSM710 confocal microscope (Zeiss).
For live imaging, 3-d-old seedlings were placed on glass bottom dishes with a drop of water and covered with a piece of solid medium. Once in the inverted LSM510 confocal microscope (Zeiss), 100 μL of 10µM FM4-64 was injected under the solid medium. Movies were edited using Fiji, and photobleaching was corrected with the same software.
Analysis of Confocal Images: Positioning of Mitotic Figures in the Root Meristem
Epidermal cell files containing at least one mitotic figure were selected for analysis. Cell length of each cell in the file was measured. Meristem boundaries were considered the first cell in focus in the epidermis plane, near the root tip (−1 on the y axis) and the first rapidly elongated cell (0 on the y axis). In this analysis, the cell marking the end of the meristem will be at position −1, the next cell toward the root tip at position −2, and so on. Once the end of the meristem was localized and the position of cells in mitosis was identified, the relative position was calculated according to: relative position = position in the file of cell i/cell number in the file.
EdU Labeling
Col-0 seedlings (4 d after sowing) were transferred to 0.5× liquid MSS containing 20 µM EdU for 15 min, washed, and incubated for different chase time periods in 0.5× MSS supplemented with 50 µM TTP. Plants were then fixed in 4% paraformaldehyde in microtubule stabilizing buffer and permeabilized as described (Lauber et al., 1997). EdU was developed with the Click-it EdU staining kit (C10340; Life Technologies), nuclei stained with DAPI, and roots imaged by confocal microscopy with a Zeiss LSM 710.
Flow Cytometry of Roots and Hypocotyls
Seeds from all the GFP-tagged histone lines and wild-type Columbia (Col-0) were grown for 5 d under the usual conditions. As most cells in the root are differentiated, root tips (∼5 mm) were cut to enrich the sample in apical tissues and avoid the masking of possible differences between cell populations. This resulted in two samples per line, the apical tissue sample (enriched in root tips), and the differentiated tissue sample (depleted of root tips).
Tissues were chopped in 250 μL cold nuclei isolation buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM 4-MOPS, pH 7.0, and 0.1% Triton X-100; Galbraith et al., 1991). Nuclei were then stained with DAPI (2 µg/mL) and kept on ice until the samples were run in the flow cytometer (FACSCanto II; Becton Dickinson). In each experiment, 10,000 events were measured, and each experiment was repeated three times. All analyses were performed with FlowJo software (LLC Data Analysis Software).
For hypocotyl cell ploidy measurements, seeds were sown, placed in the growth chamber in the dark, and analyzed at various times afterwards (3 to 10 d). The aerial part of the seedlings was chopped and nuclei were stained using the same conditions described for roots.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database under the following accession numbers: HTR3, At3g27360; HTR4, At4g40030; HTR5, At4g40040; and HTR13, At5g10390.
Supplemental Data
Supplemental Figure 1. Characterization of fluorescently labeled H3 protein-expressing plants.
Supplemental Figure 2. Expression pattern of various fluorescently labeled H3 lines used in this study.
Supplemental Figure 3. Relative position of mitotic figures in trichoblast and atrichoblast cell layers.
Supplemental Figure 4. Analysis of ploidy levels of H3-tagged nuclei.
Supplemental Figure 5. H3.1-GFP labeling during initiation of lateral roots.
Supplemental Table 1. Chromatin and cell cycle genes downregulated from slice 1 + 2 to slice 4 (fold change > 1.5) of the Arabidopsis root.
Supplemental Table 2. Primers used in this study.
Supplemental Movie 1. Time-lapse microscopy (60 min) of a root of Arabidopsis expressing H3.1-GFP and H3.3-mRFP visualizing H3.1 eviction in G2 and H3.1 incorporation during S-phase, as well as mitotic figures labeled with H3.3 alone.
Supplementary Material
Acknowledgments
We thank Fred Berger for the HTR13-GFP and HTR5-GFP seeds, T. Nakagawa, T. Suzuki, and R. Tsien for pGWB vectors, R. Martienssen and Y. Jacob for the htr1,2,3,9 seeds, E. Martinez-Salas for comments, and members of C.G.’s laboratory for discussions. We also thank the Microscopy and Flow Cytometry Services of CBMSO and V. Mora-Gil for technical assistance. S.O. was the recipient of a JAE Predoctoral Fellowship from CSIC. This research was supported by Grants BFU2012-34821 and BFU2015-68396-R and by an institutional grant from Fundación Ramón Areces to the Centro de Biologia Molecular Severo Ochoa.
AUTHOR CONTRIBUTIONS
S.O. and B.D. performed the experiments. R.P. analyzed cell cycle and chromatin genes. C.G., B.D., and S.O. designed and analyzed the experiments. C.G. and B.D. supervised the work. C.G. wrote the manuscript with the active participation of the rest of authors, who read and approved the final version.
Glossary
- RAM
root apical meristem
- QC
quiescent center
- DAPI
4′,6-diamidino-2-phenylindole
References
- Adrian J., et al. (2015). Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell 33: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I., et al. (2010). Shootward and rootward: peak terminology for plant polarity. Trends Plant Sci. 15: 593–594. [DOI] [PubMed] [Google Scholar]
- Beemster G.T., Baskin T.I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Hung C.Y., Dolan L., Schiefelbein J. (1998). Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194: 235–245. [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blythe S.A., Wieschaus E.F. (2015). Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 160: 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošković A., Eid A., Pontabry J., Ishiuchi T., Spiegelhalter C., Raghu Ram E.V., Meshorer E., Torres-Padilla M.E. (2014). Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 28: 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Burton A., Torres-Padilla M.E. (2014). Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15: 723–734. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Desvoyes B., Fernández-Marcos M., Sequeira-Mendes J., Otero S., Vergara Z., Gutierrez C. (2014). Looking at plant cell cycle from the chromatin window. Front. Plant Sci. 5: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow G.J., Bergmann D.C. (2014). Patterning and processes: how stomatal development defines physiological potential. Curr. Opin. Plant Biol. 21: 67–74. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Zielke N., Gutierrez C. (2014). Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Exner V., Taranto P., Schönrock N., Gruissem W., Hennig L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133: 4163–4172. [DOI] [PubMed] [Google Scholar]
- Forzani C., Aichinger E., Sornay E., Willemsen V., Laux T., Dewitte W., Murray J.A. (2014). WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24: 1939–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Knapp S. (1991). Systemic endopolyploidy in Arabopsis thaliana. Plant Physiol. 96: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A.D., et al. (2010). Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard-Levin Z.A., Quivy J.P., Almouzni G. (2014). Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83: 487–517. [DOI] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Sabatini S. (2014). Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 15: 301–312. [DOI] [PubMed] [Google Scholar]
- Heyman J., Kumpf R.P., De Veylder L. (2014). A quiescent path to plant longevity. Trends Cell Biol. 24: 443–448. [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Vandenbussche F., Heyndrickx K.S., Van Leene J., Vercauteren I., Vanderauwera S., Vandepoele K., De Jaeger G., Van Der Straeten D., De Veylder L. (2013). ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342: 860–863. [DOI] [PubMed] [Google Scholar]
- Ingouff M., Berger F. (2010). Histone3 variants in plants. Chromosoma 119: 27–33. [DOI] [PubMed] [Google Scholar]
- Ingouff M., Rademacher S., Holec S., Soljić L., Xin N., Readshaw A., Foo S.H., Lahouze B., Sprunck S., Berger F. (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 20: 2137–2143. [DOI] [PubMed] [Google Scholar]
- Ivanov V.B., Dubrovsky J.G. (2013). Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends Plant Sci. 18: 237–243. [DOI] [PubMed] [Google Scholar]
- Jacob Y., Bergamin E., Donoghue M.T., Mongeon V., LeBlanc C., Voigt P., Underwood C.J., Brunzellem J,S., Michaelsm S.D., Reinberg D., Couture J.F., Martienssen R.A. (2014). Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343: 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kundaje A., et al.; Roadmap Epigenomics Consortium (2015). Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jürgens G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139: 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A., Almouzni G. (2007). Marking histone H3 variants: how, when and why? Trends Biochem. Sci. 32: 425–433. [DOI] [PubMed] [Google Scholar]
- Maze I., Noh K.M., Soshnev A.A., Allis C.D. (2014). Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 15: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Otero S., Desvoyes B., Gutierrez C. (2014). Histone H3 dynamics in plant cell cycle and development. Cytogenet. Genome Res. 143: 114–124. [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Winter C.M., Benfey P.N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J., Wagner D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10: 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007a). The many faces of chromatin assembly factor 1. Trends Plant Sci. 12: 570–576. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007b). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S., Ntoukakis V., Ohmido N., Pendle A., Abranches R., Shaw P. (2014). Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell 26: 4821–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. (2007). Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8: 345–354. [DOI] [PubMed] [Google Scholar]
- Scheres B., Benfey P., Dolan L. (2002). Root development. Arabidopsis Book 1: e0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J., Aragüez I., Peiró R., Mendez-Giraldez R., Zhang X., Jacobsen S.E., Bastolla U., Gutierrez C. (2014). The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Wang J., Hong F., Spector D.L., Fang Y. (2011). Four amino acids guide the assembly or disassembly of Arabidopsis histone H3.3-containing nucleosomes. Proc. Natl. Acad. Sci. USA 108: 10574–10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Otero S., Desvoyes B., Ramírez-Parra E., Jacobsen S.E., Gutierrez C. (2012). Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 5370–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H., Holec S., Alden K., Clarke N.D., Jacques P.E., Berger F. (2012). Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 8: e1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Shermoen A.W., O’Farrell P.H. (2014). Illuminating DNA replication during Drosophila development using TALE-lights. Curr. Biol. 24: R144–R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaballos M.A., Cantero W., Azpiazu N. (2015). The TALE transcription factor homothorax functions to assemble heterochromatin during Drosophila embryogenesis. PLoS One 10: e0120662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.